A New Data Model for Time Sequences
MOE-基于结构的药物设计及在药物发现方面的应用

药物发现/设计
基于大分子结构的 基于小分子配体的 基于片段的
上市
转化医学
药代动力学 吸收/分布/排除 代谢 毒性检测 先导化合物优化 临床数据管理 2010 © CloudScientific All Rights Reserved 临床数据统计分析
药物开发 (临床前及临床试验)
about moe
2010 © CloudScientific All Rights Reserved
我们代理如下公司产品在中国的业务
ChemAxon
2010 © CloudScientific All Rights Reserved
我们提供的解决方案
生物学
生物信息学 基因组学 蛋白组学 结构生物学 信号转导通路
about cloudscientific
2010 © CloudScientific All Rights Reserved
Best Solutions for Best Research
公司介绍
上海康昱盛信息科技有限公司
2010 © CloudScientific All Rights Reserved
2010 © CloudScientific All Rights Reserved
我们能为您提供以下方面的价值
• •
产品: 世界领先的生命科学软件产品 解决方案:针对客户的专业需求,提供专业的产品组合建议和科学应 用方面咨询,并可做客户化的定制开发
•
培训: 不仅为客户提供产品定制的专业培训,并且定期邀请行业专家 提供专业知识和行业进展的高级培训
2010 © CloudScientific All Rights Reserved
Molecular Operating Environment MOETM
5 Linear Time Series Model

1
2015/3/19
Assumptions for Unbiasedness
Still assume a model that is linear in parameters: yt = 0 + 1xt1 + . . .+ kxtk + ut Still need to make a zero conditional mean assumption: E(ut|X) = 0, t = 1, 2, …, n Note that this implies the error term in any p y given period is uncorrelated with the explanatory variables in all time periods
14
Detrending (continued)
An advantage to actually detrending the d ( data (vs. adding ddi a trend) d) involves i l the h calculation of goodness of fit Time-series regressions tend to have very high R2, as the trend is well explained The R2 from a regression on detrended data better reflects how well the xt’s explain yt
6
Assumptions (continued)
Still need to assume that no x is constant, and that there is no perfect collinearity Note we have skipped the assumption of a random sample The key impact of the random sample assumption is that each ui is independent Our strict exogeneity assumption takes care of it in this case
Native Instruments MASCHINE MK3 用户手册说明书

The information in this document is subject to change without notice and does not represent a commitment on the part of Native Instruments GmbH. The software described by this docu-ment is subject to a License Agreement and may not be copied to other media. No part of this publication may be copied, reproduced or otherwise transmitted or recorded, for any purpose, without prior written permission by Native Instruments GmbH, hereinafter referred to as Native Instruments.“Native Instruments”, “NI” and associated logos are (registered) trademarks of Native Instru-ments GmbH.ASIO, VST, HALion and Cubase are registered trademarks of Steinberg Media Technologies GmbH.All other product and company names are trademarks™ or registered® trademarks of their re-spective holders. Use of them does not imply any affiliation with or endorsement by them.Document authored by: David Gover and Nico Sidi.Software version: 2.8 (02/2019)Hardware version: MASCHINE MK3Special thanks to the Beta Test Team, who were invaluable not just in tracking down bugs, but in making this a better product.NATIVE INSTRUMENTS GmbH Schlesische Str. 29-30D-10997 Berlin Germanywww.native-instruments.de NATIVE INSTRUMENTS North America, Inc. 6725 Sunset Boulevard5th FloorLos Angeles, CA 90028USANATIVE INSTRUMENTS K.K.YO Building 3FJingumae 6-7-15, Shibuya-ku, Tokyo 150-0001Japanwww.native-instruments.co.jp NATIVE INSTRUMENTS UK Limited 18 Phipp StreetLondon EC2A 4NUUKNATIVE INSTRUMENTS FRANCE SARL 113 Rue Saint-Maur75011 ParisFrance SHENZHEN NATIVE INSTRUMENTS COMPANY Limited 5F, Shenzhen Zimao Center111 Taizi Road, Nanshan District, Shenzhen, GuangdongChina© NATIVE INSTRUMENTS GmbH, 2019. All rights reserved.Table of Contents1Welcome to MASCHINE (25)1.1MASCHINE Documentation (26)1.2Document Conventions (27)1.3New Features in MASCHINE 2.8 (29)1.4New Features in MASCHINE 2.7.10 (31)1.5New Features in MASCHINE 2.7.8 (31)1.6New Features in MASCHINE 2.7.7 (32)1.7New Features in MASCHINE 2.7.4 (33)1.8New Features in MASCHINE 2.7.3 (36)2Quick Reference (38)2.1Using Your Controller (38)2.1.1Controller Modes and Mode Pinning (38)2.1.2Controlling the Software Views from Your Controller (40)2.2MASCHINE Project Overview (43)2.2.1Sound Content (44)2.2.2Arrangement (45)2.3MASCHINE Hardware Overview (48)2.3.1MASCHINE Hardware Overview (48)2.3.1.1Control Section (50)2.3.1.2Edit Section (53)2.3.1.3Performance Section (54)2.3.1.4Group Section (56)2.3.1.5Transport Section (56)2.3.1.6Pad Section (58)2.3.1.7Rear Panel (63)2.4MASCHINE Software Overview (65)2.4.1Header (66)2.4.2Browser (68)2.4.3Arranger (70)2.4.4Control Area (73)2.4.5Pattern Editor (74)3Basic Concepts (76)3.1Important Names and Concepts (76)3.2Adjusting the MASCHINE User Interface (79)3.2.1Adjusting the Size of the Interface (79)3.2.2Switching between Ideas View and Song View (80)3.2.3Showing/Hiding the Browser (81)3.2.4Showing/Hiding the Control Lane (81)3.3Common Operations (82)3.3.1Using the 4-Directional Push Encoder (82)3.3.2Pinning a Mode on the Controller (83)3.3.3Adjusting Volume, Swing, and Tempo (84)3.3.4Undo/Redo (87)3.3.5List Overlay for Selectors (89)3.3.6Zoom and Scroll Overlays (90)3.3.7Focusing on a Group or a Sound (91)3.3.8Switching Between the Master, Group, and Sound Level (96)3.3.9Navigating Channel Properties, Plug-ins, and Parameter Pages in the Control Area.973.3.9.1Extended Navigate Mode on Your Controller (102)3.3.10Navigating the Software Using the Controller (105)3.3.11Using Two or More Hardware Controllers (106)3.3.12Touch Auto-Write Option (108)3.4Native Kontrol Standard (110)3.5Stand-Alone and Plug-in Mode (111)3.5.1Differences between Stand-Alone and Plug-in Mode (112)3.5.2Switching Instances (113)3.5.3Controlling Various Instances with Different Controllers (114)3.6Host Integration (114)3.6.1Setting up Host Integration (115)3.6.1.1Setting up Ableton Live (macOS) (115)3.6.1.2Setting up Ableton Live (Windows) (116)3.6.1.3Setting up Apple Logic Pro X (116)3.6.2Integration with Ableton Live (117)3.6.3Integration with Apple Logic Pro X (119)3.7Preferences (120)3.7.1Preferences – General Page (121)3.7.2Preferences – Audio Page (126)3.7.3Preferences – MIDI Page (130)3.7.4Preferences – Default Page (133)3.7.5Preferences – Library Page (137)3.7.6Preferences – Plug-ins Page (145)3.7.7Preferences – Hardware Page (150)3.7.8Preferences – Colors Page (154)3.8Integrating MASCHINE into a MIDI Setup (156)3.8.1Connecting External MIDI Equipment (156)3.8.2Sync to External MIDI Clock (157)3.8.3Send MIDI Clock (158)3.9Syncing MASCHINE using Ableton Link (159)3.9.1Connecting to a Network (159)3.9.2Joining and Leaving a Link Session (159)3.10Using a Pedal with the MASCHINE Controller (160)3.11File Management on the MASCHINE Controller (161)4Browser (163)4.1Browser Basics (163)4.1.1The MASCHINE Library (163)4.1.2Browsing the Library vs. Browsing Your Hard Disks (164)4.2Searching and Loading Files from the Library (165)4.2.1Overview of the Library Pane (165)4.2.2Selecting or Loading a Product and Selecting a Bank from the Browser (170)4.2.2.1[MK3] Browsing by Product Category Using the Controller (174)4.2.2.2[MK3] Browsing by Product Vendor Using the Controller (174)4.2.3Selecting a Product Category, a Product, a Bank, and a Sub-Bank (175)4.2.3.1Selecting a Product Category, a Product, a Bank, and a Sub-Bank on theController (179)4.2.4Selecting a File Type (180)4.2.5Choosing Between Factory and User Content (181)4.2.6Selecting Type and Character Tags (182)4.2.7List and Tag Overlays in the Browser (186)4.2.8Performing a Text Search (188)4.2.9Loading a File from the Result List (188)4.3Additional Browsing Tools (193)4.3.1Loading the Selected Files Automatically (193)4.3.2Auditioning Instrument Presets (195)4.3.3Auditioning Samples (196)4.3.4Loading Groups with Patterns (197)4.3.5Loading Groups with Routing (198)4.3.6Displaying File Information (198)4.4Using Favorites in the Browser (199)4.5Editing the Files’ Tags and Properties (203)4.5.1Attribute Editor Basics (203)4.5.2The Bank Page (205)4.5.3The Types and Characters Pages (205)4.5.4The Properties Page (208)4.6Loading and Importing Files from Your File System (209)4.6.1Overview of the FILES Pane (209)4.6.2Using Favorites (211)4.6.3Using the Location Bar (212)4.6.4Navigating to Recent Locations (213)4.6.5Using the Result List (214)4.6.6Importing Files to the MASCHINE Library (217)4.7Locating Missing Samples (219)4.8Using Quick Browse (221)5Managing Sounds, Groups, and Your Project (225)5.1Overview of the Sounds, Groups, and Master (225)5.1.1The Sound, Group, and Master Channels (226)5.1.2Similarities and Differences in Handling Sounds and Groups (227)5.1.3Selecting Multiple Sounds or Groups (228)5.2Managing Sounds (233)5.2.1Loading Sounds (235)5.2.2Pre-listening to Sounds (236)5.2.3Renaming Sound Slots (237)5.2.4Changing the Sound’s Color (237)5.2.5Saving Sounds (239)5.2.6Copying and Pasting Sounds (241)5.2.7Moving Sounds (244)5.2.8Resetting Sound Slots (245)5.3Managing Groups (247)5.3.1Creating Groups (248)5.3.2Loading Groups (249)5.3.3Renaming Groups (251)5.3.4Changing the Group’s Color (251)5.3.5Saving Groups (253)5.3.6Copying and Pasting Groups (255)5.3.7Reordering Groups (258)5.3.8Deleting Groups (259)5.4Exporting MASCHINE Objects and Audio (260)5.4.1Saving a Group with its Samples (261)5.4.2Saving a Project with its Samples (262)5.4.3Exporting Audio (264)5.5Importing Third-Party File Formats (270)5.5.1Loading REX Files into Sound Slots (270)5.5.2Importing MPC Programs to Groups (271)6Playing on the Controller (275)6.1Adjusting the Pads (275)6.1.1The Pad View in the Software (275)6.1.2Choosing a Pad Input Mode (277)6.1.3Adjusting the Base Key (280)6.1.4Using Choke Groups (282)6.1.5Using Link Groups (284)6.2Adjusting the Key, Choke, and Link Parameters for Multiple Sounds (286)6.3Playing Tools (287)6.3.1Mute and Solo (288)6.3.2Choke All Notes (292)6.3.3Groove (293)6.3.4Level, Tempo, Tune, and Groove Shortcuts on Your Controller (295)6.3.5Tap Tempo (299)6.4Performance Features (300)6.4.1Overview of the Perform Features (300)6.4.2Selecting a Scale and Creating Chords (303)6.4.3Scale and Chord Parameters (303)6.4.4Creating Arpeggios and Repeated Notes (316)6.4.5Swing on Note Repeat / Arp Output (321)6.5Using Lock Snapshots (322)6.5.1Creating a Lock Snapshot (322)6.5.2Using Extended Lock (323)6.5.3Updating a Lock Snapshot (323)6.5.4Recalling a Lock Snapshot (324)6.5.5Morphing Between Lock Snapshots (324)6.5.6Deleting a Lock Snapshot (325)6.5.7Triggering Lock Snapshots via MIDI (326)6.6Using the Smart Strip (327)6.6.1Pitch Mode (328)6.6.2Modulation Mode (328)6.6.3Perform Mode (328)6.6.4Notes Mode (329)7Working with Plug-ins (330)7.1Plug-in Overview (330)7.1.1Plug-in Basics (330)7.1.2First Plug-in Slot of Sounds: Choosing the Sound’s Role (334)7.1.3Loading, Removing, and Replacing a Plug-in (335)7.1.3.1Browser Plug-in Slot Selection (341)7.1.4Adjusting the Plug-in Parameters (344)7.1.5Bypassing Plug-in Slots (344)7.1.6Using Side-Chain (346)7.1.7Moving Plug-ins (346)7.1.8Alternative: the Plug-in Strip (348)7.1.9Saving and Recalling Plug-in Presets (348)7.1.9.1Saving Plug-in Presets (349)7.1.9.2Recalling Plug-in Presets (350)7.1.9.3Removing a Default Plug-in Preset (351)7.2The Sampler Plug-in (352)7.2.1Page 1: Voice Settings / Engine (354)7.2.2Page 2: Pitch / Envelope (356)7.2.3Page 3: FX / Filter (359)7.2.4Page 4: Modulation (361)7.2.5Page 5: LFO (363)7.2.6Page 6: Velocity / Modwheel (365)7.3Using Native Instruments and External Plug-ins (367)7.3.1Opening/Closing Plug-in Windows (367)7.3.2Using the VST/AU Plug-in Parameters (370)7.3.3Setting Up Your Own Parameter Pages (371)7.3.4Using VST/AU Plug-in Presets (376)7.3.5Multiple-Output Plug-ins and Multitimbral Plug-ins (378)8Using the Audio Plug-in (380)8.1Loading a Loop into the Audio Plug-in (384)8.2Editing Audio in the Audio Plug-in (385)8.3Using Loop Mode (386)8.4Using Gate Mode (388)9Using the Drumsynths (390)9.1Drumsynths – General Handling (391)9.1.1Engines: Many Different Drums per Drumsynth (391)9.1.2Common Parameter Organization (391)9.1.3Shared Parameters (394)9.1.4Various Velocity Responses (394)9.1.5Pitch Range, Tuning, and MIDI Notes (394)9.2The Kicks (395)9.2.1Kick – Sub (397)9.2.2Kick – Tronic (399)9.2.3Kick – Dusty (402)9.2.4Kick – Grit (403)9.2.5Kick – Rasper (406)9.2.6Kick – Snappy (407)9.2.7Kick – Bold (409)9.2.8Kick – Maple (411)9.2.9Kick – Push (412)9.3The Snares (414)9.3.1Snare – Volt (416)9.3.2Snare – Bit (418)9.3.3Snare – Pow (420)9.3.4Snare – Sharp (421)9.3.5Snare – Airy (423)9.3.6Snare – Vintage (425)9.3.7Snare – Chrome (427)9.3.8Snare – Iron (429)9.3.9Snare – Clap (431)9.3.10Snare – Breaker (433)9.4The Hi-hats (435)9.4.1Hi-hat – Silver (436)9.4.2Hi-hat – Circuit (438)9.4.3Hi-hat – Memory (440)9.4.4Hi-hat – Hybrid (442)9.4.5Creating a Pattern with Closed and Open Hi-hats (444)9.5The Toms (445)9.5.1Tom – Tronic (447)9.5.2Tom – Fractal (449)9.5.3Tom – Floor (453)9.5.4Tom – High (455)9.6The Percussions (456)9.6.1Percussion – Fractal (458)9.6.2Percussion – Kettle (461)9.6.3Percussion – Shaker (463)9.7The Cymbals (467)9.7.1Cymbal – Crash (469)9.7.2Cymbal – Ride (471)10Using the Bass Synth (474)10.1Bass Synth – General Handling (475)10.1.1Parameter Organization (475)10.1.2Bass Synth Parameters (477)11Working with Patterns (479)11.1Pattern Basics (479)11.1.1Pattern Editor Overview (480)11.1.2Navigating the Event Area (486)11.1.3Following the Playback Position in the Pattern (488)11.1.4Jumping to Another Playback Position in the Pattern (489)11.1.5Group View and Keyboard View (491)11.1.6Adjusting the Arrange Grid and the Pattern Length (493)11.1.7Adjusting the Step Grid and the Nudge Grid (497)11.2Recording Patterns in Real Time (501)11.2.1Recording Your Patterns Live (501)11.2.2The Record Prepare Mode (504)11.2.3Using the Metronome (505)11.2.4Recording with Count-in (506)11.2.5Quantizing while Recording (508)11.3Recording Patterns with the Step Sequencer (508)11.3.1Step Mode Basics (508)11.3.2Editing Events in Step Mode (511)11.3.3Recording Modulation in Step Mode (513)11.4Editing Events (514)11.4.1Editing Events with the Mouse: an Overview (514)11.4.2Creating Events/Notes (517)11.4.3Selecting Events/Notes (518)11.4.4Editing Selected Events/Notes (526)11.4.5Deleting Events/Notes (532)11.4.6Cut, Copy, and Paste Events/Notes (535)11.4.7Quantizing Events/Notes (538)11.4.8Quantization While Playing (540)11.4.9Doubling a Pattern (541)11.4.10Adding Variation to Patterns (541)11.5Recording and Editing Modulation (546)11.5.1Which Parameters Are Modulatable? (547)11.5.2Recording Modulation (548)11.5.3Creating and Editing Modulation in the Control Lane (550)11.6Creating MIDI Tracks from Scratch in MASCHINE (555)11.7Managing Patterns (557)11.7.1The Pattern Manager and Pattern Mode (558)11.7.2Selecting Patterns and Pattern Banks (560)11.7.3Creating Patterns (563)11.7.4Deleting Patterns (565)11.7.5Creating and Deleting Pattern Banks (566)11.7.6Naming Patterns (568)11.7.7Changing the Pattern’s Color (570)11.7.8Duplicating, Copying, and Pasting Patterns (571)11.7.9Moving Patterns (574)11.7.10Adjusting Pattern Length in Fine Increments (575)11.8Importing/Exporting Audio and MIDI to/from Patterns (576)11.8.1Exporting Audio from Patterns (576)11.8.2Exporting MIDI from Patterns (577)11.8.3Importing MIDI to Patterns (580)12Audio Routing, Remote Control, and Macro Controls (589)12.1Audio Routing in MASCHINE (590)12.1.1Sending External Audio to Sounds (591)12.1.2Configuring the Main Output of Sounds and Groups (596)12.1.3Setting Up Auxiliary Outputs for Sounds and Groups (601)12.1.4Configuring the Master and Cue Outputs of MASCHINE (605)12.1.5Mono Audio Inputs (610)12.1.5.1Configuring External Inputs for Sounds in Mix View (611)12.2Using MIDI Control and Host Automation (614)12.2.1Triggering Sounds via MIDI Notes (615)12.2.2Triggering Scenes via MIDI (622)12.2.3Controlling Parameters via MIDI and Host Automation (623)12.2.4Selecting VST/AU Plug-in Presets via MIDI Program Change (631)12.2.5Sending MIDI from Sounds (632)12.3Creating Custom Sets of Parameters with the Macro Controls (636)12.3.1Macro Control Overview (637)12.3.2Assigning Macro Controls Using the Software (638)12.3.3Assigning Macro Controls Using the Controller (644)13Controlling Your Mix (646)13.1Mix View Basics (646)13.1.1Switching between Arrange View and Mix View (646)13.1.2Mix View Elements (647)13.2The Mixer (649)13.2.1Displaying Groups vs. Displaying Sounds (650)13.2.2Adjusting the Mixer Layout (652)13.2.3Selecting Channel Strips (653)13.2.4Managing Your Channels in the Mixer (654)13.2.5Adjusting Settings in the Channel Strips (656)13.2.6Using the Cue Bus (660)13.3The Plug-in Chain (662)13.4The Plug-in Strip (663)13.4.1The Plug-in Header (665)13.4.2Panels for Drumsynths and Internal Effects (667)13.4.3Panel for the Sampler (668)13.4.4Custom Panels for Native Instruments Plug-ins (671)13.4.5Undocking a Plug-in Panel (Native Instruments and External Plug-ins Only) (675)13.5Controlling Your Mix from the Controller (677)13.5.1Navigating Your Channels in Mix Mode (678)13.5.2Adjusting the Level and Pan in Mix Mode (679)13.5.3Mute and Solo in Mix Mode (680)13.5.4Plug-in Icons in Mix Mode (680)14Using Effects (681)14.1Applying Effects to a Sound, a Group or the Master (681)14.1.1Adding an Effect (681)14.1.2Other Operations on Effects (690)14.1.3Using the Side-Chain Input (692)14.2Applying Effects to External Audio (695)14.2.1Step 1: Configure MASCHINE Audio Inputs (695)14.2.2Step 2: Set up a Sound to Receive the External Input (698)14.2.3Step 3: Load an Effect to Process an Input (700)14.3Creating a Send Effect (701)14.3.1Step 1: Set Up a Sound or Group as Send Effect (702)14.3.2Step 2: Route Audio to the Send Effect (706)14.3.3 A Few Notes on Send Effects (708)14.4Creating Multi-Effects (709)15Effect Reference (712)15.1Dynamics (713)15.1.1Compressor (713)15.1.2Gate (717)15.1.3Transient Master (721)15.1.4Limiter (723)15.1.5Maximizer (727)15.2Filtering Effects (730)15.2.1EQ (730)15.2.2Filter (733)15.2.3Cabinet (737)15.3Modulation Effects (738)15.3.1Chorus (738)15.3.2Flanger (740)15.3.3FM (742)15.3.4Freq Shifter (743)15.3.5Phaser (745)15.4Spatial and Reverb Effects (747)15.4.1Ice (747)15.4.2Metaverb (749)15.4.3Reflex (750)15.4.4Reverb (Legacy) (752)15.4.5Reverb (754)15.4.5.1Reverb Room (754)15.4.5.2Reverb Hall (757)15.4.5.3Plate Reverb (760)15.5Delays (762)15.5.1Beat Delay (762)15.5.2Grain Delay (765)15.5.3Grain Stretch (767)15.5.4Resochord (769)15.6Distortion Effects (771)15.6.1Distortion (771)15.6.2Lofi (774)15.6.3Saturator (775)15.7Perform FX (779)15.7.1Filter (780)15.7.2Flanger (782)15.7.3Burst Echo (785)15.7.4Reso Echo (787)15.7.5Ring (790)15.7.6Stutter (792)15.7.7Tremolo (795)15.7.8Scratcher (798)16Working with the Arranger (801)16.1Arranger Basics (801)16.1.1Navigating Song View (804)16.1.2Following the Playback Position in Your Project (806)16.1.3Performing with Scenes and Sections using the Pads (807)16.2Using Ideas View (811)16.2.1Scene Overview (811)16.2.2Creating Scenes (813)16.2.3Assigning and Removing Patterns (813)16.2.4Selecting Scenes (817)16.2.5Deleting Scenes (818)16.2.6Creating and Deleting Scene Banks (820)16.2.7Clearing Scenes (820)16.2.8Duplicating Scenes (821)16.2.9Reordering Scenes (822)16.2.10Making Scenes Unique (824)16.2.11Appending Scenes to Arrangement (825)16.2.12Naming Scenes (826)16.2.13Changing the Color of a Scene (827)16.3Using Song View (828)16.3.1Section Management Overview (828)16.3.2Creating Sections (833)16.3.3Assigning a Scene to a Section (834)16.3.4Selecting Sections and Section Banks (835)16.3.5Reorganizing Sections (839)16.3.6Adjusting the Length of a Section (840)16.3.6.1Adjusting the Length of a Section Using the Software (841)16.3.6.2Adjusting the Length of a Section Using the Controller (843)16.3.7Clearing a Pattern in Song View (843)16.3.8Duplicating Sections (844)16.3.8.1Making Sections Unique (845)16.3.9Removing Sections (846)16.3.10Renaming Scenes (848)16.3.11Clearing Sections (849)16.3.12Creating and Deleting Section Banks (850)16.3.13Working with Patterns in Song view (850)16.3.13.1Creating a Pattern in Song View (850)16.3.13.2Selecting a Pattern in Song View (850)16.3.13.3Clearing a Pattern in Song View (851)16.3.13.4Renaming a Pattern in Song View (851)16.3.13.5Coloring a Pattern in Song View (851)16.3.13.6Removing a Pattern in Song View (852)16.3.13.7Duplicating a Pattern in Song View (852)16.3.14Enabling Auto Length (852)16.3.15Looping (853)16.3.15.1Setting the Loop Range in the Software (854)16.4Playing with Sections (855)16.4.1Jumping to another Playback Position in Your Project (855)16.5Triggering Sections or Scenes via MIDI (856)16.6The Arrange Grid (858)16.7Quick Grid (860)17Sampling and Sample Mapping (862)17.1Opening the Sample Editor (862)17.2Recording Audio (863)17.2.1Opening the Record Page (863)17.2.2Selecting the Source and the Recording Mode (865)17.2.3Arming, Starting, and Stopping the Recording (868)17.2.5Using the Footswitch for Recording Audio (871)17.2.6Checking Your Recordings (872)17.2.7Location and Name of Your Recorded Samples (876)17.3Editing a Sample (876)17.3.1Using the Edit Page (877)17.3.2Audio Editing Functions (882)17.4Slicing a Sample (890)17.4.1Opening the Slice Page (891)17.4.2Adjusting the Slicing Settings (893)17.4.3Live Slicing (898)17.4.3.1Live Slicing Using the Controller (898)17.4.3.2Delete All Slices (899)17.4.4Manually Adjusting Your Slices (899)17.4.5Applying the Slicing (906)17.5Mapping Samples to Zones (912)17.5.1Opening the Zone Page (912)17.5.2Zone Page Overview (913)17.5.3Selecting and Managing Zones in the Zone List (915)17.5.4Selecting and Editing Zones in the Map View (920)17.5.5Editing Zones in the Sample View (924)17.5.6Adjusting the Zone Settings (927)17.5.7Adding Samples to the Sample Map (934)18Appendix: Tips for Playing Live (937)18.1Preparations (937)18.1.1Focus on the Hardware (937)18.1.2Customize the Pads of the Hardware (937)18.1.3Check Your CPU Power Before Playing (937)18.1.4Name and Color Your Groups, Patterns, Sounds and Scenes (938)18.1.5Consider Using a Limiter on Your Master (938)18.1.6Hook Up Your Other Gear and Sync It with MIDI Clock (938)18.1.7Improvise (938)18.2Basic Techniques (938)18.2.1Use Mute and Solo (938)18.2.2Use Scene Mode and Tweak the Loop Range (939)18.2.3Create Variations of Your Drum Patterns in the Step Sequencer (939)18.2.4Use Note Repeat (939)18.2.5Set Up Your Own Multi-effect Groups and Automate Them (939)18.3Special Tricks (940)18.3.1Changing Pattern Length for Variation (940)18.3.2Using Loops to Cycle Through Samples (940)18.3.3Using Loops to Cycle Through Samples (940)18.3.4Load Long Audio Files and Play with the Start Point (940)19Troubleshooting (941)19.1Knowledge Base (941)19.2Technical Support (941)19.3Registration Support (942)19.4User Forum (942)20Glossary (943)Index (951)1Welcome to MASCHINEThank you for buying MASCHINE!MASCHINE is a groove production studio that implements the familiar working style of classi-cal groove boxes along with the advantages of a computer based system. MASCHINE is ideal for making music live, as well as in the studio. It’s the hands-on aspect of a dedicated instru-ment, the MASCHINE hardware controller, united with the advanced editing features of the MASCHINE software.Creating beats is often not very intuitive with a computer, but using the MASCHINE hardware controller to do it makes it easy and fun. You can tap in freely with the pads or use Note Re-peat to jam along. Alternatively, build your beats using the step sequencer just as in classic drum machines.Patterns can be intuitively combined and rearranged on the fly to form larger ideas. You can try out several different versions of a song without ever having to stop the music.Since you can integrate it into any sequencer that supports VST, AU, or AAX plug-ins, you can reap the benefits in almost any software setup, or use it as a stand-alone application. You can sample your own material, slice loops and rearrange them easily.However, MASCHINE is a lot more than an ordinary groovebox or sampler: it comes with an inspiring 7-gigabyte library, and a sophisticated, yet easy to use tag-based Browser to give you instant access to the sounds you are looking for.What’s more, MASCHINE provides lots of options for manipulating your sounds via internal ef-fects and other sound-shaping possibilities. You can also control external MIDI hardware and 3rd-party software with the MASCHINE hardware controller, while customizing the functions of the pads, knobs and buttons according to your needs utilizing the included Controller Editor application. We hope you enjoy this fantastic instrument as much as we do. Now let’s get go-ing!—The MASCHINE team at Native Instruments.MASCHINE Documentation1.1MASCHINE DocumentationNative Instruments provide many information sources regarding MASCHINE. The main docu-ments should be read in the following sequence:1.MASCHINE Getting Started: This document provides a practical approach to MASCHINE viaa set of tutorials covering easy and more advanced tasks in order to help you familiarizeyourself with MASCHINE.2.MASCHINE Manual (this document): The MASCHINE Manual provides you with a compre-hensive description of all MASCHINE software and hardware features.Additional documentation sources provide you with details on more specific topics:▪Controller Editor Manual: Besides using your MASCHINE hardware controller together withits dedicated MASCHINE software, you can also use it as a powerful and highly versatileMIDI controller to pilot any other MIDI-capable application or device. This is made possibleby the Controller Editor software, an application that allows you to precisely define all MIDIassignments for your MASCHINE controller. The Controller Editor was installed during theMASCHINE installation procedure. For more information on this, please refer to the Con-troller Editor Manual available as a PDF file via the Help menu of Controller Editor.▪Online Support Videos: You can find a number of support videos on The Official Native In-struments Support Channel under the following URL: https:///NIsupport-EN. We recommend that you follow along with these instructions while the respective ap-plication is running on your computer.Other Online Resources:If you are experiencing problems related to your Native Instruments product that the supplied documentation does not cover, there are several ways of getting help:▪Knowledge Base▪User Forum▪Technical Support▪Registration SupportYou will find more information on these subjects in the chapter Troubleshooting.1.2Document ConventionsThis section introduces you to the signage and text highlighting used in this manual. This man-ual uses particular formatting to point out special facts and to warn you of potential issues. The icons introducing these notes let you see what kind of information is to be expected:This document uses particular formatting to point out special facts and to warn you of poten-tial issues. The icons introducing the following notes let you see what kind of information can be expected:Furthermore, the following formatting is used:▪Text appearing in (drop-down) menus (such as Open…, Save as… etc.) in the software and paths to locations on your hard disk or other storage devices is printed in italics.▪Text appearing elsewhere (labels of buttons, controls, text next to checkboxes etc.) in the software is printed in blue. Whenever you see this formatting applied, you will find the same text appearing somewhere on the screen.▪Text appearing on the displays of the controller is printed in light grey. Whenever you see this formatting applied, you will find the same text on a controller display.▪Text appearing on labels of the hardware controller is printed in orange. Whenever you see this formatting applied, you will find the same text on the controller.▪Important names and concepts are printed in bold.▪References to keys on your computer’s keyboard you’ll find put in square brackets (e.g.,“Press [Shift] + [Enter]”).►Single instructions are introduced by this play button type arrow.→Results of actions are introduced by this smaller arrow.Naming ConventionThroughout the documentation we will refer to MASCHINE controller (or just controller) as the hardware controller and MASCHINE software as the software installed on your computer.The term “effect” will sometimes be abbreviated as “FX” when referring to elements in the MA-SCHINE software and hardware. These terms have the same meaning.Button Combinations and Shortcuts on Your ControllerMost instructions will use the “+” sign to indicate buttons (or buttons and pads) that must be pressed simultaneously, starting with the button indicated first. E.g., an instruction such as:“Press SHIFT + PLAY”means:1.Press and hold SHIFT.2.While holding SHIFT, press PLAY and release it.3.Release SHIFT.Unlabeled Buttons on the ControllerThe buttons and knobs above and below the displays on your MASCHINE controller do not have labels.。
pandas 填空题

pandas 的填空题1. pandas is a ______ library for data analysis and manipulation in Python.(数据分析和处理)2. The main data structures used in pandas are ______ and ______.(序列和数据帧)3. DataFrame is a two-dimensional data structure consisting of a collection of ______ objects.(列)4. Series is a one-dimensional labeled array with ______.(标签)5. The ______ method can be used to convert a dictionary into a DataFrame object.(from_dict)6. The ______ attribute can be used to view the column names in a DataFrame.(columns)7. The ______ method can be used to view the first n rows of data in a DataFrame.(head)8. The ______ method can be used to view the last n rows of data in a DataFrame.(tail)9. The ______ method can be used to query a specific column in a DataFrame.(loc)10. The ______ method can be used to sort the data in a DataFrame.(sort_values)11. The ______ method can be used to fill or drop missing values in a DataFrame.(fillna)12. The ______ method can be used to perform statistical analysis on the data in a DataFrame.(describe)13. The ______ method can be used to filter the data in a DataFrame.(query)14. The ______ method can be used to perform grouping operations on the data in a DataFrame.(groupby)15. The ______ method can be used to merge the data in a DataFrame.(merge)16. The ______ method can be used to reshape the data in a DataFrame.(pivot)17. The ______ method can be used to set or reset the index in a DataFrame.(set_index/reset_index)18. The ______ method can be used to perform pivot table operations on the data in a DataFrame.(pivot_table)19. The ______ method can be used to handle time series data in a DataFrame.(resample)20. The ______ method can be used to save a DataFrame asa CSV file.(to_csv))。
mega使用教程
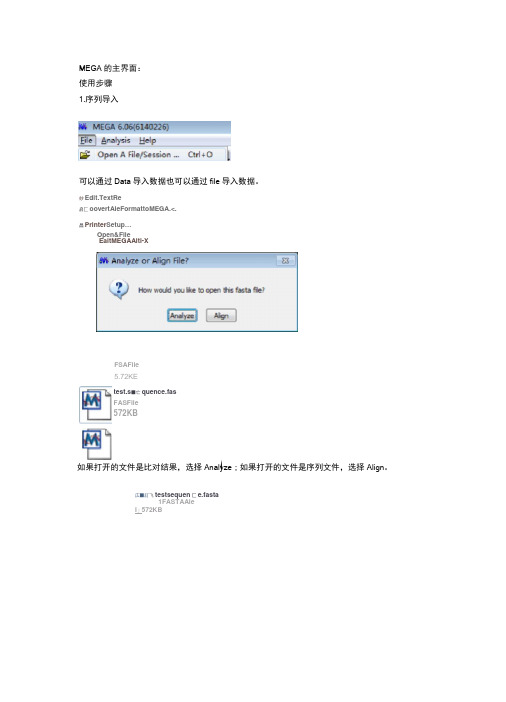
MEGA 的主界面:使用步骤1.序列导入可以通过Data 导入数据也可以通过file 导入数据。
妙Edit.TextRe启匚oovertAleFormattoMEGA.<. 昌Printer Setup …EaitMEGAAlti-X如果打开的文件是比对结果,选择Analyze ;如果打开的文件是序列文件,选择Align 。
広■訂飞testsequen 匚e.fasta1FASTAAleI 」572KBOpen&Filetest.s ■亡quence.fas FASFile572KBFSAFile5.72KE另外双击这些后缀名文件即可自动导入序列,导入后会弹出MEGA比对界面。
如果fasta序列导入报错,多是因为序列长度不同导致:fPTFTCN 黜aVLRLFPGERrHFTDEELWQYLSREVCSSPLPA5IIPEETJVCHMPWDLPS-NLEKEaYrFSTElEJiKYPWRTRSNRftlSSGYWKSLrGLEK^imSHGNQlVGL fbQLSLPPGFRnPTDEfLLVQYLCm^GYHrSL^IGDI&LYKr&FWDLPS^GTCFTin^GEYKCT-YLGKrt^rSEKEKyrFSPRDRKYPHGSRPHRVAGWW IkQ15IZETl3yRETFTE>EELm B Ef :LCR33iASHI?F5:LQLIA£:IDL¥KEI>E r WVi^5KaXFGEKEWYFi _£E !RB5i K¥F3G5S:PNHVAGSG¥HK5L HGTDKV]^TE :EREWZ A-PF-SraFHFrDEELILHYLI^t^SSSPVPLSIIftDVDIYKSDPWDLPAKAFrGEKEWYrFSPRD^PWGMPSrRAAASSYWKATSTDKLIAVPNGEGTHE^I百UFF 囲皿民卞亍诜丘1孙爬匸址m-Pt 即虑陀EFT 玳咗bPW 血塞自垃空已MFFM 亍匪站HPM 曲a 服迂弟品YIgm 』買茁二虽買IM 』玄品£MDBTJKLVKNGVIiRLPPGFRFHPTDEELWQYLKRKVCSSPLPASIIPEFDVCRAT-PtfDIxPGNLEKERYFFSTRE TDHIMEEYRLSSSPPSSMGPTQNWV1CRIFLKKRAGNKNDDODGESRNLRHJiNNNN D SDQIEIITT DQTDDKTK>AT4G274LQMGVREKDPLAQLSLPPGFRFYPTDEELLVQYLCRKVAGYHFSLQVIGDIDLYKFDPrJDLPSKQTCFTFVGEYNC GTKKA1VFYAGKAPKGTKTNWIMHEYRLIEHSRSHGSSRLDDWVLCRIYKRTSGSQFtQAVTPVQACREEHSTNGAPTNGLPSYGGYDAFRAAZGEAESGH^ZWRQQNS5GLTQSFGYSSSGFGV5G<JTETFRQ>AT3G1550QMGLQE.LDPLAQLSLPPGFREYPTDEELMVEYLCRKAAGHDFSLQLIAEIDLifKFDP»VLP£KALrGEKEWYFFSKTNWIMHE YRLIEPSRRNGSTKLDDWVLCRIYKKQTSAQKQAYNNLMTSGREYSi™STSSSSHQYDDVLESLELGLSRNVPSIRYGDGGT QQQTEGIPRFNNNSDVSftWQGFSVDPVNGFGYSGQQSS^FeFI如果序列长度不同,可以采用新建文件,将序列文件导入的方法 步骤:Align f Edit/BuildAlignment f createanewalignment f Data f open f RetrievesequencesfromFile将复制输入的序列另存输出看看 步骤:datafExportalignmentffastaformat■•■Fas-tapm 注■riTor^Ewpectedserjuenc -E-characleir,twt fou<id mSequencesaredIffereinilengihE.0**fihec5 EFVGIKK M TPOrarHFF DEELVLMT :5TEGQR¥C:|'—Irr ■TJ t.:iiZ3曰MAC-3|$-f*£3|kA A-criinH-fl,EepIcrKri'LJ::|WEditsearchgmffflfcWsbheflLienwilLP □匚r»阳触I*Q S Qp«nRacnpar9匚1口“PTylC甘BFMIK斷]MM^iiSE匚fed»Sfc-E®»rlAJ^nrw*e*rtD N*S HMJBW:* PjotewSe^jenceEi',lTranqlriBi/L-nlTAnilfliw■^vlmEOririhraLiTi^rT^lilri* JndrattlMi匚CIl'IjiJtaniSiiE■a R总昌国⑪甜段匙呈髙居屍〒$口>fiT5M37*»riL-q Ml.r'1t±iicii i i HniKD-ii>i I IHI ba■Qim■Lx■KiqQin r i'HMHad.Lki MI*I od I.uIDE SDLVIiFHPhe L FEMrt.¥CEKE?VF FSHRDRtf1FHCERFW%ft*CTn¥M^TGWH卩I GH Hl.I KI I1TH-E IRII YHInWlFII^M:!I FIHIIEF-H«LUUWWI H A:I VFIHHfa I IR KTIF~H4HI M~r I KViri DHWUC^EFKU[K.E.»fiLEIlFI«&TENFS5ECLLQKiAFUFqFQVI^SDFliD-^FOWF EQKP I■-liT^Ll^liiiHhrAiiij.tita NUiriLHLE屮I*HP!纯呂対II MSIJFLI I I WM^IMWLP&NHHEIHVl TU汕VMKiiT;I D*IASUTEIUH]>IUCLKKTLVFYKCHPPIHSiltTDtf[«HEWLSiSF H朋旬朋切沖4UDs.|tHLkl«Hiwi>^Ult I[I I Irk I IrWIHKll-1U卜摘IQ J I M-N,LLrS£D£LTSSilMfSiFA-■■-、口TM空片WHLUEEKSPLAHLELPPCF RFVPTBE[LLUqYLCRKIABYHFELqUJCO I DL Y轴hI引PL P5H L I l;lHi Wil^I'HPH.lYFHIi^UJ'rflCilAGkKYLMA IG K t>M]I mtUIHIII;jKHJI i>i¥HI;KHI'PStiCS^KLRDUULiCRIVKKirTCEiH lAUlTFVq^iCREEH5TH€S¥S5^SSqL D4ULKF PEI Hrn nMn<\iHEI Hrirt nr IHZI i'^vsiEWhAii IMAI EI AI^EIIUMH q^HNKCi ignisiwNs^u^fil3l;l^r.fllfeH[±QELbPI.A4L£LRf£FRFVPT@EfLHI£¥LCRHRACHDFSLQLJAE LDL^FbFWLPSHPmilEHl'BFRLARFinVLHAT l;T"HU I髯W CZIWUni KKII UIV I ILKAP Ifi H HIHUI HMI VRII B-Pkko FSA4k^!rM.MrS;£F£tfSwt£tmfX£HlQyhtOLESLHE EfeHft^LCFflRC^HALPHiAlignByMuscle AAA.AJ&AProteinSequencesMarlc/UnmarkSiteCtrl+M Specie a/Atibrv2+WP011931144.12+CAA44467H2氛A^N16207H1 4.WP_01229^898a AAAK1«222.1B Wr_D1203^132.1 7,KDP92253.1DeleteGap-OnlySites|A|A|AA|A|AAIVALAAA-LLL^VAI AjA|Aft|A|AA||__._.-|A|A|A|AA|AA||VV||V|A|L VL|L序列长度都被用横线补齐了2.池MHAlEgnmentExploreritesLsequEncz&fm硏DataEditSearch|AlignmentWebSequencerDi^pl^yHelpIV Signby匚tustalW选择muscle或者clustalw进行比对:clustalw—般用于DNA,muscle多用于蛋白。
桑树桑黄与杨树桑黄的分子辨别

桑树桑黄与杨树桑黄的分子辨别SONG Ji-Ling;LU Na;WANG Wei-Ke;YUAN Wei-Dong;LI Hai-Bo;CHENG Jun-Wen;KANG Xue-Ping;YAN Jing【摘要】为准确鉴别桑黄(Sanghuangporus)近缘种桑树桑黄(S.sanghuang)与杨树桑黄(S.vaninii)、暴马桑黄(S.baumii),本研究基于核糖体基因rDNA内转录间隔区(internal tran-scribed spacer,ITS)序列分析技术,对桑黄真菌进行了系统发育分析与近缘种的分子辨别研究.结果表明,在N J系统发育树上,23个桑树桑黄、11个杨树桑黄和6个暴马桑黄菌株各自以很高的Bootstrap支持率聚为了三个独立的分支,种间差异明显.依据桑树桑黄和杨树桑黄的rDNA ITS序列差异,设计两对引物Sv_U1/Sv_L和Sv_U2/Sv_L,均可特异性地扩增杨树桑黄478 bp和651 bp的ITS 片段,而不扩增桑树桑黄的ITS片段,因而可用于两个桑黄近缘种的快速分子辨别.本研究为探讨桑黄孔菌属真菌的系统发育关系提供了参考,并为桑黄种间的准确辨别提供了一种有效的分子辅助手段.【期刊名称】《四川大学学报(自然科学版)》【年(卷),期】2019(056)004【总页数】6页(P765-770)【关键词】桑黄;鉴别;分类;内转录间隔区;特异性引物【作者】SONG Ji-Ling;LU Na;WANG Wei-Ke;YUAN Wei-Dong;LI Hai-Bo;CHENG Jun-Wen;KANG Xue-Ping;YAN Jing【作者单位】;;;;;;;【正文语种】中文【中图分类】Q9491 引言桑黄是一种珍贵的多年生大型药用真菌,隶属于真菌界Fungi、担子菌门Basidiomycota、伞菌纲Agaricomycetes、锈革孔菌目Hymenochaetaceae、锈革孔菌科Hymenochaetaceae、桑黄属sanghuangporus,因其生长在桑属(Morus)植物上且子实体呈黄褐色而得名[1,2].在《药性论》、《本草纲目》和《神农本草经》等历代本草著作中均有桑黄及其药效的明确记载[3,4].现代研究表明,桑黄具有显著的抗肿瘤、抗氧化、增强免疫力和消炎抗菌等作用,是国际公认的生物抗癌天然产品[5,6],已成为国内外医药制剂和保健品行业研发的热点.目前报道的桑黄属真菌有12种,其中7个种在中国有分布,分别为高山桑黄S.alpinus、暴马桑黄S.baumii、小孔忍冬桑黄S.lonicericola、桑树桑黄S.sanghuang、杨树桑黄S.vaninii、锦带花桑黄S.weigelae和环区桑黄S.zonatus[4].此类真菌的子实体均呈马蹄形,只是在形态特征和颜色上存在细微差别,仅仅依赖这些形态特征很难准确鉴定菌株的分类归属[7].尤其是对野外采集标本的鉴定上,由于桑黄子实体颜色常常易受环境因子、子实体生长期等的影响而导致主观判断出现偏差.对于长期保藏的干标本,仅凭子实体形态特征与颜色更是难以准确区分.因此,利用分子技术手段辅助鉴定桑黄真菌具有重要意义.真菌核糖体基因rDNA的转录间隔区(Internal Transcribed Spacers,ITS)序列由于进化速率快、稳定性好和丰富的信息位点,被广泛应用于真菌的系统发育分析和物种间的分子鉴定研究[8-11].在本研究中,我们对收集的36个桑黄菌株进行ITS测序,基于桑树桑黄、杨树桑黄和爆马桑黄的ITS序列进行系统发育树构建,为深入探讨桑黄孔菌属的系统发育关系提供证据;并进一步通过ITS序列数据的比对分析,设计ITS特异引物,为桑黄真菌的种间辨别提供可靠便捷的分子辅助手段.2 材料与方法2.1 供试材料供试菌株共36个,其中23个收集自我国各主要保藏单位和栽培地区,13份采集自浙江杭州地区.菌种保存于杭州市农业科学研究院(表1).表1 用于本研究的36个供试桑黄菌株Tab.1 Thirty-six Sanghuangporus strains used in this study序号Number菌株编号Strain Number来源Source1SH1采集于桐庐保安,杭州2SH2采集于桐庐后岩,杭州3SH3采集于临安,杭州4SH4采集于淳安,杭州5SH5采集于桐庐凤川,杭州6SH6采集于淳安临歧,杭州7SH7采集于淳安临歧,杭州8SH8采集于淳安梓桐,杭州9SH9采集于淳安梓桐,杭州10SH10采集于淳安姜家,杭州11SH11采集于衢州开化,杭州12SH12采集于淳安,杭州13SH13采集于淳安,杭州14SH14引进于淳安微生物研究所15SH15引进于淳安微生物研究所16SH16引进于四川省农业科学研究院17SH17引进于韩国18SH18引进于延边特产研究院19SH19引进于福建农业大学20SH20引进于福建农业大学21SH21引进于福建农业大学22SH22引进于福建农业大学23SH23引进于中国农业微生物菌种保藏中心24SH24引进于中国农业微生物菌种保藏中心25SH25引进于四川省农业科学研究院26SH26引进于吉林27SH27引进于黑龙江伊春28SH28引进于丽水市农业科学研究院29SH29引进于安徽30SH30引进于安徽31SH31引进于安徽32SH32引进于六安,安徽33SH33引进于安徽34SH34引进于浙江工业大学35SH35引进于延边特产研究院36SH36引进于福建2.2 方法2.2.1 菌丝体培养所有供试菌株转接于PDA平板.从在PDA平板上培养7 d的菌落边缘取2 mm×2 mm的菌丝块,接种于PDA液体培养基中,在27 ℃培养10 d,收集菌丝,无菌水漂洗两次,用无菌滤纸吸干水分,于-80 ℃保存备用.2.2.2 基因组DNA提取取0.5 g菌丝体,采用新型快速植物基因组DNA提取试剂盒(BioTeke,北京)提取DNA.提取后的DNA经NanoDropTM 2000(Nanodrop Technologies,South Logan,Utah,USA)测定浓度,并经1.5%琼脂糖凝胶电泳检测,于-20℃保存备用.2.2.3 rDNA ITS扩增20μL ITS-PCR扩增反应体系为:2×TSINGKE Master Mix 10 μL,ITS引物对[12]各1 μL,20 ng·μL-1模板DNA 3 μL,ddH2O 5 μL.扩增反应在Life ECO型扩增仪(Bioer,杭州博日科技)上进行,94 ℃预变性6 min后;94 ℃变性40 s,54.8 ℃退火40 s,72 ℃延伸2 min,共30个循环;最后于72 ℃补平7 min,终止温度为4 ℃.2.2.4 rDNA ITS序列的克隆和测序采用普通琼脂糖凝胶DNA回收试剂盒(TIANGE,北京天根)从1.5%的琼脂糖凝胶中回收ITS片段.ITS片段与pGEM-Teasy 载体连接,4 ℃过夜.连接产物转化于大肠杆菌感受态细胞DH5α中,经蓝白斑筛选后,每个样品均随机挑选5个阳性克隆交付杭州擎科生物技术公司完成测序.2.2.5 系统发育分析 36个本次测序的ITS序列连同16个从GenBank下载的ITS 序列,共52个序列用于系统发育分析.16个ITS序列为戴玉成等[13]发表的桑黄属真菌基源物种(S.sanghuang、S.vaninii 和S.baumii)的rDNA序列,包括S.sanghuang 序列6个(Accession nos.AF534064.1、JN642576.1、JN642577.1、JN642586.1、JN642589.1和JN794061.1)、S.vaninii序列4个(Accession nos.HM584811.1、HM584811.1、JN642591.1和JN642593.1)、S.baumii序列6个(Accession nos.HM584807.1、JN642565.1、JN642567.1、JN642568.1、JN642570.1和JX 069836.1).采用Clustal X v.1.8[14]对52个ITS序列进行多重对位排列(multiplealignments).用MEGA v.5.0[15]进行系统发育分析,以Kimura 2-parameter模型计算遗传距离,所有对位排列结果中的空位作完全删除处理.用NJ法(Neighbor-joining method)构建系统进化进化树,Bootstrap testing为1000 replic-ates.2.2.6 ITS特异引物设计与PCR扩增根据桑黄真菌S.sanghuang和S.vaninii的ITS序列比对结果,设计可用于鉴别S.sanghuang和S.vaninii的ITS特异引物.引物设计采用Oligo 6.54软件(MBI,USA),引物合成由杭州擎科梓熙生物技术有限公司完成.利用ITS特异引物对桑黄真菌的PCR扩增参照“1.4 rDNA ITS扩增”.通过逐步提高 PCR 反应的退火温度,取得对ITS特异引物对扩增的最佳退火温度(表2).表2 辨别S. sanghuang与S. vaninii 的ITS引物序列及其最适退火温度Tab.2 Sequence of ITS primers used for distinguishing S.sanghuang fromS.vaninii and its optimal annealing temperatureITS引物ITS primer序列Sequence(5′-3′)退火温度Annealing temperature(℃)扩增S.vaninii的片段Amplified fragment from S.vaninii(bp)Sv_U1GAGTGCGTCGGGTGAAGACTSv_LAGGAGGTAACAACAACTCCTT5 4.8S.vaninii(478)Sv_U2TGCGCATGTGCACGGCCTTSv_LAGGAGGTAACAACAACTCCTT54. 8S.vaninii (651)注:字母U和L分别代表上、下游ITS特异引物,字母Sv代表杨树桑黄Note:The letters U and L refer to the specific upstream and downstream primer for the ITS region,respectively.The letter Sv refer to S.vaninii3 结果与分析3.1 基于rDNA ITS序列的系统发育分析基于52个桑黄属真菌的ITS序列对位形成的数据集(Dataset)总共包含了744个碱基位点,其中保守位点(conserved)370个,变异位点(variable)337个,简约信息位点(parsimony informative)104个,单突变位点(singleton)226个.NJ系统发育树(图1)显示,49个菌株构成了3个独立的类群Group Ⅰ、Group Ⅱ和Group Ⅲ,分别包含了29个S.sanghuang 菌株(供试菌株SH1~SH23)、14个S.vaninii 菌株(供试菌株SH24~SH33)、6个S.baumii 菌株和3个桑黄真菌以外的菌株(供试菌株SH34~SH36,分别为Sanghuangporus spp、Cordyceps militaris和Trametes hirsuta),Group Ⅰ、Group Ⅱ和Group Ⅲ的Bootstrap支持率分别为99%、99%和98%.这表明S.sanghuang、S.vaninii和S.baumii这三个桑黄真菌的种间存在明显差异,且S.sanghuang与S.vaninii的亲缘关系较S.baumii接近.这与形态特征上三种真菌的存在的些许差别、以及寄生树种的不同具有一致性. 3.2 ITS 特异引物设计对6个S.vaninii菌株(SH24、SH26、SH27、SH28、SH30、SH32)和6个S.sanghuang菌株(SH1、SH2、SH14、SH16、SH18、SH23)测序获得的12个ITS序列经多重对位排列产生了820个碱基位点.根据对位碱基位点的差异比较,设计出了可特异性扩增S.vaninii菌株而不扩增S.sanghuang的两对ITS引物Sv_U1/Sv_L和Sv_U2/Sv_L.图2显示了上游引物Sv_U1所在的第241~300位点区段、上游特异引物Sv_U2所在的第1~60位点区段,以及下游引物SS_L所在的第721~780位点区段.两对ITS特异引物的碱基序列、退火温度及扩增的ITS片段长度详见表2.图1 基于桑黄属真菌rDNA ITS序列构建的系统发育树采用Kimura two-parameter distance mode;完全删除;bootstrap=1000;节间数值代表bootstrap 值(低于50的数值不显示)Fig.1 Neighbor-joining phylogenetic tree generated for Sanghuangporus species based on rDNA ITS sequencesNucleotide:Kimura two-parameter distance model,completedeletion,bootstrap=1000;the numbers in each node represent bootstrap support values (those lower than 50 are not shown).图2 辨别S.sanghuang与S.vaninii 的ITS特异引物位点箭头所指分别为上下游引物Sv_U1、Sv_U2和 Sv_LFig.2 Localization and specificity of the specific ITS primers used for distinguishing S.sanghuang from S.vaniniiThe upstream and downstream primers (Sv_U1,Sv_U2 andSv_L)are indicated by arrows in the sequence3.3 S.sanghuang与S.vaninii 的分子鉴别分别以10个S.vaninii菌株和23个S.sanghuang菌株的基因组DNA为模板,以表2所示的ITS特异引物对进行PCR扩增.在最适退火温度下,两对特异引物Sv_U1/Sv_L和Sv_U2/Sv_L分别从10个S.vaninii菌株中扩增出了唯一的一条ITS条带,长度分别为478 bp和651 bp,而没有从23个S.sanghuang菌株中扩增出任何条带(图3).这一结果表明,本研究基于S.vaninii和S.sanghuang的ITS序列差异设计的两对特异引物可以用于S.vaninii与S.sanghuang的分子鉴别.图3 S.sanghuang与S.vaninii 菌株的ITS特异引物扩增图谱A:Sv_U1/Sv_L;B:Sv_U2/Sv;M:DNA分子量标准;白色箭头所指为目的条带;泳道1~23对应表1所列的桑黄菌株SH1~23;泳道24~33对应表1所列的杨黄菌株SH24~33Fig.3 The PCR amplification pattern of S.sanghuang and S.vaninii strains with specific ITS primersA:Sv_U1/Sv_L;B:Sv_U2/Sv;M:DNA marker;The white arrow shows the location of target band;Lanes 1~23 and Lanes 24~33 correspond to the S.sanghuang strains SH1~23 and the S.vaninii strains SH24~33 listed in Table 1,respectively.4 讨论随着分子生物学技术的发展,位于18S rDNA及28S rDNA间的rDNA内转录间隔区(internal transcribed spacer,ITS)序列分析技术由于进化速率快、稳定性好和测序方便,已广泛运用到多种真菌种属水平的分类鉴定及遗传多样性的研究[16-19].利用ITS序列分析技术,我们鉴定了杭州、衢州及安徽等地采集到的36个桑黄样品,根据ITS序列聚类分析,将36个桑黄样品明显区分为3个类群.其中,杭州地区采集的23个桑黄菌株为一组,为S.sanghuang,序列长度814 bp;收集于吉林、四川等地的10个桑黄菌株为一组,为S.vaninii,序列长度为773 bp;其余的3个菌株为一组,为sanghuangporus以外的其他菌株.核糖体基因rDNA ITS区段的序列差异比较被广泛用于近缘物种的辨别.在实际应用中,通常需要ITS区段的克隆测序和序列的同源性比对,比较耗时耗力.通过ITS特异引物设计,使得近缘物种的差异直观地表现为ITS特异扩增条带的有无,可非常便捷地辨别近缘物种,例如美味牛肝菌Boletus edulis Bull.与铜色牛肝菌B.aereus Bull.、褐红牛肝菌B.pinophilus Pilát &Dermek、夏生牛肝菌B.aestivalis (Paulet)Fr.[20,21]、牛肝菌属卷边组Boletus sect.Appendiculati 物种[10]、以及松材线虫Bursaphelenchus xylophilus 与拟松材线虫B.mucronatus 伪伞真滑刃线虫B.fraudulentus等[22].本文通过ITS特异引物的开发,可以进一步从PCR 电泳图上直观地反映了两个相似种在基因组上的差异,也验证了ITS特异引物用于S.vaninii与S.sanghuang分子鉴定的可靠性.该技术手段适用于野外采集的新鲜样本、陈年保存的干标本、古老标本或菌丝体样本,对于仅含有微量DNA的样本也可以快速完成鉴别.在研究过程中发现,由于S.sanghuang与S.vaninii的ITS1-5.8S-ITS2序列分别为773 bp和814 bp,两者的ITS序列差异很小,很难设计出用于鉴定S.sanghuang 的特异引物,因此,进一步设计用于鉴定S.sanghuang的特异引物也是今后值得尝试的技术思路.此外,在本研究的基础上,我们将收集收集更多的桑黄种质资源,包括暴马桑黄S.baumii、大孔忍冬桑黄S.lonicerinus和小孔忍冬桑黄S.lonicericola 等,进一步开发ITS特异引物用于更多相似种间的分子识别,为桑黄真菌各物种的准确鉴别提供可靠便捷的分子技术手段.参考文献:【相关文献】[1] 张小青,戴玉成.中国真菌志 [M].北京:科学出版社,2005:117.[2] Zhou L W,Vlasák J,Decock C,et al.Global diversity and taxonomy of the Inonotus linteus complex (Hymenochaetales,Basidiomycota):Sanghuangporus gen.nov.,Tropicoporus excentrodendri and T.guanacastensis gen.et spp.nov.,and 17 new combinations [J].Fungal Divers,2016,1:335.[3] 孙培龙,徐双阳,杨开,等.珍稀药用真菌桑黄的国内外研究进展 [J].微生物学通报,2006,33:119.[4] 吴声华,黄贯中,陈愉瓶,等.桑黄的分类及开发前景 [J].菌物研究,2016,14:187.[5] 朱琳,崔宝凯.药用真菌桑黄的研究进展 [J].菌物研究,2016,14:201.[6] 梁晓薇,刘远超,黄龙花,等.桑黄孔菌属的研究进展 [J].微生物学杂志,2017,37:98.[7] 戴玉成,崔宝凯.药用真菌桑黄种类研究 [J].北京林业大学学报,2014,36:1.[8] 张文娟,康帅,魏锋,等.基于ITS序列分析鉴别冬虫夏草与古尼虫草 [J].药物分析杂志,2015,35:1551.[9] 李莹,李莉,刘艳玲,等.基于ITS序列分析对杏鲍菇菌种的鉴定 [J].微生物学杂志,2014,34:62.[10] 魏海龙,李海波,王丽玲,等.牛肝菌属卷边组Boletus sect.Appendiculati物种的分子识别 [J].菌物学报,2014,3:242.[11] 黄丽洋,石卉,王晓婷,等.野生桑黄菌株的分离、鉴定和次生代谢物分析 [J].中草药,2013,44:3394.[12] White T J,Bruns T,Lee S,et al.Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics[A]//Innis M A,Gelfand D H,Sninsky J J,et al.PCR protocols:a guide to methods and applications.New York:Academic Press,1990:315.[13] Tian X M,Yu H Y,Zhou L W.Phylogeny and taxnomy of the Inonotus linteus complex [J].Fungal Divers,2013 ,58:159.[14] Thompson J D,Gibson T J,Plewniak F,et al.The Clustal X windows interface:flexible strategies for multiple sequence alignment aided by quality analysis tools [J].Nucleic Acids Res,1997,25:4876.[15] Tamura K,Peterson D,Peterson N,et al.MEGA5:molecular evolutionary genetics analysis using maximum likelihood,evolutionary distance,and maximum parsimonymethods [J].Mol Biol Evol,2011,28:2731.[16] 谢丽源,张勇,彭金华,等.桑黄真菌分子鉴定及遗传多样性分析 [J].菌物学报,2010,29:347.[17] Lim Y W,Lee J S,Jung H S.Type studies on Phellinus baumii and Phellinus linteus [J].Mycotaxon,2003,85:201.[18] 高凯,杜明,吕英华,等.10株桑黄菌基于rDNA ITS序列的分子鉴定 [J].蚕业科学,2010,36:0584.[19] 张文隽,吴亚召,雷萍,等.秦巴山区野生桑黄rDNA ITS序列及亲缘关系分析 [J].中国食用菌,2015,34:50.[20] Mello A,Ghignone S,Vizzini A,et al.ITS primers for the identification of marketable boletes [J].J Biotechnol,2006,121:318.[21] Lian B,Zang J P,Hou W G,et al.PCR-based sensitive detection of the edible fungus Boletus edulis from rDNA ITS sequences [J].Electronic J Biotechn,2008,11:1.[22] Filipiak A,Wieczorek P,Tomalak M.Multiplex polymerase chain reaction for simultaneous detection and identification of Bursaphelenchus xylophilus,B.mucronatus and B.fraudulentus-three closely related species within the xylophilus group[J].Nematology,2017,19:1.。
UVM1.1应用指南及源代码分析_20111211版

而后半部分(第 10 到第 19 章)则介绍 UVM 背后的工作原理,用户群相对稀少。 通常来说,一般的用户只要看懂前半部分就可以了。但是我想,世上总有像我一样 有好奇心的人,不满足知其然再不知其所以然,会有人像我一样,会因为一个技术 问题而彻夜难眠,如果你是这样的人,那么恭喜,这本书的后半部分就是为你准备 的。
UVM1.1 应用指南及 源代码分析
UVM1.1 Application Guide and Source Code Analysis
张强 著
在这里,读懂 UVM
序
写这本书的难度超出了我的预料。从 8 月初开始写,一直到现在,4 个多月的 时间,从刚开始的满含激情,到现在的精疲力尽。现在写出来的东西,距离我心目 中的作品差距十万八千里,有太多的地方没有讲述清楚,有太多的地方需要仔细斟 酌,有太多的语句需要换一种表述方式。
8. register model的使用 ..............................................................................................125
8.1. register model简介...................................................................................125
写这本书,只是想把自己会的一点东西完全的落于纸上。在努力学习 UVM 的 过程中,自己花费了很多时间和精力。我只想把学习的心得记录下来,希望能够给 后来的人以启发。如果这本书能够给一个人带来一点点的帮助,那么我的努力就不 算是白费。
这本书的前半部分(第 1 到第 9 章)介绍了 UVM 的使用,其用户群较为广泛;
DS2208数字扫描器产品参考指南说明书

-05 Rev. A
6/2018
Rev. B Software Updates Added: - New Feedback email address. - Grid Matrix parameters - Febraban parameter - USB HID POS (formerly known as Microsoft UWP USB) - Product ID (PID) Type - Product ID (PID) Value - ECLevel
-06 Rev. A
10/2018 - Added Grid Matrix sample bar code. - Moved 123Scan chapter.
-07 Rev. A
11/2019
Added: - SITA and ARINC parameters. - IBM-485 Specification Version.
No part of this publication may be reproduced or used in any form, or by any electrical or mechanical means, without permission in writing from Zebra. This includes electronic or mechanical means, such as photocopying, recording, or information storage and retrieval systems. The material in this manual is subject to change without notice.
英语技术写作试题及答案

英语技术写作试题及答案一、选择题(每题2分,共20分)1. The term "API" stands for:A. Application Programming InterfaceB. Artificially Programmed IntelligenceC. Advanced Programming InterfaceD. Automated Programming Interface答案:A2. Which of the following is not a common data type in programming?A. IntegerB. StringC. BooleanD. Vector答案:D3. In technical writing, what is the purpose of using the term "shall"?A. To indicate a requirement or obligationB. To suggest a recommendationC. To express a possibilityD. To denote a future action答案:A4. What does the acronym "GUI" refer to in the context of computing?A. Graphical User InterfaceB. Global User InterfaceC. Generalized User InterfaceD. Graphical Unified Interface答案:A5. Which of the following is a correct statement regarding version control in software development?A. It is used to track changes in software over time.B. It is a type of software testing.C. It is a method for encrypting code.D. It is a way to compile code.答案:A6. What is the primary function of a compiler in programming?A. To debug codeB. To execute codeC. To translate code from one language to anotherD. To optimize code for performance答案:C7. In technical documentation, what does "RTFM" commonly stand for?A. Read The Frequently Asked QuestionsB. Read The Full ManualC. Read The File ManuallyD. Read The Final Message答案:B8. Which of the following is a common method for organizing code in a modular fashion?A. LoopingB. RecursionC. EncapsulationD. Inheritance答案:C9. What is the purpose of a "pseudocode" in programming?A. To provide a detailed step-by-step guide for executing codeB. To serve as a preliminary version of code before actual codingC. To act as an encryption for the codeD. To be used as a substitute for actual code in production答案:B10. What does "DRY" stand for in software development?A. Don't Repeat YourselfB. Data Retrieval YieldC. Database Record YieldD. Dynamic Resource Yield答案:A二、填空题(每空2分,共20分)1. The process of converting a high-level code into machine code is known as _______.答案:compilation2. In programming, a _______ is a sequence of characters that is treated as a single unit.答案:string3. The _______ pattern in object-oriented programming is a way to allow a class to be used as a blueprint for creating objects.答案:prototype4. A _______ is a type of software development methodology that emphasizes iterative development.答案:agile5. The _______ is a set of rules that defines how data is formatted, transmitted, and received between software applications.答案:protocol6. In technical writing, the term "should" is used toindicate a _______.答案:recommendation7. The _______ is a type of software that is designed to prevent, detect, and remove malicious software.答案:antivirus8. A _______ is a variable that is declared outside the function and hence belongs to the global scope.答案:global variable9. The _______ is a programming construct that allows you to execute a block of code repeatedly.答案:loop10. In software development, the term "branch" in version control refers to a _______.答案:separate line of development三、简答题(每题10分,共40分)1. Explain the difference between a "bug" and a "feature" in software development.答案:A "bug" is an unintended behavior or error in a software program that causes it to behave incorrectly or crash. A "feature," on the other hand, is a planned and intentional part of the software that provides some functionality or capability to the user.2. What is the significance of documentation in technical writing?答案:Documentation in technical writing is significant as it serves to provide detailed information about a product or system, making it easier for users, developers, and other stakeholders to understand its workings, usage, and maintenance. It is crucial for training, troubleshooting, and future development.3. Describe the role of a software architect in a software development project。
tfautomodelforsequenceclassification类 -回复

tfautomodelforsequenceclassification类-回复[tfautomodelforsequenceclassification类] 是一个基于TensorFlow 框架的自然语言处理模型类,用于序列分类任务。
它是TensorFlow 模型库中的一个关键类,提供了构建和训练序列分类模型的核心功能。
本文将通过一步一步的回答,介绍tfautomodelforsequenceclassification 类的重要特性和用法,以及如何使用它来构建和训练序列分类模型。
我们还将探讨tfautomodelforsequenceclassification 类背后的原理和关键概念,以帮助读者更好地理解该类的工作原理。
首先,让我们来了解一下什么是序列分类任务。
在自然语言处理领域,序列分类是指根据文本序列的内容对其进行分类。
例如,情感分析任务就是一种典型的序列分类任务,目标是根据输入的句子判断其情感倾向是积极的、消极的还是中性的。
tfautomodelforsequenceclassification 类就是为了解决这类序列分类任务而设计的。
tfautomodelforsequenceclassification 类的最大特点之一是其灵活性。
该类提供了多种模型架构的选择,可以根据任务的特点和需求选择合适的模型。
例如,我们可以选择使用BERT、Electra、GPT-2 等预训练模型来构建序列分类模型。
这些预训练模型能够通过大规模的数据进行预训练,具有优秀的语言表示能力。
tfautomodelforsequenceclassification 类还支持对预训练模型进行微调,以适应特定的序列分类任务。
在开始构建和训练序列分类模型之前,我们需要准备好训练数据。
tfautomodelforsequenceclassification 类要求输入数据以特定的格式提供。
通常情况下,我们将训练数据拆分为训练集和验证集,用于模型的训练和评估。
青少年创意编程大赛题库

青少年创意编程大赛题库English Answer:1. Explain the difference between a variable and a constant.Variable: A variable is a container that can store data and change its value over time.Constant: A constant is a container that stores data that cannot change its value.2. What is the difference between a function and a procedure?Function: A function is a block of code that performs a specific task and returns a value.Procedure: A procedure is a block of code that performs a specific task but does not return a value.3. What are the different data types in programming?Integers: Whole numbers, such as 1, 2, and -3.Floats: Decimal numbers, such as 1.23, 4.56, and -7.89.Strings: A sequence of characters, such as "hello", "world", and "Python".Booleans: True or false values.Lists: A collection of items in a specific order.Tuples: A collection of items in a specific order that cannot be changed.Dictionaries: A collection of key-value pairs.4. What is the purpose of loops in programming?Loops: Loops allow you to execute a block of codemultiple times based on a specific condition.5. What are the different types of loops in programming?For loops: Execute a block of code a specific numberof times.While loops: Execute a block of code while a condition is true.Do-while loops: Execute a block of code at least once, and then repeat it while a condition is true.6. What are the different control structures in programming?Conditional statements: Control the flow of execution based on specific conditions.If statements: Execute a block of code if a conditionis true.Else statements: Execute a block of code if acondition is false.Elif statements: Execute a block of code if acondition is true and other specific conditions are false.7. What are the different types of operators in programming?Arithmetic operators: Perform arithmetic operations, such as addition, subtraction, multiplication, and division.Assignment operators: Assign a value to a variable.Comparison operators: Compare two values to determineif they are equal, not equal, greater than, less than, greater than or equal to, or less than or equal to.Logical operators: Combine multiple Boolean conditions to create a single Boolean expression.8. What is the difference between a library and aframework?Library: A collection of reusable code that can be added to a program to extend its functionality.Framework: A collection of libraries and tools that provide a specific set of functionality and can be used to build applications.9. What is the purpose of documentation in programming?Documentation: Provides information about a program, its components, and how to use it.10. What are the different types of documentation in programming?In-line documentation: Comments within the code that explain what the code does.External documentation: User manuals, tutorials, and other documents that provide information about the program.中文回答:1. 变量和常量的区别是什么?变量,变量是一个可以存储数据的容器,并且可以在运行期间改变其值。
fastapi basemodel 时间参数

FastAPI BaseModel 时间参数引言FastAPI 是一个现代高性能的 Python web 框架,它提供了许多强大的功能和特性,以帮助我们快速构建高效的 API。
其中之一就是BaseModel类,它是 FastAPI 框架中一个非常重要的概念。
BaseModel允许我们定义数据模型,以及对数据进行验证和转换。
在这篇文章中,我们将重点讨论如何在BaseModel中使用时间参数,以及如何处理时间相关的操作。
BaseModel 概述在使用 FastAPI 构建 API 的过程中,BaseModel是一个非常重要的概念。
它是pydantic 库的一部分,用于数据模型的定义和验证。
通过继承BaseModel类,我们可以定义一个具有验证和转换功能的数据模型。
在 FastAPI 中,我们可以在请求体中使用BaseModel来自动解析请求数据,并进行验证和转换,从而大大简化了 API 的开发过程。
时间参数概述在很多 API 中,我们经常需要处理时间相关的操作。
例如,我们可能需要接受时间戳作为参数,或者返回一些时间相关的信息。
在 FastAPI 中,我们可以使用datetime模块来处理时间相关的操作。
datetime模块提供了许多有用的类和函数,用于处理日期、时间和时间间隔。
在 BaseModel 中使用时间参数在BaseModel中,我们可以使用datetime模块中的datetime类来表示时间参数。
下面是一个使用时间参数的示例:from datetime import datetimefrom pydantic import BaseModelclass Item(BaseModel):created_at: datetime在上面的示例中,我们定义了一个Item类,它继承自BaseModel。
Item类有一个属性created_at,它的类型被指定为datetime。
这样,我们就定义了一个具有时间参数的数据模型。
运动会啦啦队的描写

运动会啦啦队的描写英文回答:The sports meet is here, and I am thrilled to be a part of the cheerleading squad. As a member of the cheerleading team, our role is to boost the spirit and energy of the audience and the athletes. We are the ones who get everyone on their feet, clapping, and cheering for their favorite teams.Our cheerleading routine is a combination of dance, gymnastics, and stunts. We practice for hours to perfect our moves and synchronize our steps. The routine is filled with flips, jumps, and high-energy dance sequences. We also incorporate catchy chants and cheers to engage the crowd and motivate the athletes.During the sports meet, we perform in between the events to keep the momentum going. When it's our turn to take the center stage, we enter the field with confidenceand enthusiasm. The crowd roars as we start our routine, and we feed off their energy. We execute our flips and jumps flawlessly, and our synchronized dance moves impress the audience.One of the most memorable moments was when we performed a pyramid stunt. We formed a human pyramid with the base consisting of strong and sturdy cheerleaders, and the smaller ones climbed up to the top. It was a nerve-wracking experience, but the feeling of accomplishment when we successfully completed the stunt was indescribable. The crowd erupted in cheers and applause, and we felt like superstars.Another highlight of being a cheerleader is interacting with the crowd. We encourage them to participate by clapping, chanting, and doing the wave. We also throw out small souvenirs, such as mini pom-poms or wristbands, to the crowd as a token of appreciation. It's amazing to see the smiles on their faces when they catch these souvenirs.Being a cheerleader is not just about performing, butalso about being a positive role model. We cheer for both the winning and losing teams, and we always maintain good sportsmanship. We believe in the power of encouragement and support, and our goal is to create a fun and uplifting atmosphere for everyone.中文回答:运动会来了,我非常激动地成为啦啦队的一员。
MELODIC-FslWiki

MELODIC-FslWikiResearch OverviewSingle-session ICA: This will perform standard 2D ICA on each of the input files. The input data will each be represented as a 2D time x space matrix. MELODIC then de-composes each matrix separately into pairs of time courses and spatial maps. The original data is assumed to be theThis approach does not assume that the temporal response pattern is the same across the population, though the final web report will contain the first Eigenvector of all different temporal responses as a summary time course. Access to all time courses is available: the time series plot is linked to a text file (tXX.txt) which contains the first Eigenvector, the best model fit in case a time series design was specified and all different subject/session-specific time courses as columns. For each component the final mixing matrix melodic_mix contains the temporal response of all different data sets concatenated into a single column vector. The final reported time course will be the best rank-1 approximation to these different responses. Multi-session Tensor-ICA: This will perform a 3D Tensor-ICA decomposition of the data. All individual data sets will be represented as a single time x space x sessions/subjects block of data. Tensor-ICA will decompose this block of data into triplets of time courses, spatial maps and session/subject modes, which - for each component - characterise the signal variation across the temporal, spatial and subject/session domain.CategoryFunctional CategoryMELODICMELODIC (17:02:49 16-07-2013由MarkJenkinson编辑)。
绣球‘杜丽’AP3基因克隆与基因编辑载体构建

㊀Guihaia㊀Feb.2024ꎬ44(2):257-266http://www.guihaia-journal.comDOI:10.11931/guihaia.gxzw202204002李童ꎬ王月莹ꎬ赵惠恩ꎬ2024.绣球 杜丽 AP3基因克隆与基因编辑载体构建[J].广西植物ꎬ44(2):257-266.LITꎬWANGYYꎬZHAOHEꎬ2024.AP3genecloningandgene ̄editingvectorconstructionofHydrangeamacrophylla Dooley [J].Guihaiaꎬ44(2):257-266.绣球 杜丽 AP3基因克隆与基因编辑载体构建李㊀童ꎬ王月莹ꎬ赵惠恩∗(花卉种质创新与分子育种北京市重点实验室ꎬ林木花卉遗传育种教育部重点实验室ꎬ国家花卉工程技术研究中心ꎬ城乡生态环境北京实验室ꎬ北京林业大学园林学院ꎬ北京100083)摘㊀要:绣球(Hydrangeamacrophylla)是以花序为主要观赏部位的园林植物ꎬ多用作切花装饰和景观营造ꎬ在亚洲㊁美洲㊁欧洲广泛栽培ꎮ为探究AP3基因在绣球花萼形成过程中的功能ꎬ加快重瓣绣球新品种培育进程ꎬ该研究以绣球 杜丽 为材料ꎬ克隆其MADS ̄boxB类基因HmAP3ꎬ并结合生物信息学方法预测基因功能ꎻ根据HmAP3序列信息ꎬ筛选出高特异性编辑靶点并构建CRISPR/Cas9基因编辑载体ꎬ通过农杆菌转化法将载体整合到绣球基因组中ꎮ结果表明:(1)克隆到1段HmAP3基因的cDNA序列ꎬ其序列全长546bpꎬ共编码181个氨基酸ꎬ测序结果表明其氨基酸序列与参考序列一致性为100%ꎬ与拟南芥AtAP3相似度为58.8%ꎮ(2)不同属植物AP3氨基酸序列差异较大ꎬ在同属不同物种中ꎬAP3蛋白主要结构较为保守ꎬ仅在少数基序上存在差异ꎮ(3)在HmAP3中共鉴定到2个高特异性靶点ꎬ并成功构建2个单靶点CRISPR/Cas9基因编辑载体ꎮ(4)该研究共获得5株基因组内含有Cas9序列的抗性芽ꎬ但其靶点均未突变ꎬ在抗性芽中没有检测到Cas9表达ꎮ该研究探讨了AP3基因在重瓣绣球育种中的价值ꎬ对绣球的CRISPR/Cas9基因编辑技术进行了初探ꎬ为绣球优良品种繁育工作奠定了基础ꎮ关键词:绣球ꎬMADS ̄box家族ꎬAP3ꎬCRISPR/Cas9ꎬ载体构建中图分类号:Q943㊀㊀文献标识码:A㊀㊀文章编号:1000 ̄3142(2024)02 ̄0257 ̄10AP3genecloningandgene ̄editingvectorconstructionofHydrangeamacrophylla DooleyLITongꎬWANGYueyingꎬZHAOHuien∗(BeijingKeyLaboratoryofOrnamentalPlantsGermplasmInnovation&MolecularBreedingꎬKeyLaboratoryofGeneticsandBreedinginForestTreesandOrnamentalPlantsofMinistryofEducationꎬNationalEngineeringResearchCenterforFloricultureꎬBeijingLaboratoryofUrbanandRuralEcologicalEnvironmentꎬCollegeofLandscapeArchitectureꎬBeijingForestryUniversityꎬBeijing100083ꎬChina)Abstract:HydrangeamacrophyllaisagardenplantwidelycultivatedinAsiaꎬAmericaꎬandEuropewithitsinflorescenceasmainornamentalfeature.Itiscommonlyusedininteriordecorationandlandscapecreation.To收稿日期:2022-05-20基金项目:国家林业和草原局引进国际先进林业科学技术项目(2015-4-15)ꎮ第一作者:李童(1997-)ꎬ硕士ꎬ主要从事花卉物种质资源创新与育种研究ꎬ(E ̄mail)130****5858@ꎮ∗通信作者:赵惠恩ꎬ博士ꎬ教授ꎬ研究方向为花卉种质资源创新与育种ꎬ(E ̄mail)zhaohuien@bjfu.edu.cnꎮinvestigatetheroleofAP3geneinhydrangeaduringcalyxformationꎬH.macrophylla Dooley wasusedasthematerial.TheMADS ̄boxClassBgeneHmAP3wasclonedꎬanditsgenefunctionwaspredictedbybioinformaticsanalysis.Toexploremethodsforquickerbreedingnewvarietiesꎬhighly ̄specificeditingtargetswerescreenedandCRISPR/Cas9gene ̄editingvectorswereconstructed.ThevectorsequencewasintegratedintotheH.macrophyllagenomebyagrobacterium ̄mediatedtransformation.Theresultswereasfollows:(1)ThecDNAsequencefulllengthofHmAP3was546bpꎬencoding181aminoacids.Itsaminoacidsequencewas100%similartothereferencesequenceand58.8%similartoArabidopsisthaliana.(2)AP3differedgreatlyindifferentgenera.WithinthesamegenusꎬthemainstructureofAP3proteinwasconservedanddifferedonlyinafewmotifs.(3)ThereweretwohighlyspecifictargetsinHmAP3.Sequencingresultsindicatedthattwosingle ̄targetCRISPR/Cas9gene ̄editingvectorswereconstructedsuccessfully.(4)TherewerefiveresistantbudswithCas9sequencesintheirgenomes.HoweverꎬtheirtargetsequencesdidnotchangeduetotheabsenceofCas9expression.InthisstudyꎬthepotentialofAP3geneinthebreedingworkofdoubleflowerphenotypewasinvestigatedꎬandapreliminaryexplorationofCRISPR/Cas9gene ̄editingtechnologyforHydrangeamacrophyllawasconducted.TheseresultsprovideabasisforthebreedingofH.macrophylla.Keywords:HydrangeamacrophyllaꎬMADS ̄boxfamilyꎬAP3ꎬCRISPR/Cas9ꎬvectorconstruction㊀㊀绣球(Hydrangeamacrophylla)ꎬ虎耳草科绣球属ꎬ又名八仙花ꎬ在庭院景观中的应用历史悠久ꎬ是一种具有较高观赏价值的园林植物ꎬ作为世界流行的切花深受大众喜爱ꎮ目前绣球主要有蕾丝帽形和圆球形两类花序ꎬ其花序中的不育花具有大而艳丽的花瓣状萼片ꎬ是绣球的主要观赏组织ꎮ绣球不育花有单瓣和重瓣之分ꎬ其中单瓣类只有一轮观赏性萼片ꎬ重瓣类则具有多轮观赏性萼片ꎮ相比之下ꎬ重瓣绣球具有更高的观赏和经济价值ꎬ是绣球新品种培育的重要方向(Suyamaetal.ꎬ2015)ꎮ目前ꎬ国内外的绣球育种方式以杂交育种为主ꎬ其育种效率低㊁周期长ꎬ难以适应日益增长的市场需求(Wuetal.ꎬ2021)ꎬ需要探索更快捷㊁高效的育种方式ꎮCRISPR/Cas9技术是一种新兴的基因编辑技术ꎬ能够定向改变植物的观赏性状ꎬ如改造花型和花色ꎬ延长观赏周期等ꎬ在园林植物新品种繁育工作中具有极大的发展潜力和经济价值(Kauretal.ꎬ2021)ꎮCRISPR/Cas9基因编辑系统由Cas9核酸酶和单引导RNA(singleguideRNAꎬsgRNA)构成(Jineketal.ꎬ2012)ꎬ二者在植物细胞内转录后形成复合体ꎬ识别植物基因组中的间区序列邻近基序(protospaceradjacentmotifꎬPAM)前端约20nt核苷酸序列并结合ꎬCas9核酸酶切割该序列形成DNA双链缺口(DNAdouble ̄strandbreaksꎬDSBs)ꎬ引发植物自身损伤修复机制ꎬ产生随机的碱基缺失(Hsuetal.ꎬ2013)ꎮ与其他园艺作物相比ꎬCRISPR/Cas9技术在园林植物中的应用较少ꎬ仅应用于毛白杨(Fanetal.ꎬ2015)㊁矮牵牛(Zhangetal.ꎬ2016ꎻSun&Kaoꎬ2018ꎻXuetal.ꎬ2020ꎻYuetal.ꎬ2021)㊁菊花(Kishi ̄Kaboshietal.ꎬ2017)㊁铁皮石斛(Kuietal.ꎬ2017)㊁百合(Yanetal.ꎬ2019)㊁牵牛花(Shibuyaetal.ꎬ2018ꎻWatanabeetal.ꎬ2018)㊁蓝猪耳(Nishiharaetal.ꎬ2018)与蝴蝶兰(Tongetal.ꎬ2020ꎻSemiartietal.ꎬ2020)ꎮ花器官由花瓣㊁花萼㊁雄蕊和心皮4个部分组成ꎬ其基因表达调控机制可以用ABCDE模型来解释ꎮ在ABCDE模型中ꎬB类基因主要负责与A类基因共同调控花瓣的形成ꎬ以及与C类基因共同调控雄蕊的形成(Coen&Meyerowitzꎬ1991)ꎻ除A类基因中的AP2属于AP2/ERF家族外ꎬ该模型中的其余基因均属于MADS ̄box基因家族(王莹等ꎬ2021)ꎮMADS ̄boxB类基因亚家族成员广泛存在于现存植物的基因组中ꎬ在裸子植物小孢子叶球与被子植物花瓣和雄蕊中均有表达ꎬ在植物发育过程中具有重要地位(Albertetal.ꎬ1998)ꎮB类基因包含APETALA3(AP3)和PITILLATA(PI)两个谱系ꎬ其中AP3谱系主要调控花瓣和花萼的形成(Jaramillo&Kramerꎬ2004)ꎮAP3蛋白中含有保守的K ̄BOX结构域ꎬ该结构域能够引导AP3蛋白与PI㊁SEP3㊁AP1蛋白形成四聚体ꎬ诱导花瓣原基形成(Melzer&Theißenꎬ2009ꎻTheißenetal.ꎬ2016)ꎮ在观赏植物中ꎬ已经发现AP3基因沉默能够导致矮牵牛(vanderKrolꎬ1993)㊁兰花(Mondragón ̄Palomino&Theißenꎬ2009)与耧斗菜(Zhangetal.ꎬ852广㊀西㊀植㊀物44卷2013)等发生从花瓣向花萼的同源异型转变ꎮ因此ꎬ本研究对绣球 杜丽 的MADS ̄boxB类基因HmAP3进行了克隆和生物信息学分析ꎻ同时结合组内前期绣球 杜丽 再生体系建立基础ꎬ借助CRISPR/Cas9基因编辑系统ꎬ构建了2个HmAP3单靶点载体ꎬ转化获得抗性芽ꎬ拟探讨以下问题:(1)HmAP3氨基酸序列保守结构域特征与蛋白结构分析ꎻ(2)HmAP3系统进化关系及其生物学功能预测ꎻ(3)探究影响绣球CRISPR/Cas9基因编辑工作成功率的因素ꎮ以期为绣球的性状改良和新品种繁育工作提供实践参考和技术支撑ꎮ1㊀材料与方法1.1试验材料和试剂绣球 杜丽 种植于北京植物园(116ʎ28ᶄE㊁40ʎ00ᶄN)ꎮ于4月选取翠绿㊁无病虫害的叶片作为试验材料ꎬ将叶片与叶柄一并剪下ꎬ放入干净的蒸馏水中转移至实验室ꎮ试验所用的试剂盒包括植物总RNA提取试剂盒[天根生化科技(北京)有限公司ꎬDP432]ꎬcDNA反转录试剂盒(TaKaRaꎬRR047A)ꎬDNA凝胶回收试剂盒(北京擎科生物科技股份有限公司ꎬGE0101)ꎬOnestepZTOPO ̄Blunt/TA零背景快速克隆试剂盒(北京庄盟生物科技有限公司ꎬZC206)ꎬSE无缝克隆和组装试剂盒(北京庄盟生物科技有限公司ꎬZC231)ꎬ限制性内切酶BsaI[纽英伦生物技术(北京)有限公司]ꎮ1.2绣球 杜丽 HmAP3基因克隆依据在NCBI上查找到的绣球 BlueSky (H.macrophylla BlueSky )AP3基因(GenBank:AF230702.1)的CDS序列进行引物设计(表1)并合成高特异性引物ꎮ按照试剂盒说明书提取试验材料RNAꎬ并将RNA反转录成cDNAꎮ以cDNA为模板ꎬ用KODOne高保真DNA聚合酶(TOYOBOꎬKMM ̄101)ꎬ以HmAP3 ̄F1/R1为引物(表1)进行PCR扩增ꎮ扩增产物利用琼脂糖凝胶电泳进行纯化ꎬ将目的片段所处区域凝胶切割下来ꎬ按照DNA凝胶回收试剂盒说明书回收ꎮ纯化后的PCR产物连接T载体后转入DH5α大肠杆菌感受态涂布平板培养12hꎬ选取单菌落送至测序公司(北京擎科生物科技股份有限公司)进行质粒提取和测序工作ꎬ获得HmAP3基因的CDS序列ꎮ1.3绣球 杜丽 HmAP3基因生物信息学分析使用Cell ̄PLoc(http://www.csbio.sjtu.edu.cn/bioinf/Cell ̄PLoc ̄2/)对HmAP3进行亚细胞定位预测(Chou&Shenꎬ2010)ꎮ利用NCBI ̄BLAST(https://blast.ncbi.nlm.nih.gov/Blast.cgi)比对HmAP3氨基酸序列相似性ꎬ下载比对结果中排名在前列的其他植物AP3氨基酸序列ꎬ同时下载拟南芥AtAP3氨基酸序列ꎬ通过MEGA ̄X软件的邻接法(neighbor ̄joiningmethodꎬNJ)构建系统进化树(Zhangetal.ꎬ2019)ꎮ利用MEME(http://meme ̄suite.org/tools/meme/)预测HmAP3氨基酸序列保守基序ꎮ通过ProrParam(https://web.expasy.org/protparam/)分析HmAP3蛋白的理化性质(Lietal.ꎬ2020)ꎮ分别使用蛋白二级结构预测工具SOPMA(https://npsa ̄prabi.ibcp.fr/cgi ̄bin/npsa_automat.pl?page=npsa_sopma.html/)和蛋白三级结构预测工具Swissmodel(https://swissmodel.expasy.org/)对HmAP3氨基酸序列进行分析ꎮ1.4CRISPR/Cas9基因编辑载体构建和转化使用CRISPR靶点设计网站CRISPRdirect(http://crispr.dbcls.jp/)根据HmAP3基因的CDS序列ꎬ选择PAM位点和GC含量在40%~60%之间的高特异性靶点ꎮ以pCAMBIA1300 ̄sgRNA/Cas9载体质粒为模板ꎬ以HmAP3 ̄F2/R2㊁HmAP3 ̄F3/R3为引物(表1)ꎬ进行PCR扩增获得带有黏性末端的目的片段ꎮ使用内切酶BsaⅠ酶切获得pCAMBIA1300 ̄sgRNA/Cas9线性载体ꎬ用无缝克隆试剂盒(北京庄盟生物科技有限公司ꎬZC231)连接载体和目的片段ꎬ获得重组质粒ꎮ构建好的质粒转入DH5α大肠杆菌感受态涂布平板培养12hꎬ选取单菌落送至测序公司(北京擎科生物科技股份有限公司)进行质粒提取和测序工作ꎬ回收构建成功的载体质粒ꎮ将构建好的载体pCAMBIA1300::HmAP3利用冻融法转入GV3101农杆菌感受态中ꎬ在2抗LB培养基(50mg L ̄1卡那霉素+50mg L ̄1利福平)中28ħ培养2dꎬ挑取单菌落在LB液体培养基扩繁ꎮ离心收集扩繁的农杆菌菌体ꎬ加入适量侵染液(MS+30g L ̄1蔗糖+200μmol L ̄1乙酰丁香酮)调至OD600=0.4ꎮ将绣球 杜丽 叶片剪切成1cmˑ1cm的小块ꎬ在叶背划3~4刀ꎬ放入上述配制好的侵染液中9522期李童等:绣球 杜丽 AP3基因克隆与基因编辑载体构建浸泡侵染10minꎬ转接到共培养培养基(MS+2.0mg L ̄16 ̄BA+0.1mg L ̄1IBA)上暗培养2dꎬ再转移到筛选培养基(MS+2.0mg L ̄16 ̄BA+0.1mg L ̄1IBA+2mg L ̄1潮霉素+200mg L ̄1头孢霉素)中至获得抗性再生芽ꎮ1.5抗性芽检测和鉴定取抗性芽叶片ꎬ使用基因组提取试剂盒获取叶片DNAꎬ再依次使用总RNA提取试剂盒㊁cDNA反转录试剂盒获取叶片cDNAꎮ分别以叶片DNA和cDNA为模板ꎬ以Cas9 ̄F/R为引物扩增Cas9序列ꎬ扩增片段长度为764bpꎮ以叶片DNA为模板ꎬ以HmAP3 ̄F1/R1为引物扩增抗性芽HmAP3序列ꎬ扩增产物送至测序公司(北京擎科生物科技股份有限公司)测序ꎮ使用DNAMAN比对测序结果与野生型序列差异ꎮ2㊀结果与分析2.1绣球 杜丽 AP3基因克隆与序列分析参考绣球 BlueSky AP3基因CDS序列ꎬ利用HmAP3 ̄F1/HmAP3 ̄R1引物(表1)ꎬ在绣球 杜丽 cDNA文库中克隆到了一段完全一致的核苷酸序列ꎮ克隆到的基因序列全长546bpꎬ共编码181个氨基酸ꎬ利用NCBI分析其氨基酸序列ꎬ发现在30~123bp处包含1个K ̄BOX保守结构域(图1)ꎮ该氨基酸序列C端含有PI基序和euAP3基序ꎬ符合MADS ̄box家族特征ꎬ命名为HmAP3ꎮ将HmAP3与拟南芥AtAP3氨基酸序列比对ꎬ其相似度为58.8%ꎻHmAP3与绣球 BlueSky AP3的DNA序列相似度为100%ꎮ因此ꎬ推测该基因为绣球 杜丽 AP3基因ꎮ亚细胞定位预测结果显示HmAP3在细胞核中表达ꎮ2.2绣球 杜丽 AP3蛋白理化性质与结构分析以拟南芥MADS ̄box类蛋白三级结构为模型ꎬ预测HmAP3蛋白三级结构ꎬ可见该结构中含有2条长α ̄螺旋ꎬ螺旋间纽结为90ʎꎻ其预测结果GMQE(全球模型质量估计)值为0.32ꎬQMEAN得分为0.74ʃ0.05ꎬ模型可信度和质量较高(图2)ꎮ绣球 杜丽 的HmAP3蛋白分子式为C927H1462N268O287S7ꎬ分子量为21177.85Dꎮ该蛋白共包含181个氨基酸ꎬ不稳定系数为38.79ꎬ属稳定蛋白ꎮ蛋白带负电荷残基总数(Asp+Glu)为28ꎬ带正电荷残基总数(Arg+Lys)为25ꎬ理论等电点为6.17ꎮ蛋白脂肪指数为79.12ꎬ亲水性(GRAVY)为-0.791ꎬ为亲水性蛋白ꎮHmAP3蛋白二级结构中α ̄螺旋占比最高ꎬ为64.09%ꎻ其他结构占比由高到低依次为无规则卷曲(22.65%)㊁延伸链(8.84%)㊁β ̄折叠(4.42%)(图3)ꎮ2.3绣球 杜丽 AP3蛋白系统进化与motif分析将HmAP3氨基酸序列提交到NCBI进行BLAST比对ꎬ在比对结果中选取下载与该序列相似度较高的其他植物AP3氨基酸序列ꎬ在MEGA ̄X软件上用邻接法构建系统进化树(图4)ꎮ从整体上来看ꎬ绣球等蔷薇亚纲菊超目植物被聚为同一大支ꎬ说明AP3蛋白在系统进化过程中具有一定保守性ꎮ从各小分支来看ꎬ不同物种间的AP3序列存在一定差异ꎬ而同物种间的序列相似度则较高ꎮ相对而言ꎬ绣球与神秘果(Synsepalumdulcificum)㊁洒金桃叶珊瑚(Aucubajaponicavar.borealis)和欧洲枸骨(Ilexaquifolium)亲缘关系最近ꎮ在模式植物中ꎬ绣球与烟草(Nicotianatabacum)的亲缘关系比拟南芥(Arabidopsisthaliana)更近ꎮ因此ꎬ使用烟草基因组作为预测绣球基因编辑靶点的参考基因组更为适宜ꎮ在MEME ̄motifsuite工具上对上述氨基酸序列进行分析后ꎬ获得了15个motif及其在序列中的相对位置(图4)ꎮ大部分植物AP3序列包含7个motifꎬ其中有8个AP3序列包含8个motifꎬ1个AP3序列包含9个motifꎮ所有AP3序列C端较为保守ꎬ均含有motif2㊁motif4㊁motif5㊁motif6和motif7ꎬ而N端多含motif3ꎮ与其他植物相比ꎬ绣球AP3序列中含有特有的motif12ꎬ此外仅洒金桃叶珊瑚AP3序列中含有这一基序ꎬ说明HmAP3相对其他植物AP3蛋白可能会有更多功能ꎮ对同源基因来说ꎬ其序列的motif大体相似ꎬ然而在不同物种间仍存在一定的差异ꎬ这些差异导致了不同物种间同源基因的功能差异ꎮ2.4CRISPR/Cas9基因编辑载体构建利用CRISPRdirect在HmAP3上共选取到2个特异性强的靶点ꎬ分别命名为HmAP3 ̄Taget1(5ᶄ ̄GATCTGTACCAGACGACAAT+GGG ̄3ᶄ)和HmAP3 ̄Taget2(5ᶄ ̄TGAACGAAAGTATCGAGTAC+CGG ̄3ᶄ)ꎬ其GC含量分别为45%和40%ꎬ其与PAM位点相邻的12bp在参考基因组(烟草)中均仅比对到1个位点ꎬ证明该靶点具有较强特异性ꎮ用引物HmAP3 ̄F2/R2㊁HmAP3 ̄F3/R3在质粒上扩增含有062广㊀西㊀植㊀物44卷表1㊀本研究中所使用的引物序列Table1㊀Primersequencesusedinthisstudy引物名称Primername序列(5ᶄң3ᶄ)Sequence(5ᶄң3ᶄ)退火温度Annealingtemperature(ħ)用途PurposeHmAP3 ̄F1HmAP3 ̄R15ᶄ ̄ATGTTCTCCACTACCAACAAACT ̄3ᶄ5ᶄ ̄CTAATCGAGCAATGCATACGTAG ̄3ᶄ56HmAP3全长扩增HmAP3full ̄lengthamplificationHmAP3 ̄F2HmAP3 ̄R25ᶄ ̄ACAGCTAGAGTCGAAGTAGTGATTGGATCTGTACCAGACGACAATGTTTTAGAGCTAGAAATAGC ̄3ᶄ5ᶄ ̄TTCTGCAGACAAATGGCCCCCATTCGGAGTTTTTGTATCT ̄3ᶄ58CRISPR/Cas9载体构建 ̄靶点1CRISPR/Cas9vectorconstruction ̄Target1HmAP3 ̄F3HmAP3 ̄R35ᶄ ̄ACAGCTAGAGTCGAAGTAGTGATTGGTACTCGATACTTTCGTTCAGTTTTAGAGCTAGAAATAGC ̄3ᶄ5ᶄ ̄TTCTGCAGACAAATGGCCCCCATTCGGAGTTTTTGTATCT ̄3ᶄ58CRISPR/Cas9载体构建 ̄靶点2CRISPR/Cas9vectorconstruction ̄Target2Cas9 ̄FCas9 ̄R5ᶄ ̄CAAGTTCATCAAGCCCATCC ̄3ᶄ5ᶄ ̄GTCCTCGTTTTCCTCATTGTC ̄3ᶄ52抗性芽Cas9序列检测Cas9sequencedetectioninresistantbuds∗表示终止子ꎮ∗indicatesterminator.图1㊀HmAP3的序列及其结构域分析Fig.1㊀SequenceandstructuraldomainanalysisofHmAP3黏性末端的目的片段ꎬ与线性载体连接ꎬ用大肠杆菌转化后挑取单菌落测序ꎬ测序结果表明目的片段已成功插入载体ꎬ插入片段与载体结构如图5所示ꎮ绣球对潮霉素敏感性很高ꎬ经2mg L ̄1潮霉素筛选后ꎬ在侵染约2000枚叶片后仅培育出9株抗性芽(图6:A)ꎮ以抗性芽叶片DNA为模板ꎬ1622期李童等:绣球 杜丽 AP3基因克隆与基因编辑载体构建图2㊀HmAP3蛋白三级结构Fig.2㊀TertiarystructureofHmAP3protein分别使用靶基因序列扩增引物HmAP3 ̄F1/HmAP3 ̄R1和Cas9序列扩增引物Cas9 ̄F/Cas9 ̄R对抗性芽进行鉴定ꎮ扩增和测序结果表明ꎬ9株抗性芽叶片基因组中有5株可克隆到Cas9序列ꎬ但其靶点序列均未发生突变(图6:B)ꎮ提取抗性芽叶片总RNA后反转录获得cDNAꎬ再次克隆Cas9序列ꎬ发现所有样品均无法扩增出条带ꎮ该现象说明虽然载体序列已成功整合到载体基因组上ꎬ但是Cas9蛋白并未成功转录和表达ꎬ因此ꎬ编辑靶点的序列没有改变ꎮ3㊀讨论与结论本研究克隆了绣球 杜丽 的HmAP3基因ꎬ并对其核苷酸序列与氨基酸序列进行了生物信息学分析ꎮ研究发现HmAP3与模式植物拟南芥(Yangetal.ꎬ2003)及园艺作物绿竹(朱龙飞ꎬ2013)㊁葡萄(胡晓燕等ꎬ2021)㊁菠萝(郑雪文等ꎬ2021)在特有的K ̄BOX结构域上长度相近㊁结构相似ꎬ表明此结构域在不同物种的AP3中高度保守ꎮ理化性质分析结果表明HmAP3是稳定的亲水性蛋白ꎬ与郑雪文等(2021)的研究结果一致ꎮ在HmAP3蛋白三级结构模型中ꎬK ̄BOX形成长α ̄螺旋结构ꎬ与植物MADS ̄box家族中K ̄BOX结构域特征一致ꎬ该结构在AP3与其他蛋白结合形成四聚体的过程中发挥关键作用(Yang&Jackꎬ2004)ꎮHmAP3蛋白系统进化树表明AP3基因在植物系统进化过程中的保守性ꎬ其中绣球与金鱼草㊁烟草和番茄AP3基因聚类在同一大分支ꎬ亲缘关系较近ꎬ与Viaene等(2009)研究结果相似ꎮMartino等(2006)和Liu等(2004)研究发现ꎬ番茄SlAP3与烟草NtAP3基因沉默后代表现出花萼轮数增多㊁花瓣消失的性状ꎮ通过同源比对ꎬ推测绣球 杜丽 HmAP3基因与其同源基因SlAP3㊁NtAP3功能相似ꎬ可能负责调控绣球花器官中花萼和花瓣的形成ꎮ本研究构建了2个绣球HmAP3单靶点载体ꎬ并在转化获得的抗性芽基因组中检测到载体序列ꎬ但在抗性芽中未检测到Cas9序列的表达和编辑位点序列突变ꎮ本研究与Ren等(2013)的研究结果相似ꎬ他推测基因编辑效率与启动子活性有关ꎬ当CRISPR/Cas9基因编辑载体中的启动子从nos ̄mini变为U6b启动子时ꎬ遗传突变率可从0提高至3.2%ꎮ在观赏植物中ꎬKishi ̄Kaboshi等(2019)的研究也佐证了这一观点ꎬ系统性比较了Ubiqutin㊁CaMV35S和CmActin2启动子的表达活性差异后ꎬ发现CaMV35S和Ubiqutin启动子在菊花愈伤组织中活性均低于菊花CmActin2启动子ꎮ本研究使用的Cas9序列启动子为Ubiqutin启动子ꎬ猜想其在绣球组织中的表达活性极低ꎬ导致Cas9序列在抗性芽中未表达ꎮ在下一步绣球基因编辑工作中ꎬ可将载体中启动子更换为绣球本源启动子ꎬ进一步探究启动子活性对基因编辑成功率的影响ꎮ本研究发现绣球对潮霉素的高敏感性也是影响CRISPR/Cas9基因编辑效率的重要因素ꎮ本研究使用2mg L ̄1潮霉素浓度对绣球抗性芽进行筛选ꎬ再生率仅为0.45%ꎬ与苹果(贾东杰等ꎬ2013)等在潮霉素筛选下的再生情况相符ꎬ推测绣球野生型基因组内不含有潮霉素抗性基因ꎮ甘煌灿等(2018)提出可通过在再生筛选过程中逐渐增加潮霉素浓度的方法ꎬ提高抗性芽的成活率ꎮ后续的绣球抗性芽的筛选条件可通过调整不同再生阶262广㊀西㊀植㊀物44卷蓝色表示α ̄螺旋ꎬ紫色表示无规则卷曲ꎬ绿色表示β ̄折叠ꎬ红色表示延伸链ꎮBlueindicatesα ̄helixꎬpurpleindicatesrandomcoilꎬgreenindicatesβ ̄angleꎬandredindicatesextendedstrand.图3㊀HmAP3蛋白二级结构Fig.3㊀SecondarystructureofHmAP3protein图4㊀HmAP3蛋白系统进化和motif分析Fig.4㊀PhylogeneticandmotifanalysisofHmAP3protein3622期李童等:绣球 杜丽 AP3基因克隆与基因编辑载体构建A.pCAMBIA1300::HmAP3载体结构ꎻB.载体中目的片段测序结果ꎮA.StructureofpCAMBIA1300::HmAP3vectorꎻB.Sequencingresultsofthetargetfragmentinthevector.图5㊀pCAMBIA1300::HmAP3载体图谱及目的片段插入情况Fig.5㊀pCAMBIA1300::HmAP3vectormappingandtargetfragmentinsertionA.绣球 杜丽 抗性芽的生长状况ꎻB.抗性芽基因组中Cas9序列扩增(M.DL2000DNAMarkerꎻWT.野生型ꎻ1~9.绣球 杜丽 抗性芽)ꎮA.ResistantbudsgrowthofHydrangeamacrophylla Dooley ꎻB.Cas9sequenceamplificationinthegenomeofresistantbuds(M.DL2000DNAMarkerꎻWT.Wildtypeꎻ1-9.ResistantbudsofH.macrophylla Dooley ).图6㊀绣球 杜丽 抗性芽检测Fig.6㊀DetectionofresistantbudsofHydrangeamacrophylla Dooley462广㊀西㊀植㊀物44卷段潮霉素浓度的方式来进一步优化ꎬ以提高基因编辑效率ꎮ本研究在绣球 杜丽 中克隆到1个HmAP3基因ꎬcDNA序列全长546bpꎬ共编码181个氨基酸ꎬ为稳定的亲水性蛋白ꎬ氨基酸序列结构分析证明其具有MADS ̄boxB类基因亚家族特征ꎬ系统进化分析表明HmAP3与烟草㊁番茄㊁金鱼草亲缘关系较近ꎬ基序组成结构保守ꎻ以HmAP3为靶点ꎬ成功构建2个以Cas9基因㊁sgRNA㊁潮霉素抗性基因为骨架的CRISPR/Cas9基因编辑载体ꎬ并将载体序列整合到绣球基因组中ꎮ上述研究结果为进一步研究HmAP3基因功能奠定了理论基础ꎬ为重瓣绣球基因编辑辅助育种工作提供技术支撑ꎮ参考文献:ALBERTVAꎬGUSTAFSSONMHGꎬLAURENZIOLDꎬ1998.Ontogeneticsystematicsꎬmoleculardevelopmentalgeneticsꎬandtheangiospermpetal[M]//MolecularSystematicsofplantsⅡ.Boston:Springer:349-374.CHOUKCꎬSHENHBꎬ2010.Cell ̄PLoc2.0:animprovedpackageofweb ̄serversforpredictingsubcellularlocalizationofproteinsinvariousorganisms[J].NatSciꎬ2(10):1090.COENESꎬMEYEROWITZEMꎬ1991.Thewarofthewhorls:geneticinteractionscontrollingflowerdevelopment[J].Natureꎬ353(6339):31-37.FANDꎬLIUTTꎬLICFꎬetal.ꎬ2015.EfficientCRISPR/Cas9 ̄mediatedtargetedmutagenesisinPopulusinthefirstgeneration[J].SciRepꎬ5(1):1-7.GANHCꎬLAICCꎬPANHꎬetal.ꎬ2018.Plantexpressionvectorconstructionandjasmine[Jasminumsambac(Linn.)Aiton]callustransformationofDFRgenefromspinegrape(VitisdavidiiFoëx.)[J].ChinJTropCropꎬ39(6):1128-1136.[甘煌灿ꎬ赖呈纯ꎬ潘红ꎬ等ꎬ2018.刺葡萄DFR基因植物表达载体构建及转化茉莉花愈伤组织的研究[J].热带作物学报ꎬ39(6):1128-1136.]HSUPDꎬSCOTTDAꎬWEINSTEINJAꎬetal.ꎬ2013.DNAtargetingspecificityofRNA ̄guidedCas9nucleases[J].NatBiotechnolꎬ31(9):827-832.HUXYꎬGUOCLꎬWANGLꎬetal.ꎬ2021.CloningandfunctionanalysisofVvMADS46inredglobeandthompsonseedlessforseedlessregulation[J].JFruitSciꎬ38(8):1231-1239.[胡晓燕ꎬ郭春磊ꎬ王莉ꎬ等ꎬ2021.无核白及红地球葡萄VvMADS46基因的克隆及其无核调控功能分析[J].果树学报ꎬ38(8):1231-1239.]JARAMILLOMAꎬKRAMEREMꎬ2004.APETALA3andPISTILLATAhomologsexhibitnovelexpressionpatternsintheuniqueperianthofAristolochia(Aristolochiaceae) [J].EvolDevꎬ6(6):449-458.JIADJꎬFANLMꎬSHENJLꎬetal.ꎬ2013.GenetictransformationandexpressingofastaxanthinbiosynthesisgenesBKTinto BrookfieldGala appletree[J].ActaHorticSinꎬ40(1):21-31.[贾东杰ꎬ樊连梅ꎬ沈俊岭ꎬ等ꎬ2013.虾青素合成关键酶基因BKT在 BrookfieldGala 苹果中的遗传转化及表达[J].园艺学报ꎬ40(1):21-31.]JINEKMꎬCHYLINSKIKꎬFONFARAIꎬetal.ꎬ2012.Aprogrammabledual ̄RNA ̄guidedDNAendonucleaseinadaptivebacterialimmunity[J].Scienceꎬ337(6096):816-821.KAURHꎬPANDEYDKꎬGOUTAMUꎬetal.ꎬ2021.CRISPR/Cas9 ̄mediatedgenomeeditingisrevolutionizingtheimprovementofhorticulturalcrops:recentadvancesandfutureprospects[J].SciHort ̄Amsterdamꎬ289(110476):1-13.KISHI ̄KABOSHIMꎬAIDARꎬSASAKIKꎬ2017.Generationofgene ̄editedChrysanthemummorifoliumusingmulticopytransgenesastargetsandmarkers[J].PlantCellPhysiolꎬ58(2):216-226.KISHI ̄KABOSHIMꎬAIDARꎬSASAKIKꎬ2019.ParsleyubiquitinpromoterdisplayshigheractivitythantheCaMV35SpromoterandthechrysanthemumActin2promoterforproductiveꎬconstitutiveꎬanddurableexpressionofatransgeneinChrysanthemummorifolium[J].BreedSciꎬ69(3):19036.KUILꎬCHENHTꎬZHANGWXꎬetal.ꎬ2017.Buildingageneticmanipulationtoolboxfororchidbiology:identificationofconstitutivepromotersandapplicationofCRISPR/Cas9intheorchidꎬDendrobiumofficinale[J].FrontPlantSciꎬ7(2036):1-13.LIUYLꎬNAKAYAMANꎬSCHIFFMꎬetal.ꎬ2004.VirusinducedgenesilencingofadeficiensorthologinNicotianabenthamiana[J].PlantMolBiolꎬ54(5):701-711.MARTINOGDꎬPANIꎬEMMANUELEꎬetal.ꎬ2006.FunctionalanalysesoftwotomatoAPETALA3genesdemonstratediversificationintheirrolesinregulatingfloraldevelopment[J].PlantCellꎬ18(8):1833-1845.MELZERRꎬTHEIßENGꎬ2009.Reconstitutionof floralquartets invitroinvolvingclassBandclassEfloralhomeoticproteins[J].NuclAcidsResꎬ37(8):2723-2736.MONDRAGÓN ̄PALOMINOMꎬTHEIßENGꎬ2009.Whyareorchidflowerssodiverse?ReductionofevolutionaryconstraintsbyparaloguesofclassBfloralhomeoticgenes[J].AnnBotꎬ104(3):583-594.NISHIHARAMꎬHIGUCHIAꎬWATANABEAꎬetal.ꎬ5622期李童等:绣球 杜丽 AP3基因克隆与基因编辑载体构建2018.ApplicationoftheCRISPR/Cas9systemformodificationofflowercolorinToreniafournieri[J].BmcPlantBiolꎬ18(1):1-9.RENXJꎬSUNJꎬHOUSDENBEꎬetal.ꎬ2013.OptimizedgeneeditingtechnologyforDrosophilamelanogasterusinggermline ̄specificCas9[J].ProcNatlAcadSciUSAꎬ110(47):19012-19017.SEMIARTIEꎬNOPITASARISꎬSETIAWATIYꎬetal.ꎬ2020.ApplicationofCRISPR/Cas9genomeeditingsystemformolecularbreedingoforchids[J].IndonesJBiotechnolꎬ25(1):61-68.SHIBUYAKꎬWATANABEKꎬONOMꎬ2018.CRISPR/Cas9 ̄mediatedmutagenesisoftheEPHEMERAL1locusthatregulatespetalsenescenceinJapanesemorningglory[J].PlantPhysiolBiochemꎬ131(36):53-57.SUNLHꎬKAOTHꎬ2018.CRISPR/Cas9 ̄mediatedknockoutofPiSSK1revealsessentialroleofS ̄locusF ̄boxprotein ̄containingSCFcomplexesinrecognitionofnon ̄selfSRNasesduringcross ̄compatiblepollinationinself ̄incompatiblePetuniainflata[J].PlantReprodꎬ31(2):129-143.SUYAMATꎬTANIGAWATꎬYAMADAAꎬetal.ꎬ2015.Inheritanceofthedouble ̄floweredtraitindecorativehydrangeaflowers[J].HorticJꎬ84(3):253-260.THEIßENGꎬMELZERRꎬRÜMPLERFꎬ2016.MADS ̄domaintranscriptionfactorsandthefloralquartetmodelofflowerdevelopment:linkingplantdevelopmentandevolution[J].Developmentꎬ143(18):3259-3271.TONGCGꎬWUFHꎬYUANYHꎬetal.ꎬ2020.High ̄efficiencyCRISPR/Cas ̄basededitingofPhalaenopsisorchidMADSgenes[J].PlantBiotechnolJꎬ18(4):889-891.VANDERKROLꎬBRUNELLEAꎬTSUCHIMOTSꎬetal.ꎬ1993.FunctionalanalysisofpetuniafloralhomeoticMADSboxgenepMADS1[J].GeneDevꎬ7(7a):1214-1228.VIAENETꎬVEKEMANSDꎬIRISHVFꎬetal.ꎬ2009.Pistillata duplicationsasamodeforfloraldiversificationin(Basal)asterids[J].MolBiolEvolꎬ26(11):2627-2645.WANGYꎬMUYXꎬWANGJꎬ2021.AdvancesintheregulationofplantfloralorgandevelopmentbytheMADS ̄boxgenefamily[J].ActaAgricZhejiangꎬ33(6):1149-1158.[王莹ꎬ穆艳霞ꎬ王锦ꎬ2021.MADS ̄box基因家族调控植物花器官发育研究进展[J].浙江农业学报ꎬ33(6):1149-1158.]WATANABEKꎬODA ̄YAMAMIZOCꎬSAGE ̄ONOKꎬetal.ꎬ2018.AlterationofflowercolourinIpomoeanilthroughCRISPR/Cas9 ̄mediatedmutagenesisofcarotenoidcleavagedioxygenase4[J].TransgenicResꎬ27(1):25-38.WUXBꎬHULSE ̄KEMPAMꎬWADLPAꎬetal.ꎬ2021.Genomicresourcedevelopmentforhydrangea[Hydrangeamacrophylla(Thunb.)Ser.] Atranscriptomeassemblyandahigh ̄densitygeneticlinkagemap[J].Horticulturaeꎬ7(25):1-13.XUJPꎬKANGBCꎬNAINGAHꎬetal.ꎬ2020.CRISPR/Cas9 ̄mediatededitingof1 ̄aminocyclopropane ̄1 ̄carboxylateoxidase1enhancesPetuniaflowerlongevity[J].PlantBiotechnolJꎬ18(1):287-297.YANRꎬWANGZPꎬRENYMꎬetal.ꎬ2019.EstablishmentofefficientgenetictransformationsystemsandapplicationofCRISPR/Cas9genomeeditingtechnologyinLiliumpumilumDC.Fisch.andLiliumlongiflorumWhiteHeaven[J].IntJMolSciꎬ20(12):2920YANGYZꎬFANNINGLꎬJACKTꎬ2003.TheKdomainmediatesheterodimerizationoftheArabidopsisfloralorganidentityproteinsꎬAPETALA3andPISTILLATA[J].PlantJꎬ33(1):47-59.YANGYZꎬJACKTꎬ2004.DefiningsubdomainsoftheKdomainimportantforprotein ̄proteininteractionsofplantMADSproteins[J].PlantlBiolꎬ55(1):45-59.YUJꎬTULHꎬSUBBURAJSꎬetal.ꎬ2021.SimultaneoustargetingofduplicatedgenesinPetuniaprotoplastsforflowercolormodificationviaCRISPR ̄Cas9ribonucleoproteins[J].PlantCellRepꎬ40(6):1037-1045.ZHANGBꎬYANGXꎬYANGCPꎬetal.ꎬ2016.ExploitingtheCRISPR/Cas9systemfortargetedgenomemutagenesisinPetunia[J].SciRepꎬ6(1):1-8.ZHANGJJꎬYANGEDꎬHEQꎬetal.ꎬ2019.Genome ̄wideanalysisoftheWRKYgenefamilyindrumstick(MoringaoleiferaLam.)[J].PeerJꎬ7(7093):1-20.ZHANGRꎬGUOCCꎬZHANGWEꎬetal.ꎬ2013.DisruptionofthepetalidentitygeneAPETALA3 ̄3ishighlycorrelatedwithlossofpetalswithinthebuttercupfamily(Ranunculaceae) [J].ProcNatlAcadSciꎬ110(13):5074-5079.ZHENGXWꎬOUYANGYWꎬPANXLꎬetal.ꎬ2022.AnalysisoncloningofAcMADS14geneanditsexpressionduringflowerdevelopmentofpineapple[J].GuangdongAgricSciꎬ49(1):42-50.[郑雪文ꎬ欧阳嫣惟ꎬ潘晓璐ꎬ等ꎬ2022.菠萝AcMADS14基因的克隆及其在花发育中的表达分析[J].广东农业科学ꎬ49(1):42-50.]ZHULFꎬ2013.CloningandpreliminaryfunctionanalysisofBclassgenesinBambusaoldhamii[D].Hangzhou:ZhejiangA&FUniversity.[朱龙飞ꎬ2013.绿竹B类基因克隆与功能初步分析[D].杭州:浙江农林大学.](责任编辑㊀周翠鸣)662广㊀西㊀植㊀物44卷。
白来航鸡STING基因克隆、生物信息学及组织表达特性分析

中国畜牧兽医 2024,51(4):1372-1381C h i n aA n i m a lH u s b a n d r y &V e t e r i n a r y Me d i c i n e 白来航鸡S T I N G 基因克隆㊁生物信息学及组织表达特性分析王艳,张 颖,赵雅妮,易 林,史 珍,周长明,周宛蓉,姜莉莉,樊兆斌(菏泽学院药学院,菏泽274015)摘 要:ʌ目的ɔ克隆白来航鸡干扰素基因刺激因子基因(s t i m u l a t o ro f i n t e r f e r o n g e n e s ,S T I N G )C D S 区序列并进行生物信息学和组织表达分析,为阐明S T I N G 基因在抗病毒免疫应答中的作用奠定基础㊂ʌ方法ɔ采用P C R 扩增并克隆白来航鸡S T I N G 基因C D S 区,测序后对其编码氨基酸序列进行相似性比对及系统进化树构建,利用生物信息学预测S T I N G 蛋白的理化特性及结构功能,并利用实时荧光定量P C R 技术检测S T I N G 基因在鸡心脏㊁肝脏等14个组织中的表达情况㊂ʌ结果ɔ白来航鸡S T I N G 基因C D S 区序列全长1140b p ,编码379个氨基酸㊂相似性比对和系统进化树分析结果表明,白来航鸡S T I N G 基因与原鸡的相似性最高(99.7%),亲缘关系最近,与冠小嘴乌鸦亲缘关系最远㊂S T I N G 蛋白为酸性㊁亲水性蛋白,分子质量为42.625k u ,等电点(pI )为6.67,不稳定系数为69.26,脂肪系数为105.01㊂该蛋白大部分在线粒体和内质网上合成,含有跨膜结构,不含信号肽㊂S T I N G 蛋白二级结构包括α-螺旋(54.62%)㊁延伸链(10.29%)㊁β-转角(3.43%)及无规则卷曲(31.66%)㊂蛋白互作分析表明,白来航鸡S T I N G 蛋白与N F K B 1㊁D D X 41㊁c G A S ㊁T B K 1等蛋白存在相互作用㊂实时荧光定量P C R 结果显示,S T I N G 基因在白来航鸡组织中广泛表达,其中在肺脏中表达量最高,且显著高于其他组织(P <0.05);在胸肌中表达量最低㊂ʌ结论ɔ本研究成功克隆了白来航鸡S T I N G 基因,其C D S 区序列全长1140b p ,编码379个氨基酸㊂白来航鸡S T I N G 蛋白为酸性㊁亲水性蛋白,含有跨膜结构㊂S T I N G 基因在白来航鸡肺脏中表达量最高㊂研究结果为深入探究白来航鸡S T I N G 基因编码蛋白功能提供了材料㊂关键词:白来航鸡;S T I N G 基因;克隆;生物信息学;表达中图分类号:S 831;Q 78文献标识码:AD o i :10.16431/j.c n k i .1671-7236.2024.04.005 开放科学(资源服务)标识码(O S I D ):收稿日期:2023-10-20基金项目:山东省自然科学基金项目(Z R 2019M C 036㊁Z R 2023Q C 302);菏泽学院大学生创新训练项目;菏泽学院新兽药工程重点实验室联系方式:王艳,E -m a i l :1722804585@q q .c o m ;张颖,E -m a i l :3082291255@q q.c o m ㊂王艳和张颖对本文具有同等贡献,并列为第一作者㊂通信作者姜莉莉,E -m a i l :l y j l l f x y l 024@163.c o m ;樊兆斌,E -m a i l :438321212@q q.c o m C l o n i n g ,B i o i n f o r m a t i c s a n dT i s s u eE x pr e s s i o nC h a r a c t e r i s t i c s o f S T I N G G e n e i n W h i t eL e gh o r nC h i c k e n s WA N G Y a n ,Z H A N G Y i n g ,Z H A O Y a n i ,Y IL i n ,S H I Z h e n ,Z H O U C h a n g m i n g,Z H O U W a n r o n g,J I A N GL i l i ,F A NZ h a o b i n (C o l l e g e o f P h a r m a c y ,H e z eU n i v e r s i t y ,H e z e 274015,C h i n a )A b s t r a c t :ʌO b j e c t i v e ɔI nt h i ss t u d y ,t h eC D Sr e g i o ns e qu e n c eo f s t i m u l a t o ro f i n t e r f e r o n g e n e s (S T I N G )g e n e i n W h i t eL e g h o r nc h i c k e n sw a s c l o n e da n da n a l y z e db y bi o i n f o r m a t i c s a n d t i s s u e e x p r e s s i o n ,w h i c h l a i da f o u n d a t i o nf o re l u c i d a t i n g th er o l eo f S T I N G g e n e i na n t i v i r a l i m m u n e r e s p o n s e .ʌM e t h o d ɔT h eC D S r e g i o n o f S T I N G g e n e i n W h i t eL e g h o r n c h i c k e n sw a s a m p l i f i e db y P C Ra n d c l o n e d .A f t e r s e q u e n c i n g ,t h es i m i l a r i t y o f t h ee n c o d e da m i n oa c i ds e qu e n c eo f S T I N G g e n ew a sc o m p a r e da n dt h e p h y l o g e n e t i ct r e e w a sc o n s t r u c t e d .T h e p h y s i c o c h e m i c a l p r o pe r t i e s a n d s t r u c t u r a lf u n c t i o no f S T I N G p r o t e i nw e r e p r e d i c t e db y b i o i n f o r m a t i c s ,a n d t h e e x pr e s s i o no f4期王艳等:白来航鸡S T I N G基因克隆㊁生物信息学及组织表达特性分析S T I N G g e n e i n14t i s s u e ss u c ha sh e a r t a n d l i v e ro f W h i t eL e g h o r nc h i c k e n sw e r ed e t e c t e db y R e a l-t i m e q u a n t i t a t i v eP C R.ʌR e s u l tɔT h e s e q u e n c eo f t h eC D Sr e g i o no f S T I N G g e n e i n W h i t e L e g h o r nc h i c k e n sw a s1140b p i n t o t a l l e n g t h,e n c o d i n g379a m i n o a c i d s.S i m i l a r i t y a l i g n m e n t a n d p h y l o g e n e t i c t r e e a n a l y s i s s h o w e d t h a t S T I N G g e n e i n W h i t eL e g h o r nc h i c k e n sh a d t h eh i g h e s t s i m i l a r i t y w i t h G a l l u s g a l l u s a n d t h e c l o s e s t g e n e t i cr e l a t i o n s h i p,a n d t h ef a r t h e s t g e n e t i c r e l a t i o n s h i p w i t h C o r v u s c o r n i xc o r n i x.S T I N G p r o t e i n i n W h i t eL e g h o r n c h i c k e n sw a s a n a c i d i c,h y d r o p h i l i c p r o t e i n w i t ha m o l e c u l a rw e i g h to f42.625k u,a n i s o e l e c t r i c p o i n t(p I)o f6.67,a ni n s t a b i l i t y c o e f f i c i e n t o f69.26,a n da f a t c o e f f i c i e n to f105.01.S T I N G p r o t e i n w a ss y n t h e s i z e d m o s t l y o nm i t o c h o n d r i a a n de n d o p l a s m i c r e t i c u l u m,c o n t a i n e da t r a n s m e m b r a n e s t r u c t u r e,a n dn o s i g n a l p e p t i d e.T h es e c o n d a r y s t r u c t u r e o fS T I N G p r o t e i ni n c l u d e d a l p h a h e l i x(54.62%), e x t e n d e dc h a i n(10.29%),b e t at u r n(3.43%)a n dr a n d o m c o i l(31.66%).P r o t e i ni n t e r a c t i o n a n a l y s i s s h o w e d t h a t t h e r ew e r e i n t e r a c t i o n sb e t w e e nS T I N Ga n dN F K B1,D D X41,c G A S,T B K1 a n do t h e r p r o t e i n s.T h er e s u l t so fR e a l-t i m e q u a n t i t a t i v eP C R s h o w e dt h a t S T I N G g e n e w a s w i d e l y e x p r e s s e d i n t h e t i s s u e s o fW h i t eL e g h o r o n c h i c k e n s,w i t h t h e h i g h e s t e x p r e s s i o n i n l u n g, w h i c hw a s s i g n i f i c a n t l y h i g h e r t h a n t h a t i no t h e r t i s s u e s(P<0.05),a n d t h e l o w e s t e x p r e s s i o n i n p e c t o r a l i s m u s c l e.ʌC o n c l u s i o nɔI nt h i ss t u d y,S T I N G g e n ei n W h i t e L e g h o r nc h i c k e n s w a s s u c c e s s f u l l y c l o n e d,t h e t o t a l l e n g t ho f t h eC D Sr e g i o nw a s1140b p,e n c o d i n g379a m i n oa c i d s. S T I N G p r o t e i n i n W h i t e L e g h o r n c h i c k e n s w a s a n a c i d i c,h y d r o p h i l i c p r o t e i n w i t h a t r a n s m e m b r a n e s t r u c t u r e.T h e r e w a st h eh i g h e s te x p r e s s i o no f S T I N G g e n ei nl u n g o f W h i t e L e g h o r o n c h i c k e n s.T h er e s u l t s p r o v i d e d m a t e r i a l s f o r t h e i n-d e p t hs t u d y o f t h e f u n c t i o no f t h e p r o t e i ne n c o d e db y S T I N G g e n e i n W h i t eL e g h o r nc h i c k e n s.K e y w o r d s:W h i t eL e g h o r n c h i c k e n s;S T I N G g e n e;c l o n i n g;b i o i n f o r m a t i c s;e x p r e s s i o n先天性免疫是机体抵御病原微生物的第一道防线㊂宿主编码模式识别受体(P R R s)对来自病原体核酸的识别启动了复杂的信号转导途径,最终产生Ⅰ型干扰素(I F N-Ⅰ)和促炎细胞因子,并激活机体的天然免疫反应[1]㊂环鸟腺苷酸合成酶(c G A S)作为D N A识别受体,能识别并结合胞质双链D N A(d s D N A)[2]㊂当胞质中存在异常d s D N A时,c G A S能催化合成第二信使2 ,3 -c G AM P(c G AM P),结合并有效激活干扰素基因刺激因子基因(s t i m u l a t o r o f i n t e r f e r o n g e n e s,S T I N G),随后S T I N G招募T A N K结合激酶(T B K1)和I K B 激酶(I K K)使其磷酸化,激活导致产生I F N-Ⅰ和干扰素调节因子3(I R F3)㊁核因子κB(N F-κB)等细胞因子的表达[3]㊂S T I N G的功能主要体现在c G A S-S T I N G-I F N 信号通路中㊂c G A S-S T I N G通路除了构成有效的防御组织损伤和病原体入侵的监测系统外,还在自噬㊁翻译㊁代谢稳态㊁细胞凝聚㊁D N A损伤修复㊁衰老和细胞死亡中发挥作用[4]㊂然而c G A S-S T I N G通路的异常或过度激活可导致原发性发病机制和多种自身免疫性疾病[5]㊂家禽马立克病病毒(M D V)和鸡新城疫病毒(N D V)均靶向S T I N G接头蛋白,从而阻碍c G A S-S T I N G通路介导抗病毒天然免疫的应答[6-7]㊂禽痘病毒(F W P V)可以刺激鸡巨噬细胞中的c G A S-S T I N G通路,上调I F N相关因子表达,使宿主有效防御F W P V感染;禽白血病病毒(A L V)通过其编码的P15蛋白抑制激活c G A S-S T I N G通路,从而有助于病毒复制和持续感染[8]㊂S T I N G在细胞因子诱导㊁自噬诱导㊁代谢调节和内质网应激等方面发挥重要功能[9],且在健康和疾病中发挥着至关重要的作用,广泛参与各种细胞过程㊂近年来,靶向S T I N G的激动剂成为国内外抗肿瘤药物研发的新热点,S T I N G激动剂在乳腺癌㊁结肠癌㊁肝癌㊁黑色素瘤及淋巴瘤等肿瘤模型中都展现出了强有力的抗肿瘤活性[10]㊂目前国内外对鸡S T I N G在抗病毒免疫中的研究鲜有报道㊂本研究以白来航鸡为研究对象,克隆S T I N G基因C D S 区,运用在线软件对该基因编码蛋白进行生物信息学分析,并采用实时荧光定量P C R技术检测S T I N G基因在白来航鸡不同组织中的表达特征,以期为深入探究S T I N G在抗病毒免疫应答中的作用提供参考依据㊂3731中 国 畜 牧 兽 医51卷1 材料与方法1.1 材料供试动物选取同一批次的3只3周龄健康的白来航鸡(菏泽市某养殖场)㊂颈部放血处理后,无菌采集每只白来航鸡的心脏㊁肝脏㊁脾脏㊁胰脏㊁肺脏㊁肾脏㊁脑㊁胆㊁腺胃㊁十二指肠㊁回肠㊁盲肠㊁直肠㊁胸肌共14个组织置于超低温备用㊂T r i z o l ㊁P r i m e S c r i p t T MR T R e a g e n t K i t w i t h gD N AE r a s e r ㊁D L 2000D N A M a r k e r ㊁限制性内切酶E c o R Ⅰ㊁H i n d Ⅲ均购自宝日医生物技术(北京)有限公司;S Y B R P r e m i x E x T a q T M (2ˑ)购自T a K a R a 公司;T 4D N A 连接酶㊁pE T -28a (+)载体㊁大肠杆菌D H 5α感受态细胞㊁T I A N p r e p Mi n i P l a s m i dK i t 均购自天根生化科技(北京)有限公司㊂1.2 方法1.2.1 引物设计及合成 根据G e n B a n k 中原鸡S T I N G 基因序列(登录号:K P 893157.1),使用P r i m e rP r e m i e r 5.0软件设计普通P C R (下划线处为E c o R Ⅰ和H i n d Ⅲ酶切位点)和实时荧光定量P C R 引物,以G A P DH 为内参基因,引物信息见表1㊂引物均由生工生物工程(上海)股份有限公司合成㊂表1 引物序列信息T a b l e 1 P r i m e r s e q u e n c e i n f o r m a t i o n 基因G e n e s引物序列P r i m e r s e qu e n c e s (5'ң3')退火温度A n n e a l i n gt e m pe r a t u r e /ħ产物长度P r o d u c tl e n g t h /b p 用途A p p l i c a t i o n S T I N G F :C C G G A A T T C A T G C C C C A G G A C C C G T C A A C C 62.81140普通P C R R :C C C A A G C T T C T G G G C T C A G G G G C A G T C A C T S T I N G F :G C C C C A G G A C C C G T C A A C C A G 60.0114实时荧光定量P C R R :A G C A C C A C G A A G C A C A C A G C C AG A P DHF :G A C G T G C A G C A G G A A C A C T A 60.0122实时荧光定量P C RR :A T G G C C A C C A C T T G G A C T T T1.2.2 总R N A 提取及反转录 利用T r i z o l 法提取各组织总R N A ,溶于D N a s e /R N a s e -f r e ed d H 2O ,利用紫外分光光度计及琼脂糖凝胶电泳检测R N A 浓度及纯度,将R N A 反转录为c D N A ,―20ħ保存备用㊂1.2.3 S T I N G 基因C D S 区克隆及测序 以白来航鸡脾脏组织c D N A 为模板进行P C R 扩增㊂P C R 反应体系20μL :c D N A 模板0.5μL ,上㊁下游引物(10μm o l /L )各0.5μL ,P C R M a s t e r M i x10μL ,d d H 2O8.5μL ㊂P C R 反应程序:94ħ预变性5m i n ;94ħ变性30s ,62.8ħ退火30s ,72ħ延伸75s ,共35个循环;72ħ延伸7m i n ㊂P C R 产物用1.0%琼脂糖凝胶电泳检测,取鉴定正确的扩增产物回收纯化后连接p E T -28a (+)载体,转化大肠杆菌D H 5α感受态细胞,均匀涂布于含卡那霉素的L B 固体培养基上,过夜培养14h 后筛选出圆滑白色单菌落并进行菌液P C R 鉴定,将挑选得到的阳性克隆菌液送生工生物工程(上海)股份有限公司测序㊂1.2.4 生物信息学分析 使用M e g A l i g n 软件将白来航鸡与G e n B a n k 数据中已公布的原鸡(G a l l u s ga l l u s ,登录号:N P _001292081.2)㊁盔珠鸡(N u m i d am e l e a g r i s ,登录号:X P _021265823.1)㊁火鸡(M e l e a g r i s g a l l o pa v o ,登录号:X P _010717095.1)㊁白尾雷鸟(L a g o pu s l e u c u r a ,登录号:X P _042749793.1)㊁日本鹌鹑(C o t u r n i x j a po n i c a ,登录号:X P _015731739.1)㊁凤头朱鹮(N i p po n i a n i p po n ,登录号:K F Q 92075.1)㊁冠小嘴乌鸦(C o r v u s c o r n i xc o r n i x ,登录号:X P _039415878.1)的S T I N G 基因氨基酸序列进行相似性比对,并利用M e ga 7.0软件构建系统进化树㊂利用P r o t P a r a m 在线软件(h t t ps :ʊw e b .e x p a s y .o r g /p r o t pa r a m /)分析S T I N G 蛋白理化性质;利用P r o t S c a l e 在线软件(h t t p s :ʊw eb .e x p a s y .o r g /pr o t s c a l e /)分析S T I N G 蛋白亲/疏水性;利用S i g n a l P4.1(h t t ps :ʊs e r v i c e s .h e a l t h t e c h .d t u .d k /s e r v i c e s /S i g n a l P -4.1/)㊁T MHMM (h t t ps :ʊs e r v i c e s .h e a l t h t e c h .d t u .d k /)在线软件分别预测S T I N G 蛋白信号肽与跨膜区;利用S M A R T (h t t p :ʊs m a r t .e m b l -h e i d e l b e r g .d e /s m a r t /b a t c h .p l )和P S P R TⅡ(h t t p :ʊp s o r t .h g c .j p/f o r m 2.h t m l )在线软件分别预测S T I N G 蛋白结构域和亚细胞定位;利用47314期王艳等:白来航鸡S T I N G基因克隆㊁生物信息学及组织表达特性分析N e t P h o s3.1在线软件(h t t p s:ʊs e r v i c e s.h e a l t h t e c h.d t u.d k/se r v i c e.p h p?N e t P h o s-3.1)预测S T I N G 蛋白磷酸位点;利用Y i n O Y a n g1.2(h t t p s:ʊs e r v i c e s.h e a l t h t e c h.d t u.d k/s e r v i c e s/Y i n O Y a n g-1.2/)和N e t N G l y c1.0(h t t p s:ʊs e r v i c e s.h e a l t h t e c h.d t u.d k/s e r v i c e s/N e t N G l y c-1.0/)分别预测S T I N G蛋O-糖基化和N-糖基化位点;通过P r a b i S O P MA(h t t p s:ʊn p s a.l y o n.i n s e r m.f r/c g i-b i n/s e c p r e d_s o p m a.p l)和S W I S S M O D E L(h t t p s:ʊs w i s s m o d e l.e x p a s y.o r g/i n t e r a c t i v e)在线软件分别预测S T I N G蛋白二级结构和三级结构;利用S T R I N G12.0在线软件(h t t p s:ʊv e r s i o n-12-0. s t r i n g-d b.o r g/)和C y t o s c a p e软件进行蛋白互作分析㊂1.2.5白来航鸡S T I N G基因组织表达特性检测以鸡14个组织c D N A为模板,利用实时荧光定量P C R检测S T I N G基因在白来航鸡各组织中的相对表达情况,以G A P DH作为内参基因㊂P C R 反应体系10μL:c D N A0.5μL,上㊁下游引物(10μm o l/L)各0.3μL,S Y B R P r e m i x E x T a q T M (2ˑ)5μL,d d H2O3.9μL㊂P C R反应程序:95ħ2m i n;95ħ15s,60ħ1m i n,共45个循环;熔解程序:95ħ15s,60ħ1m i n;95ħ15s㊂每个组织样品设3个重复,采用2―әәC t法计算相对表达量㊂1.3数据统计分析通过S P S S26.0软件中单因素方差分析对基因表达量进行显著性检测,采用L S D和邓肯式法进行多重比较,利用G r a p h P a dP r i s m8.0软件作图㊂结果用平均值ʃ标准差表示,P<0.05表示差异显著㊂2结果2.1白来航鸡S T I N G基因C D S区序列克隆1.0%琼脂糖凝胶电泳显示,在约1140b p附近出现清晰条带(图1),与预期条带大小符合㊂应用D N AMA N软件分析测序结果显示,本试验克隆的白来航鸡S T I N G基因C D S区片段长为1140b p,编码379个氨基酸,碱基组成分别为: A(17.54%)㊁G(30.35%)㊁T(18.16%)㊁C(33.95%), G C含量高于A T含量,说明白来航鸡S T I N G基因C D S区的D N A双链较稳定㊂M,D L2000D N A M a r k e r;1~3,S T I N G基因P C R扩增产物;4,阴性对照M,D L2000D N A M a r k e r;1-3,P C Ra m p l i f i c a t i o n p r o d u c t s o f S T I N G g e n e;4,N e g a t i v e c o n t r o l图1白来航鸡S T I N G基因P C R扩增结果电泳图F i g.1E l e c t r o p h o r e t i c m a p o fP C Ra m p l i f i c a t i o nr e s u l t so f S T I NG g e n e i n W h i t eL e i g h o r n c h i c k e n s2.2生物信息学分析2.2.1相似性比对及系统进化树构建相似性比对结果显示,白来航鸡S T I N G基因编码氨基酸序列与原鸡㊁灰胸竹鸡㊁环颈雉㊁盔珠鸡㊁火鸡㊁白尾雷鸟㊁日本鹌鹑㊁朱鹮和冠小嘴乌鸦的相似性分别为99.7%㊁93.7%㊁89.1%㊁87.3%㊁90.2%㊁87.3%㊁83.6%㊁63.9%和62.0%(图2)㊂采用M e g a7.0软件中M L法构建系统进化树,结果显示,白来航鸡与原鸡之间的亲缘关系最近,与灰胸竹鸡的亲缘关系次之,朱鹮和冠小嘴乌鸦形成另一个分支,与冠小嘴乌鸦亲缘关系最远(图3)㊂2.2.2理化特性分析白来航鸡S T I N G蛋白分子式为C1897H3032N530O541S22,分子质量为42.625k u,理论等电点(p I)为6.67,推测该蛋白为酸性蛋白㊂组成白来航鸡S T I N G蛋白的20种氨基酸中,L e u 所占比例最高(17.2%),而A s n和M e t含量最低(1.3%),其中带负电荷的氨基酸(A s p和G l u) 42个,带正电荷的氨基酸(A r g和L y s)40个㊂在哺乳动物网织红细胞内的半衰期为30h,脂肪系数为105.01㊂S T I N G蛋白的不稳定系数为69.26,高于阈值40,表明该蛋白不稳定㊂2.2.3亲/疏水性预测白来航鸡S T I N G蛋白在第34位氨基酸处分值最高(3.500),在第257位氨基酸处分值最低(―2.789)(图4),总平均亲水性G R A V Y为―0.041,即S T I N G为亲水性蛋白㊂5731中 国 畜 牧 兽 医51卷图2 不同物种S T I N G 基因氨基酸序列相似性比对F i g .2 S i m i l a r i t y a l i g n m e n t o f a m i n o a c i d s e q u e n c e o f S T I NG g e n e a m o n g d i f f e r e n t s pe c i es 图3 基于S T I N G 基因氨基酸序列构建的系统进化树F i g .3 P h y l o g e n e t i c t r e e b a s e do na m i n o a c i d s e qu e n c e o f S T I N G g e ne 图4 白来航鸡S T I N G 蛋白亲/疏水性预测F i g .4 H y d r o p h i l i c i t y a n dh y d r o p h o b i c i t yp r e d i c t i o no f S T I NG p r o t e i n i n W h i t eL e gh o r n c h i c k e n s 67314期王 艳等:白来航鸡S T I N G 基因克隆㊁生物信息学及组织表达特性分析2.2.4 信号肽及跨膜区预测 白来航鸡S T I N G 蛋白不含信号肽(图5),推测该蛋白不是分泌型蛋白;该蛋白含4个跨膜区,分别位于第27―42㊁57―73㊁101―112及124―141位氨基酸处(图6),属于跨膜蛋白㊂图5 白来航鸡S T I N G 蛋白信号肽预测F i g .5 S i g n a l p e p t i d e p r e d i c t i o no fS T I NG p r o t e i ni n W h i t e L e gh o r n c h i c k e ns 图6 白来航鸡S T I N G 蛋白跨膜区预测F i g .6 T r a n s m e m b r a n e r e g i o n p r e d i c t i o no f S T I NG p r o t e i n i n W h i t eL e gh o r n c h i c k e n s 2.2.5 结构域预测及亚细胞定位 预测结果显示,白来航鸡S T I N G 蛋白具有1个保守结构域T M E M 173,位于第50―342位氨基酸处㊂白来航鸡S T I N G 蛋白大部分在线粒体和内质网上合成,在细胞核中合成较多,在细胞质中合成最少㊂2.2.6 修饰位点预测 白来航鸡S T I N G 蛋白丝氨酸㊁酪氨酸㊁苏氨酸磷酸化位点分别有25㊁3及5个(图7);存在43个O -糖基化磷酸位点(图8),不存在N -糖基化磷酸位点㊂图7 白来航鸡S T I N G 蛋白磷酸化位点预测F i g .7 P h o s p h o r y l a t i o n s i t e p r e d i c t i o no f S T I NG p r o t e i n i n W h i t eL e gh o r n c h i c k e n s 2.2.7 二级结构及三级结构预测 白来航鸡S T I N G 蛋白二级结构包括α-螺旋(54.62%)㊁延伸链(10.29%)㊁β-转角(3.43%)㊁无规则卷曲(31.66%)(图9)㊂白来航鸡S T I N G 蛋白三级结构主要以α-螺旋为主(图10),与二级结构预测结果一致㊂2.2.8 蛋白互作分析 将白来航鸡S T I N G 蛋白序列导入S T R I N G12.0在线数据库,结果显示,该蛋白与N F K B 1㊁D D X 41㊁c G A S ㊁T B K 1等蛋白存在互作(图11)㊂7731中 国 畜 牧 兽 医51卷图8 白来航鸡S T I N G 蛋白O -糖基化位点预测F i g .8 O -g l y c o s y l a t i o n s i t e p r e d i c t i o no f S T I N G p r o t e i n i n W h i t eL e gh o r n c h i c k e ns h ,α-螺旋;e ,延伸链;t ,β-转角;c ,无规则卷曲h ,A l ph ah e l i x ;e ,E x t e n d e d c h a i n ;t ,B e t a t u r n ;c ,R a n d o mc o i l 图9 白来航鸡S T I N G 蛋白二级结构F i g .9 S e c o n d a r y s t r u c t u r e p r e d i c t i o no f S T I NG p r o t e i n i n W h i t eL e gh o r n c h i c k e ns 图10 白来航鸡S T I N G 蛋白三级结构F i g .10 T e r t i a r y s t r u c t u r e p r e d i c t i o no fS T I NG p r o t e i ni n W h i t eL e gh o r n c h i c k e ns 图11 白来航鸡S T I N G 蛋白互作分析F i g .11 P r o t e i ni n t e r a c t i o n a n a l y s i so fS T I NG p r o t e i ni n W h i t eL e gh o r n c h i c k e n s 87314期王 艳等:白来航鸡S T I N G 基因克隆㊁生物信息学及组织表达特性分析2.3 白来航鸡S T I N G 基因组织表达分析通过实时荧光定量P C R 检测S T I N G 基因在白来航鸡14个组织中的表达情况,结果显示,S T I N G 基因在白来航鸡肺脏中的表达量最高,且显著高于其他组织(P <0.05);在胸肌中的表达量最低(图12)㊂肩标不同字母表示差异显著(P <0.05);肩标相同字母表示差异不显著(P >0.05)V a l u e sw i t hd i f f e r e n t l e t t e r s u p e r s c r i p t sm e a n s i g n i f i c a n t d i f f e r e n c e (P <0.05);W h i l ew i t h t h e s a m e l e t t e r s u p e r s c r i p t sm e a n n o s i gn i f i c a n t d i f f e r e n c e (P >0.05)图12 S T I N G 基因在白来航鸡不同组织中的相对表达量F i g .12 T h e r e l a t i v e e x p r e s s i o no f S T I NG g e n e i nd i f f e r e n t t i s s u e s o fW h i t eL e gh o r n c h i c k e n s 3 讨 论S T I N G 是固有免疫系统I F N -Ⅰ信号通路中的一个重要接头蛋白,定位在内质网上,可激活T B K I ㊁I K K ㊁I R F 3及N F -κB 通路,强烈刺激I F N -Ⅰ和白介素等免疫炎症因子的产生[11]㊂I F N -β在S T I N G 通路中分泌量最高,它不仅能够消灭癌细胞,还能通过促使树突细胞成熟实现抗原递呈,从而将固有免疫应答与获得性免疫应答相联系[12]㊂S T I N G 基因缺失使小鼠胚胎成纤维细胞(S T I N G -/-M E F s)极易受到负链病毒感染,如水疱性口炎病毒(V S V )和1型单纯疱疹病毒(H S V -1)[13]㊂本研究成功克隆白来航鸡S T I N G 基因C D S 区序列,为后期研究禽类S T I N G 蛋白抗病毒作用机制提供基础数据㊂系统进化树显示,白来航鸡与原鸡亲缘关系最近,与冠小嘴乌鸦亲缘关系最远,表明该基因在不同物种间保守性具有一定差异㊂白来航鸡S T I N G 蛋白为酸性㊁亲水性蛋白㊂以上结果表明该基因在不同物种的发育进化过程中表现出了种内相似性和种间差异性㊂白来航鸡S T I N G 蛋白无信号肽,说明该蛋白不是分泌蛋白,结合蛋白的亚细胞定位推测其主要存在于细胞质或线粒体中㊂另外,白来航鸡S T I N G 蛋白具有4个跨膜结构域㊂在S T I N G 二聚体中,8个T M 螺旋被分成由T M 2和T M 4组成的中间层和由T M 1和T M 3组成的外围层,其中1个S T I N G 分子的T M 1和另1个分子的T M 2㊁T M 3和T M 4被堆积在一起,形成结构域包裹的结构,这种保守的结构特征对蛋白的稳定性和功能至关重要[14]㊂白来航鸡S T I N G 蛋白存在T M E M 173保守结构域,预测结果显示其可促进先天免疫信号传导,与刘健新等[15]对山羊S T I N G 蛋白结构域的预测结果一致㊂白来航鸡S T I N G 蛋白中含有43个O -糖基化磷酸位点,这些修饰位点可能影响S T I N G 蛋白的空间构象㊁合成㊁运输㊁定位等生物过程,不存在N -糖基化磷酸位点,从而对该蛋白的不稳定性产生影响㊂S T I N G 蛋白共有33个潜在磷酸化位点,该蛋白本身的磷酸化是下游信号转导所必需的,其中含有的2个保守的丝氨酸和苏氨酸位点是T B K 1的磷酸化靶点㊂另外,S T I N G 磷酸化有助于其从细胞质到高尔基体的适当易位,开启免疫反应,且还参与防止S T I N G 过度激活和维持体内平衡[16]㊂蛋白质棕榈酰化是一种功能强大且保守的翻译后修饰,脂肪酸链通过可逆的硫酯键与蛋白质的半胱氨酸残基连接[17]㊂在高尔基体中,S T I N G 的2个半胱氨酸残基(C ys 88和9731中国畜牧兽医51卷C y s91)被棕榈酰化,激活其下游信号通路,这是S T I N G激活所必需的翻译后修饰[18]㊂蛋白互作预测结果显示,S T I N G蛋白与N F K B1㊁D D X41㊁c G A S㊁T B K1等蛋白存在相互作用㊂S T I N G蛋白可与c G A S结合形成第二信使c G A M P,使得S T I N G招募并激活T B K1,进而诱导I F N-Ⅰ和其他调节因子[19]㊂研究表明,当树突状细胞在感染H S V-1或腺病毒后,D D X41与S T I N G直接相互作用介导T B K1㊁N F-κB和I R F3的激活[20]㊂推测S T I N G 通过与这些蛋白互作,从而参与免疫及抗病毒过程㊂通过对白来航鸡S T I N G基因的组织表达特性分析可知,S T I N G基因的分布具有组织特异性,其中在肺脏中表达量最高,而石树端等[21]研究发现鸭S T I N G基因在腺胃中表达水平最高,表明家禽中m R N A组织分布存在一定差异;其次是脾脏,提示S T I N G基因广泛分布于宿主的免疫器官;此外, S T I N G基因在回肠㊁十二指肠等肠道组织中也有表达,因为肠道中的微生物菌群是启动免疫的重要因子,肠道微生物与宿主的互作建立机体的适应性免疫系统,可有效维持免疫防御作用[22];在胸肌中表达量最低,与金洁[7]研究结果一致㊂研究表明, S T I N G基因表达量降低可以提示肝癌和胃癌患者的预后不良[23-24],而注射S T I N G激动剂可使人乳头瘤病毒相关肿瘤组织消退[25]㊂另外,通过抑制S T I N G通路可以阻断衰老细胞释放促炎信号,从而改善老年神经退行性疾病[26]㊂因此,抑制或激活S T I N G信号通路,可为疾病治疗提供新的途径和特异性治疗靶点[21]㊂本试验通过对白来航鸡S T I N G 基因编码蛋白进行生物信息学和组织表达特性分析,为揭示S T I N G基因在固有免疫系统中的作用提供数据,并为后期探究S T I N G编码蛋白的生物学功能提供切实的理论基础㊂4结论本研究成功克隆了白来航鸡S T I N G基因C D S 区序列,大小为1140b p,编码379个氨基酸㊂白来航鸡与原鸡的亲缘关系最近,与冠小嘴乌鸦亲缘关系最远㊂S T I N G蛋白属于酸性㊁亲水性蛋白,存在跨膜结构,无信号肽,主要在细胞质和线粒体中发挥作用㊂S T I N G基因在白来航鸡肺脏中表达量最高,在胸肌中表达最低㊂参考文献(R e f e r e n c e s):[1] G U O Y,J I A N GF,K O N G L,e t a l.O T U D5p r o m o t e si n n a t e a n t i v i r a l a n d a n t i t u m o r i m m u n i t y t h r o u g hd e u b i q u i t i n a t i n g a n ds t a b i l i z i n g S T I N G[J].C e l l u l a r&M o l e c u l a r I m m u n o l o g y,2021,18(8):1945-1955.[2]J I A N G M,C H E N P,W A N G L,e t a l.c G A S-S T I N G,a ni m p o r t a n t p a t h w a y i n c a n c e r i m m u n o t h e r a p y[J].J o u r n a lo f H e m a t o l o g y&O n c o l o g y,2020,13(1):81.[3] Z HA N G R,Q I N X,Y A N G Y,e t a l.S T I N G1i se s s e n t i a lf o ra n R N A-v i r u st r ig g e r e da u t o ph a g y[J].A u t o p h a g y,2022,18(4):816-828.[4] E R G U N S L,L IL.S t r u c t u r a l i n s i g h t si n t oS T I N Gs i g n a l i n g[J].T r e n d s i n C e l lB i o l o g y,2020,30(5):399-407.[5] Z H O UJ,Z HU A N GZ,L I J,e t a l.S i g n i f i c a n c eo f t h ec G A S-S T I N G p a t h w a y i n h e a l t h a nd d i se a s e[J].I n t e r n a t i o n a l J o u r n a lo f M o l e c u l a rS c i e n c e s,2023,24(17):13316.[6]刘永振.鸡马立克氏病病毒调控D N A受体信号通路逃避宿主天然免疫反应的分子机制[D].北京:中国农业科学院,2019.L I U Y Z.A v i a n M a r e k sd i s e a s ev i r u sm e d i a t e st h eD N A-s e n s i n gp a t h w a y t oe s c a p eh o s t i n n a t e i m m u n er e s p o n s e[D].B e i j i n g:C h i n e s e A c a d e m y o fA g r i c u l t u r a l S c i e n c e s,2019.(i nC h i n e s e)[7]金洁.鸡S T I N G基因的克隆㊁表达及其抗病毒功能的研究[D].雅安:四川农业大学,2017.J I N J.M o l e c u l a r c h a r a c t e r i z a t i o n,e x p r e s s i o n a n df u n c t i o n a la n a l y s i s o fc h i c k e n S T I N G[D].Y a a n:S i c h u a nA g r i c u l t u r a lU n i v e r s i t y,2017.(i nC h i n e s e) [8]何慧芬,张璐,秦爱建,等.c G A S-S T I N G信号通路在畜禽疾病中的研究进展[J].微生物学通报,2022,49(12):5331-5341.H E H F,Z HA N G L,Q I N AJ,e t a l.R o l eo f c G A S-S T I N G s i g n a l i n g p a t h w a y i nl i v e s t o c k a n d p o u l t r yd i se a s e s:A r e v i e w[J].M i c r o b i o l o g y C h i n a,2022,49(12):5331-5341.(i nC h i n e s e)[9] Z HA N GZ,Z H O U H,O U A N G X,e t a l.M u l t i f a c e t e df u n c t i o n so fS T I N G i n h u m a n h e a l t h a n d d i s e a s e:F r o m m o l e c u l a r m e c h a n i s mt ot a r g e t e ds t r a t e g y[J].S i g n a lT r a n s d u c t i o na n d T a r g e t e d T h e r a p y,2022,7(1):394.[10]李玺源.新型靶向S T I N G抗肿瘤药物的研发及机制研究[D].北京:中国科学院大学,2023.L I X Y.N o v e la n t i-t u m o r d r u g d e v e l o p m e n t a n dm e c h a n i s m s t u d y b y t a r g e t i n g S T I N G[D].B e i j i n g:U n i v e r s i t y o fC h i n e s eA c a d e m y o fS c i e n c e s,2023.(i nC h i n e s e)[11]曹艳杰,何怡宁,唐宁,等.三黄鸡S T I N G胞外区基因克隆及原核表达[J].南方农业学报,2016,08314期王艳等:白来航鸡S T I N G基因克隆㊁生物信息学及组织表达特性分析47(8):1401-1405.C A O Y J,H E Y N,T A N G N,e t a l.C l o n i n g a n dp r o k a r y o t i ce x p r e s s i o no fS a n h u a n g c h i c k e n S T I N Ge x t r a c e l l u l a rd o m a i n g e n e[J].J o u r n a lof S o u t h e r nA g r i c u l t u r e,2016,47(8):1401-1405.(i nC h i n e s e)[12] MO TWA N IM,P E S I R I D I SS,F I T Z G E R A L D K A.D N As e n s i n g b y t h e c G A S-S T I N G p a t h w a y i nh e a l t ha n d d i s e a s e[J].N a t u r e R e v i e w s G e n e t i c s,2019,20(11):657-674.[13]I S H I K A W A H,B A R B E R G N.S T I N G i s a ne n d o p l a s m i c r e t i c u l u m a d a p t o r t h a tf a c i l i t a t e s i n n a t ei m m u n e s i g n a l l i n g[J].N a t u r e,2008,455(7213):674-678.[14] S HA N G G,Z HA N G C,C H E N ZJ,e t a l.C r y o-E Ms t r u c t u r e s o f S T I N G r e v e a l i t s m e c h a n i s m o fa c t i v a t i o nb yc y c l i c GM P-AM P[J].N a t u r e,2019,567(7748):389-393.[15]刘健新,彭斐,黎振标,等.山羊S T I N G的克隆及其在O F T u细胞的表达[J].中国兽医学报,2018,38(5):1013-1018.L I UJX,P E N GF,L I ZB,e t a l.C l o n e a n d e x p r e s s i o no fs t i m u l a t o ro fi n t e r f e r o n g e n e so f g o a ti n O F T uc e l l s[J].C h i n e s e J o u r n a l o f V e t e r i n a r y S c i e n c e,2018,38(5):1013-1018.(i nC h i n e s e) [16] P A NJ,F E ICJ,HU Y,e t a l.C u r r e n tu n d e r s t a n d i n go ft h ec G A S-S T I N G s i g n a l i n g p a t h w a y:S t r u c t u r e,r e g u l a t o r y m e c h a n i s m s,a n d r e l a t e d d i s e a s e s[J].Z o o l o g i c a l R e s e a r c h,2023,44(1):183. [17] WA N G Y,L U H,F A N GC,e t a l.P a l m i t o y l a t i o na s as i g n a lf o r d e l i v e r y[J].A d v a n c e si n E x p e r i m e n t a lM e d i c i n e a n dB i o l o g y,2020,1248:399-424. [18]胡燕萍,许锦霞,徐阿慧,等.宿主c G A S-S T I N G信号通路调控机制的研究进展[J].中国预防兽医学报,2022,44(12):1344-1351.HU YP,X UJX,X U A H,e t a l.R e c e n t a d v a n c e s i nt h e r e g u l a t i o n m e c h a n i s m o f h o s t c G A S-S T I N Gs i g n a l i n gp a t h w a y[J].C h i n e s e J o u r n a l o f V e t e r i n a r yM e d i c i n e,2022,44(12):1344-1351.(i nC h i n e s e) [19] Z HA N GC,S HA N G G,G U IX,e t a l.S t r u c t u r a l b a s i so f S T I N G b i n d i n g w i t h a n d p h o s p h o r y l a t i o n b yT B K1[J].N a t u r e,2019,567(7748):394-398.[20] MA T ZK M,G U Z MA N R M,G O O D MA N A G.T h er o l eo fn u c l e i ca c i ds e n s i n g i nc o n t r o l l i n g m i c r o b i a la n da u t o i m m u n ed i s o r d e r s[J].I n t e r n a t i o n a lR e v i e wo f C e l l a n d M o l e c u l a rB i o l o g y,2019,345:35-136.[21]石树端,高全新,程玉强,等.S T I N G基因荧光定量检测方法的建立及其在鸭组织中的分布[J].畜牧与兽医,2017,49(7):69-73.S H I S D,G A O Q X,C H E N G Y Q,e t a l.E s t a b l i s h m e n t o f R e a l-t i m e P C R f o r d e t e c t i o n o fS T I N G g e n ea n di t st i s s u ed i s t r i b u t i o ni nd u c k[J].A n i m a lH u s b a n d r y&V e t e r i n a r y M e d i c i n e,2017,49(7):69-73.(i nC h i n e s e)[22]李淑珍,常文环,刘国华,等.肉鸡肠道微生物及其调控[J].动物营养学报,2020,32(7):2989-2996.L IS Z,C HA N G W H,L I U G H,e t a l.I n t e s t i n a lm i c r o b i o t ao fb r o i l e rc h i c k e n sa n di t sr e g u l a t i o n[J].C h i n e s e J o u r n a l o f A n i m a lN u t r i t i o n,2020,32(7):2989-2996.(i nC h i n e s e)[23] B U Y,L I UF,J I A Q A,e t a l.D e c r e a s e d e x p r e s s i o no fTM E M173p r e d i c t s p o o r p r o g n o s i si n p a t i e n t s w i t hh e p a t o c e l l u l a r c a r c i n o m a[J].P L o S O n e,2016,11(11):e0165681.[24] S O N G S,P E N G P,T A N G Z,e t a l.D e c r e a s e de x p r e s s i o n of S T I N G p r e d i c t s p o o r p r og n o s i s i np a t i e n t s w i t h g a s t r i cc a n c e r[J].S c i e n t i f i cR e p o r t s,2017,7:39858.[25]张展,刘朝晖.S T I N G信号通路在H P V相关恶性肿瘤中的作用[J].国际妇产科学杂志,2023,50(1):30-34.Z HA N G Z,L I U Z H.R o l e o f S T I N G s i g n a l i n gp a t h w a y i n H P V-r e l a t e d m a l i g n a n t t u m o r s[J].J o u r n a l o f I n t e r n a t i o n a lO b s t e t r i c s a n dG y n e c o l o g y,2023,50(1):30-34.(i nC h i n e s e)[26] G U L E N M F,S AM S O N N,K E L L E R A,e t a l.c G A S-S T I N G d r i v e s a g e i n g-r e l a t e d i n f l a m m a t i o n a n dn e u r o d e g e n e r a t i o n[J].N a t u r e,2023,620(7973):374-380.(责任编辑晋大鹏)1831。
AS 400和S 390高性能打印协议转换器说明书

Axis Protocol Converters• Any printer resolution supported*GSA pricing available.Call for more information.©1998 Axis Communications AB.Axis is a registered trademark,and AXIS Pocket Cobra and AXIS Cobra+ are trademarks of Axis Communications AB.All other company names and products are trademarks or registered trademarks of their respective companies.Pocket Cobra Height: 1.2 in/2.9 cm Width: 2.2 in/5.5 cm Depth: 4.0 in/10 cm Weight:0.3 lb/0.15 kg Power 9V AC,min 300mA,provided by external power supply T emperature:40–105°F/5–40°C Humidity:10%–95% RHG non-condensing Cobra +Height: 1.0 in/2.5 cm Width: 3.5 in/9.0 cm Depth: 4.0 in/10.0 cm Weight:.55 lb/0.25 kg Power 9V AC — 12V DC provided by external power supply T emperature:50–105°F/5–40°C Humidity:20%–80% RHG non-condensing AFP Height: 1.3 in/3.2 cm Width:7.7 in/19.5 cm Depth: 5.2 in/13.3 cm Weight: 1.8 lbs/0.8 kg Power 12–15V AC or DC,min.500 mA provided by external power supply T emperature:40–105°F /5–40°C Humidity:10%–95% RHG non-condensing HP-MIO and OKI-500Units fit into internal slot provided by printer Power:Provided by printer T emperature:32–110°F/0–43°C Humidity:0%–90% RHG non-condensing APPROVALS Pocket Cobra,Cobra+,HP-MIO,AFP EMC:FCC Class A 12:EN 55022/1987EN 50082-1/1992Safety:EN 6095,UL,CSA OKI-500EMC:FCC Class A 12:EN 50081-1EN 50082-1Safety:UL,CSA,and TUV/GS Axis Protocol ConvertersAxis CommunicationsLund +46 46 270 18 00Boston +1 800-444-AXIS (2947)Paris +33 1 49 69 15 50London +44 1715 539200Munich +49 89 95 93 96 0T okyo +81 3 3545 8282Singapore +65 250 8077Hong Kong +852 2836 0813Beijing +86 10 6510 2705Shanghai +86 21 6445 4522T aipei +886 2 8780 5972Seoul +82 2 780 9636C X 001/U S /R 1。
tfautomodelforsequenceclassification类 -回复

tfautomodelforsequenceclassification类-回复首先,我们将介绍TF-AutoModelForSequenceClassification类的基本概念和作用。
然后,我们将深入探讨其在序列分类任务中的应用和工作原理。
最后,我们将讨论一些使用该类时需要考虑的关键因素和使用示例。
TF-AutoModelForSequenceClassification类是TensorFlow库中的一个类,用于处理序列分类任务。
序列分类是指根据输入的序列数据对其进行分类或标记。
这种类型的任务在自然语言处理(NLP)中特别常见,例如情感分析、文本分类和垃圾邮件检测。
作为一个通用的序列分类模型,TF-AutoModelForSequenceClassification类可以应用于不同的语言和不同的数据集。
它的作用是根据输入的文本序列,自动选择合适的预训练模型,并使用这个模型进行分类任务。
TF-AutoModelForSequenceClassification的使用非常简单,只需要三个步骤即可完成。
首先,我们需要导入需要的库和模块,并加载数据集。
其次,我们创建一个实例并配置模型。
最后,我们训练模型并进行预测。
对于第一步,我们需要导入TensorFlow库中相应的模块。
例如,导入tensorflow、tensorflow_datasets和tensorflow_text等模块。
然后,我们需要下载和加载适合我们任务的数据集。
TensorFlow库提供了许多常用的数据集,例如IMDB情感分析数据集和MNIST手写数字数据集。
接下来,我们创建一个TF-AutoModelForSequenceClassification的实例。
首先,我们需要指定模型的架构,例如BERT、RoBERTa、ALBERT 等。
我们还可以选择不同的超参数,如学习率、批量大小和训练时的迭代次数。
然后,我们将数据集分为训练集和测试集,以便进行模型性能的评估。
NovAtel学习记录

NovAtel学习记录1、信号类型:二进制、ASCII、缩写ASCII。
信号类型由信息名字后附加的字母来表示。
“A”表示ASCII,“B”表示二进制,无符号表示缩写ASCII。
2、字段类型:Char字符型,1字节(8bit比特);UChar无符号字符型,1字节;Short短整型,2字节;UShort无符号短整型,2字节;Long长整型,4字节;ULong无符号长整型,4字节;Double双精度浮点型,8字节;Float单精度浮点型,4字节;Enum枚举型,4字节(4字节枚举型以0开始,相当于ULong,在二进制中,枚举型数据直接输出,在ASCII和缩写ASCII中,枚举型数据被拼写出来);GPSec,4字节,有两个独立的格式,二进制输出时是毫秒、长整型的,ASCII输出时是秒、浮点型的;Hex,n 字节,二进制时是一个打包的、固定长度的字节阵,ASCII或缩写ASCII时则被转换成2字符的16进制对;String,n字节,二进制情况下是一个以null结尾的变长度字节阵,另外的填充字节则是为了保持4字节属性,每个字符域的最大字节长度在记录或命令表里的行上标出的。
3、字节布置:所有的数据送入或读出都是从最低位开始,数据从接收器的最低位(LeastSignificant Bit)LSB开始存储。
字符型数据里,LSB是0,最高位MSB(Most Significant Bit)则是7。
ASCII信息结构4、ASCII信息结构:(1)每条记录的先行代码标识符是“#”;(2)依据数据的量和形式,每条记录都是变长度的;(3)所有数据域由逗号隔开,不过有两个例外,一个是头域,以分号结尾,标志数据信息的开始,另外一个例外就是最后一个数据域,它以心号*开头,标志信息数据的结束;(4)每条记录都以十六进制结束,十六进制前面有一个*,后面有回车和换行,*1234ABCD[CR][LF],记录里的所有字节是一个32位的CRC,包括#标识符和四个校验和数字之前的*;(5)ASCII字符串是一个域,由双引号括起来,如果引号里面有逗号,字符串仍然是一个域,逗号将被忽略,字符串内不允许有双引号;(6)如果接受器在解析输入信息时检测到错误,它会返回一个错误提示。
sequence函数

sequence函数sequence函数是一个重要的Python函数,可以用来实现列表、元组、字符串的分解和组合。
它可以让处理数据变得更加容易,可以大大简化我们的编程代码。
sequence函数的基本语法如下:seq_function(iterable, start = 0, step = 1)其中,iterable为可迭代的参数,start为起始位置,step为步长。
首先,我们来看一下sequence函数的应用场景:(1)将列表、元组或字符串分解为多个单独的元素:seq_function([a b c d) #出:(a b c dseq_function(abcde #出:(a b c d e(2)从多个元素中组成一个序列:seq_function((a b c d #出:[a b c dseq_function(abcde #出:abcde(3)迭代字典项:dic = {a1, b2, c3, d4}for i in seq_function(dic):print(item i, value dic[i])输出:Item a value 1Item b value 2Item c value 3Item d value 4(4)让元素按照一定的顺序进行排列:seq_function(abcde start = 3, step = 2) #出:(d b a 另外,sequence函数还可以用来进行元素替换:seq_function(abcde start = 2, step = 2, replacement = [x y) #出:(a x c y e以上就是sequence函数的主要用法,只要掌握了它,就可以大大简化我们编写程序的代码。
那么它底层到底是如何实现的?其实它的实现方式比较简单,它的实现核心在于:1、通过start和step参数计算出当前元素的位置,并使用iterable获取当前位置的元素;2、使用replacement参数替换当前元素所以,sequence函数最核心的功能就是分解和组合元素,可以说它已经成为我们编写程序的一部分了,在很多地方它都可以发挥作用。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
A New Data Model for Time SequencesDmitry V. Maslov11 Nayanova University, Samara, Russiadmaslov@industrialauto.ruAbstract. In this paper we analyze the requirements for logical modeling of or-dered data (sequences). We examine the shortcomings of existing models andpresent a new one. Our model inherits the advantages of different existing ap-proaches and has some new features. Its main contributions are: a) integrationof data quality information into the model; b) "transform" operator, allowing tochange the calendar of the sequence to any specified calendar; c) extended ag-gregation operators, that can use the ordering information.1 IntroductionData ordered over time (time sequences) can be found in many diverse financial, in-dustrial and scientific applications. That's why a lot of people face the problem of storage and preliminary processing of sequences in DBMS.Today relational data model is the most prevalent and universal. However rela-tional data model is not well suited for modeling of sequences. The most important reason for this is that a relation – the main structural element of relational model – is an unordered set of tuples. Consequently, although sequence data can be stored in relational database, it cannot be manipulated effectively. For example, it is very diffi-cult (nearly impossible) to query 5-day moving average of some stock price using relational algebra. It is also difficult to transform a sequence with granularity of one day to a sequence with granularity of one month, or to make a join of two sequences with different calendars. The problems of modeling sequence data in relational data-base are more carefully described in [8, 14]. In [14] professor Dennis Shasha formu-lates the requirements for financial time sequences model, in [1, 15] the requirements for industrial time sequences model are formulated.Because of mentioned problems a lot of specialized data models that deal with se-quences were proposed during last 15 years. However, as shown in section 3, all of them have shortcomings. One of the tasks of this research is to determine structural and functional requirements for time sequences data model (or, moreover, arbitrary sequences data model). We propose a new data model that inherits the advantages of all existing approaches. The main contributions of our model are:· integration of data quality information into a model: the domain of the function that is modeled by the sequence, the area where the sequence approximates the function and the quality (mistake of approximation);· extended transform operator;· extended aggregation operators.The rest of this paper is organized as follows: in section 2 the requirements for time sequences data model are described; in section 3 the advantages and shortcomings of existing data models are analyzed; section 4 presents a new data model; section 5 contains the conclusions and the direction for future work.2 Requirements for Time Sequences Data Model2.1 Structural RequirementsHigh Abstraction Level. The model should deal with as wide class of objects as possible.Independent (Separated from Data) Representation of Calendar. Calendar is an ordered set of time points at which the values of the sequence are defined. The model of sequences should have the structural element representing calendar with following functions [2]: a) defining time points at which time sequences have values; b) describing sets of time points (e.g., in queries); c) defining constraints; etc.Data Quality Information. Time sequence is a way to describe some external process (function). That's why the model should include the information about this function's domain and the quality of approximation.2.2 Functional RequirementsAlgebra of Sequences. According to [4], algebra of sequences doesn't have to include all statistical and other functions for time series analysis. There are a lot of specialized software packages with such capabilities. The algebra of sequences should include basic retrieving and transformation capabilities: "transform" operator, changing a calendar of a sequences to a new arbitrary calendar; temporal aggregation; moving window aggregation; calculations that use one or more sequences (possibly of different types). The algebra should be closed, i.e. the result of any operator should be a time sequence.Integration with Relational Data. The model should allow the co-existence of relational and sequence data in one query. All relational operators should be extended to deal with new structural elements of the model.3 Related Research WorkThe model, described in [6], is general for temporal objects (time sequences) and spatial objects. But this model doesn't have the structural element representing calen-dar, and doesn't pay attention to the algebra of sequences.The model for time series, proposed in [15], is a sort of valid-time temporal data model and doesn't address the problems of sequence data modeling.The model of Time Series Management System, presented in [3, 4, 5], contains ba-sic structural elements for modeling of time sequences: events, calendars, time series and groups of time series; and basic operations: calendar transformation, filtration, aggregation. However this is not a formal logical model, but the concrete implementa-tion of Time Series Management System. Moreover, the model doesn't have the ability to integrate relational and sequence data.In the model [2, 9, 10] the concepts of time sequence and time sequence collection are defined. The notion of the type of time sequence (i.e. the type of the function it describes) is presented. This model presents a general form for all operators manipu-lating time sequences, and some specific operators. It also defines the algebra of cal-endars. However this model doesn't include the operation of arbitrary calendar trans-formation. Another drawback of this model is that time sequence exists not as individ-ual object, but as a part of time sequence collection which is modeled as a relation.The SRQL query language [7] defines the minimal extension to the set of relational operators to formulate sequence queries over relational data. The advantage of such approach is the simplicity of implementation and the natural integration of relational and sequence data; the drawback is the limited functionality.SEQ model, described in [11, 12, 13], is the most general logical data model for sequences. It is designed for any kind of ordered data. However it doesn't include the operation of arbitrary calendar transformation (with many-to-many relation between source and destination calendars). Temporal aggregation ("g-collapse") doesn't pre-vent the ordering information inside the group (e.g. it is impossible to use aggregation function that selects n-th element in each group).To the best of our knowledge, no time sequence data model includes data quality information.4 A New Data Model for Time Sequences4.1 FundamentalsLet O be a set (finite or infinite), - full ordering relationship defined on this set.p Definition 1. {}j c =C , m j ,1= is a calendar if the following properties hold:·O c j j Î"|;·k j c c k j k j p Þ<"|,.An empty set Æ is also a calendar (empty calendar ).Calendar has dual nature. On one hand it is a succession, on the other hand it is aset. That's why standard set operations are applicable to the calendars.We define a sequence schema as a list of named attributes ,where each attribute A ),...,,(21n A A A A =i has the type T i , i.e. its values belongs to some set T i . The event of schema A is an element . Event contains the values of the sequence's attributes at some time point.n n T T T a a a a ´´´Î=...),...,,(2121Let T i * be the set of all non-empty subsets of T i . We define an approximate event ofschema A as an element , where each a **2*1**2*1*...),...,,(n n T T T a a a a ´´´Î=i * is some set of values of type T i , reflecting an approximate value of attribute A i .We say that a * is an approximation of a (), if *a a »*|,1i i a a n i Î=".Let be a set of all possible events and letbe a set of all possible approximateevents, where v n T T T E ´´´=...21{...**2null n v T T È´´´}{}*1*uncertain v T E È=null and v uncertain are some special values. Then each element of E * is either some approximate value or special value (v uncertain or v null ). The value v null de-notes the fact that the event is not defined at particular time point, v uncertain denotes the fact that the event is unknown at particular time point (and, moreover, it is unknown if the event is defined or not).Let be some function. For each element that belongs to its domainD it defines an event belonging to . Let a be a value of F at some point c ; and let aE O DF ®Í|n T T T ´´´...21* be an approximation of a . Then a * approximates the function F at point c . We assume that v null approximates any function at the points where the function is not defined.Now we are ready to define a time sequence as a collection of approximate events,corresponding to consecutive time points and approximating the functionE O DF ®Í|.Definition 2. A sequence H of schema A , approximating the function , is a 5-tuple , where{}j c C =E O D F ®Í|),,,,(.in M f D M V C · , m j ,1= is a calendar of the sequence;· {}j v V =, m j ,1= is a collection of values of the sequence, and the following properties hold:o ; *|E v j j Î"o if then approximates F at point с|j "uncertain j v v ¹j v j ;· is a set, where sequence H approximates the function F (which meansthat we know the points inside M , where the function F is defined, and usinginterpolation function f O M Íin we can calculate the approximation of F at any ofthese points);· is a set of the points inside M where F is defined (in generalcase we do not know such points outside M );M D D M Ç=·is an interpolation function that defines an ap-proximate event, approximating the function F , at each point**2*1...),,(n in T T T c V C f ´´´®C c D c M ÏÎ,. Let ℋ be the set of all possible sequences.So, a sequence for each time point of the calendar defines an approximate event.Depending on we can define different types of sequences. Let us note the most important of them:in M f D M F ,,,· Discrete – if , i.e. inside M the function F is defined only at the timepoints of calendar of the sequence. In this case the interpolation function is notused;C D M Í· Step-wise constant – if inside M the function F changes its value only at time points of calendar of the sequence;· Linear – if F is a numerical function and f in is a linear function, approximatingF with an error not greater than some e .Example 1. An example of discrete sequence is an information about some goods sales. To each time point of the calendar an information about a sale that took place at this time point corresponds.Example 2. Suppose some measurements periodically received from some sensor. Let d be the relative error of the sensor and e - the maximum absolute error of linear approximation. Then each value of the sequence is an interval (, where v is a measured value. The interpolation function of the sequence produces the intervals . The set M contains the interval of sensor's normal operation (the intervals of sensor's faults or system faults are excluded from M ). Outside M we cannot calculate the value of F using interpolation.),d d ×+×-v v v v ),(d n e d e ×++×--in in v v v Definition 3. Let R be a relational schema, A – sequence schema. Then an extended relation with schema (R.A) is a set of elements, consisting of a tuple of schema R and a sequence of schema A .Extended relation is an object providing the integration of relational and sequencedata in one model. In extended relation each tuple is extended with sequence, which is viewed not as an attribute of special type, but as an individual object with its own algebra. The paper [12] has already proposed a few operators for such objects. One of the future tasks of our research is to define the algebra for extended relations.Such approach to the integration of relational and sequence data has following ad-vantages:· The ability to add and remove sequences using DML-operations (that is im-possible if each sequence is modeled as a relation);· The ability to select several sequences by some criteria (that is impossible if each sequence is modeled as a relation);· The ability to combine relational and sequence data in one query, e.g. join se-quence and relation (which is impossible if the sequences are modeled as at-tributes of abstract data type);· In degenerate case (if sequence schema contains no attributes) extended rela-tion becomes classical relation, which means that proposed data model con-tains relational data model.Example 3. Suppose that some database contains an information about different companies' stocks sales. Such information can be modeled using extended relation. Each tuple of this extended relation contains general info about the company (relational part) and the history of stocks sales (sequence part).4.2 Algebra of SequencesCalendar Transformation Operator. Assume that the sequence , where , , approximates the function and let be some point. Then the value of H at point c is defined as follows:E O DF ®Í|ì=$|if ,c j v c ),,,,(in M f D M V C H ={}O c C j Í=m j ,1=O c Î)ïïîïïíÏÎÏ=cases other in the ),,,(,if ,if ,)(c V C f D c M c v M c v c H inM null uncertainj j Obviously, if , then approximates F at the point c.uncertain v c H ¹)((c H Let , O c C k Í=}'{'l k ,1= be some calendar. The calendar transformation opera-tor is designed to derive from H a sequence that has approximate values of F at the points of calendar C'.Definition 4. transform , where)',,,','(')',(in M def f D M V C H C H ==·, )}'({'k c H V =l k ,1=; ·, where j is such interpolation function transformation that H' approximates F inside M .)',('C H f in j = Operator j depends upon the implementation of the model. The simplest exampleof possible j is an operator that always return "undefined" interpolation function f undef ,which for all points, that don't belong to , returns (approximateevent that for each attribute contains the set of all its possible values).'C ),...,,(**2*1*n T T T a =Proposed calendar transformation operator has the following advantages:· The result of the operator is always a sequence (which, by definition, approxi-mates the function F );· Being a sequence, the result of the operator contains the information about the quality of approximation (the set of possible values of all attributes).Temporal Aggregation Operator. Like previous operator, temporal aggregation operator changes the calendar of the sequence. But, in contrast to "transform" operator, the result of temporal aggregation is a sequence that is result of computations that use the values of function F .Definition 5. Assume that the sequence , where ),,,,(in M f D M V C H =, , approximates the function . LetO' be some ordered set; y - some function. C' is used to denote C , E O D F ®Í|'{)('c C ==y '|O O ®l k ,1=m l £aggregatio , .)',',''in M f D ,,(H n y aggr {}O c C j Í=m j ,1='}O k Í,','(')_def M V C H func ==, where· , }'{'k v V =l k ,1=E , , whereis a calendar that consists of points such that y ;aggr_func | ℋ ® is an aggregation function that for input sequence de-fines an approximate event; )))'(,((_'1k k c H transform func aggr v -=y C c Î)'(1k c -y k c c ')(=*·; },')(,|''{'M c c c c C c M Î=$Î=y ·;},')(,|''{''M M D c c c c C c D Î=$Î=y · f'in is not used, because D'M' Ì C' (H' is discrete). The operator of temporal aggregation divides the input sequence into non-intersecting parts; to each part an aggregation function is applied. The input of aggre-gation function is a sequence (not a set), that is why it can use the ordering informa-tion. For example, aggregation function can select n-th element from each subse-quence, which is impossible with usual aggregation functions.Now let's define another, simplified, form of temporal aggregation operator. As-sume , and let be some calendar. We define the function y as fol-lows: y . Then O O =')('c C O C Î'min 'c or c =p 'C )'(',''c c c C c Î=)_,,()_,',('*func aggr H n aggregatio func aggr C H n aggregatio C defy =.Positional Operators. In the paper [12] a group of "positional" operators was defined: unary and binary positional operators, and moving window aggregation. Now we define positional operators for sequences defined in this model.Let and be the sets of all possible events with corresponding schemas, let G be some function,11211..n n T T T E ´´´=22221..k k T T T E ´´´=n E |k E ®k i T i ,1,2=®(G E g n i | - its coordinate functions. Let be someapproximate event. Then is defined as follows:***2),...,n n E a Î*1,(a a =*a )*a · Gnull null v v G =)(;uncertain uncertain v v =)(;·if then G , where ; )(|*G dom a a a λ"(|{)2*a g x T x a i i =Î=))(),..,(),(()(**2*1*a g a g a g a k =}*),(a a g i »·if then ; )(|*G dom a a a Ï»"null v a G =)(*· in the other cases .uncertain v a G =)(*Obviously, if a and G then .*a »uncertain v a ¹)(*)()(*a G a G »Definition 6. Assume that the sequence , where),,,,(in M f D M V C H =, , approximates the function . , whereD F Í|)',,('in M f C ({v G ',','D M V )}j ')(def H H G =='V =m ,1j ={}O c C j Í=m j ,1=E O ®·, ; ·; }))((|{'uncertain v c H G c M M =-=·; )}(),(|{)'(''G dom a c H a c M D D M M Ï»"-Ç=·, where j - is such interpolation function transformation that H' approximates G(F) inside M'.),('G H f in j =Let , , be the sets of all possible events with corresponding schemas, let esome function, 1121111..n n T T T E ´´´=2222122..n n T T T E ´´´=33231..k k T T T E ´´´=n n E E G ®´21k E | b k i T i n ,1,32=®***2*122),...,,(n n E b b b Î=**E E g n i |1´***2*111),...,,n n E a a Î*b ),(**b a G ** - its coordinate functions. Let, be some approximate events.Thenis defined as follows: *(a a =·if or , then G ; null v a =*null v b =*null v b a =),(*·if and b , or b and , then ; uncertain v a =uncertain v b a =),(***null v ¹uncertain v =null v a ¹*G *· if ", then, where;)(),(|,G dom b a b b a a λ"»),..,,(),,((),(**2**1**g b a g b a g b a =),,(|{),(3**a b a g x T x b a i i »=Î=**)),(**b a G k },**b b a g i »*·if , then G ; )(),(|,G dom b a b b a a Ï»"»"**null v b a =),(*· in the other cases .uncertain v b a G =),(***Obviously, if a , b , and G , then .*a »b »uncertain v b a ¹),(),(),(**b a G b a G »Definition 7. Assume that , where ),,,,(111111in M f D M V C H ={}O c j Í=C , , approximates the function ,, approximates the function . , where11n E O ®Í22n E O ®Í(2C H =,(1H H G 2in M,',',(')2def M V C H ==)},({'21j j v v G V =2|D F )','in M f 'D m j =,1m j ,1=1|D F ),,,,2222f D M V ·, ; ·; }))(),((|{)('2121uncertain v c H c H G c M M M =-Ç=·; )}(),(),(),(|{)'('2121'21G dom b a c H b c H a c M D D D M M M Ï»"»"-ÇÇ=·, where j - is such interpolation function transformation that H' approximates G inside M'. ),,('21G H H f in j =),(21F FDefinition 8. Assume that the sequence , where ),,,,(in M f D M V C H =, , approximates the function Letaggr_window(с) be some function that for each point defines a calendar insideO .O D F ®ÍO c Îdef {}O c C j Í=m j ,1=E |.')_,_,(_H func aggr window aggr H n aggregatio window =)',',',',(''in M f D M V C =, where , where H · , }'{'j v V =m j ,1=, ,where aggr_func | ℋ ® is an aggregation function that for input se-quence defines an approximate event;)))(_,((_'j j c window aggr H transform func aggr v =*E · Æ};¹ÇÎ=M c window aggr C c M )(_|{'·Æ};¹ÇÎ=M M D c window aggr C c D )(_|{''·f'in is not used, because D'M' Ì C' (H' is discrete). 5 Conclusions and Future WorkProposed data model is a general data model for all kinds of ordered data. Althoughwe call it "A Model for Time Sequences", we never used the assumption that the or-dering domain is time.Our data model integrates the data quality information. We have defined operatorsof flexible calendar transformation and temporal aggregation. Proposed algebra ofsequences is closed, which means we can use the output of any operator as the input ofanother operator, thus constructing complicated expressions.We are looking to the following areas for the further research:· Definition of algebra of extended relations and further development of se-quences algebra;· Development of optimization techniques for operators of sequences algebra; · Development of effective storage methods for sequence data.References1. Bonnet, P., Gehrke, J., Seshadri, P.: Towards Sensor Database Systems. Mobile Data Man-agement (2001) 3-142. Chandra, R., Segev, A.: Managing Temporal Financial Data in an Extensible Database.Proceedings of VLDB (1993) 302-3133. Dreyer, W., Kotz, A.D., Schmidt, D.: An Object-Oriented Data Model for a Time SeriesManagement System. Proceedings of SSDBM (1994) 186-1954. Dreyer, W., Kotz, A.D., Schmidt, D.: Research Perspectives for Time Series ManagementSystems. SIGMOD Record, Vol. 23, No. 1 (1994) 10-155. Dreyer, W., Kotz, A.D., Schmidt, D.: Using the CALANDA Time Series Management Sys-tem. Proceedings of ACM SIGMOD International Conference on Management of Data (1995) 4896. Erwig, M., Schneider, M., Gting, R.H.: Temporal and Spatio-Temporal Data Models andTheir Expressive Power. FernUniversitt Hagen, Informatik-Report 225 (1997) /erwig97temporal.html7. Ramakrishnan, R., Donjerkovic, D., Ranganathan, A., Beyer, K.S., Krishnaprasad, M.:SRQL: Sorted Relational Query Language. Proceedings of SSDBM (1998) 84-958. Schmidt, D., Dittrich, K.A., Dreyer, W., Marti, R..: Time Series, a Neglected Issue in Tem-poral Database Research?. Proceedings of the Int. Workshop on Temporal Databases (1995) 214-2329. Segev, A., Shoshani, A.: A Temporal Data Model Based on Time Sequences. In: TanselA.U. et al.: Temporal Databases – Theory, Design and Implementation. The Benja-min/Cummings Publ. Comp. (1993) 248 - 26910. Segev, A., Shoshani, A.: Logical Modeling of Temporal Data. Proceedings of ACMSIGMOD (1987) 454-46611. Seshadri, P.: Management of Sequence Data. Ph.D. Thesis. University of Wisconsin, CS-Dept (1996)12. Seshadri, P.: SEQ: A Model for Sequence Databases (1995)/2302.html13. Seshadri, P., Livny, M., Ramakrishnan, R.: The Design and Implementation of a SequenceDatabase System. Proceedings of VLDB (1996) 99-11014. Shasha, D.: Time Series in Finance: the array database approach./cs/faculty/shasha/papers/jagtalk.html15. Wolski, A., Kuha, J., Luukkanen, T., Pesonen, A.: Design of RapidBase - An Active Meas-urement Database System. IDEAS (2000) 75-82。
