Valrubicin_56124-62-0_DataSheet_MedChemExpress
VARIANT IIβ-地贫试剂

更换新的分析柱
选择CDM中的Run/Worklist屏幕来启动机器。 按开始键启动工作菜单。 运行结束后,注意第二个校准液管(6号管) 的血红蛋白HbA2的滞留时间。最佳时间是 3.65分钟(参考试剂盒中分析柱说明书)。 若HbA2的滞留时间测量值的变化幅度超过 0.05分钟,建议进行温度调节
样本测试
样本测试
选择CDM中的Run/Worklist屏幕来启动机器。 按开始键启动工作菜单。详细信息参看手 册第4.0章节。 每次分析校准物结束后,都将自动计算出 A2和F的反应因子,该因子将用于计算所有 后面样本的A2和F面积的百分比值
结果分析
结果分析
在对结果进行解释和说明时应遵循如下原则: HbA2 的滞留时间是3.65 ± 0.10,HbA2 或HbF 的反应因子需在0.7—1.3之间的范围内。 HbA2 的正常范围一般通常占全部血红蛋白的 1.75%到3.25%,而ß-海洋性贫血的范围则在 4.0% 到 9.0%之间。 通常HbF的正常范围一般占全部血红蛋白的不到 1%,杂合子和纯合子状态的ß-海洋性贫血的病人 其HbF可分别达到1-5%和80-100%。 每例分析的全面积波动在1,000,000 到3,000,000 µv.秒之间。若超出此范围产生的结果则不应报告。
试剂准备
Hb A2/F 校准液 在每安瓶Hb A2/F校准液中加入10ml校准液 稀释液以对其进行配置。 轻摇以使之彻底溶解和混合。 配置好的Hb A2/F 校准液15-30 °C下静置 10分钟。在2-8 °C下储存可保存10天。若 药片发黄或瓶体破裂,请勿使用。 取1ml校准液于贴好标签的管中
期望值
在使用本程序双重测定时,使用了68名正 常的加州男女样本。血红蛋白A2 的正常值 是2.8%. 其 95%可信区间是2.3% 到3.3%。 在确定血红蛋白F时双重测定使用了46例加 州正常男女的样本,其均值是<1.0%,95% 可信区间是<1.0%。 每个实验室均应根据自己的情况和贫血类 型确定自己的范围
SAS-1
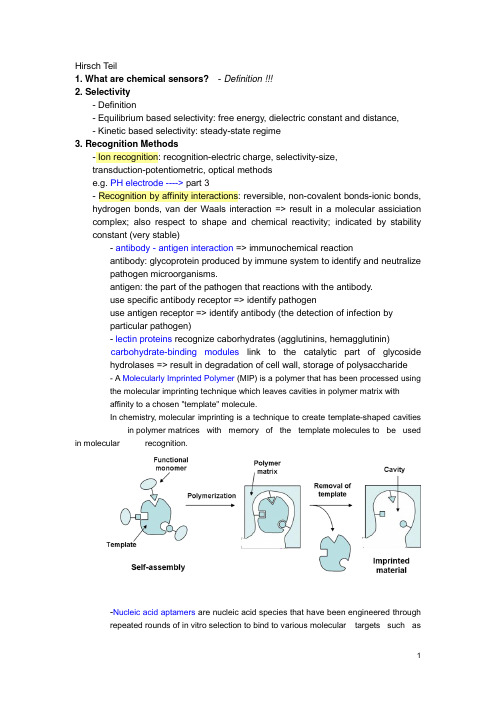
Hirsch Teil1. What are chemical sensors?- Definition !!!2. Selectivity- Definition- Equilibrium based selectivity: free energy, dielectric constant and distance,- Kinetic based selectivity: steady-state regime3. Recognition Methods- Ion recognition: recognition-electric charge, selectivity-size,transduction-potentiometric, optical methodse.g. PH electrode ----> part 3- Recognition by affinity interactions: reversible, non-covalent bonds-ionic bonds, hydrogen bonds, van der Waals interaction => result in a molecular assiciation complex; also respect to shape and chemical reactivity; indicated by stability constant (very stable)- antibody - antigen interaction => immunochemical reactionantibody: glycoprotein produced by immune system to identify and neutralizepathogen microorganisms.antigen: the part of the pathogen that reactions with the antibody.use specific antibody receptor => identify pathogenuse antigen receptor => identify antibody (the detection of infection byparticular pathogen)- lectin proteins recognize caborhydrates (agglutinins, hemagglutinin)carbohydrate-binding modules link to the catalytic part of glycosidehydrolases => result in degradation of cell wall, storage of polysaccharide- A Molecularly Imprinted Polymer (MIP) is a polymer that has been processed usingthe molecular imprinting technique which leaves cavities in polymer matrix withaffinity to a chosen "template" molecule.In chemistry, molecular imprinting is a technique to create template-shaped cavities in polymer matrices with memory of the template molecules to be used in molecular recognition.-Nucleic acid aptamers are nucleic acid species that have been engineered throughrepeated rounds of in vitro selection to bind to various molecular targets such assmall molecules, proteins, nucleic acids, and even cells, tissues and organisms.Aptamers are useful in biotechnological and therapeutic a pplications as they offer molecular recognition properties that rival that of the commonly used bimolecular antibodies.- Recognition by nucleic acids: hydrogen bonds between two distinct pairs of nucleobases => two complementary nucleic acids form a double strand association complex => called hybridizationnucleic acid sensors: short single strand NA as receptor to recognize a particular NA sequence in the analyte NA => detection of genetic anomalies and pathogen mircoorganism- Recognition by enzyme: dynamic processEnzyme: protein compound that function as catalysts in chemical reaction occurring in living system.- Recognition by cells and tissues: advantages of enzyme incorporated in biological materials => in their natural environmentsee part 3, Wegener - Recognition by gases and vapors: based on sorption at solid material => surface-adsorption, inner-absorption; purely physical phenomenon or chemical reaction.4. Transduction MetohdosChemical transduction: monitoring the change of chemical composition of the sensing element in response to the recognition process. => change in concentration/amount is measured => detect primary product -> secondary product or coreagent -> labeling productLABEL can be a simple molecular species or nanoparticals that can be detected by available physiochemical methods => enzyme, fluorescent dyes, luminescent dyes, electroactive compoundsPhysical transduction: a specific physical property of the sensing element that is affected by its interaction with the analyte is monitored. => mass, reflective index, dielectric properties, electrical resistivity => LABEL-FREE- Thermometric transductionRecognition of the analyte leads to change in temperature => only catalytical processes generate sufficient heat to the measurement => application: combustible gases react with O2 at the surface of a catalyst.- Transduction based on mechanical effectsRecognition leads to change in mass of the sensing element => monitored by mass tranducer based on quartz crystal microbalance (QCM)----------------------------------------------------------------------------------------------------------------- QCM, correct name: Thickness shear modePiezoelectric effect:generation of electrical charges on the surface of a solid by strain, pressure or torsion (mechanical deformation of solid) =>electricity resulting from pressureI nverse piezoelectric effect:application of charges to surfaces of piezoelectricsolid generates mechanical deformation (elongation, contraction, torsion)QCM is based on Inverse piezoelectric effect!# AT cut => 35`15`=> minimum temperature coefficient at 50~70 CIt makes the AT-cut well suited to applications requiring high degree of frequency stability over wide temperature ranges.## Electrodes are applied on both sides, and AC voltage applied.DC cannot flow across the crystal because it consists of an insulator material;however the crystal somewhat behaves as capacitor and allow an AC current to f low along the left-hand loop.AC voltage applied => leads to shear oscillation of crystal => when the voltage frequency matches the intrinsic vibration frequency of the crystal => the vibration amplitude is at maximal => the resonant => resonant frequency (f0) => depend on crystal thickness (e.g. d q= 330 um, f0= 5MHZ), density and elasticity of piezoelectric material### AT-cut resonator: thickness: ~0.2 mm, diameter of the active area: 5~20 mm #### Deposition of a homogenous mass film (a rigid overlay)Sauerbrey equation:Cf indicate sensitivity of QCMcondition of this equation: rigid deposited mass; △m<2% of crystal mass;operated in vacuum or in gaseous atmopphereIn liquid: the liquid breaks the vibration by friction => lessen f0Thickness of the layer must be greater than the wave decay lengththat is of 250 nm of 5 MHz resonator at water. ----> part 2!!!##### QCM in practice => see p.41----------------------------------------------------------------------------------------------------------------- - Resistive and capacitive transductionRecognition leads to changes in the electrical property of this materialResistive transduction: gases interact with MOS => change in electrical resistivity Capactive transduction => dielectric constant- Electrochemical transductionsee part 2, Matysik - Optical transductionOptical transduction can be based on light emission or light absorption, also by physical quantity (reflective index) and light scattering.5. Sensor Configuration and Fabrication- Lateral flow assayA typical test strip consists of the following components:1. Sample pad – an absorbent pad onto which the test sample is applied2. Conjugate pad –this contains antibodies specific to the target analyte;conjugated to coloured particles (e.g. gold nanoparticles)3. Reaction membrane –typically a hydrophobic nitrocellulose or celluloseacetate membrane onto which anti-target analyte antibodies are immobilized in a line across the membrane as a capture zone or test line, and a control zonecontaining antibodies specific for the conjugate antibodies.4. Wicking pad –a further absorbent pad designed to draw the sample acrossthe reaction membrane by capillary action and collect it.Double antibody sandwich assays: the sample migrates from the sample pad through the conjugate pad where any target analyte present will bind to the c onjugate.=> The sample then continues to migrate across the membrane until it reaches the test line where the target or conjugate complex will bind to the immobilized antibodies producing a visible line on the membrane. => The sample then migrates further along the strip until it reaches the control line, where excess conjugate will bind and producea second visible line on the membrane.This control line indicates that the sample has migrated across the membrane as intended. Two clear lines on the membrane is a positive result. A single line in the control zone is a negative result. Double antibody sandwich assays are most suitable for larger analytes, such as bacterial pathogens and viruses, with multiple antigenic sites. 6. Methods and Material in Sensor Preparation- Immobilization at solid surface => integration of a transducer with the receptor Physical adsorption at a solid supportNon covalent immobilization at solid surface => hydrophobic interaction, hydrogen bonding, electrostatic attraction; monolayer; no restrict access; not stable; Langmuir isotherm -> equilibrium interactionSupport material: silica, cellulose acetate, PVCCovalent bonding to the solid supportCovalent conjugation => stable, covalent bond, time consuming, expensiveCommon reactive group: -OH, -NH2, -C=O, -SH- Carboxylic acid with DCC- Glutaraldehyde reacts with the a.a. of lysine in protein => widely used Support: porous material => high specific area, high density of immobilized compounds => hydrogel: immobilized by entrapment/covalent corsslink - Natural polymers: Cellulose, Dextran- Synthetic polymers: Polystyrene- Active polymers: Epoxide (without preliminary activation) -->DNA array !!!- Inactive Polymers: Vicinal hydroxyls actived by CNBr- Inorganic support: Silica, AL2O3, TiO2 => stable at extreme PH- Metal support: noble metals, thiols on golds --> self assembled monolayers!Affinity reaction: avidin-biotin !!!Thin molecular layers: one or several molecular layers in solid support - Self-assembly of amphiphilic compounds: preparation of liposome andmicelles; liposome can be used of entrapment of molecular- Bilipid layer membranes: Langmuir-Blodgett technique- Layer by Layer assembly- Sol-Gel chemistry methods: silica gel => -O-Si-O-- Hydrogels: Xerogel, aerogel- Conducting polymers: Polyacetylene, polyaniline --> gas senor based on CP (----> part 3 !!!); also as entrapment matrix for biological receptors- Mesoporous materials: porous materials with pore (diameter: 2-50 nm,close to protein) => enzyme immobilization by entrapment (crosslinking withglutaraldehyde)- Deposition of polymers onto solid surfaces: dip coating, drop coating, spin coating ----> part 2 !!!Perm-selective memberanes: Nafin ----> Clark oxygen electrode Support-free crosslinkingEntrapment in a polymer networkEncapsulation7. Microfabrication Methodes- Spot Arraying: Contact-based & Noncontact-based; DNA microarray !!!!!Pros & Cons- Thick-film Technology: screen-printing technique (5-50 um thick layer)- Thin-film Technology: Photolithography (2 um)- Softlithography ----> experiment !!!!- Microcontact printing ----> experiment !!!!8. Optical Sensors- Electromagnetic RadiationOptical sensor => interaction of electromagnetic radiation with sensor layer - frequency; wavelength; photon energy (definition)- Structure: integration with wavelength-selection (optical filters) device and light sources (lasers), light detectors (phototransistors)- Optical Waveguides- Optical FibersOptical fibers' structuretotal internal reflection => evanescent wave- Spectrochemical Transduction MethodsSpectrochemical method analysis => light absorption or emission by sample => optical label performs absorption or emission (organic dye or metal complexes) - Light absorption: absorbance => concentration; sensitivity => thickness, absorpyivity, absorptivity => wavelength- Diffuse reflectance spectrometry: refelctance => concentration; suitable forsolid in near IR- Luminescence: Fluorescence spectromerty => fluorophore (label, organic dye or metal complexes, luminescent nanparticle ); steady-statefluorescence measurement, Time-resolved fluormetry; fluorescencequenching; resonance energy transfer (FRET); chemical- andbioluminescence => luminol; electrochemicaluminescence; Ramanspetrometry- Surface Plasmon Resonance Spectroscopy (SPR)。
hss-p-5.75.09 - hyaluronic acid derivatives说明书

5.75.09Section:Prescription DrugsEffective Date: April 1, 2020 Subsection: Neuromuscular Drugs Original Policy Date: June 9, 2011 Subject:Hyaluronic Acid DerivativesPage:1 of 7Last Review Date:March 13, 2020Hyaluronic Acid DerivativesDescriptionDurolane, Euflexxa, GelSyn-3, GenVisc 850, Hyalgan , SodiumHyaluronate, Supartz , Synojoynt*, Triluron, TriVisc, Visco-3 (sodium hyaluronate)Gel-ONE , Hymovis, Monovisc, Orthovisc (hyaluronan)Synvisc, Synvisc-One (hylan G-F 20)Bolded medications are the preferred products*These medications are included in this policy but are not available in the market as of yetBackgroundOsteoarthritis of the knee is a disease in which the elastoviscous property of the synovial fluid in the knee joint becomes diminished, resulting in less protection and shock absorption. Durolane, Euflexxa, Gel-One, GelSyn-3, GenVisc 850, Hyalgan, Hymovis, Monovisc, Orthovisc, Sodium Hyaluronate, Synvisc, Synvisc-One, Supartz, Synojoynt, Triluron, TriVisc, Visco-3 are hyaluronan derivatives that are injected into the knee joints to increase the elastoviscous properties of arthritic joint fluid and slow its outflow from the joint . The goal of therapy is torestore the viscoelasticity in the affected joints, thereby decreasing pain, improving mobility, and restoring the natural protective functions (1).The American College of Rheumatology (ACR) updated its guidelines for the treatment of osteoarthritis (OA) of the knee in 2012. In mild symptomatic OA, treatment may be limited toFederal Employee Program® 1310 G Street, N.W.Washington, D.C. 20005 202.942.1000Fax 202.942.1125Section: Prescription Drugs Effective Date: April 1, 2020 Subsection: Neuromuscular Drugs Original Policy Date: June 9, 2011 Subject: Hyaluronic Acid Derivatives Page: 2 of 7patient education, physical and occupational therapy and other non-pharmacologic modalities. Nonpharmacologic modalities strongly recommended for the management of knee OA were aerobic, aquatic, and/or resistance exercises as well as weight loss for overweight patients. Nonpharmacologic modalities conditionally recommended for knee OA included medial wedge insoles for valgus knee OA, subtalar strapped lateral insoles for varus knee OA, medially directed patellar taping, manual therapy, walking aids, thermal agents, tai chi, self-management programs, and psychosocial interventions. Pharmacologic modalities conditionally recommended for the initial management of patients with knee OA included acetaminophen, oral and topical NSAIDs, tramadol, and intraarticular corticosteroid injections (1).Regulatory StatusFDA-approved indication: Hyaluronic acid derivatives are indicated for the treatment of pain in osteoarthritis (OA) of the knee in patients who have failed to respond adequately to conservative non-pharmacologic therapy, simple analgesics (e.g., acetaminophen), NSAIDs, tramadol, or intra-articular steroid injections (2-18).The hyaluronic acid derivatives are contraindicated for use in patients with known hypersensitivity to hyaluronan (sodium hyaluronate) preparations. Orthovisc lists hypersensitivity to gram positive bacterial proteins as an additional contraindication (4). Caution should be exercised when Gel-One, Hyalgan, Visco-3, Synvisc, Synvisc-One, Supartz, and Triluron are administered to patients with allergies to avian proteins, feathers, and egg products (3-8, 18).Hyaluronic acid derivatives are contraindicated to treat patients with knee joint infections, infections or skin diseases in the area of the injection site (2-17).A treatment cycle for most of the hyaluronan derivatives typically involves multiple weekly injections. Euflexxa, GelSyn-3, Sodium Hyaluronate, Synvisc, Triluron, TriVisc, and Visco-3 are given for a total of three injections. Orthovisc is given for three or four injections. GenVisc 850, Supartz and Hyalgan are given for a total of three or five injections. Durolane, Gel-One, Synojoynt, and Synvisc-One differ from the other hyaluronan derivatives in that it only requires one injection. Repeat courses of hyaluronan derivatives may be administered if symptoms return (2-18).Upon the basis of high quality supporting evidence, the American Academy of Orthopedic Surgeons cannot recommend using hyaluronic acid for patients with symptomatic osteoarthritis of the knee (19).Related policiesSection: Prescription Drugs Effective Date: April 1, 2020 Subsection: Neuromuscular Drugs Original Policy Date: June 9, 2011 Subject: Hyaluronic Acid Derivatives Page: 3 of 7Hyaluronate PowderPolicyThis policy statement applies to clinical review performed for pre-service (Prior Approval, Precertification, Advanced Benefit Determination, etc.) and/or post-service claims.Hyaluronic acid derivatives may be considered medically necessary for the treatment of osteoarthritis of the knee and if the conditions indicated below are met.Hyaluronic acid derivatives may be considered investigational for all other indications.Prior-Approval RequirementsAge18 years or older (22 or older for Synvisc, Synvisc-One, and TriVisc)DiagnosisPatient must have the following:Osteoarthritis of the kneeAND ALL of the following:1. Inadequate response to TWO or more of the following conservative non-pharmacologic therapy:a. Cardiovascular (aerobic) activity, such as: walking, biking, stationarybike, aquatic exerciseb. Resistance exercisec. Weight reduction (for persons who are overweight)d. Participation in self-management programse. Wear of medially directed patellar tapingf. Wear of wedged insolesg. Thermal agentsh. Walking aidsi. Physical therapyj. Occupational therapy2. Inadequate response, intolerance, or contraindication to TWO or more of thefollowing:Section: Prescription Drugs Effective Date: April 1, 2020 Subsection: Neuromuscular Drugs Original Policy Date: June 9, 2011 Subject: Hyaluronic Acid Derivatives Page: 4 of 7a. Acetaminophenb. Oral NSAIDsc. Topical NSAIDs3. Inadequate response, intolerance, or contraindication to intra-articularsteroid injections in which efficacy lasted less than 8 weeks4. Radiologic confirmation of Kellgren-Lawrence Scale score of grade 2 orgreater5. NO dual therapy with another hyaluronic acid injectable6. Non-preferred medications only: Patient MUST have tried at least TWO ofthe preferred products unless the patient has a valid medical exception (e.g.inadequate treatment response, intolerance, contraindication)Prior – Approval Renewal RequirementsAge18 years or older (22 or older for Synvisc, Synvisc-One, and TriVisc)DiagnosisPatient must have the following:Osteoarthritis of the kneeAND ALL of the following:1. Documentation of improvement in pain with previous course of treatment2. At least 12 months has elapsed since last injection of the prior treatmentcycle3. Documentation of reduction of dosing of NSAIDs or other analgesicsduring the 12 month period following the last injection of the prior treatmentcycle4. NO dual therapy with another hyaluronic acid injectable5. Non-preferred medications only: Patient MUST have tried at least TWOof the preferred products unless the patient has a valid medical exception(e.g. inadequate treatment response, intolerance, contraindication) Policy GuidelinesPre - PA AllowanceNoneSection: Prescription Drugs Effective Date: April 1, 2020 Subsection: Neuromuscular Drugs Original Policy Date: June 9, 2011 Subject: Hyaluronic Acid Derivatives Page: 5 of 7Prior - Approval LimitsDuration12 monthsQuantity One course of therapy for each kneePrior – Approval Renewal LimitsSame as aboveRationaleSummaryOsteoarthritis of the knee is a disease in which the elastoviscous property of the synovial fluid in the knee joint becomes diminished, resulting in less protection and shock absorption. Durolane, Euflexxa, Gel-One, GelSyn-3, GenVisc 850, Hyalgan, Hymovis, Monovisc, Orthovisc, Sodium Hyaluronate, Synvisc, Synvisc-One, Supartz, Synojoynt, Triluron, TriVisc, Visco-3 are hyaluronan derivatives that are injected into the knee joints to increase the elastoviscous properties of arthritic joint fluid and slow its outflow from the joint. The goal of therapy is to restore the viscoelasticity in the affected joints, thereby decreasing pain, improving mobility, and restoring the natural protective functions (1-18).Prior approval is required to ensure the safe, clinically appropriate and cost effective use of the hyaluronic acid derivatives while maintaining optimal therapeutic outcomes.References1. American College of Rheumatology, Subcommittee on Osteoarthritis Guidelines.Recommendations for the medical management of osteoarthritis of the hip and knee:2012 update. Arthritis Care & Research 2012; 64(4):465-474.2. Euflexxa [package insert]. Parsippany, NJ: Ferring Pharmaceuticals Inc.; July 2016.3. Hyalgan [package insert]. Parsippany, NJ: Fidia Pharma USA Inc.; May 2014.4. Orthovisc [package insert]. Woburn, MA: Anika Therapeutics; June 2005.5. Supartz [package insert]. Durham, NC: Bioventus LLC; April 2015.6. Synvisc [package insert]. Ridgefield, NJ: Genzyme Corp.; December 2014.7. Synvisc-One [package insert]. Ridgefield, NJ: Genzyme Corp.; September 2014;8. Gel-One [package insert]. Warsaw, IN: Zimmer Inc.; May 2011.9. Monovisc [package insert]. Bedford, MA: Anika Therapeutics; December 2013.10. Hymovis [package insert]. Parsippany, NJ: O Fidia Pharma USA Inc.; October 2015.Section: Prescription Drugs Effective Date: April 1, 2020 Subsection: Neuromuscular Drugs Original Policy Date: June 9, 2011 Subject: Hyaluronic Acid Derivatives Page: 6 of 711. GenVisc 850 [package insert]. Doylestown, PA: OrthogenRx Inc.; January 2015.12. GelSyn-3 [package insert]. Durham, NC: Bioventus LLC; January 2016.13. Durolane [package insert]. Durham, NC: Bioventus LLC; November 2017.14. Visco-3 [package insert]. Warsaw, IN: Zimmer, Inc.; May 2017.15. Sodium Hyaluronate [package insert]. North Wales, PA: Teva Pharmaceuticals USA,Inc.; March 2019.16. Synojoynt [package insert]. North Wales, PA: Teva Pharmaceuticals USA, Inc.;September 2019.17. TriVisc [package insert]. Doylestown, PA: OrthogenRx, Inc.; September 2018.18. Triluron [package insert]. Florham Park, NJ: Fidia Pharma USA Inc.; March 2019.19. American Academy of Orthopaedic Surgeons. Treatment of osteoarthritis of the knee.Evidence-based guideline 2nd edition. May 2013.Policy HistoryDate Action ReasonJanuary 2012 Added minimum age - only approved for adultsDecember 2012 Annual editorial review and reference updateDecember 2013 Annual editorial review and reference updateMarch 2014 Annual editorial reviewAddition of examples of non-pharmacological agents and agents of priorfailure medications.April 2014 Line-Addition of Monovisc to PAMarch 2015 Annual criteria review and reference updateMarch 2016 Change from one tried and failed to two tried and failed non-pharmacologic and pharmacologic therapies and addition of the tried and failed of intra-articular steroid and radiologic confirmation of Kellgren-Lawrence Scalescore of grade 2 or greaterAddition of HymovisPolicy # change from 5.11.04 to 5.75.09May 2016 Addition of GelSyn-3 and GenVisc 850December 2016 Annual editorial review and reference updateAdded: no dual therapy with another hyaluronic acid injectableMarch 2017 Bolded preferred products in the title pageJuly 2017 GelSyn-3 has been changed to preferredSeptember 2017 Annual reviewDecember 2017 Addition of Durolane and Visco-3March 2018 Annual editorial reviewRemoval of Tramadol from the T/F listSeptember 2019 Annual review and reference update. Addition of Sodium Hyaluronate,Synojoynt, and TriViscSection: Prescription Drugs Effective Date: April 1, 2020 Subsection: Neuromuscular Drugs Original Policy Date: June 9, 2011 Subject: Hyaluronic Acid Derivatives Page: 7 of 7December 2019 Annual review. Addition of requirement to trial preferred products January 2020 Addition of TriluronMarch 2020 Annual reviewKeywordsThis policy was approved by the FEP® Pharmacy and Medical Policy Committee on March 13, 2020 and is effective on April 1, 2020.。
质粒图谱查询方法

3.google scholar: / 有些质粒是经过改造的,所以通过上述方法不能查询到相应信息。这时,可以在google scholar中输入质粒名称,可以直观地看哪些学者在何文章中使用了该质粒,从而可了解到质粒的来源;或者籍此向作者咨询或索取。 4.尝试从各大生物公司,例如invitrogen网站查询. 5. 这个网站收录了大量图谱: http://www.embl-hamburg.de/~geerlof/webPP/vectordb/bact_vectors/table.html
Pe
te
rX u
file:///D|/中科院/Selective Serotonin Transporter/质粒信息/质粒图谱查询方法.txt
file:///D|/中科院/Selective Serotonin Transporter/质粒信息/质粒图谱查询方法.txt(第 2/6 页)[2011/8/4 18:39:52]
By
0099--pGE-1—Stratagene--RNAi载体 0100--pSUPER.p53—OligoEngine--RNAi载体 0101--palter-ex1--promega 0102--pACYCDuet-1--NOVAGEN 0103--pEX lox(+) Vector—NOVAGEN--原核表达 0104--质粒名称:pBACgus-8 Transfer Plasmid—NOVAGEN--CHUANSUO 0105--pSCREEN?-1b(+) Vector Map—novagen--筛选 0106--PGEX-2T--BD Co--pDsRed2--Clontech 0107--pbgal-Basic—Clontech--mammalian reporter vector 0108—pBI—Clontech--express two genes of interest from a bidirectional tet-responsive promoter 0109--质粒名称:pbgal-Control—Clontech--mammalian reporter vector 0110-- pGEX-5X-1--原核表达 0111--pBI-EGFP—Clontech--pBI-EGFP-- coexpress 0112--pBI-G—Clontech--pBI-G--express b-galactosidase 0113--pBI-GL—Clontech--pBI-GL --express luciferase and b-galactosidase 0114--pCMS-EGFP—Clontech--mammalian expression vector 0115--pd2EYFP-1—Clontech--启动子测定 0116--质粒名称--pd2EYFP-N1—Clontech--融合表达 0117--pd4EGFP-Bid—Clontech--融合表达 Bid 0118--pDNR-CMV—Clontech--pDNR-CMV 0119--pDNR-EGFP Vector—Clontech 0120--pDNR-LacZ –Clontech 0121--pECFP-Endo—Clontech--真核表达0122--pECFP-ER—Clontech--真核表达0123--pEGFP-Actin—Clontech--真核表达0124--pGAD GH--Clontech--酵母表达 0125--pGADT7-Rec –Clontech--酵母表达 0126--pGADT7-RecAB—Clontech--酵母表达 0127--pGADT7-Rec2—Clontech--酵母表达 0128--pGBKT7—Clontech--酵母表达 0129--pHAT 10/11/12—Clontech 0130--pHAT20—Clontech 0131—pHygEGFP—Clontech 0132—pLacZi—Clontech 0133—pM—Clontech--pM is used to generate a fusion of the GAL4 DNA-BD 0134--pPKCa-EGFP—Clontech 0135--pPKCb-EGFP—Clontec 0136--pSIREN-DNR Vector—Clontech--RNAi 0137--pSIREN-DNR-DsRed-Express Vector—Clontech--RNAi 0138--pSIREN-RetroQ—Clontech--RNAi 0139--pIRES-EYFP—Clontech--RNAi 0140--pSRE-Luc—Clontech--RNAi 0141--pTK-neo—novagen--原核表达 0142--pZsGreen Vector—Clontech--pZsGreen is a pUC19-derived prokaryotic expression vector 0143--pTandem-1—novagen--原核表达 0144--pZsGreen1-C1Vector—Clontech----真核表达 0145--质粒名称:M13mp18—novagen--原核表达 0146--pZsGreen1-DR Vector—Clontech--真核表达 0147--PZsGreen1-N1 Vector—Clontech --真核表达 0148--T7Select415-1b—novagen----真核表达 0149--pZsYellow Vector—Clontech --真核表达 0150—pTimer—Clontech --真核表达 0151--pTA-Luc—Clontech --真核表达 0152--pTAL-Luc—Clontech --真核表达 0153--pTA-SEAP—Clontech --真核表达 0154--pTAL-SEAP—Clontech --真核表达 0155--pTet-On—Clontech --真核表达 0156--pTet-Off—Clontech --真核表达 0157--pTet-ATF—Clontech --真核表达 0158--pTet-CREB—Clontech --真核表达
双荧光素酶报告基因检测试剂盒

注意事项:
1) Fassay Buffer I和Fassay Substrate I应避免反复冻熔,可分装成合适体积分次使用。 Rassay Substrate II溶液应盖严存放,避免蒸发。配制好未用完的Fassay Reagent I和 Rassay Reagent II可在-20℃保存1月左右。
自动发光测定:
配制好的Fassay Reagent I和Rassay Reagent II置于测定仪内并连接好对应管道,Fassay Reagent I接第一注射管道,Rassay Reagent II接第二注射管道。各待测样品20 μl分别加 入测定管/板孔底部,启动自动测量程序。记录Firefly luciferase和Ranilla luciferase的发光 单位(RLU)。
测定前,在室温待Fassay Buffer I、Fassay Substrate I和Rassay Buffer II溶化,混匀(注意 避光)。按20/1比例用Fassay Buffer I稀释Fassay Substrate I,按50/1比例用Rassay Buffer II 稀释Rassay Substrate II,分别配制所需体积的Fassay Reagent I和Rassay Reagent II(注意 避光)。
2) 细胞裂解液一般在当天测定。如需隔日测定,应将样品于-20℃保存。长期保存应 在-80℃。测定样品量可为10~30μl个样品的两种试剂加入时间间 隔一致。
4) Rassay Reagent II可用于直接测定样品的Ranilla luciferase。需要注意的是,Rassay Reagent II直接测量的RLU要比双荧光素酶顺序检测获得的RLU高一些(反应体积 等因素的影响)。
ColiComplete 产品说明书

Page 1 of 2 ColiComplete ®AOAC Official Method 992.30General DescriptionColiComplete ® contains 5-bromo-4-chloro-3-indolyl-ß-Dgalactopyranoside (X-Gal) and 4-methyl umbelliferyl-ß-D-glucuronide (MUG). Discs are added to LST inoculated with selected dilutions of samples. Samples are incubated at 35–37 °C and examined after 24 and 48 ±2 h for confirmed total coliforms and after 30 ±2 h for confirmed E. coli results. ß-Galactosidase, from coliforms present in samples, cleaves X-Gal into 5-bromo-4-chloro-indoxyl intermediate which undergoes oxidation to yield water-insoluble blue dimer, visually detectable on disc or in surrounding medium as confirmed positive result for total coliform activity. ß-Glucuronidase, from E. coli present in samples, cleaves MUG into glucuronide and methyl umbelliferone which fluoresces under long wave UV light (366 nm) as confirmed positive result for E. coli presence.NOTE : As E. coli O157:H7 does not produce ß-glucuronidase, ColiComplete ® is not suitable for the detection of E. coli O157:H7.A. Sample PreparationPrepare appropriate serial dilutions as indicated in FDA Bacteriological Analytical Manual (BAM), or AOAC Official Methods of Analysis according to sample type.B. InoculationInoculate LST tubes with appropriate sample dilution series selected to determine MPN levels or presence/absence of total coliforms and E. coli in sample. Aseptically add a single ColiComplete ® disc to each tube. Incubate at 35–37 °C.C. Reading ColiComplete ®a. For total coliforms — After at least 24 h incubation, examine each tube for visually detectable blue color on disc or in surrounding medium. Presence of blue color indicates confirmed positive result for total coliforms.NOTE: A wide range of blue color intensity may be expected, depending on sample composition and microflora. All blue reactions are positive regardless of intensity of color.Reincubate at 35–37 °C. After additional 24 ±2 h re-examine. Continued absence of blue indicates negative result; presence of blue indicates confirmed positive result for total coliforms. Read and record the MPN code or presence/absence of total coliforms in the sample.b. For E.coli — After 30 ±2 h from start of initial incubation, examine tubes under long-wave UV light (366 nm). Fluorescent tubes indicate confirmed positive result for E. coli. Read and record the MPN code or presence/absence of E. coli in the sample.D. CONTROLSPositive and negative controls should be used to facilitate interpretation of MUG fluorescent reaction. Use one known positive E. coli tube and two negative controls - one non -E. coli /coliform tube (e.g., Klebsiella spp.) and one uninoculated media tube.NOTE: Use borosilicate glass tubes, flint glass gives fluorescence that may be misinterpreted for a positive result.Lit. No. MK_UG4655EN Merck KGaAFrankfurter Strasse 25064293 DarmstadtGermanyPage 2 of 2 E. Method Modification for Certain JuicesApplicable to juice products/processors which rely on treatments that do not come into direct contact with all parts of the juice, as contained in 21 CFR Part 120: Rules and Regulations. Hazard Analysis and Critical Control Point (HAACP); Procedures for the Safe and Sanitary Processing and Importing of Juice; Final Rule. Vol 66 No. 13. 6137-6202. Use the modified method “Analysis for Escherichia coli in Citrus Juices - Modifi cation of AOAC Official Method 992.30” as stated in Section 120.25 (a).F. StorageStore unused discs at 2–8 °C (36–46 °F) in a sealed container, with desiccant.G. DisposalAfter use, all tubes must be steam-sterilized at 121 °C for at least 30 min before discarding. For in-vitro diagnostic use only.Manufacturing EntityBioControl Systems, Inc, 12822 SE 32nd St, Bellevue, WA 98005, USA.BioControl Systems, Inc is an affiliate of Merck KGaA, Darmstadt, Germany.。
LabVolt系列雷达水位传感器(HART)产品说明书

LabVolt Series DatasheetRadar Level Transmitter (HART)589125 (46931-10)* The product images shown in this document are for illustration purposes; actual products may vary. Please refer to the Specifications section of each product/item for all details. Festo Didactic reserves the right to change product images and specifications at any time without notice.Festo Didactic en12/2023Radar Level Transmitter (HART), LabVolt SeriesTable of ContentsGeneral Description_________________________________________________________________________________3 List of Manuals_____________________________________________________________________________________3 Table of Contents of the Manual(s)_____________________________________________________________________3 Specifications______________________________________________________________________________________3Radar Level Transmitter (HART), LabVolt Series•••General DescriptionThe Radar Level Transmitter is a level measurement device using electromagnetic waves to detect the level of liquid in the column of the Pressure, Flow, Level, and Temperature Process Training Systems. The Radar Level Transmitter includes a horn antenna to direct the signal and a transmitter supporting either the HART orFOUNDATION Fieldbus communication protocols. The device can be configured manually via its alphanumeric display and push-buttons, but as it is difficult to efficiently program the transmitter's advanced functions on the device's display, acquiring a Software Configurator (Model 46982) is required.The Radar Level Transmitter is a noncontact sensor, in the industry it is usually used in corrosive environments. Changes in the density, conductivity, and composition of the process fluid do not affect this sensor.Available Radar Level Transmitters:46931-1: Radar Level Transmitter (HART)46931-D: Radar Level Transmitter (FOUNDATION Fieldbus)List of ManualsDescriptionManual numberRadar Level Transmitters (Workbook) __________________________________________________589760 (52200-00)Radar Level Transmitters (Workbook (Instructor)) ________________________________________589762 (52200-10)Table of Contents of the Manual(s)Radar Level Transmitters (Workbook) (589760 (52200-00))1 Fundamentals of Radar Level TransmittersSpecificationsParameterValueModelMicropilot FMR51Communication Protocol HARTRatings Power Input 24 V dc Operating Frequency ~26 GHzMeasured Variable Level (via time-of-flight)Accuracy±2 mmSensor Operating Temperature -40°C to 80°C (-40°F to 176°F)Process Temperature -196°C to 450°C (-321°F to 842°F)Sensor Operating Pressure Vacuum to 16000 kPa (Vacuum to 2320 psi)Blocking Distance200 mm (7.9 in)Radar Level Transmitter (HART), LabVolt Series Reflecting the commitment of Festo Didactic to high quality standards in product, design, development, production, installation, and service, our manufacturing and distribution facility has received the ISO 9001 certification.Festo Didactic reserves the right to make product improvements at any time and without notice and is not responsible for typographical errors. Festo Didactic recognizes all product names used herein as trademarks or registered trademarks of their respective holders. © Festo Didactic Inc. 2023. All rights reserved.Festo Didactic SERechbergstrasse 373770 DenkendorfGermanyP. +49(0)711/3467-0F. +49(0)711/347-54-88500Festo Didactic Inc.607 Industrial Way WestEatontown, NJ 07724United StatesP. +1-732-938-2000F. +1-732-774-8573Festo Didactic Ltée/Ltd675 rue du CarboneQuébec QC G2N 2K7CanadaP. +1-418-849-1000F. +1-418-849-1666。
PrepSEQ Nucleic Acid Extraction Kit使用手册说明书
PrepSEQ ™ Nucleic Acid Extraction KitTotal Nucleic Acid (DNA and RNA) ExtractionCatalog Numbers 4480466 and 4428176Pub. No. MAN0019336 Rev. C.0Product descriptionThe PrepSEQ ™Nucleic Acid Extraction Kit is designed for preparation of high-quality total nucleic acid (NA) from tissue, liquid, and swab samples. Magnetic beads allow efficient DNA and RNA capture and sample washing.This user guide describes the following methods:•“Isolate total nucleic acid from tissue samples” on page 2•“Isolate total nucleic acid from liquid samples” on page 3•“Isolate total nucleic acid from swab samples (manual method)” on page 4•“Isolate total nucleic acid from swab samples (automated method with the KingFisher ™ mL Food Protection Purification System)” on page 5Kit contents and storage[1]Refer to the product label for the expiration date.[2]Add ~35 mL of 100% isopropanol to the empty bottle before use.[3]Add 74 mL of 95% ethanol before use.Note: Kit components may ship separately depending on configuration and storage conditions.Required materials not suppliedUnless otherwise indicated, all materials are available through . "MLS" indicates that the material is available from or another major laboratory supplier.Isolate total nucleic acid from tissue samplesa.Place up to 100 mg of solid (tissue) sample in a 1.5-mL microcentrifuge tube.b.Add 300 μL of PK Buffer and 10 μL of Proteinase K.c.Incbate for 60 minutes at 45℃ and 1000 rpm in the thermomixer.d.Centrifuge for 2 minutes ar 10,000 x g , then transfer the supernatant to a new 1.5-mL centrifugetube.e.Add 200 μL of Lysis Buffer, then vortex for 15 seconds.1Treat the samples with proteinase K and perform cell lysisVortex the Magnetic Particles until complete resuspension (approximately 5 seconds).a.Add 35 μL of Magnetic Particles to the sample.b.Vortex for 10 seconds at low speed.c.Add 350 μL of Binding Solution, then vortex for 5 seconds.d.Incubate for 10 minutes at room temperature shaking continuously.e.Vortex for 10 seconds at low speed, then place the tube in the DynaMag ™‑2 Magnet.f.Let the tube rest in the DynaMag ™‑2 Magnet until complete separation occurs (approximately 1-2minutes).g.Carefully discard the liquid phase without disturbing the Magnetic Particles pellet.2Bind the nucleic acid to the magnetic beadsa.Add 300 μL of Wash Solution to the tube, then vortex at medium speed for 5 seconds, or untilthe pellet is completely resuspended.b.Place the tube in the DynaMag ™‑2 Magnet, then let it rest for 30 seconds.c.Carefully discard the liquid phase without disturbing the Magnetic Particles pellet.d.Repeat the two last steps two more times.3Wash the nucleic acida.Air-dry the Magnetic Particles in the DynaMag ™‑2 Magnet with the lid open for 5 minutes.b.Add 50 μL of Elution Buffer.c.Close the lid, then vortex the tube at medium speed for 5 seconds.d.Incubate the tube for 5 minutes at 45℃.e.Vortex the tube at medium speed for 2 seconds, then place the tube in the DynaMag ™‑2 Magnet.f.Let the tube rest in the DynaMag ™‑2 Magnet for at least 1 minute.g.Transfer the liquid phase containing the total NA to a new tube for storage.4Elute the nucleic acid Isolate total nucleic acid from liquid samplesa.Place 250 μL of liquid sample in a 1.5-mL microcentrifuge tube.b.Add 50 μL of PK Buffer and 10 μL of Proteinase K, then vortex for 15 seconds.c.Incbate for 25 minutes at 45℃ and 1000 rpm in the thermomixer.d.Centrifuge for 2 minutes ar 10,000 x g , then transfer the supernatant to a new 1.5-mL centrifugetube.e.Add Lysis Buffer up to 500 μL of total volume, then vortex for 15 seconds.1Treat the samples with proteinase K and perform cell lysisVortex the Magnetic Particles until complete resuspension (approximately 5 seconds).a.Add 35 μL of Magnetic Particles to the sample.b.Vortex for 10 seconds at low speed.c.Add 350 μL of Binding Solution, then vortex for 5 seconds.d.Incubate for 10 minutes at room temperature shaking continuously.e.Vortex for 10 seconds at low speed, then place the tube in the DynaMag ™‑2 Magnet.f.Let the tube rest in the DynaMag ™‑2 Magnet until complete separation occurs (approximately 1-2minutes).g.Carefully discard the liquid phase without disturbing the Magnetic Particles pellet.2Bind the nucleic acid to the magnetic beadsa.Add 300 μL of Wash Solution to the tube, then vortex at medium speed for 5 seconds, or untilthe pellet is completely resuspended.b.Place the tube in the DynaMag ™‑2 Magnet, then let it rest for 30 seconds.c.Carefully discard the liquid phase without disturbing the Magnetic Particles pellet.d.Repeat the two last steps two more times.3Wash the nucleic acida.Air-dry the Magnetic Particles in the DynaMag ™‑2 Magnet with the lid open for 5 minutes.b.Add 50 μL of Elution Buffer.c.Close the lid, then vortex the tube at medium speed for 5 seconds.d.Incubate the tube for 5 minutes at 45℃.e.Vortex the tube at medium speed for 2 seconds, then place the tube in the DynaMag ™‑2 Magnet.f.Let the tube rest in the DynaMag ™‑2 Magnet for at least 1 minute.g.Transfer the liquid phase containing the total NA to a new tube for storage.4Elute the nucleic acid Isolate total nucleic acid from swab samples (manual method)a.Place the swab sample in a 1.5-mL microcentrifuge tube.b.Add 650 μL of Lysis Buffer, then vortex for 15 seconds.c.Incbate for 25 minutes at 45℃ and 1000 rpm in the thermomixer.d.Centrifuge for 2 minutes ar 10,000 x g , then transfer 500 μL of supernatant to a new 1.5-mLcentrifuge tube.1Perform cell lysis Vortex the Magnetic Particles until complete resuspension (approximately 5 seconds).a.Add 35 μL of Magnetic Particles to the sample.b.Vortex for 10 seconds at low speed.c.Add 350 μL of Binding Solution, then vortex for 5 seconds.d.Incubate for 10 minutes at room temperature shaking continuously.e.Vortex for 10 seconds at low speed, then place the tube in the DynaMag ™‑2 Magnet.f.Let the tube rest in the DynaMag ™‑2 Magnet until complete separation occurs (approximately 1-2minutes).g.Carefully discard the liquid phase without disturbing the Magnetic Particles pellet.2Bind the nucleic acid to the magnetic beadsa.Add 300 μL of Wash Solution to the tube, then vortex at medium speed for 5 seconds, or untilthe pellet is completely resuspended.b.Place the tube in the DynaMag ™‑2 Magnet, then let it rest for 30 seconds.c.Carefully discard the liquid phase without disturbing the Magnetic Particles pellet.d.Repeat the two last steps two more times.3Wash the nucleic acida.Air-dry the Magnetic Particles in the DynaMag ™‑2 Magnet with the lid open for 5 minutes.b.Add 50 μL of Elution Buffer.c.Close the lid, then vortex the tube at medium speed for 5 seconds.d.Incubate the tube for 5 minutes at 45℃.e.Vortex the tube at medium speed for 2 seconds, then place the tube in the DynaMag ™‑2 Magnet.4Elute the nucleic acid4Elute the nucleic acid (continued)f.Let the tube rest in the DynaMag™‑2 Magnet for at least 1 minute.g.Transfer the liquid phase containing the total NA to a new tube for storage.Isolate total nucleic acid from swab samples (automated method with the KingFisher™ mL Food Protection Purification System)For more information about using the KingFisher™ mL Food Protection Purification System, see Thermo Scientific™ KingFisher™ mL User Manual (Pub. No. 1508260).•Ensure that the PSNA_mL_300ul script has been downloaded from the product page and loadedonto the KingFisher™ mL Food Protection Purification System.•Ensure that a water bath or heating block is heated to 83°C.•Label the following consumables for each sample to be processed and the negative extractioncontrol:–One tube strip–Two 1.5‑mL microcentrifuge tubes (nuclease free)Note: Up to 14 samples and 1 negative extraction control can be processed at a time on theKingFisher™ mL Food Protection Purification System.1Before you beginVortex the Magnetic Particles until complete resuspension (approximately 5 seconds).a.For the number of required reactions, prepare the Binding Mix according to the following table:[1]Include 10% overage when making for multiple reactions.b.Invert the Binding Mix 5 times gently to mix, then add 700 µL to Tube A of each tube strip.Include tube strips for each sample and negative extraction control.Note: Remix the Binding Mix by inversion frequently during pipetting to ensure even distributionof beads to all samples or wells. The Binding Mix is viscous, so pipet slowly to ensure that thecorrect amount is added. DO NOT reuse pipette tips to add Binding Mix to the samples, as thehigh viscosity will cause variations in the volumes added.c.Add 300 µL of Wash Buffer to Tube B and 300 µL of Wash Buffer to Tube C of each tube strip.d.Add 100 µL of Elution Buffer to Tube D of each tube strip.e.Add 1 µL of Total RNA Control (Human) to Tube A of each tube strip.f.Vortex the swab sample tubes for 30 seconds.g.Add 300 µL of a sample to Tube A of the corresponding, pre‑labeled tube strip. Repeat for theremaining samples and tube strips.h.Add 300 µL of Nuclease-free Water (not DEPC-Treated) to Tube A of the Negative ExtractionControl tube strip.2Set up processing tubesa.Load the prepared tube strips into the tray, then place the tray in the KingFisher ™mL FoodProtection Purification System.b.Fully insert the tip combs into the tip comb slots.c.Select the PSNA_mL_300ul script, then press Start .d.When prompted by the instrument, remove the tube ‑strip tray from the instrument.e.For each tube strip, transfer the elution buffer (100 µL) from Tube D into one of thecorresponding pre ‑labeled microcentrifuge tubes.f.Cap the microcentrifuge tubes, then incubate at 83°C for 4 minutes.g.Transfer the elution buffer from each microcentrifuge tube back into Tube D of thecorresponding tube strip.h.Load the tube ‑strip tray into the instrument, then restart the run.i.After the run is complete, immediately remove the tube ‑strip tray from the instrument.j.For each tube strip, transfer the elution buffer (100 μL) from Tube D into the second pre ‑labeled microcentrifuge tube.Place the microcentrifuge tubes on ice for immediate use in real-time PCR. The extracted samples can be stored at -70°C for long ‑term storage (up to one year).3Process the samples on the instrumentLimited product warrantyLife Technologies Corporation and/or its affiliate(s) warrant their products as set forth in the Life Technologies' General Terms and Conditions of Sale at /us/en/home/global/terms-and-conditions.html . If you have any questions, please contact Life Technologies at /support .Life Technologies Ltd | 7 Kingsland Grange | Woolston, Warrington WA1 4SR | United KingdomFor descriptions of symbols on product labels or product documents, go to /symbols-definition .The information in this guide is subject to change without notice.DISCLAIMER : TO THE EXTENT ALLOWED BY LAW, THERMO FISHER SCIENTIFIC INC. AND/OR ITS AFFILIATE(S) WILL NOT BE LIABLE FOR SPECIAL, INCIDENTAL, INDIRECT,PUNITIVE, MULTIPLE, OR CONSEQUENTIAL DAMAGES IN CONNECTION WITH OR ARISING FROM THIS DOCUMENT, INCLUDING YOUR USE OF IT.Important Licensing Information : These products may be covered by one or more Limited Use Label Licenses. By use of these products, you accept the terms and conditions of all applicable Limited Use Label Licenses.©2020 Thermo Fisher Scientific Inc. All rights reserved. All trademarks are the property of Thermo Fisher Scientific and its subsidiaries unless otherwise specified. Thermomixer ™is a trademark of Eppendorf./support | /askaquestion 。
Sodium-Valproate-5571[英国药典BP2009]
![Sodium-Valproate-5571[英国药典BP2009]](https://img.taocdn.com/s3/m/67847223af45b307e87197eb.png)
Sodium Valproate General Notices(Ph Eur monograph 0678)IDENTIFICATIONA. Infrared absorption spectrophotometry (2.2.24).Comparison sodium valproate CRS.If the spectra obtained in the solid state show differences, record new spectra using discs prepared by placing 50 µl of a 100 g/l solution in methanol R on a disc of potassium bromide R and evaporating the solvent in vacuo. Examine immediately.B. Examine the chromatograms obtained in the test for related substances.Results The principal peak in the chromatogram obtained with test solution (b) is similar in retention time to the principal peak in the chromatogram obtained with reference solution (b).C. 2 ml of solution S (see Tests) gives reaction (a) of sodium (2.3.1) .TESTSSolution SDissolve 1.25 g in 20 ml of distilled water R in a separating funnel, add 5 ml of dilute nitric acid R and shake. Allow the mixture to stand for 12 h. Use the lower layer.Appearance of solutionThe solution is not more opalescent than reference suspension II (2.2.1) and not more intensely coloured than reference solution Y6(2.2.2, Method II).Dissolve 2.0 g in water R and dilute to 10 ml with the same solvent.Acidity or alkalinityDissolve 1.0 g in 10 ml of water R. Add 0.1 ml of phenolphthalein solution R . Not more than 0.75 ml of 0.1 M hydrochloric acid or 0.1 M sodium hydroxide is required to change the colour of the indicator.Related substancesGas chromatography (2.2.28).Internal standard solution Dissolve 10 mg of butyric acid R in heptane R and dilute to 200 ml with the same solvent.Test solution (a) Dissolve 0.500 g of the substance to be examined in 10 ml of water R . Add 5 ml of dilute sulphuric acid R and shake with 3 quantities, each of 20 ml, of heptane R. Add 10.0 ml of the internal standard solution to the combined upper layers, shake with anhydrous sodium sulphate R , filter and evaporate the filtrate, at a temperature not exceeding 30 °C, using a rotary evaporator. Take up the residue with heptane R and dilute to 10.0 ml with the same solvent. Dilute 1.0 ml of this solution to 10.0 ml with heptane R .Test solution (b) Dissolve 40 mg of the substance to be examined in 100 ml of water R . To 10 ml of this solution add 0.5 ml of dilute sulphuric acid R and shake with 3 quantities, each of 5 ml, of heptane R . Shake with anhydrous sodium sulphate R, filter and evaporate the filtrate, at a temperature not exceeding 30 °C, to a volume of about 10 ml, using a rotaryevaporator.Reference solution (a) Dissolve 20 mg of 2-(1-methylethyl)pentanoic acid CRS (impurity C) in 5.0 ml of test solution (b) and dilute to 10 ml with heptane R . Dilute 1 ml of this solution to 10 ml with heptane R.Reference solution (b) Prepare as prescribed for test solution (b), using sodium valproate CRS instead of the substance to be examined.Column:— material: wide-bore fused silica;— size: l = 30 m, Ø = 0.53 mm;— stationary phase: macrogol 20 000 2-nitroterephthalate R (film thickness 0.5 µm). Carrier gas helium for chromatography R.Flow rate 8 ml/min.Temperature:Detection Flame ionisation.Injection 1 µl.System suitability Reference solution (a):— resolution: minimum 3.0 between the peaks due to impurity C and valproic acid.Limits Test solution (a):— any impurity: for each impurity, not more than the area of the peak due to the internal standard (0.1 per cent);— total: not more than 3 times the area of the peak due to the internal standard (0.3 per cent);— disregard limit: 0.1 times the area of the peak due to the internal standard (0.01 per cent).Chlorides (2.4.4)Maximum 200 ppm.To 5 ml of solution S add 10 ml of water R.Sulphates (2.4.13)Maximum 200 ppm, determined on Solution S.Heavy metals (2.4.8)Maximum 20 ppm.1.0 g complies with test C. Prepare the reference solution using 2 ml of lead standard solution (10 ppm Pb) R.Loss on drying (2.2.32)Maximum 2.0 per cent, determined on 1.000 g by drying in an oven at 105 °CASSAYDissolve 0.1500 g in 25 ml of anhydrous acetic acid R . Titrate with 0.1 M perchloric acid, determining the end-point potentiometrically (2.2.20) .1 ml of 0.1 M perchloric acid is equivalent to 16.62 mg of C8H15NaO2.STORAGEIn an airtight container.IMPURITIESA. R = R′ = H: pentanoic acid (valeric acid),B. R = H, R′ = CH2-CH3: (2RS)-2-ethylpentanoic acid,C. R = H, R′ = CH(CH3)2: (2RS)-2-(1-methylethyl)pentanoic acid,D. R = R′ = CH2-CH2-CH3: 2,2-dipropylpentanoic acid,E. R = R′ = H: pentanamide (valeramide),F. R = H, R′ = CH2-CH2-CH3: 2-propylpentanamide,G. R = R′ = CH2-CH2-CH3: 2,2-dipropylpentanamide,Soft SoapGeneral NoticesPreparationSoap SpiritDEFINITIONSoft Soap is soap made by the interaction of potassium hydroxide or sodium hydroxide with a suitable vegetable oil or oils or with fatty acids derived there from. It yields not less than44.0% of fatty acids. It may be coloured with chlorophyll or not more than 0.015% of a suitable green soap dye.CHARACTERISTICSA yellowish white to green or brown, unctuous substance.Soluble in water and in ethanol (96%).TESTSChlorides and other ethanol-insoluble substancesDissolve 5 g in 100 ml of hot ethanol (96%) previously neutralised to phenolphthalein solution R1, filter through a dried and tared filter, wash the residue thoroughly with hot neutralised ethanol (96%) and dry to constant weight at 105°. The residue weighs not more than 0.15 g. Free fatty acid or alkali hydroxideBoil 250 ml of ethanol (96%) to remove carbon dioxide, add 0.5 ml of phenolphthalein solution R1, allow to cool to 70° and neutralise, if necessary, with 0.1M sodium hydroxide VS or 0.05M sulphuric acid VS. To 100 ml of the neutral ethanol add 10 g of the substance being examined and dissolve it as quickly as possible by heating under a reflux condenser. Cool to 70° and, if the solution is not pink, titrate at 70° with 0.1M sodium hydroxide VS; not more than 0.2 ml is required. If the solution is pink add, in a thin stream, 5 ml of hot barium chloride solution previously neutralised to phenolphthalein solution R1, mix thoroughly and titrate with 0.1M hydrochloric acid VS until the pink colour disappears; not more than 1.0 ml is required.Total free alkaliTo 100 ml of the neutral ethanol prepared as described in the test for Free fatty acid or alkali hydroxide add 10 g of the substance being examined and dissolve it as quickly as possible by heating under a reflux condenser. Add immediately 3 ml of 0.5M sulphuric acid VS and boil under a reflux condenser on a water bath for at least 10 minutes. If the solution is not pink, cool to 70° and titrate with 1M sodium hydroxide VS until a pink colour is produced. The volume of 0.5M sulphuric acid VS neutralised by the substance being examined is not more than 1.0 ml.Unsaponifiable matter and unsaponified neutral fatDissolve 5 g in 80 ml of a mixture of 50 ml of ethanol (96%) and 100 ml of water, without。
7. Endotoxin LALTests

Charles River Endosafe
2
Woo Jung BSC Inc.
August 25, 2003
LAL Discoveries by Bang and Levin
Described role of endotoxin in coagulation of Limulus blood
Prepared Endotoxin - responsive lysate from Amoebocytes
Endotoxicity
ENDOTOXIN CAUSES HUMAN TISSUE TO RELEASE INFLAMMATORY MEDIATORS INFLAMMATION INDUCES A VARIETY OF TISSUE DAMAGE SHOCK and MULTIPLE ORGAN DYSFUNCTION MAY OCCUR
Endotoxins and Pyrogens
Pyrogens are fever-inducing agents in humans and animals
include endotoxin, gram + cell debris, fungi
Endotoxins are components from the outer membrane of gram-negative bacteria
Clotting Enzyme
Liquid Coagulogen
M++ pH=7.2
Clotted Coagulin Gel
Summary of Gel Clot Test
Endpoint sought by 180 inversion of sample tube
475150-69-7_BAN ORL 24相关参数MedBio

192322-50-2
50mg
≥98%
品牌
货号
中文名称
英文名称
CAS
包装
纯度
MedBio
MED12697
CRF (human, rat)
CRF (human, rat)
86784-80-7
5mg
≥98%
品牌
货号
中文名称
英文名称
CAS
包装
纯度
MedBio
MED13424
MED12760
Neurotensin
Neurotensin
39379-15-2
20mg
≥98%
中文名称
英文名称
CAS
包装
纯度
MedBio
MED12718
RS 100329 hydrochloride
RS 100329 hydrochloride
1215654-26-4
50mg
≥98%
品牌
货号
中文名称
英文名称
CAS
包装
纯度
MedBio
MED12752
GRP (human)
GRP (human)
93755-85-2
1mg
≥98%
品牌
货号
中文名称
英文名称
CAS
包装
纯度
MedBio
MED12866
SB 328437
SB 328437
247580-43-4
10mg
≥98%
品牌
货号
中文名称
英文名称
CAS
包装
纯度
MedBio
SMARTer PCR cDNA Synthesis Kit User Manual

SMARTer™ PCR cDNA Synthesis Kit User Manual Cat. Nos. 634925 & 634926United States/Canada 800.662.2566Asia Pacific +1.650.919.7300Europe +33.(0)1.3904.6880Japan +81.(0)77.543.6116SMART er™ PCR cDNA Synthesis Kit User ManualT able of ContentsI. List of Components (3)II. Additional Materials Required (4)III. Introduction & Protocol Overview (5)IV. RNA Preparation & Handling (7)A. General Precautions (7)B. RNA Isolation (7)C. RNA Purity (8)D. Assessing the Quality of the RNA Template (8)V. SMART er cDNA Synthesis (9)A. General Considerations (10)B. PRoToCol: First-Strand cDNA Synthesis (10)C. PRoToCol: cDNA Amplification by lD PCR (12)VI. Analysis of cDNA Amplification Results (16)VII. T roubleshooting Guide (17)VIII. References (18)Appendix A: Protocols for PCR-Select™ (19)A. Additional Materials Required (19)B. PRoToCol: cDNA Amplification by lD PCR (19)C. PRoToCol: Column Chromatography (22)D. PRoToCol: RsaI Digestion (23)E. PRoToCol: Purification of Digested cDNA (23)F. Controls for PCR-Select cDNA Subtraction (25)G. Analysis of Results of SMARTer PCR cDNA Synthesis for PCR-Select cDNA Subtraction (25)H. T roubleshooting (27)Appendix B: Virtual Northern Blots (28)Appendix C: Protocol for Non-Directional Cloning of SMART er cDNA (29)A. Additional Materials Required (29)B. PRoToCol: ds cDNA Polishing (29)List of FiguresFigure 1. Flowchart of SMARTer cDNA synthesis (5)Figure 2. Guide to using the SMARTer cDNA synthesis protocol for PCR-Select cDNA Subtraction,Virtual Northerns, Non-Directional Cloning & library Construction, and other applications. (9)Figure 3. optimizing PCR parameters for SMARTer cDNA synthesis. (15)Figure 4. Analysis for optimizing PCR parameters (16)Figure 5. optimizing PCR parameters for SMARTer cDNA synthesis for use withClontech PCR-Select (21)Figure 6. Virtual Northern blot analysis of cDNA fragments expressed in cells producing γ-globin. (28)List of T ablesTable I: Guidelines for Setting Up PCR Reactions (12)Table II: Cycling Guidelines Based on Starting Material (13)Table III: T roubleshooting Guide for First-Strand cDNA Synthesis & SMARTer PCR Amplification (17)Table IV: T roubleshooting Guide for Preparing SMARTer cDNA for Subtraction (27)SMART er™ PCR cDNA Synthesis Kit User Manual I. List of ComponentsSMART er PCR cDNA Synthesis KitCat. No.Cat. No. 634925634926 10 rxns20 rxns Box 110 µl20 µl • SMART er II A Oligonucleotide (12 µM)5'–AAGCAGTGGTATCAACGCAGAGTACXXXXX–3' Rsa I(X = undisclosed base in the proprietary SMARTer oligo sequence) 5 µl5 µl • Control Mouse Liver T otal RNA (1 µg/µl)Box 210 µl20 µl • 3’ SMART CDS Primer II A (12 µM)5’–AAGCAGTGGTATCAACGCAGAGTACT (30)N -1N–3’Rsa I (N = A, C, G, or T ; N -1 = A, G, or C)200 µl 400 µl • 5’ PCR Primer II A (12 µM)40 µl 80 µl • 5X First-Strand Buffer (RNase-Free)250 mM T ris-HCl (pH 8.3)375 mM KCl30 mM MgCl 2100 µl 200 µl • dNTP Mix (dATP , dCTP , dGTP , and dTTP , each at 10 mM)50 µl 50 µl • Dithiothreitol (DTT ; 100 mM)10 µl 10 µl • RNase Inhibitor (40 U/µl)12 µl 25 µl • SMARTScribe™ Reverse T ranscriptase (100 U/µl)1 ml 1 ml • Deionized H 2O Box 310 20 • CHROMA SPIN™+TE-1000 ColumnsStorage ConditionsStore Control Mouse Liver T otal RNA and SMARTer II A Oligonucleotide at –70°C.• Store the CHROMA SPIN +TE-1000 Columns at room temperature.• Store all other reagents at –20°C.• Licensing InformationFor important information about the use of SMART technology, please see the Notice to Purchaser at theend of this user manual.要稀释本页已使用福昕阅读器进行编辑。
安捷伦产品目录

15
Real-Time PCR
16
Mx3000P QPCR System
17
Brilliant III Ultra-Fast SYBR Green QPCR and QRT-PCR Reagents
18
Brilliant III Ultra-Fast QPCR and QRT-PCR Reagents
Agilent / STRATAGENE
Agilent website: /genomics
Welgene | Agilent Stratagene
威健股份有限公司 | Stratagene 總代理
Table of Content
Table of Contents
/ XL1-Red Competent Cells SoloPack Gold Supercompetent Cells
/ TK Competent Cells Specialty Cells
/ Classic Cells / Fine Chemicals For Competent Cells
適用於 UNG 去汙染或 bisulphite
sequencing
適用於 TA Cloning
最高敏感性
取代傳統 Taq 的好選擇
-
2
威健股份有限公司 | Stratagene 總代理
PCR Enzyme & Instrument
Agilent SureCycler 8800
市場上領先的 cycling 速度和 sample 體積 10 ~ 100 μL 簡易快速可以選擇 96 well 和 384 well 操作盤 優秀的溫控設備讓各個 well 都能保持溫度的穩定 七吋的高解析度觸控螢幕讓操作上更為簡便 可以透過網路遠端操控儀器及監控儀器 Agilent 專業的技術支援可以幫助您應對各種 PCR 的問題
Bioanalytical Method ValidationGuidance for Indust

Guidance for Industry Bioanalytical Method ValidationU.S. Department of Health and Human ServicesFood and Drug AdministrationCenter for Drug Evaluation and Research (CDER)Center for Veterinary Medicine (CVM)May 2001BPGuidance for Industry Bioanalytical Method ValidationAdditional copies are available from:Drug Information Branch (HFD-210)Center for Drug Evaluation and Research (CDER)5600 Fishers Lane, Rockville, MD 20857 (Tel) 301-827-4573Internet at /cder/guidance/index.htmorCommunications Staff (HFV-12)Center for Veterinary Medicine (CVM)7500 Standish Place, Rockville, MD 20855 (Tel) 301–594-1755Internet at /cvmU.S. Department of Health and Human ServicesFood and Drug AdministrationCenter for Drug Evaluation and Research (CDER)Center for Veterinary Medicine (CVM)May 2001BPTable of ContentsI.INTRODUCTION (1)II.BACKGROUND (1)A.F ULL V ALIDATION (2)B.P ARTIAL V ALIDATION (2)C.C ROSS-V ALIDATION (3)III.REFERENCE STANDARD (4)IV.METHOD DEVELOPMENT: CHEMICAL ASSAY (4)A.S ELECTIVITY (4)B.A CCURACY, P RECISION, AND R ECOVERY (5)C.C ALIBRATION/S TANDARD C URVE (5)D.S TABILITY (6)E.P RINCIPLES OF B IOANALYTICAL M ETHOD V ALIDATION AND E STABLISHMENT (8)F.S PECIFIC R ECOMMENDATIONS FOR M ETHOD V ALIDATION (10)V.METHOD DEVELOPMENT: MICROBIOLOGICAL AND LIGAND-BINDING ASSAYS (11)A.S ELECTIVITY I SSUES (11)B.Q UANTIFICATION I SSUES (12)VI.APPLICATION OF VALIDATED METHOD TO ROUTINE DRUG ANALYSIS (13)A CCEPTANCE C RITERIA FOR THE R UN (15)VII.DOCUMENTATION (16)A.S UMMARY I NFORMATION (16)B.D OCUMENTATION FOR M ETHOD E STABLISHMENT (17)C.A PPLICATION TO R OUTINE D RUG A NALYSIS (17)D.O THER I NFORMATION (19)GLOSSARY (20)GUIDANCE FOR INDUSTRY1Bioanalytical Method ValidationI.INTRODUCTIONThis guidance provides assistance to sponsors of investigational new drug applications (INDs), new drug applications (NDAs), abbreviated new drug applications (ANDAs), and supplements in developing bioanalytical method validation information used in human clinical pharmacology, bioavailability (BA), and bioequivalence (BE) studies requiring pharmacokinetic (PK) evaluation. This guidance also applies to bioanalytical methods used for non-human pharmacology/toxicology studies and preclinical studies. For studies related to the veterinary drug approval process, this guidance applies only to blood and urine BA, BE, and PK studies.The information in this guidance generally applies to bioanalytical procedures such as gas chromatography (GC), high-pressure liquid chromatography (LC), combined GC and LC mass spectrometric (MS) procedures such as LC-MS, LC-MS-MS, GC-MS, and GC-MS-MS performed for the quantitative determination of drugs and/or metabolites in biological matricessuch as blood, serum, plasma, or urine. This guidance also applies to other bioanalytical methods, such as immunological and microbiological procedures, and to other biological matrices, such as tissue and skin samples.This guidance provides general recommendations for bioanalytical method validation. The recommendations can be adjusted or modified depending on the specific type of analytical method used. II.BACKGROUND1 This guidance has been prepared by the Biopharmaceutics Coordinating Committee in the Center for Drug Evaluation and Research (CDER) in cooperation with the Center for Veterinary Medicine (CVM) at the Food and Drug Administration.This guidance has been developed based on the deliberations of two workshops: (1) Analytical Methods Validation: Bioavailability, Bioequivalence, and Pharmacokinetic Studies (held on December 3B5, 19902 ) and (2) Bioanalytical Methods Validation C A Revisit With a Decade of Progress (held on January 12B14, 20003).Selective and sensitive analytical methods for the quantitative evaluation of drugs and their metabolites (analytes) are critical for the successful conduct of preclinical and/or biopharmaceutics and clinical pharmacology studies. Bioanalytical method validation includes all of the procedures that demonstrate that a particular method used for quantitative measurement of analytes in a given biological matrix, such as blood, plasma, serum, or urine, is reliable and reproducible for the intended use. The fundamental parameters for this validation include (1) accuracy, (2) precision, (3) selectivity, (4) sensitivity, (5) reproducibility, and (6) stability. Validation involves documenting, through the use of specific laboratory investigations, that the performance characteristics of the method are suitable and reliable for the intended analytical applications. The acceptability of analytical data corresponds directly to the criteria used to validate the method.Published methods of analysis are often modified to suit the requirements of the laboratory performing the assay. These modifications should be validated to ensure suitable performance of the analytical method. When changes are made to a previously validated method, the analyst should exercise judgment as to how much additional validation is needed. During the course of a typical drug development program, a defined bioanalytical method undergoes many modifications. The evolutionary changes to support specific studies and different levels of validation demonstrate the validity of an assay’s performance. Different types and levels of validation are defined and characterized as follows:A.Full Validation•Full validation is important when developing and implementing a bioanalytical method for the first time.•Full validation is important for a new drug entity.• A full validation of the revised assay is important if metabolites are added to an existing assay for quantification.B.Partial ValidationPartial validations are modifications of already validated bioanalytical methods. Partial validation can range from as little as one intra-assay accuracy and precision determination to a nearly full2 Workshop Report: Shah, V.P. et al., Pharmaceutical Research: 1992; 9:588-592.3 Workshop Report: Shah, V.P. et al., Pharmaceutical Research: 2000; 17:in press.validation. Typical bioanalytical method changes that fall into this category include, but are not limited to:•Bioanalytical method transfers between laboratories or analysts•Change in analytical methodology (e.g., change in detection systems)•Change in anticoagulant in harvesting biological fluid•Change in matrix within species (e.g., human plasma to human urine)•Change in sample processing procedures•Change in species within matrix (e.g., rat plasma to mouse plasma)•Change in relevant concentration range•Changes in instruments and/or software platforms•Limited sample volume (e.g., pediatric study)•Rare matrices•Selectivity demonstration of an analyte in the presence of concomitant medications•Selectivity demonstration of an analyte in the presence of specific metabolitesC.Cross-ValidationCross-validation is a comparison of validation parameters when two or more bioanalytical methods are used to generate data within the same study or across different studies. An example of cross-validation would be a situation where an original validated bioanalytical method serves as thereference and the revised bioanalytical method is the comparator. The comparisons should be done both ways.When sample analyses within a single study are conducted at more than one site or more than one laboratory, cross-validation with spiked matrix standards and subject samples should be conducted at each site or laboratory to establish interlaboratory reliability. Cross-validation should also be considered when data generated using different analytical techniques (e.g., LC-MS-MS vs.ELISA4) in different studies are included in a regulatory submission.All modifications should be assessed to determine the recommended degree of validation. The analytical laboratory conducting pharmacology/toxicology and other preclinical studies for regulatory submissions should adhere to FDA=s Good Laboratory Practices (GLPs)5 (21 CFR part 58) and to sound principles of quality assurance throughout the testing process. The bioanalytical method for human BA, BE, PK, and drug interaction studies must meet the criteria in 21 CFR 320.29. The analytical laboratory should have a written set of standard operating procedures (SOPs) to ensure a complete system of quality control and assurance. The SOPs should cover all aspects of analysis from the time the sample is collected and reaches the laboratory until the results of the analysis are reported. The SOPs also should include record keeping, security and chain of sample custody4 Enzyme linked immune sorbent assay5 For the Center for Veterinary Medicine, all bioequivalence studies are subject to Good Laboratory Practices.(accountability systems that ensure integrity of test articles), sample preparation, and analytical tools such as methods, reagents, equipment, instrumentation, and procedures for quality control and verification of results.The process by which a specific bioanalytical method is developed, validated, and used in routine sample analysis can be divided into (1) reference standard preparation, (2) bioanalytical method development and establishment of assay procedure, and (3) application of validated bioanalytical method to routine drug analysis and acceptance criteria for the analytical run and/or batch. These three processes are described in the following sections of this guidance.III.REFERENCE STANDARDAnalysis of drugs and their metabolites in a biological matrix is carried out using samples spiked with calibration (reference) standards and using quality control (QC) samples. The purity of the reference standard used to prepare spiked samples can affect study data. For this reason, an authenticated analytical reference standard of known identity and purity should be used to prepare solutions of known concentrations. If possible, the reference standard should be identical to the analyte. When this is not possible, an established chemical form (free base or acid, salt or ester) of known purity can be used. Three types of reference standards are usually used: (1) certified reference standards (e.g., USP compendial standards); (2) commercially supplied reference standards obtained from a reputable commercial source; and/or (3) other materials of documented purity custom-synthesized by an analytical laboratory or other noncommercial establishment. The source and lot number, expiration date, certificates of analyses when available, and/or internally or externally generated evidence of identity and purity should be furnished for each reference standard.IV.METHOD DEVELOPMENT: CHEMICAL ASSAYThe method development and establishment phase defines the chemical assay. The fundamental parameters for a bioanalytical method validation are accuracy, precision, selectivity, sensitivity, reproducibility, and stability. Measurements for each analyte in the biological matrix should be validated. In addition, the stability of the analyte in spiked samples should be determined. Typical method development and establishment for a bioanalytical method include determination of (1) selectivity, (2) accuracy, precision, recovery, (3) calibration curve, and (4) stability of analyte in spiked samples.A.SelectivitySelectivity is the ability of an analytical method to differentiate and quantify the analyte in thepresence of other components in the sample. For selectivity, analyses of blank samples of theappropriate biological matrix (plasma, urine, or other matrix) should be obtained from at leastsix sources. Each blank sample should be tested for interference, and selectivity should be ensured at the lower limit of quantification (LLOQ).Potential interfering substances in a biological matrix include endogenous matrix components, metabolites, decomposition products, and in the actual study, concomitant medication and other exogenous xenobiotics. If the method is intended to quantify more than one analyte, each analyte should be tested to ensure that there is no interference.B.Accuracy, Precision, and RecoveryThe accuracy of an analytical method describes the closeness of mean test results obtained by the method to the true value (concentration) of the analyte. Accuracy is determined by replicate analysis of samples containing known amounts of the analyte. Accuracy should be measured using a minimum of five determinations per concentration. A minimum of three concentrations in the range of expected concentrations is recommended. The mean value should be within 15% of the actual value except at LLOQ, where it should not deviate by more than 20%. The deviation of the mean from the true value serves as the measure of accuracy.The precision of an analytical method describes the closeness of individual measures of an analyte when the procedure is applied repeatedly to multiple aliquots of a single homogeneous volume of biological matrix. Precision should be measured using a minimum of five determinations per concentration. A minimum of three concentrations in the range of expected concentrations is recommended. The precision determined at each concentration level should not exceed 15% of the coefficient of variation (CV) except for the LLOQ, where it should not exceed 20% of the CV. Precision is further subdivided into within-run, intra-batch precision or repeatability, which assesses precision during a single analytical run, and between-run, inter-batch precision or repeatability, which measures precision with time, and may involve different analysts, equipment, reagents, and laboratories.The recovery of an analyte in an assay is the detector response obtained from an amount of the analyte added to and extracted from the biological matrix, compared to the detector response obtained for the true concentration of the pure authentic standard. Recovery pertains to the extraction efficiency of an analytical method within the limits of variability. Recovery of the analyte need not be 100%, but the extent of recovery of an analyte and of the internal standard should be consistent, precise, and reproducible. Recovery experiments should be performed by comparing the analytical results for extracted samples at three concentrations (low, medium, and high) with unextracted standards that represent 100% recovery.C.Calibration/Standard CurveA calibration (standard) curve is the relationship between instrument response and known concentrations of the analyte. A calibration curve should be generated for each analyte in thesample. A sufficient number of standards should be used to adequately define the relationship between concentration and response. A calibration curve should be prepared in the same biological matrix as the samples in the intended study by spiking the matrix with known concentrations of the analyte. The number of standards used in constructing a calibration curve will be a function of the anticipated range of analytical values and the nature of theanalyte/response relationship. Concentrations of standards should be chosen on the basis of the concentration range expected in a particular study. A calibration curve should consist of a blank sample (matrix sample processed without internal standard), a zero sample (matrix sample processed with internal standard), and six to eight non-zero samples covering the expected range, including LLOQ.1.Lower Limit of Quantification (LLOQ)The lowest standard on the calibration curve should be accepted as the limit ofquantification if the following conditions are met:C The analyte response at the LLOQ should be at least 5 times the responsecompared to blank response.C Analyte peak (response) should be identifiable, discrete, and reproducible witha precision of 20% and accuracy of 80-120%.2.Calibration Curve/Standard Curve/Concentration-ResponseThe simplest model that adequately describes the concentration-response relationshipshould be used. Selection of weighting and use of a complex regression equation should be justified. The following conditions should be met in developing a calibration curve:C#20% deviation of the LLOQ from nominal concentrationC#15% deviation of standards other than LLOQ from nominal concentrationAt least four out of six non-zero standards should meet the above criteria, including the LLOQ and the calibration standard at the highest concentration. Excluding thestandards should not change the model used.D.StabilityDrug stability in a biological fluid is a function of the storage conditions, the chemical properties of the drug, the matrix, and the container system. The stability of an analyte in a particular matrix and container system is relevant only to that matrix and container system and should not be extrapolated to other matrices and container systems. Stability procedures should evaluate the stability of the analytes during sample collection and handling, after long-term (frozen at theintended storage temperature) and short-term (bench top, room temperature) storage, and after going through freeze and thaw cycles and the analytical process. Conditions used in stability experiments should reflect situations likely to be encountered during actual sample handling and analysis. The procedure should also include an evaluation of analyte stability in stock solution.All stability determinations should use a set of samples prepared from a freshly made stock solution of the analyte in the appropriate analyte-free, interference-free biological matrix. Stock solutions of the analyte for stability evaluation should be prepared in an appropriate solvent at known concentrations.1.Freeze and Thaw StabilityAnalyte stability should be determined after three freeze and thaw cycles. At least three aliquots at each of the low and high concentrations should be stored at the intendedstorage temperature for 24 hours and thawed unassisted at room temperature. Whencompletely thawed, the samples should be refrozen for 12 to 24 hours under the sameconditions. The freeze–thaw cycle should be repeated two more times, then analyzedon the third cycle. If an analyte is unstable at the intended storage temperature, thestability sample should be frozen at -700C during the three freeze and thaw cycles.2.Short-Term Temperature StabilityThree aliquots of each of the low and high concentrations should be thawed at roomtemperature and kept at this temperature from 4 to 24 hours (based on the expectedduration that samples will be maintained at room temperature in the intended study) and analyzed.3.Long-Term StabilityThe storage time in a long-term stability evaluation should exceed the time between the date of first sample collection and the date of last sample analysis. Long-term stabilityshould be determined by storing at least three aliquots of each of the low and highconcentrations under the same conditions as the study samples. The volume of samples should be sufficient for analysis on three separate occasions. The concentrations of allthe stability samples should be compared to the mean of back-calculated values for the standards at the appropriate concentrations from the first day of long-term stabilitytesting.4.Stock Solution StabilityThe stability of stock solutions of drug and the internal standard should be evaluated at room temperature for at least 6 hours. If the stock solutions are refrigerated or frozenfor the relevant period, the stability should be documented. After completion of thedesired storage time, the stability should be tested by comparing the instrumentresponse with that of freshly prepared solutions.5.Post-Preparative StabilityThe stability of processed samples, including the resident time in the autosampler, should be determined. The stability of the drug and the internal standard should be assessedover the anticipated run time for the batch size in validation samples by determiningconcentrations on the basis of original calibration standards.Although the traditional approach of comparing analytical results for stored samples with those for freshly prepared samples has been referred to in this guidance, other statistical approaches based on confidence limits for evaluation of an analyte=s stability in abiological matrix can be used. SOPs should clearly describe the statistical method andrules used. Additional validation may include investigation of samples from dosedsubjects.E.Principles of Bioanalytical Method Validation and Establishment•The fundamental parameters to ensure the acceptability of the performance of a bioanalytical method validation are accuracy, precision, selectivity, sensitivity,reproducibility, and stability.• A specific, detailed description of the bioanalytical method should be written. This can be in the form of a protocol, study plan, report, and/or SOP.•Each step in the method should be investigated to determine the extent to which environmental, matrix, material, or procedural variables can affect the estimation of analyte in the matrix from the time of collection of the material up to and including the time ofanalysis.•It may be important to consider the variability of the matrix due to the physiological nature of the sample. In the case of LC-MS-MS-based procedures, appropriate steps should be taken to ensure the lack of matrix effects throughout the application of the method,especially if the nature of the matrix changes from the matrix used during method validation.• A bioanalytical method should be validated for the intended use or application. All experiments used to make claims or draw conclusions about the validity of the methodshould be presented in a report (method validation report).•Whenever possible, the same biological matrix as the matrix in the intended samples should be used for validation purposes. (For tissues of limited availability, such as bone marrow, physiologically appropriate proxy matrices can be substituted.)•The stability of the analyte (drug and/or metabolite) in the matrix during the collection process and the sample storage period should be assessed, preferably prior to sampleanalysis.•For compounds with potentially labile metabolites, the stability of analyte in matrix from dosed subjects (or species) should be confirmed.•The accuracy, precision, reproducibility, response function, and selectivity of the method for endogenous substances, metabolites, and known degradation products should beestablished for the biological matrix. For selectivity, there should be evidence that thesubstance being quantified is the intended analyte.•The concentration range over which the analyte will be determined should be defined in the bioanalytical method, based on evaluation of actual standard samples over the range,including their statistical variation. This defines the standard curve.• A sufficient number of standards should be used to adequately define the relationship between concentration and response. The relationship between response and concentration should be demonstrated to be continuous and reproducible. The number of standards used should be a function of the dynamic range and nature of the concentration-responserelationship. In many cases, six to eight concentrations (excluding blank values) can define the standard curve. More standard concentrations may be recommended for nonlinear than for linear relationships.•The ability to dilute samples originally above the upper limit of the standard curve should be demonstrated by accuracy and precision parameters in the validation.•In consideration of high throughput analyses, including but not limited to multiplexing, multicolumn, and parallel systems, sufficient QC samples should be used to ensure control of the assay. The number of QC samples to ensure proper control of the assay should be determined based on the run size. The placement of QC samples should be judiciously considered in the run.•For a bioanalytical method to be considered valid, specific acceptance criteria should be set in advance and achieved for accuracy and precision for the validation of QC samples over the range of the standards.F.Specific Recommendations for Method Validation•The matrix-based standard curve should consist of a minimum of six standard points, excluding blanks, using single or replicate samples. The standard curve should cover the entire range of expected concentrations.•Standard curve fitting is determined by applying the simplest model that adequately describes the concentration-response relationship using appropriate weighting and statistical tests for goodness of fit.•LLOQ is the lowest concentration of the standard curve that can be measured with acceptable accuracy and precision. The LLOQ should be established using at least five samples independent of standards and determining the coefficient of variation and/orappropriate confidence interval. The LLOQ should serve as the lowest concentration on the standard curve and should not be confused with the limit of detection and/or the low QC sample. The highest standard will define the upper limit of quantification (ULOQ) of an analytical method.•For validation of the bioanalytical method, accuracy and precision should be determined using a minimum of five determinations per concentration level (excluding blank samples).The mean value should be within ±15% of the theoretical value, except at LLOQ, where it should not deviate by more than ±20%. The precision around the mean value should not exceed 15% of the CV, except for LLOQ, where it should not exceed 20% of the CV.Other methods of assessing accuracy and precision that meet these limits may be equally acceptable.•The accuracy and precision with which known concentrations of analyte in biological matrix can be determined should be demonstrated. This can be accomplished by analysis ofreplicate sets of analyte samples of known concentrations C QC samples C from anequivalent biological matrix. At a minimum, three concentrations representing the entire range of the standard curve should be studied: one within 3x the lower limit of quantification (LLOQ) (low QC sample), one near the center (middle QC), and one near the upperboundary of the standard curve (high QC).•Reported method validation data and the determination of accuracy and precision should include all outliers; however, calculations of accuracy and precision excluding values that are statistically determined as outliers can also be reported.•The stability of the analyte in biological matrix at intended storage temperatures should be established. The influence of freeze-thaw cycles (a minimum of three cycles at twoconcentrations in triplicate) should be studied.•The stability of the analyte in matrix at ambient temperature should be evaluated over a time period equal to the typical sample preparation, sample handling, and analytical run times.•Reinjection reproducibility should be evaluated to determine if an analytical run could be reanalyzed in the case of instrument failure.•The specificity of the assay methodology should be established using a minimum of six independent sources of the same matrix. For hyphenated mass spectrometry-basedmethods, however, testing six independent matrices for interference may not be important.In the case of LC-MS and LC-MS-MS-based procedures, matrix effects should beinvestigated to ensure that precision, selectivity, and sensitivity will not be compromised.Method selectivity should be evaluated during method development and throughout methodvalidation and can continue throughout application of the method to actual study samples.•Acceptance/rejection criteria for spiked, matrix-based calibration standards and validation QC samples should be based on the nominal (theoretical) concentration of analytes.Specific criteria can be set up in advance and achieved for accuracy and precision over therange of the standards, if so desired.V.METHOD DEVELOPMENT: MICROBIOLOGICAL AND LIGAND-BINDING ASSAYSMany of the bioanalytical validation parameters and principles discussed above are also applicable to microbiological and ligand-binding assays. However, these assays possess some unique characteristics that should be considered during method validation.A.Selectivity IssuesAs with chromatographic methods, microbiological and ligand-binding assays should be shown to be selective for the analyte. The following recommendations for dealing with two selectivity issues should be considered:1.Interference From Substances Physiochemically Similar to the Analyte•Cross-reactivity of metabolites, concomitant medications, or endogenouscompounds should be evaluated individually and in combination with the analyteof interest.•When possible, the immunoassay should be compared with a validated reference method (such as LC-MS) using incurred samples and predetermined criteria foragreement of accuracy of immunoassay and reference method.。
ViraSafe Lentiviral Packaging System (Ecotropic) 产

Product ManualViraSafe™ Lentiviral Packaging System, EcotropicCatalog NumberkitVPK-205 1FOR RESEARCH USE ONLYNot for use in diagnostic proceduresIntroductionLentivirus vector based on the human immunodeficiency virus-1 (HIV-1) has become a promising vector for gene transfer studies. The advantageous feature of lentivirus vector is the ability of gene transfer and integration into dividing and non-dividing cells. Lentivirus pseudotyped with the MLV ecotropic envelope glycoprotein will only transduce mouse and rat cells with high efficiency. Lentiviral vectors have been shown to deliver genes to neurons, lymphocytes and macrophages, cell types that previous retrovirus vectors could not be used. Lentiviral vectors have also proven to be effective in transducing brain, liver, muscle, and retina in vivo without toxicity or immune responses. Recently, the lentivirus system is widely used to integrate siRNA efficiently in a wide variety of cell lines and primary cells both in vitro and in vivo.Lentivirus particles are produced from 293T cells through transient transfection of plasmids that encode for the components of the virion (Figure 1). Due to safety concerns regarding the infectious nature of HIV-1, recent lentiviral packaging systems have separated the viral components into 3 or 4 plasmids. However, these systems still present a small chance of generating replication-competent lentivirus upon recombination. In addition, most commercial lentiviral packaging systems provide plasmids containing the viral structure proteins in a premixed formulation, making it nearly impossible to optimize the ratio of the various plasmids for your particular experiment and host cell.Cell Biolabs’ ViraSafe™ Lentiviral Packaging System provides a much safer method to package lentivirus, while still providing high viral titers. In addition, each plasmid is provided separately rather than in a packaging mixture. This allows you the flexibility to amplify individual plasmids and optimize the ratio of plasmids for your experiment.Key Features of ViraSafe™ Lentiviral Packaging System:1.Packaging Plasmids: Improve the packaging plasmid to increase performance and reduce thelikelihood of recombination between vector components.a. Minimize HIV sequences – no accessory proteins, Tat or Rev, or LTRsb. Prevent overlap with vector SM by codon wobbling Gag sequencesc. Boost particle production by incorporating adenovirus VA I element2.Flexible: All vectors including packaging vectors are provided separately to allow end-user tooptimize the vector ratio for maximal lentivirus production.Figure 1. Lentivirus Production in 293T CellsRelated Products1.VPK-206: ViraSafe™ Lentiviral Packaging System, Pantropic2.VPK-107: QuickTiter™ Lentivirus Titer Kit (Lentivirus-Associated HIV p24)3.VPK-108: QuickTiter™ Lentivirus Quantitation Kit4.VPK-090: ViraBind™ Lentivirus Concentration and Purification Kit5.LTV-200: ViraDuctin™ Lentivirus Transduction Kit6.LTV-100: 293LTV Cell LineUnique Elements of the ViraSafe™ Lentivirus Packaging SystemKit Components1.pRSV-Rev Packaging Vector (Part No. 320022): One 40 µL vial at 0.25 mg/mL.2.pCMV-Eco Envelope Vector (Part No. 320026): One 40 µL vial at 0.25 mg/mL.3.pCgpV Packaging Vector (Part No. 320024): One 40 µL vial at 0.25 mg/mL.Figure 2: pRSV-Rev Packaging Vector (4180 bp, Ampicillin-resistant). EcoRI Digestion: 300 bp + 3880 bpFigure 3: pCMV-Eco Envelop Vector (6763 bp, Ampicillin-resistant). BamHI Digestion: 777 bp + 5986 bp.Figure 4: pCgpV Packaging Vector (9118 bp, Ampicillin-resistant). Pst I Digestion: 927 bp + 1424 bp + 6767 bp.Materials Not Supplied1.Lentiviral Transfer Vector2.293T cells: we recommend 293LTV Cell Line (Cat. # LTV-100) for high titer production oflentivirus.3.Cell Culture Medium4.Transfection ReagentsStorageUpon receipt, store all other kit components at -20ºC until their expiration dates.Safety ConsiderationsRemember that you will be working with samples containing infectious virus. Follow the recommended NIH guidelines for all materials containing BSL-2 organisms. The ViraSafe™ Universal Lentiviral Expression System is designed to minimize the chance of generating replication-competent lentivirus, but precautions should still be taken to avoid direct contact with viral supernatants.Lentivirus Production1.One day before transfection, plate sufficient 293T cells or 293LTV cells (cat.# LTV-100) toachieve 70-80% confluence on the day of transfection.2.Transfect cells by Calcium Phosphate or other transfection reagents.Note: We suggest transfecting cells with FuGENE® Transfection Reagent (Roche Applied Science) or Lipofectamine™ Plus (Invitrogen). We recommend the ratio of vectors at 3:1:1:1 (transfer vector: pCMV-Eco:pRSV-REV:pCgpV).3.Harvest lentiviral supernatant 36-72 hours after transfection. Supernatant can be harvested 2 or3 times, every 12 hours. Keep it at 4ºC over the collecting period.4.Pool the collected supernatants, centrifuge 5 minutes at 1500 rpm to remove cell debris andfiltrate on 0.22 µm.5.Supernatants can be used directly or purified/concentrated if needed. For long term storage,store supernatant at -80ºC in aliquots.Post-Packaging ConsiderationsPackaging your lentivirus is only the first step to ensuring successful expression of your gene. The following steps should be considered prior to infection of your host cell:1.Concentration and purification of your lentivirus: Because of the latent nature of lentivirus,it is imperative that your virus be highly concentrated before infecting your host cell. Also,impurities from your viral supernatant can decrease the efficiency of infection. We recommend using Cell Biolabs’ ViraBind™ Lentivirus Concentration and Purification Kit (Catalog # VPK-090).2.Measure the titer of your lentivirus: This is an important step to ensure consistent viraltransduction into your host cell. However, QPCR or stable clone counting can take as much as 1-2 weeks to perform. Traditional p24 ELISA kits can greatly overestimate your lentiviral titer.Our advanced p24 ELISA, QuickTiter™ Lentivirus Titer Kit (Catalog # VPK-107), usesexclusive technology that eliminates free p24 from your supernatant, giving you much moreaccurate lentiviral titers. Results are obtained in 6-18 hours.e transduction reagents to increase infection efficiency: Many cells are difficult to infectwith lentivirus, and without supplemental reagents transduction efficiencies can be low.Reagents such as Polybrene® can help, but are often insufficient. Cell Biolabs’ proprietaryreagents in our ViraDuctin™ Lentivirus Transduction Kit (Catalog # LTV-200) form a super-complex with your virus to increase transduction efficiencies by promoting virus and cellinteraction.References1.Chen, M. et al. (2002). Nature Genetics32(4): 670-675.2.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono(1996) Science272:263-267.3.Verma, I. M., and N. Somia (1997) Nature 389:239-2424.Kahl C. A., Marsh J., Fyffe J., Sanders D. A., and K. Cornetta (2004) J Virol.78:1421-30.5.White S. M., Renda M., Nam N. Y., Klimatcheva E., Zhu Y., Fisk J., Halterman M., Rimel B. J.,Federoff H., Pandya S., Rosenblatt J. D., and V. Planelles (1999) J Virol.73:2832-40.6.Kafri T., van Praag H., Ouyang L., Gage F. H., and I. M. Verma (1999) J Virol.73:576-84. Notice to PurchaserThis product is sold for research and development purposes only and is not to be incorporated into products for resale without written permission from Cell Biolabs. The patented technology is covered by a license from University of Southern California. By the use of this product you accept the terms and conditions of all applicable Limited Use Label Licenses. You may contact our Business Development department at ********************** for information on sublicensing this technology. WarrantyThese products are warranted to perform as described in their labeling and in Cell Biolabs literature when used in accordance with their instructions. THERE ARE NO WARRANTIES THAT EXTEND BEYOND THIS EXPRESSED WARRANTY AND CELL BIOLABS DISCLAIMS ANY IMPLIED WARRANTY OF MERCHANTABILITY OR WARRANTY OF FITNESS FOR PARTICULAR PURPOSE. CELL BIOLABS’ sole obligation and purchaser’s exclusive remedy for breach of this warranty shall be, at the option of CELL BIOLABS, to repair or replace the products. In no event shall CELL BIOLABS be liable for any proximate, incidental or consequential damages in connection with the products.Contact InformationCell Biolabs, Inc.7758 Arjons DriveSan Diego, CA 92126Worldwide: +1 858-271-6500USA Toll-Free: 1-888-CBL-0505E-mail: ********************2009: Cell Biolabs, Inc. - All rights reserved. No part of these works may be reproduced in any form without permissions in writing.。
荷尔登生物医学检测产品说明书
1mL
R
1mL
SST, R, GL
1mL
No capillary
1mL
collection
Order of Draw: 1. BLOOD CULTURES 2. LT BLUE 3. RED 4. SST 5. PST/LH GRN 6. LAV 7. GRAY
M2909 (1-19)
Microtainer Microtainer 1 mL
2 Microtainers Microtainer
2 mL FILL TO LINE 1 mL 1 mL Microtainer 1 mL 1 mL Microtainer
SST, R, GL
STAT to lab in 10 min
Also known as CMP SST, R, GL SST, R, GL
TOBRAMYCIN TOTAL PROTEIN TROPONIN TSH
TYPE AND CROSSMATCH
TYPE AND SCREEN URIC ACID
VANCOMYCIN VALPROIC ACID VITAMIN D
FEA
6LA LHC 6LIP
6LIVER 6LIPID LIA 6MG 6METX 6MONO PHNOC 6PFA 6PHOS 6PBNP 6PCAL 6KA 6PTIMEN PSAC 6PTT 6RETCT 6RENAL WHRIG 6SAL SYPHB TBSATA TT3C T4C TESTOC THEOC
CHEM8
QUICK CHEM 8 CBC/AUTO DIFF CKMB
COMP CREATININE CREATININE X-RAY EVALUATION D-DIMER DIGOXIN DILANTIN ELECTROLYTES FERRITIN FSH GLUCOSE
1,3-二磷酸甘油酸ELISA检测试剂盒说明书

1,25-二羟基维生素D3检测试剂盒说明书上海西格生物科技有限公司本试剂盒仅供研究使用检测范围:1.5ng/mL~48ng/mL1,3-二磷酸甘油酸 elisa检测试剂盒主要用于定量测定血清、血浆、细胞培养上清或其他相关生物液体中的含量。
特色服务:1,3-二磷酸甘油酸 ELISA检测试剂盒使用说明书免费代测可上门取样,可免费申领24T 免费试用装elisa试剂盒检测范围:人、绵羊、小鼠、大鼠、猪、兔、山羊、牛、马、猪、其它动物细胞因子、植物细胞因子、骨代谢、细胞凋亡、激素内分泌、活性多肽、肝纤维化、自身抗体、血栓与止血、自身抗体科研elisa检测试剂盒。
产品保存:2-8℃、有效期6个月,我司1,3-二磷酸甘油酸 ELISA检测试剂盒使用说明书均为最新批次,您可放心选购。
规格:48T/96T性状:液体运输方式:快递发货有效期:6个月品特点:灵敏性高,高效性,吸附均匀,吸附性好,回收利用率高,是一款可以让您放心选购的1,3-二磷酸甘油酸 ELISA检测试剂盒使用说明书产品。
使用范围:此产品供科研实验使用,不得用于临床。
用途:用于测定人血清、血浆、组织及相关液体样本中的含量或活性。
1,3-二磷酸甘油酸 ELISA检测试剂盒使用说明书特点1.专一性强。
抗原与抗体的免疫反应是专一反应,而免疫酶技术以免疫反应为基础,所检测的对象是抗原(或抗体),使用的抗体除标记了酶以外,与普通抗体的免疫反应特性并无多大差别。
2.灵敏度高。
由于抗体联结上了酶,因此,借助于酶与底物的显色反应,显示抗原与抗体的结合,大大提高了检测的灵敏性,使检测水平接近放射免疫测定法。
3.样品易保存。
经过酶反应显示的有色产物大多比较稳定,因此有利于样品的保存。
4.结果易观察。
对检测结果即可用肉眼观察,又可用显微镜观察,也可以用分光光度计进行比色测定,还可用显微镜观察。
这是因为某些酶反应产物能使电子密度发生改变,从而引起被检测物的显示。
5.可以定量测定。
