CLUSTERING VIA DIMENSIONAL REDUCTION METHOD FOR THE PROJECTION PURSUIT BASED ON THE ICSA
Document Clustering Using Locality Preserving Indexing

Xiaofei He Department of Computer Science The University of Chicago 1100 East 58th Street, Chicago, IL 60637, USA Phone: (733) 288-2851 xiaofei@
Jiawei Han Department of Computer Science University of Illinois at Urbana Champaign 2132 Siebel Center, 201 N. Goodwin Ave, Urbana, IL 61801, USA Phone: (217) 333-6903 Fax: (217) 265-6494 hanj@
document clustering [28][27]. They model each cluster as a linear combination of the data points, and each data point as a linear combination of the clusters. And they compute the linear coefficients by minimizing the global reconstruction error of the data points using Non-negative Matrix Factorization. Thus, NMF method still focuses on the global geometrical structure of document space. Moreover, the iterative update method for solving NMF problem is computational expensive. In this paper, we propose a novel document clustering algorithm by using Locality Preserving Indexing (LPI). Different from LSI which aims to discover the global Euclidean structure, LPI aims to discover the local geometrical structure. LPI can have more discriminating power. Thus, the documents related to the same semantics are close to each other in the low dimensional representation space. Also, LPI is derived by finding the optimal linear approximations to the eigenfunctions of the Laplace Beltrami operator on the document manifold. Laplace Beltrami operator takes the second order derivatives of the functions on the manifolds. It evaluates the smoothness of the functions. Therefore, it can discover the non-linear manifold structure to some extent. Some theoretical justifications can be traced back to [15][14]. The original LPI is not optimal in the sense of computation in that the obtained basis functions might contain a trivial solution. The trivial solution contains no information and thus useless for document indexing. A modified LPI is proposed to obtain better document representations. In this low dimensional space, we then apply traditional clustering algorithms such as k -means to cluster the documents into semantically different classes. The rest of this paper is organized as follows: In Section 2, we give a brief review of LSI and LPI. Section 3 introduces our proposed document clustering algorithm. Some theoretical analysis is provided in Section 4. The experimental results are shown in Section 5. Finally, we give concluding remarks and future work in Section 6.
SPSS术语中英文对照

SPSS术语中英文对照【常用软件】SPSS术语中英文对照Absolute deviation, 绝对离差Absolute number, 绝对数Absolute residuals, 绝对残差Acceleration array, 加速度立体阵Acceleration in an arbitrary direction, 任意方向上的加速度Acceleration normal, 法向加速度Acceleration space dimension, 加速度空间的维数Acceleration tangential, 切向加速度Acceleration vector, 加速度向量Acceptable hypothesis, 可接受假设Accumulation, 累积Accuracy, 准确度Actual frequency, 实际频数Adaptive estimator, 自适应估计量Addition, 相加Addition theorem, 加法定理Additivity, 可加性Adjusted rate, 调整率Adjusted value, 校正值Admissible error, 容许误差Aggregation, 聚集性Alternative hypothesis, 备择假设Among groups, 组间Amounts, 总量Analysis of correlation, 相关分析Analysis of covariance, 协方差分析Analysis of regression, 回归分析Analysis of time series, 时间序列分析Analysis of variance, 方差分析Angular transformation, 角转换ANOVA (analysis of variance), 方差分析ANOVA Models, 方差分析模型Arcing, 弧/弧旋Arcsine transformation, 反正弦变换Area under the curve, 曲线面积AREG , 评估从一个时间点到下一个时间点回归相关时的误差ARIMA, 季节和非季节性单变量模型的极大似然估计Arithmetic grid paper, 算术格纸Arithmetic mean, 算术平均数Arrhenius relation, 艾恩尼斯关系Assessing fit, 拟合的评估Associative laws, 结合律Asymmetric distribution, 非对称分布Asymptotic bias, 渐近偏倚Asymptotic efficiency, 渐近效率Asymptotic variance, 渐近方差Attributable risk, 归因危险度Attribute data, 属性资料Attribution, 属性Autocorrelation, 自相关Autocorrelation of residuals, 残差的自相关Average, 平均数Average confidence interval length, 平均置信区间长度Average growth rate, 平均增长率Bar chart, 条形图Bar graph, 条形图Base period, 基期Bayes' theorem , Bayes定理Bell-shaped curve, 钟形曲线Bernoulli distribution, 伯努力分布Best-trim estimator, 最好切尾估计量Bias, 偏性Binary logistic regression, 二元逻辑斯蒂回归Binomial distribution, 二项分布Bisquare, 双平方Bivariate Correlate, 二变量相关Bivariate normal distribution, 双变量正态分布Bivariate normal population, 双变量正态总体Biweight interval, 双权区间Biweight M-estimator, 双权M估计量Block, 区组/配伍组BMDP(Biomedical computer programs), BMDP统计软件包Boxplots, 箱线图/箱尾图Breakdown bound, 崩溃界/崩溃点Canonical correlation, 典型相关Caption, 纵标目Case-control study, 病例对照研究Categorical variable, 分类变量Catenary, 悬链线Cauchy distribution, 柯西分布Cause-and-effect relationship, 因果关系Cell, 单元Censoring, 终检Center of symmetry, 对称中心Centering and scaling, 中心化和定标Central tendency, 集中趋势Central value, 中心值CHAID -χ2 Automatic Interac tion Detector, 卡方自动交互检测Chance, 机遇Chance error, 随机误差Chance variable, 随机变量Characteristic equation, 特征方程Characteristic root, 特征根Characteristic vector, 特征向量Chebshev criterion of fit, 拟合的切比雪夫准则Chernoff faces, 切尔诺夫脸谱图Chi-square test, 卡方检验/χ2检验Choleskey decomposition, 乔洛斯基分解Circle chart, 圆图Class interval, 组距Class mid-value, 组中值Class upper limit, 组上限Classified variable, 分类变量Cluster analysis, 聚类分析Cluster sampling, 整群抽样Code, 代码Coded data, 编码数据Coding, 编码Coefficient of contingency, 列联系数Coefficient of determination, 决定系数Coefficient of multiple correlation, 多重相关系数Coefficient of partial correlation, 偏相关系数Coefficient of production-moment correlation, 积差相关系数Coefficient of rank correlation, 等级相关系数Coefficient of regression, 回归系数Coefficient of skewness, 偏度系数Coefficient of variation, 变异系数Cohort study, 队列研究Column, 列Column effect, 列效应Column factor, 列因素Combination pool, 合并Combinative table, 组合表Common factor, 共性因子Common regression coefficient, 公共回归系数Common value, 共同值Common variance, 公共方差Common variation, 公共变异Communality variance, 共性方差Comparability, 可比性Comparison of bathes, 批比较Comparison value, 比较值Compartment model, 分部模型Compassion, 伸缩Complement of an event, 补事件Complete association, 完全正相关Complete dissociation, 完全不相关Complete statistics, 完备统计量Completely randomized design, 完全随机化设计Composite event, 联合事件Composite events, 复合事件Concavity, 凹性Conditional expectation, 条件期望Conditional likelihood, 条件似然Conditional probability, 条件概率Conditionally linear, 依条件线性Confidence interval, 置信区间Confidence limit, 置信限Confidence lower limit, 置信下限Confidence upper limit, 置信上限Confirmatory Factor Analysis , 验证性因子分析Confirmatory research, 证实性实验研究Confounding factor, 混杂因素Conjoint, 联合分析Consistency, 相合性Consistency check, 一致性检验Consistent asymptotically normal estimate, 相合渐近正态估计Consistent estimate, 相合估计Constrained nonlinear regression, 受约束非线性回归Constraint, 约束Contaminated distribution, 污染分布Contaminated Gausssian, 污染高斯分布Contaminated normal distribution, 污染正态分布Contamination, 污染Contamination model, 污染模型Contingency table, 列联表Contour, 边界线Contribution rate, 贡献率Control, 对照Controlled experiments, 对照实验Conventional depth, 常规深度Convolution, 卷积Corrected factor, 校正因子Corrected mean, 校正均值Correction coefficient, 校正系数Correctness, 正确性Correlation coefficient, 相关系数Correlation index, 相关指数Correspondence, 对应Counting, 计数Counts, 计数/频数Covariance, 协方差Covariant, 共变Cox Regression, Cox回归Criteria for fitting, 拟合准则Criteria of least squares, 最小二乘准则Critical ratio, 临界比Critical region, 拒绝域Critical value, 临界值Cross-over design, 交叉设计Cross-section analysis, 横断面分析Cross-section survey, 横断面调查Crosstabs , 交叉表Cross-tabulation table, 复合表Cube root, 立方根Cumulative distribution function, 分布函数Cumulative probability, 累计概率Curvature, 曲率/弯曲Curvature, 曲率Curve fit , 曲线拟和Curve fitting, 曲线拟合Curvilinear regression, 曲线回归Curvilinear relation, 曲线关系Cut-and-try method, 尝试法Cycle, 周期Cyclist, 周期性D test, D检验Data acquisition, 资料收集Data bank, 数据库Data capacity, 数据容量Data deficiencies, 数据缺乏Data handling, 数据处理Data manipulation, 数据处理Data processing, 数据处理Data reduction, 数据缩减Data set, 数据集Data sources, 数据来源Data transformation, 数据变换Data validity, 数据有效性Data-in, 数据输入Data-out, 数据输出Dead time, 停滞期Degree of freedom, 自由度Degree of precision, 精密度Degree of reliability, 可靠性程度Degression, 递减Density function, 密度函数Density of data points, 数据点的密度Dependent variable, 应变量/依变量/因变量Dependent variable, 因变量Depth, 深度Derivative matrix, 导数矩阵Derivative-free methods, 无导数方法Design, 设计Determinacy, 确定性Determinant, 行列式Determinant, 决定因素Deviation, 离差Deviation from average, 离均差Diagnostic plot, 诊断图Dichotomous variable, 二分变量Differential equation, 微分方程Direct standardization, 直接标准化法Discrete variable, 离散型变量DISCRIMINANT, 判断Discriminant analysis, 判别分析Discriminant coefficient, 判别系数Discriminant function, 判别值Dispersion, 散布/分散度Disproportional, 不成比例的Disproportionate sub-class numbers, 不成比例次级组含量Distribution free, 分布无关性/免分布Distribution shape, 分布形状Distribution-free method, 任意分布法Distributive laws, 分配律Disturbance, 随机扰动项Dose response curve, 剂量反应曲线Double blind method, 双盲法Double blind trial, 双盲试验Double exponential distribution, 双指数分布Double logarithmic, 双对数Downward rank, 降秩Dual-space plot, 对偶空间图DUD, 无导数方法Duncan's new multiple range method, 新复极差法/Duncan新法Effect, 实验效应Eigenvalue, 特征值Eigenvector, 特征向量Ellipse, 椭圆Empirical distribution, 经验分布Empirical probability, 经验概率单位Enumeration data, 计数资料Equal sun-class number, 相等次级组含量Equally likely, 等可能Equivariance, 同变性Error, 误差/错误Error of estimate, 估计误差Error type I, 第一类错误Error type II, 第二类错误Estimand, 被估量Estimated error mean squares, 估计误差均方Estimated error sum of squares, 估计误差平方和Euclidean distance, 欧式距离Event, 事件Event, 事件Exceptional data point, 异常数据点Expectation plane, 期望平面Expectation surface, 期望曲面Expected values, 期望值Experiment, 实验Experimental sampling, 试验抽样Experimental unit, 试验单位Explanatory variable, 说明变量Exploratory data analysis, 探索性数据分析Explore Summarize, 探索-摘要Exponential curve, 指数曲线Exponential growth, 指数式增长EXSMOOTH, 指数平滑方法Extended fit, 扩充拟合Extra parameter, 附加参数Extrapolation, 外推法Extreme observation, 末端观测值Extremes, 极端值/极值F distribution, F分布F test, F检验Factor, 因素/因子Factor analysis, 因子分析Factor Analysis, 因子分析Factor score, 因子得分Factorial, 阶乘Factorial design, 析因试验设计False negative, 假阴性False negative error, 假阴性错误Family of distributions, 分布族Family of estimators, 估计量族Fanning, 扇面Fatality rate, 病死率Field investigation, 现场调查Field survey, 现场调查Finite population, 有限总体Finite-sample, 有限样本First derivative, 一阶导数First principal component, 第一主成分First quartile, 第一四分位数Fisher information, 费雪信息量Fitted value, 拟合值Fitting a curve, 曲线拟合Fixed base, 定基Fluctuation, 随机起伏Forecast, 预测Four fold table, 四格表Fourth, 四分点Fraction blow, 左侧比率Fractional error, 相对误差Frequency, 频率Frequency polygon, 频数多边图Frontier point, 界限点Function relationship, 泛函关系Gamma distribution, 伽玛分布Gauss increment, 高斯增量Gaussian distribution, 高斯分布/正态分布Gauss-Newton increment, 高斯-牛顿增量General census, 全面普查GENLOG (Generalized liner models), 广义线性模型Geometric mean, 几何平均数Gini's mean difference, 基尼均差GLM (General liner models), 一般线性模型Goodness of fit, 拟和优度/配合度Gradient of determinant, 行列式的梯度Graeco-Latin square, 希腊拉丁方Grand mean, 总均值Gross errors, 重大错误Gross-error sensitivity, 大错敏感度Group averages, 分组平均Grouped data, 分组资料Guessed mean, 假定平均数Half-life, 半衰期Hampel M-estimators, 汉佩尔M估计量Happenstance, 偶然事件Harmonic mean, 调和均数Hazard function, 风险均数Hazard rate, 风险率Heading, 标目Heavy-tailed distribution, 重尾分布Hessian array, 海森立体阵Heterogeneity, 不同质Heterogeneity of variance, 方差不齐Hierarchical classification, 组内分组Hierarchical clustering method, 系统聚类法High-leverage point, 高杠杆率点HILOGLINEAR, 多维列联表的层次对数线性模型Hinge, 折叶点Histogram, 直方图Historical cohort study, 历史性队列研究Holes, 空洞HOMALS, 多重响应分析Homogeneity of variance, 方差齐性Homogeneity test, 齐性检验Huber M-estimators, 休伯M估计量Hyperbola, 双曲线Hypothesis testing, 假设检验Hypothetical universe, 假设总体Impossible event, 不可能事件Independence, 独立性Independent variable, 自变量Index, 指标/指数Indirect standardization, 间接标准化法Individual, 个体Inference band, 推断带Infinite population, 无限总体Infinitely great, 无穷大Infinitely small, 无穷小Influence curve, 影响曲线Information capacity, 信息容量Initial condition, 初始条件Initial estimate, 初始估计值Initial level, 最初水平Interaction, 交互作用Interaction terms, 交互作用项Intercept, 截距Interpolation, 内插法Interquartile range, 四分位距Interval estimation, 区间估计Intervals of equal probability, 等概率区间Intrinsic curvature, 固有曲率Invariance, 不变性Inverse matrix, 逆矩阵Inverse probability, 逆概率Inverse sine transformation, 反正弦变换Iteration, 迭代Jacobian determinant, 雅可比行列式Joint distribution function, 分布函数Joint probability, 联合概率Joint probability distribution, 联合概率分布K means method, 逐步聚类法Kaplan-Meier, 评估事件的时间长度Kaplan-Merier chart, Kaplan-Merier图Kendall's rank correlation, Kendall等级相关Kinetic, 动力学Kolmogorov-Smirnove test, 柯尔莫哥洛夫-斯米尔诺夫检验Kruskal and Wallis test, Kruskal及Wallis检验/多样本的秩和检验/H检验Kurtosis, 峰度Lack of fit, 失拟Ladder of powers, 幂阶梯Lag, 滞后Large sample, 大样本Large sample test, 大样本检验Latin square, 拉丁方Latin square design, 拉丁方设计Leakage, 泄漏Least favorable configuration, 最不利构形Least favorable distribution, 最不利分布Least significant difference, 最小显著差法Least square method, 最小二乘法Least-absolute-residuals estimates, 最小绝对残差估计Least-absolute-residuals fit, 最小绝对残差拟合Least-absolute-residuals line, 最小绝对残差线Legend, 图例L-estimator, L估计量L-estimator of location, 位置L估计量L-estimator of scale, 尺度L估计量Level, 水平Life expectance, 预期期望寿命Life table, 寿命表Life table method, 生命表法Light-tailed distribution, 轻尾分布Likelihood function, 似然函数Likelihood ratio, 似然比line graph, 线图Linear correlation, 直线相关Linear equation, 线性方程Linear programming, 线性规划Linear regression, 直线回归Linear Regression, 线性回归Linear trend, 线性趋势Loading, 载荷Location and scale equivariance, 位置尺度同变性Location equivariance, 位置同变性Location invariance, 位置不变性Location scale family, 位置尺度族Log rank test, 时序检验Logarithmic curve, 对数曲线Logarithmic normal distribution, 对数正态分布Logarithmic scale, 对数尺度Logarithmic transformation, 对数变换Logic check, 逻辑检查Logistic distribution, 逻辑斯特分布Logit transformation, Logit转换LOGLINEAR, 多维列联表通用模型Lognormal distribution, 对数正态分布Lost function, 损失函数Low correlation, 低度相关Lower limit, 下限Lowest-attained variance, 最小可达方差LSD, 最小显著差法的简称Lurking variable, 潜在变量Main effect, 主效应Major heading, 主辞标目Marginal density function, 边缘密度函数Marginal probability, 边缘概率Marginal probability distribution, 边缘概率分布Matched data, 配对资料Matched distribution, 匹配过分布Matching of distribution, 分布的匹配Matching of transformation, 变换的匹配Mathematical expectation, 数学期望Mathematical model, 数学模型Maximum L-estimator, 极大极小L 估计量Maximum likelihood method, 最大似然法Mean, 均数Mean squares between groups, 组间均方Mean squares within group, 组内均方Means (Compare means), 均值-均值比较Median, 中位数Median effective dose, 半数效量Median lethal dose, 半数致死量Median polish, 中位数平滑Median test, 中位数检验Minimal sufficient statistic, 最小充分统计量Minimum distance estimation, 最小距离估计Minimum effective dose, 最小有效量Minimum lethal dose, 最小致死量Minimum variance estimator, 最小方差估计量MINITAB, 统计软件包Minor heading, 宾词标目Missing data, 缺失值Model specification, 模型的确定Modeling Statistics , 模型统计Models for outliers, 离群值模型Modifying the model, 模型的修正Modulus of continuity, 连续性模Morbidity, 发病率Most favorable configuration, 最有利构形Multidimensional Scaling (ASCAL), 多维尺度/多维标度Multinomial Logistic Regression , 多项逻辑斯蒂回归Multiple comparison, 多重比较Multiple correlation , 复相关Multiple covariance, 多元协方差Multiple linear regression, 多元线性回归Multiple response , 多重选项Multiple solutions, 多解Multiplication theorem, 乘法定理Multiresponse, 多元响应Multi-stage sampling, 多阶段抽样Multivariate T distribution, 多元T分布Mutual exclusive, 互不相容Mutual independence, 互相独立Natural boundary, 自然边界Natural dead, 自然死亡Natural zero, 自然零Negative correlation, 负相关Negative linear correlation, 负线性相关Negatively skewed, 负偏Newman-Keuls method, q检验NK method, q检验No statistical significance, 无统计意义Nominal variable, 名义变量Nonconstancy of variability, 变异的非定常性Nonlinear regression, 非线性相关Nonparametric statistics, 非参数统计Nonparametric test, 非参数检验Nonparametric tests, 非参数检验Normal deviate, 正态离差Normal distribution, 正态分布Normal equation, 正规方程组Normal ranges, 正常范围Normal value, 正常值Nuisance parameter, 多余参数/讨厌参数Null hypothesis, 无效假设Numerical variable, 数值变量Objective function, 目标函数Observation unit, 观察单位Observed value, 观察值One sided test, 单侧检验One-way analysis of variance, 单因素方差分析Oneway ANOVA , 单因素方差分析Open sequential trial, 开放型序贯设计Optrim, 优切尾Optrim efficiency, 优切尾效率Order statistics, 顺序统计量Ordered categories, 有序分类Ordinal logistic regression , 序数逻辑斯蒂回归Ordinal variable, 有序变量Orthogonal basis, 正交基Orthogonal design, 正交试验设计Orthogonality conditions, 正交条件ORTHOPLAN, 正交设计Outlier cutoffs, 离群值截断点Outliers, 极端值OVERALS , 多组变量的非线性正规相关Overshoot, 迭代过度Paired design, 配对设计Paired sample, 配对样本Pairwise slopes, 成对斜率Parabola, 抛物线Parallel tests, 平行试验Parameter, 参数Parametric statistics, 参数统计Parametric test, 参数检验Partial correlation, 偏相关Partial regression, 偏回归Partial sorting, 偏排序Partials residuals, 偏残差Pattern, 模式Pearson curves, 皮尔逊曲线Peeling, 退层Percent bar graph, 百分条形图Percentage, 百分比Percentile, 百分位数Percentile curves, 百分位曲线Periodicity, 周期性Permutation, 排列P-estimator, P估计量Pie graph, 饼图Pitman estimator, 皮特曼估计量Pivot, 枢轴量Planar, 平坦Planar assumption, 平面的假设PLANCARDS, 生成试验的计划卡Point estimation, 点估计Poisson distribution, 泊松分布Polishing, 平滑Polled standard deviation, 合并标准差Polled variance, 合并方差Polygon, 多边图Polynomial, 多项式Polynomial curve, 多项式曲线Population, 总体Population attributable risk, 人群归因危险度Positive correlation, 正相关Positively skewed, 正偏Posterior distribution, 后验分布Power of a test, 检验效能Precision, 精密度Predicted value, 预测值Preliminary analysis, 预备性分析Principal component analysis, 主成分分析Prior distribution, 先验分布Prior probability, 先验概率Probabilistic model, 概率模型probability, 概率Probability density, 概率密度Product moment, 乘积矩/协方差Profile trace, 截面迹图Proportion, 比/构成比Proportion allocation in stratified random sampling, 按比例分层随机抽样Proportionate, 成比例Proportionate sub-class numbers, 成比例次级组含量Prospective study, 前瞻性调查Proximities, 亲近性Pseudo F test, 近似F检验Pseudo model, 近似模型Pseudosigma, 伪标准差Purposive sampling, 有目的抽样QR decomposition, QR分解Quadratic approximation, 二次近似Qualitative classification, 属性分类Qualitative method, 定性方法Quantile-quantile plot, 分位数-分位数图/Q-Q图Quantitative analysis, 定量分析Quartile, 四分位数Quick Cluster, 快速聚类Radix sort, 基数排序Random allocation, 随机化分组Random blocks design, 随机区组设计Random event, 随机事件Randomization, 随机化Range, 极差/全距Rank correlation, 等级相关Rank sum test, 秩和检验Rank test, 秩检验Ranked data, 等级资料Rate, 比率Ratio, 比例Raw data, 原始资料Raw residual, 原始残差Rayleigh's test, 雷氏检验Rayleigh's Z, 雷氏Z值Reciprocal, 倒数Reciprocal transformation, 倒数变换Recording, 记录Redescending estimators, 回降估计量Reducing dimensions, 降维Re-expression, 重新表达Reference set, 标准组Region of acceptance, 接受域Regression coefficient, 回归系数Regression sum of square, 回归平方和Rejection point, 拒绝点Relative dispersion, 相对离散度Relative number, 相对数Reliability, 可靠性Reparametrization, 重新设置参数Replication, 重复Report Summaries, 报告摘要Residual sum of square, 剩余平方和Resistance, 耐抗性Resistant line, 耐抗线Resistant technique, 耐抗技术R-estimator of location, 位置R估计量R-estimator of scale, 尺度R估计量Retrospective study, 回顾性调查Ridge trace, 岭迹Ridit analysis, Ridit分析Rotation, 旋转Rounding, 舍入Row, 行Row effects, 行效应Row factor, 行因素RXC table, RXC表Sample, 样本Sample regression coefficient, 样本回归系数Sample size, 样本量Sample standard deviation, 样本标准差Sampling error, 抽样误差SAS(Statistical analysis system ), SAS统计软件包Scale, 尺度/量表Scatter diagram, 散点图Schematic plot, 示意图/简图Score test, 计分检验Screening, 筛检SEASON, 季节分析Second derivative, 二阶导数Second principal component, 第二主成分SEM (Structural equation modeling), 结构化方程模型Semi-logarithmic graph, 半对数图Semi-logarithmic paper, 半对数格纸Sensitivity curve, 敏感度曲线Sequential analysis, 贯序分析Sequential data set, 顺序数据集Sequential design, 贯序设计Sequential method, 贯序法Sequential test, 贯序检验法Serial tests, 系列试验Short-cut method, 简捷法Sigmoid curve, S形曲线Sign function, 正负号函数Sign test, 符号检验Signed rank, 符号秩Significance test, 显著性检验Significant figure, 有效数字Simple cluster sampling, 简单整群抽样Simple correlation, 简单相关Simple random sampling, 简单随机抽样Simple regression, 简单回归simple table, 简单表Sine estimator, 正弦估计量Single-valued estimate, 单值估计Singular matrix, 奇异矩阵Skewed distribution, 偏斜分布Skewness, 偏度Slash distribution, 斜线分布Slope, 斜率Smirnov test, 斯米尔诺夫检验Source of variation, 变异来源Spearman rank correlation, 斯皮尔曼等级相关Specific factor, 特殊因子Specific factor variance, 特殊因子方差Spectra , 频谱Spherical distribution, 球型正态分布Spread, 展布SPSS(Statistical package for the social science), SPSS统计软件包Spurious correlation, 假性相关Square root transformation, 平方根变换Stabilizing variance, 稳定方差Standard deviation, 标准差Standard error, 标准误Standard error of difference, 差别的标准误Standard error of estimate, 标准估计误差Standard error of rate, 率的标准误Standard normal distribution, 标准正态分布Standardization, 标准化Starting value, 起始值Statistic, 统计量Statistical control, 统计控制Statistical graph, 统计图Statistical inference, 统计推断Statistical table, 统计表Steepest descent, 最速下降法Stem and leaf display, 茎叶图Step factor, 步长因子Stepwise regression, 逐步回归Storage, 存Strata, 层(复数)Stratified sampling, 分层抽样Stratified sampling, 分层抽样Strength, 强度Stringency, 严密性Structural relationship, 结构关系Studentized residual, 学生化残差/t化残差Sub-class numbers, 次级组含量Subdividing, 分割Sufficient statistic, 充分统计量Sum of products, 积和Sum of squares, 离差平方和Sum of squares about regression, 回归平方和Sum of squares between groups, 组间平方和Sum of squares of partial regression, 偏回归平方和Sure event, 必然事件Survey, 调查Survival, 生存分析Survival rate, 生存率Suspended root gram, 悬吊根图Symmetry, 对称Systematic error, 系统误差Systematic sampling, 系统抽样Tags, 标签Tail area, 尾部面积Tail length, 尾长Tail weight, 尾重Tangent line, 切线Target distribution, 目标分布Taylor series, 泰勒级数Tendency of dispersion, 离散趋势Testing of hypotheses, 假设检验Theoretical frequency, 理论频数Time series, 时间序列Tolerance interval, 容忍区间Tolerance lower limit, 容忍下限Tolerance upper limit, 容忍上限Torsion, 扰率Total sum of square, 总平方和Total variation, 总变异Transformation, 转换Treatment, 处理Trend, 趋势Trend of percentage, 百分比趋势Trial, 试验Trial and error method, 试错法Tuning constant, 细调常数Two sided test, 双向检验Two-stage least squares, 二阶最小平方Two-stage sampling, 二阶段抽样Two-tailed test, 双侧检验Two-way analysis of variance, 双因素方差分析Two-way table, 双向表Type I error, 一类错误/α错误Type II error, 二类错误/β错误UMVU, 方差一致最小无偏估计简称Unbiased estimate, 无偏估计Unconstrained nonlinear regression , 无约束非线性回归Unequal subclass number, 不等次级组含量Ungrouped data, 不分组资料Uniform coordinate, 均匀坐标Uniform distribution, 均匀分布Uniformly minimum variance unbiased estimate, 方差一致最小无偏估计Unit, 单元Unordered categories, 无序分类Upper limit, 上限Upward rank, 升秩Vague concept, 模糊概念Validity, 有效性VARCOMP (Variance component estimation), 方差元素估计Variability, 变异性Variable, 变量Variance, 方差Variation, 变异Varimax orthogonal rotation, 方差最大正交旋转Volume of distribution, 容积W test, W检验Weibull distribution, 威布尔分布Weight, 权数Weighted Chi-square test, 加权卡方检验/Cochran检验Weighted linear regression method, 加权直线回归Weighted mean, 加权平均数Weighted mean square, 加权平均方差Weighted sum of square, 加权平方和Weighting coefficient, 权重系数Weighting method, 加权法W-estimation, W估计量W-estimation of location, 位置W估计量Width, 宽度Wilcoxon paired test, 威斯康星配对法/配对符号秩和检验Wild point, 野点/狂点Wild value, 野值/狂值Winsorized mean, 缩尾均值Withdraw, 失访Youden's index, 尤登指数Z test, Z检验Zero correlation, 零相关Z-transformation, Z变换。
k-medoids 聚类公式字母公式

k-medoids 聚类算法是一种常用的基于距离的聚类方法,它主要用于将数据集中的数据点划分为若干个类别,使得同一类别内的数据点之间的相似度较高,不同类别之间的相似度较低。
与k-means 算法不同的是,k-medoids 算法使用代表性的数据点(medoids)来代表每个类别,从而使得对噪声和异常值更加稳健。
在k-medoids 聚类算法中,我们首先需要确定聚类的数量k,然后从数据集中随机选择k个数据点作为初始的medoids。
接下来的步骤是不断地迭代,直至收敛为止。
具体的迭代过程如下:1. 初始化:随机选择k个数据点作为初始的medoids。
2. 分配数据点:对于每个数据点,计算它与各个medoids 的距离,并将其分配到距离最近的medoids 所代表的类别中。
3. 更新medoids:对于每个类别,选择一个新的medoids 来代表该类别,使得该类别内所有数据点到新medoids 的距离之和最小。
4. 判断收敛:检查新的medoids 是否与旧的medoids 相同,若相同则停止迭代,否则继续进行迭代。
在k-medoids 聚类算法中,距离的计算可以使用各种不同的距离度量方式,例如欧氏距离、曼哈顿距离等。
对于大规模的数据集,k-medoids 算法可能会比k-means 算法更具有优势,因为它在每次迭代时只需要计算medoids 之间的距离,而不需要计算所有数据点之间的距离,从而可以减少计算量。
k-medoids 聚类算法是一种有效且稳健的聚类方法,它在处理一些特定情况下可以取得比k-means 更好的聚类效果。
通过对数据进行有效的分组和分类,k-medoids 聚类算法在数据挖掘和模式识别领域具有广泛的应用前景。
K-medoids clustering algorithm is a widely used distance-based clustering method for partitioning the data points in a dataset into several categories, in which the similarity of data points within the same category is relatively high, while the similarity between different categories is relatively low. Unlike the k-means algorithm, the k-medoids algorithm uses representative data points (medoids) to represent each category, making it more robust to noise and outliers.In the k-medoids clustering algorithm, the first step is to determine the number of clusters, denoted as k, and then randomly select k data points from the dataset as the initial medoids. The following steps involve iterative processes until the algorithm converges.The specific iterative process is as follows:1. Initialization: randomly select k data points as the initial medoids.2. Data point assignment: for each data point, calculate its distance to each medoid and assign it to the category represented by the nearest medoid.3. Update medoids: for each category, select a new medoid to represent the category, so that the sum of the distances from all data points in the category to the new medoid is minimized.4. Convergence check: check whether the new medoids are the same as the old medoids. If they are the same, stop the iteration; otherwise, continue the iteration.In the k-medoids clustering algorithm, various distance metrics can be used for distance calculation, such as Euclidean distance, Manhattan distance, etc. For large-scale datasets, the k-medoids algorithm may have advantages over the k-means algorithm because it only needs to calculate the distance betweenmedoids at each iteration, rather than calculating the distance between all data points, which can reduce theputational workload.In conclusion, the k-medoids clustering algorithm is an effective and robust clustering method that can achieve better clustering results than the k-means algorithm in certain situations. By effectively grouping and classifying data, the k-medoids clustering algorithm has wide application prospects in the fields of data mining and pattern recognition.Moreover, the k-medoids algorithm can be further extended and applied in various domains, such as customer segmentation in marketing, anomaly detection in cybersecurity, and image segmentation inputer vision. In marketing, k-medoids clustering can be used to identify customer segments based on their purchasing behavior, allowingpanies to tailor their marketing strategies to different customer groups. In cybersecurity, k-medoids can help detect anomalies by identifying patterns that deviate from the norm in network traffic or user behavior. Inputer vision, k-medoids can be used for image segmentation to partition an image into different regions based on similarity, which is useful for object recognition and scene understanding.Furthermore, the k-medoids algorithm can also bebined with other machine learning techniques, such as dimensionality reduction, feature selection, and ensemble learning, to improve its performance and scalability. For example, using dimensionality reduction techniques like principalponent analysis (PCA) can help reduce theputational burden of calculating distances in high-dimensional data, while ensemble learning methods like boosting or bagging can enhance the robustness and accuracy of k-medoids clustering.In addition, research and development efforts can focus on optimizing the k-medoids algorithm for specific applications and datasets, such as developing parallel and distributed versions of the algorithm to handle big data, exploring adaptive and dynamic approaches to adjust the number of clusters based on the data characteristics, and integrating domain-specific knowledge or constraints into the clustering process to improve the interpretability and usefulness of the results.Overall, the k-medoids clustering algorithm is a powerful tool for data analysis and pattern recognition, with a wide range of applications and potential for further advancements andinnovations. Its ability to handle noise and outliers, its flexibility in distance metrics, and its scalability to large-scale datasets make it a valuable technique for addressing real-world challenges in various domains. As the field of data science and machine learning continues to evolve, the k-medoids algorithm will likely remain an important method for uncovering meaningful insights fromplex data.。
k-Means-Clustering

合肥工业大学—数学建模组k-Means ClusteringOn this page…Introduction to k-Means Clustering Create Clusters and Determine Separation Determine the Correct Number of Clusters Avoid Local MinimaIntroduction to k-Means Clusteringk-means clustering is a partitioning method. The function kmeans partitions data into k mutuallyexclusive clusters, and returns the index of the cluster to which it has assigned each observation. Unlike hierarchical clustering, k-means clustering operates on actual observations (rather than the larger set of dissimilarity measures), and creates a single level of clusters. The distinctions mean that k-means clustering is often more suitable than hierarchical clustering for large amounts of data.kmeans treats each observation in your data as an object having a location in space. It finds apartition in which objects within each cluster are as close to each other as possible, and as far from objects in other clusters as possible. You can choose from five different distance measures, depending on the kind of data you are clustering.Each cluster in the partition is defined by its member objects and by its centroid, or center. The centroid for each cluster is the point to which the sum of distances from all objects in that clusteris minimized. kmeanscomputes cluster centroids differently for each distance measure, tominimize the sum with respect to the measure that you specify.kmeans uses an iterative algorithm that minimizes the sum of distances from each object to itscluster centroid, over all clusters. This algorithm moves objects between clusters until the sum cannot be decreased further. The result is a set of clusters that are as compact and well-separated as possible. You can control the details of the minimization using several optional inputparameters to kmeans, including ones for the initial values of the cluster centroids, and for themaximum number of iterations.Create Clusters and Determine SeparationThe following example explores possible clustering in four-dimensional data by analyzing the results of partitioning the points into three, four, and five clusters.Note Because each part of this example generates random numbers sequentially, i.e., without setting a new state, you must perform all steps in sequence to duplicate the results shown. If you perform the steps out of sequence, the answers will be essentially the same, but the intermediate results, number of iterations, or ordering of the silhouette plots may differ.王刚合肥工业大学—数学建模组 First, load some data:rng('default'); % For reproducibility load kmeansdata; size(X) ans =560 4 Even though these data are four-dimensional, and cannot be easily visualized, kmeans enables you to investigate whether a group structure exists in them. Call kmeans with k, the desired number of clusters, equal to 3. For this example, specify the city block distance measure, and usethe default starting method of initializing centroids from randomly selected data points.idx3 = kmeans(X,3,'distance','city');To get an idea of how well-separated the resulting clusters are, you can make a silhouette plotusing the cluster indices output from kmeans. The silhouette plot displays a measure of howclose each point in one cluster is to points in the neighboring clusters. This measure ranges from +1, indicating points that are very distant from neighboring clusters, through 0, indicating points that are not distinctly in one cluster or another, to -1, indicating points that are probably assignedto the wrong cluster. silhouette returns these values in its first output. [silh3,h] = silhouette(X,idx3,'city'); set(get(gca,'Children'),'FaceColor',[.8 .8 1]) xlabel('Silhouette Value') ylabel('Cluster')王刚合肥工业大学—数学建模组From the silhouette plot, you can see that most points in the second cluster have a large silhouette value, greater than 0.6, indicating that the cluster is somewhat separated from neighboring clusters. However, the third cluster contains many points with low silhouette values, and the first contains a few points with negative values, indicating that those two clusters are not well separated.Determine the Correct Number of ClustersIncrease the number of clusters to see if kmeans can find a better grouping of the data. This time, use the optional 'display' parameter to print information about each iteration.idx4 = kmeans(X,4, 'dist','city', 'display','iter');iter phasenumsum115602077.4321511778.643131771.14201771.1Best total sum of distances = 1771.1Notice that the total sum of distances decreases at each iteration as kmeans reassigns pointsbetween clusters and recomputes cluster centroids. In this case, the second phase of the algorithm did not make any reassignments, indicating that the first phase reached a minimum after five iterations. In some problems, the first phase might not reach a minimum, but the second phase always will.A silhouette plot for this solution indicates that these four clusters are better separated than the three in the previous solution.[silh4,h] = silhouette(X,idx4,'city'); set(get(gca,'Children'),'FaceColor',[.8 .8 1]) xlabel('Silhouette Value') ylabel('Cluster')王刚合肥工业大学—数学建模组A more quantitative way to compare the two solutions is to look at the average silhouette values for the two cases.cluster3 = mean(silh3) cluster4 = mean(silh4) cluster3 =0.5352 cluster4 =0.6400Finally, try clustering the data using five clusters.idx5 = kmeans(X,5,'dist','city','replicates',5); [silh5,h] = silhouette(X,idx5,'city'); set(get(gca,'Children'),'FaceColor',[.8 .8 1]) xlabel('Silhouette Value') ylabel('Cluster') mean(silh5) ans =0.5266王刚合肥工业大学—数学建模组This silhouette plot indicates that this is probably not the right number of clusters, since two of the clusters contain points with mostly low silhouette values. Without some knowledge of howmany clusters are really in the data, it is a good idea to experiment with a range of values for k.Avoid Local MinimaLike many other types of numerical minimizations, the solution that kmeans reaches often depends on the starting points. It is possible for kmeans to reach a local minimum, wherereassigning any one point to a new cluster would increase the total sum of point-to-centroid distances, but where a better solution does exist. However, you can use theoptional 'replicates' parameter to overcome that problem. For four clusters, specify five replicates, and use the 'display' parameter to print out the finalsum of distances for each of the solutions.[idx4,cent4,sumdist] = kmeans(X,4,'dist','city',... 'display','final','replicates',5);Replicate 1, 4 iterations, total sum of distances = 1771.1. Replicate 2, 7 iterations, total sum of distances = 1771.1. Replicate 3, 8 iterations, total sum of distances = 1771.1. Replicate 4, 5 iterations, total sum of distances = 1771.1. Replicate 5, 6 iterations, total sum of distances = 1771.1. Best total sum of distances = 1771.1王刚合肥工业大学—数学建模组In this example, kmeans found the same minimum in all five replications. However, even forrelatively simple problems, nonglobal minima do exist. Each of these five replicates began from adifferent randomly selected set of initial centroids, so sometimes kmeans finds more than one local minimum. However, the final solution that kmeans returns is the one with the lowest totalsum of distances, over all replicates.sum(sumdist) ans =1.7711e+03王刚。
降次求值的基本手法

降次求值的基本手法English Answer:Dimensional reduction is a fundamental technique in mathematics and physics that involves reducing the number of variables or dimensions in a system while preserving as much information as possible. It is widely used in various fields, including optimization, machine learning, and data analysis. There are several basic techniques for performing dimensional reduction, including:1. Projection: Projection involves finding a lower-dimensional subspace that captures the most important information in the original data. This can be achieved through methods such as principal component analysis (PCA) or singular value decomposition (SVD).2. Linear Transformation: Linear transformations can be used to reduce the dimensionality of a system by applying a linear map that transforms the original variables into anew set of variables with a reduced number of dimensions.3. Sampling: Sampling involves selecting a subset of the original data that is representative of the entire dataset. This can be used to reduce the dimensionality of the data while preserving the essential characteristics.4. Clustering: Clustering algorithms can be used to group similar data points together, which can then be represented by a single representative point. This can lead to a reduction in dimensionality while preserving the underlying structure of the data.5. Manifold Learning: Manifold learning techniques assume that the data lies on a lower-dimensional manifold embedded in a higher-dimensional space. By identifying this manifold, the dimensionality of the data can be effectively reduced while preserving its intrinsic properties.The choice of the appropriate dimensional reduction technique depends on the specific problem and the desired outcome. By carefully selecting and applying thesetechniques, it is possible to reduce the complexity of complex systems, improve computational efficiency, and gain new insights into the underlying data.中文回答:降维求值是数学和物理学中的一种基本技术,它涉及在保持尽可能多信息的情况下减少系统中的变量或维数。
AI专用词汇

AI专⽤词汇LetterAAccumulatederrorbackpropagation累积误差逆传播ActivationFunction激活函数AdaptiveResonanceTheory/ART⾃适应谐振理论Addictivemodel加性学习Adversari alNetworks对抗⽹络AffineLayer仿射层Affinitymatrix亲和矩阵Agent代理/智能体Algorithm算法Alpha-betapruningα-β剪枝Anomalydetection异常检测Approximation近似AreaUnderROCCurve/AUCRoc曲线下⾯积ArtificialGeneralIntelligence/AGI通⽤⼈⼯智能ArtificialIntelligence/AI⼈⼯智能Associationanalysis关联分析Attentionmechanism注意⼒机制Attributeconditionalindependenceassumption属性条件独⽴性假设Attributespace属性空间Attributevalue属性值Autoencoder⾃编码器Automaticspeechrecognition⾃动语⾳识别Automaticsummarization⾃动摘要Aver agegradient平均梯度Average-Pooling平均池化LetterBBackpropagationThroughTime通过时间的反向传播Backpropagation/BP反向传播Baselearner基学习器Baselearnin galgorithm基学习算法BatchNormalization/BN批量归⼀化Bayesdecisionrule贝叶斯判定准则BayesModelAveraging/BMA贝叶斯模型平均Bayesoptimalclassifier贝叶斯最优分类器Bayesiandecisiontheory贝叶斯决策论Bayesiannetwork贝叶斯⽹络Between-cla ssscattermatrix类间散度矩阵Bias偏置/偏差Bias-variancedecomposition偏差-⽅差分解Bias-VarianceDilemma偏差–⽅差困境Bi-directionalLong-ShortTermMemory/Bi-LSTM双向长短期记忆Binaryclassification⼆分类Binomialtest⼆项检验Bi-partition⼆分法Boltzmannmachine玻尔兹曼机Bootstrapsampling⾃助采样法/可重复采样/有放回采样Bootstrapping⾃助法Break-EventPoint/BEP平衡点LetterCCalibration校准Cascade-Correlation级联相关Categoricalattribute离散属性Class-conditionalprobability类条件概率Classificationandregressiontree/CART分类与回归树Classifier分类器Class-imbalance类别不平衡Closed-form闭式Cluster簇/类/集群Clusteranalysis聚类分析Clustering聚类Clusteringensemble聚类集成Co-adapting共适应Codin gmatrix编码矩阵COLT国际学习理论会议Committee-basedlearning基于委员会的学习Competiti velearning竞争型学习Componentlearner组件学习器Comprehensibility可解释性Comput ationCost计算成本ComputationalLinguistics计算语⾔学Computervision计算机视觉C onceptdrift概念漂移ConceptLearningSystem/CLS概念学习系统Conditionalentropy条件熵Conditionalmutualinformation条件互信息ConditionalProbabilityTable/CPT条件概率表Conditionalrandomfield/CRF条件随机场Conditionalrisk条件风险Confidence置信度Confusionmatrix混淆矩阵Connectionweight连接权Connectionism连结主义Consistency⼀致性/相合性Contingencytable列联表Continuousattribute连续属性Convergence收敛Conversationalagent会话智能体Convexquadraticprogramming凸⼆次规划Convexity凸性Convolutionalneuralnetwork/CNN卷积神经⽹络Co-oc currence同现Correlationcoefficient相关系数Cosinesimilarity余弦相似度Costcurve成本曲线CostFunction成本函数Costmatrix成本矩阵Cost-sensitive成本敏感Crosse ntropy交叉熵Crossvalidation交叉验证Crowdsourcing众包Curseofdimensionality维数灾难Cutpoint截断点Cuttingplanealgorithm割平⾯法LetterDDatamining数据挖掘Dataset数据集DecisionBoundary决策边界Decisionstump决策树桩Decisiontree决策树/判定树Deduction演绎DeepBeliefNetwork深度信念⽹络DeepConvolutionalGe nerativeAdversarialNetwork/DCGAN深度卷积⽣成对抗⽹络Deeplearning深度学习Deep neuralnetwork/DNN深度神经⽹络DeepQ-Learning深度Q学习DeepQ-Network深度Q⽹络Densityestimation密度估计Density-basedclustering密度聚类Differentiab leneuralcomputer可微分神经计算机Dimensionalityreductionalgorithm降维算法D irectededge有向边Disagreementmeasure不合度量Discriminativemodel判别模型Di scriminator判别器Distancemeasure距离度量Distancemetriclearning距离度量学习D istribution分布Divergence散度Diversitymeasure多样性度量/差异性度量Domainadaption领域⾃适应Downsampling下采样D-separation(Directedseparation)有向分离Dual problem对偶问题Dummynode哑结点DynamicFusion动态融合Dynamicprogramming动态规划LetterEEigenvaluedecomposition特征值分解Embedding嵌⼊Emotionalanalysis情绪分析Empiricalconditionalentropy经验条件熵Empiricalentropy经验熵Empiricalerror经验误差Empiricalrisk经验风险End-to-End端到端Energy-basedmodel基于能量的模型Ensemblelearning集成学习Ensemblepruning集成修剪ErrorCorrectingOu tputCodes/ECOC纠错输出码Errorrate错误率Error-ambiguitydecomposition误差-分歧分解Euclideandistance欧⽒距离Evolutionarycomputation演化计算Expectation-Maximization期望最⼤化Expectedloss期望损失ExplodingGradientProblem梯度爆炸问题Exponentiallossfunction指数损失函数ExtremeLearningMachine/ELM超限学习机LetterFFactorization因⼦分解Falsenegative假负类Falsepositive假正类False PositiveRate/FPR假正例率Featureengineering特征⼯程Featureselection特征选择Featurevector特征向量FeaturedLearning特征学习FeedforwardNeuralNetworks/FNN前馈神经⽹络Fine-tuning微调Flippingoutput翻转法Fluctuation震荡Forwards tagewisealgorithm前向分步算法Frequentist频率主义学派Full-rankmatrix满秩矩阵Func tionalneuron功能神经元LetterGGainratio增益率Gametheory博弈论Gaussianker nelfunction⾼斯核函数GaussianMixtureModel⾼斯混合模型GeneralProblemSolving通⽤问题求解Generalization泛化Generalizationerror泛化误差Generalizatione rrorbound泛化误差上界GeneralizedLagrangefunction⼴义拉格朗⽇函数Generalized linearmodel⼴义线性模型GeneralizedRayleighquotient⼴义瑞利商GenerativeAd versarialNetworks/GAN⽣成对抗⽹络GenerativeModel⽣成模型Generator⽣成器Genet icAlgorithm/GA遗传算法Gibbssampling吉布斯采样Giniindex基尼指数Globalminimum全局最⼩GlobalOptimization全局优化Gradientboosting梯度提升GradientDescent梯度下降Graphtheory图论Ground-truth真相/真实LetterHHardmargin硬间隔Hardvoting硬投票Harmonicmean调和平均Hessematrix海塞矩阵Hiddendynamicmodel隐动态模型H iddenlayer隐藏层HiddenMarkovModel/HMM隐马尔可夫模型Hierarchicalclustering层次聚类Hilbertspace希尔伯特空间Hingelossfunction合页损失函数Hold-out留出法Homo geneous同质Hybridcomputing混合计算Hyperparameter超参数Hypothesis假设Hypothe sistest假设验证LetterIICML国际机器学习会议Improvediterativescaling/IIS改进的迭代尺度法Incrementallearning增量学习Independentandidenticallydistributed/i.i.d.独⽴同分布IndependentComponentAnalysis/ICA独⽴成分分析Indicatorfunction指⽰函数Individuallearner个体学习器Induction归纳Inductivebias归纳偏好I nductivelearning归纳学习InductiveLogicProgramming/ILP归纳逻辑程序设计Infor mationentropy信息熵Informationgain信息增益Inputlayer输⼊层Insensitiveloss不敏感损失Inter-clustersimilarity簇间相似度InternationalConferencefor MachineLearning/ICML国际机器学习⼤会Intra-clustersimilarity簇内相似度Intrinsicvalue固有值IsometricMapping/Isomap等度量映射Isotonicregression等分回归It erativeDichotomiser迭代⼆分器LetterKKernelmethod核⽅法Kerneltrick核技巧K ernelizedLinearDiscriminantAnalysis/KLDA核线性判别分析K-foldcrossvalidationk折交叉验证/k倍交叉验证K-MeansClusteringK–均值聚类K-NearestNeighb oursAlgorithm/KNNK近邻算法Knowledgebase知识库KnowledgeRepresentation知识表征LetterLLabelspace标记空间Lagrangeduality拉格朗⽇对偶性Lagrangemultiplier拉格朗⽇乘⼦Laplacesmoothing拉普拉斯平滑Laplaciancorrection拉普拉斯修正Latent DirichletAllocation隐狄利克雷分布Latentsemanticanalysis潜在语义分析Latentvariable隐变量Lazylearning懒惰学习Learner学习器Learningbyanalogy类⽐学习Learn ingrate学习率LearningVectorQuantization/LVQ学习向量量化Leastsquaresre gressiontree最⼩⼆乘回归树Leave-One-Out/LOO留⼀法linearchainconditional randomfield线性链条件随机场LinearDiscriminantAnalysis/LDA线性判别分析Linearmodel线性模型LinearRegression线性回归Linkfunction联系函数LocalMarkovproperty局部马尔可夫性Localminimum局部最⼩Loglikelihood对数似然Logodds/logit对数⼏率Lo gisticRegressionLogistic回归Log-likelihood对数似然Log-linearregression对数线性回归Long-ShortTermMemory/LSTM长短期记忆Lossfunction损失函数LetterM Machinetranslation/MT机器翻译Macron-P宏查准率Macron-R宏查全率Majorityvoting绝对多数投票法Manifoldassumption流形假设Manifoldlearning流形学习Margintheory间隔理论Marginaldistribution边际分布Marginalindependence边际独⽴性Marginalization边际化MarkovChainMonteCarlo/MCMC马尔可夫链蒙特卡罗⽅法MarkovRandomField马尔可夫随机场Maximalclique最⼤团MaximumLikelihoodEstimation/MLE极⼤似然估计/极⼤似然法Maximummargin最⼤间隔Maximumweightedspanningtree最⼤带权⽣成树Max-P ooling最⼤池化Meansquarederror均⽅误差Meta-learner元学习器Metriclearning度量学习Micro-P微查准率Micro-R微查全率MinimalDescriptionLength/MDL最⼩描述长度Minim axgame极⼩极⼤博弈Misclassificationcost误分类成本Mixtureofexperts混合专家Momentum动量Moralgraph道德图/端正图Multi-classclassification多分类Multi-docum entsummarization多⽂档摘要Multi-layerfeedforwardneuralnetworks多层前馈神经⽹络MultilayerPerceptron/MLP多层感知器Multimodallearning多模态学习Multipl eDimensionalScaling多维缩放Multiplelinearregression多元线性回归Multi-re sponseLinearRegression/MLR多响应线性回归Mutualinformation互信息LetterN Naivebayes朴素贝叶斯NaiveBayesClassifier朴素贝叶斯分类器Namedentityrecognition命名实体识别Nashequilibrium纳什均衡Naturallanguagegeneration/NLG⾃然语⾔⽣成Naturallanguageprocessing⾃然语⾔处理Negativeclass负类Negativecorrelation负相关法NegativeLogLikelihood负对数似然NeighbourhoodComponentAnalysis/NCA近邻成分分析NeuralMachineTranslation神经机器翻译NeuralTuringMachine神经图灵机Newtonmethod⽜顿法NIPS国际神经信息处理系统会议NoFreeLunchTheorem /NFL没有免费的午餐定理Noise-contrastiveestimation噪⾳对⽐估计Nominalattribute列名属性Non-convexoptimization⾮凸优化Nonlinearmodel⾮线性模型Non-metricdistance⾮度量距离Non-negativematrixfactorization⾮负矩阵分解Non-ordinalattribute⽆序属性Non-SaturatingGame⾮饱和博弈Norm范数Normalization归⼀化Nuclearnorm核范数Numericalattribute数值属性LetterOObjectivefunction⽬标函数Obliquedecisiontree斜决策树Occam’srazor奥卡姆剃⼑Odds⼏率Off-Policy离策略Oneshotlearning⼀次性学习One-DependentEstimator/ODE独依赖估计On-Policy在策略Ordinalattribute有序属性Out-of-bagestimate包外估计Outputlayer输出层Outputsmearing输出调制法Overfitting过拟合/过配Oversampling过采样LetterPPairedt-test成对t检验Pairwise成对型PairwiseMarkovproperty成对马尔可夫性Parameter参数Parameterestimation参数估计Parametertuning调参Parsetree解析树ParticleSwarmOptimization/PSO粒⼦群优化算法Part-of-speechtagging词性标注Perceptron感知机Performanceme asure性能度量PlugandPlayGenerativeNetwork即插即⽤⽣成⽹络Pluralityvoting相对多数投票法Polaritydetection极性检测Polynomialkernelfunction多项式核函数Pooling池化Positiveclass正类Positivedefinitematrix正定矩阵Post-hoctest后续检验Post-pruning后剪枝potentialfunction势函数Precision查准率/准确率Prepruning预剪枝Principalcomponentanalysis/PCA主成分分析Principleofmultipleexplanations多释原则Prior先验ProbabilityGraphicalModel概率图模型ProximalGradientDescent/PGD近端梯度下降Pruning剪枝Pseudo-label伪标记LetterQQuantizedNeu ralNetwork量⼦化神经⽹络Quantumcomputer量⼦计算机QuantumComputing量⼦计算Quasi Newtonmethod拟⽜顿法LetterRRadialBasisFunction/RBF径向基函数RandomFo restAlgorithm随机森林算法Randomwalk随机漫步Recall查全率/召回率ReceiverOperatin gCharacteristic/ROC受试者⼯作特征RectifiedLinearUnit/ReLU线性修正单元Recurr entNeuralNetwork循环神经⽹络Recursiveneuralnetwork递归神经⽹络Referencemodel参考模型Regression回归Regularization正则化Reinforcementlearning/RL强化学习Representationlearning表征学习Representertheorem表⽰定理reproducingke rnelHilbertspace/RKHS再⽣核希尔伯特空间Re-sampling重采样法Rescaling再缩放Residu alMapping残差映射ResidualNetwork残差⽹络RestrictedBoltzmannMachine/RBM受限玻尔兹曼机RestrictedIsometryProperty/RIP限定等距性Re-weighting重赋权法Robu stness稳健性/鲁棒性Rootnode根结点RuleEngine规则引擎Rulelearning规则学习LetterS Saddlepoint鞍点Samplespace样本空间Sampling采样Scorefunction评分函数Self-Driving⾃动驾驶Self-OrganizingMap/SOM⾃组织映射Semi-naiveBayesclassifiers半朴素贝叶斯分类器Semi-SupervisedLearning半监督学习semi-SupervisedSupportVec torMachine半监督⽀持向量机Sentimentanalysis情感分析Separatinghyperplane分离超平⾯SigmoidfunctionSigmoid函数Similaritymeasure相似度度量Simulatedannealing模拟退⽕Simultaneouslocalizationandmapping同步定位与地图构建SingularV alueDecomposition奇异值分解Slackvariables松弛变量Smoothing平滑Softmargin软间隔Softmarginmaximization软间隔最⼤化Softvoting软投票Sparserepresentation稀疏表征Sparsity稀疏性Specialization特化SpectralClustering谱聚类SpeechRecognition语⾳识别Splittingvariable切分变量Squashingfunction挤压函数Stability-plasticitydilemma可塑性-稳定性困境Statisticallearning统计学习Statusfeaturefunction状态特征函Stochasticgradientdescent随机梯度下降Stratifiedsampling分层采样Structuralrisk结构风险Structuralriskminimization/SRM结构风险最⼩化S ubspace⼦空间Supervisedlearning监督学习/有导师学习supportvectorexpansion⽀持向量展式SupportVectorMachine/SVM⽀持向量机Surrogatloss替代损失Surrogatefunction替代函数Symboliclearning符号学习Symbolism符号主义Synset同义词集LetterTT-Di stributionStochasticNeighbourEmbedding/t-SNET–分布随机近邻嵌⼊Tensor张量TensorProcessingUnits/TPU张量处理单元Theleastsquaremethod最⼩⼆乘法Th reshold阈值Thresholdlogicunit阈值逻辑单元Threshold-moving阈值移动TimeStep时间步骤Tokenization标记化Trainingerror训练误差Traininginstance训练⽰例/训练例Tran sductivelearning直推学习Transferlearning迁移学习Treebank树库Tria-by-error试错法Truenegative真负类Truepositive真正类TruePositiveRate/TPR真正例率TuringMachine图灵机Twice-learning⼆次学习LetterUUnderfitting⽋拟合/⽋配Undersampling⽋采样Understandability可理解性Unequalcost⾮均等代价Unit-stepfunction单位阶跃函数Univariatedecisiontree单变量决策树Unsupervisedlearning⽆监督学习/⽆导师学习Unsupervisedlayer-wisetraining⽆监督逐层训练Upsampling上采样LetterVVanishingGradientProblem梯度消失问题Variationalinference变分推断VCTheoryVC维理论Versionspace版本空间Viterbialgorithm维特⽐算法VonNeumannarchitecture冯·诺伊曼架构LetterWWassersteinGAN/WGANWasserstein⽣成对抗⽹络Weaklearner弱学习器Weight权重Weightsharing权共享Weightedvoting加权投票法Within-classscattermatrix类内散度矩阵Wordembedding词嵌⼊Wordsensedisambiguation词义消歧LetterZZero-datalearning零数据学习Zero-shotlearning零次学习。
improve the accuracy

Ensembles based on random projections to improve the accuracy of clustering algorithmsAlberto Bertoni and Giorgio ValentiniDSI,Dipartimento di Scienze dell’Informazione,Universit`a degli Studi di Milano,Via Comelico39,20135Milano,Italia.{bertoni,valentini}@dsi.unimi.itAbstract.We present an algorithmic scheme for unsupervised clusterensembles,based on randomized projections between metric spaces,bywhich a substantial dimensionality reduction is obtained.Multiple clus-terings are performed on random subspaces,approximately preservingthe distances between the projected data,and then they are combinedusing a pairwise similarity matrix;in this way the accuracy of each“base”clustering is maintained,and the diversity between them is improved.The proposed approach is effective for clustering problems characterizedby high dimensional data,as shown by our preliminary experimentalresults.1IntroductionSupervised multi-classifiers systems characterized the early development of en-semble methods[1,2].Recently this approach has been extended to unsupervised clustering problems[3,4].In a previous work we proposed stability measures that make use of random projections to assess cluster reliability[5],extending a previous approach[6] based on an unsupervised version of the random subspace method[7].In this paper we adopt the same approach to develop cluster ensembles based on random projections.Unfortunately,a deterministic projection of the data into relatively low dimensional spaces may introduce relevant distortions,and,as a consequence,the clustering in the projected space may results consistently dif-ferent from the clustering in the original space.For these reasons we propose to perform multiple clusterings on randomly chosen projected subspaces,approxi-mately preserving the distances between the examples,and then combining them to generate thefinal”consensus”clustering.The next section introduces basic concepts about randomized embeddings between metric spaces.Sect.3presents the Randomized embedding clustering (RE-Clust)ensemble algorithm,and Sect.4show the results of the application of the ensemble method to high dimensional synthetic data.The discussion of the results and the outgoing developments of the present work end the paper.2Randomized embeddings 2.1Randomized embeddings with low distortion.Dimensionality reduction may be obtained by mapping points from a high to a low-dimensional space:µ:R d →R d ,with d <d ,approximately preserving some characteristics,i.e.the distances between points In this way,algorithms whose results depend only on the distances ||x i −x j ||could be applied to the compressed data µ(X ),giving the same results,as in the original input space.In this context randomized embeddings with low distortion represent a key concept.A randomized embedding between R d and R d with distortion 1+ ,(0< ≤1/2)and failure probability P is a distribution probability on the linear mapping µ:R d →R d ,such that,for every pair p,q ∈R d ,the following property holds with probability ≥1−P :11+ ≤||µ(p )−µ(q )||||p −q ||≤1+ (1)The main result on randomized embedding is due to Johnson and Linden-strauss [8],who proved the following:Johnson-Lindenstrauss (JL)lemma :Given a set S with |S |=n there exists a 1+ -distortion embedding into R d with d =c log n/ 2,where c is a suitable constant.The embedding exhibited in [8]consists in random projections from R d into R d ,represented by matrices d ×d with random orthonormal vectors.Similar results may be obtained by using simpler embeddings [9],represented throughrandom d ×d matrices P =1/√ r ij ),where r ij are random variables such that:E [r ij ]=0,V ar [r ij ]=1For sake of simplicity,we call random projections even this kind of embeddings.2.2Random projections.Examples of randomized maps,represented trough d ×d matrices P such that the columns of the ”compressed”data set D P =P D have approximately the same distance are:1.Plus-Minus-One (PMO)random projections:represented by matrices P =1/√d (r ij ),where r ij are uniformly chosen in {−1,1},such that P rob (r ij =1)=P rob (r ij =−1)=1/2.In this case the JL lemma holds with c 4.2.Random Subspace (RS)[7]:represented by d ×d matrices P = r ij ),where r ij are uniformly chosen with entries in {0,1},and with exactly one ”1”per row and at most one ”1”per column.Even if RS subspaces can be quickly computed,the do not satisfy the JL lemma .3Randomized embedding cluster ensemblesConsider a data set X ={x 1,x 2,...,x n },where x i ∈R d ,(1≤i ≤n );a subset A ⊆{1,2,...,n }univocally individuates a subset of examples {x j |j ∈A }⊆X .The data set X may be represented as a d ×n matrix D ,where columns correspond to the examples,and rows correspond to the ”components”of the examples x ∈X .A k-clustering C of X is a list C =<A 1,A 2,...,A k >,with A i ⊆{1,2,...,n }and such that A i ={1,...,n }.A clustering algorithm C is a procedure that,having as input a data set X and an integer k ,outputs a k-clustering C of X :C (X,k )=<A 1,A 2,...,A k >.The main ideas behind the proposed cluster ensemble algorithm RE-Clust (acronym for Randomized Embedding Clustering)are based on data compres-sion,and generation and combination of multiple ”base”clusterings.Indeed at first data are randomly projected from the original to lower dimensional sub-spaces,using projections described in Sect 2.2in order to approximately preserve the distances between the examples.Then multiple clusterings are performed on multiple instances of the projected data,and a similarity matrix between pairs of examples is used to combine the multiple clusterings.The high level pseudo-code of the ensemble algorithm scheme is the following:RE-Clust algorithm :Input :–a data set X ={x 1,x 2,...,x n },represented by a d ×n D matrix.–an integer k (number of clusters)–a real >0(distortion level)–an integer c (number of clusterings)–two clustering algorithms C and C com–a procedure that realizes a randomized map µbegin algorithm (1)d =2· 2log n +log c 2(2)For each i,j ∈{1,...,n }do M ij =0(3)Repeat for t =1to c(4)P t =Generate projection matrix (d,d )(5)D t =P t ·D(6)<C (t )1,C (t )2,...,C (t )k >=C (D t ,k )(7)For each i,j ∈{1,...,n }M (t )ij =1k k s =1I (i ∈C (t )s )·I (j ∈C (t )s )end repeat (8)M =Pc t =1M (t )c (9)<A 1,A 2,...,A k >=C com (M,k )end algorithm .Output :–the final clustering C =<A 1,A 2,...,A k >In thefirst step of the algorithm,given a distortion level ,the dimension d for the compressed data is computed according to the JL lemma.At each iteration of the main repeat loop(step3-7),the procedure Generate projection matrix outputs a projection matrix P t according to the randomized embeddingµ,and a projected data set D t=P t·D is generated;the corresponding clustering<C(t)1,C(t)2,...,C(t)k >is computed by calling C,and a M(t)similarity matrix is built.The similarity matrix M(t)associated to a clustering C=<C(t)1,C(t)2,...,C(t)k>is a n×n matrix such that:M(t)ij =1kks=1I(i∈C(t)s)·I(j∈C(t)s)(2)where I is is the characteristic function of the set C s.After step(8),M ij denotes the frequency by which the examples i and j occur in the same cluster across multiple clusterings.Thefinal clustering is performed by applying the clustering algorithm C com to the main similarity matrix M.Choosing different random projections we may generate different RE-Clust ensembles(e.g.PMO and RS cluster ensembles).4Experimental resultsIn this section we present some preliminary experimental results with the RE-Clust ensemble algorithm.The Ward’s hierarchical agglomerative clustering al-gorithm[10]has been applied as”base”clustering algorithm.4.1Experimental environmentSynthetic data generation We experimented with2different sample gen-erators,whose samples are distributed according to different mixtures of high dimensional gaussian distributions.Sample1is a generator for5000-dimensional data sets composed by3clusters. The elements of each cluster are distributed according to a spherical gaussian with standard deviation equal to3.Thefirst cluster is centered in0,that is a 5000-dimensional vector with all zeros.The other two clusters are centered in 0.5e and−0.5e,where e is a vector with all1.Sample2is a a generator for6000-dimensional data sets composed by5clus-ters of data normally distributed.The diagonal of the covariance matrix for all the classes has its element equal to1(first1000elements)and equal to2(last 5000elements).Thefirst1000variables of thefive clusters are respectively cen-tered in0,e,−e,5e,−5e.The remaining5000variables are centered in0for all clusters.For each generator,we considered30different random samples each respec-tively composed by60,100examples(that is,20examples per class).1.05 1.10 1.15 1.20 1.25 1.30 1.35 1.400.00.10.20.3PMO ensembleRS ensemblesingledistortionE r r o r (a)1.10 1.15 1.20 1.25 1.30 1.35 1.40 1.450.000.020.040.060.08PMO ensemble RS ensemble singleE r r o rdistortion(b)parison of mean errors between single hierarchical clustering,PMO and RS ensembles with different 1+ distortions.For ensembles,error bars for the 99%confidence interval are represented,while for single hierarchical clustering the 99%confidence interval is represented by the dotted lines above and below the horizontal dash-dotted line.(a)Sample1data set (b)sample2Experimental setup We compared classical single hierarchical clustering al-gorithm with our ensemble approach considering PMO and RS random projec-tions (Sect.2.2).We used 30different realizations for each synthetic data set,using each time 20clusterings for both PMO and RS ensembles.For each PMO and RS ensemble we experimented with different distortions,corresponding to ∈[0.06,0.5].We implemented the ensemble algorithms and the scripts used for the exper-iments in the R language (code is freely available from the authors).4.2ResultsWith sample1(Fig.1(a))for1.10distortion,that corresponds to projections from the original5000into a3407dimensional subspace,RE-Clust ensembles per-form significantly better than single clustering.Indeed PMO ensembles achieve a0.017±0.010mean error over30different realizations from sample1,and RS ensembles a0.018±0.011mean error against a0.082±0.015mean error for single hierarchical clustering.Also with an estimated1.20distortion(with a corresponding subspace dimension equal to852)we obtain significantly better results with both PMO and RS ensembles.With sample2(Fig.1(b))the difference is significant only for1.10distortion, while for larger distortions the difference is not significant and,on the contrary, with1.4distortion RE-Clust ensembles perform worse than single clustering. This may be due both to the relatively high distortion induced by the randomized embedding and to the loss of information due to the random projection to a too low dimensional space.Anyway,with all the high dimensional synthetic data sets the RE-Clust ensembles achieve equal or better results with respect to a ”single”hierarchical clustering approach,at least when the distortions predicted by the JL lemma are lower than1.30.5ConclusionsExperimental results with synthetic data(Sect.4.2)show that RE-Clust ensem-bles are effective with high dimensional data,even if we need more experiments to confirm these results.About the reasons why RE-Clust outperforms single clustering,we suspect that RE-Clust ensembles can reduce the variance component of the error,by ”averaging”between different multiple clusterings,and we are planning to per-form a bias-variance analysis of the algorithm to investigate this topic,using the approach proposed in[11]for supervised ensembles.To evaluate the performance of RE-Clust with other”base”clustering al-gorithms,we are experimenting with Partitioning Around Medoids(PAM)and fuzzy-c-mean algorithms.AcknowledgementThe present work has been developed in the context of the CIMAINA Center of Excellence,and it was partially funded by the italian COFIN project Linguaggi formali ed automi:metodi,modelli ed applicazioni.References[1]Dietterich,T.:Ensemble methods in machine learning.In Kittler,J.,Roli,F.,eds.:Multiple Classifier Systems.First International Workshop,MCS2000,Cagliari, Italy.Volume1857of Lecture Notes in Computer Science.,Springer-Verlag(2000) 1–15[2]Valentini,G.,Masulli,F.:Ensembles of learning machines.In:Neural Nets WIRN-02.Volume2486of Lecture Notes in Computer Science.Springer-Verlag(2002)3–19[3]Strehl,A.,Ghosh,J.:Cluster Ensembles-A Knowledge Reuse Framework forCombining Multiple Partitions.Journal of Machine Learning Research3(2002) 583–618[4]Hadjitodorov,S.,Kuncheva,L.,Todorova,L.:Moderate Diversity for BetterCluster rmation Fusion(2005)[5]Bertoni,A.,Valentini,G.:Random projections for assessing gene expressioncluster stability.In:IJCNN2005,The IEEE-INNS International Joint Conference on Neural Networks,Montreal(2005)(in press).[6]Smolkin,M.,Gosh,D.:Cluster stability scores for microarray data in cancerstudies.BMC Bioinformatics4(2003)[7]Ho,T.:The random subspace method for constructing decision forests.IEEETransactions on Pattern Analysis and Machine Intelligence20(1998)832–844 [8]Johnson,W.,Lindenstrauss,J.:Extensions of Lipshitz mapping into Hilbertspace.In:Conference in modern analysis and probability.Volume26of Contem-porary Mathematics.,Amer.Math.Soc.(1984)189–206[9]Bingham,E.,Mannila,H.:Random projection in dimensionality reduction:Ap-plications to image and text data.In:Proc.of KDD01,San Francisco,CA,USA, ACM(2001)[10]Ward,J.:Hierarchcal grouping to optimize an objective function.J.Am.Stat.Assoc.58(1963)236–244[11]Valentini,G.:An experimental bias-variance analysis of SVM ensembles basedon resampling techniques.IEEE Transactions on Systems,Man and Cybernetics-Part B:Cybernetics35(2005)。
Data Mining Techniques

Data Mining Techniquesrefer to a set of methodologies and algorithms used to extract useful information from large datasets. In today's data-driven world, where massive amounts of data are generated every day, it is crucial to effectively analyze and extract valuable insights from this data. play a key role in this process by enabling organizations to uncover hidden patterns, trends, and relationships within their data that can be used to make informed business decisions.One of the most commonly used data mining techniques is clustering, which involves grouping similar data points together based on certain characteristics. This technique is helpful in identifying natural groupings within a dataset and can be used for customer segmentation, anomaly detection, and pattern recognition.Another important data mining technique is classification, which involves creating models that can predict the class or category to which new data instances belong. Classification algorithms, such as decision trees, support vector machines, and neural networks, are widely used in applications such as spam filtering, credit scoring, and medical diagnosis.Association rule mining is another popular data mining technique that is used to discover relationships between different items in a dataset. This technique is commonly used in market basket analysis to identify patterns in customer purchasing behavior and to make recommendations for cross-selling and upselling.Regression analysis is another useful data mining technique that is used to predict the value of a continuous target variable based on one or more input variables. This technique is commonly used in financial forecasting, sales prediction, and risk analysis.Text mining is a data mining technique that is used to analyze unstructured text data, such as emails, social media posts, and customer reviews. Text mining techniques, such as sentiment analysis, topic modeling, and named entity recognition, are used to extractuseful information from text data to understand customer sentiments, identify key topics, and extract important entities.Other data mining techniques include anomaly detection, feature selection, and dimensionality reduction, which are used to identify outliers in data, select the most relevant features for analysis, and reduce the complexity of high-dimensional data, respectively.In conclusion, data mining techniques are powerful tools that can help organizations gain valuable insights from their data and make informed business decisions. By using a combination of clustering, classification, association rule mining, regression analysis, text mining, and other techniques, organizations can unlock the full potential of their data and drive business growth.。
预测蛋白转录因子的方法

预测蛋白转录因子的方法英文回答:Predicting protein transcription factors is a crucial task in understanding gene regulation and cellular processes. Various computational methods have been developed to identify potential transcription factors based on their sequence and structural features. These methods utilize machine learning algorithms, feature engineering techniques, and domain-specific knowledge to make predictions.One common approach is to train supervised machine learning models using a dataset of known transcription factors and non-transcription factors. The models are trained on a set of features extracted from protein sequences, such as amino acid composition, sequence motifs, and structural properties. Once trained, these models can predict the likelihood of a new protein being a transcription factor.Another approach involves unsupervised learning techniques, such as clustering and dimensionality reduction. These methods identify patterns and relationships withinthe data to group proteins with similar characteristics. By analyzing the clusters or reduced-dimensional representations, researchers can identify potential transcription factors based on their similarity to known factors.Sequence-based methods rely on the assumption that transcription factors share conserved sequence motifs or patterns. These methods scan protein sequences for known transcription factor binding sites or use sequencealignment techniques to identify homologous regions. By identifying these sequence features, they can predict proteins with a high probability of being transcription factors.Structural-based methods consider the three-dimensional structure of proteins to identify potential transcription factors. These methods analyze the protein's shape, surfaceproperties, and interactions with DNA or other proteins. By understanding the structural features associated with transcription factor activity, these methods can predict proteins with the necessary structural characteristics.In addition to these computational methods, experimental approaches, such as chromatin immunoprecipitation sequencing (ChIP-seq) and DNA affinity purification sequencing (DAP-seq), can also be used to identify transcription factors that bind to specific regions of DNA. These experimental techniques providedirect evidence of protein-DNA interactions and can be used to validate predictions made by computational methods.中文回答:预测蛋白质转录因子是一种了解基因调控和细胞过程的关键方法。
what is principal component analysis
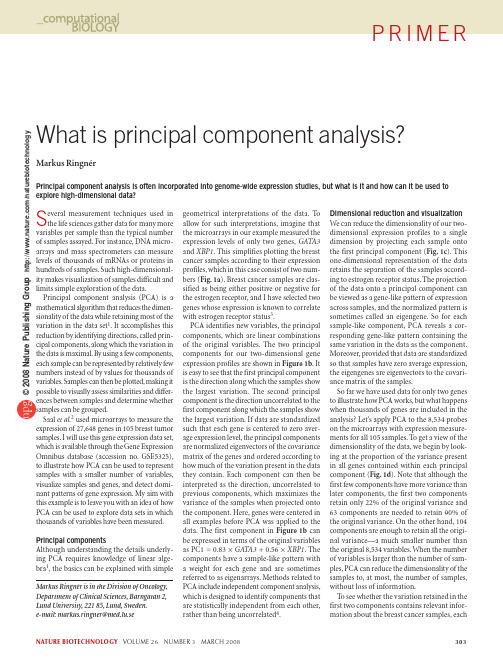
NATURE BIOTECHNOLOGY VOLUME 26 NUMBER 3 MARCH 2008 303What is principal component analysis?Markus RingnérPrincipal component analysis is often incorporated into genome-wide expression studies, but what is it and how can it be used to explore high-dimensional data?Several measurement techniques used in the life sciences gather data for many more variables per sample than the typical number of samples assayed. For instance, DNA micro-arrays and mass spectrometers can measure levels of thousands of mRNAs or proteins in hundreds of samples. Such high-dimensional-ity makes visualization of samples difficult and limits simple exploration of the data.Principal component analysis (PCA) is a mathematical algorithm that reduces the dimen-sionality of the data while retaining most of the variation in the data set 1. It accomplishes this reduction by identifying directions, called prin-cipal components, along which the variation in the data is maximal. By using a few components, each sample can be represented by relatively few numbers instead of by values for thousands of variables. Samples can then be plotted, making it possible to visually assess similarities and differ-ences between samples and determine whether samples can be grouped.Saal et al.2 used microarrays to measure the expression of 27,648 genes in 105 breast tumor samples. I will use this gene expression data set, which is available through the Gene Expression Omnibus database (accession no. GSE5325), to illustrate how PCA can be used to represent samples with a smaller number of variables, visualize samples and genes, and detect domi-nant patterns of gene expression. My aim with this example is to leave you with an idea of how PCA can be used to explore data sets in which thousands of variables have been measured.Principal componentsAlthough understanding the details underly-ing PCA requires knowledge of linear alge-bra 1, the basics can be explained with simplegeometrical interpretations of the data. To allow for such interpretations, imagine that the microarrays in our example measured the expression levels of only two genes, GATA3 and XBP1. This simplifies plotting the breast cancer samples according to their expression profiles, which in this case consist of two num-bers (Fig. 1a ). Breast cancer samples are clas-sified as being either positive or negative for the estrogen receptor, and I have selected two genes whose expression is known to correlate with estrogen receptor status 3.PCA identifies new variables, the principal components, which are linear combinations of the original variables. The two principal components for our two-dimensional gene expression profiles are shown in Figure 1b . It is easy to see that the first principal component is the direction along which the samples show the largest variation. The second principal component is the direction uncorrelated to the first component along which the samples show the largest variation. If data are standardized such that each gene is centered to zero aver-age expression level, the principal components are normalized eigenvectors of the covariance matrix of the genes and ordered according to how much of the variation present in the data they contain. Each component can then be interpreted as the direction, uncorrelated to previous components, which maximizes the variance of the samples when projected onto the component. Here, genes were centered in all examples before PCA was applied to the data. The first component in Figure 1b can be expressed in terms of the original variables as PC1 = 0.83 × GATA3 + 0.56 × XBP1. The components have a sample-like pattern with a weight for each gene and are sometimes referred to as eigenarrays. Methods related to PCA include independent component analysis, which is designed to identify components that are statistically independent from each other, rather than being uncorrelated 4.Dimensional reduction and visualization We can reduce the dimensionality of our two-dimensional expression profiles to a single dimension by projecting each sample onto the first principal component (Fig. 1c ). This one-dimensional representation of the data retains the separation of the samples accord-ing to estrogen receptor status. The projection of the data onto a principal component can be viewed as a gene-like pattern of expression across samples, and the normalized pattern is sometimes called an eigengene. So for each sample-like component, PCA reveals a cor-responding gene-like pattern containing the same variation in the data as the component. Moreover, provided that data are standardized so that samples have zero average expression, the eigengenes are eigenvectors to the covari-ance matrix of the samples.So far we have used data for only two genes to illustrate how PCA works, but what happens when thousands of genes are included in the analysis? Let’s apply PCA to the 8,534 probes on the microarrays with expression measure-ments for all 105 samples. To get a view of the dimensionality of the data, we begin by look-ing at the proportion of the variance present in all genes contained within each principal component (Fig. 1d ). Note that although the first few components have more variance than later components, the first two components retain only 22% of the original variance and 63 components are needed to retain 90% of the original variance. On the other hand, 104 components are enough to retain all the origi-nal variance—a much smaller number than the original 8,534 variables. When the number of variables is larger than the number of sam-ples, PCA can reduce the dimensionality of the samples to, at most, the number of samples, without loss of information.To see whether the variation retained in the first two components contains relevant infor-mation about the breast cancer samples, eachP R I M E RMarkus Ringnér is in the Division of Oncology, Department of Clinical Sciences, Barngatan 2, Lund University, 221 85, Lund, Sweden. e-mail: markus.ringner@med.lu.se©2008 N a t u r e P u b l i s h i n g G r o u p h t t p ://w w w .n a t u r e .c o m /n a t u r e b i o t e c h n o l o g y304 VOLUME 26 NUMBER 3 MARCH 2008 NATURE BIOTECHNOLOGYsample is projected onto these components in Figure 1e . The result is that the dimensional-ity can be reduced from the number of genes down to two dimensions, while still retaining information that separates estrogen recep-tor–positive from estrogen receptor–negative samples. Estrogen receptor status is knownto have a large influence on the gene expres-sion profiles of breast cancer cells 3. However, note that PCA did not generate two separate clusters (Fig. 1e ), indicating that discover-ing unknown groups using PCA is difficult. Moreover, gene expression profiles can also be used to classify breast cancer tumors according to whether they have gained DNA copies of ERBB2 or not 3 and this informa-tion is lost when reducing this data set to the first two principal components (Fig. 1f ). This reminds us that PCA is designed to identify directions with the largest variation and not directions relevant for separating classes of samples. Also, it is important to bear in mind that much of the variation in data from high-throughput technologies may be due to systematic experimental artifacts 5–7, result-ing in dominant principal components that correlate with artifacts.As the principal components have a sam-ple-like pattern with a weight for each gene, we can use the weights to visualize each gene in the PCA plot 8. Most genes will be close to the origin in such a biplot of genes and samples, whereas the genes having the larg-est weights for the displayed components will extend out in their respective direc-tions 9. Biplots provide one way to use the correspondence between the gene-like and sample-like patterns revealed by PCA to identify groups of genes having expression levels characteristic for a group of samples. As an example, two genes with large weights are displayed in Figure 1e .Applications in computational biology An obvious application of PCA is to explore high-dimensional data sets, as outlined above. Most often, three-dimensional visualizations are used for such explorations, and samples are either projected onto the components, as in the examples here, or plotted according to their correlation with the components 10. As much information will typically be lost in two- or three-dimensional visualizations, it is important to systematically try differentcombinations of components when visual-izing a data set. As the principal components are uncorrelated, they may represent different aspects of the samples. This suggests that PCA can serve as a useful first step before clustering or classification of samples. However, decid-ing how many and which components to use in the subsequent analysis is a major chal-lenge that can be addressed in several ways 1. For example, one can use components that correlate with a phenotype of interest 9 or use enough components to include most of the variation in the data 11. PCA results depend critically on preprocessing of the data and on selection of variables. Thus, inspecting PCA plots can potentially provide insights into different choices of preprocessing and variable selection.PCA is often implemented using the sin-gular value decomposition (SVD) of the data matrix 1. The sample-like eigenarray and the gene-like eigengene patterns are both uncovered simultaneously by SVD 10,12. Many applications beyond dimensional reduction, classification and clustering have taken advantage of global representations of expression profiles generated by this decom-position. Applications include identifying patterns that correlate with experimental artifacts and filtering them out 6, estimating missing data, associating genes and expres-sion patterns with activities of regulators and helping to uncover the dynamic archi-tecture of cellular phenotypes 7,10,12. The rapid growth in technologies that generate high-dimensional molecular biology data will likely provide many new applications for PCA in the years to come.ACKNOWLEDGMENTSI wish to thank the Swedish Foundation for Strategic Research for support through the Lund Strategic Centre for Clinical Cancer Research (CREATE Health).1. Jolliffe, I.T. Principal Component Analysis (Springer,New York, 2002).2. Saal, L.H. et al. Proc. Natl. Acad. Sci. USA 104, 7564–7569 (2007).3. Perou, C.M. et al. Nature 406, 747–752 (2000).4. Comon, P . Signal Process. 36, 287–314 (1994).5. Coombes, K.R. et al. Nat. Biotechnol. 23, 291–292(2005).6. Nielsen, T.O. et al. Lancet 359, 1301–1307 (2002).7. Li, C.M. & Klevecz, R.R. Proc. Natl. Acad. Sci. USA 103,16254–16259 (2006).8. Gabriel, K.R. Biometrika 58, 453–467 (1971).9. Landgrebe, J. Wurst, W. & Welzl, G. Genome Biol. 3,RESEARCH0019 (2002).10. Alter, O., Brown, P .O. & Botstein, D. Proc. Natl. Acad.Sci. USA 97, 10101–10106 (2000).11. Khan, J. et al. Nat. Med. 7, 673–679 (2001).12. Holter, N.S. et al. Proc. Natl. Acad. Sci. USA 97, 8409–8414 (2000).Figure 1 Principal component analysis (PCA) of a gene expression data set. (a ) Each dot represents a breast cancer sample plotted against its expression levels for two genes. (In a–c , e, samples are colored according to estrogen receptor (ER) status: ER +, red; ER –, black). (b ) PCA identifies the two directions (PC1 and PC2) along which the data have the largest spread. (c ) Samples plotted in onedimension using their projections onto the first principal component (PC1) for ER +, ER – and all samples separately. (d ) The variance of the principal components when PCA is applied to all 8,534 genes with expression levels for all samples. (e ) PCA biplot with samples plotted in two dimensions using their projections onto the first two principal components, and two genes plotted using their weights for the components (green points). The scale shown is for the samples; for the genes, the scale should be divided by 950. (f ) Samples plotted as in e but colored according to ERBB2 status (blue, ERBB2+; brown, ERBB2–; green, unknown).GATA3X B P 1abc0fdER +ER –All Principal componentProjection onto PC1Projection onto PC12Projection onto PC1X B P 1GATA3P r o p o r t i o n o f v a r i a n c e (%)P r o j e c t i o n o n t o P C 2P R I M E R ©2008 N a t u r e P u b l i s h i n g G r o u p h t t p ://w w w .n a t u r e .c o m /n a t u r e b i o t e c h n o l o g y。
子空间变换法

子空间变换法Subspace Transformation Method子空间变换法The Subspace Transformation Method is a mathematical approach that involves the transformation of data from its original space into a new, lower-dimensional subspace.子空间变换法是一种数学方法,涉及将数据从其原始空间转换到一个新的、低维的子空间。
This technique is often employed in various fields such as machine learning, computer vision, and signal processing, where it can effectively reduce the complexity of data and enhance its analysis.这种方法常应用于机器学习、计算机视觉和信号处理等多个领域,它能够有效降低数据的复杂性并加强其分析。
By projecting the data onto a smaller subspace, the Subspace Transformation Method allows for the extraction of important features while eliminating redundant or noisy information.通过将数据投影到较小的子空间上,子空间变换法能够提取重要的特征,同时消除冗余或噪声信息。
One of the key advantages of this method is its ability to preserve the essential structure and relationships within the data while reducing its dimensionality.这种方法的一个关键优势在于,它能够在降低数据维度的同时,保留数据的本质结构和关系。
Clustering
25
Graph Cut
26
Graph Cut
27
Graph Cut
28
1
0.8 0.6
0.1
5
0.8 0.8
Graph Cut
6
2
0.2 0.8
3
4
0.7
0 0.8 ������ = 0.6 0 0.1 0
0.8 0.6 0 0.1 0 0 0.8 0 0 0 0.8 0 0.2 0 0 0 0.2 0 0.8 0.7 0 0 0.2 0 0 0 0 0.7 0.8 0
Spectral clustering techniques make use of the spectrum (eigenvalues) of the similarity matrix of the data to perform dimensionality reduction before clustering in fewer dimensions. Based on spectral graph theory, spectral clustering is in essence the problem of optimal graph cut.
14
Implementation of k-means Initialize k, u k=2; 0 2 u= 2 −1
������������������������������ ������ : = argmin ������������ − ������������
������ 2
(1) (2)
Unsupervised —clustering (e.g., k-means, mixture models, hierarchical clustering); hidden Markov models,
模式识别英文版

Figure 3.2 (a) Globular (b) filamentary datasets for comparison of clustering methods.
The vertical icicle plot represents the hierarchical clustering tree and must be inspected bottom-up. Figure 3.3 b shows the clustering schedule graph. These usually correspond to a plateau before a high jump in the distance measure.
average of the distances of all possible pairs of patterns, as if they formed a single cluster:
d (ωi ,ω j ) = 1 ∑,ωx)− y C(ni + n j ,2) x, y∈(ωi j
(3-3c)
Multidimensional scaling is another method of representing data in a smaller number of dimensions preserving as much as
possible the similarity structure of the data. The following quadratic error measure known as stress is iteratively minimized:
Figure 3.1 Cross data with Euclidian clustering
CICA包:集群独立成分分析说明书

Package‘CICA’July17,2023Type PackageTitle Clusterwise Independent Component AnalysisVersion1.0.1Date2023-06-27Depends ica,RNifti,R(>=2.10)Imports mclust,plotly,multiway,methods,magrittr,neurobase,oro.nifti,servr,htmltoolsAuthor Jeffrey Durieux[aut,cre],Tom Wilderjans[aut],Juan Claramunt Gonzalez[ctb]Maintainer Jeffrey Durieux<*************************>DescriptionClustering multi-subject resting state functional Magnetic Resonance Imaging data.This meth-ods enables the clustering of subjects based on multi-subject resting state functional Mag-netic Resonance Imaging data.Objects are clustered based on similarities and differ-ences in cluster-specific estimated components obtained by Independent Component Analysis. License GPL(>=3)Encoding UTF-8RoxygenNote7.2.0URL https:///science/article/pii/S0165027022002448, https:///jeffreydurieux/CICANeedsCompilation noRepository CRANDate/Publication2023-07-1708:50:02UTCR topics documented:CICA (2)computeRVmat (4)embed_papaya (5)FindRationalStarts (5)GenRanStarts (7)12CICA GenRatStarts (8)get_papaya_version (9)loadNIfTIs (9)matcher (10)matcher.CICA (11)mpinv (12)papaya (12)papaya_div (13)pass_papaya (13)plot.CICA (14)plot.ModSel (15)SequentialScree (15)Sim_CICA (16)Sr_to_nifti (17)summary.CICA (18)summary.MultipleCICA (19)update_papaya_build (20)Index21 CICA CICA:Clusterwise Independent Component AnalysisDescriptionMain function to perform Clusterwise Independent Component AnalysisUsageCICA(DataList,nComp,nClus,RanStarts,RatStarts=NULL,pseudo=NULL,pseudoFac,userDef=NULL,userGrid=NULL,scalevalue=1000,center=TRUE,maxiter=100,verbose=TRUE,ctol=1e-06,checks=TRUE)CICA3ArgumentsDataList a list of matricesnComp number or vector of ICA components per clusternClus number or vector of clustersRanStarts number of random startsRatStarts Generate rational starts.Eiter’all’or a specific linkage method name(e.g.,’complete’).Use NULL to indicate that Rational starts should not be used.pseudo percentage value for perturbating rational starts to obtain pseudo rational starts pseudoFac factor to multiply the number of rational starts(7in total)to obtain pseudora-tional startsuserDef a user-defined starting seed stored in a data.frame,if NULL no userDef starting partition is useduserGrid user supplied data.frame for multiple model CICA.First column are the re-quested components.Second column are the requested clusters scalevalue desired sum of squares of the block scaling procedurecenter mean center matricesmaxiter maximum number of iterations for each startverbose print loss information to consolectol tolerance value for convergence criterionchecks boolean parameter that indicates whether the input checks should be run(TRUE) or not(FALSE).ValueCICA returns an object of class"CICA".It contains the estimated clustering,cluster specific com-ponent matrices and subject specific time course matricesP partitioning vector of size length(DataList)Sr list of size nClus,containing cluster specific independent componentsAis list of size length(DataList),containing subject specific time coursesLoss loss function value of the best startFinalLossDiff value of the loss difference between the last two iterations of the algorithm.IndLoss a vector with containing the individual loss function valuesLossStarts loss function values of all startsIterations Number of iterationsstarts dataframe with the used starting partitionsAuthor(s)Jeffrey Durieux4computeRVmatExamples##Not run:CICA_data<-Sim_CICA(Nr=15,Q=5,R=4,voxels=100,timepoints=10,E=0.4,overlap=.25,externalscore=TRUE)multiple_output=CICA(DataList=CICA_data$X,nComp=2:6,nClus=1:5,userGrid=NULL,RanStarts=30,RatStarts=NULL,pseudo=c(0.1,0.2),pseudoFac=2,userDef=NULL,scalevalue=1000,center=TRUE,maxiter=100,verbose=TRUE,ctol=.000001)summary(multiple_output$Q_5_R_4)plot(multiple_output$Q_5_R_4)##End(Not run)computeRVmat Compute modified RV matrixDescriptionThis function computes a NxN modified RV matrixUsagecomputeRVmat(DataList=DataList,dist=TRUE,verbose=TRUE)ArgumentsDataList a list with matricesdist boolean if TRUE distance object is returnedverbose boolean if TRUE progressbar is printed to the consoleValueRVsS a square similarity matrix of class matrix or distance object of class dist containing the pairwise modified RV valuesExamples##Not run:CICA_data<-Sim_CICA(Nr=15,Q=5,R=4,voxels=100,timepoints=10,E=0.4,overlap=.25,externalscore=TRUE)#Compute single subject ICAs(nClus equals length(ExampleData))output<-CICA(DataList=CICA_data$X,nStarts=1,nComp=5,nClus=9,verbose=FALSE)embed_papaya5RV<-computeRVmat(DataList=output$Sr,dist=TRUE,verbose=FALSE)#apply hierarchical clustering on RV outputhcl<-hclust(RV)plot(hcl)#low dimensional visualisation using Classical Multidimensional Scalingmds<-cmdscale(RV)plot(mds)##End(Not run)embed_papaya Embed images with PapayaDescriptionWrites temporary images out from nifti objects or passes characterfilenames of images to papaya JS viewerUsageembed_papaya(images,outdir=NULL)Argumentsimages characterfilenames or nifti objects to be viewedoutdir output directory for index and all to goValueOutput htmlFindRationalStarts Plot method for rstarts objectDescriptionPlot method for rstarts object6FindRationalStartsUsageFindRationalStarts(DataList,RatStarts="all",nComp,nClus,scalevalue=NULL,center=TRUE,verbose=TRUE,pseudo=NULL,pseudoFac=NULL)##S3method for class rstartsplot(x,type=1,mdsdim=2,nClus=NULL,...)ArgumentsDataList a list of matricesRatStarts type of rational start.’all’computes all types of hclust methodsnComp number of ICA components to extractnClus Number of clusters for rectangles in dendrogram,default NULL is based on number of clusters present in the objectscalevalue scale each matrix to have an equal sum of squarescenter mean center matricesverbose print output to consolepseudo percentage value for perturbating rational starts to obtain pseudo rational starts pseudoFac how many pseudo starts per rational startx an object of class rstartstype type of plot,1for a dendrogram,2for a multidimensional scaling configuration mdsdim2for two dimensional mds configuration,3for a three dimensional configuration ...optional arguments passed to hclust functionValuedataframe with(pseudo-)rational and dist object based on the pairwise modified RV values ReferencesDurieux,J.,&Wilderjans,T.F.(2019).Partitioning subjects based on high-dimensional fMRI data: comparison of several clustering methods and studying the influence of ICA data reduction in big data.Behaviormetrika,46(2),271-311.Examples##Not run:CICA_data<-Sim_CICA(Nr=15,Q=5,R=4,voxels=100,timepoints=10,E=0.4,overlap=.25,externalscore=TRUE)rats<-FindRationalStarts(DataList=CICA_data$X,nComp=5,nClus=4,verbose=TRUE,pseudo=.2) plot(rats,type=1,method= ward.D2 )plot(rats,type=2,method= ward.D2 )plot(rats,type=2,method= ward.D2 ,mdsdim=3)##End(Not run)##Not run:CICA_data<-Sim_CICA(Nr=15,Q=5,R=4,voxels=100,timepoints=10,E=0.4,overlap=.25,externalscore=TRUE)Out_starts<-FindRationalStarts(DataList=CICA_data$X,nComp=5,nClus=4,scalevalue=1000) plot(Out_starts)plot(Out_starts,type=2)plot(Out_starts,type=2,mdsdim=3,method= ward.D2 )##End(Not run)GenRanStarts Generate random startsDescriptionGenerate random startsUsageGenRanStarts(RanStarts,nClus,nBlocks,ARIlim=0.2,itmax=1000,verbose=FALSE)ArgumentsRanStarts number of randomstarts to generatenClus number of clustersnBlocks number of objectsARIlim maximal value of adjusted Rand Indexitmax maximum number of iterations used tofind suitable random startsverbose boolean that indicates whether the output should be printed on the console Valuea list where thefirst element is a matrix with random starts,second element all pairwise ARIs#’GenRatStarts TitleDescriptionTitleUsageGenRatStarts(DataList,RatStarts,nComp,nClus,scalevalue,center,verbose,pseudo,pseudoFac)ArgumentsDataList DataListRatStarts Type of rational startnComp number of componentsnClus number of clustersscalevalue value for blockscaling procedurecenter centerverbose verbosepseudo percentage used for perturbation rational starts(between0)pseudoFac multiplication factor for pseudo rational startsValueoutget_papaya_version9 get_papaya_version Get Papaya VersionDescriptionReads the papaya.jsfile installed and determines version and buildUsageget_papaya_version()ValueList of build and version,both charactersloadNIfTIs Load Niftifiles from directoryDescriptionLoad Niftifiles from directoryUsageloadNIfTIs(dir,toMatrix=TRUE)Argumentsdir Input directory containing niftifilestoMatrix logical if TRUE nifti’s are converted to matricesValuelist object containing V oxel by Time course matricesExamples##Not run:nifs<-loadNIfTIs( <FolderPath> ,toMatrix=T)outnif<-CICA(DataList=nifs,RanStarts=2,nComp=10,nClus=2)##End(Not run)10matcher matcher Match components between cluster specific spatial mapsDescriptionMatch components between cluster specific spatial mapsUsagematcher(x,reference,RV=FALSE,...)Argumentsx object of class CICAreference integer cluster index that serves as the reference.If nifti path is supplied,clusters will be matched to this templateRV compute modified-RV between cluster components...other argumentsValueoutExamples##Not run:CICA_data<-Sim_CICA(Nr=15,Q=5,R=4,voxels=100,timepoints=10,E=0.4,overlap=.25,externalscore=TRUE)multiple_output=CICA(DataList=CICA_data$X,nComp=2:6,nClus=1:5,userGrid=NULL,RanStarts=30,RatStarts=NULL,pseudo=c(0.1,0.2),pseudoFac=2,userDef=NULL,scalevalue=1000,center=TRUE,maxiter=100,verbose=TRUE,ctol=.000001)matcher(multiple_output$Q_5_R_4,reference=1,RV=TRUE)##End(Not run)matcher.CICA11 matcher.CICA Match components between cluster specific spatial mapsDescriptionMatch components between cluster specific spatial mapsUsage##S3method for class CICAmatcher(x,reference=1,RV=FALSE,...)Argumentsx object of class CICAreference integer cluster index that serves as the reference.If nifti path is supplied,clusters will be matched to this templateRV compute modified-RV between cluster components...other argumentsValueoutExamples##Not run:CICA_data<-Sim_CICA(Nr=15,Q=5,R=4,voxels=100,timepoints=10,E=0.4,overlap=.25,externalscore=TRUE)multiple_output=CICA(DataList=CICA_data$X,nComp=2:6,nClus=1:5,userGrid=NULL,RanStarts=30,RatStarts=NULL,pseudo=c(0.1,0.2),pseudoFac=2,userDef=NULL,scalevalue=1000,center=TRUE,maxiter=100,verbose=TRUE,ctol=.000001)matcher(multiple_output$Q_5_R_4,reference=1,RV=TRUE)##End(Not run)12papaya mpinv Moore Penrose inverseDescriptionMoore Penrose inverseUsagempinv(X)ArgumentsX input matrixValuemp Moore Penrose inverse of matrix Xpapaya View images with PapayaDescriptionWrites temporary images out from nifti objects or passes characterfilenames of images to papaya JS viewerUsagepapaya(images,outdir=NULL,...)Argumentsimages characterfilenames or nifti objects to be viewedoutdir output directory for index and all to go...Options to be passed to pass_papayaValueOutput directory where index.html,js,and copied nii.gzfilespapaya_div13Examples##Not run:library(neurobase)x=nifti(img=array(rnorm(100^3),dim=rep(100,3)),dim=rep(100,3),datatype=16) thresh=datatyper(x>1)index.file=papaya(list(x,thresh))##End(Not run)papaya_div Papaya Div element outputDescriptionGet the necessary div output for embedding a papaya imageUsagepapaya_div()ValueCharacter stringExamplespapaya_div()pass_papaya View images with PapayaDescriptionWrites temporary images out from nifti objects or passes characterfilenames of images to papaya JS viewerUsagepass_papaya(L=NULL,outdir=NULL,daemon=FALSE,close_on_exit=TRUE,sleeper=3,version="0.8",build="982")14plot.CICAArgumentsL list of arguments passed to papaya using paramsoutdir output directory for index and all to godaemon Argument passed to server_configclose_on_exit Should the server close once the functionfinishes?sleeper Time in seconds to sleep if close_on_exit=TRUE.This allows the server to start up.version Version of papaya.js and papaya.css to usebuild Build of papaya.js and papaya.css to useplot.CICA Plot method for CICADescriptionPlot method for CICA.This function shows the cluster specific independent components in an interactive viewer using the papayar packageUsage##S3method for class CICAplot(x,brain="auto",cluster=1,...)Argumentsx Object of class CICAbrain autocluster Components of cluster to plot.Only used when non fMRI related data is used ...other argumentsExamples##Not run:CICA_data<-Sim_CICA(Nr=15,Q=5,R=4,voxels=100,timepoints=10,E=0.4,overlap=.25,externalscore=TRUE)multiple_output=CICA(DataList=CICA_data$X,nComp=2:6,nClus=1:5,userGrid=NULL,RanStarts=30,RatStarts=NULL,pseudo=c(0.1,0.2),pseudoFac=2,userDef=NULL,scalevalue=1000,center=TRUE,maxiter=100,verbose=TRUE,ctol=.000001)plot(multiple_output$Q_5_R_4,cluster=2)##End(Not run)plot.ModSel15 plot.ModSel Plot method for sequential model selectionDescriptionPlot method for the sequential model selection option for CICAUsage##S3method for class ModSelplot(x,...)Argumentsx Object of class ModSel...other argumentsExamples##Not run:CICA_data<-Sim_CICA(Nr=15,Q=5,R=4,voxels=100,timepoints=10,E=0.4,overlap=.25,externalscore=TRUE)multiple_output=CICA(DataList=CICA_data$X,nComp=2:6,nClus=1:5,userGrid=NULL,RanStarts=30,RatStarts=NULL,pseudo=c(0.1,0.2),pseudoFac=2,userDef=NULL,scalevalue=1000,center=TRUE,maxiter=100,verbose=TRUE,ctol=.000001)ModSelOutput<-SequentialScree(multiple_output)plot(ModSelOutput)##End(Not run)SequentialScree Sequential Model Selection for Multiple CICA modelDescriptionSequential Model Selection for Multiple CICA modelUsageSequentialScree(x)16Sim_CICA Argumentsx an object of class MultipleCICAValuea list objectExamples##Not run:CICA_data<-Sim_CICA(Nr=15,Q=5,R=4,voxels=100,timepoints=10,E=0.4,overlap=.25,externalscore=TRUE)multiple_output=CICA(DataList=CICA_data$X,nComp=2:6,nClus=1:5,userGrid=NULL,RanStarts=30,RatStarts=NULL,pseudo=c(0.1,0.2),pseudoFac=2,userDef=NULL,scalevalue=1000,center=TRUE,maxiter=100,verbose=TRUE,ctol=.000001)ModSelOutput<-SequentialScree(multiple_output)plot(ModSelOutput)##End(Not run)Sim_CICA Simulate CICA dataDescriptionSimulate CICA dataUsageSim_CICA(Nr,Q,R,voxels,timepoints,E,overlap=NULL,externalscore=FALSE)Sr_to_nifti17ArgumentsNr number of subjects per clusterQ number of componentsR number of clustersvoxels number of voxelstimepoints number of time pointsE proportion of independent gaussian noiseoverlap amount of overlap between S across clusters.Smaller value means more overlap externalscore add simulated external score(default is FALSE)Valuea list with simulated CICA dataExamples##Not run:#Use set.seed(1)to obtain the dataset used in the article"Clusterwise#Independent Component Analysis(CICA):an R package for clustering subjects#based on ICA patterns underlying three-way(brain)data"Xe<-Sim_CICA(Nr=15,Q=5,R=4,voxels=100,timepoints=10,E=0.4,overlap=.25,externalscore=TRUE)##End(Not run)Sr_to_nifti Convert Cluster specific independent components to NIFTI formatDescriptionConvert Cluster specific independent components to NIFTI formatUsageSr_to_nifti(x,write=FALSE,...)Argumentsx an object of class CICAwrite if TRUE,NIfTIfiles are written to current working directory...other arguments passed to RNifti::writeNifti18summary.CICAValuea list with niftiImagefilesExamples##Not run:nifs<-loadNIfTIs( <FolderPath> ,toMatrix=T)outnif<-CICA(DataList=nifs,RanStarts=2,nComp=10,nClus=2)test<-Sr_to_nifti(outnif,write=T,datatype= int16 ,version=2)##End(Not run)summary.CICA Summary method for class CICADescriptionSummarize a CICA analysisUsage##S3method for class CICAsummary(object,...)Argumentsobject Object of the type produced by CICA...Additional argumentsValuesummary.CICA returns an overview of the estimated clustering of a CICA analysisPM Partitioning matrixtab tabulation of the clusteringLoss Loss function value of the solutionExamples##Not run:CICA_data<-Sim_CICA(Nr=15,Q=5,R=4,voxels=100,timepoints=10,E=0.4,overlap=.25,externalscore=TRUE)multiple_output=CICA(DataList=CICA_data$X,nComp=2:6,nClus=1:5,userGrid=NULL,RanStarts=30,RatStarts=NULL,pseudo=c(0.1,0.2),pseudoFac=2,userDef=NULL,scalevalue=1000,center=TRUE,maxiter=100,verbose=TRUE,ctol=.000001)summary.MultipleCICA19summary(multiple_output$Q_5_R_4)##End(Not run)summary.MultipleCICA Summary method for class MultipleCICADescriptionSummarize a CICA analysisUsage##S3method for class MultipleCICAsummary(object,...)Argumentsobject Object of the type produced by CICA...Additional argumentsValuesummary.MultipleCICA returns an overview of the estimated clustering of a CICA analysisPM Partitioning matrixtab tabulation of the clusteringLoss Loss function value of the solutionExamples##Not run:CICA_data<-Sim_CICA(Nr=15,Q=5,R=4,voxels=100,timepoints=10,E=0.4,overlap=.25,externalscore=TRUE)multiple_output=CICA(DataList=CICA_data$X,nComp=2:6,nClus=1:5,userGrid=NULL,RanStarts=30,RatStarts=NULL,pseudo=c(0.1,0.2),pseudoFac=2,userDef=NULL,scalevalue=1000,center=TRUE,maxiter=100,verbose=TRUE,ctol=.000001)summary(multiple_output$Q_5_R_4)##End(Not run)20update_papaya_build update_papaya_build Update Papaya build version from GitHubDescriptionUpdates the papaya version in the papayar package to the most current on GitHubUsageupdate_papaya_build(type=c("standard","minimal","nodicom","nojquery","standard-with-atlas-local", "standard-with-atlas"),verbose=TRUE)Argumentstype Type of release.Standard is defaultverbose Should download progress be shown?ValueResult of get_papaya_version after downloadingIndexCICA,2,18,19class,3computeRVmat,4dist,4embed_papaya,5FindRationalStarts,5GenRanStarts,7GenRatStarts,8get_papaya_version,9,20loadNIfTIs,9matcher,10matcher.CICA,11matrix,4mpinv,12papaya,12papaya_div,13pass_papaya,12,13plot.CICA,14plot.ModSel,15plot.rstarts(FindRationalStarts),5 SequentialScree,15server_config,14Sim_CICA,16Sr_to_nifti,17summary.CICA,18summary.MultipleCICA,19update_papaya_build,2021。
基于图像单元对比度与统计特性的显著性检测

第39卷第10期自动化学报Vol.39,No.10 2013年10月ACTA AUTOMATICA SINICA October,2013基于图像单元对比度与统计特性的显著性检测唐勇1,2杨林1,2段亮亮1摘要根据视觉注意机制,提出一种基于图像单元对比度与空间统计特性的可靠显著性区域检测方法.通过自适应的图像分割构造图像单元结构,以图像单元为基础,分别利用颜色对比度和空间统计特性两种模型进行显著性区域检测,最后,将两种模型的检测结果通过高斯模型进行结合,得到最终的显著性区域检测的结果.实验表明,该检测方法与现有的方法比较,具有更好的精度和召回率,能明显抑制复杂纹理和噪声,去除复杂背景的影响.关键词显著性区域检测,自适应图像分割,颜色对比度,空间统计特性引用格式唐勇,杨林,段亮亮.基于图像单元对比度与统计特性的显著性检测.自动化学报,2013,39(10):1632−1641DOI10.3724/SP.J.1004.2013.01632Image Cell Based Saliency Detection via Color Contrast and DistributionTANG Yong1,2YANG Lin1,2DUAN Liang-Liang1Abstract According to biological visual attention mechanism,a salient region detection method is proposed in this paper,which is based on image cell contrast and space statistical characteristics.By constructing image cell structure with an adaptive image segmentation,based on image cell,it makes a salient region detection using both the color contrast model and space statistical characteristics model.In the end,two models detection results combine by Gaussian model to get thefinal salient region detection results.Experiments show that this detection method has a higher precision and recall rate,which can not only resist the complex texture and the noise but also remove the influence of the complex background.Key words Salient region detection,adaptive image segmentation,color contrast,space statistical characteristics Citation Tang Yong,Yang Lin,Duan Liang-Liang.Image cell based saliency detection via color contrast and distribu-tion.Acta Automatica Sinica,2013,39(10):1632−1641图像的显著性区域检测是根据人类视觉注意机制,在自然图像中选出最能吸引人类关注的区域.显著性区域检测对于分析图像内容,优先选择重要部分,合理安排资源有重要意义,能够广泛应用于视频检测[1]、图像检索[2]、目标检测[3]、目标识别[4]、图像语义理解[5]等领域.目前,流行的显著性区域检测方法主要分为生物驱动模型与数据驱动模型两种.生物驱动模型由Koch等首先提出[6],但只是提出了生物视觉模型,并没有将其应用到计算机视觉方面,Itti等在生物视觉模型的基础上定义了图像显著性,提出了基于收稿日期2012-11-12录用日期2013-03-27Manuscript received November12,2012;accepted March27, 2013国家自然科学基金(60970073),河北省自然科学基金(F2012203084)资助Supported by National Natural Science Foundation of China (60970073)and Natural Science Foundation of Hebei Province (F2012203084)本文责任编委周杰Recommended by Associate Editor ZHOU Jie1.燕山大学信息科学与工程学院秦皇岛0660042.河北省计算机虚拟技术与系统集成重点实验室秦皇岛0660041.College of Information Science and Engineering,Yan-shan University,Qinhuangdao0660042.Key Laboratory for Computer Virtual Technology and System Integration of Hebei Province,Qinhuangdao066004生物视觉注意机制的显著性区域检测方法(IT方法),利用颜色、边缘、方向等低层信息,通过多尺度高斯差分方式计算显著性区域[7],作为一种最早提出的显著性区域检测算法,无论在检测效果或是运行效率上,IT方法都处于一个较低的水平.Harel 等通过IT方法构建特征图,采用基于图像统计的归一化方法[8],实现显著性区域计算(Graph-based, GB),该方法利用图像的统计特性,使得检测效果相对于IT方法有所提升.Goferman等和Wang等提出通过构造图像单元[9−10],计算图像单元间的差异(Context-aware,CA),能够体现出良好的全局特性,检测效果有明显提升,但这种方式运用多尺度图像金字塔进行融合,计算复杂度高,运行缓慢,而且只适合应用在低分辨率图像上.最近,Cheng等引入区域对比度的显著性区域检测方法[11](Region contrast,RC),这种方式可以在全分辨率图像上检测,提高了检测精度和运行效率,是目前相对可靠的检测方式.但是这些生物模型的方法过分强调面积较小、颜色纯粹的局部特征,可能会导致结果模糊、轮廓不清晰.数据驱动模型更依赖于计算机视觉应用的计算,主要分为两类,一类方法利用图像的局部特征,另一类利用图像的全局特征.第一类方法中,10期唐勇等:基于图像单元对比度与统计特性的显著性检测1633Ma等采用局部像素的对比度与模糊增长的方式来确认图像的显著性区域(Ma and Zhang,MZ)[12],这种方式速度慢,检测精度低.Achanta等考虑图像在多尺度空间图像子区域的像素平均特征向量与近邻的像素平均特征向量的差来衡量显著性值[13],输出全分辨率图像(Average contrast,AC),由于在多尺度空间下检测,使得检测效果有所提升,但是运行效率依然较低.这些只使用局部特征的方式,能有效地突出显著性对象的轮廓,但是检测到的显著性区域精度相对较低,显著性区域与物体实际位置相差较大,而且对纹理和噪声十分敏感.另一类数据驱动模型的方法主要利用图像的全局特征,Hou等总结图像的频域特征[14],对图像背景进行建模,在频域内进行计算频谱残留,最后转换到空间域中的显著性区域(Spectral resid-ual,SR),这种方式首先提出了在频域上计算图像显著性区域,根据频域特征计算,使得计算效率有很大提升,但是只能输出64×64像素的检测结果,不能处理全分辨率图像,而且得到的结果非常不清晰.Zhai等把像素级的显著性值定义为全图像素点间的对比度[15],在Lab颜色空间中只使用了单通道的亮度信息进行计算(L-channel contrast, LC),该方法使检测结果的精度有所提升,但依然较低.Achanta等提出了频率调谐的显著性区域检测方法[16](Frequency-tuned,FT),通过计算每个像素与全图颜色平均值的色差来直接确定显著性值, Cheng等量化全图像素数量[11],通过直方图对比度来计算显著性区域(Histogram contrast,HC).这两种方式都是统计并利用图像的全局颜色特征,得到的检测结果的精度有了很大提升.与单纯采用局部特征相比,这些总结并利用全局特征的方法可以提升运行效率,使检测结果更加清晰,提高了检测精度.以上检测方法,能够大致确定显著性区域的位置,但是检测结果的精度依然有进一步提升的空间,但是依然存在显著性区域轮廓不清晰、对高纹理区域和噪声抵抗能力差等问题.本文提出一种基于图像单元的对比度与空间统计特性的显著性区域检测方法(Cell contrast and statistics,CCS),构造图像单元结构,分别采用计算图像单元的颜色对比度,与图像单元聚类后颜色的空间统计特性,两种方式来确定图像的显著性区域.最后将两种方式的检测结果通过高斯模型结合起来,同时考虑显著性区域中心的空间信息,进一步增强显著性区域,减少非显著性物体的影响.实验证明,相比从前的方式,本方法能够使得检测结果的精度有进一步提升,而且能够有效抵抗噪声与纹理区域的影响.1划分图像结构单元以单个通道的像素为单位计算图像中的显著性区域[13],则操作易受到图像纹理、噪声等因素的影响,若对图像所有通道的全部像素操作,将会导致计算量巨大,无法分析复杂多变的自然图像.为克服以上缺点,本文首先构造以区域为基础的图像单元,将一幅图像划分成单元结构,并以图像的单元结构为基础,计算图像的显著性区域.由于图像分割算法能在保留图像信息的基础上高效地将图像划分成为多个非重叠区域,因此,在构造图像单元的过程中,本文使用一种自适应的Mean-shift图像分割算法.通过给出一组多维数据点,在图像中像素点的维数是(x,y),每个像素点由(r,g,b)三基色构成.Mean-shift算法可以用一个窗口扫描空间来找到数据密度最高的多维数据点,即数据峰值,当窗口移动时,经过窗口变换后收敛到数据峰值的点都会连通起来并属于该峰值,实现分割. Mean-shift算法能够综合考虑多通道颜色[17]、空间距离、区域大小等因素,能将颜色相同或相似、空间分布集中的像素划分到同一区域,同时将对比度大或者空间距离较远的像素区分到不同区域,除此之外,还能够保留轮廓,约束图像中的物体形状,在颜色域和空间上都具有良好特性.由于空间变量(x,y)的变化范围与颜色的变化范围有极大的不同,所以,Mean-shift算法对不同的维数要用不同的窗口半径,分别选择颜色特征向量的带宽h c与空间带宽h r,对图像进行分割,构造图像单元结构,如图1(b)所示.因此,全图被划分成为图像单元{Cell i}i=1,···,N,其中,N表示图像单元个数,计算每个Cell i内所有像素的平均颜色f i(r,g,b)作为Cell i的颜色特征向量.根据带宽参数与多元正态分布协方差矩阵的关系[18],采用以下方法选择带宽参数.首先,设定初始颜色带宽H c=6,空间带宽H r=7.计算分割结果每个Cell i的5×5协方差矩阵i:i=(cc)i(cr)i(rc)i(rr)i(1)其中,3×3的子矩阵是在颜色空间(r,g,b)的协方差矩阵,2×2的子矩阵是空间坐标(x,y)的协方差矩阵,3×2的子矩阵与2×3的子矩阵是颜色与位置的协方差.计算颜色或空间分布的平均方差:h c=1M13tri(cc)h r=1M12tri(rr)(2)1634自动化学报39卷其中,M 表示分割结果Cell 的数目,空间带宽h r 是根据计算x 与y 方向的方差得到,颜色带宽h c 是根据计算在RGB 颜色空间的方差得到.通过这种方式,能够在图像上获得良好的分割结果,文献[19]已经成功采用这种带宽计算方式.但是如果每次处理一幅图像前都进行以上操作就会导致运行速度严重降低,为了能达到好的分割效果同时提高运行效率,在实际操作前,从数据集中选择100幅图像分别计算其空间带宽h r 和颜色带宽h c ,实验发现,计算所得的每组h r 、h c 相差不大,计算h r 与h c 的平均值,分别为7与6.5.因此在实际操作时,选择这对平均值h r =7、h c =6.5进行操作,这样既可以保证分割效果,又能提升运行效率.(a)原图(a)Originalimage(b)单元结构图像(b)Image cell structure 图1构造图像单元结构Fig.1Constructing image cell structure通过Mean-shift 构造图像单元,综合考虑了图像的颜色特征与空间特征,以Cell 为处理单元进行操作,降低后续显著性区域检测的计算量.2基于图像单元的显著性区域检测经典的显著性区域检测方法可以大体上分为全局特征和局部特征两种检测方式,但大都是单独操作,彼此没有联系.本文通过构造图像单元结构,将图像单元的对比度这种局部特性,与图像单元颜色特征向量空间统计特性这种全局特性有机的结合,提高显著性区域检测结果的精确性.2.1基于图像单元的对比度的显著性区域检测一幅图像中,与周围对比度大的区域容易受到关注,因此,区域对比度这种局部特性可以作为显著性区域检测的重要依据之一.除此之外,空间位置的分布对人类的视觉注意机制也有很大影响,相邻两个区域的不同往往更容易引起注意,两个相邻Cell 的高对比度比距离较远的高对比度更容易获得视觉注意.由于第1节中采用Mean-shift 算法对图像进行分割,构造了图像单元结构,即Cell 结构,因此,以Cell 为基础将对比度和空间距离相结合,首先计算不同Cell 间的对比度,再利用每个Cell 与其他Cell 对比度的加权来确定显著性值,同时为每个加权的对比度分配权重,权重由空间距离决定.计算每个Cell 与图像中其他Cell 的对比度,公式如下:Contrast [i ][j ]=D (f i (r,g,b ),f j (r,g,b )))(3)其中,i =j ,D (·,·)为两个Cell 颜色特征f i (r,g,b )的欧氏距离.计算Cell i 的显著性值,将空间信息引入,与对比度结合,距离越近,影响越大,分配给对比度的权重越高,反之,则分配较小权重.同时将Cell i 包含的像素数目引入,增大较大区域的影响,计算如下:S contrast (Cell i )=Nj =1,j =iContrast [i ][j ]Num (j )exp −Dist [i ][j ]λ(4)其中,Num (j )表示Cell j 所包含的像素数目,Dist [i ][j ]表示Cell i 与Cell j 的空间距离,通过计算两个Cell 的中心坐标的欧氏距离得到,λ为权重参数,用来控制空间权值强度,λ越大,空间权值的控制力越弱,会导致距离较远区域的对比度对当前区域的显著性值做出较大贡献.检测结果如图2(a)所示.在确定权重参数的取值时,为了能够达到更好的实验效果,选择一系列不同的对从数据集中随机选取的100幅图像操作,计算图像的平均精度、召回率与F β值.计算结果如图3所示.10期唐勇等:基于图像单元对比度与统计特性的显著性检测1635(a)图像单元对比度检测模型(a)Image cell constrast model(b)图像单元统计特性检测模型(b)Image cell statistics models图2两种检测模型结果Fig.2The detection results by two models由图3可知,在λ=0.4计算所得的Fβ值最大,精度与召回率在该点也有良好的表现.因此在实际操作中选择λ=0.4.计算结果归一化到[0,1]之间.基于图像单元的对比度的显著性区域检测结果如图2(b)所示,利用第二部分构造的图像单元结构,以Cell为处理单元,根据Cell间的对比度这一局部特征,同时综合计算空间距离和Cell的大小,得到显著性图.这种方法的优势是局部特性好,能够准确定位出显著性区域的位置.文章使用精度,召回率与Fβ作为评价一种显著性区域检测算法优劣的标准[8].精度表示在检测到的显著性区域中,属于真实显著性区域部分所占的比例.召回率表示在真实的显著性区域中检测到的部分所占的比例.精度和召回率的计算公式如下:P recision=x(G xR x)xR xRecall=x(G xR x)xG x(5)其中,G x表示真实的显著性区域,R x表示检测到的显著性区域,x表示对应图像的序号.在计算显著性区域时,精度和召回率往往相互影响,一种检测方法的精度提升时,召回率可能会有所下降,因此,文献[8]中提出一种新的评价参数Fβ作为标准.Fβ的计算公式如下:Fβ=(1+η2)P recision×Recallη2×P recision+Recall(6)其中,根据原文献选择η2=0.3.目前,国际上评价一个显著性区域检测算法的好坏大都采用精度、召回率与Fβ作为标准.2.2基于空间分布统计特性的显著性区域检测观察发现,人类视觉总是优先的聚焦在目标物体上,而目标物体的颜色往往接近或者连续,同时,人类视觉摄取到图像中的非目标区域的颜色的分布往往相对离散,非目标区域总是被忽略.根据这一特性,统计一般图像颜色的全局空间分布特性,前景物体的颜色相对接近,在图像中的空间分布总是相对集中,而背景颜色总是均匀地分布在整幅图像中,具体来说,图像中属于前景部分的颜色,空间分布的方(a)λ-Precision曲线(a)λ-Precision curve(b)λ-Recall曲线(b)λ-Recall curve(c)λ-Fβ曲线(c)λ-Fβcurve图3权重参数λ的precision,recall,Fβ曲线Fig.3Curves of precision,recall,Fβby differentλ1636自动化学报39卷差较小,背景颜色空间分布的方差大.根据这一发现,本文将Cell颜色的空间分布的统计特征作为显著性区域检测的另一种方式,属于显著性区域的Cell总是集中在图像中的某一区域,空间分布的方差较小,属于非显著性区域的Cell总是离散的分布在全图上,空间分布的方差较大,之前的显著性检测方法都没有有效利用这一特性.2.2.1聚类计算主要颜色要将Cell的空间分布的统计特性引入显著性检测中,就是计算所有Cell颜色特征向量空间分布的方差,但是在构造图像单元结构后,大多图像的Cell数量都很大,如果计算所有的Cell颜色的空间统计特性是不现实的,因此,本文选择对图像中所有Cell的颜色特征进行聚类操作,减少Cell颜色特征的种类,得到全图的主要颜色.由于RGB颜色空间由于是非均匀分布,而Lab是均匀分布空间,更适合计算图像的颜色距离.因此,在聚类前将图像的由RGB颜色空间转到Lab颜色空间,能达到更好地效果.相对于一般图像内部的颜色差异来说,属于显著性区域的Cell与属于非显著性区域的Cell的颜色特征向量总是有很大差异,而属于显著性区域的不同Cell的颜色总是十分相近.因此,对图像中所有Cell的颜色特征进行聚类,总能有效的将显著性区域的Cell划分到同一类型中,同时将非显著性区域的Cell划分到其他的类别中.选择K-mean算法进行聚类操作,该算法的处理速度快,在处理大数据方面有很大优势,最终解的质量取决于初始聚类中心,为此,本文使用Arthur等提出的方式[20]选择初始聚类中心.聚类操作数据样本的构造如下:Sample=f1(l,a,b),···,f1(l,a,b)Num(1)...f i(l,a,b),···,f i(l,a,b)Num(i)...f N(l,a,b),···,f N(l,a,b)Num(N)(7)其中,f i(l,a,b)表示Cell i的颜色特征向量, Num(i)表示Cell i所包含的像素数目,聚类样本数目与全图的像素数相等.选择最多迭代次数和迭代精度作为迭代结束的条件.聚类操作完成后,全图划分为K种颜色,每一个Cell的颜色特征向量都能用K种聚类颜色中相应的一种表示,与此同时,属于显著性区域的Cell 与属于非显著性区域的Cell能够被划归到不同类别中.使用K种聚类颜色更新计算Cell颜色特征向量,这样方便下一步计算Cell颜色特征的空间统计特性.在选择K的取值时,根据聚类后基本不影响图像的视觉效果聚类时间较短,这两条标准,同时为了能尽量降低后续的计算量,应使K的取值尽量小.为了衡量图像聚类对视觉效果的影响,引入失真度J来计算全图像素到各自聚类中心的距离,从而衡量聚类到K种颜色对图像的视觉效果的影响,分别K=3,4,5,···,12选择进行试验,对随机选择的100幅300×400像素图像的全部像素聚类,计算失真度J如下:J=Nn=1Kk=1r nk x n−µk 2(8)其中,r nk是指示函数,表示像素n是否属于类别k, x n−µk 2表示像素n到其聚类中心的欧氏距离.根据实验结果绘制的折线图,如图4所示.(a)K-失真度曲线(a)K-compactnesscurve(b)K-聚类时间曲线(b)K-clustering time curve图4K值影响曲线Fig.4K values effect curves试验表明,在K的取值小于6时,图像失真严重,聚类后严重影响了图像的视觉效果,当K的取10期唐勇等:基于图像单元对比度与统计特性的显著性检测1637值大于6时,聚类时间会随着的增大而迅速增大.在K的取值为6时,既能保证图像的整体视觉效果不受影响又能控制聚类时间较短,因此本文选择K=6进行实验操作.2.2.2计算空间统计特性由于显著性区域的颜色分布相对集中,方差较小,反之,属于非显著性区域的颜色分布的方差较大.根据聚类操作的计算结果,计算Cell颜色特征向量的空间统计特性,即Cell的颜色特征向量所代表的某一种聚类颜色k的空间分布的方差,计算如下:D(k)=E(X k2)−(E(X k)2 2+E(X k2)−(E(X k)22(9)其中,1≤k≤K表示第k种聚类颜色,E(·)表示某种聚类颜色的在图像中横坐标或纵坐标的期望.需要计算图像中所有Cell的聚类颜色的空间分布期望,则第k种聚类颜色空间分布坐标的期望为E(L k)=1nL m,Cluster Cellm=k(10)其中,Cluster Cellm表示Cell m所属的聚类颜色,当Cell m的颜色特征向量属于聚类颜色k时,对Cell m 坐标进行加权,像素坐标归一化到[0,1]之间,n表示聚类颜色k的加权次数.2.2.3根据空间统计特性计算显著性区域通过上述算法,能够得到每种聚类颜色的方差,能够反映聚类颜色的空间分布的离散度,方差越小,该颜色的空间分布越集中,反之越分散.根据区域显著性与颜色空间分布集中度成正比的原则,某种聚类颜色方差越小,显著性值越高.具体到Cell,每个Cell的颜色特征向量的空间分布方差越小,该Cell 的显著性值越高,反之Cell的显著性值越低.首先,根据显著性值与颜色的空间分布方差成反比的原理,计算聚类颜色的对应显著性值,公式如下:S variance(k)=log1D(k)(11)其中,D(k)表示聚类颜色k空间分布的方差.之后,根据Cell i的颜色特征向量,把相应的显著性赋给Cell i,Cell i的显著性值计算公式如下:S Distributuion(i)=S variance(Cluster Celli=k)(12)其中,Cluster Cell i表示Cell i所属的聚类颜色,将式(12)的计算结果赋给相应的Cell i,计算结果归一化到[0,1]之间.根据图像单元空间分布的统计特性方法计算的出图像显著区域,如图2(b)所示,该显著性图是根据全图的颜色空间分布集中度的全局特性计算得到的,显著性区域检测的结果轮廓清晰,拥有良好的全局特性.3显著性区域联合赋值与增强根据第2节使用的两种方式得到的显著性区域检测结果,分别具有各自的优势,综合考虑这两种检测结果的优点进行联合赋值,总结显著性区域与非显著性区域的空间分布规律,引入显著性区域中心的概念,进一步提高结果的精度、召回率以及抗噪能力.3.1显著性区域联合赋值通过图像单元对比度计算得到的显著性区域局部特性好,精度较高,而根据图像单元颜色特征向量空间分布统计特性计算所得的显著性区域检测结果,轮廓清晰,拥有良好的全局特性.选用高斯函数模型将两种检测结果以不同的权重进行结合,可以的达到更好地效果.以图像的Cell为基本操作单元,将每个Cell通过对比度计算得到的显著性值与通过颜色特征向量空间分布统计特性计算所得的显著性进行结合,计算Cell i的显著性值.函数公式如下:Sal(Cell i)=exp(α·S Distributuion(i)+β·S Contrast)(13)其中,α与β是权重控制参数,分别表示两种显著性检测方式对最终结果的影响.通过联合赋值计算所得的显著性图、精度、召回率,都超过了任何单独的一种方式.在选择权重控制参数α与β的具体值时,为了能达到更好地实验效果,选择一系列不同取值的(α,β)进行测试,从实验数据集中随机选取100幅图像作为试验样本,综合评价不同取值的(α,β)对检测结果的影响,分别选择α=1,2,3,4,5,β=1,2,3,4,5,共25组权重控制参数计算精度、召回率与Fβ的平均值,计算结果如图5所示.由图5可以看出精度与召回率相互影响,因此选择Fβ作为首要的评价标准,(α=1,β=2)时, Fβ能得到最高的值,精度与召回率在该点也有良好的表现,故在实际中选择(α=1,β=2)进行操作.3.2基于像素的显著性区域增强观察发现,对一般图像,引起人类视觉注意的物体总是集中在图像某一块区域,距离这块区域越远的区域,越容易被忽略.具体来讲,距离显著性区域1638自动化学报39卷越远的像素,显著性值越小.根据这一原理,本文通过引入与图像显著性区域中心的距离,来对第3.1节计算所得的显著性区域进行增强操作,之所以使用像素而不是Cell进行操作,计算像素间的空间距离的时间复杂度相对较低,距离也更加精确.(a)(α,β)-Precision三维图(a)(α,β)-Precision3D image(b)(α,β)-Recall三维图(b)(α,β)-Recall3D image(c)(α,β)-Fβ三维图(c)(α,β)-Fβ3D image图5权重控制参数(α,β)的Precision,Recall,Fβ曲线Fig.5The Precision,Recall,Fβcurves by differen t(α,β)首先通过以下算式,确定显著性区域中心点的坐标:x=np=1x p S pnp=1S p,y=np=1y p S pnp=1S p(14)其中,x p表示像素p的横坐标,y p表示像素y的纵坐标,x p表示像素p的显著性值,表示全图的像素总数,计算的全图的显著性区域的中心坐标.然后,通过计算每个像素与图像显著性区域中心的距离,对图像显著性区域进行增强操作如下: Saliency(p)=Sal(p)·exp−D iε(15)其中,D i表示像素p的坐标与图像显著性中心的欧式距离,表示距离权重控制参数,ε在进行增强操作时,能够根据与显著性区域中心的距离,对不同位置的显著性值进行调整,ε实际上代表了高斯函数的方差,它能够控制不同位置到显著性中心的距离对该位置显著性值的影响,ε的取值越小,空间距离的影响越大,保证归一化后,接近显著性中心位置,Cell 的显著性值增大,相反,则减小.为了达到更好的检测结果,选择一系列不同的ε对从数据集中随机选取的100幅图像操作,计算检测结果的平均精度、召回率与Fβ值.计算结果图6所示.(a)ε-Precision曲线(a)ε-Precision curve(b)ε-Recall曲线(b)ε-Recall curve(c)ε-Fβ曲线(c)ε-Fβcurve图6距离权重参数ε的Precision,Recall,Fβ曲线Fig.6The Precision,Recall,Fβcurves by differentε。
DDRTree包的说明说明书

Package‘DDRTree’October12,2022Type PackageTitle Learning Principal Graphs with DDRTreeVersion0.1.5Date2017-4-14Author Xiaojie Qiu,Cole Trapnell,Qi Mao,Li WangDepends irlbaImports RcppLinkingTo Rcpp,RcppEigen,BHMaintainer Xiaojie Qiu<***********>Description Provides an implementation of the framework of reversed graph embed-ding(RGE)which projects data into a reduced dimensional space while constructs a princi-pal tree which passes through the middle of the data simultaneously.DDRTree shows superior-ity to alternatives(Wishbone,DPT)for inferring the ordering as well as the intrinsic struc-ture of the single cell genomics data.In general,it could be used to reconstruct the temporal pro-gression as well as bifurcation structure of any datatype.License Artistic License2.0RoxygenNote6.0.1SystemRequirements C++11NeedsCompilation yesRepository CRANDate/Publication2017-04-3020:54:17UTCR topics documented:DDRTree (2)get_major_eigenvalue (5)pca_projection_R (6)sqdist_R (6)Index71DDRTree Perform DDRTree constructionDescriptionPerform DDRTree constructionThis is an R and C code implementation of the DDRTree algorithm from Qi Mao,Li Wang et al.Qi Mao,Li Wang,Steve Goodison,and Yijun Sun.Dimensionality Reduction via Graph Struc-ture Learning.The21st ACM SIGKDD Conference on Knowledge Discovery and Data Mining (KDD’15),2015/citation.cfm?id=2783309to perform dimension reduction and principal graph learning simultaneously.Please cite this pack-age and KDD’15paper if you found DDRTree is useful for your research.UsageDDRTree(X,dimensions=2,initial_method=NULL,maxIter=20,sigma=0.001,lambda=NULL,ncenter=NULL,param.gamma=10,tol=0.001,verbose=F,...)ArgumentsX a matrix with D×N dimension which is needed to perform DDRTree construc-tiondimensions reduced dimensioninitial_method a function to take the data transpose of X as input and then output the reduced dimension,row number should not larger than observation and column numbershould not be larger than variables(like isomap may only return matrix on validsample sets).Sample names of returned reduced dimension should be preserved.maxIter maximum iterationssigma bandwidth parameterlambda regularization parameter for inverse graph embeddingncenter number of nodes allowed in the regularization graphparam.gamma regularization parameter for k-means(the prefix of’param’is used to avoid name collision with gamma)tol relative objective differenceverbose emit extensive debug output...additional arguments passed to DDRTreeValuea list with W,Z,stree,Y,history W is the orthogonal set of d(dimensions)linear basis vector Z isthe reduced dimension space stree is the smooth tree graph embedded in the low dimension space Y represents latent points as the center of ZIntroductionThe unprecedented increase in big-data causes a huge difficulty in data visualization and down-stream analysis.Conventional dimension reduction approaches(for example,PCA,ICA,Isomap, LLE,etc.)are limited in their ability to explictly recover the intrinisic structure from the data as well as the discriminative feature representation,both are important for scientific discovery.The DDRTree algorithm is a new algorithm to perform the following three tasks in one setting:1.Reduce high dimension data into a low dimension space2.Recover an explicit smooth graph structure with local geometry only captured by distancesof data points in the low dimension space.3.Obtain clustering structures of data points in reduced dimensionDimensionality reduction via graph structure learningReverse graph embedding is previously applied to learn the intrinisic graph structure in the original dimension.The optimization of graph inference can be represented as:min f g∈Fmin{z1,...,z M}(V i,V j)∈Eb i,j||f g(z i)−f g(z j)||2where f g is a function to map the instrinsic data space Z={z1,...,z M}back to the input data space(reverse embedding)X={x1,...,x N}.V i is the the vertex of the instrinsic undirected graph G=(V,E).b ij is the edge weight associates with the edge set E.In order to learn the intrinsic structure from a reduced dimension,we need also to consider a term which includes the error during the learning of the instrinsic structure.This strategy is incorporated as the following:min G∈ˆG b minf g∈Fmin{z1,...,z M}Ni=1||x i−f g(z i)||2+λ2(V i,V j)∈Eb i,j||f g(z i)−f g(z j)||2whereλis a non-negative parameter which controls the tradeoff between the data reconstruction error and the reverse graph embedding.Dimensionality reduction via learning a treeThe general framework for reducing dimension by learning an intrinsic structure in a low dimen-sion requires a feasible setˆG b of graph and a mapping function f G.The algorithm uses minimum spanning tree as the feasible tree graph structure,which can be solved by Kruskal’algoritm.A linear projection model f g(z)=Wz is used as the mapping function.Those setting results in thefollowing specific form for the previous framework:minW ,Z ,BN i =1||x i −Wz i ||2+λ2i,jb i,j ||Wz i −Wz j ||2where W =[w 1,...,w d ]∈R D ×d is an orthogonal set of d linear basis vectors.We can group tree graph B ,the orthogonal set of linear basis vectors and projected points in reduced dimension W ,Z as two groups and apply alternative structure optimization to optimize the tree graph.This method is defined as DRtree (Dimension Reduction tree)as discussed by the authors.Discriminative dimensionality reduction via learning a treeIn order to avoid the issues where data points scattered into different branches (which leads to lose of cluster information)and to incorporate the discriminative information,another set of points {y k }K k =1as the centers of {z i }Ni =1can be also introduced.By so doing,the objective functions of K-means and the DRtree can be simulatenously minimized.The author further proposed a soft partition method to account for the limits from K-means and proposed the following objective function:minW ,Z ,B ,Y ,R Ni =1||x i −Wz i ||2+λ2 k,kb k,k ||Wy k −Wy k ||2+γ K k =1N i =1r i,k ||z i −y k ||2+σΩ(R ) s.t.W T W =I ,B ∈B ,K k =1r i,k =1,r i,k ≤0,∀i,∀kwhere R ∈R N ×N ,Ω(R )= N i =1 kk =1r i,k log r i,k is the negative entropy regularization which transforms the hard assignments used in K-means into soft assignments and σ>0is the reg-ulization parameter.Alternative structure optimization is again used to solve the above problem by separately optimize each group W ,Z ,Y ,B ,R until convergence.The actual algorithm of DDRTree1.Input :Data matrix X ,parameters λ,σ,γ2.Initialize Z by PCA3.K =N,Y =Z4.repeat :5.d k,k =||y k −y k ||2,∀k,∀k6.Obtain B via Kruskal’s algorithm7.L =diag (B1)−Bpute R with each element9.τ=diag (1TR )10.Q =11+γI +R (1+γγ(λγL+τ)−R T R )−1RT11.C =XQX T12.Perform eigen-decomposition on C such that C =U ∧U T and diag (∧)is sorted in a descend-ing order13.W =U (:,1:d )14.Z =W T XQ15.Y =ZR (λγL +τ)−116.Until Convergenceget_major_eigenvalue5 Implementation of DDRTree algorithmWe implemented the algorithm mostly in Rcpp for the purpose of efficiency.It also has extensive optimization for sparse input data.This implementation is originally based on the matlab code provided from the author of DDRTree paper.Examplesdata( iris )subset_iris_mat<-as.matrix(t(iris[c(1,2,52,103),1:4]))#subset the data#run DDRTree with ncenters equal to species numberDDRTree_res<-DDRTree(subset_iris_mat,dimensions=2,maxIter=5,sigma=1e-2,lambda=1,ncenter=3,param.gamma=10,tol=1e-2,verbose=FALSE)Z<-DDRTree_res$Z#obatain matrixY<-DDRTree_res$Ystree<-DDRTree_res$streeplot(Z[1,],Z[2,],col=iris[c(1,2,52,103), Species ])#reduced dimensionlegend("center",legend=unique(iris[c(1,2,52,103), Species ]),cex=0.8,col=unique(iris[c(1,2,52,103), Species ]),pch=1)#legendtitle(main="DDRTree reduced dimension",col.main="red",font.main=4)dev.off()plot(Y[1,],Y[2,],col= blue ,pch=17)#center of the Ztitle(main="DDRTree smooth principal curves",col.main="red",font.main=4)#run DDRTree with ncenters equal to species numberDDRTree_res<-DDRTree(subset_iris_mat,dimensions=2,maxIter=5,sigma=1e-3,lambda=1,ncenter=NULL,param.gamma=10,tol=1e-2,verbose=FALSE)Z<-DDRTree_res$Z#obatain matrixY<-DDRTree_res$Ystree<-DDRTree_res$streeplot(Z[1,],Z[2,],col=iris[c(1,2,52,103), Species ])#reduced dimensionlegend("center",legend=unique(iris[c(1,2,52,103), Species ]),cex=0.8,col=unique(iris[c(1,2,52,103), Species ]),pch=1)#legendtitle(main="DDRTree reduced dimension",col.main="red",font.main=4)dev.off()plot(Y[1,],Y[2,],col= blue ,pch=2)#center of the Ztitle(main="DDRTree smooth principal graphs",col.main="red",font.main=4)get_major_eigenvalue Get the top L eigenvaluesDescriptionGet the top L eigenvaluesUsageget_major_eigenvalue(C,L)6sqdist_RArgumentsC data matrix used for eigendecompositionL number for the top eigenvaluespca_projection_R Compute the PCA projectionDescriptionCompute the PCA projectionUsagepca_projection_R(C,L)ArgumentsC data matrix used for PCA projectionL number for the top principal componentssqdist_R calculate the square distance between a,bDescriptioncalculate the square distance between a,bUsagesqdist_R(a,b)Argumentsa a matrix with D×N dimensionb a matrix with D×N dimensionValuea numeric value for the different between a and bIndexDDRTree,2DDRTree-package(DDRTree),2get_major_eigenvalue,5pca_projection_R,6sqdist_R,67。
single cell best practices

Single-cell best practices typically refer to the best methods and techniques for working with single-cell data, which is data generated from individual cells of a sample, such as a tissue biopsy or a blood sample. Single-cell data analysis has become increasingly important in many areas of research, including cancer biology, immunology, and neuroscience.Here are some best practices for working with single-cell data:1.Understand the technology: Single-cell data analysis often involves complex techniques such as single-cell sequencing, which can generate large amounts of data. It's important to understand the limitations and potential biases of the technology, as well as how to effectively analyze and interpret the data.2.Standardize and quality control: Standardized protocols and quality control measures are essential for generating reliable single-cell data. This includes sample preparation, cell isolation, library preparation, and sequencing.3.Normalize the data: Normalizing single-cell data is a crucial step to remove technical biases and normalize gene expression levels across cells. There are several normalization methods available, such as size factor normalization or regularized log transformation.4.Identify cell types and populations: It's important to identify and characterize the different cell types and populations present in the sample. This can be achieved through unsupervised clustering methods or by using reference datasets for comparison.5.Perform dimensionality reduction: Dimensionality reduction techniques such as t-SNE or principal component analysis can help visualize high-dimensional data and identify cell subpopulations or clusters.6.Analyze gene expression: Gene expression analysis can provide insights into the functional states of cells and identify markers for different cell types. Differential expression analysis can identify genes that are significantly expressed in different cell populations.7.Integrate multiple datasets: Integrating single-cell data from multiple sources (e.g., different patients or experimental conditions) can provide more comprehensive insights into cell populations and their dynamics. This requires appropriate preprocessing and normalization steps to ensure consistency across datasets.8.Validate results: It's essential to validate single-cell data analysis results using orthogonal techniques such as flow cytometry or immunohistochemistry. This helps ensure the accuracy and reliability of the findings.9.Follow best practices for data sharing: Sharing single-cell datawith the research community enables reproducibility and facilitates collaborative research efforts. Following best practices for data sharing, such as depositing data in public repositories or usingcommunity-supported data standards, ensures that the data are accessible and reusable by other researchers.。
基于深度学习和改进K-means聚类算法的电网无功电压快速分区研究

DOI: 10.19783/ki.pspc.201124
电力系统保护与控制
Power System Protection and Control
Vol.49 No.14 Jul. 16, 2021
基于深度学习和改进 K-means 聚类算法的电网 无功电压快速分区研究
赵晶晶,贾 然,陈凌汉,朱天天
(上海电力大学电气工程学院,上海 200090)
摘要:随着电网规模的不断扩大,对整个大电网进行统一的电压调控变得越发困难。提出一种基于深度学习和改 进 K-means 聚类算法的电网无功电压快速分区方法。首先建立电耦合强度矩阵反映系统节点间的电气耦合关系的 强弱。然后采用深度学习中的稀疏自编码器,通过训练实现对输入的高维矩阵进行特征提取和降维。最后基于改 进的 K-means 聚类算法用以对降维后的特征序列进行聚类分析,通过检验电气模块度值来确定最终的分区。以电 气模块度、无功储备校验两个评价指标对电网分区质量进行评估。对 IEEE39 节点和 IEEE118 节点系统进行仿真 分析,验证了所提方法在保证连通性以及充足的无功储备的的基础上,具有较高的电气模块度。 关键词:电耦合强度;稀疏自编码器;改进 K-means 聚类算法;电网分区;电气模块度
0 引言
近年来,世界各地相继发生了严重的电网大停 电事故,造成了重大的经济损失,影响了居民的生 活。电压不稳定事件已被确定为最近几次全球大停
基金项目:国家重点研发计划项目资助(2018YFB0905105)
电的促因[1-2]。由此可见,稳定的电压是电网安全运 行的基础,随着电网规模的不断扩大,对大规模电 力系统进行统一的电压调控变得越发困难,分区电 压控制的思想应运而生。由法国提出的分级电压控 制模式在世界范围内得到了广泛的应用,其关键是 将整个电网划分为“区内耦合,区间解耦”的小电 网,由小电网在各自区内单独进行电压调节。由
二度人脉聚类算法设计

二度人脉聚类算法设计张保龙;黄海燕【摘要】针对整个复杂CLASS全属性聚类的聚类算法在聚类算法中有较为复杂的实现要求,试图对社交软件中较为复杂的CLASS⁃USER进行整体聚类计算,难度在于将其复杂属性体系整合成高维度变量进行降维处理。
通过多次连续的数据整理,特别使用了二维模糊矩阵与排序算法实现快速降维,将高达13维的高维度变量进行降维处理,最终形成一维变量,最后使用常见的K⁃means聚类算法对该一维变量进行聚类分析。
%Since the whole complex CLASS full⁃attribute clustering algorithm in clustering algorithms has complicated imple⁃mentation requirement,the overall clustering computation for the more complex CLASS⁃USER in social software is tried to carry out,which is difficult to integrate the complex attribute system into the high dimensional variables for dimension reduction pro⁃cessing. The ranking algorithm oftwo⁃dimensional fuzzy matrix is particularly used to fast reduce the dimension by means of re⁃peatedly continuous data processing. The dimension reduction processing for the high dimensional variables with 13 dimensions is conducted to form the one⁃dimensional variable. And then the cluster analysis for the one⁃dimensional variable is conducted with common K⁃means clustering algorithm.【期刊名称】《现代电子技术》【年(卷),期】2016(039)009【总页数】3页(P126-127,132)【关键词】全属性聚类;社交软件;聚类算法;人脉分析【作者】张保龙;黄海燕【作者单位】郑州科技学院,河南郑州 450064;郑州科技学院,河南郑州450064【正文语种】中文【中图分类】TN911-34现阶段,人脉分析功能已经成为当前社交软件中的必备功能[1]。
表格题英语作文

表格题英语作文Title: Analyzing Tabular Data: A Comprehensive Approach。
In today's data-driven world, the ability to analyzeand interpret tabular data is becoming increasingly crucial. From business decisions to scientific research, tabulardata serves as a cornerstone for deriving insights and making informed judgments. In this essay, we will delveinto the methodologies and strategies for effectively analyzing tabular data.To begin with, it is essential to understand the structure and components of tabular data. Typically,tabular data consists of rows and columns, where each row represents a unique observation or entity, and each column represents a specific attribute or variable. Theseattributes could be quantitative, such as numerical values, or qualitative, such as categories or labels.One of the primary steps in analyzing tabular data isdata cleaning and preprocessing. This involves handling missing values, removing duplicates, and standardizing formats to ensure data quality and consistency. Furthermore, outliers and anomalies may need to be identified and addressed to prevent them from skewing the analysis results.Once the data is preprocessed, the next step is exploratory data analysis (EDA). EDA involves visualizingthe data using various graphical techniques such as histograms, box plots, and scatter plots. These visualizations help in understanding the distribution of variables, identifying patterns, and detectingrelationships between variables.After gaining insights from EDA, statistical analysis techniques can be applied to quantify relationships andtest hypotheses. Descriptive statistics, such as mean, median, and standard deviation, provide summaries of the data distribution. Inferential statistics, such as hypothesis testing and regression analysis, allow for making inferences and predictions based on the observed data.In addition to traditional statistical methods, machine learning algorithms play a significant role in analyzing tabular data. Supervised learning algorithms, such aslinear regression and decision trees, can be used for predictive modeling tasks, where the goal is to predict an outcome variable based on input features. Unsupervised learning algorithms, such as clustering and dimensionality reduction, are useful for pattern recognition and data segmentation.Furthermore, advancements in artificial intelligence have led to the development of deep learning modelstailored for tabular data analysis. Deep learning architectures like deep neural networks (DNNs) can effectively handle complex tabular data with high-dimensional features, enabling more accurate predictions and insights extraction.Moreover, the interpretation of analysis results is crucial for deriving actionable insights and making informed decisions. Visualizations, such as heatmaps andinteractive dashboards, facilitate the communication of findings to stakeholders effectively. Additionally, sensitivity analysis and scenario planning help in assessing the robustness of the analysis results under different conditions.In conclusion, analyzing tabular data is a multifaceted process that involves data cleaning, exploratory analysis, statistical modeling, machine learning, and interpretation of results. By employing a comprehensive approach encompassing various methodologies and techniques, analysts can unlock valuable insights from tabular data, driving innovation and informed decision-making across diverse domains.。
