DISCRIMINATION AND BIOPHYSICAL CHARACTERIZATION OF CERRADO PHYSIOGNOMIES WITH EO-1 HYPERSPE
biochemistry名词解释

生物化学(Biochemistry)是研究生物体内化学物质和生物化学过程的学科。
它涵盖了分子层面的生物学,关注生物体内分子的结构、组成和相互作用,以及这些分子如何参与细胞功能、代谢过程和遗传传递等生物学活动。
以下是一些与生物化学相关的术语的详细解释:1.分子生物学(Molecular Biology):研究生物体内分子的结构、功能和相互作用。
它涉及到DNA、RNA、蛋白质和其他生物分子的合成、调控和相互作用。
2.基因(Gene):是生物体内携带遗传信息的DNA序列。
基因编码了蛋白质的合成所需的信息,并通过转录和翻译过程将这些信息转化为蛋白质。
3.蛋白质(Protein):是生物体内由氨基酸组成的大分子。
蛋白质具有多种功能,包括酶催化、结构支持、信号传导、运输物质和免疫防御等。
4.酶(Enzyme):是一类特殊的蛋白质,能够催化生物体内化学反应的进行。
酶通过降低反应的活化能,加速化学反应速率。
5.代谢(Metabolism):是生物体内化学反应的总体过程。
它包括分解有机物和合成新的分子,以获得能量和构建细胞组分。
代谢过程还涉及调节物质转运和维持生物体内稳态。
6.基因表达(Gene Expression):是指基因信息的转录和翻译过程,将DNA序列转化为蛋白质。
这一过程包括转录(DNA转化为RNA)和翻译(RNA转化为蛋白质)两个主要步骤。
7.核酸(Nucleic Acid):是生物体内的重要分子,包括DNA(脱氧核糖核酸)和RNA(核糖核酸)。
DNA储存遗传信息,而RNA在基因表达过程中起着重要的中间信使和催化作用。
8.代谢途径(Metabolic Pathway):是一系列相互关联的化学反应,共同完成特定代谢功能。
代谢途径通常涉及多个酶的参与,以完成特定的生物合成和分解过程,以及能量的转化和物质的转运。
9.能量代谢(Energy Metabolism):是生物体内转化和利用能量的过程。
能量代谢包括产生能量的反应(如细胞呼吸和光合作用),以及利用能量的反应(如肌肉收缩和细胞运输)。
EMEA-包括生物技术衍生制品在内的药品工艺变更可比性研究-质量事项(翻译)

包括生物技术衍生蛋白作为活性物质(生物技术衍生制品)在内的药品可比性研究质量事项目录1.简介 (2)1.1目的 (2)1.2法规框架 (2)1.3范围 (2)1.4可比性研究 (2)2.给定产品生产工艺变更时的可比性研究 (2)2.1可比性研究实施时的考虑要点 (3)2.1.1引入变更的阶段 (3)2.1.2 质量标准考虑 (3)2.1.3现有分析方法的适用性 (3)2.1.4.. 安全性和有效性标准的考虑 (4)2.2生产工艺变更的可比性研究策略 (4)2.2.1不影响质量标准的变更(过程控制、活性物质和/或成品质量标准) (4)2.2.2影响过程控制但不影响活性物质或者成品标准的变更 (4)2.2.3影响产品质量标准(过程控制和活性物质、成品标准)但预期不影响安全性和有效性的变更 .. 42.2.4影响产品质量标准(过程控制和活性物质、成品标准)且预期会影响安全性和有效性的变更 .. 43. 仿制药的可比性研究 (5)4.结论 (5)1.简介1.1目的众所周知,生物技术药物、重组DNA和杂交瘤技术制备的蛋白药物都经常会发生工艺变更(活性成分和/或制剂)。
这些变更的主要原因为提高产品质量、增大产量和全球生产力,降低运营成本等。
这些变更可能发生在研发阶段或者上市之后。
无论这些变更发生在在什么阶段,都有必要比较变更前后的产品来确定变更的引入没有改变产品的理化性质和生物特性。
这些特性(可能体现在现有的过程控制或者放行标准)极为重要,是证明产品质量、安全性和有效性的基础。
这些特性的改变都可能导致产品安全性和有效性属性的改变。
因此,某个(a given product)药品工艺发生变更后,应该考虑做可比性研究。
该指南不涵盖在研发初期(即在做非临床研究和初期临床研究评价其初步安全性之前)引入的变更。
此外,当生产商进行生物仿制药的上市申请时,则应该考虑做可比性研究的必要性。
无论何种情况,做可比性研究的思路(逐步法)都很类似。
中医药名词英文翻译06(中药学)

06.001草药herb我国局部地区、某些人群或民间习用,加工炮制欠规范的中药。
06.002药材crude medicine ; crude drug初步加工处理的中药原料药。
06.003道地药材genuine regional drug特定产地的特定品种,且质量、疗效优良的药材。
06.004鲜药fresh medicine; fresh crudedrug 鲜、活应用的药物。
06.005天然药物natural medicine; naturalcrude drug 没有经过加工的有药用价值的天然动、植、矿物药。
06.006采制collection and preparation包括采集、收获、加工、干燥等制备商品中药材的传统技术。
06.007采收期collection period采集药用动、植物的适宜时期。
06.008产地加工processing in production place在产地将鲜药初步加工,使之成为药材的过程。
06.009萌发期germination period植物发芽的阶段。
06.010枯萎期withering period植物失去水分或失去生机的阶段。
06.011贮藏storage药物的贮积保存。
06.012干燥drying药材中水分气化蒸发的过程。
06.013晒干drying in sunshine将药材曝晒使干燥。
06.014阴干drying in shade将药材在通风不见阳光的地方自然干燥。
06.015烘干drying by baking将药材加温干燥,包括焙干、烤干等。
06.016虫蛀rotten due to insect bites仓虫蛀蚀药材使其受损或变质的现象。
06.017霉变mildew and rot中药生霉、腐败、腐烂等现象的统称。
06.018泛油extensive diffusion of oil药材及饮片所含油质溢出表面呈油浸润状态,使质变软、色泽变深黯,味变哈喇的现象。
基于两种卫星传感器的不透水面提取指数比较

第42卷第2期航天返回与遥感2021年4月SPACECRAFT RECOVERY & REMOTE SENSING139基于两种卫星传感器的不透水面提取指数比较马羽赫 赵牡丹 周鹏 王建(西北大学城市与环境学院,西安 710127)摘要不透水面是城市化过程和环境品质影响评价的重要指标,快速、精确地从遥感影像上提取不透水面信息对分析城市扩张、地表结构变化和评价生态环境品质具有重要意义。
前人已经提出了多种指数模型来识别不透水面信息,但是,裸土和沙地的干扰始终是困扰不透水面提取精度的一个主要因素。
文章以分布有大量裸土和沙地信息的银川市部分区域为研究区,使用归一化建筑指数(Normalized Difference Built-up Index,NDBI)、比值居民地指数(Ratio Resident-area Index,RRI)、遥感建筑用地指数(Index Based Built-up Index,IBI)、垂直不透水层指数(Perpendicular Impervious Index,PII)和增强型不透水面指数(Enhancement Normalized Difference Impervious Surface Index,ENDISI)这5种目前常用的不透水面提取指数以及2个中等分辨率的传感器(Landsat8 OLI和Sentinel-2 MSI)数据,系统对比了10种不同指数模型和传感器组合下的不透水面提取结果,进而分析5种指数模型的差异性、对不透水面和裸土沙地的区分程度以及对不同传感器数据的响应效果。
研究表明:5种不透水面指数在2种数据源下均可以基本提取出不透水面信息,其中指数RRI的提取结果总体精度最高;指数PII和RRI区分不透水面和裸土沙地的效果最好;指数NDBI、IBI和RRI对Sentinel-2 MSI的响应效果较好,指数ENDISI 和PII对两种数据响应效果较一致。
东北地理所在盐碱地改良为水稻田对区域气候的影响研究中获进展

大连化物所提出食品中兽药及其代谢物非靶向筛查新方法近日,中国科学院大连化学物理研究所高分辨分离分析及代谢组学研究组研究员许国旺团队在食品中风险物质非靶向筛查技术研究方面取得新进展,通过系统研究兽药及其相应代谢物的质谱碎裂特征,构建了复杂食品基质中兽药及其代谢物的非靶向筛查策略,可为食品中风险物的发现提供技术手段。
食品安全关系国计民生,不断出现的未知/新型风险物质给食品安全带来挑战。
针对未知风险物识别的难题,该团队在前期工作中先后建立了两种非靶向筛查策略,可实现对有空白样本(Anal Chem.,2016)和无空白样本(Anal Chem.,2018)的食品中潜在风险物质的筛查。
考虑到风险物质在体内会被代谢并以多种形式存在于食品中,团队于近期构建了包含3710种兽药及其相应代谢物的质谱数据库,研究、归纳了共有或独有的质谱碎裂特征,并基于质谱碎裂特征及智能检索程序,开发出针对复杂食品基质中已知/未知兽药及其代谢物的非靶向筛查方法。
科研人员利用该方法在蛋类样本中进行了示范性应用,证明了其在食品安全风险物筛查中具有应用潜力。
相关研究成果以Nontargeted Screening Meth⁃od for Veterinary Drugs and Their Metabolites Based on Fragmentation Characteristics from Ultra⁃high-Performance Liquid Chromatography–High-Resolution Mass Spectrometry为题,发表在《食品化学》(Food Chemistry)上。
研究工作得到国家重点研发计划、国家自然科学基金等的资助。
(来源:大连化学物理研究所)东北地理所等在大豆基因组解析研究中获进展中国科学院东北地理与农业生态研究所研究员冯献忠课题组等选择1份东北野生大豆、7份我国代表性栽培种,结合平均50×的三代测序和Hi-C测序,组装了高质量大豆参考基因组。
生物活性导向分离海洋真菌Alternaria sp.MNP801次级代谢产物

( 1 . 浙 江 工业 大 学 药 学 院 , 浙江 杭州 3 1 0 0 3 2 ; 2 . 浙江工业大学 生物与环境工程学院, 浙江 杭州 3 1 0 0 3 2 )
摘要: 生物 活性 导 向分 离海 洋微 生物 次级代 谢 产物是 海 洋天然 产物研 究 的热点 , 对 海洋链 格孢属 真
Ab s t r a c t :Bi o a c t i v i t y gu i d e i s ol a t i o n o f s e c o nd a r y me t a bo l i t e s f r om ma r i ne mi c r o or g a ni s m i s a ho t s po t o f ma r i ne n a t u r a l p r od u c t r e s e a r c h. Th e me t h a no l e xt r a c t s o f myc e l i u m f r o m ma r i ne
(工) ,x a n t h o n e (1 I) ,s t e a r i c a c i d (m ) . Th e c o mp o u n d 工 wa s i s o l a t e d f r o m ma r i n e f u n g u s
f u n g u s Al t e r n a r i a s p .M NP8 0 1 h a v e b e e n t e s t e d b y a n t i o x i d a t i v e( DP PH )a n d a n t i t u mo r( M TT)
f r o m ma r i n e f u ng u s Al t e r n ar i a s p. M NP8 0 1
非等位基因

非等位基因概述非等位基因是指同一基因座上的不同等位基因。
等位基因是指在某个给定的基因座上,可以存在多种不同的变体。
每个个体继承了一对等位基因,一对等位基因可能会导致不同的表型表达。
非等位基因的存在使得遗传学研究更加复杂,因为不同的等位基因会对个体的表型产生不同的影响。
背景在生物学中,基因座是指染色体上一个特定的位置,该位置上的基因决定了某个特征的表达方式。
每个基因座上可以有多种不同的等位基因。
等位基因是指在某个特定基因座上的不同基因变体。
每个个体都会继承一对等位基因,通过这对等位基因的不同组合,决定了个体的表型。
然而,并非所有基因座上的等位基因都具有相同的表现型。
非等位基因的影响非等位基因的存在导致不同等位基因会对个体表型产生不同的影响。
有些非等位基因会表现出显性效应,也就是说,当个体继承了一个突变的等位基因时,即使同时继承了一个正常的等位基因,但显性效应会使得突变的等位基因的表型表达得到体现。
相反,有些非等位基因会表现出隐性效应,当个体继承了两个突变的等位基因时,才会表现出突变的表型。
除了显性和隐性效应之外,非等位基因还可能发生两种其他类型的表型效应。
一种是共显效应,当个体继承了两个不同的突变等位基因时,在表型表达上会表现出一种新的特征,这个特征并不是单个突变等位基因所能导致的。
另一种是部分显性效应,当个体继承了两个不同的突变等位基因时,表型表达将介于两个单独突变等位基因的表型之间。
重组和非等位基因重组是指两个不同的染色体交换部分基因序列的过程。
在重组的过程中,非等位基因可能会发生改变,导致新的等位基因组合形成。
这一过程使得非等位基因的表型效应更加复杂,因为新的等位基因可能将不同基因座的效应组合起来。
非等位基因的重要性非等位基因对生物的适应性和多样性起着重要作用。
通过对等位基因的各种组合的研究,人们可以更好地理解基因与表型之间的关系,并揭示遗传变异对物种适应环境的重要性。
总结非等位基因是指同一基因座上的不同等位基因。
博士复试英文PPT

3. PTBP1 enhances exon11a skipping of Mena premRNA in lung cancer cells
Results
1. PTBP1 is highly expressed in lung adenocarcinoma (LUAD) tissues and 95-D cells and upregulation of PTBP1 is associated with EMT progress
2. PTBP1 promotes migration and invasion of lung cancer cells
Master Research
PTBP1 enhances exon11a skipping in Mena premRNA to promote migration and invasion in lung
carcinoma cells
Background Objectives Technology Methods Results Conclusions
5. PTBP1-mediated migration and invasion of 95-D cells are partially dependent on MenaINV
Results
2.1. Overexpressed PTBP1 promotes levels of EMT-related proteins in lung cancer cells
Technology Methods
Results
1. PTBP1 is highly expressed in lung adenocarcinoma (LUAD) tissues and 95-D cells and upregulation of PTBP1 is associated with EMT progress
世界绵羊(Ovisaries)球虫种类与地理分布

·综述·Chinese Journal of Animal Infectious Diseases中国动物传染病学报摘 要:为了解绵羊(Ovis aries )球虫的种类及在世界各地的分布情况,本文收集整理了世界上报道绵羊球虫的相关资料,按照虫种学名的字母为序,介绍了寄生于绵羊的17种艾美耳球虫(Eimeria ),其中3种在中国发现与命名的球虫为卵状艾美耳球虫(Eimeria oodeus Hu and Yan, 1990)、厚膜艾美耳球虫(Eimeria pachmenia Hu and Yan, 1990)、固原艾美耳球虫(Eimeriaguyuanensis Xiao, 1992)。
每种球虫包括中文名称与学名、宿主名称、寄生部位、地理分布,部分种类有同物异名,地理分布按照国家或地区的英文或拼音字母为序。
在收录的56个国家中,记录球虫种类最多的国家为中国(17种),其次为巴西(13种),记录11~12种球虫的国家占32.14%,6~10种球虫的国家占35.71%,1~5种球虫的国家占28.57%。
在17种绵羊球虫中,分布最广的球虫为小型艾美耳球虫(49个国家),其次为类绵羊艾美耳球虫(46个国家),分布40~45个国家的球虫种类占29.41%,20~39个国家的球虫种类占23.53%,1~19个国家的球虫种类占35.29%。
本文基本能反映出绵羊球虫在世界范围的分布状况,为全面了解绵羊球虫的地理分布提供了一份较为完整的基础资料。
关键词:绵羊;球虫;艾美耳球虫;种类;分布中图分类号:S858.26文献标志码: A文章编号:1674-6422(2022)02-0181-17收稿日期:2021-11-17基金项目: 国家寄生虫资源库(2019-194-30)作者简介:黄兵,男,学士,研究员,主要从事球虫生物学与球虫病防治研究;董辉,男,博士,研究员,主要从事球虫免疫学与分子生物学研究通信作者: 黄兵,E-mail:***********.cn世界绵羊(Ovis aries )球虫种类与地理分布黄 兵,董 辉,赵其平,韩红玉,于 珏,朱顺海(中国农业科学院上海兽医研究所 农业农村部动物寄生虫学重点实验室,上海200241)2022,30(2): 181-197Abstract: In order to comprehensively understand the coccidia species of sheep (Ovis aries ) and their distribution in different countries, we reviewed the literatures and collected references of sheep coccidia published so far in the world. A total of 17 sheep Eimeria species were introduced with the alphabetical order of their scientific names, of which E . oodeus Hu and Yan, 1990, E . pachmenia Hu and Yan, 1990 and E . guyuanensis Xiao, 1992 were firstly described and named in China. Each specie was listed and introduced with its Chinese name, scientific name, host name, parasitic site and geographical distribution. Some species also had their synonyms. The geographical distribution was listed with the alphabetical order according to English or Chinese name of a country or region. Among 56 countries listed, the country with the most reported coccidia species was China (17 species), followed by Brazil (13 species). 32.14%, 35.71% and 28.57% countries had 11-12, 6-10 and 1-5 coccidia species, respectively. Among 17 sheep coccidia species discovered, the most widely distributed coccidia species was E. parva (49 countries), followed by E . ovinoidalis (46 countries). 29.41%, 23.53% and 35.29% species were distributed in 40-45, 20-39 and 1-19 countries, respectively. This review collected the basic distribution of sheep coccidia in the world and provided necessary information for a comprehensively understanding of the species and geography of sheep coccidia.Key words: Sheep; coccidia; Eimeria ; species; distributionCoccidia Species and World Distribution of Sheep (Ovis aries )HUANG Bing, DONG Hui, ZHAO Qiping, HAN Hongyu, YU Yu, ZHU Shunhai(Key Laboratory of Animal Parasitology of Ministry of Agriculture and Rural Affairs, Shanghai Veterinary Research Institute, CAAS,Shanghai 200241, China)· 182 ·中国动物传染病学报2022年4月M o u s s u和M a r o t e l于1901年首次在绵羊(Ovis aries)检出球虫,当时归为球虫属未定种(Coccidium sp.),并于1902年将其命名为福氏球虫(Coccidium faurei)。
遗传育种相关名词中英文对照

中英文对照的分子育种相关名词3'untranslated region (3'UTR) 3'非翻译区5'untranslated region (5; UTR) 5'非翻译区A chromosome A 染色体AATAAA 多腺苷酸化信号aberration 崎变abiogenesis 非生源说accessory chromosome 副染色体accessory nucleus 副核accessory protein 辅助蛋白accident variance 偶然变异Ac-Ds system Ac-Ds 系统acentric chromosome 无着丝粒染色体acentric fragment 无着丝粒片段acentric ring 无着丝粒环achromatin 非染色质acquired character 获得性状acrocentric chromosome 近端着丝粒染色体acrosyndesis 端部联会activating transcription factor 转录激活因子activator 激活剂activator element 激活单元activator protein( AP)激活蛋白activator-dissociation system Ac-Ds 激活解离系统active chromatin 活性染色质active site 活性部位adaptation 适应adaptive peak 适应高峰adaptive surface 适应面addition 附加物addition haploid 附加单倍体addition line 附加系additive effect 加性效应additive gene 加性基因additive genetic variance 加性遗传方差additive recombination 插人重组additive resistance 累加抗性adenosine 腺昔adenosine diphosphate (ADP )腺昔二鱗酸adenosine triphosphate( ATP)腺昔三憐酸adjacent segregation 相邻分离A-form DNA A 型DNAakinetic chromosome 无着丝粒染色体akinetic fragment 无着丝粒片断alien addition monosomic 外源单体生物alien chromosome substitution 外源染色体代换alien species 外源种alien-addition cell hybrid 异源附加细胞杂种alkylating agent 焼化剂allele 等位基因allele center 等位基因中心allele linkage analysis 等位基因连锁分析allele specific oligonucleotide(ASO)等位基因特异的寡核苷酸allelic complement 等位(基因)互补allelic diversity 等位(基因)多样化allelic exclusion 等位基因排斥allelic inactivation 等位(基因)失活allelic interaction 等位(基因)相互作用allelic recombination 等位(基因)重组allelic replacement 等位(基因)置换allelic series 等位(基因)系列allelic variation 等位(基因)变异allelism 等位性allelotype 等位(基因)型allohaploid 异源单倍体allopatric speciation 异域种alloploidy 异源倍性allopolyhaploid 异源多倍单倍体allopolyploid 异源多倍体allosyndesis 异源联会allotetraploid 异源四倍体alloheteroploid 异源异倍体alternation of generation 世代交替alternative transcription 可变转录alternative transcription initiation 可变转录起始Alu repetitive sequence, Alu family Alu 重复序列,Alu 家族ambiguous codon 多义密码子ambisense genome 双义基因组ambisense RNA 双义RNAaminoacyl-tRNA binding site氨酰基tRNA接合位点aminoacyl-tRNA synthetase 氨酰基tRNA连接酶amixis 无融合amorph 无效等位基因amphipolyploid 双多倍体amplicon 扩增子amplification 扩增amplification primer 扩增引物analysis of variance 方差分析anaphase (分裂)后期anaphase bridge (分裂)后期桥anchor cell 锚状细胞androgamete 雄配子aneuhaploid 非整倍单倍体aneuploid 非整倍体animal genetics 动物遗传学annealing 复性antibody 抗体anticoding strand 反编码链anticodon 反密码子anticodon arm 反密码子臂anticodon loop 反密码子环antiparallel 反向平行antirepressor 抗阻抑物antisense RNA 反义RNAantisense strand 反义链apogamogony 无融合结实apogamy 无配子生殖apomixis 无融合生殖arm ratio (染色体)臂比artificial gene人工基因artificial selection 人工选择asexual hybridization 无性杂交asexual propagation 无性繁殖asexual reproduction 无性生殖assortative mating 选型交配asynapsis 不联会asynaptic gene 不联会基因atavism 返祖atelocentric chromosome 非端着丝粒染色体attached X chromosome 并连X 染色体attachment site 附着位点attenuation 衰减attenuator 衰减子autarchic gene 自效基因auto-alloploid 同源异源体autoallopolyploid 同源异源多倍体autobivalent 同源二阶染色体auto-diploid 同源二倍体;自体融合二倍体autodiploidization 同源二倍化autoduplication 自体复制autogenesis自然发生autogenomatic 同源染色体组autoheteroploidy 同源异倍性autonomous transposable element 自主转座单元autonomously replicating sequence(ARS)自主复制序列autoparthenogenesis 自发单性生殖autopolyhaploid 同源多倍单倍体autopolyploid 同源多倍体autoradiogram 放射自显影图autosyndetic pairing 同源配对autotetraploid 同源四倍体autozygote 同合子auxotroph 营养缺陷体B chromosome B 染色体B1,first backcross generation 回交第一代B2,second backcross generation 回交第二代back mutation 回复突变backcross 回交backcross hybrid 回交杂种backcross parent 回交亲本backcross ratio 回交比率background genotype 背景基因型bacterial artification chromosome( BAC )细菌人工染色体Bacterial genetics 细菌遗传学Bacteriophage 噬菌体balanced lethal 平衡致死balanced lethal gene 平衡致死基因balanced linkage 平衡连锁balanced load 平衡负荷balanced polymorphism 平衡多态现象balanced rearrangements 平衡重组balanced tertiary trisomic 平衡三级三体balanced translocation 平衡异位balancing selection 平衡选择band analysis 谱带分析banding pattern (染色体)带型basal transcription apparatus 基础转录装置base analog 碱基类似物base analogue 类減基base content 减基含量base exchange 碱基交换base pairing mistake 碱基配对错误base pairing rules 碱基配对法则base substitution 减基置换base transition 减基转换base transversion 减基颠换base-pair region 碱基配对区base-pair substitution 碱基配对替换basic number of chromosome 染色体基数behavioral genetics 行为遗传学behavioral isolation 行为隔离bidirectional replication 双向复制bimodal distribution 双峰分布binary fission 二分裂binding protein 结合蛋白binding site 结合部位binucleate phase 双核期biochemical genetics 生化遗传学biochemical mutant 生化突变体biochemical polymorphism 生化多态性bioethics 生物伦理学biogenesis 生源说bioinformatics 生物信息学biological diversity 生物多样性biometrical genetics 生物统计遗传学(简称生统遗传学) bisexual reproduction 两性生殖bisexuality 两性现象bivalent 二价体blending inheritance 混合遗传blot transfer apparatus 印迹转移装置blotting membrane 印迹膜bottle neck effect 瓶颈效应branch migration 分支迁移breed variety 品种breeding 育种,培育;繁殖,生育breeding by crossing 杂交育种法breeding by separation 分隔育种法breeding coefficient 繁殖率breeding habit 繁殖习性breeding migration 生殖回游,繁殖回游breeding period 生殖期breeding place 繁殖地breeding population 繁殖种群breeding potential繁殖能力,育种潜能breeding range 繁殖幅度breeding season 繁殖季节breeding size 繁殖个体数breeding system 繁殖系统breeding true 纯育breeding value 育种值broad heritability 广义遗传率bulk selection 集团选择C0,acentric 无着丝粒的Cl,monocentric 单着丝粒C2, dicentric双着丝粒的C3,tricentric 三着丝粒的candidate gene 候选基因candidate-gene approach 候选基因法Canpbenmodel坎贝尔模型carytype染色体组型,核型catabolite activator protein 分解活化蛋白catabolite repression 分解代谢产物阻遏catastrophism 灾变说cell clone 细胞克隆cell cycle 细胞周期cell determination 细胞决定cell division 细胞分裂cell division cycle gene(CDC gene) 细胞分裂周期基因ceU division lag细胞分裂延迟cell fate 细胞命运cell fusion 细胞融合cell genetics 细胞的遗传学cell hybridization 细胞杂交cell sorter细胞分类器cell strain 细胞株cell-cell communication 细胞间通信center of variation 变异中心centimorgan(cM) 厘摩central dogma 中心法则central tendency 集中趋势centromere DNA 着丝粒DNAcentromere interference 着丝粒干扰centromere 着丝粒centromeric exchange ( CME)着丝粒交换centromeric inactivation 着丝粒失活centromeric sequence( CEN sequence)中心粒序列character divergence 性状趋异chemical genetics 化学遗传学chemigenomics 化学基因组学chiasma centralization 交叉中化chiasma terminalization 交叉端化chimera异源嵌合体Chi-square (x2) test 卡方检验chondriogene 线粒体基因chorionic villus sampling 绒毛膜取样chromatid abemition染色单体畸变chromatid break染色单体断裂chromatid bridge 染色单体桥chromatid interchange 染色单体互换chromatid interference 染色单体干涉chromatid tetrad 四分染色单体chromatid translocation 染色单体异位chromatin agglutination 染色质凝聚chromosomal aberration 染色体崎变chromosomal assignment 染色体定位chromosomal banding 染色体显带chromosomal disorder 染色体病chromosomal elimination 染色体消减chromosomal inheritance 染色体遗传chromosomal interference 染色体干扰chromosomal location 染色体定位chromosomal locus 染色体位点chromosomal mutation 染色体突变chromosomal pattern 染色体型chromosomal polymorphism 染色体多态性chromosomal rearrangement 染色体质量排chromosomal reproduction 染色体增殖chromosomal RNA 染色体RNA chromosomal shift 染色体变迁,染色体移位chromosome aberration 染色体畸变chromosome arm 染色体臂chromosome banding pattern 染色体带型chromosome behavior 染色体动态chromosome blotting 染色体印迹chromosome breakage 染色体断裂chromosome bridge 染色体桥chromosome coiling 染色体螺旋chromosome condensation 染色体浓缩chromosome constriction 染色体缢痕chromosome cycle 染色体周期chromosome damage 染色体损伤chromosome deletion 染色体缺失chromosome disjunction 染色体分离chromosome doubling 染色体加倍chromosome duplication 染色体复制chromosome elimination染色体丢失chromosome engineering 染色体工程chromosome evolution 染色体进化chromosome exchange 染色体交换chromosome fusion 染色体融合chromosome gap 染色体间隙chromosome hopping 染色体跳移chromosome interchange 染色体交换chromosome interference 染色体干涉chromosome jumping 染色体跳查chromosome knob 染色体结chromosome loop 染色体环chromosome lose染色体丢失chromosome map 染色体图chromosome mapping 染色体作图chromosome matrix 染色体基质chromosome mutation染色体突变chromosome non-disjunction染色体不分离chromosome paring染色体配对chromosome polymorphism 染色体多态性chromosome puff染色体疏松chromosome rearrangement染色体质量排chromosome reduplication 染色体再加倍chromosome repeat染色体质量叠chromosome scaffold 染色体支架chromosome segregation 染色体分离chromosome set 染色体组chromosome stickiness染色体粘性chromosome theory of heredity 染色体遗传学说chromosome theory of inheritance 染色体遗传学说chromosome thread 染色体丝chromosome walking 染色体步查chromosome-mediated gene transfer 染色体中介基因转移chromosomology 染色体学CIB method CIB法;性连锁致死突变出现频率检测法circular DNA 环林DNAcis conformation 顺式构象cis dominance 顺式显性cis-heterogenote顺式杂基因子cis-regulatory element 顺式调节兀件cis-trans test 顺反测验cladogram 进化树cloning vector 克隆载体C-meiosis C减数分裂C-metaphase C 中期C-mitosis C有丝分裂code degeneracy 密码简并coding capacity 编码容量coding ratio 密码比coding recognition site 密码识别位置coding region 编码区coding sequence 编码序列coding site 编码位置coding strand 密码链coding triplet 编码三联体codominance 共显性codon bias 密码子偏倚codon type 密码子型coefficient of consanguinity 近亲系数coefficient of genetic determination 遗传决定系数coefficient of hybridity 杂种系数coefficient of inbreeding 近交系数coefficient of migration 迁移系数coefficient of relationship 亲缘系数coefficient of variability 变异系数coevolution 协同进化coinducer 协诱导物cold sensitive mutant 冷敏感突变体colineartiy 共线性combining ability 配合力comparative genomics 比较基因组学competence 感受态competent cell感受态细胞competing groups 竞争类群competition advantage 竞争优势competitive exclusion principle 竞争排斥原理complementary DNA (cDNA)互补DNA complementary gene 互补基因complementation test 互补测验complete linkage 完全连锁complete selection 完全选择complotype 补体单元型composite transposon 复合转座子conditional gene 条件基因conditional lethal 条件致死conditional mutation 条件突变consanguinity 近亲consensus sequence 共有序列conservative transposition 保守转座constitutive heterochromatin 组成型染色质continuous variation 连续变异convergent evolution 趋同进化cooperativity 协同性coordinately controlled genes 协同控制基因core promoter element 核心启动子core sequence 核心序列co-repressor协阻抑物correlation coefficient相关系数cosegregation 共分离cosuppression 共抑制cotranfection 共转染cotranscript共转录物cotranscriptional processing共转录过程cotransduction 共转导cotransformation 共转化cotranslational secrection 共翻译分泌counterselection 反选择coupling phase 互引相covalently closed circular DNA(cccDNA)共价闭合环状DNA covariation 相关变异criss-cross inheritance 交叉遗传cross 杂交crossability 杂交性crossbred 杂种cross-campatibility 杂交亲和性cioss-infertility 杂交不育性crossing over 交换crossing-over map 交换图crossing-over value 交换值crossover products 交换产物crossover rates 交换率crossover reducer 交换抑制因子crossover suppressor 交换抑制因子crossover unit 交换单位crossover value 值crossover-type gamete 交换型配子C-value paradox C 值悖论cybrid 胞质杂种cyclin 细胞周期蛋白cytidme 胞苷cytochimera 细胞嵌合体cytogenetics 细胞遗传学cytohet 胞质杂合子cytologic 细胞学的cytological map 细胞学图cytoplasm细胞质cytoplasmic genome 胞质基因组cytoplasmic heredity 细胞质遗传cytqplasmic incompatibility 细胞质不亲和性cytoplasmic inheritance 细胞质遗传cytoplasmic male sterility 细胞质雄性不育cytoplasmic mutation 细胞质突变cytofdasmic segregation 细胞质分离cytoskeleton 细胞骨架Darwin 达尔文Darwinian fitness 达尔文适合度Darwinism 达尔文学说daughter cell 子细胞daughter chromatid 子染色体daughter chromosome 子染色体deformylase 去甲酰酶degenerate code 简并密码degenerate primer 简并引物degenerate sequence 简并序列degenerated codon 简并密码子degeneration 退化degree of dominance 显性度delayed inheritance 延迟遗传deletant 缺失体deletion 缺失deletion loop 缺失环deletion mapping 缺失作图deletion mutation 缺失突变denatured DNA 变性DNA denatured protein 变性蛋白denaturing gel 变性胶denaturing gel electrophoresis 变性凝胶电泳denaturing gradient polyacrylamide gel 变性聚丙稀酰胺凝胶density gradient centrifugation 密度梯度离心density gradient separation 密度梯度分离deoxyribonucleic acid-dependent DNA polymerase 依赖于DNA的DNA聚合酶derived line 衍生系derived type 衍生类型developmental genetics 发育遗传学developmental pathway 发育途径dicentric bridge 双粒染色体桥dicentric chromosome 双着丝粒染色体differential staining technique 显带技术differentiation center 分化中心dihaploid 双单倍体,dihybrid 双因子杂种dihybrid cross 双因子杂交dimorphism 二态性diploidization 二倍化diploidize 二倍化diploidized haploid 二倍化的单倍体direct cross 正交direct repeat 同向重复(序列)direct selection 正选择directed mutagenesis 正向突变discontinuous variation 不连续变异distant hybrid 远缘杂种distant hybridization 远缘杂交diversity center 多样性中心diversity curve 多样性曲线diversity gene ( D gene) D 基因diversity indices 多样性指数diversity of species 种的多样性diversity region ( D region) D 区;多变区DNA alkylation DNA 烧化DNA amplification DNA 扩增DNA amplification in vitro DNA 体外扩增DNA amplification polymorphism DNA 扩增多态性DNA breakage DNA 断裂DNA database DNA 数据库DNA degradation DNA 降解DNA denaturation DNA 变性DNA detection DNA 检测DNA distortion DNA 变形DNA duplex DNA 双链体DNA duplicase DNA 复合酶DNA element DNA 单元DNA evolution DNA 进化DNA fingerprint DNA 指纹DNA fingerprinting DNA 指纹分析DNA homology DNA 同源性DNA hybridization DNA 杂交DNA jumping technique DNA 跳查技术DNA melting DNA 解链DNA methylation DNA 甲基化DNA modification DNA 修饰DNA modification restriction system DNA 修饰限制系统DNA nicking DNA 切口形成DNA oxidation DNA 氧化DNA packaging DNA 包装DNA pairing DNA 配对DNA pitch DNA 螺距DNA polymorphism DNA 多态性DNA probe DNA 探针DNA puff DNA 泡DNA purification DNA 纯化DNA recombination DNA 重组DNA redundant 多余DNADNA repair DNA 修复DNA replication DNA 复制DNA replication enhancer DNA 复制增强子DNA replication origin DNA 复制起点DNA replication site DNA 复制点DNA sealase DNA 连接酶DNA sequence analysis DNA 序列分析DNA sizing gene DNA大小决定基因DNA strand exchange DNA 链交换DNA strand separation DNA 链分离DNA strand transfer protein DNA 链转移蛋白DNA template DNA 模板DNA thermal cycler DNA 热循环仪DNA topoisomerase DNA 拓扑异构酶DNA transcript DNA 转录物DNA transposon DNA 转座子DNA twist DNA 扭曲DNA typing DNA 分型DNA untwisting DNA 解旋DNA unwinding enzyme DNA 解旋酶DNA unwinding protein DNA 解旋蛋白DNA-agar technique DNA 琼脂技术DNAase I footprinting DNA 酶I 足迹法DNAase-free reagent 无DNA 酶试剂DNA-binding domain DNA 结合域DNA-binding motif DNA 结合基序DNA-binding protein DNA 结合蛋白DNA-polymerase DNA 聚合酶DNA-protein complex DNA -蛋白质复合体DNA-protein interaction DNA _ 蛋白质相互作用DNA-restriction enzyme DNA 限制酶DNA-RNA hybrid DNA-RNA 杂交体DNase-free 不含DNA 酶的dominance 显性dominance type 优势型dominance variance 显性方差dominant allele 显性等位基因dominant effect 显性效应dominant gene 显性基因dominant gene mutation 显'性基因突变dominant lethal 显性致死dominant phenotype 显性表型donor DNA 供体DNAdonor organism 供体生物dosage compensation 剂量补偿作用dotting blotting 点溃法double crossing over 双交换double fertilization 汉受精duplicate genes 重复基因duplication重复duplicon 重复子dyad 二分体dynamic selection 动态选择ecological genetics 生态遗传学ecological isolation 生态隔离ecological niche 生态小境ectopic expression 异位表达ectopic integration 异位整合effective population size 有效群体大小embryoid 胚状体embryonic stem cells( ES cells)胚胎干细胞endocrine signal 内分泌信号endogamy 近亲繁殖endomitosis 核内有丝分裂endonuclease 内切核酸酶endopolyploidy 核内多倍体environment 环境environmental variance 环境方差environmental variation 环境变异epigenesis 后成说epigenetic inheritance 后生遗传epigenetically silenced 后生沉默episome 附加体epistasis 上位性epistatic dominance 超显性epistatic gene 上位基因equal segregation 均等分离equational division 均等分裂equilibrium population 平衡群体Expressed Sequence Tag(EST)表达序列标签euchromatin 常染色质euchromatin常染色质eugenics 优生学euhaploid 整单倍体eukaryote 真核生物eukaryotic chromosome 真核染色体eukaryotic cell 真核细胞eukaryotic organism 真核生物eukaryotic vector 真核载体euphenics 优型学euploid 整倍体evolutional load 进化负荷evolutionary divergence 进化趋异evolutionary genetics 进化遗传学evolutionaiy rate 进化速率excision repair 切除修复exconjugant 接合后体excretion vector 分泌型载体exit site 萌发点exogenote 外基因子exogenous gene 外源基因exonuclease 外切核酸酶expression cloning 表达克隆expression library 表达文库expression mutation 表达突变expression plasmid 表达质粒expression product 表达产物expression screening 表达筛选extinguisher loci 消失基因座,灭绝基因座extirpated species 绝迹种extrachromosomal inheritance 染色体外遗传extra-chromosome超数染色体,额外染色体extranuclear inheritance 核外遗传F1 generation F1代,子一代F2 generation F2 代,子二代facultative heterochromatin 兼性异染色质familial trait 家族性状family selection 家系选择feedback suppression 反馈抑制female gamete 雌配子fertility factor 致育因子filial generation 子代fingerprint 指纹finite population 有限群体first division segregation 第一次分裂分离first division segregation pattern 第一次分裂分离模式flanking sequence 侧翼序列flow cytometry 流式细胞仪fluorescence in situ hybridization ( FISH )荧光原位杂交fluorescent primer 荧光引物fluorescent probe 荧光探针formyl methionine (fMet)甲酰甲硫氨酸foot printing 足迹法foreign DNA 外源DNAforward genetics 正向遗传学forward mutation 正向突变forward primer 正向引物founder effect 建立者效应four strand double crossing over 四线双交换full-sib 全同胞functional genomics 功能基因组学functional RNA 功能RNAgain-of-function mutation 功能获得性突变gamete 配子gametic 配子的gametic incompatibility 配子不亲和性gametic lethal 配子致死gametic linkage 配子连锁gametic meiosis 配子减数分裂gametic ratio 配子分离比gametoclonal variation 配子无性系变异gametophyte 配子体G-band G带;中期染色体带GC box GC 框GC tailing GC 加尾gel electrophoresis 凝胶电泳gemetic sterility 配子不育gene activation 基因激活gene activity 基因活性gene amplification 基因扩增gene analysis 基因分析gene arrangement 基因排列gene balance 基因平衡gene basis 基因基础gene batteries 基因群gene block 基因区段gene carrier 基因携带者gene center theory 基因中心学说gene cluster 基因簇gene combination 基因重组gene complex 基因复合体gene content 基因含量gene conversion 基因转换gene distribution 基因分布gene diversity 基因多样性gene dosage 基因剂量gene dosage compensation 基因剂量补偿gene dosage effect 基因剂量效应gene duplication 基因重复gene element 基因元件gene exchange 基因交流gene expression 基因表达gene expression system 基因表达系统gene family 基因家族gene fixation 基因固定gene flow 基因流gene frequency 基因频率gene fusion 基因融合gene inactivation 基因失活gene inoculation 基因接种gene interaction 基因相互作用gene isolation 基因分离gene knockout 基因敲除gene knock-out 基因失效法gene linkage 基因连锁gene localization 基因定位gene location 基因位置gene locus 基因位点gene magnification 基因扩增gene manipulation 基因操作gene map 基因图谱gene mapping 基因作图gene multiplication 基因重复gene mutation 基因突变gene mutation rate 基因突变频率gene order 基因次序gene organization 基因组构gene pool 基因库gene position effect 基因位置效应gene probe 基因探针gene product 基因产物gene rearrangement 某因重排gene reassortment 基因重新配对gene replication 基因复制gene repression 基因抑制gene resortment 基因重配gene silencing 基因沉默gene splicing 基因剪接gene string 基因线gene structure 基因结构gene substitute 基因置换gene substitution 基因置换gene suppression 基因抑制gene synthesis 基因合成gene tagged 基因标签gene tagging 基因标签gene targeting 基因导向,基因寻靶gene transfer 基因转移gene transfer agent 基因传递因子gene transfer vector 基因转移载体gene transposition 基因转座genealogical classification 系谱分类genera 属general transcription factor ( GTF )通用转录因子generalized transduction 普遍性转导generation 世代generative cell 生殖细胞generative reproduction 有性繁殖generic coefficient 种属系数generic cross 属间杂交generic name 属名genes in common 共同基因gene-specific transcription factor 基因特异性转录因子genetic ablation 基因缺损genetic advance 遗传进度genetic algebra 遗传代数genetic analysis 遗传分析genetic background 遗传背景genetic balance 遗传平衡genetic block 遗传性阻碍genetic compensation 遗传补偿genetic complementation 遗传互补genetic composition 遗传组成genetic continuity 遗传连续性genetic control 遗传控制genetic covariance 遗传协方差genetic cross 杂交genetic database 遗传数据库genetic death 遗传性死亡genetic deficiency 遗传缺损genetic deformity 基因变型genetic determinant 遗传决定因子genetic dimorphism 遗传二型现象genetic distance 遗传距离genetic divergence 遗传趋异genetic diversity 遗传多样性genetic dominance 遗传优势genetic donor 基因供体genetic drift 遗传漂变genetic element遗传因子,遗传成分genetic engineering 遗传工程genetic equilibrium 遗传平衡genetic erosion 遗传冲刷,遗传蚀变genetic expression 遗传表达genetic extinction 遗传灭绝genetic facilitation 遗传促进作用genetic factor 遗传因子genetic feedback 遗传反馈genetic fingerprint 遗传指纹genetic fingerprinting 遗传指纹分析genetic fitness 遗传适合度genetic flexibility 遗传可塑性genetic gain 遗传获得量genetic heterogeneity 遗传异质性genetic homology 遗传同源genetic immunity 遗传免疫genetic imprinting 遗传印记genetic inertia 遗传惰性genetic information 遗传信息genetic inoculation 基因接种genetic instability 遗传不稳定性genetic continuity 遗传连续性genetic control 遗传控制genetic covariance 遗传协方差genetic cross 杂交genetic database 遗传数据库genetic death 遗传性死亡genetic deficiency 遗传缺损genetic deformity 基因变型genetic determinant 遗传决定因子genetic dimorphism 遗传二型现象genetic distance 遗传距离genetic divergence 遗传趋异genetic diversity 遗传多样性genetic dominance 遗传优势genetic donor 基因供体genetic drift 遗传漂变genetic element遗传因子,遗传成分genetic engineering 遗传工程genetic equilibrium 遗传平衡genetic erosion 遗传冲刷,遗传蚀变genetic expression 遗传表达genetic extinction 遗传灭绝genetic facilitation 遗传促进作用genetic factor 遗传因子genetic feedback 遗传反馈genetic fingerprint 遗传指纹genetic fingerprinting 遗传指纹分析genetic fitness 遗传适合度genetic flexibility 遗传可塑性genetic gain 遗传获得量genetic heterogeneity 遗传异质性genetic homology 遗传同源genetic immunity 遗传免疫genetic imprinting 遗传印记genetic inertia 遗传惰性genetic information 遗传信息genetic inoculation 基因接种genetic instability 遗传不稳定性genetic interaction 遗传相互作用genetic isolating factor 遗传隔离因子genetic isolation 遗传隔离genetic knock-out experiment 基因失效试验genetic linkage 遗传连锁genetic linkage map 遗传连锁图谱genetic load 遗传负荷genetic manipulation 遗传操作genetic map 遗传图谱genetic mapping 遗传作图genetic marker 遗传标记genetic masking 基因组掩饰genetic material 遗传物质genetic mobilization 遗传转移genetic modification 遗传修饰genetic module 遗传组件genetic nomenclature 遗传命名法genetic parameter 遗传参数genetic polarity 遗传极性genetic polymorphism 遗传多样性genetic population 遗传群体genetic potential 遗传潜力genetic process 遗传过程genetic property 遗传特'性genetic ratio 遗传比genetic reactivation 遗传复活genetic reassortment 遗传重排genetic recipient 基因受体genetic recombination 遗传重组genetic regulation 遗传调节genetic relationship 亲缘关系genetic repair mechanism 遗传修复机制genetic replication 遗传复制genetic risk 遗传危险性genetic screening 遗传筛查genetic segregation 遗传分离genetic selection 遗传选择genetic sex 遗传性别genetic shift 遗传漂移genetic stability 遗传稳定性genetic sterility 遗传性不育genetic strain 遗传品系genetic suppression 遗传抑制genetic switch 遗传开关genetic system 遗传体系genetic transcription 遗传转录genetic transformation 遗传转换genetic translation 遗传翻译genetic transmission 遗传传递genetic typing 遗传分型genetic unit 遗传单位genetic value 遗传值genetic variability 遗传变异性genetic variance 遗传方差genetic vulnerability 遗传易损性genetic“hot spot” 遗传“热点”genetical marker 遗传标记genetical non-disjunction 遗传不分离genetical population 遗传群体genetically heterogeneous 遗传异质的genetically modified organism 基因修饰生物genetics correction 遗传修正genetics of resistance 抗性遗传genetype 基因型genic balance 基因平衡genome allopolyploid 基因组异质多倍体genome amplification 基因组扩增genome evolution 基因组进化genome mapping 基因组作图genome project 基因组计划genome rearrangement 基因组重排genome sequencing 基因组测序genomic exclusion 基因组排斥genomic fingerprinting 基因组指纹分析genomic footprinting 基因组足迹分析genomic imprinting 基因组印记genomic instability 基因组不稳定性genomic library 基因组文库genomic walking 基因组步查genotypic frequency 基因型频率genotypic ratio 基因型比值genotypic value 基因型值genotypic variance 基因型方差geographic speciation 地理型新种形成geographical isolation 地理隔离geographical polymorphism 地理多态现象germ layer 胚层germ line 种系germ nucleus 生殖核germ plasm 种质germinal mutation 生殖细胞突变germ-line gene therapy 种系基因治疗giant chromosome 巨型染色体global homology 总体同源性global region 全局调节子globular protein 球蛋白group selection 集团选择growth factor 生长因子GT-AG rule mRNA剪接识别信号规则gynandromorphy 雌雄嵌合体hairpin loop 发夹环hairpin structure 发夹结构half life 半寿期half sib mating 半同胞交配haplogenotypic 单倍基因型的haploid 单倍体haploidization 单倍体化haplotype 单元型hapostatic gene 下位基因Hardy-Weinberg equilibrium 哈迪-温伯格平衡heat shock gene 热激基因heat sock protein 热激蛋白heavy chain 重链helical structure 螺旋结构。
Decision and biodegradation characteristic kiranto
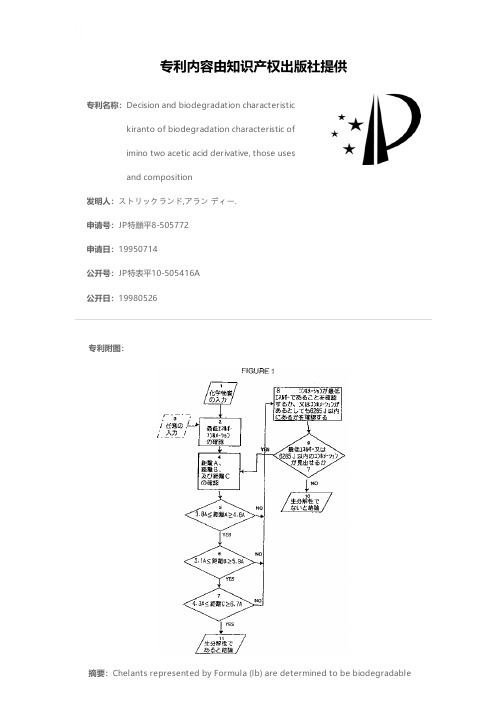
专利名称:Decision and biodegradation characteristickiranto of biodegradation characteristic ofimino two acetic acid derivative, those usesand composition发明人:ストリックランド,アラン ディー.申请号:JP特願平8-505772申请日:19950714公开号:JP特表平10-505416A公开日:19980526专利内容由知识产权出版社提供专利附图:摘要:Chelants represented by Formula (Ib) are determined to be biodegradablewhen Distance A is from 3.8 x 10<-10> to 4.6 x 10<-10> m, Distance B is from 5.1 x 10<-10> to 5.9 x 10<-10> m, and Distance C is from about 4.3 to 6.7 x 10<-10> m. Compounds meeting these criteria are referred to as compounds of Formula (2). In addition to methods of analysis and computer systems therefor, the invention includes a method of chelating a metal ion to form a chelate and biodegrading the chelate comprising step (a) contacting the metal ion with at least one compound of Formula (2) to form a chelate and (b) contacting the resultant chelate or chelant with microbes of the type specified in ASTM 2667-89, ISO 5815 or effective enzymes thereof under conditions and for a time sufficient for biodegradation wherein either (i) the chelate formed in step (a) is formed in a process of removing deposits, scale, or rust; cleaning or washing; controlling metal initiated or catalyzed oxidation or deterioration including spoilage, discoloration, or rancidification; textile treatment; paper making; stabilization of polymers or phosphates; or petroleum drilling, production or recovery; or (ii) the chelate formed in step (a) is before step (b) used as a redox catalyst, in a process of photographic bleaching, bleach-fixing or developing; in electroless deposition or plating; in removing acid or oxide gases including H2S, NOx, SOx, CO and CO2; or in providing agricultural nutrients.申请人:ザ ダウ ケミカル カンパニー地址:アメリカ合衆国,ミシガン 48640,ミッドランド,アボット ロード,ダウ センター 2030国籍:US代理人:石田 敬 (外3名)更多信息请下载全文后查看。
一株墨鱼干源枝芽胞杆菌的分离鉴定与生长特性

Hans Journal of Food and Nutrition Science 食品与营养科学, 2019, 8(1), 45-51Published Online February 2019 in Hans. /journal/hjfnshttps:///10.12677/hjfns.2019.81006Isolation Identification of a Virgibacillussp. from Dried Cuttlefish and Its GrowthCharacteristicsJing Fu1, Pan Chen1, Maohua Chen1, Aiqing Xu1,2*1School of Life Science, Hunan University of Science and Technology, Xiangtan Hunan2Key Laboratory of Ecological Remediation and Safe Utilization of Heavy Metal-Polluted Soils, College of Hunan Province, Xiangtan HunanReceived: Jan. 4th, 2019; accepted: Jan. 16th, 2019; published: Jan. 23rd, 2019AbstractThis paper is aimed to investigate the microflora of the white plaque existing at the surface of dried cuttlefish, and to isolate salt-tolerated microbial resources. The commercially available dried cuttlefish with white plaque were sampled, and a salt-tolerated bacterium, strain NYJ2, was isolated and purified on the Beef-extract-peptone Plate containing 5% of NaCl. The morphological characteristics of the colonies and cells, and several biochemical and physiological characteristics of the strain were detected, and partial sequence of the 16S rDNA was sequenced and phylogenetic analysis was performed. The results showed that a Virgibacillus sp. NYJ2 was isolated and identi-fied from the white plaque, and it can grow in Beef-extract-peptone broth containing 0.5%~15% of NaCl, and optimum at 1%~10% of NaCl, belonging to moderately halophilic bacteria. Its growth occurs at pH 5~pH 8, 10˚C~46˚C and the optimum temperature ranges from 25˚C to 42˚C.Low-temperature below 10˚C, or 0.05% of Nisin could be used to inhibit the growth and reproduc-tion of the strain NYJ2.KeywordsDried Cuttlefish, Moderately Halophilic Bacterium, Virgibacillus, Growth Characteristics一株墨鱼干源枝芽胞杆菌的分离鉴定与生长特性伏靖1,陈盼1,陈茂华1,许爱清1,2*1湖南科技大学生命科学学院,湖南湘潭*通讯作者。
次生代谢物中英文对照表

次生代谢物中英文对照表
English:
Secondary metabolites are organic compounds produced by plants, fungi, and bacteria that are not directly involved in the normal growth, development, or reproduction of the organism. Instead, these compounds often serve in defense against herbivores, pathogens, and competing organisms, as well as in attraction of pollinators and seed dispersers. Some examples of secondary metabolites include alkaloids, phenolics, terpenoids, and polyketides. These compounds have diverse functions and can provide important ecological and pharmacological benefits.
中文翻译:
次生代谢物是由植物、真菌和细菌产生的有机化合物,不直接参与生物体的正常生长、发育或繁殖。
相反,这些化合物通常在防御草食动物、病原体和竞争性生物方面发挥作用,同时也在吸引传粉者和种子传播者方面发挥作用。
一些次生代谢物的例子包括生物碱、酚类化合物、萜类化合物和多酮类化合物。
这些化合物具有多种功能,可以提供重要的生态和药理学益处。
初中生物知识与概念之生物进化论简介

初中生物知识与概念之生物进化论简介一、生物进化论概述1.1 生物进化论的定义生物进化论是描述生物种群随时间变化而发生的遗传性状和物种多样性变化的科学理论。
它揭示了生物种群如何通过自然选择、遗传变异和遗传漂变等机制,逐步适应环境,发展出更加复杂和多样的生命形态。
Evolutionary theory is the scientific theory that describes the changes in genetic traits and species diversity that occur in biological populations over time. It reveals how biological populations gradually adapt to their environments and develop more complex and diverse life forms through mechanisms such as natural selection, genetic variation, and genetic drift.1.2 生物进化论的重要性生物进化论对于理解生物多样性、生物适应性和生物演化的过程具有重要意义。
它为我们揭示了生命起源和演化的奥秘,帮助我们认识生物世界的复杂性和多样性,为生物学研究提供了重要的理论框架和指导思想。
The theory of biological evolution is crucial for understanding biodiversity, biological adaptability, and the process of biological evolution. It reveals the mysteries of the origin and evolution of life, helps us recognize the complexity and diversity of the biological world, and provides an important theoretical framework and guiding ideology for biological research.二、生物进化论的主要观点和机制2.1 自然选择自然选择是生物进化论的核心机制之一。
Biosensors and Bioelectronics

Biosensors and Bioelectronics Biosensors and bioelectronics have revolutionized the field of medical diagnostics and healthcare, offering a wide range of applications in disease detection, environmental monitoring, and food safety. These innovative technologies have the potential to significantly improve the quality of life for individuals around the world. However, they also present a number of challenges and ethical considerations that must be carefully addressed. From a scientific perspective, biosensors and bioelectronics represent a remarkable advancement in the ability to detect and analyze biological molecules with high sensitivity and specificity. These devices utilize biological components such as enzymes, antibodies, or nucleic acids to recognize and bind to target molecules, generating a measurable signal that can be translated into valuable information. This capability has opened up new possibilities for rapid and accurate diagnosis of diseases, monitoring of physiological parameters, and detection of environmental contaminants. In the medical field, biosensors have the potential torevolutionize the way diseases are diagnosed and monitored. For example, glucose biosensors have transformed the management of diabetes by enabling individuals to monitor their blood sugar levels at home, reducing the need for frequent visits to healthcare facilities. Additionally, biosensors for cardiac markers, infectious diseases, and cancer biomarkers hold great promise for early detection and personalized treatment strategies. These technologies have the potential to improve patient outcomes and reduce healthcare costs. Furthermore, biosensors and bioelectronics have significant implications for environmental monitoring and food safety. These technologies can be used to detect pollutants, toxins, and pathogens in air, water, and food samples, providing valuable information for public health and regulatory purposes. By enabling rapid and on-site analysis, biosensors have the potential to prevent outbreaks of foodborne illnesses and minimize environmental contamination. Despite these promising applications, biosensors and bioelectronics also present several challenges that need to be carefully considered. One of the primary concerns is the need for rigorous validation and standardization of these technologies to ensure their reliability and accuracy. The complex nature of biological systems and the potential for interference fromexternal factors can pose significant challenges for the development anddeployment of biosensors in real-world settings. Ethical considerations also come into play when discussing the use of biosensors and bioelectronics. The collection and analysis of biological data raise important questions about privacy, consent, and data security. As these technologies become more integrated into healthcareand everyday life, it is essential to establish clear guidelines and regulationsto protect individuals' rights and ensure responsible use of sensitive information. In addition, the accessibility and affordability of biosensors in resource-limited settings are important factors to consider. While these technologies have the potential to improve healthcare in both developed and developing countries, thereis a need to address barriers to adoption, such as cost, infrastructure, and training. Ensuring equitable access to biosensors is crucial for maximizing their impact on global health and well-being. In conclusion, biosensors andbioelectronics hold great promise for transforming healthcare, environmental monitoring, and food safety. These technologies have the potential torevolutionize disease diagnosis, enable personalized medicine, and enhance public health efforts. However, it is important to address scientific, technical, and ethical challenges to realize the full potential of biosensors while ensuringtheir responsible and equitable use. By navigating these complexities, we can harness the power of biosensors to improve the lives of people around the world.。
营养素在生物化学反应的个体差异(BiochemicalIndividuality)狄迪

营养素在生物化学反应的个体差异(BiochemicalIndividuality)狄迪——生物化学反应的个体差异(Biochemical Individuality)在营养素的需求方面,每个个体都是不同的,不同人之间存在着比较大的差异,同一个人处于不同的生理条件或者不同的环境条件下,需要量也是不同的,这就是营养素需要的个性化差异。
营养医学或者称为分子矫正医学最关心的问题就是营养的个性化,尊重营养个性化的特点,给身体细胞最佳的营养,保证细胞优质的生命活动,是保持长期健康、远离慢性疾病困扰和享受优质生命质量的根本。
Linus Pauling先生在他的书中引用了几个科学研究结果来说明营养个性化的问题。
今天,我想跟大家分享此书中几个科学实验的过程和结果。
1967年,Williams和Deason采用Guinea Pigs(豚鼠)作为实验动物,采用8组(每组10-15只)豚鼠,其中一组饲喂不含抗坏血酸(维生素C)的饲料,其它组饲喂不同剂量的维生素C的饲料(分别是0.5、1.0、2.0、4.0、8.0、16.0和32.0毫克/kg体重),观察它们罹患坏血病的情况。
研究结果表明,80%饲喂小于0.5毫克/kg体重剂量和25%饲喂1-4毫克/kg体重剂量维生素C的豚鼠在饲喂一段时间后患了坏血病,当维生素C的饲喂剂量大于8毫克/kg体重剂量时,就不会有坏血病产生。
其中有两只饲喂1.0毫克/kg体重剂量的豚鼠,生长状况超过了高剂量组,而且没有患白血病,而另外7只饲喂8-16毫克/kg体重维生素C的豚鼠却表现出生长非常缓慢的情况,当增加它们的维生素C的饲喂剂量时,他们的体重增加表现非常明显,由原来的10天增重12克增加到10天增重72克。
研究表明,有7/30的饲喂8-32毫克/kg体重的豚鼠需要更多的维生素C才能让它们生长得更好。
由此研究结果表明:对于豚鼠来说,维生素C的个体差异在20倍左右。
而对于人类来讲,Williams和Deason猜测维生素C的需要量差异在100倍左右。
phylogenomic意思

phylogenomic意思Phylogenomics 是一种将基因组学和系统发生学相结合的研究领域,其目标是通过研究物种之间的基因组序列差异来推断它们之间的系统进化关系。
通过将大量基因组数据与系统发生学方法相结合,Phylogenomics 可以提供更具亲缘关系的类群之间的系统进化关系,并提供更准确的进化历史推断。
Phylogenomics 的基本原理是在整个基因组水平上比较不同物种之间的基因序列差异。
相比于传统的研究方法,如酶电泳或单个基因的比对,Phylogenomics 可以提供更多的基因数据,并利用这些数据来构建更准确的系统发生学树。
Phylogenomics 主要依赖于两个核心步骤:物种选择和基因组分析。
在物种选择阶段,研究者需要确定要研究的物种,这些物种通常源自于一棵进化树上的一个或多个分支。
确定了研究物种后,就可以进行基因组分析了。
基因组分析包括两个主要过程:基因组测序和基因组比对。
在基因组测序阶段,研究者需要从研究物种中提取DNA 样本,并使用高通量测序技术将其转化为 DNA 序列。
这个步骤产生了大量的原始序列数据,这些数据需要经过一系列的质控和预处理步骤,以去除噪音和错误,并生产高质量的 DNA 序列数据。
接下来,研究者需要对基因组数据进行比对并提取出有用的信息。
这个步骤通常涉及基因组序列的比对和突变位点的分析。
通过将每个物种的基因组序列与已知的参考物种进行比对,研究者可以确定每个物种之间的序列差异和突变。
一旦基因组数据和突变位点被提取出来,就可以使用系统进化学的方法来构建系统进化树。
这些方法包括最大似然法、贝叶斯推断和邻接法等。
这些方法可以使用基因组序列差异数据来恢复物种之间的亲缘关系和进化历史,从而提供更深入的洞察力和理解。
Phylogenomics 在许多不同的领域有着广泛的应用。
在生物多样性研究中,Phylogenomics 可以帮助确定濒危物种的亲缘关系和进化历史,从而指导保护决策。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
DISCRIMINATION AND BIOPHYSICAL CHARACTERIZATION OF CERRADO PHYSIOGNOMIES WITH EO-1 HYPERSPECTRAL HYPERIONT OMOAKI M IURA 1A LFREDO R. H UETE 1L AERTE G. F ERREIRA 2E DSON E. S ANO 31 Terrestrial Biophysics and Remote Sensing LaboratoryDepartment of Soil, Water and Environmental ScienceUniversity of Arizona, Tucson, AZ 85721, USA{tomoaki, ahuete}@2 UFG - Universidade Federal de GoiásIESA - Instituto de Estudos Sócio-AmbientaisCaixa Postal 131 - 74.001-970 - Goiânia - GO, Brazillaerte@iesa.ufg.br3 EMBRAPA CerradosCaixa Postal 08.223 - 73.301-970 - Planaltina – DF, Brazilsano@cpac.embrapa.brAbstract.The Brazilian savanna, locally known as "cerrado", is the most intensely stressed biome with both natural- and human-induced pressures. In this study, we aimed to improve discrimination and characterization of the Brazilian cerrado physiognomies using hyperspectral Hyperion imagery, the first space-borne imaging spectrometer onboard the NASA's Earth Observing-1 (EO-1) platform. A Hyperion image was acquired over the Brasilia National Park (BNP) on July 20, 2001. Various optical measures which took a full advantage of hyperspectral remote sensing were employed and applied to the atmospherically-corrected Hyperion data. These included the 1st-order derivative-based green vegetation index (1st-DGVI), ligno-cellulose absorption index, spectral albedo, and shortwave-infrared (SWIR) spectral unmixing. All these optical measures were correlated well with field-estimates of landscape component cover fractions. Especially, the SWIR spectral unmixing results simultaneously biophysically-characterized and discriminated the cerrado physiognomies. These preliminary analyses showed a great potential of hyperspectral remote sensing in characterizing/discriminating the land covers in the Brazilian cerrado.Keywords: hyperspectral remote sensing, cerrado, land cover characterization, land cover discrimination, EO-1, Hyperion.1. IntroductionThe savanna, typically found in the sub-tropics and seasonal tropics, are the dominant vegetation biome type in the southern hemisphere, covering approximately 45 % of the South America. In Brazil, the savanna, locally known as "cerrado", is the most intensely stressed biome with natural environmental pressures (e.g., the strong seasonality in weather, extreme soil nutrient impoverishment, and widespread fire occurrences) and rapid/aggressive land conversions (Skole et al., 1994; Ratter et al., 1997). Better characterization and discrimination of cerrado physiognomies are needed in order to improve understanding of cerrado dynamics and its impact on carbon storage, nutrient dynamics, and the prospect for sustainable land use in the Brazilian cerrado biome.In this study, we aimed to improve discrimination of the Brazilian cerrado physiognomies through biophysical characterization with hyperspectral remote sensing. We used Hyperion,the first satellite-based hyperspectral imager, onboard the Earth Observing-1 (EO-1) platform.2. Materials and MethodsOur study sites were located in the Brasilia National Park (BNP) in the northern Federal District, Brazil (S 15o 40', W 48o 02'). This preserved area contains several of the major "core" cerrado vegetation associations (physiognomies), including cerrado grassland (camp limpo), shrub cerrado (campo sujo), wooded cerrado (cerrado ralo), cerrado woodland (cerrado tipico), and gallery forest (mata de galeria) in the order of increasing arboreous cover (Ribeiro and Walter, 1998).The first four major cerrado physiognomies described above were structurally characterized using ground transect surveys. At each site (physiognomy), landscape were vertically stratified into an arboreous (shrubs/trees) overstory layer and herbaceous understory layer dominated by grasses, in addition to a background soil/litter layer. Component cover fractions of each layer were then measured using a pin-point technique along a randomly-chosen 100 m transect. The landscape components considered in the measurements were: photosynthetic vegetation (PV), woody materials, and crown covers for the overstory, PV and standing litter for the understory, and soil and surface litter for the background surface layer.Hyperspectral Hyperion data were acquired over the field sites on July 20, 2001 during the field measurement campaign. Hyperion collected full range spectral data (400-2400 nm) in 10 nm intervals (full-width at half maximum = 10nm) at a 30 m ground spatial resolution. The data were preprocessed and radiometrically-calibrated into a Level 1A product at the TRW Hyperion data processing facility. The data were further processed to correct for several known artifacts in the Level 1A products and then converted to ground reflectances using a MODTRAN4-based atmospheric radiative transfer code (ACORN4, /acorn/intro.asp). The atmospherically-corrected Hyperion data were compared to airborne spectrometer (Analytical Spectral Devices, Inc., Boulder, CO, USA) data collected at a large dry pasture field in the north of BNP near the Hyperion overpass time, but on the next day. An aircraft was flown "below the atmosphere" at 150 m AGL. The airborne data were calibrated to ground reflectances by taking a ratio to the readings made over a calibrated Spectralon white reference panel before and after the flight. The Hyperion and airborne spectrometer data were statistically similar, indicating good accounting of atmospheric constituents in the Hyperion correction.Pixels over the field sites were extracted from the atmospherically-corrected Hyperion image. GPS coordinates of the sites and low-altitude aerial photos were used to locate the sites in the image. In addition, another set of pixels were extracted over gallery forest, cultivated pasture, and lake water (Santa Maria Lake at the center of BNP) for comparisons.3. ResultsField Measurements. Measured landscape component cover fractions of the four cerrado physiognomies as well as the field site locations are summarized in Table 1. The herbaceous layers were dominated by senescent tissues, while the shrub/tree layer were still green at the time of this field campaign. As used for the basis on many cerrado classification schemes (e.g., Ribeiro and Walter, 1998), the crown cover fractions increased from the cerrado grassland to cerrado woodland sites with a discrete increase between the shrub cerrado and wooded cerrado sites. There was a general increase (decrease) in the PV (NPV) cover fractions with an increase in the crown covers, except for the wooded cerrado site. Two of the four sites, the wooded cerrado and shrub cerrado sites, were dominated by the species that grow quickly after burnings and that remained green, which resulted in a larger green cover in the wooded cerrado site than the cerrado woodland site. Nearly no soils were exposed at any of the sites.Table 1. Landscape Component Cover Fractions of the Cerrado Physiognomies Measured in the Field SitesSite Name (Physiognomy) Site Location(Lat./Lon.)Crown (%)Green (PV)(%)NPV (%)Soil (%)Cerrado Grassland N15o39'55"/ W48o01'52" 1 18 82 < 1 Shrub Cerrado N15o35'20"/ W48o00'25" 3 23 76 < 1 Wooded Cerrado N15o36'26"/ W48o01'47" 10 34 63 3 Cerrado Woodland N15o43'58"/ W48o00'11" 13 30 69 < 1 Hyperion Reflectance Data. The Hyperion hyperspectral signatures clearly depicted the differences between pasture, gallery forest, and the other four cerrado physiognomies (Figure 1). Spectral signatures in the visible and near-infrared (NIR) regions for the cerrado physiognomies showed small differences, but with the red-NIR reflectance contrast corresponding well with green cover fractions (Figure 1, Table 1). The reflectance values atSWIR2 spectral region (2000-2400 nm), and performed a correlative comparison with coverFigure 2. First-DGVI plotted against field estimates of PV cover fractions for cerradophysiognomies.fractions. We employed various optical measures to take the full advantages of hyperspectral remote sensing, including the 1st-order derivative-basedgreen vegetation index derived from local baseline (1st-DGVI) (Elvidge and Chen, 1995),SWIR2 spectral albedo (an integral of reflectancesfrom 2100-2350 nm), ligno-cellulose absroption index (Elvidge, 1988), and 3-endmembers SWIRspectral unmixing (Asner and Lobell, 2000). Details of these methods were provided in thecorresponding references.In Figure 2, the 1st-DGVI values were plotted against the green cover fractions. The 1st-DGVI and green (PV) cover fractions correlated very well. Similarly, the spectral albedo of the SWIR region (2100-2350 nm) and ligno-cellulose absorption indices for 2090 and 2280 nm werecorrelated very well with the crown and NPV cover fractions, respectively (Figures 3, 4). The ligno-cellulose absorption indices, however, had large standard deviations (Figure 4) due most likely to the low signal-to-noise ratios of the Hyperion sensor in this wavelength region (< 30:1).Figure 4. Ligno-cellulose absorption depths plotted against field estimates of NPV cover fractions.Finally, the SWIR unmixing results from the Hyperion image were compared to an existing vegetation map for BNP (Macedo,1992). The regional fractional estimates were consistent with the vegetation map (Figure 5). Largest values of green vegetation fractions corresponded well to the occurrences of gallery forest along stream lines. Similarly, relatively large values of green vegetation fractions corresponded spatially well with wooded cerrado and cerrado woodland, while cerrado grassland and shrub cerrado areas were consistent with large NPV fractions.Figure 3. SWIR spectral albedo plotted against field estimates of crown cover fractions.4. Conclusions and DiscussionsIn this study, we evaluated the utility of hyperspectral remote sensing in biophysical characterization and discrimination of cerrado physiognomies by taking an advantage of a newly available satellite-based imaging spectrometer, EO-1 Hyperion. The atmospherically-corrected hyperspectral reflectance of Hyperion clearly depicted such diagnostic absorption features of vegetation as the red edge, red-NIR transition, and ligno-cellulose absorptions. Likewise, these spectral features were found to be corresponding well with biophysical characteristics (i.e., landscape component cover fractions) of cerrado physiognomies. As the cerrado physiognomic classes are based on differences in the proportion of a grass understory and tree/shrub overstory layer, the cover component fractional estimates of green vegetation, NPV, and soil with the SWIR spectral unmixing resulted in not only biophysically characterizing, but also discriminating cerrado physiognomies. These preliminary analyses showed a great potential of hyperspectral remote sensing in biophysical characterization as well as discrimination of the land covers in the Brazilian cerrado.Figure 5. Color composite of green vegetation (red), total litter (green), and bare soil (blue) fractions in comparison to a vegetation map adapted from Macedo (1992) for the Brasilia National Park.Acknowledgement.The authors would like to thank personnel of EMBRAPA-Cerrado, University of Brasilia, and the LBA project office for their logistical support and assistance in the field data collections. This work was supported by a NASA MODIS contract NAS5-31364 and NASA EO-1 grant NCC5-478.ReferencesAsner, G. P. and Lobel, D. B. A biogeophysical approach for automated SWIR unmixing of soils and vegetation. Remote Sensing of Environment, v. 74, p. 99-112, 2000.Elvidge, C. D. Examination of the spectral features of vegetation in 1987 AVIRIS data. Proceedings of the First AVIRIS Performance Evaluation Workshop, Jet Propulsion Laboratory Publication 88-38, Pasadena, CA, USA, p. 97-101, 1988.Elvidge, C. D. and Chen, Z. Comparison of broad-band and narrow-band red and near-infrared vegetation indices. Remote Sensing of Environment, v. 54, p. 38-48, 1995.Macedo, J. Soil studies from the Brasilia National Park. Technical Report for Subsidiaring the Review of the Brasilia National Park Monitoring Plan, Appendix 2, 39p., 1992. (in Portugese)Ratter, J. A., Riberio, J. F., and Bridgewater, S. The Brazilian cerrado vegetation and threats to its biodiversity. Annals of Botany, v. 80, p. 223-230, 1997.Ribeiro, J. F. and Walter, T. M. B. The major physiognomy in the Brazilian Cerrado region. Cerrado: Ambiente e Flora (ed. by Sano, S. M. and Almeida, S. P.), EMBRAPA-CPAC, Planaltina, DF, chap. 3, p. 169-188, 1998. Skole, D. L., Chomentowiski, W. H., Salas, W. A., and Nobre, C. A. Physical and human dimensions of deforestation in Amazonia. Bioscience, v. 44, p. 314-322, 1994.。
