OpenMP Dual Population...(IJIEEB-V7-N1-8)
并行程序设计实验报告-OpenMP 进阶实验

实验2:OpenMP 进阶实验1、实验目的掌握生产者-消费者模型,具备运用OpenMP相关知识进行综合分析,可实现实际工程背景下生产者-消费者模型的线程级负责均衡规划和调优。
2、实验要求1)single与master语句制导语句single 和master 都是指定相关的并行区域只由一个线程执行,区别在于使用master 则由主线程(0 号线程)执行,使用single 则由运行时的具体情况决定。
两者还有一个区别是single 在结束处隐含栅栏同步,而master 没有。
在没有特殊需求时,建议使用single 语句。
程序代码见程序2-12)barrier语句在多线程编程中必须考虑到不同的线程对同一个变量进行读写访问引起的数据竞争问题。
如果线程间没有互斥机制,则不同线程对同一变量的访问顺序是不确定的,有可能导致错误的执行结果。
OpenMP中有两种不同类型的线程同步机制,一种是互斥机制,一种是事件同步机制。
其中事件同步机制的设计思路是控制线程的执行顺序,可以通过设置barrier同步路障实现。
3)atomic、critical与锁通过critical 临界区实现的线程同步机制也可以通过原子(atomic)和锁实现。
后两者功能更具特点,并且使用更为灵活。
程序代码见程序2-2、2-3、2-44)schedule语句在使用parallel 语句进行累加计算时是通过编写代码来划分任务,再将划分后的任务分配给不同的线程去执行。
后来使用paralle for 语句实现是基于OpenMP 的自动划分,如果有n 次循环迭代k 个线程,大致会为每一个线程分配[n/k]各迭代。
由于n/k 不一定是整数,所以存在轻微的负载均衡问题。
我们可以通过子句schedule 来对影响负载的调度划分方式进行设置。
5)循环依赖性检查以对π 的数值估计的方法为例子来探讨OpenMP 中的循环依赖问题。
圆周率π(Pi)是数学中最重要和最奇妙的数字之一,对它的计算方法也是多种多样,其中适合采用计算机编程来计算并且精确度较高的方法是通过使用无穷级数来计算π 值。
openmp用法

openmp用法OpenMP是一种支持共享内存多线程编程的标准API。
它提供了一种简单而有效的方法,用于在计算机系统中利用多核和多处理器资源。
本文将逐步介绍OpenMP的用法和基本概念,从简单的并行循环到复杂的并行任务。
让我们一步一步来学习OpenMP吧。
第一步:环境设置要开始使用OpenMP,我们首先需要一个支持OpenMP的编译器。
常见的编译器如GCC、Clang和Intel编译器都支持OpenMP。
我们需要确保在编译时启用OpenMP支持。
例如,在GCC中,可以使用以下命令来编译包含OpenMP指令的程序:gcc -fopenmp program.c -o program第二步:并行循环最简单的OpenMP并行化形式是并行循环。
在循环的前面加上`#pragma omp parallel for`指令,就可以让循环被多个线程并行执行。
例如,下面的代码演示了如何使用OpenMP并行化一个简单的for循环:c#include <stdio.h>#include <omp.h>int main() {int i;#pragma omp parallel forfor (i = 0; i < 10; i++) {printf("Thread d: d\n", omp_get_thread_num(), i);}return 0;}在上面的例子中,`#pragma omp parallel for`指令会告诉编译器将for 循环并行化。
`omp_get_thread_num()`函数可以获取当前线程的编号。
第三步:数据共享与私有变量在并行编程中,多个线程可能会同时访问和修改共享的数据。
为了避免数据竞争和不一致的结果,我们需要显式地指定哪些变量是共享的,哪些变量是私有的。
我们可以使用`shared`和`private`子句来指定。
`shared`子句指定某个变量为共享变量,对所有线程可见。
OpenMP

OpenMP是一种针对共享内存的多线程编程技术,由一些具有国际影响力的大规模软件和硬件厂商共同定义的标准.它是一种编译指导语句,指导多线程、共享内存并行的应用程序编程接口(API)OpenMP是一种面向共享内存以及分布式共享内存的多处理器多线程并行编程语言,OpenMP是一种能被用于显示指导多线程、共享内存并行的应用程序编程接口.其规范由SGI发起.OpenMP具有良好的可移植性,支持多种编程语言.OpenMP能够支持多种平台,包括大多数的类UNIX以及WindowsNT系统.OpenMP最初是为了共享内存多处理的系统结构设计的并行编程方法,与通过消息传递进行并行编程的模型有很大的区别.因为这是用来处理多处理器共享一个内存设备这样的情况的.多个处理器在访问内存的时候使用的是相同的内存编址空间.SMP是一种共享内存的体系结构,同时分布式共享内存的系统也属于共享内存多处理器结构,分布式共享内存将多机的内存资源通过虚拟化的方式形成一个同意的内存空间提供给多个机子上的处理器使用,OpenMP对这样的机器也提供了一定的支持.OpenMP的编程模型以线程为基础,通过编译指导语句来显示地指导并行化,为编程人员提供了对并行化的完整控制.这里引入了一种新的语句来进行程序上的编写和设计.OpenMP的执行模型采用Fork-Join的形式,Fork-Join执行模式在开始执行的时候,只有一个叫做“主线程“的运行线程存在.主线程在运行过程中,当遇到需要进行并行计算的时候,派生出线程来执行并行人物,在并行执行的时候,主线程和派生线程共同工作,在并行代码结束后,派生线程退出或者是挂起,不再工作,控制流程回到单独的主线程中。
OpenMP的功能由两种形式提供:编译指导语句和运行时库函数,并通过环境变量的方式灵活控制程序的运行.OpenMP和MPI是并行编程的两个手段,对比如下:∙OpenMP:线程级(并行粒度);共享存储;隐式(数据分配方式);可扩展性差;∙MPI:进程级;分布式存储;显式;可扩展性好。
基于MPI与OpenMP混合并行计算技术的研究

基于MPI与OpenMP混合并行计算技术的研究李苏平,刘羽,刘彦宇(桂林理工大学信息科学与工程学院,广西桂林541004)摘要:针对多核机群系统的硬件体系结构特点,提出了节点间MPI消息传递、节点内部OpenMP共享存储的混合并行编程技术。
该编程模型结合了两者的优点,更为有效地利用了多核机群的硬件资源。
建立了单层混合并行的Jacobi求对称矩阵特征值算法。
实验结果表明,与纯MPI算法相比,混合并行算法能够取得更好的加速比。
关键词:混合编程模型;多核机群;MPI; OpenMPTP312:A:1672-7800(2010)03-0050-031 MPI与OpenMP混合模型MPI( Message Passing Interface)是消息传递并行编程模型的代表和事实标准,可以轻松地支持分布存储和共享存储拓扑结构;OpenMP是为共享存储环境编写并行程序而设计的一个应用编程接口,是当前支持共享存储并行编程的工业标准。
在SMP机群系统中,混合编程模型已经有一些成功的应用,对于多核PC机群的混合编程模型研究才开始起步。
在多核PC 机群中,结合MPI与OpenMP技术,充分利用这两种编程模型的优点,在付出较小的开发代价基础上,尽可能获得较高的性能。
按照在MPI进程间消息传递方式和时机,即消息何时由哪个或哪些线程在MPI进程间传递进行分类,混合模型可以分为以下两种:(1)单层混合模型(Hybrid master- only)。
MPI调用发生在应用程序多线程并行区域之外,MPI实现进程间的通信由主线程执行。
该混合模型编程易于实现,即在基于MPI模型程序的关键计算部分加上OpenMP循环命令#pramgma omp par-allel- -即可。
(2)多层混合模型(Hybrid multiple)。
MPI调用可以发生在应用程序多线程并行区域内,进程间通信的可由程序任何区域内的任何一个或一些线程完成。
在该模型中,当某些线程进行通信时,其它的非通信线程同时进行计算,实现了通信与计算的并行执行,优化了进程间的通信阻塞问题。
Inference of Population Structure Using Multilocus Genotype Data

Copyright©2000by the Genetics Society of AmericaInference of Population Structure Using Multilocus Genotype DataJonathan K.Pritchard,Matthew Stephens and Peter DonnellyDepartment of Statistics,University of Oxford,Oxford OX13TG,United KingdomManuscript received September23,1999Accepted for publication February18,2000ABSTRACTWe describe a model-based clustering method for using multilocus genotype data to infer populationstructure and assign individuals to populations.We assume a model in which there are K populations(where K may be unknown),each of which is characterized by a set of allele frequencies at each locus.Individuals in the sample are assigned(probabilistically)to populations,or jointly to two or more popula-tions if their genotypes indicate that they are admixed.Our model does not assume a particular mutationprocess,and it can be applied to most of the commonly used genetic markers,provided that they are notclosely linked.Applications of our method include demonstrating the presence of population structure,assigning individuals to populations,studying hybrid zones,and identifying migrants and admixed individu-als.We show that the method can produce highly accurate assignments using modest numbers of loci—e.g.,seven microsatellite loci in an example using genotype data from an endangered bird species.The softwareused for this article is available from /فpritch/home.html.I N applications of population genetics,it is often use-populations based on these subjective criteria representsa natural assignment in genetic terms,and it would beful to classify individuals in a sample into popula-tions.In one scenario,the investigator begins with a useful to be able to confirm that subjective classifications sample of individuals and wants to say something aboutare consistent with genetic information and hence ap-the properties of populations.For example,in studies propriate for studying the questions of interest.Further, of human evolution,the population is often consideredthere are situations where one is interested in“cryptic”to be the unit of interest,and a great deal of work has population structure—i.e.,population structure that isdifficult to detect using visible characters,but may be focused on learning about the evolutionary relation-ships of modern populations(e.g.,Cavalli et al.1994).significant in genetic terms.For example,when associa-In a second scenario,the investigator begins with a settion mapping is used tofind disease genes,the presence of predefined populations and wishes to classify individ-of undetected population structure can lead to spurious uals of unknown origin.This type of problem arisesassociations and thus invalidate standard tests(Ewens in many contexts(reviewed by Davies et al.1999).A and Spielman1995).The problem of cryptic population standard approach involves sampling DNA from mem-structure also arises in the context of DNAfingerprint-bers of a number of potential source populations and ing for forensics,where it is important to assess thedegree of population structure to estimate the probabil-using these samples to estimate allele frequencies inity of false matches(Balding and Nichols1994,1995; each population at a series of unlinked ing theForeman et al.1997;Roeder et al.1998).estimated allele frequencies,it is then possible to com-Pritchard and Rosenberg(1999)considered how pute the likelihood that a given genotype originated ingenetic information might be used to detect the pres-each population.Individuals of unknown origin can beence of cryptic population structure in the association assigned to populations according to these likelihoodsmapping context.More generally,one would like to be Paetkau et al.1995;Rannala and Mountain1997).able to identify the actual subpopulations and assign In both situations described above,a crucialfirst stepindividuals(probabilistically)to these populations.In is to define a set of populations.The definition of popu-this article we use a Bayesian clustering approach to lations is typically subjective,based,for example,ontackle this problem.We assume a model in which there linguistic,cultural,or physical characters,as well as theare K populations(where K may be unknown),each of geographic location of sampled individuals.This subjec-which is characterized by a set of allele frequencies at tive approach is usually a sensible way of incorporatingeach locus.Our method attempts to assign individuals diverse types of information.However,it may be difficultto populations on the basis of their genotypes,while to know whether a given assignment of individuals tosimultaneously estimating population allele frequen-cies.The method can be applied to various types ofmarkers[e.g.,microsatellites,restriction fragment Corresponding author:Jonathan Pritchard,Department of Statistics,length polymorphisms(RFLPs),or single nucleotide University of Oxford,1S.Parks Rd.,Oxford OX13TG,United King-dom.E-mail:pritch@ polymorphisms(SNPs)],but it assumes that the marker Genetics155:945–959(June2000)946J.K.Pritchard,M.Stephens and P.Donnellyloci are unlinked and at linkage equilibrium with one observations from each cluster are random draws another within populations.It also assumes Hardy-Wein-from some parametric model.Inference for the pa-berg equilibrium within populations.(We discuss these rameters corresponding to each cluster is then done assumptions further in background on clusteringjointly with inference for the cluster membership of methods and the discussion.)each individual,using standard statistical methods Our approach is reminiscent of that taken by Smouse(for example,maximum-likelihood or Bayesian et al.(1990),who used the EM algorithm to learn about methods).the contribution of different breeding populations to aDistance-based methods are usually easy to apply and sample of salmon collected in the open ocean.It is alsoare often visually appealing.In the genetics literature,it closely related to the methods of Foreman et al.(1997)has been common to adapt distance-based phylogenetic and Roeder et al.(1998),who were concerned withalgorithms,such as neighbor-joining,to clustering estimating the degree of cryptic population structuremultilocus genotype data(e.g.,Bowcock et al.1994). to assess the probability of obtaining a false match atHowever,these methods suffer from many disadvan-DNAfingerprint loci.Consequently they focused ontages:the clusters identified may be heavily dependent estimating the amount of genetic differentiation amongon both the distance measure and graphical representa-the unobserved populations.In contrast,our primarytion chosen;it is difficult to assess how confident we interest lies in the assignment of individuals to popula-should be that the clusters obtained in this way are tions.Our approach also differs in that it allows for themeaningful;and it is difficult to incorporate additional presence of admixed individuals in the sample,whoseinformation such as the geographic sampling locations genetic makeup is drawn from more than one of the Kof individuals.Distance-based methods are thus more populations.suited to exploratory data analysis than tofine statistical In the next section we provide a brief descriptioninference,and we have chosen to take a model-based of clustering methods in general and describe someapproach here.advantages of the model-based approach we take.TheThefirst challenge when applying model-based meth-details of the models and algorithms used are given inods is to specify a suitable model for observations from models and methods.We illustrate our method witheach cluster.To make our discussion more concrete we several examples in applications to data:both onintroduce very briefly some of our model and notation simulated data and on sets of genotype data from anhere;a fuller treatment is given later.Assume that each endangered bird species and from humans.incorpo-cluster(population)is modeled by a characteristic set rating population information describes how ourof allele frequencies.Let X denote the genotypes of the method can be extended to incorporate geographicsampled individuals,Z denote the(unknown)popula-information into the inference process.This may betions of origin of the individuals,and P denote the useful for testing whether particular individuals are mi-(unknown)allele frequencies in all populations.(Note grants or to assist in classifying individuals of unknownthat X,Z,and P actually represent multidimensional origin(as in Rannala and Mountain1997,for exam-vectors.)Our main modeling assumptions are Hardy-ple).Background on the computational methods usedWeinberg equilibrium within populations and complete in this article is provided in the appendix.linkage equilibrium between loci within populations.Under these assumptions each allele at each locus ineach genotype is an independent draw from the appro-BACKGROUND ON CLUSTERING METHODSpriate frequency distribution,and this completely speci-Consider a situation where we have genetic data fromfies the probability distribution Pr(X|Z,P)(given later a sample of individuals,each of whom is assumed toin Equation2).Loosely speaking,the idea here is that have originated from a single unknown population(nothe model accounts for the presence of Hardy-Weinberg admixture).Suppose we wish to cluster together individ-or linkage disequilibrium by introducing population uals who are genetically similar,identify distinct clusters,structure and attempts tofind population groupings and perhaps see how these clusters relate to geographi-that(as far as possible)are not in disequilibrium.While cal or phenotypic data on the individuals.There areinference may depend heavily on these modeling as-broadly two types of clustering methods we might use:sumptions,we feel that it is easier to assess the validityof explicit modeling assumptions than to compare the 1.Distance-based methods.These proceed by calculatingrelative merits of more abstract quantities such as dis-a pairwise distance matrix,whose entries give thetance measures and graphical representations.In situa-distance(suitably defined)between every pair of in-tions where these assumptions are deemed unreason-dividuals.This matrix may then be represented usingable then alternative models should be built.some convenient graphical representation(such as aHaving specified our model,we must decide how to tree or a multidimensional scaling plot)and clustersperform inference for the quantities of interest(Z and may be identified by eye.2.Model-based methods.These proceed by assuming that P).Here,we have chosen to adopt a Bayesian approach,947Inferring Population Structureby specifying models(priors)Pr(Z)and Pr(P),for both Assume that before observing the genotypes we haveZ and P.The Bayesian approach provides a coherent no information about the population of origin of eachframework for incorporating the inherent uncertainty individual and that the probability that individual i origi-of parameter estimates into the inference procedure nated in population k is the same for all k,and for evaluating the strength of evidence for the in-Pr(z(i)ϭk)ϭ1/K,(3) ferred clustering.It also eases the incorporation of vari-ous sorts of prior information that may be available,independently for all individuals.(In cases where somesuch as information about the geographic sampling lo-populations may be more heavily represented in thecation of individuals.sample than others,this assumption is inappropriate;itHaving observed the genotypes,X,our knowledge would be straightforward to extend our model to dealabout Z and P is then given by the posterior distribution with such situations.)We follow the suggestion of Balding and Nichols Pr(Z,P|X)ϰPr(Z)Pr(P)Pr(X|Z,P).(1)(1995)(see also Foreman et al.1997and Rannala While it is not usually possible to compute this distribu-and Mountain1997)in using the Dirichlet distri-tion exactly,it is possible to obtain an approximate bution to model the allele frequencies at each locus sample(Z(1),P(1)),(Z(2),P(2)),...,(Z(M),P(M))from Pr(Z,within each population.The Dirichlet distributionP|X)using Markov chain Monte Carlo(MCMC)meth-D(1,2,...,J)is a distribution on allele frequenciesods described below(see Gilks et al.1996b,for more pϭ(p1,p2,...,p J)with the property that these frequen-general background).Inference for Z and P may then cies sum to1.We use this distribution to specify the be based on summary statistics obtained from this sam-probability of a particular set of allele frequencies pkl·ple(see Inference for Z,P,and Q below).A brief introduc-for population k at locus l,tion to MCMC methods and Gibbs sampling may befound in the appendix.pkl·فD(1,2,...,J l),(4)independently for each k,l.The expected frequency of MODELS AND METHODS allele j is proportional toj,and the variance of thisfrequency decreases as the sum of thej increases.We We now provide a more detailed description of ourtake1ϭ2ϭ···ϭJ lϭ1.0,which gives a uniform modeling assumptions and the algorithms used to per-distribution on the allele frequencies;alternatives are form inference,beginning with the simpler case wherediscussed in the discussion.each individual is assumed to have originated in a singleMCMC algorithm(without admixture):Equations2, population(no admixture).3,and4define the quantities Pr(X|Z,P),Pr(Z),and The model without admixture:Suppose we genotypePr(P),respectively.By settingϭ(1,2)ϭ(Z,P)and N diploid individuals at L loci.In the case without admix-letting(Z,P)ϭPr(Z,P|X)we can use the approach ture,each individual is assumed to originate in one ofoutlined in Algorithm A1to construct a Markov chain K populations,each with its own characteristic set ofwith stationary distribution Pr(Z,P|X)as follows: allele frequencies.Let the vector X denote the observedAlgorithm1:Starting with initial values Z(0)for Z(by genotypes,Z the(unknown)populations of origin ofdrawing Z(0)at random using(3)for example),iterate the the individuals,and P the(unknown)allele frequenciesfollowing steps for mϭ1,2,....in the populations.These vectors consist of the follow-ing elements,Step1.Sample P(m)from Pr(P|X,Z(mϪ1)).(x(i,1)l,x(i,2)l)ϭgenotype of the i th individual at the l th locus,Step2.Sample Z(m)from Pr(Z|X,P(m)).where iϭ1,2,...,N and lϭ1,2,...,L;z(i)ϭpopulation from which individual i originated;Informally,step1corresponds to estimating the allele p kljϭfrequency of allele j at locus l in population k,frequencies for each population assuming that the pop-where kϭ1,2,...,K and jϭ1,2,...,J l,ulation of origin of each individual is known;step2 where J l is the number of distinct alleles observed at corresponds to estimating the population of origin of locus l,and these alleles are labeled1,2,...,J l.each individual,assuming that the population allele fre-Given the population of origin of each individual,quencies are known.For sufficiently large m and c,(Z(m), the genotypes are assumed to be generated by drawing P(m)),(Z(mϩc),P(mϩc)),(Z(mϩ2c),P(mϩ2c)),...will be approxi-alleles independently from the appropriate population mately independent random samples from Pr(Z,P|X). frequency distributions,The distributions required to perform each step aregiven in the appendix.Pr(x(i,a)lϭj|Z,P)ϭp z(i)lj(2)The model with admixture:We now expand ourmodel to allow for admixed individuals by introducing independently for each x(i,a)l.(Note that p z(i)lj is the fre-a vector Q to denote the admixture proportions for each quency of allele j at locus l in the population of originof individual i.)individual.The elements of Q are948J.K.Pritchard,M.Stephens and P.Donnellyq (i )k ϭproportion of individual i ’s genome thattion of origin of each allele copy in each individual isknown;step 2corresponds to estimating the population originated from population k.of origin of each allele copy,assuming that the popula-It is also necessary to modify the vector Z to replace the tion allele frequencies and the admixture proportions assumption that each individual i originated in some are known.As before,for sufficiently large m and c ,unknown population z (i )with the assumption that each (Z (m ),P (m ),Q (m )),(Z (m ϩc ),P (m ϩc ),Q (m ϩc )),(Z (m ϩ2c ),P (m ϩ2c ),observed allele copy x (i ,a )l originated in some unknown Q (m ϩ2c )),...will be approximately independent random population z (i ,a )l :samples from Pr(Z ,P ,Q |X ).The distributions required to perform each step are given in the appendix.z (i ,a )l ϭpopulation of origin of allele copy x (i ,a )l .Inference:Inference for Z,P,and Q:We now discuss how We use the term “allele copy”to refer to an allele carried the MCMC output can be used to perform inference on at a particular locus by a particular individual.Z ,P ,and Q.For simplicity,we focus our attention on Q ;Our primary interest now lies in estimating Q.We inference for Z or P is similar.proceed in a manner similar to the case without admix-Having obtained a sample Q (1),...,Q (M )(using suitably ture,beginning by specifying a probability model for large burn-in m and thinning interval c )from the poste-(X ,Z ,P ,Q ).Analogues of (2)and (3)arerior distribution of Q ϭ(q 1,...,q N )given X using the MCMC method,it is desirable to summarize the Pr(x (i ,a )l ϭj |Z ,P ,Q )ϭp z (i ,a )l lj(5)information contained,perhaps by a point estimate of andQ.A seemingly obvious estimate is the posterior meanPr(z (i ,a )l ϭk |P ,Q )ϭq (i )k ,(6)E (q i |X )≈1M ͚M m ϭ1q (m )i .(8)with (4)being used to model P as before.To complete our model we need to specify a distribution for Q ,which However,the symmetry of our model implies that the in general will depend on the type and amount of admix-posterior mean of q i is (1/K ,1/K ,...,1/K )for all i ,ture we expect to see.Here we model the admixturewhatever the value of X.For example,suppose that there proportions q (i )ϭ(q (i )1,...,q (i )K )of individual i using are just two populations and 10individuals and that the the Dirichlet distributiongenotypes of these individuals contain strong informa-tion that the first 5are in one population and the second q (i )فD (␣,␣,...,␣)(7)5are in the other population.Then eitherindependently for each individual.For large values of ␣(ӷ1),this models each individual as having allele q 1...q 5≈(1,0)and q 6...q 10≈(0,1)(9)copies originating from all K populations in equal pro-orportions.For very small values of ␣(Ӷ1),it models each individual as originating mostly from a single popu-q 1...q 5≈(0,1)and q 6...q 10≈(1,0),(10)lation,with each population being equally likely.As with these two “symmetric modes”being equally likely,␣→0this model becomes the same as our model leading to the expectation of any given q i being (0.5,without admixture (although the implementation of the 0.5).This is essentially a problem of nonidentifiability MCMC algorithm is somewhat different).We allow ␣caused by the symmetry of the model [see Stephens to range from 0.0to 10.0and attempt to learn about ␣(2000b)for more discussion].from the data (specifically we put a uniform prior on In general,if there are K populations then there will ␣[0,10]and use a Metropolis-Hastings update step be K !sets of symmetric modes.Typically,MCMC to integrate out our uncertainty in ␣).This model may schemes find it rather difficult to move between such be considered suitable for situations where little is modes,and the algorithms we describe will usually ex-known about admixture;alternatives are discussed in plore only one of the symmetric modes,even when run the discussion.for a very large number of iterations.Fortunately this MCMC algorithm (with admixture):The following does not bother us greatly,since from the point of algorithm may be used to sample from Pr(Z ,P ,Q |X ).view of clustering all the symmetric modes are the same Algorithm 2:Starting with initial values Z (0)for Z (by drawing Z (0)at random using (3)for example),iterate the [compare the clusterings corresponding to (9)and following steps for m ϭ1,2,....(10)].If our sampler explores only one symmetric mode then the sample means (8)will be very poor estimates Step 1.Sample P (m ),Q (m )from Pr(P ,Q |X ,Z (m Ϫ1)).of the posterior means for the q i ,but will be much better Step 2.Sample Z (m )from Pr(Z |X ,P (m ),Q (m )).estimates of the modes of the q i ,which in this case turn Step 3.Update ␣using a Metropolis-Hastings step.out to be a much better summary of the information in the data.Ironically then,the poor mixing of the Informally,step 1corresponds to estimating the allele MCMC sampler between the symmetric modes gives frequencies for each population and the admixture pro-portions of each individual,assuming that the popula-the asymptotically useless estimator (8)some practical949Inferring Population Structure value.Where the MCMC sampler succeeds in moving Simulated data:To test the performance of the clus-tering method in cases where the “answers”are known,between symmetric modes,or where it is desired to combine results from samples obtained using different we simulated data from three population models,using standard coalescent techniques (Hudson 1990).We as-starting points (which may involve combining results corresponding to different modes),more sophisticated sumed that sampled individuals were genotyped at a series of unlinked microsatellite loci.Data were simu-methods [such as those described by Stephens (2000b)]may be required.lated under the following models.Inference for the number of populations:The problem of Model 1:A single random-mating population of con-inferring the number of clusters,K ,present in a data stant size.set is notoriously difficult.In the Bayesian paradigm the Model 2:Two random-mating populations of constant way to proceed is theoretically straightforward:place a effective population size 2N.These were assumed to prior distribution on K and base inference for K on the have split from a single ancestral population,also of posterior distributionsize 2N at a time N generations in the past,with no subsequent migration.Pr(K |X )ϰPr(X |K )Pr(K ).(11)Model 3:Admixture of populations.Two discrete popu-However,this posterior distribution can be peculiarly lations of equal size,related as in model 2,were fused dependent on the modeling assumptions made,even to produce a single random-mating population.Sam-where the posterior distributions of other quantities (Q ,ples were collected after two generations of random Z ,and P ,say)are relatively robust to these assumptions.mating in the merged population.Thus,individuals Moreover,there are typically severe computational chal-have i grandparents from population 1,and 4Ϫi lenges in estimating Pr(X |K ).We therefore describe an grandparents from population 2with probability alternative approach,which is motivated by approximat-(4i )/16,where i {0,4}.All loci were simulated inde-ing (11)in an ad hoc and computationally convenient pendently.way.We present results from analyzing data sets simulated Arguments given in the appendix (Inference on K,the under each model.Data set 1was simulated under number of populations )suggest estimating Pr(X |K )usingmodel 1,with 5microsatellite loci.Data sets 2A and 2B Pr(X |K )≈exp(Ϫˆ/2Ϫˆ2/8),(12)were simulated under model 2,with 5and 15microsatel-lite loci,respectively.Data set 3was simulated under wheremodel 3,with 60loci (preliminary analyses with fewer loci showed this to be a much harder problem than ˆϭ1M ͚M m ϭ1Ϫ2log Pr(X |Z (m ),P (m ),Q (m ))(13)models 1and 2).Microsatellite mutation was modeled by a simple stepwise mutation process,with the mutation andparameter 4N set at 16.0per locus (i.e.,the expected variance in repeat scores within populations was 8.0).ˆ2ϭ1M ͚Mm ϭ1(Ϫ2log Pr(X |Z (m ),P (m ),Q (m ))Ϫˆ)2.We did not make use of the assumed mutation model in analyzing the simulated data.(14)Our analysis consists of two phases.First,we consider We use (12)to estimate Pr(X |K )for each K and substi-the issue of model choice—i.e.,how many populations tute these estimates into (11)to approximate the poste-are most appropriate for interpreting the data.Then,rior distribution Pr(K |X ).we examine the clustering of individuals for the inferred In fact,the assumptions underlying (12)are dubious number of populations.at best,and we do not claim (or believe)that our proce-Choice of K for simulated data:For each model,we dure provides a quantitatively accurate estimate of the ran a series of independent runs of the Gibbs sampler posterior distribution of K.We see it merely as an ad for each value of K (the number of populations)be-hoc guide to which models are most consistent with the tween 1and 5.The results presented are based on runs data,with the main justification being that it seems of 106iterations or more,following a burn-in period of to give sensible answers in practice (see next section for at least 30,000iterations.To choose the length of the examples).Notwithstanding this,for convenience we burn-in period,we printed out log(Pr(X |P (m ),Q (m ))),and continue to refer to “estimating”Pr(K |X )and Pr(X |K ).several other summary statistics during the course of a series of trial runs,to estimate how long it took to reach (approximate)stationarity.To check for possible prob-APPLICATIONS TO DATAlems with mixing,we compared the estimates of P (X |K )and other summary statistics obtained over several inde-We now illustrate the performance of our method on both simulated data and real data (from an endangered pendent runs of the Gibbs sampler,starting from differ-ent initial points.In general,substantial differences be-bird species and from humans).The analyses make use of the methods described in The model with admixture.tween runs can indicate that either the runs should950J.K.Pritchard,M.Stephens and P.DonnellyTABLE 1Estimated posterior probabilities of K ,for simulated data sets 1,2A,2B,and 3(denoted X 1,X 2A ,X 2B ,and X 3,respectively)K log P (K |X 1)P (K |X 2A )P (K |X 2B )P (K |X 3)1ف1.0ف0.0ف0.0ف0.02ف0.00.210.999ف1.03ف0.00.580.0009ف0.04ف0.00.21ف0.0ف0.05ف0.0ف0.0ف0.0ف0.0The numbers should be regarded as a rough guide to which models are consistent with the data,rather than accurate esti-mates of posterior probabilities.Figure 1.—Summary of the clustering results for simulated data sets 2A and 2B,respectively.For each individual,webe longer to obtain more accurate estimates or that computed the mean value of q (i )1(the proportion of ancestry independent runs are getting stuck in different modes in population 1),over a single run of the Gibbs sampler.Thein the parameter space.(Here,we consider the K !dashed line is a histogram of mean values of q (i )1for individuals from population 0;the solid line is for individuals from popula-modes that arise from the nonidentifiability of the K tion 1.populations to be equivalent,since they arise from per-muting the K population labels.)We found that in most cases we obtained consistent and Q estimating the number of grandparents from estimates of P (X |K )across independent runs.However,each of the two original populations,for each individual.when analyzing data set 2A with K ϭ3,the Gibbs sampler Intuitively it seems that another plausible clustering found two different modes.This data set actually con-would be with K ϭ5,individuals being assigned to tains two populations,and when K is set to 3,one of clusters according to how many grandparents they have the populations expands to fill two of the three clusters.from each population.In biological terms,the solution It is somewhat arbitrary which of the two populations with K ϭ2is more natural and is indeed the inferred expands to fill the extra cluster:this leads to two modes value of K for this data set using our ad hoc guide [the of slightly different heights.The Gibbs sampler did not estimated value of Pr(X |K )was higher for K ϭ5than manage to move between the two modes in any of our for K ϭ3,4,or 6,but much lower than for K ϭ2].runs.However,this raises an important point:the inferred In Table 1we report estimates of the posterior proba-value of K may not always have a clear biological inter-bilities of values of K ,assuming a uniform prior on K pretation (an issue that we return to in the discussion ).between 1and 5,obtained as described in Inference for Clustering of simulated data:Having considered the the number of populations.We repeat the warning given problem of estimating the number of populations,we there that these numbers should be regarded as rough now examine the performance of the clustering algo-guides to which models are consistent with the data,rithm in assigning particular individuals to the appro-rather than accurate estimates of the posterior probabil-priate populations.In the case where the populations ities.In the case where we found two modes (data set are discrete,the clustering performs very well (Figure 2A,K ϭ3),we present results based on the mode that 1),even with just 5loci (data set 2A),and essentially gave the higher estimate of Pr(X |K ).perfectly with 15loci (data set 2B).With all four simulated data sets we were able to The case with admixture (Figure 2)appears to be correctly infer whether or not there was population more difficult,even using many more loci.However,structure (K ϭ1for data set 1and K Ͼ1otherwise).the clustering algorithm did manage to identify the In the case of data set 2A,which consisted of just 5population structure appropriately and estimated the loci,there is not a clear estimate of K ,as the posterior ancestry of individuals with reasonable accuracy.Part probability is consistent with both the correct value,K ϭof the reason that this problem is difficult is that it is 2,and also with K ϭ3or 4.However,when the number hard to estimate the original allele frequencies (before of loci was increased to 15(data set 2B),virtually all of admixture)when almost all the individuals (7/8)are the posterior probability was on the correct number of admixed.A more fundamental problem is that it is diffi-populations,K ϭ2.cult to get accurate estimates of q (i )for particular individ-Data set 3was simulated under a more complicated uals because (as can be seen from the y -axis of Figure model,where most individuals have mixed ancestry.In 2)for any given individual,the variance of how many this case,the population was formed by admixture of two populations,so the “true”clustering is with K ϭ2,of its alleles are actually derived from each population。
基于OpenMP的分子动力学并行算法的性能分析与优化

基于OpenMP的分子动力学并行算法的性能分析与优化作者:白明泽程丽豆育升孙世新来源:《计算机应用》2012年第01期文章编号:1001-9081(2012)01-0163-04 doi:10.3724/SP.J.1087.2012.00163摘要:为提高分子动力学模拟在共享内存式服务器上的计算速度,对基于OpenMP的分子动力学并行算法(Critical方法)进行了性能分析与优化。
通过在多核服务器上的测试,以及加速比和并行效率的计算分析了Critical方法的并行性能,进而提出优化的三角形方法。
所提方法中每个线程所计算的粒子数固定,且粒子数目呈阶梯状上升,使得各线程能够错时到达临界区。
从而使程序在临界区的闲置时间比Critical方法减半,加速比明显提高。
关键词:分子动力学;并行计算;多核中央处理器;OpenMP;临界区中图分类号: TP399; O641 文献标志码:AAbstract: To enhance the computing speed of the molecular dynamics simulations on the shared memory servers, the performance of parallel molecular dynamics program based on Open Multi-Processing (OpenMP) approach with the critical section method was analyzed and improved. After testing performance on a multi-core server, as well as the calculations of speedup and parallel efficiency, an optimized triangle method was developed. In this method, stationary atom sets were assigned to threads respectively, and the number of atoms increased stepwise, which made the threads arrive at critical sections at different time. The triangle method can efficiently halve the idle time in critical sections and therefore can significantly enhance the parallel performance.Key words: molecular dynamics; parallel computing; multi-core Central Processing Unit (CPU); Open Multi-Processing (OpenMP); critical section0 引言分子动力学(Molecular Dynamics, MD)是一种应用广泛并可在粒子级别模拟固态、液态物质的主要计算方法之一。
基于OpenMP的Multi-Critical分子动力学并行算法优化

段 振华 ,白明泽 , 。 豆育升 。
(. 1 重庆 邮 电大学 高性能 计算 与应 用研 究所 ,重庆 4 0 6 ; .D p.o P yia S ine ,Nc o s t eU ies 0 0 5 2 e t f h s l cecs i l a nvri c h lS t . t,L 0 1 3 y A 7 3 0; .电子 科技 大 学 计算机 科 学与 工程 学院 ,成都 6 1 3 ) 17 1
Si c Tcn l yo C ia hn d 104,C ia c ne& e oo hn ,C eg u6 05 e h gf hn )
Ab t a t n o d rt r v h o u i gs e d o lc l r y a c i lt n o t — oe s ae mo s r e ,t i sr c :I r e i o et e c mp t p e fmoe u a n mi ss o mp n d mua i n mu i c r h r d me  ̄ e v r h s o l p p rp o o e l — r ia lo t m a e n e i i gp rl lag r h a e r p s d Mut C t l g r h b s d o xs n a al l o i m.T i ag r h u e n a iiin f re marx i i c a i t e t h s lo i m s d ma u ldv s oc t , t o i t k t p et r a se tr df r n rt a s cin wi i e e tn me n h t o ft e s p r o i o lc p i o ma e mu i l h e d n e i e e tci c l e t t d f r n a .a d t e me h d o h u ep s in b o k o t l i i o h i t . mie a al l l o t m ,i r v d t e p r l l f ce c .T e e p rme t l e u s s o h t o a e ot e p e iu r i zd p rl gr h ea i mp o e h aa l i i n y h x e i n a s h h w t a ,c mp r d t h r vo sc i — ee r t
基于OpenMP的压缩感知多描述并行处理算法

s e ns i n g,t h i s pa pe r p r o po s e d a mul t i p l e de s c r i p t i o n o f c o mp r e s s e d s e ns i n g p a r a l l e l p r o c e s s i ng a l g o r i t h m o n t h e b a s i s o f a c a r e f u l a n a l y s i s .I n o r d e r t o i mp r o v e t he s p e e d a n d t h e qu a l i t y o f r e c o n s t r u c t i o n i ma g e,di v i de d t he c o e ic f i e nt s a f t e r s pa r s e t r a ns f o r m
像 重建质 量 差的 问题 。 关 键词 :压缩 感知 ;多描 述 ;O p e n MP;并行 ;交 织抽 取 ;加速 比 中图分类号 j T P 3 0 1 . 6 ; T P 3 9 1 . 4; T N 9 1 1 . 7 3
d o i : 1 0 . 3 9 6 9 / j . i s s n . 1 0 0 1 - 3 6 9 5 . 2 0 1 3 . 0 4 . 0 8 7
基于avx与openmp的libsvm并行优化研究

大约在 300ms 左右,这对性能要求比较高的系统,
为解决支持向量机(SVM)分类算法性能不能满足实时性要求的问题,提出了一种使用 Intel 高级矢量扩展指令
集(AVX)对 SVM 分类算法进行并行加速的计算方法。首先以 LIBSVM 的串行版本作为算法优化的基准,分析了 LIBSVM 训
练阶段产生的模型文件读入内存后的布局特点,给出了满足 AVX 指令集操作的内存布局优化方案,接着在满足分类结果精
the optimized layout suitable for AVX is proposed. In order to improve the parallelism of the algorithm,the double float is replaced
by the single float based on the accuracy of classification results. Finally,the OpenMP technology is used to optimize the SIMD algo⁃
2504
田林琳等:基于 AVX 与 OpenMP 的 LIBSVM 并行优化研究
第 47 卷
维目标识别、人脸识别、文本图像分类等实际问题
的解决过程中都能看到 SVM 的影子[5]。
SVM 算法是典型的计算时间长的算法,它的主
要过程分为训练和分类。训练过程计算开销很大,
图1
因为参数优化算法本身需要若干次迭代。SVM 的
基金项目:国家自然科学基金项目(编号:61603262);辽宁省教育厅科学研究一般项目(编号:L2015380)资助。
作者简介:田林琳,女,硕士,副教授,研究方向:虚拟现实、并行计算、高性能计算。刘业峰,男,博士,副教授,研究方
基于OpenMP编程模型的多线程程序性能分析

基于OpenMP编程模型的多线程程序性能分析李梅【期刊名称】《电子设计工程》【年(卷),期】2014(22)23【摘要】并行化程序的出现大大提高了应用程序的执行效率,多核程序设计时需要对程序的性能进行考虑。
本文重点讨论OpenMP编程模型中多核多线程程序在并行化开销、负载均衡、线程同步开销方面对程序性能的影响。
%The emergence of the parallel program has greatly increased the application execution efficiency, multi-coreprogram design needs to consider the performance of the program. This article focuses on OpenMP multi-core multi-hread program in the programming model in parallelization overhead, load balancing, thread synchronization overhead aspects effect the performance of the program.【总页数】4页(P42-44,50)【作者】李梅【作者单位】西安欧亚学院陕西西安 710065【正文语种】中文【中图分类】TP311【相关文献】1.基于MPI+OpenMP混合编程模型的并行声纳信号处理技术研究 [J], 胡银丰;孔强2.基于OpenMP的电磁场FDTD并行程序性能分析 [J], 李正浩;周俊;刘大刚3.基于OpenMP/MPI并行编程模型的N体问题的优化实现 [J], 祝永志;续士强;禹继国4.基于 OpenMP 并行编程模型与性能优化的稀疏矩阵操作研究 [J], 蔡文海;陈洺均5.基于多核集群的MPI+OpenMP混合并行编程模型研究 [J], 谷克宏;黄岷;何江银因版权原因,仅展示原文概要,查看原文内容请购买。
美森电源分布块说明书

E P.M E R S E N.CO MPD 4D© 2019 Mersen. All rights reserved. Mersen reserves the right to change, update,or correct, without notice, any information contained in this datasheet.Mersen power distribution blocks provide a safe and easy method of splicing cables, splitting primary power into secondary circuits and fulfilling requirements for fixed junction tap-off points. Unless noted otherwise, all blocks are UL and CSA approved while meeting spacing requirements for feeder and branch circuits in conjunction with UL508A and the National Electrical Code®. PDB options include single or dual conductor primary inputs and up to 30 secondary outputs. Specialty blocks are available allowing for up to 7 primary inputs. The MPDB series is offered in three size categories: miniature (MPDB62 and MPDB63 series), intermediate (MPDB66 and MPDB67 series), and large (MPDB68 and MPDB69 series), in both aluminum and copper.E AT U R E S /B E N EF I T Adder Poles: All sizes have optional adder poles for increasedMPDB SeriesOpen-Style Power Distribution BlocksP OW E R D I S T R I B U T I O N B LO C K STHE NEXT GENERATIONPOWER DISTRIBUTION BLOCK (PDB)E P.M E R S E N.C O MPD 5P OW E R D I S T R I B U T I O N B LO C K S P DPA R T S E L E C T I O N N O T E SMPDBs in each size category come in one, two, and three pole configurations (ending in -1, -2, and -3 accordingly). Users also have the ability to field install additional poles, end barriers, and safety covers.Adder Pole Snap-on Adder poles to fully assembled units to add additional poles in the field. Adder pole catalog numbers in all.Adder Pole Field assemble Adder poles to form multi-pole units.Safety CoverOptional, snap-on, hinged safety coverMPDBC6263Miniature Series MPDBC6667Intermediate Series MPDBC6869Large SeriesEnd BarrierSnap-on to Adder pole to complete assemblyMPDBE6263Miniature Series MPDBE6667Intermediate Series MPDBE6869Large Series F E AT U R E S /B E N E F I T S (C O N T I N U E D ):•Insulators: Insulators are virtually unbreakable, made of glass-filled polycarbonate. “See-through,” hinged safety covers are optional and provide a greater degree of safety and shock resistance where required. Hinged covers can be installed without tools.• Spacings: 1 inch through air and 2 inches over surface between uninsulated live parts of opposite polarity meets requirements for feeder and branch circuit applications of UL508A.•Safety Covers: Polycarbonate safety covers provide dead-front protection. One cover is needed for each pole. Each cover has a test probe hole in the center for circuit checking. Covers are optional accessories and catalog numbers can be found in the catalog selection tables for each size block.A D D I T I O N A L S P E C I F I C AT I O N S :Wire Type: Copper Blocks: 60/75ºC Solid/Stranded CU; Aluminum Blocks: 60/75/90ºC Solid/Stranded AL and CUConnector:Copper Blocks: Highly conductive tin-plated copper; Aluminum Blocks: Highly conductive tin-plated aluminumInsulating Material: Glass-filled polycarbonate with verified dielectric strength in excess of 2500V Flammability: UL 94-V0Mounting: Direct panel mount Environmental:RoHS compliant, Lead FreeM P D BOpen-Style Power Distribution BlocksP OW E R D I S T R I B U T I O NB LOC K SPD C ATA L O G N U M BE R S,M I N I AT U R E A L U M I N U M M P D B s, B O X-B O X C O NF IG U R AT I O NC ATA L O G N U M B E R S,M I N I AT U R E A L U M I N U M M PD B s, B O X-S T U D C O N F I G U R AT I O NC ATA L O G N U M B E R S,M I N I AT U R E C O P P E R M PD B s,B O X-B O XC O N F I G U R AT I ONC ATA L O G N U M B E R S,M I N I AT U R E C O P P E R M PD B s,S T U D-S T U D C O N F I G U R AT I O NEnd Barrier for MPDB62 and MPDB63 series: Catalog Number MPDBE6263M P D B62A N D M P D B63Open-Style Power Distribution BlocksE P.M E R S E N.CO MPD 6E P.M E R S E N.C O MPD 7P OW E R D I S T R I B U T I O N B LO C K S PDC ATA L O G N U M B E R S , I N T E R M ED I ATE A L U M I N U M M P D B s , B O X -B O X C O NF IG U R AT I ON(M) Indicates connection UL approved for use with multiple conductors in the same opening. Quantities and sizes of wires are as follows:#2-#14 Openings (4) #14(4) #12(2) #104/0-#6 Openings (2) #2(2) #3(2) #4(2) #6200-#4 Openings (2) #4(2) #3(2) #2(2) #1(2) 1/0(2) #2/0(2) 3/0End Barrier for MPDB66 and MPDB67 series: Catalog Number MPDBE6667C ATA L O G N U M B E R S , I N T E R M ED I ATE A L U M I N U M M P D B s , B O X -S T U D C O NF IG U R AT I O NM P D B 66 A N D M P D B 67Open-Style Power Distribution BlocksP OW E R D I S T R I B U T I O NB LOC K SP D C ATA L O G N U M B E R S,I N T E R M E D I AT E C O P P E R M P D B s, B O X-B O X C O N F I G U R AT I O NC ATA L O G N U M B E R S,I N T E R M ED I ATE C O P P E R M P D B s, S T U D-S T U D C O NF IG U R AT I O NHinged Safety Cover for MPDB66 and MPDB67 series: Catalog number MPDBC6667End Barrier for MPDB66 and MPDB67 series: Catalog Number MPDBE6667M P D B66A N D M P D B67Open-Style Power Distribution BlocksE P.M E R S E N.CO MPD 8E P.M E R S E N.C O MPD 9P OW E R D I S T R I B U T I O N B LO C K S PD(DLO) Indicates Ampere Rating or Wire Range applicable to Copper DLO class wire(M) Indicates connection UL approved for use with multiple conductors in the same opening. Quantities and sizes of wires are as follows:#2-#14 Openings (4) #14(4) #12(2) #104/0-#6 Openings (2) #2(2) #3(2) #4(2) #6200-#4 Openings (2) #4(2) #3(2) #2(2) #1(2) 1/0(2) #2/0(2) 3/0C ATA L O G N U M B E R S , L A R G E A L U M I N U M M PD B s , B O X -B O X C O N F I G U R AT I ONC ATA L O G N U M B E R S , L A R G E A L U M I N U M M PD B s , B O X -S T U D C O N F I G U R AT I O NEnd Barrier for MPDB68 and MPDB69 series: Catalog Number MPDBE6869M P D B 68 A N D M P D B 69Open-Style Power Distribution BlocksP OW E R D I S T R I B U T I O NB LOC K SP D C ATA L O G N U M B E R S,L A R G E C O P P E R M P D B s, B O X-B O X C O N F I G U R AT I ONC ATA L O G N U M B E R S,L A R G E C O P P E R M PD B s, S T U D-B O X C O N F I G U R AT I O NHinged Safety Cover for MPDB68 and MPDB69 series: Catalog number MPDBC6869End Barrier for MPDB68 and MPDB69 series: Catalog Number MPDBE6869C ATA L O G N U M B E R S,L A R G E C O P P E R M PD B s, S T U D-S T U D C O N F I G U R AT I O NM P D B68A N D M P D B69Open-Style Power Distribution BlocksE P.M E R S E N.CO MPD 10E P.M E R S E N.C O MPD 11P OW E R D I S T R I B U T I O N B LO C K S P DD O U B LE W I D E A L U M I N U M C ATA L O G N U M B E R S , B O X -B O X C O NF IG U R AT I O NThe MPDB double-wide series are designed for custom applications where large ampacities are required. Double-wide blocks are not UL or CSA certified unless otherwise noted. All double-wide blocks are Mersen self-certifiedand approved.D O U B LE W I D E C O P P E R C ATA L O G N U M B E R S ,B O X -B O XC O N F I G U R AT I OND O U B LE W I D E C O P P E R C ATA L O G N U M B E R S , B O X -S T U D C O NF IG U R AT I ONM P D B D O U B L E -W I D EOpen-Style Power Distribution BlocksP OW E R D I S T R I B U T I O NB LOC K SP D D I M E N S I O N SMiniature (MPDB63133 shown for reference) Intermediate (MPDB67563 shown for reference) Large (MPDB69123 shown for reference)M P D BOpen-Style Power Distribution BlocksE P.M E R S E N.CO MPD 12E P.M E R S E N.C O MPD 13P OW E R D I S T R I B U T I O N B LO C K S P DD I ME N S I O N S (C O N T I N U E D )Double-Wide (MPDB69331 shown for reference)Triple-Wide (MPDB800061 shown for reference)M P D BOpen-Style Power Distribution Blocks。
关于Open MP:一个并行编程接口

关于Open MP:一个并行编程接口
王玉红;刘振中;任健
【期刊名称】《哈尔滨商业大学学报(自然科学版)》
【年(卷),期】2003(019)004
【摘要】OpenMP是一个公认的共享存储系统的并行编程接口.它由一些语言指导(directives)及库函数组成,并建立在Fortran 或者C、C++语言的基础上.优点是简单、通用,有利于快速开发并行程序.介绍了它的发展历史、执行模型以及它的三个组成部份, 即语言指导(一些在Fortran、C或C++基础上增加的注释语句)、运行库函数(共有10个与执行环境有关的运行库函数)和环境变量(设置该OpenMP 执行时所需的线程总数).文中还提供了已公开发布的OpenMP应用程序的情况, 并讨论了它将来的发展趋势.OpenMP的推广,还需要解决它的可扩展性问题.
【总页数】6页(P439-444)
【作者】王玉红;刘振中;任健
【作者单位】哈尔滨市投资专科学校,黑龙江,哈尔滨,150001;哈尔滨市环境检测中心站,黑龙江,哈尔滨,150076;黑龙江大学,计算机学院,黑龙江,哈尔滨,150086
【正文语种】中文
【中图分类】TP311
【相关文献】
1.基于OpenMP/MPI并行编程模型的N体问题的优化实现 [J], 祝永志;续士强;禹继国
2.MPI+OpenMP混合并行编程模型应用研究 [J], 冯云;周淑秋
3.MPX5100压力传感器直接与微处理器接口的一个测量系统 [J], 赵敏
4.MPI+OpenMP混合并行编程的分析 [J], 孙秋实;王移芝
5.基于多核集群的MPI+OpenMP混合并行编程模型研究 [J], 谷克宏;黄岷;何江银
因版权原因,仅展示原文概要,查看原文内容请购买。
基于共享内存的高效OpenMP并行多层快速多极子算法

基于共享内存的高效OpenMP并行多层快速多极子算法潘小敏;皮维超;盛新庆
【期刊名称】《北京理工大学学报》
【年(卷),期】2012(32)2
【摘要】提出并实现了一种基于共享内存并行平台的OpenMP并行多层快速多极子算法.结合OpenMP并行算法开发的要点和多层快速多极子算法数据分布的特性,对多层快速多极子的填充矩阵模块、矩阵向量相乘中的远相互作用部分进行了OpenMP并行化设计.在分析调度方式和循环次序对计算效率的影响的基础上,提出了一种高效的OpenMP并行多层快速多极子方案.数值实验表明,并行算法与串行精度一致,OpenMP并行算法具有较好的并行效率.
【总页数】6页(P164-169)
【关键词】多层快速多极子(MLFMA);并行;OpenMP;雷达散射截面积
【作者】潘小敏;皮维超;盛新庆
【作者单位】北京理工大学信息与电子学院电磁仿真中心
【正文语种】中文
【中图分类】TN454
【相关文献】
1.自适应多层快速多极子算法及其并行算法 [J], 袁军;邱扬;刘其中;郭景丽;谢拥军
2.一种基于矢量有限元与多层快速多极子技术的电磁散射快速并行算法 [J], 袁军;刘其中;郭景丽
3.一种多层快速多极子的高效并行方案 [J], 潘小敏;盛新庆
4.多层快速多极子算法并行实现的数据划分策略 [J], 胡悦;童维勤;龚治勋
5.并行多层快速多极子算法的最细层处理改进 [J], 刘战合;姬金祖;蒋胜矩;李洁因版权原因,仅展示原文概要,查看原文内容请购买。
基于OpenMP的湍流场中颗粒碰撞聚合的并行数值模拟

基于OpenMP的湍流场中颗粒碰撞聚合的并行数值模拟雷洪;赫冀成
【期刊名称】《东北大学学报(自然科学版)》
【年(卷),期】2009(030)011
【摘要】为了快速求解10 000个粒子的湍流碰撞聚合问题,采用OpenMP对Smoluchowki方程的FORTRAN求解程序进行了并行处理,数值结果表明:在不改变串行程序结构的情况下,仅对循环体部分进行并行处理,并行效率可高达80%,且串行程序与并行程序的计算结果完全吻合.对于大计算量循环体的并行计算,采用全部处理器进行并行计算时耗时最小.但是对于小计算量循环体的并行计算,采用全部处理器进行并行计算时耗时不一定最小.
【总页数】4页(P1602-1605)
【作者】雷洪;赫冀成
【作者单位】东北大学,材料电磁过程研究教育部重点实验室,辽宁,沈阳,110004;东北大学,材料电磁过程研究教育部重点实验室,辽宁,沈阳,110004
【正文语种】中文
【中图分类】TF701;TP319
【相关文献】
1.基于OpenMP的凝固数值模拟并行计算 [J], 黄江林;陈立亮
2.基于OpenMP的流化床颗粒堆积过程三维并行数值模拟 [J], 李斌;姚路;焦明月;周遵凯
3.钢液中Al2O3夹杂物颗粒布朗碰撞聚合的三维可视化数值模拟研究 [J], 王耀;李宏;郭洛方
4.OpenMP在CO2地质储存数值模拟并行计算中的应用 [J], 杨艳林;许天福;靖晶
5.潮汐环境中垂向湍射流流场的数值模拟 [J], 杨志峰;周雪漪;许协庆
因版权原因,仅展示原文概要,查看原文内容请购买。
编译器指导的OpenMP Fortran程序数据分布

编译器指导的OpenMP Fortran程序数据分布
周虎成;黄春;赵克佳
【期刊名称】《南京大学学报:自然科学版》
【年(卷),期】2005(41)5
【摘要】数据分布是提高分布存储系统上OpenMP程序性能的主要方法之一.基于两阶段分析方法,提出了一个面向OpenMP程序的自动数据分布框架及算法并实现其于CCRGOpenMP编译器之中.第一阶段,编译器分析程序中数据访问模式,结合OpenMP程序中DO指导命令提供的任务调度信息,为每次数组访问产生分布方式候选;第二阶段,采用多面体作为迭代空间及数组空间的几何模型,提出自动计算有界多面体中整数点个数以衡量通信量之多少的方法,并且用Ehrhart多项式表示其结果以更便于符号比较和最优分布方式的选取.实验表明,在最终选取的分布方式下,程序性能明显优于其他候选分布方式.
【总页数】7页(P562-568)
【关键词】OpenMP;数据分布;空间多面体;Ehrhart多项式
【作者】周虎成;黄春;赵克佳
【作者单位】国防科技大学计算机学院
【正文语种】中文
【中图分类】TP314
【相关文献】
1.OpenMP数据分布子句自动生成算法 [J], 黄品丰;赵荣彩;韩林;刘晓娴
2.OpenMP并行程序的编译器优化 [J], 张平;李清宝;赵荣彩
3.OpenMP Fortran程序中死锁的静态检测 [J], 王昭飞;黄春
4.纳格资讯推出最先进的Fortran编译器NAG Fortran Builder
5.2版 [J],
5.OpenMP Fortran程序中的未指定行为的静态检测 [J], 王昭飞;黄春;赵克佳因版权原因,仅展示原文概要,查看原文内容请购买。
openmp详解教程
Open Multi-Processing的缩写,是一个应用程序接口(API),可用于显式指导多线程、共享内存的并行性。
在项目程序已经完成好的情况下不需要大幅度的修改源代码,只需要加上专用的pragma来指明自己的意图,由此编译器可以自动将程序进行并行化,并在必要之处加入同步互斥以及通信。
当选择忽略这些pragma,或者编译器不支持OpenMp时,程序又可退化为通常的程序(一般为串行),代码仍然可以正常运作,只是不能利用多线程来加速程序执行。
OpenMP提供的这种对于并行描述的高层抽象降低了并行编程的难度和复杂度,这样程序员可以把更多的精力投入到并行算法本身,而非其具体实现细节。
对基于数据分集的多线程程序设计,OpenMP是一个很好的选择。
OpenMP支持的语言包括C/C++、Fortran;而支持OpenMP的编译器VS、gcc、clang等都行。
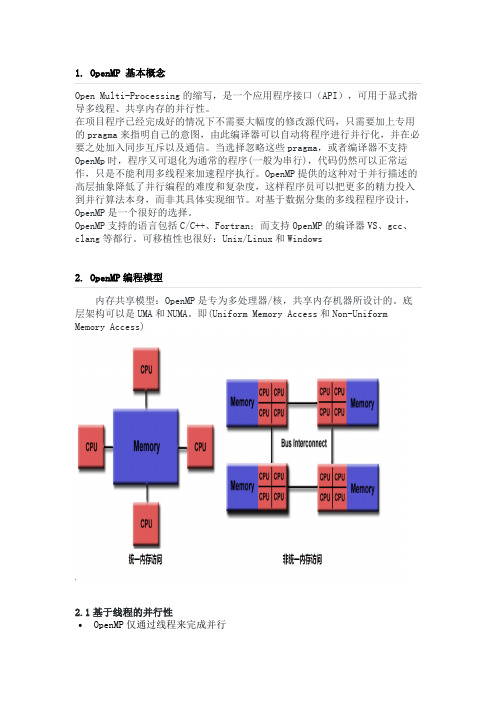
可移植性也很好:Unix/Linux和Windows内存共享模型:OpenMP是专为多处理器/核,共享内存机器所设计的。
底层架构可以是UMA和NUMA。
即(Uniform Memory Access和Non-Uniform Memory Access)2.1基于线程的并行性•OpenMP仅通过线程来完成并行•一个线程的运行是可由操作系统调用的最小处理单•线程们存在于单个进程的资源中,没有了这个进程,线程也不存在了•通常,线程数与机器的处理器/核数相匹配,然而,实际使用取决与应用程序2.2明确的并行•OpenMP是一种显式(非自动)编程模型,为程序员提供对并行化的完全控制•一方面,并行化可像执行串行程序和插入编译指令那样简单•另一方面,像插入子程序来设置多级并行、锁、甚至嵌套锁一样复杂2.3 Fork-Join模型•OpenMP就是采用Fork-Join模型•所有的OpenML程序都以一个单个进程——master thread开始,master threads按顺序执行知道遇到第一个并行区域•Fork:主线程创造一个并行线程组•Join:当线程组完成并行区域的语句时,它们同步、终止,仅留下主线程2.4 数据范围•由于OpenMP时是共享内存模型,默认情况下,在共享区域的大部分数据是被共享的•并行区域中的所有线程可以同时访问这个共享的数据•如果不需要默认的共享作用域,OpenMP为程序员提供一种“显示”指定数据作用域的方法2.5嵌套并行•API提供在其它并行区域放置并行区域•实际实现也可能不支持2.6动态线程•API为运行环境提供动态的改变用于执行并行区域的线程数•实际实现也可能不支持3.openmp使用需要使用openmp就需要引入omp.h库文件。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
M. S. Joshi
Department of Computer Engineering, Jawaharlal Nehru Engineering College, Aurangabad, MS, India madhuris.joshi@ P. D. Sheth Department of Information Technology, Walchand College of Engineering, Sangli, 416-416, India Email: pranalisheth@ Abstract—Dual Population Genetic Algorithm is an effective optimization algorithm that provides additional diversity to the main population. It deals with the premature convergence problem as well as the diversity problem associated with Genetic Algorithm. But dual population introduces additional search space that increases time required to find an optimal solution. This large scale search space problem can be easily solved using all available cores of current age multi-core processors. Experiments are conducted on the problem set of CEC 2006 constrained optimization problems. Results of Sequential DPGA and OpenMP DPGA are compared on the basis of accuracy and run time. OpenMP DPGA gives speed up in execution. Index Terms—Dual Population Genetic Algorithm (DPGA), Open Multiprocessing (OpenMP), Constrained Optimization Problems (COPs), High Performance Computing (HPC), Meta-heuristic Algorithms, Function Optimization. I. INTRODUCTION Dual Population Genetic algorithms (DPGA) is population based search algorithm that obtains solution to optimization problems. It is a diversity based algorithm that addresses diversity as well as premature convergence problems of Genetic Algorithm (GA). DPGA uses a reserve population along with the main population. This provides additional diversity to the main population. With the existence of two populations, execution time required for evolution is further increased. Recent advancement in High Performance Computing (HPC) includes programming for multi-core processors. These processors can be employed in parallel for faster evolution of the reserve as well as the main population. Information exchange between populations takes place through interpopulation crossbreeding [1]. In this paper, Maximum Constraints Satisfaction Method (MCSM) along with DPGA to solve Constrained Copyright © 2015 MECS II. LITERATURE REVIEW Dual Population Genetic Algorithm for solving COPs is an open research problem. Therefore we studied literature about evolution of DPGA. We have also surveyed how other Evolutionary Algorithms applied to solve COPs. Park and Ruy (2006) [4] introduced DPGA. It has two distinct populations with different evolutionary objectives: The Prospect (main) population works like population of the regular genetic algorithm which aims to optimize the objective function. The Preserver (reserve) population serves to maintain the diversity. Park and Ruy (2007) [5] proposed DPGA-ED that is an improved design-DPGA. Unlike DPGA, the reserve population of DPGA-ED evolves by itself. Park and Ruy (2007) [6] proposed a method to dynamically set the distance between the Optimization Problems (COPs) are used. MCSM is a novel technique which tries to satisfy maximum constrains first and then it tries to optimize an objective function. OpenMP is an API for writing shared-memory parallel applications in C, C++, and FORTRAN. With the release of API specifications in 1997 uses of multiple cores of multi-core CPU for parallel computing has become easy [2]. The basic execution unit of OpenMP is a thread. OpenMP is an implementation of multithreading. A master thread forks a specified number of slave threads and a task is distributed among them. The designers of OpenMP developed a set of compiler pragmas, directives, and environment variables, function calls which are platform-independent [3]. In application exactly where and how to insert threads are explicitly instructed by these constructs. Section II gives a brief literature review of DPGA and COPs. Section III describes the proposed algorithm DPGA for solving COPs using OpenMP. Section IV presents experimental results and discussion. Section V gives some conclusions and exhibits future scope.
I.J. Information Engineering and Electronic Business, 2015, 1, 59-65 Published Online January 2015 in MECS (/) DOI: 10.5815/ijieeb.2015.01.08
OpenMP Dual Population Genetic Algorithm for Solving Constrained Optimization Problems
A. J. Umbarkar
Department of Information Technology, Walchand College of Engineering, Sangli, 416-416, India Email: anantumbarkar@
populations using the distance be
