胆固醇代谢过程
胆固醇的代谢

胆固醇是一种含有27个碳原子并高度修饰的生物小分子。
它是脊椎动物细胞膜的重要成分,也是脂蛋白的组成成分。
胆固醇的衍生物胆酸盐、维生素D和类固醇激素在脂类消化中和动物的生长、发育过程中都具有重要的作用。
生物体内的胆固醇主要来源于两个方面,一方面是自身合成;另一方面是从外界摄入。
膳食中摄入的胆固醇被小肠吸收后,通过血液循环进入肝代谢。
当外源胆固醇摄入量增高时,可抑制肝内胆固醇的合成,所以在正常情况下体内胆固醇量维持动态平衡。
各种因素引起胆固醇代谢紊乱都可使血液中胆固醇水平增高,从而引起动脉粥样硬化,因此高胆固醇血症患者应注意控制膳食中胆固醇的摄入量。
除成年动物脑组织和成熟红细胞外,其他组织和细胞均可以合成胆固醇,其中肝是合成胆固醇的主要场所,机体内70%~80的胆固醇是由肝合成,其他如小肠、皮肤、肾上腺皮质、性腺和动脉血管壁均能合成少量胆固醇。
合成胆固醇的酶系存在于细胞液和滑面内质网膜上。
采用14C和13C标记乙酸的甲基碳及羧基碳,研究结果表明,乙酸分子中的2个碳原子都参与了胆固醇的合成。
其中有15个胆固醇中的碳原子来自乙酸的甲基,12个来自于乙酸的羧基。
合成胆固醇的原料是乙酰CoA,它可以经过“柠檬酸–丙酮酸循环”从线粒体转运至细胞液中。
由于乙酰CoA也可用于脂肪酸的合成,因此它是胆固醇和脂肪酸这两种脂类物质合成途径的分支点。
鲨烯(squalene)是胆固醇生物合成的中间代谢物,它由5个异戊二烯单位行成,胆固醇的合成可以归纳四个阶段:乙酰CoA3-甲基-35-二羟基戊酸异戊烯焦磷酸酯鲨烯胆固醇27C30C5C6C2C CoA3353–甲基–35–二羟戊酸(mevalonic acid,MVA)简称甲羟戊酸(mevalonate)。
由2分子乙酰CoA缩合成乙酰乙酰CoA,然后再与1分子乙酰CoA缩合成3–羟基–3–甲基戊二酸单酰CoA(HMG–CoA),该化合物是合成胆固醇和酮体的重要中间产物。
在线粒体中,HMG–CoA裂解后生成酮体;而在细胞液中,HMG–CoA则在内质网HMG–CoA还原酶催化下,由NADPH提供氢,还原形成甲羟戊酸。
胆固醇的合成与代谢途径

胆固醇的合成与代谢途径胆固醇是体内一种重要的脂质物质。
它的主要作用是构建细胞膜,制造激素和胆汁,同时还可能对心血管系统产生影响。
人体内胆固醇的合成和代谢途径复杂,下面让我们来了解一下。
一、胆固醇的合成胆固醇的合成主要发生在肝脏和小肠中,包括以下几个步骤:1、乙酰辅酶A的转化:在肝细胞中乙酰辅酶A首先转化为乙酰丙酮酸。
2、缬氨酸和丙酮酸的合成:乙酰丙酮酸接下来与缬氨酸结合,经过一系列催化反应后生成羟甲基戊二酸。
3、胆固醇的前体物质:羟甲基戊二酸接下来通过一系列催化反应分解成异戊二烯丙酰辅酶A,然后再转化为色氨酸,接着是一系列的反应后最终合成甲基戊二酸。
4、甲基戊二酸的转化:甲基戊二酸经过一系列的化学反应,最终合成出胆固醇。
这一过程主要需要受到3-羟基-3-甲基戊二酸的介导。
二、胆固醇的代谢胆固醇的代谢过程十分复杂。
从摄入膳食中的胆固醇开始,到最终经过多次转化变成胆酸和排泄,期间经历了以下几个步骤:1、膳食胆固醇的吸收:大部分的膳食胆固醇被小肠粘膜吸收,进入小肠细胞内部。
小部分的膳食胆固醇经过酯化反应后和其他脂质物质一起被吸收,形成胆固醇酯。
2、肝脏中胆固醇的代谢:膳食中的胆固醇在被吸收后,需要经过肝脏代谢后才能达到其他细胞。
肝脏将血液中的胆固醇提取出来,一部分被转化为胆汁酸,一部分是胆固醇酯储存在肝细胞里面。
3、胆汁中胆固醇的排泄:肝脏将胆汁酸和胆固醇酯合成胆汁排放进入肠道,一部分胆固醇被肠道吸收,剩下的胆固醇排泄出体外。
4、胆固醇的运输:胆固醇主要通过低密度脂蛋白和高密度脂蛋白进行运输,在血液中达到目的组织后,才能被细胞吸收利用。
总的来说,胆固醇的合成和代谢途径非常复杂,需要多种物质的参与。
人体内胆固醇合成与代谢的平衡关系对我们的健康有重要的影响,合理的饮食和生活习惯能够有效地影响胆固醇平衡关系。
胆固醇代谢

.
10
.
11
胆固醇合成的第二个阶段的反应
第三阶段:6个异戊二烯单位→鲨
烯
此阶段主要包括三步反应,首先在二甲
烯丙基转移酶的催化下,一个二甲烯丙
基与一个异戊二烯焦磷酸头尾缩合成牻
牛儿焦磷酸;随后在牻牛儿转移酶催化
下,牻牛儿焦磷酸与另一个异戊二烯焦
磷酸头尾缩合成法尼焦磷酸;最后,在
法尼转移酶或鲨烯合酶催化下,两个法
26
:)
HDL
合成与肝细胞和小肠 含有卵磷脂-胆固醇脂酰基转移酶(LCAT)
的激活剂 参与胆固醇的逆向运输 (好胆固醇) 通过受体介导的内吞被肝细胞吸收
.
27
胆固醇代谢的调节
胆固醇合成的限速酶是HMG-CoA还原酶 调节的方式包括
(1)“蛋白质的可逆磷酸化” (2)酶的降解 (3)酶基因的表达调控
.
28
“蛋白质的可逆磷酸化”调节.HMG-CoA还原酶的活性
29
HMG-CoA还原酶基因表达的调节机制
.
30
胆固醇的降解
环结构不能代谢 体内胆固醇的清除是将其转变成胆汁
酸,在分泌到小肠以后,部分随粪便 排出。
.
31
主要 成分
脂肪
VLD L
IDL
0.95~1.006
1.006~ 1.019
30~80 25~35
肝
脂肪
VLDL
脂肪,
ห้องสมุดไป่ตู้
部分降解 胆固醇
主要载 脂蛋白
A,B-48, CI,II,III, E B-100, C-I,II,III, E
B-100, E
主要功能
运输食物中的脂肪和 胆固醇 运输内源的脂肪
胆固醇代谢的生理学

胆固醇代谢的生理学胆固醇是人体中一种关键的脂类物质,它在人体内起着重要的生理功能。
然而,当胆固醇水平过高时,尤其是低密度脂蛋白胆固醇(LDL-C) 的增加,会成为心血管疾病的危险因素。
因此,了解胆固醇的代谢过程对于了解和预防与胆固醇相关的疾病至关重要。
一、胆固醇的来源1.内源性合成:胆固醇的70%-80%是由肝脏合成的。
在肝脏中,乙酰辅酶A (Acetyl-CoA) 被合成为胆固醇的起始物质。
经过一系列酶的催化作用,最终生成胆固醇。
2.外源性摄入:剩余的20%-30%胆固醇来自于我们食物的摄入。
胆固醇主要存在于富含动物脂肪的食物如肉类、乳制品和蛋黄中。
二、胆固醇代谢的过程胆固醇代谢主要包括吸收、转运、合成、转化和排泄五个主要环节。
1.吸收:胆固醇主要在小肠中被吸收。
在肠道内,胆固醇与胆汁酸结合形成胆固醇酯,通过肠道上皮细胞的摄取,与胆酸结合在微绒毛上,然后包裹在混合胆汁中的胆固醇颗粒中形成混合胆固醇颗粒。
2.转运:胆固醇在肠道内以混合胆固醇颗粒的形式被转运至肝脏。
在肝脏内,混合胆固醇颗粒与其他脂质结合成为低密度脂蛋白(LDL) 。
3.合成:在肝脏中合成的胆固醇主要以三酸甘油酯 (TG) 的形式储存,也可以通过转运脂蛋白将其分布到其他组织器官。
4.转化:胆固醇可以通过胆汁酸的合成转化为胆酸,以促进脂类的消化和吸收。
同时,胆固醇也可以经过脱饰反应转化为类固醇激素、维生素D和胆固醇醇醚等物质。
5.排泄:胆固醇的排泄主要通过肝脏和肠道进行。
肝脏将胆固醇排泄至胆汁中,然后通过肠道排出体外。
三、胆固醇调控的机制1.内源性调控:胆固醇自身可以通过负反馈机制调控合成,即当胆固醇水平升高时,它会抑制胆固醇合成酶的活性,从而减少胆固醇的合成。
2.外源性调控:体内的胆固醇水平也受到脂质摄入的影响。
当体内胆固醇水平不足时,肠道会通过增加胆固醇的吸收和合成来增加其供应量。
相反,当胆固醇水平过高时,肠道会降低对胆固醇的吸收量。
胆固醇代谢ce变为fc解毒原理

胆固醇代谢ce变为fc解毒原理1. 胆固醇代谢ce变为fc解毒原理的基础知识胆固醇是人体中一种重要的脂类物质,它在维持细胞膜的完整性、合成激素和胆汁酸等方面具有重要作用。
但当体内胆固醇浓度过高时,就会增加患心血管疾病的风险。
这时,身体会通过代谢ce变为fc的方式将多余的胆固醇转化为更容易排出体外的成分,以达到解毒的目的。
2. 胆固醇代谢过程中的转化机理胆固醇代谢ce变为fc的过程,主要是通过肝脏中的胆固醇酯酶参与转化。
胆固醇酯酶能够将胆固醇酯水解为游离的胆固醇和游离的脂肪酸。
而后,游离的胆固醇可以被转化为胆汁酸,经过胆汁酸排泄进而排出体外。
这一过程中,胆固醇代谢ce变为fc的转化机理是非常关键的,它能够有效地减少体内胆固醇的积累,从而保护心血管和其他重要器官的健康。
3. 对胆固醇代谢ce变为fc解毒原理的个人见解胆固醇代谢ce变为fc的解毒原理,是人体自身具有的一种重要的生理调节机制。
通过这一机制,人体能够有效地调控胆固醇的代谢,防止其在体内过量积累,从而减少心血管疾病等慢性病的风险。
对于我来说,深入了解胆固醇代谢ce变为fc的解毒原理,不仅有助于我更好地保护自己的健康,也能够帮助我更好地理解人体内部复杂的生化代谢过程。
总结回顾:通过本文的介绍,我们深入了解了胆固醇代谢ce变为fc的解毒原理。
我们了解了胆固醇代谢的转化机理,以及这一过程在维持身体健康方面的重要作用。
通过对这一主题的深入探讨,我们不仅能够增加对人体生理代谢的理解,也能更好地保护自己的健康。
在未来的生活中,我会继续关注这一领域的最新研究成果,以便不断更新对胆固醇代谢ce变为fc解毒原理的理解和应用。
以上就是一篇关于胆固醇代谢ce变为fc解毒原理的文章。
希望对你有所帮助。
生物化学领域的研究显示,胆固醇代谢ce变为fc的解毒原理,是人体内部非常重要的一种生理调节机制。
通过这一机制,人体能够有效地调控胆固醇的代谢,并防止其在体内过量积累,从而减少心血管疾病等慢性病的风险。
胆固醇代谢教学过程论文

胆固醇代谢的教学过程摘要:胆固醇代谢是生物化学课程教学重点内容之一,上好这节课具有非常重要的意义。
笔者通过教材分析、学情分析得出适合本节课的教学方法,并结合多媒体教学手段的使用,使本堂课达到一个很好的教学效果。
关键词:tc ldl【中图分类号】622 【文献标识码】a 【文章编号】胆固醇代谢是生物化学课程教学中脂类代谢的重要内容之一,而脂类代谢又是代谢生化的一个重点,所以讲好这节课具有非常重要的意义。
我首先针对所教学生特点来设计我的教学过程。
我所任课班级为中专护理一年级学生,他们理论基础较差,学习缺乏自信,不太喜欢枯燥的纯讲授法,但他们同时也具有思维活跃,敢于发表不同意见,活泼好动等优点。
因此对于这节课我采用案例分析结合多媒体演示以及讨论等多种教学方法相结合来讲解。
本节课一开始,我用多媒体课件向学生展示一张近期我校教职工体检时的异常血脂化验单,告诉学生单子上有两项tc(总胆固醇)、ldl(低密度脂蛋白胆固醇)结果明显升高,并指出,众所周知,胆固醇升高是动脉粥样硬化发生的高危因素,那么人体内胆固醇是从哪里来的,为什么会升高,怎样才能使升高的胆固醇下降呢?这些问题的提出就导入了我们要学习的内容—胆固醇代谢。
我通过向学生展示我校教职工体检单,目的是让学生感到自己熟悉的人出现异常,出于关注的心态,从而产生一种想探究其真相的心里。
这样的设计,让学生带着问题和兴趣去学习,达到一种“课伊始,趣已生”的效果。
紧接着我给学生展示今天的教学内容:胆固醇的概述、胆固醇的合成、胆固醇的转化。
并标注出今天学习的重点(合成部位、原料及限速酶)和难点(胆固醇合成的调节),引起学生注意,从而让他们学习的目的性更明确,注意力更集中。
在胆固醇概述中我首先介绍胆固醇名字的由来,指出因它最早是在动物胆石中提取出来的固体醇类化合物,故取名胆固醇。
这样讲目的是引起学生学习兴趣,引导他们顺着老师思路往下进行。
接着讲解胆固醇的生理功能和来源。
胆固醇代谢

精品课件
生理功能
➢ 是生物膜的重要成分,对控制生物膜的 流动性有重要作用;
➢ 是合成胆汁酸、类固醇激素及维生素D
等生理活性物质的前体。
精品课件
合成代谢
15个碳原子来自于乙酰CoA 的甲基, 12个碳原子来自于乙酰CoA 的羧基
精品课件
合成代谢
(二)合成原料
1
• 发生在细胞质,由四个阶段的反应组成
本
1. 内质网
特
2. 细胞液
点
• 起始物质是乙酰CoA (18个),共27个碳原子
• 共29步反应,四大步骤
• 单向不可逆反应
• 耗能反应,共消耗18个ATP和16个还原能力 (NAD精P品H课•件H+)
胆固醇的调节
• 限速酶——HMG-CoA还原酶(羟甲基戊二酸COA)
精品课件
thanks!
精品课件
➢ 酶的活性具有昼夜节律性 (午夜最高,中午最低) ➢ 可被磷酸化而失活,脱磷酸可恢复活性 ➢ 受胆固醇的反馈抑制作用 ➢ 胰岛素、甲状腺素能诱导肝HMG-COA还原酶的合成
精品课件
胆固醇的调节
• 饥饿与饱食 ➢ 饥饿与禁食可抑制肝合成胆固醇。 ➢ 摄取高糖、高饱和脂肪膳食后,胆固醇的合 成增加。
18乙酰CoA + 36ATP + 16(NADPH+H+)
葡萄糖有氧氧化
磷酸戊糖途径
精品课件
合成代谢
胆 固 醇 合 成 的 四 个 阶 段 反 应
精品课件
胆固醇合成精品的课件第一个阶段的反应
胆固醇合成的精第品二课件个阶段的反应
胆固醇合成的第精三品个课件阶段的反应
胆固醇合成精品的课第件 四个阶段的反应
胆固醇彻底分解产物 -回复

胆固醇彻底分解产物-回复胆固醇是一种脂质,它在人体中发挥着重要的生理功能,但高水平的胆固醇会增加心血管疾病的风险。
因此,了解如何彻底分解胆固醇成为降低心血管疾病风险的重要一步。
本文将一步一步回答有关胆固醇彻底分解产物的主题。
第一步:胆固醇的基本概念胆固醇是一种脂质,主要由肝脏合成,也可通过饮食摄入。
它在人体中发挥着结构和功能上的重要作用,是细胞膜的重要组成成分,同时也是合成类固醇激素和维生素D的前体。
然而,高水平的胆固醇会导致胆固醇在血液中沉积,形成动脉粥样硬化的根源,从而增加心血管疾病的风险。
第二步:胆固醇代谢的基本过程胆固醇在人体中的代谢过程大致分为三个主要步骤:摄取、吸收以及分解。
首先,通过饮食,人们摄取的胆固醇会进入肠道。
在肠道中,胆固醇会与胆汁中的溶解物结合,并通过胆盐的辅助帮助吸收。
这个过程中,一些细菌会将胆固醇转化成胆固醇代谢物,进一步影响其结构和特性。
然后,吸收的胆固醇会进入肝脏,部分用于再合成胆汁酸,而另一部分则在肝脏中处理。
肝脏中,胆固醇可以被进一步分解、合成或转运给其他器官以满足需要。
第三步:胆固醇分解产物在胆固醇的分解过程中,存在一些重要的产物。
其中,胆汁酸是胆固醇代谢的主要产物之一。
胆汁酸是由肝脏合成的,其作用是在肠道中帮助消化脂类,并促进胆固醇的排泄。
通过胆汁酸的合成和胆固醇的排泄,人体可以维持胆固醇平衡。
另外,胆固醇酯是另一个重要的胆固醇分解产物。
它是一种酯化合物,由胆固醇和脂肪酸合并而成。
胆固醇酯主要存在于人体脂肪组织中,并且在血液中以脂蛋白的形式存在。
胆固醇酯的形成和代谢受到多种因素的影响,包括饮食摄入的脂肪酸类型和数量,以及胆固醇酯酶的活性等。
第四步:胆固醇分解与心血管疾病的关系高水平的胆固醇与心血管疾病的发生密切相关。
当胆固醇摄入量超过人体需要,或者胆固醇代谢过程中出现异常,会导致胆固醇在血液中过量沉积。
这些过量的胆固醇可以沉积在动脉壁上,形成胆固醇斑块,最终导致动脉粥样硬化。
脂肪酸与胆固醇的合成与代谢

脂肪酸与胆固醇的合成与代谢脂肪酸与胆固醇是人体内重要的脂类物质,在维持人体健康和正常生理功能方面具有重要作用。
本文将探讨脂肪酸与胆固醇的合成与代谢过程,以及它们在人体内的功能和调节机制。
一、脂肪酸的合成与代谢1.1 脂肪酸的合成在人体内,脂肪酸的合成主要发生在肝脏和脂肪组织中。
脂肪酸的合成过程被称为脂肪酸合成途径,主要依赖于一系列酶的参与。
首先,由葡萄糖酶磷酸化酶(glucokinase)催化的葡萄糖经糖酵解途径产生的乙酰辅酶A(Acetyl-CoA)是脂肪酸合成的前体物质。
然后,乙酰辅酶A与二氢异戊酸(Malonyl-CoA)在脂肪酸合成酶复合物的催化下,通过一系列反应逐渐合成长链脂肪酸。
最后,脂肪酸通过脂肪酸合成酶的催化与甘油酯结合形成三酰甘油(Triglyceride),被储存在脂肪细胞中。
1.2 脂肪酸的代谢脂肪酸代谢包括脂肪酸的氧化和合成反应。
脂肪酸氧化是指将脂肪酸分解为乙酰辅酶A,进而通过TCA循环(三羧酸循环)产生能量。
脂肪酸氧化发生在线粒体内,经过一系列酶的催化作用,脂肪酸被解体为乙酰辅酶A分子,并进入TCA循环产生能量。
此外,脂肪酸还可通过β氧化作用进一步分解为辅酶A和乙酰辅酶A,提供更多的能量供给。
二、胆固醇的合成与代谢2.1 胆固醇的合成胆固醇合成主要发生在肝脏和肠道上皮细胞中。
胆固醇合成是复杂的生物合成过程,主要依赖于多个酶的催化。
胆固醇的合成途径包括两个主要过程:前体物质戊二酸(Acetyl-CoA)通过一系列反应转化为甲酰辅酶A(Methylmalonyl-CoA),然后合成胆固醇。
胆固醇的合成过程是一个调控严格的代谢途径,受到多个酶和转运蛋白的调控。
2.2 胆固醇的代谢胆固醇的代谢主要通过两条途径进行:胆固醇酯化和胆汁酸合成。
胆固醇酯化是指将胆固醇与脂肪酸结合形成胆固醇酯,以便在脂蛋白颗粒中进行转运。
胆固醇酯可以被转运到组织细胞中,也可以通过胆固醇酯酶的催化重新释放为游离胆固醇。
胆固醇代谢的转化

胆固醇代谢转化
1、转化1)胆固醇在肝中转化成胆汁酸是胆固醇在体内代谢的主要去路,基本步骤为:
胆酸胆固醇7α-羟化酶7α-羟胆固醇
甘氨酸或牛磺酸结合型胆汁酸
NADPH鹅脱氧胆酸
胆酸 肠道细菌 7-脱氧胆酸
甘氨酸牛磺酸 鹅脱氧胆酸 石胆酸
2)转化为类固醇激素胆固醇是肾上腺皮质、睾丸,卵巢等内分泌腺合成及分泌类固醇激素的原料,如睾丸酮、皮质醇、雄激素、雌二醇及孕酮等。
3)转化为7-脱氢胆固醇在皮肤,胆固醇可氧化为7-脱氢胆固醇,后者经紫外光照射转变为维生素D。
胆固醇的合成与调控

胆固醇的合成与调控胆固醇是一种重要的脂类化合物,广泛存在于人体细胞中。
它在机体内起着不可或缺的生理功能,并参与多种代谢途径。
然而,高胆固醇水平与心血管疾病之间存在紧密的联系,因此了解胆固醇的合成与调控机制显得尤为重要。
一、胆固醇的合成过程胆固醇的合成主要发生在肝脏和小肠上皮细胞中。
这个过程通常被称为内源性胆固醇合成,约占全身胆固醇的80%。
具体的合成过程可以分为几个关键步骤。
首先是醋酸与乙酰辅酶A的缩合,形成一种6碳酮体——羟甲戊二酮。
随后,羟甲戊二酮进一步转化为甲戊二酮,并通过一系列酶的作用逐步合成甾体内异构酶。
此过程中,乙酰辅酶A与洛尔氏反应的关键酶物质是HMG-CoA还原酶和HMG-CoA合酶。
在胆固醇的合成过程中,HMG-CoA还原酶与HMG-CoA合酶等酶发挥着重要的调控作用。
同时,还受到许多内外因素的影响,例如胰岛素、糖皮质激素和雌激素等。
二、胆固醇的调控机制胆固醇的合成和代谢是一个复杂的过程,其中存在许多调控机制,以维持胆固醇的正常水平。
主要的调控机制包括内源性和外源性的胆固醇调控。
内源性胆固醇调控主要通过转录因子SREBP(内质网膜蛋白SREBP)调控。
在细胞内,SREBP通过结合内质网上的SCAP (SREBP切割激活蛋白)而转移到高尔基体,并被S1P(SREBP切割蛋白)和S2P(SREBP切割激活蛋白)酶切割,释放出活性的SREBP。
活性的SREBP进入细胞核,并与基因组中调控胆固醇合成的基因启动子结合,从而促进胆固醇合成。
外源性胆固醇调控主要通过LDL受体介导。
人体通过食物摄入胆固醇,肠道细胞合成胆盐以帮助胆固醇吸收。
胆盐通过胆汁流向小肠,并与胆固醇形成混合胆汁。
混合胆汁经过肝脏重新吸收胆固醇,将其转运至体内需要的部位。
当体内胆固醇水平过高时,LDL受体的合成和表达会受到调控。
LDL受体可以识别血液中载脂蛋白LDL(低密度脂蛋白)上的胆固醇,并介导其从血液中清除。
因此,LDL受体的增加可以帮助降低体内胆固醇水平。
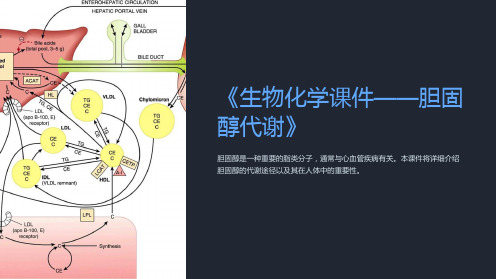
《生物化学课件-胆固醇代谢》
3 胆固醇吸收抑制剂
通过抑制肠道胆固醇的吸收,减少胆固醇水平。
结语
了解胆固醇的代谢过程对于维持健康非常重要。通过健康饮食和适度运动, 我们可以维持良好的胆固醇水平,促进身体的健康。
增加血浆胆固醇水平,提高罹患心血管疾病的风 险。
高水平的HDL有助于胆固醇从组织中转运到肝脏, 起到保护心血管系统的作用。
高水平的LDL胆固醇可导致动脉粥样硬化的发生。
胆固醇的药物干预
1 他汀类药物
通过抑制胆固醇合成酶,降 低胆固醇水平。
2 胆汁酸结合树脂
通过结合胆汁酸,促使胆固 醇排泄,减少胆固醇水平。
胆固醇的调节
肝脏与胆固醇代谢
肝脏是胆固醇代谢的主要器官, 通过调节胆固醇合成和吸收控制 其水平。
脂蛋白与胆固醇运输
脂蛋白参与胆固醇的运输,包括 LDL和HDL。
固醇调节元件结合蛋白
SREBPs是胆固醇代谢的主要调 节因子,有助于胆固醇的合成和 吸收的平衡。
高胆固醇与L) 低密度脂蛋白(LDL)
《生物化学课件——胆固 醇代谢》
胆固醇是一种重要的脂类分子,通常与心血管疾病有关。本课件将详细介绍 胆固醇的代谢途径以及其在人体中的重要性。
胆固醇的生物合成
1
异戊二烯酸(Acetyl-CoA)
胆固醇合成的起始物质。
2
戊二酰辅酶A(Mevalonate)
异戊二烯酸经过一系列酶催化反应转化而成。
3
脱羟胆甾酮(Cholestrol)
戊二酰辅酶A经过调控酶的作用转化为胆固醇,用于细胞膜构建及激素合成。
胆固醇的吸收和运输
肠道吸收
脂肪酸与胆盐结合形成的混合物通过肠道黏膜进 入体内。
冠状动脉内壁
胆固醇在血管内壁沉积可能导致动脉粥样硬化。
高中生物校本课程-胆固醇代谢

细胞定位:细胞质、光面内质网
(二)合成原料
1分子胆固醇 18乙酰CoA + 36ATP + 16(NADPH+H+)
葡萄糖有氧氧化
磷酸戊糖途径
乙酰CoA通过柠檬酸-丙酮酸循环出线粒体
(三)合成基本过程
线粒体中 乙酰CoA
原料
柠檬酸-丙 酮酸循环
入胞质
HMG-CoA
2.引起AS的脂蛋白
脂蛋白残粒
CM和VLDL经LPL水解生成CM残粒与IDL, 并转变成富含胆固醇酯和ApoE的颗粒沉积于血 管壁,经清道夫受体介导摄取进入巨噬细胞引起 AS的增强作用。
变性LDL
乙酰LDL 、 氧化LDL 、 糖化LDL 经修饰的LDL,如OX-LDL会激活巨噬细胞,使 巨噬细胞摄取乙酰LDL而转变成泡沫细胞,促进 AS形成。
HDL2在CETP介导下,与VLDL、LDL进行CE交换,同时也转 运TG,以VLDL、LDL形式经肝脏摄取,最终使末梢组织的FC输
送到肝脏。HDL主要功能是参与胆固醇逆转运(RCT) (胆固醇
逆转运)
三、动脉粥样硬化(AS)
1.概述
AS是指动脉内膜的脂质、血液成分的沉积,平滑肌 细胞及胶原纤维增生,伴有坏死及钙化等不同程度病变 的一类慢性进行性病理过程。AS主要损伤动脉内壁膜。
限速酶
HMG-CoA 还原酶
胆固醇酯
胆固醇
鲨烯
甲基羟戊酸 (MVA)
胆固醇合成的调节
限速酶
饱食 胰岛素 甲状腺素
HMG-CoA还原酶
胆固醇 饥饿禁食
胰高血糖素
➢酶的活性具有昼夜节律性 (午夜最高,中午最低)
胆固醇的生理作用和代谢途径

胆固醇的生理作用和代谢途径胆固醇是一种有机化合物,是人体内重要的基础物质之一,广泛存在于细胞膜、激素、维生素D等生物分子中。
在人们的膳食中,胆固醇的多寡是一个备受关注的问题。
一、胆固醇的生理意义胆固醇在人体内具有非常重要的生理作用。
它是细胞膜的重要组成部分之一,能够维持细胞膜的稳定性和透过性,对细胞的生长、代谢和信号传递起着至关重要的作用。
此外,胆固醇还是合成性激素、胆汁酸和维生素D等物质的前体,是维持正常生理功能的必需物质。
二、胆固醇的代谢途径胆固醇主要是由肝脏和肠道合成。
在肝脏中,胆固醇主要是通过血液中的脂蛋白转运到各个组织细胞中,并参与到细胞膜的合成和代谢中。
同时,肝脏也是胆固醇代谢和排泄的重要器官之一。
在肠道中,胆固醇主要是由膳食中的胆固醇和肝脏合成的胆汁酸组成,通过肝胆循环参与到脂质代谢中。
在肠道中,一部分胆固醇会被肠道内的细菌代谢,而另一部分则会被小肠黏膜上皮细胞摄取。
摄取的胆固醇随后会与脂蛋白一起转运到肝脏,然后进入胆汁中再次进入肠道参与到脂质代谢中。
三、胆固醇相关的代谢紊乱胆固醇在人体内的代谢存在一系列的调控机制,能够维持正常的代谢平衡。
然而,在人们长期高脂、高热量饮食、缺乏运动、肥胖等不良生活方式的影响下,胆固醇的代谢平衡可能会被打破,出现一系列代谢紊乱。
其中最常见的就是高胆固醇血症,即血液中胆固醇浓度过高的一种病理状态。
高胆固醇血症是导致冠心病、中风等心血管疾病的主要危险因素之一。
此外,胆固醇还存在着多种脂质代谢紊乱病症,如脂蛋白代谢紊乱等。
四、降低胆固醇的方法减少膳食中的胆固醇摄入、保持适量的运动、控制体重,是降低胆固醇的关键措施之一。
此外,膳食纤维、植物固醇等营养素也可以对降低胆固醇有一定的帮助。
至此,我们可以看出胆固醇在人类的健康中扮演了非常重要的角色,同时也需要注意,不良的生活方式可能会导致胆固醇的代谢紊乱,出现相关的健康问题。
生物化学第五节 胆固醇代谢

第五节胆固醇代谢2015-07-07 71752 0一、体内胆固醇来自食物和内源性合成胆固醇有游离胆固醇( free cholesterol,FC),亦称非酯化胆固醇(unesterified cholesterol)和胆固醇酯( cholesterol ester)两种形式,广泛分布于各组织,约1/4分布在脑及神经组织,约占脑组织20%。
肾上腺、卵巢等类固醇激素分泌腺,胆固醇含量达1% ~5%。
肝、肾、肠等内脏及皮肤、脂肪组织,胆固醇含量约为每100g组织200~500mg,以肝最多。
肌组织含量约为每100g组织100~200mg。
(一)体内胆固醇合成的主要场所是肝除成年动物脑组织及成熟红细胞外,几乎全身各组织均可合成胆固醇,每天合成量为lg左右。
肝是主要合成器官,占自身合成胆固醇的70%~80%,其次是小肠,合成10%。
胆固醇合成酶系存在于胞质及光面内质网膜。
(二)乙酰CoA和NADPH是胆固醇合成基本原料14C及13C标记乙酸甲基碳及羧基碳,与肝切片孵育证明,乙酸分子中的2个碳原子均参与构成胆固醇,是合成胆固醇唯一碳源。
乙酰CoA是葡萄糖、氨基酸及脂肪酸在线粒体的分解产物,不能通过线粒体内膜,需在线粒体内与草酰乙酸缩合生成柠檬酸,通过线粒体内膜载体进入胞质,裂解成乙酰CoA,作为胆固醇合成原料。
每转运1分子乙酰CoA,由柠檬酸裂解成乙酰CoA时消耗1分子ATP。
胆固醇合成还需NADPH供氢、ATP供能。
合成1分子胆固醇需18分子乙酰CoA、36分子AIP及16分子NADPH。
(三)胆固醇合成由以HMG-CoA还原酶为关键酶的一系列酶促反应完成胆固醇合成过程复杂,有近30步酶促反应,大致可划分为三个阶段(图7_9)。
1.由乙酰CoA合成甲羟戊酸2分子乙酰CoA在乙酰乙酰CoA硫解酶作用下,缩合成乙酰乙酰CoA;再在HMG-CoA合酶作用下,与1分子乙酰CoA缩合成HMG-CoA。
在线粒体中,HMG-CoA被裂解生成酮体;而胞质生成的HMG-CoA,则在内质网HMG-CoA还原酶(HMG-CoA reductase)作用下,由NADPH供氧,还原生成甲羟戊酸(mevalonic acid,MVA)。
胆固醇的代谢

胆固醇的代谢
胆固醇是生物结构和功能的重要组分,它对于动物细胞和内分泌有重要作用。
它还是胆囊分泌液的主要组成成分,参与脂质和脂肪的合成。
因此,胆固醇的代谢对于维持生命功能至关重要。
胆固醇的主要来源是动物摄食,不仅包括动物的肉类,还包括动物的内脏,以及人工制造的食物,如奶油和奶酪等。
除此之外,还有一些植物源的胆固醇,如橄榄油和大豆油等,这些也是胆固醇的主要来源。
胆固醇在人体内的代谢主要分为三个步骤:脂肪酸合成、胆固醇衍生物的合成和吸收。
1. 脂肪酸合成:胆固醇被酶酸化,分解为脂肪酸,其中包括较小的乙醇酸和较大的乙醛酸。
这些脂肪酸随着尿液排出体外,排出量占胆固醇总量的50%。
2. 胆固醇衍生物的合成:经过酶酸化后,胆固醇的衍生物形成,包括胆碱、胆酸、胆酐和胆汁酸。
这些物质可以用于脂质的合成和清除,同时有助于调节血液中胆固醇的含量。
3. 吸收:将摄入的胆固醇酯通过胆囊系统及小肠结肠系统吸收到血液中,其中约有20%被复合物结合,形成体外循环的胆固醇。
然后,胆固醇随着血液流动,到达肝脏,开始参与体内代谢。
胆固醇的代谢不仅是供给身体所必需的物质,同时也为身体排除有害物质提供了重要途径,因此胆固醇的代谢对于维持身体健康十分重要。
高密度脂蛋白胆固醇的代谢与心脑血管健康

高密度脂蛋白胆固醇的代谢与心脑血管健康一、引言心血管疾病是目前全球范围内主要的健康问题之一,而高密度脂蛋白(HDL)被广泛认为是维护心脑血管健康的保护因子之一。
本文将重点讨论高密度脂蛋白胆固醇(HDL-C)的代谢过程以及它们与心脑血管健康之间的关系。
二、HDL胆固醇代谢1. HDL合成和分泌HDL是由肝脏和小肠上皮细胞合成并分泌到循环系统中。
在肝脏中,Apolipoprotein A-I(ApoA-I)被合成,并与磷脂、游离胆固醇和其他辅助载体结合形成初始HDL颗粒。
2. 转运途径HDL通过循环系统转运到外周组织,负责收集体内过剩的游离胆固醇并带回肝脏进行排泄。
这个转运途径包括反式胆固醇转运和胆固醇酯转运。
在反式胆固醇转运过程中,HDL通过接触组织和细胞膜上的ATP结合盒A1 (ABCA1)来促进胆固醇从外周组织与细胞中释放出来并与HDl颗粒结合。
而在胆固醇酯转运过程中,HDL通过交换蛋白及硫化物转移酶等多种蛋白质调控,将游离胆固醇与肝脏合成的甘油三酯互相交换。
3. HDL代谢途径HDL颗粒可以通过多个途径代谢。
一种途径是被肝脏特异性受体识别并清除。
另外一种途径是被肠道内泡状半隐孢子球体(enterocytic chylomicron remnants)夹带入肝脏,并参与到动态平衡过程中。
三、HDL-C与心脑血管健康1. HDL-C的关联性研究高密度的脂蛋白具有明显的抗动脉粥样硬化作用,HDL-C水平则被广泛认为是心脑血管疾病的一个重要危险因子。
多项流行病学和临床研究表明,HDL-C对冠心病和中风等心脑血管事件具有显著的保护作用。
2. HDL-C与动脉粥样硬化HDL-C通过多种机制参与动脉粥样硬化的发生和进展过程。
首先,HDL-C可以促进胆固醇从动脉壁中去除,并促进其返回到肝脏进行代谢。
其次,HDL-C能够抑制胆固醇氧化、减少低密度脂蛋白(LDL)被氧化的程度,从而减轻血管内皮损伤。
此外,HDL-C可通过抗炎、抗凝血和抗氧化作用来改善心血管健康状态。
胆固醇代谢

乙酰CoA通过柠檬酸-丙酮酸循环出线粒体 (三)胆固醇合成由以HMG-CoA还原酶为关键酶的一系列酶促反应完成
1. 由乙酰CoA合成甲羟戊酸
合成胆固醇 的关键酶
• 甲羟戊酸经15碳化合物转变成30 碳鲨烯
• 鲨烯环化为羊毛固醇后转变为胆固醇
(四)胆固醇合成通过HMG-CoA还原酶调节
• 关键酶——HMG-CoA还原酶
(一)体内胆固醇合成的主要场所是肝 : 组织定位 除成年动物脑组织及成熟红细胞外,几乎全身各组织均可合成,以肝、小肠为主。 : 细胞定位 胞质、光面内质网膜
(二)乙酰CoA和NADPH是胆固醇合成基本原料
1分子胆固醇
18乙酰CoA + 36ATP + 16(NADPH+H+)
葡萄糖有氧氧化
磷酸戊糖途径
(三)胆固醇可转化为维生素D3的前体
7-脱氢胆固醇
Thanks
➢ 酶的活性具有昼夜节律性 (午夜最高,中午最低) ➢ 可被磷酸化而失活,脱磷酸可恢复活性 ➢ 受胆固醇的反馈抑制作用 ➢ 胰岛素、甲状腺素能诱导肝HMG-COA还原酶的合成
• 饥饿与饱食
➢ 饥饿与禁食可抑制肝合成胆固醇。 ➢ 摄取高糖、高饱和脂肪膳食后,胆固醇的合成增加。
• 胆固醇
➢ 胆固醇可反馈抑制肝胆固醇的合成。它主要抑制HMG-CoA还原 酶的合成。
• 激素
➢ 胰岛素及甲状腺素能诱导肝HMG-CoA还原酶的合成,从而增加胆固醇 的合成。
➢ 胰高血糖素及皮质醇则能抑制HMG-CoA还原酶的活性,因而减少胆固 醇的合成。
➢ 甲状腺素还促进胆固醇在肝转变为胆汁酸。
二、转化成胆汁酸是胆固醇的主要去路
胆固醇的母核——环戊烷多氢菲在体内不能被降解,但侧链可被氧化、还原或降解,实现 胆固醇的转化。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Fig. 2. Proposed pathway for cholesterol degradation under aerobic conditions. Cholest-4-en-3-one or any of the subsequent metabolites from degradation of the side-chain up to (and including) AD may undergo a dehydrogenation reaction to introduce a double bond in the position 1, leading to compound cholest-1,4-diene-3-one in the case of cholest-4-en-3-one, or to the corresponding 1,2-dehydro derivatives for other molecules. The side-chain degradation of this compounds will be identical to that of the cholest-4-en-3-one to the common intermediate 9a-hydroxyandrosta-1,4-diene-3,17-dione. The microorganisms from which enzymes implicated in different steps are indicated by numbers. Numbers in brackets are assigned arbitrarily to
facilitate the compound identification in
Fig. 3. Proposed b-oxidation-like reactions for cholesterol side-chain degradation. The Fad proteins have been assigned according to the nomenclature of the E. coli genes involved in the b-oxidation of fatty acids. LiuE is the name assigned to 3-hydroxy-3-methylglutaryl-coenzymelyases.
Fig. 4. Organization of the main gene clusters implied or suggested to be involved in the degradation of cholesterol in M. smegmatis mc2155.The identity number for each MSMEG gene is indicated within the arrows. The name of some genes of interest is written above them.
Numbers below genes indicate the number of bp between adjacent genes; numbers in brackets indicate separation and numbers in parentheses indicate overlap.
Numbers above diagonal lines indicate the genomic position in kb.
Orange: mce cluster. Green: genes suggested and/or proved to participate in the side-chain degradation. Blue: genes suggested and/or proved to participate in the central or lower catabolic pathway.
Yellow: genes coding the transcriptional repressors KstR and KstR2.
Genes surrounded by a dashed line are controlled by KstR2, the rest of the genes showed in this figure are controlled by KstR (except for MSMEG_5905, 5909, 5910, 5912, 5916, 5917, 5924, 5926, 5928, 5936, 5938, 6005, 6006, 6007, 6010, 6034,
which could not be proved to be controlled by any of both repressors)。
