1-s2.0-S1673852711001305-main
ASCII码表
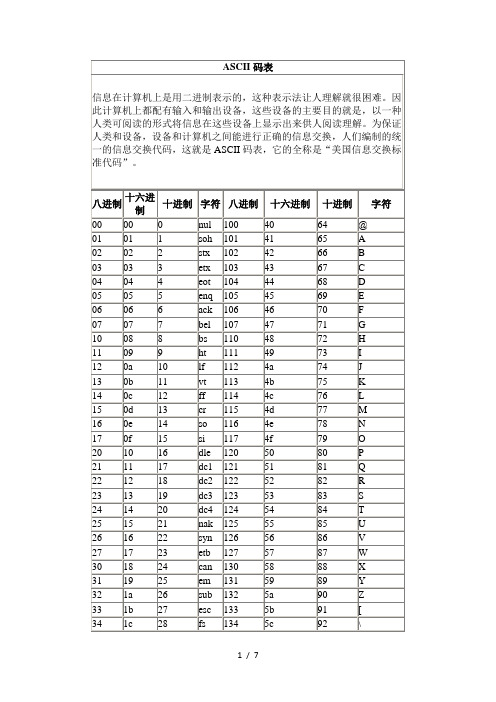
ASCII码对照表II2012-09-12 11:18:48分类:WINDOWS(ASCII = American Standard Code for Information Interchange)Decimal Octal Hex Binary Value------- ----- --- ------ -----000 000 000 00000000 NUL (Null char.)001 001 001 00000001 SOH (Start of Header)002 002 002 00000010 STX (Start of Text)003 003 003 00000011 ETX (End of Text)004 004 004 00000100 EOT (End of Transmission)005 005 005 00000101 ENQ (Enquiry)006 006 006 00000110 ACK (Acknowledgment)007 007 007 00000111 BEL (Bell)008 010 008 00001000 BS (Backspace)009 011 009 00001001 HT (Horizontal Tab)010 012 00A 00001010 LF (Line Feed)011 013 00B 00001011 VT (Vertical Tab)012 014 00C 00001100 FF (Form Feed)013 015 00D 00001101 CR (Carriage Return)014 016 00E 00001110 SO (Shift Out)015 017 00F 00001111 SI (Shift In)016 020 010 ******** DLE (Data Link Escape)017 021 011 00010001 DC1 (XON) (Device Control 1)018 022 012 00010010 DC2 (Device Control 2)019 023 013 00010011 DC3 (XOFF)(Device Control 3)020 024 014 00010100 DC4 (Device Control 4)021 025 015 00010101 NAK (Negative Acknowledgement)022 026 016 00010110 SYN (Synchronous Idle)023 027 017 00010111 ETB (End of Trans. Block)024 030 018 00011000 CAN (Cancel)025 031 019 00011001 EM (End of Medium)026 032 01A 00011010 SUB (Substitute)027 033 01B 00011011 ESC (Escape)028 034 01C 00011100 FS (File Separator)029 035 01D 00011101 GS (Group Separator)030 036 01E 00011110 RS (Request to Send)(Record Separator)031 037 01F 00011111 US (Unit Separator)032 040 020 ******** SP (Space)033 041 021 ******** ! (exclamation mark)034 042 022 ******** " (double quote)035 043 023 ******** # (number sign)036 044 024 ******** $ (dollar sign)037 045 025 ******** % (percent)038 046 026 00100110 & (ampersand)039 047 027 ******** ' (single quote)040 050 028 ******** ( (left/opening parenthesis)041 051 029 ******** ) (right/closing parenthesis)042 052 02A 00101010 * (asterisk)043 053 02B 00101011 + (plus)044 054 02C 00101100 , (comma)045 055 02D 00101101 - (minus or dash)046 056 02E 00101110 . (dot)047 057 02F 00101111 / (forward slash)048 060 030 00110000 0049 061 031 00110001 1050 062 032 00110010 2051 063 033 00110011 3052 064 034 00110100 4053 065 035 00110101 5054 066 036 00110110 6055 067 037 00110111 7056 070 038 00111000 8057 071 039 00111001 9058 072 03A 00111010 : (colon)059 073 03B 00111011 ; (semi-colon)060 074 03C 00111100 < (less than)061 075 03D 00111101 = (equal sign)062 076 03E 00111110 > (greater than)063 077 03F 00111111 ? (question mark)064 100 040 01000000 @ (AT symbol)065 101 041 01000001 A066 102 042 01000010 B067 103 043 01000011 C068 104 044 01000100 D069 105 045 01000101 E070 106 046 01000110 F071 107 047 01000111 G072 110 048 01001000 H073 111 049 01001001 I074 112 04A 01001010 J075 113 04B 01001011 K076 114 04C 01001100 L077 115 04D 01001101 M078 116 04E 01001110 N079 117 04F 01001111 O080 120 050 01010000 P081 121 051 01010001 Q082 122 052 01010010 R083 123 053 01010011 S084 124 054 01010100 T085 125 055 01010101 U086 126 056 01010110 V087 127 057 01010111 W088 130 058 01011000 X089 131 059 01011001 Y090 132 05A 01011010 Z091 133 05B 01011011 [ (left/opening bracket) 092 134 05C 01011100 \ (back slash)093 135 05D 01011101 ] (right/closing bracket) 094 136 05E 01011110 ^ (caret/circumflex)095 137 05F 01011111 _ (underscore)096 140 060 01100000 `097 141 061 01100001 a098 142 062 01100010 b099 143 063 01100011 c100 144 064 01100100 d101 145 065 01100101 e102 146 066 01100110 f103 147 067 01100111 g104 150 068 01101000 h105 151 069 01101001 i106 152 06A 01101010 j107 153 06B 01101011 k108 154 06C 01101100 l109 155 06D 01101101 m110 156 06E 01101110 n111 157 06F 01101111 o112 160 070 01110000 p113 161 071 01110001 q114 162 072 01110010 r115 163 073 01110011 s116 164 074 01110100 t117 165 075 01110101 u118 166 076 01110110 v119 167 077 01110111 w120 170 078 01111000 x121 171 079 01111001 y122 172 07A 01111010 z123 173 07B 01111011 { (left/opening brace)124 174 07C 01111100 | (vertical bar)125 175 07D 01111101 } (right/closing brace)126 176 07E 01111110 ~ (tilde)127 177 07F 01111111 DEL (delete)------------------------------------------------------------------0 1 2 3 4 5 6 7 8 9 A B C D E F0 NUL SOH STX ETX EOT ENQ ACK BEL BS HT LF VT FF CR SO SI1 DLE DC1 DC2 DC3 DC4 NAK SYN ETB CAN EM SUB ESC FS GS RS US2 SP ! " # $ % & ' ( ) * + , - . /3 0 1 2 3456789:;<=> ?4 @ A B C D E F G H I J K L M N O5 P Q R S T U V W X Y Z [ \ ] ^ _6 ` a b c d e f g h i j k l m n o7 p q r s t u v w x y z { | } ~ DEL------------------------------------------------------------------[文档可能无法思考全面,请浏览后下载,另外祝您生活愉快,工作顺利,万事如意!]。
最全ASCII码对照表

最全ASCII码对照表Bin Dec Hex 缩写/字符解释0000 0000 0 00 NUL (null) 空字符0000 0001 1 01 SOH (start of handing) 标题开始0000 0010 2 02 STX (start of text) 正文开始0000 0011 3 03 ETX (end of text) 正文结束0000 0100 4 04 EOT (end of transmission) 传输结束0000 0101 5 05 ENQ (enquiry) 请求0000 0110 6 06 ACK (acknowledge) 收到通知0000 0111 7 07 BEL (bell) 响铃0000 1000 8 08 BS (backspace) 退格0000 1001 9 09 HT (horizontal tab) 水平制表符0000 1010 10 0A LF (NL line feed, new line) 换行键0000 1011 11 0B VT (vertical tab) 垂直制表符0000 1100 12 0C FF (NP form feed, new page) 换页键0000 1101 13 0D CR (carriage return) 回车键0000 1110 14 0E SO (shift out) 不用切换0000 1111 15 0F SI (shift in) 启用切换0001 0000 16 10 DLE (data link escape) 数据链路转义0001 0001 17 11 DC1 (device control 1) 设备控制1 0001 0010 18 12 DC2 (device control 2) 设备控制2 0001 0011 19 13 DC3 (device control 3) 设备控制3 0001 0100 20 14 DC4 (device control 4) 设备控制4 0001 0101 21 15 NAK (negative acknowledge) 拒绝接收0001 0110 22 16 SYN (synchronous idle) 同步空闲0001 0111 23 17 ETB (end of trans. block) 传输块结束0001 1000 24 18 CAN (cancel) 取消0001 1001 25 19 EM (end of medium) 介质中断0001 1010 26 1A SUB (substitute) 替补0001 1011 27 1B ESC (escape) 溢出0001 1100 28 1C FS (file separator) 文件分割符0001 1101 29 1D GS (group separator) 分组符0001 1110 30 1E RS (record separator) 记录分离符0001 1111 31 1F US (unit separator) 单元分隔符0010 0000 32 20 空格0010 0001 33 21 !0010 0010 34 22 "0010 0011 35 23 #0010 0100 36 24 $0010 0101 37 25 %0010 0110 38 26 &0010 0111 39 27 "0010 1001 41 29 ) 0010 1010 42 2A * 0010 1011 43 2B + 0010 1100 44 2C , 0010 1101 45 2D - 0010 1110 46 2E . 0010 1111 47 2F / 0011 0000 48 30 0 0011 0001 49 31 1 0011 0010 50 32 2 0011 0011 51 33 3 0011 0100 52 34 4 0011 0101 53 35 5 0011 0110 54 36 6 0011 0111 55 37 7 0011 1000 56 38 8 0011 1001 57 39 9 0011 1010 58 3A : 0011 1011 59 3B ; 0011 1100 60 3C < 0011 1101 61 3D = 0011 1110 62 3E > 0011 1111 63 3F ? 0100 0000 64 40 @0100 0001 65 41 A 0100 0010 66 42 B 0100 0011 67 43 C 0100 0100 68 44 D 0100 0101 69 45 E 0100 0110 70 46 F 0100 0111 71 47 G 0100 1000 72 48 H 0100 1001 73 49 I 0100 1010 74 4A J 0100 1011 75 4B K 0100 1100 76 4C L 0100 1101 77 4D M 0100 1110 78 4E N 0100 1111 79 4F O 0101 0000 80 50 P 0101 0001 81 51 Q 0101 0010 82 52 R0101 0100 84 54 T 0101 0101 85 55 U 0101 0110 86 56 V 0101 0111 87 57 W 0101 1000 88 58 X 0101 1001 89 59 Y 0101 1010 90 5A Z 0101 1011 91 5B [ 0101 1100 92 5C \ 0101 1101 93 5D ] 0101 1110 94 5E ^ 0101 1111 95 5F _ 0110 0000 96 60 `0110 0001 97 61 a 0110 0010 98 62 b 0110 0011 99 63 c 0110 0100 100 64 d 0110 0101 101 65 e 0110 0110 102 66 f 0110 0111 103 67 g 0110 1000 104 68 h 0110 1001 105 69 i 0110 1010 106 6A j 0110 1011 107 6B k 0110 1100 108 6C l 0110 1101 109 6D m 0110 1110 110 6E n 0110 1111 111 6F o 0111 0000 112 70 p 0111 0001 113 71 q 0111 0010 114 72 r 0111 0011 115 73 s 0111 0100 116 74 t 0111 0101 117 75 u 0111 0110 118 76 v 0111 0111 119 77 w 0111 1000 120 78 x 0111 1001 121 79 y 0111 1010 122 7A z 0111 1011 123 7B { 0111 1100 124 7C | 0111 1101 125 7D }0111 1111 127 7F DEL (delete) 删除ESC键VK_ESCAPE (27)回车键:VK_RETURN (13)TAB键:VK_TAB (9)Caps Lock键:VK_CAPITAL (20)Shift键:VK_SHIFT ()Ctrl键:VK_CONTROL (17)Alt键:VK_MENU (18)空格键:VK_SPACE (/32)退格键:VK_BACK (8)左徽标键:VK_LWIN (91)右徽标键:VK_LWIN (92)鼠标右键快捷键:VK_APPS (93)Insert键:VK_INSERT (45)Home键:VK_HOME (36)Page Up:VK_PRIOR (33)PageDown:VK_NEXT (34)End键:VK_END (35)Delete键:VK_DELETE (46)方向键(←):VK_LEFT (37)方向键(↑):VK_UP (38)方向键(→):VK_RIGHT (39)方向键(↓):VK_DOWN (40)F1键:VK_F1 (112)F2键:VK_F2 (113)F3键:VK_F3 (114)F4键:VK_F4 (115)F5键:VK_F5 (116)F6键:VK_F6 (117)F7键:VK_F7 (118)F8键:VK_F8 (119)F9键:VK_F9 (120)F10键:VK_F10 (121)F11键:VK_F11 (122)F12键:VK_F12 (123)Num Lock键:VK_NUMLOCK (144)小键盘0:VK_NUMPAD0 (96)小键盘1:VK_NUMPAD0 (97)小键盘2:VK_NUMPAD0 (98)小键盘3:VK_NUMPAD0 (99)小键盘4:VK_NUMPAD0 (100)小键盘5:VK_NUMPAD0 (101)小键盘6:VK_NUMPAD0 (102) 小键盘7:VK_NUMPAD0 (103) 小键盘8:VK_NUMPAD0 (104) 小键盘9:VK_NUMPAD0 (105) 小键盘.:VK_DECIMAL (110) 小键盘*:VK_MULTIPLY (106) 小键盘+:VK_MULTIPLY (107) 小键盘-:VK_SUBTRACT (109) 小键盘/:VK_DIVIDE (111) Pause Break键:VK_PAUSE (19) Scroll Lock键:VK_SCROLL (145)。
ASSIC码对照表

ASSIC码对照表编码对应字符:✔:\u2714✘:\u2718<script type="text/javascript">var aaa = "\u2718";document.write(aaa);</script>测试ASCII码的⽅法:在记事本中,按住ALT键,同时⽤⼩键盘输⼊⼗进制的ASCII码,然后松⼿,就可以看到效果了!ASCII值控制字符ASCII值控制字符ASCII值控制字符ASCII值控制字符0NUT32(space)64@96、1SOH33!65A97a2STX34”66B98b3ETX35#67C99c4EOT36$68D100d5ENQ37%69E101e6ACK38&70F102f7BEL39,71G103g8BS40(72H104h9HT41)73I105i10LF42*74J106j11VT43+75K107k12FF44,76L108l13CR45-77M109m14SO46.78N110n15SI47/79O111o16DLE48080P112p17DCI49181Q113q18DC250282R114r19DC351383S115s20DC452484T116t21NAK53585U117u22SYN54686V118v23TB55787W119w24CAN56888X120x25EM57989Y121y26SUB58:90Z122z27ESC59;91[123{28FS60<92\124|29GS61=93]125}30RS62>94^126~31US63?95—127DELNUL VT 垂直制表SYN 空转同步SOH 标题开始FF ⾛纸控制ETB 信息组传送结束STX 正⽂开始CR 回车CAN 作废ETX 正⽂结束SO 移位输出EM 纸尽EOY 传输结束SI 移位输⼊SUB 换置ENQ 询问字符DLE 空格ESC 换码ACK 承认DC1 设备控制1FS ⽂字分隔符BEL 报警DC2 设备控制2GS 组分隔符BS 退⼀格DC3 设备控制3RS 记录分隔符HT 横向列表DC4 设备控制4US 单元分隔符LF 换⾏NAK 否定DEL 删除键盘常⽤ASCII码ESC键VK_ESCAPE (27)回车键:VK_RETURN (13)TAB键:VK_TAB (9)Caps Lock键:VK_CAPITAL (20)Shift键:VK_SHIFT ($10)Ctrl键:VK_CONTROL (17)Alt键:VK_MENU (18)空格键:VK_SPACE ($20/32)退格键:VK_BACK (8)左徽标键:VK_LWIN (91)右徽标键:VK_LWIN (92)⿏标右键快捷键:VK_APPS (93)Insert键:VK_INSERT (45)Home键:VK_HOME (36)Page Up:VK_PRIOR (33) PageDown:VK_NEXT (34)End键:VK_END (35)Delete键:VK_DELETE (46)⽅向键(←):VK_LEFT (37)⽅向键(↑):VK_UP (38)⽅向键(→):VK_RIGHT (39)⽅向键(↓):VK_DOWN (40)F1键:VK_F1 (112)F2键:VK_F2 (113)F3键:VK_F3 (114)F4键:VK_F4 (115)F5键:VK_F5 (116)F6键:VK_F6 (117)F7键:VK_F7 (118)F8键:VK_F8 (119)F9键:VK_F9 (120)F10键:VK_F10 (121)F11键:VK_F11 (122)F12键:VK_F12 (123)Num Lock键:VK_NUMLOCK (144)⼩键盘0:VK_NUMPAD0 (96)⼩键盘1:VK_NUMPAD0 (97)⼩键盘2:VK_NUMPAD0 (98)⼩键盘3:VK_NUMPAD0 (99)⼩键盘4:VK_NUMPAD0 (100)⼩键盘5:VK_NUMPAD0 (101)⼩键盘5:VK_NUMPAD0 (101)⼩键盘6:VK_NUMPAD0 (102)⼩键盘7:VK_NUMPAD0 (103)⼩键盘8:VK_NUMPAD0 (104)⼩键盘9:VK_NUMPAD0 (105)⼩键盘.:VK_DECIMAL (110)⼩键盘*:VK_MULTIPLY (106)⼩键盘+:VK_MULTIPLY (107)⼩键盘-:VK_SUBTRACT (109)⼩键盘/:VK_DIVIDE (111)Pause Break键:VK_PAUSE (19)Scroll Lock键:VK_SCROLL (145)ASCII码中:第0~32号及第127号是控制字符,常见的控制符如:007 = 07 = U+0007 : BELL 转义符:\a 响铃 008 = 08 = U+0008 : BACKSPACE 转义符:\b 退格键 009 = 09 = U+0009 : HORIZONTAL TABULATION 转义符:\t Tab键 010 = 0A =U+000A : LINE FEED 转义符:\n 换⾏符 011 = 0B = U+000B : VERTICALTABULATION 转义符:\v 垂直 Tab 符 012 = 0C = U+000C : FORMFEED 转义符:\f 换页符 013 = 0D = U+000D : CARRIAGERETURN 转义符:\r 回车键 027 = 1B = U+001B : ESCAPE 转义符:\e Esc 键第33~126号是字符,其中第48~57号为0~9⼗个阿拉伯数字;65~90号为26个⼤写英⽂字母,97~122号为26个⼩写英⽂字母,其余的是⼀些标点符号、运算符号等。
常用一维条形码编码规则汇总

常用一维条形码139码(CODE39)39码可以包含数字及英文字母。
除了超市、零售业的应用中使用UPC/EAN码外,几乎在其他饿应用环境中,都是使用39码。
39码是目前使用最广泛的条码规格,支持39码的软硬件设备也最齐全。
1.1 特征◆能表示44个字符,A-Z、0-9、SPACE、-、.、$、/、+、%、*◆分散式,条码组之间使用细白条分隔◆两种宽度◆自我检查◆有扩展模式《Full ASCII Mode》◆检查码字符可有可无,视需求而定1.2 组成◆各个字符有9条黑白相间,粗细不同的线条组成,其中6条为黑白细条3条黑白粗条◆一串字符必须在头尾加上起始字符和结束字符“*”1.3 校验方法找到输入字符串每个字符对应值,求和,除以43,取余数。
1.4 条码说明1.5 编码表P.S.在程序中可以使用“11”表示宽黑条,‘1’表示细黑条,“00”表示宽白条,“0”表示细白条。
那么字符1就可以表示为110100101011。
使用此方法建立一个编码表,每个字符可以长度为12的“01”字符串来表示。
1.6 典型CODE39条码1.7 CODE39的扩展码扩展码表同CODE93。
但是扩展方式不同,39码使用$,/,+.%与其26个大写字母组合,表示ASCII码表中的其他字符。
条空表示方式和校验方式与标准39码相同。
93码中使用的控制码与26个大写字母的组合。
293码(CODE93)2.1 组成◆字母:A-Z,数字:0-9,符号:SPACE, - , . , $ , / , +, %, 控制码:$ , / , +, %,起始结束码:□◆每个字由9个模组成,包括3条粗细黑条及3条粗细白条。
每一黑条或白条有可能为1.2.3.4模组成2.2 特征◆用4个控制码$, %, /, + 组合其他字母或符号,可编程FULL ASCII字母,读码器读到上面4个控制码的组合时候,送出的字尾所对应的ASCII。
◆有2个检验码C和K。
2.3 校验方法◆先查出资料所对应值,对应值的表如下顺序号作为权值,分别乘以对应值,求和,除以47,取余数◆检查码K由C位用1-15顺序排列,若资料差偶偶15位,再从1-15起算,顺序号作为权值,分别乘以对应值,求和,除以47,取余数◆举例:资料C O D E SP 9 3 “C”“K”资料对应值12 24 13 14 38 9 3C 排列顺序7 6 5 4 3 2 1K 排列顺序8 7 6 5 4 3 2 1(1x3) + (2x9) + (3x38) + (4x14) + (5x13) + (6x24) + (7x12) = 484C = 484 ÷47 = 10……14 (余数)则 C = 14 = E (对应值)(1x14) + (2x3) + (3x9) + (4x38) + (5x14) + (6x13) + (7x24) + (8x12)= 611K = 611 ÷47 = 13……0 (余数)則K = 0 = 0 (对应值)2.4 条码说明2.5 编码表P.S.程序编码中,结束符号模块比起始符号多一个“1”;使用控制符组合字母所表示的字符,编码时需要分解成控制符和大写字母两个模块。
asiii码表阿斯克码表大全之欧阳地创编

121
51
81
Q
22
12
18
dc2
122
52
82
R
23
13
19
dc3
123
53
83
S
24
14
20
dc4
124
54
84
T
25
15
21
nak
125
55
85
U
26
16
22
syn
126
56
86
V
27
17
23
etb
127
57
87
W
30
18
24
can
130
58
88
X
31
19
25
em
131
59
89
Y
32
1a
时间:2021.03.04
创作:欧阳地
76
118
v
67
37
55
7
167
77
119
w
70
38
56
8
170
78
120
x
71
39
57
9
171
79
121
y
72
3a
58
:
172
7a
122
z
73
3b
59
;
173
7b
123
{
74
3c
60
<
174
7c
124
|
75
3d
61
对中纠偏型号对照表

对中纠偏型号对照表光电式我公司型号 EMG 型号 说明 LSE 480/01 LSE 770/01 LSE 1075/01 LSE 1375/01LIC 480/01 LIC 770/01 LIC 1075/01 LIC 1375/01光源LR13.01 LS13.01测量光电接收头 LR14.01 LS14.01参考光电接收头 RCT2.11.1 EVK2.11.XEVK2.12.X X 表示可选数字光电接收调节装置中的控制电路板ORAD1-CP/250 ORAD1-CP/400 ORAD1-CP/600 ORAD1-CP/800 ORAD1-CP/1000 EVK2-CP/250 EVK2-CP/400 EVK2-CP/600 EVK2-CP/800 EVK2-CP/1000 光电接收调节装置(单头对边整机) ORAD2-CP/550 ORAD2-CP/750ORAD2-CP/1000 ORAD2-CP/1300 ORAD2-CP/1650 EVM2-CP/550 EVM2-CP/750EVM2-CP/1000 EVM2-CP/1300 EVM2-CP/1650光电接收调节装置 (双头对中整机)电感式我公司型号 EMG 型号 说明ISI300S/E ISI500S/E ISI800S/E IM300S02/E01 IM500S02/E01 IM800S02/E01 普通电感式检测头(线圈)ISH300 ISH500 ISH800IMP300.01 IMP500.02 IMP800.02高精度电感式检测头(线圈) ISB/01 PIB04.0XX 表示可选数字高精度线圈驱动盒(发射)IEB/01 PIB04.04高精度线圈放大盒(接收)IEA.01BMI2.01 BMI2.02电感式信号处理电路板(模拟)IED.01 BMI2.11.XX 表示可选数字电感式信号处理电路板(数字带CANBUS )IND2-CP BMI2-CP普通电感式整机 INDH-CP BMH-CP高精度电感式整机 IFM普通电感式框架 HFM高精度电感式框架 FRC玻璃钢盖板 框架尺寸按需要定制,价格根据尺寸定,玻璃钢盖板按米计价。
ASCII码表完整版(带16进制)

q
18
DC2
50
2
82
R
114
r
19
DC3
51
3
83
X
115
s
20
DC4
52
4
84
T
116
t
21
NAK
53
5
85
U
117
ll
22
SYN
54
6
86
V
118
V
23
TB
55
7
87
W
119
W
24
CAN
56
8
88
X
120
X
25
EM
57
9
89
Y
121
y
26
SUB
58
90
Z
122
Z
27
ESC
59
I
5
91
[
123
被选区域结束
88
HTS
水平制表符集
89
HTJ
对齐的水平制表符集
8A
VTS
垂直制表符集
8B
PLD
部分行向下
8C
PLU
部分行向上
8D
RI
反向索引
8E
SS2
单移2
8F
SS3
单移3
90
DCS
设备控制字符串
91
PU1
专用1
92
PU2
专用2
93
STS
设置传输状态
94
CCH
取消字符
95
MW
消息等待
[计算机]ASCII码对照表-精品文档
![[计算机]ASCII码对照表-精品文档](https://img.taocdn.com/s3/m/3cc83c5801f69e31433294fc.png)
ASCII码对照表2009-04-23 19:43ASCII(American Standard Code for Information Interchange)定义从 0 到127 的共128个数字所代表的英文字母或一样的结果与意义。
由于使用7个位(bit)就可以表示从0到127的数字,大部分的电脑都使用8个位来存取字元集(character set),所以从128到255之间的数字可以用来代表另一组128个符号,称为extended ASCII。
目前计算机中用得最广泛的字符集及其编码,是由美国国家标准局(ANSI)制定的ASCII码(American Standard Code for Information Interchange,美国标准信息交换码),它已被国际标准化组织(ISO)定为国际标准,称为ISO 646标准。
适用于所有拉丁文字字母,ASCII码有7位码和8位码两种形式。
因为1位二进制数可以表示(21=)2种状态: 0、1;而2位二进制数可以表示(22)=4种状态:00、01、10、11;依次类推,7位二进制数可以表示(27=)128种状态,每种状态都唯一地编为一个7位的二进制码,对应一个字符(或控制码),这些码可以排列成一个十进制序号0~127。
所以,7位ASCII码是用七位二进制数进行编码的,可以表示128个字符。
第0~32号及第127号(共34个)是控制字符或通讯专用字符,如控制符:LF(换行)、CR(回车)、FF(换页)、DEL(删除)、BEL(振铃)等;通讯专用字符:SOH(文头)、EOT(文尾)、ACK(确认)等;第33~126号(共94个)是可打印字符,其中第48~57号为0~9十个阿拉伯数字;65~90号为26个大写英文字母,97~122号为26个小写英文字母,其余为一些标点符号、运算符号等。
注意:在计算机的存储单元中,一个ASCII码值占一个字节(8个二进制位),其最高位(b7)用作奇偶校验位。
asiii码表阿斯克码表大全之欧阳数创编

创作:欧阳数
dc1
121
51
81
Q
22
12
18
dc2
122
52
82
R
23
13
19
dc3
123
53
83
S
24
14
20
dc4
124
54
84
T
25
15
21
nak
125
55
85
U
26
16
22
syn
126
56
86
V
27
17
23
127
57
87
W
30
18
24
can
130
58
88
X
31
19
25
em
131
59
89
Y
32
1a
26
sub
132
5a
90
Z
33
1b
27
esc
133
5b
91
[
34
1c
28
fs
134
5c
92
\
35
1d
29
gs
135
5d
93
]
36
1e
30
re
136
5e
94
^
37
1f
31
us
137
5f
95
_
40
20
32
sp
140
60
96
'
一维条码概述

最近意外找到以前曾整理的一維條碼資料, 自己都完全遺忘了. 於是便拿出來po一下, 提供給有需要的人參考吧.-一維條碼by青衫(邱奕南)一維條碼可分為39碼、EAN-13碼、EAN-8碼、UPC-A碼、UPC-E碼、交錯式25碼、CODABAR碼(又稱NW-7碼)、128碼等多種,以下便說明一下各種條碼的意義,以及編碼方式:(一)39碼39碼發展於1974年,由於限制少,又支援文數字,因此多應用於一般管理軟體。
其編碼特性如下:1. 資料碼可以是0~9、A~Z、-、.、Space、$、/、+、%等43種,資料長度不限,各資料編號次序依上列次序編號0~42。
2. 編碼結構為〔起始碼+資料碼+檢查碼+終止碼〕,其中檢查碼可有可無,且檢查碼亦會被視為資料輸入。
3. 起始碼與終止碼均固定為*。
4. 檢查碼為資料編號值(0~42)累加後(Sum)除以43取餘數,並將該值視為相對應資料編號值的資料。
5. 編碼資料如下,每碼有9條,其中3條為粗,各佔12線。
以下各位元奇數位為黑,要畫線,偶數位為白,不畫線,0為細,1為粗:0 0001101001 1001000012 0011000013 1011000004 0001100015 1001100006 0011100007 0001001018 1001001009 001100100A 100001001B 001001001C 101001000D 000011001E 100011000F 001011000G 000001101H 100001100I 001001100J 000011100K 100000011L 001000011M 101000010N 000010011O 100010010P 001010010Q 000000111R 100000110S 001000110T 000010110U 110000001V 011000001W 111000000X 010010001Y 110010000Z 011010000- 010000101. 110000100Space 011000100$ 010101000/ 010100010+ 010001010% 000101010* 010010100各資料碼之間必須再空開一條白線。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
DNA methylation changes detected by methylation-sensitive amplified polymorphism in two contrasting rice genotypes under salt stressWensheng Wang a ,b ,1,Xiuqin Zhao a ,1,Yajiao Pan a ,Linghua Zhu a ,Binying Fu a ,*,Zhikang Li a ,baInstitute of Crop Sciences,National Key Facility for Crop Gene Resources and Genetic Improvement,Chinese Academy of Agricultural Sciences,Beijing 100081,ChinabInternational Rice Research Institute,DAPO Box 7777,Metro Manila,PhilippinesReceived 8April 2011;revised 19July 2011;accepted 20July 2011AbstractDNA methylation,one of the most important epigenetic phenomena,plays a vital role in tuning gene expression during plant development as well as in response to environmental stimuli.In the present study,a methylation-sensitive amplified polymorphism (MSAP)analysis was performed to profile DNA methylation changes in two contrasting rice genotypes under salt stress.Consistent with visibly different phenotypes in response to salt stress,epigenetic markers classified as stable inter-cultivar DNA methylation differences were determined between salt-tolerant FL478and salt-sensitive IR29.In addition,most tissue-specific DNA methylation loci were conserved,while many of the growth stage-dependent DNA methylation loci were dynamic between the two genotypes.Strikingly,salt stress induced a decrease in DNA methylation specifically in roots at the seedling stage that was more profound in IR29than in the FL478.This result may indicate that demethylation of genes is an active epigenetic response to salt stress in roots at the seedling stage,and helps to further elucidate the implications of DNA methylation in crop growth and development.Keywords:DNA methylation;MSAP;Salt tolerance;Rice1.IntroductionCrop growth and development is greatly influenced by various abiotic stresses,such as salinity,cold,heat and drought.Rice is an agronomically important cereal crop and it is considered to be salt-sensitive,with expressing different responses at different growth stages to salt stress.At the seedling stage,salt stress can cause a significant reduction in the germination index and seedling vigor (Shannon et al.,1998).The different responses of rice varieties have been described (Lutts et al.,1995),and previous efforts to dissect the genetic basis of salt tolerance in rice indicate that it is a quantitative trait with generally low heritability and expres-sivity (Ammar et al.,2007).A number of quantitative trait loci(QTLs)have been mapped (Takehisa et al.,2004;Hu et al.,2010),and several genes were identified to be related to salt tolerance (Ren et al.,2005;Hu et al.,2008).However,there have been only a few studies aimed at understanding the relationship between epigenetics and the regulation of gene expression under salt stress in rice.DNA methylation is an important epigenetic modification that may contribute to environmentally-induced phenotypic variations by modifying gene expression in a tissue-specific or a developmental stage-dependent manner (Angers et al.,2010;Zhang et al.,2010).Abiotic stress is one such example that results in altered gene expression by DNA hypo-methylation or hyper-methylation.In maize roots,it was reported that cold stress treatment induced DNA hypo-methylation in the nucle-osome core,resulting in up-regulation of ZmM11gene (Steward et al.,2002).Studies on leaves show that aluminum,salt,cold and oxidative stresses induced DNA demethylation in the coding sequence of NtGPDL gene within 1h,concomitant with NtGDPL gene expression (Choi and Sano,2007).There is*Corresponding author.Tel:þ861082106698,fax:þ861082108559.E-mail address:fuby@ (B.Fu).1These authors contributed equally to thiswork.Available online at Journal of Genetics and Genomics 38(2011)419e4241673-8527/$-see front matter Copyright Ó2011,Institute of Genetics and Developmental Biology,Chinese Academy of Sciences,and Genetics Society of China.Published by Elsevier Limited and Science Press.All rights reserved.doi:10.1016/j.jgg.2011.07.006an evidence to suggest that alterations in DNA methylation are required for environmental stress protection.High increases in CNG methylation level were observed in Mesembryan-themum crystallinum genome under high salinity conditions (Dyachenko et al.,2006).Furthermore,DNA methylation may endow plants with a better ability to respond to envi-ronmental stresses.For example,Wada et al.(2004)describes the activation of a set of specific stress response genes following the down-regulation of Met1-induced genomic hypo-methylation.This study sought to investigate DNA methylation changes in salt-stressed roots and leaves at various developmental stages using two rice genotypes,FL478and IR29,which share approximately50%of their genome yet have different degrees of salt tolerance(Gregorio et al.,1997).The effect of salt stress on the spatial and temporal DNA methylation changes revealed in this study will provide information for further dissecting the molecular mechanism of rice adaptation to salt stress.2.Materials and methods2.1.Plant materials and growth conditionsTwo indica rice genotypes,FL478and IR29,were used in this study.FL478is a recombinant inbred line developed at the International Rice Research Institute(IRRI)using Pokkali (salt-tolerant donor)and IR29.IR29is an improved indica cultivar currently used as a salt-sensitive standard(Bonilla et al.,2002).Previous studies showed the contrasting morphological and physiological changes of those two culti-vars under salt stress(Gregorio et al.,2002;Suriya-arunroj et al.,2004;Walia et al.,2005).All seeds were sterilized in0.1%NaClO(v/v)and then germinated at37 C in an incubator.The germinated seeds were planted in a seedling nursery.2.2.Salt stress treatment and morphological analysis at seedling stageRice plants with four leaves were transplanted into tanks,as described by Gregorio et al.(1997)and watered with Yoshida solution(Yoshida et al.,1976)in the greenhouse.Salt stress was applied by adding NaCl to afinal electrical conductivity (EC)of12dS mÀ1.The EC of control tanks was around 1.0dS mÀ1.The pH of the nutrient solution was adjusted daily to5.5by adding sulfuric acid and was refreshed every week. Rice plants were exposed to salt stress for13days.Roots and leaves under stress and control plants were collected and snap-frozen for DNA extraction.Three replicates were prepared from each sample for DNA methylation analysis.After13days of salt stress,the Modified Standard Evalua-tion System for rice was used to rate the visual symptoms of salt injury,as described by Gregorio et al.(1997)(Supplementary Table1).The plant height(cm)was measured from the base of the stem to the tip of the topmost leaf of the plants.Root length(cm)was measured for each plant.2.3.Salt stress treatment and morphological analysis at tillering and booting stagesRice plants were grown in large potsfilled with mixed soil (50N,25P and25K mg KgÀ1)and irrigated with fresh water. Salinity stress treatment was performed by adding NaCl to the stress tank until the EC was8dS mÀ1,and adjusting the EC with fresh water or NaCl every two days;the EC of the control tank was1dS mÀ1by adding fresh water.Roots and leaves were harvested and snap-frozen for DNA extraction after15 days of stress treatment.Three replicates were prepared from each sample for DNA methylation analysis.After15days of salt stress,the plant height(cm)was measured,and the tiller number was counted for each plant. Each treatment was replicated three times.The SAS program GLM was used to perform ANOV A analysis(SAS Institute, 1999).2.4.Methylation-sensitive amplified polymorphism (MSAP)analysisTotal genomic DNA was extracted using the improved CTAB method.MSAP(methylation-sensitive amplified poly-morphism)analysis was performed as described by Xiong et al.(1999),with minor modifications.First,we used several primers to identify the robustness of the three repli-cates for each sample;the results showed identical MSAP bands among three replicates(data not shown).These repli-cates were then mixed to provide a pool sample for the following MSAP analysis.Two enzyme combinations were used:Eco R I/Msp I and Eco R I/Hpa II;the sequence infor-mation for the adapters and primers of pre-amplification and selective amplification are provided in Supplementary Table2. The MSAP PCR products were separated on the6% sequencing gels and visualized with silver staining.The MSAP pattern for displaying the DNA fragments resulting from digestions with the isoschizomers were divided into the following four types(Supplementary Table3):Type I bands, present for both enzyme combinations;Type II bands,present only for Eco R I/Hpa II;Type III bands,present for Eco R I/ Msp I;and Type IV bands,absent from both enzyme combi-nations.Here,Type II represents semi-methylated bands, whereas Type III and IV bands represent semi-or full meth-ylation,respectively(Chen et al.,2009).2.5.Cloning and characterization of the differentiallyamplified DNA fragmentsDifferentially amplified fragments were selected,isolated, reamplified and purified with Wizard SV gel and the PCR clean-up system(Promega,Madison,USA).Briefly,the polymorphic bands were excised from the gel using a steril-ized surgical blade,hydrated in20m L of TE buffer(pH8.0)in an eppendorf tube and incubated in boiling water for5min. The supernatant was recovered by centrifugation and used for the re-amplification.The reamplified DNA fragments were purified and cloned with T-vector(TaKaRa,Dalian,China)for420W.Wang et al./Journal of Genetics and Genomics38(2011)419e424sequencing,and the sequences obtained were analyzed by NCBI BLAST (/Blast.cgi )and EMBL BLAST (/Tools/blast/).3.Results3.1.Phenotypic differences between FL478and IR29under salt stressThree developmental stages were used to test the various effects of salinity on rice growth:seedling,tillering and booting stages.The two rice genotypes,FL478and IR29,showed extreme variability in their reaction to salt stress at the seedling stage (Supplementary Fig.1).While FL478had a high degree of salinity tolerance (mean SES score of 2),IR29was very sensitive to salt stress (mean SES score of 7).In both genotypes,salt stress significantly reduced the plant height at the seedling stage,but the root lengths were less affected (Table 1).The plant height and tiller number of both lines were also comparatively analyzed at the tillering stage under salt stress and control conditions.The plant height of IR29was significantly reduced by salt stress treatment,but there was no significant reduction of plant height detected in FL478under salt stress (Table 1and Supplementary Fig.2).However,the tiller numbers of FL478and IR29were not affected by salinity (Table 1and Supplementary Fig.2).At the booting stage,no significant difference in plant height or tiller number were detected between salt-stressed and control plants in both lines (Table 1).3.2.MSAP analysis of DNA methylation in FL478and IR29Thirty-eight primer combinations were used to detect cytosine methylation at CCGG sequences in the two rice genotypes.During the three different developmental stages,971e 996clear and reproducible fragments were amplified in at least one sample of leaves or roots of FL478and IR29under high salinity conditions.Based on the MSAP profiles,the number of methylated (hemi-methylated and fully-methylated)DNA bands was determined.The results showed that 212e 269bands were polymorphic (Type II þType III þType IV bands),accounting for 21.8%e 27.0%of the total bands.Thesefindings indicate distinct DNA methylation levels in the different samples (Table 2).3.3.DNA methylation status in leaves or roots of FL478and IR29under normal growth conditions at three developmental stagesThe DNA methylation levels were assessed in two tissues of the FL478and IR29rice genotypes.Overall,the number of methylated DNA bands observed in the leaves or roots of FL478(230e 268)was higher than that of IR29(212e 249),comprising 23.1%e 26.9%and 21.8%e 25.6%of the total bands,respec-tively (Table 2).Among these,28bands (2.8%of the total MSAP bands)were found to be polymorphic between FL478and IR29,with 15and 10bands specific to FL478and IR29,respectively;this indicated an epigenetic difference between these two lines.All 28of these polymorphic bands were stably independent of tissue,growth stage and environmental condi-tions (Supplementary Table 4).We detected 17bands (1.7%of the total MSAP bands)that were differentially amplified between the leaf and root tissues.Of these,16were commonly tissue-specific in both FL478and IR29,with only one band specific to the leaves of IR29(Supplementary Table 5).At each of the developmental stages,a number of DNA-methylated bands were detected to be stage-specific.DNA methylation levels in leaves at the seedling and tillering stages of FL478(26.4%)and IR29(24.9%)were relatively higher than those in leaves at the booting stage (24.4%and 24.5%,respectively)under control conditions.However,in the roots,the DNA methylation level at the tillering stage was higher than the levels observed at the seedling and booting stages (Table 2).In FL478,54DNA-methylated bands were identi-fied to be stage-specific.Among them,19and 8bands were present in leaves and roots at the booting stage,respectively;but only 21and 4bands were present in roots at the seedling stage and tillering stage,respectively.As for IR29,33stage-specific DNA-methylated bands were identified.At the seed-ling,tillering and booting stages,14,4and 9bands were amplified in roots,respectively,whereas 5bands were present in leaves at the booting stage.The full data are provided as supplementary material (Supplementary Table 6).Table 1The mean performances of FL478and IR29for phenotypic traits under salinity stress and control conditions evaluated at different development stages.Stage Genotype Treatment Plant height a Root length a Tiller number a SeedlingFL478Control 52.77Æ2.8011.06Æ0.80e Stress 39.72Æ0.80***12.96Æ1.50e IR29Control 39.15Æ1.7012.81Æ0.70e Stress 26.33Æ1.30***14.41Æ0.70eTilleringFL478Control 98.23Æ4.60e 10.67Æ1.50Stress 94.33Æ4.50e 9.67Æ2.10IR29Control 87.87Æ2.38e 12Æ1.00Stress 77.27Æ2.91**e 12.67Æ1.20BootingFL478Control 114Æ2.60e 8.67Æ0.60Stress 108.33Æ6.50e 9.33Æ1.50IR29Control 100.00Æ1.00e 9.67Æ1.20Stress98.67Æ1.50e10.00Æ1.00**,***denote P <0.01and P <0.001,respectively.aResults are mean values of three replicates ÆSD.Each replicate was derived from three plants.421W.Wang et al./Journal of Genetics and Genomics 38(2011)419e 4243.4.DNA methylation changes of FL478and IR29induced by salt stressDNA methylation levels of FL478and IR29under salt-stressed and control conditions were compared by MSAP profiles(Supplementary Fig.3).As shown in Table2,FL478 leaves at the booting stage showed a slightly enhanced proportion of DNA methylation after salt stress,from244 (control)to251(stressed)bands.However,no obvious changes were detected in the FL478leaves at the seedling and tillering stages under salt stress.However,in FL478roots,salt stress decreased DNA methylation levels at the seedling stage, with the total DNA methylation bands under control and stress conditions were252and230,respectively(25.3%and23.1% of the total bands),whilst no obvious differences were found at either the tillering or booting stages.The IR29variety also showed a tendency toward the changes in DNA methylation by salt stress.Strikingly,in IR29 roots,DNA methylation levels decreased from24.6%to 21.8%at the seedling stage under salt stress,a far greater decrease than we observed in the FL478.However,at the tillering and booting stages and in both leaves and roots,DNA methylation was unaffected by salt stress.The changes in DNA methylation banding patterns(hemi-or full methylation)induced by salt stress were further analyzed.The results showed that,in both rice lines,all of these changed bands were from type IV fully-methylated DNA fragments(Table2).This indicates that salt stress induced the decrease in DNA methylation in seedling roots in both lines, and the changes resulted from the full-methylated cytosine on both DNA strands.3.5.Sequence analysis of the differentially methylatedDNA bandsTwenty-seven differentially methylated DNA bands were cloned and sequenced.The resulting sequences were blasted against the databases at NCBI and EBI websites.As shown in Table3,the range of fragment sizes was82e202bp,with an average of150bp.Among these27DNA sequences,three (S1,S2and S3)were specifically methylated during the booting stage in both lines and unaffected by salt stress.Their sequences are homologous to genes encoding GYF domain-containing proteins,histone-lysine N-methyltransferases and OsWRKY transcription factors with WRKY and zincfinger domains.Five sequences(T1to T5)were present only in leaf or root tissue and not affected by salt stress.Of them,T1and T2are leaf-specifically methylated and highly homologous to cytochrome b6-f complex subunit4,Ser/Thr phosphatase family of proteins and NB-ARC domain-containing proteins.In contrast,T3,T4and T5were root tissue-specific and homolo-gous to photosystem I assembly protein,retrotransposon and NADPH quinine oxidoreductase.The remaining nineteen DNA sequences,coded as P1e P19, were de-methylated or methylated in response to salt stress in FL478and IR29.Three of these(P1,P2and P10)were identified as genes encoding putative proteins.The rest of the sequences were found to be homologous to genes encoding hydrolases(P3,P13and P14),cytochrome subunits(P4and P6),transcription factors(P8and P15),kinases(P9and P12), and genes involved in redox regulation,such as Acetyl-CoA, peroxidases and ABC transporters.Together these results confirm that salt stress affected a large amount of diverse genes by DNA methylation changes.4.DiscussionEpigenetic processes,such as DNA methylation,can be influenced by environmental stimuli(Angers et al.,2010).In this study,the MSAP approach was used to characterize the spatial and temporal DNA methylation changes in rice caused by salt stress.The two genotypes,FL478and IR29,share at least50%of their genome,as IR29is one of the parents used to develop the RIL population,including FL478(Gregorio et al.,1997);however,these two varieties were previously identified as differing in salt tolerance.The phenotypic results in this study confirmed that these two lines have strikingly different responses to salt stress,especially at the seedlingTable2DNA methylation changes in leaves and roots at three stages under control and stress conditions.Genotype FL478IR29Tissue Leaves Roots Leaves RootsStage Seedling Tillering Booting Seedling Tillering Booting Seedling Tillering Booting Seedling Tillering Booting MSAP band types C S C S C S C S C S C S C S C S C S C S C S C SI733734733733752745744766728727732733729729729729734733732759725722729729 II777777777977777777777877III160159159159159159157157157157157157159159159159159159156156156156156156 IV969697977885886610410310099767676767172764983857979 Total methylatedbands a263262263263244251252230268269264263242242242242237238239212246249242242MSAP(%)26.426.326.426.424.525.225.323.126.92726.526.424.924.924.924.924.424.524.621.825.325.624.924.9 Fully-methylatedbands b256255256256237244245223261260257256235235235235230231232205239241235235Full methylationratio(%)25.725.625.725.723.824.524.622.426.226.125.825.724.224.224.224.223.723.823.921.124.624.824.224.2C and S indicate the control and stress conditions,respectively.a Total methylated bands¼IIþIIIþIV;b Fully-methylated bands¼IIIþIV.422W.Wang et al./Journal of Genetics and Genomics38(2011)419e424stage.Correspondingly,we detected a number of salt stress-induced DNA methylation changes that were genotype-and organ/tissue-dependent.Previous results indicated that DNA methylation differences might occur among distinct cultivars (Ashikawa,2001;Wang et al.,2004;Takata et al.,2005).In this study,we found that small but distinct changes to DNA methylation in FL478and IR29rice varieties.These stable DNA methylation differences may be epigenetic markers that are potentially linked to phenotypic variations between closely related strains (Takata et al.,2005).Previously,studies showed that tissue-specific DNA methylation was related to the differential expression of genes that constitute transcription networks regulating tissue specificity (Aceituno et al.,2008;Lu et al.,2008).We identi-fied a few common leaf-or root-specific DNA methylation bands in both FL478and IR29.These conserved,inter-cultivar,tissue-specific DNA methylation loci could be related to diverse functional differentiations of leaves and roots in rice.Additionally,a large amount of differentially methylated DNA bands was confirmed to be dependent on the growth stage in FL478and IR29,indicating the dynamic nature of cytosine methylation in different developmental stages of rice plants.Strikingly,in both lines,we found distinct decreases in DNA methylation in roots at the seedling stage induced by salt stress,but the salt-sensitive genotype IR29showed a greater decrease in DNA methylation under high-salt conditions.Further analysis indicated that the majority of salt-induced cytosine methylation changes occurred symmetrically on both DNA strands.Our results are consistent with the previ-ously reported induction of a large number of genes by salinity stress in sensitive genotypes (Walia et al.,2005,2007),con-firming that DNA methylation can fine-tune gene expression in response to salt stress.Although DNA demethylation may be either an indirect effect of salt stress or a protectiveTable 3BLAST results of 27polymorphic methylated DNA fragments.MSAP fragment Accession no./locus nameNucl.Id (%)Sequence homologyName Primer (E/HM)Size (bp)S19/3883AT5G08430.166SWIB complex BAF60b domain-containing protein/GYF domain-containing protein S210/312139LOC_Os11g3890066Histone-lysine N-methyltransferas,H3lysine-9specific SUVH1,putative,expressed S38/32115LOC_Os02g26430.163OsWRKY42e Super family of TFs having WRKY and zinc finger domains,expressed T110/312193LOC_Os09g3629079Ser/Thr protein phosphatase family protein,putative,expressed T29/32145LOC_Os05g1504065NB-ARC domain-containing protein,expressed T36/31196LOC_Os10g3821695Photosystem I assembly protein ycf3,putativeT46/315137LOC_Os12g2907072Retrotransposon protein,putative,Ty3-gypsy subclass LOC_Os01g72430j 65NADPH quinone oxidoreductase,putative,expressed T57/3890LOC_Os01g3142074Retrotransposon protein,putative,Ty3-gypsy subclass P15/35110LOC_Os04g1437074Hypothetical proteinP21/32105AC116369.5100OSJNBb0113I20genomic sequenceP39/315147AT3G60480.169Hydrolase,acting on carbon-nitrogen (but not peptide)bonds LOC_Os07g0971069OsFBX220-F-box domain-containing protein,expressedP48/313202CD718353.193An expressed sequence tag for abiotic stressed leaves of Vitis vinifera var.Chardonnay Vitis viniferaLOC_Os04g1684496Cytochrome b 6-f complex subunit 4,putative LOC_Os09g28740100Gibberellin receptor GID1L2,putative,expressedLOC_Os09g3629072Ser/Thr protein phosphatase family protein,putative,expressed P53/33128LOC_Os11g3922066Acyl-coenzyme A oxidase,putative,expressed P62/31199LOC_Os10g3703480Cytochrome P450,putativeLOC_Os07g3636082OsFBO19w F-box and other domain-containing protein,expressed LOC_Os02g2601483Terpene synthase,putative,expressed LOC_Os07g4449975Peroxidase,putative,expressedCB096663.180IRRI Drought Stress Panicle Library,Oryza sativa L Indica Group cDNA clone P710/38145LOC_Os03g2240067Ulp1protease family protein,putative,expressedP85/33143LOC_Os02g4045063DEAD/DEAH box helicase domain-containing protein,expressed P95/33198LOC_Os03g5549061Casein kinase II subunit alpha-2,putative,expressed P105/33143LOC_Os11g2538070Retrotransposon protein,putative,Ty3-gypsy subclassP115/33148LOC_Os05g1459065MCM6-Putative minichromosome maintenance MCM complex subunit 6,expressed P125/33116LOC_Os08g1029074SHR5-receptor-like kinase,putative,expressed P133/3898AT2G42990.165GDSL-motif lipase/hydrolase family protein AT4G33520.364HMA6,metal-transporting P-type ATPase 1P148/3282LOC_Os02g0962088GDSL-like lipase/acylhydrolase,putative,expressedP158/32160LOC_Os02g5296068PHD-finger domain-containing protein,putative,expressed LOC_Os04g0547062Protein kinase,putativeP168/36129LOC_Os04g5493067ABC transporter,ATP-binding protein,putative,expressedP177/38132LOC_Os09g3252666Peptidyl-prolyl cis-trans isomerase,FKBP-type,putative,expressed P186/31187LOC_Os10g3821699Photosystem I assembly protein ycf3,putativeCI435299.1100O.sativa L leaf of seedling 50ppm CuSO4for 3days,Japonica Group cDNA P1910/31291AT4G39680.172SAP domain-containing proteinLOC_Os04g54360.168Glycosyl transferase 8domain-containing protein,putative,expressed423W.Wang et al./Journal of Genetics and Genomics 38(2011)419e 424mechanism by tuning gene expression must be further eluci-dated.Most importantly,this result may suggest that deme-thylation of genes in roots at the rice seedling stage could be an active epigenetic response to salt stress.Many differential changes in DNA methylation were found in this study,and salt stress has induced extensive alterations, such as hypo-methylation or hyper-methylation,at multiple genomic loci across the rice genome.The BLAST results of the differentially methylated DNA sequences showed a similar frequency of methylation in the coding and non-coding regions of the genome,suggesting that DNA methylation changes occur throughout the whole rice genome in response to salt stress.In addition,several genes related to biotic and abiotic stress responses were differentially methylated,sug-gesting that DNA methylation is widely involved in salt stress response by regulating these genes at the transcription level.Our results may point to gene demethylation in seedling roots as an active epigenetic response to salt stress,providing further clues for unraveling the molecular mechanism of rice adaptation to salt stress.AcknowledgementsThis work was supported by the grants from the Central Public-interest Scientific Institution Basal Research Fund of China and The National High Technology Research and Development Program of China(863Program,No. 2007AA10Z191).Supplementary dataSupplementary data related to this article can be found online at doi:10.1016/j.jgg.2011.07.006.ReferencesAceituno,F.F.,Nick,M.,Seung,Y.R.,Rodrigo,A.G.,2008.The rules of gene expression in plants:organ identity and gene body methylation are key factors for regulation of gene expression in Arabidopsis thaliana.BMC Genomics9,438.Ammar,M.H.M.,Singh,R.K.,Singh,A.K.,Mohapatra,T.,Sharma,T.R., Singh,N.K.,2007.Mapping QTLs for salinity tolerance at seedling stage in rice(Oryza sativa L.).Afr.Crop Sci.Proc.8,617e620.Angers,B.,Castonguay,E.,Massicotte,R.,2010.Environmentally induced phenotypes and DNA methylation:how to deal with unpredictable conditions until the next generation and after.Mol.Ecol.19,1283e1295. Ashikawa,I.,2001.Surveying CpG methylation at50-CCGG in the genomes of rice cultivars.Plant Mol.Biol.45,31e39.Bonilla,P.,Dvorak,J.,Mackill,D.,Deal,K.,Gregorio,G.,2002.RFLP and SSLP mapping of salinity tolerance genes in chromosome1of rice(Oryza sativa L.)using recombinant inbred lines.Philipp.J.Agric.Sci.85,68e76. Chen,X.Q.,Ma,Y.,Chen,F.,Song,W.Q.,Zhang,L.,2009.Analysis of DNA methylation patterns of PLBs derived from Cymbidium hybridium based on MSAP.Plant Cell Tissue Organ Cult.98,67e77.Choi,C.S.,Sano,H.,2007.Abiotic-stress induces demethylation and tran-scriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants.Mol.Genet.Genome277,589e600.Dyachenko,O.V.,Zakharchenko,N.S.,Shevchuk,T.V.,Bohnert,H.J., Cushman,J.C.,Buryanov,Y.I.,2006.Effect of hypermethylation of CCWGG sequences in DNA of Mesembryanthemum crystallinum plants on their adaptation to salt stress.Biochemistry(Mosc)71,461e465. Gregorio,G.B.,Senadhira, D.,Mendoza,R.D.,1997.Screening rice for salinity tolerance.In:IRRI Discussion Paper Series,no.22.International Rice Research Institute,Manila,Philippines.Gregorio,G.B.,Senadhira,D.,Mendoza,R.D.,Manigbas,N.L.,Roxas,J.P., Guerta,C.Q.,2002.Progress in breeding for salinity tolerance and asso-ciated abiotic stresses in rice.Field Crops Res.76,91e101.Hu,H.H.,You,J.,Fang,Y.J.,Zhu,X.Y.,Qi,Z.Y.,Xiong,L.Z.,2008.Char-acterization of transcription factor gene SNAC2conferring cold and salt tolerance in rice.Plant Mol.Biol.67,169e181.Hu,T.T.,Liu,C.,Wang,J.K.,Ding,C.W.,Guo,R.L.,Wu,Y.L.,Xu,J.A., Wang,Y.S.,2010.Progress of genetic and breeding on salt tolerance in rice.Mol.Plant Breed.7,110e116.Lu,Y.L.,Rong,T.Z.,Cao,M.J.,2008.Analysis of DNA methylation in different maize tissues.J.Genet.Genomics35,41e48.Lutts,S.,Kinet,J.M.,Bouharmont,J.,1995.Changes in plant response to NaCl during development of rice(Oryza sativa L.)varieties differing in salinity resistance.J.Exp.Bot.46,1843e1852.Ren,Z.H.,Gao,J.P.,Li,L.G.,Cai,X.L.,Wei,H.,Chao,D.Y.,Zhu,M.Z., Wang,Z.Y.,Luan,S.,Lin,H.X.,2005.A rice quantitative trait locus for salt tolerance encodes a sodium transporter.Nat.Genet.37,1141e1146. SAS Institute,1999.SAS/Stat User’s Guide,Version8.2.SAS Inst.,Cary,NC, USA.Shannon,M.C.,Rhoades,J.D.,Draper,J.H.,Scardaci,S.C.,Spyres,M.D., 1998.Assessment of salt tolerance in rice cultivars in response to salinity problems in California.Crop Sci.38,394e398.Steward,N.,Ito,M.,Yamaguchi,Y.,Koizumi,N.,Sano,H.,2002.Periodic DNA methylation in maize nucleosomes and demethylation by environ-mental stress.J.Biol.Chem.277,37741e37746.Suriya-arunroj,D.,Supapoj,N.,Toojinda,T.,Vanavichit,A.,2004.Relative leaf water content as an efficient method for evaluating rice cultivars for tolerance to salt stress.ScienceAsia30,411e415.Takata,M.,Yuji,K.,Yoshio,S.,2005.DNA methylation polymorphisms in rice and wild rice strains:detection of epigenetic markers.Breed.Sci.55, 57e63.Takehisa,H.,Shimodate,T.,Fukuta,Y.,Ueda,T.,Yano,M.,Yamaya,T., Kameya,T.,Sato,T.,2004.Identification of quantitative trait loci for plant growth of rice in paddyfieldflooded with salt water.Field Crops Res.89, 85e95.Wada,Y.,Miyamoto,K.,Kusano,T.,Sano,H.,2004.Association between up-regulation of stress-responsive genes and hypomethylation of genomic DNA in tobacco plants.Mol.Genet.Genome271,658e666.Walia,H.,Wilson, C.,Condamine,P.,Liu,X.,Ismail, A.M.,Zeng,L., Wanamaker,S.I.,Mandal,J.,Xu,J.,Cui,X.,Close,T.J.,parative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage.Plant Physiol.139,822e835. Walia,H.,Wilson,C.,Zeng,L.,Ismail,A.M.,Condamine,P.,Close,T.J., 2007.Genome-wide transcriptional analysis of salinity stressed japonica and indica rice genotypes during panicle initiation stage.Plant Mol.Biol.63,609e623.Wang,Y.M.,Lin,X.Y.,Dong,B.,Wang,Y.D.,Liu,B.,2004.DNA methylation polymorphism in a set of elite rice cultivars and its possible contribution to intercultivar differential gene expression.Cell.Mol.Biol.Lett.9,543e556. Xiong,L.Z.,Xu,C.G.,Maroof,S.,Zhang,Q.F.,1999.Patterns of cytosine methylation pattern in an elite rice hybrid and its parental lines detected bya methylation sensitive amplification polymorphism technique.Mol.Gen.Gene261,439e446.Yoshida,S.,Forna,D.A.,Cock,J.H.,Gomez,K.A.,boratory Manual for Physiological Studies of Rice.International Rice Research Institute, Los Ban˜os,Philippines.Zhang,M.S.,Kimatu,J.N.,Xu,K.Z.,Liu,B.,2010.DNA cytosine methylation in plant development.J.Genet.Genomics37,1e12.424W.Wang et al./Journal of Genetics and Genomics38(2011)419e424。
