Section I Gene library and screening
基因文库的构建

为了最大限度保证度的断裂。制备的
DNA分子规方法制备的染色体 DNA的长度一般在100 kb左右 如果先将细胞固定在低融点凝 构建流程
随机引物引导的 cDNA 合成是采用 6-10 个随机 碱基的寡核苷酸短片段来锚定 mRNA 并作为反转录的 起点。 由于随机引物可能在一条 mRNA 链上有多个结合 位点而从多个位点同时发生反转录,比较容易合成特 长的 mRNA 分子的 5‘端序列。 随机引物 cDNA 合成的方法不适合构建 cDNA 文 库,一般用于克隆特定 mRNA 的 5' 末端,如 RTPCR 和物体中,不同组织和细胞在不同时段的mRNA
种类不同(即基因
① 细胞总 RNA 的提取和 mRNA 分离; ② 第一链 cDNA 合成; ③ 第二链 cDNA 合成; ④ 双链 cDNA 克隆
Total RNA的提取 mRNA的分离 cDNA双链的合成 载体的制备 cDNA双链和载体的连接 转化或转染 快速鉴定 pBlueScript cDNA库扩增
5’
AAAAAAAAAAAAAAOH 3’ TTTTTTTTTTTTTTp 5’ T4-DNA ligase AAAAAAAAAAAAAAOH 3’ TTTTTTTTTTTTTTp 5’
5’ 3’
cDNA第二链的合成
引导合成法:获得的双链cDNA 能保留完整的5’端序列
G 5‘ppp’5 G
AAAAAOH 3’ TTTTTp 5’ TdT dCTP AAAAACCCCCCCOH 3’ TTTTTp 5’ NaOH TTTTTp 5’
A A A A
白酶K、RNaseA的缓冲液中浸泡,可获得 >100 kb大小的DNA片段基因的构建程序③基因组DNA的切割
基因克隆,表达及功能研究

更多生物医学资源下载:
Lac Z
本资源由药智网收集整理: A plasmid vector for gene expression
Expression vectors: allowing the exogenous DNA to be inserted, stored and expressed. 1. Promoter and terminator for RNA transcription are required. 2. Intact ORF and ribosomal binding sites (RBS) are required for translation. 3. Include:(1) bacterial expression vectors, (2) yeast expression vectors, (3) mammalian expression vector
本资源由药智网收集整理:
Recombinant plasmid (contain insert)
更多生物医学资源下载:
back
Multiple cloning sites 本资源由药智网收集整理:
更多生物医学资源下载:
本资源由药智网收集整理:
本资源由药智网收集整理:
Cosmid vectors
1. Utilizing the properties of the phage l cos sites in a plasmid vector. 2. A combination of the plasmid vector and the COS site which allows the target DNA to be inserted into the l head. 3. The insert can be 37-52 kb
Daudi细胞系特异性Fab噬菌体抗体库的构建_筛选与初步鉴定

#论著#文章编号:1007-8738(2009)02-0150-05Daudi 细胞系特异性Fab 噬菌体抗体库的构建、筛选与初步鉴定申咏梅1,2*,杨晓春3,董宁征4,白 霞4(苏州大学附属第二医院:1放免中心,2检验教研室,3磁共振室,江苏苏州215004;4江苏省血液研究所,苏州大学附属第一医院,江苏苏州215006)收稿日期:2008-06-30; 接受日期:2008-09-08基金项目:国家自然科学基金资助项目(30400111);江苏省自然科学基金资助项目(BK2004041)作者简介:申咏梅(1969-),女,重庆人,副教授,医学博士Te:l 0512-********;E-m ai:l szs y m@yahoo .co *Corres pond i ng authorG enerati on ,screeni ng and prelim i naryi dentification of specific Fab phage an -ti body li brary agai nst D audi cell strai nSHE N Yong-m ei 1,2*,YANG X iao -chun 3,DONG N ing-zheng 4,BAI X ia41R adi o i m munoassay Cente r ,2D epart m ent of C linical L abo rato ry ,3D epart m ent o f M agnetic R esonance ,Second A ffiliated H osp ita,lSoocho w U niversity ,Suzhou 215004;4Ji ang su Institutie o fH em a -to l ogy ,F irst A ffiliated H ospita ,lSoochow U n i ve rsity ,Suzhou215006,Ch i na[Abstract] A I M:To gene ra t e and screen the specificFab p hage an ti b ody library aga inst human Daud i ce ll stra in in B-ly mphoma and i d en tify t he positive c l o nes .M ETH-OD S :BA LB /c m ice we re m i mun ized w it h Daud i ce lls ,and an tisera we re titra ted by EL I SA .Fo llo w ing t he demonstra -tion o f su fficien t an tibody tit e r ,to t a l RNA was extract ed from sp l e n ic ly mphocytes o f the m i m unized m i c e and RT -PCR w as used to am p lify J ligh t cha in and Fd fragmen ts o f heavy chain .A ft e r re strictive d igestion w ith S a c I/Xba I and X ho I/S pe ,I the J ligh t cha in and the Fd fragm ents were succes -sive ly inse rt ed in t o the p hagem id vector pComb 3H -SS and then e l e ctropo ra t ed in t o E.co li XL 1-B l u e .The spe cific Fab phage antibody li b ra ry aga inst Daud i ce ll stra i n in hu man B-l y m pho m a was constructed by in f ec tion o f he l p e r phage VC -SM 13.Fo llo w ing six rounds of b iopanning w ith Daud i ce lls ,the antigen b inding acti v ities o f rando m clone s we re t ested by EL I S A to se lect the positive c l o nes ,wh ich we re further DNA sequenced ,expre ssed i n E.co li X L 1-B lue and i d en t -i fi e d byW estern b lo.t RE SULTS :The Fab phage an tibody l -i b ra ry w it h 3.13@107size was construct ed and f our positive c lones wh ich spec ifica lly recogn ized Daudi ce ll stra in w ere -i so l a t ed .In am i n o ac i d seq uences ,the var i a b le heavy do -m ains (V H )we re found to be 80%-94%and va riab l e ligh tdom a i n s (V L )88%-95%homo logous w ith respective m u -r i n e ge r m line genes in GenBank .Fu rthe r mo re ,so lub l e Fab antibod ies o f t he po sitive clone s we re successfu ll y ex -p ressed in E.c o l i XL 1-B l u e and t he reactivity w ith t he memb rane p ro t e ins o f Daud i ce lls wa s demonstra t ed by W estern b l o .t CONCLUSI ON :Fab phage an ti b ody library is succe ssfu lly constructed and specific an tibod ies aga instmemb rane antigens in Daud i ce lls are ob ta ined ,wh ich pro -vides an expe rm i en t a l f ounda tion for the further i n ve stiga tion o f B -ly m phoma m i munotherapy .[K ey w ords]B-ly mphoma ;Fab ;Phage anti b ody li b ra ry ;Daud i ce ll stra i n ;pComb 3H -SS vector[摘要] 目的:构建针对B 细胞淋巴瘤D aud i 细胞系噬菌体抗体库,从中筛选特异性抗体,并对所筛选出的阳性克隆进行鉴定。
基因工程外文翻译(中英对照)(可编辑)

基因工程外文翻译(中英对照)Retrovirus-mediated gene transfer and expression cloning: Powerful tools in functional genomics Most of the human genome has now been sequenced and about 30,000 potential open reading frames have been identified, indicating that we use these 30,000 genes to functionally organize our biologic activities. However, functions of many genes are still unknown despite intensive efforts using bioinformatics as well as transgenic and knockout mice. Retrovirus-mediated gene transfer is a powerful tool that can be used to understand gene functions. We have developed a variety of retrovirus vectors and efficient packaging cell lines that have facilitated the development of efficient functional expression cloning methods. In this review, we describe retrovirus-mediated strategies used for investigation of gene functions and function-based screening strategies 2003 International Society for Experimental Hematology. Published by Elsevier Inc.摘要:人类基因组的大部分现在已经测序完成,大约30,000潜在的开放阅读框已经确定,表明我们使用这30,000个基因管理我们的生物学活和功能性。
全基因组CRISPRCas9高通量筛选人体细胞中的功能基因组学

全基因组CRISPRCas9高通量筛选人体细胞中的功能基因组学点击左上方蓝字“HACS”!全基因组CRISPR/Cas9高通量筛选人体细胞中的功能基因组学英文名:《Genome-Wide CRISPR/Cas9 Screening for High-Throughput Functional Genomics in Human Cells》简单说就是利用CRISPR/Cas9技术对人类的基因组(23对染色体上的全部基因,大约两万个)进行筛查,看看这些基因到底有哪些功能。
摘要基因作为生命的本质,基因的具体功能一直是科学家的研究方向。
我们知道人体的基因组大约有两万个基因,研究一个基因的工作量就已经很大,这意味我们不可能一个一个地研究每一个基因。
如果能以高通量的方式鉴定基因的功能,那么我们就可能进行全基因组基因功能研究,而近几年来的CRISPR/Cas9系统已经可以满足这样的需求,这也是目前研究的热点。
在这里,我们介绍一种可以产生覆盖全人类基因组的慢病毒单链引导RNA (sgRNA)文库的通用方法。
CRISPR/Cas9系统有两个重要组成部分,包括Cas9蛋白并sgRNA,前者可以切割DNA,相当于一个士兵;后者起到引导作用,将CRISPR/Cas9这个编辑器引导到靶位点,相当于指挥官;sgRNA可以覆盖全基因组就意味着CRISPR/Cas9这个编辑器可以对全基因组进行编辑。
因为可以覆盖全基因组,而且具有普适性,这个实验方法将成为各种模式动物,各种生物活动相关基因研究的有力工具。
关键词 CRISPR-Cas9系统,高通量,敲除,筛选,sgRNA。
一. 背景介绍CRISPR/Cas9系统是古菌和细菌常用的一种防御系统[1]。
不过这并不重要,毕竟英雄不问出处。
目前,应用最最广泛的工程设计CRISPR/Cas体系由Cas9核酸酶和单链引导RNA (sgRNA)组成。
引导 RNA (sgRNA) 是一段长度为20bp的核糖核酸序列。
genecard的all section -回复

genecard的all section -回复GENECARDS: A Comprehensive Resource for Genetic InformationIntroduction:In the field of genetics, the quest to understand the functions and roles of individual genes and their associated proteins is of paramount importance. With the advent of high-throughput sequencing technologies, the accumulation of genetic data has skyrocketed over the past few decades. However, the challenge lies in the effective analysis and interpretation of this vast amount of genetic information. This is where GeneCards, a comprehensive genetic database, plays a crucial role by providing a user-friendly platform for accessing and understanding genetic information. In this article, we will explore the various sections of GeneCards and how they contribute to our understanding of genes and proteins.Section 1: Gene SummaryThe Gene Summary section of GeneCards provides a concise overview of the gene of interest. It includes information such as gene name, location, and aliases. Additionally, it provides information on the protein encoded by the gene, its function, and its involvement in various biological processes. This section isespecially useful for researchers and clinicians looking to quickly gather pertinent information about a gene of interest.Section 2: Gene FunctionThe Gene Function section of GeneCards delves deeper into the molecular and cellular functions of the gene and its associated protein. It provides detailed information on the protein's involvement in signaling pathways, metabolic processes, and cellular localization. GeneCards also highlights any known protein-protein interactions and the roles of the gene in disease pathways. This section aids researchers in understanding the intricate functions and pathways in which the gene is involved.Section 3: Gene ExpressionUnderstanding the tissue-specific expression patterns of genes is crucial for comprehending their roles in physiological and pathological processes. The Gene Expression section of GeneCards provides information on the expression levels of genes across various tissues and cell lines. This data is obtained fromhigh-throughput transcriptomic studies, allowing researchers to identify the organs or tissues in which a gene is predominantly expressed. This section aids in elucidating the normal physiologicalroles of genes and their potential involvement in diseases.Section 4: Gene OntologyGene Ontology (GO) is a widely accepted system for categorizing gene functions into standardized terms. The Gene Ontology section of GeneCards utilizes this framework to classify the functions, processes, and cellular components associated with a gene's protein product. By providing a standardized vocabulary, researchers can easily compare and analyze the functions of different genes. This section also aids in identifying potential gene-gene interactions and cross-talk within biological processes.Section 5: PathwaysThe Pathways section of GeneCards provides a comprehensive overview of the various signaling pathways in which the gene of interest is involved. It includes information on both canonical and non-canonical pathways, enabling researchers to gain insights into the complex regulatory networks governing gene function. This section also highlights the gene's involvement in disease pathways, facilitating the identification of therapeutic targets and potential biomarkers.Section 6: VariantsGenetic variation plays a critical role in human health and disease. The Variants section of GeneCards catalogs known genetic variants associated with the gene of interest. It includes single nucleotide polymorphisms (SNPs), insertions, deletions, and other types of structural variations. This section provides information on the frequency of these variants in different populations and their potential impact on gene function. It is an invaluable resource for researchers studying the genetic basis of diseases and the impact of genetic variation on drug response.Section 7: PublicationsThe Publications section of GeneCards compiles information on scientific papers that have mentioned the gene of interest. It includes links to these publications, allowing researchers to delve deeper into the literature and explore the findings in more detail. This section serves as a valuable resource for researchers looking to stay up-to-date with the latest advancements in the field.Conclusion:GeneCards serves as a comprehensive resource for genetic information, providing researchers and clinicians with auser-friendly platform to access and understand the functions and roles of individual genes and their associated proteins. Its various sections, such as Gene Summary, Gene Function, Gene Expression, Gene Ontology, Pathways, Variants, and Publications, collectively contribute to our understanding of the genetic landscape. By harnessing the power of high-throughput sequencing and integrating diverse sources of data, GeneCards plays a pivotal role in advancing our knowledge of genetic processes, disease mechanisms, and potential therapeutic targets.。
(高考生物)生物英语证书考试(PEC)生物化学词汇

(生物科技行业)生物英语证书考试 (PEC) 生物化学词汇生物英语证书考试(PEC) - 生物化学常用词汇nucleoside核苷Okazakifragment冈崎片段oncogene癌基因,原癌基因onecarbonunit一碳单位operator操控基因operon操控子oroticacid乳清酸ossification成骨作用oxaloaceticacid草酰乙酸oxidases氧化酶类oxidativephosphorylation氧化磷酸化oxidoreductase氧化复原酶palindrome回文构造pancreaticlipase胰脂肪酶pantothenicacid遍多酸pentose戊糖pentosephosphatepathway磷酸戊糖门路pepsin胃蛋白酶pepsinogen胃蛋白酶原peptide肽peptidebond肽键peptidylsite肽基位或P 位peroxidase过氧化物酶phenylalanine苯丙氨酸phosphatidicacid磷脂酸phosphogluconate磷酸葡萄糖酸phospholipase磷脂酶plasmid质粒polycistron多作用子polypeptide多肽porphyrin卟啉precipitation积淀preproalbumin前清蛋白原primarystructure一级构造primase引起酶primer引物glucogenicaminoacid生糖氨基酸glucokinase葡萄糖激酶gluconeogenesis糖 (原 )异生作用glutamicacid谷氨酸glutaminase谷氨酰胺酶glutamine谷氨酰胺glutathione谷胱甘肽glycerol甘油glycine甘氨酸glycogen糖原glycogenphosphorylase糖原磷酸化酶glycogensynthase糖原合成酶glycolysis糖酵解guanosine鸟苷helicase解链酶(解旋酶)heme血红素heteroduplex杂化双链hexokinase己糖激酶histamine组胺histidine组氨酸housekeepinggene管家基因hybridization杂交hydrogenbond氢键hydrolase水解酶类hydroperoxidases氢过氧化酶类hydrophobicbond(hydrophobicinteraction)疏水键hydroxyapatite羟磷灰石hydroxymethylglutarylCoAcleavageenzymeHMGCoA裂解酶hydroxymethylglutarylCoAsynthetaseHMGCoA合酶Hydroxyproline羟脯氨酸acceptorsite受位acetone丙酮activator激活蛋白,激活剂,活化物adenine(A)腺嘌呤adenosine腺苷aerobicdehydrogenase需氧脱氢酶alanine丙氨酸albumin白蛋白,清蛋白allopurinol别嘌呤醇allostericeffect别构 (位 )效应allostericenzyme变构酶,别位酶allostericregulation别构调理amine胺aminoacylsiteA位,氨酰基位anticodon反密码子arginine精氨酸ascorbicacid 抗坏血酸 (维生素 C) asparagine天冬酰胺asparticacid天冬氨酸asymmetrictranscription不对称转录attenuator衰减子base 碱基basepairing碱基配对bilepigment胆色素biotin生物素biotransformation生物转变calcitriol1,25二羟胆骨化醇(钙三醇)calciumdependentproteinkinaseCa依靠性蛋白激酶,蛋白激酶C(C 激酶 ) Calmodulincarbohydrate糖carnitine肉毒碱catalase触酶,过氧化氢酶cephalin脑磷脂denovosynthesis从头合成degradation降解denaturation变性deoxycholicacid脱氧胆酸deoxyribonucleotide脱氧核糖核苷酸dialysis透析dihydroxyacetonephosphate磷酸二羟丙酮disulfidebond二硫键DNApolymeraseDNA聚合酶domain 域 ,构造域 ,功能区donorsite给位doublehelix双螺旋effector 效应器 ,效应物elongation延伸endopeptidase内肽酶enhancer加强子enolphosphopyruvate磷酸烯醇式丙酮酸enzyme酶essentialaminoacid必要氨基酸essentialfattyacid必要脂肪酸exon外显子exopeptidase外肽酶fat 脂肪feedbackinhibition反应克制作用feritin铁蛋白ferrochelatase亚铁螯合酶folicacid叶酸freefattyacid游离脂肪酸freeradicals自由基fructosediphosphatase果糖二磷酸酶genecloning基因克隆geneexpression基因表达genelibrary基因文库genetransfer基因导入 ,转基因geneticcode遗传密码geneticengineering基因工程geneticrecombination基因重组genome染色体基因,基因组globin珠蛋白hypocalcemia低钙血症induction引诱initiatorcodon起动信号 ,开端密码子intermediarymetabolism中间代谢ionicbond离子键isocitratedehydrogenase异柠檬酸脱氢酶isoleucine异亮氨酸isomerase异构酶类isozyme同工酶jaundice黄疸ketogenicaminoacid生酮氨基酸keyenzyme重点酶kinase激酶lactate乳酸盐lecithin卵磷脂leucine亮氨酸ligase连结酶linoleate亚油酸linolenate亚麻酸lipoicacid硫酸楚lipoid类脂lipoprotein脂蛋白lithocholicacid石胆酸lyases 裂合酶类malate苹果酸malateaspartateshuttle苹果酸天冬氨酸穿越metabolicregulation代谢调理mitogenactivatedproteinkinase分裂原活化蛋白激酶mixedfunctionoxidase混淆功能氧化酶molecularcloning分子克隆moleculardisease分子病monooxygenase单加氧酶monooxygenasesystem单加氧酶系统nicotinamide烟酰胺,尼克酰胺nitrogenbalance氮均衡pyruvatecarboxylase丙酮酸羧化酶pyruvatedehydrogenasecomplex丙酮酸脱氢酶复合体pyruvatekinase丙酮酸激酶quaternarystructure四级构造recombinantDNA重组 DNA geneticengineering基因工程regulatorygene调理基因renaturation复性repair修复replication复制repression隔绝residue残基respiratorychain呼吸链restrictionendonuclease限制性内切核酸酶retinol 视黄醇 ( 维生素 A) reversetranscriptase逆转录酶reversetranscription逆转录作用saltingout盐析salvagepathway挽救 ( 从头利用 )门路screening挑选secondarystructure二级构造semiconservativereplication半保存复制sensestrand存心义链sequence序列serine丝氨酸signalrecognitionparticle信号肽辨别颗粒silencer克制子simpleprotein纯真蛋白质specificity特异性splicing剪接作用squalene鲨烯stagespecificity阶段特异性stercobilinogen粪胆素原stress 应激structuralgene构造基因substrate作用物substratelevelphosphorylation作用物 ( 底物 )水平磷酸化subunit亚单位,亚基succinatedehydrogenase琥珀酸脱氢酶supersecondarystructure超二级构造Taurine牛磺酸telomerase端粒酶telomere 端区 (端粒 )templatestrand模板链termination停止terminator停止子terminatorcodon停止信号tertiarystructure三级构造thiamine 硫胺素 (维生素 B1) threonine苏氨酸thymidine胸苷,胸腺嘧啶核苷thymine(T)胸腺嘧啶tocopherol生育酚proalbumin清蛋白原processing加工proenzyme酶原proline脯氨酸promoter 启动基因 (启动子 ),催化剂prostheticgroup辅基protease蛋白酶pyridoxal吡哆醛pyridoxamine吡哆胺。
英语作文基因改造过程

英语作文基因改造过程Genetic Modification Process。
Genetic modification is the process of altering the genetic makeup of an organism by introducing foreign DNAinto its genome. The process involves several steps, including isolation of the desired gene, cloning of the gene, and insertion of the gene into the target organism.The first step in genetic modification is the identification and isolation of the desired gene. This is done using a variety of techniques, including PCR (polymerase chain reaction), which amplifies the gene of interest, and restriction enzymes, which cut the gene outof the DNA sequence. Once the gene has been isolated, it is cloned using a vector, such as a plasmid or a virus.The next step is to insert the cloned gene into the target organism. This is done using a variety of techniques, including electroporation, which uses an electric field tocreate temporary pores in the cell membrane, allowing the foreign DNA to enter the cell. Another technique is microinjection, which involves injecting the foreign DNA directly into the nucleus of the target cell.Once the foreign DNA has been inserted into the target organism, it must be integrated into the genome. This is done through a process called homologous recombination, which involves the replacement of a segment of the target DNA with the foreign DNA. This process is facilitated by the use of selectable markers, which allow scientists to identify cells that have successfully integrated the foreign DNA.After the foreign DNA has been integrated into the genome, the modified organism must be screened to ensure that the desired trait has been expressed. This is done using a variety of techniques, including PCR, which can be used to detect the presence of the foreign DNA, and phenotypic analysis, which involves the observation of physical traits.In conclusion, genetic modification is a complexprocess that involves several steps, including isolation of the desired gene, cloning of the gene, insertion of thegene into the target organism, integration of the foreign DNA into the genome, and screening for the desired trait. Despite the controversy surrounding genetic modification,it has the potential to revolutionize medicine, agriculture, and industry.。
人民大2023农林国际学术交流英语 PPTUnit 1 Gene Editing

G__M5O__
It is a plant, animal, microorganism or other organisms whose genetic makeup has been modified in a laboratory using genetic engineering or transgenic technology.
2. Can you arrange them in chronological order?
B-C-A
Part I History of Gene Editing
Timeline Method and Technique selective breeding discovery of DNA
Detailed Information It can strengthen ____u_s_e__f1u_l_t_r_a_i_t_s___ in plants and animals, but never truly understand ___h_o2w___i_t. worked
Part II The Working Principles of CRISPR-Cas9
Can you retell the working principles of CRISPR after watching the video clip?
Part II The Working Principles of CRISPR-Cas9
This creates combinations of plant, animal, bacterial and virus genes that do not occur in nature or through traditional crossbreeding methods.
定点突变和定向进化的流程

定点突变和定向进化的流程英文回答:Site-Directed Mutagenesis.Site-directed mutagenesis (SDM) is a method used to introduce specific mutations into a DNA sequence at a predetermined location. This technique allows researchers to study the effects of specific mutations on gene function and regulation. SDM is commonly used in molecular biology, genetics, and protein engineering.The basic principle of SDM is to use a short oligonucleotide primer that is complementary to the target DNA sequence, except for a single nucleotide change at the desired mutation site. The primer is designed to bind to the target DNA and extend by DNA polymerase, incorporating the desired mutation into the newly synthesized DNA strand.Several methods are commonly used for SDM, including:PCR mutagenesis: The target DNA sequence is amplified using PCR with the mutagenic primer. The amplified DNA is then treated with restriction enzymes and ligated to create a plasmid containing the desired mutation.Cassette mutagenesis: A synthetic DNA fragment containing the desired mutation is inserted into a plasmid that contains the target DNA sequence. The plasmid is then transformed into a suitable host organism.Oligonucleotide-directed mutagenesis: A synthetic oligonucleotide primer containing the desired mutation is hybridized to the target DNA sequence. The primer is then extended by DNA polymerase to incorporate the mutation into the newly synthesized DNA strand.Directed Evolution.Directed evolution is a method for evolving proteins or other molecules to improve their properties or functions. This technique involves iteratively selecting and mutatinga population of molecules and screening for individuals with the desired traits. Directed evolution is commonly used in protein engineering, enzyme engineering, and antibody engineering.The basic principle of directed evolution is to start with a population of molecules with diverse genetic sequences. This population is then subjected to selection pressure to identify individuals with the desired traits. The selected individuals are then mutated to create a new population with an increased frequency of the desired traits. This cycle of selection and mutation is repeated until a population with the desired properties is obtained.Several methods are commonly used for directed evolution, including:Random mutagenesis: The starting population is subjected to random mutagenesis using techniques such as error-prone PCR or chemical mutagenesis.Site-directed mutagenesis: Specific mutations areintroduced into the starting population using techniques such as SDM.DNA shuffling: DNA fragments from differentindividuals are recombined to create a new population witha higher diversity of genetic sequences.中文回答:定点突变。
Section I-基因文库与筛选
Second strand synthesis
对第一链的3’端 加尾” 对第一链的 端“加尾”, 再用与之互补的引物合成
第二链,保证获得全长 第二链,保证获得全长cDNA。(Fig 1.1) 。
Molecular Biology Course
Section I Gene libraries es and screes
remove protein, lipids and other unwanted macromolecules by protease digestion and phase extraction.
extracted DNA directly from cells
Prokaryotes
I1 Genomic libraries
I2 cDNA libraries
Three methods to isolate mRNA
1.传统的提取方法是把总 RNA 通过寡聚 (dT)-纤维素 传统的提取方法是把总 纤维素 柱。 2.直接将结合了寡聚 (dT)的磁珠加入到细胞裂解液中, 直接将结合了寡聚 的磁珠加入到细胞裂解液中, 直接将结合了 的磁珠加入到细胞裂解液中 利用磁性将吸附了mRNA的磁珠分离出来后,再用溶 的磁珠分离出来后, 利用磁性将吸附了 的磁珠分离出来后 剂把mRNA从磁珠上洗脱。 从磁珠上洗脱。 剂把 从磁珠上洗脱 3. 利用蔗糖梯度,从细胞裂解液中制备 利用蔗糖梯度,从细胞裂解液中制备mRNA-核 - 糖体复合物,再提取mRNA。 糖体复合物,再提取 。
动物实验手册说明书

A Alligators.......................................................80,219,225, 226,229,230,243,247–262 Amphibians..............................................v,1,3,4,17–30, 163,222,295,303,393,394,420,423,424 Animal welfareanesthesia (351)animal facilities.....................407,408,413,415,416 ethics.....................................407,409,410,412,421 guidelines...........................................6,272,407–429 organizations.................................407,411,412,414 protocols......................408,412,415,422–424,426 regulatory compliance..................409,412,416,426 Antibodiesalpha-tubulin................................................20,25,28 bromodeoxyuridine.......................227,231,237,238flag.....................................................................64,109 gamma-tubulin....................................................20,28 IgM...................................................................69,324 MAPK (64)monoclonal antibodies..................20,64,69,72,184 phosphotyrosine..................................................64,68 PLCgamma. (64)pMAPK (64)proliferating cell nuclear antigen(PCNA) (236)Src1/2 (64)SSEA-1...................................................319,321,323 tyrosine phosphorylation (68)uroplakin III (68)vimentin (236)Antisensemorpholino.............................2,8,77,328,339,347 oligonucleotide.......................2,6,77,328,339,347 Avian.............................................99–109,248,317–325BBalbiani body(Bb)manual isolation....................................267,269,271 percoll gradient purification..................266,268–271 proteomics......................................................295–302 visualization.........................................................v,278C Calcium.....................................................41–56,85,101, 220,222,228,239,241,242,272Capillary tubes,see Micro-tubes Chick......................v,75–80,88,99,100,102,106,184 Chromatin immunoprecipitation(ChIP)............99–102, 105,109Cloning,see Somatic cell nuclear transfer(SCNT)Co-immunoprecipitation(Co-IP).......................99–101, 106,107Clustered regularly interspaced short palindromic repeats (CRISPR-Cas9)CHOPCHOP................................380,382,383,388 designing guide RNA...........................378,379,386 propagation of INDELS.. (388)screening insertions/deletions(INDELS).................................................377–391 synthesis of guide RNA.........................379–381,384 Cultured cells.................................64,70,217,296,365DDNApreparation.......................................................36,150 plasmid cloning (34)template preparation...............................36,151,157, 186,194,195vector construction.............................78,82,88,150EEggactivation.................................................3,41–43,61, 63,68,71,360,367,368,370,372,373 animal-vegetal polarity........................................69,70 chorion removal............................331,332,339,362 defolliculation.................................................6,47,48 dejellying.................................21,307,308,311,312 enucleation....................................................353,362, 363,366,367,370extrusion............................................9,119,125,304 laying........................................6,9,11,78,119–121, 123,125,249,306–308,310–312,314,325Francisco J.Pelegri(ed.),Vertebrate Embryogenesis:Embryological,Cellular,and Genetic Methods,Methods in Molecular Biology, vol.1920,https:///10.1007/978-1-4939-9009-2,©Springer Science+Business Media,LLC,part of Springer Nature2019431Embryosasters...........................................................18,22,394 bead implantation.. (77)blastomeres............................................266,393,394 bleaching embryos..............................................22,24 cell-free extracts..................................................37,47, 48,50,52,67,69,270,271cell implantation (167)cell shape.................................................393–395,404 chromatin fragmentation. (103)clearing............................................................213,225 crosslinking.....................................................102,104 cytoplasmic collection......................................49,164 dechorionation.....................121,332,334,337,365 dissociation...........................102,104,334,339,365 electroporation............................................77,78,80, 83,90,91,95,106,248,252,254,256 ex ovo culture..............................................77,79,83, 84,90,96,248,250,252,255,261explant culture....................................................77,79, 82–86,96,100,102,166,248,255explant processing............................................78,251 ex utero culture..............................................163–179 incubation....................................................77,78,86, 207,211,324in ovo culture................................77–79,82,83,248 live imaging.....................................................18,112, 169,170,172–175,179,296,393lysis..............................................................35,37,103 membrane microdomains(MD)........................67–70 microtubules........................................................17–30 mitotic spindle...................................17,28,126,394 mounting...................................................26,27,148, 149,153,156,165nucleic acid electroporation.............................78,165 RNA extraction (34)spindle positioning.........................................394,398 tissue sectioning....................................133,140,216 Expression systemsß-galactosidase (205)fluorescent protein............................33,78,165,183 luciferase.. (33)nanoLuc (34)reporter constructs.............................................33,82, 183,205,217,261,286,344tomo-seq (130)FFertilizationin vitro fertilization.......................................6,11,18, 63,67,68,112,114,119,125,353–355,360,361,363–365,368–370,372,374,419 mating systems...............................................354,363 strain selection.........................................................353GGeckoanesthesia...............................................222,234,242 biopsy.....................................................222,225,234 husbandry.............................221–224,228,232,233 perfusion................................................222,233,242 sex determination...........................................221,223 Gene expression librariesamplified RNA (136)cDNA synthesis.............................130,131,134–137 gene library preparation........................131,137,138 Gene functiongain of function............................................78,82,88 genetic screen. (123)knockdown......................................77,78,82,87,88 Genome editing,see CRISPR-Cas9Germ cellsblastomere transplant (329)germ cell transplant.......................................317–325, 331,334,335germ line chimera..........................................327–329 host embryo sterilization.......................328–330,332 host-transfer..................................................324–325, 334–338,346–349isolation..........................................................296,334 migration........................................................328,329 Germplasm..............................................2,265–274,295 IInterspecies hybrid (vi)In vitro transcription(IVT)...........35,36,131,136,137 LLabelingAlcian blue.............................................225,229,234 Alizarin red............................................225,229,234 alkaline phosphatase (200)bromodeoxyuridine (227)calcium indicators................................................48–52 DAPI.................................................................29,286 DiOC6...........................................281,286,287,292 DNA dyes............................................................20,25 Fixation...............................................21–23,91,191, 196–198,202–204,206–209,234–238,256–259,288–290,323Harris hematoxylin counterstain (236)immunofluorescence............225,230,231,236,237 immunohistochemistry.........................184,278,286 in situ hybridization.............184,186,210,248,286 MitoTracker....................................................286,292 TO-PRO-3...................................................20,25,29 vital dyes...................................9,286,299,324,337 YO-PRO-1..........................................................20,29432V ERTEBRATE E MBRYOGENESIS:E MBRYOLOGICAL,C ELLULAR,AND G ENETIC M ETHODS IndexMMaternal genes....................................1–3,277,343,344 Micro-computed tomography(microCT).................248, 250,252,255,257,261 Microinjection...................................................5,8,9,12, 35–37,49–51,94,145,158,248,297,298,300,319,324,330–333,339,345,347,350,356,366,367,369,370Micro-tubescalibration (152)injection..............................................8,36,152,331, 332,334,337,349–350transplantation in adults..........................10,346–349 transplantation in embryos....................324,332–338 Mount.........................................................19,21,22,26, 27,49,147,159,184,186,187,189,191,193,196–198,200,201,203–209,211,216,217,235–237,324Mousepostimplantation embryo.............................167,168, 170,174–176,178preimplantation embryo...............................166,167, 169,172–174mRNA microinjection (36)mRNA sequencing(RNA-Seq) (129)NNeedles,see Micro-tubesOOocyteculture..........................................................1–13,273, 279,281,284,286,291,292,296,298 dissociation............................................279,285,286 endoplasmic reticulum..........................277,286,295 extract..........................................................18,38,47, 48,52,53,55,281follicle.........................................................7,8,12,46, 54,284–286,347,350histology (286)immunohistochemistry.................................278,282, 286,287,289,292maturation...............................................9,12,42,60, 61,71,72,284,291,351mitochondria.................................................265,267, 270,272,273,277,281,286–288,295 ovary dissection..............................................284,285 sorting....................................................279,281,286 transplantation......................3–5,317,345–347,351 Oogenesis............................................................1,59–72, 265,269,277,344,357Optogenetics........................................................143–161 Ovaries.............................................................5–8,12,28, 46,59,270,278,281,284–287,289,291,292,297,298,300,344–346,348,350,351PPloidygynogenesis (112)gynogenetic diploid.......................................121,122 haploid...................................................112,117,120 heat shocked (123)tetraploid.......................................113,117,120–122 triploid (303)Ploidy analysischromosome spreads.....................113,117,121,122fluorescence-assisted cell sorting(FACS).............113, 118,122,123genetic markers (371)Polymerase chain reaction(PCR).................................88, 117,122,127,130,131,138,140,147,151,156,157,194,195,257,320,323,325,371,379–381,385,387,388,390,420Primordial germ cell,see Germ cellProtein analysisCoomassie Brilliant Blue (68)immunoblotting (69)mass-spectrometry (70)SDS-PAGE..........................................................68–70 silver staining.......................................................68,70 two-dimensional gel electrophoresis (70)Protein-DNA interactions.............................99–109,377 Protein-protein interactions...................................99–109R Rehydration.............................................20,22,213,251 Reptilia (219)RNA(mRNA)concentration..................................................37,136, 137,152,158,386–388microinjection..........................................36,333,347 purification. (66)synthesis.........................................................147,150, 152,157,212,260,379–381,384–386,388,390 S Sectioning.....................................................19,130,131, 133,140,191,203,205,211,216,235–237 Signal transduction (43)Somatic cell nuclear transfer(SCNT)egg enucleation (366)microinjection...............................356,366,367,369V ERTEBRATE E MBRYOGENESIS:E MBRYOLOGICAL,C ELLULAR,AND G ENETIC M ETHODSIndex433Somatic cell nuclear transfer(SCNT)(cont.)preparation of nucleus-donor cells........................355, 356,365,366,369Spermextraction (359)in vitro fertilization............................................63,67, 68,112,369polyspermy (61)UV-inactivation....................120,121,123–125,127 Src............................................................................42,45, 54,63,64,68,71,72Subcellular protein localization...........................143–161 T3D modelinginput imaging stack........................................397–399 Mathworks Matlab.. (394)simulations.............................................394,398,401 surface evolver................................................394–396 Time-lapse...........................................................161,164, 166,169,175 Transfection.............................................................64,66, 78,88,248,254,261Transgenic..................................................118,123,124,127,146,183,205,286,318,348,357,365,369,373,411,419Turtles......................................v,219–221,240,247–262 XXenopus laevis...................................................v,4,17–19, 41,44,59–72,265–274,303Xenopus tropicalis...................................................19,303 Y Yolk.......................................................18,25,37,47,59,67,76–79,82–84,90,93,95,96,102,121,122,126,179,220,249,298,332,333,337,364,386,393,398,400–402,404Yolk clearing (25)ZZebrafish......................................................v,1,111–127,131,133,143,146,152,154,155,158,163,248,277–293,295–302,327–340,343–351,353–374,377–391,411,417,419,420,426434V ERTEBRATE E MBRYOGENESIS:E MBRYOLOGICAL,C ELLULAR,AND G ENETIC M ETHODS Index。
genecloning

cDNA I
• Eukaryotic DNA differs from bacterial (prokaryotic) DNA in that it has introns (intervening sequences) and exons (expressed or translated sequences). • In order for a eukaryotic gene to be expressed, the introns are ‘edited’ out of mRNA after transcription
• Bacteria can’t deal with introns, so in cases where a product (e.g. insulin) is to be expressed by the bacteria, an uninterrupted coding sequence is needed. • Also, since introns can account for up to 90% of an eukaryotic gene, and cloning long fragments is difficult, it is sometimes desirable to work only with the expressed sequences (exons)
Gene Cloning
& Creating DNA Libraries
Gene Cloning
• What does the term cloning mean? • What is gene cloning? How does it differ from cloning an entire organism? • Why is gene cloning done? • How is gene cloning accomplished ? • What is a DNA ‘Library’? A cDNA Library? • What are some of the ethical considerations regarding gene cloning?
4-目的基因的制备(龙敏南)

5’ 3’
cDNA第一链 cDNA第二链
去掉发卡结构 用核酸酶S1可以切掉发卡结构(但这会 导致cDNA中有用的序列被切掉!)。 5’ 3’ cDNA第一链 cDNA第二链 核酸酶S1
5’ 3’
cDNA第一链 cDNA第二链
3’ 5’
(4)cDNA与载体连接 在双链cDNA末端接上人工接头,即可与 载体连接,转入受体菌。
• 分子杂交法
(三)、双抗体免疫法分 离编码蛋白的基因
适用于某一真核基因的蛋白质已被分离纯 化,且足以产生特定抗体。通过抗体和多 聚核糖体结合,从而将含有特定mRNA的 多聚核糖体从总多聚核糖体中分离,再分 离mRNA和合成cDNA。
(四)、利用反转录直接从特 定mRNA分离基因
适用于合成分子质量较大,转录产物 mRNA易分离的目的基因,由mRNA 反转录为cDNA。
亚磷酸三酯法、 自动化合成法。
磷酸三酯法、
固相合成法、
第三节 目的基因的分离
从可能存在目的基因的大量片段中筛 选含目的基因的克隆,进行基因序列、 功能、定位和转录产物等研究
1、已知特异蛋白的氨基酸序列分离目的基因
可设计简并引物进行扩增
2、基因表达系列分析
(Serial Analysis of Gene Expression, SAGE)
3、目的基因片段的差别杂交
富集特定基因的mRNA或cDNA模板
探针柱分离特异mRNA 根据已知的基因序列合成探针,结合到纤维 素柱上,用来分离纯化该基因的mRNA。
Total mRNA过柱
探针DNA 纤 探针DNA 维 探针DNA 柱 探针DNA
探针DNA 纤 探针DNA 特异 维 mRNA 探针DNA 柱 探针DNA 洗脱
Biomics CRISPR Cas9 基因敲除技术
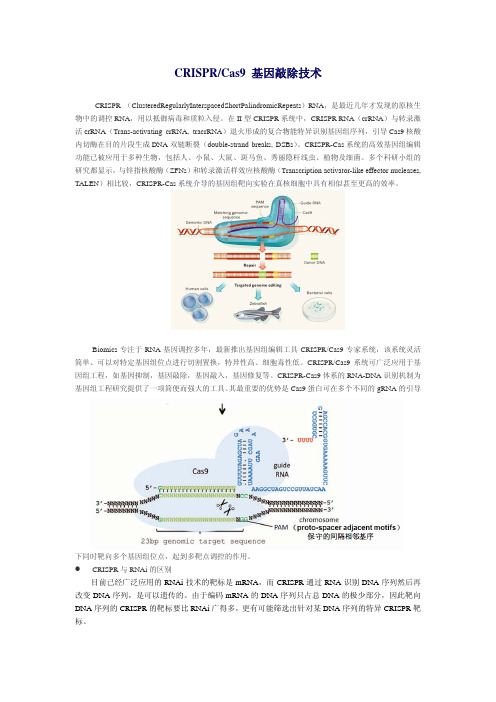
CRISPR/Cas9 基因敲除技术CRISPR (ClusteredRegularlyInterspacedShortPalindromicRepeats)RNA,是最近几年才发现的原核生物中的调控RNA,用以抵御病毒和质粒入侵。
在II型CRISPR系统中,CRISPR RNA(crRNA)与转录激活crRNA(Trans-activating crRNA, tracrRNA)退火形成的复合物能特异识别基因组序列,引导Cas9核酸内切酶在目的片段生成DNA双链断裂(double-strand breaks, DSBs)。
CRISPR-Cas系统的高效基因组编辑功能已被应用于多种生物,包括人、小鼠、大鼠、斑马鱼、秀丽隐杆线虫、植物及细菌。
多个科研小组的研究都显示,与锌指核酸酶(ZFNs)和转录激活样效应核酸酶(Transcription activator-like effector nucleases, TALEN)相比较,CRISPR-Cas系统介导的基因组靶向实验在真核细胞中具有相似甚至更高的效率。
Biomics专注于RNA基因调控多年,最新推出基因组编辑工具CRISPR/Cas9专家系统,该系统灵活简单、可以对特定基因组位点进行切割置换,特异性高、细胞毒性低。
CRISPR/Cas9系统可广泛应用于基因组工程,如基因抑制,基因敲除,基因敲入,基因修复等。
CRISPR-Cas9体系的RNA-DNA识别机制为基因组工程研究提供了一项简便而强大的工具。
其最重要的优势是Cas9蛋白可在多个不同的gRNA的引导下同时靶向多个基因组位点,起到多靶点调控的作用。
z CRISPR与RNAi的区别目前已经广泛应用的RNAi技术的靶标是mRNA,而CRISPR通过RNA识别DNA序列然后再改变DNA序列,是可以遗传的。
由于编码mRNA的DNA序列只占总DNA的极少部分,因此靶向DNA序列的CRISPR的靶标要比RNAi广得多,更有可能筛选出针对某DNA序列的特异CRISPR靶标。
生物工程专业英语词汇总结

专业英语词汇总结第一章bacteria 细菌yeast 酵母fungi 真菌mammalian 哺乳动物细胞molecular biology 分子生物学biomass 生物体enzyme 酶antibiotic 抗生素catabolic 分解代谢的glucose 葡萄糖anabolic 合成代谢的exergonic 放能的endergonic 吸能的vaccine 疫苗interferon 干扰素date back into 始于…fermentative 发酵的sterile 无菌的;不生育的acetic acid 醋酸butanol 丁醇acetone 丙酮polysaccharide 多糖citric acid 柠檬酸genetic engineering 基因工程genome 基因组mutation 突变geneticist 遗传学家recombinant DNA 重组DNA gene 基因hybird 杂交物insulin 胰岛素protoplast fusion 原生质体融合monoclonal antibody 单克隆抗体tissue culture 组织培养immobilize 固定化amino acid 氨基酸septic 感染的protein 蛋白质lipid 脂质hormone 激素第二章exploit 利用divert 转移,使转向matrix 发源地divergent 分歧的nucleic acid 核酸carbohydrate 碳水化合物;糖类aerobic 需氧的anaerobic 厌氧的anhydride 酐adenine 腺嘌呤striking 显著的mould 霉菌hydrocarbon 烃methanol 甲醇precursor 前体hydride 氢化物modulate 调整lactobacilli 乳酸杆菌tricarboxylic acid cycle 三羧酸循环cytochrome 细胞色素oxidant 氧化剂reductant 还原剂lyase 裂解酶hydrolyze 水解commence 开始ketone 酮gluconeogenesis 葡糖异生作用oxidoreductase 氧化还原酶nucleotide 核苷酸glycoside 糖苷ribosome 核糖体code 密码messenger RNA 信使RNARNA polymerase RNA聚合酶transcription 转录codon 密码子translation 转译transfer RNA(tRNA)转移RNA regulatory gene 调节基因operator gene 操纵基因structural gene 结构基因operon 操纵子promotor 启动基因.glycogen 糖原feedback inhibition 反馈抑制isoenzyme 同工酶proteolytic enzyme 蛋白(水解)酶cell cycle 细胞周期germ cell 生殖细胞duplication 成倍mutagen 诱变剂lethal 致死的adverse 不利的entail 必需inclination 倾向(于某事,某种状态)competence 能力第三章genetics 遗传学breeding 育种mutagenesis 诱变作用vial 病毒slime 粘质物strain (菌)株screen,screening 筛选transformation 转化hybridization 杂交transduction (基因)转导gene cloning 基因克隆genotype 基因型dispersal 分散conjugation 接合sexual cycle 有性循环parasexual cycle 准性循环gene library 基因文库shake-flask culture 摇瓶培养spore 孢子phenotype 表型phenotypic 表型的degeneration 退化culture maintenance 菌种保藏subculturing 移种;接种freeze drying 冷冻干燥mutator gene 增变基因antimutator gene 减变基因mutator strain 增变(菌)株complete medium 完全培养基minimal medium 基本培养基offspring 后代context 背景,环境sexual hybridization 有性杂交haploid 单倍体meiosis 减数分裂homologous recombinant 同源重组life cycle 生命周期chromatid 染色单体breeding system 繁殖系统outbreeding 远交,远系繁殖inbreeding 近郊,同系交配self-fertilization 自体受精vegetation cell 营养细胞gamete 配子zygote 合子vector 载体homogenic 同种的,同基因的heterogenic 异种的,异基因的incompatibility 不相容的translocation 移位denude 使裸露encapsulate 用囊状物包,封装differentiate 分化somatic 体细胞的erythrocyte 红细胞vertebrate animal 脊椎动物plasma cell 浆细胞lymphocyte 淋巴细胞antisera 抗血清proliferate 增殖transfusion 输血tissue typing 组织定型propagation 繁殖amplification 扩大heritable 可遗传的restriction endonuclease 限制性核酸内切酶express 表达DNA ligase DNA连接酶chimeric DNA 嵌合DNAprobe 探针competent cell 感受态细胞replicon 复制子.transformant 转化体transposon 转位子,转座子annealing 退火digestion (酶切)消化transfection 转染reverse transcriptase 逆转录酶intron 内含子chimera 嵌合体cosmid 粘粒autoradiography 放射自显影术site-directed mutation 定点突变signal peptide 信号肽第四章beverage 饮料sauerkraut 泡菜alkaloid 生物碱perfume 香水flavour 调味品perfumery 香料(总称)embryo 胚trickle 滴流fed-batch culture 半连续培养respiratory quotient 呼吸商doubling time 倍增时间generation time 传代时间balanced growth 平衡生长exponential growth 指数生长exponential phase 指数(生长)期deceleration phase 降速期stationary phase 稳定期death phase 死亡期residence time 停留时间dilution rate 稀释率wash-out 洗出continuous stirred tank reactor(CSTR)连续搅拌釜式反应器loop bioreactor 环路式(循环式)生物反应器tap water 自来水starch 淀粉sensor 传感器controller 控制器on-line 在线off-line 离线cholesterol 胆固醇indigenous 在当地生长的microflora 微生物区系vegetation 植物(总称)hyphae 菌丝【复】第五章inanimate 无生命的urease 脲酶hydrolase 水解酶glucose oxidase 葡萄糖氧化酶lipase 脂肪酶catalage 过氧化氢酶built-in 固有的idiophase 繁殖期trophophase 生长期synthetase 合成酶allergenic 变应原的,引起变态反应的extraneous 外来的mycotoxin 真菌毒素antigenicity 抗原性toxicological 毒理学的carcinogenic 致癌的subacute 亚急性的fertility 能育性immobilized enzyme 固定化酶encapsulation 胶囊封装collagen 胶原sweetener 甜味剂ampicillin 氨苄青霉素unfold 展开cell homogenate 细胞匀浆液【下载本文档,可以自由复制内容或自由编辑修改内容,更多精彩文章,期待你的好评和关注,我将一如既往为您服务】.。
《基因工程》 第七章 重组子选择筛选与分析
第十八页,共四十四页。
③ 卡那霉素 (kanamycin, Kn或 Kan): 含Kan抗性基因菌体转译一种能修饰Kn的 酶,阻碍Kn对核糖体的干扰(gānrǎo)。 四环素(tetracymic, Tc或 Tet): 含Tc抗性基因的菌体转译一种能改变细菌膜的蛋白, 防止Tc 进入细胞后干扰细菌蛋白质的合成。
⑥ 链霉素(strentomycin, Sm或 Str): 含Str 抗性基因的菌体转译一种能修 饰Sm的酶,抑制Sm与核糖体结合。
第三页,共四十四页。
1.根据载体选择标记初步筛选转化子
载体DNA分子上通常携带了一定的选择遗传标记基因,可进行转化 子或重组子初步筛选。做法(zuòfǎ)是将转化处理后的菌液(包括对照处理) 适量涂布在选择培养基上,在最适生长温度条件下培养一定时间,观察 菌落生长情况,即可挑选出转化子。
对于受体细胞而言,通常用LB培养基,在LB 培养基中加入适量的某种 选择物,即为选择培养基。选择物是由载体DNA分子上携带的选择标记基因 所决定。
3) Immunological screening for expressed genes (Western blotting)
4) Analysis of cloned genes (Restriction mapping, DNA sequencing)
2
第二页,共四十四页。
第一节 转基因筛选(shāixuǎn)和选择的方法
(1)抗药性筛选法 (2)插入失活筛选法 (3)插入表达筛选法
第四页,共四十四页。
(1)抗药性筛选
这是利用载体DNA分子上的抗药性筛选标记进行筛选的方法。主要用于 重组质粒DNA分子的转化子筛选。重组质粒DNA分子携带特定的抗药性 选择标记基因,在含有相应选择药物的选择培养基上正常生长。 ① 氨苄青霉素(ampicillin,Ap 或Amp): 含bla基因的菌体能转译 -丙酰胺酶 (-lactarnase),可降解Ap。 ② 氯霉素 (chloramphenicol, Cm 或 Cmp): 含 cat基因的菌体能转译氯霉素乙酰 转酰酶 (chloramphenicol acetyltransferase),使Cm乙酰化而失效。
第五章目的基因的获取

第五章目的基因的获取第五章目的基因的获取1976年H.G. Khorana 提出了用化学方法合成基因的设想,并于1979年在science 上发表了率先成功地合成大肠杆菌酪氨酸tRNA 基因的论文。
第一节化学合成目的基因目前常用的方法一、目前常用的方法磷酸二酯法、磷酸三酯法、亚磷酸三酯法、固相合成法自动化合成法。
①保护dNTP 的5’端P 或3’端-OH (1)原理1. 磷酸二酯法保证合成反应的定向进行。
然后再用酸②带保护的单核苷酸连接或碱脱去保护基团带5’保护的单核苷酸与带3’保护的另一个单核苷酸以磷酸二酯键连接起来。
然后去掉一个保护,使其循环往复连接。
(2)合成过程5’端保护的单核苷酸可以同3’端保护的二核苷酸聚合。
保护连接去保护连接原理与磷酸二酯法一样。
只是参加反应的单核苷酸都是在3’端磷酸和5’-OH 上都先连接了一个保护基团。
2. 磷酸三酯法固相磷酸三酯合成法将第一个核苷酸的3’-OH 端固定在固相支持物上。
(第一个核苷酸不需要保护)固相磷酸酯合成法是目前通用的合成方法。
化学合成的DNA 片断一般在200bp 以内。
需要把几个正确片段连接起来二、化学合成DNA 片断的组装1. 互补连接法(1)互补配对用T4多核苷酸激酶使各个片段的5’端带上磷酸。
预先设计合成的片断之间都有互补区域,不同片断之间的互补区域形成有断点的完整双链。
(2)5’端磷酸化T4多核苷酸激酶使5’-OH磷酸化(3)连接酶连成完整双链T4 DNA连接酶完整的DNA双链2. 互补延伸连接法预先设计的片断之间有局部互补区,可以相互作为另一个片断延长的引物,用DNA聚合酶延伸成完整的双链。
3’5’5’3’5’3’T4DNA连接酶Klenow片段引物1. 直接合成基因三、寡聚核苷酸化学合成的优点(1)有些组织特异性mRNA的含量很低,很难用cDNA方法克隆)有些基因比较短化学合成费用较低2. 合成测序或PCR用的引物(20bp左右)(2)有些基因比较短,化学合成费用较低3. 合成探针序列以筛选特定的基因4. 定点突变合成合成带有定点突变的基因片段。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Section I Gene library and screening
Molecular Biology
Gene libraries and screening
I1 Genomic libraries I2
cDNA libraries
I3 Screening procedures
Molecular Biology
Expression screening One-sixth of the clones in a cDNA expression libraries will produce their encoded polypeptide as a fusion protein with the vector-encodeed βgalactosidase. Antibidies to the desired protein can be used to screen the library in a process similar to plaque hybridization to obtain particular clone. Hybrid arrest and release cDNA clones can be hybridized to mRNA preparations to prevent, or arrest, the subsequent translation of some mRNA species. Alternatively, those mRNA hybridized to the cDNA clones can be purified, released from the cDNA and translated to identify the polypeptides encoded. Chromosome walking This is the repeated screening of genomic libraries to obtain overlapping clones and hence build up a collection of clones covering part of a chromosome. It involves using the ends of the inserts as probes for subquent rounds of screening. Chromosome jumping is a similar process.
Molecular Biology
பைடு நூலகம்
Gene libraries and screening
I2
cDNA libraries
mRNA isolation, purification and fractionation
mRNA can be readily isolated from lysed eukaryotic cells by adding magnetic beads which have oligo(dT) covalently attached. The mRNA binds to the oligo(dT) via its poly(A) tail and thus be isolated from the solution. The integrity of an mRNA preparation can be checked by translation in a wheat germ extract or reticulocyte lysate and then visualizing the translation products by polyacrylaminde gel electrophoresis. Integrity can also be studied using gel electrophoresis, which allows the mRNA to be size fractionated by recovering chosen regions of the gel lane. Specific sequences can be removed from the mRNA by hybridization.
Molecular Biology
Fig. 1. cDNA cloning. (a) First and second strand synthesis; (b) end preparation and linker addition to duplex cDNA.
Molecular Biology
Molecular Biology
Gene libraries and screening
I1 Genomic libraries
Representative gene libraries Gene libraries made from genomic DNA are called genomic libraries and those made from complementary DNA are known as cDNA libraries. The latter lack nontranscribed genomic sequences(repetitive sequences, etc.). Good gene libraries are representative of the starting material and have not lost certain sequences due to cloning artifacts. Size of library A gene library must contain a certain number of recombinant for there to be a high probability of it containing any particular sequence. This value can be calculated if the genome size and the average size of the insert in the vector are known. Genomic DNA For making libraries, genomic DNA, usually prepared by protease digestion and phase extraction, is fragmented randomly by physical shearing or restriction enzyme digestion to give a size range appropriate for the chosen vector. Often combination of restriction enzymes are used to partially digest the DNA. Vectors Plasmids, λ pahge, cosmid, BAC or yeast artificial chromosome vectors can be used to construct genomic libraries, the choice depending on the genome size. The upper size limit of these vectors is about 10,23,45,350 and 1000 kb respectively. The genomic DNA fragments are ligated to the prepared vector molecule using T4 DNA ligase.
Molecular Biology Synthesis of cDNA In the first synthesis, reverse transcriptase is used to make a cDNA copy of the mRNA by exetending a primer, usually oligo(dT), by the addition of deoxyribounucletides to 3’-end. Synthesis can be detected by trace labeling. 3’-Tailing of the first strand cDNA using terminal transferase makes full-length second strand synthesis easier. Reverse transcriptase or Klenow enzyme can extend a primer [e.g.oligo(dG)] annealed to a homopolymeric tail [e.g.oligo(dC)] to synthesize second strand cDNA. Treatment of cDNA ends To avoid blunt end ligation of cDNA to vector, linkers are usually added to the cDNA after the ends have been repaired(blunted) using a single strand-specific nuclease followed by Klenow enzyme. The cDNA may also be methylated to keep it from being digested when the added linkers are cleaved by a restriction enzyme. Adaper molecules can be used as an alternative to linkers. Ligation to vector The vector is usually dephosphorylated using alkaline phosphatase to prevent self-ligation, and so promote the formation of recombinant molecules. Plasmid or phage vectors can be used to make cDNA libraries, but the phage λgt11 is preferred for the cnstruction of expression libraries.
