Crastin CE2056
Light Vision 清晰视界系列眼镜说明书

Light Vision ™11356-00000-10Clear AF Lens, Grey Frame,Dual LED LightsSolus ™ 1000S1101SGAFClear Scotchgard ™ AF Lens,Blue/Black Frame S1102SGAFGrey Scotchgard ™ AF Lens,Blue/Black Frame S1103SGAFAmber Scotchgard ™ AF Lens,Blue/Black Frame S1107SGAF Indoor/Outdoor Grey Scotchgard ™ AF LensBlue/Black Frame S1101SGAF-KTClear Scotchgard ™ AF Lens,Blue/Black Frame, Foam Gasket and StrapS1102SGAF-KTGrey Scotchgard ™ AF Lens,Blue/Black Frame, Foam Gasket and StrapS1107SGAF-KTIndoor/Outdoor Grey Scotchgard ™AF Lens, Blue/Black Frame, Foam Gasket and StrapS1201SGAFClear Scotchgard ™ AF Lens,Green/Black FrameS1202SGAFGrey Scotchgard ™ AF Lens,Green/Black FrameS1203SGAFAmber Scotchgard ™ AF Lens,Green/Black FrameS1207SGAF Indoor/Outdoor Grey Scotchgard ™ AF LensGreen/Black Frame S1201SGAF-KTClear Scotchgard ™ AF Lens,Green/Black Frame, Foam Gasket and StrapS1202SGAF-KTGrey Scotchgard ™ AF Lens,Green/Black Frame, Foam Gasket and Strap S1201SGAF-SKTClear Scotchgard ™ AF Lens, Green/Black Frame, Foam Gasket and Strap (temples not included)S1202SGAF-SKTGrey Scotchgard ™ AF Lens, Green/Black Frame, Foam Gasket and Strap (temples not included)S1207SGAF-SKTIndoor/Outdoor Grey Scotchgard ™ AF Lens, Green/Black Frame, Foam Gasketand Strap (temples not included)Solus-foamSolus Accessories Foam GasketSolus-strapSolus Accessories StrapSmart Lens ™13407-00000-5Photochromic Lens, Black FramePREMIUM PROTECTIVE EYEWEARThese eye or face protection products help provide limited eye and face protection. Misuse or failure to follow warning and instruction may result in serious potential injury, including blindness or death. For proper use, selection, and applications against flying particles, optical radiation and/or splash, see supervisor, read User Instructions and warning on the package or call 3M PSD Technical Service in the USA at 1-800-243-4630. In Canada, call 1-800-267-4414.WARNING!3Personal Safety Division 3M CenterBuilding 235-2W-70St. Paul, MN 55144-1000For more information:In U.S.Technical Assistance 1-800-243-4630 Customer Care Center 1-800-328-1667/WorkerSafety In CanadaTechnical Assistance 1-800-267-4414 Customer Care 1-800-364-35773M.ca/Safety3M PSD products are occupational use only.3M and all other trademarks used herein are trademarks of 3M Company, used under license in Canada. Please recycle. Printed in U.S.A. © 3M 2018. All rights reserved. 70-0715-6919-1Rev. 01/201811476-00000-10Clear AF Lens, Grey Frame,Dual LED LightsLight Vision ™ 2Fuel ™11640-00000-10Red Mirror Lens, Metallic Sand Frame11641-00000-20Blue Mirror Lens, Silver Frame11664-00000-10Blue HC Mirror Lens, White Frame11650-0000-10Red Mirror Lens, Metallic Sand Frame11654-00000-10Grey AF Lens, Black Rubberized FrameSecureFit ™ 600Safety SunwearSS1330AS-GYellow Mirror AS Lens, Grey FrameSS1428AS-SBlue Mirror AS Lens, Silver Alum FrameSS1502AF-BGrey AF Lens, Black Frame w/GasketSS1502AF-WGrey AF Lens, White Frame w/GasketSS1514AS-SSilver Mirror AS Lens, Silver Black Frame w/GasketSS1511AF-BGrey Polarize AF Lens, Black Frame w/GasketSS1514AS-BSilver Mirror AS Lens, Black/Grey Frame w/GasketSS1629AS-BRed Mirror AS Lens, Black FrameSF601SGAFClear Scotchgard ™ AF LensSF601SGAF-FMClear Scotchgard ™ AF Lens, Foam GasketSF602SGAFGrey Scotchgard ™ AF LensSF602SGAF-FMGrey Scotchgard ™ AF Lens, Foam GasketSF603SGAFAmber Scotchgard ™ AF LensSF607SGAFIndoor/Outdoor Grey Scotchgard ™ AF LensSF601RASClear Rugged Anti-Scratch LensSF602RASGrey Rugged Anti-Scratch LensSF617ASLow IR R1.7, AS LensSF630ASShade W3.0, AS LensSF650AS W5, AS Lens SF611ASPolarized, AS LensSF613ASPhotochromic, AS LensLight Vision ™ OTG11489-00000-10Clear AF Lens, Dual LED Lights, OTG16617-00000-10Clear Lens, Black Frame w/Strap, Medium16618-00000-10Clear Lens, Black Frame w/Strap, LargeLexa ™Dust GoggleGear™Lexa ™ Splash GoggleGear ™16644-00000-10 Clear Lens, Medium, D3 D416645-00000-10 Clear Lens, Large, D3 D416400-00000-10Clear Lens, Elastic Strap, Medium16408-00000-10Clear Lens, Black Adjustable Temple,Medium16412-00000-10Clear Lens, Elastic Strap, Large16420-00000-10Clear Lens, Black Adjustable Temple,LargeFectoggles ™40300-00000-10Clear Lens, Impact Goggle40301-00000-10Clear AF Lens, Impact Goggle40304-00000-10Clear Lens, Splash Goggle, D3 D440305-00000-10Clear AF Lens, Splash Goggle, D3 D4Centurion ™Maxim ™ 2x2 Goggle40686-00000-10Clear Lens, Black Frame w/Strap, Temples40687-00000-10Grey Lens, Black Frame w/Strap, Temples40696-00000-10Clear Lens, Black Frame, Side Venting40698-00000Clear AF Lens, Black Strap, Air Flow Goggle40699-00000Grey AF Lens, Black Strap, Air Flow Goggle332 Impact Goggle40650-00000-10Clear Lens 40651-00000-10 Clear AF Lens334 Splash Goggle40660-00000-10 Clear Lens, D3 D440661-00000-10 Clear AF Lens, D3 D4GoggleGear ™ 500GG501SGAFClear Scotchgard ™ AF Lens, D3 D4,Cloth Strap GG501NSGAFClear Scotchgard ™ AF Lens, D3 D4,Neoprene Strap GG500-PI Prescription InsertGG500-NeoStrapReplacementNeoprene Strap GG500-ClthStrap Replacement Cloth StrapSAFETY GOGGLESACCESSORIESMaxim ™ Splash GoggleModul-R ™40671-00000-10Clear AF Lens, O-T-G, D3 D440658-00000-10Clear AF Lens, Chin ProtectorThe Complete Line of Protective Eyewear Products11215-00000-20 Grey AF Lens, Black Frame11216-00000-20I/O Mirror Lens, Black FrameMoon Dawg ™11532-10000-20Clear AF Lens, Blue Frame11554-00000-20Clear AF Lens, Bronze Frame11555-00000-20Bronze AF Lens, Bronze Frame11556-00000-20Blue Mirror HC Lens, Bronze FrameMetaliks ™ GTHIE6 Protective EyewearAttaches directly to 3M ™ Hard Hat SuspensionHIE601AF - Clear AF Lens HIE602AF - Amber AF Lens HIE603AF - Grey AF Lens3M ™ Protective Eyewear Slip-On Side Shields23451-00030-20 ClearReplacement Foam GasketSF-FOAM (SF400)SF600FI (SF600)GoggleGear ™ 2890 SeriesGG2891-SGAFClear Scotchgard ™ AF Lens,Indirect Vent3M ™ Lens Cleaner83803-00000Lens Cleaning Fluid 83745-00000Lens Cleaning T owelettesQX Privo 12261-00000-20Clear AF Lens, Black/Orange Frame12262-00000-20Grey AF Lens, Black/Orange Frame12263-00000-20Amber AF Lens, Black/Orange Frame12264-00000-20I/O Mirror Lens, Black/Orange Frame12265-00000-20Clear AF Lens, Silver/Red Frame12266-00000-20SF401SGAF-REDClear Scotchgard Anti-Fog LensRed/Gray Frame SF401SGAF-BLUClear Scotchgard Anti-Fog LensBlue/Gray Frame SF401SGAF-BLU-FClear Scotchgard Anti-Fog Lens with FoamGasket, Blue/Gray Frame SF402SGAF-BLU12100-10000-20 Clear Lens, Black Temple12101-10000-20Grey Lens, Black Temple 12109-10000-20Clear Lens, Black Temple, Soft Nose12110-10000-20Grey Lens, Black Temple, Soft Nose12115-10000-20Clear Lens, Black Sport Grip Temple, Soft Nose12180-10000-2011380-00000-20 Clear AF Lens, Silver Frame11381-00000-20Grey AF Lens, Silver Frame11471-00000-20Clear AF Lens, Blue Frame11472-00000-20I/O Mirror Lens, Blue Frame11523-00000-20Light Blue AF Lens, Blue Frame11477-00000-10Clear AF Lens, Grey Frame, +1.5 Diopter11478-00000-10Clear AF Lens, Grey Frame, +2.0 Diopter11479-00000-10Clear AF Lens, Grey Frame, +2.5 Diopter11374-00000-20Clear Lens, Silver Frame, +1.5 Diopter11375-00000-20Clear Lens, Silver Frame, +2.0 Diopter11376-00000-20Clear Lens, Silver Frame, +2.5 Diopter11377-00000-20Grey Lens, Silver Frame, +1.5 Diopter11378-00000-2011457-00000-10Clear AF Lens, Silver Frame, Dual +1.5 Diopter11458-00000-10Clear AF Lens, Silver Frame, Dual +2.0 Diopter11459-00000-10Clear AF Lens, Silver Frame, Dual +2.5 DiopterBX ReadersMetaliks Sport11343-10000-20Clear AF Lens, Brushed Nickel Frame11344-10000-20Grey AF Lens, Brushed Nickel Frame11345-10000-20I/O Mirror HC Lens, Brushed Nickel Frame11540-10000-20Blue Mirror HC Lens, Brushed Nickel FrameLexa ™15100-00000-20Clear AF Lens, Black Temple, Large15200-00000-20Clear AF Lens, Black Temple, Medium15154-00000-100Clear AF Lens, Black Temple, Large15204-00000-20Grey AF Lens, Black Temple, Medium15152-00000-100Clear AF Lens, Black Temple, Medium14246-00000-20Clear AF Lens, Metallic Grey/Black Frame14247-00000-20Grey AF Lens, Metallic Grey/Black Frame14248-00000-20 I/O Mirror Lens, Metallic Grey/Black FrameSF401AF Clear AF Lens SF402AF Grey AF Lens SF410ASIndoor/Outdoor Mirror LensSF401AF-FM Clear AF Lens, Foam SF402AF-FM Grey AF Lens, Foam SF410AS-FMSecureFit 400OX。
转基因植物35S基因核酸检测试剂盒说明书

转基因植物35S基因核酸检测试剂盒说明书转基因植物35S基因核酸检测试剂盒说明书试剂盒简介货为了适应新加坡石斑鱼虹彩病毒染料法荧光定量PCR试剂盒快速检测和疫病研究的需要,本公司参照 OIE国际标准中规定的引物序列,经多次实验及系统优化,开发生产了本试剂盒。
应用本试剂盒进行检测具有快速、灵敏、特异、准确、安全操作简单、应用广泛和高通量检测等特点及优点。
试剂盒组成及试剂配制:酶联板(Assay plate):一块(96孔)。
2.标准品(Standard):2瓶(冻干品)。
3.样品稀释液(Sample Diluent):1×20ml/瓶4.生物su标记抗体稀释液(Biotinantibody Diluent):1×10ml/瓶。
5.辣根过氧化物酶标记亲和素稀释液(HRPavidin Diluent):1×10ml/瓶。
6.生物su标记抗体(Biotinantibody):1×120μl/瓶(1:100)7.辣根过氧化物酶标记亲和素(HRPavidin):1×120μl/瓶(1:100)8.底物溶液(TMB Substrate):1×10ml/瓶。
9.浓洗涤液(Wash Buffer):1×20ml/瓶,使用时每瓶用蒸馏水稀释25倍。
10.终止液(Stop Solution):1×10ml/瓶(2N H2SO4)。
样本处理及要求:1.血清:全血标本请于室温放置2小时或4℃过夜后于1000g离心20分钟,取上清即可检测,或将标本放于20℃或80℃保存,但应避免反复冻融。
2. 血浆:可用EDTA或肝素作为抗凝剂,标本采集后30分钟内于2 8°C 1000g离心20分钟,或将标本放于20℃或80℃保存,但应避免反复冻融。
3. 组织匀浆:用预冷的PBS (0.01M, pH=7.4)冲洗组织,去除残留血液(匀浆中裂解的红细胞会影响测量结果),称重后将组织剪碎。
酪蛋白肽锌螯合物的制备及体外消化分析

周桂成,肖珊,王波,等. 酪蛋白肽锌螯合物的制备及体外消化分析[J]. 食品工业科技,2023,44(23):270−279. doi:10.13386/j.issn1002-0306.2023020002ZHOU Guicheng, XIAO Shan, WANG Bo, et al. Preparation and in Vitro Digestive Analysis of Casein-Derived Peptide-Zinc Chelates[J]. Science and Technology of Food Industry, 2023, 44(23): 270−279. (in Chinese with English abstract). doi:10.13386/j.issn1002-0306.2023020002· 分析检测 ·酪蛋白肽锌螯合物的制备及体外消化分析周桂成1,2,肖 珊2,王 波2, *,王际辉2,*(1.大连工业大学生物工程学院,辽宁大连 116034;2.东莞理工学院生命健康技术学院,广东东莞 523808)摘 要:为开发安全高效且易吸收的补锌剂,利用碱性蛋白酶酶解和乳酸菌发酵相结合的方法制备生物活性肽,并以此多肽制备了酪蛋白肽锌螯合物。
采用光谱法对螯合物结构进行表征,利用体外消化模型和Caco-2细胞实验对其胃肠消化特性及生物安全性进行评价。
结果表明,制备酪蛋白肽的最优条件为酶解pH 为9、碱性蛋白酶添加量为0.3%(w/v ),乳酸菌发酵时间为12 h ,此时反应体系中多肽含量为142.39±0.95 mg/g ,对锌的螯合率为31.41%±0.97%。
与锌螯合后,酪蛋白肽表面的致密结构遭到破坏,形成疏松的状态;光谱学分析表明,Zn 2+能与酪蛋白肽上的活性基团进行结合,螯合位点为羧基氧、羟基氧和氨基。
体外模拟消化结果显示,酪蛋白肽锌螯合物在消化过程中锌溶解性优于硫酸锌;在胃肠消化后酪蛋白肽锌螯合物DPPH 和ABTS +自由基的清除能力分别提升了26.19%±3.30%和71.96%±7.06%,而铁还原力下降了36.26%±2.80%;同时,在消化过程中肽锌螯合物的β-转角与无规则卷曲含量减少,β-折叠结构增加,Zn 2+起到了维持多肽结构的作用。
IGCS19-0405 : 产品说明书

abnormal vaginal bleeding requiring intervention had no statis-tical difference between VP and WVP patients group (p=0.3074)as other complications as well(table1).Median of related days of vaginal bleeding after the procedure were 7.4days(SD8.75)in VP group and7.34days(SD8.52)in WVP group,with no statistical difference(p=0.912). Conclusions Insert a vaginal pack or not,after LEEP,do not affect the number of postoperative gynecologic intervention due to vaginal bleeding or the amount of postoperative bleed-ing days.Previous pregnancies,hormonal status,cytology or LEEP specimen characteristics did not affect the disclosure. We also could not find any risk factor associated to abnormal bleeding.Based on that,the use of vaginal pack can be omit-ted with no further complications.IGCS19-0405382LATERALLY EXTENDED ENDOPELVIC RESECTION(LEER) AND NEOVAGINE,PATIENT WITH RECTALADENOCARCINOMA AND RECURRENCE IN CERVIX,VAGINA AND PELVIC WALL:A PURPOSE OF A CASE1J Torres*,2J Saenz,3O Suescun,3M Medina,4L Trujillo.1Especialista en entrenamiento–Universidad Militar Nueva Granada–Instituto Nacional de Cancerologia,Department of Gynecologic Oncology,Bogota D.C.,Colombia;2Especialista en entrenamiento–Universidad Militar Nueva Granada–Instituto Nacional de Cancerologia,Department of Gynecologic Oncology,Bogota D.C,Colombia;3Instituto Nacional de Cancerologia, Department of Gynecologic Oncology,Bogota D.C,Colombia;4Instituto Nacional de Cancerologia,Department of Gynecologic Oncology,Bogota D.C.,Colombia10.1136/ijgc-2019-IGCS.382Objectives Exenteration is used to treat cancers of the lower and middle female genital tract in the irradiated pelvis. Höckel described laterally extended endopelvic resection (LEER)as an approach in which the resection line extends to the pelvic side wall.Methods A49-year-old patient diagnosed with rectal adenocar-cinoma10years ago,managed with chemotherapy plus radio-therapy.T umor relapse at3years,management with low abdominoperineal resection and definitive colostomy.Second relapse4years later,compromising the posterior aspect of the coccyx and right side of the pelvis with irresecability criteria, management was decided with chemotherapy with capecita-bine,oxaliplatin and bevacizumab.New relapse at2years in the cervix,vagina and pelvic wall.Images without distance disease,type LEER management with extension of pelvic floor margins and resection of muscle pubococcygeus and right lat-eral iliococcygeus with neovagina(Singapore flap)and non-continent urinary derivation with bilateral cutaneous ureteros-tomy,achieving adequate lateral margin with curative intent. During follow-up with favorable evolution.Results LEER combines at least two procedures:total mesorec-tal excision,total mesometrial resection or total mesovesical resection.It may even require resection of the pelvic wall, internal obturator muscle,pubococcygeus,iliococcygeus,coccy-geus or internal iliac vessels.In combination with neovagina, it would offer better results in non-gynecological cancer relapses.Conclusions LEER with neovagina can be offered as a new therapy to a selected subset of patients with relapse in adja-cent gynecological organs with good oncological,functional and aesthetic results.Symptom Management–Supportive Cancer CareIGCS19-0706383PHOTOBIOMODULATION AND MANUAL LYMPHDRAINAGE FOR NIPPLE NECROSIS TREATMENT INBREAST CANCER:A CASE REPORT1J Baiocchi,2L Campanholi,3G Baiocchi*.1Oncofisio,Physical Therapy,Sao Paulo,Brazil;2CESCAGE,Physical Therapy,Ponta Grossa,Brazil;3AC Camargo Cancer Center, Gynecologic Oncology,Sao Paulo,Brazil10.1136/ijgc-2019-IGCS.383Objectives Recently,breast reconstruction after mastectomywith nipple preservation became an option of breast cancer surgery.Despite its efficacy and aesthetic superiority,the nip-ple preservation is associated with several complications in the postoperative period.The photobiomodulation therapy,for-merly known as low-intensity laser therapy,demonstrated tis-sue promotion repair by cellular repair biostimulation, angiogenesis and anti-inflammatory effects.These characteris-tics suggest a potential role for repair of chronic wounds andmay be applicable in necrosis treatment.Our aim was toreport the effects of the physiotherapeutic intervention through photobiomodulation therapy in a patient with nipple necrosis after risk reducing mastectomy.Methods We report a case of a breast cancer surgery with nip-ple necrosis treated with low-level laser therapy.The patientwas a36-year-old women who developed skin nipple necrosisin the right breast after bilateral reconstructive mastectomy.She had6sessions of low-level laser therapy.Results A female subject developed a nipple necrosis of morethan40%on the right breast after mastectomy and recon-struction.She was referred to Physical Therapy(PT)and thePT sessions were composed by manual lymph drainage,man-ual therapy for de AWS,exercises of strength and flexibility, followed by LLLT with laser660nm,2joules per point atevery1cm.Therapy was implemented for12times in total,from May2016to June2016.A re-evaluation was performed monthly from July13,2016to November2017.After18 months of follow-up,the sustained effects of LLLT were found.Conclusions Low-level laser therapy is effective for the skin cicatrization after nipple necrosis.IGCS19-0446384CONTRACEPTION AND FERTILITY COUNSELING INPATIENTS RECEIVING CHEMOTHERAPY1A Elnaggar*,2A Calfee,1LB Daily,2T Hasley,1T Tillmanns.1West Cancer Center and Research Institute,Gynecologic Oncology,Memphis,USA;2University of Tennessee Health Science Center,Obstetrics and Gynecology,Mempis,USA10.1136/ijgc-2019-IGCS.384Objectives Cancer care advances allow more patients to pursue fertility.Unfortunately,treatments may have detrimental effectson fertility and fetus should pregnancy occur.This study examines physician documentation and patient perceptions of fertility and contraception counseling. on December 24, 2023 by guest. Protected by copyright./ Int J Gynecol Cancer: first published as 10.1136/ijgc-2019-IGCS.384 on 18 September 2019. Downloaded fromMethods IRB approval obtained for a cross-sectional study of men and women,ages18–50,with newly diagnosed malig-nancy between May2017and2018.Prior sterilization,secon-dary or synchronous cancer,or prior chemotherapy were exclusionary.Consented patients received a survey regarding perception on receipt and quality of,counseling.Demographic, sexual,and social information was obtained.Differences were evaluated using chi-square tests.Results Fifty-three of179patients identified participated. Majority were women(75v25%).Patients were more likely to have perceived counseling for contraception and fertility than documented.The majority perceived counseling as suffi-cient regarding contraception and fertility.Men were more likely than women to be perceive counsel-ing regarding fertility(85v43%,p=0.010).However,both felt fertility counseling to be sufficient with similar rates of documentation.Caucasians were more likely to perceive receipt of fertility counseling(68v29%)and to perceive it to be sufficient(70v40%),then African Americans,with the same rate of documentation(35%).Conclusions Significant discrepancies in perception counsel-ing regarding contraception and fertility were seen.Gen-der and race were important factors for the perception of fertility counseling,while only race was a factor to qual-ity of perceived counseling.These differences occurred despite equal rates of physician documentation,across all groups.IGCS19-0430385WHO ARE YOU CALLING OLD?PRACTICE PATTERNS AND MANAGEMENT OF NONAGENARIANS PRESENTINGTO A GYNECOLOGIC ONCOLOGIST FOR INITIALCONSULTATIONE Ryan*,B Margolis,B Pothuri.New York University Langone Health,Obstetrics and Gynecology,New York,USA10.1136/ijgc-2019-IGCS.385Objectives T o describe the practice patterns and treatment of nonagenarians who initiated care with a gynecologic oncologist.Methods Retrospective chart review of women aged90or older who presented to a gynecologic oncologist between10/ 09and12/18at an urban academic medical center.Descrip-tive statistics utilized for variables of interest.Results We identified34nonagenarians(median age92,range 90–98):10(29%)had benign disease,8(24%)pre-malignancy or suspected malignancy,and16(47%)malignancy.Of these, 79%had age and/or functional status discussed in the care plan.Of the8with suspected malignancy,5declined further workup.The cancer distribution revealed5(31%)vulvar,5 (31%)uterine,4(25%)ovarian,1(6%)vaginal and1(6%) cervical bined,37%had stage I disease;6% stage3;6%stage4;13%recurrent;and25%unstaged.All received treatment plans:7(47%)with palliative intent and8 (53%)with curative intent.In the curative group,7under-went surgery(1adjuvant chemotherapy)and1chemotherapy/radiation.In the palliative group,4underwent radiation,1 chemotherapy and2declined/unknown.Overall,13(87%) completed the proposed treatment.T reatment-related complica-tions included1superficial skin infection and1thirty-day readmission.Conclusions Nonagenarians often presented with vulvar or endometrial cancer and87%successfully completed treatmentwith minimal adverse effects or toxicity.Age and/or functionalstatus were considered in the care plan for79%of women,but it did not preclude treatments that had the potential to preserve meaningful quality of life and/or cure patients oftheir disease.IGCS19-0646386RISK FACTORS COMPREHENSIVE GERIATRICASSESSMENT FOR EARLY DEATH IN ELDERLY PATIENTSWITH GYNECOLOGICAL CANCER.A PROSPECTIVECOHORT STUDY1J Sales*,2C Azevedo,2C santos,3L sales,4M Bezerra,5G Bezerra,4Z cavalcanti,6MJ Mello.1IMIP,Geriatric Oncology,Recife,Brazil;2IMIP,Oncology,Recife,Brazil;3FPS,Medical Course,Recife,Brazil;4IMIP,geriatric,Recife,Brazil;5HMV,oncology,caruaru,Brazil;6IMIP,post graduation,Recife,Brazil10.1136/ijgc-2019-IGCS.386Objectives T o determine risk factors for early death identifiedthe Comprehensive Geriatric Assessment(CGA)in elderly patients with gynecological cancer(EPGC).Methods Prospective cohort study.Participants with a recent diagnosis of cancer were from eight community hospitals andone cancer center in Northeast Brazil and were recruited dur-ing their first medical appointment at the outpatient oncologic clinic.A basal CGA was done before the treatment decision (ADL,Charlson Comorbidity Index-CCI,Karnofsky Perform-ance status–KPS,GDS15,IPAQ,MMSE,MNA,MNA-SF,PS,PPS,Polipharmacy,TUG).During the follow up of12 months,information about the treatments performed,the tar-geted interventions and early death was collected.Overall sur-vival was estimated using the Kaplan–Meier method,and survival curves were compared using the Log rank test for cat-egorical variables.A multivariate Cox proportional hazardsmodel was used.Results From2015–2017,84EPGC,mean age69,6±7,9;range60–96),were enrolled,25%were metastatic disease.tumor site:40,4%cervical uterine,36,9%endometrial,20,2%ovary and2,3vulva.Nine(10.7%)ECP died in less than12 months of follow-up.In our multivariate model,controlled byage,site of cancer and cancer stage,the remaining significantrisk factors were malnutrition/nonutrition determined byMNA-SF(HR3.70,95%CI1.81–5.99,p<0.001),Katz index(HR 3.60,CI 1.56–3.81,p<0.001)CCI>2(HR2,74,CI1.0.74–10.20,p=0.013)and Polipharmacy(HR2.65,CI0.71–9.81,p<0.001).Conclusions The CGA at admission identified risk factors (Nutritional risk,polypharmacy,functionality for Katz indexand comorbidity index)for premature death in EPGC.They can help to plan a personalized care. on December 24, 2023 by guest. Protected by copyright./ Int J Gynecol Cancer: first published as 10.1136/ijgc-2019-IGCS.384 on 18 September 2019. Downloaded from。
英语词根词缀记忆大全-第五部分

第五部分:abol=do away with(消除) ablish v 废除accoutr=dress (穿着)accoutrement n 装备acm=top (顶端)acme n 顶峰adip=fat (肥)adipose a 肥胖的adjut=assist (帮助)adjutant 副手,副官advant=ahead (前面)advantage n 利益,好处avant n 先峰agger=heap (堆积)ecaggerate v 夸张alacr=swift (迅速)alacrity n 迅速,敏捷ald=old (老)alderman n 市政议员alesc=grow (成长)coalesce v 结合,联合amen=pleasant (愉快)amenity n 舒服,愉快amenable a 通情达理的amic=friend (朋友)amicable a 友好的angu,anx=distress (苦恼)amguish a 苦恼的anxious a 焦虑的anth = flower (花)antholigy n 选集chrysanthemum n 菊花(chrys黄色anthem 花+um) ap=bee (蜜蜂)apiary n 养蜂场aper=open (开)aperture n 开口,孔April 四月(开花季节) apex=aummit (顶峰)aquil=eagie (鹰)aquiline a 鹰的,似鹰的ar=plow (犁地)arable a 可耕种的ar=dry (干)arid a 干旱的arefavtion n 弄干arbor=tree (树)arboreous a 树木的arboriculture n 种树业arch=chief (主要的)architect n 建筑师archipei ago n 群岛arch=bear (熊)arctic a 北极的(在大熊星座下) antarctic a 南极的(ant相反+arctic) ard=burn (燃烧)ardent a 热情的arson v 纵火(ars=ard)ardu=difficult (困难)arduous a 劳累的aren=sand (沙)arena 角斗场(以沙铺地) arom=apice n (香料) aroma n 芳香aromatic a 芳香的arter=pipe (管道)artery n 动脉articul=joint (关节)artivulate v 关节结合;咬字清楚asin=ass (驴子)asinine a 像驴在的;愚蠢的asper=rough (粗糙)asperity n 粗暴exasperate v 激怒;使恶化asthm=panting (喘气) asthma n 气喘病athl=contest (比赛) athlete n 运动员atmo=vapor (蒸汽) atmosphere n 大气层atroc =cruel (残酷) atrocious a 残暴的atrocity n 残暴aur=gold (金)aureate v 镀金auriferous a 含金的;产金的aur = ear (耳朵) aueiform a 耳状的auricular a 耳的;听觉的auster=severe (严重的) austere a 严肃的;朴素的austr=aouth (南方)austeal a 南面的Australia n 澳大利亚(在南半球)auxili=help (帮助)auxiliary a 辅助的aval=kownward (向下)1/9avalanche v 雪崩(aval+lanche滑—向下滑)avar=greedy (贪婪)avarice n 贪婪axi=worthy (有价值)axiom n 公理,真理badin=jest (说笑)badinage v 打趣,逗笑bank=bench (长椅)bank n 银行(由柜台转化而来)banktupt n 破产者(bank椅子—柜台+rupt断—柜台断了—破产)bard=beard (胡子)barber n 理发师(兼修理胡子)bard n 倒钩;刺人的话(像胡子一样刺人) barbar=stammer (胡说) barbarian a 野蛮的(野蛮人说的话听不懂)barr=bar (栅栏)barrier n 障碍embargo v 禁运(em进入+bar禁止+go状态beat=blessed (有福的)beatify v 祝福beatific a 快乐的;天使般的bell=fine (美好)embellish v 装饰(em使+bell+使—美好)belle n 美女bever=drink (喝)beverage n 饮料bey=expect (期待)abeyance n 暂停,中止(a在+bey+ance—在期待中—观望,暂停)bias=slant (斜)bias n 偏见bib=drink (喝)imbibe v 喝入,饮bibulous a 吸水的;喜喝酒的blas=bamage (毁坏)blasphemy v 亵渎神灵(blas+phemy神的显示) blem=stain (弄脏) blemish v 玷污bombast=cotton (棉花)bombast n 吹牛的话(像棉花一样膨胀)bon=good (好)bonus n 奖金boon n 恩惠bor=dweller (居民)neighbor n 邻居(neigh附近+bor人)bord=side (边)border n 边境transborder a 横穿边境的bosc=feed (喂)proboscis n 象鼻子;吻针(pro前面+boscis,象用鼻子喂食) botan=erb (草)botany n 植物学bouch=mouth (嘴)debouch v 流出;走出bov=ox (牛)boving a 牛的brack=vomit (呕)brackish a 令人恶心的broch=pierce (刺,缝)brochure n 手册(用线缝在一起的纸)broach n 钻孔器v开瓶塞;提出问题bry=sprout (发芽)embryo n 胚胎,萌芽burl=mockery (嘲笑)burlespue n 滑稽戏(burl+espue反缀,如grotespue古怪的)byss=borrom (底)abyss n 深渊(a没有+byss—没有底) cadaver=corpse (尸体) cadaverous a 死灰色的,苍白的cal=beautiful (美丽)calligraphy n 书法kaleidoscope n 万花筒calam=misfortune (不幸)calamity n 灾难,不幸calend=first day (每月第一天) calendar n 日历call=vhard skin (硬皮)callous a 冷漠的,无感觉的can=dog (狗)canine a 狗的capr=goat (山羊)caprice v 突变;任性caper v 欢蹦,跳跃carcer=prison (监狱)caric=load (过分)aricature n 漫画;讽刺文caten=chain (链)concatenare v 把—连在一起cathar=pure (纯洁)cathartic n 泻药catharsis v 导泻;精神发泄cathol=general (普通的)catholic a 普遍的,一般的caul=stem (根,杆)cauliflower n 花菜(从根上长出花来)2/9caust=bun (烧)caustic a 苛刻的;腐蚀性的holocaust n 大火灾(holo全部+caust—全部烧光)caval=horse (马)cavalier n 骑士cavalry n 骑兵ceal=hide (藏)concea v 隐藏cel=heaven (天空)celestia a 天的,天空的ceiling n 天花板(ceil+cel) celebr=honor (荣誉)celebrate 庆祝celebrity n 名人celib=single (单个)celibacy n 独身celibate n 独身生活的人a独身的cem=sleep (睡)cemetery n 墓地cephal=head (头)acephalous a 没有头的ceram=earth (陶土)eramic n 陶瓷erebr=brain (脑)cerebrum n 大脑cess=stop (停止)cessatinon n 停止,中止incessant a 无穷无尽的chame=ground (地)chameleon n 变色龙(chame+leon狮子—地上的狮子—变色龙)chondr=cartilage (软骨)h软骨ypochondria n 臆想病症(hypo在下面+chondr+ia病—在软骨下的病—人的臆想病)chtys=golden (金色)chrysanthemum n 菊花ciner=ash (灰)incinerate v 焚烧inerary a 骨灰的cinct=bins (捆)succinct a 简洁的cist=box (盒子)cistern n 贮水桶cla=break (打破)iconoclast n 反对崇拜偶像者clandestin=secret (秘密)clandestine a 秘密的clys=dash (猛冲)cataclysm n 洪水(cata向下+clysm)col=strain (排水)cpercolart v 渗滤,渗透colander n 漏勺oll=neck (脖子)collar n 衣领accolade v 奖励,授骑士士爵位礼(把奖品挂在脖子上) com=sleep (睡) coma n 昏迷,昏睡cintamin=pollute (污染)contaminate v 弄脏,污染cop=abundance (丰富)copious a 大量的cornucopia n 丰富;丰饶角(corn角+u+cop+ia——象征丰收的角——丰饶角) cori=skin (皮)excoriate v 剥皮;严厉批评corrig=correct (改正)incorrigible a 不能改正的corusc=glitter (发光)coruscate v 闪耀,闪烁cost=side (旁边)accost v 向人搭话;调情cras=mixing (混合)idiosyncras (人的)特性crastin=tomorrow (明天)procrastinate v 拖延(明日复明日)creas=flesh (肉)pancreas n 胰腺(pan接近+creas——像肉的东西——胰腺) crepit=burst (爆裂)becrepit a 衰老的(脾气不再火爆)crimin=ceime (罪)crimina n 罪犯recriminate v 反责;反诉(crud=raw (生的)crude a 粗糙的;原始的recrudesce (病等)复发(re再+crud+esce——再次变粗糙——复发)crust=shell (外壳)crustscean a 甲壳纲的(动物)culin=kitchen (厨房)culinary a 厨房的culmin=top (顶)culminate v 到达顶峰cmul=heap (堆积)accumulate v 积累cumulus n 积云cup=desire (渴望)cpidity n 贪财,贪心curt=short (短)curt (说话)短而粗暴的curtail v 截短;缩短(讲话、节目等)3/9custod=guard (看护)custody v 监护cylind=roll (卷)cylinder n 圆柱体dama=conpuer (征服)adamant a 坚定的,坚固的(a不+dama+ant——不可征服的) de=god (神) deity n 神diefy v 神化deb=owe (欠债)debenture n 借据;债券dibit n 借方;负债debt n 债务dibil=weak (衰弱)dibility n 衰弱debilitate v 使衰弱deleter=destroyer (毁坏者) deleteriois a 有害的delect=delight (愉快) delectable a 愉快的delir=mad (疯)delirious a 神志失常的,妄想的deterior=worse (更坏) deteriorate 恶化didact=teach (教)didactic 教训的,教导的digit=finger (手指)digitate 有指的,指状的dogm=opinion (观点)domga 教条dogmatic 教条主义的domin=lord (主人)dominate 统治dominion 领地;统治domit=tame (驯服)indomitable 不可征服的dors=back (背)dorsal 背部的indorse 背书;同意(在背后签子)dot=give (给)antidote 解毒药(anti对抗毒)anecdote 轶事;短故事(an不+ec 出+dote——not published——没出版的小故事)dra=perform ( 表演)drastic 剧烈的drama 戏剧dubi=doubtful (怀疑)dubilus 怀疑的indubitable 明确的,无可怀疑的ebri=drunken (醉) inebriate 陶醉的,酒鬼eleg=lament (悲伤)elegy 哀歌emul=equal (平等)emulate 模仿;超越et=being (存在)entity 存在nonentity 不存在;无名之人entom=insect (昆虫)entomology 昆虫学equ=horse (马)equine 马的ero=love (爱)erotic 色情的ert=erect (直)alert 警惕的(al+ert站直了看——警惕的) escal=ladder (梯子) escalade 攀登escalate 逐步上升esoter=inner (内在)esoteric 秘传的estim=value (价值)estimate 估计,估量esteem (尊敬)estimable 可尊敬的;可估计的ethn=nation (民族)ethnology 人种学ethnic 种族的etymo=true (真的)etymology 词源学(词的真实来源)fam=hunger (饿)famine 饥荒famish 挨饿fan=temple (庙)fanatic 狂热的,盲信的(在庙里盲目信仰) fascin=enchant (迷住) fascinate 使---入迷fatig=weary (疲倦)fatigue 疲劳indefatigable 不知疲倦的fatu=silly (笨)fatuous 昏庸的infatuate 使糊涂,使迷醉febr=fever (烧)febrile 发烧的fecund=fruit (果实)fecundity 肥沃;多产(果实很多)felic=happy (幸福)4/9felicity 幸福;得体femin=woman (女人) feminine 女人的;温柔的fer=wild (野)ferocity 凶猛feral 野生的;凶猛的ferr=iron (铁)ferreous 铁的,含铁的fisc=pruse (钱包)fiscal 财政的confiscate 没收,把---充公fiss=split (分裂) fissure 裂缝,分裂fissile 易分裂的flagell=whip (鞭子) flagellate 鞭打fluctu=wave (波浪)fluctuate 波动for=bore (打孔)perforate 打孔fratern=brother (兄弟)fraternity 兄弟情谊fraternize 亲如兄弟;友善fraterm=fresh (新鲜)fresco 壁画(用新鲜颜料画的画) fresc=shine (果实) fructufy 使多产;结果实fulg=shine (发光)effulgence 光辉,灿烂refulgent 光辉的fulmin=thunder (雷声)fulminate 大声怒吼fulminous 怒喝的furt=steal (偷)furtive 偷偷摸摸的furc=fork (分叉)bifurcate 分成两叉furcal 叉状的fusc=dark (黑的)obfuscate 混淆;弄黑暗fuscous 暗褐色的;深色的gain =against (反对) gainsay 否认against 反对galax=milk (乳)galaxy 欢蹦,跳跃(用腿蹦跳) garr=chatter (唠叨) garrulous 唠叨的gastr=stomach (胃)gastric 胃的gasreitis 胃炎genu=knee (膝盖)genuflection 曲膝ger=bear (带有)belligerent 好斗的(bell战争)glomer=ball (球)conglomerate 合并,联合glomerulus 小球glut=devour (吞吃)glut 吞吃glutton 饱食者(gluttony 暴饮暴食)gorg=throat (喉)qorge 吞咽,峡谷gorgeous 壮观的;美丽的gross=great (大)engross 正式写成(议案、决议等);大量(或全部)收买,独占gross 总共的;重大的gymn=naked 裸体的gymnasium 体育馆(脱衣服锻炼的地方)harm=fitting (适合)harmony 和谐charm 魅力吸引力(harm的变体) haught=high (高)haughty 高傲的hein=odious (可恨的)heinous 憎恨的hemo=blood (血)hemorrhage 大出血(hemo=rrhage流出) hemer=day (一天)eohemeral 转瞬即逝的(一天就消失了) hom=man (人)homicide 杀人者(homi+cide杀) homage 尊敬,崇敬(把别人当人看)host =enemy (敌人)hostile 故意的hostility 敌对hum=moist (湿)humid 潮湿的icon=image (形象) iconoclast 反对崇拜偶像者iconography 肖像画法ident=same (相同) identical 一样的identify 认出,识别ign=fire (点火)ignite 点燃igneous 火的,似火的5/9incip=begin (开始) incipient 开始的intim=inmost (内心的) intimate 亲密的暗示isol=island (岛)isolate 孤立isolable 可隔离的isthm=narrow pass (狭道)isthmus 地狭jac=lie (躺)adjacent 邻近的,毗连的circumjacent 周围的jacul=javelin (标枪)ejaculate 突然说出(像把标枪扔出去一样) jaculate 把—向前掷去jubil=shout of joy (欢呼)jubilation 欢庆,欢腾jubilant 欢快的laver=torn (撕开)lacerate 割碎;伤害lass=weaty (倦)lassitude 疲倦,无精打采lat=wide (宽)latitude 纬度;自由laud=praise (赞扬)laudable 值得赞美的laudatory 表示赞美的,褒扬的lemma=argument (争论)dilemma 进退两难(di表示二,二种争论——进退两难)lent=loose (松)relent 放松;宽厚(re再+lent——再放松)relentless 无情的leth=oblivion (遗忘)lethargy 昏睡(leth原意是“忘川”,喝了忘川水,一切都忘记) lethal 致命的(忘了,死了)lig=bind (捆绑)ligament 韧带;纽带ligature 绷带lign=wood (木头)ligneous 木质的lignivorous (昆虫的幼虫)食木的loft=sky (天空)aloft 在高处,在空中lofty 极高的;崇高的lop=run (跑)elope 私奔interloper 入侵者,干涉者(跑入二者之间)lubric=slip (滑) lubricate 使润滑lubricant 润滑油luc=light (灯)lucubration 灯下工作lucid 清晰的lug=mourn (悲伤)lugubrious 悲伤的,伤心的(lug+ubr工作+ious——悲伤起作用) lup=wolf (狼)lupine 狼的lyr=lute (笛子)lyric 抒情诗maci=lean (瘦) emaciate 使憔悴;瘦弱macilent 瘦的macul=spot (斑点) maculate 弄上污点immacvulate 无缺点的mall=hammer (锤子) malleable 可锻造的,可塑的mallet 小锤子mamm=breast (奶) mammal 哺乳动物mascul =male (雄性) masculine 男子气的emasculate 阉割mastic=chew (嚼)masticate 咀嚼;三思matin=morning (早上)matinee 日场戏me=go (走)permeate 渗透medit=ponder (思考)meditate 思考premeditate 预先思考medl=mic (混合)medley混合(品)mell=honey (蜜)mellifluous (声音)甜美的melan=black (黑)melancholy 忧郁(的);令人伤感(的)(原指伤心得胆汁发黑) melior=better (更好)ameliorate 改善,改良mendic=beg (乞讨)mendicant 乞丐mer=lake (湖)mermaid 美人鱼(maid女人)6/9mol=meal (饭)immolate 焚祭,作祭品(给神吃饭)mol=heap (堆)molecule分子demolish 破坏,摧毁(de去掉+mol+ish,把堆起来的东西毁掉) morb=disease (病)morbid 病态的myth=fable (寓言)mythology 神话学narc=numbxx 神话学narcotic 麻醉的nebul=cloud (云)nebula 星云状的nebulous 星云的;朦胧的nec=kill (杀)internecine 自相残杀的negat=deny (否认)negative 否认的abnegation 否认;自我克制nom=pasture (牧场)nomad 游牧成员nomadic 游牧的;流浪的nu=nod (点头)innuendo 暗示,含沙射影(点头表示) nugator=trifle (烦事)nugatory 无价值的numism=coin (钱币)numismatist 钱币学家nupti=wedding (婚礼)unptial 婚姻的;婚礼的obed=obey (服从)obedient 服从的obeisance 敬礼;敬意obes=fat (肥)obese 肥胖的obit=death (死)obituary 讣告obliv=forget (忘记) oblivion 遗忘oblivious 疏忽的obsol=decay (腐烂) obsolete 过时的obsolescent 逐渐被废弃的ocul=eye (眼睛)ocular 眼睛的oculist 眼科医生od=road (道路)odometer 里程表(meter仪表)odi=hate (恨)odium 憎恨odious 可憎的odyn=pain (痛)anodyne 止痛药(an没有+odyne痛)ole=oil (油)petroleum 石油oleaginous 产油的;油质的olfact=scent (味)olfaction 嗅觉olfactoty 嗅觉的omin=omen (预兆)ominous 凶兆的abominate 厌恶,憎恶(ab离开+omin+ate—离开凶兆—憎恨凶兆) opl=armor (盔甲)panoply 全副武装(pan全部,全身都是盔甲)ornith=bird (鸟)ornithology 鸟类学oscul=kiss (吻)osculate 亲吻;接触ostens=appear (出现)ostensible 表面的,明显的ostentation 夸示的;虚饰的ostrac=tile (瓦片)ostracize 流放;放(古希腊人投瓦片表示放入出境)ov=shout (叫喊)ovation 大声欢迎;鼓掌pachy=thick (厚)pachyderm 厚皮动物;脸皮厚的人pan=bread (面包)pantry 面包房company 同伴;公司(共同有面包吃)pat=walk (走)peripatetic 巡回走动的(peri来回+pat+etic) phem=speech (讲话) blasphemy 亵渎(blas坏+phem+y—讲坏话)euphemism 委婉的说法(eu好+phem+is—m—讲好话) phen=show (显示) phenomenon 现象(phen+omen预兆+on—显示预兆—现象)pher=bring (带来)periphery 周边,周围环境(peri四周)ping=fasten (系紧)impinge 撞击;侵犯(im进入+ping+e—紧迫进入—侵犯)pinn=peak (顶)pinnacle 顶峰,尖顶plais=please (高兴)7/9complaisant 讨好的plas=mold (模式)plaster 膏药plastic 可塑的protoplast 原物型pleb=people (人们)plebeian 普通民众的pleth=crowdx (挤,多)plethora 多余,供过于求supplicant 恳求者 a.哀求的(跪下去求别人) plum=feather (羽毛) plumage 羽毛plume 整理—羽毛;骄傲pneumon=lung (肺)pneumonia 肺炎pol=smooth (光滑)polish 抛光,擦亮plite 有礼貌的postul=demand (要求)postulate 要求;假定存在expostulate 规劝,忠告pragm=deed (行为)pragmatic 实用主义的(以行为为最重要)psych=soul (灵魂)psychology 心理学psychical 心理学;灵魂的pud=shame (害羞)imoudent 无礼的,不知羞耻的pudency 羞怯,害羞puer=boy (男孩)puerile 孩子气的,幼稚的pulver=dust (灰尘)pulverize 把—弄粉碎pusill=mean (卑微的)pusillanimous 胆小的,胆怯的(anim生命)pyr=fire (火)pyre 火葬柴堆empyrean 苍天,太空(em进入+pyr+ean,进入火—进入太阳—天空) quarant=forty (四十)quarantine 隔离检查(被隔离40天) quer=complain (抱怨) querulous 抱怨的quarrel 争吵rab=rage (怒)rabid 狂怒的,狂热rabies 狂犬病radi =ray (光线)radiant 发光的irradiate 发光,(光)漫射ram=branch (枝)ramify 分叉,分枝ramiform 枝状的reg=king (国王)regnant 国王的,皇家的regicide 杀害国王regn=reign (王朝)regnant 统治的,占优势的intrerregunm 无王时期remn=remain (留下)remnant 剩余物rest=stay (停留)arrest 阻止,逮捕redtless 不安的;得不到休息的(和restive 是同一意思) retic=silent (安静)reticent 沉默的rever=awe (敬畏)revere 尊敬reverent 尊敬的,敬畏的rhe=flow (流)diarrhea 痢疾(dia对穿—对穿流过—拉肚子) rheum 泪水rhin=nose (鼻子)rhinoceros 犀牛riv=stream (河流)rivulet 小河(let表示“小”)derive 派生,得出rug=wrinkle (皱)corrugate 起皱(cor一起,皱到一起)sacchar=sugar (糖)saccharin 糖精salu=health (健康)salubrious 有益健康的(salubr=salu) salutary 有益健康的sanat=taste (味道)sapid 美味的,有趣的insipis 无味的;无吸引力的sarc=flesh (肉)sarcasm 尖刻讽刺(尖刻的话像咬人一块肉一样) sarcophagous 食肉的scintill=spark (火花)sembl=be like (相像)resemble 和—相像dissemble 假装,掩盖(dis不,和本性不一样,假8/9装)sept=rotten (烂)antiseptic 防腐的septic 腐败的;败血病的sibil=hiss (咝咝声)sibilant 发嘶声的sider=star (星星)sidereal 星星的,星空的consider 考虑(研究星象引申而来)stetho=chest (胸)stethoscope 听诊器succ=juice (果汁)succulent 多汁的succus 液,汁sud=sweat (汗)sudatory 发汗的exude 渗出,慢慢流出(由“出汗”引申出来) sumptu=expense (花费) sumptuous 奢侈的,豪华的sumptuary 规定个人费用的tabul=plank (平板)tabular 表格的;平坦的tabulate 把—列成表格taph=tomb (墓)epitaph 墓志铭(epi的旁边,在墓边的“墓志铭”) tauto=same (相同) tautology 同义反复,累赘templ=tmple (庙)contemplate 静思,深思(像庙中人一样深思) temple 寺庙torp=numb (麻木)torpid 呆板的torpify 使麻木;使迟钝torr=dry (干)torrid 酷热的torrefy 焙,烤trepid=trembling (颤抖)trepidation 胆怯intrepid 大无畏的tum=swell (肿)tumid 肿胀的intumescence 开始发胀uber=fruitful (果实)exuberant 茂盛的;多产的uberty 丰饶,多产,生育力ucor=wife (老婆)uxorious 怕老婆的,宠爱老婆的uxoricide 杀妻vale=farewell (再见)valediction 告别演说veh=carry (带来)vehicle 车辆vehement (感情等)猛烈的(猛烈状态带来后果) velop=cover (盖上) envelop 把—包住,裹住develop 发展verd=green (绿)erdant 碧绿的erdigris 铜绿,铜绣;碱性veter=old (老)veteran 老兵inveterate 根深蒂固的viti=fault (错误) itiate 污染;使堕落vituper=blame (责备) vituperate 严厉责备zym=ferment (发酵) enzyme 酶zymology 发酵学9/9。
格瑞普莱特 工具和射钉对照表说明书

Grip-Rite ®, the leader in fasteners, is committed to making your collated fastener purchase simple and easy. Use this c ross-reference guide to quickly identify which tools are compatible with each type of Grip-Rite collated fastener, and vice versa (which Grip-Rite collated fasteners will fit your tools).*Model numbers change often as new or updated tools are introduced. Please use the icon system when possible or call 800 676-7777 for assistance.APPLICATION F FEATURE THE ICON MATCHING SYSTEMAND MODELS ARE L ISTED WITH EACH TYPE OF GRIP-RITE COLLATE D FASTENERSPACKAGING CLEARLY SHOWCASE THE FASTENER ICONFASTENER CATEGORIES AND TYPES OF COLLATION FRAMING21° Plastic StripRound Head NailsGrip-Rite: GRTFR83ROOFINGSIDING/FENCING15° COILR OOFING NAILS0° COILS HEET NAILS15° PLASTIC S HEET NAILS15° WIRE W ELD NAILSLASER WELDED PLASTIC CAPS AND STAPLESGrip-Rite: GRTCR175, GRTRN45Bostitch: RN45, RN45B, RN46, RN46-1, BRN175ADeWalt: DW45RN, DCN45RN Duo-Fast: DRN-45, RN-175H itachi/Metabo HPT:NV45AB2, NV45A M akita:AN451, AN453, AN454Milwaukee: 7120-21, 7220-20, 7220-80 Max: CN445R, CN445R2, CN450R, CN445R3 P aslode:R-175C, 3175/44RCUPorter-Cable: 134R, RN175, RN175A, RN175C R idgid:R175RNESenco: SNC40R, SCN200R,ROOFPRO 450, ROOFPRO 455XPGrip-Rite: GRC58, GRC58A Pneu Tools: RC-58, RC-58II Senco: BC58Grip-Rite: GRTCS250ZDuo-Fast: DF225C, 502950 Pneu Tools: CN65ZThe list of tools for each fastener is not exclusive. Refer to tool manuals for fastener size compatibility.Grip-Rite is a trademark of PrimeSource Building Products, Inc.O ther product and company names listed may be trademarks of their respective owners.Grip-Rite: GRTCS250Bostitch: N63CP, N63CP-1, N66C, N66C-1DeWalt: DW66C-1Hitachi/Metabo HPT: NV65AH,N V75AG, NV50A1, NV65AH2, NV65ANMakita: AN611Max: CN565D, CN565S, CN890S Senco: SCN49, SCN55S, SCN56Grip-Rite: GRTCS250Bostitch: N50C, N55C, N65CP, N65CP-2, N66C-1 DeWalt: DW66C-1Hitachi/Metabo HPT: NV65AB, NV65AE, NV65AH, NV75AG, NV50A1, NV65AH2, NV65AN M akita: AN611Max: CN55, CN550S, CN565S, CN665, CN890II , CN665D, CN565S3Senco: SCN45, SCN49, SCN55S, SCN56Tool/FastenerCompatibility GuideFRAMINGJOIST HANGER33° PAPER T APE JOIST H ANGER NAILS33° PLASTIC STRIP JOIST HANGER NAILS28° PLASTIC STRIP CLIPPED HEAD NAILSThe list of tools for each fastener is not exclusive. Refer to tool manuals for fastener size compatibility.Grip-Rite is a trademark of PrimeSource Building Products, Inc.O ther product and company names listed may be trademarks of their respective owners.Bostitch:DeWalt:Duo-Fast:H NV65ANMakita:M ax:CN890F2Paslode:P orter-Cable:S enco:Grip-Rite: GR150, GR250, GRSB150, GRSB250Bostitch: MCN150, MCN250 DeWalt: DWMC150Hitachi/Metabo HPT: NR65AK, NR65AK(S),NR38AKM Max: SN438JPaslode: 5250/65S PP, 5250S PP, F250S-PP, PF150S-PPPneu Tools: RN150, RN250, RNS150, RNS250Senco: HN150, HN250, JoistPro 150/250Grip-Rite: GR150, GR250 Bostitch: MCN150, MCN250 Pneu Tools: RN150, RN250 Senco: HN150, HN250Milwaukee: 7110-20Max: SN883CH/34, SN890CH/34, SNH890CH2/34P aslode: 5300, 900420, IMPULSE 35,CF325XP, PF250S, PF350S, 513000, F350S POWER MASTER PLUSPorter-Cable: FC350, FC350A, FC350B Ridgid: R350CHA, R350CHE S enco:FRAMEPRO 601/651,FRAMEPRO 701XP/751XP, SN901XP, SN951XPBostitch: D eWalt:Duo-Fast:C N-350B,H NR90AEPR, NR83A3, NR83A3(S), NR83A5, NR83A5(S), NR90AES, NR90AES1, NR90A5 Husky: DPFR2190Makita: AN922, AN923, AN8300, AN924 M ax: GS683RH-EX, SN80, SN883RH2, SN890RHMilwaukee: 274-22, 7200-20Paslode: 5325/SRH, 5350/SRH-20, F350-21 P orter-Cable: FR350BRidgid: R350RHA, R350RHE, R350RHF S enco:FRAMEPRO 502, FRAMEPRO 650, SN60, SN65, SN902XP, GT90FRHDuo-Fast: NSPM-325FFINISHFLOORINGCONCRETE15-1/2 GAUGE FLOORING STAPLES1/2" Crown“L” HARDWOOD FLOORING CLEATSCONCRETE T-NAILS16 GAUGE STRAIGHT FINISH NAILSBostitch:DA1564KD eWalt:D uo-Fast:NT65MA4Makita: M ax:P R idgid:Ryobi:Senco: P orter-Cable: PIN100 R idgid: R138HPASenco: FINISHPRO 10, FINISHPRO 11, SHP10Bostitch: Max:Grip-Rite: GR200FSB ostitch: MIIIFSDuo-Fast: FLOOR MASTER 200-SGrip-Rite: GR200LCNBostitch: MFN200, MFN201 Porter-Cable: FCN200Senco: SHF10, SHF15, SHF50Bostitch: MIII812CNCT Porta-Nails: 460ASpotnails: MT9764, XT8664 Grex: 2564 Senco: GT40CPP aslode:Senco: FUSION F-16A Milwaukee: 2742-21CTGrip-Rite: GRTFN250D eWalt:D51257K, DWFP71917, DCN660D1 D uo-Fast:SURE SHOT 764 Hitachi/Metabo HPT: NT65GS Makita: AF601 Max: NF565/16Milwaukee: 2741-21CT, 2741-20, 7145-21 Paslode: 902000, T250S-F16, 916000, 515500 Porter-Cable: FN250CRidgid: R250SFE, R250SFF Ryobi: YN250FSD, P325S enco:FUSION F-16S, FINISHPRO 32, FINISHPRO 16XPThe list of tools for each fastener is not exclusive. Refer to tool manuals for fastener size compatibility.TACKER STAPLESSTAPLES“A11” STYLE T ACKER STAPLES3/8" Crown“54”-STYLET ACKER STAPLES3/8" Crown“50”-STYLETACKER STAPLES1/2" Crown“A19”-STYLE TACKER STAPLES3/8" Crown“STCR”-STYLE TACKER STAPLES7/16" Crown“SHCR”-STYLE T ACKER STAPLES7/16" CrownDuo-Fast: N3804AB3 Makita: Max: Milwaukee:P orter-Cable:NS150BRidgid: Ryobi:Senco: Bostitch: SX150, SB2N1Grip-Rite: GRR11C Rapid: 54Grip-Rite: GR50BLD uo-Fast:HT-550, SURESHOT 5020Grip-Rite: GRR19 Rapid: A19, R19Bostitch: H2BGrip-Rite: GRSTCR5019B ostitch: H30-8, PC2K, PC4000, T6-8 Rapid: 31Grip-Rite: GRR11Arrow: T-50, HT-50, HT-55Bostitch: PHT150C, TR100, TR200, TRE500Duo-Fast: SLAPSHOT R apid:A11, R11Senco: PC0705, 85000, 85060Bostitch: S4Duo-Fast: M akita:Max:DeWalt: Makita: M ax:Porter-Cable: Senco: 800-676-7777 。
《2024年向日葵列当寄生过程关键基因OcEXPA6的功能验证及siRNA特征分析》范文

《向日葵列当寄生过程关键基因OcEXPA6的功能验证及siRNA特征分析》篇一一、引言向日葵列当是一种常见的植物寄生生物,它通过寄生在向日葵等植物上获取营养,从而对植物的生长和发育产生重要影响。
近年来,随着分子生物学和基因编辑技术的发展,关于向日葵列当寄生的相关研究也越来越多。
本篇文章主要介绍在列当寄生过程中起关键作用的基因OcEXPA6的功能验证以及相关siRNA特征的分析。
二、材料与方法1. 材料本实验所使用的材料包括向日葵列当、向日葵植物、以及相关的生物试剂和仪器等。
2. 方法(1)基因克隆与表达分析:通过PCR技术克隆出OcEXPA6基因,并构建表达载体,进行表达分析。
(2)功能验证:利用基因编辑技术,对OcEXPA6基因进行敲除或过表达,观察其对向日葵列当寄生过程的影响。
(3)siRNA特征分析:通过高通量测序技术,对向日葵列当中与OcEXPA6相关的siRNA进行鉴定和特征分析。
三、实验结果1. 基因克隆与表达分析结果成功克隆出OcEXPA6基因,并构建了表达载体。
通过实时荧光定量PCR技术,发现OcEXPA6基因在向日葵列当寄生过程中表达量显著上升。
2. 功能验证结果通过对OcEXPA6基因进行敲除或过表达,发现该基因在向日葵列当寄生过程中具有重要作用。
敲除OcEXPA6基因后,向日葵列当的寄生能力明显减弱;而过表达OcEXPA6基因则能显著增强向日葵列当的寄生能力。
这表明OcEXPA6基因是向日葵列当寄生过程中的关键基因。
3. siRNA特征分析结果通过高通量测序技术,鉴定出与OcEXPA6基因相关的siRNA。
这些siRNA在向日葵列当中的表达量较高,且具有明显的时空特异性。
进一步分析发现,这些siRNA可能通过调控OcEXPA6基因的表达,从而影响向日葵列当的寄生过程。
四、讨论本实验结果表明,OcEXPA6基因在向日葵列当寄生过程中具有重要作用,且与相关siRNA的表达密切相关。
鬼笔环肽标记系列产品说明书

鬼笔环肽标记系列产品说明书保存:-20℃干燥、避光保存,有效期一年。
若配制成水溶液,请小量分装保存,避免反复冻融。
注意:本产品为冻干粉形式,使用前请瞬时离心,加适当溶剂溶解后使用。
产品介绍:鬼笔环肽是从致命的伞形毒蕈蘑菇中分离出来的一种毒素。
它是特异性结合于F-肌动蛋白的双环肽(1)。
因此用荧光染料标记的鬼笔环肽可以非常方便的研究F-肌动蛋白的分布。
鬼笔环肽内部,在半胱氨酸和色氨酸之间含有不常见的硫醚桥形成内环结构。
在pH 升高时,该硫醚被裂解,鬼笔环肽失去对肌动蛋白的亲和力。
荧光标记的鬼笔环肽可在纳摩尔水平染色F-肌动蛋白(1-3)。
在各种植物细胞或动物细胞中,标记的鬼笔环肽对大、小细丝具有相似的亲和力,平均每个肌动蛋白亚基结合一个鬼笔环肽分子。
不同于抗体,鬼笔环肽与肌动蛋白的结合亲和力在不同物种间没有显着变化。
非特异性染色可以忽略不计,染色和未染色区域之间的对比度非常大。
鬼笔环肽将单体/聚合物的平衡转向聚合状态,将聚合临界浓度降低至30倍(3,4)。
Phallotoxins 可通过抑制细胞松弛素的解聚,碘化钾和升高的温度,稳定F-肌动蛋白。
因为鬼笔环肽缀合物很小,大约直径12-15埃,分子量<2000道尔顿,多种肌动蛋白结合蛋白,包括肌球蛋白,原肌球蛋白和后肌钙蛋白依然可以和鬼笔环肽标记的肌动蛋白结合。
更重要的是,鬼笔环肽标记的肌动蛋白丝保持功能,标记甘油肌纤维仍然收缩,标记的肌动蛋白丝仍然可以继续移动(5,6)。
而且荧光标记的鬼笔环肽也可用于对细胞中F-肌动蛋白进行定量研究(7,8)。
实验方法:1.储液制备:荧光染料标记的鬼笔环肽:取适量甲醇或无菌水溶解棕色管中冻干的粉末,制备成200U/mL 的储液(300T 规格染料加入1.5mL 的液体)。
荧光标记的标记鬼笔环肽的一个单位(T )的定义是染色一个加载细胞的载玻片所用染料的量。
使用时的推荐稀释比例为1:40-1:200,一个单位相当于200µL 总染色体积中加入1-5µL 200U /mL 储备溶液。
Bosch 家用电器产品说明

Touch controls with LCD display.
600 CFM centrifugal integrated blower included.
Four speed touch controls with LCD display.
Dishwasher safe filters with stainless steel covers.
36" Glass Canopy Chimney Hood
Benchmark® Series – Stainless Steel HCG56651UC
HCG56651UC Stainless Steel
Wall mounted chimney hoods add style and sophistication to the kitchen and gives you greater freedom in your kitchen planning.
Damper included
Yes
Grease filter material
Washable stainless steel
Grease filter type
Multilayer cassette
Technical Details
Watts
443 W
Volts
120 V
Frequency
60 Hz
Working Speed
460
160
Sones
High Speed
Working Speed
11.0
2.0
Instpper ue duct B Lower ue duct C Screws D Angle bracket
PMMA Disc安全数据说明书

PMMA Disc Page 1 of 5SECTION 1: Identification of the substance/mixture and of the company/undertaking1.1. Product identifierPMMA Disc1.2. Relevant identified uses of the substance or mixture and uses advised against1.3. Details of the supplier of the safety data sheetCompany Name:Address:City, State, Zip Code:Telephone:email address:Website:SECTION 2: Hazards identification VinciSmile Group LLC 16453 Old Valley BlvdLa Puente, CA, 91744, USA +1 626 283 5808************************ 2.1. Product classificationMay cause sensitisation by skin contact.2.2. Health HazardThe product presents no toxic hazard when used according the manufacturer’s instructions. If a patient is known to beallergic to any of the ingredients of the teeth, the product should not be used.2.3. Additional dangers to humans and the environmentSee point 12.SECTION 3: Composition/information on ingredients3.2. MixturesChemical characterizationsolidHazardous componentsPoly methyl methacrylate with high molecular weight Methyl methacrylatePigments: Iron oxide and Titanium dioxideHazardous Components C.A.S. # EINECS # Substance Classification WT %Poly(methyl methacrylate)9011-14-7 232-674-9 Not Applicable >90SECTION 4: First aid measures4.1. Description of first aid measuresGeneral informationNone.PMMA DiscPage 2 of 5 After inhalationIn case of trouble after inhaling abrasive dust, seek medical advice.After eye contactI n case of mechanical eye irritation, rinse thoroughly with large amounts of water. Consult a doctor if irritation persists. After skin contactIn case of dust contact with skin, wash off with soap and water. Consult a doctor if skin irritation persists.After swallowingIn case of swallowing, seek medical advice.Cautions for the doctorNo specific antidote. Effect support therapy. Care should be effected according doctor’s evaluation with respect to thepatient’s reactions.Most important symptoms of exposureThis product is a non-hazardous polymer solid. Dust generated from polishing or grinding may cause eye or respiratorytract irritation.SECTION 5: Firefighting measures5.1. Extinguishing mediaSuitable extinguishing agentsThese products are not classified as flammable or combustible but will burn under fire conditions. Water spray jet, foam,dry powder, carbon dioxideFor safety reasons unsuitable extinguishing agentsWater with full jet.Protective equipment for fire-fightingUse self-contained respirator (breathing apparatus). -Wear full protection suit.SECTION 6: Accidental release measures6.1 Person-related safety precautionsAvoid build up of dust. Use breathing protection (Fine dust mask FFP) when exposed to dust. Use personal protectiveequipment (overalls, protective glasses and protective gloves).6.2 Environmental PrecautionsReport releases as required by local and national authorities.6.3 Methods and Materials for Containment and Clean-up:Pick up teeth and return to container for use. For dust from grinding or polishing products: Contain spills, sweep orgather spilled material in a manner that minimizes the generation of airborne dusts, and transfer to a suitable containerfor disposal.SECTION 7: Handling and storage7.1. Precautions for safe handlingAvoid generating dust. Avoid breathing dust. Use adequate ventilation for polishing or grinding operations. Wash handsthoroughly after polishing or grinding teeth. Use good housekeeping to minimize accumulation of dust.7.2. Conditions for safe storage, including any incompatibilitiesNo special storage required.Safety Data SheetPMMA DiscPage 3 of 5SECTION 8: Exposure controls/personal protection8.1 Exposure informationNone.8.2 Technical devicesNone.8.3 Personnel protective equipmentNone.8.4 Respiratory ProtectionNo needed with normal usage. When product is grinded or polished, use localized aspiration of dusts and/or protectivemask.8.5 Skin protectionNone particular precaution should be taken with normal usage.8.6 Eye and visage protectionNone particular precaution should be taken with normal usage. When the product is grinded or polished, use protectiveglasses.SECTION 9: Physical and chemical propertiesPhysical state at 20°C solidColor variousOdor unscentedSpecific weight (at 20°C) --Solubility in water (at 20°C) insolublePH (at 20°C) --Casting temperature --Boiling temperature --Auto ignition temperature --Inflammability temperature --Air deflagration limits --SECTION 10: Stability and reactivity10.1 Dangerous reactionsNo dangerous reactions known.10.2 Chemical StabilityStable.10.3 Conditions to AvoidStable in normal conditions.10.4 Hazardous Decomposition ProductsDecomposition may release oxides of carbon, methyl methacrylate, and irritating smoke and fumes.10.5 Materials to avoidNone in particular.10.6 Hazardous decomposition productsNone when correctly utilized.PMMA DiscPage 4 of 5SECTION 11: Toxicological information11.1. Information on toxicological effectsSensitisationAllergic reactions were reported among humans. The product contains small quantities of sensitising substances(methyl methacrylate ). Intensive skin contact, especially with the dissolved product, can trigger an allergic reaction inpersons previously sensitised.Further informationThe biocompatibility tests carried out on the teeth, in accordance with EN ISO 10993, regarding cytotoxicity, toxicitytest, stimulate or intradermal reaction, acute systemic toxicity and delayed type, did not result in any negative effects,under the test conditions, on the cell materials used.The fines which appear during sharpening can lead to mechanical irritations of the skin, eyes, and mucous membranes.Skin and eye contact with the product should be avoided. The inhalation of product dusts should be avoided.SECTION 12: Ecological information12.1 DegradationThe material is known to be stable in environment. Due to the law biodegradability the product should not reach wateror ground.12.2 Aquatic toxicityInsoluble. No dangers for environment are known.SECTION 13: Disposal considerations13.1.RecommendationThe waste is not dangerous and can be disposed together with household garbage.Disposal area must comply with the environment and national safety standard.SECTION 14: Transport informationUN number: n.a.class: n.a.packaging group: n.a.ocean harmful substances: n.a.other relevant information: no hazardous goodSECTION 15: Regulatory information15.1. Safety, health and environmental regulations/legislation specific for the substance or mixtureThe product is subject to the regulations of the Medical Device Directive 93/42/EEC, as well as to the national MedicalDevice Act and the Chemicals ActPMMA DiscPage 5 of 5 SECTION 16: Other informationNo further technical informationThe present data sheet contains technical-scientific information processed at best of our knowledge. We recommendverifying national and regional regulations applicable to the specific utilize field as well as regulations relative hygienicand safety on work and environment worship.All information contained in the present data sheet is correct and processed in good faith. However they do not involveany obligation, guarantee and patent concession.The characteristics mentioned in the following document do not constitute contractual specifications.The data for the hazardous ingredients were taken respectively from the last version of the sub-contractor's safety data sheet.。
SMARTer_RACE_5_3__Kit_User_Manual_022714

Clontech Laboratories, Inc.SMARTer® RACE 5’/3’Kit User ManualCat. No(s). 634858, 634859(022714)Clontech Laboratories, Inc.A Takara Bio Company1290 Terra Bella Avenue, Mountain View, CA 94043, USAU.S. Technical Support: tech@United Asia Pacific Europe Japan Page 1 of 30I. Introduction (4)II. List of Components (7)III. Additional Materials Required (8)IV. Primer Design (9)A. Primer Sequence (9)B. Additional Considerations for Design (10)C. Location of Primer Sequences within Genes (10)D. Nested Primers (10)V. Generating RACE-Ready cDNA (11)A. General Considerations (11)B. Preparation and Handling of Total and Poly A+ RNA (11)C. Assessing RNA Template Quality (12)D. Protocol: First-Strand cDNA Synthesis (13)VI. Rapid Amplification of cDNA Ends (RACE) (15)A. Things You Should Know Before Starting RACE PCR Reactions (15)B. Protocol: Rapid Amplification of cDNA Ends (RACE) (15)VII. Characterization of RACE Products (17)A. Protocol: Gel Extraction with the NuceloSpin Gel and PCR Clean-Up Kit (17)B. Protocol: In-Fusion Cloning of RACE Products (18)C. Sequencing RACE Products (19)VIII. References (20)Appendix A. Troubleshooting Guide (21)A. Troubleshooting Touchdown PCR (21)B. Multiple Band RACE Products (23)C. Other Specific Problems (25)Ap pendix B. Detailed Flow Chart of 5’ RACE (27)Appendix C. Detailed Flow Chart of 3’ RACE (28)Appendix D. 5’-RACE cDNA Amplification with Random Primers (29)A. Protocol: First-Strand cDNA Synthesis with Random Priming (29)Figure 1. Mechanism of SMARTer cDNA synthesis (4)Figure 2. Overview of the SMARTer RACE procedure.. (6)Figure 3. The relationship of gene-specific primers to the cDNA template. (9)Figure 4. 5'- and 3'-RACE sample results (22)Figure 5. Detailed mechanism of the 5'-RACE reactions. (27)Figure 6. Detailed mechanism of the 3'-RACE reactions. (28)Table of TablesTable 1. Additional 5'-RACE Sequence Obtained with SMART Technology (5)Table 2. Setting up 5'- and 3'-RACE PCR Reactions (15)Table 3. Troubleshooting Guide: Other Specific Problems (25)I. IntroductionThe SMARTer RACE 5’/3’ Kit provides a method for performing both 5’- and 3’-rapid amplification of cDNA ends (RACE). The SMARTer RACE 5’/3’ Kit includes our SMARTer II A Oligonucleotide and SMARTScribe™Reverse Transcriptase, which provides better sensitivity, less background and higher specificity than previouskits. This powerful system allows you to amplify the complete 5’ sequence of your target transcript from as little as 10 ng of total RNA. The cornerstone of SMARTer RACE cDNA synthesis is SMART™ technology, which eliminates the need for problematic adaptor ligation and lets you use first-strand cDNA directly in RACE PCR, a benefit that makes RACE far less complex and much faster (Chenchik et al., 1998). Additionally, the SMARTer RACE Kit exploits Clontech’s technology for suppression PCR & step-out PCR to increase the sensitivity andreduce the background of the RACE reactions. You can use either poly A+ or total RNA as starting material for constructing full-length cDNAs, even of very rare transcripts.The SMARTer RACE 5’/3’ Kit is an improved version of our original SMARTer RACE cDNA Amplification Kit, designed to accommodate larger RNA input volumes and perform more efficiently on challenging targets(e.g., those that are long, GC-rich, etc.). RACE PCR products are amplified with our highly robust SeqAmp™DNA Polymerase, and cloned into the linearized pRACE vector with In-Fusion® HD Cloning. The In-Fusion HD Cloning Kit, NucleoSpin Gel and PCR Clean-Up Kit, and Stellar™ Competent Cel ls are included for yourconvenience in cloning RACE products.SMART technology provides a mechanism for generating full-length cDNAs in reverse transcription reactions (Zhu et al., 2001). This is made possible by the joint action of the SMARTer II A Oligonucleotide andSMARTScribe Reverse Transcriptase. When the SMARTScribe RT reaches the 5’ end of the RNA, its terminal transferase activity adds a few additional nucleotides to the 3’ end of the first-strand cDNA (Figure 1).Figure 1. Mechanism of SMARTer cDNA synthesis.First-strand cDNA synthesis is primed using a modified oligo (dT) primer. After SMARTScribe Reverse Transcriptase (RT) reaches the end of the mRNA template, it adds several nontemplated residues. The SMARTer IIA Oligonucleotide anneals to the tail of the cDNA and serves as an extended template for SMARTScribe RT.The SMARTer II A Oligonucleotide contains a terminal stretch of modified bases that anneal to the extended cDNA tail, allowing the oligo to serve as a template for the RT. SMARTScribe RT switches templates from the mRNA molecule to the SMARTer oligo, generating a complete cDNA copy of the original RNA with the additional SMARTer sequence at the end. Since the template switching activity of the RT occurs only when the enzyme reaches the end of the RNA template, the SMARTer sequence is typically only incorporated into full-length, first-strand cDNAs. This process guarantees that the use of high quality RNA will result in the formation of a set of cDNAs that have a maximum amount of 5’ sequence (Table I).Table 1. Additional 5'-RACE Sequence Obtained with SMART TechnologyHuman gene Size of mRNA(kb)Additional sequence(bp)*Matches genomicsequencesPiccolo presynaptic cytomatrix protein 20.29 +59 yesDynein, cytoplasmic 1, heavy chain 1 14.36 +36 yesPolycystic kidney disease 1 14.14 +21 yesSolute carrier family 1 12.02 +73 yes Microtubule-associated protein 1A 10.54 +13 yesSpectrin, beta, non-erythrocytic 10.24 +32 yesTransferrin receptor 5.0 +25 yesInterferon-α receptor 2.75 +17 yesSmooth muscle g-actin 1.28 +31 yesFollowing reverse transcription, SMART technology allows first-strand cDNA to be used directly in 5’- and3’-RACE PCR reactions. Incorporation of universal primer binding sites in a single-step during first-strand cDNA synthesis eliminates the need for tedious second-strand synthesis and adaptor ligation. This simple and highly efficient SMARTer cDNA synthesis method ensures higher specificity in amplifying your target cDNA. Suppression PCR & step-out PCR techniques are used in combination with SMARTer technology to decrease background amplification in RACE PCR.Requirements for SMARTer RACE cDNA AmplificationThe only requirement for SMARTer RACE cDNA amplification is that you know at least 23–28 nucleotides (nt) of sequence information in order to design gene-specific primers (GSPs) for the 5’- and 3’-RACE reactions. (Additional sequence information will facilitate analysis of your RACE products.) This limited requirement makes SMARTer RACE ideal for characterizing genes identified through diverse methods, including cDNA subtraction, differential display, RNA fingerprinting, ESTs, library screening, and more.Uses of SMARTer RACE cDNA AmplificationSMARTer RACE cDNA amplification is a flexible tool—many researchers use this kit in place of conventional kits to amplify just the 5’ or 3’ end of a particular cDNA. Others perform both 5’- and 3’-RACE, and many then go on to clone full-length cDNAs using one of the two methods described in the latter part of this protocol. In many cases, researchers obtain full-length cDNAs without ever constructing or screening a cDNA library.Figure 2. Overview of the SMARTer RACE procedure.Detailed flow charts of the SMARTer RACE mechanisms can be found in Appendices B & C. Alternatively, you can obtain the sequences of the extreme ends of the transcript by sequencing the 5’ end of the 5’ product and the 3’ end of the 3’ product. Using this information, you can design 5’ and 3’ gene-specific primers to use in LD PCR with the 5’-RACE-Ready cDNA as template to generate the full-length cDNA.Note that with the cloned RACE fragments you can use a restriction site in an overlapping region to construct a full-length cDNA by subcloning, or design new GSPs to generate PCR products compatible with In-Fusion cloning.II. List of ComponentsThis section lists the components for Cat. No. 634858, a 10 reaction kit. The larger, 20 reaction kit (Cat. No.634859) contains two of every item listed below.SMARTer RACE 5’/3’ Kit Components (Cat. No. 634860) (Not sold separately)Store SMARTer II A Oligonucleotide and Control Mouse Heart Total RNA at –70°C. Store all other components at –20°C.∙First-Strand cDNA Synthesiso10 µl SMARTer II A Oligonucleotide (24 μM)o10 µl 3' RACE CDS Primer A (12 μM)o10 µl 5' RACE CDS Primer A (12 μM)o10 µl 10X Random Primer Mix (20 μM)o40 µl 5X First-Strand Buffer (RNAse-Free)o 5 µl Dithiothreitol (DTT) (100 mM)o 1 ml Deionized H2Oo10 µl RNase Inhibitor (40 U/µl)o20 µl SMARTScribe Reverse Transcriptase (100 U/µl)o10 µl dNTP mix (20 mM)∙5’- and 3’-RACE PCRo400 µl 10X Universal Primer A Mix (UPM)o50 µl Universal Primer Short (10 µM)o 5 µl Control Mouse Heart Total RNA (1 µg/µl)o25 µl Control 5'-RACE TFR Primer (10 µM; designed for compatibility with In-Fusion cloning)o25 µl Control 3'-RACE TFR Primer (10 µM; designed for compatibility with In-Fusion cloning) ∙In-Fusion Cloningo10 µl Linearized pRACE (50 ng/µl)∙General Reagentso 2 tubes Tricine-EDTA Buffer (1 ml each)SeqAmp DNA Polymerase (Cat. No. 638504)Store all components at –20°C.∙50 μl SeqAmp DNA Polymerase∙ 1.25 ml SeqAmp PCR Buffer (2X)In-Fusion HD Cloning Kit (Cat. No. 639648) (Not sold separately)Store all components at –20°C.∙20 μl 5X In-Fusion HD Enzyme Premix∙ 5 μl pUC19 Control Vector, linearized (50 ng/μl)∙10 μl 2 kb Control Insert (40 ng/μl)NucleoSpin Gel and PCR Clean-Up Kit (Cat. No. 740609.10) (Not sold separately)Store all components at room temperature∙10 ml Binding Buffer NTI∙ 6 ml Wash Buffer NT3 (concentrate)∙ 5 ml Elution Buffer NE (5 mM Tris/HCl, pH 8.5)∙10 NucleoSpin Gel and PCR Clean-Up Columns (yellow rings)∙10 Collection Tubes (2 ml)Stellar Competent Cells (Cat. No. 636763)Store Stellar Competent Cells at –70°C. Store all other components at –20°C.∙10 tubes Stellar Competent Cells (100 µl/tube)∙10 tubes SOC Medium (1 ml/tube)∙10 µl pUC19 Vector (0.1 ng/µl)III. Additional Materials RequiredIf your RNA template is from a non-eukaryotic organism and lacks a polyadenylated tail, you can add one prior to first-strand 3’-cDNA synthesis using the following enzyme:∙Poly(A) Polymerase (Takara Bio Cat. No. 2180A)The following materials are required for In-Fusion cloning and transformation, but not supplied: ∙Ampicillin (100 mg/ml stock) or other antibiotic required for plating the In-Fusion reaction∙LB (Luria-Bertani) medium (pH 7.0)∙LB/antibiotic plates∙SOC mediumThe following material is required for the NucleoSpin Gel and PCR Clean-Up Kit, but not supplied: ∙96–100% ethanolIV. Primer DesignA. Primer SequenceGene-Specific Primers (GSPs) should:∙be 23–28 nt to ensure specific annealing∙be 50–70% GC∙have a T m≥65°C; best results are obtained if T m >70°C, which enables the use of touchdown PCR.(T m should be calculated based upon the 3’ (gene-specific) end of the primer, NOT the entire primer.) ∙not be complementary to the 3’-end of the Universal Primer MixLong primer = 5’–TAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT–3'Short primer = 5’–CTAATACGACTCACTATAGGGC–3’∙be specific to your gene of interest∙both have 15 bp overlaps with the vector at their 5’ end s (i.e., add the sequenceGATTACGCCAAGCTT to the 5’ end s of both GSPs’ sequences; see details below) The relationship of the primers used in the SMARTer RACE reactions to the template and resultingRACE products are shown in detail in Figure 3.For the complete SMARTer RACE protocol, you will need at least two GSPs: an antisense primer for the5’-RACE PCR and a sense primer for the 3’-RACE PCR. If you are performi ng only 5’- or 3’-RACE,you will only need one GSP. In our experience, longer GSPs with annealing temperatures above 70°Cgive more robust amplification in RACE, particularly from difficult samples; however, there is generallyno advantage to using primers with gene-specific sequence longer than 30 nt.Successful In-Fusion cloning requires a 15 bp overlap with the linearized vector. Given this, you will needto add the sequence GATTACGCCAAGCTT to the 5’-end of your 5’ and 3’ GSPs to facilitate In-Fusioncloning of your RACE PCR products. This specific sequence is in addition to the 22 nt gene-specificsequence described above. The provided linearized pRACE vector already contains this overlap with theUniversal Primer A Mix included for PCR, and adding this sequence to the 5’-end of your GSPs willcomplete the necessary overlap for the cloning reaction. Please note that the In-Fusion User Manualcontains only general primer recommendations that should not be used for this particular protocol.Figure 3. The relationship of gene-specific primers to the cDNA template.This diagram shows a generalized first-strandcDNA template. This RNA/DNA hybrid does not precisely represent either the 5’- or 3’-RACE-Ready cDNAs. For a detailedlook at those structures, see Appendices B & C. Note that the gene-specific primers designed here contain tails with In-Fusionhomology, and also produce overlapping RACE products. This overlap permits the use of the primers together in a control PCRreaction. Additionally, if a suitable restriction site is located within this region, it will be possible to construct the full-lengthcDNA by subcloning.B. Additional Considerations for DesignThe primers shown in Figure 3 will create overlapping 5’- and 3’-RACE products. If a suitable restriction site is located in the region of overlap, the fragments can subsequently be joined by restriction digestion and ligation to create the full-length cDNA. If no suitable restriction sites are available, you canalternately design new GSPs suitable for multi-fragment In-Fusion cloning. By designing primers thatgive a 100–200-bp overlap in the RACE products, you will also be able to use the primers together as an internal positive control for the PCR reactions. However, it is not absolutely necessary to use primers that give overlapping fragments. In the case of large and/or rare cDNAs, it may be better to use primers that are closer to the ends of the cDNA and therefore do not create overlapping fragments. The primersthemselves can overlap (i.e., be complementary).C. Location of Primer Sequences within GenesWe have had good success using the SMARTer RACE Kit to amplify 5’ and 3’ cDNA fragments thatextend up to 6.5 kb from the GSP binding sites. Nevertheless, for optimum results, we recommendchoosing your primers so that the 5’- and 3’-RACE products will range from 1–3 kb in length. If you are working with an annotated genome, we suggest using NCBI’s Primer-BLAST to aid in your design for each transcript.D. Nested PrimersWe recommend that you do not use nested PCR in your initial experiments. The UPM Primer and a GSP will usually generate a good RACE product with a low level of nonspecific background. However, nested PCR may be necessary in some cases where the level of background or nonspecific amplification in the 5’- or 3’-RACE reaction is too high with a single GSP. In nested PCR, a primary amplification isperformed with the outer primers and, if a smear is produced, an aliquot of the primary PCR product is re-amplified using the inner primers. The SMARTer RACE protocols include optional steps indicatingwhere nested primers can be used. The Universal Primer Short (provided with the kit) can be used forbo th 5’- and 3’-RACE with nested primers.Nested gene specific primers (NGSP) should be designed according to the same guidelines discussedabove. If possible, nested primers should not overlap with the outer gene-specific primers; if they mustoverlap due to limited sequence information, the 3’ end of the inner primer should have as much unique sequence as possible. Additionally, your nested primers should also contain the 15 bp overlap required for In-Fusion cloning.V. Generating RACE-Ready cDNAPLEASE READ THE ENTIRE PROTOCOL BEFORE STARTINGA. General Considerations∙We recommend using the Tricine-EDTA Buffer provided in the kit to resuspend and dilute your cDNA samples throughout the protocols in this user manual, because Tricine buffers maintain theirpH at high temperature better than Tris-based buffers. Tris-based buffers can lead to low pHconditions that degrade DNA.∙Resuspend pellets and mix reactions by gently pipetting the solution up and down or by flicking the bottom of the tube. Always spin tubes briefly prior to opening to collect the contents at the bottom ofthe tubes.∙Perform all reactions on ice unless otherwise indicated.∙Add enzymes to reaction mixtures last.∙Ethidium bromide (EtBr) is a carcinogen. Use appropriate precautions when handling and disposing of this reagent. For more information, see Molecular Cloning: A Laboratory Manual by Sambrook &Russell (2001).B. Preparation and Handling of Total and Poly A+ RNA1. General PrecautionsThe integrity and purity of your total or poly A+ RNA starting material is an important element inhigh-quality cDNA synthesis. The following precautions will help you avoid contamination anddegradation of your RNA:∙Have a separate bench and/or pipette set dedicated to RNA work, free of RNasecontamination.∙Wear gloves throughout to protect your RNA samples from nucleases.∙Use freshly deionized (e.g., MilliQ-grade) H2O directly, without treatment with DEPC(diethyl pyrocarbonate). Takara Bio also offers RNase-Free Water (Cat. No. 9012).∙Use only single-use plastic pipettes and pipette tips. Filter tips are recommended.2. RNA IsolationClontech® offers several kits for isolating total or poly A+ RNA from a variety of sources:Purified Product Starting Material Product Cat. #Total RNA Plant or fungal samples NucleoSpin RNA Plant 740949.50mRNA Total RNA derived fromcultured cells or animal tissuesMagnosphere UltraPuremRNA Purification Kit9186Many procedures are available for the isolation of poly A+ RNA (Farrell, 1993; Sambrook et al., 2001).3. RNA PurityThe purity of RNA is the key factor for successful cDNA synthesis and SMARTer RACE. Thepresence of residual organics, metal ions, salt or nucleases in your RNA sample can have a largeimpact on downstream enzymatic applications by inhibiting enzymatic activity or degrading theRNA. We strongly recommend checking the stability of your RNA to ensure that it is free ofcontaminants. Impurities such as salt or organic contaminants can be removed by repeatedethanol precipitation, subsequent washing with 80% ethanol and the complete removal of allremaining ethanol.Since RNA stability is a good indicator of RNA purity, we strongly recommend checking thestability of your RNA to ensure that it is free of contaminants.Incubate a small portion of your RNA at 37°C for 2 hours, then compare the sample to a duplicatecontrol stored at –70°C. If the sample incubated at 37°C shows a lower 28S:18S ratio than thecontrol or a significant downward shift on a formaldehyde agarose gel, the RNA may havenuclease contaminants (see Section V.C., below, for methods for assessing RNA quality).If your RNA template is from a plant or some other species with high pigment levels, pleasepay special attention to polysaccharide/pigment contamination. Polysaccharides/pigments arehard to remove and can’t be detected on the agarose gel. These glycoproteins might interfere withprimer binding sites of RNA during the first-strand cDNA synthesis leading to reduced cDNAyield.C. Assessing RNA Template Quality1. Methods for Assessing Total RNA Integrity∙Detection with the Agilent 2100 BioAnalyzer (Agilent Technologies, CA):This microfluidics-based technology, which provides an alternative to traditional gel-based analysis, requires only 2–7 ng of RNA per analysis. We recommend using RNAsamples with an RNA Integrity Number (RIN) of 7 or higher. In addition to assessingRNA quality, this automated system provides a good estimate of RNA concentration.∙If you do not have access to an Agilent 2100 BioAnalyzer, you can visualize your RNA on a denaturing formaldehyde agarose gel under UV light. The theoretical 28S:18S ratiofor eukaryotic RNA is approximately 2:1. If the 28S:18S ratio of your RNA is less than 1,your RNA template is not suitable for SMARTer RACE. When visualizing RNA usingEtBr, you need at least 0.5–1 µg of total RNA. Alternatively, SYBR® Green II or SYBRGold dyes (Molecular Probes; Eugene OR), allow you to detect as little as 1 or 2 ng ofRNA on your gel, respectively.2. Methods for Assessing mRNA IntegrityAll of the methods mentioned above can be used to assess the quality of your mRNA. However,because mRNA does not contain strong ribosomal bands, the assessment of its quality will besomewhat subjective. Typically, mRNA appears as a smear between 0.5 kb to 6 kb, with an areaof higher intensity around 1.5 and 2 kb. This size distribution may be tissue or species-specific. Ifthe average size of your mRNA is less than 1.5 kb, it could be an indication of degradation.D. Protocol: First-Strand cDNA SynthesisThe two 20 µl reactions described in the protocol convert 10 ng–1 µg of total or poly A+ RNA intoRACE-Ready first-strand cDNAWe recommend that you use poly A+ RNA whenever possible. However, if you have less than 50 µg of total RNA we do not recommend purification of poly A+ RNA because the final yield will be too small to effectively analyze the RNA quantity and quality.We strongly recommend that you perform a positive control cDNA synthesis using the includedMouse Heart Total RNA in addition to your experimental reactions.NOTE: If your RNA template is from a non-eukaryotic organism and/or lacks a polyadenylated tail,follow the protocol for 5’-first-strand cDNA synthesis with random primers in Appendix D.For 3’-first-strand cDNA synthesis, add a poly(A) tail using Poly(A) Polymerase (Takara Cat. No.2180A), and proceed with the following protocol.IMPORTANT:∙Prior to cDNA synthesis, please make sure that your RNA is intact and free of contaminants (see Section V.C. Assessing the Quality of the RNA Template).∙Do not change the volume of any of the reactions. All components have been optimized for the volumes specified.1.Prepare enough of the following Buffer Mix for all of the 5’- and 3’-RACE-Ready cDNA synthesisreactions plus 1 extra reaction to ensure sufficient volume. Mix the following reagents and spinbriefly in a microcentrifuge, then set aside at room temperature until Step 6:4.0 µl 5X First-Strand Buffer0.5 µl DTT (100 mM)1.0 µl dNTPs (20 mM)5.5 µl Total Volumebine the following reagents in separate microcentrifuge tubes:For preparation of 5’-RACE-Ready cDNA For preparation of 3’-RACE-Ready cDNA1.0–10 µl RNA* 1.0–11 µl RNA*1.0 µl 5’-CDS Primer A 1.0 µl 3’-CDS Primer A0–9 µl Sterile H2O 0–10 µl Sterile H2O11 µl Total Volume 12 µl Total Volume*For the control reactions, use 1 µl of Control Mouse Heart Total RNA (1 µg/µl).3.Mix contents and spin the tubes briefly in a microcentrifuge.4.Incubate tubes at 72°C for 3 minutes, then cool the tubes to 42°C for 2 minutes. After cooling, spinthe tubes briefly for 10 seconds at 14,000 x g to collect the contents at the bottom.NOTE: This step can be performed in a thermal cycler. While the tubes are incubating, you can prepare the Master Mix in Step 6.5.To just the 5’-RACE cDNA synthesis reaction(s), add 1 µl of the SMARTer II A Oligonucleotideper reaction.6.Prepare enough of the following Master Mix for all 5’- and 3’-RACE-Ready cDNA synthesisreactions. Mix these reagents at room temperatures in the following order:5.5 µl Buffer Mix from Step 10.5 µl RNase Inhibitor (40 U/µl)2.0 µl SMARTScribe Reverse Transcriptase (100 U)8.0 µl Total Volume7.Add 8 µl of the Master Mix from Step 6 to the denatured RNA from Step 4 (3’-RACE cDNA) andStep 5 (5’-RACE cDNA), for a total volume of 20 µl per cDNA synthesis reaction.8.Mix the contents of the tubes by gently pipetting, and spin the tubes briefly to collect the contents atthe bottom.9.Incubate the tubes at 42°C for 90 minutes in an air incubator or a hot-lid thermal cycler.NOTE: Using a water bath for this incubation may reduce the volume of the reaction mixture (due to evaporation), and therefore reduce the efficiency of first-strand cDNA synthesis.10.Heat tubes at 70°C for 10 minutes.11.Dilute the first-strand cDNA synthesis reaction product with Tricine-EDTA Buffer:∙Add 10 µl if you started with <200 ng of total RNA.*∙Add 90 µl if you started with >200 ng of total RNA.*∙Add 240 µl if you started with poly A+ RNA.*The copy number of your gene of interest should be the determining factor for diluting your sample.If you have 200 ng of total RNA but your gene of interest has low abundance, dilute with 10 µl. If you have 200 ng of total RNA and the gene of interest is highly abundant, dilute with 90 µl.12.You now have 3’- and 5’-RACE-Ready cDNA samples. Samples can be stored at –20°C for up tothree months.VI. Rapid Amplification of cDNA Ends (RACE)PLEASE READ THE ENTIRE PROTOCOL BEFORE STARTING.At this point, you have 3’- and 5’-RACE-Ready cDNA samples. The RACE reactions in this section use only a fraction of this material for each RNA of interest. There is sufficient single-stranded cDNA for PCR amplification of multiple genes.A. Things You Should Know Before Starting RACE PCR ReactionsIf you intend to use LD PCR to construct your full-length cDNA after completing 5’- and 3’-RACE, besure to set aside an aliquot of the 5’-RACE-Ready cDNA to use as a template in the PCR reaction.Please note that the efficiency of RACE PCR depends on the abundance of the mRNA of interest in yourRNA sample. Additionally, different primers will have different optimal annealing/extensiontemperatures. Refer to the Troubleshooting Guide (Appendix A) for suggestions on optimizing PCRconditions.NOTE: This is a RACE-specific protocol. It differs from the general SeqAmp protocol in many regards.B. Protocol: Rapid Amplification of cDNA Ends (RACE)This procedure describes the 5’-RACE and 3’-RACE PCR reactions that generate the 5’ and 3’ cDNAfragments. We recommend that you also perform positive control 5’- and 3’-RACE using the TFRprimers and UPM. Although the Universal Primer Short (UPM short) is provided, nested PCR isgenerally not necessary in SMARTer RACE reactions.1.Prepare enough PCR Master Mix for all of the PCR reactions plus one extra reaction to ensuresufficient volume. The same Master Mix can be used for both 5’- and 3’-RACE reactions. For each50 µl PCR reaction, mix the following reagents:15.5 µl PCR-Grade H2O25.0 µl 2X SeqAmp Buffer1.0 µl SeqAmp DNA Polymerase41.5 µl Total Volume2.Prepare PCR reactions as shown below in Table 2. Add the components to 0.5 ml PCR tubes in theorder shown and mix gently.Table 2. Setting up 5'- and 3'-RACE PCR ReactionsComponent 5’- or 3’-RACESampleUPM only(– control)GSP only(– control)5’- or 3’-RACE-Ready cDNA(experimental)2.5 µl 2.5 µl 2.5 µl 10X UPM 5 µl 5 µl —5’ or 3’ GSP (10 µM) 1 µl — 1 µl H2O — 1 µl 5 µl Master Mix (Step 1) 41.5 µl 41.5 µl 41.5 µl Total Volume 50 µl 50 µl 50 µl。
Crafter’s Choice黑樱桃莓香油(7820)说明书
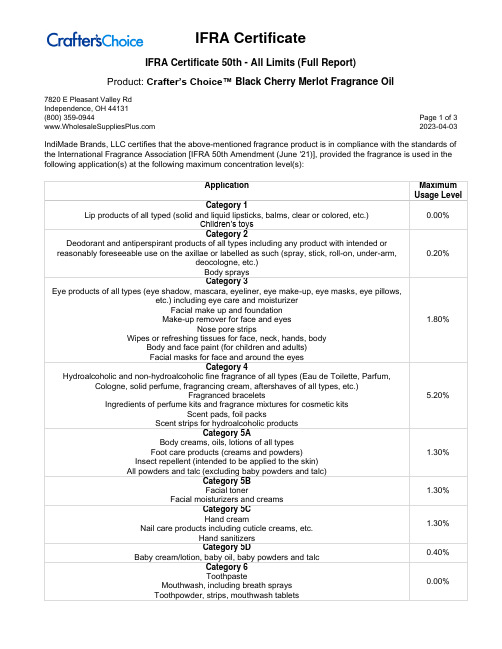
Product: Crafter’s Choice™ Black Cherry Merlot Fragrance Oil7820 E Pleasant Valley RdIndependence, OH 44131(800) 359-0944 Page 1 of 3 2023-04-03 IndiMade Brands, LLC certifies that the above-mentioned fragrance product is in compliance with the standards of the International Fragrance Association [IFRA 50th Amendment (June '21)], provided the fragrance is used in the following application(s) at the following maximum concentration level(s):Product: Crafter’s Choice™ Black Cherry Merlot Fragrance Oil7820 E Pleasant Valley RdIndependence, OH 44131(800) 359-0944 Page 2 of 3 2023-04-03Product: Crafter’s Choice™ Black Cherry Merlot Fragrance Oil7820 E Pleasant Valley RdIndependence, OH 44131(800) 359-0944 Page 3 of 3 2023-04-03For all other applications, or use at higher concentration levels, a new evaluation will be required.The IFRA standards regarding use restrictions are based on safety assessments by the Research Institute for Fragrance Materials (RIFM) Expert Panel (REXPAN) and are enforced by the IFRA Scientific Committee. Evaluation of individual fragrance materials is made according to the safety standards contained in the relevant section of the IFRA Code of Practice.It is the ultimate responsibility of the customer to ensure the safety of the final product containing this fragrance, by further testing, if necessary.The above-mentioned fragrance product contains ingredients which are NOT considered GRAS, Generally Regarded as Safe as a Flavor Ingredient.。
ViralSEQ Lentivirus Physical Titer Kit说明书

ViralSEQ™ Lentivirus Physical Titer KitCatalog Numbers A52597 and A52598Pub. No. MAN0026127 Rev. A.0Note: For safety and biohazard guidelines, see the “Safety” appendix in the ViralSEQ™ Lentivirus Titer Kits User Guide(Pub. No. MAN0026126). Read the Safety Data Sheets (SDSs) and follow the handling instructions. Wear appropriate protective eyewear, clothing, and gloves.Product descriptionThe Applied Biosystems™ ViralSEQ™ Lentivirus Physical Titer Kit is a TaqMan™-based RT-qPCR kit. The kit measures viral count based on highly sensitive viral RNA quantitation from the supernatants of cell-based, bioproduction systems. Viral titers of 104 to 1011 viral particles (VP) per mL can be quantitated using a standard curve generated from the synthetic RNA control included with the kit. Lentivirus quantitation by RT-qPCR is accurate, sensitive, and reproducible.The ViralSEQ™ Lentivirus Physical Titer Kit is compatible with the PrepSEQ™ Nucleic Acid Sample Preparation Kit (Cat. A50485), which offers both a manual and automated sample preparation workflow. For real‑time PCR, the ViralSEQ™ Lentivirus Physical Titer Kit has been validated on the Applied Biosystems™ 7500 Fast Real-Time PCR System and the Applied Biosystems™ QuantStudio™ 5 Real‑Time PCR System. Data analysis is streamlined using AccuSEQ™ Real‑Time PCR Software that provides accurate quantitation and security, audit, and e-signature capabilities to help enable 21 CFR Pt 11 compliance.For more information about reagent use, see the ViralSEQ™ Lentivirus Titer Kits User Guide (Pub. No. MAN0026126).Treat samples with DNase I, RNase‑free (1 U/µL)DNase I, RNase‑free (1 U/µL) treatment is used to digest double-stranded DNA.Thaw all reagents on ice. Invert the DNase I, RNase‑free (1 U/µL) several times to mix, then centrifuge briefly. All other reagents should be vortexed, then centrifuged briefly before use.1.Set up the DNase I, RNase‑free (1 U/µL) reactions in a MicroAmp™ Optical 96-Well Reaction Plate (0.2 mL).[1]Mix gently by pipetting 3-5 times when adding.2.Mix the reactions by gently pipetting up and down 5 times, then seal the reaction plate with MicroAmp™ Clear Adhesive Film.3.Centrifuge the plate at 1,000 x g for 2 minutes.4.Load the reactions onto the VeritiPro™ 96-well Thermal Cycler, then start the DNase I treatment.Set cover temperature: 105℃Set reaction volume: 18 µL[1]Do not hold for more than 5 minutes. Proceed immediately to DNase I inactivation.5.Centrifuge the plate at 1,000 x g for 2 minutes.CAUTION! The plate is in contact with the heated lid. Remove carefully.6.Gently remove the MicroAmp™ Clear Adhesive Film, then discard.IMPORTANT! Do not touch wells when removing the MicroAmp™ Clear Adhesive Film. Contamination can lead to inaccurate results.7.Add 2 µL of 50mM EDTA to each reaction well. Mix by gently pipetting 5 times with a P10/P20 pipettor set to 10 µL.8.Seal the reaction plate with MicroAmp™ Clear Adhesive Film, then centrifuge the plate at 1,000 x g for 2 minutes.9.Load the reactions onto the VeritiPro™ 96-well Thermal Cycler, then start the DNase I inactivation.Set cover temperature: 105℃Set reaction volume: 20 µL[1]Do not hold for more than 5 minutes.10.Centrifuge the plate at 1,000 x g for 2 minutes.CAUTION! The plate is in contact with the heated lid. Remove carefully.IMPORTANT! Do not vortex.Place the plate on ice until use.Prepare the serial dilutionsThaw the Physical Titer RNA Control (2 ✕1010 copies/µL) on ice. Vortex at medium speed for 5 seconds, briefly centrifuge, then place on ice until use.bel nonstick 1.5‑mL microfuge tubes: NTC, SD1, SD2, SD3, SD4, SD5, and SD6 [used for limit of detection (LOD)].2.Add 35 µL of RNA Dilution Buffer (RDB) to the NTC (no template control) tube. Place the tube on ice.3.Perform the serial dilutions.When dispensing RNA, pipette up and down gently. After each transfer, vortex for 7 seconds, then centrifuge briefly.Table 1 Standard curve dilutions (ViralSEQ™ Lentivirus Physical Titer Kit)Store the standard curve dilution tubes at 4°C or on ice. Use the dilutions within 6 hours for RT-qPCR.Prepare the kit reagents and premix solutionThaw all kit reagents on ice. Vortex the reagents for 5 seconds, briefly centrifuge, then place the reagents on ice until use.bel a microcentrifuge tube for the Premix Solution.2.Prepare the Premix Solution according to the following tables.IMPORTANT! Use a separate pipette tip for each component.Table 2 Premix Solution[1]Includes 10% excess to compensate for pipetting loss.3.Vortex the Premix Solution for 10 seconds to mix, then briefly centrifuge.Store the Premix Solution at 4°C or on ice until use.Prepare the PCR reactionsPlace the plate containing DNase I-treated samples on a MicroAmp™ 96-Well Base, then gently remove the MicroAmp™ Clear Adhesive Film. Gently pipette up and down 3 times to mix the samples.1.Dispense the following into the appropriate wells of a MicroAmp™ Fast Optical 96-Well Reaction Plate, 0.1 mL, gently pipetting at thebottom of the well.Figure 1 Recommended plate layout2.Seal the plate with MicroAmp ™Optical Adhesive Film.3.Vortex the reaction plate for 10 seconds, then centrifuge at 1,000 x g for 2 minutes.Note: Ensure there are no bubbles in the reaction wells. If present, tap the well gently to remove bubbles, then re-centrifuge.Proceed immediately to “Start the run (QuantStudio ™ 5 Real ‑Time PCR Instrument)”.Create a ViralSEQ ™ templateCreate a new template in the (Home) screen of the AccuSEQ ™Real ‑Time PCR Software v3.1.1.ClickCreate New on the home screen.Create New ExperimentpaneFactory default/Admin Defined Templates —List of existing default or Admin Defined templates. These templates can beused as templates for new experiments.My Templates —List of templates available to the user that is signed in. These templates can be used as templates for newexperiments.Create New —Used to create an experiment or template with nopre-existing settings.Create My Template —Used to create a new template (stored locally in My Templates).Create Admin Defined Template —Used to create a new template (Administrator only).2.Select Create My Template or Create Admin Defined Template .3.Edit the Experiment Properties as required.a.In the Template Name field, modify the template name. For example, LV Titer template.b.(Optional ) Enter information in the Comments field.c.In the Setup tab, select:•Experiment Type —Quantitation-Standard Curve •Chemistry —TaqMan ® Reagents•Ramp Speed —Standard-2hrs •Block Type —96-Well 0.1mL Blockd.(Optional ) Select Is Locked to lock the template. If locked, users are unable to edit the template.4.ClickAnalysis settings to change the default C t Settings and Flag Settings.a.In the C t Settings tab, click Edit Default Settings .b.Deselect Automatic Threshold , then enter 0.200.c.Ensure that Automatic Baseline is selected.d.Click Save Changes .e.Deselect Default Settings , then click Applyto save any changes before closing the window.2C tSettingsFlag SettingsEdit Default SettingsbuttonDefault Settingscheckbox Apply buttonf.In the Flag Settings tab, deselect the following flags.•CQCONF —Low Cq confidence •EXPFAIL —Exponential algorithm failed •NOAMP —No amplification •NOSIGNAL —No signal in wellNote: Use the scrollbar on the right to scroll down the list of flags.g.Click Apply to save any changes before closing the window.5.Click Next .Template name cannot be changed after this step.The qPCR Method screen is displayed.Edit the run method and optical filter selectionThis section provides general procedures to edit the run method and optical filter selection in the qPCR Method. To edit the default run method, see the AccuSEQ™ Real‑Time PCR Software v3.1 User Guide (Pub. No. 100094287).1.Set the reaction volume to 25 µL.2.Edit Step 1 of the Hold Stage to 45℃ for 30 minutes.3.Set Step 2 of the Hold Stage to 95℃ for 10 minutes.4.Set Step 1 of the PCR Stage to 95℃ for 15 seconds.5.Edit Step 2 of the PCR Stage to 60℃ for 45 seconds.6.Set the cycle number to 40.7.Ensure that Data Collection occurs after Step 2.123Figure 2 Lentivirus Physical Titer Run MethodReaction volume- set to 25µLStageCycle number- set to 40 cycles8.(Optional) Click (Optical Filter Settings) to view the default filter settings.•The default optical filter selection is suitable for the ViralSEQ™ Lentivirus Physical Titer Kit.•The ViralSEQ™ Lentivirus Physical Titer Kit requires the QuantStudio™ 5 System to be calibrated for FAM™, VIC™, and ROX™.•For more information about system dyes and their calibration and optical filter selection, see QuantStudio™ 3 and 5 Real‑Time PCR Systems Installation, Use, and Maintenance Guide (Pub. No. MAN0010407).9.Click Next.Assign plate and well attributesNote: This section provides general procedures to set up the plate.For specific instructions for each assay type, see the corresponding chapter in this guide. Do not change Targets for default assay templates.TargetsSamplesPlate setup toolbarSelect Item to highlight (Sample, Target, or Task).Select Item. For example, Sample 1. Sample 1 replicates arehighlighted.Define& setup Standard(View Legend)(Print Preview)View (Grid View or Table View)1.In Plate Setup screen, click or click‑drag to select plate wells in the (Grid View) of the plate.2.Assign the well attributes for the selected wells. Each well should have a Sample Name under Samples, as well as the appropriateTargets under Targets. Reporters should be FAM™ dye for Lentivirus Physical Titer, and VIC™ dye for internal positive control (IPC).a.To add new Samples or Targets, click Add in the appropriate column on the left of the screen, then edit the new Name andother properties as required. The new sample or target is then selectable within the wells of the plate.b.For each sample (e.g. DNase-treated lentivirus sample, standard curve dilution, or NTC), two targets should be included.•Select the FAM™ dye reporter for Lentivirus Physical Titer detection.•Select the VIC™ dye for IPC detection.c.Select NFQ-MGB as the quencher for both targets.d.For standard curve dilution samples (SD1 to SD5), the Task for Lentivirus Physical Titer target should be indicated as “S” forStandard, with the appropriate copy number written under Quantity. For instance, the quantity for SD1 is 1E9 copies. Change the Task by clicking on the field and using the drop-down menu. Copy numbers can be indicated using scientific notation (e.g.“1E9”) and the program will convert it to numerical format.e.For DNase-treated samples and SD6, set the Task for Lentivirus Physical Titer target to U for Unknown.f.For NTC wells, set the Task for Lentivirus Physical Titer target to N for NTC.g.For IPC wells, set the Task for Lentivirus Physical Titer target to U for Unknown.h.To change sample names, click the name in the Name column, then type the new name. To change Reporters and Quenchers,click the dye, then select from the dropdown list.When a Sample or Targetname are edited, two entries are added to the Audit trail (one for Delete, and another for Create).23AddbuttonCheckbox—Select Targets and Samples to go in the selectedwell.Textbox—Click the name to edit.Scrollbar—Use to scroll to additional properties.•Use the plate setup toolbar (above the plate) to make edits to the plate.–Click View to show/hide the Sample Name, Sample Color, or Target from the view.•To add consecutive samples (with the same Target ), select a well, then click ‑drag the dark blue box to the right.3.(Optional ) Double ‑click a well to enter comments for the selected well.4.Select ROX ™dye from the Passive Reference drop-down list (bottom left of screen).5.Click Save to save the template.This template can then be used to create experiments.Start the run (QuantStudio ™ 5 Real ‑Time PCR Instrument)Ensure that the plate is loaded in the QuantStudio ™5 Real ‑Time PCR Instrument.™A message stating Run has been started successfully is displayed when the run has started.Review the resultsAfter the qPCR run is finished, use the following general procedure to analyze the results. For more detailed instructions see theAccuSEQ ™Real ‑Time PCR Software v3.1 User Guide (Pub. No. 100094287).1.In the AccuSEQ ™Real ‑Time PCR Software, open your experiment, then navigate to the Result tab.ResulttabAnalysis SettingsPlot horizontal scrollbar Analyze button2.In the Result Analysis tab, select individual targets, then review the Amplification Curve plots for amplification profiles in thecontrols, samples, and the standard curve. Ensure that threshold is set to 0.200 with an automatic baseline.3.In the Result Analysis tab, review the QC Summary for any flags in wells.4.In the Result Analysis tab, review the Standard Curve plot. Verify the values for the Slope, Y ‑intercept, R 2, and Efficiency are withinacceptable limits.Note: The Standard Curve efficiency should be between 90-110% and the R 2>0.99. If these criteria are not met, up to two points,not in the same triplicate, can be removed from the standard curve data, and the analysis repeated.5.In Table View, ensure that C t values are within the standard curve range.•Samples with C t values that exceed the upper limit of quantitation (109 copies) of the standard curve should be diluted and re-run.•Samples with C t values that exceed the lower limit of detection (LoD of 10 copies) and IPC shows no signs of PCR inhibition,suggests the absence of lentivirus.6.(Optional ) Outliers can be excluded from the results. To exclude, select the well, then click Omit/Include , then reanalyze by clickingAnalyze .7.(Optional ) Select File 4Print Report to generate a hard copy of the experiment, or click Print Preview to view and save the report asa PDF or HTML file.8.Export the results.a.Navigate to the Report tab.b.Check all boxes under Contents .c.Select Export Data in One File .d.Select the XLS format, then click Export .Calculate the titer (VP/mL)1.Download the Physical Titer Calculation Tool .a.Go to .b.Search for the ViralSEQ ™Lentivirus Physical Titer Kit.c.Download the tool from the Documents section.2.Open the tool, then follow the instructions in the tool to calculate the titer.Calculate lentivirus titers from qPCR dataTo determine the number of lentivirus RNA copies per mL in the original sample, the copy numbers obtained from the qPCR must be multiplied by the dilution factor of the sample during extraction and DNase I treatment. Since there are 2 copies of RNA/target per lentivirus particle, the number of viral particles per mL (VP/mL) is 0.5x the number of lentivirus RNA copies.Viral particles per mL =qPCR copies x sample dilution factor x 0.5Volume of sample used (mL)For help in determining the qPCR copy numbers, see the QuantStudio ™Design and Analysis Desktop Software User Guide (Pub. No. MAN0010408).For example, if the following parameters were used,•10 µL of lentivirus culture was extracted with the KingFisher ™Flex Purification System with 96 Deep-Well Head and eluted in 200 µL •10 µL of this eluate (20x dilution) was treated with DNase I, RNase ‑free (1 U/µL) in a total volume of 20 µL.• 5 µL of the DNase-treated sample (4x dilution) was used for the qPCR reaction.Lentivirus sample10 μL Sample Extraction Elute: 200 μL DNaseI Treat Total: 20 μL reaction RT-qPCRTotal: 25 μL reactionUse 10 μL (1:20 dilution)Use 5 μL (1:4 dilution)then, the calculation would be:Viral particles per mL =qPCR copies x (20x4) x 0.50.01 (mL)Note: qPCR can only determine the number of physical particles in a virus culture. To determine the numbers of infectious units,cell-based transduction experiments must be carried out. The titers of physical particles are often higher than infectious titers by 10-1000fold, depending on the purity of the lentivirus preparation and the levels of infectious particles within the culture.Limited product warrantyLife Technologies Corporation and/or its affiliate(s) warrant their products as set forth in the Life Technologies' General Terms and Conditions of Sale at /us/en/home/global/terms-and-conditions.html . If you have any questions, please contact Life Technologies at /support .Life Technologies Ltd | 7 Kingsland Grange | Woolston, Warrington WA1 4SR | United KingdomThe information in this guide is subject to change without notice.DISCLAIMER : TO THE EXTENT ALLOWED BY LAW, THERMO FISHER SCIENTIFIC INC. AND/OR ITS AFFILIATE(S) WILL NOT BE LIABLE FOR SPECIAL, INCIDENTAL, INDIRECT,PUNITIVE, MULTIPLE, OR CONSEQUENTIAL DAMAGES IN CONNECTION WITH OR ARISING FROM THIS DOCUMENT, INCLUDING YOUR USE OF IT.Important Licensing Information : These products may be covered by one or more Limited Use Label Licenses. By use of these products, you accept the terms and conditions of all applicable Limited Use Label Licenses.©2022 Thermo Fisher Scientific Inc. All rights reserved. All trademarks are the property of Thermo Fisher Scientific and its subsidiaries unless otherwise specified. Vortex-Genie is a trademark of Scientific Industries. Windows and Excel are trademarks of Microsoft Corporation. TaqMan is a registered trademark of Roche Molecular Systems, Inc., used under permission and license./support | /askaquestion 23 August 2022。
2016年出口卷心菜用药

★采收前12天,尽量避免使用农药。★以上所列农药均符合有关标准,但在实际应 用过程中,以客户需要为准,仅供客户选择。★病虫防治时间应掌握在害虫1-2龄 期、病害发生前或初期进行。具体以公司植保技术员视田间实际情况而定。
漳州市大地农业科技有限公司 技术部0596-8321ቤተ መጻሕፍቲ ባይዱ32、13695939786 2016年8月4日
日本 呋虫胺(Dinotefuran) 20%呋虫胺 2500 2
施定康 上海生农 吡丙醚•吡虫啉 吡丙醚•吡虫啉 1500 0.7 暂定基准值: 0.5 七、防治棉铃虫药剂; 商品名: 全攻 厂家: 安徽华星 成份(英文): 杀螟丹(Carbamic acid) 包装照片 有效成分(含量): 98%杀螟丹 稀释倍数: 1000 暂定基准值: 一律 商品名: 厂家: 成份(英文): 包装照片 有效成分(含量): 稀释倍数: 暂定基准值: 商品名: 厂家: 成份(英文): 包装照片 有效成分(含量): 稀释倍数: 暂定基准值: 除虫脲 上海生农 除虫脲(DIFLUBENZURON) 20%除虫脲 1000 1 凯恩 美国杜邦 茚虫威(Indoxacarb) 15%茚虫威 2500 1
包装照片 稀释倍数: 1500 暂定基准值: 0.7 商品名: 厂家: 成份(英文): 包装照片 有效成分(含量): 稀释倍数: 暂定基准值: 商品名: 厂家: 成份(英文): 包装照片 有效成分(含量): 稀释倍数: 暂定基准值: 商品名: 厂家: 成份(英文): 包装照片 有效成分(含量): 稀释倍数: 暂定基准值: 商品名: 厂家: 成份(英文): 包装照片 有效成分(含量): 稀释倍数: 暂定基准值: 商品名: 厂家: 成份(英文): 包装照片 有效成分(含量): 稀释倍数: 暂定基准值: 六、防治粉虱、蚜虫药剂; 商品名: 厂家: 成份(英文): 包装照片 有效成分(含量): 稀释倍数: 暂定基准值: 商品名: 厂家: 成份(英文): 包装照片 有效成分(含量): 稀释倍数: 暂定基准值: 稳敌 上海生农
FS FORTH-SYSTEME GmbH Mod520C_2 产品说明书

Mod520C_2P.O. Box 11 03l D-79200 Breisach, Germany Kueferstrasse 8l D-79206 Breisach, Germany (+49 (7667) 908-0 l Fax +49 (7667) 908-200 l e-mail:****************Mod520C_2© Copyright 2004:FS FORTH-SYSTEME GmbHPostfach 1103, D-79200 Breisach a. Rh., GermanyRelease of Document:May 27, 2004Filename:Mod520C_2.docAuthor:Hans-Peter SchneiderAll rights reserved. No part of this document may be copied or reproduced in any form or by any means without the prior written consent of FS FORTH-SYSTEME GmbH.2Mod520C_2 Table of Contents1.Introduction (4)2.Features (5)3.Functional Description (6)3.1.1.CPU AMD ÉlanSC520 (6)3.1.2.SDRAM stage (6)3.1.3.ROM stage (6)3.1.4.SRAM stage (6)3.1.5.32 I/O Ports (7)3.1.6.256 Byte EEPROM for BIOS and Applications on PIO30,31 (7)3.1.7.On-board Power Supply (7)3.1.8.Voltage Supervision, RESET Generation (7)3.1.9.Serial Ports (7)3.1.10.Fast Ethernet Controller Stage (8)3.1.11.Dual CAN Controller Stage (8)3.1.12.GP Bus used for ISA Bus (9)4.Connectors Of MOD520C (10)4.1.System Connector X2 (10)4.2.System Connector X4 (12)5.Application Notes (14)5.1.Power Supply (14)5.2.Important Signals (14)5.2.13.PCICLKRTN PCICLK PCICLKETHER (14)5.2.14.ISA-Bus Signals (14)5.2.15.CAN-Interrupt IRQ11 (15)6.Members of the MOD520C family (16)3Mod520C_21. IntroductionThe module MOD520C with its integrated and optional peripherals, based on the 32 Bit AMD ÉlanSC520 microcontroller, is designed for medium to high performance applications in telecommunication, data communication and information appliances on the general embedded market. It can easily be designed in customized boards.The AMD ÉlanSC520 microcontroller combines a low voltage 586 CPU running on 133 MHz, including FPU (Floating Point Unit) with a set of integrated peripherals: 32 Bit PCI controller, SDRAM controller for up to 256 MByte, GP (General Purpose) bus with programmable timing and ROM/Flash controller. Enhanced PC compatible peripherals like DMA controller, two UARTs and battery buffered RTC and CMOS, watchdog and software timers make this device a very fast system for both real time and PC/AT compatible applications. Insyde Software’s Mobile BIOS is available which offers serial and parallel remote features (video, keyboard, floppy). Furthermore FS FORTH-SYSTEME has adapted Datalight Sockets (TCP/IP Stack), ROM-DOS and the Flash File System FlashFX to this environment.The MOD520C offers the software engineer the possibility to reduce the time-to-market phase even more. FS FORTH-SYSTEME added several features on-board as SDRAM (up to 64 MByte), PCI Fast Ethernet controller to facilitate networking and remote control. A Strata-FLASH for booting and data is included on board. Two CAN Ports are additionally available for communication. 512 Kbyte SRAM is available for battery buffered data. The enhanced JTAG port for low-cost debugging is supported. This allows instruction tracing during execution. FS FORTH-SYSTEME has adapted Windows CE 3.0 to this platform and offers drivers and support. With Ethernet debugging the software designer has powerful means for fast debugging his applications.Due to the 16 MByte FLASH it is possible to build larger, complete systems on this module like Linux, QNX or VxWorks.4Mod520C_252. Features• 16 MByte STRATA-FLASH or 2 MByte AMD FLASH • 64 MByte or 16 MByte SDRAM • 512 KByte battery buffered SRAM• PCI Ethernet controller with EEPROM. Rx and Tx signals are providedon the System Connectors • Two CAN-Buses.• Enhanced JTAG port available on System Connector.• GP-Bus signals available on System Connector • PCI-Bus signals available on System Connector• BIOS for ÉlanSC520 by Insyde Software Inc. Including serial or parallelremote features (Video, Keyboard, Floppy).Mod520C_23. Functional Description3.1.1. CPU AMD ÉlanSC520The CPU AMD ÉlanSC520 is powered with 2.5V (core and analog path) and 3.3V (all other voltages) except VRTC, which is powered with about 3V either from battery or from on-board 3.3V. This voltage is limited to 3.3V, the other 3.3V power planes have a limit of 3.6V.The CPU is clocked with a 32.768 kHz quartz. An internal PLL derives from this frequency the RTC clock and DRAM refresh clock and the clocks for PC/AT compatible PIT (1.1882 MHz) and UARTs (18.432 MHz). All other stages (CPU, PCI, GP bus, GP DMA, ROM, SSI, timers) are fed from the second clock generator driven by a 33.33 MHz clock oscillator. SDRAM is clocked with 66.66 MHz.3.1.2. SDRAM stageThe SDRAM (up to 128 MByte on-board) has its own DRAM bus containing memory addresses MA0..12, memory data MD0..31 and control signals for up to four banks. Due to small load no buffering of clocks and signals is necessary.3.1.3. ROM stageROM or FLASH are driven by the general purpose address bus GPA0..25. It has three programmable chip selects with each up to 64 MByte range. The ROM Data bus is either the 32 bit general purpose bus GPD0..31 or the memory data bus MD0..31. Configuration pins decide, which bus at boot time is used. The bus size is selectable with 8, 16 or 32 bit. The MOD520C has a FLASH IC for up to 16 MByte 16 bit ROM or FLASH selected by BOOTCS# connected to MD0..15. 3.1.4. SRAM stageThe SRAM (512 KByte on-board) is buffered by VBAT. The Memory Location is defined in the System BIOS. ROMCS1# is used to access the SRAM.6Mod520C_23.1.5. 32 I/O PortsThe ÉlanSC520 CPU has 32 I/O ports. They have alternate functions. Most of them are control signals for GP bus (PIO0..26) used as ISA-bus. PIO27 (GPCS0#) is used as a programmable external chip select and PIO28,29 are not connected. PIO30,31 are used to drive a serial parameter EEPROM on-board. 3.1.6. 256 Byte EEPROM for BIOS and Applications on PIO30,31An on-board serial EEPROM with 256 byte and I2C bus is controlled by PIO30 (I2CDAT) and PIO31 (I2CCLK). 128 byte are used for non-volatile BIOS defaults, the remaining range may contain application specific data and parameters. The BIOS contains calls to read and write to this memory (see BIOS documentation).3.1.7. On-board Power SupplyThe 2.5V on-board voltage is generated from +5V.An external battery may be connected to the signal VBATIN. Battery status is controlled by BBATSEN, which sets a power fail bit in a status register for RTC, if BBATSEN is low at power-up.3.1.8. Voltage Supervision, RESET GenerationThree voltages are used on board: +2.5V, +3.3V and +5V. U2 controls +5V and U10 controls +3.3V. LBOUT or PWRGOOD will become low, if these voltages are out of tolerance. An external SRESET# is wired or-ed to U10. It can also be activated from extended JTAG signal SRESET# via X1. The wired OR of 1RESET# and 2RESET# control U10. Its output PWRGOOD is low (not active), if either the signals described above from U10 are low or +5V is out of tolerance. Typical length of PWRGOOD low is longer than 1 sec (minimum 790 msec).3.1.9. Serial PortsThe AMD ÉlanSC520 CPU has two internal asynchronous ports and one synchronous serial port. Both of this ports are available at the System Connectors.7Mod520C_23.1.10. Fast Ethernet Controller StageConnected to the PCI bus device 0 (REQ/GNT0#) of the Élan SC520 CPU, a Fast Ethernet Controller U7 supports 10/100Mbps transfer depending on driver software. X3 is a JST B5B-PH-SM3 5 pin connector. Parameters as physical address and power down modes are stored in a 64X16 bit Serial EEPROM controlled by U7. 2 status LEDs LE1, LE2 show the state of the Ethernet connection. For a more detailed hardware and software description see Intel 82559ER manual.3.1.11. Dual CAN Controller StageThe CAN Controller is selected via ROMCS1# and the Memory location is selectable in the BIOS Setup Screen. The CAN Interrupt provided by 82C900 is inverted by the onboard Lattice CPLD. Since the Interrupt asserted by the 82C900 is only a low active pulse of 0.2µs the CPLD holds the interrupt active until the software accesses the Memory at the location CAN-Base+1xxh. The BIOS routes this interrupt to IRQ11.8Mod520C_2 3.1.12. GP Bus used for ISA BusThe AMD ÉlanSC520 CPU contains an 8/16bit General Purpose Bus (GP bus) with 26 address lines (GPA0..25), 16 data lines (GPD0..15) and different control lines using PIO ports in their alternate GP bus function. Its timing is programmable for speeds up to 33MHz. This bus is to emulate a 16 bit ISA bus (PC/104) running with 8 MHz. ISA bus signal are connected without buffers directly to the lines of the CPU due to the 5V tolerance of the 3.3V signals.8 bit signals SMEMRD# and SMEMWR# (active only at addresses beyond 1 MByte) are not supported (GPMEM_RD# and GPMEM_WR# used).The AMD ÉlanSC520 CPU has only 4 DMA channels on GP bus. DMA channel 2 is used for Super-I/O (U2) on EVAMOD520. All four channels are connected to edge connector X2.Not supported ISA bus signals 0WS#, IOCHK#, IRQ15, REFRESH#, 8MHz and 14.318 MHz clocks, MASTER#.9Mod520C_24. Connectors Of MOD520C4.1. System Connector X2Pin Function I/O Pin Function I/O1+3.3V power2GND power 3+3.3V power4GND power 5GPD0I/O6TDP O7GPD1I/O8TDN O9GPD2I/O10not connected11GPD3I/O12RDP I13GPD4I/O14RDN I15GPD5I/O16not connected17GPD6I/O18DRQ0I19GPD7I/O20DRQ2I21GPD8I/O22DRQ5I23GPD9I/O24DRQ7I25GPD10I/O26DACK0#O27GPD11I/O28DACK2#O29GPD12I/O30DACK5#O31GPD13I/O32DACK7#O33GPD14I/O34GND power 35GPD15I/O36GPRESET O37GND power38GPIORD#O39GPA0O40GPIOWR#O41GPA1O42GPALE O43GPA2O44GPBHE#O45GPA3O46GPRDY I47GPA4O48GPAEN O49GPA5O50GPTC O51GPA6O52GPDBUFOE#O53GPA7O54GPIO_CS16#O55GPA8O56GPMEM_CS16#O57GPA9O58GPCS0#O59GPA10O60GPMEM_RD#O61GPA11O62GPMEM_WR#O63GPA12O64GND power 10Pin Function I/O Pin Function I/O65GPA13O66EXTRES#I67GPA14O68PWRGOOD O69BUFA15O70CLKTEST71BUFA16O72PRG_RESET73BUFA17O74GND power75BUFA18O76GPCS1#O77BUFA19O78GPCS2#O79BUFA20O80GPCS3#O81BUFA21O82GPCS4#O83BUFA22O84GPCS5#O85BUFA23O86GPCS6#O87BUFA24O88GPCS7#O89BUFA25O90VBATIN power91GND power92GND power93IRQ1I94RSTLD0I95IRQ3I96RSTLD1I97IRQ4I98RSTLD2I99IRQ5I100RSTLD3I101IRQ6I102RSTLD4I103IRQ7I104RSTLD5I105IRQ9I106RSTLD6I107IRQ10I108RSTLD7I109CANINT I110DBGDIS I111IRQ12I112INSTRC I113IRQ14I114DBGENTR I115SPEAKER O116not connected117+5V power118GND power119+5V power120GND power114.2. System Connector X4Pin Function I/O Pin Function I/O1PCICLKRTN I2GND power 3PCICLK O4PCICLKETHER I5AD0I/O6CBE0#I/O7AD1I/O8CBE1#I/O9AD2I/O10CBE2#I/O11AD3I/O12CBE3#I/O13AD4I/O14not connected15AD5I/O16not connected17AD6I/O18not connected19AD7I/O20not connected21AD8I/O22GND power 23AD9I/O24RXD1I25AD10I/O26TXD1O27AD11I/O28CTS1#I29AD12I/O30DCD1#I31AD13I/O32DSR1#I33AD14I/O34RIN1#I35AD15I/O36DTR1#O37AD16I/O38RTS1#O39AD17I/O40RXD2I41AD18I/O42TXD2O43AD19I/O44CTS2#I45AD20I/O46DCD2#I47AD21I/O48DSR2#I49AD22I/O50RIN2#I51AD23I/O52DTR2#O53AD24I/O54RTS2#O55AD25I/O56CANH1/TXD157AD26I/O58CANL1/RXD159AD27I/O60CANH2/TXD261AD28I/O62CANL2/RXD263AD29I/O64GND power 12Pin Function I/O Pin Function I/O65AD30I/O66SRESET#I67AD31I/O68GPRESET#O69GND power70TCK O71INTA#I72TMS73INTB#I74TDI I75INTC#I76TDO O77INTD#I78TRST#I79REQ0#I80CMDACK81REQ1#I82BR/TC83REQ2#I84GND Power85REQ3#I86STOP/TX87REQ4#I88TRIG/TRACE89GNT0#O90not connected91GNT1#O92ACTLED#O93GNT2#O94LILED#O95GNT3#O96SPEEDLED#O97GNT4#O98not connected99GND power100GND power101PAR I/O102SSI_CLK O103PERR#I/O104SSI_DO O105SERR#I106SSI_DI I107FRAME#I/O108not connected109TRDY#I/O110ISP_TDI I111IRDY#I/O112ISP_TDO O113STOP#I/O114ISP_TMS115DEVSEL#I/O116ISP_TCK I117RST#O118BSCAN#119GND power120GND power135. Application Notes5.1. Power SupplyThe MOD520C needs +3.3V and 5V power supply.3.3V worst case supply current is 1130 mA5V worst case supply current is 550 mABe sure to design your power supply for this current including large load transients.5.2. Important Signals5.2.13. PCICLKRTN PCICLK PCICLKETHERPCICLK is the clock source for the PCI-Bus. PCICLKETHER is the clock input for the Ethernet Controller. PCICLKRTN is the clock input of the AMD Élan Sc520. This pin is used to synchronize the CPU with the external PCI-Bus. Therefore it is important that all clock traces have the same length to provide each PCI-Target with the clock at the same time. Trace length of each of this clocks is 68mm on the module. If you do not plan to connect an additional PCI-Target on your board you just add a serial resistor 33R between PCICLK and PCICLKRTN and one between PCICLK and PCICLKETHER.5.2.14. ISA-Bus SignalsIf you use the ISA-Bus Signals you have to add some resistors to the following Signals:GPD0 – GPD15 4k7 pull upGPRDY1k pull downIRQ´s10k pull upDRQ´s10k pull down145.2.15. CAN-Interrupt IRQ11IRQ11 is used for the CAN-Controller 82C900 and is not sharable. Since the interrupt provided by 82C900 is a low active pulse with a length of 0.2µs the CPLD inverts this signal and holds it until the software acknowledges the interrupt. To acknowledge this interrupt the software has to access a memory location with the offset 1xxh to the CAN-Base. This memory access is just an access of the system memory, not an access of the CAN-Controller.156. Members of the MOD520C familyNumbe r Variant Flash SDRAM SRAM CAN CAN-DriverTemp.320MOD520C_0_V018M*16, Strata(=16 Mbyte)1*TM014452*4M*16(=16 Mbyte)2*TM01519512k*8Yes Yes0..70°321MOD520C_0_V021M*16, AMD(=2 Mbyte)1*TM015202*4M*16(=16 Mbyte)2*TM01519512k*8Yes Yes0..70°322MOD520C_0_V038M*16, Strata(=16 Mbyte)1*TM014452*16M*16(=64 Mbyte)2*TM01249512k*8Yes Yes0..70°334MOD520C_1_V01MOD520C_2_V018M*16, Strata(=16 Mbyte)1*TM014452*4M*16(=16 Mbyte)2*TM01519512k*8TM0Yes no0..70°335MOD520C_1_V02MOD520C_2_V021M*16, AMD(=2 Mbyte)1*TM015202*4M*16(=16 Mbyte)2*TM01519512k*8TM0Yes no0..70°336MOD520C_1_V03MOD520C_2_V038M*16, Strata(=16 Mbyte)1*TM014452*16M*16(=64 Mbyte)2*TM01249512k*8TM0Yes no0..70°184MOD520C_1_V048M*16, Strata(=16 Mbyte)1*TM014452*4M*16(=16 Mbyte)2*TM01519No No No0..70°16。
HGPT-35-A-B并行手掌机器人产品说明书
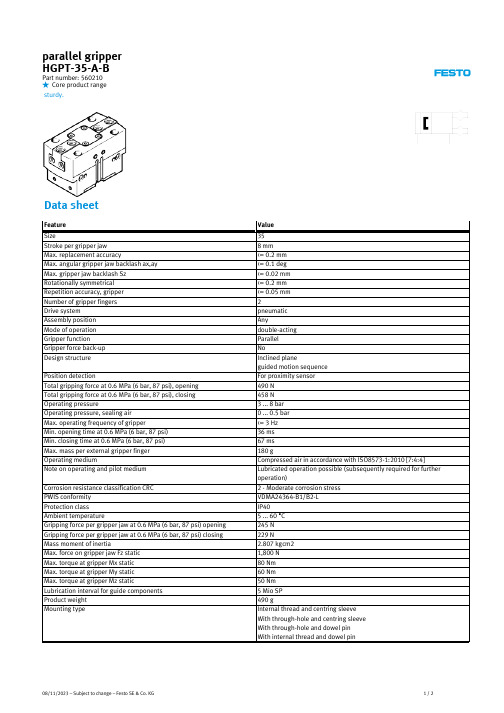
Part number: 560210 Core product range
sturdy.
Data sheet
Feature
Size Stroke per gripper jaw Max. replacement accuracy Max. angular gripper jaw backlash ax,ay Max. gripper jaw backlash Sz Rotationally symmetrical Repetition accuracy, gripper Number of gripper fingers Drive system Assembly position Mode of operation Gripper function Gripper force back-up Design structure
2/2
Value
Optional M5 M5 Conforms to RoHS High alloy steel, non-corrosive Anodised aluminium Steel, hardened
08/11/2023 – Subject to change – Festo SE & Co. KGFra bibliotekValue
35 8 mm <= 0.2 mm <= 0.1 deg <= 0.02 mm <= 0.2 mm <= 0.05 mm 2 pneumatic Any double-acting Parallel No Inclined plane guided motion sequence For proximity sensor 490 N 458 N 3 ... 8 bar 0 ... 0.5 bar <= 3 Hz 36 ms 67 ms 180 g Compressed air in accordance with ISO8573-1:2010 [7:4:4] Lubricated operation possible (subsequently required for further operation)
金蝉止痒胶囊中盐酸小檗碱的溶出曲线测定

金蝉止痒胶囊中盐酸小檗碱的溶出曲线测定发布时间:2021-09-24T08:22:20.078Z 来源:《中国医学人文》2021年21期作者:曾凡华1 梅勇1龙涛1 王文娟1 李建华1杨莉1[导读] 目的:建立高效液相色谱法测定金蝉止痒胶囊中盐酸小檗碱的溶出曲线。
曾凡华1 梅勇1龙涛1 王文娟1 李建华1杨莉1重庆希尔安药业有限公司重庆 401520摘要:目的:建立高效液相色谱法测定金蝉止痒胶囊中盐酸小檗碱的溶出曲线。
方法:采用中国药典2015年版四部0931第二法(桨法),以900ml pH4.0、pH6.8、pH7.4磷酸盐缓冲溶液和水作为溶出介质,转速50转/分,分别在5、10、15、30和45分钟取样,采用高效液相色谱法测定盐酸小檗碱的含量。
结果:盐酸小檗碱在测定浓度0.9μg/ml~1.5μg/ml范围内,不同溶出介质条件下,浓度与峰面积线性良好。
在溶出介质水条件下线性方程为Y=0.0000205X-0.002(R= 0.9999);在溶出介质pH4.0条件下线性方程为Y=0.0000214X-0.003(R=1.0000);在溶出介质pH6.8条件下线性方程为Y=0.0000211X-0.006(R=1.0000);在溶出介质pH7.4条件下线性方程为Y=0.0000217X-0.001(R=0.9999)。
个条件下盐酸小檗碱的平均加样回收率均大于98.0%,RSD分别均小于1.0%。
结论:该高效液相色谱法专属性强、准确度高,重复性好,可作为金蝉止痒胶囊的溶出曲线测定。
关键词金蝉止痒胶囊;高效液相色谱法;盐酸小檗碱;溶出度曲线金蝉止痒胶囊是重庆希尔安药业有限公司开发用于治疗夏季皮炎,丘疹性荨麻疹等皮肤瘙痒症状的中药组合物胶囊[1](批准文号为国药准字Z20090396)。
其主要成份包括金银花、黄柏、黄芩、蛇床子、栀子、苦参、龙胆、白芷、白鲜皮、蝉蜕、连翘、地肤子、地黄、青蒿、广藿香、甘草等十六味中药组成,药理学研究表明其具有镇静、抗过敏、抗炎、解热、抑菌、利尿、解毒等作用,并对非免疫性接触性荨麻疹疗效明确,对多种原因导致的小鼠皮肤瘙痒症状具有良好的止痒作用,临床报道金蝉止痒胶囊在治疗神经因素、内分泌等因素引起的瘙痒,慢性荨麻疹,湿热型肛周湿疹,热型湿疹等疗效显著。
