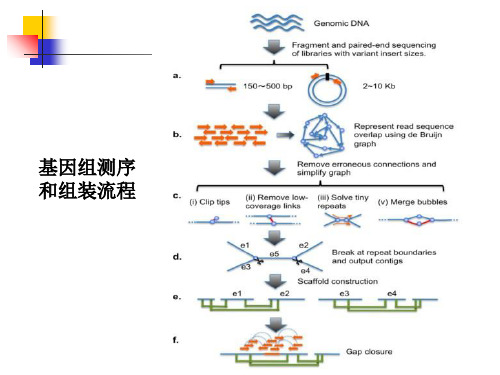
gene_families
细菌全基因组测序 ppt课件
基因家族(gene family) 和基因簇(gene cluster)分析
基因组中来源相同,结构和功能相关的基因 聚集在一起形成基因家族。
基因家族的各个成员紧密成簇排列成大段的串联 重复单位,分布在某一条染色体的特殊区域
genefamily.xls
基因家族聚类结果
genefamily.stat
各基因家族统计信息
培养条件① 培养条件②
或活性较低
测定转录 组mRNA
细菌全基因组测序
比较 新 差异 基因
其他方面的应用研究
❖ 应用NMR、FTIR、UV, 14C标记的木质 素降解机理方面的研究; ❖农药残留物以及其他一些难降解有机物的 降解; ❖ 重金属有机物化合物的降解。
② 木质素降解过程中涉及到的细胞外酶主要有:木质素过氧化物酶
(LiP)和锰过氧化物酶(MnP),以及漆酶(Lac)。此外,一 些附属酶参与过氧化氢的产生,乙二醛氧化酶(glyoxal oxidase, 缩写作GLOX)和芳基醇氧化酶(aryl alcohol oxidase,缩写作 AAO)属于这类酶。
对4株菌的亲缘关系进行分析,确定菌株之间的相互关 系;
通过对4株菌进行进化分析,判定是否为古菌或新的菌 种。
细菌全基因组测序
基因分离
下一步的实验安排
对已注释出的基因进行验证
载体
酶切
酶切
连接
转化
筛选 表达
细菌全基因组测序
未注释出功能的基因鉴定,挖掘新基因
DNA 转录 RNA 翻译 Protein
细菌全基因组测序
“一个物种基因组计划的完成, 就意味着这一物种学科和产业 发展的新开端”
向仲怀院士
谢谢!!
细菌全基因组测序
基因家族开题报告研究计划

基因家族开题报告研究计划(中英文实用版)Title: Research Plan for the Study of Gene Families基因家族研究计划旨在深入探索基因家族的演变、功能及调控机制。
本计划将采用多种研究方法,包括生物信息学分析、实验验证和功能研究,力求揭示基因家族在生物体生长发育、疾病发生和进化过程中的重要作用。
The research plan for the study of gene families aims to delve into the evolution, function, and regulatory mechanisms of gene families.This plan will employ a variety of research methods, including bioinformatics analysis, experimental validation, and functional studies, to reveal the crucial roles of gene families in the growth, development, disease occurrence, and evolution of organisms.研究将首先通过生物信息学方法对基因家族进行系统分析,挖掘其成员、结构及进化关系。
随后,我们将对筛选出的关键基因家族进行实验验证,通过基因敲除、过表达等手段研究其功能。
此外,我们还将探讨基因家族在不同生物过程中的调控机制,如信号传导、转录调控等。
The research will first employ bioinformatics methods to systematically analyze gene families, uncovering their members, structures, and evolutionary relationships.Subsequently, we will validate the selected key gene families through experimental methods, such as gene knockout and overexpression, to study their functions.In addition, we will explore the regulatory mechanisms of gene families in variousbiological processes, such as signal transduction and transcriptional regulation.基因家族研究计划将有助于揭示生物体的基因调控网络,为疾病诊断、治疗和预防提供新的靶点。
基因家族、管家基因的概念

基因家族、管家基因的概念
1、基因家族概念:基因家族(gene family),是来源于同一个祖先,由一个基因通过基因重复而产生两个或更多的拷贝而构成的一组基因,它们在结构和功能上具有明显的相似性,编码相似的蛋白质产物,同一家族基因可以紧密排列在一起,形成一个基因簇,但多数时候,它们是分散在同一染色体的不同位置,或者存在于不同的染色体上的,各自具有不同的表达调控模式。
2、管家基因的概念:管家基因是指所有类型组织细胞在任何时候都需要表达的基因。
由于管家基因是生命活动必需的基因,表达相对稳定,差异小。
所以在基因芯片技术中根据各芯片的管家基因可以得出标准化系数进行标准化较正;管家基因在所有的细胞中都有表达,因此有关管家基因的概念有助于分析差异表达基因的表达情况,进而进行差异表达基因的克隆;通过管家基因,能比较不同样本中某种mRNA的水平。
大麦VQ_基因家族鉴定及表达分析

54卷大麦VQ基因家族鉴定及表达分析倪守飞1,母景娇1,耿梓瀚1,王孜逸2,丛钰莹1,王月雪1,刘梦迪1,蔡倩1,赵彦宏1*,王艳芳2*(1鲁东大学农学院,山东烟台264025;2鲁东大学生命与科学学院,山东烟台264025)摘要:【目的】鉴定大麦VQ基因家族成员并进行表达分析,为大麦VQ基因的功能挖掘提供理论依据。
【方法】从大麦基因组中鉴定VQ基因家族成员,利用生物信息学方法对其结构特征及编码蛋白序列进行分析,基于转录组测序数据及实时荧光定量PCR方法进行大麦组织表达模式、盐胁迫和生物胁迫分析。
【结果】在大麦基因组中鉴定出29个HvVQ 基因(HvVQ1~HvVQ29),HvVQ蛋白序列平均长度较短(214aa),多数HvVQ蛋白为碱性或偏中性蛋白,HvVQ基因不均地分布在大麦染色体上,定位于细胞核中。
29个HvVQ蛋白均含有保守基序FxxxVQxhTG,近90%的HvVQ基因不含内含子。
进化分析将大麦、拟南芥与水稻的VQ基因家族成员分为7个亚族(Ⅰ~Ⅶ),HvVQs基因不均地分布在Ⅱ~Ⅶ亚族中。
大麦与水稻的共线性基因对数(17对)远多于与拟南芥的共线性基因对数(1对),种内共线性分析发现1对共线性基因对,非同义替换率/同义替换率(Ka/Ks)计算发现HvVQ蛋白主要处于纯化选择状态。
HvVQ基因启动区富含生长发育作用元件、非生物胁迫反应元件和激素反应元件,种类及分布均呈多样性。
对蛋白网络预测分析推断其与HvWRKY的2类亚族(Ⅱ-c和Ⅲ)存在互作关系。
大多数HvVQ基因在组织中表达,HvVQ19在受到盐胁迫时表达量明显上调,在根尖和根伸长区表达量分别上调1.40和1.10倍;对其中10个HvVQ基因进行实时荧光定量PCR检测,HvVQ2基因在蚜虫和黄矮病毒胁迫下表达量均显著下调(倍数变化<0.5为显著抑制,>2.0为显著诱导),HvVQ7和HvVQ15基因在蚜虫和黄矮病毒胁迫下表达量上调最显著,其他7个HvVQ基因也均表现出差异表达。
名词解释-5道

名词解释5道(基因组(同源……);基因突变;蛋白质降解;表达调控)基因组(第二章)Genome (基因组):一种生物细胞内全部遗传物质的总和,包括构成基因和基因之间区域的所有DNA;C值:基因组中的全部DNA量称为C值。
Families of genes(基因家族):同一物种中结构与功能相似,进化起源上密切相关的一组基因。
多基因家族(multi gene family)指由某一祖先基因经过倍增和变异所产生的一组基因。
假基因(pseudo gene):在多基因家族中,某些基因并不产生有功能的基因产物,这些基因称为假基因(pseudo gene)(来源:突变或来自RNA的逆转录;重新插入基因组)经典的多基因家族:成员的序列相等或近乎相等,人们认为多基因家族成员来自祖先基因的倍增(e.g.rRNA )“复合”多基因家族:序列相似,编码产物特性上有差异orthologs直系同源基因: genes in two separate species that derive from the same ancestral gene in the last common ancestor of those two species.paralogs旁系同源基因: related genes that have resulted from a gene duplication event within a single genome — likely to have diverged in their function—Homologs:Genes that are related by descent in either way are called homologs, a general term used to cover both types of relationshipgene superfamily: sometimes it is possible to see relationships not only within a single gene family but also between different families.(e.g.the α- and β-globin families )Operon:(操纵子)(为原核生物所特有)a group of genes that are located adjacent to one another in the genome, with perhaps just one or two nucleotides between the end of one gene and the start of the next.all the genes in an operon are expressed as a single unit.蛋白质降解(第四章)熔球(molten globule) 包含了二级结构的大部分元件,其结构已接近于蛋白质的最终结构。
基因家族生信分析报告

基因家族生信分析一、什么是基因家族概念:是来源于同一个祖先,有一个基因通过基因重复而产生两个或更多的拷贝而构成的一组基因,他们在结构和功能上具有明显的相似性,编码相似的蛋白质产物。
划分:按功能划分:把一些功能类似的基因聚类,形成一个家族。
按照序列相似程度划分:一般将同源的基因放在一起认为是一个家族。
1.常见基因家族:WRKY基因家族:是植物前十大蛋白质基因家族之一,大量研究表明,WRKY 基因家族的许多成员参与调控植物的生长发育,形态建成与抗病虫。
NBS-LRR抗病基因家族:是植物中最大类抗病基因家族之一。
MADS-BOX基因家族:是植物体的重要转录因子,它们广泛地调控着植物的生长、发育和生殖等过程。
在植物中参与花器官的发育,开花时间的调节,在果实,根,茎,叶的发育中都起着重要的作用。
热激蛋白70家族(HSP70)是一类在植物中高度保守的分子伴侣蛋白,在细胞中协助蛋白质正确折叠。
二、基因家族分析流程:●利用蛋白保守域结构提取号在Pfam数据库提取其隐马尔科夫模型矩阵文件(*.hmm)●在数据库(Ensemble 、JGI、NVBI)下载你所需要的物种的基因组数据(*.fa,*.gff)●在虚拟机中Bio-Linux中的hummsearch程序,用隐马尔科夫模型矩阵文件在蛋白序列文件中搜索含有该保守结构域的蛋白●将蛋白序列导入MEGA软件构建进化树(可以阐明成员之间系统进化关系,从进化关系上揭示其多样性)●利用MEME搜索蛋白质的保守结构域利用MEME搜索基因家族成员的motif可以揭示基因家族在物种的多样化及其功能,如果他们都含有相同的motif表明其功能具有相似性,如果部分家族成员含有其他不同的motif,很可能这些成员有其他特异功能,或者可以归分为一个亚族●绘制基因染色体位置图从*.gff文件中抽取我们搜索到的基因位置信息,http://mg2c.iask.in/mg2c_v2.0/在线绘制基因染色体位置图通过染色体位置分布,可以了解基因主要分布字哪条染色体上,及是否能形成基因簇(被认为是通过重组与错配促进基因交流)●基因结构分析从gff文件中抽取基因的结构信息,绘制转录本结构图。
基于基因家族大小的比较研究脊椎动物的适应性进化

Hereditas (Beijing) 2019年2月, 41(2): 158―174收稿日期: 2018-08-06; 修回日期: 2018-12-13作者简介: 孟玉,硕士研究生,专业方向:遗传学。
E-mail: m1994yu@通讯作者:杨若林,教授,博士生导师,研究方向:进化遗传学和生物信息学。
E-mail: desert.ruolin@ DOI: 10.16288/j.yczz.18-225网络出版时间: 2019/1/14 13:15:21URI: /kcms/detail/11.1913.R.20190114.1315.004.html 研究报告基于基因家族大小的比较研究脊椎动物的适应性进化孟玉,杨若林西北农林科技大学生命科学学院,杨凌712100摘要:同源基因家族的拷贝数在不同物种间普遍存在差异,这种差异是由不同的基因得失速率引起。
众所周知,基因拷贝数变异是特定物种表型创新的可能原因。
本研究选取具有代表性的脊椎动物主要类群并跨约6亿年进化时间的64个物种,鉴定了它们的同源基因家族,揭示了脊椎动物基因家族大小的进化模式。
结果表明:在推断的存在于脊椎动物最近共同祖先的6857个基因家族中,有6712个都在至少一个种系中发生了大小的变化,而且基因家族在大多数种系中都是收缩的;其中,霍氏树懒(Choloepus hoffmanni)中有最高的基因家族收缩水平,而在斑马鱼(Danio rerio)中则相反。
基于脊椎动物基因家族大小进化的高度动态性,本研究从基因家族大小变化的角度鉴定了一些可能与特定脊椎动物类群进化有关的基因组信号。
结果观察到在现存真骨鱼类最近共同祖先基因组中出现了可能因全基因组复制所导致的高比例的基因家族扩增现象,随后在后裔物种中发生基因收缩事件。
此外,本研究还发现了硬骨鱼特异性的orphan基因可能对这些鱼类在水生环境中的适应性进化有所贡献的证据,如在有些硬骨鱼中orphan基因与鳍、尾巴、肾脏等发育有关。
2.2 基因组(多基因家族)

Characteristics of globin gene expression
(1) the development stage-specific expression 5’——3’ in order (beginning or closing) (2) the tissue-specific expression
ε-crystallin, found in the eye lenses of some birds and crocodiles, is also the enzyme lactate dehydrogenase (LDH).
• In vertebrates are those in the Hox family. For invertebrates, like Drosophila. • The homeotic (同源异型 ) gene complex (HOM).
• In the mouse, each of its four clusters is located on a different chromosome and extends for over 100kb.
• Amphioxus (文昌鱼 ) has a single cluster of at least 10 Hox gene (spanning 270 kb), each of which is homologous to a different Hox gene in vertebrates, so that the origin of the vertebrates coincided with s series of gene duplications.
Pseudogenes usually have mutations that would produce stop codons within what would normally be the coding region, thus only fragments of the protein they seem to code for would be produced if they were expressed. It is not known if pseudogenes are expressed in any way.
植物ppr基因家族的鉴定与进化分析

摘要随着近年来DNA测序技术的快速发展,越来越多的物种已完成基因组测序,因此从全基因组层面鉴定和研究基因家族的分类、序列特点、进化特征及分子功能预测等已成为生物学领域需要关注的重要问题和研究手段。
本文主要关注植物进化分支中的Pentatricopeptide repeat proteins (PPR)基因。
此基因家族作为植物基因组中最庞大的家族之一(占总基因数目百分比,1-2%),及具有非常多样性的对植物表型的影响和特殊的定向绑定、调控靶标基因的分子机制及潜在的巨大生物工程应用价值,近年来正吸引大量研究者从不同角度及方式研究此基因家族,而基因、蛋白结构域的鉴定就自然成为了后续研究的重中之重。
随着大量植物基因组数据的公布,准确、自动化、高效的计算机预测方法成为急需的研究部分。
优秀的鉴定方法、高质量的结果直接影响到基因家族后续的比较进化分析、靶标基因的预测。
在本研究中,首先在18个代表性植物基因组中,用生物信息方法准确、高效鉴定PPR 家族基因及其内部一级序列结构;再借用比较基因组学、系统分子发育学方法,比较PPR 基因及其所含蛋白结构域的结构特征、序列变化、及基因家族的进化等。
研究目标首先是,在大范围的植物分支中,为此领域其他研究者,提供最全面、可靠的PPR基因、所含蛋白结构域的注释信息;接着,展示此类基因家族的基本概况、数量分布、基因结构、序列特征及进化历程;同时,期望能解释PPR家族在高等植物中的拷贝数量巨大的原因;最后,结合上述各种分析、结果,进一步推测其蕴含的分子生物学意义,深化人们对该蛋白家族的认识,为进一步研究PPR蛋白的分子功能及其作为调控靶标基因表达的工具在生物技术领域的应用提供参考。
研究结果:建立了一个高效、准确的生物信息学鉴定流程。
通过比较分析,发现了此基因家族在整个植物领域中快速膨胀现象,但出人意料PLS、E1、E2、E+、DYW亚族仅仅存在于高等植物中。
再通过物种之间、物种内部对此基因家族的比较分析,我们可以勾画出PPR基因家族可能的进化路;同时揭开此基因家族迅速扩张的可能原因:基因的反转录复制事件,而后产生大量的单外显子拷贝。
The R2R3-MYB gene family in Arabidopsis thaliana

447MYB factors represent a family of proteins that include the conserved MYB DNA-binding domain. In contrast to animals, plants contain a MYB-protein subfamily that is characterised by the R2R3-type MYB domain. ‘Classical’ MYB factors, which are related to c-Myb, seem to be involved in the control of the cell cycle in animals, plants and other higher eukaryotes. Systematic screens for knockout mutations in MYB genes, followed by phenotypic analyses and the dissection of mutants with interesting phenotypes, have started to unravel the functions of the 125 R2R3-MYB genes in Arabidopsis thaliana. R2R3-type MYB genes control many aspects of plant secondary metabolism, as well as the identity and fate of plant cells. AddressessMax-Planck-Institute for Plant Breeding Research, Carl-von-Linné-Weg 10, D-50829Köln, Germany*e-mail: weisshaa@mpiz-koeln.mpg.deCurrent Opinion in Plant Biology2001, 4:447–4561369-5266/01/$ — see front matter© 2001 Elsevier Science Ltd. All rights reserved.AbbreviationsAGI Arabidopsis Genome InitiativeAS1ASYMMETRICAL LEAVES1CDC5cell division cycle5GL1GLABROUS1KNOX KNOTTEDPAP1PRODUCTION OF ANTHOCYANIN PIGMENT1PCR polymerase chain reactionPHAN PHANTASTICATT2TRANSPARENT TESTA2WER WEREWOLFIntroductionRegulation of gene expression at the level of transcription controls many crucial biological processes. A number of different factors are required for the process of transcription. These include factors required for chromatin remodelling and DNA unwinding, as well as proteins of the pre-initiation complex and the RNA polymerase II complex. In addition to this general inventory, other factors control promoter strength. These factors are referred to as ‘transcription factors’, a term usually used to describe proteins that recognise DNA in a sequence-specific manner and that regulate the frequency of initiation of transcription upon binding to specific sites in the promoter of target genes. T ranscription factors—which can be activators, repressors, or both—display a modular structure. On the basis of similarities in one of the modules, namely the DNA-binding domain, transcription factors have been classified into families [1]. In plants, MYB factors comprise one of the largest of these families [2,3•].The first MYB gene identified was the ‘oncogene’v-Myb derived from the avian myeloblastosis virus [4]. Evidence obtained from sequence comparisons indicates that v-Myb may have originated from a vertebrate gene, which mutated once it became part of the virus. Many vertebrates contain three genes related to v-Myb—c-Myb, A-Myb and B-Myb [5]—and other similar genes have been identified in insects, plants, fungi and slime moulds [6]. The encoded proteins are crucial to the control of proliferation and differentiation in a number of cell types, and share the con-served MYB DNA-binding domain. This domain generally comprises up to three imperfect repeats, each forming a helix-turn-helix structure of about 53 amino acids. Three regularly spaced tryptophan residues, which form a trypto-phan cluster in the three-dimensional helix-turn-helix structure, are characteristic of a MYB repeat [7,8]. The three repeats in c-Myb are referred to as R1, R2 and R3; and repeats from other MYB proteins are categorised according to their similarity to either R1, R2 or R3. Here, we discuss the MYB gene inventory of the Arabidopsis thaliana genome, and review the conservation of sequence and function among A. thaliana MYB proteins by including several well-known examples from other species.MYB genes in plantsMYB proteins can be classified into three subfamilies depending on the number of adjacent repeats in the MYB domain (one, two or three) [9,10]. We refer to MYB-like proteins with one repeat as ‘MYB1R factors’, with two as ‘R2R3-type MYB’ factors, and with three repeats as ‘MYB3R’ factors. The MYB-like proteins with a single repeat(or sometimes just a partial one)are fairly divergent and include factors that bind the consensus sequence of plant telomeric DNA (TTTAGGG) [11]. It has also been shown that MYB1R factors (e.g.MYBST1 or StMYB1R1) can act as transcriptional activators [12], and some are associated closely with the activity of the circadian clock (A1 [CIRCADIAN CLOCK ASSOCIATED1] and LHY [LATE ELONGATED HYPOCOTYL]) [13]. CCA1 and LHY1 bind DNA, indicating that they might act by modulating transcription [14,15].Primarily through genome sequencing, genes encoding three Myb repeats have been detected in A. thaliana[16]. MYB3R genes have also been detected in all major lineages of land plants [17]. These data show that the genes encoding R2R3-MYB factors (see below) are not the equivalents of c-Myb from animals, as was initially thought before the detection of plant MYB3R genes.A.thaliana contains five AtMYB3R genes (see T able1). It has been shown recently that plant MYB3R factors similar to MYB proteins in animals are involved in controlling the cell cycle [18]. Like B-MYB in blood cells [19], MYB3R factors are involved in regulating the transcription of cyclin genes via MYB recognition elements in cyclin promoters [18]. This may indicate functional conservation amongThe R2R3-MYB gene family in Arabidopsis thaliana Ralf Stracke, Martin Werber and Bernd Weisshaar*448Cell signalling and gene regulationThe R2R3-MYB gene family in Arabidopsis thaliana Stracke, Werber and Weisshaar 449MYB3R genes from plants to humans. However, the exact contribution of MYB3R factors and proteins such as AtMYBCDC5 [20] in cell cycle control in plants requires further elucidation. AtMYBCDC5 is related to the yeast cell cycle protein CDC5+ and the human regulator of mitotic entry HsCDC5. These are potentially multifunctional MYB proteins that are involved in transcript splicing [21] and transcriptional regulation[22].We have so far been unable to find knockout alleles for AtMYB3R genes, although we have screened several of the available insertion-mutagenesis populations. AtMYB3R genes may play a role in cell cycle control, and so, it is possible that mutations in these genes are lethal, thus the respective mutant alleles are under-represented in knockout populations. The map positions of the A.thaliana MYB3R genes were compared with those of embryonic lethal mutations (emb), but candidates for mutant alleles were not revealed conclusively. This might be because of the high number of emb genes and the difficulties involved in integrating physical and genetic maps [23].450Cell signalling and gene regulationR2R3-MYB genes in A.thalianaMYB genes containing two repeats (i.e. R2R3-MYB) con-stitute the largest MYB gene family in plants. The large size of this gene family was apparent from the work of Romero et al.[2] in A.thaliana and was also confirmed in Zea mays[24]. About 80 different A.thaliana genes were described initially [2], and this number increased to 97 through the combined efforts of six European laboratories in an European Community funded consortium.Our listing herein is based on the systematic names previ-ously assigned to R2R3-MYB genes [2,25,26]. In some instances, synonyms are listed for genes bearing different names in the literature. The list includes AtGL1,the first R2R3-MYB gene identified in A.thaliana as AtMYB0(the designation ‘AtMYB1’ was assigned to the first A.thaliana R2R3-type MYB gene identified by polymerase chain reac-tion [PCR]-based methods) [25]. As new MYB genes were disclosed through sequencing by the Arabidopsis Genome Initiative (AGI), we generated Genbank/EMBL database entries for the deduced cDNAs of AtMYB and AtMYB3R genes (see T able 1). Upon inspection, and in some cases correction, of the predicted splicing patterns, we con-firmed the existence of the respective transcript by reverse transcription (RT)-PCR using gene-specific primers, resulting in an amplicon spanning at least one intron posi-tion (R Stracke, B Weisshaar, unpublished data). In an attempt to complete the listing, we screened the TIGR and MIPS whole-genome datasets (version 03202001) with various sequence-similarity search tools. T able1 lists 125 distinct AtMYB genes of the R2R3 type that were detected on the basis of the (near) complete sequence ofThe R2R3-MYB gene family in Arabidopsis thaliana Stracke, Werber and Weisshaar 451452Cell signalling and gene regulationA.thaliana[27•]. The systematic designations, and the gene identifiers (e.g. At3g27920) assigned to genes in the AGI annotation, may help to avoid the confusion that often results when multiple names are used for the same gene in such a large gene family.There are two unusual genes encoding MYB proteins with two or more repeats: AtMYBCDC5and AtMYB4R1. AtMYBCDC5 contains a MYB domain consisting of two repeats that are only distantly related to those of the R2R3-type MYB domain (having 31% identity to a typical R2R3-type MYB domain, whereas R2R3-type MYB domains generally display at least 40% identity to the consensus); AtMYB4R1 (T able1) is a putative MYB protein containing four R1R2-like repeats.Riechmann et al.[3•] calculated that of the almost 26,000 A.thaliana genes, about 1600 genes (6%) encode transcription factors, and classified 131 of them as factors of the R2R3-MYB type. The InterPro MYB domain signature patterns—PS00037 (W-[ST]-x[2]-E-[DE]-x[2]-[LIV]) and PS00334 (W-x[2]-[LI]-[SAG]-x[4,5]-R-x[8]-[YW]-x[3]-[LIVM]; accession IPR001005)[28] —used to detect MYB factors in the Riechmann study do, as already stated in the ‘Supplementary Material’ of that study, retrieve ‘false-positives’ resulting in an over-estimation of the total number of AtMYB genes. We have tried to exclude such false positives by manual inspection of the deduced amino-acid sequence of every gene shown in T able1. This may explain the difference between the number of R2R3-type MYB genes detected (131 versus 125). Clarification of this discrepancy will require information, which is currently unavailable,on which open reading frames were considered by Riechmann et al.[3•]. We cannot exclude, however, the existence of a small number of additional MYB proteins with two or more repeats in A.thaliana. This is because not all of the genome has been sequenced yet, and because a potential candidate gene with an unusual gene structure and/or very small exons might have escaped our detection. All 125 A.thaliana R2R3-type MYB domain sequences were used to deduce a consensus sequence and to determine the frequency of the most prevalent amino acids at each position within the repeats. Figure1 shows clearly the high level of conservation not only of the tryptophan residues but also of the phenylalanine residue that replaces the first tryptophan in the R3 repeat. In R2, 25 (of 53) positions are occupied by a single, or two very similar, residues in more than 80% of the proteins. For R3, the equivalent number is 31 (out of 51), indicating that R3 is slightly more conserved than R2. It is worth noting that there are five additional amino-acid positions conserved immediately amino-terminal to R2 (consensus: CDKAG) of the R2R3-type MYB domain (M Werber, B Weisshaar, unpublished data). However, it is not known what function this sequence might have.R2R3-MYB genes mainly regulateplant-specific processesNo functional data are available for most of the 125 R2R3-type AtMYB genes. However, systematic searches for knockouts have been initiated recently [29] and the number of AtMYB genes for which functional information has become available has grown significantly during the past year. R2R3-type MYB genes have been shown to regulate phenylpropanoid metabolism in A.thaliana. Overexpression of AtMYB75/PAP1 (PRODUCTION OF ANTHOCYANIN PIGMENT1) and AtMYB90/PAP2[30•] results in accumulation of anthocyanins, and AtMYB4 represses the synthesis of sinapoyl malate [31••]. The analysis of AtMYB4 also demonstrated that R2R3-type MYB proteins can act as transcriptional activators as well as repressors. Recently, the TRANSPARENT TESTA2(TT2) gene has been shown to encode an R2R3-type MYB factor (AtMYB123; N Nesi, L Lepiniec, personal communication). This is not a surprise as several R2R3-type MYB genes, such as ZmMYBC1from Zea mays[32] or PhMYBAN2from Petunia hybrida [33] (which also control phenylpropanoid metabolism [9,34]), are known to be derived from other plant species. AtMYB34/ATR1(ALTERED TRYPTOPHAN REGULATION1) is a regulator of tryptophan biosynthesis, which demonstrates that pathway control by such factors is not limited to secondary metabolism[35].Another important function for R2R3-type MYB factors is the control of development and determination of cell fate and identity. AtMYB0/GLABROUS 1(GL1) [36] and AtMYBB66/WEREWOLF(WER) [37•] are involved in epidermal cell patterning. In fact, both genes encode very similar proteins (subgroup 15, Figure2; [26]). The classical A.thaliana genetic locus ASYMMETRICAL LEAVES1 (AS1)was shown to encode a MYB factor—AtMYB91/AS1 [38•]—and is orthologous to the cell differentiation genes AmMYBPHAN[39] from Anthirrhinum majus and ZmMYBRS2 [40,41]. AtMYB0/GL1 interacts with a basic helix-loop-helix (bHLH) factor encoded by AtbHLH1/GL3[42••] —similar to the the combined action of ZmMYBC1 and ZmR, which together regulate anthocyanin production in Zea mays[43]. This evidence suggests that R2R3-type MYB factors often are involved in the combinatorial interactionThe R2R3-MYB gene family in Arabidopsis thaliana Stracke, Werber and Weisshaar 453of transcription factors for the generation of highly specific expression patterns.R2R3-type MYB factors also participate in plant responses to environmental factors and in mediating hormone actions, examples of which have been discussed in detail recently[9]. In A.thaliana, AtMYB2 has been found to regulate the AtADH1(ALCOHOL DEHYDROGENASE1) gene promoter, and it might also be involved in the response to low oxygen [44]. AtMYB30expression is strongly correlated with cell death during the hypersensitive response upon pathogen attack or elicitor treatment [45•]. Elicitor-responsive R2R3-type MYB genes have also been described from Nicotiana tabacum[46], and an AtMYB78-related MYB gene from Oryza sativa has been shown to be expressed in response to fungal attack [47]. T aken as a whole, it seems that R2R3-type MYB genes are involved predominantly in controlling ‘plant-specific’ processes[9,34]. This finding is especially interesting in the light of the observation that,to the best of our current knowledge,MYB genes of the R2R3 type are only present in plants [3•]. It is tempting to speculate that R2R3-type MYB genes have, during evolution, contributed to plant speciation.None of the known mutations in R2R3-type MYB genes, including those detected so far in various knockout pop-ulations [29], result in strong phenotypes that are expected for essential genes. More data are required to determine if any R2R3-type MYB genes are absolutely essential, or whether functional redundancy often masks the effects of mutations in crucial R2R3-type MYB genes. Complexity of the R2R3-MYB gene familyThe R2R3-type MYB factors encoded by the AtMYB genes have been categorised into 22 subgroups on the basis of conserved amino-acid sequence motifs present carboxy-terminal to the MYB domain [26]. Re-evaluation of these motifs, using the newly extended dataset, confirmed most of these subgroups (Figure2). Despite the divergence of the amino-acid sequence outside of the MYB domain, there are some conserved motifs that may contribute to function. Evidence for the importance of these motifs comes from AtMYB0/GL1 and the nonfunctional gl1 protein encoded by the gl1-2allele that lacks the motif-defining subgroup15 [26]. The conserved motifs may facilitate the identification of functional domains outside of the DNA-binding domain of R2R3-type MYB factors.It is also interesting to note that there are several cases of functional conservation of genes that cluster together in the dendrogram. AtMYB91/AS1and AmMYBPHAN have both been shown to negatively regulate KNOX(KNOTTED) expression in organ primordia [38•]. AtMYB66/WER and AtMYB0/GL1, both clustering together in subgroup15 (Figure2), can functionally complement each other and display different biological functions only because of their different spatial expression patterns [48••]. AtMYB23 is closely related to AtMYB66/WER and AtMYB0/GL1, and it would be interesting to see if the AtMYB23 gene is also able to complement wer and gl1 mutants when expressed under the control of the respec-tive promoter. A third case for correlation between sequence similarity and function might be AtMYB123, because this factor is sequence-related to ZmMBC1 and is identical to the R2R3-type MYB factor encoded by TT2 (see above). Both, AtMYB123/TT2 and ZmMYBC1, con-trol the pigmentation of seed or kernals, respectively. AtMYB75/PAP1 and AtMYB90/PAP2 are also closely related to ZmMYBC1 (Figure2) and appear to have a somehow related function[30•].ConclusionsThe clear examples of functional conservation described above support the optimistic view that data from the model plant A.thaliana will contribute to a better understanding of MYB gene functions in other species, and vice versa. Part of the apparent redundancy observed among the MYB genes at the amino-acid sequence level is due to genes that have similar molecular functions but still display different biological phenotypes when mutated because of differential temporal and/or spatial expression characteristics. Consequently, the genes are not redundant in terms of the developmental biology of the plant. This concept can explain the existence, and maintenance during evolution, of genes encoding a number of closely related factors in the same species. When transferred to the level of the target genes, this interpretation suggests that a single target gene is regulated by a fairly high number of factors from the same gene family, which all transmit their signal via the same cis-acting element. There is no evidence that any of the 125 R2R3-type MYB genes might represent a pseudogene, inferring that selection might maintain intact MYB genes. This indicates that there must be a phenotype associated with any mutation on which selection can act. The challenge is to find the correct conditions and screens to force the plant to display this phenotype, thereby helping to reveal the function of the remaining 90% of the MYB genes in A.thaliana. AcknowledgementsWe would like to thank all members of the EC MYB consortium, particularly Javier Paz-Ares, Chiara T onelli, Mike Bevan, Sjef Smeekens and Cathie Martin, for stimulating discussions on plant MYB proteins; Loic Lepiniec and Masaki Ito for communicating results prior to publication; AGI for sequencing and annotation; and Heiko Schoof for updating gene identifiers in MATDB. We also thank Norddeutsche Pflanzenzucht (NPZ), Hohenlieth, for funding the PhD work of MW; John Doonan, Imre Somssich and Cathie Martin for critical reading of the manuscript; and Klaus Hahlbrock and Francesco Salamini for continuous support and laboratory space. References and recommended readingPapers of particular interest, published within the annual period of review, have been highlighted as:•of special interest••of outstanding interest1.Pabo CO, Sauer RT: Transcription factors: structural families andprinciples of DNA recognition.Annu Rev Biochem 1992,61:1053-1095.2.Romero I, Fuertes A, Benito MJ, Malpical JM, Leyva A, Paz-Ares J:More than 80 R2R3-MYB regulatory genes in the genome ofArabidopsis thaliana.Plant J 1998, 14:273-284.454Cell signalling and gene regulation3.Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, •Adam L, Pineda O, Ratcliffe OJ, Samaha RR et al.: Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes.Science 2000, 290:2105-2110.The authors evaluated the sequenced eukaryotic genomes in terms of transcription factor genes and gene families. About 45% of the approximately 1600 Arabidopsis transcription factors seem to belong to families that are specific to plants.4.Klempnauer KH, Gonda TJ, Bishop JM: Nucleotide sequence of theretroviral leukemia gene v-myb and its cellular progenitor c-myb: the architecture of a transduced oncogene.Cell 1982,31:453-463.5.Weston K: Myb proteins in life, death and differentiation.Curr OpinGenet Dev 1998, 8:76-81.6.Lipsick JS: One billion years of Myb.Oncogene 1996, 13:223-235.7.König P, Giraldo R, Chapman L, Rhodes D: The crystal structure ofthe DNA-binding domain of yeast RAP1 in complex with telomeric DNA.Cell 1996, 85:125-136.8.Ogata K, Hojo H, Aimoto S, Nakai T, Nakamura H, Sarai A, Ishii S,Nishimura Y: Solution structure of a DNA-binding unit of Myb: ahelix-turn-helix-related motif with conserved tryptophans forminga hydrophobic core.Proc Natl Acad Sci USA 1992, 89:6428-6432.9.Jin H, Martin C: Multifunctionality and diversity within the plantMYB-gene family.Plant Mol Biol 1999, 41:577-585.10.Rosinski JA, Atchley WR: Molecular evolution of the Myb family oftranscription factors: evidence for polyphyletic origin.J Mol Evol1998, 46:74-83.11.Yu EY, Kim SE, Kim JH, Ko JH, Cho MH, Chung IK:Sequence-specific DNA recognition by the Myb-like domain ofplant telomeric protein RTBP1.J Biol Chem 2000, 275:24208-24214.12.Baranowskij N, Frohberg C, Prat S, Willmitzer L: A novel DNAbinding protein with homology to Myb oncoproteins containingonly one repeat can function as a transcriptional activator.EMBO J 1994, 13:5383-5392.13.Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E:Microarray analysis of diurnal and circadian-regulated genes inArabidopsis.Plant Cell 2001, 13:113-123.14.Wang ZY, Kenigsbuch D, Sun L, Harel E, Ong MS, Tobin EM:A Myb-related transcription factor is involved in the phytochromeregulation of an Arabidopsis Lhcb gene.Plant Cell 1997,9:491-507.15.Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA,Coupland G: The late elongated hypocotyl mutation ofArabidopsis disrupts circadian rhythms and the photoperiodiccontrol of flowering.Cell 1998, 93:1219-1229.16.Braun EL, Grotewold E: Newly discovered plant c-myb-like genesrewrite the evolution of the plant myb gene family.Plant Physiol1999, 121:21-24.17.Kranz H, Scholz K, Weisshaar B: c-MYB oncogene-like genesencoding three MYB repeats occur in all major plant lineages.Plant J 2000, 21:231-235.18.Ito M, Araki S, Matsunaga S, Itoh T, Nishihama R, Machida Y,Doonan JH, Watanabe A: G2/M-phase-specific transcription during the plant cell cycle is mediated by c-Myb-like transcription factors.Plant Cell 2001, 13:in press.19.Muller C, Yang R, Idos G, Tidow N, Diederichs S, Koch OM,Verbeek W, Bender TP, Koeffler HP: c-myb transactivates thehuman cyclin A1 promoter and induces cyclin A1 geneexpression.Blood 1999, 94:4255-4262.20.Hirayama T, Shinozaki K: A cdc5+ homolog of a higher plant,Arabidopsis thaliana.Proc Natl Acad Sci USA 1996,93:13371-13376.21.Burns CG, Ohi R, Krainer AR, Gould KL: Evidence that Myb-relatedCDC5 proteins are required for pre-mRNA splicing.Proc Natl Acad Sci USA 1999, 96:13789-13794.22.Lei XH, Shen X, Xu XQ, Bernstein HS: Human Cdc5, a regulator ofmitotic entry, can act as a site-specific DNA binding protein.J Cell Sci 2000, 113:4523-4531.23.Meinke DW, Cherry JM, Dean C, Rounsley SD, Koornneef M:Arabidopsis thaliana: a model plant for genome analysis.Science 1998, 282:662-682.24.Rabinowicz P, Braun E, Wolfe A, Bowen B, Grotewold E: Maize R2R3Myb genes: sequence analysis reveals amplification in the higher plants.Genetics 1999, 153:427-444.25.Shinozaki K, Yamaguchi-Shinozaki K, Urao T, Koizumi M: Nucleotidesequence of a gene from Arabidopsis thaliana encoding a mybhomologue.Plant Mol Biol 1992, 19:493-499.26.Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, Meissner RC,Petroni K, Urzainqui A, Bevan M, Martin C et al.: Towards functional characterisation of the members of the R2R3-MYB gene familyfrom Arabidopsis thaliana.Plant J 1998, 16:263-276.27.The Arabidopsis Genome Initiative: Analysis of the genome •sequence of the flowering plant Arabidopsis thaliana.Nature 2000, 408:796-815.This paper summarises years of highly coordinated work from laboratories all over the world to obtain the first complete genome sequence of a plant. Within 115.4 Megabases of sequenced DNA, 25,500 genes were identified and annotated.28.Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M,Bucher P, Cerutti T, Corpet F, Croning MDR et al.: The InterProdatabase, an integrated documentation resource for protein families, domains and functional sites.Nucleic Acids Res 2001, 29:37-40. 29.Meissner RC, Jin H, Cominelli E, Denekamp M, Fuertes A, Greco R,Kranz HD, Penfield S, Petroni K, Urzainqui A et al.: Function search ina large transcription factor gene family in Arabidopsis: assessingthe potential of reverse genetics to identify insertional mutations in R2R3 MYB genes.Plant Cell 1999, 11:1827-1840.30.Borevitz JO, Xia YJ, Blount J, Dixon RA, Lamb C: Activation tagging •identifies a conserved MYB regulator of phenylpropanoid biosynthesis.Plant Cell 2000, 12:2383-2393.An A.thaliana mutant line with intensive purple pigmentation in many vegetative organs was identified from an activation-tagging population. The dominant phenotype resulted from overexpression of AtMYB75/PAP1, which caused a massive activation of phenylpropanoid biosynthetic genes and enhanced accumulation of lignin, hydroxycinnamic acid esters, and purple anthocyanins. AtMYB75/PAP1, and the closely related factor AtMYB90/PAP2, are both similar to Petunia hybrida AtMYBAN2.31.Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, ••Weisshaar B, Martin C: Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis.EMBO J 2000, 19:6150-6161.The R2R3-type MYB factor AtMYB4 was shown to act as a negative regulator of cinnamate 4-hydroxylase gene expression. It also negatively regulates other steps of phenylpropanoid metabolism in a dose-dependent way. This is the first example of a plant MYB protein that acts as a transcriptional repressor. Functionally, AtMYB4 controls the synthesis of sinapate esters, which provide tolerance to UV-B irradiation.32.Paz-Ares J, Wienand U, Peterson PA, Saedler H: Molecular cloningof the c1 locus of Zea mays: a locus regulating the anthocyaninpathway.EMBO J 1986, 5:829-834.33.Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N,Mol J, Koes R: Molecular analysis of the anthocyanin2gene ofpetunia and its role in the evolution of flower color.Plant Cell1999, 11:1433-1444.34.Martin C, Paz-Ares J: MYB transcription factors in plants.TrendsGenet 1997, 13:67-73.35.Bender J, Fink GR: A Myb homologue, ATR1, activates tryptophangene expression in Arabidopsis.Proc Natl Acad Sci USA 1998,95:5655-5660.36.Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD:A MYB gene required for leaf trichome differentiation inArabidopsis is expressed in stipules.Cell 1991, 67:483-493.37.Lee MM, Schiefelbein J: WEREWOLF, a MYB-related protein in •Arabidopsis, is a position-dependent regulator of epidermal cell patterning.Cell 1999, 24:473-483.The authors isolated the AtMYB66/WER gene. Plants mutated at WER display a hairy root phenotype. AtMYB66/WER is required for normal epidermal-cell patterning, and regulates the position-dependent expression of GL2.38.Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, •Martienssen RA: Asymmetric leaves1mediates leaf patterning and stem cell function in Arabidopsis.Nature 2000,408:967-971.The paper describes the cloning of AtMYB91/AS1, which was found to be a homologue of AmMYBPHAN from A.majus and ZmMYBRS2 from Z.mays. Molecular and genetic analyses define conserved interactions between AS1 and the KNOX homeobox genes.The R2R3-MYB gene family in Arabidopsis thaliana Stracke, Werber and Weisshaar 455。
分子生物学课件 第3章 基因与基因组

实际应用中“基因组”这个词既可以特指储存在细胞核中 的整套DNA(即核基因组),也可以指储存在细胞器中的 整套DNA(即线粒体基因组或叶绿体基因组),还可以指 一些非染色体的遗传元件,如病毒基因组、质粒基因组和 转座元件等。
不同基因家族各成员之间的序列 相似度也不同:
序列高度相似:经典的基因家族,如rRNA基因家族和组蛋 白基因家族。 保守性较低,但是编码产物具有大段的高度保守的氨基酸 序列。
序列保守性很低,编码产物之间也只有很短的保守氨基酸 序列,但通常由于具有保守的结构和功能区域,因而编码产 物具有相似的功能。
基因家族的成员在染色体上 的分布形式不同:
成簇存在的基因家族(clustered gene family)或称基因簇 (gene cluster),如人类类α链基因簇和类β链基因簇。 散布的基因家族(interspersed gene family),如肌动蛋白 基因家族和微管蛋白基因家族。
基因间隔区较短且内含子较少,基因排列紧密。
3.2.7 沉默基因
沉默基因( Silent Gene)也叫隐蔽基因(Cryptic gene), 是处于不表达状态的基因。它可能是假基因,也可能是被关闭的 基因。这些基因以隐性的方式埋藏在染色体中,但遇到特殊因子 的刺激,有可能解除关闭变成显性基因。
3.2.8 RNA基因
tRNA、rRNA; 核仁小分子RNA(small nucleolar RNA, snoRNA) 微小分子RNA(microRNA, miRNA); 小分子干扰RNA(small interfering RNA, siRNA); 核内小分子RNA(small nuclear RNA, snRNA);
humann2 genefamilies文件转化为go、kegg注释

humann2 genefamilies文件转化为go、kegg注释摘要:1.介绍humann2 genefamilies 文件2.文件转化为go、kegg 注释的意义3.详细步骤和方法4.结果展示与分析5.总结与展望正文:Humann2 是一种广泛应用于代谢组学数据分析的生物信息学工具,能够对代谢物进行注释和代谢途径分析。
在分析过程中,我们通常会使用humann2 genefamilies 文件,这个文件包含了所有与代谢物相关的基因家族信息。
然而,这个文件通常以一种特定的格式存储,不便于我们直接进行后续的分析。
因此,我们需要将humann2 genefamilies 文件转化为go、kegg 注释格式。
文件转化为go、kegg 注释的意义在于,这两种格式是生物信息学领域广泛接受的标准格式,可以方便地进行后续的功能注释和代谢途径分析。
GO (Gene Ontology)是一种对基因进行功能注释的标准方法,它将基因分为三个层次:生物学过程、细胞组分和分子功能。
KEGG(Kyoto Encyclopedia of Genes and Genomes)则是一种代谢途径数据库,提供了大量的代谢途径信息。
具体的转化步骤如下:1.安装humann2 软件包:首先,我们需要安装humann2 软件包,以便于我们处理humann2 genefamilies 文件。
可以通过运行以下命令进行安装:`conda install -c bioconda humann2`。
2.读取humann2 genefamilies 文件:使用humann2 软件包中的`read_humann2_genefamilies()`函数读取humann2 genefamilies 文件。
例如:`humann2_genefamilies <-read_humann2_genefamilies("path/to/humann2_genefamilies_file.txt")`。
gene family名词解释

gene family名词解释Gene family是指基因家族,是生物学中一组与其它基因有高度同源性的基因。
基因家族拥有类似的序列和结构,包括DNA序列、外显子和内含子等。
它们可能位于同一染色体内,也可能散布在染色体不同位置上。
基因家族可以出现在所有生物的基因组中,可以是多个基因副本,还可以是一组基因序列中的变化导致不同类型的基因。
在不同物种之间,基因家族的数量和特征也有所不同。
下面是关于基因家族的一些基本知识点:1.基因家族的分类基因家族可以分为四类:同感家族、重复基因家族、多基因家族和保守基因家族。
同感家族是指在一个物种的基因组中由一个基因产生的蛋白质分子,可以正常地识别与与其它基因在物种中共有的序列相似。
重复基因家族是指具有高度相似DNA序列并以相似方式表达的基因。
这是基因家族中最常见的类型之一。
多基因家族是指包含多个相同或相似的基因,这些基因通常来自基因组中不同位置的基因复制和分化。
保守基因家族是指与特定功能相关的基因家族,这些基因在不同物种中的结构和序列比较相似。
这种家族成员通常是形成蛋白质的基本功能酶或结构蛋白质。
2.基因家族的重要性基因家族的形成是在基因进化方面发挥了重要的作用。
不同种的组织和不同的细胞具有不同的蛋白质需要。
因此,如果一个基因家族具有不同的副本,就可以产生各种不同类型的蛋白质来适应各种不同的细胞。
同样的,基因家族的变异可以允许新功能的产生,从而对进化产生影响。
3.检测和分析基因家族来自不同基因家族的基因序列可以通过DNA序列分析软件比较。
这些分析可以帮助确定家族成员的数量和DNA序列的关系。
有很多计算工具和数据库,可以帮助生物学家快速比较大量的DNA序列,以便于更全面的了解基因家族之间的逻辑关系。
简言之,基因家族是指具有高度同源性的基因,并且能够实现类似的生物学功能。
基因家族的分类、作用和检测方法都包含了生物医学领域的重要知识点。
医学遗传学英文名词

原癌基因(proto-oncogene) 原癌基因 抑癌基因(tumor suppressor gene) 抑癌基因 基因诊断(gene diagnosis) 基因诊断 基因治疗(gene therapy) 基因治疗 基因替代( 基因替代(gene replacement) ) 遗传咨询(genetic counseling) 遗传咨询 新生儿筛查(neonatal screening) 新生儿筛查
“医学遗传学”英语名词范围(共30个) 医学遗传学”英语名词范围( 个
其中前部分15个 其中前部分 个: 遗传病( disease) 遗传病(genetic disease) 基因家族( family) 基因家族(gene family) 断裂基因( gene) 断裂基因(split gene) 移码突变( mutation) 移码突变(frame shift mutation) 遗传印记( imprinting) 遗传印记(genomic imprinting) 基因突变( mutation) 基因突变(Gene mutation) 动态突变( 动态突变(dynamic mutation) 单基因病( disease) 单基因病(monogenic disease)
其中后部分 个: 其中后部分15个
X染色质(X chromatin) 染色质( 染色质 核型(karyotype) 核型(karyotype) 异染色质( 异染色质(heterochromatin) ) 罗伯逊易位( translocation) 罗伯逊易位(Robertsonian translocation) 染色体病( 染色体病(chromosome disease) 标记染色体( chromosome) 标记染色体(marker chromosome) 癌基因(oncogene) 癌基因 肿瘤的遗传易感性(tumor susceptability) 肿瘤的遗传易感性
基因家族的概念

基因家族的概念基因家族的概念1. 引言基因家族(gene family)是指一组相似或相关的基因,它们在生物体中以多个拷贝的形式存在。
基因家族的出现和演化是通过基因重复事件而产生的,这些事件包括基因复制、基因转座和基因重组等。
基因家族在进化过程中发挥了重要的作用,它们对生物体的适应性和多样性起到了关键的推动作用。
本文将深入探讨基因家族的概念,其重要性以及相关领域的研究进展。
2. 基因家族的形成和演化基因家族是由基因重复事件导致的,这些事件可以是基因复制(gene duplication)、基因转座(gene transposition)或基因重组(gene recombination)等。
在基因复制过程中,某个基因的拷贝通过突变和选择的作用逐渐发展出新的功能和表达模式。
基因转座则是指基因在染色体上的位置发生了改变,从而产生了新的基因。
基因重组则是指两个不同的基因在染色体上发生了交换,从而形成了新的组合。
3. 基因家族的重要性基因家族在生物的进化和多样性中起到了重要的作用。
基因家族提供了基因多样性的基础。
通过基因的多次重复和演化,新的基因产生了新的功能和表达模式,进而促进了生物体对环境的适应和进化。
基因家族在维持生物体的稳定性上也起到了关键的作用。
由于基因家族中的基因具有相似的序列和结构,它们通常会在相同条件下被调控,在某种程度上实现了对基因的冗余。
这种冗余性使得基因家族中的某个基因发生突变或缺失时,其他基因可以起到补偿作用。
4. 基因家族的研究进展近年来,基因家族的研究在生物学领域取得了显著的进展。
通过高通量测序技术和生物信息学方法,研究人员可以对基因家族的成员进行鉴定和分类,并分析它们的进化关系和功能。
人们还发现了许多具有重要生物学功能的基因家族,例如免疫球蛋白基因家族和微小RNA基因家族等。
这些研究为我们深入理解基因家族提供了重要的线索。
5. 总结与展望基因家族是生物进化和多样性的重要基础,它们通过基因重复事件的发生和演化推动了生物体的适应性和进化。
遗传学英语

遗传学英语
遗传学英语是一门研究基因及其变异、遗传信息的传递和表达方式的学科。
在遗传学英语中,常使用一些专业术语来描述基因的结构、功能、表达方式以及遗传变异等内容。
遗传学英语中一些常用术语包括:gene(基因)、allele(等位基因)、mutation(突变)、genotype(基因型)、phenotype(表型)、inheritance(遗传)、dominant(显性)、recessive(隐性)、heterozygous(杂合)、homozygous(纯合)等等。
研究遗传学英语不仅可以帮助我们更好地理解基因的结构、功能和表达方式,还可以使我们了解基因的遗传模式和遗传疾病等方面的知识。
这对于遗传咨询、基因检测和基因治疗等领域都有着重要的意义。
因此,对于从事遗传学相关工作的人员,学习遗传学英语是必不可少的。
同时,对于广大科学爱好者来说,学习遗传学英语也可以增加我们对生命科学的认识和理解。
- 1 -。
家族族谱简介英语作文

Family Genealogy IntroductionFamily genealogy, also known as a family tree, is a record of a family's history and lineage. It typically includes information about ancestors, descendants, and relatives, providing a comprehensive overview of the family's roots and connections.The importance of family genealogy lies in its ability to preserve and pass down family history from one generation to the next. By documenting the names, dates, and relationships of family members, genealogy helps to maintain a sense of identity and belonging within the family.In addition to serving as a historical record, family genealogy can also facilitate connections between relatives. Discovering shared ancestors and distant cousins can bring families closer together, fostering a sense of unity and shared heritage.In the modern era, technological advancements have made it easier than ever to research and compile family genealogy. Online databases, DNA testing, and genealogy software have revolutionized the process, enabling individuals to uncover new information about their ancestry and connect with relatives around the world.In conclusion, family genealogy plays a vital role in preserving family history, fostering connections between relatives, and maintaining a sense of heritage and identity. By documenting and sharing our family's genealogy, we can ensure that future generations have a deep understanding of their roots and the interconnectedness of their family tree.家族族谱简介家族族谱,又称家谱,是一份家庭历史和血统的记录。
高中英语词汇5-gene-出生

*gene-*genə-, also *gen-, Proto-Indo-European root meaning "give birth, beget," with derivatives referring to procreation and familial and tribal groups.engine,gender,gene,general,generate,generation,generic, generous,genesis,genius,genre,gentle,genuine,oxygen2019全国I卷阅读理解D 篇32. What sort of girl was the author in her early years of elementary school?A. Unkind.B. Lonely.C. Generous.D. Cool.七选五Is Fresh Air Really Good for You?We all grew up hearing people tell us to "go out and get some fresh air". 36 According to recent studies, the answer is a big YES, if the air quality in your camping area is good.37 If the air you’re breathing is clean — which it would be if you’re away from the smog of cities — then the air is filled with life-giving, energizing oxygen. If you exercise out of doors, your body will learn to breathe more deeply, allowing even more oxygen to get to your muscles(肌肉)and your brain.2018年听力第2题2. What can we say about the woman?A. She’s generous.B. She’s curious.C. She’s helpful.阅读理解C篇第3段At present, the world has about 6,800 languages. The distribution of these languages is hugely uneven. The general rule is that mild zones have relatively few languages, often spoken by many people, while hot, wet zones have lots, often spoken by small numbers.阅读理解D篇第2段This method provided a readout for how home energy use has evolved since the early 1990s. Devices were grouped by generation.七选五第4段Medium color choices are generally furniture pieces such as sofas, dinner tables or bookshelves.核心词1:engineengine [ˈendʒɪn] n. 引擎,发动机;机车,火车头;工具en-一般形成动词,表使动+-gin-(gen-的变体)+ -e 动词或名词后缀c. 1300, "mechanical device," especially one used in war; "manner of construction," also "skill, craft, innate ability;deceitfulness, trickery," from Old French engin "skill, wit, cleverness," also "trick, deceit, stratagem,strategy; war machine" (12c.), from Latin ingenium "innate qualities, ability; inborn character," in Late Latin "a war engine, battering ram" (Tertullian, Isidore of Seville); literally "that which is inborn," from in-"in" (from PIE root *en "in") + gignere, from PIE *gen(e)-yo-, suffixed form of root *gene- "give birth, beget." Sense of "device that converts energy to mechanical power" is 18c.; in 19c. especially of steam engines. Middle English also had ingeny (n.) "gadget, apparatus, device," directly from Latin ingenium.Battering ram,ram 撞车,破城槌1.1 engineer [ˌendʒɪˈnɪr]n. 工程师;工兵;火车司机vt. 设计;策划;精明地处理vi. 设计;建造engine+-er →engineer n. →名词动用1.2 engineering [ˌendʒɪˈnɪrɪŋ] n. 工程,工程学v.engineer+-ing→engineering n. 名词动用civil engineering 土木工程the branch of engineering dealing with the design and construction of highways, bridges, tunnels, waterworks, harbors, etc.civil engineer n.核心词2:gender[ˈdʒendər] n. 性别gen-出生+-d-(with euphonious[juːˈfoʊniəs] -d- in French)+-er名词后缀,表人或物→出生的人,男的女的?→出生决定性别c. 1300, "kind, sort, class, a class or kind of persons or things sharing certain traits," from Old French gendre, genre "kind, species; character; gender" (12c., Modern French genre), from stem of Latin genus (genitive generis) "race, stock, family; kind, rank, order; species," also "(male or female) sex," from PIE root *gene-"give birth, beget," with derivatives referring to procreation and familial and tribal groups.Also used in Latin to translate Aristotle's Greek grammatical term genos. The grammatical sense is attested in English fromlate 14c. The unetymological -d- is a phonetic accretion in Old French2.1 gender['dʒendəlis] a. 无性的asexual , sexlessa- prefix①与---相反,缺乏---, 不;没有showing an opposite or the absence of something; not; withoutsexual, asexual; moral, amoral; typical, atypical② in the condition or state of 处于某种状况,以某种方式afloat,alive,asleep, aloud一般只能做表语,不能做定语③(old use 旧)in, to, at, or on somethingabed, in bed;afar, far away2.2engender(ɪnˈdʒɛndə) V.en-表使动+gender生→使---发生1. (transitive)to bring about or give rise to; produce or cause使产生;造成2. to be born or cause to be born; bring or come into being产生;引起early 14c., "beget, procreate," from Old French engendrer (12c.) "give birth to, beget, bear; cause, bring about," from Latin ingenerare "to implant, engender, produce," from in-"in" (from PIE root *en "in") + generare "bring forth, beget, produce," from genus "race, kind" (from PIE root *gene- "give birth, beget," with derivatives referring to procreation and familial and tribal groups). With euphonious[juːˈfoʊniəs] -d- in French. Also from early 14c. engendered was used in a theological sense, with reference to Jesus, "derived (from God)." Meaning "cause, produce" is mid-14c. Related: Engendering.核心词3:gene [dʒiːn] n. [遗] 基因,遗传因子1911, from German Gen, coined 1905 by Danish scientist Wilhelm Ludvig Johannsen (1857-1927), from Greek genea "generation, race," from PIE root *gene-"give birth, beget."3.1 genetic 遗传的;基因的;起源的gene+-tic(形容词后缀,a suffix, equivalent in meaning to -ic, occurring in adjectives of Greek origin ( analytic))3.2 genetical 遗传的;起源的;创始的genetic +-al形容词或名词后缀核心词4:general[ˈdʒenrəl]adj. 一般的,普通的;综合的;大体的n. 一般;将军,上将;常规of wide application, generic, affecting or involving all" (as opposed to special or specific), from Old French general (12c.) and directly from Latin generalis "relating to all, of a whole class, generic" (contrasted with specialis), from genus (genitive generis) "stock, kind" (from PIE root *gene- "give birth, beget," with derivatives referring to procreation and familial and tribal groups).4.1 generally ad.4.2 generalize [ˈdʒenrəlaɪz] vt. 概括;推广;使...一般化vi. 形成概念4.3 generality [ˌdʒenəˈræləti] n. 概论;普遍性;大部分4.4 generalization, generalisation n.核心词5:generation[ˌdʒenəˈreɪʃn]n. 一代;产生;一代人;生殖early 14c., "body of individuals born about the same period" (historically 30 years but in other uses as few as 17), on the notion of "descendants at the same stage in the line of descent," from Old French generacion "race, people, species; progeny [ˈprɒdʒəni], offspring; act of procreating" (12c., Modern French génération) and directly from Latin generationem (nominative generatio) "generating, generation," noun of action from past-participle stem of generare "bring forth, beget, produce," from genus "race, kind" (from PIE root *gene- "give birth, beget," with derivatives referring to procreation and familial and tribal groups).From late 14c. as "act or process of procreation; process of being formed; state of being procreated; reproduction; sexual intercourse;" also "that which is produced, fruit, crop; children; descendants, offspring of the same parent." Generation gap first recorded 1967; generation x is 1991, by author Douglas Coupland (b.1961) in the book of that name;generation y attested by 1994. Adjectival phrase first-generation, second-generation, etc. with reference to U.S. immigrant families is from 1896. Related: Generational.5.1 generational [ˌdʒenəˈreɪʃənl] adj. 一代的;生育的5.2 regeneration [rɪˌdʒenəˈreɪʃn]n. [生物][化学][物] 再生,重生;重建5.3 generate v.5.4 generable a.5.5 generator [ˈdʒenəreɪtər] n. 发电机;发生器;生产者5.6 generative [ˈdʒenərətɪv] a. 生殖的;生产的;有生殖力的;有生产力的generate +-ive n./a.后缀→generative5.7 intergenerational a.核心词6:generic[dʒəˈnerɪk] a. 类的;一般的;属的;非商标的"of a general kind, not special. In reference to manufactured products, "not special; not brand-name; in plain, cheap packaging,"gener出生-+-ic 形容词或名词后缀from Latin gener-, stem of genus "race, kind", "give birth, beget,"6.1 generically ad.generic+-al+-ly→generically核心词7:generous [ˈdʒenərəs] a. 有雅量的,宽宏大量的,慷慨的,大方的;"of noble birth," from Middle French généreux (14c.), from Latin generosus "of noble birth," Secondary senses of "unselfish" (1690s) and "plentiful" (1610s) in English were present in French and in Latin.gener+-ous形容词后缀→-ous, suffix in adjectives构成形容词, describes something that causes or has a particular quality具有---的;有---特征的dangerous, full of danger; spacious, with much space, etc.7.1 generosity[ˌdʒenəˈrɑːsəti] n. 宽宏大量,慷慨,大方generous+ -ity抽象名词后缀→ generosity n.例如: curious + -ity→curiosity n.7.2 generously ad.7.3 generousness n.核心词8:genesis [ˈdʒenəsɪs] n. 发生;起源Genesis 创世纪from Latin genesis "generation, nativity,"E.g.The Bible begins with Genesis.核心词9:genius [ˈdʒiːniəs] n. 天才,天赋;精神from Latin genius "guardian deity or spirit which watches over each person from birth; spirit, incarnation; wit, talent;" also "prophetic skill; the male spirit of a gens," originally "generative power" (or "inborn nature"), from PIE *gen(e)-yo-, from root *gene- "give birth, beget,"根据古希腊神话,每一个人出生时都有一位介于神人之间的守护神被分派来终生守护在人的身边,而守护神的希腊文为" daimon ",拉丁文则为" genius "(birth ),即发生、起源、诞生之意。
植物F-box基因家族的研究进展

植物F-box基因家族的研究进展许克恒;张云彤;张莹;王彬;王法微;李海燕【期刊名称】《生物技术通报》【年(卷),期】2018(034)001【摘要】F-box基因家族是植物中最大的基因家族之一,由于其数量巨多,根据其蛋白C末端结构域的不同被分为不同的亚家族.F-box基因编码的蛋白能够调节多种多样的生命活动,如延缓植物衰老、调控植物开花以及响应生物胁迫、干旱和盐等逆境胁迫.近年来,随着全基因组测序的不断完善,越来越多物种的F-box基因被分析鉴定出来.已经鉴定出功能的F-box基因编码的蛋白大多能够和结合蛋白Skp1、骨架蛋白Cullin 1及Rbx1形成SCF复合体,进而参与泛素-蛋白酶途径(UPP)而发挥作用;少部分F-box蛋白以非SCF复合体形式发挥作用.泛素-蛋白酶途径(UPP)是机体重要的调节机制之一,大多数细胞内蛋白都是经过这一途径降解.主要对其蛋白结构,作用途径以及生物学功能进行概述,探讨F-box基因参与的生命活动,旨为F-box的深入研究奠定基础.%The F-box gene family is one of the largest gene families in plants. According to its protein C-terminal domain,F-box genes are divided into different subfamilies,for its number is large. Proteins encoded by F-box gene can regulate a variety of life activities,such as delaying plant senescence,regulating plant flowering and response to biological stress,drought and salt stress. In recent years,with the improvement of whole genome sequencing,more and more F-box genes of different species were identified. Most of the F-box proteins encoded by the F-box gene can form SCF complexes by binding with proteins Skp1,theskeletal proteins Cullin 1 and Rbx1,which then participates in the ubiquitin-protease pathway(UPP). A small number of F-box proteins play a role in non-SCF complexes. UPP is one of the key regulatory mechanisms,and through which the most of the intracellular proteins are degraded. This review mainly summarizes the protein structures, function pathways and biological functions of F-box gene,and explores the life activities involved by F-box gene,aiming at laying the foundation for intensively investigating F-box.【总页数】7页(P26-32)【作者】许克恒;张云彤;张莹;王彬;王法微;李海燕【作者单位】吉林农业大学生命科学学院生物反应器与药物开发教育部工程研究中心,长春 130118;吉林农业大学生命科学学院生物反应器与药物开发教育部工程研究中心,长春 130118;吉林农业大学生命科学学院生物反应器与药物开发教育部工程研究中心,长春 130118;吉林农业大学生命科学学院生物反应器与药物开发教育部工程研究中心,长春 130118;吉林农业大学生命科学学院生物反应器与药物开发教育部工程研究中心,长春 130118;吉林农业大学生命科学学院生物反应器与药物开发教育部工程研究中心,长春 130118【正文语种】中文【相关文献】1.F-box蛋白在植物生长发育中的功能研究进展 [J], 吴丹;唐冬英;李新梅;李丽;赵小英;刘选明2.植物SHMT基因家族研究进展 [J], 倪弦之;柏浩东;韩进财;罗丁峰;李祖任;胡一鸿;金晨钟3.MADS-box基因家族调控植物花器官发育研究进展 [J], 王莹;穆艳霞;王锦4.F-box蛋白参与植物逆境胁迫研究进展 [J], 许涛;夏冬健;万菁;姜书涵;宋江华5.植物转录因子HD-Zip基因家族参与逆境胁迫的研究进展 [J], 罗少;唐翠明;戴凡炜;王振江;冀宪领;吴福安;罗国庆因版权原因,仅展示原文概要,查看原文内容请购买。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
• gene families typically encode functionally related proteins, and sometimes the term gene families is refer to proteins that the genes encode
其它
乙酰胆碱 6, 8,
(M1, 9)
M2,
M3,
M4,
M5)
·
多巴胺
(D1,
D2,
D3,
D4,
D5)
·
组织胺
(H1,
H2,
H3,
H4)
·
褪黑素
(1A,
1B,
1C)
·
痕量胺相关受体
(1,
2,
3,
5,
花生酸 半胱氨酰白三烯素 (1, 2) · LTB4 (1, 2) · FPRL1 · 酮基花生酸 · 前列腺素 (D型 (1, 2) , E型 (1, 2, 3, 4) , FP) · 前列腺环素 · 血栓素
B2) 2)
· ·
趋化因子 · 胆囊收缩素 生长素 · 转移抑素 ·
(A, B) · 内皮 黄体生成素/绒毛膜促
孤儿受体
G蛋白耦合受体 63, 65, 68,
(1, 75,
3, 4, 6, 12, 15, 17, 18, 19, 20, 21, 22, 23, 77, 78, 81, 82, 83, 84, 85, 87, 88, 92, 101,
• Storeage and recall tools
https:///info/genome/compara/index.html
G-protein-coupled receptors (GPCR)
• The G protein–coupled receptor is activated by an external signal in the form of a ligand or other signal mediator.
opsin 3 opsin 4
opsin 5 retinal G protein coupled receptor rhodopsin retinal pigment epithelium-derived rhodopsin homolog
Unclassfied families
how to build a family
• get annotated genes from public databases:
o the key words of your interested gene family: (wikipedia, paper) o understand the hierarchy level of your gene families: (HGNC, InterPro) o get gene id or gene names belong to the families: (InterPro, Pfam) o get gene sequences from databases (database tools always contain them) o remove very similar sequences
Gene families main categories
•14-3-3 protein family •Homeobox (Hox gene family) •Achaete-scute complex (Neuroblast formation) •Krüppel-type zinc finger (ZNF) •MADS-box Gene Family
• This creates a conformational change in the receptor, causing activation of a G protein.
• Further effect depends on the type of G protein. • G proteins are subsequently inactivated by GTPase activating
what is gene families
• A gene family is a set of several similar genes, formed by duplication of a single original gene, and generally with similar biochemical functions
类型F:
卷曲 卷曲受体 (1, 2, 3, 4, 5, 6, 7, 8, 9, 10)
卷曲/平滑受体
平滑 平滑受体
https:///cgi-bin/genefamilies/set/139
opsin receptor family
• The visual sense: The opsins use a photoisomerization reaction to translate electromagnetic radiation into cellular signals. Rhodopsin, for example, uses the conversion of 11-cis-retinal to all-trans-retinal for this purpose
味觉 TAS1R(甜味) (1, 2, 3) · TAS2R(苦味) (1, 3, 4, 5, 8, 9, 10, 12, 13, 14, 16, 19, 20, 30, 31, 38, 39, 40, 41, 42, 43, 45, 46, 50, 60)
代谢型谷氨酸受体/信息素 其它 钙敏感受体 · γ-氨基丁酸B (1, 2) · 谷氨酸受体 (代谢型 (1, 2, 3, 4, 5, 6, 7, 8)) · G蛋白耦合受体C族6组A型 · GPR (156, 158, 179) · 维甲酸诱导 (1, 2, 3, 4)
Gene families collection
YCC
Contents
• 1. what is gene families • 2. gene families category • 3. source of databases • 4. build self gene families database
•Myosin •Kinesin •Dynein
•G-proteins •MAP Kinase •Olfactory Receptor •Receptor tyrosine kinases
Regulatory protein gene families
Motor proteins
Signal transducing proteins
proteins, known as RGS proteins.
G-protein-coupled receptors (GPCR)
细胞表面受体: G蛋白偶联受体
肾上腺素 α1 (A, B, D) · α2 (A, B, C) · β1 · β2 · β3
类型A: 视紫质类
神经递质
代谢物及
信号分子
类型B: 分泌素受体类 类型C:
孤儿受体 G蛋白耦合受体 (56, 64, 97, 98, 110, 111, 112, 113, 114, 115, 116, 123, 124, 125, 126, 128, 133, 143, 144, 155, 157)
其它
脑血管生成抑制受体 (1, 2, 3) · 钙粘蛋白 (1, 2, 3) · 降钙素 · 降钙素受体类似受体 · CD97 · 促肾上腺皮质激素释放激素 (1, 2) · 内嵌表皮生长因子粘蛋白类激素 (1, 2, 3) · 胰高血糖激素 (胰高血糖激素, 抑胃多肽, 胰高血糖激素类肽1, 胰高血糖激素类肽2) · 生长激素释放激素 · PACAPR1 · GPR · 蛛毒素 (1, 2, 3, 表皮生长因子/蛛毒素/7跨膜域集
•ABC transporters •Antiporter •Aquaporins
Transporters
•Major histocompatibility complex (MHC) •Immunoglobulin superfamily
Immune system proteins
•Expansin Gene Family •FH2 protein (formin) Gene Family •Ion channels •SNARE (protein) family •Protocadherin Gene Family
5,
6)
·
神经肽 B/W (1, 2) · FF (1, 2) · S · Y (1, 2, 4, 5) · 神经调节肽 (B, U (1, 2)) · 神经降压素 (1, 2)
其它
过敏毒素 素 (A,
(C3a, B) ·
C5a) · 血管紧张素 甲酰肽 (1, 2, 3)
(1, 2) · 爱佩琳 · 铃蟾肽 (BRS3, 胃泌素释素, NMBR) · 缓激肽 (B1, · 促卵泡激素 · 甘丙肽 (1, 2, 3) · γ-羟丁酸 · 促性腺激素释素 (1,
• get unannotated genes from newly published genomes:
o scan multiple genomes in two methods: • tblastn: • hmm scan: TreeFam HMM/InterProscan/Pfamscan
