Macro12Micro-foundation and the Behavior Analysis(宏观经济学-南开大学,龚刚)
纳米酶的应用及其伦理问题

优势以及核 心技术
该材料应用于免疫组化检测时将显色步骤减少为1步,时间 缩短为1小时,检测结果不仅可以对肿瘤性质和患者预后做出 判断,而且可以指导医师选择合适的药物进行治疗,是免疫组 化领域革命性和创新性的技术,2012年获得国家自然科学奖二 等奖。
该核心技术是在重组的24聚体Ferritin纳米小体内部包入磁 颗粒,合成一种外径12纳米,内核8纳米的双功能纳米材料, 利用这种纳米材料的蛋白外壳在癌症细胞上的特异受体 (TFR1)实现肿瘤的靶向性,同时利用该材料磁纳米颗粒内 核的酶活性催化底物显、色,从而区别正常组织和肿瘤组织,实 现癌症的及早诊断和判断。
纳米酶及纳米材料所涉及的伦理问题
科学家们因纳米技术可能对人类健康和生态环境造成消极影响而忧心忡忡。 很多纳米粒子被吸入肺中会有何影响以及纳米材料是否有毒等都成为了人 们思考的问题。目前人造纳米材料已经广泛应用到医药工业、染料、涂料、 食品、化妆品、环境污染治理等传统或新兴产业中人们在研究、生产、生 活中接触到纳米材料的机会越来越多纳米技术的应用可能引发相应的环境 伦理问题。这些问题包括:为了追求经济利益,某些机构可能在对纳米技术的 环境安全评估尚不充分的时候就推行纳米技术的产业化、肆意排放纳米废 物从而污染环境、危及他人利益和健康、造成代内不公正;对纳米技术的滥 用可能从微观层次破坏生态系统,并且这种破坏造成的危害很可能是无法挽 回的,这便违背了环境伦理学的代际公正原则。显然纳米技术有积极作用的 同时也具有一些隐患。
纳米酶及纳米材料所涉及的伦理问题
生命伦理学是指对生命科学和卫生保健领域中人类行为的系统研究, 用道德价值和原则检验此范围内人的行为。纳米技术为我们提供了一 种对医学乃至生命科学的全新认识方法和实践方法。纳米器件直接作 用于患处,纳米微粒药物可以全面渗透患处等大大提高了我们的医疗 水平,但如果不加限制地滥用,也会引发一系列的伦理问题。威胁人 类健康 由于纳米微小的身材,导致它及其不易控制。散发到空气中 的纳米微粒,被人体吸收后将会产生特殊生物机制,作用于人体心肺, 严重威胁人体健康。 纳米药物的使用不当也会产生严重后果。“是药 三分毒”,何况纳米药物特性的神秘不可预测。例如,当用纳米颗粒 作为蛋白质的载体,它一旦注入体内,纳米颗粒也会与体内的天然蛋 白质发生结合,从而干扰血液和细胞中蛋白质的功能。
结构生物学

发展
结构生物学的发展经过以下几个阶段:结构生物学起源于上世纪五十年代众所周知的Waston Crick发现DNA 双螺旋结构,建立DNA的双螺旋模型。60年代当时的开文迪许实验室的ew用X-射线晶体衍射技术获得了球蛋白的 结构。由于X射线晶体衍射技术的应用,使我们可以在晶体水平研究大分子的结构,在分子原子基础上解释了大分 子,由于他们开创性的工作,Waston Crick获得了1962年的诺贝尔生理学或医学奖,M.Pertt和J.Kendrew获得 了同年的化学奖。从那时起,技术的发展就成为结构生物学发展最重要的决定因素。60到70年代,在同一实验室 的他们又发展了电子晶体学技术,当时的研究对象主要是有序的,对称性高的生物体系,如二维的晶体和对称性 很高的三维晶体。70-80年代,多维核磁共振波谱学的发明使得在水溶液中研究生物大分子成为可能,水溶液中 的生物大分子更接近于生理状态。最近二十年,80年代到本世纪初,冷冻电镜的发明,这种技术的发明使我们不 仅能够研究生物大分子在晶体状态和溶液状态的结构,而且能够研究研究复杂的大分子体系(molecular complex)超分子体系,这就是细胞器和细胞。可见结构生物学的发展过程经历了从结晶到溶液再到大分子体系, 超分子体系,如核糖体(ribosome),病毒,溶酶体(lysosome),线粒体等。
定义
什么是结构生物学?生物大分子要发挥功能,必须满足两个条件。第一,凡要发挥功能和活性的生物大分子 必须具有特定的,自身特有,相对稳定的三级结构。第二,结构运动。没有稳定的三级结构和结构运动,生物大 分子是很难发挥生物功能或活性的。那么,结构生物学研究什么呢?按我的看法,是以生物大分子三级结构的确 定作为手段,研究生物大分子的结构功能关系,将探讨生物大分子的作用机制和原理作为研究目的。结构生物学 是近代生物学发展过程中,定量阐明生命现象的一门科学,这个是我对结构生物学的看法。生物大分子的三级结 构和结构功能研究的结构生物学已经成为生命科学当前的前沿和带头学科。
Curli Biogenesis and Function

Curli Biogenesis and FunctionMichelle M.Barnhart and Matthew R.ChapmanDepartment of Molecular,Cellular,and Developmental Biology,University of Michigan,Ann Arbor,Michigan 48109;email:chapmanm@Annu.Rev.Microbiol.2006.60:131–47First published online as a Review in Advance on May 3,2006The Annual Review of Microbiology is online at This article’s doi:10.1146/annurev.micro.60.080805.142106Copyright c2006by Annual Reviews.All rights reserved0066-4227/06/1013-0131$20.00Key Wordsamyloid,cellulose,enteric,biofilm,extracellular matrixAbstractCurli are the major proteinaceous component of a complex extra-cellular matrix produced by many Enterobacteriaceae .Curli were first discovered in the late 1980s on Escherichia coli strains that caused bovine mastitis,and have since been implicated in many physiologi-cal and pathogenic processes of E.coli and Salmonella spp.Curli fibers are involved in adhesion to surfaces,cell aggregation,and biofilm formation.Curli also mediate host cell adhesion and invasion,and they are potent inducers of the host inflammatory response.The structure and biogenesis of curli are unique among bacterial fibers that have been described to date.Structurally and biochemically,curli belong to a growing class of fibers known as amyloids.Amyloid fiber formation is responsible for several human diseases including Alzheimer’s,Huntington’s,and prion diseases,although the process of in vivo amyloid formation is not well understood.Curli provide a unique system to study macromolecular assembly in bacteria and in vivo amyloid fiber formation.Here,we review curli biogenesis,regulation,role in biofilm formation,and role in pathogenesis.131A n n u . R e v . M i c r o b i o l . 2006.60:131-147. D o w n l o a d e d f r o m w w w .a n n u a l r e v i e w s .o r g b y O h i o S t a t e U n i v e r s i t y L i b r a r y o n 11/07/13. F o r p e r s o n a l u s e o n l y .CR:Congo red csg :curli specific geneContentsINTRODUCTION.................132CURLI BIOGENESIS..............132CURLI GENE REGULATION.. (135)CURLI ARE PART OF THE BACTERIALEXTRACELLULAR MATRIX...137THE ROLE OF CURLI INBIOFILM FORMATION........137CURLI ARE A BACTERIALAMYLOID ......................138ROLE OF CURLI INPATHOGENESIS (140)ROLE OF CURLI IN ATTACHMENT ANDINVASION OF HOST CELLS..141CONCLUSIONS...................141INTRODUCTIONBacteria are able to integrate and survive in a remarkably diverse collection of environ-ments.In recent years,bacterial communi-ties have been better appreciated as an inte-gral part of most microbial lifestyles.These communities,or biofilms,are prominent dur-ing infections and are generally characterized by an extracellular matrix that can help sculpt three-dimensional structures,which promote the survival of its inhabitants in the face of environmental stresses (20,21).Community behavior is complex and can involve many genetic loci.Genetic loci involved in com-munity behaviors often encode extracellu-lar factors that promote surface colonization or cell-cell contact.Enteric bacteria such as Escherichia coli and Salmonella spp.express pro-teinaceous extracellular fibers called curli that are involved in surface and cell-cell contacts that promote community behavior and host colonization (3,31,32,88,89).Understand-ing the biogenesis of structures that promote biofilm formation,such as curli,is a prereq-uisite to the development of therapeutics that can attenuate biofilm formation and host col-onization.Here,we discuss curli regulation and biogenesis and the role of these fibers in the lifestyle of enteric bacteria such as E.coli .CURLI BIOGENESISCurliated bacteria stain red when grown on plates supplemented with the diazo dye Congo red (CR),providing a convenient way to identify genes important for curli produc-tion (17).At least six proteins,encoded by the divergently transcribed csgBA and csgDEFG operons,are dedicated to curli formation in E.coli (33)(Figure 1).Homologous oper-ons have been identified in Salmonella spp.and are called agfBA and agfDEFG (16,67).Salmonella typhimurium agf genes can com-plement mutations in E.coli csg genes (67).In S.typhimurium the fibers encoded by the agf operons are called T afi(thin aggregative fimbrae).For the purposes of this review we refer to structures encoded by the agf and csg operons as curli.The csgBA operon encodes the major structural subunit,CsgA,and the nucleator protein CsgB (33,34).A third gene,csgC ,is in the csgBA operon,but no transcript for csgC has been detected and there is no re-ported role for CsgC in curli biogenesis (16,33).CsgA and CsgB are proteins of identical predicted size,share 30%sequence identity,and are built up of similar repeat motifs (34).In the absence of CsgB,curli are not assem-bled and the major subunit protein,CsgA,is secreted from the cell in an unpolymerized form (14,34).CsgA and CsgB do not have to be expressed from the same cell for curli assembly to occur.During a process called in-terbacterial complementation,a csgB mutant cell secretes soluble CsgA that can be assem-bled on the surface of a cell expressing only csgB (34)(Figure 2a ).Interbacterial comple-mentation is best illustrated with established nomenclature in which the strain that secretes soluble CsgA is the donor,and the strain that presents CsgB on its cell surface is the ac-ceptor (Figure 2a ).In E.coli ,interbacterial complementation can work when strains are132Barnhart·ChapmanA n n u . R e v . M i c r o b i o l . 2006.60:131-147. D o w n l o a d e d f r o m w w w .a n n u a l r e v i e w s .o r g b y O h i o S t a t e U n i v e r s i t y L i b r a r y o n 11/07/13. F o r p e r s o n a l u s e o n l y .Figure 1Model of curli assembly.A schematic diagram of the two curli gene operons is shown (bottom ).CsgD is a positive transcriptional regulator of the csgBA operon.All the proteins encoded by the csg operons,except for CsgD,contain sec signal sequences for translocation into the periplasm.CsgG is an outer membrane protein required for the secretion of the two curli structural subunits CsgA and CsgB.CsgA is secreted outside of the cell where CsgB nucleates it into a fiber.CsgE and CsgF both interact with CsgG and are required for efficient curli assembly.grown within a few millimeters of each other.However,in Salmonella enterica interbacterial complementation was not observed between donor and acceptor strains (86),suggesting an alternative mechanism for curli assembly.When mutations are made in lipopolysaccha-ride (LPS)O polysaccharide in S.enterica,in-terbacterial complementation can occur (87).The K-12E.coli strains that were first used to study interbacterial complementation are LPS O polysaccharide deficient.Interbacterial complementation has led to the hypothesis that curli assemble via the nu-cleation precipitation pathway (7,47).Nucle-ation precipitation is based on the idea that CsgA is secreted into the extracellular milieu and nucleated into a fiber by CsgB.However,LPS:lipopolysaccharide Wt:wild type OM:outer membraneuntil recently there was no evidence that CsgA is secreted from wild-type (Wt)cells in an unpolymerized form.Cherny and colleagues (65)showed that small,rationally designed peptides could abrogate curli formation when added to the media.These peptides are pre-dicted to block reactive surfaces on CsgA that would prevent it from assembling into a fiber.Because the peptides are not thought to enter the cell,the assumption is that CsgA polymer-ization was blocked at the cell surface.The csgDEFG operon encodes four ac-cessory proteins required for curli assembly (33).CsgD is a positive transcriptional reg-ulator of the csgBA operon and is discussed in more detail in the section on curli gene regulation.The roles of CsgE,CsgF ,and •Curli Biogenesis and Function133A n n u . R e v . M i c r o b i o l . 2006.60:131-147. D o w n l o a d e d f r o m w w w .a n n u a l r e v i e w s .o r g b y O h i o S t a t e U n i v e r s i t y L i b r a r y o n 11/07/13. F o r p e r s o n a l u s e o n l y .Figure 2Phenotype of csg mutant strains after growth on CR-indicator plates.(a )A CR-indicator plate demonstrating interbacterial complementation.csgA −/csgB +acceptor and csgA +/csgB −donor strains were struck from top to bottom of aCR-indicator plate.csgA −and csgB −strains were struck from left to right on the same CR-indicator plate.When a csgA −strain is cross-streaked with a csgB −strain,CR binding occurs.A CR-indicator plate of wild-type (Wt),csgA −,csgE −,and csgF −strains is shown after (b )30and (c )48h of growth at 26◦C.CsgG are just beginning to be elucidated.CsgG is an outer membrane (OM)lipopro-tein that is required for the stability and se-cretion of CsgA and CsgB (14,47).When streaked on CR-indicator plates,csgG mutants stain white and no fibers are visualized by electron microscopy (EM).csgG mutants do not act as acceptors or donors during inter-bacterial complementation,suggesting that no functional CsgA or CsgB is produced by these cells (64).CsgG interacts with itself and purified CsgG visualized by high-resolution EM forms oligomeric,ring-shaped complexes (Figure 3).These structures are analogous to those formed by other OM channel-forming proteins (10,77,78).Overexpression of CsgG is also corre-lated with pore formation in the OM.Nor-mally,gram-negative bacteria are resistant to erythromycin because this 741-Da antibi-otic does not efficiently cross the OM.How-ever,if the integrity of the OM is breached,erythromycin can access the bacterial cyto-plasm and poison translation.Expression of CsgG renders E.coli sensitive to erythromycin (64),suggesting that CsgG is able to per-meabilize the OM.The possibility remains that CsgG only modifies the activity of an-other,yet unidentified,protein that is ulti-mately responsible for curli subunit transloca-tion across the OM.Because CsgG has been purified to homogeneity,in vitro liposome swelling or voltage-gating experiments should determine whether CsgG could form pores in the absence of other proteins.The CsgG-mediated secretion of CsgA is dependent on the N-terminal 22amino acids of the mature CsgA protein.These residues are not predicted to be an integral part of the curli fiber (19).However,these residues are required for CsgA to be secreted and assem-bled into a fiber (64).When the N-terminal 22amino acids of CsgA are fused to PhoA (alka-line phosphatase),the resulting CsgA/PhoA chimera forms a complex with CsgG at the OM.This suggests that the N-terminal 22amino acids on CsgA provide specificity for secretion of the major curli subunit.134Barnhart·ChapmanA n n u . R e v . M i c r o b i o l . 2006.60:131-147. D o w n l o a d e d f r o m w w w .a n n u a l r e v i e w s .o r g b y O h i o S t a t e U n i v e r s i t y L i b r a r y o n 11/07/13. F o r p e r s o n a l u s e o n l y .CsgE is a periplasmic protein and csgE mu-tants are defective in curli assembly as visual-ized by their inability to bind CR when grown on CR-indicator plates (14)(Figure 2b ,c ).CsgA and CsgB stability is greatly reduced in csgE mutants,which do not act as donors or acceptors during interbacterial complementa-tion.csgE mutants produce a few CsgA fibers,but these fibers are morphologically distinct from those produced by Wt cells (14).CsgE also physically interacts with CsgG at the OM (64).csgE and csgG mutants have simi-lar phenotypes and these two proteins likely work together to promote curli assembly,al-though the molecular role of CsgE is currently unknown.CsgF is a periplasmic protein that also interacts with CsgG in the OM (64),but csgF mutants have a phenotype different from that of csgE mutants.csgF mutants stain pink when streaked onto CR-indicator plates (14)(Figure 2b ,c ).csgF mutants are phenotypi-cally similar to csgB mutants because they se-crete soluble,unpolymerized CsgA (14)and therefore act as donors,but not acceptors,during interbacterial complementation.CURLI GENE REGULATIONThe regulation of curli gene expression is extraordinarily complex and is responsive to many environmental cues (28).The inter-genic region between the csgDEFG and the csgBA operons is one of the largest in E.coli .At the center of the curli regulatory network is the CsgD protein,which is a transcriptional regulator in the FixJ/UhpA family (33).CsgD positively regulates the csgBA operon (33),but unlike most transcriptional regulators it does not regulate its own expression (69).CsgD contains an N-terminal receiver domain and a C-terminal helix-turn-helix DNA binding domain.Although CsgD is proposed to reg-ulate the csgBA promoter directly,there is no experimental evidence to demonstrate CsgD DNA binding activity.It is also unclear what stimulates CsgD expression and/or activity,but activation of CsgD is thought toresultFigure 3High-resolution EM of purified CsgG.Purified CsgG was visualized by high-resolution EM (left ).At least two shapes of CsgG were observed:donut-like structures,which might indicate a top or bottom view of CsgG oligomers,and cylinders,which might represent a side view of the CsgG oligomeric complex (right ).EM:electron microscopyfrom phosphorylation of a conserved aspar-tic acid residue in the N-terminal receiver domain (69).Because CsgD is absolutely re-quired for csgBA promoter activity,it is not surprising that regulators of CsgD expression influence csgBA expression.One of the first conditions recognized to promote curli gene expression was growth at temperature below 30◦C (2,54).For most lab-oratory strains of E.coli and most Salmonella strains,curli expression is best at temperatures below 30◦C.However,it has been demon-strated that many clinical strains of E.coli ,including sepsis isolates,can express curli at 37◦C (6).Furthermore,mutations in the csgD promoter can result in strains that express curli regardless of temperature (70,81).In addition to temperature,other envi-ronmental conditions also influence curli ex-pression.Curli expression occurs maximally in media without salt (70).Nutrient limita-tion (nitrogen,phosphate,and iron)stimu-lates curli gene expression (27,70).Oxygen tension also plays a role in curli expression,with microaerophilic conditions resulting in maximal csgD transcription (27,67). •Curli Biogenesis and Function135A n n u . R e v . M i c r o b i o l . 2006.60:131-147. D o w n l o a d e d f r o m w w w .a n n u a l r e v i e w s .o r g b y O h i o S t a t e U n i v e r s i t y L i b r a r y o n 11/07/13. F o r p e r s o n a l u s e o n l y .Table 1Regulators of curli gene expression Regulator P csgBA a P csgDEFG aReference(s)CsgD +33OmpR/EnvZ +62,67,85RpoS +1,9,52,60Crl +CpxA/R −−23,39Rcs −−24,39,84MlrA +11IHF +26HN-S+or −b1,26,52aActivation or repression of the promoter is indicated by a +or −,respectively.No +or −indicates that no effect on the promoter has been demonstrated.A bold +or −indicates that direct binding to the promoter was observed.bThe role of HN-S in curli gene expression is dependent on the strain background.In E.coli K-12HN-S has a negative role (1,52),whereas in S.typhimurium HN-S positively influences curli gene expression (26).A number of regulatory systems con-tribute to the expression of the curli operons (Table 1).RpoS,the stationary-phase sigma factor,plays a key role in curli gene regulation both directly and indirectly (1,52).Curli genes are maximally expressed during station-ary phase and their expression is dependent upon RpoS (1).Crl interacts with RpoS to fa-cilitate RpoS binding to the csgBA promoter region,and therefore Crl is required in most strains for curli expression (2).However,some strains express curli independent of Crl (63).Crl and RpoS cooperatively regulate other stationary-phase-induced genes (60).Crl was proposed to be the thermal sensor that maxi-mized curli operon expression at low temper-atures (2),and it was recently shown that the Crl protein is more stable at lower temper-atures (9).Selective stability of Crl at lower temperatures may explain the propensity of curli fibers to be maximally expressed below 30◦C,although thermal regulation of curli expression occurs in strains that do no have the Crl protein (M.M.Barnhart &M.R.Chapman,unpublished observations).RpoS also modulates curli gene expression by ac-tivating MlrA expression,which is a positive transcriptional regulator of csgD (11).Three two-component regulatory systems regulate curli gene expression:OmpR/EnvZ,CpxA/R,and Rcs (24,39,62,70,84,85).Of these two-component regulatory systems the OmpR/EnvZ has the most dramatic effect on curli gene regulation.The OmpR/EnvZ sys-tem responds to changes in osmolarity and regulates the porins OmpF and OmpC (59).EnvZ is the sensor kinase that senses a sig-nal it transmits to the OmpR response regula-tor,which modulates gene expression.OmpR works by positively regulating csgD expres-sion,and in an ompR mutant there is no csgD transcription (70).The ompR234allele,which has a leucine-to-arginine change at position 43,constitutively activates curli gene expres-sion and promotes biofilm formation in strains that normally do not make biofilms (85).The CpxA/R system is activated in re-sponse to envelope stress and/or misfolded periplasmic proteins,resulting in the upreg-ulation of many periplasmic chaperones and proteases (36).CpxA is the sensor kinase and CpxR is the response regulator that modulates gene expression.CpxA/R negatively regulates both curli operons.Overexpression of csgA in the absence of a corresponding overexpres-sion of csgG results in the activation of the Cpx pathway (23,61,62).In fact,it has been diffi-cult to experimentally detect the curli subunit proteins as they pass through the periplasm during curli assembly,suggesting that they are only transiently present in the periplasm.The Cpx pathway also regulates the P pilus system.P pilus subunits that misfold in the periplasm are recognized by the Cpx system and are rapidly degraded by the proteases present in the Cpx regulon (36,44,71).It is tempting to speculate that the Cpx system is at least partly responsible for the apparent instability of curli subunits in the periplasm.Similar to Cpx,the Rcs pathway responds to membrane stress,specifically OM stress,and is best known for its positive regulatory effect on capsule synthesis (49,50).The Rcs pathway negatively regulates csgD expression (24,39,84).The Rcs pathway is also required for biofilm formation (24).Thus,regulation136Barnhart·ChapmanA n n u . R e v . M i c r o b i o l . 2006.60:131-147. D o w n l o a d e d f r o m w w w .a n n u a l r e v i e w s .o r g b y O h i o S t a t e U n i v e r s i t y L i b r a r y o n 11/07/13. F o r p e r s o n a l u s e o n l y .of the curli operons seems to be tightly cou-pled to the ability to form biofilms (23).It is interesting to note that both Rcs and Cpx negatively regulate curli operon expression.Therefore,curli may be important only dur-ing the initial stages of biofilm formation,pos-sibly for initial adhesion,and then turned off as the Cpx and Rcs pathways become active during biofilm maturation.T wo global regulatory proteins,histone-like protein (HN-S)and IHF (integration host factor),have been implicated in curli gene expression.Both of these proteins modulate DNA architecture.In S.typhimurium deletion of ihf reduced csgD transcription and there-fore curli production (28).The role of HN-S in curli gene expression is more complicated.In S.typhimurium deletion of hns results in a decrease in csgD transcription,suggesting that HN-S is a positive regulator of curli gene ex-pression (26).However,in E.coli K-12strains hns mutants cause an increase in csgA tran-scription,suggesting that HN-S negatively affects curli gene expression (1).These re-sults underscore the complexity of curli gene expression.CURLI ARE PART OF THEBACTERIAL EXTRACELLULAR MATRIXRecently,it has been shown that many enteric bacteria express different morphotypes,which correspond to differences in the extracellular matrix that they produce (66,69,88).S.ty-phimurium and E.coli produce an extracellu-lar matrix that features curli as the major pro-teinaceous component.Cellulose is a second component of the matrix,and a third polysac-charide component is proposed to be present,although its identity is unknown (87).Differ-ent morphotypes can be visualized by grow-ing bacteria on CR-indicator plates.The four described morphotypes are rdar (red,dry,and rough;curli and cellulose),pdar (pink,dry,and rough;cellulose only),bdar (brown,dry,and rough;curli only),and saw (smooth and white;neither curli nor cellulose)(88,89)(Figure 4).MC4100,the K-12E.coli strain that has been used to study curli assembly,does not produce cellulose,whereas pathogenic and commensal E.coli isolates can produce cellulose,curli,or both (8,88).CsgD plays an integral role in ex-tracellular matrix production because it regu-lates curli gene expression and indirectly reg-ulates cellulose production by activating adrA (33,69,89).AdrA synthesizes cyclic-di-GMP ,which is required to produce cellulose (72),but does not regulate cellulose gene expres-sion (89).Recently it has been demonstrated that cyclic-di-GMP is an important signaling molecule in bacteria,but exactly how it works to modulate gene expression has yet to be elucidated (22,68).THE ROLE OF CURLI IN BIOFILM FORMATIONBiofilms,communities of bacteria that live together for the benefit of the group,are char-acterized by water channels,complex three-dimensional structures,and an increased re-sistance to environmental stresses.Curli are important for biofilm development in both E.coli and Salmonella spp.Biofilm formation is a multi-step developmental process that in-cludes at least five distinguishable steps:(a )re-versible attachment,(b )irreversible attach-ment and production of adhesive molecules such as exopolysaccharides and adhesions,(c )biofilm development characterized by a dis-tinct mushroom shape,(d )biofilm maturation,and (e )biofilm dispersal.Many bacterial sur-face structures,including curli,flagella,pili,Figure 4Morphotypes of Salmonella typhimurium grown on CR-indicator plates for 48h at 26◦C.Figure was kindly provided by Ute Romling. •Curli Biogenesis and Function137A n n u . R e v . M i c r o b i o l . 2006.60:131-147. D o w n l o a d e d f r o m w w w .a n n u a l r e v i e w s .o r g b y O h i o S t a t e U n i v e r s i t y L i b r a r y o n 11/07/13. F o r p e r s o n a l u s e o n l y .ThT:thioflavin Tand exopolysaccharide,play roles in various aspects of biofilm development (83).Biofilms can be problematic in the food industry and hospital settings.Curli allow Salmonella enteriditis to adhere to T eflon and stainless steel,which can lead to biofilm formation and contamination of surfaces often used in the food industry (3).In a screen used to identify genes that allow a nonadherent strain of E.coli to form biofilms,an allele of ompR called ompR234(L 43R)that activated curli gene expression was iden-tified (85).The ability of this strain to form a biofilm was dependent on csgA .These results suggested that curli were important in the ini-tial stages of biofilm development during the attachment phase.Recent work has suggested that biofilms formed by curli-proficient strains have a morphology different from biofilms formed by curli-deficient strains.Curli-deficient strains form flat biofilms on polyurethane sheets,compared with the ma-ture biofilms produced by curli-expressing strains (40).During biofilm formation,E.coli K-12is able to produce curli at 37◦C,even though on agar plates or in static broth it is able to express curli at only 26◦C (40).CURLI ARE A BACTERIAL AMYLOIDThe biochemical and structural properties of curli are fascinating.Curli share many dis-tinguishing biochemical and structural prop-erties with eukaryotic amyloid fibers.Amy-loid fiber formation is traditionally associated with human diseases including Alzheimer’s,Parkinson’s,and prion diseases;however,curli are part of a growing number of functional amyloids (15).Like eukaryotic amyloid fibers,curli are nonbranching (14,54)(Figure 5),β-sheet rich fibers that are resistant to pro-tease digestion and 1%sodium dodecyl sulfate (17–19).Regardless of their origin,allamyloidFigure 5Electronmicrographs of curli.Negative-stain EMs of (a )a bacterium-expressing curli and (b )bacteria not expressing curli are shown.(c )High-resolution EM of purified CsgA fibers.138Barnhart·ChapmanA n n u . R e v . M i c r o b i o l . 2006.60:131-147. D o w n l o a d e d f r o m w w w .a n n u a l r e v i e w s .o r g b y O h i o S t a t e U n i v e r s i t y L i b r a r y o n 11/07/13. F o r p e r s o n a l u s e o n l y .Figure 6Model of CsgA and CsgB structure.(a )A model of the predictedstrand-loop-strand motif of CsgA (19).A similar prediction was made for CsgB (86).(b )CsgA and (c )CsgB are composed of five repeating units (R1–R5),and each repeating unit is the equivalent of one strand-loop-strand.Residues that are conserved within the repeating units are colored the same.The red residues in R5of (c )CsgBindicate residues that differ from R1–R4of CsgB.fibers cause a red shift when mixed with CR (42,43)and cause fluorescence when mixed with thioflavin T (ThT)(45,46).These two dyes have been used as a diagnostic for amy-loid formation.Curli cause a red shift when mixed with CR and a 10-to 20-fold increase in ThT fluorescence (13,14).The hallmark of amyloid fibers is the conserved cross β-strand structure,in which condensed β-sheets are stacked parallel to the fiber axis and in-dividual β-strands are perpendicular to the fiber axis (75,76).Computer modeling of the two curli subunits CsgA and CsgB predicts that they form a similar structure composed of five repeating strand-loop-strand motifs (19,86)(Figure 6).Each repeating unit is composed of conserved glycines,glutamines,and asparagines (Figure 6).Many eukaryotic amyloids are also rich in glutamine and as-paragine residues (51,56).The glutamine and asparagine residues are predicted to form a hy-drogen bond network that might contribute to the extreme stability of these fibers (57,58).Curli represent a novel twist to amyloid for-mation because mammalian amyloid forma-tion is generally considered an off-pathway protein-folding event,but curli are the prod-uct of a highly regulated and directed process.In addition to curli,there is a growing list of functional amyloids that have been described •Curli Biogenesis and Function139A n n u . R e v . M i c r o b i o l . 2006.60:131-147. D o w n l o a d e d f r o m w w w .a n n u a l r e v i e w s .o r g b y O h i o S t a t e U n i v e r s i t y L i b r a r y o n 11/07/13. F o r p e r s o n a l u s e o n l y .AA:amyloid protein At-PA:tissue type plasminogen activatorin yeast,fungi,and mammals (7a,25a,76a,79a).Therefore,the amyloid fold is not just a biological mishap,but an important part of cellular physiology.Westermark and colleagues (48)recently reported that injection of curli fibers into mice resulted in increased polymerization of the disease-associated amyloid protein A (some-times called secondary amyloid protein,or AA).AA fibril formation is a manifestation of chronic inflammatory disease,the result of which is severe tissue damage and morbidity.Like other amyloid-associated diseases,a key question in the pathogenesis of AA is,What are the underlying causes of the conversion of AA from a soluble protein to an amyloid fiber?The work of Westermark suggests that one possibility could be that heterologous amy-loid fibers such as curli act as a seed to drive AA polymerization.E.coli and other enteric bacteria that express curli are found as part of our normal gut flora and many of these bacte-ria can also cause diseases such as sepsis,which might allow the direct interaction of AA and curli fibers.ROLE OF CURLI IN PATHOGENESISMany extracellular surface fibers produced by bacteria are important in pathogenesis.A uni-fying role of curli in pathogenesis has not been elucidated,but several lines of evidence sug-gest that curli are important during the infec-tious process.Curli have been implicated inTable 2Proteins that interact with curliProtein FunctionReference(s)Fibronectin Extracellular matrix protein 17,54Laminin Extracellular matrix protein 52MHC class I Antigen-presenting molecule 55TLR2Innate immune system activation 80Plasminogen Serine protease that degrades soft tissue when activated to plasmin 73t-PA Converts plasminogen to plasmin H-Kininogen Contact-phase proteins5,35,53Fibrinogen Factor XIIthe attachment and invasion of host cells,in-teraction with host proteins,and activation of the immune system.Curli bind to the extra-cellular matrix proteins fibronectin (54)and laminin (52).However,the role of curli dur-ing host colonization may not be fully appre-ciated,because it was thought that curli were expressed only at temperatures below 30◦C (54).It is now known that curli expression is strain and condition specific and that many enterics express curli under conditions found in the host.Curli are expressed in biofilms at 37◦C (40)and mutations in the csgD promoter region can also result in curli expression at 37◦C (70,81,82).Furthermore,many clinical isolates express curli at 37◦C.Curli bind to many host proteins (Table 2).Many of the proteins with which curli interact are proposed to facilitate bacte-rial dissemination through the host (6).Curli bind to the tissue-degrading enzyme plas-minogen.Plasminogen is a serine protease that degrades fibrin and soft tissue and must be activated from its proenzyme form (12).Tissue type plasminogen activator (t-PA)activates plasminogen to plasmin.Curli bind to plasminogen and t-PA simultaneously,resulting in the activation of plasminogen to plasmin (73).By activating plasminogen,curliated bacteria might gain an advantage inside the host because this enzyme degrades soft tissue,which would allow the bacteria to gain access to deeper tissue.Curliated bacteria and curli also bind to human contact-phase proteins including H-kininogen,fibrinogen,and factor XII (5,35,53).By binding to the contact-phase proteins,curliated bacteria slow clotting,which could increase the spread of bacteria to surround-ing tissue (35).Curli have been implicated in sepsis because antibodies to CsgA were present in the sera from sepsis patients (6).Furthermore,bacteria isolated from these pa-tients were capable of expressing curli at 37◦C.Because curli bind to contact-phase proteins and plasminogen/t-PA,curliated bacteria may have an advantage in spreading throughout the body and thus might play a role in sepsis.140Barnhart·ChapmanA n n u . R e v . M i c r o b i o l . 2006.60:131-147. D o w n l o a d e d f r o m w w w .a n n u a l r e v i e w s .o r g b y O h i o S t a t e U n i v e r s i t y L i b r a r y o n 11/07/13. F o r p e r s o n a l u s e o n l y .。
人类基因组-转化医学-精准医学

克隆与表达(蛋白质) 建立筛选实验方法 纯 化 生物筛选
新药研发国家实验室 药物所、复旦药学院 筛选天然产物 组合化学库 传统医学 植物化学组
中医 民族医药学
先导化合物
Phytochemomics
中草药中心
张浦 江东 : 国 家 「 药 谷
传统药物化学 组合化学 代谢物组学 化学基因组学 药物基因组学
生物学数据: 内在结构 Pre-designed 生物学数据: 非标准+高噪声 标准化+QA 真态 Realness Noise+Incomplete+ Biased+Anomaly
动态 Dynamics High speed +Real time
生物学数据: 整合、管理 交叉、共享
量态 Quantitation Mass data (EB)
MTB: Molecular Tumor Board
2A Yes AKT1 AR EBV FGFR1 FGFR2 FGFR3 FGFR4 HRAS JAK1 KDR MAP2K1 MAP2K2 MET NF1 FOLR1 NRAS PDGFR PIK3CA PIK3CD PIK3R1 RET SMO SRC STK11 TOP1 TSC1 TSC2
系统生物学
健康医学
动物
生科院 及高校研究基础
动物模型(比较医学)
B2B2B
转化型医学研究
临床医院 及社区样本基地
4P:
Personalized Predictive
(个体化) (预测)
Preemptive
(干预)
Participatory
(参与)
六十而耳顺:生物大数据
状态 Status Complex+ Non-structural
仿生润湿阵列微芯片开发和生物医学应用

仿生润湿阵列微芯片开发和生物医学应用下载提示:该文档是本店铺精心编制而成的,希望大家下载后,能够帮助大家解决实际问题。
文档下载后可定制修改,请根据实际需要进行调整和使用,谢谢!本店铺为大家提供各种类型的实用资料,如教育随笔、日记赏析、句子摘抄、古诗大全、经典美文、话题作文、工作总结、词语解析、文案摘录、其他资料等等,想了解不同资料格式和写法,敬请关注!Download tips: This document is carefully compiled by this editor. I hope that after you download it, it can help you solve practical problems. The document can be customized and modified after downloading, please adjust and use it according to actual needs, thank you! In addition, this shop provides you with various types of practical materials, such as educational essays, diary appreciation, sentence excerpts, ancient poems, classic articles, topic composition, work summary, word parsing, copy excerpts, other materials and so on, want to know different data formats and writing methods, please pay attention!引言仿生润湿阵列微芯片是一种结合仿生学原理与微技术的创新产物,其在生物医学领域具有广阔的应用前景。
高通量自动化类器官芯片研究进展

天津医药2024年1月第52卷第1期1专题研究·类器官与器官芯片编者按:类器官与器官芯片作为近年来兴起的一种新型强大研究手段,在实现干细胞体外增殖与分化、模拟器官内细胞间互作方式、重现来源脏器形态学特征与功能、优化精准药物筛选方案和建立罕见病个体化治疗等方面发挥重要作用,已引起国内外研究人员、临床工作者和药物研发公司的广泛关注。
目前已应用于包括肺、脑、肠在内的多种脏器的病理生理学研究与转化医学研究。
未来结合工程学、基因编辑等手段进行类器官与器官芯片个性化、大规模、均质化、标准化和自动化的完善与优化将给个体化医学带来深远影响。
我受《天津医药》编辑部邀约,特组织国内类器官与器官芯片研究领域的知名专家共同就肺、脑和肠这3种研究进展最快的类器官与器官芯片的构建及其在炎症反应、病毒感染、抗肿瘤药物筛选和罕见病等方面的应用进行阐述,并对目前关于该技术待解决的关键问题进行深入讨论。
本专题旨在加深各学科基础研究、临床和药物研发同道对类器官与器官芯片的认识,共同推动该技术的深入应用,为疾病机制研究、临床诊断与目标药物发现提效增速。
由于篇幅限制,本专题未涉及包括肝、肾、胃和胰腺等其他脏器的类器官与器官芯片的研究进展,但是这方面的研究同样值得关注。
最后,本人对参与和支持本专题工作的全体作者和编审专家所付出的辛勤工作表示由衷感谢!陈怀永(专题主编)高通量自动化类器官芯片研究进展孟繁露,韩益明,修继冬,黄建永△摘要:类器官是利用干细胞的自更新和自组织能力在体外构建的三维多细胞培养物,其复现了对应器官或组织的关键结构和功能特征,为发育生物学、再生医学、疾病建模和药物开发等领域提供了理想的体外模型和研究平台。
然而,传统的类器官培养体系依赖于手动操作,培养流程较为繁琐,且培养物个体差异和批次差异较大,成为限制类器官转化和应用的重要原因之一。
因此,工程化类器官培养体系通过引入微流控芯片技术来提升类器官培养体系的通量和自动化程度,对于实现类器官大规模、均质化、标准化培养具有重要意义。
(2008)Dimensionality reduction: A comparative review

L.J.P. van der Maaten ∗ , E.O. Postma, H.J. van den Herik
MICC, Maastricht University, P.O. Box 616, 6200 MD Maastricht, The Netherlands.
22 February 2008
the number of techniques and tasks that are addressed). Motivated by the lack of a systematic comparison of dimensionality reduction techniques, this paper presents a comparative study of the most important linear dimensionality reduction technique (PCA), and twelve frontranked nonlinear dimensionality reduction techniques. The aims of the paper are (1) to investigate to what extent novel nonlinear dimensionality reduction techniques outperform the traditional PCA on real-world datasets and (2) to identify the inherent weaknesses of the twelve nonlinear dimenisonality reduction techniques. The investigation is performed by both a theoretical and an empirical evaluation of the dimensionality reduction techniques. The identification is performed by a careful analysis of the empirical results on specifically designed artificial datasets and on the real-world datasets. Next to PCA, the paper investigates the following twelve nonlinear techniques: (1) multidimensional scaling, (2) Isomap, (3) Maximum Variance Unfolding, (4) Kernel PCA, (5) diffusion maps, (6) multilayer autoencoders, (7) Locally Linear Embedding, (8) Laplacian Eigenmaps, (9) Hessian LLE, (10) Local Tangent Space Analysis, (11) Locally Linear Coordination, and (12) manifold charting. Although our comparative review includes the most important nonlinear techniques for dimensionality reduction, it is not exhaustive. In the appendix, we list other important (nonlinear) dimensionality reduction techniques that are not included in our comparative review. There, we briefly explain why these techniques are not included. The outline of the remainder of this paper is as follows. In Section 2, we give a formal definition of dimensionality reduction. Section 3 briefly discusses the most important linear technique for dimensionality reduction (PCA). Subsequently, Section 4 describes and discusses the selected twelve nonlinear techniques for dimensionality reduction. Section 5 lists all techniques by theoretical characteristics. Then, in Section 6, we present an empirical comparison of twelve techniques for dimensionality reduction on five artificial datasets and five natural datasets. Section 7 discusses the results of the experiments; moreover, it identifies weaknesses and points of improvement of the selected nonlinear techniques. Section 8 provides our conclusions. Our main conclusion is that the focus of the research community should shift towards nonlocal techniques for dimensionality reduction with objective functions that can be optimized well in practice (such as PCA, Kernel PCA, and autoencoders).
转基因动物在microRNA研究中的应用

·1096·
生物化学与生物物理进展
Prog. Biochem. Biophys.
2009; 36 (9)
Stefan 等应用转基因技术建立了 miR-155 过表达的 小鼠模型 . [24] 2007 年 Zhao 等[25]应用基因打靶技术 建 立 了 miR-1-2 的 基 因 敲 除 模 型 . 近 三 年 , 与
“miRNA 和转基因动物”相关的研究数量更是不断 增长,建立策略不断丰富. miRNA 转基因动物模 型正成为 miRNA 功能研究的有力工具(表 1).
Table 1 The functions of microRNAs revealed by using transgenic mouse 表 1 应用转基因小鼠证实的 microRNA 功能
从“转基因”表达水平的结果上看,转基因动 物模型可分为:过表达(overexpression)、基因敲除 (knockout, KO) 及 基 因 敲 减 (knockdown). 通 常 , “转基因动物”也可单指外源基因“过表达”的动 物. 基因不同水平的表达导致其产物生成量的差 异,最终动物出现不同表型(phenotype),从而为不 同研究提供材料.
MicroRNA 的表达具有阶段和组织特异性,有 些 microRNA 在胚胎发育过程中起着关键作用,有 些 microRNA 只在特定组织表达. 这完全适合运用 条件基因打靶(conditional gene targeting)技术对其进 行功能研究. 在传统基因打靶过程中,microRNA 从发育的开始就在其全部脏器内被“敲除”,这会 影响胚胎发育,甚至造成胎死(embryonic lethality) 现象. 而条件基因敲除的应用很好地解决了这一问 题. 目前,重组酶 Cre-LoxP 系统已在 microRNA 基因敲除动物中有所应用. 2007 年,van Rooij 等[41] 报道了心肌细胞条件敲除 miR-208(图 1c)对 琢MHC 基因表达的影响,进一步揭示了其对心脏发育的作 用. 该技术需要一种含组织特异性表达 Cre 重组酶 的转基因动物,同时经同源重组建立另一种含 LoxP 重组酶位点的基因打靶动物,二者杂交产 生的后代,就可产生组织特异性的 miRNA 敲除的 动物.
细胞龛中造血干细胞和祖细胞的活体示踪

活体动物位于其龛中的造血干细胞/祖细胞的追踪Abstract干细胞留驻在一个特化的,受调控的环境中,这个环境称为龛,龛控制着干细胞如何产生,维持及修复组织(1,2)。
我们之前记录了移植的造血干细胞和祖细胞群局限在骨髓微血管的亚区域中,那里的化学因子CXCL12特别丰富(3)。
将高精度共聚焦显微镜与双光子成像技术联合起来,持续观察活体小鼠颅盖骨骨髓的个体造血细胞,我们在受辐射及c-Kit-受体缺失容受小鼠中(recipient mice),检查了造血干细胞和祖细胞与血管,成骨细胞以及它们定居及移入的骨内膜表面之间的关系。
成骨细胞陷入微血管中,而干细胞/祖细胞在这个复杂组织中的相对位置不是随机的,并且是动态的。
细胞自主因素和非自主因素都影响了细胞的原始局限化。
不同的造血细胞亚群局限在完全不同的位置,这取决于分化的时期。
当生理上的激发事件驱动细胞移入或扩张时,骨髓干细胞/祖细胞占据了(assumed)最接近骨和成骨细胞的位置。
我们的分析使得我们可以在单细胞水平上实时观察某种过程,之前对这种过程只能在机体水平上研究其长期结果。
哺乳动物干细胞龛在很大程度上是基于干细胞的局限化而被定义的,还有少数有限的定义是关于龛内异源细胞的,这种细胞调控干细胞的功能。
还没有人在活体上检查过这些龛的生理内容。
造血干细胞(HSC)龛已经被研究过,办法是改变骨的组成部分,诱导干细胞功能的变化(4~7),或是通过免疫组化方式(8~10)。
前者不能精确定义干细胞的局限化,后者则不能定义其功能。
一些研究已经记录了成骨细胞在HSC命运上的调控作用(4,7,8,11~13);但是,其它研究显示,从免疫表型上来说最像HSCs的那些细胞被观察到邻近于骨髓中的脉管系统(9)。
骨髓脉管系统包括以有规律的间隔穿入密质骨的动脉,分叉成毛细血管覆盖中央窦(central sinus)(14)。
长骨的CD31免疫染色可以使人们观察到邻近骨内膜表面的这种脉管网络,特别是骨小梁区域,这是已知的HSCs局限的地方(Fig.1a and data not shown)(7,10)。
与基质芯片相关的方法和组合物[发明专利]
![与基质芯片相关的方法和组合物[发明专利]](https://img.taocdn.com/s3/m/d112ef6602d276a201292e0d.png)
专利名称:与基质芯片相关的方法和组合物
专利类型:发明专利
发明人:安德鲁·J·曼尼托斯,罗伯特·弗伯格,卡拉·瓦尼纳吉申请号:CN200480037140.0
申请日:20041013
公开号:CN101389753A
公开日:
20090318
专利内容由知识产权出版社提供
摘要:本发明涉及用于评估细胞间和细胞与基质材料间相互作用的设备和方法,其中由于这些相互作用形成的细胞分布模式是对细胞入侵潜能的指示物。
此外,这些设备和方法可以提供对入侵性细胞转移的优选位点的指示;对使用到这些细胞上的抗癌药物的功效的指示;以及对试剂促进或提高肿瘤生长或转移的潜能的指示。
申请人:伊利诺伊大学评仪会
地址:美国伊利诺伊州
国籍:US
代理机构:北京金信立方知识产权代理有限公司
代理人:黄威
更多信息请下载全文后查看。
锌指蛋白KOX12单克隆抗体的制备及在肿瘤组织中的表达

参考 文献
[ ] i r , a i y I K l r H, t . d oet d i r 1 Kz 皿 B rl , u e L e a A i n i a s o c - e za J l 1 p c n n rk f o
斑块的分解破裂与 炎性损 伤有密 切的关 系。多项 关于 内皮 细胞 、 单核巨噬 细胞 以及 血管平 滑肌 的研究 已经证 实 C P R 在动脉粥样硬化形成中有着重要 的作 用 。而 h—R sC P检 测提供 了一种观察极低水平炎症信 息的方法 。
本文 结 果 中, P 含 量 Ct 组 低 于 粥 样 硬 化 组 AN I D ( O0 ) 粥样硬 化组低 于正 常组 ( 0 0 ) C D组 内 P< .5 , P< . 5 ; H
[ ] l aJ , hg e A t a , t . ycr ai a t n e - 4 Ap T y n e S s e K, nm nE e a M o d ln r o d 1 a i fc r e i
矗 e _ cl ̄s8 ou e tf e on erpa oiyo a il A ndao l nu c m n it uoensce C r oo f l d ot j h t f d  ̄/ ・
脉粥样硬化 的形成 、 发展 过程 ; 动脉粥样 硬化 的进 展和粥样
Me b 2 0 , 3 9 35 -3 4 t , 0 8 9 ( ): 373 6 . a [ ] t k ,S g a ,K j ,e a ls d oet vl 2 Os aF ui maS omaS t1 u y i .Pamaai nci l e p ne s
miR-126 Regulates Angiogenic Signaling and Vascular Integrity
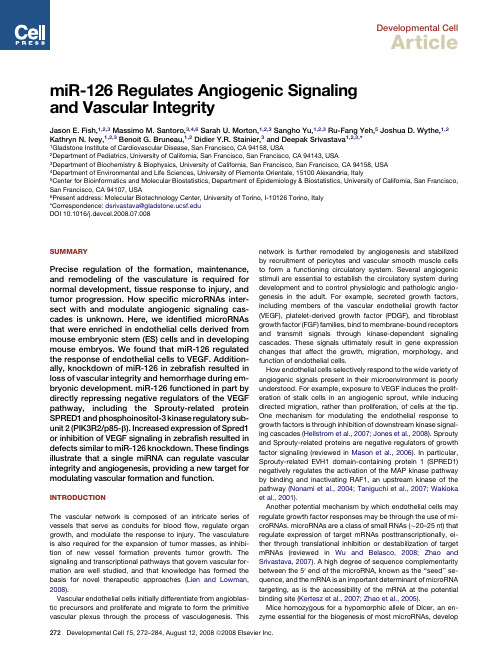
Developmental CellArticlemiR-126Regulates Angiogenic Signalingand Vascular IntegrityJason E.Fish,1,2,3Massimo M.Santoro,3,4,6Sarah U.Morton,1,2,3Sangho Yu,1,2,3Ru-Fang Yeh,5Joshua D.Wythe,1,2 Kathryn N.Ivey,1,2,3Benoit G.Bruneau,1,2Didier Y.R.Stainier,3and Deepak Srivastava1,2,3,*1Gladstone Institute of Cardiovascular Disease,San Francisco,CA94158,USA2Department of Pediatrics,University of California,San Francisco,San Francisco,CA94143,USA3Department of Biochemistry&Biophysics,University of California,San Francisco,San Francisco,CA94158,USA4Department of Environmental and Life Sciences,University of Piemonte Orientale,15100Alexandria,Italy5Center for Bioinformatics and Molecular Biostatistics,Department of Epidemiology&Biostatistics,University of California,San Francisco, San Francisco,CA94107,USA6Present address:Molecular Biotechnology Center,University of Torino,I-10126Torino,Italy*Correspondence:dsrivastava@DOI10.1016/j.devcel.2008.07.008SUMMARYPrecise regulation of the formation,maintenance, and remodeling of the vasculature is required for normal development,tissue response to injury,and tumor progression.How specific microRNAs inter-sect with and modulate angiogenic signaling cas-cades is unknown.Here,we identified microRNAs that were enriched in endothelial cells derived from mouse embryonic stem(ES)cells and in developing mouse embryos.We found that miR-126regulated the response of endothelial cells to VEGF.Addition-ally,knockdown of miR-126in zebrafish resulted in loss of vascular integrity and hemorrhage during em-bryonic development.miR-126functioned in part by directly repressing negative regulators of the VEGF pathway,including the Sprouty-related protein SPRED1and phosphoinositol-3kinase regulatory sub-unit2(PIK3R2/p85-b).Increased expression of Spred1 or inhibition of VEGF signaling in zebrafish resulted in defects similar to miR-126knockdown.Thesefindings illustrate that a single miRNA can regulate vascular integrity and angiogenesis,providing a new target for modulating vascular formation and function. INTRODUCTIONThe vascular network is composed of an intricate series of vessels that serve as conduits for bloodflow,regulate organ growth,and modulate the response to injury.The vasculature is also required for the expansion of tumor masses,as inhibi-tion of new vessel formation prevents tumor growth.The signaling and transcriptional pathways that govern vascular for-mation are well studied,and that knowledge has formed the basis for novel therapeutic approaches(Lien and Lowman, 2008).Vascular endothelial cells initially differentiate from angioblas-tic precursors and proliferate and migrate to form the primitive vascular plexus through the process of vasculogenesis.This network is further remodeled by angiogenesis and stabilized by recruitment of pericytes and vascular smooth muscle cells to form a functioning circulatory system.Several angiogenic stimuli are essential to establish the circulatory system during development and to control physiologic and pathologic angio-genesis in the adult.For example,secreted growth factors, including members of the vascular endothelial growth factor (VEGF),platelet-derived growth factor(PDGF),andfibroblast growth factor(FGF)families,bind to membrane-bound receptors and transmit signals through kinase-dependent signaling cascades.These signals ultimately result in gene expression changes that affect the growth,migration,morphology,and function of endothelial cells.How endothelial cells selectively respond to the wide variety of angiogenic signals present in their microenvironment is poorly understood.For example,exposure to VEGF induces the prolif-eration of stalk cells in an angiogenic sprout,while inducing directed migration,rather than proliferation,of cells at the tip. One mechanism for modulating the endothelial response to growth factors is through inhibition of downstream kinase signal-ing cascades(Hellstrom et al.,2007;Jones et al.,2008).Sprouty and Sprouty-related proteins are negative regulators of growth factor signaling(reviewed in Mason et al.,2006).In particular, Sprouty-related EVH1domain-containing protein1(SPRED1) negatively regulates the activation of the MAP kinase pathway by binding and inactivating RAF1,an upstream kinase of the pathway(Nonami et al.,2004;Taniguchi et al.,2007;Wakioka et al.,2001).Another potential mechanism by which endothelial cells may regulate growth factor responses may be through the use of mi-croRNAs.microRNAs are a class of small RNAs($20–25nt)that regulate expression of target mRNAs posttranscriptionally,ei-ther through translational inhibition or destabilization of target mRNAs(reviewed in Wu and Belasco,2008;Zhao and Srivastava,2007).A high degree of sequence complementarity between the50end of the microRNA,known as the‘‘seed’’se-quence,and the mRNA is an important determinant of microRNA targeting,as is the accessibility of the mRNA at the potential binding site(Kertesz et al.,2007;Zhao et al.,2005).Mice homozygous for a hypomorphic allele of Dicer,an en-zyme essential for the biogenesis of most microRNAs,developgross abnormalities during blood vessel development in the em-bryo and in the yolk sac (Yang et al.,2005).While several broadly expressed microRNAs regulate in vitro endothelial cell behavior,including proliferation,migration,and the ability to form capillary networks (Chen and Gorski,2008;Kuehbacher et al.,2007;Lee et al.,2007a;Poliseno et al.,2006;Suarez et al.,2007),the in vivo functions of endothelial-specific microRNAs,or their tar-gets,have yet to be described.By microRNA profiling of ES cell-derived endothelial cells,we identified a group of endothelial-enriched microRNAs,including miR-126,miR-146,miR-197,and miR-625.These microRNAs were also enriched in endothe-lial cells of developing mouse embryos.Through modulation of the expression of miR-126in vitro and in vivo,we demonstrate that miR-126positively regulates the response of endothelial cells to VEGF.Furthermore,we show that miR-126regulates VEGF-dependent PI3kinase and MAP kinase signaling by directly targeting PI3KR2(p85-b )and SPRED1,respectively,two negative regulators of the VEGF signaling pathway.RESULTSmiR-126Is the Most Highly Enriched MicroRNA in Endothelial CellsTo determine when the endothelial lineage first appears in dif-ferentiating mouse ES cells in the embryoid body (EB)model,extensive mRNA expression profiling was performed for endo-thelial marker expression using quantitative reverse transcrip-tase PCR (qRT-PCR).Oct4,a marker of pluripotent ES cells,was rapidly downregulated during differentiation (data not shown).At day 4(d4)of EB formation,Flk1/Vegfr2was dra-matically induced,but other endothelial markers,such as Tie2/Tek and CD31/Pecam1,were unchanged (Figure 1A).This population of Flk1-positive cells at d4contains vascular precursor cells,as evidenced by the ability of isolated Flk1-positive cells to differentiate into the endothelial lineage in the presence of VEGF (data not shown).By d6of EB forma-tion,endothelial markers,including CD31and Tie2,wereFigure 1.Identification of microRNAs Enriched in Endothelial Cells(A)Gene expression changes were monitored by qRT-PCR during differentiation of mouse ES cells in an embryoid body (EB)model.Flk1is expressed in vascular progenitors and mature endothelial cells,while CD31and Tie2are markers of mature endothelial cells.Expression was normalized to Tata-binding protein (Tbp )levels.The average of multiple experiments is shown.(B)Endothelial cells were isolated from day 7(d7)EBs by cell sorting with anti-CD31antibodies and microRNA arrays were performed.microRNAs enriched more than 1.5-fold relative to miR-16in CD31+cells compared to CD31Àcells are shown.(C)Enrichment of microRNAs identified in (B)in CD31+endothelial cells sorted from E10.5mouse embryos assayed by qRT-PCR.(D)Expression of Egfl7,miR-126and miR-126*in EBs assayed by qRT-PCR.(E)miR-126was enriched in sorted vascular progenitors (Flk1+)from d4EBs and in endothelial cells (CD31+)from d7EBs compared to ES cells.Tie2expression was used to assess mature endothelial gene expression in sorted cells used in (B)and (E).Developmental CellmiR-126Regulates Angiogenic Signalingrobustly expressed and remained elevated even after14days of differentiation.We isolated CD31-positive and CD31-negative cells from d7 EBs byfluorescence-activated cell sorting(FACS)and profiled microRNA expression by microarray.While several microRNAs, including miR-146b,miR-197,miR-615,and miR-625,were enriched more than1.5-fold in CD31-positive cells,miR-126 was the most highly enriched microRNA(Figure1B).MicroRNA arrays were also performed at d14of EB formation,and the same subset of microRNAs was enriched(data not shown),sug-gesting that relatively few microRNAs are enriched in endothelial cells.To determine if these microRNAs were also enriched in endo-thelial cells in vivo,we used FACS to isolate CD31-positive cells from E10.5mouse embryos.qRT-PCR with RNA from these cells confirmed the enrichment of the above microRNAs,with the ex-ception of miR-615,in CD31-positive cells compared to CD31-negative cells from the same embryos(Figure1C).Additionally, miR-126*,expressed from the opposite strand of the miR-126 pre-miRNA,was also highly enriched in endothelial cells in vivo,as was miR-146a,which differs from miR-146b by only two nucleotides near the30end of the mature microRNA (Figure1C).miR-126is located in an intron of Egfl7,a gene that is highly expressed in endothelial cells(Parker et al.,2004).The expres-sion of Egfl7largely mirrored that of endothelial markers during EB formation(Figure1D).Interestingly,expression of Egfl7, miR-126,and miR-126*was induced at d4of EB formation and further increased at d6,when endothelial markers were robustly expressed.miR-126(Figure1E)and miR-126*(data not shown) were highly enriched in Flk1-positive vascular progenitors sorted at d4and were also enriched in mature CD31-expressing endothelial cells at d7.miR-126Does Not Control Endothelial Lineage CommitmentWe have previously shown that the muscle-specific microRNA miR-1controls cell fate decisions of multipotent cells(Ivey et al.,2008).Because of the early induction of miR-126in vascular progenitors,we investigated whether this microRNA might regu-late differentiation toward the endothelial lineage.We created stable mouse ES cell lines that expressed miR-126under control of the ubiquitously expressed EF1-a promoter(mES miR-126)and confirmed miR-126overexpression(Figure S1A,available on-line).The expression of several endothelial genes,including Flk1,eNOS/NoS3,Tie2,and CD31,was not affected by miR-126 overexpression during ES cell differentiation(Figure S1B),and the number of CD31-positive endothelial cells at d7was not al-tered(Figure S1C).This suggests that while miR-126is enriched in vascular progenitors,it is not sufficient to promote differentia-tion of pluripotent cells toward the endothelial lineage.miR-126Modulates Endothelial Phenotype In VitroTo study the loss-of-function of miR-126in endothelial cells, a morpholino(MO)antisense to miR-126that spanned the miR-12650Dicer cleavage site of the miR-126pricursor was in-troduced into human umbilical vein endothelial cells(HUVECs). These cells expressed high levels of miR-126(Figure S2A).Intro-duction of the MO resulted in decreased levels of both mature miR-126and miR-126*and an increase in miR-126pricursor,be-ginning at24hr(h)posttransfection(Figure2A and Figure S2B). While both miR-126and miR-126*were reduced to a similar ex-tent,the absolute basal level of miR-126was much higher than miR-126*in endothelial cells(Figure S2C).Importantly,levels of spliced EGFL7mRNA,detected by qRT-PCR with primers spanning the intron containing miR-126,and protein levels of EGFL7,were unaffected by introduction of this MO (Figure2A).Endothelial cells with reduced levels of miR-126were pheno-typically similar to control MO-transfected cells but had an elevated rate of proliferation(Figure S3A).The endothelial pheno-type was further studied in an in vitro wound closure or scratch assay,in which the rate of migration of cells into a denuded area of a confluent monolayer was monitored.Modulating miR-126levels had no effect on cell migration when complete me-dium was used(Figure S3B).However,VEGF-(Figure2B)and bFGF-induced(Figure S3C)migration was inhibited in miR-126 knockdown cells compared to control MO-transfected cells. Conversely,in cells transfected with miR-126mimic,which ex-press50-fold more miR-126,there was a trend toward increased migration in response to VEGF stimulation(Figure2B).Examina-tion of the actin cytoskeleton revealed a reduction in large actin filaments and more diffuse actin staining in many miR-126 knockdown cells(Figure2C).Stimulation of endothelial cells with VEGF normally results in the reorganization of the cytoskel-eton,but this rearrangement was defective in miR-126knock-down cells(Figure S3D).Consequently,in scratch assays,there was a reduction in cell protrusions into the denuded area in knockdown cells(Figure2D).These data suggest that endothe-lial cell migration is regulated by miR-126abundance.The effect of miR-126on the formation and stability of capillary tubes on matrigel was also assessed.While initial formation of tubes appeared normal,the capillary tubes were less stable and appeared thin,with dissociation of many tubes after24hr (Figure2E).Assessment of cell adhesion by measuring the kinet-ics of endothelial cell attachment(Figure S3E)and cell-cell junc-tion formation by VE-cadherin immunostaining(data not shown) revealed no obvious defects in miR-126knockdown cells,sug-gesting that the instability of tubes was not attributable to de-fects in cell adhesion.To determine if there were survival defects in cells with reduced levels of miR-126,cells were serum starved in the presence or absence of VEGF.The number of control se-rum-starved cells increased with addition of VEGF,but miR-126 knockdown cells were refractory to this increase in cell number in response to VEGF(Figure2F).TUNEL staining demonstrated that VEGF treatment of control serum-starved cells resulted in a decrease in apoptosis,but this effect was absent in miR-126 knockdown cells(Figure2F).miR-126Regulates Blood Vessel Integrity In Vivo Considering the dramatic effects of miR-126on the behavior of human endothelial cells in vitro,we assessed the in vivo function of miR-126.For this purpose,we used zebrafish as a model sys-tem in which a functioning cardiovascular system is not required for viability through relatively advanced stages of embryogene-sis.The mature forms of zebrafish miR-126and miR-126*are identical to their human orthologs.FACS isolation of GFP-posi-tive cells from the endothelial cell-specific zebrafish reporterDevelopmental CellmiR-126Regulates Angiogenic Signalingline Tg(flk1:GFP )s843(Jin et al.,2007)demonstrated that miR-126and miR-126*were highly enriched in zebrafish endothelial cells (Figure 3A).As in human endothelial cells,miR-126was more abundant than miR-126*in zebrafish embryos (Figure 3B).miR-126expression was induced in 24hr post fertilization (hpf)embryos,was further increased at 48hpf,and remained elevated between 72and 96hpf (Figure 3C).We decreased miR-126ex-pression during zebrafish development by injecting two uniquemorpholinos targeting pri-miR-126(miR-126MO1and MO2[Figure S4A])into fertilized eggs.Injection of these MOs blocked processing of pri-miR-126,resulting in a profound decrease in mature levels of miR-126and miR-126*(Figure 3D).Importantly,levels of egfl7,which hosts one of the two copies of zebrafish miR-126,and regulates tubulogenesis in zebrafish (Parker et al.,2004),were not appreciably altered by the miR-126MOs (Figure 3D).Figure 2.miR-126Regulates Endothelial Phenotype In Vitro(A)Relative levels of mature miR-126,spliced EGFL7mRNA across the miR-126-containing intron,and EGFL7protein (immunoblot)were measured 72hr after HUVECs were electroporated with miR-126morpholinos (MOs)or control (con)MO.Densitometric analysis of EGFL7protein levels is indicated above immunoblot.(B)The effect of miR-126knockdown (MO)or overexpression (mimic)on endothelial cell migration in response to VEGF in the presence or absence of a VEGF receptor inhibitor was determined by generating a ‘‘scratch’’in a confluent monolayer of endothelial cells and measuring the degree of ‘‘wound closure’’after 8and 24hr.Dashed lines indicate width of ‘‘wound.’’Percent wound closure is shown as the mean ±SEM of five scratches from one representative experiment.*p <0.05compared to control.(C)Actin cytoskeletal structure was observed by phalloidin staining of control and miR-126knockdown HUVECs.(D)Actin staining revealed a decrease in cytoskeletal reorganization and cytoplasmic extensions into the denuded area in a scratch assay.(E)Capillary tube formation of endothelial cells transfected with control or miR-126MOs and seeded onto Matrigel.Endothelial cells transfected with miR-126MOs formed capillary tubes,but these tubes were unstable,exhibiting frequent dissociation of tubular structures compared to control.Shown is a representative image at 24hr after plating.Arrows indicate examples of broken tubes.Quantification of the average number of tubes per field of view at 4hr and the number of complete and broken tubes at 24hr from a representative experiment is shown below.(F)VEGF-mediated changes in cell number and cell death by TUNEL were assessed in control and miR-126knockdown cells.VEGF treatment increased total cell number and decreased the percent of serum-starved cells that were TUNEL positive,but this effect was absent in cells with reduced levels of miR-126.*denotes a significant difference between VEGF treated and non-treated cells (p <0.05).Developmental CellmiR-126Regulates Angiogenic SignalingThrough the use of Tg(flk1:GFP )s843;Tg(gata1:dsRed )sd2zebrafish,which express GFP in the vasculature,and dsRed in blood cells,we assessed the effect of miR-126knockdown on vascular development and circulation.No differences in gross morphology (Figure 3E,top panel)or vascular patterning (Fig-ure 3E,middle panel)were evident between control and miR-126morpholino-injected (morphant)embryos.Additionally,FACS quantification revealed no significant difference in the percent-age of Tg(flk1:GFP )s843-expressing endothelial or Tg (gata1:dsRed )sd2-expressing blood cells in control,miR-126MO1,or miR-126MO2-injected zebrafish (Figure S4B).However,severalabnormalities were evident in the circulation and vessel mor-phology.These defects occurred in 70%±5%(n =60per experiment,3independent experiments)of the miR-126MO-injected embryos and were similar with either MO.While circula-tion of blood occurred normally between 24and 36hpf,the presence of Tg(gata1:dsRed )sd2-expressing blood cells in the head vasculature,intersomitic vessels (ISVs),dorsal aorta (DA),and primary cardinal vein (PCV)was reduced between 36and 72hpf (Figures 3E,bottom panel,and 3F;data not shown).Blood cells were made and were visible in the heart at these time points (Figures 3E and 3F).Severe cranial hemorrhageswereFigure 3.miR-126Regulates Vascular Integrity and Lumen Maintenance In Vivo(A)miR-126and miR-126*enrichment (qRT-PCR)in GFP +endothelial cells from 72hpf Tg(flk1:GFP)s843zebrafish compared to GFP Àcells.(B)Relative levels of miR-126and miR-126*in 72hpf zebrafish embryos.(C)miR-126expression monitored by qRT-PCR during zebrafish development.(D)Levels of miR-126/126*or egfl7(across intron containing miR-126)quantified by qRT-PCR in 72hpf zebrafish injected with miR-126MOs relative to control.(E)Lateral views of control and miR-126MO-injected Tg(flk1:GFP)s843;Tg(gata1:dsRed)sd2zebrafish (72hpf).Brightfield microscopy (top)revealed no major changes in gross morphology,while flk1:GFP showed normal blood vessel patterning (middle).Presence of blood cells (gata1:dsRed )in the intersomitic vessels (isv),dorsal aorta (da)and primary cardinal vein (pcv)was greatly reduced in morphants (bottom).y,yolksac;h,head;ht,heart.(F)miR-126morphants (48hpf)had normal vessel patterning (flk1:GFP ),but developed cranial hemorrhages (gata1:dsRed ;arrow)in the head.baa,branchial arch arteries.(G)Ventral view of baa suggested smaller luminal size in miR-126morphants.(H)Transverse section of control or miR-126MO-treated zebrafish revealed that the da and pcv of morphants had a smaller lumen than controls;higher magnification (right panels)of boxed area shows collapsed da and small pcv in morphants.flk1:GFP ,green;ZO-1,an epithelial marker,red.Developmental CellmiR-126Regulates Angiogenic Signalingalso present in20%±3%(n=60per experiment,3independent experiments)of embryos,evidenced by the accumulation of dsRed-positive cells(Figure3F).Importantly,cardiac contractile function was not grossly affected by miR-126MO at either48 or72hpf.We observed that some Tg(gata1:dsRed)sd2-expressing cells were trapped in the ISVs of miR-126morphants(data not shown),indicating that blood vessel integrity might be compro-mised.Indeed,branchial arch vessels(Figures3F and3G) appeared to have a reduced lumen diameter in morphants.To better characterize these defects,we analyzed the integrity of endothelial tubes by examining the DA and PCV in miR-126mor-phants by confocal microscopy(Figure3H).These experiments revealed collapsed lumens and compromised endothelial tube organization in miR-126morphants,suggesting that miR-126 expression was required to maintain vessel integrity and caliber during zebrafish vascular development.Identification of Genes Regulated by miR-126by MicroarrayAlthough miR-126morphants had severe defects in vessel integ-rity,the number of endothelial cells was not altered.We took advantage of this phenotype and isolated Tg(flk1:GFP)s843-expressing endothelial cells by FACS from control and miR-126 MO1-and MO2-injected zebrafish and analyzed mRNA expres-sion by microarray.Since similar genes were altered in miR-126 MO1-and MO2-injected zebrafish,the data sets were combined to identify dysregulated genes in miR-126morphants(see Ta-bles S1and S2for up-and downregulated genes,respectively). By gene ontology(GO)statistical analysis,the most highly dysre-gulated class of genes in the endothelium of miR-126morphants encoded transcription factors(Table S3).The homeobox(HOX) and forkhead box(FOX)family of genes were especially affected in miR-126morphants,many of which regulate endothelial cell biology,including angiogenesis(Bruhl et al.,2004;Dejana et al.,2007;Myers et al.,2000).Microarray analysis was also performed with RNA from human endothelial cells(HUVECs)in which miR-126was knocked down for72hr(see Tables S4and S5for up-and downregulated genes, respectively).The most overrepresented GO terms were related to the cell cycle and the cytoskeleton(Table S6).This observation supports ourfinding that cells with reduced levels of miR-126 proliferated more rapidly than control cells and had altered cyto-skeletal structures(Figure S3A and Figures2C and2D).Platelet-derived growth factors(PDGF)A,B,C,and D,which are impor-tant in endothelial biology,were all significantly downregulated in cells with reduced levels of miR-126(Table S6;data not shown).In addition,genes categorized as important for vascular development were highly dysregulated(Table S6).A total of61 genes were similarly altered(p<0.05)in zebrafish and human miR-126knockdown expression arrays(data not shown),sug-gesting a high conservation in the gene repertoire regulated by miR-126.To determine how many of the upregulated genes in miR-126knockdown endothelial cells might be direct miR-126 targets,we performed bioinformatic analyses of miR-126and miR-126*seed matches in the30UTRs of genes upregulated by >1.5-fold in human cells with reduced levels of miR-126.miR-126seed matches were highly enriched in the upregulated genes, while seed matches for miR-126*or an unrelated microRNA,miR-124,were not enriched(Figure S5).Surprisingly,genes contain-ing both miR-126and miR-126*seed matches were also statisti-cally overrepresented,while the combination of miR-126and miR-124seed matches was not.This suggests that miR-126 and miR-126*,which are derived from the same pri-miRNA, coordinately regulate target genes.miR-126Regulates EGFL7Expression in a Negative Feed-Back LoopEGFL7mRNA was highly upregulated on the human array(Table S4)despite our earlierfinding that levels of spliced EGFL7mRNA and protein were unchanged(Figure2A).To understand this discrepancy,we used qRT-PCR with primer sets specific for the transcriptional start sites of the three EGFL7isoforms(named here EGFL7isoform A,B,and C,all of which contain the same open reading frame[ORF]),as well as several primer sets that were common to all three isoforms.EGFL7mRNA levels were in-creased throughout the EGFL7transcriptional unit(Figure S6A), except for the spliced EGFL7mRNA surrounding the miR-126-containing intron,as we noted earlier(Figure2A).Thus,EGFL7 was upregulated in miR-126MO-treated cells,but the MO appar-ently inhibited processing of the intron containing miR-126,re-sulting in no net change in EGFL7protein levels.Only EGFL7iso-form B was induced by miR-126MO(Figure S6A).Since all three isoforms contain the same30UTR,miR-126may regulate isoform B in a30UTR-independent fashion.By performing RNA polymer-ase II(Pol II)chromatin immunoprecipitation(ChIP)experiments, we noted an increase in Pol II density at the promoter of isoform B and in the coding region(which is common to all three isoforms), but not at the promoter of isoform A,which was not induced by miR-126MO(Figure S6B).Thus,miR-126may indirectly regulate EGFL7isoform B at the transcriptional level.miR-126Represses SPRED1,VCAM1,and PIK3R2 PosttranscriptionallyTo understand the mechanisms by which miR-126regulates en-dothelial biology,we searched for potential direct mRNA targets of miR-126.Several miRNA target prediction algorithms were employed,including one developed in our lab that incorporates sequence complementarity and mRNA target site accessibility (K.Ivey and D.S.,unpublished data).Portions of the30UTR of several potential targets were cloned into the30UTR of a lucifer-ase construct,and the ability of miR-126to affect luciferase ex-pression was determined in HeLa cells,which do not normally express miR-126.Six potential targets were initially chosen based on binding sites(Figure S7)and a known role in endothe-lial cell signaling or vascular function.These included regulator of G protein signaling3(RGS3)(Bowman et al.,1998;Lu et al., 2001),SPRED1(Wakioka et al.,2001),PIK3R2(also known as p85-b)(Ueki et al.,2003),CRK(Park et al.,2006),integrin alpha-6(ITGA6),and vascular cell adhesion molecule1 (VCAM1).miR-126,but not a control miRNA,miR-1,significantly repressed the activity of luciferase derived from RNAs containing the30UTR of SPRED1,VCAM1,and PIK3R2(Figure4A). Luciferase experiments were also performed in endothelial cells in which endogenous miR-126levels were knocked down by antisense MO.The activity of luciferase from constructs that included portions of the SPRED1,VCAM1,or PIK3R230 UTR was increased upon knockdown of miR-126(Figure4B).Developmental CellmiR-126Regulates Angiogenic SignalingIn contrast,a MO directed to miR-21,which is also expressed in endothelial cells,had no effect on the activity of the constructs tested (data not shown).MicroRNAs can regulate mRNA stability or translation of target mRNAs.We quantified mRNA expression of potential miR-126targets by qRT-PCR in HUVECs that had been transfected with antisense miR-126MO or a miR-126mimic (Figure 4C).While SPRED1and PIK3R2mRNA levels were reciprocally regulated by miR-126abundance,VCAM1mRNA levels were elevated upon miR-126inhibition but were not decreased in the presence of miR-126mimic.As a control,we examined levels of RGS3,since the 30UTR of this gene did not affect luciferase activity in the presence of miR-126.RGS3expression was unchanged when miR-126levels were modulated (Figure 4C).We also as-sessed the expression of SPRED1,PIK3R2,and VCAM1proteinFigure 4.Identification of miR-126mRNA Targets(A)Relative luciferase activity of constructs con-taining the 30UTR of potential miR-126targets in-troduced into HeLa cells in the presence of miR-1or miR-126.The 30UTR was also inserted in the antisense orientation as a control (control 30UTR).Firefly luciferase activity for each construct was normalized to the co-transfected Renilla lucif-erase construct and then normalized to the change in pGL3luciferase in the presence of microRNA.For each 30UTR construct,normalized luciferase activity in the absence of microRNA was set to 1.*p <0.05compared to pGL3.(B)Relative luciferase activity of select constructs in (A)in HUVECs upon inhibition of miR-126with MOs.Normalization was performed as in (A).(C)mRNA levels of potential targets in HUVECs transfected with miR-126mimic or MO quantified by qRT-PCR.Values are relative to transfection controls.(D)Immunoblot of SPRED1,VCAM1,and PIK3R2protein in control or miR-126MO-transfected HUVECs 72hr post-transfection.GAPDH is shown as a loading control.(E)Sequence complementarity of a potential miR-126binding site in the zebrafish spred130UTR.(F and G)Luciferase assays (as in (A)and (B),respectively)using the zebrafish spred130UTR.by western blot after introduction of con-trol or miR-126MOs (Figure 4D).SPRED1and PIK3R2protein were increased when miR-126levels were decreased (Figure 4D).However,we could not de-tect VCAM1protein in either control or miR-126MO-transfected endothelial cells.This was not due to an ineffective antibody,since VCAM1protein was read-ily detectible in TNF-a -treated endothelial cells (data not shown).Indeed,VCAM1has recently been identified as a miR-126target in TNF-a -treated endothelial cells (Harris et al.,2008).The 30UTR of zebrafish spred1con-tains a highly conserved 8-mer that isperfectly complementary to nucleotides 2–9of miR-126(Figure 4E).Addition of miR-126specifically repressed the activ-ity of luciferase reporters containing this 30UTR (Figure 4F).Con-versely,knockdown of miR-126led to an increase in luciferase activity of the spred130UTR luciferase construct when trans-fected into HUVECs (Figure 4G).This suggests that miR-126tar-geting of Spred1is conserved in zebrafish.miR-126Modulates VEGF-Dependent Events by Targeting SPRED1and PIK3R2SPRED1and PIK3R2negatively regulate growth factor signaling via independent mechanisms.SPRED1functions by inhibiting growth factor-induced activation of the MAP kinase pathway (Taniguchi et al.,2007),while PIK3R2is thought to negatively regulate the activity of PI3kinase (Ueki et al.,2003).ActivationDevelopmental CellmiR-126Regulates Angiogenic Signaling。
[精华版]《跨文化交际》_名词解释
![[精华版]《跨文化交际》_名词解释](https://img.taocdn.com/s3/m/568cda0115791711cc7931b765ce0508763275e4.png)
1.Globalization is considered as a process of increasing involvment in international business operations.经济学视角中的全球化表现为不断增加的国际商务往来过程。
6.Macroculture:The term macroculture implies losing ethnic differences and forming onelarge society.宏观文化意味着种族差异的消失和一个大社会的形成。
7.Melting pot means a sociocultural assimilation of people of different backgrounds andnationlities.熔炉:不同背景和国籍的人们之间的社会文化的同化。
8.Microcultures:cultures within cultures微观文化:文化中的文化9.Intercultural communication refers to communication between people whose cultureperceptions and symbol systems are distinct enough to alter the communication event.跨文化交际:指拥有不同文化认知和符号体系的人文之间进行的交际。
Chapter 110.Culture is a learned set of shared interpretations about beliefs,values,and norms,which affectthe behavior of a relatively large group of people.文化是习得的一套关于信仰,价值观,规范的公认的解释,这些信仰,价值观,规范对相当大人类群体的行为产生影响。
含有内含子的启动子及其用途[发明专利]
![含有内含子的启动子及其用途[发明专利]](https://img.taocdn.com/s3/m/a3e92a68c950ad02de80d4d8d15abe23482f033b.png)
专利名称:含有内含子的启动子及其用途专利类型:发明专利
发明人:纪良辉,Y·刘,C·M·J·许,S·A·叶
申请号:CN201780022143.4
申请日:20170202
公开号:CN109072247B
公开日:
20220603
专利内容由知识产权出版社提供
摘要:本发明涉及分子生物学领域,且更具体地涉及用于酵母或真菌中代谢工程的启动子,以产生具有广泛应用的基于生物的化学制品。
在油积累阶段期间,具有强活性的含内含子的启动子特别适用于酵母和真菌,特别是红冬孢酵母属或红酵母属中的遗传工程。
这种启动子能够驱动红冬孢酵母属或红酵母属的物种中RNA或蛋白质的强烈表达。
申请人:淡马锡生命科学研究院有限公司
地址:新加坡新加坡
国籍:SG
代理机构:永新专利商标代理有限公司
代理人:区斌
更多信息请下载全文后查看。
用于治疗的尿石素A和其衍生物[发明专利]
![用于治疗的尿石素A和其衍生物[发明专利]](https://img.taocdn.com/s3/m/8c9bbbe8b52acfc788ebc941.png)
专利名称:用于治疗的尿石素A和其衍生物
专利类型:发明专利
发明人:V·R·贾拉,H·博杜鲁里,R·辛格,P·K·韦姆拉,S·钱德拉谢哈拉帕,A·A·希瓦莱
申请号:CN201980047329.4
申请日:20190514
公开号:CN112638889A
公开日:
20210409
专利内容由知识产权出版社提供
摘要:在本发明的一些实施方案中,公开了本发明化合物(例如,式(I)、(IA)、(II)和(III)以及尿石素衍生物)。
其它实施方案包含包括本发明化合物的组合物(例如,药物组合物)。
本发明的仍其它实施方案包含用于使用本发明化合物治疗例如某些疾病的组合物(例如,药物组合物)。
一些实施方案包含使用本发明化合物(例如,在组合物中或在药物组合物中)进行施用和治疗(例如,疾病)的方法。
另外的实施方案包含用于制备本发明化合物的方法。
本文还讨论了本发明的另外的实施方案。
申请人:路易斯维尔大学研究基金会,干细胞生物学与再生医学研究所
地址:美国肯塔基州
国籍:US
代理机构:永新专利商标代理有限公司
代理人:过晓东
更多信息请下载全文后查看。
干货第四期课程回顾打开微生物世界的秘钥『宏基因组技术』

干货第四期课程回顾打开微生物世界的秘钥『宏基因组技术』CNGBdb组学/数据库系列课程开讲啦!2019年7月25日<第四讲>上线课程名称:打开微生物世界的秘钥『宏基因组技术』讲师:宋泽伟/深圳华大生命科学研究院,宏基因组研究中心,研究员;明尼苏达大学,博士/博士后课程现场1课程概要1.什么是生态学?为什么要研究微生物生态?2.测序能干什么?3.如何开展宏基因组项目:实验、计算、分析2现场Q&AQ:目前基因测序与干细胞移植有没有相关联交叉的课题呢?A:这个问题更多的是和人的基因组相关,例如病人术后康复会和其肠道菌群密切相关,不过这主要在癌症上,即调节病人肠道菌群可以帮助其进行术后康复,具体到胚胎实验的话,这个不在我的领域范围。
Q:关于样本采集,比如研究一个湖泊的微生物,如何采样比较合理?A:这个首先需要研究者清楚自己要回答什么问题(研究目的),统计学可以计算我们需要的样本量,这需要你事先知道测定样本的变异程度(variation),所以有条件的话,我会进行预实验,如果没有条件进行预实验,建议尽可能的多采样(尽管不是那么科学)。
Q:如果用denovo来建立那个表的话,每一行是一个OTU,它们基于数据集产生,denovo产生的OTU只能是一次项目使用吗?对于同类型的其他项目是否有参考意义?如果基于目前的数据库也找不到OTU,要怎么分析呢?A:我们前期的研究显示:目前针对扩增子(16s或ITS)的数据库,已经基本能代表大家平时能想到的样品,即使对于一些非常罕见的样品,大部分序列都可以在参考数据库找到。
基于这样的研究结果,我们认为可以用refrence来代替denovo的方法。
如果想研究未知的物种怎么办?这个需要从两个方面来思考:1)解读菌落结构,利用菌落矩阵选择可靠的refrence的方法,得到一些结论;2)把已知数据库无法解读的信号单独出来再做研究。
即分成独立的两个问题来回答,而不是用同一个方法解决两个问题。
美国布鲁克海文国家实验室构建低木质素转基因植物

美国布鲁克海文国家实验室构建低木质素转基因植物
小仓红叶
【期刊名称】《生物产业技术》
【年(卷),期】2012(0)6
【摘要】2012年7月31日,美国能源部(DOE)下属的美国布鲁克海文国家实验室(Brookhaven National Laboratory,BNL)宣布,其研究团队构建了导入新酶基因的转基因植物,所导入的新酶能够有效地修饰木质素合成前体。
木质素是使植物生物质的分解变得特别困难的植物细胞壁组成成分。
【总页数】2页(P56-57)
【关键词】转基因植物;美国能源部;木质素合成;国家实验室;植物细胞壁;组成成分;酶基因;生物质
【作者】小仓红叶
【作者单位】
【正文语种】中文
【中图分类】Q943.2
【相关文献】
1.美国布鲁克海文国家实验室大观 [J], 于彬
2.美国布鲁克海文国家实验室的问题 [J], 邓寿昌
3.美国布鲁克海文国家实验室简介 [J], 赵淑权;何介薇
4.美国布鲁克海文国家实验室大观 [J], 无
5.美国能源部布鲁克海文国家实验室开发出能够分解神经毒剂的新型催化剂 [J], 宫学源
因版权原因,仅展示原文概要,查看原文内容请购买。
Microsatellites的名词解释

Microsatellites的名词解释在现代生物学领域中,Microsatellites是一种常见的遗传学工具,被广泛用于基因组学和遗传学研究中。
它们是一类由重复的短DNA序列组成的特殊序列标记。
这些序列通常由1到9个碱基重复单元构成,例如CA、GC、CT等。
由于其短小的长度和高度多态性,Microsatellites已成为基因组间的标记点和遗传变异的探针。
Microsatellites的结构和特点使其成为一种极具变异性的遗传标记。
重复的DNA单元导致了不同个体之间的长度差异,这被称为allele size polymorphism。
这种多态性是由于Microsatellites的不稳定性引起的。
通常情况下,重复单元越多,其长度就越长,而反之亦然。
这使得Microsatellites成为一种高度可变的分子标记,能够检测不同个体和群体之间微小的遗传差异。
Microsatellites的应用广泛且多样化。
首先,它们被广泛用于遗传多样性研究。
通过比较个体或种群之间Microsatellites的差异,我们可以了解到不同个体或种群之间的基因流动或遗传分化。
这对于保护濒危物种、种群遗传监测和遗传学进化研究都具有重要意义。
其次,Microsatellites还被广泛应用于个体识别和亲子鉴定上。
由于其高度多态性的特点,通过分析Microsatellites的组合模式,我们可以确定个体之间的关系和亲缘关系。
这在野生动物保护、畜牧业和人类学研究中都有重要的应用。
此外,Microsatellites还可以用于构建遗传图谱和基因定位。
通过在基因组中特定位置上定位Microsatellites标记,我们可以了解到这些标记与某个特定性状或疾病之间的关联。
这对于研究复杂的遗传疾病、农作物育种和生命科学研究都具有重要意义。
值得注意的是,Microsatellites虽然是一种独特的遗传标记,但它们并不适用于所有的研究对象。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
II. Irving Fisher’s Inter-temporal Choice of Consumption
The Inter-temporal Budget Constraint (continued)
Express S in (B) in terms of (A), we obtain C2 = (1+r)(Y1 - C1) + Y2 or C1+C2/(1+r) = Y1 + Y2/(1+r) This budget constraint can be graphed as in the figure of the next page.
IV. Friedman’s Permanent Income Hypothesis
The Basic Hypothesis
The total income of a consumer is composed of two parts:
• the permanent income Yp and • the transitory income Yt
II. Irving Fisher’s Inter-temporal Choice of Consumption
Consumer Preference (continued)
II. Irving Fisher’s Inter-temporal Choice of Consumption
Decision Problem
Macroeconomics
Lecture 12
Micro-foundation and the Behavior Analysis on Consumption
I. Introduction
The Critique to Keynesian Economics
The lacking in micro-foundation; Unable to explain growth; Unable to explain and deal with stagflation.
III. Modigliani’s Life Cycle Hypothesis
The Implication
Since b = R/T < 1, the model indicates that saving allows the consumer to move income from the periods when income is high to the periods when income is low.
III. Modigliani’s Life Cycle Hypothesis
The Model (continued)
Thus, the life time income (when ignoring interest rate) is equal to W+RY.
III. Modigliani’s Life Cycle Hypothesis
II. Irving Fisher’s Inter-temporal Choice of Consumption
The Effects of Interest Rate Change (continued)
II. Irving Fisher’s Inter-temporal Choice of Consumption
II. Irving Fisher’s Inter-temporal Choice of Consumption
The Effect of Income Change (continued)
1 1+r 0 C2
(1+r)Y1+Y2
II. Irving Fisher’s Inter-temporal Choice of Consumption
I. Introduction
The objective of this lecture
is to dealing with the first critique—the lacking of micro-foundation by introducing
• some models of behavior analysis on the consumption derived from optimization. • The bounded rationality, which may regarded to be a model of output determination in New Keynesian analysis.
The Implication
Consumption is not only related to the current income, but also to the interest rate and to the future income the consumer expects. There is no sense of saving over the life of the consumer.
The Effects of Interest Rate Change
Increasing interest rate r will increase C2 while reduce C1 yet whether the utility will increase or decrease is not clear (see the figure in the next page)
III. Modigliani’s Life Cycle Hypothesis
The Model
Let T: the years the consumer expect to live R: the years from now to retire Y: income expect earned from now to retire W: the current wealth
III. Modigliani’s Life Cycle Hypothesis
The Basic Hypothesis
Income is varied systematically over a consumer’s life. The consumer pursues a relatively smoothed consumption over its life.
II. Irving Fisher’s Inter-temporal Choice of Consumption
The Inter-temporal Budget Constraint
Suppose a consumer’s life can be divided into 2 periods, and therefore we denote Y Y1: income in period 1; Y2: income in period 2; C1: consumption in period 1; C2: consumption in period 2.
C1
1 1+r 0 C2
(1+r)Y1+Y2
II. Irving Fisher’s Inter-temporal Choice of Consumption
The Effect of Income Change
Increasing income Y1 and Y2 will increase both C1 and C2 (see the figure in the next page).
MRS between C1 and C2 is equal to 1/(1+r) (see the figure in the next page)
II. Irving Fisher’s Inter-temporal Choice of Consumption
Solution to the Decision Problem (continued)
S = Y1 - C1 (A)
II. Irving Fisher’s Inter-temporal Choice of Consumption
The Inter-temporal Budget Constraint (continued)
This indicates that
(B)
C2 = (1+r)S+Y2 where r is the interest rate. Note that here S can also be regarded as borrowing (dis-saving).
The Model (continued)
The Decision Property: Since consumer pursues a smoothed consumption sequence over his life, the consumption in each period could be written as C = (1/T)W+(R/T)Y or C = aW + bY
Choose C1 and C2 such that
Ma43;r) = Y1 + Y2/(1+r)
II. Irving Fisher’s Inter-temporal Choice of Consumption
Solution to the Decision Problem
