pEGFP质粒图谱
质粒图谱大全

〔转载〕Pllp-OmpA, pllp-STII, pMBP-P, pMBP-C,pET-GST, pET-Trx, pET-His, pET-CKS, pET-DsbApTZ19RDNApUC57DNAPMD18TPQE30pUC18pUC19pTrcHisApTrxFuspRSET-ApRSET-BpVAX1PBR322pbv220pBluescriptIIKS( )L4440pCAMBIA-1301pMAL-p2XpGD926ProteinExpression?ProkaryoticExpression?pETDsbFusionSystems39band40b ProteinExpression?ProkaryoticExpression?pETExpressionSystem33b ProteinExpression?ProkaryoticExpression?pETExpressionSystems ProteinExpression?ProkaryoticExpression?pETExpressionSystemsplusCompetentCells ProteinExpression?ProkaryoticExpression?pETGSTFusionSystems41and42 ProteinExpression?ProkaryoticExpression?pETVectorDNAProteinPurification?PurificationSystems?Strep?TactinResinsandPurificationKitsTEcoR?pGEX-1I/BAPpGEX-2TpGEX-2TKpGEX-3XpGEX-4T-1pGEX-4T-2pGEX-4T-3pGEX-5X-1pGEX-5X-2pGEX-5X-3pGEX-6P-1pGEX-6P-2pGEX-6P-3PTYB1PTYB2PTYB11PTYB12pCDNA3.1(-)pCDNA3.1( )pPICZalphaApGAPZαApBI121pEGFP-N1pEGFP-C1pPIC9K如何阅读分析质粒图谱载体主要有病毒和非病毒两大类,其中质粒DNA是一种新的非病毒转基因载体。
质粒图谱查询方法

3.google scholar: / 有些质粒是经过改造的,所以通过上述方法不能查询到相应信息。这时,可以在google scholar中输入质粒名称,可以直观地看哪些学者在何文章中使用了该质粒,从而可了解到质粒的来源;或者籍此向作者咨询或索取。 4.尝试从各大生物公司,例如invitrogen网站查询. 5. 这个网站收录了大量图谱: http://www.embl-hamburg.de/~geerlof/webPP/vectordb/bact_vectors/table.html
Pe
te
rX u
file:///D|/中科院/Selective Serotonin Transporter/质粒信息/质粒图谱查询方法.txt
file:///D|/中科院/Selective Serotonin Transporter/质粒信息/质粒图谱查询方法.txt(第 2/6 页)[2011/8/4 18:39:52]
By
0099--pGE-1—Stratagene--RNAi载体 0100--pSUPER.p53—OligoEngine--RNAi载体 0101--palter-ex1--promega 0102--pACYCDuet-1--NOVAGEN 0103--pEX lox(+) Vector—NOVAGEN--原核表达 0104--质粒名称:pBACgus-8 Transfer Plasmid—NOVAGEN--CHUANSUO 0105--pSCREEN?-1b(+) Vector Map—novagen--筛选 0106--PGEX-2T--BD Co--pDsRed2--Clontech 0107--pbgal-Basic—Clontech--mammalian reporter vector 0108—pBI—Clontech--express two genes of interest from a bidirectional tet-responsive promoter 0109--质粒名称:pbgal-Control—Clontech--mammalian reporter vector 0110-- pGEX-5X-1--原核表达 0111--pBI-EGFP—Clontech--pBI-EGFP-- coexpress 0112--pBI-G—Clontech--pBI-G--express b-galactosidase 0113--pBI-GL—Clontech--pBI-GL --express luciferase and b-galactosidase 0114--pCMS-EGFP—Clontech--mammalian expression vector 0115--pd2EYFP-1—Clontech--启动子测定 0116--质粒名称--pd2EYFP-N1—Clontech--融合表达 0117--pd4EGFP-Bid—Clontech--融合表达 Bid 0118--pDNR-CMV—Clontech--pDNR-CMV 0119--pDNR-EGFP Vector—Clontech 0120--pDNR-LacZ –Clontech 0121--pECFP-Endo—Clontech--真核表达0122--pECFP-ER—Clontech--真核表达0123--pEGFP-Actin—Clontech--真核表达0124--pGAD GH--Clontech--酵母表达 0125--pGADT7-Rec –Clontech--酵母表达 0126--pGADT7-RecAB—Clontech--酵母表达 0127--pGADT7-Rec2—Clontech--酵母表达 0128--pGBKT7—Clontech--酵母表达 0129--pHAT 10/11/12—Clontech 0130--pHAT20—Clontech 0131—pHygEGFP—Clontech 0132—pLacZi—Clontech 0133—pM—Clontech--pM is used to generate a fusion of the GAL4 DNA-BD 0134--pPKCa-EGFP—Clontech 0135--pPKCb-EGFP—Clontec 0136--pSIREN-DNR Vector—Clontech--RNAi 0137--pSIREN-DNR-DsRed-Express Vector—Clontech--RNAi 0138--pSIREN-RetroQ—Clontech--RNAi 0139--pIRES-EYFP—Clontech--RNAi 0140--pSRE-Luc—Clontech--RNAi 0141--pTK-neo—novagen--原核表达 0142--pZsGreen Vector—Clontech--pZsGreen is a pUC19-derived prokaryotic expression vector 0143--pTandem-1—novagen--原核表达 0144--pZsGreen1-C1Vector—Clontech----真核表达 0145--质粒名称:M13mp18—novagen--原核表达 0146--pZsGreen1-DR Vector—Clontech--真核表达 0147--PZsGreen1-N1 Vector—Clontech --真核表达 0148--T7Select415-1b—novagen----真核表达 0149--pZsYellow Vector—Clontech --真核表达 0150—pTimer—Clontech --真核表达 0151--pTA-Luc—Clontech --真核表达 0152--pTAL-Luc—Clontech --真核表达 0153--pTA-SEAP—Clontech --真核表达 0154--pTAL-SEAP—Clontech --真核表达 0155--pTet-On—Clontech --真核表达 0156--pTet-Off—Clontech --真核表达 0157--pTet-ATF—Clontech --真核表达 0158--pTet-CREB—Clontech --真核表达
质粒图谱

)质粒图谱登记号:00012)质粒名称:pIRES3)来源:BD Co4)用途:真核双表达5)是否可以提供更详细资料:可以6)是否可以共享:7)联系方式:PM)质粒图谱登记号:00022)质粒名称:pECFP-C13)来源:BD Co4)用途:检测真核表达5)是否可以提供更详细资料:可以6)是否可以共享:7)联系方式:pm1)质粒图谱登记号:00032)质粒名称:pShuttle3)来源:4)用途:5)是否可以提供更详细资料:6)是否可以共享:7)联系方式:2)pShuttle MCS很多人用pEGFP-C1,我也来发一个)质粒图谱登记号:00042)质粒名称:pSBR322、pUC183)来源:4)用途:5)是否可以提供更详细资料:6)是否可以共享:7)联系方式:1)质粒图谱登记号:00052)质粒名称:pcDNA3.1(+)/CAT3)来源:invitrogen4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:无偿7)联系方式:PM1)质粒图谱登记号:00062)质粒名称:pQEx3)来源:Qiagen4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM2)1)质粒图谱登记号:00072)质粒名称:pIVEX2.33)来源:Rocho4)用途:体外转录翻译5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM质粒图谱登记号:0008质粒名称:pIRES-EGFP来源:用途:是否可以提供更详细资料:是否可以共享:否联系方式:1)质粒图谱登记号:00092)质粒名称:pET-28a(+)3)来源:Novagen4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM1)质粒图谱登记号:00112)质粒名称:pET-32a(+)3)来源:novagen4)用途:原核表达5)是否可以提供更详细资料:6)是否可以共享:7)联系方式:pm1)质粒图谱登记号:00132)质粒名称:pcDNA3.1/Zeo (+)3)来源:invitrogen4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)质粒图谱登记号:00132)质粒名称:pEGFP-N33)来源:clontech4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)质粒图谱登记号:00142)质粒名称:pcDNA33)来源:invitrogen4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)质粒图谱登记号:00152)质粒名称:pfastbac13)来源:invitrogene4)用途:昆虫表达5)是否可以提供更详细资料:不可以6)是否可以共享:不可以7)联系方式:PM)质粒图谱登记号:00162)质粒名称:pEGFP-C33)来源:clontech4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PMwangjun2002274 edited on 2004-06-22 00:041)质粒图谱登记号:00172)质粒名称:pSecTag23)来源:Invitrogen4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)1质粒图谱登记号:00192)质粒名称:pET20b3)来源:NOVAGEN4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM1)质粒图谱登记号:00222)质粒名称:pThioHisA3)来源:invitrogen4)用途:原核表达5)是否可以提供更详细资料:4.365kb , HP-thioredoxin fusion proteinexpressionvector, trc promoter, Ampr, a EK cleavage site lies between HP-thioredoxin and MCS宿主菌TOP10(基因型为:F-mcrA △(mrr-hsd RMS-mcrBC)Ф80 lacZ M15 △lacX74 deoR recAl araD139 △(ara-leu)7697 galU galK rpsL endAl nupG6)是否可以共享:交换或其它7)联系方式:PM2)3)1)质粒图谱登记号:00242)质粒名称:pcDNA3.1-Myc-His-A-3)来源:invitrogen4)用途:真核核表达4))质粒图谱登记号:00252)质粒名称:pSUPER.neo3)来源:4)用途:siRNA5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)质粒图谱登记号:00312)质粒名称:pSilencer1.0-siRNA3)来源:Ambion4)用途:RNAi5)是否可以提供更详细资料:/techlib/Documents.html?fkResSxn=7&fkSub Sxn=236)是否可以共享:实验结束后,可提供含shRNA模板的质粒7)联系方式:pm5) )质粒图谱登记号:00322)质粒名称:pSilencer2.0-U6siRNA 3)来源:Ambion4)用途:RNAi ,与1.0相比,可以建立稳转株5)更详细资料:/techlib/prot/fm_7209.pdf 6)是否可以共享:交换 7)联系方式:pm6)会员名:mlluoE-mail:*************可提供试验资源名称和简要介绍:pSilencer 3.1-H1 neo Vector,是Ambion公司目前最高版本的shRNA 载体,为扩增此载体我已插入目的片段,如有战友需要,将此片段双酶切再连上自己的片段即可。
质粒图谱大全

(转载)一. 九种表达载体Pllp-OmpA, pllp-STII, pMBP-P, pMBP-C,pET-GST, pET-Trx, pET-His, pET-CKS, pET-DsbA二. 克隆载体pTZ19RDNApUC57DNAPMD18TPQE30pUC18pUC19pTrcHisApTrxFuspRSET-ApRSET-BpVAX1PBR322pbv220pBluescriptIIKS( )L4440pCAMBIA-1301pMAL-p2XpGD926三.PET 系列表达载体ProteinExpression?ProkaryoticExpression?pETDsbFusionSystems39band40b ProteinExpression?ProkaryoticExpression?pETExpressionSystem33b ProteinExpression?ProkaryoticExpression?pETExpressionSystems ProteinExpression?ProkaryoticExpression?pETExpressionSystemsplusCompetentCells ProteinExpression?ProkaryoticExpression?pETGSTFusionSystems41and42 ProteinExpression?ProkaryoticExpression?pETNusAFusionSystems43.1and44 ProteinExpression?ProkaryoticExpression?pETVectorDNAProteinPurification?PurificationSystems?Strep?TactinResinsandPurificationKits四.PGEX 系列表达载体TEcoR?pGEX-1I/BAPpGEX-2TpGEX-2TKpGEX-3XpGEX-4T-1pGEX-4T-2pGEX-4T-3pGEX-5X-1pGEX-5X-2pGEX-5X-3pGEX-6P-1pGEX-6P-2pGEX-6P-3五.PTYBsystemPTYB1PTYB2PTYB11PTYB12六. 真核表达载体pCDNA3.1(-)pCDNA3.1( )pPICZalphaApGAPZα APYES2.0pBI121pEGFP-N1pEGFP-C1pPIC9KpPIC3.5K如何阅读分析质粒图谱载体主要有病毒和非病毒两大类, 其中质粒DNA是一种新的非病毒转基因载体。
pEGFP-HPV16E7重组融合蛋白质粒的构建及其表达

pEGFP-HPV16E7重组融合蛋白质粒的构建及其表达左泽华;李辉;伍欣星【期刊名称】《中国病毒学》【年(卷),期】2003(018)004【摘要】应用基因重组技术,构建增强绿色荧光蛋白(EGFP)与人乳头瘤病毒16型E7(HPV16E7)的重组融合表达质粒,经限制性内切酶酶切鉴定和PCR分析后,用基因转染技术将其导入小鼠肝癌细胞,荧光显微镜下观察融合蛋白的表达.酶切鉴定和PCR分析证实重组质粒中插入目的基因片段的大小、方向和插入位点均正确,在转染的小鼠肝癌细胞中观察到绿色荧光蛋白的表达.构建的pEGFP-HPV16E7融合表达质粒能直观地反映转染细胞中EGFP-HPV16E7融合蛋白的表达.由于转化率与表达率融为一体,故有利于对转染细胞的筛选,缩短转染细胞在体外的筛选的时间适用于对HPV16E7分子生物学特性、致瘤机理及APC提呈等的研究.为建立表达HPV16E7的实体瘤动物模型奠定了基础.【总页数】4页(P344-347)【作者】左泽华;李辉;伍欣星【作者单位】武汉大学医学院病毒学研究所分子生物学研究室,湖北武汉,430071;武汉大学医学院病毒学研究所分子生物学研究室,湖北武汉,430071;武汉大学医学院病毒学研究所分子生物学研究室,湖北武汉,430071【正文语种】中文【中图分类】R373【相关文献】1.重组融合蛋白Tumstatin-TNF-α分泌型真核表达载体的构建及其在中国仓鼠卵巢细胞中的表达 [J], 赵亮;姚丽娟;孔建新;濮跃晨;孙安源;罗以勤;张林杰2.人IL-6和TNF突变体重组融合蛋白表达质粒的构建及在大肠杆菌中的表达 [J], 刘丽;欧阳应斌;黄培堂;刘及3.组织蛋白酶B过表达质粒和特异性siRNA表达质粒的构建与转染及对血管平滑肌细胞作用初步探讨 [J], 张涛;羊镇宇;薄小萍;王强;陈茂华;李坚;郭素峡;冯健;尤华彦4.含弓形虫ROP1基因真核表达重组质粒DNA免疫小鼠的研究Ⅴ.IFN-γ基因真核表达重组质粒的构建及其在DNA免疫中基因佐剂的作用 [J], 郭虹;陈观今;郑焕钦5.弓形虫ROP1基因真核表达重组质粒DNA免疫小鼠的研究Ⅰ.pcDNA3-ROP1真核表达重组质粒的构建 [J], 郭虹;陈观今;周永安;郑焕钦;刘彦文因版权原因,仅展示原文概要,查看原文内容请购买。
质粒pECFP-N1图谱
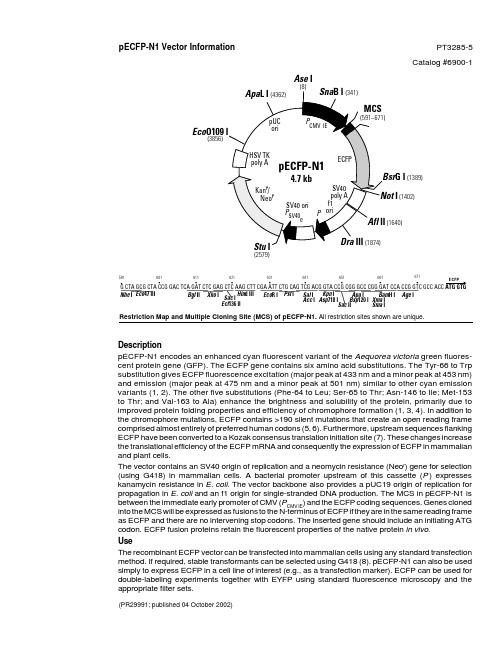
Restriction Map and Multiple Cloning Site (MCS) of pECFP-N1. All restriction sites shown are unique.pECFP-N1 Vector Information PT3285-5Catalog #6900-1(PR29991; published 04 October 2002)DescriptionpECFP-N1 encodes an enhanced cyan fluorescent variant of the Aequorea victoria green fluores-cent protein gene (GFP). The ECFP gene contains six amino acid substitutions. The Tyr-66 to Trp substitution gives ECFP fluorescence excitation (major peak at 433 nm and a minor peak at 453 nm)and emission (major peak at 475 nm and a minor peak at 501 nm) similar to other cyan emission variants (1, 2). The other five substitutions (Phe-64 to Leu; Ser-65 to Thr; Asn-146 to Ile; Met-153to Thr; and Val-163 to Ala) enhance the brightness and solubility of the protein, primarily due to improved protein folding properties and efficiency of chromophore formation (1, 3, 4). In addition to the chromophore mutations, ECFP contains >190 silent mutations that create an open reading frame comprised almost entirely of preferred human codons (5, 6). Furthermore, upstream sequences flanking ECFP have been converted to a Kozak consensus translation initiation site (7). These changes increase the translational efficiency of the ECFP mRNA and consequently the expression of ECFP in mammalian and plant cells.The vector contains an SV40 origin of replication and a neomycin resistance (Neo r ) gene for selection (using G418) in mammalian cells. A bacterial promoter upstream of this cassette (P ) expresses kanamycin resistance in E. coli . The vector backbone also provides a pUC19 origin of replication for propagation in E. coli and an f1 origin for single-stranded DNA production. The MCS in pECFP-N1 is between the immediate early promoter of CMV (P CMV IE ) and the ECFP coding sequences. Genes cloned into the MCS will be expressed as fusions to the N-terminus of ECFP if they are in the same reading frame as ECFP and there are no intervening stop codons. The inserted gene should include an initiating ATG codon. ECFP fusion proteins retain the fluorescent properties of the native protein in vivo .UseThe recombinant ECFP vector can be transfected into mammalian cells using any standard transfection method. If required, stable transformants can be selected using G418 (8). pECFP-N1 can also be used simply to express ECFP in a cell line of interest (e.g., as a transfection marker). ECFP can be used for double-labeling experiments together with EYFP using standard fluorescence microscopy and the appropriate filter sets.G CTA GCG CTA CCG GAC TCA GAT CTC GAG CTC AAG CTT CGA ATT CTG CAG TCG ACG GTA CCG CGG GCC CGG GAT CCA CCG GTC GCC ACC 591•611•601•621•631•641•661•651•671•Hin d III Xho I Apa I Bsp 120 I Kpn I Asp 718 I Bam H I ECFPAge I Xma I Sma ISac II Sal I Acc I Sac I Ecl 136 II Eco R I Pst I Eco 47 IIIBgl II Nhe II (1402) (1640)Eco (3856)(2579)Ase IG I (1389)Location of features•Human cytomegalovirus (CMV) immediate early promoter: 1–589Enhancer region: 59–465TATA box: 554–560Transcription start point: 583C→G mutation to remove Sac I site: 569•MCS: 591–671•Enhanced cyan fluorescent protein (ECFP) geneKozak consensus translation initiation site: 672–682Start codon (ATG): 679–681; stop codon: 1396–1398Insertion of Val at position 2: 682–684ECFP mutations:Phe-64 to Leu, Ser-65 to Thr, and Tyr-66 to Trp: 871–879Asn-146 to Ile: 1117–1119Met-153 to Thr: 1138–1140Val-163 to Ala: 1168–1170His-231 to Leu mutation (A→T): 1373•SV40 early mRNA polyadenylation signalPolyadenylation signals: 1552–1557 & 1581–1586mRNA 3' ends: 1590 & 1602•f1 single-strand DNA origin: 1649–2104(Packages the noncoding strand of ECFP.)•Bacterial promoter for expression of Kan r gene:–35 region: 2166–2171; –10 region: 2189–2194Transcription start point: 2201•SV40 origin of replication: 2445–2580•SV40 early promoterEnhancer (72-bp tandem repeats): 2278–2349 & 2350–242121-bp repeats: 2425–2445, 2446–2466 & 2468–2488Early promoter element: 2501–2507Major transcription start points: 2497, 2535, 2541 & 2546•Kanamycin/neomycin resistance geneNeomycin phosphotransferase coding sequences:Start codon (ATG): 2629–2631; stop codon: 3421–3423G→A mutation to remove Pst I site: 2811C→A (Arg to Ser) mutation to remove Bss H II site: 3157•Herpes simplex virus (HSV) thymidine kinase (TK) polyadenylation signalPolyadenylation signals: 3659–3664 & 3672–3677•pUC plasmid replication origin: 4008–4651Primer Locations•EGFP-N Sequencing Primer (#6479-1): 745–724•EGFP-C Sequencing Primer (#6478-1): 1332–1353Propagation in E. coli•Suitable host strains: DH5α, HB101, and other general-purpose strains. Single-stranded DNA production requires a host containing an F plasmid such as JM101 or XL1-Blue.•Selectable marker: plasmid confers resistance to kanamycin (30 µg/ml) in E. coli hosts.• E. coli replication origin: pUC;Copy number: ~500•Plasmid incompatibility group: pMB1/Col E1References1.Heim, R.,& Tsien, R. Y. (1996) Curr. Biol. 6:178–182.2.Heim, R., et al. (1994) Proc. Natl. Acad. Sci. USA 91:12501–12504.3.Cormack, B., et al. (1996) Gene 173:33–38.4.Heim, R., et al. (1995) Nature373:663–664.5.Yang, T. T., et al. (1996) Nucleic Acids Res.24:4592–4593.6.Haas, J., et al. (1996) Curr. Biol.6:315–324.7.Kozak, M. (1987) Nucleic Acids Res.15:8125–8148.8.Gorman, C. (1985) In DNA cloning: A practical approach, vol. II. Ed. D. M. Glover. (IRL Press, Oxford, U.K.), pp. 143–190.BD Biosciences Clontech Protocol # PT3285-5 2Version # PR29991Protocol # PT3285-5 BD Biosciences ClontechVersion # PR299913Notice to PurchaserUse of BD Biosciences Clontech’s Living Colors™ products containing DNA sequences coding for mutant Aequorea victoria green fluorescent protein (GFP) variants or proteins thereof requires a license from Amersham Biosciences under U.S. Patent Nos. 5,625,048; 5,777,079; 6,054,321and other pending U.S. and foreign patent applications. In addition, certain BD Biosciences Clontech products are made under U.S. Patent No.5,804,387 licensed from Stanford University.Not-For-Profit research institutes or entities are granted an automatic license with the purchase of this product for use in non-commercial internal research purposes, the terms of which are disclosed in detail in the license that accompanies the shipment of this product. Such license specifi-cally excludes the right to sell or otherwise transfer this product or its components to third parties.For-Profit research institutes or entities must obtain a license from Amersham Biosciences. E-mail: gfp@Please contact BD Biosciences Clontech directly for any other assistance, including purchasing and technical support. All companies and institutions purchasing Living Colors™ products will be included in a quarterly report to Aurora Biosciences, as required by the BD Biosciences Clontech/Aurora Biosciences license agreement.This product is intended to be used for research purposes only. It is not to be used for drug or diagnostic purposes nor is it intended for human use. BD Biosciences Clontech products may not be resold, modified for resale, or used to manufacture commercial products without written approval of BD Biosciences Clontech.© 2002, Becton, Dickinson and Company Note: The attached sequence file has been compiled from information in the sequence databases, published literature, and other sources, together with partial sequences obtained by BD Biosciences Clontech. This vector has not been completely sequenced.。
质粒图谱的阅读PPT课件

CHENLI
51
2021/3/7
CHENLI
52
2021/3/7
CHENLI
53
2021/3/7
CHENLI
54
2021/3/7
CHENLI
55
免疫法筛选
• 外源基因在细胞内表达出的蛋白质,可 与特异性的抗体结合。
2021/3/7
CHENLI
56
利用抗体筛选
原理 目的基因编码的蛋白质作为抗原,用特异
2021/3/7
CHENLI
33
氨苄青霉素 抗性基因
2021/3/7
CHENLI
复制起点
34
2021/3/7
CHENLI
35
2021/3/7
CHENLI
36
2.利用颜色筛选
插入失活现象
X-gal :5-溴-4-氯-3-吲哚-B-D-半乳糖苷
2021/3/7
CHENLI
37
多 克 隆 位 点
CHENLI
7
一.目的基因与载体的连接
Link of cohesive end
2021/3/7
粘末端连接
CHENLI
8
Link of blunt end
平末端连接
2021/3/7
CHENLI
9
平末端连接
vector
1
T4 DNA ligase
Recombinant DNA
2021/3/7
CHENLI
2021/3/7
CHENLI
40
2021/3/7
CHENLI
表示外源 基因插入 编码半乳 糖苷酶的 基因区段
41
Target gene
人源pEGFP-C1-p38γ真核表达质粒的构建及其功能研究

・97 •安徽医科大学学报 Acta Unwersitath Medicinalit An,aui 2028 Jo ;56(8)网络出版时间:2020 -10 -9 O :/) 网络出版地址:https ://kus. cnkd uePkcms/dxWl/34. 1065. R ; 20201208. 091). 033. html人源pEGFP-C1-2337真核表达质粒的构建及其功能研究胡 爽*8,杨 莉8,陈 晨8,潘林鑫2,李良云8,杨俊发8,周 焕6,徐 涛82225 -08 -10 接收基金项目:国家自然科学基金(编号:81779522-安徽省自然科学基金(编号:00085 MH235)作者单位:安徽医科大学8药学院、、生命科学学院,合肥2369348蚌埠医学院第一附属医院国家药物临床试验机构,蚌埠233794作者简介:胡爽,女,硕士研究生;周焕,男,副教授,硕士生导师,责任作者,E-mail : zhhuhuankest@ 163. com ;徐 涛,男,副教授,硕士生导师,责任作者,E-mail : xxtoc@ abmu. eXu. cn摘要目的构建人源pEGFP-L)-p33Y 表达质粒,并观察其对乙醇刺激的L0-2肝细胞的增殖和凋亡及炎症因子分泌的影响。
方法 在人源L0O 肝细胞中提取RNA 并且逆转录 为cDNA,同时将引物稀释到10 pnol/L ,其余作为储液备用。
采用PCR 技术扩增p 38y 并鉴定,使用AxyPrep DNA 凝胶回收试剂盒进行PCR 产物的纯化回收,再对PCR 产物及载体进行酶切回收,酶切片段连接后,进行连接产物的转化、 挑菌、摇菌、质粒小抽。
酶切鉴定后,进行测序鉴定。
将构建好的质粒转染至乙醇刺激的人源L0-2细胞中,通过MTT 实验和流式细胞术检测对其增殖和凋亡的影响,并用Westernblot 技术检测炎症因子白细胞介素O((P-9)和肿瘤坏死因子-c(TNF-c )在L0C 肝细胞中的表达。
质粒图谱大全

.(转载)一.九种表达载体Pllp-OmpA, pllp-STII, pMBP-P, pMBP-C,pET-GST, pET-Trx, pET-His, pET-CKS, pET-DsbA二.克隆载体pTZ19RDNApUC57DNAPMD18TPQE30pUC18pUC19pTrcHisApTrxFuspRSET-ApRSET-BpVAX1PBR322pbv220pBluescriptIIKS( )L4440pCAMBIA-1301pMAL-p2XpGD926三.PET系列表达载体ProteinExpression?ProkaryoticExpression?pETDsbFusionSystems39band40b ProteinExpression?ProkaryoticExpression?pETExpressionSystem33b ProteinExpression?ProkaryoticExpression?pETExpressionSystems ProteinExpression?ProkaryoticExpression?pETExpressionSystemsplusCompetentCells ProteinExpression?ProkaryoticExpression?pETGSTFusionSystems41and42 ProteinExpression?ProkaryoticExpression?pETNusAFusionSystems43.1and44 ProteinExpression?ProkaryoticExpression?pETVectorDNAProteinPurification?PurificationSystems?Strep?TactinResinsandPurificationKits四.PGEX系列表达载体TEcoR?pGEX-1I/BAP.pGEX-2TpGEX-2TKpGEX-3XpGEX-4T-1pGEX-4T-2pGEX-4T-3pGEX-5X-1pGEX-5X-2pGEX-5X-3pGEX-6P-1pGEX-6P-2pGEX-6P-3五.PTYBsystemPTYB1PTYB2PTYB11PTYB12六.真核表达载体pCDNA3.1(-)pCDNA3.1( )pPICZalphaApGAPZαAPYES2.0pBI121pEGFP-N1pEGFP-C1pPIC9KpPIC3.5K如何阅读分析质粒图谱载体主要有病毒和非病毒两大类,其中质粒DNA是一种新的非病毒转基因载体。
质粒图谱——精选推荐

1)质粒图谱登记号:00012)质粒名称:pIRES3)来源:BD Co4)用途:真核双表达5)是否可以提供更详细资料:可以6)是否可以共享:7)联系方式:PM)质粒图谱登记号:00022)质粒名称:pECFP-C13)来源:BD Co4)用途:检测真核表达5)是否可以提供更详细资料:可以6)是否可以共享:7)联系方式:pm1)质粒图谱登记号:00032)质粒名称:pShuttle3)来源:4)用途:5)是否可以提供更详细资料:6)是否可以共享:7)联系方式:2)pShuttle MCS很多人用pEGFP-C1,我也来发一个)质粒图谱登记号:00042)质粒名称:pS BR322、pUC183)来源:4)用途:5)是否可以提供更详细资料:6)是否可以共享:7)联系方式:1)质粒图谱登记号:00052)质粒名称:pcDNA3.1(+)/CAT3)来源:invitrogen4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:无偿7)联系方式:PM1)质粒图谱登记号:00062)质粒名称:pQEx3)来源:Qiagen4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM2)1)质粒图谱登记号:00072)质粒名称:pIVEX2.33)来源:Rocho4)用途:体外转录翻译5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM质粒图谱登记号:0008质粒名称:pIRES-EGFP来源:用途:是否可以提供更详细资料:是否可以共享:否联系方式:1)质粒图谱登记号:00092)质粒名称:pET-28a(+)3)来源:Novagen4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM1)质粒图谱登记号:00112)质粒名称:pET-32a(+)3)来源:novagen4)用途:原核表达5)是否可以提供更详细资料:6)是否可以共享:7)联系方式:pm1)质粒图谱登记号:00132)质粒名称:pcDNA3.1/Zeo (+)3)来源:invitrogen4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)质粒图谱登记号:00132)质粒名称:pEGFP-N33)来源:clontech4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)质粒图谱登记号:00142)质粒名称:pcDNA33)来源:invitrogen4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)质粒图谱登记号:00152)质粒名称:pfastbac13)来源:invitrogene4)用途:昆虫表达5)是否可以提供更详细资料:不可以6)是否可以共享:不可以7)联系方式:PM)质粒图谱登记号:00162)质粒名称:pEGFP-C33)来源:clontech4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PMwangjun2002274 edited on 2004-06-22 00:041)质粒图谱登记号:00172)质粒名称:pSecTag23)来源:Invitrogen4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)1质粒图谱登记号:00192)质粒名称:pET20b3)来源:NOVAGEN4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM1)质粒图谱登记号:00222)质粒名称:pThioHisA3)来源:invitrogen4)用途:原核表达5)是否可以提供更详细资料:4.365kb , HP-thioredoxin fusion proteinexpressionvector, trc promoter, Ampr, a EK cleavage site lies between HP-thioredoxin and MCS宿主菌TOP10(基因型为:F-mcrA △(mrr-hsd RMS-mcrBC)Ф80 lacZ M15 △lacX74 deoR recAl araD139 △(ara-leu)7697 galU galK rpsL endAl nupG6)是否可以共享:交换或其它7)联系方式:PM2)3)1)质粒图谱登记号:00242)质粒名称:pcDNA3.1-Myc-His-A-3)来源:invitrogen4)用途:真核核表达4))质粒图谱登记号:00252)质粒名称:pS UPER.neo3)来源:4)用途:siRNA5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)质粒图谱登记号:00312)质粒名称:pSilencer1.0-siRNA3)来源:Ambion4)用途:RNAi5)是否可以提供更详细资料:/techlib/Documents.html?fkResSxn=7&fkSub Sxn=236)是否可以共享:实验结束后,可提供含shRNA模板的质粒7)联系方式:pm5))质粒图谱登记号:00322)质粒名称:pSilencer2.0-U6siRNA3)来源:Ambion4)用途:RNAi,与1.0相比,可以建立稳转株5)更详细资料:/techlib/prot/fm_7209.pdf6)是否可以共享:交换7)联系方式:pm6)会员名:mlluoE-mail:*************可提供试验资源名称和简要介绍:pSilencer 3.1-H1 neo Vector,是Ambion公司目前最高版本的shRNA 载体,为扩增此载体我已插入目的片段,如有战友需要,将此片段双酶切再连上自己的片段即可。
pEGFP_N1_BMP_2真核表达质粒的构建与鉴定

基金项目:高等学校博士点专项科研基金(编号:20060183050)3通讯作者文章编号:1007-4287(2009)12-1683-03pEGFP 2N1/BMP 22真核表达质粒的构建与鉴定张明磊,常 非,高忠礼,柳 扬,刘光耀3(吉林大学中日联谊医院骨科,吉林长春130033)摘要:目的 构建pEG FP 2N1/BMP 22真核表达质粒,并进行鉴定。
方法 利用RT 2PCR 方法从组织中提取骨形态发生蛋白(BMP 22)的基因片段,与pM D182T 载体连接,构建pM D182T/BMP 22重组质粒,酶切后与pEG FP 2N1真核表达载体连接,构建pEG FP 2N1/BMP 22真核表达质粒,进行测序、酶切鉴定,表明质粒构建成功。
结果 通过测序、酶切鉴定,证明pM D182T/BMP 22质粒构建成功。
结论 本实验成功构建了pEG FP 2N1/BMP 22真核表达质粒,为进一步研究利用BMP 22基因修饰骨组织工程骨种子细胞提供实验基础。
关键词:pEG FP 2N1;骨形成蛋白2;基因中图分类号:R34文献标识码:AConstruction and identification of euk aryotic expression vector pEGFP 2N 1/BMP 22 ZH ANG Ming 2lei ,CH ANG fei ,G AO Zhong 2li ,et al.(China 2Japan Union Hospital o f Jilin Univer sity ,Changchun 130033,China )Abstract :Objective T o construct a eukary otic expression vector carrying BMP 22gene.Methods BMP 22gene was cloned from tissue by using RT 2PCR method and was inserted into pM D182T to constrcuct pM D182T/BMP 22plasmid ,and to construct pEG FP 2N1/BMP 22plasmid by using restriction enzyme.T o identify the plasmid by sequencing and restriction enzyme.R esults The eukary otic expression vector pEG FP 2N1/BMP 22was constructed and identified.Conclusion T o construct the expression vector suc 2cess fully ,BMP 22gene was able to was for bone engineering.K ey w ords :pEG FP 2N1;BMP 22;gene(Chin J Lab Diagn ,2009,13:1683) 在新生骨组织的形成过程中,多种相关细胞因子参与其中并起到十分重要的作用,通过细胞因子的浓度发生改变,继而通过募集祖细胞,发挥调节细胞的增殖和分化等作用。
质粒图谱大全

(转载)一.九种表达载体Pllp-OmpA, pllp-STII, pMBP-P, pMBP-C,pET-GST, pET-Trx, pET-His, pET-CKS, pET-DsbA二.克隆载体pTZ19RDNApUC57DNAPMD18TPQE30pUC18pUC19pTrcHisApTrxFuspRSET-ApRSET-BpVAX1PBR322pbv220pBluescriptIIKS( )L4440pCAMBIA-1301pMAL-p2XpGD926三.PET系列表达载体ProteinExpression?ProkaryoticExpression?pETDsbFusionSystems39band40b ProteinExpression?ProkaryoticExpression?pETExpressionSystem33b ProteinExpression?ProkaryoticExpression?pETExpressionSystems ProteinExpression?ProkaryoticExpression?pETExpressionSystemsplusCompetentCells ProteinExpression?ProkaryoticExpression?pETGSTFusionSystems41and42 ProteinExpression?ProkaryoticExpression?pETNusAFusionSystems43.1and44 ProteinExpression?ProkaryoticExpression?pETVectorDNAProteinPurification?PurificationSystems?Strep?TactinResinsandPurificationKits四.PGEX系列表达载体TEcoR?pGEX-1I/BAPpGEX-2TpGEX-2TKpGEX-3XpGEX-4T-1pGEX-4T-2pGEX-4T-3pGEX-5X-1pGEX-5X-2pGEX-5X-3pGEX-6P-1pGEX-6P-2pGEX-6P-3五.PTYBsystemPTYB1PTYB2PTYB11PTYB12六.真核表达载体pCDNA3.1(-)pCDNA3.1( )pPICZalphaApGAPZαAPYES2.0pBI121pEGFP-N1pEGFP-C1pPIC9KpPIC3.5K如何阅读分析质粒图谱载体主要有病毒和非病毒两大类,其中质粒DNA是一种新的非病毒转基因载体。
质粒图谱查询方法

0037--pQE9--Qiagen--原核表达--jpsgf
0038--pCANTAB5E--Phamcia--噬菌体抗库--yanBaggio
0039--pET-24a--Novagene--原核表达--yanBaggio
0006--pQEx--Qiagen--原核表达--wangjun2002274
0007--pIVEX2.3--Roche--体外转录翻译--wangjun2002274
0008--pIRES-EGFP--/--/--luoqingli2003
0009--pET-28a(+)--Novagen--原核表达--wangjun2002274
0014--pcDNA3--invitrogen--真核表达--kras
0015--pfastbac1--invitrogene--昆虫表达--cox2wj
0016--pEGFP-C3--clontech--真核表达--smilely
0017--pSecTag2--invitrogen--真核表达--kenmed
0018--PYX212--/--真核表达--Gmail
0019--pET20b--Novagen--原核表达--Gmail
0020--pEGFP-1--BD BIO--转录调控--wyh
0021--pEGFP/U6 --/--siRNA--songbinn
0022--pThioHisA--invitrogen--原核表达--xuezhishui
0081--pcDNA5/FRT/CAT—INVITROGEN--真核表达--renke3333
pEGFP-N1质粒图谱

pEGFP-N1 Vector Information PT3027-5GenBank Accession #U55762Catalog #6085-1Restriction Map and Multiple Cloning Site (MCS) of pEGFP-N1 Vector. (Unique restriction sites are in bold.) The Not I site follows the EGFP stop codon. The Xba I site (*) is methylated in the DNA provided by CLONTECH. If you wish to digest the vector with this enzyme, you will need to transform the vector into a dam – and make fresh DNA.G I (1389) I (1402)I * (1412) (1640)Eco (3856)(2579)Ase IDescription:pEGFP-N1 encodes a red-shifted variant of wild-type GFP (1–3) which has been optimized for brighter fluorescence and higher expression in mammalian cells. (Excitation maximum = 488 nm;emission maximum = 507 nm.) pEGFP-N1 encodes the GFPmut1 variant (4) which contains the double-amino-acid substitution of Phe-64 to Leu and Ser-65 to Thr. The coding sequence of the EGFP gene contains more than 190 silent base changes which correspond to human codon-usage preferences (5). Sequences flanking EGFP have been converted to a Kozak consensus translation initiation site (6) to further increase the translation efficiency in eukaryotic cells. The MCS in pEGFP-N1 is between the immediate early promoter of CMV (P CMV IE ) and the EGFP coding sequences. Genes cloned into the MCS will be expressed as fusions to the N-terminus of EGFP if they are in the same reading frame as EGFP and there are no intervening stop codons. SV40polyadenylation signals downstream of the EGFP gene direct proper processing of the 3' end of the EGFP mRNA. The vector backbone also contains an SV40 origin for replication in mammalian cells expressing the SV40 T antigen. A neomycin-resistance cassette (Neo r ), consisting of the SV40 early promoter, the neomycin/kanamycin resistance gene of Tn5, and polyadenylation signals from the Herpes simplex virus thymidine kinase (HSV TK) gene, allows stably transfected eukaryotic cells to be selected using G418. A bacterial promoter upstream of this cassette expresses kanamycin resistance in E. coli . The pEGFP-N1 backbone also provides a pUC origin of replication for propagation in E. coli and an f1 origin for single-stranded DNA production.G CTA GCG CTA CCG GAC TCA GAT CTC GAG CTC AAG CTT CGA ATT CTG CAG TCG ACG GTA CCG CGG GCC CGG GAT CCA CCG GTC GCC ACC 591•611•601•621•631•641•661•651•671•Hin d III Xho I Apa I Bsp 120 I Kpn I Asp 718 I Bam H I EGFPAge I Xma I Sma ISac II Sal I Acc I Sac I Ecl136 II Eco R I Pst I Eco 47 IIIBgl II Nhe I (PR93634; published 19 March 1999)Use:Fusions to the N terminus of EGFP retain the fluorescent properties of the native protein allowing the localization of the fusion protein in vivo . The target gene should be cloned into pEGFP-N1 so that it is in frame with the EGFP coding sequences, with no intervening in-frame stop codons. The inserted gene should include the initiating ATG codon. The recombinant EGFP vector can be transfected into mammalian cells using any standard transfection method. If required, stable transformants can be selected using G418 (7). pEGFP-N1 can also be used simply to express EGFP in a cell line of interest (e.g., as a transfection marker).Location of features:•Human cytomegalovirus (CMV) immediate early promoter: 1–589Enhancer region:59–465; TATA box: 554–560Transcription start point: 583C→G mutation to remove Sac I site: 569•MCS: 591–671•Enhanced green fluorescent protein (EGFP) geneKozak consensus translation initiation site: 672–682Start codon (ATG): 679–681; Stop codon: 1396–1398Insertion of Val at position 2: 682–684GFPmut1 chromophore mutations (Phe-64 to Leu; Ser-65 to Thr): 871–876His-231 to Leu mutation (A→T): 1373•SV40 early mRNA polyadenylation signalPolyadenylation signals: 1552–1557 & 1581–1586; mRNA 3' ends: 1590 & 1602•f1 single-strand DNA origin: 1649–2104 (Packages the noncoding strand of EGFP.)•Bacterial promoterfor expression of Kan r gene:–35 region: 2166–2171; –10 region: 2189–2194Transcription start point: 2201•SV40 origin of replication: 2445–2580•SV40 early promoterEnhancer (72-bp tandem repeats): 2278–2349 & 2350–242121-bp repeats: 2425–2445, 2446–2466 & 2468–2488Early promoter element: 2501–2507Major transcription start points: 2497, 2535, 2541 & 2546•Kanamycin/neomycin resistance geneNeomycin phosphotransferase coding sequences: start codon (ATG): 2629–2631; stop codon: 3421–3423 G→A mutation to remove Pst I site: 2811C→A (Arg to Ser) mutation to remove Bss H II site: 3157•Herpes simplex virus (HSV) thymidine kinase (TK) polyadenylation signalPolyadenylation signals: 3659–3664 & 3672–3677•pUC plasmid replication origin: 4008–4651Primer Locations:•EGFP-N Sequencing Primer (#6479-1): 745–724•EGFP-C Sequencing Primer (#6478-1): 1332–1353Propagation in E. coli:•Suitable host strains: DH5a, HB101 and other general purpose strains. Single-stranded DNA production requires a host containing an F plasmid such as JM101 or XL1-Blue.•Selectable marker: plasmid confers resistance to kanamycin (30 µg/ml) to E. coli hosts.• E. coli replication origin: pUC•Copy number: ≈500•Plasmid incompatibility group: pMB1/ColE1References:1.Prasher, D. C. et al. (1992) Gene 111:229–233.2.Chalfie, M. et al. (1994) Science263:802–805.3.Inouye, S. & Tsuji, F. I. (1994) FEBS Letters 341:277–280.4.Cormack, B. et al. (1996) Gene 173:33–38.5.Haas, J., et al. (1996) Curr. Biol. 6:315–324.6.Kozak, M. (1987) Nucleic Acids Res. 15:8125–8148.7.Gorman, C. (1985). In DNA cloning: A practical approach, vol. II. Ed. D.M. Glover. (IRL Press, Oxford, U.K.) pp. 143–190.Notice to PurchaserUse of CLONTECH’s Living Colors® products containing DNA sequences coding for mutant Aequorea victoria green fluorescent protein (GFP) variants or proteins thereof requires a license from Aurora Biosciences Corporation under U.S. Patent Nos. 5,625,048 and 5,777,079 and other pending U.S. and foreign patent applications. In addition, certain CLONTECH products are made under U.S. Patent No. 5,804,387 licensed from Stanford University.Not-For-Profit research institutes or entities are granted an automatic license with the purchase of this product for use in non-commercial internal research purposes, the terms of which are disclosed in detail in the license that accompanies the shipment of this product. Such license specifically excludes the right to sell or otherwise transfer this product or its components to third parties.For-Profit research institutes or entities that wish to use this product in non-commercial applications are required to obtain a license from CLONTECH prior to purchasing these reagents or using them for any purpose. For information on the terms of this license, or to obtain information on approved applications, you can download a copy of the license agreement, without payment terms, at / license/.Any For-Profit research institute that wishes to use this product for commercial applications must obtain a license from Aurora Biosciences Corporation. For commercial license information only contact: Court Turner at 619-404-8416 or Fax 619-404-6743 or . Please contact CLONTECH directly for any other assistance, including purchasing and technical support.All companies and institutions purchasing Living Colors® products will be included in a quarterly report to Aurora Biosciences Corporation, as required by the CLONTECH/Aurora license agreement.The attached sequence file has been compiled from information in the sequence databases, published literature, and other sources, together with partial sequences obtained by CLONTECH. This vector has not been completely sequenced.This product is intended to be used for research purposes only. It is not to be used for drug or diagnostic purposes nor is it intended for human use. CLONTECH products may not be resold, modified for resale, or used to manufacture commercial products without written approval of CLONTECH.© 1999, CLONTECH Laboratories, Inc.。
