遗传算法的C语言程序案例
遗传算法旅行商问题c语言代码

遗传算法是一种模拟自然选择过程的优化算法,可以用于解决各种复杂的组合优化问题。
其中,旅行商问题是一个经典的组合优化问题,也是一个典型的NP难题,即寻找最优解的时间复杂度是指数级的。
在本文中,我们将讨论如何使用遗传算法来解决旅行商问题,并给出相应的C语言代码实现。
我们将介绍旅行商问题的数学模型,然后简要介绍遗传算法的原理,最后给出C语言代码实现。
旅行商问题是指一个旅行商要拜访n个城市,恰好拜访每个城市一次,并返回出发城市,要求总路程最短。
数学上可以用一个n*n的距离矩阵d[i][j]表示城市i到城市j的距离,问题可以形式化为求解一个排列p={p1,p2,...,pn},使得目标函数f(p)=Σd[p[i]][p[i+1]]+d[p[n]][p[1]]最小。
这个问题是一个组合优化问题,其搜索空间是一个n维的离散空间。
遗传算法是一种基于生物进化过程的优化算法,主要包括选择、交叉、变异等操作。
在使用遗传算法解决旅行商问题时,可以将每个排列p看作一个个体,目标函数f(p)看作个体的适应度,通过选择、交叉和变异等操作来搜索最优解。
以下是遗传算法解决旅行商问题的C语言代码实现:1. 我们需要定义城市的距离矩阵和其他相关参数,例如城市的数量n,种裙大小pop_size,交叉概率pc,变异概率pm等。
2. 我们初始化种裙,即随机生成pop_size个排列作为初始种裙。
3. 我们进入遗传算法的迭代过程。
在每一代中,我们首先计算种裙中每个个体的适应度,然后通过选择、交叉和变异操作来更新种裙。
4. 选择操作可以采用轮盘赌选择法,即根据个体的适应度来进行选择,适应度越高的个体被选中的概率越大。
5. 交叉操作可以采用部分映射交叉方法,即随机选择两个个体,然后随机选择一个交叉点,将交叉点之后的基因片段进行交换。
6. 变异操作可以采用变异率为pm的单点变异方法,即随机选择一个个体和一个位置,将该位置的基因值进行随机变异。
7. 我们重复进行迭代操作,直到达到停止条件(例如达到最大迭代次数或者适应度达到阈值)。
用混合遗传算法解决单机调度问题c语言程序

用混合遗传算法解决单机调度问题c语言程序#include#include#include#include#include#include "ceshi.h"void main(){s_individual *temp=NULL;long run_start,run_finish,run_sum=0;float run_avg=0;int sum_k=0;srand((unsigned)time(NULL));/*将当前时间设置成随机函数的种子,所以每次产生的数都不一样*///printf("%d\n",time(NULL));//srand((unsigned)1398759277);if((outfp = fopen("newfile1","w")) == NULL){fprintf(stderr,"Cannot open output file %s\n","file1");exit(-1);}init_parameter();//初始化工件参数print_parameter();//打印工件参数for(run=1; run<=maxruns; run++){run_start=clock();initialize();//初始化包括初始参数设置initmalloc();//分配全局空间gererate_pop();//产生初始种群statistics(oldpop);//统计种群数据,得到最优个体init_temperature=(min-max)/log(preceive); temperature_k=init_temperature;while(k{generation();//交叉select_better(oldpop,newpop);//保留最佳个体newpop popsize_copy(oldpop,newpop);gen_mu(newpop);//变异保留最佳个体select_better(oldpop,newpop);popsize_copy(oldpop,newpop);CSA(newpop);//模拟退火操作定步长抽样select_better(oldpop,newpop);statistics(newpop);run_finish=clock();if (last_fitness==bestfit.fitness){last_maxgen++;}else{last_maxgen=0;last_runtime=run_finish;}last_fitness=bestfit.fitness;k++;//退温temperature_k=temperature_k*ptemper;temp=oldpop;oldpop=newpop;newpop=temp;}sum_k=sum_k+k;run_time=last_runtime-run_start;run_sum=run_sum+run_time;print_popsize();ceshi_popstatic();freeall();}run_avg=(float)run_sum/float(maxruns);fprintf(outfp,"\nmax=%d min=%d avg=%d run_sum=%f avg_k=%d\n",pop_max,pop_min,pop_sum/maxruns,run_avg,su m_k);fclose(outfp);}。
解TSP问题的遗传算法C语言程序

解TSP问题的遗传算法C语言程序#include<stdio.h>#include<stdlib.h>#include<math.h>#include<alloc.h>#include<conio.h>#include<float.h>#include<time.h>#include<graphics.h>#include<bios.h>#define maxpop 100#define maxstring 100struct pp{unsigned char chrom[maxstring];float x,fitness;unsigned int parent1,parent2,xsite;};struct pp *oldpop,*newpop,*p1;unsigned int popsize,lchrom,gem,maxgen,co_min,jrand;unsigned int nmutation,ncross,jcross,maxpp,minpp,maxxy;floatpcross,pmutation,sumfitness,avg,max,min,seed,maxold,oldrand[maxstring];unsigned char x[maxstring],y[maxstring]; float*dd,ff,maxdd,refpd,fm[201]; FILE *fp,*fp1;float objfunc(float);void statistics();int select();int flip(float);int crossover();void generation();void initialize();void report();float decode();void crtinit();void inversion();float random1();void randomize1();main(){unsigned int gen,k,j,tt;char fname[10];float ttt;clrscr();co_min=0;if((oldpop=(struct pp *)farmalloc(maxpop*sizeof(struct pp)))==NULL) {printf("memory requst fail!\n");exit(0);} if((dd=(float*)farmalloc(maxstring*maxstring*sizeof(float)))==NULL){printf("memory requst fail!\n");exit(0);} if((newpop=(struct pp *)farmalloc(maxpop*sizeof(struct pp)))==NULL){printf("memory requst fail!\n");exit(0);} if((p1=(struct pp*)farmalloc(sizeof(struct pp)))==NULL){printf("memory requst fail!\n");exit(0);} for(k=0;k<maxpop;k++) oldpop[k].chrom[0]='\0'; for(k=0;k<maxpop;k++) newpop[k].chrom[0]='\0'; printf("Enter Result Data Filename:"); gets(fname);if((fp=fopen(fname,"w+"))==NULL){printf("cannot open file\n");exit(0);}gen=0;randomize();initialize();fputs("this is result of the TSP problem:",fp);fprintf(fp,"city: %2d psize: %3d Ref.TSP_path:%f\n",lchrom,popsize,refpd);fprintf(fp,"Pc: %f Pm: %f Seed: %f\n",pcross,pmutation,seed);fprintf(fp,"X site:\n");for(k=0;k<lchrom;k++){if((k%16)==0) fprintf(fp,"\n"); fprintf(fp,"%5d",x[k]);}fprintf(fp,"\n Y site:\n"); for(k=0;k<lchrom;k++){if((k%16)==0) fprintf(fp,"\n"); fprintf(fp,"%5d",y[k]);}fprintf(fp,"\n");crtinit();statistics(oldpop);report(gen,oldpop);getch();maxold=min;fm[0]=100.0*oldpop[maxpp].x/ff; do {gen=gen+1;generation();statistics(oldpop);if(max>maxold){maxold=max;co_min=0;}fm[gen%200]=100.0*oldpop[maxpp].x/ff; report(gen,oldpop);gotoxy(30,25);ttt=clock()/18.2;tt=ttt/60;printf("Run Clock: %2d: %2d: %4.2f",tt/60,tt%60,ttt-tt*60.0);printf("Min=%6.4fNm:%d\n",min,co_min); }while((gen<100)&&!bioskey(1)); printf("\n gen= %d",gen);do{gen=gen+1;generation();statistics(oldpop);if(max>maxold){maxold=max;co_min=0;}fm[gen%200]=100.0*oldpop[maxpp].x/ff; report(gen,oldpop);if((gen%100)==0)report(gen,oldpop); gotoxy(30,25);ttt=clock()/18.2;tt=ttt/60;printf("Run Clock: %2d: %2d: %4.2f",tt/60,tt%60,ttt-tt*60.0);printf("Min=%6.4fNm:%d\n",min,co_min); }while((gen<maxgen)&&!bioskey(1));getch();for(k=0;k<lchrom;k++) {if((k%16)==0)fprintf(fp,"\n");fprintf(fp,"%5d",oldpop[maxpp].chrom[k]);}fprintf(fp,"\n");fclose(fp);farfree(dd);farfree(p1);farfree(oldpop);farfree(newpop);restorecrtmode();exit(0);}/*%%%%%%%%%%%%%%%%*/float objfunc(float x1) {float y;y=100.0*ff/x1;return y;}/*&&&&&&&&&&&&&&&&&&&*/void statistics(pop) struct pp *pop;{int j;sumfitness=pop[0].fitness; min=pop[0].fitness; max=pop[0].fitness; maxpp=0;minpp=0;for(j=1;j<popsize;j++) {sumfitness=sumfitness+pop[j].fitness;if(pop[j].fitness>max){max=pop[j].fitness;maxpp=j;}if(pop[j].fitness<min){min=pop[j].fitness;minpp=j;}}avg=sumfitness/(float)popsize;}/*%%%%%%%%%%%%%%%%%%%%*/void generation(){unsigned int k,j,j1,j2,i1,i2,mate1,mate2;float f1,f2;j=0;do{mate1=select();pp:mate2=select();if(mate1==mate2)goto pp;crossover(oldpop[mate1].chrom,oldpop[mate2].chrom,j);newpop[j].x=(float)decode(newpop[j].chrom);newpop[j].fitness=objfunc(newpop[j].x); newpop[j].parent1=mate1;newpop[j].parent2=mate2;newpop[j].xsite=jcross;newpop[j+1].x=(float)decode(newpop[j+1].chrom);newpop[j+1].fitness=objfunc(newpop[j+1].x); newpop[j+1].parent1=mate1;newpop[j+1].parent2=mate2;newpop[j+1].xsite=jcross;if(newpop[j].fitness>min){for(k=0;k<lchrom;k++)oldpop[minpp].chrom[k]=newpop[j].chrom[k];oldpop[minpp].x=newpop[j].x;oldpop[minpp].fitness=newpop[j].fitness;co_min++;return;}if(newpop[j+1].fitness>min){for(k=0;k<lchrom;k++)oldpop[minpp].chrom[k]=newpop[j+1].chrom[k];oldpop[minpp].x=newpop[j+1].x;oldpop[minpp].fitness=newpop[j+1].fitness;co_min++;return;}j=j+2;}while(j<popsize);}/*%%%%%%%%%%%%%%%%%*/void initdata(){unsigned int ch,j;clrscr();printf("-----------------------\n");printf("A SGA\n");printf("------------------------\n");/*pause();*/clrscr();printf("*******SGA DATA ENTRY AND INITILIZATION *******\n");printf("\n");printf("input pop size");scanf("%d",&popsize); printf("input chrom length");scanf("%d",&lchrom); printf("input maxgenerations");scanf("%d",&maxgen); printf("input crossoverprobability");scanf("%f",&pcross); printf("input mutationprob");scanf("%f",&pmutation); randomize1();clrscr();nmutation=0;ncross=0;}/*%%%%%%%%%%%%%%%%%%%%*/void initreport(){int j,k;printf("pop size=%d\n",popsize);printf("chromosome length=%d\n",lchrom);printf("maxgen=%d\n",maxgen);printf("pmutation=%f\n",pmutation); printf("pcross=%f\n",pcross);printf("initial generation statistics\n"); printf("ini pop maxfitness=%f\n",max); printf("ini pop avr fitness=%f\n",avg); printf("ini pop min fitness=%f\n",min); printf("ini pop sum fit=%f\n",sumfitness); } void initpop(){unsigned char j1;unsigned int k5,i1,i2,j,i,k,j2,j3,j4,p5[maxstring];float f1,f2;j=0;for(k=0;k<lchrom;k++)oldpop[j].chrom[k]=k;for(k=0;k<lchrom;k++)p5[k]=oldpop[j].chrom[k];randomize();for(;j<popsize;j++){j2=random(lchrom);for(k=0;k<j2+20;k++){j3=random(lchrom);j4=random(lchrom);j1=p5[j3];p5[j3]=p5[j4];p5[j4]=j1;}for(k=0;k<lchrom;k++)oldpop[j].chrom[k]=p5[k]; }for(k=0;k<lchrom;k++)for(j=0;j<lchrom;j++)dd[k*lchrom+j]=hypot(x[k]-x[j],y[k]-y[j]);for(j=0;j<popsize;j++) {oldpop[j].x=(float)decode(oldpop[j].chrom); oldpop[j].fitness=objfunc(oldpop[j].x);oldpop[j].parent1=0;oldpop[j].parent2=0;oldpop[j].xsite=0;}}/*&&&&&&&&&&&&&&&&&*/void initialize(){int k,j,minx,miny,maxx,maxy; initdata();minx=0;miny=0;maxx=0;maxy=0;for(k=0;k<lchrom;k++){x[k]=rand();if(x[k]>maxx)maxx=x[k]; if(x[k]<minx)minx=x[k]; y[k]=rand();if(y[k]>maxy)maxy=y[k]; if(y[k]<miny)miny=y[k]; }if((maxx-minx)>(maxy-miny)){maxxy=maxx-minx;}else {maxxy=maxy-miny;}maxdd=0.0;for(k=0;k<lchrom;k++)for(j=0;j<lchrom;j++){dd[k*lchrom+j]=hypot(x[k]-x[j],y[k]-y[j]);if(maxdd<dd[k*lchrom+j])maxdd=dd[k*lchrom+j];}refpd=dd[lchrom-1];for(k=0;k<lchrom;k++)refpd=refpd+dd[k*lchrom+k+2]; for(j=0;j<lchrom;j++)dd[j*lchrom+j]=4.0*maxdd; ff=(0.765*maxxy*pow(lchrom,0.5)); minpp=0; min=dd[lchrom-1];for(j=0;j<lchrom-1;j++){if(dd[lchrom*j+lchrom-1]<min){min=dd[lchrom*j+lchrom-1];minpp=j;}}initpop();statistics(oldpop);initreport();}/*&&&&&&&&&&&&&&&&&&*/void report(int l,struct pp *pop){int k,ix,iy,jx,jy;unsigned int tt;float ttt;cleardevice();gotoxy(1,1);printf("city:%4d para_size:%4d maxgen:%4d ref_tour:%f\n",lchrom,popsize,maxgen,refpd);printf("ncross:%4d Nmutation:%4d Rungen:%4d AVG=%8.4f MIN=%8.4f\n\n",ncross,nmutation,l,avg,min);printf("inpath:%6.4f Minpath length:%10.4f Ref_co_tour:%f\n",pop[maxpp].x/maxxy,pop[maxpp].x,ff); printf("Co_minpath:%6.4f Maxfit:%10.8f",100.0*pop[maxpp].x/ff,pop[maxpp].fitness); ttt=clock()/18.2;tt=ttt/60;printf("Run clock:%2d:%2d:%4d.2f\n",tt/60,tt%60,ttt-tt*60.0); setcolor(1%15+1);for(k=0;k<lchrom-1;k++){ix=x[pop[maxpp].chrom[k]];iy=y[pop[maxpp].chrom[k]]+110;jx=x[pop[maxpp].chrom[k+1]];jy=y[pop[maxpp].chrom[k+1]]+110;line(ix,iy,jx,jy);putpixel(ix,iy,RED);}ix=x[pop[maxpp].chrom[0]];iy=y[pop[maxpp].chrom[0]]+110;jx=x[pop[maxpp].chrom[lchrom-1]];jy=y[pop[maxpp].chrom[lchrom-1]]+110; line(ix,iy,jx,jy); putpixel(jx,jy,RED);setcolor(11);outtextxy(ix,iy,"*");setcolor(12);for(k=0;k<1%200;k++){ix=k+280;iy=366-fm[k]/3;jx=ix+1;jy=366-fm[k+1]/3;line(ix,iy,jx,jy);putpixel(ix,iy,RED);}printf("GEN:%3d",l);printf("Minpath:%f Maxfit:%f",pop[maxpp].x,pop[maxpp].fitness); printf("Clock:%2d:%2d:%4.2f\n",tt/60,tt%60,ttt-tt*60.0);}/*###############*/float decode(unsigned char *pp) {int j,k,l;float tt;tt=dd[pp[0]*lchrom+pp[lchrom-1]]; for(j=0;j<lchrom-1;j++){tt=tt+dd[pp[j]*lchrom+pp[j+1]];} l=0;for(k=0;k<lchrom-1;k++)for(j=k+1;j<lchrom;j++){if(pp[j]==pp[k])l++;}return tt+4*l*maxdd;}/*%%%%%%%%%%%%%%%%%%*/ void crtinit(){int driver,mode;struct palettetype p;driver=DETECT;mode=0;initgraph(&driver,&mode,""); cleardevice();}/*$$$$$$$$$$$$$$$$$$$$*/ int select(){double rand1,partsum; float r1;int j;partsum=0.0;j=0;rand1=random1()*sumfitness; do{partsum=partsum+oldpop[j].fitness;j=j+1;}while((partsum<rand1)&&(j<popsize));return j-1;}/*$$$$$$$$$$$$$$$*/int crossover(unsigned char *parent1,unsigned char *parent2,int k5) {int k,j,mutate,i1,i2,j5;int j1,j2,j3,s0,s1,s2; unsigned charjj,ts1[maxstring],ts2[maxstring];float f1,f2;s0=0;s1=0;s2=0;if(flip(pcross)){jcross=random(lchrom-1); j5=random(lchrom-1); ncross=ncross+1;if(jcross>j5){k=jcross;jcross=j5;j5=k;}}else jcross=lchrom; if(jcross!=lchrom) {s0=1;k=0;for(j=jcross;j<j5;j++) {ts1[k]=parent1[j]; ts2[k]=parent2[j]; k++; }j3=k;for(j=0;j<lchrom;j++) {j2=0;while((parent2[j]!=ts1[j2])&&(j2<k)){j2++;}if(j2==k){ts1[j3]=parent2[j];j3++;}}j3=k;for(j=0;j<lchrom;j++){j2=0;while((parent1[j]!=ts2[j2])&&(j2<k)){j2++;}if(j2==k){ts2[j3]=parent1[j];j3++;}}for(j=0;j<lchrom;j++){newpop[k5].chrom[j]=ts1[j];newpop[k5+1].chrom[j]=ts2[j]; }}else{for(j=0;j<lchrom;j++){newpop[k5].chrom[j]=parent1[j]; newpop[k5+1].chrom[j]=parent2[j]; } mutate=flip(pmutation);if(mutate){s1=1;nmutation=nmutation+1;for(j3=0;j3<200;j3++){j1=random(lchrom);j=random(lchrom);jj=newpop[k5].chrom[j];newpop[k5].chrom[j]=newpop[k5].chrom[j1];newpop[k5].chrom[j1]=jj;}}mutate=flip(pmutation);if(mutate){s2=1;nmutation=nmutation+1;for(j3=0;j3<100;j3++){j1=random(lchrom);j=random(lchrom);jj=newpop[k5+1].chrom[j];newpop[k5+1].chrom[j]=newpop[k5+1].chrom[j1];newpop[k5+1].chrom[j1]=jj;}}}j2=random(2*lchrom/3);for(j=j2;j<j2+lchrom/3-1;j++)for(k=0;k<lchrom;k++){if(k==j)continue;if(k>j){i2=k;i1=j;}else{i1=k;i2=j;}f1=dd[lchrom*newpop[k5].chrom[i1]+newpop[k5].chrom[i2]]; f1=f1+dd[lchrom*newpop[k5].chrom[(i1+1)%lchrom]+newpop[k5].chrom[(i2+1)%lchrom]];f2=dd[lchrom*newpop[k5].chrom[i1]+newpop[k5].chrom[(i1+1)%lchrom]];f2=f2+dd[lchrom*newpop[k5].chrom[i2]+newpop[k5].chrom[(i2+1)%lchrom]];if(f1<f2){inversion(i1,i2,newpop[k5].chrom);}}j2=random(2*lchrom/3);for(j=j2;j<j2+lchrom/3-1;j++)for(k=0;k<lchrom;k++){if(k==j)continue;if(k>j){i2=k;i1=j;}else{i1=k;i2=j;}f1=dd[lchrom*newpop[k5+1].chrom[i1]+newpop[k5+1].chrom[i2]];f1=f1+dd[lchrom*newpop[k5+1].chrom[(i1+1)%lchrom]+newpop[k5+1].chrom[(i2+1)%lchrom]];f2=dd[lchrom*newpop[k5+1].chrom[i1]+newpop[k5+1].chrom[(i1+1)%lchrom]];f2=f2+dd[lchrom*newpop[k5+1].chrom[i2]+newpop[k5+1].chrom[(i2+1)%lchrom]];if(f1<f2){inversion(i1,i2,newpop[k5+1].chrom);} }return 1;}/*$$$$$$$$$$$$$$$*/void inversion(unsigned int k,unsigned int j,unsigned char *ss) {unsigned int l1,i;unsigned char tt;l1=(j-k)/2;for(i=0;i<l1;i++){tt=ss[k+i+1];ss[k+i+1]=ss[j-i];ss[j-i]=tt;}}/*%%%%%%%%%%%%%%%*/void randomize1(){int i;randomize();for(i=0;i<lchrom;i++) oldrand[i]=random(30001)/30000.0;jrand=0;}/*%%%%%%%%%%%*/float random1(){jrand=jrand+1;if(jrand>=lchrom){jrand=0;randomize1();}return oldrand[jrand]; }/*%%%%%%%%%%*/int flip(float probability) {float ppp;ppp=random(20001)/20000.0; if(ppp<=probability)return 1; return 0;}%TSP问题(又名:旅行商问题,货郎担问题)遗传算法通用matlab程序 %D是距离矩阵,n为种群个数,建议取为城市个数的1~2倍, %C为停止代数,遗传到第 C 代时程序停止,C的具体取值视问题的规模和耗费的时间而定%m为适应值归一化淘汰加速指数 ,最好取为1,2,3,4 ,不宜太大 %alpha为淘汰保护指数,可取为0~1之间任意小数,取1时关闭保护功能,最好取为0.8~1.0 %R为最短路径,Rlength为路径长度function [R,Rlength]=geneticTSP(D,n,C,m,alpha)[N,NN]=size(D);farm=zeros(n,N);%用于存储种群for i=1:nfarm(i,:)=randperm(N);%随机生成初始种群endR=farm(1,:);%存储最优种群len=zeros(n,1);%存储路径长度fitness=zeros(n,1);%存储归一化适应值counter=0;while counter<Cfor i=1:nlen(i,1)=myLength(D,farm(i,:));%计算路径长度endmaxlen=max(len);minlen=min(len);fitness=fit(len,m,maxlen,minlen);%计算归一化适应值rr=find(len==minlen);R=farm(rr(1,1),:);%更新最短路径FARM=farm;%优胜劣汰,nn记录了复制的个数nn=0;for i=1:nif fitness(i,1)>=alpha*rand nn=nn+1;FARM(nn,:)=farm(i,:);endendFARM=FARM(1:nn,:);[aa,bb]=size(FARM);%交叉和变异while aa<nif nn<=2nnper=randperm(2);elsennper=randperm(nn);endA=FARM(nnper(1),:);B=FARM(nnper(2),:);[A,B]=intercross(A,B); FARM=[FARM;A;B]; [aa,bb]=size(FARM);endif aa>nFARM=FARM(1:n,:);%保持种群规模为nendfarm=FARM;clear FARMcounter=counter+1endRlength=myLength(D,R);function [a,b]=intercross(a,b) L=length(a); if L<=10%确定交叉宽度W=1;elseif ((L/10)-floor(L/10))>=rand&&L>10W=ceil(L/10);elseW=floor(L/10);endp=unidrnd(L-W+1);%随机选择交叉范围,从p到p+W for i=1:W%交叉x=find(a==b(1,p+i-1)); y=find(b==a(1,p+i-1)); [a(1,p+i-1),b(1,p+i-1)]=exchange(a(1,p+i-1),b(1,p+i-1));[a(1,x),b(1,y)]=exchange(a(1,x),b(1,y));endfunction [x,y]=exchange(x,y) temp=x;x=y;y=temp;% 计算路径的子程序function len=myLength(D,p) [N,NN]=size(D);len=D(p(1,N),p(1,1)); for i=1:(N-1)len=len+D(p(1,i),p(1,i+1));end%计算归一化适应值子程序function fitness=fit(len,m,maxlen,minlen) fitness=len;for i=1:length(len)fitness(i,1)=(1-((len(i,1)-minlen)/(maxlen-minlen+0.000001))).^m;end。
(完整版)遗传算法c语言代码

遗传算法代码# iiiclude<stdio.h>#mclude<stnng.h> #mclude<stdlib.h> #mclude<math.h> #mclude<tmie.h>^define cities 10 〃城市的个数 ^define MAXX ]00//迭代次数 #define pc 0.8 〃交配概率 #define pm 0.05 〃变异概率 ^define num 10〃种群的人小 int bestsolution;//最优染色体int distance[cities] [cities];// 城市之间的距离stmct group //染色体的结构{int city [cities];// 城市的顺序int adapt;// 适应度 double p 〃在种群中的幸存概率} group [num] ,grouptemp [num];〃随机产生10个城市之间的相互距离 voidinit(){intij ;meniset(distance.0.sizeof(distance)); srand((uiisigned)tuue(NULL)); fbr(i=O ;i<cities;i++){fbr(j=i+l ;j<cities J++){distance [i] [j]=rand()% 100; distance[j][i]=distance[i]Ij];} }fbr(i=O;i<cities;i++)printf( ”************ 城市的距离矩阵如下************\ii n );pruitf(M%4d H,distance[i][j]);}}〃随机产生初试群void groupproduceQ{mt i j 丄k,flag;fbi(i=O ;i<num; i++) //初始化for(j=OJ<citiesj++) group[i].city[j]=-l;srand((uiisigned)tuue(NULL));fbi(i=O ;i<num: i++)血(J=Oj<citi 亡s;){ t=rand()%cities;flag=l;for(k=0;k<j;k++){if(group[i] .city[k]=t){flag=O;break:}} if(flag){group[i].city|j]=t; J++;}}}pnntfC************ 初始种群如下^***************^);fbi(i=0;i<num: i++)血(J=0 J <citi 亡s;j++) pimtf(M%4d,\gioup[i].city[j]);〃评价函数,找岀最优染色体void pmgjiaQint ij;iiit nl,ii2;mt sumdistance5biggestsum=O; double biggestp=O;fdr(i=O;i<num; i++){sumdistance=O;{nl=group[i].city|j-l];n2=group[i].city[j]; sumdistance4-=distance[nl][n2];}group [1] .adapt=sumd istaiice; 〃每条染色体的路径总和biggestsum+=sumdistance; 〃种群的总路径}fbi(i=O ;i<num: i++){group [i].p= 1 -(double)gioup(i] .adapt/(double)biggestsum;biggestp+=group[i].p;}fbi(i=O ;i<num: i++)gioup[i] .p=gioup[i] .p./biggestp;〃求最佳路劲bestsolution=0;fbi(i=O ;i<num: i++) if(gioup[i].p>gioup[bestsolution].p) bestsolution=i; }〃选择void xuanzeQ{mt ij^temp;double gradient[num]^/梯度概率double xuaiize[num];//选择染色体的随机概率mt xuaii[num];//选择了的染色体〃初始化梯度概率fbi(i=O ;i<num: i++)gradient[i]=O.O; xuaiize[i]=O.O;}gradient[0]=gioup[0].p;fbr(i= 1 ;i<num:i++)gradient[i]=gradient[i-1 ]+group[i] .p; srand((uiisigned)tune(NULL));〃随机产生染色体的存活概率fdr(i=O;i<num: i++){xuanze[i]=(rand()% 100); xuaiize[i]/=100;}〃选择能生存的染色体fdr(i=0;i<num: i++){{if(xu aiize [i] <gradient [j ]){xuan[i]=j; //第i个位置存放第j个染色体break;}}}〃拷贝种群fdr(i=0;i<num: i++){grouptenip [i]. adapt=gioup [i].adapt;giouptemp[i] .p=group[i] .p;fbi(j=0 j <cities;j ++)grouptenip [i].citv|j]=group [i]. c ity[j ];}〃数据更新fdr(i=0;i<num: i++){temp=xuan[i];groupfi] .adapt=giouptemp[temp] .adapt;group [1] .p=giouptemp [temp] .p;fbi(j=0 j <cities;j ++)group[i].city[j]=grouptemp[tenip].city[j];〃变异void bianyiQ{intij;mt t;mt temp 1 ,temp2.point;double buinyip[num]; 〃染色体的变异概率mtbianyiflag[num];//染色体的变异情况fbi(i=O ;i<num: i++)〃初始化bianyiflag[i]=O;〃随机产生变异概率srand((uiisigned)tune(NULL));fbi(i=O ;i<num: i++){bianyip[i]=(rand()% 100); bianyip[i]/=100;}〃确定可以变异的染色体t=0;for(i=0 ;i<num; i++){if(biaiivip[i]<pm){ biaiiviflag[i]=l; t++;}}〃变异操作,即交换染色体的两个节点srand((iuisigned)tiine(NULL));for(i=0 ;i<num; i++){if(biaiiviflag[i]== 1){templ=rand()%10;temp2=rand()% 10;pomt=group[i]・ city[temp 1 ]; group [i]. city [temp1 ]=gioup [1]. city [temp2 ]; group[i] .city[temp2]=pomt;o e :a【n d(XXVINV£3n¥fo C T bB U T doQonpo】ddno・bb O -S Hg o q o ・Teqo「(£2】o o y )U S S A S()UTCU 二UTo lunp 」f (A R §o q o )2wl^20q o ^.・o %w uc o sXUTPsqsns^******** ***************^f l 】0 A)近士定—障尉QTry f ******************5-:************* **********⑧ 琛c>^建翠 唳********************=)七.s】d宀(dcl.mdno乩z'uxpt%-®^^・・)七宀「(曰目。
遗传算法的C语言实现(二)-----以求解TSP问题为例

遗传算法的C语⾔实现(⼆)-----以求解TSP问题为例上⼀次我们使⽤遗传算法求解了⼀个较为复杂的多元⾮线性函数的极值问题,也基本了解了遗传算法的实现基本步骤。
这⼀次,我再以经典的TSP问题为例,更加深⼊地说明遗传算法中选择、交叉、变异等核⼼步骤的实现。
⽽且这⼀次解决的是离散型问题,上⼀次解决的是连续型问题,刚好形成对照。
⾸先介绍⼀下TSP问题。
TSP(traveling salesman problem,旅⾏商问题)是典型的NP完全问题,即其最坏情况下的时间复杂度随着问题规模的增⼤按指数⽅式增长,到⽬前为⽌还没有找到⼀个多项式时间的有效算法。
TSP问题可以描述为:已知n个城市之间的相互距离,某⼀旅⾏商从某⼀个城市出发,访问每个城市⼀次且仅⼀次,最后回到出发的城市,如何安排才能使其所⾛的路线最短。
换⾔之,就是寻找⼀条遍历n个城市的路径,或者说搜索⾃然⼦集X={1,2,...,n}(X的元素表⽰对n个城市的编号)的⼀个排列P(X)={V1,V2,....,Vn},使得Td=∑d(V i,V i+1)+d(V n,V1)取最⼩值,其中,d(V i,V i+1)表⽰城市V i到V i+1的距离。
TSP问题不仅仅是旅⾏商问题,其他许多NP完全问题也可以归结为TSP问题,如邮路问题,装配线上的螺母问题和产品的⽣产安排问题等等,也使得TSP问题的求解具有更加⼴泛的实际意义。
再来说针对TSP问题使⽤遗传算法的步骤。
(1)编码问题:由于这是⼀个离散型的问题,我们采⽤整数编码的⽅式,⽤1~n来表⽰n个城市,1~n的任意⼀个排列就构成了问题的⼀个解。
可以知道,对于n个城市的TSP问题,⼀共有n!种不同的路线。
(2)种群初始化:对于N个个体的种群,随机给出N个问题的解(相当于是染⾊体)作为初始种群。
这⾥具体采⽤的⽅法是:1,2,...,n作为第⼀个个体,然后2,3,..n分别与1交换位置得到n-1个解,从2开始,3,4,...,n分别与2交换位置得到n-2个解,依次类推。
遗传算法案例及源代码

计算智能作业三:遗传算法计算问题1.问题描述:求下述二元函数的最大值:222121),(m ax x x x x f +=S.t. }7,6,5,4,3,2,1{1∈x }7,6,5,4,3,2,1{2∈x2.程序结构:(1)变量:C :是一个1*6数组,每个数组里面是一个6位二进制数,它是遗传算法中的染色体。
new_c:每一轮的新变量c 。
first_c:初始群体矩阵。
sur_value :个体适应值的概率值,为0-1之间的数,所有概率值和为1。
survived :经过选择运算后产生的个体基因型组合。
intersect_c :经过交叉运算后产生的个体基因型组合。
mutation_c :经过变异运算后产生的个体基因型组合。
f :最后计算得到的最大值 (2)程序里面的方程function out = value_function( ci ):价值函数(自适应度函数),即222121),(x x x x f +=。
function [ sur_value ] = calc_value( c ):计算群体中每一个个体的适应度的值function survived = surviver( sur_value ):利用概率选择函数 function [ intersect_c ] = intersect( new_c ):交叉运算function [ mutation_c ,mutation_value] = mutation( intersect_c ):变异运算3.源程序(1)遗传算法的主程序主程序包括初始群体产生,最终结果展示,即各函数之间的调用关系。
个体编码遗传算法的运算对象是表示个体的符号串,所以必须把变量 x1, x2 编码为无符号二进制整数。
这个二进制整数位个体的基因型。
因为x1, x2 为 0 ~ 7之间的整数,所以分别用3位无符号二进制整数来表示,将它们连接在一起所组成的6位无符号二进制数就形成了个体的基因型,表示一个可行解。
用遗传算法求解多目标函数优化c++程序

遗传算法是一种优化搜索方法,它模拟了自然选择和遗传学中的一些概念,如基因突变、交叉和选择。
这种方法可以用于解决多目标优化问题,其中多个目标之间可能存在冲突。
以下是一个使用C++和OpenCV库实现遗传算法的基本示例。
这个例子解决的是一个简单的多目标优化问题,目标是找到一个最优的图像分割方案,使得两个目标(分割的精度和计算的效率)同时最大化。
注意:这个示例是为了演示遗传算法的基本概念,并不一定适用于所有问题。
你可能需要根据你的具体需求来调整遗传算法的参数和约束条件。
```cpp#include <iostream>#include <vector>#include <algorithm>#include <opencv2/opencv.hpp>// 多目标函数优化struct ObjectiveFunction {std::vector<double> values;void operator()(const std::vector<double>& x) const {// 这里应该根据你的具体问题来定义函数的具体形式// 这里只是一个简单的示例,只考虑了分割精度和计算效率两个目标values.resize(x.size(), 0); // 初始化所有目标值为0values[0] = 1.0; // 精度目标values[1] = 1.0; // 效率目标}};class GeneticAlgorithm {public:GeneticAlgorithm(int populationSize, int generations, double crossoverRate, double mutationRate) : populationSize(populationSize), generations(generations), crossoverRate(crossoverRate), mutationRate(mutationRate) {} std::vector<std::vector<double>> optimize(const std::vector<std::vector<double>>& inputs) {std::vector<std::vector<double>>bestSolution(inputs.size(),std::vector<double>(populationSize, 0)); // 初始化最优解double bestScore = -1; // 初始最佳分数为-1,通常需要先运行一次算法以找到初始最佳分数for (int generation = 0; generation <generations; ++generation) {std::vector<std::vector<double>>population(populationSize,std::vector<double>(populationSize, 0)); // 初始化种群for (int i = 0; i < populationSize; ++i) { std::vector<double>randomSolution(inputs.size(), 0); // 随机生成解for (int j = 0; j < inputs.size(); ++j) {randomSolution[j] = inputs[j][rand() % inputs[j].size()]; // 在输入范围内随机选择一个数作为解}population[i] = randomSolution; // 将随机解加入种群}while (!population.empty()) { // 当种群不为空时继续迭代std::sort(population.begin(), population.end(), [](const std::vector<double>& a, const std::vector<double>& b) { // 对种群进行排序,根据适应度进行排序(这里适应度是解的分数)return ObjectiveFunction()(a) > ObjectiveFunction()(b); // 如果分数更高,则适应度更好,优先选择这个解作为下一代解的一部分});std::vector<double>nextGeneration(population[0]); // 选择当前种群中的第一个解作为下一代解的一部分for (int j = 1; j < populationSize; ++j) { // 对剩余的解进行交叉和变异操作,生成下一代解if (rand() / double(RAND_MAX) < crossoverRate) { // 如果满足交叉条件,则进行交叉操作for (int k = 0; k < inputs.size(); ++k) { // 将两个解的部分基因进行交叉操作,生成新的基因序列nextGeneration[k] = population[j][k]; // 将两个解的部分基因复制到下一代解中if (rand() / double(RAND_MAX) < mutationRate) { // 如果满足变异条件,则对部分基因进行变异操作,增加种群的多样性nextGeneration[k] = nextGeneration[k] * (1 - mutationRate) + population[j][k] * mutationRate; // 对部分基因进行变异操作,增加种群的多样性}}} else { // 如果不满足交叉条件,则直接复制当前解作为下一代解的一部分for (int k = 0; k < inputs.size(); ++k) { // 将当前解的部分基因复制到下一代解中 nextGeneration[k] = population[。
遗传算法求解函数极值C语言代码

#include "stdio.h"#include "stdlib.h"#include "conio.h"#include "math.h"#include "time.h"#define num_C 12 //个体的个数,前6位表示x1,后6位表示x2 #define N 100 //群体规模为100#define pc 0.9 //交叉概率为0.9#define pm 0.1 //变异概率为10%#define ps 0.6 //进行选择时保留的比例#define genmax 2000 //最大代数200int RandomInteger(int low,int high);void Initial_gen(struct unit group[N]);void Sort(struct unit group[N]);void Copy_unit(struct unit *p1,struct unit *p2);void Cross(struct unit *p3,struct unit *p4);void Varation(struct unit group[N],int i);void Evolution(struct unit group[N]);float Calculate_cost(struct unit *p);void Print_optimum(struct unit group[N],int k);/* 定义个体信息*/typedef struct unit{int path[num_C]; //每个个体的信息double cost; //个体代价值};struct unit group[N]; //种群变量groupint num_gen=0; //记录当前达到第几代int main(){int i,j;srand((int)time(NULL)); //初始化随机数发生器Initial_gen(group); //初始化种群Evolution(group); //进化:选择、交叉、变异getch();return 0;}/* 初始化种群*/void Initial_gen(struct unit group[N]){int i,j;struct unit *p;for(i=0;i<=N-1;i++) //初始化种群里的100个个体{p=&group[i]; //p指向种群的第i个个体for(j=0;j<12;j++){p->path[j]=RandomInteger(0,9); //end }Calculate_cost(p); //计算该种群的函数值}//end 初始化种群}/* 种群进化,进化代数由genmax决定*/void Evolution(struct unit group[N]){int i,j;int temp1,temp2,temp3,temp4,temp5;temp1=N*pc/2;temp2=N*(1-pc);temp3=N*(1-pc/2);temp4=N*(1-ps);temp5=N*ps;for(i=1;i<=genmax;i++){//选择for(j=0;j<=temp4-1;j++){ Copy_unit(&group[j],&group[j+temp5]); }//交叉for(j=0;j<=temp1-1;){Cross(&group[temp2+j],&group[temp3+j]);j+=2;}//变异Varation(group,i);}Sort(group);Print_optimum(group,i-1); //输出当代(第i-1代)种群}/* 交叉*/void Cross(struct unit *p3,struct unit *p4){int i,j,cross_point;int son1[num_C],son2[num_C];for(i=0;i<=num_C-1;i++) //初始化son1、son2{son1[i]=-1;son2[i]=-1;}cross_point=RandomInteger(1,num_C-1); //交叉位随机生成//交叉,生成子代//子代1//子代1前半部分直接从父代复制for(i=0;i<=cross_point-1;i++) son1[i]=p3->path[i];for(i=cross_point;i<=num_C-1;i++)for(j=0;j<=num_C-1;j++) //补全p1{son1[i]=p4->path[j];}//end 子代1//子代2//子代1后半部分直接从父代复制for(i=cross_point;i<=num_C-1;i++) son2[i]=p4->path[i];for(i=0;i<=cross_point-1;i++){for(j=0;j<=num_C-1;j++) //补全p1{son2[i]=p3->path[j];}}//end 子代2//end 交叉for(i=0;i<=num_C-1;i++){p3->path[i]=son1[i];p4->path[i]=son2[i];}Calculate_cost(p3); //计算子代p1的函数值Calculate_cost(p4); //计算子代p2的函数值}/* 变异*/void Varation(struct unit group[N],int flag_v){int flag,i,j,k,temp;struct unit *p;flag=RandomInteger(1,100);//在进化后期,增大变异概率if((!flag>(flag_v>100))?(5*100*pm):(100*pm)){i=RandomInteger(0,N-1); //确定发生变异的个体j=RandomInteger(0,num_C-1); //确定发生变异的位k=RandomInteger(0,num_C-1);p=&group[i]; //变异temp=p->path[j];p->path[j]=p->path[k];p->path[k]=temp;Calculate_cost(p); //重新计算变异后的函数值}}/* 将种群中个体按函数值从小到大排序*/void Sort(struct unit group[N]){int i,j;struct unit temp,*p1,*p2;for(j=1;j<=N-1;j++) //排序总共需进行N-1轮{for(i=1;i<=N-1;i++){p1=&group[i-1];p2=&group[i];if(p1->cost>p2->cost) //值大的往后排{Copy_unit(p1,&temp);Copy_unit(p2,p1);Copy_unit(&temp,p2);}}//end 一轮排序}//end 排序}/* 计算某个个体的函数值*/float Calculate_cost(struct unit *p){double x1,x2;x1=0;if(p->path[0]>5){x1=p->path[1]+p->path[2]*0.1+p->path[3]*0.01+p->path[4]*0.001+p->path[5]*0.0001;}else if(p->path[0]<6){x1=0-(p->path[1]+p->path[2]*0.1+p->path[3]*0.01+p->path[4]*0.001+p->path[5]*0.0001);}x2=0;if(p->path[6]>5){}else if(p->path[6]<6){x2=0-(p->path[7]+p->path[8]*0.1+p->path[9]*0.01+p->path[10]*0.001+p->path[11]*0.0001);}p->cost=20+x1*x1+x2*x2-10*(cos(2*3.14*x1)+cos(2*3.14*x2));return(p->cost);}/* 复制种群中的p1到p2中*/void Copy_unit(struct unit *p1,struct unit *p2){int i;for(i=0;i<=num_C-1;i++)p2->path[i]=p1->path[i];p2->cost=p1->cost;}/* 生成一个介于两整型数之间的随机整数*/int RandomInteger(int low,int high){int k;double d;k=rand();k=(k!=RAND_MAX)?k:(k-1); //RAND_MAX是VC中可表示的最大整型数d=(double)k/((double)(RAND_MAX));k=(int)(d*(high-low+1));return (low+k);}/* 输出当代种群中的最优个体*/void Print_optimum(struct unit group[N],int k){struct unit *p;double x1,x2;x1=x2=0;p=&group[0];if(p->path[0]>5){x1=p->path[1]+p->path[2]*0.1+p->path[3]*0.01+p->path[4]*0.001+p->path[5]*0.0001;}else if(p->path[0]<6){}x2=0;if(p->path[6]>5){x2=p->path[7]+p->path[8]*0.1+p->path[9]*0.01+p->path[10]*0.001+p->path[11]*0.0001;}else if(p->path[6]<6){x2=0-(p->path[7]+p->path[8]*0.1+p->path[9]*0.01+p->path[10]*0.001+p->path[11]*0.0001);}printf(" 当x1=%f x2=%f\n",x1,x2);printf(" 函数最小值为:%f \n",p->cost);}。
TSP的遗传算法程序实验报告
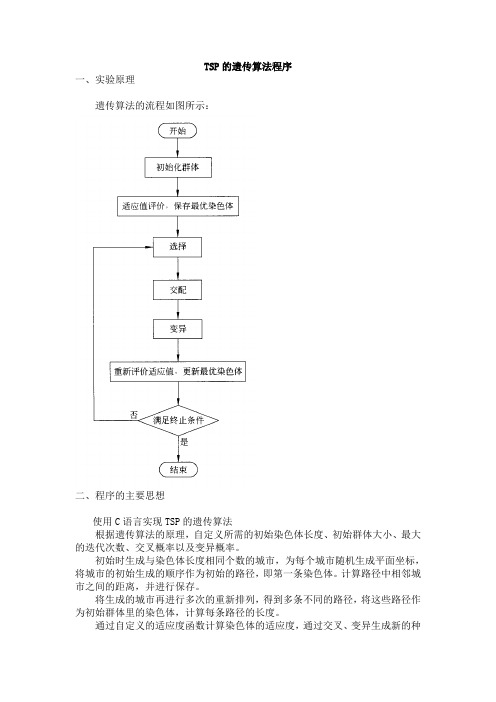
TSP的遗传算法程序一、实验原理遗传算法的流程如图所示:二、程序的主要思想使用C语言实现TSP的遗传算法根据遗传算法的原理,自定义所需的初始染色体长度、初始群体大小、最大的迭代次数、交叉概率以及变异概率。
初始时生成与染色体长度相同个数的城市,为每个城市随机生成平面坐标,将城市的初始生成的顺序作为初始的路径,即第一条染色体。
计算路径中相邻城市之间的距离,并进行保存。
将生成的城市再进行多次的重新排列,得到多条不同的路径,将这些路径作为初始群体里的染色体,计算每条路径的长度。
通过自定义的适应度函数计算染色体的适应度,通过交叉、变异生成新的种群。
对新种群继续迭代操作,直到达到初始定义的迭代次数,获得最终的路径及路径图。
三、程序的主要步骤①染色体初始化的子函数1)void initialize()2){int k,j,minx,miny,maxx,maxy;3)initdata();4)minx=0;5)miny=0;6)maxx=0;maxy=0;7)for(k=0;k<lchrom;k++)8){x[k]=rand();9)if(x[k]>maxx)maxx=x[k];10)if(x[k]<minx)minx=x[k];11)y[k]=rand();12)if(y[k]>maxy)maxy=y[k];13)if(y[k]<miny)miny=y[k];14)}15)if((maxx-minx)>(maxy-miny))16){maxxy=maxx-minx;}17)else {maxxy=maxy-miny;}18)maxdd=0.0;19)for(k=0;k<lchrom;k++)20)for(j=0;j<lchrom;j++)21){dd[k*lchrom+j]=hypot(x[k]-x[j],y[k]-y[j]);22)if(maxdd<dd[k*lchrom+j])maxdd=dd[k*lchrom+j];23)}24)refpd=dd[lchrom-1];25)for(k=0;k<lchrom;k++)26)refpd=refpd+dd[k*lchrom+k+2];27)for(j=0;j<lchrom;j++)28)dd[j*lchrom+j]=4.0*maxdd;29)ff=(0.765*maxxy*pow(lchrom,0.5));30)minpp=0;31)min=dd[lchrom-1];32)for(j=0;j<lchrom-1;j++)33){if(dd[lchrom*j+lchrom-1]<min)34){min=dd[lchrom*j+lchrom-1];35)minpp=j;36)}37)}38)initpop();39)statistics(oldpop);40)initreport();41)}7) for(k=0;k<lchrom;k++)8) {x[k]=rand();9) if(x[k]>maxx)maxx=x[k];10)if(x[k]<minx)minx=x[k];11)y[k]=rand();12)if(y[k]>maxy)maxy=y[k]13)if(y[k]<miny)miny=y[k];14)}此段程序是初始化多个城市的坐标值(x,y),其中(x[k],y[k])代表第k+1个城市的坐标值,也相当于初始染色体的第k+1个基因值。
C++实现简单遗传算法

C++实现简单遗传算法本⽂实例讲述了C++实现简单遗传算法。
分享给⼤家供⼤家参考。
具体实现⽅法如下://遗传算法 GA#include<iostream>#include <cstdlib>#include<bitset>using namespace std;const int L=5; //定义编码的长度int f(int x) //定义测设函数f(x){int result;result=x*x*x-60*x*x+900*x+100;return result;}int main(int argc,char *argv[]){int a(0),b(32); //定义x的定义域范围const int pop_size=8; //定义种群⼤⼩// int L; //指定编码的长度const int NG=20; //指定种群最⼤的繁殖的代数int t=0; //当前繁殖的代数int p[pop_size]; //定义种群int q[pop_size]; //定义繁殖种群即种群的下⼀代srand(6553); //定义随机数⽣成的种⼦double sum; //适值总和double avl_sum; //适度平均值double p_probability[pop_size]; //适值概率double pp[pop_size];double pro; //定义随机⽣成的概率float pc=0.90; //定义交叉的概率float pm=0.05; //定义变异的概率cout<<"初始的种群 ";for(int i=0;i<pop_size;i++) //⽣成初始的第0代种群{p[i]=rand()%31;cout<<p[i]<<" ";}cout<<endl;cout<<endl;void Xover(int &,int &); //声明交叉函数//当停⽌准则不满⾜即繁殖代数没到最⼤代数 ,继续繁殖while(t<=NG){cout<<"繁殖的代数:t="<<t<<endl;sum=0.0;for(int i=0;i<pop_size;i++){q[i]=p[i];cout<<q[i]<<" ";}cout<<endl;for(int i=0;i<pop_size;i++) //计算sumsum +=f(p[i]);avl_sum=sum/pop_size;cout<<"sum="<<sum<<endl;cout<<"适度平均值="<<avl_sum<<endl;for(int i=0;i<pop_size;i++) //计算适值概率{p_probability[i]=f(p[i])/sum;if(i==0){pp[i]=p_probability[i];cout<<"pp"<<i<<"="<<pp[i]<<endl;}else{pp[i]=p_probability[i]+pp[i-1];cout<<"pp"<<i<<"="<<pp[i]<<endl;}//cout<<"p_probability"<<i<<"="<<p_probability[i]<<endl;}//选择双亲for(int i=0;i<pop_size;i++)pro=rand()%1000/1000.0;if(pro>=pp[0]&&pro<pp[1])p[i]=q[0];else if(pro>=pp[1]&&pro<pp[2])p[i]=q[1];else if(pro>=pp[2]&&pro<pp[3])p[i]=q[2];else if(pro>=pp[3]&&pro<pp[4])p[i]=q[3];else if(pro>=pp[4]&&pro<pp[5])p[i]=q[4];elsep[i]=q[5];}//杂交算⼦int r=0;int z=0;for(int j=0;j<pop_size;j++){pro=rand()%1000/1000.0;if(pro<pc){++z;if(z%2==0)Xover(p[r],p[j]);elser=j;}}//变异算⼦for(int i=1;i<=pop_size;i++)for(int j=0;j<L;j++){pro=rand()%1000/1000.0; //在【0,1】区间产⽣随机数if(pro<pm){bitset<L>v(p[i]);v.flip(j);p[i]=v.to_ulong();}}t++;cout<<endl; //种群繁殖⼀代}cout<<"最终结果:";for(int i(0);i<pop_size;i++) //算法结束,输出结果{cout<<p[i]<<" ";}cout<<endl;return 0;}//定义杂交操作void Xover(int &a,int &b){int pos; //随机⽣成杂交点即第⼏个分量进⾏相互交换pos=rand()%5+1; //在n个分量中,随机确定第pos个分量int j,k;j=pos;k=pos;bitset<L>e(a);bitset<L>f(b); //前pos个分量进⾏相互交换bitset<L>g;bitset<L>h;for(int i=0;i<pos;i++){if(e[i]==1)g.set(i);}for(int i=0;i<pos;i++){if(f[i]==1)h.set(i);}for(j;j<L;j++)if(f[j]==1)g.set(j);}for(k;k<L;k++){if(e[k]==1)h.set(k);}a=g.to_ulong();b=h.to_ulong();}希望本⽂所述对⼤家的C++程序设计有所帮助。
遗传算法 c语言代码

以下是一个简单的遗传算法的C语言代码示例:c#include <stdio.h>#include <stdlib.h>#include <time.h>#include <math.h>#define POPULATION_SIZE 100#define GENE_LENGTH 10#define MAX_GENERATIONS 1000#define MUTATION_RATE 0.01#define CROSSOVER_RATE 0.8typedef struct Individual {char genes[GENE_LENGTH];double fitness;} Individual;double calculate_fitness(Individual* individual) {// 计算适应度函数,这里使用简单的二进制字符串中1的个数作为适应度 int count = 0;for (int i = 0; i < GENE_LENGTH; i++) {if (individual->genes[i] == '1') {count++;}}return count;}void initialize_population(Individual* population) {// 初始化种群for (int i = 0; i < POPULATION_SIZE; i++) {for (int j = 0; j < GENE_LENGTH; j++) {population[i].genes[j] = rand() % 2 ? '0' : '1';}population[i].fitness = calculate_fitness(&population[i]); }}void selection(Individual* population, Individual* parents) {// 选择操作,采用轮盘赌算法选择两个父代个体double total_fitness = 0;for (int i = 0; i < POPULATION_SIZE; i++) {total_fitness += population[i].fitness;}double rand1 = rand() / (double)RAND_MAX * total_fitness;double rand2 = rand() / (double)RAND_MAX * total_fitness;double cumulative_fitness = 0;int parent1_index = -1, parent2_index = -1;for (int i = 0; i < POPULATION_SIZE; i++) {cumulative_fitness += population[i].fitness;if (rand1 < cumulative_fitness && parent1_index == -1) {parent1_index = i;}if (rand2 < cumulative_fitness && parent2_index == -1) {parent2_index = i;}}parents[0] = population[parent1_index];parents[1] = population[parent2_index];}void crossover(Individual* parents, Individual* offspring) {// 交叉操作,采用单点交叉算法生成两个子代个体int crossover_point = rand() % GENE_LENGTH;for (int i = 0; i < crossover_point; i++) {offspring[0].genes[i] = parents[0].genes[i];offspring[1].genes[i] = parents[1].genes[i];}for (int i = crossover_point; i < GENE_LENGTH; i++) {offspring[0].genes[i] = parents[1].genes[i];offspring[1].genes[i] = parents[0].genes[i];}offspring[0].fitness = calculate_fitness(&offspring[0]);offspring[1].fitness = calculate_fitness(&offspring[1]);}void mutation(Individual* individual) {// 变异操作,以一定概率翻转基因位上的值for (int i = 0; i < GENE_LENGTH; i++) {if (rand() / (double)RAND_MAX < MUTATION_RATE) {individual->genes[i] = individual->genes[i] == '0' ? '1' : '0'; }}individual->fitness = calculate_fitness(individual);}void replace(Individual* population, Individual* offspring) {// 替换操作,将两个子代个体中适应度更高的一个替换掉种群中适应度最低的一个个体int worst_index = -1;double worst_fitness = INFINITY;for (int i = 0; i < POPULATION_SIZE; i++) {if (population[i].fitness < worst_fitness) {worst_index = i;worst_fitness = population[i].fitness;}}if (offspring[0].fitness > worst_fitness || offspring[1].fitness > worst_fitness) {if (offspring[0].fitness > offspring[1].fitness) {population[worst_index] = offspring[0];} else {population[worst_index] = offspring[1];}}}。
遗传算法程序正确例子

% 求下列函数的最大值%% f(x)=10*sin(5x)+7*cos(4x) x∈[0,10] %% 将x 的值用一个10位的二值形式表示为二值问题,一个10位的二值数提供的分辨率是每为(10-0)/(2^10-1)≈0.01 。
%% 将变量域[0,10] 离散化为二值域[0,1023], x=0+10*b/1023, 其中b 是[0,1023] 中的一个二值数。
%% 编程%-----------------------------------------------% 2.1初始化(编码)% initpop.m函数的功能是实现群体的初始化,popsize表示群体的大小,chromlength表示染色体的长度(二值数的长度),% 长度大小取决于变量的二进制编码的长度(在本例中取10位)。
%遗传算法子程序%Name: initpop.m%初始化function pop=initpop(popsize,chromlength)pop=round(rand(popsize,chromlength)); % rand随机产生每个单元为{0,1} 行数为popsize,列数为chromlength的矩阵,% roud对矩阵的每个单元进行圆整。
这样产生的初始种群。
% 2.2 计算目标函数值% 2.2.1 将二进制数转化为十进制数(1)%遗传算法子程序%Name: decodebinary.m%产生[2^n 2^(n-1) ... 1] 的行向量,然后求和,将二进制转化为十进制function pop2=decodebinary(pop)[px,py]=size(pop); %求pop行和列数for i=1:pypop1(:,i)=2.^(py-i).*pop(:,i);endpop2=sum(pop1,2); %求pop1的每行之和% 2.2.2 将二进制编码转化为十进制数(2)% decodechrom.m函数的功能是将染色体(或二进制编码)转换为十进制,参数spoint表示待解码的二进制串的起始位置% (对于多个变量而言,如有两个变量,采用20为表示,每个变量10为,则第一个变量从1开始,另一个变量从11开始。
遗传算法示例

实验一、过河问题一、问题描述有三个牧师和三个野人过河,只有一条能装下两个人的船,在河的任何一方或者船上,如果野人的人数大于牧师的认输,那么牧师就会有危险.找出一种按的渡河方法。
将该问题转变为:假如有n个牧师和n个野人准备渡河,但只有一条能容纳c个人的小船,为了防止野人侵犯牧师,要求无论在何处,牧师的人数不得少于野人的人数(除非牧师人数为0),且假定两种人都会划船,试设计一个算法,确定它们能否渡过河去,若能,则给出一只小船来回次数最少的最佳方案。
二、基本要求输入:牧师人数(即野人人数)n,小船一次至多载客人数c。
输出:若问题无解,则显示“渡河失败”信息,否则,输出一组最佳方案,用三组(X1,X2,X3)表示渡河过程中的状态。
并用箭头输出这些状态之间的迁移:目的状态<- <-中间状态<- <-初始状态。
例:当n=2,c=2时,输出000<-021<-211<-110<-221上述(X1,X1,X2)表示渡河过程中各个状态。
其中:X1表示始岸上牧师人数,X2表示始岸上野人人数,X3表示小船位置,(0-在目的岸,1-在起始岸)三、算法描述(1)算法基本思想的文字描述;从初始状态S(n,n,1)出发,形成的有合法且未达状态S11、S12、……、Sli。
再分别从S11、S12、……、Sli出发形成所有合法而未达状态S111、S112、……、Sli1、Sli2、Sli ……最终达到目标(0,0,0)(有解),或者找不到合法而未达状态(无解)。
若有解,则从目标返回找前趋状态,前趋状态的前趋状态……直到初始状态。
(2)判别(X1,X2,X3)为合法状态条件:X1=0或X1=n或X1=X2。
(3)数据结构:1 栈STACK ,记下“已达”状态及踪迹,并兼作队列。
2 STATE[X1][X2]=(4)算法基本思想的具体实现:1 初始化:置STATE[N+1][N+1][2]中的有状态为“未达”置队列STACK 空,cond 为当前是否已达到目标: cond= cond 置初值 2 以S (n,n,1)为始点,置STATE 为“已达”。
一个简单实用的遗传算法c程序

一个简单实用的遗传算法c程序(转载)C++ 2009-07-28 23:09:03 阅读418评论0 字号:大中小这是一个非常简单的遗传算法源代码,是由Denis Cormier (North Caroli na State University)开发的,Sita S.Raghavan (University of North Carolina at Charlotte) 修正。
代码保证尽可能少,实际上也不必查错。
对一特定的应用修正此代码,用户只需改变常数的定义并且定义评价函数”即可。
注意代码的设计是求最大值,其中的目标函数只能取正值;且函数值和个体的适应值之间没有区别。
该系统使用比率选择、精华模型、单点杂交和均匀变异。
如果用Gaussian变异替换均匀变异,可能得到更好的效果。
代码没有任何图形,甚至也没有屏幕输出,主要是保证在平台之间的高可移植性。
读者可以从,目录coe/evol中的文件prog.c中获得。
要求输入的文件应该命名为gadata.txt '系统产生的输出文件为galog.txt '输入的文件由几行组成:数目对应于变量数。
且每一行提供次序--- 对应于变量的上下界。
如第一行为第一个变量提供上下界,第二行为第二个变量提供上下界,等等。
/**************************************************************************//* This is a simple genetic algorithm implementation where the *//* evaluati on fun ctio n takes positive values on ly and the *//* fitn ess of an in dividual is the same as the value of the *//* objective fun ctio n *//**************************************************************************/#in clude <stdio.h>#in clude <stdlib.h>#in clude <math.h>/* Change any of these parameters to match your n eeds */#defi ne POPSIZE 50 #defi ne MAXGENS 1000 #defi ne NVARS 3#defi ne PXOVER 0.8#defi ne PMUTATION 0.15#defi ne TRUE 1 #defi ne FALSE 0 int gen erati on;int cur_best;/* curre nt gen erati on no.*//* best in dividual */FILE *galog; /* an output file */struct geno type /* geno type (GT), a member of the populati on */double gen e[NVARS]; /* a stri ng of variables 一个变量字符串 */ double fitn ess; /* GT's fitness 适应度 */ double upper[NVARS]; /* GT's variables upper bound 变量的上限 */ double lower[NVARS]; /* GT's variables lower bou nd 变量的下限 */double rfitn ess;/* relative fitness相对适应度 */double cfitn ess;/* cumulative fitness累计适应度 */};struct ge no type populatio n[POPSIZE+1]; struct geno type n ewpopulati on [POPSIZE+1]; /*/* populati on size *//* max. nu mber of gen erati ons *//* no. of problem variables *//* probability of crossover *//* probability of mutatio n *//* populati on */new populati on; *//* replaces the *//* old gen erati on */ /* Declarati on of procedures used by this gen etic algorithm */void in itialize(void);double ran dval(double, double);void evaluate(void);void keep_the_best(void);void elitist(void);void select(void);void crossover(void);void Xover(i nt,i nt);void swap(double *, double *);void mutate(void);void report(void);/***************************************************************//* In itializatio n fun cti on: In itializes the values of genes *//* with in the variables boun ds. It also in itializes (to zero) *//* all fitn ess values for each member of the populati on .It *//* reads upper and lower bounds of each variable from the */ /* in put file 'gadata.txt'. It ran domly gen erates values *//* betwee n these bounds for each gene of each geno type in the *//* populatio n. The format of the in put file 'gadata.txt' is *//* var1_lower_bo und var1_upper bound */ /* var2_lower_bo und var2_upper bound ... */ /***************************************************************/void in itialize(void){FILE *i nfile;int i, j;double lbo und, ubo und;if ((i nfile = fope n("gadata.txt","r"))==NULL){fprin tf(galog,"\nCannot ope n in put file!\n");exit(1);}/* in itialize variables with in the bounds */for (i = 0; i < NVARS; i++){fscanf(in file, "%lf",&l bou nd);fscan f(i nfile, "%lf",&ubou nd);for (j = 0; j < POPSIZE; j++)populati on [j].fit ness = 0;populati on [j].rfit ness = 0;populati on [j].cfit ness = 0;populati on [j].lower[i] = Ibo und;populati on [j].upper[i]= ubo und;populati on [j].ge ne[i] = ran dval(populati on [j].lower[i],populatio n[j].upper[i]);}}fclose(i nfile);}/***********************************************************//* Ran dom value gen erator: Gen erates a value within bounds *//***********************************************************/double ran dval(double low, double high){double val;val = ((double)(ra nd()%1000)/1000.0)*(high - low) + low;return(val);/* Evaluati on function: This takes a user defi ned function. */ /* Each time this is cha nged, the code has to be recompiled. *//* The curre nt fun ctio n is: x[1F2-x[1]*x[2]+x[3] */ /*************************************************************/*********************************************************void evaluate(void){int mem;int i;double x[NVARS+1];for (mem = 0; mem < POPSIZE; mem++){for (i = 0; i < NVARS; i++)x[i+1] = populati on[ mem].ge ne[i];populatio n[mem].fit ness = (x[1]*x[1]) - (x[1]*x[2]) + x[3];}}/***************************************************************//* Keep_the_best fun cti on: This fun cti on keeps track of the */ /* best member of the populati on. Note that the last entry in *//* the array Populati on holds a copy of the best in dividual*//***************************************************************/void keep_the_best(){int mem;int i;cur_best = 0; /* stores the in dex of the best in dividual */for (mem = 0; mem < POPSIZE; mem++){if (populati on[ mem].fit ness > populatio n[ POPSIZE].fit ness){cur_best = mem;populati on[ POPSIZE].fit ness = populatio n[ mem].fit ness;}}/* once the best member in the populati on is found, copy the genes */ for (i = 0; i < NVARS; i++)populati on [POPSIZE].ge ne[i] = populati on[ cur_best].ge ne[i];}/****************************************************************//* Elitist fun cti on: The best member of the previous gen erati on *//* is stored as the last in the array. If the best member of*//* the curre nt gen eratio n is worse the n the best member of the *//* previous gen erati on, the latter one would replace the worst *//* member of the curre nt populati on */ /****************************************************************/void elitist(){int i;double best, worst; /* best and worst fitn ess values */int best_mem, worst_mem; /* i ndexes of the best and worst member */best = populati on[ O].fit ness;worst = populati on[ O].fit ness;for (i = 0; i < POPSIZE - 1; ++i){if(populati on [i].fit ness > populati on [i+1].fit ness){if (populatio n[i].fit ness >= best){best = populati on [i].fit ness;best_mem = i;if (populati on [i+1].fit ness <= worst){worst = populati on [i+1].fit ness; worst_mem = i + 1;}}else{if (populati on [i].fit ness <= worst){worst = populati on [i].fit ness;worst_mem = i;}if (populati on [i+1].fit ness >= best){best = populati on [i+1].fit ness;best_mem = i + 1;}}}/* if best in dividual from the new populati on is better tha n *//* the best i ndividual from the previous populati on, the n /* copy the best from the new populati on; else replace the */ /* worst i ndividual from the curre nt populati on with the *//* best one from the previous gen erati on */if (best >= populatio n[POPSIZE].fit ness){for (i = 0; i < NVARS; i++)populati on[ POPSIZE].ge ne[i] = populatio n[ best_mem].ge ne[i]; populati on[ POPSIZE].fit ness = populati on[ best_mem].fit ness;} else{for (i = 0; i < NVARS; i++)populati on[ worst_mem].ge ne[i] = populati on[ POPSIZE].ge ne[i]; populati on[ worst_mem].fit ness = populati on[ POPSIZE].fit ness;} }**********************************************************/* Selectio n function: Stan dard proporti onal selectio n for *//* maximizati on problems in corporat ing elitist model - makes /* sure that the best member survives*/ */ */**********************************************************void select(void){int mem, i, j, k;double sum = 0;double p;/* find total fitn ess of the populati on */for (mem = 0; mem < POPSIZE; mem++){sum += populatio n[ mem].fit ness;}/* calculate relative fitn ess */for (mem = 0; mem < POPSIZE; mem++){populati on[ mem].rfit ness = populati on[ mem].fit ness/sum;}populatio n[ O].cfit ness = populati on[ O].rfit ness;/* calculate cumulative fitn ess */for (mem = 1; mem < POPSIZE; mem++){populati on[ mem].cfit ness = populati on[ mem-1].cfit ness +populati on[ mem].rfit ness;}/* fin ally select survivors using cumulative fitn ess. */for (i = 0; i < POPSIZE; i++){p = ran d()%1000/1000.0;if (p < populati on[ O].cfit ness)n ewpopulati on [i] = populati on[ 0];else{for (j = 0; j < POPSIZE;j++)if (p >= populati on [j].cfit ness && p<populati on [j+1].cfit ness)n ewpopulati on [i] = populati on [j+1];}}/* once a new populati on is created, copy it back */for (i = 0; i < POPSIZE; i++)populati on [i] = n ewpopulati on [i];}***********************************************************/* Crossover selecti on: selects two pare nts that take part in *//* the crossover. Impleme nts a sin gle point crossover */ /***************************************************************/void crossover(void){int i, mem, one;int first = 0; /* count of the nu mber of members chose n */double x;for (mem = 0; mem < POPSIZE; ++mem){x = ran d()%1000/1000.0;if (x < PXOVER){++first;if (first % 2 == 0)Xover( one, mem);elseone = mem;}**********************************************************/* Crossover: performs crossover of the two selected pare nts. */ **********************************************************void Xover(i nt on e, i nt two){int i;int point; /* crossover point *//* select crossover point */if(NVARS > 1){if(NVARS == 2)point = 1;elsepoi nt = (ran d() % (NVARS - 1)) + 1;for (i = 0; i < poi nt; i++)swap(&populati on[on e].ge ne[i], &populati on[ two].ge ne[i]);}}/*************************************************************//* Swap: A swap procedure that helps in swapp ing 2 variables *//*************************************************************/void swap(double *x, double *y) { double temp; temp = *x; *x = *y; *y = temp;}/**************************************************************//* Mutati on: Ran dom uniform mutatio n. A variable selected for */ /* mutati on is replaced by a ran dom value betwee n lower and */ /* upper bounds of this variable void mutate(void){ int i, j;double lbo und, hbo und; double x;for (i = 0; i < POPSIZE; i++)for (j = 0; j < NVARS; j++){ x = ran d()%1000/1000.0; if (x < PMUTATION){/* find the bounds on the variable to be mutated */ Ibound = populati on [i].lower[j]; hbo und = populatio*/**********************************************************n[i].upper[j];populati on [i].ge ne[j] = ran dval(lbo und, hbo un d);}}}***********************************************************/* Report fun ctio n: Reports progress of the simulati on. Data *//* dumped into the output file are separated by commas */ /***************************************************************/ void report(void) {int i;double best_val; /* best populati on fitn ess */double avg; /* avg populati on fitn ess */double stddev;/* std. deviati on of populati on fitn ess */double sum_square; /* sum of square for std. calc *//* square of sum for std. calc */sum = 0.0;sum_square = 0.0;for (i = 0; i < POPSIZE; i++){sum += populatio n[i].fit ness;double square_sum;double sum; /* total populati on fitn ess */sum_square += populati on [i].fit ness * populati on [i].fit ness;}avg = sum/(double)POPSIZE;square_sum = avg * avg * POPSIZE;stddev = sqrt((sum_square - square_sum)/(POPSIZE - 1));best_val = populatio n[POPSIZE].fit ness;fprin tf(galog, "\n%5d, %6.3f, %6.3f, %6.3f \n\n", gen eratio n,best_val, avg, stddev);}/**************************************************************//* Main fun cti on: Each gen erati on in volves select ing the best *//* members, perform ing crossover & mutatio n and the n */ /* evaluati ng the result ing populatio n, un til the term in ati ng *//* con diti on is satisfied*/ **********************************************************void mai n(void){int i;if ((galog = fope n( "galog.txt","w"))==NULL) {exit(1);}gen erati on = 0;fprin tf(galog, "\n gen erati on best average fprin tf(galog, " nu mber value fitn ess in itialize();evaluate();keep_the_best();while(ge nerati o*M AXGENS){gen erati on++;select();crossover();mutate();report();evaluate();sta ndard \n");deviati on}fprin tf(galog,"\n\n Simulatio n completed' n");fprin tf(galog,"\n Best member: \n");for (i = 0; i < NVARS; i++){fprintf (galog,"\n var(%d) = %3.3f',i,populatio n[POPSIZE].ge ne[i]);}fprin tf(galog,"\n\n Best fitn ess = %3.3f',populatio n[POPSIZE].fit ness);fclose(galog);prin tf("Success\n");}/***************************************************************/链接库文件?这个简单一般的第三方库文件有2种提供方式1.lib静态库,这样必须在工程设置里面添加。
基本遗传算法的C源程序。doc【精品毕业设计】(完整版)

/********************************************************** ********//* 基于基本遗传算法的函数最优化SGA.C *//* A Function Optimizer using Simple Genetic Algorithm *//* developed from the Pascal SGA code presented by David E.Goldberg *///********************************************************** ********/#include#include#include#include "graph.c"/* 全局变量*/struct individual /* 个体*/{unsigned *chrom; /* 染色体*/double fitness; /* 个体适应度*/double varible; /* 个体对应的变量值*/int xsite; /* 交叉位置*/int parent[2]; /* 父个体*/int *utility; /* 特定数据指针变量*/};struct bestever /* 最佳个体*/{unsigned *chrom; /* 最佳个体染色体*/double fitness; /* 最佳个体适应度*/double varible; /* 最佳个体对应的变量值*/int generation; /* 最佳个体生成代*/};struct individual *oldpop; /* 当前代种群*/ struct individual *newpop; /* 新一代种群*/ struct bestever bestfit; /* 最佳个体*/double sumfitness; /* 种群中个体适应度累计*/ double max; /* 种群中个体最大适应度*/ double avg; /* 种群中个体平均适应度*/ double min; /* 种群中个体最小适应度*/float pcross; /* 交叉概率*/float pmutation; /* 变异概率*/int popsize; /* 种群大小*/int lchrom; /* 染色体长度*/int chromsize; /* 存储一染色体所需字节数*/ int gen; /* 当前世代数*/int maxgen; /* 最大世代数*/int run; /* 当前运行次数*/int maxruns; /* 总运行次数*/int printstrings; /* 输出染色体编码的判断,0 -- 不输出, 1 -- 输出*/ int nmutation; /* 当前代变异发生次数*/int ncross; /* 当前代交叉发生次数*//* 随机数发生器使用的静态变量*/static double oldrand[55];static int jrand;static double rndx2;static int rndcalcflag;/* 输出文件指针*/FILE *outfp ;/* 函数定义*/void advance_random();int flip(float);rnd(int, int);void randomize();double randomnormaldeviate();float randomperc(),rndreal(float,float);void warmup_random(float);void initialize(),initdata(),initpop();void initreport(),generation(),initmalloc();void freeall(),nomemory(char *),report();void writepop(),writechrom(unsigned *);void preselect();void statistics(struct individual *);void title(),repchar (FILE *,char *,int);void skip(FILE *,int);int select();void objfunc(struct individual *);int crossover (unsigned *, unsigned *, unsigned *, unsigned *); void mutation(unsigned *);void initialize() /* 遗传算法初始化*/{/* 键盘输入遗传算法参数*/initdata();/* 确定染色体的字节长度*/chromsize = (lchrom/(8*sizeof(unsigned)));if(lchrom%(8*sizeof(unsigned))) chromsize++;/*分配给全局数据结构空间*/initmalloc();/* 初始化随机数发生器*/randomize();/* 初始化全局计数变量和一些数值*/ nmutation = 0;ncross = 0;bestfit.fitness = 0.0;bestfit.generation = 0;/* 初始化种群,并统计计算结果*/ initpop();statistics(oldpop);initreport();}void initdata() /* 遗传算法参数输入*/{char answer[2];setcolor(9);disp_hz16("种群大小(20-100):",100,150,20); gscanf(320,150,9,15,4,"%d", &popsize);if((popsize%2) != 0){fprintf(outfp, "种群大小已设置为偶数\n"); popsize++;};。
遗传算法简单c#程序实现(非专业)副本2

遗传算法简单c#程序实现(非专业)副本2using System;using System.Collections.Generic;using System.Linq;using System.Text;using System.Threading.Tasks;namespace 张小叶叶{//求下列元素的最大值:目标函数max=f(x1,x2)=x1*x1+x2*x2;取值范围在{1,2,3,4,5,6,7}//本算法在选择时使用:轮盘赌选择法、交叉时使用:单点交叉。
class Program{static int groupscale;//种群规模static int variation;//变异概率static int n;static int x1 = 1;//基因组数(便于对两组基因进行区分)static int x2 = 2;int result;static void Main(string[] args){Program p = new Program();Console.WriteLine("请输入种群规模?");groupscale = input();Console.WriteLine("请输入变异概率?");variation = input();Console.Write("产生随机数:\n");//产生二进制数int[,] arrnum1 = new int[groupscale, 3];//二进制随机数组x1int[,] arrnum2 = new int[groupscale, 3];//二进制随机数组x1getrand(groupscale, ref arrnum1, ref arrnum2);Console.WriteLine("二进制转十进制的结果是:");//返回一维数组就是两组,种群规模个,十进制数(转十进制便于进行适应度计算)int[] tennum1 = convert(groupscale, x1, arrnum1);//十进制随机数组x1int[] tennum2 = convert(groupscale, x2, arrnum2);//十进制随机数组x2Console.WriteLine("适应度的计算:");int[] one = max(groupscale, tennum1, tennum2,out p.result);//个体适应度int zuidazhi = promax(one);Console.WriteLine("适应度总和是:{0}",p.result);double[] sumprob = probablity(one, groupscale, p.result);//累积适应度概率double[] random = rand(groupscale);//0-1随机数的产生Console.WriteLine("选择出的群体:");int[] chooseDNA=choose(groupscale, random, sumprob);//得到被选中的数在数组中的位置int[,] answer=ans(arrnum1, arrnum2, chooseDNA);//得出选择结果Console.WriteLine("单点交叉后的群体:");int[,] jcanswer = jiaocha(groupscale, answer);shuchu(jcanswer);Console.WriteLine("变异后的群体:");int[,] varanswer = var(variation, jcanswer);shuchu(varanswer);Console.WriteLine("请输入还需循环代数?");n = input();for (int i = 0; i < n; i++){fenzu(varanswer, ref arrnum1, ref arrnum2);tennum1 = convert(groupscale, x1, arrnum1);tennum2 = convert(groupscale, x2, arrnum2);one = max(groupscale, tennum1, tennum2, out p.result);zuidazhi = promax(one);sumprob = probablity(one, groupscale, p.result);random = rand(groupscale);chooseDNA = choose(groupscale, random, sumprob);answer = ans(arrnum1, arrnum2, chooseDNA);jcanswer = jiaocha(groupscale, answer);varanswer = var(variation, jcanswer);Console.WriteLine("第{0}代产生的子种群是:",i+2);shuchu(varanswer);}Console.ReadKey();}//通用输入inputpublic static int input(){int a;while (!int.TryParse(Console.ReadLine(), out a)){Console.WriteLine("格式错误,请输入整数");}return a;}//随机产生个体(产生的二进制随机数,数组内元素个数=种群规模,并要求对产生的随机数进行编号,使用二维数组)public static void getrand(int s, ref int[,] s1, ref int[,] s2){s1 = new int[s, 3];s2 = new int[s, 3];Random rand = new Random();Console.WriteLine("第一组随机数x1\t");for (int i = 0; i < s; i++){for (int j = 0; j < 3; j++){s1[i, j] = rand.Next(0, 2);Console.Write(s1[i, j]);}Console.WriteLine("\t");}Console.WriteLine("第二组随机数x2\t");for (int i = 0; i < s; i++){for (int k = 0; k < 3; k++){s2[i, k] = rand.Next(0, 2);Console.Write(s2[i, k]);}Console.WriteLine("\t");}}//二进制编码成十进制public static int[] convert(int s, int s2, int[,] arrnum) {Console.WriteLine("第{0}组十进制数是:", s2);int[] ten = new int[s];for (int i = 0; i < s; i++)int result = 0;for (int j = 0; j < 3; j++){result += (int)Math.Pow((double)2, (double)2 - j) * arrnum[i, j];}ten[i] = result;Console.Write("{0}\t", ten[i]);}Console.WriteLine(" \n");return ten;}//适应度计算public static int[] max(int s, int[] s1, int[] s2,out int result){int[] one = new int[s];result = 0;for (int k = 0; k < groupscale; k++){one[k] = (int)Math.Pow((double)s1[k], (double)2) + (int)Math.Pow((double)s2[k], (double)2);Console.WriteLine("第{0}个初始种群适应度为:{1}", k + 1, one[k]);result += one[k];}return one;}//适应度概率计算及累积适应度概率public static double [] probablity(int[] s,int a,int sum)double [] p=new double [a];double[] sump = new double[a+1];sump[0] = 0;for (int i = 0; i{p[i] = (double)s[i] / (double)sum;sump[i+1]=p[i]+sump[i];Console.WriteLine("第{0}个初始种群适应概率为:{1:0.00},累积适应度概率为{2:0.00}",i+1,p[i],sump[i+1]);}return sump;}//0-1之间的随机数的产生public static double[] rand(int s){int[] ran= new int[s];double[] rand=new double[s];Random a = new Random();for (int i = 0; i < s; i++){ran[i] = a.Next(0, 1000);rand[i]= (double)ran[i]/ (double)1000;}return rand;}//进行选择(1,得到被选中的数在数组中的位置2,得到被选中的二进制数组)public static int[] choose(int s,double[] s1,double[] s2){int[] chooseDNA = new int[s];for (int i = 0; i < s; i++){for (int k = 1; k < s+1; k++){if (s2[k - 1] <=s1[i] && s2[k] > s1[i]){chooseDNA[i] = k;}}}return chooseDNA;}public static int[,] ans(int[,] s1,int[,] s2,int[] s) {int[,] answer = new int[s.Length, 6];for (int i = 0; i < s.Length; i++){int a = s[i] - 1;for (int j = 0; j < 3; j++){Console.Write(s1[a, j]);answer[i, j] = s1[a, j];}for (int k = 0; k < 3; k++){Console.Write(s2[a, k]);answer[i, 3 + k] = s2[a, k];}Console.WriteLine("\n");}return answer;}//单点交叉,假定交叉率为100%public static int[,] jiaocha(int s, int[,] a){//群体进行随机配对,产生随机交叉点,相互交换染色体的基因Random ran = new Random();int[] random = new int[s / 2];if (s % 2 == 0){for (int i = 0; i < s / 2; i++){random[i] = ran.Next(1, 6);for (int j = 0; j < random[i]; j++){int[,] c = new int[s, 6];c[i, j] = a[i, j];a[i, j] = a[(s / 2)+i, j];a[(s / 2) + i, j] = c[i, j];}}}else{for (int i = 0; i < s / 2; i++){random[i] = ran.Next(1, 6);for (int j = 0; j < random[i]; j++){int[,] c = new int[(s - 1) / 2, 6];c[i, j] = a[i, j];a[i, j] = a[(s - 1) / 2, j];a[(s - 1) / 2, j] = c[i, j];}}}return a;}public static void shuchu(int[,] a) {for (int i = 0; i < groupscale; i++) {for (int j = 0; j < 6; j++){Console.Write(a[i, j]);}Console.WriteLine("\n");}}//变异public static int[,] var(int a,int[,] s) {//二维转一维int[] s1 = new int[groupscale * 6]; for (int i = 0; i < groupscale; i++) {for (int j = 0; j < 6; j++){s1[i * 6 + j] = s[i, j];}}if (groupscale * 6 * a * 0.01 < 1){Console.WriteLine("变异概率过小,不发生变异");}else{int b = Convert.ToInt32(Math.Floor(groupscale * 6 * a * 0.01)); Random ran = new Random();int[] c = new int[b];for (int i = 0; i < b; i++){c[i] = ran.Next(0, groupscale * 6);if (s1[c[i]] == 0){s1[c[i]] = 1;}else{s1[c[i]] = 0;}}}//一维转二维for (int i = 0; i < groupscale; i++){for (int j = 0; j < 6; j++){s[i, j] = s1[i * 6 + j];}return s;}//进行分组public static void fenzu(int[,] s,ref int[,] s1,ref int[,] s2) {s1 = new int[groupscale, 3];s2 = new int[groupscale, 3];for (int i = 0; i < groupscale; i++){for (int k = 0; k < 3; k++){s1[i, k] = s[i, k];}for (int j = 0; j < 3; j++){s2[i, j] = s[i, j + 3];}}}//适应度最大值public static int promax(int[] a){int max = 0;for (int i = 0; i < groupscale; i++){if (a[i]>max){max = a[i];}Console.WriteLine("适应度最大值为{0}", max); return max;}}}。
遗传算法的C语言程序案例
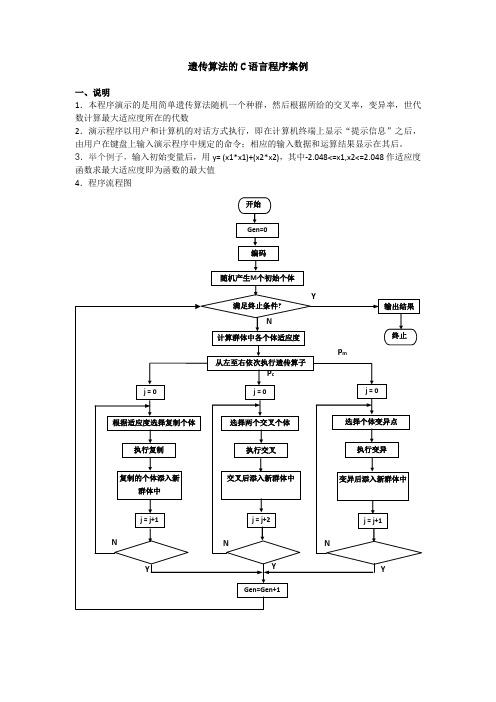
遗传算法的C语言程序案例一、说明1.本程序演示的是用简单遗传算法随机一个种群,然后根据所给的交叉率,变异率,世代数计算最大适应度所在的代数2.演示程序以用户和计算机的对话方式执行,即在计算机终端上显示“提示信息”之后,由用户在键盘上输入演示程序中规定的命令;相应的输入数据和运算结果显示在其后。
3.举个例子,输入初始变量后,用y= (x1*x1)+(x2*x2),其中-2.048<=x1,x2<=2.048作适应度函数求最大适应度即为函数的最大值4.程序流程图5.类型定义int popsize; //种群大小int maxgeneration; //最大世代数double pc; //交叉率double pm; //变异率struct individual{char chrom[chromlength+1];double value;double fitness; //适应度};int generation; //世代数int best_index;int worst_index;struct individual bestindividual; //最佳个体struct individual worstindividual; //最差个体struct individual currentbest;struct individual population[POPSIZE];3.函数声明void generateinitialpopulation();void generatenextpopulation();void evaluatepopulation();long decodechromosome(char *,int,int);void calculateobjectvalue();void calculatefitnessvalue();void findbestandworstindividual();void performevolution();void selectoperator();void crossoveroperator();void mutationoperator();void input();void outputtextreport();6.程序的各函数的简单算法说明如下:(1).void generateinitialpopulation ()和void input ()初始化种群和遗传算法参数。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
遗传算法的C语言程序案例一、说明1.本程序演示的是用简单遗传算法随机一个种群,然后根据所给的交叉率,变异率,世代数计算最大适应度所在的代数2.演示程序以用户和计算机的对话方式执行,即在计算机终端上显示“提示信息”之后,由用户在键盘上输入演示程序中规定的命令;相应的输入数据和运算结果显示在其后。
3.举个例子,输入初始变量后,用y= (x1*x1)+(x2*x2),其中-2.048<=x1,x2<=2.048作适应度函数求最大适应度即为函数的最大值4.程序流程图5.类型定义int popsize; //种群大小int maxgeneration; //最大世代数double pc; //交叉率double pm; //变异率struct individual{char chrom[chromlength+1];double value;double fitness; //适应度};int generation; //世代数int best_index;int worst_index;struct individual bestindividual; //最佳个体struct individual worstindividual; //最差个体struct individual currentbest;struct individual population[POPSIZE];3.函数声明void generateinitialpopulation();void generatenextpopulation();void evaluatepopulation();long decodechromosome(char *,int,int);void calculateobjectvalue();void calculatefitnessvalue();void findbestandworstindividual();void performevolution();void selectoperator();void crossoveroperator();void mutationoperator();void input();void outputtextreport();6.程序的各函数的简单算法说明如下:(1).void generateinitialpopulation ()和void input ()初始化种群和遗传算法参数。
input() 函数输入种群大小,染色体长度,最大世代数,交叉率,变异率等参数。
(2)void calculateobjectvalue();计算适应度函数值。
根据给定的变量用适应度函数计算然后返回适度值。
(3)选择函数selectoperator()在函数selectoperator()中首先用rand ()函数产生0~1间的选择算子,当适度累计值不为零时,比较各个体所占总的适应度百分比的累计和与选择算子,直到达到选择算子的值那个个体就被选出,即适应度为fi的个体以fi/∑fk的概率继续存在;显然,个体适应度愈高,被选中的概率愈大。
但是,适应度小的个体也有可能被选中,以便增加下一代群体的多样性。
(4)染色体交叉函数crossoveroperator()这是遗传算法中的最重要的函数之一,它是对个体两个变量所合成的染色体进行交叉,而不是变量染色体的交叉,这要搞清楚。
首先用rand ()函数产生随机概率,若小于交叉概率,则进行染色体交叉,同时交叉次数加1。
这时又要用rand()函数随机产生一位交叉位,把染色体的交叉位的后面部分交叉即可;若大于交叉概率,则进行简单的染色体复制即可。
(5)染色体变异函数mutation()变异是针对染色体字符变异的,而不是对个体而言,即个体变异的概率是一样。
随机产生比较概率,若小于变异概率,则1变为0,0变为1,同时变异次数加1。
(6)long decodechromosome(char *,int,int)本函数是染色体解码函数,它将以数组形式存储的二进制数转成十进制数,然后才能用适应度函数计算。
(7)void findbestandworstindividual()本函数是求最大适应度个体的,每一代的所有个体都要和初始的最佳比较,如果大于就赋给最佳。
(8)void outputtextreport () 输出种群统计结果输出每一代的种群的最大适应度和平均适应度,最后输出全局最大值二、运行环境本程序的开发工具是VC++,在VC++下运行。
源代码#include"stdafx.h"#include <stdio.h>#include <stdlib.h>#include <time.h>/////////////The definiton of user data 定义用户数据////#define Cmax 100 //certain maximal value#define Cmin 0 //certain minimum value#define LENGHT1 3#define LENGHT2 3//总染体长度#define CHROMLENGTH LENGHT1+LENGHT2const int MaxGeneration = 100;const int PopSize = 10;const double Pc = 0.6;const double Pm = 0.001;////////////// 数据结构定义///////////////////struct Individual{char chrom[CHROMLENGTH + 1];double value;double fitness;int generation ;int bestIndex;int worstIndex;Individual bestIndividual ;Individual worstIndividual ;// best individual by nowIndividual currentBest ;Individual population [PopSize] ;///////////////////////void generateInitialPopulation();void generateNextPopulation();void evalutePopulation();long decomdeChromosome(char*, int, int); void calculateObjectValue();void calculateFitnessValue();void findBestAndWorstIndividual();void performEvolution();void selectionOperator();void crossoverOperator();void mutationOperator();void outputTextReport();//以上为函数以及全局变量定义部分int main(){generation = 0; generateInitialPopulation(); evalutePopulation();while (generation < MaxGeneration) { generation++; generateNextPopulation(); evalutePopulation();performEvolution();outputTextReport();}system("pause");return 0;}////////////////////////////////////////////////////////////////////产生第一代样本/////void generateInitialPopulation() {int i, j;srand((unsigned)time(NULL));for (i = 0; i < PopSize; i++) {for (j = 0; j < CHROMLENGTH; j++) {population[i].chrom[j] = ((rand() % 10) < 5) ? '0' : '1';}population[i].chrom[CHROMLENGTH] ='/0';}}////////产生下一代样本//////void generateNextPopulation() {selectionOperator();crossoverOperator();mutationOperator();}//变异算子//void mutationOperator() {int i, j;double p;// bit mutationfor (i = 0; i < PopSize; i++) {for (j = 0; j < CHROMLENGTH; j++) {p = rand() % 1000 / 1000.0;if (p < Pm) {population[i].chrom[j] = (population[i].chrom[j] == '0') ? '1': '0'; }}}}//交叉算子///void crossoverOperator() {int i, j;int index[PopSize];int point, temp;double p;char ch;for (i = 0; i < PopSize; i++) {index[i] = i;}for (i = 0; i < PopSize; i++) {point = rand() %(PopSize - i);temp = index[i];index[i] = index[point + i];index[point + i] = temp;}for (i = 0; i < PopSize - 1; i+=2) {p = rand() % 1000 / 1000.0;if (p < Pc) {point = rand()% (CHROMLENGTH - 1) + 1;for (j = point; j < CHROMLENGTH; j++) {ch = population[index[i]].chrom[j];population[index[i]].chrom[j] = population[index[i + 1]].chrom[j]; population[index[i + 1]].chrom[j] = ch;}}}}///选择算子/////////////void selectionOperator() {int i, index;double p, sum = 0.0;double cfitness[PopSize];Individual newpopulation[PopSize];for (i = 0; i < PopSize; i++) {sum += population[i].fitness;}for (i = 0; i < PopSize; i++) {cfitness[i] = population[i].fitness / sum;}// calculate cumulative fitnessfor (i = 1; i < PopSize; i++) {cfitness[i] = cfitness[i] + cfitness[i - 1];}for (i = 0; i < PopSize; i++) {p = rand() % 1000 / 1000.0;index = 0;while (p > cfitness[index]) {index++;}newpopulation[i] = population[index];}for (i = 0; i < PopSize; i++) {population[i] = newpopulation[i]; /}}/////依据某些公式对样本进行评价////void evalutePopulation() {calculateObjectValue();calculateFitnessValue(); findBestAndWorstIndividual(); /}//找出到目前为止最好的个体//////void findBestAndWorstIndividual() {int i;double sum = 0.0;bestIndividual = population[0];worstIndividual = population[0];for (i = 0; i < PopSize; i++) {if (population[i].fitness > bestIndividual.fitness) { bestIndividual = population[i];bestIndex = i;} else if (population[i].fitness < worstIndividual.fitness) { worstIndividual = population[i];worstIndex = i;}sum += population[i].fitness;}if (generation == 0) {currentBest = bestIndividual;} else {if (bestIndividual.fitness > currentBest.fitness) {currentBest = bestIndividual;}}}//计算适应度///void calculateFitnessValue() {int i;long temp1, temp2;double x1, x2;for (i = 0; i < PopSize; i++) {temp1 = decomdeChromosome(population[i].chrom, 0, LENGHT1);temp2 = decomdeChromosome(population[i].chrom, LENGHT1, LENGHT2); x1 = temp1 * temp1;x2 = temp2 * temp2;population[i].fitness = x1+x2; //x1平方加上x2平方}}//计算目标值//目标函数为f(x) = x1* x1 + x2*x2void calculateObjectValue() {int i;long temp1, temp2;double x1, x2;for (i = 0; i < PopSize; i++) {temp1 = decomdeChromosome(population[i].chrom, 0, LENGHT1);temp2 = decomdeChromosome(population[i].chrom, LENGHT1, LENGHT2); x1 = temp1 * temp1;x2 = temp2 * temp2;population[i].value = x1 + x2;}}//把二进制转化为十进制long decomdeChromosome(char* string, int point, int length) {int i;long decimal = 0L;char * pointer;for(i = 0, pointer=string+point; i < length;i++,pointer++){decimal += (*pointer - '0') << (length - 1 - i);}return decimal;}//进经同时把最坏个体用目前最好个体替代///void performEvolution() {if (bestIndividual.fitness > currentBest.fitness) {currentBest = population[bestIndex];} else {population[worstIndex] = currentBest;}}//打印当前样本信息///void outputTextReport() {int i;double sum;double average;sum = 0.0;for (i = 0; i < PopSize; i++) {sum += population[i].value;}average = sum / PopSize;printf("gen=%d, avg=%f, best=%f",generation, average,currentBest.value); printf(" chromosome=");for( i = 0; i < CHROMLENGTH; i++){printf("%c", currentBest.chrom[i]);}printf("/n");}。
