毕赤酵母表达操作手册(PDF精译版)
毕赤酵母发酵手册
毕赤酵母发酵手册总览简介:毕赤酵母和酿酒酵母很相似,都非常适合发酵生长。
毕赤酵母在有可能提高总体的蛋白质产量的发酵中能够达到非常高的细胞浓度,我们建议只有那些有过发酵经验或者能得到有经验的人的指导的人参与发酵。
因为发酵的类型很多,所以我们很难为您的个人案例提高详细的过程。
下面所给出的指导是基于Mut+和Mut s两种基因型的毕赤酵母菌株在15L的台式玻璃发酵罐中发酵而成。
请在您的发酵开始前先阅读操作员手册。
下面所给出的表就发酵参数:在整个发酵过程中监测和调控下列参数非常重要。
下面的表格描述了这些参设备推荐:下面是所推荐设备的清单:·发酵罐的夹套需要在发酵过程中给酵母菌降温,尤其是在甲醇流加过程中。
你需要一个固定的来源来提供冷却水(5-10℃)。
这可能意味着你需要一个冷冻装置来保持水的冷却。
·一个泡沫探针就像消泡剂一样不可或缺。
·一个氧气的来源——空气(不锈钢的发酵罐需要1-2vvm)或者纯氧(玻璃发酵罐需要0.1-0.3vvm)。
·添加甘油和甲醇的补料泵。
·pH的自动控制。
培养基的准备:你需要准确配置下列溶液:·发酵所需的基本盐类(第11页)·PTM1补充盐类(第11页)·75ml的50%的甘油每升初始发酵液,12ml的PTM1补充盐每升甘油。
·740ml的100%的甲醇每升初始发酵液,12ml的PTM1补充盐每升甲醇。
毕赤酵母生长的测定:在不同的时间点通过测OD600的吸光值和湿细胞的重量来检测毕赤酵母的生长。
培养的代谢速率通过通过观察溶氧浓度对应于有效碳源来测定。
溶氧的测定:简介:溶解氧的浓度时指氧气在培养基中的相关比例,溶氧100%是指培养基中氧达到饱和。
毕赤酵母的生长需要消耗氧气,减少溶解氧的满度。
毕赤酵母在生长时会消耗氧气,减少溶氧的程度。
然而,因为代谢甲醇的最初阶段需要氧气,所以将溶氧浓度维持在一个适当的水平(>20%)来确保毕赤酵母在甲醇上的生长就至关重要。
毕赤酵母手册

毕赤酵母表达实验手册作者:Jnuxz 来源:丁香园时间:2007-9-5大肠杆菌表达系统最突出的优点是工艺简单、产量高、周期短、生产成本低。
然而,许多蛋白质在翻译后,需经过翻译后的修饰加工,如磷酸化、糖基化、酰胺化及蛋白酶水解等过程才能转化成活性形式。
大肠杆菌缺少上述加工机制,不适合用于表达结构复杂的蛋白质。
另外,蛋白质的活性还依赖于形成正确的二硫键并折叠成高级结构,在大肠杆菌中表达的蛋白质往往不能进行正确的折叠,是以包含体状态存在。
包含体的形成虽然简化了产物的纯化,但不利于产物的活性,为了得到有活性的蛋白,就需要进行变性溶解及复性等操作,这一过程比较繁琐,同时增加了成本。
大肠杆菌是用得最多、研究最成熟的基因工程表达系统,当前已商业化的基因工程产品大多是通过大肠杆菌表达的,其主要优点是成本低、产量高、易于操作。
但大肠杆菌是原核生物,不具有真核生物的基因表达调控机制和蛋白质的加工修饰能力,其产物往住形成没有活性的包涵体,需要经过变性、复性等处理,才能应用。
近年来,以酵母作为工程菌表达外源蛋白日益引起重视,原因是与大肠杆菌相比,酵母是低等真核生物,除了具有细胞生长快,易于培养,遗传操作简单等原核生物的特点外,又具有真核生物时表达的蛋白质进行正确加工,修饰,合理的空间折叠等功能,非常有利于真核基因的表达,能有效克服大肠杆菌系统缺乏蛋白翻译后加工、修饰的不足。
因此酵母表达系统受到越来越多的重视和利用。
[1]。
同时与大肠杆菌相比,作为单细胞真核生物的酵母菌具有比较完备的基因表达调控机制和对表达产物的加工修饰能力。
酿酒酵母(Saccharomyces.Cerevisiae)在分子遗传学方面被人们的认识最早,也是最先作为外源基因表达的酵母宿主。
1981年酿酒酵母表达了第一个外源基因----干扰素基因[2],随后又有一系列外源基因在该系统得到表达[3、4、5、6]。
干扰素和胰岛素虽然已经利用酿酒酵母大量生产并被广泛应用,当利用酿酒酵母制备时,实验室的结果很令人鼓舞,但由实验室扩展到工业规模时,其产量迅速下降。
毕赤酵母实验操作手册

毕赤酵母表达实验手册大肠杆菌表达系统最突出的优点是工艺简单、产量高、生产成本低。
然而,许多蛋白质在翻译的修饰加工,如磷酸化、糖基化、酰胺化及蛋白酶水解等过程才能转化成活性形式。
大肠杆菌缺少适合用于表达结构复杂的蛋白质。
另外,蛋白质的活性还依赖于形成正确的二硫键并折叠成高级结表达的蛋白质往往不能进行正确的折叠,是以包含体状态存在。
包含体的形成虽然简化了产物的纯的活性,为了得到有活性的蛋白,就需要进行变性溶解及复性等操作,这一过程比较繁琐,同时增与大肠杆菌相比,酵母是低等真核生物,具有细胞生长快,易于培养,遗传操作简单等原核生物的生物时表达的蛋白质进行正确加工,修饰,合理的空间折叠等功能,非常有利于真核基因的表达,菌系统缺乏蛋白翻泽后加工、修饰的不足。
因此酵母表达系统受到越来越多的重视和利用。
大肠杆菌是用得最多、研究最成熟的基因工程表达系统,当前已商业化的基因工程产品大多是通过其主要优点是成本低、产量高、易于操作。
但大肠杆菌是原核生物,不具有真核生物的基因表达调加工修饰能力,其产物往住形成没有活性的包涵体,需要经过变性、复性等处理,才能应用。
近年程菌表达外源蛋白日益引起重视,主更是因为酵母是单细胞真核生物,不但具有大肠杆菌易操作、化生产的特点,还具有真核生物表达系统基因表达调控和蛋白修饰功能,避免了产物活性低,包涵间题[1]。
与大肠杆菌相比,酵母是单细胞真核生物,具有比较完备的基因表达调控机制和对表达产物的们对酿酒酵母(Saccharomyces.Cerevisiae)分子遗传学方面的认识最早,酿酒酵母也最先作为外宿主.1981年酿酒酵母表达了第一个外源基因一干扰素基因,随后又有一系列外源基因在该系统得素和胰岛素已大量生产并在人群中广泛应用,但很大部分表达由实验室扩展到工业规模时,培养基数的选择压力消失,质粒变得不稳定,拷贝数下降,而大多数外源基因的高效表达需要高拷贝数的量下降。
同时,实验室用培养基复杂而昂贵,采用工业规模能够接受的培养基时,往往导致产量的酵母的局限,人们发展了以甲基营养型酵母(methylotrophic yeast)为代表的第二代酵母表达系甲基营养型酵母包括:Pichia、Candida等.以Pichia.pastoris(毕赤巴斯德酵母)为宿主的外源来发展最为迅速,应用也最为广泛,已利用此系统表达了一系列有重要生物学活性的蛋自质。
毕赤酵母发酵工艺手册

毕赤酵母发酵工艺手册1. 引言欢迎使用毕赤酵母发酵工艺手册。
本手册旨在介绍毕赤酵母发酵的基本原理、工艺步骤以及相关注意事项。
通过遵循本手册,您可以更好地理解和掌握毕赤酵母的发酵过程,从而在生产中取得更好的效果。
2. 毕赤酵母发酵基本原理- 毕赤酵母是一种常见的酵母菌,其发酵能力强,适用于多种发酵产品的生产。
- 发酵是指通过酵母菌对底物中的糖类进行代谢,产生酒精和二氧化碳的过程。
- 毕赤酵母在发酵过程中需要适宜的温度、pH值和营养物质等条件。
3. 毕赤酵母发酵工艺步骤1. 发酵前准备:- 准备好所需的发酵基质,包括糖类、氮源和维生素等。
- 对基质进行消毒处理,确保无害菌的存在。
2. 接种毕赤酵母:- 选择合适的毕赤酵母培养液进行接种,注意接种量的控制。
- 将毕赤酵母培养液均匀加入发酵基质中。
3. 发酵条件控制:- 控制发酵温度在合适的范围内,一般为25-30摄氏度。
- 监测发酵基质的pH值,保持在适宜的范围内。
- 提供足够的氧气供给,促进酵母的生长和代谢。
4. 发酵过程监测:- 定期对发酵过程中的温度、pH值和酵母数量等进行监测和记录。
- 根据监测结果及时调整发酵条件,确保发酵过程稳定进行。
5. 发酵结束:- 当发酵基质中的糖类被完全代谢,产物达到预期时,发酵过程结束。
- 将发酵产物经过处理和提取,得到最终的产品。
4. 注意事项- 在发酵过程中,应注意卫生和消毒,以防止杂菌的污染。
- 严格控制发酵条件,避免过高或过低的温度、pH值对发酵效果产生不利影响。
- 根据不同的发酵产品,可能需要调整发酵步骤和条件,建议根据具体要求进行调整。
- 在使用本工艺手册时,请参考其他文献和专业意见,确保准确性和可靠性。
以上是关于毕赤酵母发酵工艺手册的简要介绍。
希望本手册能对您在毕赤酵母的发酵工艺中提供帮助和指导。
如有任何问题,请随时与我们联系。
谢谢!。
GS115毕赤酵母表达菌使用说明

编号
名称
北京华越洋生物 NRR01030 GS115 毕赤酵母表达菌
基 本 信 息 :
名称:GS115 毕赤酵母表达菌
规格:300ul 甘油菌
储 存 温 度 : -‐80℃
发突变为组氨酸野生型的概率一般低于 10-‐8。GS115 毕赤酵母可以在 YPD
培养基中生长,或者在补充有组氨酸的 minimal media 中生长,但是无法
在单独的 minimal media 中生长。GS115 毕赤酵母在做质粒转化的时候,
可 采 用 电 转 化 的 方 式 将 质 粒 转 入 。
基 因 组 :
His4( 基 因 5 是毕赤酵母菌株,是巴斯德毕赤酵母的一种,属于真核细胞。
一般的针对原核生物的抗生素例如卡那和氨苄对酵母是无效的,因此为了
操作说明:
1,本品包含一份甘油菌,使用本甘油菌时可以不用完全融解,在甘油菌表
面蘸取少量涂板或进行液体培养即可。也可以完全融解后使用,但随着冻融次数
的增加,细菌的活力会逐渐下降。
2,为保证菌种纯正,避免其它细菌污染,尽量先划平板,然后再挑单克隆
菌落进行后续操作。
毕赤酵母适宜的生长温度是 28 至 30 度,温度超过 32 度对蛋白的表
达是有害的,并可能导致细胞的死亡。GS115 毕赤酵母是是组氨酸缺陷型
(His4 基因型),如果表达载体上携带有组氨酸基因,可补偿宿主菌的组
氨酸缺陷,因此可以在不含组氨酸的培养基上筛选转化子。这些受体菌自
养。细菌在 30-‐35℃培养箱中培养 24-‐48h,真菌在 23-‐28℃培养箱中培养 24-‐72h
(必要时,可适当延长培养时间)。
植物科学领域最全的“方法步骤软件”

植物科学领域最全的“⽅法步骤软件”该⽹盘所有内容也将根据⼤家的补充逐渐更新⽅法步骤1Western_blot_protocol.pdf2Yeast transformation tips.pdf3标签蛋⽩简单介绍.pdf4采⽤本⽒烟草瞬时表达系统进⾏蛋⽩质免疫共沉淀实验 Protein Immuno.. (2).pdf5超级感受态制备.pdf6超级感受态制作⽅法ULTRA.pdf7从纯化到星⾠.pdf8蛋⽩质电泳相关试剂、缓冲液的配制⽅法 - 副本.pdf9分⼦克隆实验指南第三版上册.pdf10分⼦克隆实验指南第三版下册.pdf11分⼦克隆实验指南上.pdf12分⼦⽣物学实验技术简册.pdf13分⼦⽣物学实验室⼯作⼿册.pdf14国内重点实验室分⼦⽣物学实验⽅法汇总实验室常⽤实验⽅法.pdf15核酸电泳相关试剂、缓冲液的配制⽅法 - 副本.pdf16核酸电泳相关试剂、缓冲液的配制⽅法.pdf17核算蛋⽩互算.pdf18基因northern blot⽅法.pdf19基因X 中⽂版.pdf20基因⼯程原理(吴乃虎编著).pdf21基因芯⽚⾼质量-CTAB提DNA.pdf22基于Gateway体系⼀步PCR法构建CRISPR终载体.pdf23酵母单杂说明书.pdf24酵母⾼效转化⽅法.pdf25酵母双杂交操作.pdf26酵母双杂交技术 - 副本.pdf27酵母相关实验⽅法.pdf28芥菜幼苗中超氧化物和H2O2累积的组织化学检测.pdf29抗⽣素的贮存溶液及其⼯作浓度 - 副本.pdf30类胡萝⼘素测定-Functional analysis of beta- and epsilon-ring carotenoid hydroxylases in Arabidopsis.pdf31免疫共沉淀Co-IP实验操作步骤.pdf32拟南芥的实验室⼿册.pdf33拟南芥提DNA.pdf34拟南芥叶⽚过氧化氢DAB染⾊.pdf35凝胶迁移实验(EMSA).pdf36农杆菌注射烟草叶⽚进⾏体内瞬时表达的⽅法.pdf37染⾊质免疫共沉淀(ChIP)实验.pdf38如何使⽤DNAMAN软件制作序列⽐对.pdf39实时荧光pcr中⽂.pdf40实时荧光定量PCR全⽅位解析.pdf41实验室常⽤培养基的配制⽅法 - 副本.pdf42实验室常⽤培养基的配制⽅法.pdf43实验室常⽤试剂、缓冲液的配制⽅法 - 副本.pdf44⼿把⼿教你使⽤EndNote_X8.pdf45数量遗传学、基因组学和植物分⼦育种.pdf46双酶切连接反应之全攻略.pdf47⽔稻多基因敲除系统.pdf48⽔稻原⽣质体的分离转化.pdf49体外降解.pdf50⽂献综述的写法详细.pdf51现代分⼦⽣物学第4版_朱⽟贤2013.pdf52原位杂交procotol.pdf53植物病原体感染期间的植物组织台盼蓝染⾊Bio-protocol2078.pdf 54植物组织电镜基本制样⽅法20171117.pdf55转基因⼤麦中gfp基因的染⾊体位置及其表达.pdf56转录组分析软件上机指南.pdf57综述的写法.pdf58总-常⽤逆境⽣理指标测定⽅法-LCL.pdf59[]毕⾚酵母表达操作⼿册(精译版).pdf609页的操作yeast Transformation.pdf612003 pMDC100 pDMC32 A Gateway Cloning Vector Set for High-Throughput Functional Analysis of Genes in Planta.pdf622007 pMDC100 pDMC32 Recombinational Cloning with Plant Gateway Vectors.pdf 63630491 中⽂版-单杂载体说明.pdf64 A Foxtail mosaic virus Vector for Virus-Induced Gene silencing in maize.pdf65 a loop-mediated isothermal amplification method.pdf66 A Toolkit for Illustrating Heatmap.PDF67Alexander_staining(⼀种花粉活性染⾊法).pdf68arabidopsis mesophyll bifc.pdf69Arabidopsis mesophyll protoplasts.pdf70BiFC.pdf71Bio-Rad荧光定量PCR应⽤指南.pdf72bp_clonaseii_man.pdf73CAPS_and_dCAPS.pdf74CapsaicinHPLC-UV-MS-PP.pdf75DUAL?Membrane?System?Protocol.pdf76EndNote.pdf77FISH Protocols.pdf78GAPDH酶活测定.pdf79Gateway Protocol.pdf80gateway system mannual.pdf81gateway_pdonr_vectors.pdf82GenStat统计⽅法与数据分析(1).pdf83GUS染⾊.pdf84GUS染⾊-杜长青.pdf85GV3101电击.pdf86HiTail PCR BTN_A_000112601_O_1421a.pdf87imageJ使⽤⽅法-叶⽚⾯积统计.pdf88ImpGWB manual E (pGWBnxx).pdf89infusion protocol.pdf90JoinMapML作图⽅法.pdf91Khalid Meksem, Guenter Kahl-The Handbook of Plant Genome Mapping_ Genetic and Physical Mapping (v. 1)-Wiley-VCH (2005).pdf92KOD –Plus- Neo (KOD-401).pdf93KOD-401.pdf94lr_clonase_man.pdf95LSM brief manual_final.pdf96Matchmaker Gold Yeast Two-Hybrid System User Manual (PT4084-1)_092413.pdf 97Maxent简明教程(翻译).pdf98MEGA3.1 中⽂使⽤说明.pdf99miRNA northern blot ⽅法.pdf100Molecular Plant Pathology.pdf101MultiExperiment_Viewer_Quickstart_Guide_MeV软件教程-1.pdf102Oligo的使⽤⽅法介绍.PDF103One-step enzymatic assembly of DNA-YubingHe.pdf104PCR Mutation Detection Protocols.pdf105Pcr Protocols-Methods In Molecular Biology.pdf106pDONR201.pdf107pDONR201 207 info..pdf108pgwb pdf.pdf109plant metabolite protocol and method.pdf110PrimeSTAR - 副本.pdf111R for Beginners 中⽂版.pdf112R050A PrimeSTAR? GXL DNA Polymerase.pdf113Rice Protocols_Methods in Molecular Biology, 2013, Vol. 956(Non-commercial use permitted).pdf114ROS染⾊⽅法.pdf115RPA III Kit.pdf116SDS PAGE.pdf117SPSS与多元统计分析⽅法讲义.pdf118Tai Te Wu Analytical Molecular Biology .pdf 119TAK-101_201.pdf120Vector_NTI_简体中⽂使⽤教程PDF版本.pdf 121Western blot详细操作步骤.pdf122western_blot - 副本.pdf123实验protocol⼤全124VIGS125endnote x8破解.rar.xltd.cfg126Endnote_X7实⽤教程.pptx127ij.jar128mmexport1510878447197.jpg129PastedGraphic-1.png130烟草遗传转化.jpg131可溶性糖和可溶性蛋⽩测定⽅案.wps132Adobe Acrobat XI Pro 11.0.1.rar.xltd.cfg 133SeqScannerInstall.exe.重命名134蛋⽩的相互作⽤ - 副本.ppt135蛋⽩质结构与功能预测.ppt136实验技术.zip.xltd.cfg137⽔稻花药花粉发育过程.jpg138注册账号.txt139载体构建.ppt140科研论⽂作图141⽟⽶营养液.xlsx142LI6400143protoplast_workshop.wmv144SPSS介绍与应⽤.pptx145⽂献管理教学-zd.pptx146拟南芥蜡质测定⽅法.jpg147软件148国家基础地理信息系统数据.zip149分⼦⽣物学150克隆构建载体知识积累151植物实验⽅法152转录组培训资料153补充的软件和试验⽅法154酵母双杂155实验技术156原⽣质体制备157备份word158⼦叶节培养程序2008整理.docx1594.5h Immunolocalization .doc160Arlequin操作说明.doc161change solution on August 2nc.doc162ChIP for Arabidopsis.doc163CHIP protocol.doc164CHIP实验⽅法.doc165CHIP实验⽅法2.doc166co-IP in vivo.doc167CTAB 提取DNA.doc168CTAB-96孔法⽔稻基因组DNA提取.doc169CTAB法提前DNA及其检测.doc170DNA粗提.doc171DNA提取过程中各种试剂的作⽤.doc172EMSA实验步骤.doc173FISH protocol.DOC174FISH药品配置.doc175H2O2的测定-KI法.doc176Hydrogen proxide_CN.doc177IAA treatment used for detecting IAA induced genes.doc 178In Situ hybridization in plants- protocol.doc179IPCR确定T-DNA突变体插⼊位点 - 副本.doc180Leica⽯蜡切⽚机操作规程.doc181LYM基因克隆构建PAbAi载体.doc182LYM酵母单杂流程.doc183Megazyme_总淀粉测定试剂盒翻译.doc184MS母液配制⽅法及⽤量.doc185MS培养基及配制注意事项 - 副本.doc186NEB mRNA 建库protocol.doc187N-P-K联合消煮测定.doc188PAGE胶流程.doc189PFP PFK ADH PDC enzyme activities.doc190POD 电泳⽅法.doc191Rice Leaf Protoplast Protocol-Modified by WM.doc192RT-PCR.doc193RuBP-PEP-Pr测定.doc194SEM.doc195SOD 电泳⽅法.doc196SOD、POD、CAT、MDA和可溶性蛋⽩测定.doc197VIGS实验⽅案.doc198WB实验步骤.doc199Y1H.doc200yongjiang氮代谢酶.doc201矮牵⽜转化.doc202半薄切⽚.doc203曹建康-果蔬采后⽣理⽣化实验指导.doc204测酶时需要配的溶液.doc205常⽤试剂配⽅.doc206超级感受态制备 (2).doc207超级感受态制备.doc208成熟胚⾼效转基因系统的建⽴(Ver_2011.03.21).doc209⼤肠杆菌电击感受态制备⽅法-曹⾼燚.doc210蛋⽩各组分含量测定⽅法.doc211蛋⽩酶活性的测定⽅法.doc212蛋⽩透析复性⽅法.doc213蛋⽩质胶内酶解及同位素标记实验步骤.doc214蛋⽩组学实验流程.doc215分⼦克隆技术实验操作⼿册2005.doc216分⼦⽣物学实验技术实验操作指南.doc217柑橘原⽣质体提取及转化.doc218根系活⼒测定-NRA测定.doc219过氧化氢测定⽅法.doc220花粉液体纯体外萌发⽅法.doc221花序浸染法转化拟南芥 - 副本.doc222基因枪法瞬时转化花粉.doc223甲基化⽔平重硫酸盐测序.doc224减数分裂染⾊体观察⽅法.doc225酵母感受态制备及转化.doc226磷酸盐缓冲液配制⽅法.doc227酶活测定⽅法.doc228免疫⾦⼿册.doc229茉莉酸JA提取.doc230拟南芥⾓质测定⽅法.doc231拟南芥解剖学图 - 副本.doc232拟南芥茎切⽚.doc233拟南芥原⽣质体转化系统.doc234凝胶迁移滞后实验 - 副本.doc235农杆菌感受态的制备.doc236农杆菌侵染拟南芥花序的转化⽅法 - 副本.doc237农杆菌转化番茄.doc238农杆菌转化烟草.doc239切⽚技术.doc240琼脂糖切⽚.doc241设计引物如何设计酶切位点.doc242⽯蜡包埋机(paraffin-embedding-station)Leica-EG1150H.doc 243⽯蜡切⽚制作⽇程(新).doc244实验室常⽤试剂、缓冲液的配制⽅法.doc245实验室⽣理⽣化指标实验简略步骤.doc246实验室原核表达步骤.doc247史上最全限制性内切酶酶切位点汇总.doc248树⽊DNA的提取.doc249双分⼦荧光互补实验原理 - 副本.doc250⽔稻根尖压⽚-2015-7-13.doc251⽔稻叶中CAT、POD和SOD活性的测定.doc252⽔稻营养液配⽅.doc253透明comfocs.doc254⼟壤速效钾测定.doc255⼟壤有机质测定.doc256⼟壤有效氮测定.doc257⼟壤有效磷测定.doc258⽡村单倍体诱导protocol.doc259我们实验室⽊本果树RNA提取⽅法.doc260烟草注射.doc261叶绿素含量测定.doc262液泡分离及V-PPase酶活和H 转运检测.doc263仪器操作规范-2017.11.17.doc264荧光定量引物⾃动化设计教程.doc265营养品质测定⽅法.doc266⽤于质谱鉴定的蛋⽩质凝胶样品酶解⽅法.doc267油菜转基因技术体系终结版.doc268原位.doc269中药鉴定学重点整理.doc270最新实验原理与⽅法2009.doc271[]进化树软件MEGA最新6.06说明书.docx2722个试验⽅法分享.docx2731003总结载体构建说明书.docx27420170106电击法农杆菌感受态的制备及其转化-曹⾼燚.docx275A simple and rapid method for the preparation of plant genomic DNA for PCR analysis..docx 276Benzoxazinoids extraction and quantification.docx277BSMV基因沉默相关.docx278ChIP for Plant.docx279circos画共线性图⽚.docx280CO-IP (2).docx281co-IP.docx282CoIP-Western blot.docx283CTAB法提DNA.docx284CTAB法提取⽔稻DNA.docx285Dellaporta法⼩量快速提取植物全基因组DNA.docx286EMSA protocol.docx287EST-SSR实验流程及软件使⽤.docx288FAA固定protocol.docx289FNPI-PCR⽅案.docx290GISH分析.docx291His-tag原核蛋⽩提取.docx292Histochemical detection of ROS.docx293Hoagland营养液配制⽅法.docx294IAA、 JA、 ABA、 SA 定量检测分析.docx295In vitro Pollen germination media.docx296IP MASS.docx297JASA LC-MS extraction.docx298Native chip.docx299Northern杂交.docx300PAGE 银染.docx301PCR平端加A流程.docx302Pi含量测定.docx303Protein expressed in yeast microsome.docx304Protein extracted from plant tissues__ZT.docx305Protein extraction protocol.docx306PS 、AI、CDR设计视频教程.docx307SEM实验⽅法.docx308Sls法提DNA.docx309TPS法提取植物DNA.docx310trizol法⼤量提取油菜RNA.docx311Trizol法提取RNA.docx312VIGS流程-汪汪-20161102.docx313Western Blot protocol .docx314Western Blot.docx315Western blot.docx316保护酶的测定⽅法.docx317便携式调制叶绿素荧光仪PAM-2500简明操作⼿.docx318表达蛋⽩的分离与纯化;EMSA.docx319打烟草coip或看荧光⽐较常⽤.docx320⼤肠杆菌感受态细胞制备实验(CaCl2法) _1510835367078.docx321⼤⾖蛋⽩提取.docx322⼤⾖⼦叶节遗传转化流程.docx323蛋⽩提取 CO-ip WESTERN BLOT.docx324多靶点pCRISPR载体(单⼦叶植物⽤)使⽤⽅法2014-05-19.docx325多靶点pCRISPR载体改良版(双⼦叶植物⽤)使⽤⽅法2014-10-26(P).docx 326改良的⽟⽶SLS法提取DNA.docx327柑橘DNA和RNA提取⽅法.docx328感受态细胞制备.docx329花青素原花⾊素相关protocol.docx330活性氧,胼胝质,叶⽚⽓孔的检测实验.docx331基础分⼦⽣物学实验操作指南.docx332基因枪轰击洋葱表⽪观察.docx333基因枪瞬时转化矮牵⽜花瓣.docx334简单粗暴提取拟南芥DNA-1.docx335酵母双杂筛库1.docx336酵母双杂实验⽅法总结.docx337酵母双杂最终版.docx338酵母转化⽅法及⽔稻组织培养液配⽅-1.docx339聚丙烯酰胺凝胶电泳检测PCR产物.docx340菌落PCR(⾃⼰总结).docx341可溶性糖测定.docx342临界点⼲燥.docx343拟南芥材料快速PCR鉴定.docx344拟南芥可溶性全蛋⽩的双向电泳.docx345拟南芥原⽣质体.docx346拟南芥原⽣质体的提取和转化.docx347拟南芥原⽣质体亚细胞定位Copy of Localization of procedure.docx348拟南芥原⽣质体制备.docx349拟南芥杂交.docx350拟南芥总蛋⽩简易提取.docx351凝胶迁移实验(EMSA)实验⽅法 - 副本.docx352农杆菌注射烟草顺时转化.docx353全基因组甲基化WGBS和转录组分析过程.docx354试验⽅法.docx355⽔稻缺素处理配⽅及步骤.docx356⽔稻原⽣质体分离及转化.docx357⽔稻原⽣质体提取及转化.docx358⽔稻原⽣质体制备和瞬时转化.docx359体外泛素连接酶活性测定.docx360⼟壤基本指标检测.docx361兔源抗体制备.docx362新Y1H载体构建及转化步骤.docx365亚细胞定位.docx366烟草瞬时表达.docx367叶⽚碳代谢相关物质和酶试验⽅法-棉花.docx368油菜下胚轴暗光培养转化.docx369原核表达.docx370原核表达protocol.docx371原核表达步骤.docx372原核蛋⽩的提取.docx373原⽣质体叶⾁细胞瞬时表达.docx374原位杂交.docx375真核细胞表达外源基因技术⽅法及操作步骤.docx376植物常见外援激素的作⽤.docx377植物⽣理⽣化指标.docx378植物⽯蜡切⽚.docx379植物有效磷测定.docx380植物总RNA提取与琼脂糖凝胶电泳分析.docx381注射烟草看BiFC.docx382SA测定⽅法3832007_Dolezel_NATURE_PROTOCOLS_2233.pdf384【代码M0101】常⽤溶液的配制.docx385AGROBACTERIUM COMPETENT CELLS.doc386Agrobacterium growth and transformation.doc387AutoDock&ADT教程(1).doc388cellulose content detection method.doc389CHIP protocal.docx390CRISPR-Cas9载体构建(⽔稻).docx391CTAB-96孔法⽔稻基因组DNA提取.doc392CTAB提取真菌DNA.docx393DAB染⾊,台盼蓝染⾊-钱礼超.docx394【代码M0201】PCR引物设计⽅法.docx395【代码M0202】PCR引物设计11条黄⾦原则.docx396ELISA的原理和技术要点.pdf397EMS mutagenesis.docx398Endnote简易教程-郑夏⽣.ppt399Floral dip transformation of Arabidopsis.pdf400【代码M0203】RACE技术扩增构建全长cDNA⽂库.docx 401【代码M0204】逆转录-聚合酶链式反应(RT-PCR).docx 402GPX测定⽅法.docx403【代码M0205】常规PCR.docx404GUS染⾊及GUS蛋⽩提取及GUS蛋⽩酶活性测定.doc405GUS组织化学染⾊.docx406【代码M0206】原位PCR.docx407Hoagland & Kimura营养液配⽅.docx408Hplc测定内源激素[1].doc409Image-Pro Plus官⽅简体中⽂快速⼊门指南.pdf410IN SITU AND DOUBLE IN SITU HYBRIDIZATION.doc411LIC 克隆中⽂版.docx412LIC-QLC.docx413lignin content detection method.docx414microRNA的stem-loop-Qrt-PCR引物设计.doc415mRNA的分离纯化.docx416【代码M0207】实时定量PCR.docx417【代码M0208】关于PCR的⼏个经验之谈.docx418【代码M0209】PCR扩增产物鉴定与纯化.docx419Phos-tag实验指导⼿册.pdf420Plant Genomic DNA Extraction using CTAB.docx421Poisoned Primer Extension.pdf422【代码M0301】酶切.docx42317. 扫描电镜.docx424Protocol for staining of Arabidopsis leaves with trypan blue and aniline blue.pdf425Protocol -EMSA.docx426Protocol-拟南芥转基因.docx427【代码M0302】质粒DNA的电泳检测.docx428【代码M0303】DNA⽚段回收.docx429【代码M0304】⽬的基因⽚段与载体连接.docx430SDS-PAGE胶观察.doc431AC双⾊原位杂交.doc432SOD、POD、CAT、辅氨酸测定⽅法.docx433Southern blot hybridization.doc434Southern-blotting Protocal正确.doc435AFLP实验流程.doc436staining for pollen viability.pdf437Taqman设计总结005.doc438【代码M0305】质粒DNA的转化.docx439【代码M0306】菌落PCR.docx440⼆代测序信息分析.pdf441亚细胞定位.doc442免疫荧光004.doc443全氮、有机质测定⽅法.doc444农杆菌侵染烟草叶⽚表⽪细胞实验步骤.doc445农杆菌感受态制作.docx446分⼦克隆指南(上)().pdf447分⼦克隆指南(下)().pdf448分⼦植物育种-徐云碧-中⽂版.pdf449半薄切⽚流程.doc450基因枪瞬时转化洋葱表⽪.docx451实验室常⽤分⼦⽣物学实验技术.docx452实验室常⽤培养基的配制⽅法.pdf453AsA测定⽅法.doc454实验报告RRF纯化.xls455【代码M0307】细菌的转化与平板筛选.docx456【代码M0402】蛋⽩电泳.docx457快速提取酵母基因组DNA.docx458⼿动进样HPLC-RID使⽤⽅法002.docx459拟南芥成花⽹络2015.pdf460拟南芥转基因试验流程.pdf461提取DNA和制种聚丙烯酰胺凝胶.docx462【代码M0308】E.coli感受态细胞的制备(CaCl2法).docx463⽊质素测定⽅法.docx464根系测定.doc465【代码M0405】双向电泳.docx466植物Western Blot.docx467植物分⼦遗传研究室实验技术.docx468植物病毒 dot-ELISA 检测⽅法.docx469植物组织⽯蜡切⽚.docx470⽔果果实(淀粉糖含量⾼)RNA提取⾃⼰配药品⽅法(经济性).doc 471⽔稻茎结培养操作流程.doc472泛素化蛋⽩质组.docx473液相⾊谱操作指南003.docx474烟草瞬时转化.docx475⽟⽶原⽣质体转化及原核表达⽅案.docx476⽟⽶叶⽚原⽣质体拟制备转化操作流程.doc477cDNA⽂库构建.doc478电镜切⽚制样流程.doc479直径测量.pdf480短柄草农杆菌转化流程.doc481⽯蜡切⽚制作流程.docx482⽯蜡切⽚染⾊.docx483⽯蜡切⽚的制作.doc484⽯蜡包埋切⽚操作程序.doc485碱煮法快速提取⽟⽶基因组DNA.docx486种⼦发芽流程.docx487简易CTAB法提取植物基因组DNA.docx488组培苗操作流程.doc489【代码M0406】多克隆抗体的免疫电泳技术.docx490草莓⾼效稳定转化.docx491蛋⽩实验相关操作及注意事项.doc492蛋⽩质电泳实验技术-第2版 (1).pdf493DAB和NBT染⾊.doc494超级感受态细胞的制备.docx495转基因技术.ppt496转基因拟南芥.doc497酵母单杂.docx498酵母单杂交系统及其应⽤.doc499酵母双杂交张娟娟.doc500酵母双杂交⽂库筛选流程_mating法.docx501酵母双杂实验操作.WMV502酵母⽂库筛选-Mating篇.doc503酶法催化⽣产母乳脂肪替代物001.doc504酶活测定基本⽅法.doc505醇溶蛋⽩的提取与电泳.doc506限制性内切酶酶切位点汇总.docx507⾼通量测序原理,⽂库构建及质量评价-1108.pdf508鲍鱼酶裂解海带配⼦体细胞壁步骤-卞曙光-中国科学院海洋所.doc509麦⾕蛋⽩亚基的粗提与电泳.doc510(原创)傻⽠软件进⾏简并引物设计.ppt511【代码M0401】Western Blotting.docx51215. 琼脂糖切⽚.docx51316. 叶原基离体培养和活体成像.docx514AFLP原理和操作.docx515AI_及SS活性的测定.doc516DAPI staining of mature pollen nuclei.pdf517Design degenerate primer ——Important.docx518【代码M0407】单细胞凝胶电泳标准操作.docx519H2O2的测定-KI法.doc520His标签蛋⽩纯化试剂盒(可溶性蛋⽩).pdf521【代码M0403】蛋⽩电泳后的凝胶染⾊(考马斯亮蓝、银染、铜染).docx 522mRNA差异显⽰之变性聚丙烯酰胺凝胶电泳及银染.docx523NSC测定⽅法⽐较.docx524P32放射性同位素测定注意事 (2016修改版)(1).docx525Rubisco含量测定.docx526SS,SPS,R酶,Q酶,ADPGase酶活性.docx527【代码M0408】明胶酶谱分析法.docx528第七期⽣物数据分析技能实训会.doc529多靶点pCRISPR载体改良版(单⼦叶植物⽤)使⽤⽅法2014-10-31(P).docx 530发给⼤家后⼩艾⼜修改的---⽔稻转基因实验操作.doc531⽢薯花青素提取与测定⽅法2.doc532DAB染⾊.doc533国内重点实验室分⼦⽣物学实验⽅法汇总实验室常⽤实验⽅法.pdf534GSH 测定⽅法.doc535全国Python科研应⽤专题实操班.doc536实时荧光定量PCR技术.ppt537⽔稻多基因敲除系统.pdf538⽔果抗性相关指标的测定⽅法.docx539⼀种快速鉴定转基因植物纯合体的新⽅法.pdf540软件541【代码M0404】RNA电泳(甲醛变性电泳).docx542【代码M0409】对流免疫电泳.docx543【代码M0410】Southern Blotting实验步骤.docx544【代码M0411】Northern Blotting实验步骤.docx545【代码M0501】细胞膜蛋⽩的分离.docx546【代码M0503】蛋⽩质在原核⽣物中的表达.docx547植物RNA提取⽅法资料548液相测维⽣素E549ddRT-PCR(差⽰反转录PCR).docx550ec值.pdf551GLDH含量测定.pdf552Griess试剂测NO含量.doc553H2O2测定⽅法.doc554vigs侵染⽅法.doc555Western blot(聂).doc556苯丙氨酸解氨酶(PAL)活性的测定.doc557丙酮酸的测定.pdf558酚类和⽊质素.doc559过氧化氢测定⽅法(廖).doc560抗氧化酶和MDA以及蛋⽩测定指南.doc561类胡萝⼘素提取⽅法⽂档.doc562酶联免疫测ABA .doc563台盼蓝染⾊.doc564糖含量的测定1.pdf565糖含量的测定2.pdf566叶黄素提取.doc567叶绿素⽅法.doc568载体构建⽅法.doc569MDA含量测定.doc570POD_测定⽅法.doc571POD活性测定.doc572SOD活性测定.doc573sss测定.doc574标准曲线.xls575不同光质的H2O2和超氧(1).xls576根系活⼒测定⽅法.docx577脯氨酸标准曲线.xls578脯氨酸含量.doc579⽣理实验数据(SOD,POD,DPPH,超氧).xls580植物体内氧⾃由基含量测定.docx581植物体内游离脯氨酸含量的测定.doc582本⽣烟草种植和管理.docx583病毒接毒及其后续管理.docx584植物病毒dot-ELISA检测⽅法.doc585AKTA FPLC 蛋⽩纯化操作.doc586CTAB法提取RNA.doc587CTAB提取植物DNA.docx588DNA的琼脂糖凝胶电泳.doc589Greiss试剂测定NO的⽅法.doc590变性梯度凝胶电泳.doc591实验⽅法整理 .docx592线粒体测定最终版.docx593叶绿体提取.doc594【代码M0502】细菌总蛋⽩和膜蛋⽩提取⽅法.docx 595【代码M0504】免疫复合物的纯化.docx596【代码M0602】动物组织块DNA的提取.docx597半薄切⽚技术.ppt598⽯蜡切⽚流程.doc599透射电镜切⽚的步骤.doc600荧光参数.doc601植物RNA的提取及其电泳鉴定.doc6021-1 ⽯蜡切⽚的制作.pdf603N-P-K联合消煮测定.doc604SOD、POD、CAT、MDA和可溶性蛋⽩测定.doc 605沉降值测定⽅法.doc606FISH procedure-2.doc607PCR-SSCP实验操作详细步骤.doc608Promega公司的TA载体克隆与转化.doc609shotgun_⽂库的构建.doc610southern 杂交实验流程.doc611毕业⽣离室前注意事项.doc612基因定点突变.doc613基于⽑细管电泳的荧光检测SSR实验程序.doc614拟南芥培养转化的流程[1].doc615庞斌双的蛋⽩电泳和连接转化程序.doc616实时荧光定量PCR技术.doc617⼩麦成熟胚标准技术规程.doc618⼩麦基因组DNA的提取.doc619⼩麦⽥间记载和室内考种项⽬与标准.doc620怎样记录实验结果.doc621重点实验室protocol-CWF全长cDNA⽂库构建流程.doc622重点实验室protocol-总RNA提取(Trizol法)和mRNA提取[Oligo(dT)cellulose]流程zgy.doc 623ABI3730XL测序实验流程-2007-3-15.doc624BAC⽂库构建与筛选实验程序-2007-3-10.doc625PCR产物的TA克隆(DNA的胶回收和连接).docx626基因X 中⽂版.pdf627植物中基因编辑.docx628【代码M0604】CTAB法提取植物总DNA.docx629【代码M0605】磁珠法提纯mRNA.docx630【代码M0606】连续梯度法纯化闭环DNA.docx631【代码M0607】不连续梯度法纯化闭环DNA.docx632【代码M0608】从外周⾎淋巴细胞中提取RNA.docx633【代码M0609】饱和氯化钠法提取外周⾎DNA.docx634【代码M0701】荧光原位杂交(FISH)探针的制备.docx635【代码M0801】Conditional gene knockout in C. elegans- CRISPR-Cas9.docx636【代码M0802】Target Sequence Cloning Protocol- CRISPR-Cas9.docx637【代码M0803】SAM target sgRNA cloning protocol- CRISPR-Cas9.docx638【代码M0901】酵母双杂交筛选简介和流程.docx639【代码M0902】构建于 AD 载体的 cDNA ⽂库的扩增.docx640【代码M0903】构建于 AD 载体的 cDNA ⽂库的纯化.docx641【代码M0904】pGBKT7bait ⼩规模转化酵母菌 AH109.docx642【代码M0905】钓饵蛋⽩⾃⾝激活报道基因的活性检测.docx643【代码M0906】⽂库质粒⼤规模转化已携带钓饵质粒的酵母.docx644【代码M0907】转化⼦的筛选与库质粒纯化.docx645【代码M0908】酵母质粒 DNA 的提取.docx646【代码M0909】蛋⽩相互作⽤的验证—pGBKT7bait 与 pACT2 ⼩规模共转化酵母.docx 647【代码M1001】分⼦⽣物学实验常⽤培养基的配制⽅法.docx648【代码M0601】⼩量制备质粒DNA(强碱法).docx649【代码M0603】动物组织RNA提取及纯化.docx650Southern Blot原理及实验⽅法.doc651SSR分⼦标记实验操作及其注意事项.doc652单核苷酸多态性(SNP)实验.docx653动物基因组DNA的提取.doc654甲基化检测原理及步骤.doc655降落PCR.docx656聚合酶链式反应(PCR)基本操作步骤.doc657菌落PCR.doc658⽬的基因PCR扩增产物的克隆.doc659逆转录-聚合酶链反应(RT-PCR).doc660凝胶迁移实验(EMSA).docx661普通PCR、原位PCR、反向PCR和反转录PCR的基本原理和操作步骤.doc 662染⾊质免疫共沉淀(ChIP)实验.docx663实时荧光定量PCR具体实验步骤.doc664限制性⽚段长度多态性实验(RFLP).doc665原位PCR技术基本原理.doc666质粒DNA的提取.doc667重叠延伸PCR实验.docx668重组质粒的连接、转化及筛选.doc669(原创)实时定量神器Allele ID 5破解教程.doc670分⼦⽣物学⼤实验2-L.pptx671分⼦⽣物学实验II新教案.pdf672分⼦⽣物学实验673实验技术视频674现科课件675研究⽣课件676研究⽣分⼦⽣物学实验I讲义-2017.doc677⼤肠杆菌感受态细胞的制备和转化.doc678pGEMT.pdf679TaKaRa_LA_Taq说明书.pdf680TAKARA双酶切表.doc681分⼦克隆技术实验操作⼿册.pdf682宝⽣物去磷酸化说明书.pdf683构建载体知识查询.docx684载体图谱685载体图谱序列686上海硕盟_TOYOBO说明书.pdf687分⼦克隆实验指南第三版上册.pdf688分⼦克隆实验指南第三版下册.pdf6891 PCR实验技术6902 DNA实验技术6913 RNA实验技术6924 蛋⽩质实验技术6935 补充694DH5α感受态制备⽅法.pdf695EMSA696Easy Yeast plasmid isolation kit user mannual.pdf697GM配⽅.pdf698GST pull down.pdf699word700Immobilized Glutathione.pdf701MagneGST Pull-Down System.pdf702Ni-NTA handbook.pdf703PCR704PHARMACIA_GST_Gene_Fusion_System_Handbook.pdf705Pull-Down GST kit.pdf706RNAi707RNA提取⽅法——TRIZOL.pdf708SDS_PAGE胶配⽅表.pdf709Y2H Membrane Protein Kit.pdf710activation tagging.pdf711chip EZ-ChIP_manual.pdf712chromas.rar713his tag pull down assay .pdf714iTILLING.pdf715pfu.pdf716promega_GloMax2020完整版.pdf717western breeze chromo_man.pdf718western_blot技术集锦.rar719免疫共沉淀与Western Blot.pdf720原位杂交⽅法721国内重点实验室分⼦⽣物学实验⽅法汇总实验室常⽤实验⽅法.pdf 722常⽤缓冲液的配制.pdf723普通表达载体724植物细胞组织学⾼⽼师课件725荧光定量PCR.pdf726荧光激发及吸收波长.PDF727酵母双杂交728酵母双杂交技术研究与应⽤进展_李先昆.pdf729AxyPrep_DNA_Gel_DYE_Ext_Kit_ver20090727.pdf730His Spin Trap.pdf731In-Fusion Kit User manual.pdf732P020091009585923799380.ppt733PPICZalpha.pdf734[]基因克隆研究⽅法综述.pdf735matchmaker gold yeast one-hybrid library screening system.pdf736pET28_map.pdf737ppic9k_man.pdf738分⼦克隆实验指南上册v3.pdf739分⼦克隆实验指南下册v3.pdf740拟南芥T-DNA插⼊突变纯合体的鉴定.ppt741EMSA.pdf742淀粉检测说明书.pdf743RT-PCR744DNA kit.pdf745DP441 RNAprep pure多糖多酚植物总RNA提取试剂盒.pdf746Gateway_载体构建.ppt747RNAprep Pure 多糖多酚植物总RNA提取试剂盒--天根.pdf748系统发育分析课件2017完整版.pdf749Real-time PCR750office2013完美激活.rar751[3]实时荧光定量PCR技术_天根.ppt752[4]国内重点实验室分⼦⽣物学实验⽅法汇总实验室常⽤实验⽅法.PDF 753[7]PCR技术讲座.pdf754[8]国内重点实验室分⼦⽣物学实验⽅法汇总实验室常⽤实验⽅法(1).pdf755[11]荧光定量PCR技术讲座:理论基础、引物及探针设计、体系优化、实验⽅案、数据分析、污染防控 2.ppt756R语⾔绘图-实操训练.pptx757R语⾔绘图⼊门.pptx758基因功能分析.pptx759基因聚类分析和样品相关性分析.pptx 760基因表达定量.pptx761差异表达分析.pptx762数据质控.pptx763⽆参转录组序列组装及实际操作.pptx764测序数据⽐对.pptx765测序数据统计.pptx766转录组数据库注释.pptx767转录组⽣物信息分析整体介绍.pptx下⾯是软件软件1最新版SCI-Endnote⽂献样式.zip2circos-0.69-5.tgz3hmmer-3.1b2-cygwin32.tar.gz4Adobe_Photoshop_CC.7z5BeoPlayer ⾳质最好的播放器.7z6EndNote X8.7z7genespring.7z8PRIMER 6破解版.7z9[]DPS数据处理系统.rar10[]primer5x64bit.rar11[]DNAMAN8_Registered.rar12[专业图像分析软件].ipp6.0.rar1364位系统所需要的primer5程序.rar14Adobe Acrobat XI Pro 11.0.1.rar15Adobe Photoshop CS6.rar16BeaconDesignerpjb.rar17blast.rar18CE Design V1.04.rar19chromas (2).rar20Chromas.rar21clustal.rar22clustalx.rar23Design-Expert.8.05b完全破解版.rar24DNAMAN.rar25DNAMAN7setup.rar26DNAMAN8_20150421.rar27DNAMAN8_Registered.rar28DNASTAR Lasergene v7.1.0.rarsergene.v7.1.rar30DNASTAR6.0.rar31dps7.05.rar32EditPlus_.rar33EndNote X7 批量授权中科⼤版.rar34EndNote X8.rar35EndnoteX8(含汉化⽂件).rar36EndNote教程.rar37FigTree v1.4.2.rar38GWAS常⽤软件及⼿册.rar39jionmap4.0注册版.rar40LI-6400XT OPEN6.2 中⽂操作⼿册(2014年2⽉校).rar41MapChart 2.2.rar42MapDraw.rar43MapMaker 3.0.rar44MapQTL6.03.187破解版 best.rar45MapQTL6补丁.rar46MEGA6 (2).rar47MEGA6.rar48Mindmanager15.rar49NoteExpress完美破解版.rar50Oligo.v7.53.rar51origin8.0破解版附安装图⽂教程.rar52OriginLab 8.0 绿⾊破解版.rar53PHOTOSHOP_CS_8.01_简体中⽂版.rar54POPGENE32.rar55PP600Installer.rar56Primer5.0 64wei.rar57Primer5.064.rar58Protocol.rar59Protocols.rar60python批量下载指定序列__.rar61RIP.rar62SAS Software Depot.rar63SAS.rar64ScienceSlides.rar65sigmaplot 10.0 破解版.rar66Simvector 4.5.rar67SnapGene2.3.2.rar68SnapGene各版本破解版-淼灵⽣物.rar69SPSS.rar70TBtools-crossplatform.rar71TeamViewer.rar72Trados翻译软件2014特别版(密码).rar 73treeview.rar74Uedit32.rar75Vector NTI 11.5.1 破解版.rar76Vector NTI 11.5.3破解版.rar77迅龙数据恢复.rar78[GraphPad.Prism.v5.0.注册版].prism.zip79Adobe Illustrator CS5.zip80Adobe Illustrator CS6.zip81AdobeIllustrator21_HD_win64安装包.zip82AMOS21.0.zip83BEAST.v2.3.0.Windows.zip84BioEdit (2).zip85bioedit.zip86BioMercator2-1.zip87BioXM2.6.zip88BLAST2GO - 复件.zip89BLAST2GO.zip90ChemOffice已破解解压即⽤.zip91cluster.zip92CRISPR PROTOCOL For Plant.zip93Diverge3.0B1(家族分歧软件).zip94DNAMAN.zip95DNAMAN_setup.zip96DNAMAN9_registered.zip97Dual_genomic_comparison.zip98EditPlus_3.3.0.715_XiaZaiBa.exe.zip99EMS-Mutagenesis-of-Arabidopsis.zip100Endnote X7 Windows.zip101EndNoteX7.zip102FV10-ASW_Viewer3_1.zip103Gateway Cloning Vectors.zip104GPS-SNO.zip105GraphPad Prism (2).zip106GraphPad Prism 5.zip107GraphPad Prism.zip108GraphPad.Prism.v5.01.zip109GraphPad Prism7.zip110GraphPadPrism.zip111GST_pull_down.zip112hmmer3.0_windows.zip113ICIMSetup V4.1.zip114IGV详细使⽤.zip115jionmap4.0.zip116MEGA6.zip117MrBayes-3.2.6_WIN32_x64.zip118Origin 9 32位专业破解版.zip119phylip-3.695.zip120Plant Vectors.zip121PRIMER 5.0含序列号.zip122Primer premier 5 - 32bit.zip123Primer Premier 5.zip124Primer Premier 5q.zip125Protocols.zip126python⼊门及数据分析.zip127SCI论⽂写作语⾔润⾊修改WhiteSmoke(破解).zip 128SigmaPlot_V12.5.zip129SIMCA 13.0 (破解版).zip130SIMCA 14.1 32-bit.zip131simca.14.1.crack.zip132SIMCA 13.0.zip133SnapGene 2(1).zip134snapgene 3.2.1 mac 版本.zip135SnapGene.zip136SPDBV_4.10_PC.zip137spss23 64.zip138TreeView_1.1.6r2_win.zip139UltraEdit.zip140VectorNTIAdvance11.5.1破解.zip141VIGS.zip142显微镜图像处理软件 - 复件.zip143MAGA6.0144SnapGene145EndNoteX6_16.0.0.6348_PortableSoft.zip146MEGA7.0.21_win64_setup.exe147SnapGene-.zip148SnapGene_pc().rar149Vector NTI 11.5.1 破解版.zip150Vector_NTI_简体中⽂使⽤教程PDF版本.pdf151DNAMAN汉化版.rar152software-win153⽣物信息分析软件与TCGA相关⽂献(2)154⽣物_PilotEdit_5.3.0.rar155⼀些软件 - 复件(1).rar156引物设计.rar157必应词典.zip158补充的软件和试验⽅法.zip159蛋⽩保守结构域分析与分⼦进化树构建.zip160蛋⽩三维结构处理raswin.zip161基因蛋⽩结构图绘制IBS_1.0_windows_20141228.zip 162实验protocol.zip163⽣物软件.zip164实验⼯具软件.zip165新建⽂件夹 (2).zip166英⽂⽂章语法⾃动纠错.exe167endnote x8破解.rar.xltd168ICIMSetup V4.1.exe169ij146-jdk6-64bit-setup.exe170Illustrator CS5_Lite.exe171MEGA5.05_Setup-系统树构建.exe172npp_6.9.2_Installer.exe173Pathway Builder-通路绘图.exe174Photoshop setup.exe175Photoshop_CS3.exe176R-3.4.1-win.exe177RStudio-1.0.143.exe178SETUNA.exe179iSlideTools.Setup.Online.2.4.1.exe180HemI_windows_1_0_win32_x64.exe181HemI_windows_1_0_win32_x64 (2).exe182ApE.exe183MeV_4_9_0_r2731_win(1).rar184GraphPad Prism 7 教程及软件185信号通路作图软件186winmd5free---检测数据完整性.zip187全基因组扫描-转录因⼦结合位点-⼯具.pdf188⽤origin软件绘制折线图和误差线(By 蜗⽜菌).pdf 189多序列⽐对软件clustalx-2.1-win.msi190dnasp51001.msi191Sigmaplot.rar192SPSS22.0.rar。
毕赤酵母翻译

毕⾚酵母翻译Glycoengineered(糖基化⼯程) 毕⾚ pastoris(毕⾚酵母) 中表达的重组单克隆抗体的纯化⼯艺开发摘要⼀个强健的和可扩展的净化过程被开发快速从 glycoengineered 毕⾚酵母发酵⽣成抗体的纯度⾼、数量充⾜。
蛋⽩ A 亲和层析⽤于捕获从发酵液抗体。
PH 梯度洗脱已应⽤到要防⽌抗体沉淀在低 pH 的蛋⽩ A 列。
从蛋⽩抗体⾊谱法所载⼀些产品的相关杂质,被劈重链、重链和轻链的misassembling。
它也有⼀些处理相关的杂质,包括蛋⽩ A 残留、 endotoxin(内毒素)、宿主细胞 DNA 和蛋⽩质。
Ph 值为 4.5-6.0 的最优氯化钠渐变阳离⼦交换⾊谱法有效去除这些产品和过程相关的杂质。
从 glycoengineered P.酵母抗体是其 heterotetramer 折叠、物理稳定性和约束⼒的亲和性的商业同⾏相媲美。
介绍单克隆抗体(Mab)正迅速成为关键产品的⽣物医药产业。
⼤多数商业治疗性抗体是原封 IgG 分⼦与 IgG1、 IgG2 正在共同的⼦类。
这些 IgGs 由调解抗原和效应器函数之间的联系,在免疫过程中发挥中⼼作⽤。
对⽬标细胞的特异性抗原的 igg 抗体可变域的绑定将抗体依赖的细胞毒作⽤ (ADCC) 及补体依赖性细胞毒性 (CDC) 定向杀害的⽬标单元格。
ADCC 触发由交联体 Fc 域的 IgG 抗体(Fc(Rs),尤其是 Fc (RI 和Fc (制证机对免疫效应细胞。
可能调解 ADCC 效应细胞包括⾃然杀⼿细胞、巨噬细胞和嗜中性粒细胞。
疾病预防控制中⼼是由补充组件 C1q 绑定到 IgG,触发蛋⽩⽔解的级联,以激活补充 fc 功能区启动。
IgG 型抗体有两重链和两条轻链由分⼦内的⼆硫键形成 heterotetramer 在⼀起。
重链是通过共价键固定在天冬酰胺 297 (Asn-297) 低聚糖的糖基化。
⼈免疫球蛋⽩的主要N-聚糖复杂 biantennary (⼆分⽀的) 类型,有 '核⼼' heptasaccharide(七庚糖),GlcNac2Man3GlcNac2,被称为 G0 在我们的⼯作和⽂学。
毕赤酵母表达(pichia pastoris expression )实验手册(3)

毕赤酵母表达(pichia pastoris expression )实验手册(3)液体YPD培养基可常温保存;琼脂YPD平板在4℃可保存几个月。
加入Ze ocin 100ug / ml,成为YPDZ培养基,可以4℃条件下保存1~2周。
2.4 YPDS + Zeocin 培养基(Yeast Extract Peptone Dextrose Medi um):yeast extract 1%peptone 2%dextrose (glucose) 2%sorbitol 1 M+agar 2%+ Zeocin 100 μg/ml不管是液体 YPDS培养基,还是YPDS + Zeocin 培养基,都必须存放4℃条件下,有效期1~2周。
2.5 MGYMinimal Glycerol Medium (最小甘油培养基)(34%YNB;1%甘油;4*10-5%生物素)。
将800ml灭菌水、100ml的 10* YNB母液、2ml的500*B母液和100ml的10*GY母液混匀即可,4℃保存,保存期为2个月。
2.6 MGYHMinimal Glycerol Medium + Histidine (最小甘油培养基 + 0.004%组氨酸)在1000ml的MGY培养基中加入 10ml的100*H母液混匀,4℃保存,保存期为2个月。
2.7 RDRegeneration Dextrose Medium (葡萄糖再生培养基)(含有:1mol/L的山梨醇;2%葡萄糖;1.34%YNB;4*10-5%生物素;0. 005%氨基酸)1. 将186g的山梨醇定容至700ml,高压灭菌;2. 冷却后于45℃水浴;3. 将100ml的10*D、100ml的10*YNB;2ml的500*B;10ml的100*AA等母液和88ml无菌水混匀,预热至45℃后,与步骤2 的山梨醇溶液混合。
4℃保存。
2.8 RDHRegeneration Dextrose Medium + Histidine (葡萄糖再生培养基 + 0.004%组氨酸)在RD培养基配制的第三步中,在加入10ml的100*H母液,同时无菌水的体积减少至78ml即可,其余配制方法与RD相同。
毕赤酵母表达操作手册(精译版)

毕赤酵母多拷贝表达载体试剂盒用于在含多拷贝基因的毕赤酵母菌中表达并分离重组蛋白综述:基本特征:作为真核生物,毕赤酵母具有高等真核表达系统的许多优点:如蛋白加工、折叠、翻译后修饰等。
不仅如此,操作时与E.coli及酿酒酵母同样简单。
它比杆状病毒或哺乳动物组织培养等其它真核表达系统更快捷、简单、廉价,且表达水平更高。
同为酵母,毕赤酵母具有与酿酒酵母相似的分子及遗传操作优点,且它的外源蛋白表达水平是后者的十倍以至百倍。
这些使得毕赤酵母成为非常有用的蛋白表达系统。
与酿酒酵母相似技术:许多技术可以通用:互补转化基因置换基因破坏另外,在酿酒酵母中应用的术语也可用于毕赤酵母。
例如:HIS4基因都编码组氨酸脱氢酶;两者中基因产物有交叉互补;酿酒酵母中的一些野生型基因与毕赤酵母中的突变基因相互补,如HIS4、LEU2、ARG4、TR11、URA3等基因在毕赤酵母中都有各自相互补的突变基因。
毕赤酵母是甲醇营养型酵母:毕赤酵母是甲醇营养型酵母,可利用甲醇作为其唯一碳源。
甲醇代谢的第一步是:醇氧化酶利用氧分子将甲醇氧化为甲醛,还有过氧化氢。
为避免过氧化氢的毒性,甲醛代谢主要在一个特殊的细胞器-过氧化物酶体-里进行,使得有毒的副产物远离细胞其余组分。
由于醇氧化酶与O2的结合率较低,因而毕赤酵母代偿性地产生大量的酶。
而调控产生醇过氧化物酶的启动子也正是驱动外源基因在毕赤酵母中表达的启动子。
两种醇氧化酶蛋白:毕赤酵母中有两个基因编码醇氧化酶-AOX1及AOX2。
细胞中大多数的醇氧化酶是AOX1基因产物。
甲醇可紧密调节、诱导AOX1基因的高水平表达,较典型的是占可溶性蛋白的30%以上。
AOX1基因已被分离,含AOX1启动子的质粒可用来促进编码外源蛋白的目的基因的表达。
AOX2基因与AOX1基因有97%的同源性,但在甲醇中带AOX2基因的菌株比带AOX1基因菌株慢得多,通过这种甲醇利用缓慢表型可分离Muts菌株。
表达:AOX1基因的表达在转录水平受调控。
酵母表达系统步骤

酵母表达系统步骤毕赤酵母表达系统步骤(参考Invitrogen公司说明书):一、pPICZαA、B、C质粒以及DH5α菌株的保存1取0.5μl pPICZα A、B、C质粒,热击转化DH5α,在低盐LB (含有25μg/ml Zeocin)的平板上37℃培养过夜。
2挑取转化子,甘油保存。
二、载体构建1将目的基因构建到pPICZα载体上,转化DH5α,用Zeocin筛选转化子。
2提质粒酶切鉴定或PCR鉴定3载体测序测序可用α-Factor引物或5’AOX1引物,3’AOX1引物三、线性化DNA1提取足够量的质粒DNA(一次转化至少需要5-10μg质粒)2 酶切线性化10μg构建好的载体,同时酶切空载体做对照,根据载体选择线性化酶切位点(样品分管酶切),pPICZα载体在5’AOX1区域有三个酶切位点可选择:SacI、PmeI、BstXI3 取1-2μl酶切产物跑电泳,确定是否酶切完全;4 过柱纯化线性化质粒(用50μl EB洗脱);四、线性化DNA的去磷酸化处理线性化质粒43μlCIAP Buffer 5μlCIAP酶2μl四、总体积为50μl的样品37℃ 1h,过柱纯化,用30μl ddH2O 洗脱;五、感受态细胞的制备实验前准备:无抗性YPD平板一个、无抗生素液体YPD培养基,100μg/ml Zeocin YPD 平板和液体、50ml离心管两个、500ml预冷的无菌水、20ml 1M 山梨醇(灭菌预冷的),0.2cm预冷的电击杯;1YPD平板划线培养菌,30℃培养2-3d;250ml三角瓶中,加入5ml YPD,挑取酵母单菌落,30℃培养过夜;3吸取0.5ml菌液,加入至含有200ml新鲜YPD的1L三角瓶中,30℃,225rpm/min培养至OD值1.3-1.5;41500g,4℃离心5min收集菌体;540ml冰预冷的无菌水重悬沉淀;61500g,4℃,5min;730ml无菌水重悬;81500g,4℃,5min;910ml 1M 山梨醇重悬;101500g,4℃,5min;11加入1ml山梨醇,重悬冰上放置,直接做转化,或加入灭菌甘油每管80ul分装,冻存于-80℃(长时间保存会影响转化效率);六、电击转化15-10μg线性化DNA(20μl<)与80ul上述感受态细胞混合,转移至预冷的0.2cm电击杯中(点击条件:电压1.5kV;电容25μF;电阻200Ω,电击时间为4~10msec);2冰上放置5min3电击(按生产厂商提供的适合酵母用的参数)4迅速加入1ml预冷的1M 山梨醇,转移至1.5ml EP管中530℃静置培养1-2h(如果要增加存活率,获得更多的转化克隆,可在30℃静置培养1h后,加入1mlYPD培养基,30℃200rpm培养1h后取部分涂布与不同浓度抗生素的平板)6取50、100、200ul分别涂布于含有Zeocin的YPD平板,30℃培养2-10 d至有菌落出现;7如果要筛选多拷贝转化子,将转化克隆混合在一起,涂布在Zeocin 浓度为500、1000、2000μg/ml的YPD平板,培养2-3d。
毕赤酵母表达步骤

毕赤酵母表达系统的构建1.确定转入的目的基因,并设计相应引物,进行PCR反应,获取目的基因片段。
(所需药品-高保真酶,引物,dntp,胶回收试剂盒。
)2.对目的基因及载体Ppic9k进行酶切产生粘性接头并纯化回收。
(所需药品- SalI、StuI、SacI 【用于于GS115产生His+Mut+】;BglII【用于于GS115产生His+Muts】)酶切体系酶切体系50 μL目的DNA 10 μL酶切缓冲液 5 μL限制性内切酶 1 μL超纯水34μL37 ℃酶切 1~4小时。
酶切产物进行琼脂糖凝胶电泳检测分析。
3.将酶切正确的目的片度与质粒相连(所需药品-连接试剂盒SolutionⅠ[宝生物])载体DNA0.5µL目的片段DNA 4.5µL连接试剂盒SolutionⅠ 5 µL充分混匀,置于16 ℃连接4 h或4 ℃连接过夜。
4.转入DH5α扩繁质粒(所需药品- A mp;DH5α;CaCl2)将DNA目的片段和载体的连接产物与200µL感受态细胞混匀,冰浴30min。
45℃热击45-60s,冰浴3min后加入800μL LB 液体培养基37℃震荡培养1h。
(1 )转化后,将转化混合物涂在含50-100ug/ul A mp 的LB平板上,选择A mp 抗性克隆(2)挑取10 个A mp 抗性转化子,接种含150ug/ul A mp 的培养基,37 度振荡培养过夜(3 )提取质粒进行PCR及酶切检测并送交测序。
(pPIC9k 测序时,用α-factor 引物及3’AOX1 测序引物。
将引物重悬于20ul灭菌水中,制成0.1ug/ul 溶液)4 在0.85ml 过夜培养菌液中加入0.15ml 灭菌甘油,以便保存所需克隆,涡旋混匀转入标记好的储存管中。
在液氮或干冰/酒精浴中冷冻后移入-70 度保存。
5 测序证实结构正确后,可准备转化DNA5.毕赤酵母电转化:细胞准备:毕赤酵母感受态制备:(1)取1 mL GS115过夜培养物(OD6006.0~10.0)转接于100 mL YPD液体培养基中28℃-3O℃、250—300 r/min培养至酵母菌的对数生长期(OD600 1.0~1.3)(2)取此菌1 mL分装到1.5 mL EP管中,4℃、10 000 g离心1 min,弃上清液,沉淀用无菌水(4℃预冷)洗涤,同样条件下离心,弃上清液。
毕赤酵母表达手册(详细)

毕赤酵母表达(pichia pastoris expression )实验手册2010-07-15 10:54:56| 分类:毕赤酵母| 标签:|字号大中小订阅一.毕赤酵母表达常用溶液及缓冲液的配制二.毕赤酵母表达的培养基配制三.主要试验环节的操作 3.1 酵母菌株的分离纯化 3.2 pPICZαA原核宿主菌TOP10F’的活化培养 3.3毕赤酵母表达的试验方法 3.4 毕赤酵母电转化方法 3.5 Pichia酵母表达直接PCR鉴定重组子的方法 3.6 毕赤酵母基因组提取方法 3.7 Mut+表型重组酵母的诱导表达实验关键词:酵母实验毕赤酵母表达 pichia pastoris expression 毕赤酵母酵母菌株大肠杆菌表达系统最突出的优点是工艺简单、产量高、周期短、生产成本低。
然而,许多蛋白质在翻译后,需经过翻译后的修饰加工,如磷酸化、糖基化、酰胺化及蛋白酶水解等过程才能转化成活性形式。
大肠杆菌缺少上述加工机制,不适合用于表达结构复杂的蛋白质。
另外,蛋白质的活性还依赖于形成正确的二硫键并折叠成高级结构,在大肠杆菌中表达的蛋白质往往不能进行正确的折叠,是以包含体状态存在。
包含体的形成虽然简化了产物的纯化,但不利于产物的活性,为了得到有活性的蛋白,就需要进行变性溶解及复性等操作,这一过程比较繁琐,同时增加了成本。
大肠杆菌是用得最多、研究最成熟的基因工程表达系统,当前已商业化的基因工程产品大多是通过大肠杆菌表达的,其主要优点是成本低、产量高、易于操作。
但大肠杆菌是原核生物,不具有真核生物的基因表达调控机制和蛋白质的加工修饰能力,其产物往住形成没有活性的包涵体,需要经过变性、复性等处理,才能应用。
近年来,以酵母作为工程菌表达外源蛋白日益引起重视,原因是与大肠杆菌相比,酵母是低等真核生物,除了具有细胞生长快,易于培养,遗传操作简单等原核生物的特点外,又具有真核生物时表达的蛋白质进行正确加工,修饰,合理的空间折叠等功能,非常有利于真核基因的表达,能有效克服大肠杆菌系统缺乏蛋白翻译后加工、修饰的不足。
毕赤酵母表达载体pPICzalpha手册

Recommended Pichia Host Strain
We recommend using the X-33 Pichia strain as the host for expression of recombinant proteins from pPICZα . Other Pichia strains are suitable. The X-33 Pichia strain is available from Invitrogen (see page vii for ordering information) and has the following genotype and phenotype: Genotype: Wild-type Phenotype: Mut+ Continued on next page
Shipping/Storage Reference Sources
The vectors are shipped on wet ice and should be stored at –20°C. The pPICZα A, B, and C vectors may be used with the Original Pichia Expression Kit (Cat. no. K1710-01) and are included in the EasySelect™ Pichia Expression Kit (Cat. no. K1740-01) available from Invitrogen. Additional general information about recombinant protein expression in Pichia pastoris is provided in the manuals for the Original Pichia Expression Kit and the EasySelect™ Pichia Expression Kit. The manuals can be downloaded from our Website () or obtained by calling Technical Support (see page 33). For more information about the Original Pichia Expression Kit or the EasySelect™ Pichia Expression Kit, refer to our Website or contact Technical Support. More detailed information and protocols dealing with Pichia pastoris may also be found in the following general reference (see page vii for ordering information): Higgins, D. R., and Cregg, J. M. (1998) Pichia Protocols. In Methods in Molecular Biology, Vol. 103. (J. M. Walker, ed. Humana Press, Totowaer Manual
毕赤酵母表达手册

Pichia Expression KitVersion M01110225-0043Pichia Expression KitA Manual of Methods for Expression of Recombinant Proteins in Pichia pastorisCatalog no. K1710-01tech_service@iiINDIVIDUAL PICHIA EXPRESSION KIT LICENSE AGREEMENTThe Pichia Expression Kit is based on the yeast Pichia pastoris. Pichia pastoris was developed into an expression system by scientists at Salk Institute Biotechnology/Industry Associates (SIBIA) for high-level expression of recombinant proteins. All patents for Pichia pastoris and licenses for its use as an expression system are owned by Research Corporation Technologies, Inc. Tucson, Arizona. Invitrogen has an exclusive license to sell the Pichia Expression Kit to scientists for research purposes only, under the terms described below. Use of Pichia pastoris by commercial corporations requires the user to obtain a commercial license as detailed below. Before using the Pichia Expression Kit, please read the following license a greement. If you do not agree to be bound by its terms, contact Invitrogen within 10 days for authorization to return the unused Pichia Expression Kit and to receive a full credit. If you do agree to the terms of this Agreement, please complete the User Registration Card and return it to Invitrogen before using the kit.INDIVIDUAL PICHIA EXPRESSION KIT LICENSE AGREEMENTInvitrogen Corporation (INVITROGEN) grants you a non-exclusive license to use the enclosed Pichia Expression Kit (EXPRESSION KIT) for academic research or for evaluation purposes only. The EXPRESSION KIT is being transferred to you in furtherance of, and reliance on, such license. You may not use the EXPRESSION KIT, or the materials contained therein, for any commercial purpose without a license for such purpose from RESEARCH CORPORATION TECHNOLOGIES, INC., Tucson, Arizona. Commercial purposes include the use in or sale of expressed proteins as a commercial product, or use to facilitate or advance research or development of a commercial product. Commercial entities may conduct their evaluation for one year at which time this license automatically terminates. Commercial entities will be contacted by Research Corporation Technologies during the evaluation period regarding the purchase of a commercial license.Access to the EXPRESSION KIT must be limited solely to those officers, employees and students of your institution who need access thereto in order to perform the above-described research or evaluation. You must inform each of such officer, employee and student of the provisions of this Agreement and require them to agree, in writing, to be bound by the provisions of this Agreement. You may not distribute the EXPRESSION KIT to others, even those within your own institution. You may transfer modified, altered or original material from the EXPRESSION KIT to a third party following notification of INVITROGEN such that the recipient can be licensed. You may not assign, sub-license, rent lease or otherwise transfer this License or any of the rights or obligation hereunder, except as expressly permitted.This License is effective until terminated. You may terminate it at any time by destroying all Pichia expression products in your control. It will also terminate automatically if you fail to comply with the terms and conditions of the Agreement. You shall, upon termination of the License, destroy all Pichia Expression Kits in your control, and so notify INVITROGEN in writing.This License Shall be governed in its interpretation and enforcement by the laws of the State of California.Product User Registration CardPlease complete and return the enclosed Product User Registration Card for each Pichia Expression Kit that you purchase. This will serve as a record of your purchase and registration and will allow Invitrogen to provide you with technical support and manual updates. It will also allow Invitrogen to update you on future developments of and improvements to the Pichia Expression Kit. The agreement outlined above becomes effective upon our receipt of your User Registration Card or 10 days following the sale of the Pichia Expression Kit to you. Use of the kit at any time results in immediate obligation to the terms and conditions stated in this Agreement.Technical ServicesInvitrogen provides Technical Services to all of our registered Pichia Expression Kit users. Please contact us if you need assistance with the Pichia Expression Kit.United States Headquarters:Japanese Headquarters European Headquarters:Invitrogen Corporation1600 Faraday AvenueCarlsbad, CA 92008 USATel: 1 760 603 7200Tel (Toll Free): 1 800 955 6288 Fax: 1 760 602 6500E-mail:tech_service@ Invitrogen Japan K.K.Nihonbashi Hama-Cho Park Bldg. 4F2-35-4, Hama-Cho, NihonbashiTel: 81 3 3663 7972Fax: 81 3 3663 8242E-mail: jpinfo@Invitrogen Ltd3 Fountain DriveInchinnan Business ParkPaisley PA4 9RF, UKTel (Free Phone Orders): 0800 269 210Tel (General Enquiries): 0800 5345 5345Fax: +44 (0) 141 814 6287E-mail: eurotech@iiiivTable of ContentsMaterials (vii)Purchaser Notification (x)Product Qualification (xii)Introduction (1)Overview (1)Experimental Outline (3)Recombination and Integration in Pichia (7)Methods (11)Pichia Strains (11)E. coli Strains (13)Selecting a Pichia Expression Vector (14)pHIL-D2 (16)pPIC3.5 (17)pHIL-S1 (18)pPIC9 (19)Signal Sequence Processing (20)Cloning into the Pichia Expression Vectors (21)Transformation into E. coli (26)Preparation of Transforming DNA (27)Growth of Pichia for Spheroplasting (30)Preparation of Spheroplasts (32)Transformation of Pichia (34)Screening for Mut+ and Mut S Transformants (36)PCR Analysis of Pichia Integrants (40)Expression of Recombinant Pichia Strains (42)Analysis by SDS-Polyacrylamide Gel Electrophoresis (45)Optimization of Pichia Protein Expression (47)Scale-up of Expression (49)Protein Purification and Glycosylation (51)Recipes (53)E. coli Media Recipes (53)Pichia Media Recipes (54)Appendix (59)Electroporation of Pichia (59)PEG 1000 Transformation Method for Pichia (60)Lithium Chloride Transformation Method (61)Total DNA Isolation from Pichia (62)Detection of Multiple Integration Events (63)Procedure for Total RNA Isolation from Pichia (64)β-Galactosidase Assay (65)Technical Service (67)References (69)vviMaterialsKit Contents Box 1: Spheroplast Module. Store at room temperature.Reagent Amount ComponentsSOS media 20 ml 1 M Sorbitol0.3X YPD10 mM CaCl2Sterile Water 2 x 125 ml Autoclaved, deionized waterSE 2 x 125 ml 1 M Sorbitol25 mM EDTA, pH 8.0SCE 2 x 125 ml 1 M Sorbitol10 mM Sodium citrate buffer, pH 5.81 mM EDTA1 M Sorbitol2 x 125 ml --CaS 2 x 60 ml 1 M Sorbitol10 mM Tris-HCl, pH 7.5;10 mM CaCl240% PEG 25 ml 40% (w/v) PEG 3350 (Reagent grade) in waterCaT 25 ml 20 mM Tris-HCl, pH 7.520 mM CaCl2Stab Vials: Pichia and E. coli stabs. Store at +4°C.Phenotype(Pichia only)GenotypeStrain Amountstab his4Mut+GS115 1stab arg4 his4 aox1::ARG4 Mut S, Arg+KM71 1GS115 Albumin 1 stab HIS4Mut SGS115 β-Gal 1 stab HIS4Mut+stab F´ {pro AB, lac I q, lac Z∆M15, Tn10 (Tet R)} mcr A,TOP10F´ 1∆(mrr-hsd RMS-mcr BC), φ80lac Z∆M15, ∆lac X74,deo R, rec A1, ara D139, ∆(ara-leu)7697, gal U,gal K, rps L (Str R), end A1, nup G λ-.Box 2: Spheroplast Module. Store at -20°C.ComponentsReagent AmountZymolyase 10 x 20 µl 3 mg/ml Zymolyase in water(100,000 units/g lytic activity)1 M DTT 10 x 1 ml 1 M dithiothreitol in watercontinued on next pageviiKit Contents,continuedVector Box. Store at -20°C.Reagent DescriptionpHIL-D210 µg, lyophilized in TE, pH 8.0Vector for intracellular expression in PichiapPIC3.510 µg, lyophilized in TE, pH 8.0Vector for intracellular expression in PichiapHIL-S110 µg, lyophilized in TE, pH 8.0 Vector for secreted expression in Pichia. Uses the PHO1 signal sequencepPIC910 µg, lyophilized in TE, pH 8.0 Vector for secreted expression in Pichia. Uses the α-factor signal sequencePrimer Box. Store at -20°C.5´ AOX1 sequencing primer2 µg (312 pmoles), lyophilized5´-GACTGGTTCCAATTGACAAGC-3´3´ AOX1 sequencing primer2 µg (314 pmoles), lyophilized5´-GCAAATGGCATTCTGACATCC-3´α-Factor sequencing primer2 µg (315 pmoles), lyophilized5´-TACTATTGCCAGCATTGCTGC-3´Media The following prepackaged media is included for your convenience. Instructions for use are provided on the package.Media Amount Yield YP Base Medium 2 pouches 2 liters of YP mediumYP Base Agar Medium 2 pouches 2 liters of YP mediumYeast Nitrogen Base 1 pouch 500 ml of 10X YNBFor transformation of Pichia by spheroplasting, the Pichia Spheroplast Module isavailable separately from Invitrogen (see below for ordering information).Product Reactions or Amount Catalog no.Pichia Spheroplast Module 10 spheroplast preparations(50 transformations)K1720-01continued on next pageviiiRequired Equip-ment and Supplies (not provided) • 30°C rotary shaking incubator• Water baths capable of 37°C, 45°C, and 100°C• Centrifuge suitable for 50 ml conical tubes (floor or table-top)• Baffled culture flasks with metal covers (50 ml, 250 ml, 500 ml, 1000 ml, and 3 L)• 50 ml sterile, conical tubes• 6 ml and 15 ml sterile snap-top tubes (Falcon 2059 or similar)• UVSpectrophotometer• Mini agarose gel apparatus and buffers• Polyacrylamide Gel Electrophoresis apparatus and buffers• Media for transformation, growth, screening, and expression (see Recipes, pages 53-58) • 5% SDS solution (10 ml per transformation)• Sterile cheesecloth or gauze• Breaking Buffer (see Recipes, page 58)• Acid-washed glass beads (available from Sigma)• Replica-plating equipment (optional)• BeadBreaker™ (optional)ixPurchaser NotificationIntroduction The Pichia Expression Kit is based on the yeast Pichia pastoris. Pichia pastoris wasdeveloped into an expression system by scientists at Salk Institute Biotechnology/ IndustryAssociates (SIBIA) and Phillips Petroleum for high-level expression of recombinantproteins. All patents for Pichia pastoris and licenses for its use as an expression system areowned by Research Corporation Technologies (RCT), Inc., Tucson, Arizona. Forinformation on commercial licenses, please see page x.The Nature of the Invitrogen License Invitrogen has an exclusive license to sell the Pichia Expression Kit to scientists for research purposes only, under the terms described below. Use of Pichia pastoris by commercial entities for any commercial purpose requires the user to obtain a commercial license as detailed below. Before using the Pichia Expression Kit, please read the following license agreement. If you do not agree to be bound by its terms, contact Invitrogen within 10 days for authorization to return the unused Pichia Expression Kit and to receive a full credit. If you do agree to the terms of this license agreement, please complete the User Registration Card and return it to Invitrogen before using the kit.Pichia pastoris Patents Pichia pastoris is covered by one or more of the following U.S. patents and corresponding foreign patents owned and licensed by Research Corporation Technologies:4,683,293 4,808,537 4,812,405 4,818,700 4,837,148 4,855,231 4,857,467 4,879,231 4,882,279 4,885,242 4,895,800 4,929,555 5,002,876 5,004,688 5,032,516 5,122,465 5,135,868 5,166,329Individual Pichia Expression Kit License Agreement Invitrogen Corporation ("Invitrogen") grants you a non-exclusive license to use the enclosed Pichia Expression Kit ("Expression Kit") for academic research or for evaluation purposes only. The Expression Kit is being transferred to you in furtherance of, and reliance on, such license. You may not use the Expression Kit, or the materials contained therein, for any commercial purpose without a license for such purpose from Research Corporation Technologies, Inc., Tucson, Arizona.Definition of Commercial Purpose Commercial purposes include:(a) any use of Expression Products in a Commercial Product(b) any use of Expression Products in the manufacture of a Commercial Product(c) any sale of Expression Products(d) any use of Expression Products or the Expression Kit to facilitate or advanceresearch or development of a Commercial Product(e) any use of Expression Products or the Expression Kit to facilitate or advance anyresearch or development program the results of which will be applied to thedevelopment of Commercial Products"Expression Products" means products expressed with the Expression Kit, or with the use of any vectors or host strains in the Expression Kit. "Commercial Product" means any product intended for sale or commercial use.Commercial entities may conduct their evaluation for one year at which time this license automatically terminates. Research Corporation Technologies will contact commercial entities during the evaluation period regarding their desire for a commercial license.continued on next pagexPurchaser Notification, continuedIndividual Responsibilities Access to the Expression Kit must be limited solely to those officers, employees and students of your institution who need access to perform the above-described research or evaluation. You must inform each such officer, employee and student of the provisions of this license agreement and require them to agree, in writing, to be bound by the provisions of this license agreement. You may not distribute neither the Expression Kit nor the vectors or host strains contained in it to others, even to those within your own institution. You may only transfer modified, altered, or original material from the Expression Kit to a third party following written notification of, and written approval from, Invitrogen so that the recipient can be licensed. You may not assign, sub-license, rent, lease or otherwise transfer this license agreement or any of the rights or obligation thereunder, except as expressly permitted by Invitrogen and RCT.Termination of License This license agreement is effective until terminated. You may terminate it at any time by destroying all Pichia expression products in your control. It will also terminate auto-matically if you fail to comply with the terms and conditions of the license agreement. You shall, upon termination of the license agreement, destroy all Pichia Expression Kits in your control, and so notify Invitrogen in writing.This License shall be governed in its interpretation and enforcement by the laws of the State of California.Contact for Commercial Licensing Bennett Cohen, Ph.D.Research Corporation Technologies 101 North Wilmot Road, Suite 600 Tucson, Arizona 85711-3335 Phone: (520) 748-4400Fax: (520)748-0025User Registration Card Please complete and return the enclosed User Registration Card for each PichiaExpression Kit that you purchase. This will serve as a record of your purchase and regis-tration and will allow Invitrogen to provide you with technical support and manualupdates. It will also allow Invitrogen to update you on future developments and improve-ments to the Pichia Expression Kit. The agreement outlined above becomes effectiveupon our receipt of your User Registration Card or 10 days following the sale of thePichia Expression Kit to you. Use of the kit at any time results in immediate obligation tothe terms and conditions stated in this license agreement.xiProduct QualificationIntroduction This section describes the criteria used to qualify the components in the PichiaExpression Kit.Vectors All expression vectors are qualified by restriction enzyme digestion. Restriction digests must demonstrate the correct banding pattern when electrophoresed on an agarose gel.Spheroplast Reagents The spheroplast reagents are qualified by spheroplast preparation of GS115 following the protocol provided in the Pichia Expression Kit manual. At least 70% of the Pichia pastoris cells must form spheroplasts in 30 minutes or less.Pichia Strains The Pichia strains are by demonstrating viability of the culture. Single colonies should arise within 48 hours after streaking on YPD medium from the stabPrimers Sequencing primers are lot tested by automated DNA sequencing experiments.Buffers andSolutionsAll buffers and solutions are extensively tested for sterility.Media All Pichia growth and expression media are qualified by growing the GS115 Pichiastrain.xiiIntroductionOverviewReview Articles The information presented here is designed to give you a concise overview of the Pichia pastoris expression system. It is by no means exhaustive. For further information, pleaseread the articles cited in the text along with recent review articles (Buckholz and Gleeson,1991; Cregg et al., 1993; Sreekrishna et al., 1988; Wegner, 1990). A general review offoreign gene expression in yeast is also available (Romanos et al., 1992).General Characteristics of Pichia pastoris As a eukaryote, Pichia pastoris has many of the advantages of higher eukaryotic expression systems such as protein processing, protein folding, and posttranslational modification, while being as easy to manipulate as E. coli or Saccharomyces cerevisiae. It is faster, easier, and less expensive to use than other eukaryotic expression systems such as baculovirus or mammalian tissue culture, and generally gives higher expression levels. As a yeast, it shares the advantages of molecular and genetic manipulations with Saccharomyces, and has the added advantage of 10- to 100-fold higher heterologous protein expression levels. These features make Pichia very useful as a protein expression system.Similarity to Saccharomyces Many of the techniques developed for Saccharomyces may be applied to Pichia including: • transformation by complementation• genedisruption• genereplacementIn addition, the genetic nomenclature used for Saccharomyces has been applied to Pichia. For example, the HIS4 gene in both Saccharomyces and Pichia encodes histidinol dehydrogenase. There is also cross-complementation between gene products in both Saccharomyces and Pichia. Several wild-type genes from Saccharomyces complement comparable mutant genes in Pichia. Genes such as HIS4, LEU2, ARG4, TRP1, and URA3 all complement their respective mutant genes in Pichia.Pichia pastoris as a Methylotrophic Yeast Pichia pastoris is a methylotrophic yeast, capable of metabolizing methanol as its sole carbon source. The first step in the metabolism of methanol is the oxidation of methanol to formaldehyde using molecular oxygen by the enzyme alcohol oxidase. This reaction generates both formaldehyde and hydrogen peroxide. To avoid hydrogen peroxide toxicity, methanol metabolism takes place within a specialized cell organelle called the peroxisome, which sequesters toxic by-products from the rest of the cell. Alcohol oxidase has a poor affinity for O2, and Pichia pastoris compensates by generating large amounts of the enzyme. The promoter regulating the production of alcohol oxidase drives heterologous protein expression in Pichia.Two Alcohol Oxidase Proteins The AOX1 and AOX2 genes code for alcohol oxidase in Pichia pastoris. The AOX1 gene product accounts for the majority of alcohol oxidase activity in the cell. Expression of the AOX1 gene is tightly regulated and induced by methanol to high levels, typically > 30% ofthe total soluble protein in cells grown with methanol as the carbon source. The AOX1 gene has been isolated and the AOX1 promoter is used to drive expression of the gene of interest (Ellis et al., 1985; Koutz et al., 1989; Tschopp et al., 1987a). While AOX2 is about 97% homologous to AOX1, growth on methanol is much slower than with AOX1. This slowgrowth allows isolation of Mut S strains (aox1) (Cregg et al., 1989; Koutz et al., 1989).continued on next page1Overview, continuedExpression Expression of the AOX1 gene is controlled at the level of transcription. In methanol-grown cells approximately 5% of the polyA+ RNA is from the AOX1 gene. The regulation of theAOX1 gene is a two step process: a repression/derepression mechanism plus an inductionmechanism (e.g. GAL1 gene in Saccharomyces (Johnston, 1987)). Briefly, growth onglucose represses transcription, even in the presence of the inducer methanol. For thisreason, growth on glycerol is recommended for optimal induction with methanol. Pleasenote that growth on glycerol (derepression) is not sufficient to generate even minute levelsof expression from the AOX1 gene. The inducer, methanol, is necessary for detectablelevels of AOX1 expression (Ellis et al., 1985; Koutz et al., 1989; Tschopp et al., 1987a).Phenotype of aox1 mutants Loss of the AOX1 gene, and thus a loss of most of the cell's alcohol oxidase activity, results in a strain that is phenotypically Mut S (Methanol utilization slow). This has in the past been referred to as Mut. The Mut S designation has been chosen to accurately describe the phenotype of these mutants. This results in a reduction in the cells' ability to metabolize methanol. The cells, therefore, exhibit poor growth on methanol medium. Mut+ (Methanol utilization plus) refers to the wild type ability of strains to metabolize methanol as the sole carbon source. These two phenotypes are used when evaluating Pichia transformants for integration of your gene (Experimental Outline, page 3).Intracellular and Secretory Protein Expression Heterologous expression in Pichia can be either intracellular or secreted. Secretion requires the presence of a signal sequence on the expressed protein to target it to the secretory pathway. While several different secretion signal sequences have been used successfully, including the native secretion signal present on some heterologous proteins, success has been variable. The secretion signal sequence from the Saccharomyces cerevisiaeα factor prepro peptide has been used most successfully (Cregg et al., 1993; Scorer et al., 1993).The major advantage of expressing heterologous proteins as secreted proteins is that Pichia pastoris secretes very low levels of native proteins. That, combined with the very low amount of protein in the minimal Pichia growth medium, means that the secreted heterologous protein comprises the vast majority of the total protein in the medium and serves as the first step in purification of the protein (Barr et al., 1992). Note: If there are recognized glycosylation sites (Asn-X-Ser/Thr) in your protein's primary sequence, glycosylation may occur at these sites.Posttranslational Modifications In comparison to Saccharomyces cerevisiae, Pichia may have an advantage in the glyco-sylation of secreted proteins because it may not hyperglycosylate. Both Saccharomyces cerevisiae and Pichia pastoris have a majority of N-linked glycosylation of the high-mannose type; however, the length of the oligosaccharide chains added posttranslationally to proteins in Pichia (average 8-14 mannose residues per side chain) is much shorter than those in S. cerevisiae (50-150 mannose residues) (Grinna and Tschopp, 1989; Tschopp et al., 1987b). Very little O-linked glycosylation has been observed in Pichia.In addition, Saccharomyces cerevisiae core oligosaccharides have terminal α1,3 glycan linkages whereas Pichia pastoris does not. It is believed that the α1,3 glycan linkages in glycosylated proteins produced from Saccharomyces cerevisiae are primarily responsible for the hyper-antigenic nature of these proteins making them particularly unsuitable for therapeutic use. Although not proven, this is predicted to be less of a problem for glycoproteins generated in Pichia pastoris, because it may resemble the glycoprotein structure of higher eukaryotes (Cregg et al., 1993).2Experimental OutlineSelection of Vector and Cloning To utilize the strong, highly inducible P AOX1 promoter for expression of your protein, four expression vectors are included in this kit. pHIL-D2 and pPIC3.5 are used for intracellular expression while pHIL-S1 and pPIC9 are used for secreted expression (see pages 14-19 for more information). Before cloning your insert, you must...• decide whether you want intracellular or secreted expression.• analyze your insert for the following restriction sites: Sac I, Stu I, Sal I, Not I, and Bgl II. These sites are recommended for linearizing your construct prior to Pichiatransformation. If your insert has all of these sites, see pages 28-29 for alternate sites.Transformation and IntegrationTwo different phenotypic classes of His+ recombinant strains can be generated: Mut+ and Mut S. Mut S refers to the "Methanol utilization slow" phenotype caused by the loss of alcohol oxidase activity encoded by the AOX1 gene. A strain with a Mut S phenotype has a mutant aox1 locus, but is wild type for AOX2. This results in a slow growth phenotype on methanol medium. Transformation of strain GS115 can yield both classes of transformants, His+ Mut+ and His+Mut S, while KM71 yields only His+ Mut S since the strain itself is Mut S. Both Mut+ and Mut S recombinants are useful to have as one phenotype may favor better expression of your protein than the other. Due to clonal variation, you should test 6-10 recombinants per phenotype. There is no way to predict beforehand which construct or isolate will better express your protein. We strongly recommend that you analyze Pichia recombinants by PCR to confirm integration of your construct (see page 40).Once you have successfully cloned your gene, you will then linearize your plasmid to stimulate recombination when the plasmid is transformed into Pichia. The table below describes the types of recombinants you will get by selective digestion of your plasmid. RestrictionEnzymeIntegration Event GS115 Phenotype KM71 PhenotypeSal I or Stu I Insertion at his4His+ Mut+ His+ Mut SSac I Insertion at 5´AOX1 regionHis+ Mut+ His+ Mut SNot I or Bgl II Replacement atAOX1 locusHis+ Mut SHis+ Mut+His+ Mut S (notrecommended, see page 11)Expression and Scale-up After confirming your Pichia recombinants by PCR, you will test expression of both His+Mut+ and His+ Mut S recombinants. This will involve growing a small culture of each recombinant, inducing with methanol, and taking time points. If looking for intracellular expression, analyze the cell pellet from each time point by SDS polyacrylamide gel electrophoresis (SDS-PAGE). If looking for secreted expression, analyze both the cellpellet and supernatant from each time point. We recommend that you analyze your SDS-PAGE gels by both Coomassie staining and Western blot, if you have an antibody to your protein. We also suggest checking for protein activity by assay, if one is available. Not all proteins express to the level of grams per liter, so it is advisable to check by Western blotor activity assay, and not just by Coomassie staining of SDS-PAGE gels for production of your protein.Choose the Pichia recombinant strain that best expresses your protein and optimizeinduction based on the suggestions on pages 47-48. Once expression is optimized, scale-up your expression protocol to produce more protein.continued on next page3。
裂解酶 毕赤酵母表达

裂解酶毕赤酵母表达裂解酶是一种重要的酶类,能够将高分子物质裂解为较小的分子,被广泛应用于制药、食品、化工等领域。
毕赤酵母是一种常见的酵母菌,因其生长速度快,易培养,被广泛应用于分子生物学研究领域。
本文将围绕着“裂解酶毕赤酵母表达”这一主题,进行详细阐述。
第一步,克隆裂解酶基因首先需要克隆裂解酶基因。
裂解酶基因可以从天然菌株或基因库中获得。
一般情况下,可采用PCR技术或构建文库的方法进行克隆。
PCR克隆是一种常见的将特定基因扩增的方法,需要设计引物。
而文库构建则需要将大量的DNA片段克隆到载体上,获取含有目标基因的克隆体。
第二步,构建表达载体获取裂解酶基因后,需要将其插入到表达载体中,以实现外源基因的表达。
表达载体一般包括启动子、编码区、终止序列等部分。
还需要添加适合裂解酶表达的表达基因启动子和信使RNA聚合酶结合位点。
可采用限制性内切酶、LiGA等方法,将裂解酶基因插入到表达载体中。
第三步,转化毕赤酵母将构建好的表达载体转化到毕赤酵母细胞中。
目前主要有两种转化方法,一种是化学转化,一种是电转化。
化学转化依赖于离子间的相互作用力,可将DNA引入到细胞中。
而电转化则是利用高电压作用于细胞,使其渗透性提高,DNA片段能够进入细胞。
第四步,筛选表达菌株进行转化后,将细胞涂布于含有蔗糖的SD-UF板上进行筛选。
含有蔗糖的培养基只能被表达裂解酶的菌株使用,其他菌株无法生长,通过这种方法可以筛选出表达裂解酶的毕赤酵母菌株。
第五步,表达和纯化裂解酶最后,使用相应的表达条件和纯化方法,纯化表达的裂解酶。
一般情况下,裂解酶是一种外分泌酶,可采用诱导性表达,通过添加诱导因子(如甘露醇)来促进裂解酶的分泌。
常见的纯化方法包括离子交换层析、凝胶过滤层析、亲和层析等。
总之,裂解酶毕赤酵母表达是一个复杂的过程,需要准确的基因克隆、合适的表达载体、科学的转化方法、合适的筛选条件和纯化方法等。
只有在这些步骤都正确执行的情况下,才能够得到表达裂解酶的毕赤酵母菌株,并最终纯化出高质量的裂解酶。
毕赤酵母_Pichiapastoris_分泌表达牛白细胞介素2
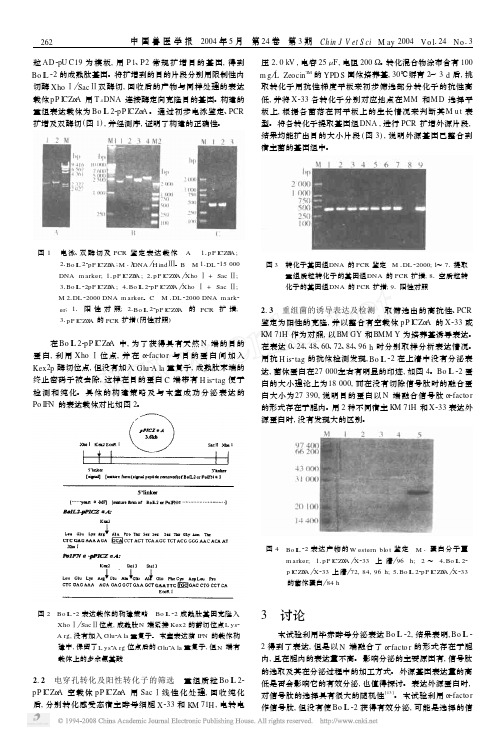
毕赤酵母(P ich ia p astor is )分泌表达牛白细胞介素2孙红立1,3,陈 芳1,3,于瑞嵩2,3,刘惠莉2,3,曹祥荣1,李 震2,33 (11南京师范大学生命科学学院,江苏南京210097; 21上海市农业科学院畜牧兽医研究所,上海201106;31上海市农业遗传育种重点实验室,上海201106;)摘要:利用含有强启动子P AOX1和Α2因子信号肽序列的巴斯德毕赤酵母载体pP I CZ ΑA ,构建出含牛白细胞介素2(Bo I L 22)基因的重组质粒Bo I L 22pP I CZ ΑA 。
线性化的重组表达载体转化到巴斯德毕赤酵母X 233及K M 71H 中,筛选Zeocin 高抗性酵母菌株,甲醇诱导目的蛋白表达。
经SD S 2PA GE 和W estern b lo t 检测表明,Bo I L 22以融合蛋白形式在胞内表达,但没能分泌到胞外。
通过Bo I L 22在巴斯德毕赤酵母中的表达,重点讨论了信号肽、基因的偏爱性等对外源基因分泌表达的影响。
关键词:牛白细胞介素2(Bo I L 22);毕赤酵母(P ich ia p astoris );分泌表达中图分类号:Q 78 文献标识码:A 文章编号:100524545(2004)0320261203 收稿日期:2003208219 基金项目:上海市科技兴农重点攻关项目(科农攻字2001第327号) 作者简介:孙红立(19792),男,硕士。
3通讯作者,T el :021*********;021*********;E 2m ail :zhenli 60@public 3.sta .net .cn 白细胞介素2(I L 22)是一种具有广泛生物活性的细胞因子,它能够有效地提高机体的免疫功能。
目前已证实,牛白细胞介素2(Bo I L 22)在牛疱疹病毒疫苗免疫中具有免疫佐剂的作用[1,2];在牛乳腺炎的预防与治疗中也具有很好的作用[3]。
毕赤酵母发酵手册

Version B Pichia Fermentation Process GuidelinesOverviewIntroduction Pichia pastoris, like Saccharomyces cerevisiae, is particularly well-suited forfermentative growth. Pichia has the ability to reach very high cell densities duringfermentation which may improve overall protein yields.We recommend that only those with fermentation experience or those who have accessto people with experience attempt fermentation. Since there are a wide variety offermenters available, it is difficult to provide exact procedures for your particular case.The guidelines given below are based on fermentations of both Mut+ and Mut S Pichiastrains in a 15 liter table-top glass fermenter. Please read the operator's manual for yourparticular fermenter before beginning. The table below provides an overview of thematerial covered in these guidelines.Step Topic Page1 Fermentationparameters 12 Equipment needed and preparation of medium 23 Measurement and use of dissolved oxygen (DO) in the culture 34 Growth of the inoculum 45 Generation of biomass on glycerol in batch and fed-batch phases 4-56 Induction of expression of Mut+ and Mut S recombinants in themethanol fed-batch phase6-77 Harvesting and lysis of cells 88 References 9-109 Recipes 11Fermentation Parameters It is important to monitor and control the following parameters throughout thefermentation process. The following table describes the parameters and the reasons for monitoring them.Parameter Reason Temperature (30.0°C) Growth above 32°C is detrimental to protein expression Dissolved oxygen (>20%) Pichia needs oxygen to metabolize glycerol andmethanolpH (5.0-6.0 and 3.0) Important when secreting protein into the medium andfor optimal growthAgitation (500 to 1500 rpm) Maximizes oxygen concentration in the mediumAeration (0.1 to 1.0 vvm*for glass fermenters)Maximizes oxygen concentration in the medium whichdepends on the vesselAntifoam (the minimumneeded to eliminate foam)Excess foam may cause denaturation of your secretedprotein and it also reduces headspaceCarbon source (variablerate)Must be able to add different carbon sources at differentrates during the course of fermentationcontinued on next pageOverview, continuedRecommended Equipment Below is a checklist for equipment recommendations.• A jacketed vessel is needed for cooling the yeast during fermentation, especially during methanol induction. You will need a constant source of cold water (5-10°C). This requirement may mean that you need a refrigeration unit to keep the water cold. • A foam probe is highly recommended as antifoam is required.• A source of O2--either air (stainless steel fermenters at 1-2 vvm) or pure O2(0.1-0.3 vvm for glass fermenters).• Calibrated peristaltic pumps to feed the glycerol and methanol.• Automatic control of pH.Medium Preparation You will need to prepare the appropriate amount of following solutions:• Fermentation Basal Salts (page 11)• PTM1Trace Salts (page 11)• ~75 ml per liter initial fermentation volume of 50% glycerol containing 12 ml PTM1 Trace Salts per liter of glycerol.• ~740 ml per liter initial fermentation volume of 100% methanol containing 12 mlPTM1Trace Salts per liter of methanol.Monitoring the Growth of Pichia pastoris Cell growth is monitored at various time points by using the absorbance at 600 nm (OD600) and the wet cell weight. The metabolic rate of the culture is monitored by observing changes in the concentration of dissolved oxygen in response to carbon availability (see next page).Dissolved Oxygen (DO) MeasurementIntroduction The dissolved oxygen concentration is the relative percent of oxygen in the mediumwhere 100% is O2-saturated medium. Pichia will consume oxygen as it grows, reducing the dissolved oxygen content. However, because oxygen is required for the first step ofmethanol catabolism, it is important to maintain the dissolved oxygen (DO) concentra-tion at a certain level (>20%) to ensure growth of Pichia on methanol. Accuratemeasurement and observation of the dissolved oxygen concentration of a culture willgive you important information about the state and health of the culture. Therefore, it isimportant to accurately calibrate your equipment. Please refer to your operator's manual.Maintaining the Dissolved Oxygen Concentration (DO) 1. Maintaining the dissolved oxygen above 20% may be difficult depending on theoxygen transfer rates (OTR) of the fermenter, especially in small-scale glassvessels. In a glass vessel, oxygen is needed to keep the DO above 20%, usually~0.1-0.3 vvm (liters of O2per liter of fermentation culture per minute). Oxygen consumption varies and depends on the amount of methanol added and the protein being expressed.2. Oxygen can be used at 0.1 to 0.3 vvm to achieve adequate levels. This can beaccomplished by mixing with the air feed and can be done in any glass fermenter.For stainless steel vessels, pressure can be used to increase the OTR. Be sure toread the operator's manual for your particular fermenter.3. If a fermenter cannot supply the necessary levels of oxygen, then the methanol feedshould be scaled back accordingly. Note that decreasing the amount of methanol may reduce the level of protein expression.4. To reach maximum expression levels, the fermentation time can be increased todeliver similar levels of methanol at the lower feed rate. For many recombinantproteins, a direct correlation between amount of methanol consumed and theamount of protein produced has been observed.Use of DO Measurements During growth, the culture consumes oxygen, keeping the DO concentration low. Note that oxygen is consumed whether the culture is grown on glycerol or methanol. The DO concentration can be manipulated to evaluate the metabolic rate of the culture and whether the carbon source is limiting. The metabolic rate indicates how healthy the culture is. Determining whether the carbon source is limiting is important if you wish to fully induce the AOX1 promoter. For example, changes in the DO concentrations (DO spikes) allow you to determine whether all the glycerol is consumed from the culture before adding methanol. Secondly, it ensures that your methanol feed does not exceed the rate of consumption. Excess methanol (> 1-2% v/v) may be toxic.Manipulation of DO If carbon is limiting, shutting off the carbon source should cause the culture to decrease its metabolic rate, and the DO to rise (spike). Terminate the carbon feed and time how long it takes for the DO to rise 10%, after which the carbon feed is turned back on. If the lag time is short (< 1 minute), the carbon source is limiting.Fermenter Preparation and Glycerol Batch PhaseInoculum Seed Flask Preparation Remember not to put too much medium in the baffled flasks. Volume should be 10-30% of the total flask volume.1. Baffled flasks containing a total of 5-10% of the initial fermentation volume ofMGY or BMGY are inoculated with a colony from a MD or MGY plate or from a frozen glycerol stock.2. Flasks are grown at 30°C, 250-300 rpm, 16-24 hours until OD600= 2-6. Toaccurately measure OD600> 1.0, dilute a sample of your culture 10-fold before reading.Glycerol Batch Phase 1. Sterilize the fermenter with the Fermentation Basal Salts medium containing 4%glycerol (see page 11).2. After sterilization and cooling, set temperature to 30°C, agitation and aeration tooperating conditions (usually maximum rpm and 0.1-1.0 vvm air), and adjust the pH of the Fermentation Basal Salts medium to 5.0 with 28% ammonium hydroxide(undiluted ammonium hydroxide). Add aseptically 4.35 ml PTM1trace salts/liter of Fermentation Basal Salts medium.3. Inoculate fermenter with approximately 5-10% initial fermentation volume from theculture generated in the inoculum shake flasks. Note that the DO will be close to 100% before the culture starts to grow. As the culture grows, it will consumeoxygen, causing the DO to decrease. Be sure to keep the DO above 20% by adding oxygen as needed.4. Grow the batch culture until the glycerol is completely consumed (18 to 24 hours).This is indicated by an increase in the DO to 100%. Note that the length of timeneeded to consume all the glycerol will vary with the density of the initial inoculum.5. Sampling is performed at the end of each fermentation stage and at least twice daily.We take 10 ml samples for each time point, then take 1 ml aliquots from this 10 mlsample. Samples are analyzed for cell growth (OD600and wet cell weight), pH, microscopic purity, and protein concentrations or activity. Freeze the cell pellets and supernatants at -80°C for later analysis. Proceed to Glycerol Fed-Batch Phase,page 5.Yield A cellular yield of 90 to 150 g/liter wet cells is expected for this stage. Recombinant protein will not yet be produced due to the absence of methanol.Introduction Once all the glycerol is consumed from the batch growth phase, a glycerol feed isinitiated to increase the cell biomass under limiting conditions. When you are ready toinduce with methanol, you can use DO spikes to make sure the glycerol is limited.Glycerol Fed-Batch Phase 1. Initiate a 50% w/v glycerol feed containing 12 ml PTM1trace salts per liter of glycerol feed. Set the feed rate to 18.15 ml/hr /liter initial fermentation volume.2. Glycerol feeding is carried out for about four hours or longer (see below). A cellularyield of 180 to 220 g/liter wet cells should be achieved at the end of this stage while no appreciable recombinant protein is produced.Note The level of expressed protein depends on the cell mass generated during the glycerolfed-batch phase. The length of this feed can be varied to optimize protein yield. A rangeof 50 to 300 g/liter wet cells is recommended for study. A maximum level of 4%glycerol is recommended in the batch phase due to toxicity problems with higher levelsof glycerol.Important If dissolved oxygen falls below 20%, the glycerol or methanol feed should bestopped and nothing should be done to increase oxygen rates until the dissolvedoxygen spikes. At this point, adjustments can be made to agitation, aeration, pressure oroxygen feeding.Proteases In the literature, it has been reported that if the pH of the fermentation medium islowered to 3.0, neutral proteases are inhibited. If you think neutral proteases aredecreasing your protein yield, change the pH control set point to 3.0 during the glycerolfed-batch phase (above) or at the beginning of the methanol induction (next page) andallow the metabolic activity of the culture to slowly lower the pH to 3.0 over 4 to 5 hours(Brierley, et al., 1994; Siegel, et al., 1990).Alternatively, if your protein is sensitive to low pH, it has been reported that inclusion ofcasamino acids also decreases protease activity (Clare, et al., 1991).Introduction All of the glycerol needs to be consumed before starting the methanol feed to fullyinduce the AOX1 promoter on methanol. However, it has been reported that a "mixedfeed" of glycerol and methanol has been successful to express recombinant proteins(Brierley, et al., 1990; Sreekrishna, et al., 1989). It is important to introduce methanolslowly to adapt the culture to growth on methanol. If methanol is added too fast, it willkill the cells. Once the culture is adapted to methanol, it is very important to use DOspikes to analyze the state of the culture and to take time points over the course ofmethanol induction to optimize protein expression. Growth on methanol also generates alot of heat, so temperature control at this stage is very important.Mut+ Methanol Fed-Batch Phase 1. Terminate glycerol feed and initiate induction by starting a 100% methanol feedcontaining 12 ml PTM1trace salts per liter of methanol. Set the feed rate to 3.6 ml/hr per liter initial fermentation volume.2. During the first 2-3 hours, methanol will accumulate in the fermenter and thedissolved oxygen values will be erratic while the culture adapts to methanol.Eventually the DO reading will stabilize and remain constant.3. If the DO cannot be maintained above 20%, stop the methanol feed, wait for theDO to spike and continue on with the current methanol feed rate. Increaseagitation, aeration, pressure or oxygen feeding to maintain the DO above 20%. 4. When the culture is fully adapted to methanol utilization (2-4 hours), and is limitedon methanol, it will have a steady DO reading and a fast DO spike time (generally under 1 minute). Maintain the lower methanol feed rate under limited conditions for at least 1 hour after adaptation before doubling the feed. The feed rate is then doubled to ~7.3 ml/hr/liter initial fermentation volume.5 After 2 hours at the 7.3 ml/hr/liter feed rate, increase the methanol feed rate to~10.9 ml/hr per liter initial fermentation volume. This feed rate is maintainedthroughout the remainder of the fermentation.6. The entire methanol fed-batch phase lasts approximately 70 hours with a total ofapproximately 740 ml methanol fed per liter of initial volume. However, this may vary for different proteins.Note: The supernatant may appear greenish. This is normal.Yield The cell density can increase during the methanol fed-batch phase to a final level of 350 to 450 g/liter wet cells. Remember that because most of the fermentation is carried out ina fed-batch mode, the final fermentation volume will be approximately double the initialfermentation volume.Fermentation of Mut S Pichia Strains Since Mut S cultures metabolize methanol poorly, their oxygen consumption is very low. Therefore, you cannot use DO spikes to evaluate the culture. In standard fermentations of a Mut S strain, the methanol feed rate is adjusted to maintain an excess of methanol in the medium which does not exceed 0.3% (may be determined by gas chromatography). While analysis by gas chromatography will insure that nontoxic levels of methanol are maintained, we have used the empirical guidelines below to express protein in Mut S strains. A gas chromatograph is useful for analyzing and optimizing growth of Mut S recombinants.continued on next pageMethanol Fed-Batch Phase, continuedMut S Methanol Fed- Batch Phase The first two phases of the glycerol batch and fed-batch fermentations of the Mut S strains are conducted as described for the Mut+ strain fermentations. The methanol induction phases of the Mut+ and Mut S differ in terms of the manner and amount in which the methanol feed is added to the cultures.1. The methanol feed containing 12 ml PTM1trace salts per liter of methanol is initiated at 1 ml/hr/liter initial fermentation volume for the first two hours. It is then increased in 10% increments every 30 minutes to a rate of 3 ml/hr which ismaintained for the duration of the fermentation.2.. The vessel is then harvested after ~100 hours on methanol. This time may be variedto optimize protein expression.Harvesting and Lysis of CellsIntroduction The methods and equipment listed below are by no means complete. The amount of cells or the volume of supernatant will determine what sort of equipment you need.Harvesting Cells and Supernatant For small fermentations (1-10 liters), the culture can be collected into centrifuge bottles (500-1000 ml) and centrifuged to separate the cells from the supernatant.For large fermentations, large membrane filtration units (Millipore) or a Sharples centrifuge can be used to separate cells from the supernatant. The optimal method will depend on whether you need the cells or the supernatant as the source of your protein and what you have available.Supernatants can be loaded directly onto certain purification columns or concentrated using ultrafiltration.Cell Lysis We recommend cell disruption using glass beads as described in Current Protocols inMolecular Biology, page 13.13.4. (Ausubel, et al., 1990) or Guide to ProteinPurification (Deutscher, 1990). This method may be tedious for large amounts of cells.For larger amounts, we have found that a microfluidizer works very well. Frenchpressing the cells does not seem to work as well as the glass beads or the microfluidizer.ReferencesIntroduction Most of the references below refer to papers where fermentation of Pichia wasperformed. Note that some of these are patent papers. You can obtain copies of patentsusing any of the following methods.• Patent Depository Libraries. U. S. patents and international patents granted underthe Patent Cooperation Treaty (PCT) are available on microfilm. These can be copiedand mailed or faxed depending on length. There is a fee for this service. The referencelibrarian at your local library can tell you the location of the nearest Patent DepositoryLibrary.• Interlibrary Loan. If you are not near a Patent Depository Library, you may request acopy of the patent through interlibrary loan. There will be a fee for this service.• U. S. Patent Office. Requests may be made directly to the Patent Office, Arlington,VA. Please call 703-557-4636 for more information on cost and delivery.• Private Library Services. There are private companies who will retrieve and sendyou patents for a fee. Two are listed below:Library Connection: 804-758-3311Rapid Patent Services: 800-336-5010Citations Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A.,Struhl, K., eds (1990) Current Protocols in Molecular Biology. GreenePublishing Associates and Wiley-Interscience, New York.Brierley, R. A., Siegel, R. S., Bussineau, C. M. Craig, W. S., Holtz, G. C., Davis, G. R.,Buckholz, R. G., Thill, G. P., Wondrack, L. M., Digan, M. E., Harpold, M. M.,Lair, S. V., Ellis, S. B., and William, M. E. (1989) Mixed Feed RecombinantYeast Fermentation. International Patent (PCT) Application. Publication No.WO 90/03431.Brierley, R. A., Bussineau, C., Kosson, R., Melton, A., and Siegel, R. S. (1990)Fermentation Development of Recombinant Pichia pastoris Expressing theHeterologous Gene: Bovine Lysozyme. Ann. New York Acad. Sci.589: 350-362.Brierley, R. A., Davis, G. R. and Holtz, G. C. (1994) Production of Insulin-Like GrowthFactor-1 in Methylotrophic Yeast Cells. United States Patent5,324,639.Clare, J. J., Romanos, M. A., Rayment, F. B., Rowedder, J. E., Smith, M. A., Payne, M.M., Sreekrishna, K. and Henwood, C. A. (1991) Production of EpidermalGrowth Factor in Yeast: High-level Secretion Using Pichia pastoris StrainsContaining Multiple Gene Copies. Gene105: 205-212.Cregg, J. M., Tschopp, J. F., Stillman, C., Siegel, R., Akong, M., Craig, W. S.,Buckholz, R. G., Madden, K. R., Kellaris, P. A., Davis, G. R., Smiley, B. L.,Cruze, J., Torregrossa, R., Veliçelebi, G. and Thill, G. P. (1987) High-levelExpression and Efficient Assembly of Hepatitis B Surface Antigen in theMethylotrophic Yeast Pichia pastoris. Bio/Technology5: 479-485.Cregg, J. M., Vedvick, T. S. and Raschke, W. C. (1993) Recent Advances in theExpression of Foreign Genes in Pichia pastoris. Bio/Technology11: 905-910.Deutscher, M. P. (1990) Guide to Protein Purification. In: Methods in Enzymology (J.N. Abelson and M. I. Simon, eds.) Academic Press, San Diego, CA.continued on next pageReferences, continuedCitations, continuedDigan, M. E., Lair, S. V., Brierley, R. A., Siegel, R. S., Williams, M. E., Ellis, S. B., Kellaris, P. A., Provow, S. A., Craig, W. S., Veliçelebi, G., Harpold, M. M. andThill, G. P. (1989) Continuous Production of a Novel Lysozyme via Secretionfrom the Yeast Pichia pastoris. Bio/Technology7: 160-164.Hagenson, M. J., Holden, K. A., Parker, K. A., Wood, P. J., Cruze, J. A., Fuke, M., Hopkins, T. R. and Stroman, D. W. (1989) Expression of Streptokinase inPichia pastoris Yeast. Enzyme Microbiol. Technol.11: 650-656.Laroche, Y., Storme, V., Meutter, J. D., Messens, J. and Lauwereys, M. (1994) High-Level Secretion and Very Efficient Isotopic Labeling of Tick AnticoagulantPeptide (TAP) Expressed in the Methylotrophic Yeast, Pichia pastoris.Bio/Technology12: 1119-1124.Romanos, M. A., Clare, J. J., Beesley, K. M., Rayment, F. B., Ballantine, S. P., Makoff,A. J., Dougan, G., Fairweather, N. F. and Charles, I. G. (1991) RecombinantBordetella pertussis Pertactin p69 from the Yeast Pichia pastoris High LevelProduction and Immunological Properties. Vaccine9: 901-906.Siegel, R. S. and Brierley, R. A. (1989) Methylotrophic Yeast Pichia pastoris Produced in High-cell-density Fermentations With High Cell Yields as Vehicle forRecombinant Protein Production. Biotechnol. Bioeng.34: 403-404.Siegel, R. S., Buckholz, R. G., Thill, G. P., and Wondrack, L. M. (1990) Production of Epidermal Growth Factor in Methylotrophic Yeast Cells. International Patent(PCT) Application. Publication No. WO 90/10697.Sreekrishna, K., Nelles, L., Potenz, R., Cruse, J., Mazzaferro, P., Fish, W., Fuke, M., Holden, K., Phelps, D., Wood, P. and Parker, K. (1989) High LevelExpression, Purification, and Characterization of Recombinant Human TumorNecrosis Factor Synthesized in the Methylotrophic Yeast Pichia pastoris.Biochemistry28(9): 4117-4125.©2002 Invitrogen Corporation. All rights reservedRecipesFermentation Basal Salts Medium For 1 liter, mix together the following ingredients:Phosphoric acid, 85% (26.7 ml)Calcium sulfate 0.93 gPotassium sulfate 18.2 gMagnesium sulfate-7H2O 14.9gPotassium hydroxide 4.13 gGlycerol 40.0g Water to 1 literAdd to fermenter with water to the appropriate volume and sterilize.PTM1 Trace Salts Mix together the following ingredients:Cupric sulfate-5H2O 6.0gSodium iodide 0.08 gManganese sulfate-H2O 3.0gSodium molybdate-2H2O 0.2gBoric Acid 0.02 g Cobalt chloride 0.5 g Zinc chloride 20.0 gFerrous sulfate-7H2O 65.0gBiotin 0.2gSulfuric Acid 5.0 mlWater to a final volume of 1 literFilter sterilize and store at room temperature.Note: There may be a cloudy precipitate upon mixing of these ingredients. Filter-sterilize as above and use.11。
毕赤酵母诱导表达实验步骤(1)

毕赤酵母诱导表达实验步骤(1)
BMGY的配制(每200 mL)
Yeast extract 2 g
Peptone 4 g
K3PO4(pH 6.0) 20 mL
丙三醇 2 mL
定容至180 mL,121,灭20 min。
使用前再加入
10*YNB 20 mL(10%)(过滤除菌)
生物素0.4 mL(0.02%)(过滤除菌)
加入后,用紫外照射10 min左右再接菌。
BMMY的配制(每200 mL)
Yeast extract 2 g
Peptone 4 g
K3PO4(pH 6.0) 20 mL
定容至180 mL,121,灭20 min。
使用前再加入
10*YNB 20 mL(10%)(过滤除菌)
生物素0.4 mL(0.02%)(过滤除菌)
甲醇0.5%
加入后,用紫外照射10 min左右再接菌。
10*YNB的配制
酵母基础氮源培养基 3.4 g
硫酸铵10 g
双蒸水80 mL
定容至100 mL,过滤除菌(用0.22 μm滤膜过滤)。
毕赤酵母诱导表达
1.配BMGY和BMMY灭菌
2.使用前向BMGY按比例加入生物素、10*YNB,紫外照10 min。
3.接菌于BMGY,250 rpm,28℃摇24 h
4.向BMMY中按比例加入生物素、10*YNB、甲醇,紫外照10
min。
5.将含有菌液的BMGY转移至50 mL离心管,4℃5000 rpm 离心5 min,弃上
清。
6.用BMMY将沉淀菌体重悬,倒回瓶中,250 rpm,28℃,摇3 d。
每24 h补
一次甲醇。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
版权声明:本站几乎所有资源均搜集于网络,仅供学习参考,不得进行任何商业用途,否则产生的一切后 果将由使用者本人承担! 本站仅仅提供一个观摩学习与交流的平台, 将不保证所提供资源的完 整性,也不对任何资源负法律责任。
所有资源请在下载后 24 小时内删除。
如果您觉得满意, 请购买正版,以便更好支持您所喜欢的软件或书籍!☆☆☆☆☆生物秀[]☆☆☆☆☆中国生物科学论坛[/bbs/]☆☆☆☆☆生物秀下载频道[/Soft/]生物秀——倾力打造最大最专业的生物资源下载平台!■■■ 选择生物秀,我秀我精彩!!■■■欢迎到生物秀论坛(中国生物科学论坛)的相关资源、软件版块参与讨论,共享您的资源,获 取更多资源或帮助。
毕赤酵母多拷贝表达载体试剂盒用于在含多拷贝基因的毕赤酵母菌中表达并分离重组蛋白综述:基本特征:作为真核生物,毕赤酵母具有高等真核表达系统的许多优点:如蛋白加工、折叠、翻译后修饰等。
不仅如此,操作时与E.coli及酿酒酵母同样简单。
它比杆状病毒或哺乳动物组织培养等其它真核表达系统更快捷、简单、廉价,且表达水平更高。
同为酵母,毕赤酵母具有与酿酒酵母相似的分子及遗传操作优点,且它的外源蛋白表达水平是后者的十倍以至百倍。
这些使得毕赤酵母成为非常有用的蛋白表达系统。
与酿酒酵母相似技术:许多技术可以通用:互补转化基因置换基因破坏另外,在酿酒酵母中应用的术语也可用于毕赤酵母。
例如:HIS4基因都编码组氨酸脱氢酶;两者中基因产物有交叉互补;酿酒酵母中的一些野生型基因与毕赤酵母中的突变基因相互补,如HIS4、LEU2、ARG4、TR11、URA3等基因在毕赤酵母中都有各自相互补的突变基因。
毕赤酵母是甲醇营养型酵母:毕赤酵母是甲醇营养型酵母,可利用甲醇作为其唯一碳源。
甲醇代谢的第一步是:醇氧化酶利用氧分子将甲醇氧化为甲醛,还有过氧化氢。
为避免过氧化氢的毒性,甲醛代谢主要在一个特殊的细胞器-过氧化物酶体-里进行,使得有毒的副产物远离细胞其余组分。
由于醇氧化酶与O2的结合率较低,因而毕赤酵母代偿性地产生大量的酶。
而调控产生醇过氧化物酶的启动子也正是驱动外源基因在毕赤酵母中表达的启动子。
两种醇氧化酶蛋白:毕赤酵母中有两个基因编码醇氧化酶-AOX1及AOX2。
细胞中大多数的醇氧化酶是AOX1基因产物。
甲醇可紧密调节、诱导AOX1基因的高水平表达,较典型的是占可溶性蛋白的30%以上。
AOX1基因已被分离,含AOX1启动子的质粒可用来促进编码外源蛋白的目的基因的表达。
AOX2基因与AOX1基因有97%的同源性,但在甲醇中带AOX2基因的菌株比带AOX1基因菌株慢得多,通过这种甲醇利用缓慢表型可分离Muts菌株。
表达:AOX1基因的表达在转录水平受调控。
在甲醇中生长的细胞大约有5%的polyA+ RNA 来自AOX1基因。
AOX1基因调控分两步:抑制/去抑制机制加诱导机制。
简单来说,在含葡萄糖的培养基中,即使加入诱导物甲醇转录仍受抑制。
为此,用甲醇进行优化诱导时,推荐在甘油培养基中培养。
注意即使在甘油中生长(去抑制)时,仍不足以使AOX1基因达到最低水平的表达,诱导物甲醇是AOX1基因可辨表达水平所必需的。
AOX1突变表型:缺失AOX1基因,会丧失大部分的醇氧化酶活性,产生一种表型为Muts的突变株(methanol utilization slow),过去称为Mut,而Muts可更精确地描述突变子的表型。
结果细胞代谢甲醇的能力下降,因而在甲醇培养基中生长缓慢。
Mut+(methanol utilization plus)指利用甲醇为唯一碳源的野生型菌株。
这两种表型用来检测外源基因在毕赤酵母转化子中的整合方式。
蛋白胞内及分泌表达:外源蛋白可在毕赤酵母胞内表达或分泌至胞外。
分泌表达需要蛋白上的信号肽序列,将外源蛋白靶向分泌通路。
几种不同的分泌信号序列已被成功应用,包括几种外源蛋白本身分制作者:陈苗商汉桥第 2 页,共 36 页泌信号序列,利用酿酒酵母α因子前原肽信号序列也获得许多成功。
分泌表达外源蛋白的最大优点是:毕赤酵母只分泌很少的自身蛋白,加上毕赤酵母最小生长培养基中只有少量的蛋白,这意味着分泌的外源蛋白是培养基中蛋白的主要组成成份,也可算作蛋白纯化的第一步。
注意,如果外源蛋白一级结构中有可识别的糖基化位点(Asn-X-Ser/Thr),则这些位点可能发生糖基化。
翻译后修饰:与酿酒酵母相比,毕赤酵母在分泌蛋白的糖基化方面有优势,因为不会使其过糖基化。
酿酒酵母与毕赤酵母大多数为N-连接糖基化高甘露糖型,然而毕赤酵母中蛋白转录后所增加的寡糖链长度(平均每个支链8-14个甘露糖残基)比酿酒酵母中的(50-150个甘露糖残基)短得多。
另外,酿酒酵母核心寡糖有末端α-1,3聚糖连接头,而毕赤酵母则没有。
一般认为酿酒酵母中糖基化蛋白的α-1,3聚糖接头与蛋白的超抗原性有关,使得这些蛋白不适于治疗应用。
虽然未经证明,但这对毕赤酵母产生的糖蛋白不构成问题,因为毕赤酵母表达蛋白与高级真核生物糖蛋白结构相似。
选择载体用于基因多拷贝整合:在某些情况下,毕赤酵母中重组基因多拷贝整合可增加所需蛋白的表达量。
该试剂盒中的三个载体均可用于在体内(pPIC3.5K, pPIC9K)或体外(pAO815)产生并分离多拷贝插入,同时可检测增加重组基因的拷贝数是否增加蛋白表达量。
体内整合可通过高遗传霉素抗性,筛选可能的多拷贝插入;而体外整合可通过连接产生外源基因的串联插入。
pPIC3.5K, pAO815用于胞内表达,而pPIC9K用于分泌表达,所有载体均利用AOX1启动子来诱导高水平表达。
多拷贝插入频率:毕赤酵母His+转化子高拷贝整合事件自发发生的概率为1-10%,体内方法可筛选可能插入多拷贝外源基因的His+转化子,体外方法可通过连接构建多拷贝子。
当选择His+转化子时,它们中插入体外构建结构多聚体的概率很高。
体内多拷贝插入的产生:Ppic3.5k及Ppic9k含有细菌kan基因,赋予毕赤酵母遗传霉素抗性,注意Kan并不赋予毕赤酵母卡那霉素抗性。
遗传霉素抗性水平主要依赖整合的kan基因的数目。
单拷贝Ppic3.5k或Ppic9k整合入毕赤酵母基因组后,赋予毕赤酵母约0.25mg/ml的遗传霉素抗性水平。
任何载体多拷贝整合可增加遗传霉素抗性水平,从0.5mg/ml(1-2拷贝)到4mg/ml(7-12拷贝)。
由于kan基因与表达盒(pAOX1及目的基因)之间有遗传连锁,可从遗传霉素高抗性推断该克隆所包含多拷贝目的基因数。
由于基因的剂量效益,蛋白的表达可能会增加。
因此,kan基因可检测转化子是否含有多拷贝目的基因。
下图显示多拷贝插入及kan基因与表达盒的连锁。
制作者:陈苗商汉桥第 3 页,共 36 页制作者:陈苗 商汉桥遗传霉素直接选择:在酵母中对遗传霉素抗性进行直接选择并不十分有效,因为新转化的细胞需要时间表达足够量的抗性因子。
由于酵母生长比细菌慢得多,大部分重组酵母在积累足够多的抗性因子以抵抗平板上抗生素之前就已经被杀死了。
最有效的筛选遗传霉素抗性及高抗性克隆的程序需要先对HIS +转化子进行选择,再进行不同水平遗传霉素抗性筛选。
虽然可以用电泳进行直接筛选,但用在遗传霉素筛选之后再进行电泳筛选,获得含高拷贝克隆的机会更大,大约可获得5-9拷贝的克隆,而直接电泳选择只能获得平均为1-3拷贝的克隆。
原生质转化时不能用遗传霉素直接选择。
体外多拷贝插入的产生:下图显示如何产生多表达盒插入载体以转化毕赤酵母。
目的基因插入独个EcoRI 位点 后,产生的表达盒(pAOX1及目的基因)上下游侧翼分别为独个的BglII 及BamHI 位点。
含目的基因的pAOX815用BglII 及BamHI 消化以分离表达盒,表达盒再插入BamHI位点以产生串联重复表达盒,重复该插入程序可产生一系列含单个HIS4基因及逐渐增加数目表达盒的载体。
用体外形成的多拷贝子转化毕赤酵母增加了多拷贝表达盒重组子出现的频率,可设计包含一特定数目多拷贝插入的毕赤酵母重组子。
转化及整合:可产生两个不同表型的His+重组菌株:质粒DNA 线性化位置不同,转化GS115后可产生两种转化子His+Mut+及His+Muts 。
KM71只产生His+Muts ,因为该菌株为Muts 表型。
两种重组子Mut +及 Muts 都是有用的,因为一个表型可能比另一个表型更有利于蛋白表达。
理想条件下,每一个表型应该检测6-10个重组子。
没有办法预测哪个结构或克隆更利于蛋白表达。
强烈推荐用PCR 分析重组子来证实整合情况。
成功将基因构建至AOX1启动子下游后,线性化质粒转化毕赤酵母时激发重组。
下图显示用不同酶消化时产生何种重组子。
限制酶插入事件 GS115表型 KM71表型 SalI 或StuI插入his4 His+Mut+ His+Muts SacI插入5’AOX1 His+Mut+ His+Muts BglII 取代AOX1 His+Muts His+Muts (不推荐)第 4 页,共 36 页表达及扩大培养:用PCR证实毕赤酵母重组后,可检测His+Mut+及His+Muts的表达。
小规模培养每个重组子,用甲醇诱导,检测时间点.如果是胞内表达,每个时间点细胞沉淀用SDS-PAGE分析;如果是分泌表达,分析每个时间点的细胞及上清。
如果有蛋白的抗体,推荐既用考马斯亮蓝染色又用western blot分析SDS-PAGE凝胶。
如果可以,建议检测蛋白活性。
因为并不是所有蛋白都能达到g/l的水平,所以建议进行western blot或活性分析,不要仅做SDS-PAGE 考马斯亮蓝染色分析。
如何选择最佳的表达蛋白毕赤酵母菌株及优化诱导见P49-50。
如表达已达最优,大规模表达以产生更多蛋白。
制作者:陈苗商汉桥第 5 页,共 36 页方法毕赤菌株表型:毕赤酵母菌GS115及KM71在组氨酸脱氢酶位点(His4)有突变,因而不能合成组氨酸,所有表达质粒都有HIS4基因可与宿主进行互补,通过不含组氨酸的培养基来选择转化子。
GS115及KM71自发回复突变到His+原养生物机率小于1/108。
KM71的亲本菌在精氨酸琥珀酸裂解酶基因(arg4)有突变,在不含精氨酸的培养基中不能生长。
用野生型ARG4基因破坏AOX1基因后,产生KM71 MutsArg+His-菌株。
GS115及KM71都可在复合培养基如YPD(YEPD)及含组氨酸的最小培养基中生长。
转化之前,GS115及KM71都不能在最小培养基中生长,因为它们是His-。
KM71结构:ARG4基因(约2kb)插入到克隆的野生型AOX1基因的BamHI(AOX1基因15/16密码子)及SalI(AOX1基因227/228密码子)位点。
