大学分子生物学经典双语课件
合集下载
大学分子生物学经典双语课件C4-Gene_mutation_and_exchange资料

Meslson-Radding Model
2 Site-specific recombination
This involves the exchange of nonhomologous
but specific pieces of DNA and is mediated by
proteins (enzyme) that recognize specific DNA
or more genes conferring antibiotic resistance.
4.1.1.3 TnA family
TnA family is about 5000bp, which is much greater than the insertion sequence. The same as composite transposon, TnA family also carries the gene who is responsible for its own transposition and other gene such as resistance gene β -amine acyl enzyme (AmpR) . It has no IS, but there are terminal repeat sequences of about 37-38bp in the end.
4.4.2 Transposons in eukaryotes
4.4.2.1 Transposons in maize
Autonomous element
Nonautonomous element
(1)Ac-Ds system
11bp Ac
Ds
transposable element 是引起玉米糊粉层花斑不稳定
大学分子生物学经典双语课件C3: DNA replication

Chapter 03: DNA Replication
3.1 The principle of DNA replication 3.2 DNA replication model 3.3 Enzymes and protein needed in DNA replication 3.4 Process of DNA replication 3.5 Telomere and Telomerase
parental duplex is unwound.
On the lagging strand, a stretch of single-stranded
parental DNA must be exposed, and then a
segment is synthesized in the reverse direction (relative to fork movement). A series of these fragments are synthesized, then they are joined together to create an intact lagging strand.
v33enzymesandproteinsneededindnareplicationdna聚合酶dna聚合酶dna聚合酶结构基因polapolbpolc亚基1410相对分子质量1030008800083000053聚合酶活性是是是35外切酶活性校正是是是53外切酶活性是否否聚合速度ntss1620402501000持续合成能力32001500500000功能切除引物修复修复复制表31大肠杆菌dna聚合酶的比较?53exonucleaseactivity
enters newly synthesized DNA in the form of
3.1 The principle of DNA replication 3.2 DNA replication model 3.3 Enzymes and protein needed in DNA replication 3.4 Process of DNA replication 3.5 Telomere and Telomerase
parental duplex is unwound.
On the lagging strand, a stretch of single-stranded
parental DNA must be exposed, and then a
segment is synthesized in the reverse direction (relative to fork movement). A series of these fragments are synthesized, then they are joined together to create an intact lagging strand.
v33enzymesandproteinsneededindnareplicationdna聚合酶dna聚合酶dna聚合酶结构基因polapolbpolc亚基1410相对分子质量1030008800083000053聚合酶活性是是是35外切酶活性校正是是是53外切酶活性是否否聚合速度ntss1620402501000持续合成能力32001500500000功能切除引物修复修复复制表31大肠杆菌dna聚合酶的比较?53exonucleaseactivity
enters newly synthesized DNA in the form of
大学分子生物学经典双语课件

2.1.2.2 Conformation polymorphism of the double helix
Alternative doublehelical structures of DNA
Base Obliquity
helix rise per base pair
bp number per turn
biological activity changed (even lost); viscosity decreased,粘度 solubility decreased,溶解度 Hyperchromicity: the absorbance of ssDNA is greater than that dsDNA.增色 concentration = 50μg/ml: dNTPs A260 = 1.60 S.S DNA A260 = 1.37 D.S DNA A260 = 1.0
2.1.3
Triplex DNA
1953, Watson & Crick proposed D.S DNA model and found many redundant hydrogen bonding donor and receptors along big grooves. 1957, Felsenfeld proposed T.S DNA concept
transferring the other dsDNA through the break.
Type I topoisomerase
Type II topoisomerase
Contents
1
2 3 4 5 6 7
Structure of DNA Denaturation, renaturation and hybridization
分子生物学全套课件96P课件精品文档

分子生物学
Molecular Biology
Presumed that you have learned “Molecular Biology” before your receiving the Bachelor’s Degree of Sciences;
Designed for helping you to become associated with the most researching interests (areas) of our university;
+
lucifCAAAACCCC
luciferase
Light + oxylufiferin
AC G TG
Pyrograph
Recorded by computer
dNTP added in serial
Sequencing by ligation: Applied Biosystems
polony
20 microns
Position overlapped one base one image color order = base adding order = sequence
Reversible terminators: Illumina
Bridge amplification of DNA fragments is randomly distributed across eight channels of a glass slide, to which high-density forward and reverse primers are covalently attached. The solid-phase amplification produces ~80 million molecule clusters (MCs) from individual ssDNA templates. A primer is annealed to the free ends of templates in each MC. The polymerase extends and then terminates DNA synthesis from a set of four reversible terminators (RTs), each labeled with a different dye. Unincorporated RTs are washed away, base identification is performed by four-colour imaging, and blocking and dye groups are removed by chemical cleavage to permit the next cycle. Colour images for a given MC provide reads of ~45 bases. Substitutions are the most common error type.
Molecular Biology
Presumed that you have learned “Molecular Biology” before your receiving the Bachelor’s Degree of Sciences;
Designed for helping you to become associated with the most researching interests (areas) of our university;
+
lucifCAAAACCCC
luciferase
Light + oxylufiferin
AC G TG
Pyrograph
Recorded by computer
dNTP added in serial
Sequencing by ligation: Applied Biosystems
polony
20 microns
Position overlapped one base one image color order = base adding order = sequence
Reversible terminators: Illumina
Bridge amplification of DNA fragments is randomly distributed across eight channels of a glass slide, to which high-density forward and reverse primers are covalently attached. The solid-phase amplification produces ~80 million molecule clusters (MCs) from individual ssDNA templates. A primer is annealed to the free ends of templates in each MC. The polymerase extends and then terminates DNA synthesis from a set of four reversible terminators (RTs), each labeled with a different dye. Unincorporated RTs are washed away, base identification is performed by four-colour imaging, and blocking and dye groups are removed by chemical cleavage to permit the next cycle. Colour images for a given MC provide reads of ~45 bases. Substitutions are the most common error type.
2024年《分子生物学》全册配套完整教学课件pptx

2024/2/29
运输功能
如载体蛋白,血红蛋白等 ,在生物体内运输各种物 质。
免疫功能
如抗体蛋白,参与生物体 的免疫应答。
18
蛋白质的功能与调控
调节功能
如激素,生长因子等,调节生物 体的生长发育和代谢过程。
2024/2/29
储存功能
如植物种子中的贮藏蛋白,动物体 内的肌红蛋白等,储存能量和营养 物质。
个性化医疗
根据患者的基因信息,制定个 性化的治疗方案。
药物基因组学
预测患者对药物的反应和副作 用,指导合理用药。
30
基因治疗的原理与应用
基因治疗的原理
通过导入正常基因或修复缺陷基因, 从而治疗由基因突变引起的疾病。
遗传性疾病的治疗
如视网膜色素变性、腺苷脱氨酶缺乏 症等。
2024/2/29
癌症治疗
利用基因编辑技术,修复或敲除癌症 相关基因,抑制肿瘤生长。
基因表达调控的层次
基因表达调控可分为转录前调控、转录水平调控、转录后调控和翻 译水平调控等多个层次。
基因表达调控的意义
基因表达调控对于生物体的生长发育、代谢、免疫应答等生理过程具 有重要意义,同时也是疾病发生发展的重要因素。
2024/2/29
22
原核生物的基因表达调控
1 2 3
原核生物基因表达调控的特点
26
DNA损伤的修复机制
直接修复
针对某些简单的DNA损伤,如碱 基错配,可通过特定的酶直接进行 修复。
碱基切除修复
通过识别并切除受损碱基,再合成 新的DNA片段进行修复。
2024/2/29
核苷酸切除修复
针对较严重的DNA损伤,如嘧啶 二聚体,通过切除一段包含受损部
运输功能
如载体蛋白,血红蛋白等 ,在生物体内运输各种物 质。
免疫功能
如抗体蛋白,参与生物体 的免疫应答。
18
蛋白质的功能与调控
调节功能
如激素,生长因子等,调节生物 体的生长发育和代谢过程。
2024/2/29
储存功能
如植物种子中的贮藏蛋白,动物体 内的肌红蛋白等,储存能量和营养 物质。
个性化医疗
根据患者的基因信息,制定个 性化的治疗方案。
药物基因组学
预测患者对药物的反应和副作 用,指导合理用药。
30
基因治疗的原理与应用
基因治疗的原理
通过导入正常基因或修复缺陷基因, 从而治疗由基因突变引起的疾病。
遗传性疾病的治疗
如视网膜色素变性、腺苷脱氨酶缺乏 症等。
2024/2/29
癌症治疗
利用基因编辑技术,修复或敲除癌症 相关基因,抑制肿瘤生长。
基因表达调控的层次
基因表达调控可分为转录前调控、转录水平调控、转录后调控和翻 译水平调控等多个层次。
基因表达调控的意义
基因表达调控对于生物体的生长发育、代谢、免疫应答等生理过程具 有重要意义,同时也是疾病发生发展的重要因素。
2024/2/29
22
原核生物的基因表达调控
1 2 3
原核生物基因表达调控的特点
26
DNA损伤的修复机制
直接修复
针对某些简单的DNA损伤,如碱 基错配,可通过特定的酶直接进行 修复。
碱基切除修复
通过识别并切除受损碱基,再合成 新的DNA片段进行修复。
2024/2/29
核苷酸切除修复
针对较严重的DNA损伤,如嘧啶 二聚体,通过切除一段包含受损部
双语分子生物学ppt

主要参考教材
• PC Turner et al. Instant Notes in Molecular Biology (Second edition). BIOS Scientific Publishers Limited, 2000
• 分子生物学(第三版)导读版(精要速览系列) • 朱玉贤,李毅,郑晓峰编著。现代分子生物学(第3版) 高等教育出版
2.2 Prokaryotic and Eukaryotic chromosome structure
2.1 Properties of nucleic acid
2.1.1 Nucleic acid structure 2.1.2 Chemical & physical properties 2.1.3 Spectroscopic (光谱学) & thermal (热力学) properties 2.1.4 DNA supercoiling
– Glycerides (甘油酯), phospholipids(磷脂),sphingolipids (鞘脂,如神经氨酰)
• Complex macromolecules
– Nucleoproteins(核蛋白), ribozyme(核糖体) – Glycoproteins – Proteoglycans (mucoproteins)蛋白多糖(粘蛋白) – Lipid-linked proteins – glycolipids
A, B and Z helices
• Z-DNA:它是左手双螺旋,与右手螺旋的不同是螺距延长 (4.5nm左右),直径变窄(1.8nm),每个螺旋含12个碱基对, 分子长链中磷原子不是平滑延伸而是锯齿形排列,有如“之” 字形一样,因此叫它Z构象,这一构象中的重复单位是二核 苷酸而不是单核苷酸;而且Z-DNA只有一个螺旋沟,它相当 于B构象中的小沟,它狭而深,大沟则不复存在。
分子生物学英文课件:molecular biotechnology
Analyze the recombinant plasmid and bacteriophage - screening DNA library.
(2) Northern blotting
❖ Similar to the southern blotting ❖ RNAs instead of DNAs ❖ No need of RE digestion ❖ Application
produce single strand DNA Transferring: transfer DNA to the NC. Immobilization: heating NC in 80℃ for 1~2 h
or uv crosslink (nylon membrane) Hybridization: NC is exposed to probe (biotin/
高等教育 > 生物学 > 分子生物学英文课件:molecular biotechnology molecularbiotechnology section molecularhybridization blottingtechnique heteroduplexfrom two complementary polynucleotide strands from different sources (dna dna;dna rna;rna molecularhybridization related conceptions/principles dnadenaturation dnarenaturation probedna rnafragment labeled radioisotope,biotin detectspecific nucleic acid sequences hybridization.?dna probe ?rna probe blottingtransfer (blot) biologic macromolecules separated fixthem nitrocellulose/nylonmembrane diffusion,electro-transferring vacuumabsorption, 1975,edwen southern blotting widelyused specificmacro- molecules (proteins, mrnas dnasequences) southernblotting genomic dna (from tissues re,separated gelelectro- phoresis nitrocellulosemembrane detectingspecific dna sequence labeledprobe. used qualitativelyanalyze genomic dna, recombinant plasmid, screening dna library. steps extraction:genomic dna from tissues digestion:cut separation:separate dna fragments denaturation:dna fragments treated p
(2) Northern blotting
❖ Similar to the southern blotting ❖ RNAs instead of DNAs ❖ No need of RE digestion ❖ Application
produce single strand DNA Transferring: transfer DNA to the NC. Immobilization: heating NC in 80℃ for 1~2 h
or uv crosslink (nylon membrane) Hybridization: NC is exposed to probe (biotin/
高等教育 > 生物学 > 分子生物学英文课件:molecular biotechnology molecularbiotechnology section molecularhybridization blottingtechnique heteroduplexfrom two complementary polynucleotide strands from different sources (dna dna;dna rna;rna molecularhybridization related conceptions/principles dnadenaturation dnarenaturation probedna rnafragment labeled radioisotope,biotin detectspecific nucleic acid sequences hybridization.?dna probe ?rna probe blottingtransfer (blot) biologic macromolecules separated fixthem nitrocellulose/nylonmembrane diffusion,electro-transferring vacuumabsorption, 1975,edwen southern blotting widelyused specificmacro- molecules (proteins, mrnas dnasequences) southernblotting genomic dna (from tissues re,separated gelelectro- phoresis nitrocellulosemembrane detectingspecific dna sequence labeledprobe. used qualitativelyanalyze genomic dna, recombinant plasmid, screening dna library. steps extraction:genomic dna from tissues digestion:cut separation:separate dna fragments denaturation:dna fragments treated p
英汉对照分子生物学导论课件Sample

Vocabulary of Day 3 (4/4)
anti-parallel base-stacking major groove minor groove
nanometer denature
denaturation absorbance
absorb adsorb
反向平行的 碱基堆积 (DNA)大沟 (DNA)小沟 纳米 变性(动词) 变性(名词) 吸收(名词) 吸收(动词) 吸附
1) Nitrogenous base / 含氮碱基
2) Sugar / 糖
No oxygen here !
Ribonucleotides and deoxyribonucleotides 核糖核苷酸 与 脱氧核糖核苷酸
3) Triphosphate / 三磷酸
NH2 65 1N
7 N
8
O
O
2.4 DNA in the Cell
2.4 细胞中的DNA
2.5 RNA (Ribonucleic Acid)
2.5 RNA(核糖核酸)
2.6 Experiments
2.6 实验研究
Vocabulary of Day 3 (1/4)
nucleic acid genetic material
inherit nucleotide nitrogenous base triphosphate
1.1
Tm
1.0 65 70 75 80 85 90 95
Temperature
Light absorbance by DNA
DNA对光的吸收
dsDNA
ssDNA
2.1 Properties of a Genetic Material
分子生物学英文课件:Eukaryote gene and genome
Exon: segments of eukaryotic gene (or of the primary transcript of that gene) that are saved as part of a functional, mature mRNA, rRNA or tRNA molecules. Intron: segments of eukaryotic gene (or of the primary transcript of that gene) that are removed by splicing during RNA processing and are not included in the mature, functional mRNA, rRNA, or tRNA.
Repetitive sequences
高度重复序列(highly repetitive sequence) 中度重复序列(moderately repetitive sequence) 单拷贝序列(single copy sequence)或低度重复序列
(一)高度重复序列(highly repetitive sequence)
➢ Have unique structure elements
基因 -N
转录起点
+1
结构基因区
调控区
mRNA 5’
AUG
翻译起点
蛋白质 N
转录 翻译
转录终点
+N
UAA
3’
翻译终点
C
Gene expression: DNA→mRNA→Protein DNA →RNA
mRNA
mRNA
转录
tRNA
rRNA
Repetitive sequences
高度重复序列(highly repetitive sequence) 中度重复序列(moderately repetitive sequence) 单拷贝序列(single copy sequence)或低度重复序列
(一)高度重复序列(highly repetitive sequence)
➢ Have unique structure elements
基因 -N
转录起点
+1
结构基因区
调控区
mRNA 5’
AUG
翻译起点
蛋白质 N
转录 翻译
转录终点
+N
UAA
3’
翻译终点
C
Gene expression: DNA→mRNA→Protein DNA →RNA
mRNA
mRNA
转录
tRNA
rRNA
分子生物学英文课件:genetic engineering

Recombinant DNA: is a form of artificial DNA that is engineered through the combination or insertion of one or more DNA strands.
DNA cloning involves separating a specific gene or segment of DNA from its larger chromosome and attaching it to a small molecule of carrier DNA, then replicating this modified DNA thousands of times. The result is a selective amplification of that particular gene or DNA segment.
Structure of pBR322: ---A common cloning vector derived from a naturally occurring plasmid ---Having antibiotic resistance genes for selection of transformants containing the plasmid ---Having unique restriction enzyme cleavage sites for insertion of foreign DNA ---Having origin of DNA replication (ori) for propagation in E. coli
Vector
Plasmid Bacteriophage
DNA cloning involves separating a specific gene or segment of DNA from its larger chromosome and attaching it to a small molecule of carrier DNA, then replicating this modified DNA thousands of times. The result is a selective amplification of that particular gene or DNA segment.
Structure of pBR322: ---A common cloning vector derived from a naturally occurring plasmid ---Having antibiotic resistance genes for selection of transformants containing the plasmid ---Having unique restriction enzyme cleavage sites for insertion of foreign DNA ---Having origin of DNA replication (ori) for propagation in E. coli
Vector
Plasmid Bacteriophage
分子生物学英文课件:CHAPTER 14 Translation

Accuracy of Aminoacylation
Correct AA
Correct tRNA
Aminoacyl-tRNA synthetases
Each of the 20 amino acids is attached to the appropriate tRNA (s) by aminoacyl-tRNA synthetases.
mRNA
Only a portion of each mRNA can be translated. The protein-coding region of the mRNA consists
of an ordered series of 3-nt-long units called codons that specify the order of amino acids.
D-arm
Composed of 3 or 4 bp stem and a loop called the Dloop (DHU-loop) usually containing the modified base dihydrouracil.
(二氢尿嘧啶)
Anticodon arm:
Consisting of a 5 bp stem and a 7 residues loop .
The recognition has to ensure two levels of accuracy: (1) each tRNA synthetase must recognize the correct set of tRNAs for a particular amino acids; (2) each synthetase must charge all of these isoaccepting tRNAs
分子生物学课件(共51张PPT)(2024)

四级结构
由两条或两条以上的多肽链组 成的一类结构,每一条多肽链
都有完整的三级结构。
21
蛋白质的功能与分类
结构蛋白:作为细胞的结构,如膜蛋白,染色体蛋白等 。 酶:催化生物体内的化学反应。
抗体:参与免疫应答。
2024/1/29
功能蛋白
激素:调节生物体的生理活动。
蛋白质的分类还可以根据其溶解度、形状等进行划分。 例如,根据溶解度可分为清蛋白、球蛋白等;根据形状 可分为纤维状蛋白和球状蛋白等。
RNA的基本组成单位是核糖核苷酸, 由磷酸、核糖和碱基组成。
磷酸二酯键
核糖核苷酸之间通过磷酸二酯键连接 形成RNA链。
碱基
RNA中的碱基主要有腺嘌呤(A)、 鸟嘌呤(G)、胞嘧啶(C)和尿嘧啶 (U)。
2024/1/29
12
RNA的种类与结构
mRNA
信使RNA,负责携带遗 传信息并指导蛋白质合
成。
翻译水平调控
通过控制翻译的起始、延伸和 终止来调控基因表达。
蛋白质水平调控
通过控制蛋白质的活性、稳定 性和相互作用来调控基因表达
。
表观遗传学调控
通过改变染色质结构和DNA 甲基化等方式来调控基因表达
。
2024/1/29
18
05
蛋白质的结构与功能
2024/1/29
19
蛋白质的分子组成
氨基酸
蛋白质的基本组成单元,共有20 种标准氨基酸。
2024/1/29
tRNA
转运RNA,负责携带氨 基酸并识别mRNA上的
遗传密码。
rRNA
其他RNA
核糖体RNA,是核糖体 的组成部分,参与蛋白
质合成。
13
如miRNA、snRNA等, 在基因表达调控等方面
由两条或两条以上的多肽链组 成的一类结构,每一条多肽链
都有完整的三级结构。
21
蛋白质的功能与分类
结构蛋白:作为细胞的结构,如膜蛋白,染色体蛋白等 。 酶:催化生物体内的化学反应。
抗体:参与免疫应答。
2024/1/29
功能蛋白
激素:调节生物体的生理活动。
蛋白质的分类还可以根据其溶解度、形状等进行划分。 例如,根据溶解度可分为清蛋白、球蛋白等;根据形状 可分为纤维状蛋白和球状蛋白等。
RNA的基本组成单位是核糖核苷酸, 由磷酸、核糖和碱基组成。
磷酸二酯键
核糖核苷酸之间通过磷酸二酯键连接 形成RNA链。
碱基
RNA中的碱基主要有腺嘌呤(A)、 鸟嘌呤(G)、胞嘧啶(C)和尿嘧啶 (U)。
2024/1/29
12
RNA的种类与结构
mRNA
信使RNA,负责携带遗 传信息并指导蛋白质合
成。
翻译水平调控
通过控制翻译的起始、延伸和 终止来调控基因表达。
蛋白质水平调控
通过控制蛋白质的活性、稳定 性和相互作用来调控基因表达
。
表观遗传学调控
通过改变染色质结构和DNA 甲基化等方式来调控基因表达
。
2024/1/29
18
05
蛋白质的结构与功能
2024/1/29
19
蛋白质的分子组成
氨基酸
蛋白质的基本组成单元,共有20 种标准氨基酸。
2024/1/29
tRNA
转运RNA,负责携带氨 基酸并识别mRNA上的
遗传密码。
rRNA
其他RNA
核糖体RNA,是核糖体 的组成部分,参与蛋白
质合成。
13
如miRNA、snRNA等, 在基因表达调控等方面
分子生物学英文课件:lecture1 Welcome to Molecular Biology(1)

1. Carefully read each Part Introduction to find
教师介绍
the connections between different chapters of
课程介绍 课程内容 知识回顾 小结
each Part, and how the contents are organized. 2. Find the organization of each chapter by
reading the Chapter Introduction. 3. Read the figures to understand the concept in
each subtitle.
Main principles: from large view to details, from
easy to difficult.
have learnt (understood and mastered), any progress? • 5. Improve your study through discussion and quiz.
• 6. Don’t be pretentious when you learn well and don’t be disappointed, either.
Robert F. Weaver. Molecular Biology, (5th Edition) 现代分子生物学(第三版)(作者: 朱玉贤 李毅 郑晓峰)高等教 育出版社
大连理工大学
Molecular 教材-Molecular Biology of the Gene, 5/E
Biology
1 导言
几丁质合酶 几丁质内切酶及几丁质外切酶 几丁质去乙酰化酶
分子生物学双语

Molecular Biology 分子生物学
分子生物学双语
Chapter 1 Welcome to Molecular Biology
➢ What is Molecular Biology? ➢ Why should we learn? ➢ How should we learn?
分子生物学双语
分子生物学双语
普通生物学
分类单位: 界门纲目科属种
分子生物学双语
微生物学
分子生物学双语
细胞生物学
分子生物学双语
生物化学
分子生物学双语
分子生物学的延伸
分子生物学
分子结构生物学 分子发育生物学 分子神经生物学 分子育种学 分子肿瘤学
分子细胞生物学 分子免疫学 分子病毒学 分子生理学 分子考古学
分子生物学双语
3)分子生物学的主要任务:
– 阐明这些生物大分子复杂的结构及结构与功能的
关系。 – 揭示生命的本质的分子机理
• Examples: Flash1
– Human immunodeficiency virus (HIV) is a lentivirus慢病毒(a member of the retrovirus 逆 转录病毒 family) that causes acquired immunodeficiency syndrome (AIDS)
分子生物学双语
4. History of Molecular Biology 分子生物学发展简史
分为三个主要阶段:
• 对DNA和遗传信息传递的认识阶段
– 确立DNA为生物遗传物质的地位
• 重组DNA技术的建立和发展阶段 • 重组DNA技术的应用和分子生物学的迅猛发展
阶段
分子生物学双语
Chapter 1 Welcome to Molecular Biology
➢ What is Molecular Biology? ➢ Why should we learn? ➢ How should we learn?
分子生物学双语
分子生物学双语
普通生物学
分类单位: 界门纲目科属种
分子生物学双语
微生物学
分子生物学双语
细胞生物学
分子生物学双语
生物化学
分子生物学双语
分子生物学的延伸
分子生物学
分子结构生物学 分子发育生物学 分子神经生物学 分子育种学 分子肿瘤学
分子细胞生物学 分子免疫学 分子病毒学 分子生理学 分子考古学
分子生物学双语
3)分子生物学的主要任务:
– 阐明这些生物大分子复杂的结构及结构与功能的
关系。 – 揭示生命的本质的分子机理
• Examples: Flash1
– Human immunodeficiency virus (HIV) is a lentivirus慢病毒(a member of the retrovirus 逆 转录病毒 family) that causes acquired immunodeficiency syndrome (AIDS)
分子生物学双语
4. History of Molecular Biology 分子生物学发展简史
分为三个主要阶段:
• 对DNA和遗传信息传递的认识阶段
– 确立DNA为生物遗传物质的地位
• 重组DNA技术的建立和发展阶段 • 重组DNA技术的应用和分子生物学的迅猛发展
阶段
分子生物学英文课件:Chapter12 Regulation of metabolism

1. Heart (aerobic oxidation) The heart uses a variety of fuels: fatty acids, ketone
bodies, lactate, glucose. 2. Brain Under normal conditions the brain only uses blood glucose. During fasting uses ketone bodies. 3. Muscle Glycogenesis and stores Phosphocreatin, Glycogenolysis, FA oxidation to provide energy lacks glucose –6-phosphatase, cannot release free Glc
Satiation → Insulin↑ glucagon↓ (1) Glucose supply energy; (2) Glucose → Glycogen (liver, muscle); (3) Glucose →TG (VLDL)
2. Fasting
(1) After eating 6~8h, Liver glycogenolysis↑
6. Kidney Gluconeogenesis
SectionⅤ. Regulation of metabolism Metabolic regulation at different levels:
at cell level (original) at hormone level at entirety level
metabolic character 4. Common metabolic pool 5. ATP is the common form for energy storage
bodies, lactate, glucose. 2. Brain Under normal conditions the brain only uses blood glucose. During fasting uses ketone bodies. 3. Muscle Glycogenesis and stores Phosphocreatin, Glycogenolysis, FA oxidation to provide energy lacks glucose –6-phosphatase, cannot release free Glc
Satiation → Insulin↑ glucagon↓ (1) Glucose supply energy; (2) Glucose → Glycogen (liver, muscle); (3) Glucose →TG (VLDL)
2. Fasting
(1) After eating 6~8h, Liver glycogenolysis↑
6. Kidney Gluconeogenesis
SectionⅤ. Regulation of metabolism Metabolic regulation at different levels:
at cell level (original) at hormone level at entirety level
metabolic character 4. Common metabolic pool 5. ATP is the common form for energy storage
分子生物学英文课件

(And some phenotypes are determined by more than one gene) (The luck of Mendel’s discovery: a simple trait)
Figure 1-2 The inheritance of flower color in the snapdragon (金鱼草).
Chapter 2: Nucleic Acids convey genetic information
The stories/efforts led to the fundamental knowledge of life, which open a new era of modern biology.
By the 1930s, geneticists began speculating as to what sort of molecules could have the kind of stability that the gene demanded, yet be capable of permanent, sudden change to the mutant forms that must provide the basis of evolution... It was generally assumed that genes would be composed of amino acids because, at that time, they appeared to be the only biomolecules with sufficient complexity to convey genetic information. This hypothesis is eventually dead from its shaking base and attractive complexity.
Figure 1-2 The inheritance of flower color in the snapdragon (金鱼草).
Chapter 2: Nucleic Acids convey genetic information
The stories/efforts led to the fundamental knowledge of life, which open a new era of modern biology.
By the 1930s, geneticists began speculating as to what sort of molecules could have the kind of stability that the gene demanded, yet be capable of permanent, sudden change to the mutant forms that must provide the basis of evolution... It was generally assumed that genes would be composed of amino acids because, at that time, they appeared to be the only biomolecules with sufficient complexity to convey genetic information. This hypothesis is eventually dead from its shaking base and attractive complexity.
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Positive supercoils
Relaxed coils Negative supercoils
Topoisomerases
•Topoisomerases: exist in cell to regulate the level
of supercoiling of DNA molecules
2.1.4
Tetraplex DNA
1958, Poly(G) X-ray photograph Ring structure of hydrogen Tetrable helix DNA
Formation condition: polyG, 4(dG)
DNA Sculpture at Disneyland
biological activity changed (even lost); viscosity decreased,粘度 solubility decreased,溶解度 Hyperchromicity: the absorbance of ssDNA is greater than that dsDNA.增色 concentration = 50μg/ml: dNTPs A260 = 1.60 S.S DNA A260 = 1.37 D.S DNA A260 = 1.0
polyA/polyU
polydA/polydT
polyd(AG)/polyd(CT)
1983, Mirkin S.M. found plasmid T.S DNA in pH=4.3 solution
Major 1963 groove Hoogsteen
Trible helix
Py:Pu:Py
Triple Helix DNA
C0t曲线
2.2.2 hybridization
• Definition:the renaturation of regions of complementarity between different nucleic acid strands(DNA or RNA) • Characteristic:sensitive、 specific
transferring the other dsDNA through the break.
Type I topoisomerase
Type II topoisomerase
Contents
1
2 3 4 5 6 7
Structure of DNA Denaturation, renaturation and hybridization
2.1.2.2 Conformation polymorphism of the double helix
Alternative doublehelical structures of DNA
Base Obliquity
helix rise per base pair
bp number per turn
DNA 复性过程遵循二级反应动力学 DNA复性过程中单链消失的速度用公式表示:
-dC/dt=kC2
C/C0=1/(1+kC0t)
其中,C是单位时间的单链DNA的浓度 C0为开始反应时变性解链的单链DNA浓度, t为复性时间 K是复性速度常数(L/mol· s),k取决于阳 离子浓度、温度、pH值、DNA片段大小。
2.1.3
Triplex DNA
1953, Watson & Crick proposed D.S DNA model and found many redundant hydrogen bonding donor and receptors along big grooves. 1957, Felsenfeld proposed T.S DNA concept
Writhing number(W) 扭曲数
L=T+W
Positive supercoils
Negative supercoils
DNA isolated from cell negatively supercoiled by ~5 turns per 100 turns of the helix. Lk / Lk = -0.05
2.1.2
secondary structure of DNA
----DNA double helix
♬ Experimental basis
X~ray photograph of DNA with high quality: DNA specimens from different species have the same results(constant width; 3.4nm); Chargaff rules:the rule of the composition of DNA Physical chemistry studies and acid and alkali titrate studies on DNA base ;
Denaturation factor
pH(>11.3或<5.0) Chemical denaturation (urea、methanal 甲醛) Thermal denaturation Low ion strength低离子强度
Characters of denatured DNA
Genome
Genetic information flow
2.1 Structure of DNA
2.1.1
primary structure of DNA Definition:the nucletide residue sequence of the polynucleotide chain; Linkage:3’,5’-phosphodiester bond; Backbone:phosphate + pentose; Direction: 5’ →3’ ;
Concept of gene Gene cluster and repetitive sequence Chromosome and nucleosome
Genome
Genetic information flow
2.2 Denaturation, renaturation and hybridization
•Type I topoisomerase: break one strand of the DNA , and change the linking number in steps of ±1 by passing the other strand through the break. •Type II topoisomerase: break both strands of the DNA , and change the linking number in steps of ±2 by
2.1.2.1 Stable factors of the double helix
• Base-stacking interaction(hydrophobic effect, the major factor); • Hydrogen bond between complementary base pairs; • electrovalent bond(between the negative charges carried on the phosphate groups and the positive charges carried on the proteins or metal ions)
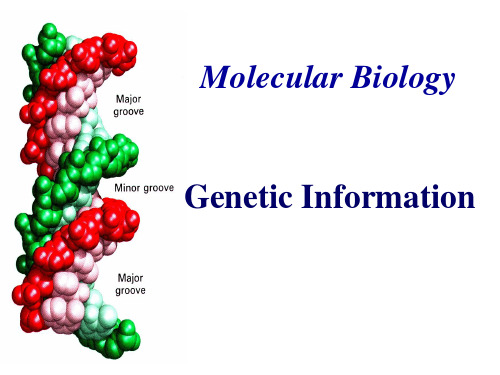
The bases lie on the inside,the sugarphosphate backbone is on the outside; The bases are flat structure, lying in pairs perpendicular to the axis
The diameter of the double helix is 2nm; There is a complete turn every 3.4nm, with 10bp per turn.
Chapter 2 Gene and Chromosome
Contents
1
2 3 4 5 6 7
Structure of DNA Denaturation, renaturation and hybridization
Concept of gene Gene cluster and repetitive sequence Chromosome and nucleosome
melting curve and Tm
• Increased temperature can bring about DNA denaturation; • Tm (melting temperature): Temperature when 50% DNA denaturation • Tm is a characteristic constant of DNA
charged PO4 on the outside
--
Hydrophobic bases inside
Pitch length
10.5 bp /turn
11Å 20Å
Fig 8-15
5’ 3’
♬ key notes of DNA double helix
Two polynucleotide chains in a DNA double helix; Along the same axis,two chains are wound around each other, resulting in a right-handed double helix; Forms a major groove and a minor groove
