基于人眼评估的灰喜鹊卵色的窝内变_省略_模拟寄生卵的拒绝行为无关_英文_Jes_
新高考新教材2025届高考生物二轮总复习专题突破练10生物与环境
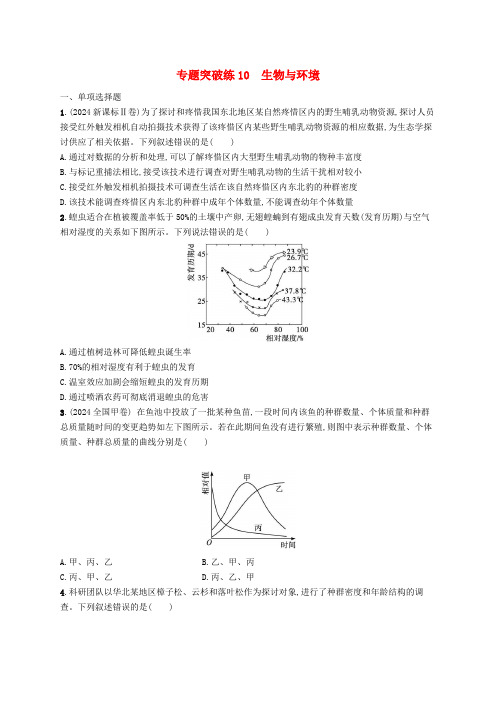
专题突破练10 生物与环境一、单项选择题1.(2024新课标Ⅱ卷)为了探讨和疼惜我国东北地区某自然疼惜区内的野生哺乳动物资源,探讨人员接受红外触发相机自动拍摄技术获得了该疼惜区内某些野生哺乳动物资源的相应数据,为生态学探讨供应了相关依据。
下列叙述错误的是( )A.通过对数据的分析和处理,可以了解疼惜区内大型野生哺乳动物的物种丰富度B.与标记重捕法相比,接受该技术进行调查对野生哺乳动物的生活干扰相对较小C.接受红外触发相机拍摄技术可调查生活在该自然疼惜区内东北豹的种群密度D.该技术能调查疼惜区内东北豹种群中成年个体数量,不能调查幼年个体数量2.蝗虫适合在植被覆盖率低于50%的土壤中产卵,无翅蝗蝻到有翅成虫发育天数(发育历期)与空气相对湿度的关系如下图所示。
下列说法错误的是( )A.通过植树造林可降低蝗虫诞生率B.70%的相对湿度有利于蝗虫的发育C.温室效应加剧会缩短蝗虫的发育历期D.通过喷洒农药可彻底消退蝗虫的危害3.(2024全国甲卷) 在鱼池中投放了一批某种鱼苗,一段时间内该鱼的种群数量、个体质量和种群总质量随时间的变更趋势如左下图所示。
若在此期间鱼没有进行繁殖,则图中表示种群数量、个体质量、种群总质量的曲线分别是( )A.甲、丙、乙B.乙、甲、丙C.丙、甲、乙D.丙、乙、甲4.科研团队以华北某地区樟子松、云杉和落叶松作为探讨对象,进行了种群密度和年龄结构的调查。
下列叙述错误的是( )树种樟子松12 58 210云杉8 46 208落叶松460 464 568A.该群落中樟子松、落叶松种群的年龄结构分别属于增长型、稳定型B.落叶松是该群落中的优势种,其种群密度为 1 492 株/hm2C.表中数据是多个样方调查数据的平均值,要求每个样方的非生物因素基本相同D.针叶树和阔叶树所占比例随海拔上升呈现不同变更,体现了群落的水平结构5.化感作用指植物通过分泌化学物质对其他植物产生影响。
现利用入侵植物薇甘菊及本地物种葛藤和鸡矢藤的叶片制备叶片水提取液(供体),然后用制备的水提取液分别处理三种植物幼苗(受体),然后计算化感作用效应指数R(R=处理组干重/空白比照组干重-1)。
中国鸟类的种内巢寄生 基于超常窝卵数的证据 贡国鸿 卢 欣

动物学报 49(6):851~853,2003A cta Zoologica S i nica Received J une 26,2003;revised Sep.04,2003 3This research was funded by a grant from National Natural Science Foundation of China (No.39800016,30170156)33Corresponding author. E 2mail :luxinwh @ B rief introduction to the f irst author Male.Research interests :avian ecology. ν2003动物学报Acta Zoologica Si nicaIntraspecif ic nest parasitism among birds in China ,evidence basedm ainly on abnorm ally large clutches 3GON G Guo 2Hong L U Xin 33(College of L if e Sciences ,W uhan U niversity ,W uhan 430072,Chi na )中国鸟类的种内巢寄生:基于超常窝卵数的证据3贡国鸿 卢 欣33(武汉大学生命科学学院,武汉 430072)摘 要 鸟类种内巢寄生(Intraspecific nest parasitism ,INP )是近年来行为生态学和进化生物学理论遇到的新问题。
自1980年以来,国外学者就开始搜集总结具有INP 的鸟种,迄今的列表已达236种。
然而,由于语言的障碍,中国鸟类学者所发现的例子未能归入列表中。
本文以超常窝卵数为标准,查阅所发表的中文鸟类学文献,发现18种发生于中国的鸟类具有INP ,其中14种未被国际学者收录。
2023年《医学科研诚信与医学研究伦理》考试题库

2023年《医学科研诚信与医学研究伦理》考试题库()对自然科学和哲学社会科学领域重大科研失信事件应加强信息通报与公开A、工信部和科技部B、科技部和教育部C、科技部和中国社科院D、科技部和公安部E、公安部和中国社科院C()分别负责统筹自然科学和哲学社会科学领域科研诚信案件的调查处理工作A、工信部和科技部B、社科院和科技部C、科技部和社科院D、科技部和工信部E、科技部和教育部C()负责全国医疗卫生机构开展的涉及人的生命科学和医学研究伦理审查监督A、国家卫生健康委B、国家中医药局C、教育部D、科技部E、公安部A()开启了医学伦理实验规范的开端A、《希波克拉底誓言》B、《纽伦堡法典》C、《医学伦理学法典》D、《日内瓦宣言》E、《东京宣言》B()是科技创新的基石A、科研诚信B、政治站位C、科研制度D、科研监管E、组织管理A()是伦理审查关注的重点A、普通人群B、特定人群C、一般人群D、男性E、女性B()要加强科技计划的科研诚信管理,建立健全以诚信为基础的科技计划监管机制,将科研诚信要求融入科技计划管理全过程A、科技计划管理部门B、教育部C、卫生健康委D、新闻出版部门E、中国科协A()负责全国高等学校开展的涉及人的生命科学和医学研究伦理审查监督,并管理教育部直属高等学校相关工作。
国家卫健委国家中医药局教育部医学科研伦理委员会国家疾控局C()负责涉及人的中医药学研究伦理审查监督国家卫健委国家中医药局药品监督管理局医学科研伦理委员会国家疾控局B()卫生健康主管部门会同有关部门制定区域伦理审查委员会的建设和管理办法县级区级市级省级各级D()以上地方人民政府卫生健康、教育等部门依据职责分工负责本辖区涉及人的生命科学和医学研究伦理审查的监督管理乡镇县级区级市级省级B()以上人民政府有关行政部门对违反本办法的机构和个人作出的行政处理,应当向社会公开乡镇县级区级市级省级B()应当牵头设立同级医学伦理专家委员会或者委托相关机构承担同级医学伦理专家委员会工作国家和省级卫生健康主管部门药品监督管理部门国家中医药局医学科研伦理委员会国家疾控局A“难发现”的科研不端行为是()A、研究设计偏见或事后修改B、剽窃C、篡改D、伪造、杜撰E、不合法的人体试验A“唯论文”不良导向指的是()A、过度看重论文数量多少B、过度看重论文影响因子高低C、忽视标志性成果的质量、贡献D、忽视标志性成果的影响E、以上均是E“在医学伦理原则指导下,医生为患者利益或他人和社会利益,对患者自主权进行干预和限制,并由医生作出决定的一种医疗伦理行为”,下列权利中符合该描述的是()患者自主权医疗干预权知情权管理权裁量权B《发表学术论文“五不准”》中,错误的是()A、不准由“第三方”代写论文B、不准由“第三方”代看论文C、不准由“第三方”对论文内容进行修改D、不准提供虚假同行评审人信息E、不准违反论文署名规范B《发表学术论文“五不准”》中,错误的是()A.不准由“第三方”代写论文B.不准由“第三方”代看论文C.不准由“第三方”对论文内容进行修改D.不准提供虚假同行评审人信息E.不准违反论文署名规范B《规范》中,医学科研人员在涉及()等研究中,要树立公共卫生和实验室生物安全意识A.传染病B.新发传染病C.不明原因疾病D.已知病原改造E.以上均是E《规范》中,医学科研人员在研究结束后,除()外都要遵循相应的生物安全和科研管理规定A.论文B.人体或动物样本C.毒害物质D.数据或资料的储存E.分享和销毁A《科研诚信案件调查处理规则(试行)》监管对象是()A、科学家B、科研诚信案件C、研究成果D、失信机构E、科研资金B《科研失信行为调查处理规则》监管对象是()A、科学家B、科研失信行为C、研究成果D、失信机构E、科研资金B《科研失信行为调查处理规则》于()印发施行A、2022年08月25日B、2022年06月25日C、2022年10月25日D、2019年10月25日E、2019年06月25日A《涉及人的生命科学和医学研究伦理审查办法》()起施行A、2022年2月18日B、2023年2月18日C、2022年6月18日D、2023年6月18日E、2021年6月18日B《涉及人的生命科学和医学研究伦理审查办法》不适用于()A、医疗机构B、卫生机构C、实验室D、科研院所E、高等学校C《涉及人的生命科学和医学研究伦理审查办法》出台目的描述有误的是()A、保护动物生命和健康B、维护人格尊严C、尊重和保护研究参与者的合法权益D、促进生命科学和医学研究健康发展E、规范涉及人的生命科学和医学研究伦理审查工作A《涉及人的生命科学和医学研究伦理审查办法》分为()章()条六章二十七条六章五十四条五章五十四条四章三十七条六章二十一条B《涉及人的生命科学和医学研究伦理审查办法》基本伦理要求不包含控制风险知情同意公平公正保护隐私权及个人信息超额赔偿E《涉及人的生命科学和医学研究伦理审查办法》坚持了《涉及人的生物医学研究伦理审查办法》的哪些基本原则和制度框架()A、坚持机构主体责任B、坚持知情同意制度C、坚持伦理审查制度D、遵循国际公认的伦理准则,坚持基本的伦理要求E、以上均是E《涉及人的生命科学和医学研究伦理审查办法》施行前,从事涉及人的生命科学和医学研究的机构已设立伦理审查委员会的,应当自本办法施行之日起()内按规定备案,并在国家医学研究登记备案信息系统上传信息A、1个月B、3个月C、6个月D、9个月E、12个月C《涉及人的生命科学和医学研究伦理审查办法》依据()制定A、《中华人民共和国民法典》B、《中华人民共和国基本医疗卫生与健康促进法》C、《中华人民共和国科学技术进步法》D、《中华人民共和国生物安全法》E、以上均是E《新加坡声明》提出的科学研究的诚信原则是()A、在研究的所有方面都要诚实B、在进行研究时负责任C、在与他人工作时保持专业的姿态与公平D、为了其他各方利益对研究进行有益的监督E、以上均是E《医学科研诚信和相关行为规范》第七条规定,医学科研人员在采集科研样本、数据和资料时要()A、客观、全面、准确B、主观、包容、及时C、客观、针对、及时D、客观、理解、包容E、及时、准确、筛选A《医学科研诚信和相关行为规范》第七条规定,医学科研人员在采集科研样本、数据和资料时要()A.主观、包容、及时B.客观、针对、及时C.客观、全面、准确D.客观、理解、包容E.及时、准确、筛选C《医学科研诚信和相关行为规范》对实施动物试验时的要求,认识错误的是()A.人道地对待动物B.实验动物饲养和使用中产生的问题不必上报C.严格按照已批准的实验方案进行实验D.遵循科研机构关于动物饲养和使用的一般措施E.实验动物饲养和使用中产生的问题必须上报B《医学科研诚信和相关行为规范》发布的目的,描述错误的是()A.践行社会主义核心价值观B.加强医学科研诚信建设C.约束科研人员行为D.预防科研不端行为E.提高医学科研人员职业道德修养C《医学科研诚信和相关行为规范》发布的目的不包括()A、践行社会主义核心价值观B、加强医学科研诚信建设C、打击犯罪D、预防科研不端行为E、提高医学科研人员职业道德修养C《医学科研诚信和相关行为规范》发布通知中突出了()A.医学科研机构科研诚信监管的责任B.规范医学科研诚信行为C.养成良好科研行为习惯D.遵守诚信原则E.加强科研诚信体制建设A《医学科研诚信和相关行为规范》明确了医学研究活动应当遵循的哪些基本规范?A.医学科研行为涵盖科研项目的申请、预实验研究、实施研究、结果报告、项目检查、执行过程管理、成果总结及发表、评估审议、验收等科研活动全流程B.医学研究要牢固树立生物安全意识,在从事致病病原研究过程做到依法合规C.医学科研活动有关记录和数据由所在单位集中保存的原则D.提出科普宣传中不得向公众传播未经科学验证的现象和观点,在疫情防控期间应当严格遵守疫情防控管理要求等准则E.以上均是E《医学科研诚信和相关行为规范》是依据哪些法条制定的?A.中华人民共和国科学技术进步法、中华人民共和国著作权法B.中华人民共和国人类遗传资源管理条例、涉及人的生物医学研究伦理审查办法C.关于进一步加强科研诚信建设的若干意见、关于进一步弘扬科学家精神加强作风和学风建设的意见D.科研诚信案件调查处理规则(试行)E.以上均是E《医学科研诚信和相关行为规范》适用于()A.从事医学研究的所有人员B.医学研究的全过程C.基础医学、临床医学D.中医学与中药学E.以上均是E《医学科研诚信和相关行为规范》突出了()科研诚信主体责任A、医学研究机构B、科研人员C、项目负责人D、导师E、领导A《医学科研诚信和相关行为规范》突出了医学研究机构作为科研诚信主体的哪些责任?A.建立健全内部管理体系,是加强内部管理制度B.加强成果管理,建立科研成果全流程追溯机制C.淡化论文发表数量、影响因子等与人员奖励奖金、临床医生考核等的关系,建立科研人员职业培训和教育体系,加强对医学科研人员诚信教育D.对特殊领域如传染病、生物安全等的研究成果进行评估E.以上均是E2021版《医学科研诚信和相关行为规范》第二条删除了()等领域A、临床医学、护理B、口腔、护理、计划生育C、中医学、计划生育D、基础医学、中药学E、计划生育、临床医学B2021版《医学科研诚信和相关行为规范》第五条增加了()A、接受伦理监督B、遵循科研伦理准则C、主动申请伦理审查D、保障受试者的合法权益E、遵循涉及人的生物医学研究伦理审查办法相关规定C2021版《医学科研诚信和相关行为规范》发布通知,相比2014年版强调了()A、医学科研机构科研诚信监管的责任B、规范医学科研诚信行为C、养成良好科研行为习惯D、遵守诚信原则E、加强科研诚信体制建设A2021版《医学科研诚信和相关行为规范》相比2014年版增加的依据规定不包括()A、《中华人民共和国人类遗传资源管理条例》B、《中华人民共和国著作权法》C、《关于进一步加强科研诚信建设的若干意见》D、《关于进一步弘扬科学家精神加强作风和学风建设的意见》E、《涉及人的生物医学研究伦理审查办法》B2021版《医学科研诚信和相关行为规范》由()共同修订A、国家卫生健康委员会、教育部、国家中医药管理局B、国家卫生健康委员会、教育部C、国家卫生健康委员会、科技部、国家中医药管理局D、国家卫生健康委员会、科技部、教育部E、科技部、教育部、国家中医药管理局C2021版《医学科研诚信和相关行为规范》自()起施行A、2021年1月27日B、2021年1月21日C、2021年6月27日D、2021年6月21日E、2021年12月21日A版本《医学科研诚信和相关行为规范》共()A.四章、34条B.五章、34条C.五章、35条D.四章、35条E.六章、34条A保护研究参与者的隐私权,应如实将研究参与者个人信息的()告知研究参与者并得到许可,未经研究参与者授权不得将研究参与者个人信息向第三方透露A、收集B、储存C、使用D、保密措施情况E、以上均是E本办法对涉及()等特定群体的研究参与者,应当予以特别保护儿童孕产妇老年人智力障碍者、精神障碍者以上都是E本办法施行前,从事涉及人的生命科学和医学研究的机构已设立伦理审查委员会的,应当自本办法施行之日起()个月内按规定备案,并在国家医学研究登记备案信息系统上传信息。
基于K-means动态聚类的鸢乌贼角质颚模式识别

标准化后的角质歆数据。
1.3 K-means聚类建模
第1步,以标准化后的150组角质额样本总量为 行,以10个形态学参数为列,形成150x10的总矩阵。 第2步,根据K-means聚类原则,将预处理完毕的角 质额数据按照采样的海区来源分为3类,即K=3。第 3步,K-means算法在数据集中随机选择1个点作为 第1个初始类的中心点,接着选择距离该点最远的点 作为第2个初始类簇中心点,最后选择距离前2个点 的最近距离最大的点作为第3个初始类簇的中心点,最 终选定3个初始聚类的中心即C15C2, C3(曾俊,2020)o 第4步,选择任意2组角质顎形态学参数 xij(i= 1,2,...,450打=1,2,..・,10)按曼哈顿距离(或欧氏
第42卷第4期 2 0 2 1年8月
渔业科学进展 PROGRESS IN FISHERY SCIENCES
Vdl.42, No.4 Aug., 2021
DOI: 10.19663/j.issn2095-9869.20200315002
/
郑芯瑜,刘必林,孔祥洪,王雪辉.基于^-means动态聚类的莺乌贼角质额模式识别.渔业科学进展,2021,42(4):6472
其具有普适性,将会提高鸯乌贼种群的识别能力。 关键词 鸯乌贼;角质频;模式识别;曼哈顿距离;欧氏距离
中图分类号S917.4 文献标识码A 文章编号2095-9869(2021)04-0064-09
头足类作为海洋食物链中重要的一环,研究其分 类状况可为其他捕食者的食性分析提供有力依据,同 时,为进一步研究海洋食物网关系、生态系统能量流
基于微卫星标记的绿背山雀遗传结构分析(英文)

Chinese Birds 2013,4(2):144–154DOI 10.5122/cbirds.2013.0017ORIGINAL ARTICLEReceived 25February 2013;accepted 16A pril 2013f (F L )f @z Microsatellites underestimate genetic divergence in the Green-backed Tit (Parus monticolus)Wenjuan WANG 1,6,Chuanyin DAI 1,2,Bailey D.MCKA Y 3,Na ZHAO 4,Shou-Hsien LI 5,Fumin LEI 1,1K ey Laboratory of Zoological Systematicsand Evolution,Institute of Zoology,Chinese A cademy of S ciences,Beijing 100101,China 2School of Chemist ry and Life Sciences,Guizhou Normal College,Guiyang 550001,China 3Department of Ornithology,American Museum of Natural History,Central Park W est at 79th Street,New Y ork,NY10024-5192,US A4K ey L aboratory of E cological Impacts of Hydraulic-Projects and Restoration of A quatic Ecosystemof Minist ry of W ater Resources,Institute of Hydroecology,Ministryof W ater Resources and Chinese A cademyof Sciences,Wuhan 430079,China 5Department of Life Science,National Taiwan Normal University,Taipei,Taiwan 6University of Chinese A cademy of S ciences,Beijing 100049,ChinaA bst ract A lthough a growing num ber of both sequence-based and microsatellite nuclear loci have been used to infer genet ic structures,their relative ef ciencies remain poorly understood.In our study,we used the Green-backed Tit (Parus monticolus)to explore the resolving ability of these two typesof m arkers.The south-western and central mitochondrial DNA (mt DNA)phylogroups were divergent to some extent in sequence-based nuclear data,while mixed together in microsatellites dat a.The F ST values among clades w ere about four t imes lower in microsatellite loci than t hose in sequence-based nuclear loci.We are of the opinion t hat size hom oplasy may have cont ributed to the inability of m icrosatellites to uncover differen-tiation.Our results suggest that sequence-based nuclear loci outperformed m icrosatellite loci in detecting population structures,especially those focused on populations w ith large effective population sizes.There was no signi cant correlation between F ST values and allelic size variability,which suggested that the ef-ciency of microsatellite loci in detect ing genetic structure may be independent of their polym orphism.F ST is bett er than R ST in detecting intraspeci c divergence due to the high variance of R S T .In agreement with sequence-based nuclear loci,microsatellite loci did resolve the genetic distinctness of t he Taiwanese phylogroup.The genetic differentiat ion betw een the Taiwanese and cont inental clades may involve allopat-ric divergence without gene ow .K eywords genetic structure,microsatellite,Parusmont icolus,sequence-based nuclear data,size hom oplasyIntroduct ionOne of the primary g oals of phylogeography is to dis-cover genetic lineag es within and among closely related species (A vise et al.,1987).At the beginning of a phy-logeographic study ,investigators are typically facedwith making a decision about which type of molecularmarker to use.The three most commonly used DNA markers in phylogeogr aphy are mitochondr ial DNA (mtDNA),sequence-based nuclear markers and mi-crosatellite markers.mtDNA has been widely used in phy logeography for almost two decades (A vise,2009).It is a leading indicator of genetic structures,especially for populations with relativ ely short periods of isolation (Zink and Barrowclough,2008).However,using mtD-N z y f ,y www.c hine se birds.ne tA uthor or correspondenc e umin ei E-mail:lei m io A alone has been critici ed recentl because o some shortcomings such as evolutionar linkage and ma-W enjuan Wang et al.Green-backed Tit diversi cation 1453B j F y U y O S yternal inheritance (Edwards and Bensch,2009).It now seems that multiple,unlinked nuclear loci are essential for better understanding the complex ev olutionary his-tor y of species (Ballard and Whitlock,2004;Brito and Edwards,2008).Still,another question that needs to be addressed is whether sequence-based nuclear mark-ers or frequency -based microsatellite markers are more suitable for detecting genetic structures.Both sequence-based nuclear markers and microsat-ellite markers have their advantages and disadv antages.Many microsatellite markers hav e high mutation rates (between 102and 105mutations per locus per genera-tion (Estoup and Angers,1998));this high mutation rate improves the power of these mark ers to distinguish populations (Schl tterer,2000).It has been suggested that fewer unlinked microsatellite markers than se-quence-based nuclear markers are needed to achiev e the same precision (Haasl and Payseur,2010).However,the extremely high mutation rate of microsatellite markers may lead to size homoplasy ,which means allelic size is identical in state but not identical by descent (Estoup et al.,2002).Size homoplasy can underestimate popula-tion differentiation (Hedrick,1999;L i et al.,2010b).Al-though a fraction of size homoplasy could be detected by single-strand conformation polymorphism (SSCP)and DNA sequencing,the detectable homoplasy could improv e only a slight underestimation of genetic differ-entiation (Adams et al.,2004).Most size homoplasy is undetectable when two alleles are identical in sequence but not identical by descent [i.e.they hav e different g e-nealog ical histories (Selkoe and T oonen,2006)].In ad-dition,the complex mutation models of microsatellite data further complicate the power to detect population str uctures (Balloux an d L ugon-Moulin,2002;Haasl and Payseur,2010).The drawbacks of allele-frequency based markers in recognizing histor ical grouping of populations,calculating g ene flow and estimating di-v erg ence times hav e led some researchers to sug gest that sequence-based markers are superior to microsatellites in evolutionar y studies (Brito and Edwards,2008;Zink,2010).Sequence-based nuclear markers have lower mu-tation rates (108to 109,e.g.Ax elsson et al.,2004)and thus lower lev els of homoplasy.Mutation models are much simpler and the exploration of g enealogical pat-terns is easier (Zhang and Hewitt,2003).Moreover,it is conv enient to compare the parameters estimated using sequence-based nuclear data with those estimated using DN (B ,),f q restricts the resolving power to discover shallower ge-netic structures.In this study ,we selected the Green-backed Tit (Parus monticolus),an East Asian endemic bird species,to re-solve which maker is more suitable for revealing genetic structures within species.Our previous work (Wang et al.,2013)revealed three mtDNA phy logroups in China and adjacent areas:a south-western phy logroup includ-ing samples from the Himalayas,southwest China and northeast India,a central phylog roup containing samples from centr al China and a T aiwanese phylogroup,re-stricted to T aiwan Island (Fig.1).A species tree analy sis including both mtDNA and sequence-based nuclear loci,revealed that the ancestral population of the Green-backed Tit,in first instance,splits into the T aiwanese phy logroup and the ancestral population of the south-western /centralphylog roup,which further splits into the south-western and the central phy logroups.The genetic diversification within the Green-backed Tit is relatively recent.A ll diversification probably occurred within the last 0.9million year s.H aplotype networks based on sequence-based nuclear loci revealed x ed base pair dif-ferences between the T aiwanese and other phylogroups.However,the genetic div ergence between the south-western and central phylogroups was shallower,with shared nuclear haplotypes.Given their higher mutation rate,microsatellite markers might resolve div ergence pat-terns within these more recently diverg ent phylogroups.Besides,previous studies hav e successfully utilized mi-crosatellite markers to rev eal genetic divergence within species when sequence-based nuclear markers failed (B runner et al.,1998;T onione et al.,2010).We used two Z-linked sequence loci and nine micro-satellite loci in this study .We rst describe the genetic structure revealed by microsatellite data.Second,we compare the g enetic structures rev ealed by sequence-based nuclear loci and microsatellite loci and address the resolving ability of these two types of markers.Third,given the genetic distinctiveness of the T aiwanese population,we also explored whether allopatric diver-gence without gene flow or isolation with migration might result in this genetic pattern.Met hodsSample collection,DNA ampli cation and microsatellite genotypingW 5f f201ei ing orestr niversit and C hina rnithologic al oc iet mt A data rito and Edwards 2008.However the low mutation rate o se uence-based nuclear markerse u sed 14samples rom 24sites across most oChinese Birds 2013,4(2):144–154146the range of the Green-backed Tit (Fig.1).Samplesincluded three of the four descr ibed subspecies (we lacked samples of P.m.legendrei,which is restricted to souther n Vietn am on the Da Lat plateau).Gross genomic DNA was extracted using the DNeasy Tissue kit (QIAGEN)following the instructions of the manu-facturer.T wo Z-linked sequence loci and nine micro-satellite loci were used in this study.The two Z-linked sequence loci were MUSK (muscle-specific tyrosine kinase)and Z-185(similar to a transient receptor po-tential cation channel,subfamily M,member 3,Shou-Hsien Li,unpublished data).These sequences were used in our previous work (Wang et al.,2013,GenBank number JQ324378–JQ324420,JQ324466–JQ324472,JQ324475,JQ324479,JQ324481–JQ324499,JQ324501,JQ 324506–JQ324513,JX849802–JX849862).Among the nine microsatellite loci,four loci (Titgata02,Tit-gata67,Titgata87,Titgata89;Wang et al.,2005)were developed specifically for the Green-backed Tit and ve additional loci (PmaGAn27,PmaT AGAn71,PmaT-GAn45,Pma48m,Pma69u;Kawano,2003;Saladin et al.,2003)for its closest relative Parus majo r (Gill et al.,2005;Dai et al.,2010).We started with the original PCR conditions descr ibed in these three papers and thenthe adjusted MgCl 2,template DNA concentrations and annealing temperatures to amplify our targ et loci.Mic-rosatellites were ampli ed with uorescent labeled for-ward primers (FAM and HEX dyes).Fragment lengths were analyzed with the internal size marker GENES-CAN-500ROX (Applied Biosystems)and scored with GENEMARKER 3.7(SoftGenetics).Data analysisFor microsatellite data,a Hardy-W einberg equilibrium (HWE)and linkage disequilibrium (LD)were tested using GENEPOP 4.0(Raymond and Rousset,1995),with a sequential Bonferroni correction for assessing statistical signi cance.Basic population genetic param-eters (number of alleles,private alleles,observed and expected heterozy gosity per locus)were estimated with MST ools (Park,2001).T o correct for sample size differ-ences,we calculated allelic richness using FSTA T 2.9.3(Goudet,2001).Null alleles were examined with Micro-Checker (V an Oosterhout et al.,2004).We used three methods to identify g enetically distinct groups in microsatellites.First,PHY LIP 3.6(Felsen-stein,1989)was utilized to construct aneighbor-joiningFig.1T opological map of East Asia showing sampling sites and the three phylogroups revealed by mitochondrial DNA (mtDNA ,data from Wang et al.,2013).Circles represent sampling sites where the size of the circles is proportional to the sample size.Dashed lines show the three mtDNA phylogroups:the south-western,central and Taiwanese phylogroups.Elevation is represented by shading:T G T f x y 3S T f f lower elevations are shown lighter and higher elevations are shown darker.he reen-backed it is distributed rom appro imatel 1000to 000m in elevation.ee able1or details o population codes.W enjuan Wang et al.Green-backed Tit diversi cation 1473B j F y U y O S ytree using Cavalli-Sforza ’s chord distance (Dc),with 1000bootstrap replicates.A Dc distance based tree was utilized because this has been suggested to be robust to null alleles (Chapuis and Estoup,2007)and is suit-able for allele frequency data (Takezaki and Nei,1996).Second,we used a Bayesian approach implemented in STRUCTURE 2.3.3(Falush et al.,2007)to assign indi-viduals to clusters without prior information.Analyses were run using an admixture and correlated allele fre-quency model.The number of groups (K)was var ied from 1to 10,with 5runs for each K v alue.The burning length and MCMC iterations were set to 500000and 5000000,respectively .We used the E vanno et al.(2005)method implemented in the program STRUCTURE HARVESTER (Earl,2012)in order to determine the most likely value of K and DISTRUCT 1.1(Rosenberg,2004)to display the results.Third,pairwise F S T (Wright,1965)and R ST (Slatkin,1995)v alues among clades were calculated for individual microsatellite loci as well as all loci combined using FST A T.We also calculated pairwise G ST (N ei,1978)an d D (Jost,2008)values amon g clades for all loci combined using GENODIVE 2.0b20(Meirmans,2009),because combined utilization of F ST with G ST and D has been recommended (Jost,2009;L eng and Zhang,2011;Meirmans and Hedrick,2011).For the two sequence-based nuclear loci,the E w-en s-Watterson (Ewens,1972;Watterson,1974)and Chakraborty ’s A malgamation (Chakraborty ,1990)tests implemented in ARL EQUIN 3.5(E xcof er et al.,2005),were used to test for selectiv e neutrality .Haplotype net-works were reconstructed using TCS 1.21(Clement et al.,2000).In order to compare the genetic differentia-tion between microsatellite and sequence-based nuclear data,we also calculated F ST v alues among clades at each locus.Result sFor microsatellite data,no evidence emerged for linkage disequilibrium between any pair of loci across all popu-lations.Locus Titgata67significantly deviated from the Hardy-Weinberg equilibrium (p <0.05)and was excluded from all further analyses.There were null al-leles for populations BSand KKS at locus Pma69u,with frequencies of 0.25and 0.18respectively .The number of alleles per locus ranged from seven in Pma48m to 22in Titgata87.Allelic richness ranged from 1.56to (T ),population.However,NT and HL,the two T aiwanese populations,exhibited high levels of private alleles,with ve and 14alleles in each population,respectiv ely .For sequence-based nuclear data,Chakraborty ’s amalgama-tion test and the Ewens-Watterson test were not sig ni -cant for any locus.The neighbor-joining tree constructed with mic-rosatellite data (Fig.2a)identified two genetic clades:a continental clade (south-western +central mtDNA phylogroups)containing samples from the Asian con-tinent and a T aiwanese clade was restricted to T aiwan.This was consistent with the STRUCTURE result (Fig.2b).The E vanno et al.(2005)method indicated the optimal number of clusters as K =2.STRUCTURE analyses did not resolve any genetic structure within the A sian continent.Allelic size frequencies were similar be-tween the south-western and central phylogroups,but signi cantly different between the T aiwanese and conti-nental clades (Fig.3).The weighted R ST values were 0.01between the south-western and central phylogroups,0.25between the central and T aiwanese phylogroups,and 0.22between the south-wester n and Taiwanese phylogroups.G ST values were 0.00between thesouth-Fig.2Neighbor-joining tree and population structure based on allelic frequencies of microsatellite data.(a)Neighbor-joining tree based on Cavalli-Sforza ’s chord dist ance (Dc).Numbers above branches are percentage bootstrap support values from 1000replicates (only values >75%shown).(b)Bayesian-based cluster analysis implemented in STRUCTURE.Each vertical bar y f (K =)201ei ing orestr niversit and C hina rnithologic al oc iet 1.79able 1.All the continental population s had limited private alleles with less than two alleles in eachrepre se nts one individual and its probabilit o being assigned toc lusters 2.Chinese Birds 2013,4(2):144–154148T able 1Summary of genetic diversity in theGreen-backed Tit (Parus monticolus )Population code Location Longitude (°)Latitude (°)n A P A A R H O H E South-western 42 3.3347.750.620.66ZM Zhangmu 85.98827.9894 3.751 1.670.660.67MY Miyi 101.89927.1275 4.130 1.670.550.67Y B Y anbian 101.53527.10613 6.001 1.680.700.68Y L Y ulong 100.26127.0972 2.630 1.670.690.67BS Baoshan 98.78324.82011 5.381 1.640.570.64BM Bomi 95.75929.8732 2.250 1.580.560.58CY Chayu 97.08428.5625 4.000 1.640.530.64Central 88 3.27129.880.670.72ZQ Zhouqu 104.17133.6972 2.381 1.560.560.56ZZ Zhouzhi 108.22134.1609 5.380 1.710.660.71FP Foping 107.76833.59610 5.750 1.720.670.72NS Ningshan 108.31833.3133 3.500 1.760.710.76HP Haoping 107.71134.08812 6.501 1.680.660.68MG Meigu 103.30828.5187 5.632 1.720.730.72DX Dingxi 110.19531.5668 5.000 1.690.730.69BQ Banqiao 108.63829.9334 3.630 1.660.700.66L JS Laojunshan 110.91831.8804 4.500 1.760.660.76L D L uding 102.27729.9206 4.500 1.700.630.70SM Shimian 102.33829.1004 3.631 1.690.590.69L L Longlin 104.87124.6661 1.630 1.630.630.63GD Guiding107.13526.5823 3.630 1.760.670.76KK S K uankuoshui 107.19027.94713 6.501 1.740.660.74L SLeishan 108.07826.3842 2.880 1.790.810.79Taiwanese 24 2.9018 6.880.600.64NT Nantou 121.18223.97714 5.135 1.560.530.58HLHualian121.38824.192105.50141.610.710.61Note:n,sample size;A,average number of alleles per locus;P A ,private allele;A R ,allelic richness;H O ,observed heterozygosity;and H E ,expectedheterozygosity.F 3z f ig.Allelic si e distributions o the eight microsatellite lociW enjuan Wang et al.Green-backed Tit diversi cation 1493B j F y U y O S ywestern and central phylogroups,0.01between the central and T aiwanese phylogroups,and 0.10between the south-western and Taiwanese phylogroups.In gener al,estimated D values were higher than the estimated G ST values.D values were 0.02between the south-western and central phylogroups,0.42between th e centr al and Taiwan ese phylogr oups,and 0.44between the south-western and T aiwanese phy logroups.F ST between the south-western and centr al clades for the eight microsatellite loci combin ed was 0.01(ranging from 0.00to 0.01).In contrast,F ST was much higher for individual sequence-based loci (MUSK:F ST =0.03,Z185:F ST =0.04).F ST between the south-western and Taiwanese clades for the eight microsatellite loci combined (average:0.17,ranging from 0.01to 0.24)was much lower than that for the two nuclear loci (MUSK:F ST =0.61,Z185:F ST =0.89),so was F ST between the central and T aiwanese clades [microsatellite:F ST =0.15(ranging from 0.03to 0.22),MUSK:F ST =0.54,Z185:F ST =0.77].The single-locus estimates of F ST among clades were similar across the eight microsatellite loci,while R ST showed high var iance (Fig.4).H aplotype networks revealed fixed base pair differences between th e Taiwan ese a nd south-western/centr al mtDNA phylogroups,while haplotypes were shared between the south-wester n and central phylogroups (Fig.5).Both south-western and central phylogroups contained pr ivate haplotypes,suggesting that the two mtDNA phy logroups also diverged to some extent in sequence-based nuclear loci.DiscussionSuitability of microsatellite markers in resolving genetic str uctur esThe mutation rates of microsatellite markers are sev-eral orders of magnitude higher than sequence-based nuclear mar kers and are expected therefore to havehigher resolving power of genetic structures (Estoup and Angers,1998).Howev er,inconsistent with the ex-pectation,the estimates of genetic div ergence among mtDNA phylogroups of Green-backed Tit were much lower in microsatellites than in sequence-based nuclear data.Speci cally ,the south-western and central mtDNA phy logroups each contained priv ate nuclear haplotypes,which suggested that the two phylogroups were also divergent to some extent in sequence-based nuclear loci (Fig.5).Nevertheless,the microsatellite data failed to detect any g enetic divergence across the Asian continent (Fig.2).The allelic size distributions between the south-wester n and centr al phylogroups were similar across the eight microsatellite loci,and limited private alleles were found in continental populations (Fig.3,Table 1).The R ST ,G ST and D values between the south-western and central phy log roups were small.F ST values among clades were about four times lower in microsatellite loci than that in sequence-based loci.Two possible reasons may explain the lower genetic str ucture in the microsatellite data:size homoplasy of microsatellites and strong selection pressure on sequence-based loci (Epperson,2005;Li et al.,2010b;Storchov áet al.,2010).The Chakraborty ’s amalgama-tion and Ewens-W atterson tests did not show signi cant differences for any sequence-based nuclear loci,which suggests that neutr al selection may not be the cause.Therefore,we think that reduced g enetic differentiation in microsatellite loci may result from size homoplasy .In agreement with this explanation,we found that ob-serv ed heterozy gosity per locus was lower than expected for most sampling sites (T able 1).Size homoplasy is closely related to effective popu-lation size (Nauta and Weissing,1996).Populations with a large effective population tend to hav e higher size homoplasy than populations with a small effective population (Estoup et al.,2002).The effective popula-tion size of the continental clade was larger than that of the Taiwanese clade.Therefore,it is likely thattheF S f F S T R S T 201ei ing orestr niversit and China rnithological ociet ig.4ingle-locus estimates o /vs.allelic richnessChinese Birds 2013,4(2):144–154150continental clade was more affected by size homoplasy and the T aiwanese clade to a lesser extent.In agreement with this expectation,the genetic distinctiveness of the T aiwanese phylogroup was resolved,while no genetic distinct group was uncovered in the Asian continent.The ranges of allele sizes larg ely ov erlapped between the south-western and central phy log roups,with only par-tial overlapping between the T aiwanese and continental clades.As well,it should be noted that the two Tai-wanese populations (NT and HL )have a much higher lev el of private alleles than the continental populations.Based on these results,we suggest that our microsatel-lite markers are less suitable than sequence-based nu-clear markers in resolving genetic divergence,especially for populations with large effectiv e population sizes.However,when effective population size is small,differ-ences among marker types are less pronounced.Besides effective population size,size homoplasy also depends on the mutation rates of microsatellite loci (E stoup et al.,2002).The large variation in the number f ,(f T ),large differences in rates of mutation.The relationship between locus variability (typically represented as allele diversity )and the magnitude of population differentia-tion is under debate.Some suggest that highly variable loci could increase the power for estimating genetic structures (Epperson,2004),while others argue that high levels of locus polymorphism may underestimate population differentiation because of size homoplasy (Estoup et al.,2002;O ’Reilly et al.,2004).Previous studies revealed a negativ e relationship between allele diversity and estimated F ST values (O ’Reilly et al.,2004;Olsen et al.,2004).Howev er,our study did not find a signi cant correlation between F S T v alues and allelic size variability (Fig.4),which indicates that the ef ciency of microsatellite loci in detecting genetic structure may be independent of their polymorphism.R ST is an F ST -analogue,but this statistic takes into ac-count the difference in repeat numbers and assumes a stepwise mutation model.It has been suggested that R ST provides less biased estimates of population divergence than F ST under the stepwise mutation model (Slatkin,1995).However,this feature no longer holds when the stepwise model is violated (Slatkin,1995).This is likely the case of microsatellite loci with rather complex mu-tation models (Anger s and Bernatchez,1997).It has also been suggested that ev en under the strict stepwise mutation model,F ST is better than R ST when sample sizes are moderate and small and the number of loci scored is low due to the high variance of R ST (Gag-giotti et al.,1999;Balloux and Lugon-Moulin,2002).In contrast to the stable F ST ,our results rev ealed that the variance of R S T across microsatellite loci was high (Fig.4).Therefore,in agreement with previous studies,our study shows that high variance restricts the suitability of R ST .Role of geographical isolation in genetic diversi cationAlthough genetic div ersi cation within the Asian con-tinent has not been resolved,our microsatellite data did recover the genetic distinctness of the T aiwanese clade.In agreement with the microsatellite results,the T ai-wanese clade had x ed base pair differences in mtDNA as well as in sequence-based nuclear loci (Wang et al.,2013).The genetic distinctiveness of the Taiwanese population has been found in other studies of birds (S ,;L ,),(G ,;,)(L L ,)Fig.5Haplotype networks of the two sequence-based nuclearloci.Each circle represents a haplotype;the size of the circle is proportional to the frequency of the haplotype.Dots represent unsampled haplotypes and dashes their corresponding mutation steps.o alleles det ected across the eight m icrosatellit e loci rom seven in Pm a48m to 22in itgata87representsong et al.2009iu et al.2012plants ao et al.2007Chou et al.2011and insects ee and in 2012.W enjuan Wang et al.Green-backed Tit diversi cation 1513B j F y U y O S yT aiwan has also been regarded as an impor tant zoogeo-graphic region (Zheng and Zhang,1959;Zhang,1999)and an area of endemism (Lei et al.,2007;L ópez-Pujol et al.,2011).All these observations sugg est the evolu-tionar y peculiarity of Taiwanese taxa.It has been sugg ested that most T aiwanese tax a colo-nized the island from the A sian continent during land bridge connections when the climate was cooler (Kano,1940).Geographical isolation has been emphasized as the most important factor for intraspecific and inter-speci c diversi cation between T aiwan and continental tax a (Grant and Grant,1998;L ei et al.,2007).However,the land bridge between T aiwan and the adjacent Asian continent formed and submerg ed intermittently during Pleistocene glacial cycles (V oris,2000).Therefore,the g eographical barrier was restricted largely to interglacial periods.When the land bridge formed during glacial periods,gene exchange between T aiwan and continen-tal taxa was possible.Recent studies,using coalescent-based methods [e.g.isolation with migration (IM;Hey ,2005)and approximate Bayesian computation (ABC;Pritchard et al.,1999)],have revealed that bidirec-tional gene flow existed in the sibling hwamei species (L i et al.,2010a)and Euphaea damsel ies species (Lee and Lin,2012)during land bridge connections.These studies suggest that the role of geographical barriers in genetic differentiation may not be as impor tant as previously sug gested.Other factors which can counter the homog enizing effect of g ene ows,such as natural selection and sexual selection,may be important driv ers of genetic diversi cation (L i et al.,2010a;Lee and Lin,2012).The interconnected area between T aiwanese and continental taxa is much larger in the Green-backed Tit than that in the hwameis and Euphaea damsel ies spe-cies.The disjunct area in both the hwameis and Eupha-ea damselflies species is restricted to the Taiwan land bridge.There is no unsuitable habitat on the Asian con-tinent.Never theless,there is a large area in southeastern China without Green-backed Tits.Therefore,it is less likely that the Taiwanese and the continental popula-tions of the Green-backed Tit would experience gene exchange during glacial periods.In addition,the eco-logical niche of the Green-back ed Tit is different from that of the hwameis and Euphaea damselflies species.The Green-backed Tit is a montane species that gener-ally occupies higher elevations,with an approximate range of 1000to 3000m (L i et al.,1982).Palaeov eg eta-f L G M x y ,semi-arid temperate woodland or scrub habitats (Har-rison et al.,2001;Ray an d Adams,2001),cur rently not suitable for the Green-backed Tit.Moreover,the results of ecological n iche modeling have suggested that there was no habitat suitable for the Green-backed Tit on the interconnected area during the Last Glacial Maximum (Wang et al.,2013).Therefore,in contrast to the divergence with our gene ow scenario,genetic differentiation in the Green-backed Tit may inv olve al-lopatric divergence without gene ows.Nevertheless,to resolv e the history of div erg ence between the T aiwanese and continental populations properly ,coalescent-based analyses are required in further inv estigations.A cknowledgments We thank Zuohua YIN,Gang SONG,Wulin LIU,Huatao LIU and Bin GAO for eld assistance and Chunlan ZHANG,Ruiying ZHANG,Y ongjie WU,Deyan GE and Baoyan LIU for assistance with data analyses.This study was supported by the National Science Fund for Distinguished Y oung Scientists (No.30925008),the Major International (Regional)Joint Re-search Project (No.31010103901),the CA S-IOZ Innovation Pro-gram (KSCX2-EW-J-2)and by a grant (No.O529YX 5105)from the K ey Laboratory of the Zoological Systematics and Evolution of the ChineseAcademy of Sciences to F.M.Lei.Reference sA dams RI,Brown KM,Hamilton MB.2004.The impact of mic-rosatellite electromorph size homoplasy on multilocus popu-lation structure estimates in a tropical tree (Corythophora alta)and an anadromous sh (Morone saxatilis).Mol Ecol,13:2579–2588.A ngers B,Bernatchez L plex evolution of a salmonid microsatellite locus and its consequences in inferring allelic divergence from size information.Mol Biol Evol,14:230–238.A vise JC,A rnold J,Ball RM,Bermingham E,Lamb T ,Neigel J E,Reeb CA,Saunders NC.1987.Intraspecific phylogeography:the mitochondrial DNA bridge between poulation genetics and systematics.Annu Rev Ecol Syst,18:489–522.A vise JC.2009.Phylogeography:retrospect and prospect.J Bio-geogr,36:3–15.Axelsson E,Smith NGC,Sun dstr m H,Berlin S,Ellegren H.2004.Male-biased mutation rate and divergence in autosomal,Z-linked and W-linked introns of chicken and turkey .Mol Biol E vol,21:1538.Ballard JWO,Whitlock MC.2004.The incomplete natural his-tory of mitochondria.Mol Ecol,13:729–744.B x F ,L M N T f ff M ,55–201ei ing orestr niversit and C hina rnithologic al oc iet tion maps o the ast lacial a imum indicate the interconnected land bridge area was covered b steppes allou ugon-oulin .2002.he estimation o populationdi erentiation with m icrosate llite marke r s.ol Ecol 11:1。
基于人眼评估的灰喜鹊卵色的窝内变_省略_模拟寄生卵的拒绝行为无关_英文_Jes_

Jesús M. Avilés and Deseada Parejo. Egg variation and discrimination in Azure-winged Magpie
303
1997; Davies, 2000). Hosts can learn how their eggs look like and recognize and reject those with an appearance different from that of their own eggs (Victoria, 1972; Rothstein, 1975; Lotem et al., 1992). Experimental studies have shown that hosts of avian brood parasites are more likely to reject parasitic eggs that look different from their own eggs (e.g. Brooke and Davies, 1988; Davies and Brooke, 1988; Soler and Møller, 1990; Moksnes et al., 1993; Lotem et al., 1995; Marchetti, 2000; Moskát and Honza, 2002; Lathi and Lathi, 2002; Rutila et al, 2002; Cassey et al., 2008; Avilés, 2008; Avilés et al., 2010; Spottiswoode and Stevens, 2010; Stoddard and Stevens, 2010), which supports the view that mimetic cuckoo egg phenotypes have evolved due to host egg rejection. As matching between parasite and host eggs improves, hosts are expected to reduce variation in the appearance of their eggs within their clutches. A low variation in egg appearance within host clutches would facilitate host’s recognition of parasite eggs (e.g. Victoria, 1972; Davies and Brooke, 1989b). In addition, a high homogeneity in appearance within clutches would lead to a higher variation in egg appearance among clutches of different host individuals (i.e. inter-clutch variation). Hence, a higher inter-clutch variation in egg appearance will prevent the evolution of a good match for the parasite (Soler and Møller, 1996). Passerine species frequently parasitized by the Common Cuckoo (Cuculus canorus) have evolved a lower degree of intraclutch variation in egg appearance, and a higher degree of inter-clutch variation than those that have not experienced such interactions (e.g. Øien et al., 1995; Soler and Møller, 1996; Stokke et al., 2002; Avilés and Møller, 2003). Recent findings with three Australian hosts of the Pallid Cuckoo (Cuculus pallidus), however, cast doubts on the generality of these processes as intra-clutch variation did not vary between populations with different exposure to parasitism (Landstrom et al., 2011). Intra-specific evidence has so far focused on reporting a link between intra-clutch variation in host egg appearance and probability of rejection of artificial eggs, and, is even more controversial. While some studies have shown that low intra-clutch variation in host egg appearance may favor parasite egg discrimination (Stokke et al., 1999; Soler et al., 2000; Moskát et al., 2008), others did not find support for the parasite recognition function of such egg traits (Karcza et al., 2003; Avilés et al., 2004; Honza et al., 2004; Stokke et al., 2004; Lovászi and Moskát, 2004; Polacikova et al., 2007; Cherry et al.,
基于视觉注意机制的UWB SAR叶簇隐蔽目标变化检测

基于视觉注意机制的UWB SAR叶簇隐蔽目标变化检测李超;李悦丽;安道祥;王广学【摘要】在超宽带合成孔径雷达叶簇隐蔽目标检测中,传统的UWB SAR图像变化检测方法易受图像灰度值起伏和成像条件变化的影响,致使现有的变化检测算法的性能下降.本文根据人类视觉系统的生理结构和认知特点,提出了一种基于视觉注意机制的叶簇隐蔽目标变化检测算法.该方法使用视觉注意模型,将图像的多尺度特征信息融合为单幅视觉显著图像,并利用图像局部邻域信息和目标的空间相关特性对视觉显著图中视觉注意焦点进行分层筛选和变化检测.实验结果表明:本文中基于视觉注意机制的变化检测方法可以有效检测多时相UWB SAR图像中的叶簇隐蔽目标,较之传统的基于统计原理的变化检测方法,其检测速度更快,且对场景复杂的UWB SAR图像亦具有鲁棒性.【期刊名称】《电子学报》【年(卷),期】2016(044)001【总页数】7页(P39-45)【关键词】低频超宽带合成孔径雷达;叶簇隐蔽目标检测;变化检测;视觉注意机制【作者】李超;李悦丽;安道祥;王广学【作者单位】国防科学技术大学电子科学与工程学院,湖南长沙410073;国防科学技术大学电子科学与工程学院,湖南长沙410073;国防科学技术大学电子科学与工程学院,湖南长沙410073;空军预警学院信息对抗系,湖北武汉430019【正文语种】中文【中图分类】TN957.51低频超宽带合成孔径雷达(Ultra Wide Bandwidth Synthetic Aperture Radar,UWB SAR)工作于VHF至UHF波段,其频段低、波长长,能够对树林遮蔽下的目标进行高分辨成像和侦察,逐步在民用与军事领域得到应用.但是,在实际UWB SAR图像中,叶簇覆盖区域除了存在车辆、坦克等隐蔽人工目标外,还有许多由粗大树干反射形成的点目标,严重影响了基于单幅UWB SAR图像叶簇区域隐蔽目标检测的准确度[1,2],除此之外,单幅低频UWB SAR提供的场景信息有限,无法有效的将隐蔽目标从场景中固有目标中完全分离出来,也是造成较高虚警率的主要原因,因此,有必要研究UWB SAR图像叶簇隐蔽目标检测的新方法. 与常规的基于单幅图像的目标检测技术不同,UWB SAR变化检测技术是一种利用同一地区不同时间UWB SAR图像之间差异来实现目标检测的方法,该方法进一步利用了同一地区叶簇区域在不同图像之间的相关性,具有更高的抑制强树干杂波的潜力,并日益受到关注.目前,基于多时相UWB SAR图像的叶簇隐蔽目标变化检测技术的难点主要存在于如何降低UWB SAR图像中相干斑的干扰;如何对UWB SAR图像杂波模型进行建模;如何实现快速的UWB SAR叶簇隐蔽目标检测等三个方面.在该领域,最初的研究多集中于基于图像像素灰度值差异的变化检测方法[3],之后,为了降低多时相UWB SAR图像灰度值起伏对于目标检测的影响,基于统计模型或统计量分析的变化检测方法逐步受到关注[4],但随着研究的深入,研究人员发现不同成像条件下获得的图像间所存在的统计分布差异制约了基于统计特性差异的检测方法的应用.此外,以上两种变化检测方法对图像配准精度的要求都很高,在处理时需要对整片树林区域进行基于像素点的逐一检测,因此存在检测效果易受图像匹配精度影响,虚警率高,运算量大等局限性.考虑到实际处理中人们所关心的目标仅占叶簇覆盖区域中很小一部分面积,如果能够选择关心的区域进行优先处理,则有利于提升检测速度和准确度.近年来Itti等人从视觉分析角度出发,模拟人类视觉系统(Human Visual System,HVS)机制构建适用于图像分析的高斯金字塔模型,该模型能够以不同的次序和力度对各个场景区域进行选择性加工,最终形成与HVS感知结果相近似的视觉显著图[5].受此启发,本文根据HVS的生理结构与认知特点,提出基于视觉注意机制的UWB SAR叶簇隐蔽目标变化检测算法(Change Detection of Foliage-Concealed Targets Based on Visual Attention,CDVA),该方法基于视觉注意机制构建视觉显著图,并利用显著图中目标的局部邻域信息和空间相关信息,以分层检测的方法来实现对叶簇隐蔽目标的变化检测.最后,基于实测UWB SAR图像的变化检测实验结果表明,本文提出的CDVA方法可以有效检测多时相UWB SAR图像中的叶簇隐蔽目标,且较之传统的基于统计原理的变化检测方法,其检测速度更快,并对场景复杂的UWB SAR图像亦具有鲁棒性.视觉注意是人类视觉的一项重要的心理调节方式,根据Treisman的特征融合理论[6],人眼的注意过程分为预注意期和注意期两个阶段:在预注意期,对视觉信息的处理过程是一个经过串行分层逐步处理的过程,而具体在每一个层次中,对视觉信息的处理则是以并行的方式进行的,在此期间,彼此独立的早期特征被抽取出来,但未进行处理;而注意期的处理是将彼此独立的特征统一的联系起来,形成对某一目标的综合描述,且随着注意焦点的选择和转移,目标之间相关性逐步清晰,整个场景逐渐被感知.参考上述过程我们将CDVA算法的处理过程分为视觉显著图的生成、视觉注意焦点的选择和变化检测三部分(如图1).其中叶簇隐蔽区域视觉显著图的生成过程是对人眼视觉注意的预注意期的模拟,该过程直接提取图像中的显著特征信息,计算图像区域中每个区域的视觉显著程度,继而生成视觉显著图.而视觉注意焦点的选择和变化检测过程则对应人眼视觉的注意期,在此过程中,视觉注意焦点因其散射强度和散射截面积不同,致使其位置和区域信息不断发生变化,这与心理学研究发现的视觉转移现象[7]不谋而合,大大增强了目标检测的准确性.在视觉注意机制的研究中Itti模型具有里程碑式的意义,它是第一个使用计算的方式模拟视觉认知过程的模型.该模型应用于UWB SAR图像处理中主要考虑灰度通道和方向通道的特征信息.在处理过程中,输入图像通过高斯金字塔实现分辨率以2为因子在水平和垂直方向依次递减的八级降采样过程,每层图像的亮度表示为I(σ),其中σ∈[0,8]代表金字塔结构中不同的层数.考虑到图像中视觉注意点与周围像素差异越大越容易引起视觉注意,因而引入中央周边差方法进行处理,即将不同分辨率的图层差值后,然后再进行点对点的相减.我们定义“Θ”为中央周边差算子,则灰度特征提取的过程表示为:其中c∈{ 2,3,4},s = c +δ,δ∈{ 3,4},表示将c尺度上的特征图差值到s尺度上的特征图来进行运算的过程.然后使用Gabor滤波器提取图像的朝向特征,二维Gabor滤波器其数学表达式为: 其中,(x0,y0)表示目标中心坐标,(ξ0,v0)表示滤波器在频域上的最优空间频率,σ表示高斯函数在x轴方向上的标准差,β表示高斯函数在y轴上的标准差.使用中央周边差方法提取方向特征信息的表达式为:其中θ为默认的四个方向,即通过式(1)、(3)的特征提取,可以形成不同尺度的亮度特征图和朝向特征图,考虑到对于叶簇隐蔽目标的检测无法事先获取先验信息,选取全局加强的合并策略来生成视觉显著图.这是一种无需先验信息的非监督的合并策略,具体的合并策略为:将各个特征图归一化到[0,N]的范围内,其中N∈(0,255).对于单幅特征图,寻找全局最大值M和除M之外的局部平均值-m.给每一幅特征图乘以权值(M --m).假设N表示全局加强合并过程,则亮度通道显著图的合并表示为:朝向通道显著图的合并表示为:将式(3)、(4)合并得到视觉显著图S,表示为:图2为使用UWB SAR对某一树林区域成像所得到的灰度图像(图2(a))和经过上述视觉注意模型处理后的视觉显著图(图2(b)).对比两幅图可以发现,经过视觉注意机制处理后,图像固有粗大树干形成的散射亮点减少,场景中目标更为突出,若能够将场景中所有目标提取出来,再进行变化检测,则会大大减少目标检测所需要的时间,为了能准确的从视觉显著图中提取场景目标,下一节我们研究UWB SAR 图像中视觉注意焦点的选择方法.人类视觉系统在面对一个复杂场景时,总会迅速选择几个感兴趣的区域进行优先处理,这些被选中的区域被称为注意焦点(Focus of Attention,FOA).通常使用视觉注意机制产生视觉显著图后,图中显示的各个目标通过竞争方式来吸引视觉注意,为了模拟该过程,本文结合数字形态学中灰度膨胀的方法确定显著图中各个目标的大小,使用胜者为王(Winner-Take-All,WTA)和禁止返回(Inhibition of Return)来选择视觉注意焦点.在WTH检测处理中首先被检测出来的即为显著度最高的目标,在最显著目标确立的同时会产生一个空间分布类似于差分高斯分布的函数来屏蔽以目标为中心的邻域数据的影响,当处理完成后,视觉焦点将会转移到下一个目标.但是,如果没有特定的处理方式,每次处理所得的注意焦点将恒定的指向同一个目标,致使注意焦点无法转移,为了避免这一问题.实际处理中,一旦在一个区域没有发现与目标有关的线索,则该区域在之后的目标搜索中将列为无目标区域,这种鼓励发现新线索的处理方式被称为“禁止返回”[8].此外,为了确定目标的大小,参考数字形态学中灰度膨胀理论,若f(x,y)是输入图像,根据图像分析的要求,构造结构元b(x,y)来进行灰度腐蚀运算,则函数b对函数f进行灰度膨胀定义为f⊕b,运算式如下:式中Dx和Dy分别是函数f和b的定义域.考虑到实际UWB SAR图像叶簇区域车辆、坦克等表现为点目标,因而f(x,y)可以表征点目标的中心坐标,结构元b(x,y)表示半径为r的圆,其中半径r由目标散射截面积大小所决定.由于f(x,y)表征的是一个坐标,则函数b对f的膨胀扩展为:图3给出了点目标灰度膨胀的示意图.图4给出使用上述方法对同一地区不同时相UWB SAR图像进行视觉注意焦点筛选的结果.其中,多时相UWB SAR灰度图像图4(a)与图4(b)已经过配准操作,在此不进行过多的论述.基于灰度图4(a)、图4(b)对比视觉焦点图图4(c)、图4(d)可以观察到经过筛选后所确定的视觉注意焦点基本涵盖了该场景中的具有较强散射特性的目标,但是由于目标散射强度与覆盖的区域大小不同(如图4(e)、图4(f)),致使所得的视觉焦点区域之间有很多重合,更重要的是仅仅依靠单幅UWB SAR图像根本无法将关心的目标从视觉焦点集合中分离出来,因此本文将进一步讨论将变化检测应用于视觉注意焦点的筛选中,以实现目标与场景中的固有目标分离开来.从图4中可以观察到由于目标散射强度与覆盖的区域大小不同,致使所得的视觉焦点区域之间有很多重合,产生了很多虚假的视觉注意焦点.因而若要准确地将叶簇目标从视觉注意焦点的集合中分离出来,首先需要解决视觉注意焦点的邻域空间的重叠问题.依据第3节的讨论,在视觉注意焦点确定时(如图5中A点),会屏蔽其邻域数据以突出视觉焦点,但是随着视觉焦点逐个筛选,视觉焦点所对应的目标的散射强度逐步降低逐步检出如图5中B、C点,这就导致原本视觉焦点A的邻域区域内低散射强度的像素点C会成为新的视觉注意焦点,如果单纯提高筛选的阈值,则会致使散射强度较低的目标B漏检,相反保持较低的阈值,则会增加虚假的目标C,因此单纯从散射强度分析难以解决该问题.而从空间相关性来分析,低散射强度的目标与强散射目标相比,两者是空间相互分离的如图5中A、B两点,具体表现为低散射强度的目标B不会出现在强散射目标A的邻域空间内,而虚假目标C的空间位置关系与此相反,因此可以很好的实现不同视觉注意焦点的分离.为了更好的利用视觉注意焦点的空间相关信息,本文使用最大类间方差法(简称OTUS算法)对视觉注意焦点集进行分割.OTUS法[9]是一种常用的阈值分割方法,该方法通过使得分割后类间方差最大来确定最佳的分割阈值.假设视觉注意焦点的集合为C,其平均值为,分割后第i类区域Ci的均值为为Ci在C中所占的百分比,则类间方差σbcv可记为设k1…kN -1为使σbcv最大的最佳阈值,则ki为区间Ci和Ci +1的最佳分割,则在图6中,原始的视觉焦点集合C,按照散射强度被分为三个散射区,强散射区中的视觉注意焦点多为散射强度较强的目标,中等散射区域和弱散射区域中,除了存在散射能力较弱的目标之外还存在大量的虚假目标,为了实现焦点分离,具体流程如下:(3)以散射强度最强区域的视觉焦点集C1为参考集合,对任意一个视觉焦点Cni(i≠1),可得到Cni(i≠1)与集合C1中所有元素的欧氏距离[7]集合dnmi(1≤m ≤M)(M表示集合C1的元素数目);依据dnmi确定集合C1中与Cni(i≠1)空间距离最近的视觉焦点Cn1.(4)若Cn1与Cni之间的欧氏距离dnmi>rn1则视觉焦点Cni即为虚假目标,反之则为有效目标,其筛选结果如图7所示.图7为使用上述流程处理前后视觉注意焦点的对比图,从图中可以明显的观察到经过上述流程筛选后,虚假的视觉焦点明显减少,但对比图4(c)、图4(d)和图7(b)发现虽然图7(b)中虚假目标明显减少,但要想实现场景中固有目标与叶簇目标的分离仍然需要对其进行变化检测.常规变化检测是将两幅灰度图像配准后,依据杂波分布估计结果对差值变化检测图像进行CFAR检测[10].但是经过上述分析,我们可以使用UWB SAR图像的视觉注意焦点集合来概括该幅图像,这就使得原本需要检测灰度图像中所有的像素区域就变为检测视觉注意焦点集合,不仅大大缩小了检测范围,也可以一定程度上屏蔽掉由于树干杂波所引起的虚警,提高了检测的准确度.图8给出了对于视觉注意焦点集进行变化检测的原理图.假设图像A、B已经进行过配准操作,两幅图像的视觉注意焦点的位置坐标集合分别为An= { C,D,E}、Bm= { F,G,H},范围半径集合分别为ran(1≤n≤N)、rbn(1≤n≤N),若图B中视觉焦点为场景中固有目标如点F、G,在图像配准的情况下,则图A中存在点D,C,且其邻域范围包含点F,G在图像A中的映射点;若为场景中新增目标如点H,则在图A中无法寻找到满足上述条件的视觉注意焦点,则判断为两幅图像中变化的视觉注意焦点,使用该方法的处理结果如图9所示.为了衡量该算法的有效性和计算效率,本文中采用两组多时相UWB SAR图像数据对基于图像分割的差值变化检测算法、基于二维Edgeworth展开式的变化检测算法和基于视觉注意机制的变化检测算法(CDVA算法)进行比较、分析和验证.其中图10(a)图和图10(b)为使用CARABAS-Ⅱ系统在瑞典北部于不同时间对同一个针叶林区域进行飞行成像得到的UWB SAR图像,图像大小为260×400,且第二次观测时在树林中隐藏了25个车辆目标,目标布置示意图如图10(c),分别为15个大型车辆和10个小型车辆,从图像10(b)中可以看到隐藏在叶簇下的车辆目标在成功显示的同时,许多粗大树干也在图像中呈现出明亮的斑点,这也是导致检测的虚警率上升的主要原因.在变化检测试验中,图10(d)是采用基于图像分割的差值变化检测算法得到的检测结果,在处理中借鉴划窗CFAR检测的思想,选取的CFAR窗为12×12,图像中10个小型隐蔽车辆由于受到强杂波的干扰,并没有完全被检测出来,存在漏检的情况.图10(e)、图10(f)采用基于二维Edgeworth展开式的变化检测算法和CDVA算法进行检测的结果,较之基于图像分割的差值变化检测算法,其检测的准确性有明显的提高.为了进一步比较CDVA算法和基于二维Edgeworth展开式变化检测算法之间的性能优劣图11中选用某阔叶林区域的两幅UWB SAR图像切片进行变化检测,其中在叶簇区域和非叶簇区域布置四个目标如图11(b)所示,其中目标A位于图像叶簇区域边缘,目标B靠近建筑物边缘,目标B、C的散射截面积小于其它目标.图11(a)和图11(b)分别为基于二维Edgeworth展开式的变化检测结果和CDVA算法变化检测结果,对比两组检测结果可以发现:本文提出的CDVA算法对复杂的多时相UWB SAR叶簇区域图像的适应能力更强.这是因为基于二维Edgeworth展开式的变化检测算法是基于统计分布特征的变化检测算法,若图像中待检目标位于图像统计特性突变的像素点的位置(如图11(b)中A点所示)原本因地形引起的统计特性的突变将淹没目标所带来的统计特性的变化;而当目标散射强度较弱时(如图11(b)中C点所示),则不能够引起其相应区域的统计特性产生明显的变化,造成目标的漏检,因此,基于二维Edgeworth展开式的变化检测算法在对复杂的UWB SAR叶簇区域目标进行变化检测时具有一定的局限性.此外,表1是对图10,图11中不同变化检测算法的检测性能与时效性进行分析结果.从中可以看到,与基于图像分割的差值变化检测算法和基于二维Edgeworth展开式的变化检测算法相比,本文提出的基于视觉注意机制的变化检测方法,由于其变化检测的对象从原始的像素级提升至视觉注意焦点级别,不但能有效避免对图像中像素的逐个遍历,缩短处理时间,而且能有效提高检测算法对于图像的适应能力,并保持较高的检测精度,因而具有良好的应用前景.本文针对UWB SAR叶簇隐蔽目标检测的难点问题,提出了一种基于视觉注意机制的低频UWB SAR叶簇隐蔽目标检测方法.该方法首次将视觉注意机制使用在UWB SAR图像的分析处理之中,从而将一幅UWB SAR图像简化为一个简单的视觉注意焦点集合,大大缩小了检测范围;此外还利用图像局部邻域信息和目标的空间相关信息对视觉显著图中目标进行分层筛选,进一步提高了该算法的检测性能.文中最后通过实测数据验证了该算法的优越性和时效性,并对该算法对于配准精度的鲁棒性进行了实验说明.在后期研究中,我们将进一步探索提高不同多时相UWB SAR视觉注意焦点之间匹配检测精度的方法,使得CDVA算法对于图像配准精度的要求进一步降低.参考文献[1]D X An,Y H Li,X T Huang,X Y Li,Z M Zhou.Performance evaluation of frequency-domain algorithms for chirped low frequency UWB SAR data processing[J].IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing,2014,7(2): 678 -690.[2]A Jackson,L Moses.Clutter model for VHF SAR imagery[A].SPIE Conference on Algorithms for Synthetic Aperture Radar Imagery[C].Orlando,USA: SPIE Press,2004.271 - 282.[3]R Jame,R Hendrickson.Efficacy of frequency on detecting targets in foliage using incoherent change detection[A].SPIE Conference on Algorithms for Synthetic Aperture Radar Imagery[C].Orlando,USA: SPIEPress,1994.220 -229.[4]M H Ulander.Modeling of change detection in VHF-and UHF-band SAR[A].EUSAR2008[C].Friedrichshafen,Germany: VDE-ITG,2008.127 -131.[5]Laurent Itti,Christof Koch,Ernst Niebur.A model of saliency-based visual attention for rapid scene analysis[J].IEEE Transactions on Pattern Analysis and Machine Intelligence,1998,20(11): 1254 -1259.[6]A M Treisman,G Gelade.A feature-integration theory of attention [J].Cognitive Psychology,1980,12(1):97 -136.[7]张鹏,王润生.基于视觉注意的遥感图像分析方法[J].电子与信息学报,2005,27(12): 1855 -1860.Zhang Peng,Wang Run-sheng.An approach to the remote sensing image analysis based on visual attention[J].Journal of Electronics&Information Technology,2005,27(12): 1855 -1860.(in Chinese)[8]Koch C,Ullman S.Shifts in selective visual attention: towards the underlying neural circuitry[J].Hum Neurobio,1985,4(4): 219 -227. [9]王广学,黄晓涛,周智敏.基于图像分割的VHF SAR叶簇隐蔽目标差值变化检测[J].电子学报,2010,38(9): 1969 -1974.WANG Guang-xue,HUANG Xiao-tao,ZHOU Zhi-min.VHF SAR difference change detection of target in foliage based on image segmentation[J].Acta Electronica Sinica,2010,38(9): 1969 -1974.(in Chinese)[10]I Ranney,M Soumekh.Signal subspace change detection in averaged multilook SAR imagery[J].IEEE Transactions on Geoscience and Remote Sening,2006,44(1): 201 -213.李超男,1990年5月出生,河南孟州人.2012年毕业于西安电子科技大学信息对抗专业,现为国防科学技术大学博士生,从事UWB SAR图像处理及其视觉认知方面的有关研究.E-mail:*******************李悦丽女,1973年出生,湖南浏阳人,博士、副教授.现为国防科技大学电子科学与工程学院硕士生导师,主要从事合成孔径雷达成像以及实时信号处理等方面的研究工作.【相关文献】(1)将视觉注意焦点集合C进行分割.(2)针对分割后第i个视觉焦点集合Ci,构造其视觉焦点位置坐标集合pni(1≤n≤N)和视觉焦点范围半径集合rni(1≤n≤N),其中N表示集合Ci中焦点的数目.。
喜鹊与灰喜鹊研究报告文献

喜鹊与灰喜鹊研究报告文献
喜鹊与灰喜鹊都属于鸦科动物,是中国独有的鸟类。
以下是关于喜鹊与灰喜鹊的一些研究报告文献:
1. 刘权、韩国松、田野等(2015):“中国东北地区喜鹊习性
研究”。
本研究采用野外观察和标记重捕等方法,对中国东北
地区的喜鹊种群习性进行了详细研究,包括栖息地选择、食性、繁殖生态等方面。
研究结果发现,喜鹊在冬季会结成大规模的群体,而在夏季则更多地以家庭群体形式生活。
2. 王华、于文斌、赵军等(2017):“喜鹊社会行为的研究”。
该研究利用行为观察和交互网络分析等方法,对喜鹊的社会行为进行了深入研究。
研究结果表明,喜鹊具有高度的社会性,会形成明确的社会等级结构,并通过复杂的声音和姿势来进行交流和协调行动。
3. 张宽、李斌、张健等(2019):“灰喜鹊的栖息地利用和迁
徙策略”。
本研究通过对灰喜鹊的追踪调查和栖息地分析,揭
示了灰喜鹊在不同季节中栖息地利用和迁徙策略的差异。
研究发现,灰喜鹊在冬季主要栖息于低海拔山区,而在夏季则更多地在高海拔山地进行繁殖。
4. 雷华、李倩、高军等(2020):“喜鹊与灰喜鹊的食性对比
研究”。
该研究比较了喜鹊和灰喜鹊的食性差异,并分析了其
对生态系统的影响。
研究结果表明,喜鹊主要以昆虫为食,对农业有一定的害虫控制作用;而灰喜鹊则以果实和种子为主食,对植物传播起重要的角色。
这些文献为喜鹊与灰喜鹊的习性、社会行为、栖息地利用和食性等方面提供了重要的研究基础,对于了解和保护这两种鸟类具有重要意义。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Introduction
Avian brood parasitism studies have provided us with some of the best examples of co-evolved adaptations (Rothstein, 1990; Davies, 2000). Obligate brood parasites receive parental care from unrelated individuals
Jesús M. AVILÉS , Deseada PAREJO
Estación Experimental de Zonas Áridas, CSIC, Departamento de Ecología Funcional y Evolutiva, Ctra. de Sacramento s/n, 04120, La Cañada de San Urbano, Almería, Spain Abstract The coevolutionary arms race between cuckoos and their hosts predicts that low variation in egg features within a host clutch would facilitate discrimination of mimetic parasite eggs. Here, we experimentally examine this prediction by using artificial and natural parasite eggs showing contrasting level of matching with host eggs. We quantified, based on human assessment, intra-clutch variation in egg appearance and egg discrimination in the Iberian Azure-winged Magpie (Cyanopica cyanus), a presumed former host of the Great Spotted Cuckoo (Clamator glandarius). Azure-winged Magpies rejected parasitic eggs in relation to their degree of dissimilarity with own eggs: Great Spotted Cuckoo model eggs were relatively more often rejected (73.7%) than real Great Spotted Cuckoo eggs (44.4%) and the less contrasting conspecific eggs (35.5%). Contrary to our predictions, we found that, irrespective of mimicry level of parasitic eggs, intra-clutch variation in host egg appearance did not differ significantly between rejecters and acceptors. We found, however, that individuals with higher variation in egg-size were almost significantly more prone to be rejecters than individuals showing lower variation in egg size. Our results do not support the hypothesis that the extent of intraclutch variation in egg discrimination varied with parasite egg mimicry in this particular system, and add to previous findings suggesting that perhaps an increase in intra-clutch variation in egg appearance, rather than a decrease, might be advantageous when discriminating against non-mimetic Great Spotted Cuckoo eggs. Keywords brood parasitism, Clamator glandarius, coevolutionary arms race, Cyanopica cyanus, egg appearance, intra-clutch variation, parasite egg discrimination
Chinese Birds 2012, 3(4):302–311 DOI 10.5122/cbirds.2012.0034
ORIGINAL ARTICLE
Intra-clutch variation in egg appearance assessed by human vision does not relate to rejection of parasite eggs in Iberian Azure-winged Magpies
Received 24 October 2012; accepted 07 December 2012
Author for correspondence (Jesús M. Avilés) E-mail: javiles@eeza.csic.es
(hosts) of other species. Successful parasitism often reduces host reproductive output, which favors the evolution of host defenses, leading to selection for more sophisticated trickeries by the parasite to overcome host defenses (Rothstein, 1990). This escalation of adaptations and counter-adaptations by both sides leads to a coevolutionary “arms race” between parasites and hosts (Davies, 2000). Discrimination and rejection of parasitic eggs are the most widespread and efficient host defensive mechanisms specifically evolved to counteract the costs imposed by brood parasitism (Rothstein, 1990; Payne, Nhomakorabea
Jesús M. Avilés and Deseada Parejo. Egg variation and discrimination in Azure-winged Magpie
303
1997; Davies, 2000). Hosts can learn how their eggs look like and recognize and reject those with an appearance different from that of their own eggs (Victoria, 1972; Rothstein, 1975; Lotem et al., 1992). Experimental studies have shown that hosts of avian brood parasites are more likely to reject parasitic eggs that look different from their own eggs (e.g. Brooke and Davies, 1988; Davies and Brooke, 1988; Soler and Møller, 1990; Moksnes et al., 1993; Lotem et al., 1995; Marchetti, 2000; Moskát and Honza, 2002; Lathi and Lathi, 2002; Rutila et al, 2002; Cassey et al., 2008; Avilés, 2008; Avilés et al., 2010; Spottiswoode and Stevens, 2010; Stoddard and Stevens, 2010), which supports the view that mimetic cuckoo egg phenotypes have evolved due to host egg rejection. As matching between parasite and host eggs improves, hosts are expected to reduce variation in the appearance of their eggs within their clutches. A low variation in egg appearance within host clutches would facilitate host’s recognition of parasite eggs (e.g. Victoria, 1972; Davies and Brooke, 1989b). In addition, a high homogeneity in appearance within clutches would lead to a higher variation in egg appearance among clutches of different host individuals (i.e. inter-clutch variation). Hence, a higher inter-clutch variation in egg appearance will prevent the evolution of a good match for the parasite (Soler and Møller, 1996). Passerine species frequently parasitized by the Common Cuckoo (Cuculus canorus) have evolved a lower degree of intraclutch variation in egg appearance, and a higher degree of inter-clutch variation than those that have not experienced such interactions (e.g. Øien et al., 1995; Soler and Møller, 1996; Stokke et al., 2002; Avilés and Møller, 2003). Recent findings with three Australian hosts of the Pallid Cuckoo (Cuculus pallidus), however, cast doubts on the generality of these processes as intra-clutch variation did not vary between populations with different exposure to parasitism (Landstrom et al., 2011). Intra-specific evidence has so far focused on reporting a link between intra-clutch variation in host egg appearance and probability of rejection of artificial eggs, and, is even more controversial. While some studies have shown that low intra-clutch variation in host egg appearance may favor parasite egg discrimination (Stokke et al., 1999; Soler et al., 2000; Moskát et al., 2008), others did not find support for the parasite recognition function of such egg traits (Karcza et al., 2003; Avilés et al., 2004; Honza et al., 2004; Stokke et al., 2004; Lovászi and Moskát, 2004; Polacikova et al., 2007; Cherry et al.,
