Lacosamide racemate_175481-26-2_DataSheet_MedChemExpress
FLIR A325sc 热成像相机说明书

FLIR A325scCámara termográfica para análisis en tiempo real CALIDAD DE IMAGEN Y SENSIBILIDAD TÉRMICA EXCELLENTFLIR A325sc está equipada con un detector microbolómetro de óxido de vanadio (VoX) de última generación sin refrigerar que genera imágenestérmicas claras de 320 x 240 píxeles. Estos píxeles generan imágenes nítidas y detalladas fáciles de interpretar con un elevado nivel de precisión. FLIR A325sc hace visibles incluso las más mínimas diferencias de temperatura de 50 mK.TRANSFERENCIA RÁPIDA DE DATOSFLIR A325sc incorpora una conexión Gigabit Ethernet RJ-45 que suministra imágenes de 320 × 240 y 14 bits a velocidades de hasta 60 Hz.COMPATIBILIDAD CON EL ESTÁNDAR GIGE VISION™GigE Vision permite una transferencia rápida de imágenes con cables de estándar de bajo coste y hasta 100 metros. Con GigE Vision, tanto el hardware como el software de otros proveedores pueden interactuar sin problemas en conexiones gigabit ethernet.COMPATIBLE CON EL PROTOCOLO GENICAM™GenICam crea una interfaz de programación de aplicaciones (API) para las cámaras independientemente de la tecnología de interfaz o las funciones que se implementen. Debido a que la API para las cámaras con GenICam siempre será la misma, las cámaras como A325sc se pueden integrar con toda facilidad en software de terceros.SOFTWARELa cámara FLIR A325sc funciona sin problemas con el software FLIR ResearchIR Max permitiendo una visualización intuitiva, grabación yprocesamiento avanzado de los datos termográficos proporcionados por la cámara. Como opción se dispone de un Kit para desarrolladores de software.MATHWORKS® MATLABControle la cámara y capture datos directamente en el software MathWorks ® MATLAB para análisis y procesamiento avanzados de las imágenes.CARACTERÍSTICAS PRINCIPALES• MICROBOLÓMETRO NO REFRIGERADO: 320 X 240 PÍXELES• INTERFAZ GIGABIT ETHERNET• LENTES DE APROXIMACIÓN Y TELEOBJETIVO DISPONIBLES • SOFTWARE RESEARCHIR MAX INCLUIDO• COMPATIBLE CON MATLABVerificación de PCBMotor de turbinaEspecificaciones de captura de imagenLas especificaciones están sujetas a cambios sin previo aviso.© Copyright 2014, FLIR Systems, Inc. Todas las demás marcas y nombres de productos son marcas registradas de sus respectivos propietarios. Las imágenes mostradas podrían no representar la resolución real de la cámara mostrada. Las imágenes son únicamente ilustrativas. (Creado en 08/14) NASDAQ: FLIRPORTLANDSede corporativa FLIR Systems, Inc.27700 SW Parkway Ave.Wilsonville, OR 97070EE. UU.Tlfn: +1 866.477.3687EUROPAFLIR Commercial Systems Luxemburgstraat 22321 Meer BélgicaTlfn: +32 (0) 3665 5100Fax : +32 (0) 3303 5624E-mail:*************ESPAÑAFLIR Commercial Systems Avenida de Bruselas, 15- 3º28108 Alcobendas (Madrid)EspañaTel. : +34 91 573 48 27Fax. : +34 91 662 97 48E-mail:*************Conector de alimentación,terminal a tornillo2 polos: 10-30 VCC, máx. <10 W Puerto Gigabit Ethernet, 1000 mB,conector RJ-45:Control y transferencia de imágenesConector de E/S digital terminal a tornillo de 6 polos:Salida digital: 2 salidas, con aislamiento óptico, suministro de 10-30 V, 100 mA.Entrada digital:2 entradas, con aislamiento óptico,10-30 V.。
Hybrid Plastics公司 Epoxycyclohexyl POSS

55 W.L. Runnels Industrial Drive; Hattiesburg, MS 39401SAFETY DATA SHEET1. IdentificationProduct Name Epoxycyclohexyl POSS® Cage Mixture Solution Product Number EP3F08.04Synonyms NACAS Number NAProduct Use VariousManufacturer Hybrid Plastics, Inc.55 Runnels DrHattiesburg, MS 39401USTelephone+1.601.544.3466Fax+1.601.545.3103EmailEmergency Telephone US & Canada: 1.800.255.3924International: +01.813.248.05852. Hazards IdentificationGHS ClassificationPhysical hazardsNoneHealth hazardsAcute toxicity, oral Classification not possibleAcute toxicity, dermal Classification not possibleAcute toxicity, inhalation Classification not possibleSkin corrosion/irritation Classification not possible Serious eye damage/irritation Classification not possible Sensitization, respiratory Classification not possibleSensitization, skin Category 1Germ cell mutagenicity Classification not possibleCarcinogenicity Classification not possibleReproductive toxicity Classification not possibleSpecific target organ toxicity, Classification not possiblesingle exposureSpecific target organ toxicity, Classification not possiblerepeated exposureAspiration hazard Classification not possibleEnvironmental hazardsHazardous to the aquatic environment, Category 3acute hazardHazardous to the aquatic environment, Classification not possiblelong-term hazardHazardous to the ozone layer Classification not possibleGHS Label ElementsSignal WordWarningHazard Statement(s)H317 May cause an allergic skin reactionH402 Hazardous to aquatic lifePrecautionary Statement(s)P261: Avoid breathing fumes/mist/vapors/spray.P272: Contaminated work clothing should not be allowed out of the workplace.P273: Avoid release to the environment.P280: Wear protective gloves.P302 + P352: IF ON SKIN: Wash with plenty of soap and water.P333 + P313: If skin irritation or rash occurs: Get medical advice/attention.P363: Wash contaminated clothing before reuse.P501: Dispose of contents/container to an approved waste disposal plant.3. Composition/Information on IngredientsChemical Identity CAS# EC# Conc. Epoxycyclohexyl Silsesquioxanes 187333-74-0 NA 65-75% 3,4-Epoxycyclohexylmethyl 3,4-epoxycyclohexanecarboxylate 2386-87-0 NA 25-35%4. First Aid MeasuresInhalationRemove to fresh air. If breathing becomes difficult, seek immediate medical attention.Skin ContactWash off with soap and water.Eye ContactFlush eyes with plenty of water.IngestionWash out mouth with water if person is conscious.5. Fire Fighting MeasuresSuitable extinguishing mediaUse water spray, carbon dioxide, dry chemical powder or alcohol-resistant foam.Special protective equipment and precaution for fire fightersFire fighters exposed to vapors should wear a self-contained breathing apparatus and full protective clothing to prevent contact with skin and eyes.Unusual Fire and Explosion HazardsFlammable liquid and vapor.Combustion ProductsIrritating or toxic substances may be emitted upon thermal decomposition. Thermal decomposition products may include oxides of carbon, silicon and nitrogen6. Accidental Release MeasuresPersonal precautionsExercise appropriate precautions to minimize direct contact with skin or eyes.Environmental precautionsDo not let product enter drains.Methods for cleaning upUse suitable absorbent, sweep up, place in bag and hold for disposal. Ventilate area and wash spill site after material pick up is complete.7. Handling and StorageHandling precautionHandle in a fume hood or in properly ventilated area. Avoid contact with eyes, skin, and clothing. Avoid prolonged or repeated exposure.Storage precautionAmbient temperatures in tightly closed containers.8. Exposure Controls/Personal ProtectionRespiratory protectionWhere respiratory protection is desired, use respirators and components tested and approved under appropriate government standards such as NIOSH (US) or CEN (EU).Hand protectionWear protective gloves. Wash thoroughly after handling.Eye protectionWear chemical safety goggles or a face shieldSkin and body protectionChoose body protection in relation to its type, to the concentration and amount of dangerous substances, and to the specific workplace. The type of protective equipment must be selected according to the concentration and amount of the dangerous substance at the specific workplace.Hygiene measuresUse common industrial hygiene practices.9. Physical and Chemical PropertiesAppearance Clear to hazy liquidOdor No data availableOdor threshold No data availablepH No data availableMelting/freezing point No data availableInitial boiling point and range > 170 °C (> 338 °F)Flash point 118 °C (244 °F) Closed CupEvaporation rate No data availableFlammability Not flammableUpper/lower flammability explosive limits No data availableVapor pressure No data availableVapor density No data availableRelative density 1.2Solubility(ies) Water < 1g/l @ 20 °C (68 °F)Partition coefficient (n-octanol/water) No data availableAutoignition temperature No data availableDecomposition temperature No data availableViscosity No data available10. Stability and ReactivityReactivityNo data availableChemical stabilityStable under recommended storage conditions.Possibility of hazardous reactionsVapors may form explosive mixture with air.Conditions to avoidSparks, heat, flamesIncompatible MaterialsStrong oxidizing agents, strong bases, strong reducing agentsHazardous decomposition productsCarbon dioxide, Carbon monoxide, Silicon Oxides11. Toxicological InformationAcute toxicityNo data availableSkin corrosion/irritationNo data availableSerious eye damage/eye irritationNo data availableRespiratory or skin sensitizationNo data availableGerm cell mutagenicityNo data availableCarcinogenicityIARC: No component of this product present at levels greater than or equal to 0.1% is identified as a probable, possible or confirmed human carcinogen by IARC.Reproductive toxicityNo data availableSpecific target organ toxicity – single exposureNo data availableSpecific target organ toxicity – repeated exposureNo data availableAspiration hazardNo data availableAdditional InformationTo the best of our knowledge the toxicological properties have not been thoroughly investigated.12. Ecological InformationToxicityNo data availablePersistence and degradabilityNo data availableBioaccumulative potentialNo data availableMobility in soilNo data availablePBT and vPvB assessmentNo data availableOther adverse effectsNo data available13. Disposal ConsiderationsProductContact a licensed waste disposal service to dispose of this material.Contaminated packagingDispose of as unused product.14. Transport InformationClassification for road and rail transport (ADR/RID)Not regulatedClassification for sea transport (IMO-IMDG)Not regulatedClassification for air transport (IATA/ICAO)Not regulated15. Regulatory InformationU.S. Federal Regulations:This product is not currently regulated by SARA/EPCRA TSCA:Not listed. R&D use onlyREACH: Not registered16. Other InformationReviewed by: Director of Commercial ProductsDate prepared: 05.03.2016The information and recommendations contained in this Safety Data Sheet are from sources believed to be reliable and to represent the most reasonable current opinion on the subject when the SDS was prepared. While the above information is believed to be accurate, no warranty, guaranty, or representation is made as to the correctness or sufficiency of the information and the information is intended only as a guide. Hybrid Plastics shall not be held liable for any damage resulting from handling or from contact with this product. The user of this product must decide what safety measures are necessary to safely use this product, either alone or in combination with other products, and determine environmental regulatory compliance obligations under any applicable laws.。
波士顿·雷克斯罗特电子控制系统简介说明书

1/26Information on available spare parts:/spcVariable-speed pressure and flow control system Sytronix DFEn 5000Type SYDFEn-2XWith axial piston variable displacement pump A10VSO.../31Size 18 to 140Component series 2XMaximum operating pressure 280 barRE 62240/12.11Replaces: 30030, onlyType SYDFEnTable of contentsFeaturesAn SYDFEn-2X control system is used for the electro-hydraulic control of swivel angle, pressure and power/torque of an axial piston variable displacement pump.The control system consists of the following components:- Axial piston variable displacement pump A10VSO.../31- VT-DFPn-2X proportional valve as pilot valve including induc-tive position transducer for valve position sensing. The pilot valve includes electronics for control of the system.- Position transducer for sensing the swivel angle- Pressure transducer with suitable signal level and dynamics (optionally HM 16, otherwise separate order)- Preload valve with integrated pressure relief function SYDZ (optional)Contents PageFeatures 1Ordering code 2Cross section 6Schematic diagrams 7Technical data 9Electrical connection 11Closed-loop control quality 12Transition function 12Unit dimensions14Unit dimensions: Combination pumps 15Hubs for through-drives 16Unit dimensions: Through-drives17Torsionally flexible couplings for attachment to a standard electric motor24Project planning information25More information about this control system25H7111_dC o u r t e s y o f C M A /F l o d y n e /H y d r a d y n e ▪ M o t i o n C o n t r o l ▪ H y d r a u l i c ▪ P n e u m a t i c ▪ E l e c t r i c a l ▪ M e c h a n i c a l ▪ (800) 426-5480 ▪ w w w .c m a f h .c o mO rdering code: Pump of the Sytronix DFEn 5000 control system SYDFEn-2X/071R -P R A 12N00-0000-…123456789See following pagesSeries1Control system with internal digital electronics, variable-speed, DFEn 5000SYDFEn-2XPump combinations (see order example page 4)SY2DFEn-2X, SY3DFE3-2X● = available - = not available Preferred program1) ANSI B92.1a-1976, 30° pressure angle, flat root, side fit, tolerance class 52) Also observe the conditions for the attachment pumps on page 16.Ordering code: Pilot and preload valve of the Sytronix DFEn 5000 control system SYDF En-2X/071R-P R A12N00-0000-A0A0F L2-* 123456789101112131415161717Further details in the plain text e.g. SO variantComment on feature 11: Valve, installation orientation of integrated electronicsClockwise direction of rotation, installation orientation 0Clockwise direction of rotation,installation orientation 2Counterclockwise direction of rotation,installation orientation 0Counterclockwise direction of rotation,installation orientation 21) With the SYDFEn control system with the additional function (feature 12 of the ordering code) "Teach-in version for cyclic operation" and with analog interfaces, the plug-in connector X2 cannot be used as actual pressure value input. Thus, a sepa-rate pressure transducer has to be used and connected to plug-in connector X1 in this case.Ordering code: Order examplesOrder example for single pump:SYDFEn-2X/100R-PSA12KC3-0000-A0A0VXXO rder example for pump combination:Both material numbers and/or type designations are to be connected by means of "+".Main pump (1st pump)+Attachment pump (2nd pump)SY2DFEn-2X/100-100/01292063+01292063SY2DFEn-2X/100-100/SYDFEN-2X/100R-PSA12KD5-0000-A0A0VX3+SYDFEN-2X/100R-PSA12KD5-0000-A0A0VX3 DoublepumpSize of the main pumpSize of the attachment pumpMaterial number without "R9" for the main pump ortype designation if material number is not knownPump combination, mounted with accessoriesMaterial number without "R9" for the attachment pump or type designation if material number is not knownExample of name plate of an SY2DFEn pump combinationWord markSY2DFEN-2X/071-071/01234567+01234567Notice:For enquiries regarding the control system, material number,production order number, serial number, and date of manu-facture are necessary.O rdering code: AccessoriesVersion 10/2011, enquire availabilityAccessories for Sytronix DFEn 5000Material number Data sheetMating connector 12-pin for central connection X1 without cable (assembly kit) R90088467108006 Mating connector 12-pin for central connection X1 with cable set 2 x 5 m R900032356Mating connector 12-pin for central connection X1 with cable set 2 x 20 m R900860399Mating connector for interface X3, M12, straight, can be connected independently,R9010769105-pin, shielded, A-coded, cable diameter 6...8 mmPressure transducer HM 12-1X measurement range 315 bar (4...20 mA)R90019987129933 Pressure transducer HM 13-1X measurement range 315 bar (0...10 V)R90017437429933 Pressure transducer HM 17-1X measurement range 315 bar (4...20 mA)R90077306530269 Pressure transducer HM 17-1X measurement range 315 bar (0.1...10 V)R90077312430269 Test device VT-PDFE-1-1X/V0/0R90075705129689-B Compact power supply unit VT-NE32-1X R90008004929929Converter USB/serial for laptops without serial interfaceR901066684VT-ZKO-USB/S-1-1X/V0/0Cable for connecting a Win-PED PC (RS232) to the X2 interface, length 3 m R901156928T connector for the simultaneous connection of a Win-PED PC (RS232) andR901117164use of the pressure transducer at connector X2More accessories PageHubs for through-drives16Torsionally flexible couplings for attachment to a standard electric motor241Swash plate 2Pilot valve 3Counter spool 4Actuating piston 5Spring6Inductive position transducer for valve position 7Swivel angle position sensor 8Proportional solenoid 9Valve spoolCross section10Spring11Integrated electronics 12Connector X113Connector X2 for connection of the HM 16 pres-sure transducer 14Mating connector X3 for connecting the CAN bus 15Drive shaft 16Connection flange17Subplate, optionally with through-driveSchematic diagram: Actuating system supplied internallyActuator(q V; p)1) When using the HM 16 pressure transducer:Installation in P (pump) or MP1 (preload valve) in connection with electronic version "actual pressure value input F". When using an external pressure transducer:Installation in the P1 line (preferably close to the actuator) and electrical connection via the central connector. When using a preload valve, the pressure transducer is to be connected to P1 or MP1.(q V ; p )Schematic diagram: Actuating system supplied externallyActuator 1) The use of a pressure relief and anti-cavitation valve (checkvalve with 0.2 bar spring) is essential in order to prevent dry-running in case of an error.I mportant notices on the external supply:– In the case of an actuating system with external supply, the pump will - in case of voltage failure - not swivel to zero stroke but to the negative stop (displacement of 100 % flow from the system to the tank).– With an active fault message, it is imperative that the machine control reacts (e.g. switching off the drive motor of the pump, interrupting the external supply of the actuating system).– Command values for pressure and flow must always be greater than zero (p Command ≥ 3 bar, αCommand ≥ 5 %), as due to drift or tolerances, there is no exact "zero" pressure or "zero" swivel angle. In the unfavorable case, smaller command value provisions may lead to cavitation.– The actual pressure value must not be less than 10 bar for more than 10 minutes (lubrication).3) Maximum pressure limitation must be provided by the customer!4) Observe upper limit for external pilot oil pressure! (seeoperating instructions), recommendation: 20 bar absolute.2)Pressure transducerMounting optionsCommentHM 16P Only in connection with actual pressure value input "F"HM 12 / HM 13 / HM 17P1Preferably close to the actuatorTechnical data (For applications outside these parameters, please consult us!)1) The values are applicable at an absolute pressure of 1 barin suction port S. With a reduction of the displacementor an increase in the inlet pressure, the speed can beincreased according to the following characteristic curve.With a reduced inlet pressure, the speed is to be reduced.2) In case of higher radial forces, please consult us3) In case of higher pressures, please consult us1,21,11,00,90,70,80,91,01,61,21,00,91,40,8Displacement Vg/Vgmax→Speedn/nmax→Inletpressurepabs[bar]→electricOperating voltage UB24 VDC +40 % –5 % Operating range (short-time operation)Upper limit UB (t)max35 VLower limit UB (t)min21 VCurrent consumption (in static control operation)Rated current IRated0.6 AMaximum current Imax1.25 AInputs Actual pressure value inputX1; pin 10 and 11U or I Parameterizable:0...20 mA; 4...20 mA;0...10 V; 0…5 V; 0.5…5 V; 0.1...10 V; 1...10 VAnalog current inputs, load RB100 ΩAnalog voltage inputs RE≥ 100 kΩDigital inputs Logic 0≤ 8 VLogic 1≥ 14 VOutputs ncommand/ UOUT1 1)UAImax±10 V2 mAαactual/ UOUT2 2)UAImax±10 V2 mADigital outputs Logic 0Ua< 1 VLogic 1Ua≥ UB– 5 V; 10 mA (short-circuit-proof)Ambient temperature range at the pump ϑ0…50 °CStorage temperature range (pump+electronics)ϑ0…70 °CElectronics design Integrated in the pilot valve (OBE) Electrical connection See page 11Protection class according to EN 60529Pump incl. pilot valve IP 65 with mounted and locked plug-in connectors Technical data (For applications outside these parameters, please consult us!)Notice:For information on the environment simulation testing for the areas of EMC (electromagnetic compatibility), climate and me-chanical load, see data sheet 30030-U.1, 2) The outputs are parameterizable, condition as supplied see page 111234567891011E lectrical connection X2: Serial interface RS232 and a selectable digital input S1/pressure transducer input for HM 16(mating connector M12)Top viewMating connectorPin Signal input Pin Signal RS2321OUT, +U B2RxD3Reference L04Analog input 0.5...5 V for HM 16 or digital input 0 V low, 10 V high (max. 12 V)Depending on additional function (feature 12 of the ordering code):– Teach-in version: Digital input "Variable-speed operation ON, S1"– Real-time version: Input as analog input for pressure trans-ducer HM 165TxDAssignment of connector or mating connector and cable set Pin Signal Description Signal direction Type ofsignal Assignment in the cable set (accessories)1+U B Voltage supplyIN 24 V DC1Supply line 3 x 1.0 mm ²20 V = L0Reference potential for the voltage supply -2PE Earth Earthing connection for the electronics-Green/yellow 3Fault Signals failures, e.g. cable break command / ac-tual values, controller monitoring (logic 0 = error)OUT Logic 24 V White Supply line 10 x 0.14 mm ² shielded (one end of the shield must be con-nected to the control!)4M0Reference potential for analog signals -Yellow 5AI2Analog input AI2Standard: Swivel angle command value IN Analog ±10 V Green 6U OUT2Analog outputStandard: Actual swivel angle value normalized OUT Analog ±10 V Violet7AI1Analog input AI1Standard: Pressure command value IN Analog 0...10 V Pink 8U OUT1Analog outputStandard: Speed command value OUTAnalog ±10 VRed9DI1Digital input DI1Depending on additional function (feature 12 ofthe ordering code):– Teach-in version: Synchronization bit DI1– Real-time version: Activate real-time operationINLogic 24 VBrown10Actual pres-sure value H Actual pressure value input: Signal level depends on feature 14 in the ordering code. INAnalog Black 11Actual pres-sure value L -AnalogBlue n.c.GrayX1: Central connectionMating connector according to EN 175201-804 (12-pin), ordering code see section Accessories on page 5Closed-loop control qualitySwivel angle controlPressure control 1)Linearity tolerance ≤ 1.0 %≤ 1.5 % (≤ 1.0 % 2)Temperature error ≤ 0.5 % / 10 K ≤ 0.5 % / 10 K Hysteresis ≤ 0.2 %≤ 0.2 %Repeatability≤ 0.2 %≤ 0.2 %1) Without considering the pump pulsation 2) Using the integrated calibration functionTransition function with pressure command value step with spool design "A"The specified curve shapes and control times refer to a drive speed of 1500 rpm and are only reached with an optimization of the pressure controller.Notices:– The specified values are only valid when using the system-related components specified in this data sheet.– At pressures < 20 bar, higher tolerances have to be anticipated due to lower actuating forces.T 95 % in ms with a connected hydraulic fluid volume (lines and actuators)Hydraulic fluid volumeT 95 %< 5 l 150 ms 5 – 10 l 200 ms 15 – 25 l250 msFor pressures up to 40 bar, the values of the response times are larger.Top view ConnectorPin Signal input Pin Signal CAN 1n.c.3CAN GND 2IN, digital IN2 (DI2)Depending on additional function (feature 12 of the ordering code):– Teach-in version: Start teach-in, S2– Real-time version: Manual speed provision active, speed isaccepted according to the real-time operation status and the setting of the R parameters.4CAN-HIGH5CAN-LOW X3: CAN bus and digital input 2 (connector M12)E lectrical connection (continued)Transition function with swivel angle command value step with spool desi gn "A"Size 100 p = 50 barSize 140 pUnit dimensions (dimensions in mm)Size 18 to 140(Valve mounting direction "0"; shaft design "S"; without through-drive "N00")with direction of rotation counterclockwise with direction of rotation clockwiseSpace required for removing Dimension A7 → installation space required for connecting the optional pressure transducer HM 16Pilot oil port "Z"Size A1A3A4A5A6A6 I A6 II A7A8 I A8 II A8 III 18120198158631786311560233125100281282081586319580115602431351154513421815863205901156025314512571146232158632541041506026715915010015123715863247100147602721641501401622501437825711014760285182150The unit dimensions of the base pump (axial piston variable displacement pump A10VSO.../31) are contained in data sheet 92711.Unit dimensions: Combination pumps (dimensions in mm)Main pump A10VSO 18A10VSO 28A10VSO 45A10VSO 71A10VSO 100A10VSO 140Attachmentpump A 1A 2A 3A 4A 1A 2A 3A 4A 1A 2A 3A 4A 1A 2A 3A 4A 1A 2A 3A 4A 1A 2A 3A 4A10VSO 18164204349399164204349399184229374424217267412462275338483533275350495554A10VSO 28164204368.5410184229393.5435217267431.5473275338502.5544275350514556A10VSO 45184229413453217267451491275338522562275350534574A10VSO 71217267484524275338555595275350567609A10VSO 100275338613664275350625679A10VSO 140275350625688A10VSO.../31 + A10VSO.../31(SYDFEn-2X/... + SYDFEn-2X/...)Main pump Attachment pumpH ubs for through-drivesHubs for the combination of single pumps or the combination of SYDFEn with other pumps. Observe that the attachment pump has a splined shaft SAE J744 with the specified diameter.To the attachment pumps listed in the table, the following conditions apply:– SYDFE and A10VSO with shaft S or R– Internal gear pump PGH with shaft R, flange U2, see data sheet 10223– Internal gear pump PGF3 with shaft J, flange U2, see data sheet 10213– External gear pump AZPF with shaft R, front cover R, see data sheet 10089Also observe that the through-drive of the main pump and the flange of the attachment pump (see ordering code page 2)are identical. Check in the current data sheet of the gear pump whether the shaft ends have the specified dimensions.Main pump SYDFE or A10VSO...Attachment pump Size 18Size 28Size 45Size 71Size 100Size 140ø shaft Pump type (examples)R902436099R902436199R902436100R902436200R902436201R9024362023/4″ 19-4(SAE A-B)SYDFE-2X, A10VSO..31Size 018 shaft SR902436098R902436084R902436083R902436101R9024361027/8″ 22-4(SAE B)SYDFE-2X, A10VSO..31Size 028 shaft RPGF3R902436103R910968921R902436105R9024362041″ 25-4(SAE B-B)SYDFE-2X, A10VSO..31Size 045 shaft RPGH4R902436085R902436086R9024361061 ¼″ 32-4(SAE C)SYDFE-2X, A10VSO..31Size 071 shaft RR910943565R9109435551 ½″ 38-4(SAE C-C)SYDFE-2X, A10VSO..31Size 100 shaft SPGH5R9109321721 ¾″ 44-4(SAE D)SYDFE-2X, A10VSO..31Size 140 shaft SR910943528R910986299R910943529R910943545R910943560R9109435515/8″ 16-4(SAE A)1PF2G2, PGF2,PGH2, PGH3, AZPFUnit dimensions: Through-drives (dimensions in mm)KD3Flange ISO 100, 2-hole for the attachment of– SYDFEn-2X (size 28 and size 45, flange A)– A10VSO..31 (size 28 and size 45, flange A, see data sheet 92711)Sectional presentation with examples for hubs(order number for hubs see page 16) Top view Hub 7/8″Hub 1″Size A1A2A3A4A5A62820441.717.8--M12; 15 right through4522941.717.946.718.4M12; 14 right through7126744.120.349.120.8M12; 20 deep1003384117.645.918.2M12; 20 deep14035041.11845.918.3M12; 20 deepUnit dimensions: Through-drives (dimensions in mm)KD5Flange ISO 125, 2-hole for the attachment of– SYDFEn-2X (size 71 and size 100, flange A)– A10VSO..31 (size 71 and size 100, flange A, see data sheet 92711)Sectional presentation with examples for hubs(order number for hubs see page 16) Top view Hub 1 ¼″Hub 1 ½″Size A1A2A3A4A5A67126758.621.8--M16; 20 right through10033856.419.563.97.9M16; 20 deep14035055.417.473.37.9M16; 24 deepUnit dimensions: Through-drives (dimensions in mm)KD7Flange ISO 180, 4-hole for the attachment of– SYDFEn-2X (size 140, flange B)– A10VSO..31 (size 140, flange B, see data sheet 92711)Sectional presentation with examples for hubs(order number for hubs see page 16) Top view Hub 1 ¾″140350758M16; 22 right throughUnit dimensions: Through-drives (dimensions in mm)KC1Flange SAE 82-2 (SAE A, 2-hole) for the attachment of– SYDFEn-2X (size 18, flange C)– A10VSO..31 (size 18, flange C, see data sheet 92711)– PGF2 (shaft J, flange U2, see data sheet 10213)– PGH2 and PGH3 (shaft R, flange U2, see data sheet 10223)– AZPF (shaft R, front cover R, see data sheet 10089)Sectional presentation with examples for hubs(order number for hubs see page 16)Top view Hub 5/8″Hub 3/4″Size A1A2A3A4181824043M10; 14.5 deep282043947M10; 16 deep4522940.553M10; 16 deep712674061M10; 20 deep1003384065M10; 20 deep1403504177M10; 17 deepUnit dimensions: Through-drives (dimensions in mm)KC3Flange SAE 101-2 (SAE B, 2-hole) for the attachment of– SYDFEn-2X (size 28 and size 45, flange C)– A10VO..31 (size 28 and size 45, flange C, see data sheet 92701)– PGF3 (shaft J, flange U2, see data sheet 10213)– PGH4 (shaft R, flange U2, see data sheet 10223)Sectional presentation with examples for hubs(order number for hubs see page 16) Top view Hub 7/8″Hub 1″Size A1A2A3A4A5A6A7282044316.547-M12; 15 deep452294216.55318.446.7M12; 18 deep712674316.56120.849.1M12; 20 deep1003384116.56510.565M12; 20 deep1403504416.57718.345.9M12; 20 deepUnit dimensions: Through-drives (dimensions in mm)KC5Flange SAE 127-2 (SAE C, 2-hole) for the attachment of– SYDFEn-2X (size 71 and size 100, flange C)– A10VO..31 (size 71 and size 100, flange C, see data sheet 92701)– PGH5 (shaft R, flange U2, see data sheet 10223)Sectional presentation with examples for hubs(order number for hubs see page 16) Top view Hub 1 ¼ ″Hub 1 ½ ″Size A1A2A3A4A5A6A77126755.517.961--M16; 18 deep1003385717.965865M16; 25 deep1403506017.977977.3M16; 32 deepUnit dimensions: Through-drives (dimensions in mm)KC6Flange SAE 152-4 (SAE D, 4-hole) for the attachment of – SYDFEn-2X (size 140, flange D)– A10VO..31 (size 140, flange D, see data sheet 92701)Sectional presentation with examples for hubs(order number for hubs see page 16)Top viewHub 1 ¾ ″Size A1A3A4A514035010.577M16; 24 deepT orsionally flexible couplings for attachment to a standard electric motor Motor SYDFEn-2XFrame size/ characteristic Shaft diameter Size 18Shaft S, 3/4 ″Size 28Shaft S or R, 7/8 ″Size 45Shaft S or R, 1 ″100/0112/028R901038012R901038017 132/038R900704699R901012344R900772898 160/042R900726977R900991864R900994283 180/048R900032918R900062159 200/055R901038026R901038025 225/060R900750847R901066409 250/065R900988348Motor SYDFEn-2XFrame size/ characteristic Shaft diameter Size 71Shaft S or R, 1 ¼″Size 100Shaft S, 1½ ″Size 140Shaft S, 1 ¾ ″160/042R900228413180/048R900240468R900242567200/055R901038021R901104689R901038048 225/060R900228375R901050508R900988121 250/065R900986404R901046864R900708084 280/075R900218487R901055216R901052451 315/080R901046894 1)R901041730 1) 315/180R9010468851) Up to 40 °CProject planning information– Always shield command and actual value lines. Observe the notices in the instructions 30014-B, section 7.6.– The distance to aerial lines or radios must be at least 1 m.– Do not lay signal lines close to power cables.– Supplementary notices on the SYDFEn control system can be found in the operating instructions (See section "More infor-mation about this control system" on this page).More information about this control systemOperating instructions for SY(H)DFEn30014-BUser manual CANopen interface for SY(H)DFEn30014-02-ZData sheet for axial piston variable displacement pump A10VSO../3192711Data sheet for pilot valve VT-DFP.-2X29016Data sheet for pump preload valve SYDZ 0001-1X29255Data sheet for swivel angle sensor VT-SWA-1-1X30268Data sheet for pressure transducer HM 12-1X and HM 13-1X29933Data sheet for pressure transducer HM 16-1X30266Data sheet for pressure transducer HM 17-1X30269Operating instructions for test device VT-PDFE29689-BCurrent information is also available on the Internet at the address /sydfe (English) or http://www.boschrexroth.de/sydfe (German).Bosch Rexroth AG HydraulicsZum Eisengießer 197816 Lohr am Main, Germany Phone +49 (0) 93 52 / 18-0 Fax +49 (0) 93 52 / 18-23 58 ***************************** www.boschrexroth.de © This document, as well as the data, specifications and other informa-tion set forth in it, are the exclusive property of Bosch Rexroth AG. It may not be reproduced or given to third parties without its consent.The data specified above only serve to describe the product. No state-ments concerning a certain condition or suitability for a certain applica-tion can be derived from our information. The information given does not release the user from the obligation of own judgment and verification. It must be remembered that our products are subject to a natural process of wear and aging.NotesBosch Rexroth AG HydraulicsZum Eisengießer 197816 Lohr am Main, Germany Phone +49 (0) 93 52 / 18-0 Fax +49 (0) 93 52 / 18-23 58 ***************************** www.boschrexroth.de © This document, as well as the data, specifications and other informa-tion set forth in it, are the exclusive property of Bosch Rexroth AG. It may not be reproduced or given to third parties without its consent.The data specified above only serve to describe the product. No state-ments concerning a certain condition or suitability for a certain applica-tion can be derived from our information. The information given does not release the user from the obligation of own judgment and verification. It must be remembered that our products are subject to a natural process of wear and aging.NotesBosch Rexroth AG HydraulicsZum Eisengießer 197816 Lohr am Main, Germany Phone +49 (0) 93 52 / 18-0 Fax +49 (0) 93 52 / 18-23 58 ***************************** www.boschrexroth.de © This document, as well as the data, specifications and other informa-tion set forth in it, are the exclusive property of Bosch Rexroth AG. It may not be reproduced or given to third parties without its consent.The data specified above only serve to describe the product. No state-ments concerning a certain condition or suitability for a certain applica-tion can be derived from our information. The information given does not release the user from the obligation of own judgment and verification. It must be remembered that our products are subject to a natural process of wear and aging.Notes。
Lacosamide_175481-36-4_MSDS_MedChemExpress

MSDS1 Composition7 Accident Release MeasureProduct Name:LacosamideChemical Name:PROCEDURE(S) OF PERSONAL PRECAUTION(S)-Wear respirator, chemical safety goggles, rubber boots, and heavyrubber gloves.METHODS FOR CLEANING UP-Sweep up, place in a bag and hold for waste disposal. Avoid raising dust. Ventilate area andwash spill site after material pickup is complete.Propanamide, 2-(acetylamino)-3-methoxy-N-(phenylmethyl)-,(2R)-CAS No.:175481-36-48 Accident Release MeasureAppearance:White to off-white(Solid)Formula:C13H18N2O39 Toxicological InformationSolubility:To the best of our knowledge, the chemical, physical, andtoxicological properties have not been thoroughly investigated.No data available.p p p p DMSO ≥48mg/mL Water ≥24mg/mL Ethanol ≥48mg/mL2 Handling and Storage10 Regulary Information3 Stability and Reactivity11Disposal ConsiderationsCLASSIFICATION- Substance not yet fully tested.SAFETY PHASES- 26-36 (In case of contact with eyes, rinse immediately with plenty of water and seek medical advice. Wear suitable protective clothing.) 36/37/38 (Irritating to eyes,respiratory system and skin.)STABILITY- Stable under normal handling conditions.HANDLING- Do not breathe dust. Avoid contact with eyes,skin,and clothing.Avoid prolonged or repeated exposure.STORAGE- Store in a properly sealed container store at -20℃,shelflife is 2 years.11 Disposal Considerations 4 Hazards Identification12 Transport Information5First Aid RID/ADR- Non-hazardous for road transport. IMDG- Non-hazardous for sea transport.IATA - Non-hazardous for air transport.As specific country, federal, state and local environmentalregulations vary and change frequently we suggest you contact a local, authorized waste disposal contractor for adequate disposal.Special indication of hazards to humans and the environment.Irritating to eyes, respiratory system and skin.MATERIALS TO AVOID- Strong oxidizing agents.REACTIVITY- May emit toxic gasses like Carbon monoxide,Carbon dioxide, Nitrogen oxides upon thermal decomposition.5 First Aid13 Other InformationThe above information is believed to be correct but does not purport to be all inclusive and shall be used only as a guide. The information in this document is based on the present state of our knowledge and is applicable to the product with regard to appropriate safety precautions. It does not represent any guarantee of the properties of the product. Medchemexpress LLC shall not be held liable for any damage resulting from h dli f t t ith th b d tINHALATION- If inhaled, remove to fresh air. If not breathing give, artificial respiration. If breathing is difficult, give oxygen.SKIN CONTACT- In case of contact, immediately wash skin withsoap and copious amounts of water.EYE CONTACT- In case of contact, immediately flush eyes withcopious amounts of water for at least 15 minutes.INGESTION- If swallowed, wash out mouth with water provided person is conscious. Call a physician.6 Fire Fighting Measureshandling or from contact with the above product.EXTINGUISHING MEDIA Water spray- Carbon dioxide, dry chemical powder, or appropriate foam.SPECIAL RISKS Specific Hazard(s)- Emits toxic fumes under fire conditions. SPECIAL PROTECTIVE EQUIPMENT FOR FIREFIGHTERS Wear self-contained breathing apparatus and protective clothing Caution: Not fully tested. For research purposes onlyMedchemexpress LLCto prevent contact with skin and eyes.18W i l k i n s o n W a y , P r i n c e t o n , N J 08540,U S AE m a i l : i n f o @m e d c h e m e x p r e s s .c o m W e b : w w w .m e d c h e m e x p r e s s .c o m。
GDC-0834 Racemate_1133432-46-8_DataSheet_MedChemExpress
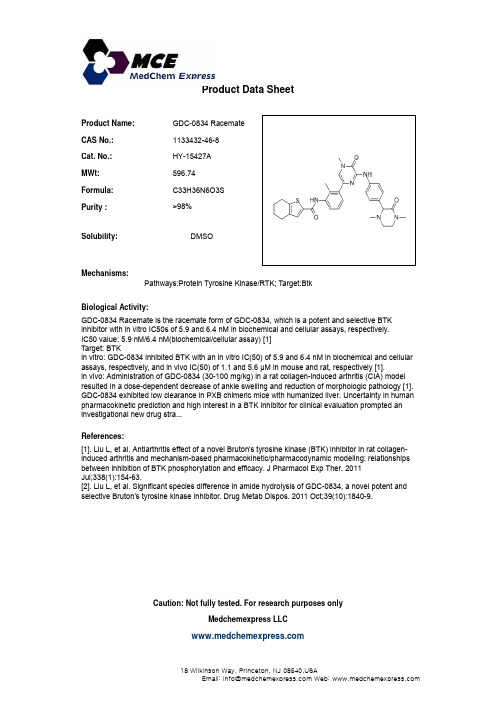
Product Name:GDC-0834 Racemate CAS No.:1133432-46-8Cat No :HY-15427AProduct Data SheetCat. No.:HY 15427A MWt:596.74Formula:C33H36N6O3S Purity :>98%Solubility:Mechanisms:Biological Activity:Pathways:Protein Tyrosine Kinase/RTK; Target:Btk DMSOGDC-0834 Racemate is the racemate form of GDC-0834, which is a potent and selective BTK inhibitor with in vitro IC50s of 5.9 and 6.4 nM in biochemical and cellular assays, respectively.IC50 value: 5.9 nM/6.4 nM(biochemical/cellular assay) [1]Target: BTK in vitro: GDC-0834 inhibited BTK with an in vitro IC(50) of 5.9 and 6.4 nM in biochemical and cellularassays, respectively, and in vivo IC(50) of 1.1 and 5.6 μM in mouse and rat, respectively [1].in vivo: Administration of GDC-0834 (30-100 mg/kg) in a rat collagen-induced arthritis (CIA) model resulted in a dose-dependent decrease of ankle swelling and reduction of morphologic pathology [1].GDC 0834exhibited low clearance in PXB chimeric mice with humanized liver Uncertainty in human References:[1]. Liu L, et al. Antiarthritis effect of a novel Bruton's tyrosine kinase (BTK) inhibitor in rat collagen-induced arthritis and mechanism-based pharmacokinetic/pharmacodynamic modeling: relationships between inhibition of BTK phosphorylation and efficacy. J Pharmacol Exp Ther. 2011J l 338(1)15463GDC-0834 exhibited low clearance in PXB chimeric mice with humanized liver. Uncertainty in human pharmacokinetic prediction and high interest in a BTK inhibitor for clinical evaluation prompted an investigational new drug stra...Jul;338(1):154-63.[2]. Liu L, et al. Significant species difference in amide hydrolysis of GDC-0834, a novel potent andselective Bruton's tyrosine kinase inhibitor. Drug Metab Dispos. 2011 Oct;39(10):1840-9.Caution: Not fully tested. For research purposes onlyMedchemexpress LLC18W i l k i n s o n W a y , P r i n c e t o n , N J 08540,U S AE m a i l : i n f o @m e d c h e m e x p r e s s .c o m W e b : w w w .m e d c h e m e x p r e s s .c o m。
Omega FL-1500流量计说明书

B-45e Exatidão de ±2% do Fundo de Escala e R epetibilidade de Fundo de Escala de ±1⁄2%e C onstrução em Vidro e Aço Inoxidável Proporcionando Compatibilidade com Vários Meios e E quações de Correlação para Uso com Diversos Fluidose Construção com Chapa Laterale Escala de 250 mm (9,85")e Escala de Fluxo de 10 a 100%e B lindado para Uso em Sistemas Pressurizados e Somente para Montagem VerticalOs medidores de vazão FL-1500 da OMEGA™ combinam exatidão de 2% e construção com chapalateral. Estes medidores apresentam construção em vidro e açoinoxidável 316, com chapas lateraisde alumínio e proteções de plástico acrílico. O desenho do flutuador oferece alta imunidade à variação de viscosidade e as conexões de rosca NPT facilitam a instalação emsistemas industriais e laboratoriais.ROTÂMETROS DE ALTA EXATIDÃOCAPACIDADES: 0,078 a 6,28 GPM para Água,FL-1501A-B, mostrado com conexões terminais de latão opcionais (o padrão é aço inox 316), em tamanho menorque o real.Vem completo, com manual do usuário.Para modelos com conexões terminais de latão, adicione o sufixo “-B” ao código do produto; consulte a engenharia de fluxos.Exemplos de Pedido: rotâmetros FL-1503A-B com conexão terminal de latão; 1,045 a 10,45 SCFM, 0,253 a 2,53 GPM.FL-1501A, conexão terminal de aço inoxidável 316, 0,317 a 3,17 SCFM, 0,078 a 0,78 GPM.ESPECIFICAÇÕESEscala: fundida no tubo medidor, 10 a 100% da escala do fluxoTubo Medidor: vidro borossilicato Flutuador e Limitadores: aço inoxidável 316Conexões Terminais:Padrão: aço inoxidável 316 Opcional: latãoAnel de Vedação: FKM-A Estrutural:Chapa Lateral: proteções de alumínio; de plástico acrílico, na frente e atrásPressão Máxima:FL-1501 e FL-1502: 20,68 bar (300 psig)FL-1503: 12,06 bar (175 psig) FL-1504: 6,9 bar (100 psig)Temperatura Máxima: 121°C (250°F)Exatidão: ±2% do fundo de escala Repetibilidade: ±0,5% do fundo de escalaSérie FL-1500A。
Colony-Forming Cell (CFC) Assays for Mouse Cells

THIS REAGENT IS FOR RESEARCH USE ONLY.NOT FOR DIAGNOSTIC OR THERAPEUTIC APPLICATIONS.Revised:January 2007PRODUCT DESCRIPTIONComponents include: 2.6% Methylcellulose Iscove’s MDMMethoCult ® methylcellulose-based media aremanufactured using tightly controlled processes and extensively pre-screened components.Each batch of MethoCult ® is sterility tested according to USP standards and Quality Control performance-tested in CFC assays using mouse BM samples. A Certificate of Analysis is available upon request.STABILITY/STORAGEStore at -20°C (-25°C to -15°C). Stable at -20°C for at least 2 years. Storage at 2-8°C is not recommended.Avoid freezing and thawing repeatedly.Contents guaranteed sterile if seal is intact.If product is received partially thawed, place immediately at -20°C or thaw and aliquot as described under ‘Handling and Directions for Use’.HANDLING AND DIRECTIONS FOR USEFor more detailed instructions, including detection of different progenitor cell types, refer to the Technical Manual for Mouse Colony-Forming Cell Assays Using MethoCult ® (Catalog #28405), available upon request and on our website at:/technical/manuals.aspThawing and Dispensing Bottles of MethoCult ®.Depending on your expected usage, prepare entire bottle or individual tubes of complete medium as required. Refer to Tables 1 and 2 for more information on recommended supplements and volumes required to prepare complete medium.To prepare 100 mL bottle of complete medium:1. Thaw provided 40 mL bottle of base medium at room temperature, or overnight under refrigeration (2-8°C).2. Add a 60 mL volume of desired supplements, including fetal bovine serum (FBS), bovine serum albumin (BSA), growth factors, and Iscove's MDM (IMDM, Catalog #36150), as required (total volume of 100 mL). Shake vigorously for 1-2 minutes and then let stand for at least 5 minutes to allow bubbles to dissipate before aliquoting.3. Using a 3 or 6 mL luer lock syringe attached to a 16gauge blunt-end needle (Catalog #28110/28120), aliquot 3 mL per tube for 1.1 mL duplicate cultures, or 4 mL per tube for 1.1 mL triplicate cultures. Tubes of complete medium can be used immediately or stored at -20°C for later use. Do not use pipettes to aliquot methylcellulose as the volume dispensed will not be accurate. Use of blunt-end needles for dispensing prevents needle-stick injuries.To prepare individual tubes of complete medium: 1. Thaw provided 40 mL bottle base medium at roomtemperature, or overnight under refrigeration (2-8°C). 2. Shake vigorously for 1-2 minutes and then let stand forat least 5 minutes to allow bubbles to dissipate before aliquotting.3. Using a 3 or 6 mL luer lock syringe attached to a 16gauge blunt-end needle (Catalog #28110/28120), aliquot base medium into tubes. For 1.1 mL duplicate cultures, aliquot 1.2 mL per tube and add desired supplements in a 1.8 mL volume (total 3.0 mL/tube). For triplicate cultures, aliquot 1.6 mL per tube and add desired supplements in a 2.4 mL volume (total 4.0mL/tube).4.Vortex tubes to mix well. Tubes can be usedimmediately or stored at -20°C for later use. RemainingRECOMMENDED FOR Colony-Forming Cell (CFC) Assays for Mouse CellsMethoCult ® M3134 is recommended as a base for thepreparation of methylcellulose-based medium for specialized applications. This incomplete medium formulation contains components that have been selected for optimal growth of mouse hematopoietic CFCs. Suitable for detection of CFU-E, mature BFU-E, BFU-E, CFU-GM, CFU-GEMM, and CFU pre-B, with the addition of appropriate growth factors and supplements. MethoCult ® M3134 is suitable for the detection and quantification of hematopoietic progenitors in mouse bone marrow (BM), spleen, peripheral blood (PB), and fetal liver (FL) samples using CFC assays. MethoCult ® M3134“Base”Methylcellulose Medium for MouseCellsCatalog # 03134 40 mL/bottleVersion 1.1.1THIS REAGENT IS FOR RESEARCH USE ONLY.NOT FOR DIAGNOSTIC OR THERAPEUTIC APPLICATIONS.Revised:January 2007base medium should be aliquoted for duplicate or triplicate cultures as required, and stored at -20°C for later use.Set-up of Mouse Colony-Forming Cell Assays1. Thaw tubes under refrigeration (2-8°C) overnight orat room temperature.2. Isolate cells from mouse BM, spleen, PB, or FL, asdescribed in detail in the Technical Manual (Catalog #28405).3. Prepare cell suspension: Unprocessed cell suspensions can beprepared directly from BM, FL, or spleen.Red blood cell depleted cell suspensions can be prepared from spleen or PB by lysis of RBC using ammonium chloride solution (Catalog #07800/07850).Lineage-depleted (Lin -) cells can be preparedfrom BM by depletion of mature lineage-committed hematopoietic cells using EasySep ® (Catalog #19756), RoboSep ® (#19756R), StemSep ® (#13056/13066), or SpinSep ® (#17056/17066) Progenitor Enrichment Kits.c-KIT + or SCA1+ cells can be prepared from BMby positive selection using EasySep ® (Catalog #18756/18757) or RoboSep ® (#18756R/18757R). For more details on StemCell’s cell separationsolutions for isolation of mouse progenitors, please contact us or visit our website at:/product_catalog/mprogenitors.asp4. Count nucleated cells using trypan blue (Catalog#07050) dye exclusion, 3% acetic acid (#07060), or automated cell counter. Methods to assay viable cells (i.e. dye exclusion) should be used for cell preparations where a decrease in cell viability may be expected (e.g. cryopreserved cells, ex vivo manipulation).5. Prepare a 10X concentrated cell suspension inIMDM (Catalog #36150). For example, prepare acell sample of 2 x 105cells per mL in IMDM for aplating concentration of 2 x 104cells per dish.6. Add 0.3 mL of cells to 3 mL of MethoCult ® or 0.4 mLof cells to 4 mL of MethoCult ®. This 1:10 v/v ratio of cells:medium gives the correct viscosity to ensure optimal CFC growth and morphology.7. Vortex tube to mix thoroughly and then let stand for2-5 minutes to allow bubbles to dissipate before dispensing.8. Using a 3 mL syringe attached to a 16 gauge blunt-end needle, dispense 1.1 mL of the MethoCult ® mixture containing cells into each of two or three 35 mm dishes (Catalog #27100/27150). Gently tilt and rotate each dish to distribute methylcellulose evenly. Dishes are pre-screened to ensure low cell adherence. Cell adherence can inhibit CFC growth.9. Add 3-4 mL of sterile water to an extra uncovered35 mm dish. For duplicate assays, place all 3 dishes into a 100 mm culture dish (Catalog#27125/27127). For triplicate assays, place 35 mm dishes in cultureware with a loose-fitting lid (e.g. 150 mm dishes, square bacterial dishes). Always provide water dishes to maintain humidity. 10. Incubate cells at 37°C, in 5% CO 2, with ≥95%humidity. Proper culture conditions are critical for optimal CFC growth. Use of water-jacketed incubators with a water pan in the chamber and routine monitoring of temperature and CO 2 levels is recommended. See Technical Manual (Catalog #28405) for information on culture conditions for different progenitor types.COUNTING AND CLASSIFICATION OF MOUSECOLONIESScoring OverviewUse a high-quality inverted microscope equipped with 2X, 4X and 10X planar objectives and stage holder for a 60 mm dish. A blue filter will enhance the contrast of erythroblasts in BFU-E and CFU-GEMM colonies. First scan the dish on low power (2X objective, 20-25X magnification) to evaluate the relative distribution of colonies. Score CFU-E and mature BFU-E on high power. Score BFU-E, CFU-GM, CFU-GEMM, and CFU pre-B on low power. Use high power to confirm colony type as required.Colony DescriptionsCFU-E: Colony-forming unit-erythroid produces a small colony containing at least 8 (8-32) erythroblasts, usually present in 1-2 clusters, detected after 2-3 days of culture. Mature BFU-E: Mature burst-forming unit-erythroidproduces a small colony containing 3 or more clusters of erythroblasts, detected after 3-4 days of culture.BFU-E: Burst-forming unit-erythroid produces a colony containing ≥30 erythroblasts, present in multiple clusters, detected after 7-14 days of culture.CFU-G: Colony-forming unit-granulocyte produces a colony containing ≥30 granulocytes, detected after 7-14 days of culture.CFU-M: Colony-forming unit-macrophage produces a colony containing ≥30 macrophages, detected after 7-14 days of culture.CFU-GM: Colony-forming unit-granulocyte, macrophage produces a colony containing ≥30 granulocytes and macrophage cells, detected after 7-14 days of culture. CFU-GEMM: Colony-forming unit-granulocyte, erythroid, macrophage, megakaryocyte produces a colony containing at least 30 cells, including granulocytes, erythroblasts, macrophages, and megakaryocytes, detected after 7-14 days of culture.CFU pre-B: Colony-forming unit-pre-B cell produces acolony containing 30 or more B-lineage cells, detected after 7-9 days of culture .THIS REAGENT IS FOR RESEARCH USE ONLY.NOT FOR DIAGNOSTIC OR THERAPEUTIC APPLICATIONS.Revised:January 2007Reference:Miller CL, Lai B: Human and Mouse Hematopoietic Colony-Forming Cell Assays, in Helgason CD, Miller CL (eds): Basic Cell Culture Protocols, Totowa, NJ, Humana Press Inc., pp 71-89, 2005 (28901)Table 1. Preparation of MethoCult ® MediaComponents VolumePer bottle: MethoCult ®M313440 mL IMDM, Supplements, Cytokines* 60 mL Total volume100 mLPer tube for 2 or (3) 1.1 mL cultures: MethoCult ® M31341.2 (1.6) mL IMDM, Supplements, Cytokines* 1.8 (2.4) mL Total volume3.0 (4.0) mL*Refer to Table 2 for recommended formulation and final concentrations.Table 2. Recommended Formulations for Mouse Myeloid CFC (BFU-E, CFU-GM and CFU-GEMM)Component Catalog # Final Concentrationrm SCF 02731 02931 50 ng/mL rm IL-3 02733 02903 10 ng/mL rm IL-6 orrh IL-6 02706 02906 02506 02606 10 ng/mL rh EPO 02625 3 U/mL FBS06200 0625015%BSA* 09300 1% Insulin* NA 10 µg/mL Transferrin* NA 200 µg/mL 2-Mercaptoethanol NA10-4ML-glutamine 07100 2 mM Methylcellulose in IMDM03134 1%*Alternatively, BIT 9500 Serum Substitute (#09500) can be usedin place of BSA, Insulin, and Transferrin (provided as a 5X concentrate for ease of use).NOTE: For other possible formulations, please refer to Technical Manual (#28405).Table 3. Recommended Cell Plating ConcentrationsCell Source Cells per 35mm dishBone Marrow (BM)* 2 x 104Peripheral Blood (PB)* 2 x 105 Spleen*2 x 105 Fetal Liver (FL)** 2 x 104 Lin - BM or FL*1 x 103 Yolk Sac5 x 104*Plating concentrations were established using cells from C57BL/6 mice at 6-12 weeks of age, in MethoCult ® medium containing SCF, IL-3, IL-6, and EPO.**Plating concentrations were established using FL cells from day 12.5-16.5 pc C57BL/6 embryos.For other strains, transgenic, or treated mice, and othercytokine combinations, plate cells at 2-3 different densities to establish optimal plating concentrations. Sufficient cells should be plated to yield 25-150 colonies per 35 mm dish (1.1 mL culture).Table 4. Recombinant Cytokines and Conditioned Media for Culture of Mouse Hematopoietic CellsCytokine Catalog # Applicationrm G-CSF orrh G-CSF02715 02915 02615 02855 growth of granulocytic progenitorsrm GM-CSF 02735 02935 growth of granulocytic and monocytic progenitors rm IL-3 02733 02903 growth of early myeloid progenitors of all lineages (in combination with other cytokines)rm IL-6 or rh IL-602706 02906 02506 02606 pleiotropic growth and differentiation ofhematopoietic progenitors rm M-CSF02751 02951 growth of monocytic progenitorsrm SCF02731 02931growth of mast cells, and promotes growth of myeloid and lymphoid progenitors (in combination with other cytokines)rm Tpo0272002920 promotes growth ofmegakaryocytic progenitors (in combination with other cytokines)rh EPO02625growth of erythroidprogenitors (in combination with other cytokines)PWM-SCCM* 02100source of colony stimulating factors*Pokeweed mitogen-stimulated spleen cell conditioned medium For a complete list of available cytokines, please visit: /product_catalog/cytokines.asp。
PASCO Force Sensor (PS-2104) User Manual
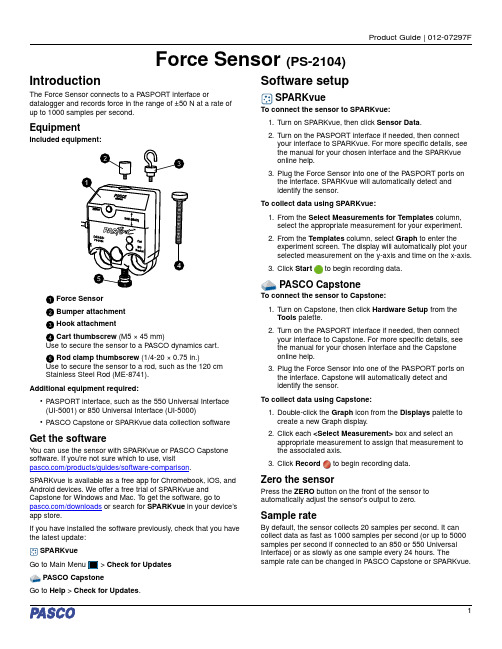
Force Sensor (PS-2104)Force Sensor Bumper attachment Hook attachmentCart thumbscrew (M5 × 45 mm)Use to secure the sensor to a P ASCO dynamics cart. Rod clamp thumbscrew (1/4-20 × 0.75 in.)Use to secure the sensor to a rod, such as the 120 cm Stainless Steel Rod (ME-8741).Additional equipment required:•P ASPORT interface, such as the 550 Universal Interface (UI-5001) or 850 Universal Interface (UI-5000)•P ASCO Capstone or SP ARKvue data collection softwareGet the softwareY ou can use the sensor with SP ARKvue or P ASCO Capstone software. If you’re not sure which to use, visit/products/guides/software-comparison .SPARKvue is available as a free app for Chromebook, iOS, and Android devices. We offer a free trial of SP ARKvue and Capstone for Windows and Mac. T o get the software, go to/downloads or search for SPARKvue in your device’s app store.If you have installed the software previously, check that you havethe latest update:SPARKvueCheck for UpdatesPASCO CapstoneGo to Help > Check for Updates .Software setup1.T urn on SP ARKvue, then click Sensor Data .2.T urn on the P ASPORT interface if needed, then connectyour interface to SP ARKvue. For more specific details, see the manual for your chosen interface and the SP ARKvue online help.3.Plug the Force Sensor into one of the P ASPORT ports on the interface. SP ARKvue will automatically detect and identify the sensor.To collect data using SPARKvue:1.From the Select Measurements for Templates column,select the appropriate measurement for your experiment.2.From the Templates column, select Graph to enter the experiment screen. The display will automatically plot your selected measurement on the y-axis and time on the x-axis.3.Click Startto begin recording data.1.T urn on Capstone, then click Hardware Setup from the Tools palette.2.T urn on the P ASPORT interface if needed, then connect your interface to Capstone. For more specific details, see the manual for your chosen interface and the Capstone online help.3.Plug the Force Sensor into one of the P ASPORT ports on the interface. Capstone will automatically detect and identify the sensor.To collect data using Capstone:1.Double-click the Graph icon from the Displays palette to create a new Graph display.2.Click each <Select Measurement> box and select an appropriate measurement to assign that measurement to the associated axis.3.Click Recordto begin recording data.Zero the sensorPress the ZERO button on the front of the sensor to automatically adjust the sensor’s output to zero.Sample rateBy default, the sensor collects 20 samples per second. It can collect data as fast as 1000 samples per second (or up to 5000samples per second if connected to an 850 or 550 Universal Interface) or as slowly as one sample every 24 hours. Thesample rate can be changed in PASCO Capstone or SP ARKvue.Product Guide | 012-07297F1Hardware setupConnect bumper and hook attachmentsScrew the bumper or hook into the sensor as illustrated below.Sensor mountingTo mount the sensor on a PASCO cart:1.Align the hole in the sensor labeled Cart with one of the threaded holes in the accessory tray of the cart.2.Insert the included cart thumbscrew through the Cart hole in the sensor.3.Screw the thumbscrew into the threaded hole on top of thecart, as shown below.To mount the sensor on a rod:1.Slide the sensor onto a rod, as shown below.2.Tighten the thumbscrew to secure the rod in place.Software helpThe SPARKvue and P ASCO Capstone Help provide additional information on how to use this product with the software. Y oucan access the help within the software or online.SPARKvueSoftware:Online:/sparkvuePASCO CapstoneSoftware: Help > P ASCO Capstone Help Online: /capstoneSpecifications and accessoriesVisit the product page at /product/PS-2104 to view the specifications and explore accessories. Y ou can also download experiment files and support documents from the product page.Experiment filesDownload one of several student-ready activities from the P ASCO Experiment Library. Experiments include editable student handouts and teacher notes. Visit /freelabs/PS-2104.Technical supportNeed more help? Our knowledgeable and friendly T echnical Support staff is ready to answer your questions or walk you through any issues.Chat Phone 1-800-772-8700 x1004 (USA)+1 916 462 8384 (outside USA) Email*****************Force Sensor | PS-21042Regulatory informationLimited warrantyFor a description of the product warranty, see the Warranty and Returns page at /legal.CopyrightThis document is copyrighted with all rights reserved. Permission is granted to non-profit educational institutions for reproduction of any part of this manual, providing the reproductions are used only in their laboratories and classrooms, and are not sold for profit. Reproduction under any other circumstances, without the written consent of P ASCO scientific, is prohibited.TrademarksP ASCO and P ASCO scientific are trademarks or registered trademarks of PASCO scientific, in the United States and in other countries. All other brands, products, or service names are or may be trademarks or service marks of, and are used to identify, products or services of, their respective owners. For more information visit /legal.Product end-of-life disposalThis electronic product is subject to disposal and recycling regulationsthat vary by country and region. It is your responsibility to recycle yourelectronic equipment per your local environmental laws and regulationsto ensure that it will be recycled in a manner that protects human healthand the environment. T o find out where you can drop off your waste equipment for recycling, please contact your local waste recycle or disposal service, or the place where you purchased the product. The European Union WEEE (Waste Electronic and Electrical Equipment) symbol on the product or its packaging indicates that this product must not be disposed of in a standard waste container.Product Guide | 012-07297F3。
CPX-4AE-T CPX-4AE-I 分析模块说明书

Terminal CPXMódulos de E/S analógicas CPX-4AE-T/CPX-4AE-IAmplio margen•CPX-4AE-T: menores costes del sistema y de almacenamiento mediante conexión directa de detectores de temperatura,termómetros de resistencia eléctrica tipos PT y N•Prescindir de costosos detecto-res con convertidores integra-dosSoluciones específicasConexiones en función de la apli-cación, de las características del módulo electrónico y del están-dar válido en la empresa.Menos es másTecnología avanzada para reducir costos y ahorrar tiempo y espa-cio.•Menores costos por canal con cuatro canales por módulo •Menos tiempos improductivos mediante diagnóstico por cana-les e indicación de fallos con LED por canal, unidad manual CPX-MMI o bus de campo /Ethernet•Terminales más compactos gra-cias a la gran cantidad de cana-les por móduloLos nuevos módulos de E/S son óptimos para una gran cantidad de canales analógicos o detectores de temperatura en la automatización de procesos. CPX-4AE-T para la detección de temperaturas desde -200°C hasta 850 °C; CPX-4AE-I para señales desde 4 hasta 20 mA.Medir temperaturas,captar señales,ahorrar espacio.210.8.PSIProduct Short InformationFesto AG &Co.KGRuiter Strasse 8273734 EsslingenInternet Tel. ++49 (0)711 347-0 Fax ++49 (0)711 347-2144E-mail service_international@270406R e s e r v a d o e l d e r e c h o d e m o d i f i c a c i ónTerminal CPXMódulos de E/S analógicas CPX-4AE-T / CPX-4AE-IAmplia modularidadMás economía mediante la utili-zación de módulos electrónicos.Medir temperaturas, caudales,presiones y distancias.CPX: diversidad de conexiones Gran cantidad de funciones eléc-tricas. El encadenamiento de la placa de alimentación, módulo electrónico y placa de alimenta-ción permite numerosas combi-naciones. Elección rápida y man-tenimiento sencillo gracias a la sustitución de módulos electróni-cos sin modificar el cableado.TipoCPX-4AE-ICPX-4AE-TMódulos de entradas analógicas Captación de señales Detección de temperaturas Cantidad de entradas 44Cantidad de salidas ––Línea característica ––Resolución12 Bit16 BitNivel de conm./Margen señales 0 ... 20 mA, 4 ... 20 mA Fuente de intensidad constante Alim. máx. de corr. por canal 40 mA En función del detector Alim. máx. de corr. por módulo 0,7 A0,7 A Detectores P . ej. sensor de presión y vacío SDE–DetectoresSensores de presión, detectores de caudal PT 100, PT 200, PT 500, PT 1000y medidores de distancias Ni 100, Ni 120, Ni 500, Ni 1000Margen de temperatura –PT estándar: -200 °C ... 850 °C DetectoresPT entorno: -120 °C ... 130 °C NI estándar: -60 °C ... 180 °CConexiones para detectores 2, 3, 4 hilos 2, 3, 4 hilos ParametrizaciónFormato de datos, valores límite, factor de escala Formato de datos, valores límite, factor de escala,detector de temperatura, comportamiento de la fuente de corriente en caso de sobrecargaDiagnóstico 4 LED para errores de canal y 1 LED para error de móduloFuncionesParametrización y diagnóstico por canal, señales del canal defectuoso a través de la red o en CPX-MMIMPA1 y MPA2Bloque de enlace CPX-GE-7/8" 5 cont.CPX-4AE-ISensor de presión SDE1Medidor de distancias SOELDetector de caudal SFE1Módulo electrónico CPX-4AE-TDetector de temperatura PT 100Datos técnicos。
拟南芥

The Conserved Splicing Factor SUA Controls Alternative Splicing of the Developmental Regulator ABI3in Arabidopsis W OAMatteo Sugliani,a Vittoria Brambilla,a Emile J.M.Clerkx,b,1Maarten Koornneef,a,b and Wim J.J.Soppe a,2a Department of Plant Breeding and Genetics,Max Planck Institute for Plant Breeding Research,50829Cologne,Germanyb Laboratory of Genetics,Wageningen University,6708PB Wageningen,The NetherlandsABSCISIC ACID INSENSITIVE3(ABI3)is a major regulator of seed maturation in Arabidopsis thaliana.We detected two ABI3 transcripts,ABI3-a and ABI3-b,which encode full-length and truncated proteins,respectively.Alternative splicing of ABI3is developmentally regulated,and the ABI3-b transcript accumulates at the end of seed maturation.The two ABI3transcripts differ by the presence of a cryptic intron in ABI3-a,which is spliced out in ABI3-b.The suppressor of abi3-5(sua)mutant consistently restores wild-type seed features in the frameshift mutant abi3-5but does not suppress other abi3mutant alleles.SUA is a conserved splicing factor,homologous to the human protein RBM5,and reduces splicing of the cryptic ABI3intron,leading to a decrease in ABI3-b transcript.In the abi3-5mutant,ABI3-b codes for a functional ABI3protein due to frameshift restoration.INTRODUCTIONSeeds are essential for the spread and survival of most plant species and constitute a major food source.Seed features like desiccation tolerance,dormancy,and the accumulation of stor-age proteins are established during seed maturation.In Arabi-dopsis thaliana,the phytohormone abscisic acid(ABA)controls seed maturation and dormancy by preventing germination and reserve mobilization.ABA signaling at this stage is concomitant with the expression of four major regulatory genes of seed maturation with partially redundant functions:LEAFY COTYLE-DON1(LEC1),LEC2,FUSCA3(FUS3),and ABSCISIC ACID INSENSITIVE3(ABI3)(Kroj et al.,2003;To et al.,2006).ABI3is a main component of the ABA signaling pathway and is highly conserved among plant species.The ABI3protein contains four functional domains(Giraudat et al.,1992;Suzuki et al.,1997). The A1domain is an acidic transcriptional activator(McCarty et al.,1991),and B1can interact with the seed-specific tran-scription factor ABI5(Nakamura et al.,2001).B2and B3are two basic DNA binding domains responsible for the ABA-dependent activation of seed maturation genes(Suzuki et al.,1997;Ezcurra et al.,2000;Nag et al.,2005).Several abi3mutant alleles were isolated in Arabidopsis.One of the most severe is abi3-5,which was originally identified by its stay-green seed phenotype.abi3-5seeds are insensitive to ABA during germination,are desiccation intolerant,and have reduced longevity,similar to other strong abi3alleles(Ooms et al.,1993).ABI3transcription is promoted by LEC1,LEC2,FUS3,ABI3(To et al.,2006),and ABA(Lopez-Molina et al.,2002).During germi-nation,ABI3is repressed by the chromatin remodeling factor PICKLE(Perruc et al.,2007)and the ABI3protein is targeted to 26S proteasome degradation by the ABI3-INTERACTING PRO-TEIN2(Zhang et al.,2005).The identification of several splice variants of ABI3homologs in monocotyledon and dicotyledon species(McKibbin et al.,2002;Fan et al.,2007;Gagete et al., 2009)implies that alternative splicing also has an important role in controlling ABI3expression.However,splicing variants of ABI3were not observed in Arabidopsis.Although alternative splicing of mRNA is an important com-ponent of posttranscriptional regulation in higher eukaryotes,its relevance and mechanisms in plants are poorly understood.In Arabidopsis,;42%of all transcripts from intron-containing genes are alternatively spliced(Filichkin et al.,2010).Alternative splicing can produce transcripts that encode for proteins with altered or lost function.Furthermore,it can lead to tissue-specific transcripts or affect mRNA stability and turnover via nonsense-mediated decay(McGlincy and Smith,2008).Splicing is directed by the spliceosome,a dynamic RNA-protein multicomponent machinery that is conserved among eukaryotes.In Arabidopsis, only a few splicing-related proteins have been characterized (Lopato et al.,1999;Ali et al.,2007;Tanabe et al.,2007;Zhang and Mount,2009),and the information on their biochemical function and their targets in relevant developmental and envi-ronmental contexts is limited.We identified SUPPRESSOR OF ABI3-5(SUA)as a novel plant splicing factor that influences seed maturation by controlling alternative splicing of ABI3.SUA is an evolutionary conserved protein that suppresses splicing of a cryptic ABI3intron.Splicing of this intron leads to a transcriptThe authors responsible for distribution of materials integral to thefindings presented in this article in accordance with the policy describedin the Instructions for Authors()are:Matteo Sugliani(sugliani@mpipz.mpg.de)and Wim J.J.Soppe(soppe@mpipz.mpg.de).1Current address:University of Applied Sciences,Hogeschool HAS DenBosch,5223DE,Hertogenbosch,The Netherlands.2Address correspondence to soppe@mpipz.mpg.de.W Online version contains Web-only data.OA Open Access articles can be viewed online without a subscription./cgi/doi/10.1105/tpc.110.074674The Plant Cell,Vol.22:1936–1946,June2010,ã2010American Society of Plant Biologiststhat encodes a truncated ABI3protein in the wild type but a functional protein in the abi3-5mutant background. RESULTSIsolation of the abi3-5sua-1Double MutantSeeds of the abi3-5glabra1(gl1)transparent testa5-1(tt5-1)triple mutant were mutagenized by g-irradiation to isolate mutants involved in ABI3signaling.The gl1and tt5-1mutations are located on both sides of the ABI3locus and were used as phenotypic markers to distinguish suppressor mutants from wild-type contaminants.A strong suppressor mutant of abi3-5 was identified in the M2generation and named suppressor of abi3-5(sua-1).The gl1-1and tt5-1mutations were removed from this line by backcrossing with its wild-type Landsberg erecta (L er)genetic background and subsequent selection for abi3-5 and sua-1in the progeny.Ripe abi3-5seeds are green due to the presence of chlorophyll,but abi3-5sua-1seeds are yellow-brown,similar to the wild type(Figure1A).In addition,abi3-5 seeds are nondormant and sensitive to desiccation,which causes reduced longevity.Seeds of abi3-5sua-1are also non-dormant,but their longevity is strongly improved and they still germinate nearly100%after10weeks of storage(Figure1B). Finally,abi3-5sua-1seeds show an increased sensitivity to ABA and cannot germinate on15m M ABA,whereas viable abi3-5mutant seeds show100%germination on30m M ABA (Figure1C).Identification of the SUA GeneInitial mapping indicated that the sua mutation is located on chromosome 3.Fine-mapping was performed using an F2 mapping population of;4000individuals derived from a cross between the abi3-5sua-1double mutant(in L er background)and Columbia(Col-0).The abi3-5sua-1double mutant was identified in this mapping population by its yellow-brown seed color trait in combination with the ability to germinate in the presence of5m m ABA.The location of the sua-1mutation was narrowed down to a region of64kb at the bottom of chromosome3between two markers located at20.056and20.120Mb.This region contains 17genes and did not show recombination in ourmappingFigure1.The sua Mutation Suppresses abi3-5Phenotypes.(A)Seeds of wild type(L er and Col-0),abi3-5(in L er background),abi3-5sua-1(in L er background),abi3-5sua-2(in L er/Col-0background)and abi3-5 sua-1PSUA:SUA:GFP#8(in L er background).(B)Germination of L er,abi3-5,and abi3-5sua-1seeds after different periods of dry storage.Harvested seeds were stored at208C and42%relative humidity.Percentages are means(6SE)of three biological replicates.(C)Germination of L er,abi3-5,abi3-5sua-1,abi3-5sua-2,and abi3-5sua-3seeds,imbibed at different ABA concentrations.Seeds were1week after-ripened and stratified4d.Percentages are means(6SE)of four biological replicates.(D)Germination of1-week-old abi3-5,abi3-5sua-1PSUA:SUA:GFP#8,and abi3-5sua-1PSUA:SUA:GFP#15seeds at different ABA concentrations. Percentages are means(6SE)of three biological replicates.(E)Germination of L er,sua-1,Col-0,and sua-2seeds,imbibed at different ABA concentrations.Seeds were6months after-ripened and4d stratified. Percentages are means(6SE)of four biological replicates.SUA Controls Alternative Splicing of ABI31937parison of sequenced candidate genes with sequences in The Arabidopsis Information Resource (Garcia-Hernandez et al.,2002)revealed a 47-bp deletion in the 15th exon of At3G54230in the abi3-5sua-1double mutant (Fig-ure 2A).The identity of At3G54230as the SUA gene was confirmed by complementation of the sua-1mutant in the abi3-5background.A construct containing the SUA cDNA,expressed from a 2711-bp putative SUA promoter and fused with a C-terminal green fluorescent protein (GFP )tag (PSUA:SUA:GFP ),was used to transform abi3-5sua-1plants.Two independent T2transform-ants,containing a single insertion event,both complemented sua-1and showed the abi3-5phenotype.One of these trans-formants,abi3-5sua-1PSUA:SUA:GFP #8,even showed an enhanced abi3-5phenotype,yielding seeds with a more intense green color and stronger ABA insensitivity (Figures 1A and 1D).Additional mutant alleles of SUA in the Col-0background (sua-2and sua-3)were obtained from the Salk insertion mutant collection and from the GABI-Kat collection.These lines contain T-DNA insertions in the fourth and the ninth intron and were named sua-2and sua-3,respectively (Figure 2A).Both alleles lack full-length SUA expression and were crossed with abi3-5.The double mutants abi3-5sua-2and abi3-5sua-3were selected in the resulting F2,and all of them showed suppression of the abi3-5phenotypes,similar to abi3-5sua-1(Figures 1A and 1C).The sua single mutants did not have any obvious visual phenotype.Detailed analysis revealed that sua-1seeds are more susceptible to ABA germination inhibition compared with wild-type L er .By contrast,sua-2seeds germinated better than wild-type Col-0in the presence of ABA (Figure 1E).SUA Encodes an RNA Binding Protein Located in the Nucleus and Expressed in All Plant TissuesSUA encodes a protein with a conserved domain architecture that suggests a function in RNA metabolism.SUA contains two RNA recognition motifs surrounding a Zinc finger domain,an octamer repeat domain,and a Gly-rich domain close to the carboxy end (Figure 2B).The Arabidopsis genome does not contain a second gene with this combination of domains.SUA homologs,however,can be found throughout the eukaryotic kingdom (Figure 2C).SUA has 45%sequence similarity with the human RNA Binding Motif Protein 5(RBM5),which was originally identified as a putative tumor suppressor gene that is part of a small gene family (Edamatsu et al.,2000).Publicly available microarray data (Zimmermann et al.,2004)show ubiquitous SUA expression in Arabidopsis ,with a moder-ate enrichment in seeds.Quantitative real-time RT-PCR analysis confirmed that the relative abundance of SUA transcripts is comparable in most Arabidopsis tissues,but highest in siliques toward the end of seed maturation (Figure 3A).The subcellular localization of the SUA protein was studied using the PSUA:SUA:GFP lines.A GFP signal was detected in the nucleus of vegeta-tive and reproductive tissues (Figure 3B).The SUA_GFP chimeric protein showed diverse patterns.Speckles of different size were observed in some nuclei,but fluorescence was diffuse and rather weak in others (Figure 3B).We did not observe a correlation between the SUA_GFP fluorescence pattern and tissue or de-velopmental stages.SUA Interacts with the Prespliceosomal Component U2AF 65RBM5,the human homolog of SUA,is a member of the prespliceosomal complex (Behzadnia et al.,2007)and interacts with U2AF 65in vivo (Bonnal et al.,2008).U2AF 65is the larger subunit of the conserved pre-mRNA splicing factor U2AF.It guides splice site selection during the formation of the spliceo-somal complex (Zamore et al.,1992;Sickmier et al.,2006).In a yeast two-hybrid GAL4assay,we detected interaction between SUA and Arabidopsis U2AF 65(BAH19725)(Domon et al.,1998;Figure 4A).To confirm the SUA-U2AF 65interaction in planta,we performed a fluorescence resonance energy transfer/fluo-rescence lifetime imaging (FRET/FLIM)assay.Arabidopsis leaf protoplasts were cotransfected with two vectors fortheFigure 2.Genetic Structure,Domain Organization,and Phylogenetic Relationships of SUA .(A)Schematic structure of the SUA gene.Triangles indicate the T-DNA insertion sites of sua-2and sua-3,and the dashed region represents the 47-bp deletion of the sua-1allele.UTRs are shown in white,exons in gray,and introns as thick lines.(B)Domain structure of the SUA protein.aa,amino acids;RRM,RNA recognition motif;Zn,zinc finger;OCRE,octamer repeat;G-p,Gly patch.(C)Phylogram of SUA and its closest related proteins.FCA is an RNA binding protein that was added to the tree to emphasize the similarity between SUA and its homologs in evolutionary distant species.Populus trichocarpa (Pt ),Vitis vinifera (Vv ),Oryza sativa (Os ),Physcomitrella patens (Pp ),Chlamydomonas reinhardtii (Cr ),Xenopus laevis (Xl ),Mus musculus (Mm ),and Homo sapiens (Hs ).Bootstrap values are shown when higher than 50.1938The Plant Celloverexpression of SUA_YFP (yellow fluorescent protein)and U2AF 65_CFP chimerical proteins.FRET/FLIM analysis of proto-plasts coexpressing SUA_YFP and U2AF 65_CFP (cyan fluores-cent protein)showed a significant reduction of the mean CFP fluorescence lifetime compared with those expressing the U2AF 65-CFP alone (Figures 4B to 4F),confirming interaction of both proteins in planta.The Suppression of abi3-5by sua-1Is Allele Specific The abi3-5mutant is one of the strongest abi3alleles,which all show reduced seed dormancy and decreased sensitivity to ABA during germination (Bies-Etheve et al.,1999).Seeds of the abi3-4and abi3-6mutants are nondormant,highly insensitive to ABA,and show reduced longevity and a high chlorophyll content similar to abi3-5.To study the suppression effect of the sua-1mutant on different abi3mutant alleles,double mutants were constructed.The ABA-insensitive abi3-4and abi3-6alleles,as well as the weak abi3-1and abi3-7alleles (Figure 5A),were combined with sua-1,sua-2,and sua-3.Surprisingly,none of these combinations showed any suppression phenotype,indicating that the suppres-sion of abi3-5by sua mutants is allele specific (Figure 5B).Detection of Functional ABI3Protein in the abi3-5sua-1Double MutantThe abi3-5mutation causes a frameshift leading to a premature stop codon after 34erroneous codons.The abi3-4mutant hasaFigure 3.SUA Is Expressed in All Tissues and Its Protein Is Localized in the Nucleus.(A)Quantitative real-time RT-PCR analysis of SUA expression in different tissues.SUA mRNA levels are normalized to ACTIN8mRNA levels.S6D,seedlings 6d after germination;R,roots;RL,rosette leaves;CL,cauline leaves;FB,flower buds;S10to S20,siliques 10,12,14,16,18,and 20d after pollination.Data are from two independent biological replicates.Error bars represent SE .(B)Confocal analysis of subcellular localization of SUA:GFP in develop-ing embryo tissue from transgenic abi3-5sua plants containing the PSUA:SUA:GFP construct.Three nuclei with different GFP patterns are shown.Bar =2mM.Figure 4.SUA Interacts with U2AF 65.(A)Interaction between SUA and U2AF 65detected with the yeast two-hybrid assay.Cotransformed yeast strains were grown on SD-L-W-H with 5mM 3AT.Snf1and Snf4are yeast proteins that strongly interact (Jiang and Carlson,1997).(B)to (E)Interaction between SUA and U2AF 65based on FRET mea-sured by FLIM.FLIM analysis of protoplasts transiently expressing U2AF 65-CFP ([B]and [C])and coexpressing U2AF 65-CFP and SUA-YFP ([D]and [E]).Intensity channel ([B]and [D])and false color code ([C]and [E]).The absence of interaction results in a long lifetime,visible as a dark-blue color.Interaction leads to a reduction in donor lifetime,visible as a shift toward orange.A representative protoplast nucleus is shown.(F)Average CFP fluorescence lifetime values for the FRET/FLIM analysis.N,number of nuclei analyzed.SUA Controls Alternative Splicing of ABI31939single nucleotide mutation that causes a stop codon at approx-imately the same position (Bies-Etheve et al.,1999;Figure 5A).Therefore,abi3-4and abi3-5produce ABI3transcripts that translate into truncated ABI3proteins with similar sizes.Never-theless,the phenotype of the abi3-5mutant is strongly sup-pressed by sua ,whereas that of abi3-4is not.To understand this discrepancy,we analyzed the ABI3protein in dry seeds of L er ,sua-1,abi3-4,abi3-5,and the double mutants abi3-4sua-1and abi3-5sua-1by immunoblotting.A specific antibody,targeted to the amino end of ABI3,was used for detection.The ABI3protein (720amino acids)migrates as a 116-kD polypeptide (Parcy et al.,1997).We detected two bands of approximately this size for the ABI3protein in L er and sua-1seeds.One of these two bands probably represents a modified version of ABI3.A truncated ABI3protein corresponding to a 428–amino acid polypeptide and migrating as a 70-kD band,was observed in the abi3-5mutant (Figure 6).A similar sized (416amino acids)highly abundant ABI3protein was found in abi3-4and abi3-4sua-1.The high abundance of the truncated ABI3protein in abi3-4seeds was previously observed by Parcy et al.(1997).In the abi3-5sua-1double mutant,two weak bands of comparable size to full-length ABI3were detected,along with the smaller truncated abi3-5mutant protein (Figure 6).The presence of full-length ABI3protein in abi3-5sua-1seeds was consistent with all the observed suppression phenotypes and predicts the pres-ence of an ABI3transcript with a restored reading frame that has lost the abi3-5premature stop codon.Identification of a Novel ABI3Splice VariantThe abi3-5transcripts were analyzed in detail by RT-PCR and sequencing.In the abi3-5sua-1double mutant,we identified,besides the expected full-length abi3-5transcript,an alterna-tively spliced novel abi3-5transcript that lacks a cryptic intron of 77nucleotides.This cryptic intron is located shortly downstream of the abi3-5mutation and includes the premature abi3-5stop codon (Figure 5C).The combination of the 1-bp abi3-5deletion and the removal of the 77-nucleotide cryptic intron results in a transcript that restores the reading frame of abi3-5after 21erroneous and 26deleted codons.We named this transcript abi3-5-b and named the transcript with the retained intron abi3-5-a .The translated abi3-5-b polypeptide (abi3-5-b )is predicted to be 694amino acids.This protein contains all four ABI3protein domains (Figure 6B),and the phenotype of the abi3-5sua-1seeds indicated that abi3-5-b largely retains the ABI3molecular functions (Figure 1).The ABI3-b transcript only encodes a functional protein in the abi3-5mutant background.In the wild type,it causes a frame-shift and codes for a truncated protein of 429amino acids.This predicted truncated polypeptide was immunodetected in the sua-1single mutant and also,at lower levels,in wild-type L er seed protein extracts.The wild-type ABI3-b protein migrates with a similar speed in the gel as the proteins encoded by abi3-4(a and b splicing forms)and abi3-5(a splicing form)mutants (Figure 6).In addition to the accumulation of the ABI3-b splice variant,the sua-1mutant also shows an overall increase in ABI3expression.The amount of ABI3-a transcript,coding for full-length ABI3,is higher in sua-1than in the wild type (Figure 7A).This could explain the increased ABA sensitivity of sua-1seeds.Instead,overall ABI3expression in sua-2seeds is similar to that in wild-type Col-0,but the portion of the transcript coding for full-length ABI3is reduced,resulting in a decrease of ABA sensitivity (Figure 1E).We tested the possibility that sua-1has again-of-functionFigure 5.sua Is an Allele-Specific Suppressor of the abi3-5Allele.(A)Schematic structure of the ABI3gene.The locations and nature of the abi3-1,abi3-4,abi3-5,abi3-6,and abi3-7mutations are indicated.UTRs are shown in white,exons in gray,and introns as thick lines.The box with diagonal stripes represents the cryptic intron.(B)Table showing the suppression of the abi3phenotype in different combinations of sua and abi3mutant alleles.A check mark indicates abi3suppression;the “x”indicates absence of abi3suppression.ND,not determined.(C)Sequence of the ABI3cryptic intron and surrounding region.The asterisk indicates the single base pair deleted in abi3-5,and the subsequent stop codon is underlined.The cryptic intron is shown in lowercase letters.1940The Plant Celleffect on ABI3expression by transforming the sua-1mutant allele,expressed from the endogenous SUA promoter,into the sua-2mutant.Indeed,the obtained transformants showed a higher ABI3expression than the sua-2mutant and had increased ABA sensitivity (see Supplemental Figure 1online).This indicates that sua-1is a gain-of-function mutant regarding ABI3expres-sion.ABI3Alternative Splicing Is Developmentally Regulated The relative abundance of ABI3-a and ABI3-b transcripts was quantified in wild-type seeds by real-time RT-PCR.Developing siliques 16d after pollination showed a very low abundance of ABI3-b transcript in L er and Col-0(1.5361.36%and 0.9560.83%,respectively,of the overall ABI3transcripts;Figure 7A).During progressive development of wild-type siliques,the ratiobetween both ABI3transcripts shifted toward ABI3-b .At 20d after pollination,the amount of ABI3-b exceeded that of ABI3-a (Figure 7B).The observed change in ratio between ABI3-a and ABI3-b transcripts during seed maturation indicates that alter-native splicing of ABI3is developmentally regulated.DISCUSSIONABI3Is Regulated by Alternative SplicingThe transcription factor ABI3regulates seed maturation and influences seed quality.The abundance of ABI3is tightly regu-lated at different levels.In addition to complex genetic interac-tions with LEC1,LEC2,and FUS3at the transcriptional level (To et al.,2006),ABI3expression is controlled posttranscriptionally.Alternative splicing of ABI3homologs in cereal species (Triticum aestivum and Oryza sativa )and dicots (Pisum sativum )(McKibbinFigure 6.Detection of Full-Length ABI3Protein in the abi3-5sua-1Double Mutant.(A)Immunoblot analysis of ABI3protein.Total protein was extracted from freshly harvested seeds and separated on a Tris-Gly SDS 4to 12%polyacrylamide gradient gel.The ABI3protein is identified as a double band of ;116kD in L er ,sua-1,and abi3-5sua-1.The truncated ABI3proteins (D ABI3)produced by abi3-4,abi3-5,and abi3-5sua-1and the novel splicing variant of ABI3are ;70kD.Asterisk indicates a nonspe-cific band that is used as loading control.Sizes of the molecular markers (in kilodaltons)are shown next to the blot.(B)Predicted ABI3protein isoforms.Gray boxes represent the con-served functional motifs of ABI3(from left to right:A1,B1,B2,and B3).Boxes with diagonal stripes represent erroneous amino acidsstretches.Figure 7.Quantification of ABI3Splicing Variants.Quantitative real-time RT-PCR analysis of ABI3-a (white)and ABI3-b (gray)expression in L er ,sua-1,abi3-5,abi3-5sua-1,Col-0,and sua-2(A)and in L er developing siliques 10to 20d after pollination (DAP)(B).For (A),mRNA was extracted from siliques 16d after pollination.ABI3mRNA levels are normalized to ACTIN8mRNA levels.Data are from two independent biological replicates.Error bars represent SE .SUA Controls Alternative Splicing of ABI31941et al.,2002;Fan et al.,2007;Gagete et al.,2009)generates multiple mis-spliced transcripts that often code for truncated polypeptides.This has been linked to reduced grain quality in rice and wheat(McKibbin et al.,2002;Fan et al.,2007).Here,we show that the ABI3gene of Arabidopsis is also regulated by alternative splicing.A77-bp cryptic ABI3intron is alternatively spliced,which leads to the occurrence of two transcripts.The ABI3-a transcript encodes a full-length ABI3protein,and the ABI3-b transcript encodes a truncated protein that contains two of the four functional domains.Splicing of the cryptic intron of ABI3is developmentally regulated,and ABI3-b accumulates only at the end of seed maturation.This probably contributes to a fast downregulation of full-length ABI3in ripe seeds,which is neces-sary to inhibit the seed maturation program in germinating seeds. Transcripts with a long39untranslated region(UTR)or with 39UTR-located introns can be detected and degraded by the nonsense-mediated decay machinery in plants(Kere´nyi et al., 2008).To distinguish a natural stop codon from a premature stop codon,nonsense-mediated decay requires a second signal that has not been identified yet in plants(van Hoof and Green,2006). The ABI3-b transcript contains a premature stop codon but probably lacks this second signal because it is not affected by nonsense-mediated decay.The protein encoded by the ABI3-b transcript contains the A1acidic transcriptional activation do-main and thefirst basic domain and might still mediate ABA signaling during late seed maturation.The prevalent model of splicing in Arabidopsis is intron defi-nition,in which intronic sequences are recognized by the spliceosomal complex.The features of a canonical plant intron are a consensus59splice site(AG/GU,where GU is the more conserved dinucleotide),a U-rich sequence,and a consensus 39splice site(CAG/G where AG is invariant)(Simpson and Filipowicz,1996;Lorkovic´et al.,2000).Arabidopsis exons con-tain on average29%U,while introns have on average42%U (Reddy,2007).It was shown that U-rich elements can function as splicing signals(Simpson et al.,2004),and short introns and introns with low AU content are more likely to be retained(Wang and Brendel,2006).The ABI3cryptic intron has sequence similarities with canonical plant introns,in particular with the consensus sequences at the two borders(Figure5C),but it has a U content of only29%,while the other ABI3introns have,on average,46%U.Because of that,the cryptic ABI3intron may not be easily recognized by the spliceosomal complex.SUA Controls Alternative Splicing of ABI3SUA suppresses splicing of the cryptic ABI3intron and thereby influences the ratio between the ABI3-a and ABI3-b transcripts. Reduced suppression of the cryptic intron in the sua mutant leads to an increased amount of ABI3-b transcript and de-creased levels of the ABI3-a transcript.However,a substantial amount of ABI3-a transcript could still be detected in the sua mutant.Other splicing factors probably act redundantly with SUA in the suppression of the cryptic ABI3intron.Alternative splicing in plants is regulated by tissue-specific developmental cues and stresses and might provide a means for optimal adaptation to the environment(Ali and Reddy,2008). Alternative splicing of ABI3could also be regulated by specific environmental conditions.In this respect,it is interesting to note that publicly available microarray data show an upregulation of SUA expression by senescence(Zimmermann et al.,2004).The water content of seeds strongly decreases during the maturation phase until;7%in mature seed(Baud et al.,2002).This process is comparable to senescence and also coincides with increased SUA mRNA levels(Figure3A).Higher SUA abundance will favor cryptic intron retention and increase the full-length ABI3protein levels during seed maturation.Consistent with that,our exper-iments showed a correlation between increased levels of SUA transcript and a reduction in ABI3-b levels in transgenic plants. The abi3-5sua-1PSUA:SUA:GFP#8line,for instance,showed increased levels of SUA transcript and reduced amounts of abi3-5-b,resulting in an enhanced abi3-5phenotype(see Supple-mental Figure2online;Figures1A and1D).SUA-mediated alternative splicing of ABI3could represent a system tofine-tune seed maturation.However,in wild-type plants,the ABI3-b transcript accumulates at the end of seed maturation when SUA is still substantially expressed.Possibly,the SUA protein is not active or degraded at the end of seed maturation.Alternatively, other factors could counteract the role of SUA in retention of the cryptic intron at this time.The sua-1single mutant showed increased ABA sensitivity during germination.This is probably caused by upregulation of ABI3expression,which does not occur in sua-2.This difference between sua-1(in a L er background)and sua-2(in a Col-0 background)could be explained by natural genetic variation between L er and Col-0that modifies the sua mutant phenotype. However,sua-2plants transformed with sua-1also showed an increased ABI3expression and enhanced ABA sensitivity. Therefore,it is more likely that sua-1is a gain-of-function allele, which is translated into a truncated protein.This predicted polypeptide includes the RNA recognition motifs and the Zn finger motif but lacks the G patch domain at the C terminus.The nonfunctional mutant sua-1protein might compete for sub-strates with other proteins of the mRNA splicing machinery and could therefore function as a dominant-negative allele.The abi3-5mutant still contains a small amount of abi3-5-b transcript,which encodes a functional ABI3protein.Conse-quently,abi3-5is not a complete loss-of-function mutant.The sua mutant can suppress abi3-5because it enhances the amount of abi3-5-b transcript.SUA Has a Conserved Role in SplicingThe conserved domain architecture of SUA and its role in the suppression of the cryptic ABI3intron indicate a function in mRNA processing.Moreover,the speckledfluorescence pat-terns observed in nuclei expressing the chimeric SUA:GFP gene are similar to those obtained with Ser/Arg-rich GFP proteins, which are involved in RNA metabolism in plants(Lorkovic´and Barta,2004).The SUA protein has two RNA recognition motifs, which are also found in many eukaryotic RNA processing pro-teins(Burd and Dreyfuss,1994).Based on its functional motifs, SUA could bind directly to specific RNA targets.However,SUA might also interact with the mRNA targets indirectly and be part of the spliceosome,which is composed of;300proteins in Arabidopsis(Reddy,2007).1942The Plant Cell。
沙多玛单体

Applications
Adhesives Plastics : PVC, ABS, PC wood coatings Glass
Isobornyl Acrylate
SR506
IBOA O O
Product Specification
Functionality Color (APHA) Viscosity(cps,25℃) Refractive index Tg 1 20 9 1.474 88
Features
Low volatile Weatherability Chemical Resistance Water Resistance
Applications
Encapsulant Photo resist Inks and Coatings
Propoxylated Diacrylates
Applications
Laminating Adhesives Wood coating Plastics : PS, PET
Difunctional Monomers
SR238NS SR508NS SR306 SR9003 SR833S BPA Type…
Alkane Diacrylates
O
O
O
Cyclic Trimethylolpropane Acrylate
SR531 CTFA
O O O O
Product Specification
Functionality Color (APHA) Viscosity(cps,25℃) Refractive index Tg 1 120 13 1.462 32
Polyethylene-glycol Di(Meth)Acrylates
CellTiter Glo Luminescent Cell Viability Assay Protocol

Promega Corporation ·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll F ree in USA 800-356-9526·Phone 608-274-4330 ·F ax 608-277-2516 ·1.Description (1)2.Product Components and Storage Conditions (4)3.Performing the CellTiter-Glo ®Assay (5)A.Reagent Preparation (5)B.Protocol for the Cell Viability Assay (6)C.Protocol for Generating an ATP Standard Curve (optional) (7)4.Appendix (7)A.Overview of the CellTiter-Glo ®Assay..............................................................7B.Additional Considerations..................................................................................8C.References............................................................................................................11D.Related Products. (12)1.DescriptionThe CellTiter-Glo ®Luminescent Cell Viability Assay (a–e)is a homogeneous method to determine the number of viable cells in culture based on quantitation of the ATP present, which signals the presence of metabolically active cells. The CellTiter-Glo ®Assay is designed for use with multiwell-plate formats, making it ideal for automated high-throughput screening (HTS) and cell proliferation and cytotoxicity assays. The homogeneous assay procedure (Figure 1) involves adding a single reagent (CellTiter-Glo ®Reagent) directly to cells cultured in serum-supplemented medium. Cell washing, removal of medium or multiple pipetting steps are not required.The homogeneous “add-mix-measure” format results in cell lysis and generation of a luminescent signal proportional to the amount of ATP present (Figure 2).The amount of ATP is directly proportional to the number of cells present in culture in agreement with previous reports (1). The CellTiter-Glo ®Assay relies on the properties of a proprietary thermostable luciferase (Ultra-Glo™ Recombinant Luciferase), which generates a stable “glow-type” luminescent signal and improves performance across a wide range of assay conditions. The luciferase reaction for this assay is shown in Figure 3. The half-life of the luminescent signal resulting from this reaction is greater than five hours (Figure 4). This extended half-life eliminates the need for reagent injectors and provides flexibility for continuous or batch-mode processing of multiple plates. The unique homogeneous format reduces pipetting errors that may be introduced during the multiple steps required by other ATP-measurement methods.CellTiter-Glo ®Luminescent Cell Viability AssayAll technical literature is available on the Internet at: /protocols/ Please visit the web site to verify that you are using the most current version of this Technical Bulletin. Please contact Promega Technical Services if you have questions on useofthissystem.E-mail:********************Figure 1. Flow diagram showing preparation and use of CellTiter-Glo ®Reagent.Promega Corporation ·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll F ree in USA 800-356-9526·Phone 608-274-4330 ·F ax 608-277-2516 ·3170M A 12_0ACellTiter-Glo CellTiter-Glo MixerLuminometer®System Advantages•Homogeneous:“Add-mix-measure” format reduces the number of plate-handling steps to fewer than that required for similar ATP assays.•Fast:Data can be recorded 10 minutes after adding reagent.•Sensitive:Measures cells at numbers below the detection limits of standard colorimetric and fluorometric assays.•Flexible:Can be used with various multiwell formats. Data can be recorded by luminometer or CCD camera or imaging device.•Robust:Luminescent signal is very stable, with a half-life >5 hours,depending on cell type and culture medium used.•Able to Multiplex:Can be used with reporter gene assays or other cell-based assays from Promega (2,3).Figure 3. The luciferase reaction.Mono-oxygenation of luciferin is catalyzed byluciferase in the presence of Mg 2+, ATP and molecular oxygen.Promega Corporation ·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll F ree in USA 800-356-9526·Phone 608-274-4330 ·F ax 608-277-2516 ·3171M A 12_0A L u m i n e s c e n c e (R L U )Cells per Well10,00060,00020,00030,00040,00050,0000R² = 0.9990.5 × 1061.0 × 1061.5 × 1062.0 × 1062.5 × 1063.0 × 1063.5 × 1064.0 × 106r² = 0.99020,00010,00030,00040,00050,000r² = 0.9900100200300400HO SN S N O S N S N OCOOH +ATP+O 2Ultra-Glo™ Recombinant Luciferase +AMP+PP i +CO 2+LightBeetle Luciferin OxyluciferinMg 2+0Figure 2. Cell number correlates with luminescent output.A direct relationship exists between luminescence measured with the CellTiter-Glo ®Assay and the number of cells in culture over three orders of magnitude. Serial twofold dilutions of HEK293cells were made in a 96-well plate in DMEM with 10% FBS, and assays wereperformed as described in Section 3.B. Luminescence was recorded 10minutes after reagent addition using a GloMax ®-Multi+ Detection System. Values represent the mean ± S.D. of four replicates for each cell number. The luminescent signal from 50HEK293 cells is greater than three times the background signal from serum-supplemented medium without cells. There is a linear relationship (r 2= 0.99)between the luminescent signal and the number of cells from 0to 50,000 cells per well.Figure 4. Extended luminescent half-life allows high-throughput batchprocessing.Signal stability is shown for three common cell lines. HepG2 and BHK-21cells were grown and assayed in MEM containing 10% FBS, while CHO-K1 cells were grown and assayed in DME/F-12 containing 10% FBS. CHO-K1, BHK-21 and HepG2 cells, at 25,000 cells per well, were added to a 96-well plate. After an equal volume of CellTiter-Glo ®Reagent was added, plates were shaken and luminescence monitored over time with the plates held at 22°C. The half-lives of the luminescent signals for the CHO-K1, BHK-21 and HepG2 cells were approximately 5.4, 5.2 and5.8hours, respectively.2.Product Components and Storage ConditionsProduct Size Cat.#CellTiter-Glo ®Luminescent Cell Viability Assay 10ml G7570Substrate is sufficient for 100 assays at 100µl/assay in 96-well plates or 400 assays at 25µl/assay in 384-well plates. Includes:• 1 × 10mlCellTiter-Glo ®Buffer • 1 vial CellTiter-Glo ®Substrate (lyophilized)Product Size Cat.#CellTiter-Glo ®Luminescent Cell Viability Assay 10 × 10ml G7571Each vial of substrate is sufficient for 100 assays at 100µl/assay in 96-well plates or 400 assays at 25µl/assay in 384-well plates (1,000 to 4,000 total assays). Includes:•10 × 10mlCellTiter-Glo ®Buffer •10 vials CellTiter-Glo ®Substrate (lyophilized)Promega Corporation ·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll F ree in USA 800-356-9526·Phone 608-274-4330 ·F ax 608-277-2516 ·R e l a t i v e L u m i n e s c e n c e (%)Time (minutes)CHO-K101020304050607080901003173M A 12_0AProduct Size Cat.# CellTiter-Glo®Luminescent Cell Viability Assay100ml G7572 Substrate is sufficient for 1,000 assays at 100µl/assay in 96-well plates or 4,000assays at 25µl/assay in 384-well plates. Includes:•1 × 100ml CellTiter-Glo®Buffer• 1 vial CellTiter-Glo®Substrate (lyophilized)Product Size Cat.# CellTiter-Glo®Luminescent Cell Viability Assay10 × 100ml G7573Each vial of substrate is sufficient for 1,000 assays at 100µl/assay in 96-well plates or4,000 assays at 25µl/assay in 384-well plates (10,000to 40,000 total assays). Includes:•10 × 100ml CellTiter-Glo®Buffer•10 vials CellTiter-Glo®Substrate (lyophilized)Storage Conditions:For long-term storage, store the lyophilized CellTiter-Glo®Substrate and CellTiter-Glo®Buffer at –20°C. For frequent use, the CellTiter-Glo®Buffer can be stored at 4°C or room temperature for 48hours without loss of activity. See product label for expiration date information. ReconstitutedCellTiter-Glo®Reagent (Buffer plus Substrate) can be stored at room temperaturefor up to 8hours with <10% loss of activity, at 4°C for 48hours with ~5% lossof activity, at 4°C for 4days with ~20% loss of activity or at –20°C for 21weekswith ~3% loss of activity. The reagent is stable for up to ten freeze-thaw cycles,with less than 10% loss of activity.3.Performing the CellTiter-Glo®AssayMaterials to Be Supplied by the User•opaque-walled multiwell plates adequate for cell culture•multichannel pipette or automated pipetting station for reagent delivery•device (plate shaker) for mixing multiwell plates•luminometer, CCD camera or imaging device capable of reading multiwell plates •optional:ATP for use in generating a standard curve (Section 3.C)3.A.Reagent Preparation1.Thaw the CellTiter-Glo®Buffer, and equilibrate to room temperature priorto use. For convenience the CellTiter-Glo®Buffer may be thawed andstored at room temperature for up to 48hours prior to use.2.Equilibrate the lyophilized CellTiter-Glo®Substrate to room temperatureprior to use.Promega Corporation·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll F ree in USA 800-356-9526·Phone 608-274-4330 ·F ax 608-277-2516 ·3.A.Reagent Preparation (continued)3.Transfer the appropriate volume (10ml for Cat.# G7570 and G7571, or 100mlfor Cat.# G7572 and G7573) of CellTiter-Glo ®Buffer into the amber bottlecontaining CellTiter-Glo ®Substrate to reconstitute the lyophilizedenzyme/substrate mixture. This forms the CellTiter-Glo ®Reagent.4.Mix by gently vortexing, swirling or inverting the contents to obtain ahomogeneous solution. The CellTiter-Glo ®Substrate should go intosolution easily in less than 1minute.3.B.Protocol for the Cell Viability AssayWe recommend that you perform a titration of your particular cells todetermine the optimal number and ensure that you are working within thelinear range of the CellTiter-Glo ®Assay. Figure 2 provides an example of sucha titration of HEK293 cells using 0 to 50,000 cells per well in a 96-well format.1.Prepare opaque-walled multiwell plates with mammalian cells in culturemedium, 100µl per well for 96-well plates or 25µl per well for 384-wellplates.Multiwell plates must be compatible with the luminometer used.2.Prepare control wells containing medium without cells to obtain a value forbackground luminescence.3.Add the test compound to experimental wells, and incubate according toculture protocol.4.Equilibrate the plate and its contents at room temperature forapproximately 30 minutes.5.Add a volume of CellTiter-Glo ®Reagent equal to the volume of cell culturemedium present in each well (e.g., add 100µl of reagent to 100µl of mediumcontaining cells for a 96-well plate, or add 25µl of reagent to 25µl ofmedium containing cells for a 384-well plate).6.Mix contents for 2 minutes on an orbital shaker to induce cell lysis.7.Allow the plate to incubate at room temperature for 10 minutes to stabilizeluminescent signal.Note:Uneven luminescent signal within standard plates can be caused bytemperature gradients, uneven seeding of cells or edge effects in multiwellplates.8.Record luminescence.Note:Instrument settings depend on the manufacturer. An integration timeof 0.25–1 second per well should serve as a guideline.Promega Corporation ·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll F ree in USA 800-356-9526·Phone 608-274-4330 ·F ax 608-277-2516 ·3.C.Protocol for Generating an ATP Standard Curve (optional)It is a good practice to generate a standard curve using the same plate onwhich samples are assayed. We recommend ATP disodium salt (Cat.# P1132,Sigma Cat.# A7699 or GE Healthcare Cat.# 27-1006). The ATP standard curveshould be generated immediately prior to adding the CellTiter-Glo®Reagentbecause endogenous ATPase enzymes found in sera may reduce ATP levels.1.Prepare 1µM ATP in culture medium (100µl of 1µM ATP solution contains10–10moles ATP).2.Prepare serial tenfold dilutions of ATP in culture medium (1µM to 10nM;100µl contains 10–10to 10–12moles of ATP).3.Prepare a multiwell plate with varying concentrations of ATP standard in100µl medium (25µl for a 384-well plate).4.Add a volume of CellTiter-Glo®Reagent equal to the volume of ATPstandard present in each well.5.Mix contents for 2 minutes on an orbital shaker.6.Allow the plate to incubate at room temperature for 10 minutes to stabilizethe luminescent signal.7.Record luminescence.4.Appendix4.A.Overview of the CellTiter-Glo®AssayThe assay system uses the properties of a proprietary thermostable luciferase toenable reaction conditions that generate a stable “glow-type” luminescentsignal while simultaneously inhibiting endogenous enzymes released duringcell lysis (e.g., ATPases). Release of ATPases will interfere with accurate ATPmeasurement. Historically, firefly luciferase purified from Photinus pyralis(LucPpy) has been used in reagents for ATP assays (1,4–7). However, it hasonly moderate stability in vitro and is sensitive to its chemical environment,including factors such as pH and detergents, limiting its usefulness fordeveloping a robust homogeneous ATP assay. Promega has successfullydeveloped a stable form of luciferase based on the gene from another firefly,Photuris pennsylvanica(LucPpe2), using an approach to select characteristics thatimprove performance in ATP assays. The unique characteristics of this mutant(LucPpe2m) enabled design of a homogeneous single-reagent-addition approachto perform ATP assays with cultured cells. Properties of the CellTiter-Glo®Reagent overcome the problems caused by factors, such as ATPases, thatinterfere with ATP measurement in cell extracts. The reagent is physicallyrobust and provides a sensitive and stable luminescent output.Promega Corporation·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll F ree in USA 800-356-9526·Phone 608-274-4330 ·F ax 608-277-2516 ·4.A.Overview of the CellTiter-Glo®Assay (continued)Sensitivity and Linearity:The ATP-based detection of cells is more sensitivethan other methods (8–10). In experiments performed by Promega scientists,the luminescent signal from 50HEK293 cells is greater than three standarddeviations above the background signal from serum-supplemented mediumwithout cells. There is a linear relationship (r2= 0.99) between the luminescentsignal and the number of cells from 0 to 50,000 cells per well in the 96-wellformat. The luminescence values in Figure 2 were recorded after 10minutes ofincubation at room temperature to stabilize the luminescent signal as describedin Section3.B. Incubation of the same 96-well plate used in the experimentshown in Figure 2 for 360minutes at room temperature had little effect on therelationship between luminescent signal and number of cells (r2= 0.99).Speed:The homogeneous procedure to measure ATP using the CellTiter-Glo®Assay is quicker than other ATP assay methods that require multiple steps toextract ATP and measure luminescence. The CellTiter-Glo®Assay also is fasterthan other commonly used methods to measure the number of viable cells(such as MTT, alamarBlue®or Calcein-AM) that require prolonged incubationsteps to enable the cells’ metabolic machinery to convert indicator moleculesinto a detectable signal.4.B.Additional ConsiderationsTemperature:The intensity and decay rate of the luminescent signal from theCellTiter-Glo®Assay depends on the luciferase reaction rate. Environmentalfactors that affect the luciferase reaction rate will change the intensity andstability of the luminescent signal. Temperature is one factor that affects therate of this enzymatic assay and thus the light output. For consistent results,equilibrate assay plates to a constant temperature before performing the assay.Transferring eukaryotic cells from 37°C to room temperature has little effect onATP content (5). We have demonstrated that removing cultured cells from a37°C incubator and allowing them to equilibrate to 22°C for 1–2 hours hadlittle effect on ATP content. For batch-mode processing of multiple assayplates, take precautions to ensure complete temperature equilibration. Platesremoved from a 37°C incubator and placed in tall stacks at room temperaturewill require longer equilibration than plates arranged in a single layer.Insufficient equilibration may result in a temperature gradient effect betweenwells in the center and at the edge of the plates. The temperature gradientpattern also may depend on the position of the plate in the stack.Promega Corporation·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll F ree in USA 800-356-9526·Phone 608-274-4330 ·F ax 608-277-2516 ·Chemicals:The chemical environment of the luciferase reaction affects theenzymatic rate and thus luminescence intensity. Differences in luminescenceintensity have been observed using different types of culture media and sera.The presence of phenol red in culture medium should have little impact onluminescence output. Assaying 0.1µM ATP in RPMI medium without phenolred resulted in ~5% increase in luminescence output (in relative light units[RLU]) compared to assays in RPMI containing the standard concentration ofphenol red, whereas assays in RPMI medium containing twice the normalconcentration of phenol red showed a ~2% decrease in luminescence.Solvents for the various test compounds may interfere with the luciferasereaction and thus the light output from the assay. Interference with theluciferase reaction can be detected by assaying a parallel set of control wellscontaining medium without cells. Dimethylsulfoxide (DMSO), commonly usedas a vehicle to solubilize organic chemicals, has been tested at finalconcentrations of up to 2% in the assay and only minimally affects light output.Plate Recommendations:We recommend using standard opaque-walledmultiwell plates suitable for luminescence measurements. Opaque-walledplates with clear bottoms to allow microscopic visualization of cells also maybe used; however, these plates will have diminished signal intensity andgreater cross talk between wells. Opaque white tape may be used to decreaseluminescence loss and cross talk.Cellular ATP Content:Different cell types have different amounts of ATP,and values reported for the ATP level in cells vary considerably (1,4,11–13).Factors that affect the ATP content of cells may affect the relationship betweencell number and luminescence. Anchorage-dependent cells that undergocontact inhibition at high densities may show a change in ATP content per cellat high densities, resulting in a nonlinear relationship between cell numberand luminescence. Factors that affect the cytoplasmic volume or physiology ofcells also will affect ATP content. For example, oxygen depletion is one factorknown to cause a rapid decrease in ATP (1).Promega Corporation·2800 Woods Hollow Road ·Madison, WI 53711-5399 USA Toll F ree in USA 800-356-9526·Phone 608-274-4330 ·F ax 608-277-2516 ·4.B.Additional Considerations (continued)Mixing:Optimal assay performance is achieved when the CellTiter-Glo®Reagent is mixed completely with the cultured cells. Suspension cell lines (e.g., Jurkat cells) generally require less mixing to achieve lysis and extract ATP than adherent cells (e.g., L929 cells). Tests were done to evaluate the effect ofshaking the plate after adding the CellTiter-Glo® Reagent. Suspension cellscultured in multiwell plates showed only minor differences in light outputwhether or not the plates were shaken after adding the CellTiter-Glo®Reagent.Adherent cells are more difficult to lyse and show a substantial differencebetween shaken and nonshaken plates.Several additional parameters related to reagent mixing include the force ofdelivery of CellTiter-Glo®Reagent, sample volume and dimensions of the well.All of these factors may affect assay performance. The degree of reagent mixing required may be affected by the method used to add the CellTiter-Glo®Reagent to the assay plates. Automated pipetting devices using a greater or lesser force of fluid delivery may affect the degree of subsequent mixing required.Complete reagent mixing in 96-well plates should be achieved using orbitalplate shaking devices built into many luminometers and the recommended2-minute shaking time. Special electromagnetic shaking devices that use aradius smaller than the well diameter may be required to efficiently mixcontents of 384-well plates. The depth of medium and geometry of themultiwell plates may have an effect on mixing efficiency. We recommend that you take these factors into consideration when performing the assay andempirically determine whether a mixing step is necessary for the individualapplication.LuminometersFor highly sensitive luminometric assays, the luminometer model and settings greatly affect the quality of data obtained. Luminometers from differentmanufacturers will vary in sensitivities and dynamic ranges. We recommend the GloMax®products because these instruments do not require gainadjustments to achieve optimal sensitivity and dynamic range. Additionally, GloMax®instruments are preloaded with Promega protocols for ease of use.If you are not using a GloMax®luminometer, consult the operating manual for your luminometer to determine the optimal settings. The limits should beverified on each instrument before analysis of experimental samples. The assay should be linear in some portion of the detection range of the instrument used.For an individual luminometer there may be different gain settings. Werecommend that you optimize the gain settings.4.C.References1.Crouch, S.P. et al.(1993) The use of ATP bioluminescence as a measure of cellproliferation and cytotoxicity. J. Immunol. Methods160, 81–8.2.Farfan, A.et al.(2004) Multiplexing homogeneous cell-based assays. Cell Notes10, 2–5.3.Riss, T., Moravec, R. and Niles, A. (2005) Selecting cell-based assays for drugdiscovery screening. Cell Notes13, 16–21.4.Kangas, L., Grönroos, M. and Nieminen, A.L. (1984) Bioluminescence of cellular ATP:A new method for evaluating cytotoxic agents in vitro. Med. Biol.62, 338–43.5.Lundin, A. et al.(1986) Estimation of biomass in growing cell lines by adenosinetriphosphate assay.Methods Enzymol. 133, 27–42.6.Sevin, B.U. et al.(1988) Application of an ATP-bioluminescence assay in human tumorchemosensitivity testing. Gynecol. Oncol.31, 191–204.7.Gerhardt, R.T.et al.(1991) Characterization of in vitro chemosensitivity ofperioperative human ovarian malignancies by adenosine triphosphatechemosensitivity assay. Am. J. Obstet. Gynecol. 165, 245–55.8.Petty, R.D. et al.(1995) Comparison of MTT and ATP-based assays for themeasurement of viable cell number. J. Biolumin. Chemilumin.10, 29–34.9.Cree, I.A. et al.(1995) Methotrexate chemosensitivity by ATP luminescence in humanleukemia cell lines and in breast cancer primary cultures: Comparison of the TCA-100assay with a clonogenic assay. AntiCancer Drugs6, 398–404.10.Maehara, Y. et al.(1987) The ATP assay is more sensitive than the succinatedehydrogenase inhibition test for predicting cell viability. Eur. J. Cancer Clin. Oncol.23, 273–6.11.Stanley, P.E. (1986) Extraction of adenosine triphosphate from microbial and somaticcells. Methods Enzymol.133, 14–22.12.Beckers, B. et al.(1986) Application of intracellular ATP determination in lymphocytesfor HLA-typing. J. Biolumin. Chemilumin.1, 47–51.13.Andreotti, P.E. et al.(1995) Chemosensitivity testing of human tumors using amicroplate adenosine triphosphate luminescence assay: Clinical correlation forcisplatin resistance of ovarian carcinoma. Cancer Res. 55, 5276–82.4.D.Related ProductsCell Proliferation ProductsProduct Size Cat.# ApoLive-Glo™ Multiplex Assay10ml G6410 ApoTox-Glo™ Triplex Assay10ml G6320 CellTiter-Fluor™ Cell Viability Assay (fluorescent)10ml G6080 CellTiter-Blue®Cell Viability Assay (resazurin)20ml G8080 CellTiter 96®AQ ueous One SolutionCell Proliferation Assay (MTS, colorimetric)200 assays G3582 CellTiter 96®AQ ueous Non-RadioactiveCell Proliferation Assay (MTS, colorimetric)1,000 assays G5421 CellTiter 96®AQ ueous MTS Reagent Powder1g G1111 CellTiter 96®Non-RadioactiveCell Proliferation Assay (MTT, colorimetric)1,000 assays G4000 Additional sizes available.Cytotoxicity AssaysProduct Size Cat.# CytoTox-Glo™ Cytotoxicity Assay (luminescent)*10ml G9290Mitochondrial ToxGlo™ Assay*10ml G8000 MultiTox-Glo Multiplex Cytotoxicity Assay(luminescent, fluorescent)*10ml G9270 MultiTox-Fluor Multiplex Cytotoxicity Assay(fluorescent)*10ml G9200 CytoTox-Fluor™ Cytotoxicity Assay (fluorescent)*10ml G9260 CytoTox-ONE™ Homogeneous MembraneIntegrity Assay (LDH, fluorometric)*200–800 assays G7890 CytoTox-ONE™ Homogeneous MembraneIntegrity Assay, HTP1,000–4,000 assays G7892 CytoTox 96® Non-Radioactive Cytotoxicity Assay1,000 assays G1780 (LDH, colorimetric)*GSH-Glo™ Glutathione Assay10ml V691150ml V6912 GSH/GSSG-Glo™ Assay10ml V661150ml V6612 *Additional sizes available.LuminometersProduct Size Cat.# GloMax®-Multi+ Detection System with Instinct™ Software:Base Instrument with Shaking 1 each E8032 GloMax®-Multi+ Detection System with Instinct™ Software:Base Instrument with Heating and Shaking 1 each E9032 GloMax®-Multi+ Luminescence Module 1 each E8041Apoptosis ProductsProduct Size Cat.# Caspase-Glo®2 Assay*10ml G0940 Caspase-Glo®6 Assay*10ml G0970 Caspase-Glo®3/7 Assay* 2.5ml G8090 Caspase-Glo®8 Assay* 2.5ml G8200 Caspase-Glo®9 Assay* 2.5ml G8210Apo-ONE®Homogeneous Caspase-3/7 Assay1ml G7792 DeadEnd™ Fluorometric TUNEL System60 reactions G3250 DeadEnd™ Colorimetric TUNEL System20 reactions G7360Anti-ACTIVE®Caspase-3 pAb50µl G7481Anti-PARP p85 Fragment pAb50µl G7341Anti-pS473Akt pAb40µl G7441 Caspase Inhibitor Z-VAD-FMK, 20mM50µl G7231125µl G7232*Additional sizes available.(a)U.S. Pat. Nos. 6,602,677 and 7,241,584, European Pat. No. 1131441, Japanese Pat. Nos. 4537573 and 4520084 and other patents pending(b)U.S. Pat. No. 7,741,067, Japanese Pat. No. 4485470 and other patents pending.(c)U.S. Pat. No. 7,700,310, European Pat. No. 1546374 and other patents pending.(d)U.S. Pat. Nos 7,083,911, 7,452,663 and 7,732,128, European Pat. No. 1383914 and Japanese Pat. Nos. 4125600 and 4275715.(e)The method of recombinant expression of Coleoptera luciferase is covered by U.S. Pat. Nos. 5,583,024, 5,674,713 and 5,700,673.© 2001–2012 Promega Corporation. All Rights Reserved.Anti-ACTIVE, Apo-ONE, Caspase-Glo, CellTiter 96, CellTiter-Blue, CellTiter-Glo, CytoTox 96 and GloMax are registered trademarks of Promega Corporation. ApoTox-Glo, ApoLive-Glo, CellTiter-Fluor, CytoTox-Fluor, CytoTox-Glo, CytoTox-ONE, DeadEnd, GSH-Glo, GSH/GSSG-Glo, Instinct, Mitochondrial ToxGlo and Ultra-Glo are trademarks of Promega Corporation. alamarBlue is a registered trademark of Trek Diagnostic Ssystems, Inc.Products may be covered by pending or issued patents or may have certain limitations. Please visit our Web site for more information.All prices and specifications are subject to change without prior notice.Product claims are subject to change. Please contact Promega Technical Services or access the Promega online catalog for the most up-to-date information on Promega products.。
英文文献汇报PPT课件
Monoclonal antibodies against P. vivax MSP-1 42 Localization of P. vivax MSP-1 42 by indirect immunofluorescence
SDS–PAGE and immunoblotting
Purification of recombinant P. vivax MSP-1 42 expressed in E. coli Purification of plasmid for immunization
IF=2.218
.
1
.
2Hale Waihona Puke 1. Introduction
Merozoite surface protein-1 of malaria parasites, present on the surface of merozoite and playing a key role in the invasion process, is one of the leading vaccine targets for both P. falciparum and P. vivax malaria. MSP-1 occurs as a 200 kDa precursor and undergoes step-wise proteolytic processing resulting in a glycosyl-phosphatidylinositol-anchored 42 kDa protein fragment (MSP-1 42 ) on the surface of free merozoite. A large number of studies have shown the protective potential of MSP-1 fragments (42 and 19 kDa) of P. falci-parum and P. yoelii .
NVIDIA nForce 590 SLI系列产品说明说明书

Características y ventajasMCP NVIDIA nForce® 590 SLIPensado para entusiastasLos procesadores de comunicaciones y contenidos multimedia (MCP) NVIDIA nForce® 590 SLI™ proporcionan todas las herramientas y el rendimiento que necesitan los frikis del PC. Si se combinan con determinadas tarjetas gráficas NVIDIA GeForce y otros componentes del sistema, la velocidad del bus aumenta de forma dinámica. Además, ofrecen funciones de overclocking y mayor rapidez en la transmisión de datos.Tecnología NVIDIA LinkBoost™El MCP nForce 590 SLI incrementa automáticamente el ancho de banda cuando detecta la presencia de determinadas tarjetas gráficas NVIDIA® GeForce®.Diseñado para NVIDIA® SLI™La tecnología NVIDIA SLI es una innovación revolucionaria que permite aumentar drásticamente el rendimiento gráfico combinando varias GPU NVIDIA en un mismo sistema dotado de un MCP NVIDIA nForce SLI.Componentes con certificación NVIDIA SLICuando se combinan ciertos componentes (como determinadas GPU NVIDIA® GeForce® y memorias del sistema) con placas basadas en el MCP nForce 590 SLI, se incrementa automáticamente la velocidad del bus del sistema. Si quieres información sobre los componentes con certificación SLI, visita \nForce.Dos enlaces PCI Express SLI x16El ancho de banda de los dos enlaces PCI Express de 16 vías garantiza el máximo rendimiento gráfico para los juegos y las GPU de última generación. Multiplica por dos el ancho de banda de las soluciones PCI Express SLI x8.Almacenamiento MediaShield™ de NVIDIAConjunto de funciones que mantiene a salvo la información digital. Siempre fiable, escalable y accesible. Incluye soporte de configuraciones RAID y unidades de disco SATA.Configuración de varios discosUn sencillo asistente ayuda a configurar fácilmente las unidades de disco para obtener mayor protección de los datos, un acceso más rápido a los discos o máxima capacidad de almacenamiento.MediaShield selecciona automáticamente la configuración RAID 0, 1, 0+1 o 5 en función de tus necesidades. Los más expertos pueden manejar las opciones RAID directamente si lo prefieren.Sistema de alerta de discosSi falla alguno de los discos, MediaShield presenta una imagen en la que señala el disco defectuoso para facilitar su identificación, sustitución y recuperación.Migración de nivel RAID (morphing)MediaShield ofrece al usuario la posibilidad de cambiar la configuración RAID existente por otra configuración en un solo paso denominado cambio de nivel o morphing. Esto elimina la necesidad de hacer la copia de seguridad de los datos y efectuar los numerosos pasos que conllevaría el proceso manual.Matriz de discos de arranqueLas funciones MediaShield permiten utilizar una matriz de discos para cargar el sistema operativo al arranque.Seis unidades de disco SATA 3Gb/sPosibilidad de combinar hasta 6 unidades SATA en un volumen para obtener configuraciones RAID más rápidas y de mayor capacidad. La presencia de más discos significa más opciones de configuración, lo que incluye, por ejemplo, dos matrices RAID 5 o 6 unidades RAID 0 (striping) para obtener máxima velocidad de acceso a los datos. Además, el soporte de unidades SATA-2 3Gb/s permite aprovechar ventajas como las funciones de conexión en caliente y reordenación de colas de comandos (Native Command Queuing y Tagged Command Queuing). Las colas de comandos nativas aumentan la eficacia del acceso a los discos en entornos multithread porque permiten mantener las operaciones de lectura/escritura en espera para ejecutarlas en el orden más conveniente.Comunicación en red con NVIDIA nForceLa tecnología de red de NVIDIA proporciona la máxima velocidad de transmisión con el menor índice de utilización de la CPU. Además de ser extraordinariamente estable y manejable, esta solución facilita la administración de la red y reduce el coste total de propiedad. Sólo NVIDIA integra este nivel de funcionalidad de red para llevar la comunicación online a otra dimensión.Gigabit Ethernet nativo de NVIDIALa máxima velocidad existente en conexiones Gigabit Ethernet. Elimina los cuellos de botella y mejora la eficacia global del sistema.Tecnología NVIDIA FirstPacket™Conviértete en el “rey del ping” con la tecnología FirstPacket de NVIDIA. Tendrás la mejor calidad de comunicación en tus llamadas telefónicas y todo el rendimiento que necesitas al jugar online.FirstPacket garantiza que los datos de los juegos, las comunicaciones de voz sobre IP (VoIP) y las transferencias de archivos de gran tamaño se gestionarán de acuerdo con las preferencias que tú establezcas a través de un sencillo asistente de configuración.Tecnología NVIDIA DualNet®Duplica la capacidad de tus comunicaciones en red con las dos conexiones Gigabit Ethernet integradas en el MCP NVIDIA nForce 500.Combinación de las dos conexiones Gigabit EthernetLa combinación de ambas conexiones permite sumar su capacidad para duplicar el ancho de banda Ethernet y, de esta forma, poder transferir grandes cantidades de datos desde el servidor de archivos a otros PC. Además, proporciona redundancia gracias al cambio automático de enlace en caso de fallo (failover).Aceleración de las funciones TCP/IPOfrece el máximo rendimiento del sistema al realizar mediante hardware el trabajo de filtrado de paquetes normalmente reservado a la CPU, lo que proporciona un entorno de red más rápido. Utilidad NVIDIA nTune™ 4.0La nueva versión de esta utilidad Windows incluye más opciones para optimizar el rendimiento. nTune permite ajustar manual o automáticamente los parámetros del sistema para conseguir el rendimiento deseado. Una vez realizada la configuración, la utilidad elige automáticamente los parámetros adecuados para la aplicación que se ejecute basándose en las reglas personales y los perfiles definidos por el usuario.PCI ExpressDiseñado para funcionar con el bus PCI Express. Este bus duplica el ancho de banda del bus AGP 8X, lo que proporciona una velocidad superior a 4 GB/s en las transferencias de datos en ambas direcciones.Audio de alta definición (HDA)El audio de alta definición introduce en el PC la calidad de sonido de los equipos electrónicos de consumo. Con HDA, los sistemas pueden proporcionar un extraordinario sonido de 192 kHz/32 bits a través de ocho canales que admiten los nuevos formatos de audio.USB 2.0Interfaz estándar que proporciona conexión inmediata con los dispositivos USB.。
PMMA Disc安全数据说明书

PMMA Disc Page 1 of 5SECTION 1: Identification of the substance/mixture and of the company/undertaking1.1. Product identifierPMMA Disc1.2. Relevant identified uses of the substance or mixture and uses advised against1.3. Details of the supplier of the safety data sheetCompany Name:Address:City, State, Zip Code:Telephone:email address:Website:SECTION 2: Hazards identification VinciSmile Group LLC 16453 Old Valley BlvdLa Puente, CA, 91744, USA +1 626 283 5808************************ 2.1. Product classificationMay cause sensitisation by skin contact.2.2. Health HazardThe product presents no toxic hazard when used according the manufacturer’s instructions. If a patient is known to beallergic to any of the ingredients of the teeth, the product should not be used.2.3. Additional dangers to humans and the environmentSee point 12.SECTION 3: Composition/information on ingredients3.2. MixturesChemical characterizationsolidHazardous componentsPoly methyl methacrylate with high molecular weight Methyl methacrylatePigments: Iron oxide and Titanium dioxideHazardous Components C.A.S. # EINECS # Substance Classification WT %Poly(methyl methacrylate)9011-14-7 232-674-9 Not Applicable >90SECTION 4: First aid measures4.1. Description of first aid measuresGeneral informationNone.PMMA DiscPage 2 of 5 After inhalationIn case of trouble after inhaling abrasive dust, seek medical advice.After eye contactI n case of mechanical eye irritation, rinse thoroughly with large amounts of water. Consult a doctor if irritation persists. After skin contactIn case of dust contact with skin, wash off with soap and water. Consult a doctor if skin irritation persists.After swallowingIn case of swallowing, seek medical advice.Cautions for the doctorNo specific antidote. Effect support therapy. Care should be effected according doctor’s evaluation with respect to thepatient’s reactions.Most important symptoms of exposureThis product is a non-hazardous polymer solid. Dust generated from polishing or grinding may cause eye or respiratorytract irritation.SECTION 5: Firefighting measures5.1. Extinguishing mediaSuitable extinguishing agentsThese products are not classified as flammable or combustible but will burn under fire conditions. Water spray jet, foam,dry powder, carbon dioxideFor safety reasons unsuitable extinguishing agentsWater with full jet.Protective equipment for fire-fightingUse self-contained respirator (breathing apparatus). -Wear full protection suit.SECTION 6: Accidental release measures6.1 Person-related safety precautionsAvoid build up of dust. Use breathing protection (Fine dust mask FFP) when exposed to dust. Use personal protectiveequipment (overalls, protective glasses and protective gloves).6.2 Environmental PrecautionsReport releases as required by local and national authorities.6.3 Methods and Materials for Containment and Clean-up:Pick up teeth and return to container for use. For dust from grinding or polishing products: Contain spills, sweep orgather spilled material in a manner that minimizes the generation of airborne dusts, and transfer to a suitable containerfor disposal.SECTION 7: Handling and storage7.1. Precautions for safe handlingAvoid generating dust. Avoid breathing dust. Use adequate ventilation for polishing or grinding operations. Wash handsthoroughly after polishing or grinding teeth. Use good housekeeping to minimize accumulation of dust.7.2. Conditions for safe storage, including any incompatibilitiesNo special storage required.Safety Data SheetPMMA DiscPage 3 of 5SECTION 8: Exposure controls/personal protection8.1 Exposure informationNone.8.2 Technical devicesNone.8.3 Personnel protective equipmentNone.8.4 Respiratory ProtectionNo needed with normal usage. When product is grinded or polished, use localized aspiration of dusts and/or protectivemask.8.5 Skin protectionNone particular precaution should be taken with normal usage.8.6 Eye and visage protectionNone particular precaution should be taken with normal usage. When the product is grinded or polished, use protectiveglasses.SECTION 9: Physical and chemical propertiesPhysical state at 20°C solidColor variousOdor unscentedSpecific weight (at 20°C) --Solubility in water (at 20°C) insolublePH (at 20°C) --Casting temperature --Boiling temperature --Auto ignition temperature --Inflammability temperature --Air deflagration limits --SECTION 10: Stability and reactivity10.1 Dangerous reactionsNo dangerous reactions known.10.2 Chemical StabilityStable.10.3 Conditions to AvoidStable in normal conditions.10.4 Hazardous Decomposition ProductsDecomposition may release oxides of carbon, methyl methacrylate, and irritating smoke and fumes.10.5 Materials to avoidNone in particular.10.6 Hazardous decomposition productsNone when correctly utilized.PMMA DiscPage 4 of 5SECTION 11: Toxicological information11.1. Information on toxicological effectsSensitisationAllergic reactions were reported among humans. The product contains small quantities of sensitising substances(methyl methacrylate ). Intensive skin contact, especially with the dissolved product, can trigger an allergic reaction inpersons previously sensitised.Further informationThe biocompatibility tests carried out on the teeth, in accordance with EN ISO 10993, regarding cytotoxicity, toxicitytest, stimulate or intradermal reaction, acute systemic toxicity and delayed type, did not result in any negative effects,under the test conditions, on the cell materials used.The fines which appear during sharpening can lead to mechanical irritations of the skin, eyes, and mucous membranes.Skin and eye contact with the product should be avoided. The inhalation of product dusts should be avoided.SECTION 12: Ecological information12.1 DegradationThe material is known to be stable in environment. Due to the law biodegradability the product should not reach wateror ground.12.2 Aquatic toxicityInsoluble. No dangers for environment are known.SECTION 13: Disposal considerations13.1.RecommendationThe waste is not dangerous and can be disposed together with household garbage.Disposal area must comply with the environment and national safety standard.SECTION 14: Transport informationUN number: n.a.class: n.a.packaging group: n.a.ocean harmful substances: n.a.other relevant information: no hazardous goodSECTION 15: Regulatory information15.1. Safety, health and environmental regulations/legislation specific for the substance or mixtureThe product is subject to the regulations of the Medical Device Directive 93/42/EEC, as well as to the national MedicalDevice Act and the Chemicals ActPMMA DiscPage 5 of 5 SECTION 16: Other informationNo further technical informationThe present data sheet contains technical-scientific information processed at best of our knowledge. We recommendverifying national and regional regulations applicable to the specific utilize field as well as regulations relative hygienicand safety on work and environment worship.All information contained in the present data sheet is correct and processed in good faith. However they do not involveany obligation, guarantee and patent concession.The characteristics mentioned in the following document do not constitute contractual specifications.The data for the hazardous ingredients were taken respectively from the last version of the sub-contractor's safety data sheet.。
AquaSnap ATP测试设备用户指南说明书

|ATP en muestras líquidasAquaSnap es un test fácil de utilizar que permite determinar la presencia deATP en muestras líquidas y es compatible con todos los luminómetros deHygiena. Este dispositivo está disponible en dos formatos: Free y Total.AquaSnap Free valora el ATP libre que está presente en la solución (ATP nomicrobiano). AquaSnap Total valora ambos, ATP libre y ATP microbianopresentes en la solución (ATP no microbiano y ATP microbiano). La diferenciaentre el ATP total y libre proporciona una indicación de la contaminaciónmicrobiana en la muestra.Ambos dispositivos ofrecen un diseño específico con un asa de inmersióncalibrada que permite recoger 100 µl asegurando una toma de muestraconsistente.Un reactivo único y estable combinado con el dispositivoofrece una precisión y reproducibilidad sin igual.Ideal para su uso en sistemas CIP, tratamiento deaguas industriales, torres de refrigeración ysistemas cerrados de aguas.Beneficios:∙Fácil de utilizar∙El diseño patentado Snap-V a l v e™ permite obtener resultados más precisos y con menosvariación∙Único reactivo líquido y estable que proporciona precisión y reproducibilidad∙Resistente a temperaturas elevadas y a los efectos de los desinfectantes∙Asa calibrada para la toma de 100 µl muestra∙Plástico 100% reciclable∙Amplia caducidad, 15 meses de vida útil en nevera (2-8 °C)∙ 4 semanas a temperatura ambiente (21-25 °C)Libre TotalSe ns ibilidad:∙Extremadamente sensible, detecta 0.1 femtomoles de ATP con EnSURE∙Detecta por debajo de 103 CFURefencia Descripción CantidadAQ-100FX A qu a Sn a p Fr ee100Productos para Alimentación y BebidasTest de ATPDescubra videos instructivos,soporte documental y más enw ww.h yg i e na.c o mVer demostración enyout ube.c o m/H yg i e na TVProcedimiento :Productos relacionados :EnSUREEl EnSure es un luminometro de alta sensibilidad que permite realizar múltiples tests decalidad en un solo instrumento tales como ATP, TVC, Coliformes, E. Coli, y otros.El EnSURE incluye el software SureTrend que permite hacer un seguimiento de todos los datos . SystemSURE PlusEl SystemSURE Plus se utiliza para realizar los controles de higiene. Este instrumento de reducidasdimensiones es fácil de utilizar, muy sensible y asequible, lo que permite a los usuarios determinarrápidamente la eficacia de la limpieza, el estado de las superficies y muestras líquidas paragarantizar la calidad del producto y reducir costes. Ultrasnap y SuperSnapUltrasnap mide ATP en superficies. SuperSnap es el test más sensible para la medida del ATP ensuperficies de Hygiena. Diseñado para trabajar con todos los luminómetros Hygiena, Ultrasnap ySuperSnap, todo en un solo dispositivo, detectan niveles extremadamente bajos de ATP. MicroSnapMicroSnap es una test rápido para la detección y enumeración de bacterias específicas. Losresultados están disponibles en 8 horas o menos, por lo que se pueden obtener resultados en elmismo día de trabajo o turno. MicroSnap está disponible en cuatro formatos: TVC ,enterobacterias , coliformes y E. coli .Refencia Descripción Cantidad Productos para Alimentación y Bebidas Test de ATPRev310315 MS-ECOLI MicroSnap E.coli 100 MS-TOTAL MicroSnap Total 100。
Vortex Shifter Engine Manual

Thank you for purchasing Shifter Vortexengines. This manual contains information tohelp you to get the best results from yournew engine. Furthermore, it will explain youhow to operate your Vortex engine safely andin a proper manner. All the information inthis manual is based on the latest experienceand product information available at the timeof writing. Vortex reserves the right to makeany kind of changes to this manual at anytime without notice and/or incurring in anyobligation.OTK KART GROUP s.r.l.Via dei Soprini 16 25080 Prevalle (Brescia) ITVORTEX FactoryVia E. Fermi 527040 Campospinoso (Pavia) IT**************************TABLE OF CONTENTChapter Information Page 1 GENERAL INFORMATIONSymbols 3Safety information 3Technical specifications 4Special technical specifications for homologated engine 5Packaging 52 ENGINE ASSEMBLY 6Engine bracket 7Coil 8Spark plug 10Fuel pomp 11Carburetor 123 ENGINE RUNNING IN 164 MAINTEANCE 17Maintenance schedule guide and adjustments 17Torque chart 17General tolerances 17Maintenance detail chart 18Carburetor cleaning 18Chancing main jet 19Spark plug cleaning and replacement 20Cylinder head cleaning 21Cylinder check and maintenance 22Piston check and maintenance 23Cleaning and/or replacing clutch 24GENERAL INFORMATIONSYMBOLSPay attention to the symbols of this manual. They alert you of dangerous situations for you or foryour engine.Personal Injury Mechanical Danger CautionIn order to perform a job, special tools are requested.SAFETY INFORMATION-Do not start the engine indoors garages, trailers etc. Start the engine in a well---ventilated areaonly. Exhaust emission are hazardous to your health.-Always wear gloves and proper clothing when working on your engine.-Use caution when handling fuel, as fuel is very flammable and explosive. When working with fuel,do not smoke or use it near fire or flames. Avoid any skin contact and inhaling fuel vapors. -Never touch moving parts when the engine is running.-During operation, both engine and muffler, become very hot. Do not touch them and do not place anything on them after operation.-Do not touch the spark plug or cable. It may provoke electrical shocks.-Understand the operation of all controls and learn how to stop the engine quickly in case of emergency.-Do not use the engine without clutch cover and chain protection.TECHNICAL SPECIFICATIONS-All sizes and measurements in this manual are expressed in metrics.-Always use original Vortex parts and proper tools when working on your engine. -Proper fuel mix is necessary for optimum engine life and performance.SPECIAL TECHNICAL SPECIFICATIONS FOR HOMOLOGATED ENGINESEvery engine is specified according to the homologation of its country. For specific rules and/or sizes refer to your country homologation filePACKAGINGYour engine will be packed in a sealed box with the Vortex logo printed on and a sticker with model and serial number attached. In a complementary box all the accessories as carburetor, muffler and more will be provided.The boxes need to have the original vortex logo.The engine box an official OTK KART USA sticker.And on the engine starter motor there needs to bean official OTK KART USA sticker too.ENGINE ASSAMBLYIn order to assemble the engine, you will need the following tools:Compressed airUnpack the engine and remove any packagingmaterial on it. Clean the engine withcompressed air and after take of the protectingPVC cups on the inlet, exhaust and spark plug.ENGINE BRACKET6mm T-wrenchLay the engine on its side and attachthe engine mount to the engine basewith four 8mm Allen screws. Engine mount and screws come with the engine.10mm Allen T-WrenchWarning: Engine comes with NO oil.Do not start the engine before filling it with oil. Remove the oil filler cap with 10mm Allen T-wrench and fill with 280gr of engine oil, viscosity W10/40 (type JASO MA-2 or API SL).Replace the oil cap.Coil10mm fixed wrench4mm T-fixed wrenchAssemble coil support and coil asper instructions. All needed partscome with the engine.Assemble the two silent blocks on the coils support plate in the engine by inserting between the plate and the silent block the ground cable.Insert coil in the two silent blocks.Insert ground cable from the coil and the other end of the ground cable in the silent block.Fix ground cables and the coil to the silent block by using M6 nut and washers provided.Be careful when assembling ground cable, the end must be in contact with the coil metal support, the other one in contact with engine crank case. Wrong assembling will result in coil failure and/or the engine will not start. Connect the coil connecter with the starter connector.SPARK PLUGUnbox the spark plug, the ignition distance must be0.8 mm like in the drawing belowSpark plug wrenchPlier Remove the PVC cap with the plier from thecylinder head.Manually tighten the spark plug into the cylinderhead.Lock and unlock with the spark plug wrench 2/3times to allow the gasket to seat properly. Now youcan tighten the spark plug properly.Insert the cable coil in the spark plug cap andtighten it.For safety, we recommend you to secure the cablecoil to the spark plug cap with a plastic strap. Placethe spark plug cap on the spark plug and press thecap fully.FUEL POMP10mm fixed wrench5mm fixed wrenchPlace the fuel pump in the specific fuel pump bracket as per scheme.All material needed comes with the engineWhen the fuel pump is placed on the engine insert the pulse line at pictured (red arrow). Later on, the other two lines will be assembled.CARBURETOR ScrewdriverFlat plier8 mm fixed wrenchRemove the plastic cap from the intakemanifold.Remove the carburetor from the box.Put the metal retaining clamp providedwith the carburetor, on the rubber intakemanifold boot.Push the carburetor firmly into intakemanifold boot.Making sure that the carburetor is in themanifold in the correct position, secure itwith the metal retaining clamp provided.Feed the throttle cable inside the register and the carburetor cover making sure to leave the cable inside.Feed the throttle cable through carburetor slide spring. Pull the slide from the carburetor.Place the ball at the end of the throttle cable through the hole in the center of the slide. Lock the throttle cable inside the slide by moving the cable to the side to the center.Place the slide and the spring in the carburetor. Replace the cap of the carburetor to the carburetoritself, screwing manually.Very carefully, by using the pliers, turn the twocarburetor vents so that they are facing up.Place the vent line supplied on each ofthe two vents. Cut half of the line in thecenter, to create a vent hole.Insert fuel line from fuel pump to carburetor. Secure fuel line with a plastic zip tie.10 mm Fixed WrenchPlace the clutch cable register. Lateron, when engine will be placed in the chassis, will serve to get a propertensioned clutch cable.5 mm Fixed Wrench Assemble gear lever before placing the engine in the chassis. Install the lever on theshaft and tighten it with the Allen screw.AFTER placing the lever in the shaft.ENGINE RUNNING INENGINE BREAK INOnly a proper break-in will insure the best performanceout of your engine in the future and guarantee its longand trouble-free life. Break-in is required when an engineis new or has undergone a major service of the engine’smain parts (piston, cylinder, connecting rod, etc.).Prepare fuel. Vortex engine works with commercialgasoline, leaded or unleaded, as well as racing fuel, withminimum 95 Octane. Mix Oil and fuel at 4% (i.e. 40cc ofoil every 1.000cc of fuel). Use high-quality synthetic oilspecifically made for kart engines. Vortex suggestsPetronas Rok Lube, however other brands with the samecomposition might be suitable.Shake the can thoroughly to mix the fuel and the oilproperly. Then fill the gas tank in your kart.A mistake in measurements could result inengine damage.Do not accelerate fully but only partially.Check that the cooling system warms up evenly; in case it warms unevenly proceed again with the bleeding of the cooling system.Once the engine is warmed up and the cooling system works properly, proceed to the track. Run the engine by alternating RPM’s a few seconds on and off the throttle at 3⁄4 maximum throttle.Do not hold the throttle at a constant speed. Continue this way for 5/6 laps and return to pits. Check everything on the kart is tightening properly.Be careful, both engine and muffler are hot.Return to the track and slowly increase the RPM and duration of the acceleration phasefor 10/15min more. Intermittently open the throttle fully and then release it.After 10/15 minutes of brake-in, your engine is ready for competition.During the break-in, nuts and bolts tend to loosen. Once the engine is cold, check thetorque of the exhaust, head, etc.MAINTENANCEGood maintenance is essential for safe, economical and trouble-free operation. Here you will find a maintenance schedule for your engine. Routine inspection procedures are very simple by using basic tools. Some service tasks that are more difficult or needs special tools must be performed by Vortex technicians or qualified mechanics.Timing schedule periods are only indicative. Extreme carburation set ups highly modify timing schedule periods.Maintenance schedule guide an adjustmentsTorque chartGeneral tolerancesMAINTENEANCE DETAIL CHARTIn the following section, you will find a detailed most important maintenance jobs to be performed.CARBURETORCLEANINGFlat Screwdriver1. Take the intake silencer off the carburetor byunscrewing the clamp.2. Disconnect the throttle cable from the carburetortogether with the spring and the guillotine slide.3. Take the carburetor off the engine and open thefloating chamber by unscrewing the special cap in themiddle of the chamber. Clean the parts, openings andpassages with compressed air.4. Unscrew the main jet by means of a flat screw driver.Replace it with another calibrated differently. Becareful, the washer must be installed in one positiononly. A wrong assembly may cause the carburetorcomplete malfunction.5. Check the plug gasket of the floating chamber is stillintact and if damaged, replace it.6. Close the floating chamber by securing the specialcap and mount the carburetor on the engine.7. Clean the inside of the intake silencer.8. Mount the intake silencer on the carburetor flange.9. Tighten it with the specific clamp.Wrong assembly will cause the loss of the intake silencer.CHANGING JETSFlat screwdriver22mm fixed wrenchWrong carburation set up could cause sever enginedamage.1. Take the intake silencer off the carburetor byunscrewing the clamp.2. Disconnect the throttle cable from the carburetortogether with the spring and the guillotine slide.3. Take the carburetor off the engine and open thefloating chamber by unscrewing the special cap in themiddle of the chamber. Clean the parts, openings andpassages with compressed air.4. Unscrew the main jet by means of a flat plier.Replace it with another calibrated differently. Becareful, the washer must be installed in one positiononly. A wrong assembly may cause the carburetorcomplete malfunction.5. Check the plug gasket of the floating chamber is stillintact and if damaged, replace it.6. Close the floating chamber by securing the specialcap and mount the carburetor on the engine.7. Clean the inside of the intake silencer.8. Mount the intake silencer on the carburetor flange.9. Tighten it with the specific clamp.Wrong assembly will cause the loss of the intake silencer.Wrong carburetor assembling may cause engine malfunction and damage.SPARK PLUG CLEANING AND REPLACEMENTSparkplug wrench Metal BrushRisk of burning: Perform this task ONLYwhen engine is cool.Oils produce carbon deposits or residues that makenecessary the spark plug to be checked andcleaned, at least every 5 hours.Remove the spark plug and clean it by using a brassmetal brush.Use a specific spark plug gap gauge to set up correctgap. Correct gap: 0.8 mm.Every 30 hours it is highly recommended to changethe spark plug.EXHAUST CLEANINGMetal BrushHeaterOils produce carbon deposits or residues that make necessary the exhaust to be checked andcleaned, at least every 10 hours.Disassemble the exhaust from the engine by removing the two springs and check the exhaust carefully.Heat the exhaust with a heater and remove all carbon deposits with a metal brush.CYLINDER HEAD CLEANING10mm Tube wrenchDynamometric wrenchSpark plugThe oil in the fuel produces carbon deposits and/orresidues that makes it necessary to check thecylinder head at least every race.Be aware, cylinder head combustionchamber volume may vary during therace. Carbon deposits may causevariations in cylinder head volume.Remove spark plug. Remove the six head nuts and relativewashers.Remove cylinder head by pulling it up carefully.Use rubber gloves.After cleaning the combustion chamber with fuel,reassemble cylinder head. Please check cooper washer, incase it is present, is located in the right position.This specific washer, assembled wrongly, will change the combustion chamber volume.Insert carefully the head on to the studs and check all O-rings are fitted in the right place. If any is damage, change it. Insert washers and nuts in the studs and tight them manually. Now, by using a dynamometric wrench tighten them alternatively at 16 Nm.CYLINDER CHECK AND MAINTENANCEDynamometric wrenchmechanic must perform inspection and honing.Remove exhaust.relative washers.Remove cylinder head by pulling it up carefully.Remove the cylinder from the crankcase slowly. Oncecylinder is separate from crankcase, hold the connectingrod with the other hand and pull the cylinder out totallyby pulling it up carefully.Every time cylinder is removed we recommend changingcylinder gasket.clearance is more than 0.19mm or whenovalization is more than 0.02 mm.Change piston (see next page)New piston cylinder/piston clearance must be 0.09mm.REASEMBLINGInsert a new gasket on to the studs carefully and place itin the crankcase surface flat.Insert now the cylinder in the studs very carefully. With the other hand close piston ring and inset the piston into the cylinder. If piston ring is not closed totally, risk of cylinder and piston ring is very high Push the cylinder down firmly. Tight four cylinder nuts with dynamometric wrench at 20Nm. Insert cooper washer. Please check cooper washer is located in the right position.This specific washer, assembled wrongly, will change the combustion chamber volume. Install, carefully, the head onto the studs and check all o-rings are fitted in the right place. If any is damaged, change it.Install washers and nuts onto the studs and tight them manually. Now, by using a dynamometric wrench tighten them alternatively at 16 Nm.Every time cylinder is honed or piston changed, the engine break-in procedure must beperformedPISTON CHECK AND MAINTENANCE10mm Tube wrenchDynamometric wrenchTo avoid engine damage, a Vortex qualifiedmechanic must perform inspection and honing.Remove exhaust.Remove spark plug. Remove the six head nutsand relative washers.Remove cylinder head by pulling it up carefully.Once cylinder is separate from crankcase, hold theconnecting rod with the other hand and pull the cylinderout totally by pulling it up carefully.Every time cylinder is removed we recommend changing cylinder gasket. Remove the circlips from the piston pin.Push the pin out of the piston.Before assembling a new piston checkdimensional sizes. Correct cylinder/pistonclearance is 0.09mm.ASSEMBLINGOil and insert the roller cage in the connecting rod.Install the new piston in the connecting rod by placingthe arrow stamped in the head of the piston facing theexhaust port. Insert now the piston pin and secure it withthe circlips.Attention, wrong circlips assembling may causeserious damage.Insert the piston ring in the piston carefully. Check bothends of the piston ring when totally closed have a gap of0.30/0.35 mm.Now you are ready to assemble cylinder and cylinderhead.Engine needs a brake-in in session when a newpiston has been placed.CHANGING CLUTCH DISCS4mm fixed wrenchDynamometric wrenchClutch is composed of disc and spacers to transmit the movement to the pinion. Even when the use of clutch is minimum disc suffer wear.Remove the clutch protection cover. Disassemble the screws thathold the disc and springs. Take screws, springs and spacers off. 2, 3,4. Now you can disassemble the cover and take the disc and spaceroff the clutch bell. Exchange the old disc with new ones.Be careful, first and last disc (A) of the series are differentthan the other ones (B). Follow the scheme to assemblediscs and spacers. Proceed backwards to reassemble. Screws mustbe tightened 0.8Nm。
LAQUAtwin系列水质测试仪操作指南说明书

CODE: GZ0000472457Instruction Manual (Operation)COMPACT WATER QUALITY METER LAQUAtwin-EC-11, LAQUAtwin-EC-22, LAQUAtwin-EC-33SpecificationsItems in packageConsumable parts sold separatelyPart Namesotherwise specified.Initial SetupAttaching/detaching the sensorInserting/removing batteriesElectrode conditioningNote●Before using the sensor for the first time or after several days of disuse, perform electrode conditioning.●Perform calibration after electrode condition-ing.1.Place some drops of the conditioning solu-tion into the measurement cell.2.Wait 10 min before use.There is no need to switch the meter ON.3.Clean the flat sensor with running water.Model LAQUAtwin-EC-112233Minimum sample volume 0.12 mL √√√Range and resolution (valid digits)Conductivity0 to 199 μS/cm:200 to 1999 μS/cm:2.00 to 19.99 mS/cm: 1 μS/cm 1 μS/cm 0.01 mS/cm √√√20.0 to 199.9 mS/cm:0.1 mS/cm √√TDS 0.0 to 99.9 ppm:100 to 999 ppm:1000 to 9990 ppm:0.1 ppm1 ppm 10 ppm√Calibration Up to 2 points√Up to 3 points√√Accuracy *1±2% full scale (for each range)√±2% full scale (0 to 19.99 mS/cm)±5% full scale (20.0 to 199.9 mS/cm)√√Temperature display 0 to 50.0︒C √√Target Electrical conductivity Measurement principle 2 electrode bipolar AC Titanium coated with Platinum black Display Custom (monochrome) digital LCD withbacklightOperating environment 5 to 40︒C, 85% or less relative humidity (no condensation)Power CR2032 batteries (⨯2)Battery life Approx. 400 h continuous operation*2Outer dimen-sions/mass 164 ⨯ 29 ⨯ 20 mm (excluding projections), Approx. 45 g (excluding batteries)Main functionTemperature compensation (2%/︒C fixed), waterproof *3, auto stable/auto hold, auto-matic power OFF*1Accuracy is the closeness of agreement betweenthe measured value and actual value of the stan-dard solution in the measurement of the same standard solution as the one used for the calibra-tion. Temperature during the calibration and mea-surement is the same. The error of standard solutions and rounding error (±1 digit) are not included.*2The life period if the meter is used in the backlightoff mode. If the backlight is used, battery life will shorten.*3IP67: no failure when immersed in water at adepth or 1 meter for 30 minutes.Please note that the meter can not be used underwater ItemsQuantitySensor S0701Meter1Storage case 1BatteriesCR20322Standard solution1413 μS/cm 112.88 mS/cm1Pipette1Conditioning solution1Instruction manual (Operation)1Instruction manual (Before use)1Items Specifications Part No.Sensor S070, COND3200459672Standard solution514-22, 1413 μS/cm 3999960110514-23, 12.88 mS/cm 3999960111Conditioning solution514-203999960114Attaching the sensor 1.Power OFF the meter.2.Confirm that the waterproofing gasket is clean and undamaged.Detaching the sensor 1.Power OFF the meter.2.Lift the sensor tongue tip and slide the sensor a little away from the meter.3.Pull out the sensor all the way from the meter. Inserting the batteries 1.Power OFF the meter.2.Slide both batteries into the battery case as shown.Be sure to use two CR2032 batteries, and put them with the plus sides (+) upwards.Removing the batteries 1.Power OFF the meter.e a ball-point pen or other tool to pry the batter-ies out from the clips as shown.Basic OperationCalibrationCalibration is required before measurement.Use standard solution within the measurement range in the specifications.Tip●Calibration values are saved even if the meter is switched OFF.●Calibration value is rewritten if calibration is repeated using the same standard solution.MeasurementNote●If a measured value is out of the specified measurement range, "Or" is displayed for upper range and "Ur" is displayed for under range.●Ambient air may cause the measured values to fluctuate. To reduce environmental interfer-ence, close the protection cover.●When you have a problem with the calibration or measurement, refer to frequently asked questions.Measurement display changeMeasurement display change is available on LAQUAtwin-EC-22 and LAQUAtwin-EC-33.The display mode switches as follows by press-ing the MEAS switch in the AS mode.●LAQUAtwin-EC-22:Between conductivity and temperature alter-nately●LAQUAtwin-pH-33:Among conductivity, TDS, and temperaturePower ON1.Press and hold the ON/OFF switch.The power is switched ON,and the meter model num-ber is displayed on the LCD. Power OFF1.Press and hold the ON/OFF switch.The power is switched OFF.Precaution on sample settingPlace an appropriate amount of a sample or stan-dard solution into the measurement cell without trapping bubbles inside. If not, the measurement may be inaccurate.Calibration pointsThe number of calibration points is dependent on the meter model.●LAQUAtwin-EC-11:Up to two-point calibration at 1413 μS/cm and 12.88 mS/cm●LAQUAtwin-EC-22 and LAQUAtwin-EC-33:Up to three-point calibration at 1413 μS/cm,12.88 mS/cm, and 111.8 mS/cmMulti-point calibration1.Open the protection cover and place some drops of the standard solution into the measurement cell.Rinsing the sensor with the standard solution beforehand will provide a more accurate cali-bration as it will reduce sample crossover con-tamination.2.Close the protection cover and press theplayed.The calibration value at 25 C is displayed for 1s and the display returns to the measurement mode automatically.3.Open the protection cover and remove the standard solution. Then remove moisture on the sensor by gently dabbing with a soft tissue.This completes the 1st point calibration.4.To perform 2nd point calibration, repeat steps 1. to 3. Calibration error If blinks and Er4 (error dis-play) appears, the calibration has failed.Perform electrode conditioning.Check that the correct standard solution is used,and repeat calibration after cleaning the sensor.If the calibration repeatedly fails when using the correct standard solution(s), the sensor may have deteriorated. Replace the sensor with newone.Sample setting1.Open the protection cover and put some drops of sample into the measurement cell.2.Close the protection cover. Measurement modeThe auto stable (AS) mode and the auto hold (AH) mode can be selected. Refer to " Mea-surement mode change" (page 4) for the opera-tion to set the measurement mode. Auto stable (AS) mode1.Confirm that the meter is in the measure-ment mode, and place a sample on the sen-sor.locked.2.appears.Auto hold (AH) modeand will not change until the MEAS switch is pressed for the next measurement.1.Confirm that the meter is in the measure-ment mode, and place a sample on the sen-sor.2.Press the MEAS switch.The auto hold function is acti-vated.blinks until the mea-sured value has stabilized.When the measured value is stable, stops blinking and the displayed value is locked with3.Document the displayed value.4.Press the MEAS switch.disappears.Be sure to perform this step before starting the next measurement. Or, you may mistake the displayed hold value for the next measured value.MaintenanceAppendixFrequently asked questions Storage1.Clean the sensor with tap water.2.Dab gently with soft tissue or cloth to remove moisture on the sensor and meter.Especially be sure to treat the sensor gently to prevent damaging it.3.Close the protection cover before storing the meter. Temperature sensor adjustmentTemperature sensor adjustment is available on LAQUAtwin-EC-22 and LAQUAtwin-EC-33.To perform accurate measurement with correc-tion for temperature effects, follow the steps below. Normally this is not necessary.1.Ready a reference thermometer, and allow the meter and reference thermometer to reach to room temperature.2.Set the display mode to temperature refer-ring to " Measurement display change"(page 2).3.Press the CAL switch.The meter displays the setting screen for tar-get temperature.4.Press the MEAS switch to adjust the dis-played temperature on the meter to match the temperature indicated by the reference thermometer.Pressing the MEAS switch increases the dis-played temperature. After the displayed tem-perature reaches 40°C, it returns to 5°C.5.Press the CAL switch again to apply the displayed value to the adjustment.The adjustment starts. The adjusted value blinks with and displayed.After the adjustment is complete, the adjusted value stops blinking with MEAS and dis-played.If Er4 (error display) appears, the adjustment has failed. Retry the above steps increasing the time spent on the step 1.If the adjustment repeatedly fails, the sensor may have deteriorated. Replace the sensor with new one.Initializing calibration dataInitialize calibration in the following cases.●To delete the calibration data●If the number of points for the last calibration is uncertain.●After the sensor is replaced.1.Press and hold the CAL and ON/OFF switches for over 3 seconds when the meter is switched OFF to Initialize calibra-tion.After a moment of all segment indication, the software version is displayed. And then, the display changes as shown right.2.Press the CAL switch.All calibration data is reset.When the initialization of cali-bration data is complete, End appears.The meter automatically switches OFF. Initializing the settingsAll setup choices are erased. The meter is reset to the factory default values.1.Press and hold the MEAS, CAL and ON/OFF switches for over 3 seconds when the meter is switched OFF to enter the initial-ization.After a moment of all segment indication, the software version is displayed. And then, the display changes as shown right.2.Press the CAL switch.All calibration data is reset.When the initialization of set-tings is complete, End appears.The meter automatically switches OFF.Er4 is dis-played during the calibra-tionPlease note that if you press the CAL switch in mV or temperature display mode, Er4 is displayed. This is because there is no calibration facility available for these modes.Er1 is dis-played soon power ON.The internal IC in the meter may be defective. Perform meter initializa-tion.If Er1 is still displayed after the initial-ization, the internal IC in the meter is defective. Replace the meter with a new one (the meter cannot be repaired).Er2 is dis-played right after power ON.The internal IC in the meter is defec-tive. Replace the meter with a new one (the meter cannot be repaired).Er3 is dis-played right after power ON.The internal IC in the meter is defec-tive. Replace the meter with a new one (the meter cannot be repaired).Question AnswerSetup ModeThe setup mode allows the user to customize the meter to his specific needs.To enter the setup mode, press and hold the MEAS and ON/OFF switches for over 3 seconds when the meter is switched OFF. All the LCD segments appear and then the meter enters the setup mode.Tip●To have the changes apply, you need to go through the entire steps from “Setup mode entry” to “Setup completion” shown below. To leave a setting as it is, just press CAL switch in the setting.●To exit the setup mode with no change of settings, press the ON/OFF switch earlier than pressing CAL switch in the last step but one, or the “Backlight setting” step.Setup mode entryUnit settingThe display units can be changed.TDS method setting (Only LAQUAtwin-EC-33)The TDS method can be selected from the following options only on LAQUAtwin-EC-33.●FACt: KCl with factor adjustable from 0.4 to 1.0 (default 0.5)●442: Myron L 442 non-linear standard curve●En: European environmental standard non-linear curve ●NACL: Non linear salinity curveThis step is bypassed on LAQUAtwin-EC-11 and LAQUAtwin-EC-22.Factor setting (Only LAQUAtwin-EC-33 with the TDS method set to FACt)This step is bypassed on LAQUAtwin-EC-11 and LAQUAtwin-EC-22, and when the TDS method is set to 442, En, or NACL on LAQUAtwin-EC-33.*The setting range is from F0.4 to F1.0.In this setting, pressing the MEAS switchincreases the displayed value. After the dis-played value reaches F1.0, it returns toF0.4. Measurement mode changeThe measurement mode can be switched.NoteThe AH (auto hold) mode is applied only to conductivity measurement.* Measurement display change is available in the AS mode. Refer to " Measurement display change" (page 2).Backlight settingThe backlight can be switched to ON or OFF.Setup completion31, Miyanonishi-cho, Kisshoin Minami-ku, Kyoto,。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Product Name:
Lacosamide racemate CAS No.:
175481-26-2Cat No :HY-13015A Product Data Sheet
Cat. No.:
HY 13015A MWt:
250.29Formula:
C13H18N2O3Purity :>98%
Solubility:Mechanisms:
Biological Activity:
Pathways:Others; Target:Others DMSO
Lacosamide racemate(Vimpat racemate; Erlosamide racemate) is a medication developed for the
adjunctive treatment of partial-onset seizures and diabetic neuropathic pain.
IC50 value:
Target: Lacosamide is a functionalized amino acid that produces activity in the maximal electroshock seizure (MES) test, that, like some other antiepileptic drugs (AEDs), are believed to act through voltage-gated sodium channels. Lacosamide enhances the slow inactivation of voltage-gated sodium channels without affecting the fast inactivation of voltage-gated sodium channels. This
inactivation prevents the channel from opening helping end the action potential Many antiepileptic References:
[1]. Rauck, Richard L. et al. Lacosamide in Painful Diabetic Peripheral Neuropathy: A Phase 2
Double-blind Placebo-controlled Study Clinical Journal of Pain: February 2007 - Volume 23 - Issue 2
- pp 150-158[2]V tti A L i G Oli i i C Z lli E Z G inactivation prevents the channel from opening, helping end the action potential. Many antiepileptic drugs, like carbamazepine or lamotrigine, slow the recovery from inactivation and hence reduce the ability of neurons to fire action potentials....
[2]. Verrotti A, Loiacono G, Olivieri C, Zulli E, Zaccara G.
Lacosamide in patients with pharmacoresistant epilepsy.
Expert Opin Pharmacother. 2012 Aug 8.
[3]. Kamel JT, Degruyter MA, D'Souza WJ, Cook MJ. Clinical experience with using lacosamide for the treatment of epilepsy in a tertiary centre.
Acta Neurol Scand. 2012 Jul 31. doi: 10.1111/j.1600-0404.2012.01704.x.
[4]. Fountain NB, Krauss G, Isojarvi J, Dilley D, Doty P , Rudd GD.Safety and tolerability of adjunctive lacosamide intravenous loading dose in lacosamide-naive patients with partial-onset seizures Caution: Not fully tested. For research purposes only
Medchemexpress LLC
patients with partial onset seizures.
Epilepsia. 2012 Jun 18. doi: 10.1111/j.1528-1167.2012.03543.x....
18W i l k i n s o n W a y , P r i n c e t o n , N J 08540,U S A
E m a i l : i n f o @m e d c h e m e x p r e s s .c o m W e b : w w w .m e d c h e m e x p r e s s .c o m。
