GSK2606414_1337531-36-8_CoA_MedChemExpress
稳定表达猪δ_冠状病毒S1_蛋白CHO_细胞系的构建与鉴定

河南农业科学,2023,52(6):131‐138Journal of Henan Agricultural Sciencesdoi :10.15933/ki.1004‐3268.2023.06.014稳定表达猪δ冠状病毒S1蛋白CHO 细胞系的构建与鉴定孙雪珂1,2,丁培阳3,王思桥1,2,刘思源2,李明慧2,常泽杰2,陈艺兰2,李瑞琪2,张改平1,2,4(1.河南农业大学生命科学学院,河南郑州450002;2.河南省农业科学院动物免疫学重点实验室,河南郑州450002;3.郑州大学生命科学学院,河南郑州450001;4.江苏高校动物重要疾病与人兽共患病防控协同创新中心,江苏扬州225009)摘要:为构建稳定表达猪δ冠状病毒(PDCoV )S1蛋白的CHO 细胞系,利用电转染技术将重组质粒pCGS3-S 1转染至CHO 细胞,通过有限稀释法筛选稳定表达重组S1蛋白的单克隆细胞系。
利用阴离子交换层析和凝胶过滤层析方法对重组S1蛋白进行纯化,采用间接ELISA 方法检测重组S1蛋白活性。
将纯化的重组S1蛋白免疫BALB/c 小鼠,通过间接ELISA 、间接免疫荧光试验(IFA )及病毒中和试验对重组S1蛋白免疫原性进行检测。
结果显示,稳定表达PDCoV S1蛋白的CHO 细胞系成功建立,并获得了纯度高于90%、产量为28.5mg/L 的重组S1蛋白;且重组S1蛋白与PDCoV 阳性血清反应性良好,具有良好的免疫原性,中和效价为1∶128。
综上,成功建立了稳定表达PDCoV S1蛋白的CHO 细胞系,且纯化获得的重组S1蛋白具有良好的生物学活性。
关键词:猪δ冠状病毒;S1蛋白;CHO 细胞;稳定表达;蛋白质纯化中图分类号:S855.3文献标志码:A文章编号:1004-3268(2023)06-0131-08收稿日期:2023-01-14基金项目:河南省重大科技专项(221100110600)作者简介:孙雪珂(1997-),女,河南新乡人,在读硕士研究生,研究方向:动物免疫学。
GSK2606414_DataSheet_MedChemExpress

Inhibitors, Agonists, Screening Libraries Data SheetBIOLOGICAL ACTIVITY:GSK2606414 is an orally available and selective PERK inhibitor with an IC 50 of 0.4 nM.IC50 & Target: IC50: 0.4 nM (PERK)In Vitro: GSK2606414 inhibits PERK activation in cells [1].In Vivo: GSK2606414 (50 and 150 mg/kg, p.o.) inhibits the growth of a human tumor xenograft in mice [1].PROTOCOL (Extracted from published papers and Only for reference)Animal Administration: GSK2606414 is formulated in vehicle (0.5% hydoxypropylmethylcellulose, 0.1% Tween 80 inwater, pH 4.8).[1]Exponentially growing BxPC3 tumor cells (10×106 cells/mouse) from cell culture are implanted subcutaneously into the right flank of female nude mice. Sixteen days after implantation, mice with –200 mm 3 tumors are randomized into various treatment groups (n = 8 mice/group). Animals are orally treated with vehicle (0.5% hydoxypropylmethylcellulose, 0.1% Tween 80 inwater, pH 4.8), compound at 50 or 150 mg/kg, b.i.d. for 21 days. Tumor volume is measured twice weekly with calipers and calculated using the following equation: tumor volume (mm 3) = (length × width 2)/2. Results are represented as percent inhibition on completion of dosing, which is 100[1–(average growth of drug–treated population)/(average growth of vehicle–treated control population)]. Statistical analysis is performed using a two–tailed t test.References:[1]. Axten JM, et al. Discovery of 7–methyl–5–(1–<[3–(trifluoromethyl)phenyl]acetyl>–2,3–dihydro–1H–indol–5–yl)–7H–pyrrolo[2,3–d]pyrimidin–4–amine (GSK2606414), a potent and selective first–in–class inhibitor of protein kinase R (PKR)–like endoplasmic reticuluProduct Name:GSK2606414Cat. No.:HY-18072CAS No.:1337531-36-8Molecular Formula:C 24H 20F 3N 5O Molecular Weight:451.44Target:PERK Pathway:Cell Cycle/DNA Damage Solubility:DMSO: ≥ 31 mg/mLCaution: Product has not been fully validated for medical applications. For research use only.Tel: 609-228-6898 Fax: 609-228-5909 E-mail: tech@ Address: 1 Deer Park Dr, Suite Q, Monmouth Junction, NJ 08852, USA。
实时定量PCR测定治疗性HBV_DNA疫苗工程菌的质粒拷贝数

广东药科大学学报Journal of Guangdong Pharmaceutical University Sep,2023,39(5)实时定量PCR测定治疗性HBV DNA疫苗工程菌的质粒拷贝数陈孟璋,何星垚,林汉森(广州广药益甘生物制品股份有限公司,广东广州511495)摘要:目的比较不同的定量方法和总DNA制备方法对实时定量PCR(real-time quantitative PCR,qPCR)测定治疗性HBV DNA疫苗工程菌的质粒拷贝数(plasmid copy number,PCN)的影响,为建立快速简便的治疗性HBV DNA 疫苗工程菌的PCN测定方法提供参考。
方法通过试剂盒提取法、Triton X-100煮沸法和培养基煮沸法分别制备总DNA样品用于qPCR检测,使用绝对定量方法和相对定量方法计算不同制备方法样品的PCN,并对测定结果进行统计学分析。
结果试剂盒提取法制备的样品经绝对定量和相对定量计算PCN,结果分别为43.10±4.02和42.92±3.98,显著低于Triton X-100煮沸法的结果(69.32±6.51和68.58±6.42)以及培养基煮沸法的结果(75.73±7.46和76.15±7.50)。
任一样品制备方法,比较其绝对定量和相对定量结果,差异均无统计学意义。
结论qPCR的绝对定量和相对定量计算方法都适用于治疗性HBV DNA疫苗工程菌PCN测定,Triton X-100煮沸法更适合于总DNA模板制备。
关键词:HBV疫苗;DNA疫苗;实时定量PCR;质粒拷贝数中图分类号:R392.1文献标识码:A文章编号:2096-3653(2023)05-0087-06DOI:10.16809/ki.2096-3653.2023051703Determination of plasmid copy number of therapeutic HBV DNA vaccine engineered strain by real-time quantitative PCRCHEN Mengzhang*,HE Xingyao,LIN Hansen(Guangzhou Guangyao Yigan Biological Products Co.,Ltd.,Guangzhou511495,China)*Corresponding author Email:**************Abstract:Objective To compare the effects of different quantitative methods and total DNA preparation methods on the plasmid copy number(PCN)determination of therapeutic HBV DNA vaccine engineered strain by real-time quantitative PCR(qPCR),so as to provide reference for the establishment of a rapid and simple method for the PCN determination of therapeutic HBV DNA vaccine engineered strain.Methods Total DNA samples were prepared for qPCR by kit extraction method,Triton X-100boiling method and medium boiling method, respectively.The PCN of samples prepared by different methods was calculated by absolute and relative quantification methods,and the results were statistically analyzed.Results The absolute and relative quantification results of PCN of samples prepared by kit extraction method were43.10±4.02and42.92±3.98, respectively,which were significantly lower than those obtained by Triton X-100boiling method(69.32±6.51) and medium boiling method(75.73±7.46).There was no significant difference between the absolute and relative quantitative results of any sample preparation method.Conclusion Both the absolute and relative quantitative methods of qPCR are suitable for the determination of PCN of therapeutic HBV DNA vaccine engineered strain, and the Triton X-100boiling method is more suitable for the preparation of total DNA template.Key words:HBV vaccine;DNA vaccine;real-time quantitative PCR;plasmid copy number收稿日期:2023-05-17基金项目:广东省重大科技专项(2012A080204009)作者简介:陈孟璋(1985-),男,硕士,制药工程师,主要从事生物医药研发和临床试验研究,Email:**************。
不同亚型CHO_宿主细胞对抗体表达的影响

生物技术进展 2023 年 第 13 卷 第 5 期 698 ~ 703Current Biotechnology ISSN 2095‑2341进展评述Reviews不同亚型CHO 宿主细胞对抗体表达的影响曹辉 , 董静 , 贾宇 , 江一帆*华北制药集团新药研究开发有限责任公司,抗体药物河北省工程研究中心,抗体药物研究国家重点实验室,石家庄 050015摘要:CHO 细胞作为宿主细胞广泛应用于生物药工业化生产中。
其中,CHO -K1、CHO -DG44和CHO -S 是最常见的3种亚型。
虽然这些亚型是从共同的原始CHO 细胞分离出来的,但在不同的实验室或生物医药公司、研究人员、培养基或培养方式下连续传代、驯化和保存,使得CHO 细胞积累了大量变异,导致宿主细胞应用于抗体药生产时会在细胞生长状态、抗体表达量及以糖型为代表的质量属性方面表现出较大差异。
综述了CHO 细胞不同亚型的染色体差异、生长状态、表达差异以及糖型差异,以期为抗体药物研发中宿主细胞的选择提供参考。
关键词:CHO 细胞;抗体;表达量;糖型DOI :10.19586/j.2095‑2341.2023.0064中图分类号:Q28, R392-33 文献标志码:AEffects of Different Sources of CHO Host Cells on Antibody ExpressionCAO Hui , DONG Jing , JIA Yu , JIANG Yifan *State Key Laboratory of Antibody Drug Development , Hebei Engineering Research Center of Antibody Medicine , New Drug Research and Development Co. Ltd , North China Pharmaceutical Corporation , Shijiazhuang 050015, ChinaAbstract :CHO cells comprise a variety of lineages including CHO -K1, CHO -DG44 and CHO -S , which have been widely used in the industrial production of biological drugs. All CHO cell lines share a common ancestor , however , during the process of cell passage cultivation , cell domesticated , and preservation by different laboratories or companies , substantial genetic heterogeneity among them has been produced , that showed great differences in cell growth state , antibody titer , glycosylation and other product quality attributes. This article reviewed the difference in chromosome , growing status and expression , and glycoform in different sources ofCHO host cells , which was expected to be helpful in host cell selection during antibody drug research and development process.Key words :CHO cells ; monoclonal antibody ; antibody titer ; glycosylation生物药物在国际医药市场中占据主导地位,截至2023年,全球范围内已有100多个抗体药物被批准上市,近1 200个抗体药物处于不同临床试验阶段[1]。
茶多酚对人脂肪来源间充质干细胞成骨分化的影响

茶多酚对人脂肪来源间充质干细胞成骨分化的影响王华1,齐玉成-杨云芳-赵艺洋2,王慧1,陈培1,杨旭芳1(1.牡丹江医学院,黑龙江牡丹江157011;2.南方医科大学第一临床医学院,广东广州510515)摘要:目的探讨茶多酚(Epigallocatechin-3-gallate,EGCG)对人脂肪间充质干细胞(human adipose-derived mesenchy^-mal stem cells,hADSCs)成骨分化的影响。
方法利用胶原酶消化法和贴壁筛选法从人脂肪组织中分离、培养及扩增hADSCs,倒置显微镜下观察各代hADSCs的形态学特点;利用流式细胞术检测各代hADSCs免疫学表型;取P3代细胞进行成骨诱导分化,实验分三组,即未诱导组、常规成骨诱导组与EGCG组(常规成骨诱导+5^mol/L EGCG),14d后,镜下观察细胞形态学改变及碱性磷酸酶(ALP)染色。
结果体外分离、培养的hADSCs形态均一;流式细胞术结果显示hADSCs具备间充质干细胞的免疫学表型,即CD44、CD73、CD105阳性;成骨诱导14d后部分细胞由长梭形变成多角形,细胞呈现聚集趋势;ALP染色显示EGCG组呈强阳性。
结论成功的从脂肪组织中分离培养出了hADSCs,EGCG能加强其成骨分化能力,这将为骨质疏松症的临床药物开发提供新的思路,亦为组织工程骨的构建提供丰富可靠的种子细胞来源。
关键词:EGCG;人脂肪来源间充质干细胞;成骨分化中图分类号:R595.2文献标识码:A文章编号:1001-7550(2021)01-0001-04Effect of EGCG on osteogenic differentiation of human adipose-derived mesenchymal stem cellsWANG Hua et al(Mudanjiang Medical University,Mudanjiang157011,China)Abstract:Objective To explore the effect of tea polyphenol EGCG on the osteogenic differentiation of human adipose-derived mesenchymal stem cells(hADSCs) .Methods To isolate,culture and amplify hADSCs from human adipose tissue by collagenase digestion and adherent screening methods, the morphological characteristics of each passage of hADSCs were observed under an inverted microscope.The immunophenotype of each generation of hADSCs was detected by flow-cytometry.P3passage cells were taken for osteogenic induction and differentiation,and were divided into three groups:non-induced group, conventional osteogenic induction group and EGCG group(conventional osteogenic induction with+5Rmol/L EGCG).After14days,morphological changes and alkaline phosphatase(ALP)staining were observed under the microscope.Results The morphology of hADSCs isolated and cultured in vitro was uni-form.The results of flow cytometry showed that hADSCs had the immunophenotype of mesenchymal stem cells,such as CD44,CD73and CD105.After14days of osteogenic induction,some cells changed from long spindle shape to polygonal shape,and the cells showed aggregation trend.ALP staining showed strong positive in EGCG group.Conclusion hADSCs have been successfully isolated and cultured from adipose tissue.EGCG can enhance the osteogenic differentiation ability of hADSCs,which will provide a new idea for the clinical drug development of osteoporosis and provide an abundant and reliable source of seed cells for the construction of tissue-engineered bone.Key words:EGCG;human adipose-derived mesenchymal stem cells;osteogenic differentiation随着人口老龄化,骨质疏松症已成为影响人们生活质量的主要因素之一⑷。
影响因子_Chem2012
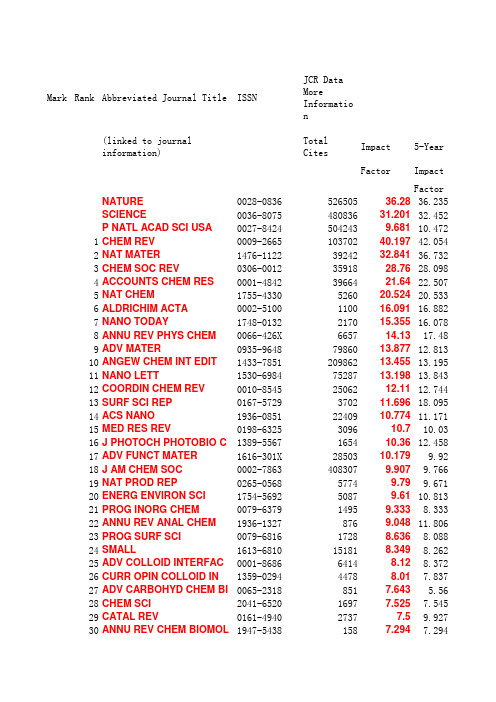
Mark Rank Abbreviated Journal Title ISSN JCR Data More Information (linked to journal information)Total Cites Impact5-YearFactor ImpactFactor NATURE0028-0836********.2836.235SCIENCE0036-807548083631.20132.452P NATL ACAD SCI USA0027-84245042439.68110.4721CHEM REV0009-266510370240.19742.0542NAT MATER1476-11223924232.84136.7323CHEM SOC REV0306-00123591828.7628.0984ACCOUNTS CHEM RES0001-48423966421.6422.5075NAT CHEM1755-4330526020.52420.5336ALDRICHIM ACTA0002-5100110016.09116.8827NANO TODAY1748-0132217015.35516.0788ANNU REV PHYS CHEM0066-426X665714.1317.489ADV MATER0935-96487986013.87712.81310ANGEW CHEM INT EDIT1433-785120986213.45513.19511NANO LETT1530-69847528713.19813.84312COORDIN CHEM REV0010-85452506212.1112.74413SURF SCI REP0167-5729370211.69618.09514ACS NANO1936-0851*******.77411.17115MED RES REV0198-6325309610.710.0316J PHOTOCH PHOTOBIO C1389-5567165410.3612.45817ADV FUNCT MATER1616-301X2850310.1799.9218J AM CHEM SOC0002-78634083079.9079.76619NAT PROD REP0265-056857749.799.67120ENERG ENVIRON SCI1754-569250879.6110.81321PROG INORG CHEM0079-637914959.3338.33322ANNU REV ANAL CHEM1936-13278769.04811.80623PROG SURF SCI0079-681617288.6368.08824SMALL1613-6810151818.3498.26225ADV COLLOID INTERFAC0001-868664148.128.37226CURR OPIN COLLOID IN1359-029444788.017.83727ADV CARBOHYD CHEM BI0065-23188517.643 5.5628CHEM SCI2041-652016977.5257.54529CATAL REV0161-494027377.59.92730ANNU REV CHEM BIOMOL1947-54381587.2947.29431CHEM MATER0897-4756699267.286 6.94932ADV ORGANOMET CHEM0065-30559517 6.63633NANO RES1998-01242017 6.977.46134CHEMSUSCHEM1864-56313040 6.8277.17135GREEN CHEM1463-926212023 6.32 6.76136TRAC-TREND ANAL CHEM0165-99366620 6.273 6.87337J PHYS CHEM LETT1948-71854695 6.213 6.217 38CHEM COMMUN1359-7345105562 6.169 6.082 39ADV SYNTH CATAL1615-415014629 6.048 5.904 40J CATAL0021-951733570 6.002 6.201 41J MATER CHEM0959-942846166 5.968 5.992 42INT REV PHYS CHEM0144-235X1533 5.967 6.977 43CHEM-EUR J0947-653954127 5.925 5.866 44NANOSCALE2040-33643187 5.914 5.914 45ORG LETT1523-706068838 5.862 5.558 46ANAL CHEM0003-270095262 5.856 5.983 47J CONTROL RELEASE0168-365926071 5.7327.112 48LAB CHIP1473-019713729 5.67 6.497 49APPL CATAL B-ENVIRON0926-337320525 5.625 6.052 50BIOSENS BIOELECTRON0956-566320029 5.602 5.637 51BIOMACROMOLECULES1525-779721280 5.479 5.646 52CARBON0008-622327798 5.378 6.008 53J MED CHEM0022-262356481 5.248 5.321 54J CHEM THEORY COMPUT1549-96188327 5.215 5.673 55PROG NUCL MAG RES SP0079-65651765 5.214 6.296 56CHEMCATCHEM1867-38801502 5.207 5.207 57FARADAY DISCUSS1364-549853725 4.687 58BIOCONJUGATE CHEM1043-180213657 4.93 5.224 59CURR MED CHEM0929-867311387 4.859 4.806 60J PHYS CHEM C1932-744760782 4.805 5.049 61CRYST GROWTH DES1528-748319082 4.72 4.877 62J CHEM INF MODEL1549-959611209 4.675 4.305 63INORG CHEM0020-166982190 4.601 4.553 64J COMPUT CHEM0192-865122334 4.583 4.795 65ANAL CHIM ACTA0003-267038343 4.555 4.144 66J CHROMATOGR A0021-967360179 4.531 4.362 67CHEM-ASIAN J1861-47284472 4.5 4.461 67ELECTROANAL CHEM0070-9778451 4.56 69J ORG CHEM0022-326398614 4.45 4.204 70SOFT MATTER1744-683X10988 4.39 4.998 71CHEM REC1527-89991256 4.377 4.161 72ANALYST0003-265414243 4.23 4.119 73PROG SOLID STATE CH0079-67861516 4.188 3.551 74LANGMUIR0743-7463103776 4.186 4.514 75CURR TOP MED CHEM1568-02664646 4.174 4.417 76PHARM RES-DORDR0724-874117160 4.093 4.668 77INT J HYDROGEN ENERG0360-319926939 4.054 4.402 78J AM SOC MASS SPECTR1044-03058794 4.002 3.746 79ORGANOMETALLICS0276-733339562 3.963 3.78680CHEMBIOCHEM1439-42279059 3.944 3.719 81APPL CATAL A-GEN0926-860X26533 3.903 3.961 82CRIT REV ANAL CHEM1040-8347849 3.902 3.956 83SENSOR ACTUAT B-CHEM0925-400527323 3.898 3.751 84MAR DRUGS1660-33971161 3.854 3.704 85CRYSTENGCOMM1466-80339036 3.842 4.023 86DALTON T1477-922637605 3.838 3.887 87TALANTA0039-914024981 3.794 3.747 88CHEM RES TOXICOL0893-228X10444 3.779 3.969 89ANAL BIOANAL CHEM1618-264218911 3.778 3.733 90J PHYS CHEM B1520-6106118812 3.696 4.061 90ORG BIOMOL CHEM1477-052015667 3.696 3.652 92ACS CHEM NEUROSCI1948-7193310 3.676 3.676 93ADV CATAL0360-05641395 3.667 5.04 94FOOD CHEM0308-814636842 3.655 4.268 95CARBOHYD POLYM0144-861715055 3.628 3.987 96PHYS CHEM CHEM PHYS1463-907631819 3.573 3.931 97EXPERT OPIN THER PAT1354-37761563 3.571 2.122 98ULTRASON SONOCHEM1350-41774594 3.567 3.83 99GOLD BULL0017-15571139 3.517 3.162 100STRUCT BOND0081-59931940 3.475 6.315 101FOOD HYDROCOLLOID0268-005X5607 3.473 3.46 102COLLOID SURFACE B0927-77658715 3.456 3.354 103CURR ORG SYNTH1570-1794665 3.434 3.842 104J CHEMINFORMATICS1758-2946179 3.419 3.419 105CHEMPHYSCHEM1439-423510354 3.412 3.553 106J COMB CHEM1520-47663046 3.408 2.893 107CATAL TODAY0920-586122303 3.407 3.584 108ACS MED CHEM LETT1948-5875511 3.355 3.355 109J INORG BIOCHEM0162-01349265 3.354 3.495 110EUR J MED CHEM0223-523411332 3.346 3.509 111EUR J ORG CHEM1434-193X18591 3.329 3.289 112ELECTROPHORESIS0173-083517540 3.303 2.917 113J BIOL INORG CHEM0949-82573803 3.289 3.488 114J NANOPART RES1388-07644770 3.287 3.574 115MICROPOR MESOPOR MAT1387-181113858 3.285 3.254 116CHEM CENT J1752-153X292 3.281 3.033 117J MASS SPECTROM1076-51745549 3.268 3.301 117PHYTOMEDICINE0944-71134731 3.268 3.3 119BIOANALYSIS1757-6180829 3.223 3.223 120J ANAL ATOM SPECTROM0267-94776969 3.22 2.966 121J PHYS CHEM REF DATA0047-26894693 3.172 3.804 122MOL DIVERS1381-19911114 3.153 3.236123CHEMMEDCHEM1860-71793364 3.151 3.445 124J NAT PROD0163-386418661 3.128 3.164 125DYES PIGMENTS0143-72086586 3.126 3.28 126MAR CHEM0304-42036836 3.074 3.551 127J COLLOID INTERF SCI0021-979742700 3.07 3.263 128CURR ORG CHEM1385-27283649 3.064 3.468 129J PHARM SCI-US0022-354916617 3.055 3.324 130EUR J INORG CHEM1434-194815553 3.049 2.904 131ADV INORG CHEM0898-******** 3.048 3.049 131MICROCHEM J0026-265X3125 3.048 2.878 133MICROCHIM ACTA0026-36724182 3.033 2.508 134TETRAHEDRON0040-402053591 3.025 3.06 135J ETHNOPHARMACOL0378-874121075 3.014 3.728 136ANAL BIOCHEM0003-269740002 2.996 3.247 137PLASMONICS1557-1955654 2.989 4.026 138CATAL COMMUN1566-73677950 2.986 3.299 139J PHARMACEUT BIOMED0731-708514454 2.967 2.979 140J MOL CATAL A-CHEM1381-116917457 2.947 3.336 141J PHYS CHEM A1089-563953462 2.946 2.941 142FUEL PROCESS TECHNOL0378-******** 2.945 3.131 143BIOORGAN MED CHEM0968-089622584 2.921 3.157 144J ELECTROANAL CHEM1572-665722586 2.905 2.731 145J CHROMATOGR B1570-023221252 2.888 3.057 146DRUG DES DEV THER1177-8881203 2.877147ELECTROANAL1040-039710069 2.872 2.856 148ANTI-CANCER AGENT ME1871-52061203 2.862149J SUPERCRIT FLUID0896-84465365 2.86 3.145 150J AGR FOOD CHEM0021-856171104 2.823 3.239 151MEDCHEMCOMM2040-2503223 2.8 2.8 152RAPID COMMUN MASS SP0951-419813453 2.79 2.782 153PURE APPL CHEM0033-454512669 2.789 2.987 154J MOL CATAL B-ENZYM1381-11774576 2.735 2.701 155J SEP SCI1615-93067275 2.733 2.725 156SYNLETT0936-521417866 2.71 2.567 157TETRAHEDRON LETT0040-403976620 2.683 2.588 158TETRAHEDRON-ASYMMETR0957-416613382 2.652 2.652 159SOLID STATE IONICS0167-273822111 2.646 3.097 160RUSS CHEM REV+0036-021X3090 2.644 2.941 161PHYTOCHEM ANALYSIS0958-03441883 2.633 2.32 162TOP CATAL1022-55284801 2.624 2.973 163SEP PURIF REV1542-2119175 2.615 3.629 164NEW J CHEM1144-05469359 2.605 2.775 165INT J MOL SCI1661-65962744 2.598 2.617166PHOTOCH PHOTOBIO SCI1474-905X3988 2.584 2.688 167ENVIRON CHEM1448-25171188 2.57 2.694 168BIOORG MED CHEM LETT0960-894X31475 2.554 2.539 169MINI-REV MED CHEM1389-55752649 2.528 2.772 170FUTURE MED CHEM1756-8919616 2.522 2.522 171BEILSTEIN J ORG CHEM1860-5397812 2.517 2.078 172PLANT FOOD HUM NUTR0921-96681448 2.505 2.889 173J ANAL APPL PYROL0165-23704356 2.487 2.687 174REACT FUNCT POLYM1381-51484207 2.479 2.727 175APPL CLAY SCI0169-13175168 2.474 3.06 176SYNTHESIS-STUTTGART0039-788118726 2.466 2.448 177J CHEM THERMODYN0021-96145451 2.422 2.173 178J PHOTOCH PHOTOBIO A1010-603012665 2.421 2.925 179MINI-REV ORG CHEM1570-193X460 2.406 2.283 180ORG PROCESS RES DEV1083-61603609 2.391 2.38 181MOL INFORM1868-1743171 2.39 2.407 182MOLECULES1420-30495118 2.386 2.411 183J ORGANOMET CHEM0022-328X22966 2.384 2.085 184CHIRALITY0899-******** 2.35 2.232 185AUST J CHEM0004-94255762 2.342 2.258 186CHEM PHYS LETT0009-261455511 2.337 2.215 187CARBOHYD RES0008-621514992 2.332 2.386 188COLLOID POLYM SCI0303-402X5510 2.331 2.077 189J ALLOY COMPD0925-838836296 2.289 2.104 190CHEM BIOL DRUG DES1747-02771504 2.282 2.477 191ADV QUANTUM CHEM0065-3276754 2.275 1.356 192CATAL LETT1011-372X9443 2.242 2.314 193COLLOID SURFACE A0927-775717780 2.236 2.359 194BIOPHYS CHEM0301-46224908 2.203 2.163 195J CHEM TECHNOL BIOT0268-25756388 2.168 2.27 196THEOR CHEM ACC1432-881X4587 2.162 2.909 197MATCH-COMMUN MATH CO0340-62531420 2.161 1.826 198J SOLID STATE CHEM0022-459617779 2.159 2.412 199PLANTA MED0032-094311107 2.153 2.338 200SUPRAMOL CHEM1061-02782323 2.145 2.021 201FLUID PHASE EQUILIBR0378-******** 2.139 2.225 202J FLUORESC1053-05092486 2.107 2.197 203APPL SURF SCI0169-433228315 2.103 2.032 204PHYTOTHER RES0951-418X7721 2.086 2.469 204SAR QSAR ENVIRON RES1062-936X715 2.086 1.874 206J FOOD COMPOS ANAL0889-15753582 2.079 3.257 207APPL ORGANOMET CHEM0268-26052774 2.061 1.849 208POLYHEDRON0277-538713245 2.057 2.118209J BIOMOL SCREEN1087-05712131 2.049 2.066 210J FLUORINE CHEM0022-11394628 2.033 1.95 211SOLVENT EXTR ION EXC0736-******** 2.024 1.81 212J ANAL TOXICOL0146-47602651 2.022 1.994 213ADSORPTION0929-560713282 2.215 214SURF SCI0039-602823707 1.994 1.661 215J ENVIRON MONITOR1464-03253800 1.991 2.245 216J IND ENG CHEM1226-086X1844 1.977 1.647 216PROG ORG COAT0300-94404252 1.977 2.167 218INORG CHEM COMMUN1387-70036187 1.972 1.975 219BIOMED CHROMATOGR0269-38792728 1.966 1.88 220J PHYS ORG CHEM0894-32302668 1.963 1.648Eigenfactor® MetricsMore Information Immediacy Articles Cited Eigenfactor®Article Influence®Index Half-life Score Score9.698419.4 1.6565820.3536.0758719.4 1.4128217.5081.87436147.8 1.6033 4.8927.1581967.90.2146413.3056.246134 4.70.2208917.8915.471314 3.20.13678.0693.4612670.10127.2865.308120 1.80.032847.94472 5.70.00328 5.2462.32437 2.80.0121 5.1443.267309.70.016877.4672.15578950.26226 4.0612.8982002 5.40.513933.372.082955 4.20.34576 5.0592.1791517.70.04173.2063.2588.70.009037.8051.631114120.12069 3.5842.12524 6.80.00678 2.7330.37516 6.60.00367 3.3871.51453340.112652.9391.86531767.50.816772.7922.61172 5.70.01594 2.9722.049548 1.90.01802 2.6940>10.00.00081 2.9341.0520 3.10.0052 3.930.51290.00413 3.9451.221430 3.20.072332.5070.633797.80.01312 2.3720.829707.40.00919 2.2640.3336>10.00.00074 1.6142.8483280.90.00453 2.4890.4297>10.00.00219 2.9011.41724 1.40.000932.9691.637673 6.40.15823 1.910.52>10.00.00074 1.8340.918122 2.30.01112 2.3841.1142012.40.01339 1.8641.09442 4.20.03184 1.4971.022138 5.60.01667 1.8071.405529 1.40.024172.072 1.5033408 5.50.24077 1.547 1.003398 4.10.04809 1.4850.842389.60.04417 1.5691.0882527 4.30.12631 1.571 1.75127.10.004032.549 1.29416973.90.1688 1.527 1.187653 1.40.01164 1.561 1.48516804.90.18767 1.411 0.93313287.70.17541 1.5470.915331 6.70.04644 1.6491.143538 3.60.05012 1.7050.858471 5.40.04732 1.4211.102704 4.20.05043 1.2040.752509 4.90.06046 1.3951.081641 6.30.06074 1.595 1.0497097.40.09638 1.3 1.145413 3.10.04017 1.716 1.55207.60.004652.345 1.049224 1.60.00601 1.352 1.3771389.20.01265 1.752 0.577300 5.50.03426 1.347 0.474403 5.20.03177 1.297 0.723166 2.90.28453 1.339 0.7237333.50.053380.912 0.595289 6.30.017020.7660.82814997.40.127630.9641.0763438.40.03476 1.516 0.597626 6.80.070850.9630.622110470.09860.9211.0453772.70.02083 1.243>10.00.00011 1.506 1.09912429.90.13784 1.05 0.8981353 2.50.05461 1.584 0.3182260.0032 1.1690.75869480.0249 1.0571.25>10.00.00172 1.053 0.711936 6.50.22306 1.179 0.706201 4.70.01384 1.168 0.7392618.50.02793 1.196 0.5911753 3.30.059170.7180.711228 5.80.02249 1.0541.013827 6.90.067680.830.819337 4.30.03418 1.204 0.5245527.10.045390.929 0.235177.20.001430.864 0.355904 5.50.057310.801 0.404171 1.90.004220.839 0.8441018 2.90.021720.749 0.831492 5.50.077660.883 0.447854 5.10.05450.81 0.68921970.02114 1.039 0.702967 4.10.062970.967 0.7151689 6.70.2465 1.159 0.8941061 3.90.051860.961 0.69669 1.40.00158 1.21405>10.00.00072 1.57 0.7321483 4.70.079720.839 0.64835 4.90.028380.744 0.91231440.11162 1.241 0.70896 2.90.005410.609 0.964197 4.70.009170.739 0.28257.30.002590.91611980.00341 1.67 0.825234 5.40.011580.756 0.716524 4.20.021180.7390.43848 4.40.002050.9771.4250 1.50.000460.805 0.71410 4.50.04139 1.1870 4.50.008370.668 0.45566870.04130.919 0.793179 1.30.00205 1.044 0.635222 6.80.015560.791 0.484672 3.50.026580.705 0.766789 4.80.049630.8180.424413 6.90.029650.6261.018111 5.60.01044 1.028 0.287669 3.80.015170.929 0.796368 5.20.03430.799 0.20986 3.20.001390.839 0.58143 6.50.013621 0.441195 5.40.009240.65 0.795171 1.60.001670.422 0.6142647.70.010820.673 0.21414>10.00.004512.295 0.734943.90.002830.6860.765226 3.30.014480.8530.4644018.60.025550.6821.052213 4.90.013750.659 0.5487>10.00.01419 1.433 0.6410467.80.070860.835 0.47215 5.80.008280.868 0.6764449.50.023950.793 0.605583 5.20.036290.663 0.310>10.00.00060.798 0.686185 5.30.005860.592 0.5226 4.60.008520.479 0.588119180.085760.724 0.344756 6.80.024870.586 0.475524>10.00.034330.912 0.276105 3.30.00338 1.185 0.46378 3.80.026420.746 0.602487 5.90.028250.647 0.496280 6.20.034720.765 0.5991610 6.30.124690.81 0.402326 5.90.013060.799 0.52978550.057240.716 0.526409>10.00.020090.672 0.3015487.20.038990.75 0.64937 1.90.000820.528362 5.80.018750.583 0.495103 3.30.005940.555200 4.80.011950.645 0.36816487.70.106370.722 0.591164 1.10.000580.7 0.56342660.032820.763 0.928139>10.00.014250.916 0.574183 5.70.0090.576 0.26453 3.80.026470.7 0.59358360.04060.66 0.51817349.50.098910.574 0.33328870.023130.591 0.29421100.028010.852 0.14854>10.00.004550.917 0.533757.10.00310.519 0.237152 5.20.014030.848 0.3339 4.70.000610.947 0.684358 6.80.01790.68 0.205611 2.60.01090.6360.62216 4.70.011730.744 0.56760 5.20.00460.935 0.5051516 5.10.073520.5710.402102 4.70.008450.7241.286112 1.70.002350.689 0.491962.10.002940.478 0.311617.50.00190.52 0.2641407.50.007770.652 0.623151 5.60.009150.618 0.30427650.011510.675 0.5575198.10.032360.622 0.531271 6.80.01050.542 0.3713267.50.020820.69 0.1503.90.001480.557 0.6211774.90.008970.574 0.37777 1.40.000560.519 0.325756 2.80.015320.517 0.658584>10.00.024370.447 0.4051265.80.006880.602 0.6922089.50.008880.572 0.451010>10.00.078030.694 0.401404>10.00.017360.539 0.51319590.007720.464 0.7552050 3.80.097330.507 0.376165 3.30.006880.645 0.66712>10.00.00110.509 0.4782497.40.017690.585 0.3366606.60.036250.587 0.5871098.60.00970.678 0.3982017.80.010420.532 0.974265 5.60.011680.86 0.575120 3.70.005470.581 0.3184918.50.029120.67 0.328274>10.00.010820.469 0.26411090.003750.484 0.4863257.70.015030.544 0.237257 5.10.005680.505 0.3831811 5.30.077130.549 0.29290 6.70.012010.475 0.2563950.001450.383 0.448163 5.60.008040.74 0.4131267.20.004260.402 0.4064617.20.019290.3870.46126 5.50.006040.602 0.2781807.30.008730.496 0.15645>10.00.001770.415 0.2139480.004840.536 0.29100 5.90.003030.562 0.511319>10.00.034280.619 0.318371 5.40.010620.649 0.2145 3.70.005160.354 0.275204 6.70.007280.481 0.454476 4.80.011950.353 0.422161 4.80.006940.423 0.466146 6.60.004980.398。
基于GEO_和TCGA_数据库对肺腺癌差异表达基因的生物信息学分析

第 49 卷第 6 期2023年 11 月吉林大学学报(医学版)Journal of Jilin University(Medicine Edition)Vol.49 No.6Nov.2023DOI:10.13481/j.1671‑587X.20230612基于GEO和TCGA数据库对肺腺癌差异表达基因的生物信息学分析叶汇, 孙哲, 周丽婷, 齐雯, 叶琳(吉林大学公共卫生学院劳动卫生与环境卫生教研室,吉林长春130021)[摘要]目的目的:采用生物信息学方法筛选影响肺腺癌(LUAD)的关键基因,分析其生物学功能及其对LUAD预后的影响。
方法方法:于高通量基因表达(GEO)数据库下载GSE118370和GSE136043芯片数据,癌症基因组图谱(TCGA)数据库筛选LUAD相关数据。
采用R软件分析共同表达的差异表达基因(DEGs)。
采用clusterProfile R包对DEGs进行基因本体(GO)功能富集分析,DAVID数据库进行京都基因与基因组百科全书(KEGG)通路富集分析,STRING数据库构建蛋白-蛋白相互作用(PPI)网络。
采用Cytoscape筛选连接度排名前10位的关键基因,GEPIA数据库和人类蛋白质图谱(HPA)数据库分析正常肺组织和LUAD组织中关键基因mRNA和蛋白表达情况及不同分期LUAD组织中关键基因表达情况。
关键基因免疫浸润分析和生存分析获取关键基因表达与患者生存期的相关关系。
结果:共筛选DEGs 428个。
GO分析,LUAD的DEGs在主要富集于上皮-间质转化(EMT)等生物过程(BP)方面、细胞基部等细胞组分(CC)方面和细胞外基质(ECM)结构形成等分子功能(MF)方面。
KEGG分析,LUAD的DEGs主要富集于细胞因子受体相互作用通路等方面。
筛选DNA拓扑异构酶Ⅱα(TOP2A)、果蝇纺锤体异常基因(ASPM)、细胞周期蛋白B1(CCNB1)、人类细胞分裂周期相关基因8(CDCA8)、含杆状病毒IAP重复序列蛋白5(BIRC5)、苏氨酸激酶(AURKA)、驱动蛋白超家族成员20A(KIF20A)、中心体相关蛋白55(CEP55)、着丝粒蛋白F(CENPF)和微管组织因子(TPX2)为关键基因。
一种细胞间通信的新的信号分子——Microvesicle运输的microRNA

一种细胞间通信的新的信号分子——Microvesicle运输的microRNA梁宏伟;陈熹;曾科;张辰宇【摘要】Microvesicle(MV)是机体内的细胞在正常和病理状态下都会分泌的直径在30~1000 nm之间的微小囊泡.细胞把特异的生物活性分子如蛋白质、mRNA 等包裹到MV中,这些生物活性分子会通过MV被运输到相应的受体细胞以调节受体细胞的生物功能.这种由MV介导的细胞间信息传递在一些生理和病理过程中扮演着十分重要的作用.近期研究表明MV中包含microRNA(miRNA),并且通过MV 的运输,这些miRNA会被运送入靶细胞以调控靶细胞相关基因的表达.本文就MV 运输的miRNA在细胞间通信的功能做一总结.【期刊名称】《分子诊断与治疗杂志》【年(卷),期】2010(002)006【总页数】7页(P417-423)【关键词】Microvesicle;MicroRNA;细胞间通信;外泌体【作者】梁宏伟;陈熹;曾科;张辰宇【作者单位】南京大学医药生物技术国家重点实验室,南京,江苏210093;南京大学医药生物技术国家重点实验室,南京,江苏210093;南京大学医药生物技术国家重点实验室,南京,江苏210093;南京大学医药生物技术国家重点实验室,南京,江苏210093【正文语种】中文【中图分类】R3Microvesicle(MV)是机体内细胞在正常和病理状态下都会分泌的直径在30~1000 nm之间的微小囊泡[1,2]。
多数研究者用MV来指代具有细胞间信号传递功能的囊胞[3,4]。
体内和体外的实验都证明MV可以由网织红细胞[5]、B细胞[6]、T细胞[7]、树突细胞[8]、肥大细胞[9]、上皮细胞[10,11]和肿瘤细胞[12,13]等多种细胞分泌。
细胞把特异的生物活性分子如蛋白质、mRNA等包裹到MV中,被运输到相应的受体细胞并调节受体细胞的生物功能。
这种由MV介导的细胞间信息传递在一些生理和病理过程中扮演着十分重要的作用[14~17]。
IgA_肾病患者肾组织、血清、尿液C4d水平与病情和疗效的关系

IgA 肾病患者肾组织、血清、尿液C 4d 水平与病情和疗效的关系王真硕1,李艳存2,李巧英31 北京积水潭医院检验科,北京100035;2 北京市大兴区中西医结合医院肾内科;3 北京市大兴区中西医结合医院急诊内科摘要:目的 探讨肾组织、血清、尿液中人补体片段4d (C 4d )水平对IgA 肾病(IgAN )患者病情、疗效及预后的评估价值。
方法 选取原发性IgAN 患者214例,采用经皮肾穿刺活检术取肾组织,免疫组化法检测C 4d 表达情况;采集患者血清和尿液,ELISA 法检测C 4d 水平。
比较肾组织、血清及尿液C 4d 不同表达水平患者的临床病理资料。
给予ACEI /ARB 药物联合免疫抑制剂治疗6~8个月后观察疗效,采用Spearman 相关分析法分析肾组织、血清、尿液C 4d 表达与治疗效果的相关性。
于随访截止日对患者进行肾功能CDK 分期,比较不同分期患者肾组织、血清、尿液C 4d 表达水平差异。
结果 肾组织C 4d 表达阳性76例、阴性138例,血清C 4d 水平为(1 270.27 ± 332.99)ng /mL ,尿液C 4d 水平为(297.53 ± 45.53)ng /mL 。
与肾组织C 4d 阴性患者比较,肾组织C 4d 阳性患者24 h 尿蛋白定量更高,血白蛋白更低(P 均<0.05);与血清C 4d 低水平患者比较,血清C 4d 高水平患者24 h 尿蛋白定量更高,血白蛋白更低(P 均<0.05);与尿液C 4d 低水平患者比较,尿液C 4d 高水平患者MAP 、24 h 尿蛋白定量、Scr 更高,eGFR 、血白蛋白更低(P 均<0.05),且病理分期更为严重。
治疗后214例患者完全缓解143例,部分缓解28例,无反应(无效+终末期肾病)43例。
相关性分析显示,肾组织、血清、尿液C 4d 水平升高与IgAN 患者的治疗效果呈负相关(r s 分别为-0.234、-0.270、-0.324,P 均<0.01),其中尿液C 4d 与治疗效果的相关性最强。
SCI_Chem 影响因子

Abbreviated JournalTitle(linked to journal information)Total CitesImpact Factor5-YearImpactFactor Immediacy Index综合1Nature 0028-0836********.59738.1599.2432Science0036-807550848931.02733.587 6.6913P Natl Acad Sci Usa 0027-84245349519.73710.583 1.893化学综合1Chem Rev 0009-266511259641.29845.79514.3352Nat Mater 1476-11224634835.74942.3768.4113Nat Nanotechnol 1748-33872192031.1736.011 5.8764Chem Soc Rev 0306-00124764624.89230.1817.9975Prog Mater Sci 0079-6425592123.19422.3337.2176Nat Chem 1755-4330865221.75723.02 5.5327Accounts Chem Res 0001-48424211220.83324.633 5.2958Nano Today 1748-0132294417.68918.1920.7849Annu Rev Mater Res 1531-7331525916.17914.4950.66710Surf Sci Rep 0167-5729411515.33322.2817.7511Adv Mater 0935-96489195214.82913.86 2.55712Mat Sci Eng R 0927-796X 485013.90218.9740.66713Angew Chem Int Edit 1433-785122989413.73413.56 2.95914Annu Rev Phys Chem 0066-426X 700213.36518.121 4.13815Nano Lett 1530-69848843113.02514.132 2.47116Acs Nano 1936-0851*******.06212.524 1.9417Energ Environ Sci 1754-56921284911.65312.462 3.08718Coordin Chem Rev 0010-85452560111.01612.257 2.29419J Am Chem Soc 0002-786343128610.67710.237 2.16420Nat Prod Rep 0265-0568660910.17810.072 2.38521Adv Energy Mater 1614-6832199510.04310.05 2.11822Adv Funct Mater 1616-301X 347589.76510.342 1.81523Med Res Rev 0198-632532979.5839.978 1.81124Npg Asia Mater 1884-40493019.0428.556 2.41425Annu Rev Anal Chem 1936-132711498.612.2830.69626Top Curr Chem 0340-102255078.456 6.205 4.27327Chem Sci 2041-652048458.3148.33 2.77128Chem Mater 0897-4756746518.2387.627 1.12329J Photoch Photobio C 1389-556720268.06911.9520.7530Small 1613-6810181377.8238.084 1.42931Prog Photovoltaics 1062-799545357.7127.023 3.22332J Control Release 0168-3659297557.6338.078 1.13633Annu Rev Chem Biomol 1947-54383317.5127.512 1.04534Int Mater Rev 0950-660822557.487.1491.188RankISSNJCR Data35Chemsuschem1864-563150567.4757.951 1.189 36Prog Solid State Ch0079-678614747.429 3.3380 37Nano Res1998-012430317.3927.8010.979 38Prog Surf Sci0079-681619047.1369.140.273 39Adv Carbohyd Chem Bi0065-23188337.133 5.8460.75 40Green Chem1463-926215554 6.828 6.992 1.269 41Adv Organomet Chem0065-3055841 6.758.9410 42Curr Opin Colloid In1359-02944670 6.6297.0360.884 43J Phys Chem Lett1948-71858575 6.585 6.651 1.301 44Top Organometal Chem1436-60021152 6.384 1.762 45Chem Commun1359-7345122728 6.378 6.226 1.53 46Catal Rev0161-49402529 6.37510.1750.889 47Trac-Trend Anal Chem0165-99367327 6.351 6.7610.92 48Nanoscale2040-33647835 6.233 6.262 1.167 49Adv Colloid Interfac0001-86867115 6.1698.01 1.32 50Org Lett1523-706073440 6.142 5.563 1.572 51Mater Today1369-70213769 6.0718.677 1.977 52Prog Nucl Mag Res Sp0079-65651993 6.022 6.065 1.389 53Crit Rev Solid State1040-8436744 5.9477.3681 54Carbon0008-622332742 5.868 6.35 1.197 55Chem-Eur J0947-653960788 5.831 5.623 1.241 56Appl Catal B-Environ0926-337323011 5.825 6.0310.965 57J Catal0021-951734516 5.787 6.249 1.025 58Wires Comput Mol Sci1759-0876570 5.738 5.738 3.518 59Lab Chip1473-019716485 5.697 6.136 1.256 60Anal Chem0003-270096794 5.695 5.7690.948 61J Med Chem0022-262359227 5.614 5.383 1.225 62Adv Synth Catal1615-415015502 5.535 5.323 1.109 63Curr Opin Solid St M1359-02862508 5.4387.3290.913 64Biosens Bioelectron0956-566322068 5.437 5.389 1.105 65J Chem Theory Comput1549-961811067 5.389 5.936 1.067 66Biomacromolecules1525-779724209 5.371 5.750.721 67Acs Catal2155-54351461 5.265 5.265 1.222 68Adv Catal0360-05641399 5.25 5.2860 69Chemcatchem1867-38802718 5.181 5.3070.941 70Mrs Bull0883-******** 5.024 5.590.569 71Acs Appl Mater Inter1944-82448635 5.008 5.040.683 72Electroanal Chem0070-977837757.50.25 73J Comb Chem1520-47662925 4.933 3.10274Int Rev Phys Chem0144-235X1524 4.92 5.595 2.231 75J Phys Chem C1932-744778595 4.814 5.1520.738 76Pharm Res-Dordr0724-874119035 4.742 5.0460.735 77Cryst Growth Des1528-748322310 4.689 4.8730.869 78Sol Energ Mat Sol C0927-024818447 4.63 5.205 1.215 79J Chromatogr A0021-967363419 4.612 4.5820.71480Inorg Chem0020-166985446 4.593 4.5510.956 81Bioconjugate Chem1043-180213900 4.58 4.7960.768 82Chem-Asian J1861-47286084 4.572 4.488 1.04683J Org Chem0022-326396723 4.564 4.135 1.101 84Anal Chim Acta0003-267039448 4.387 4.3440.684 85Chem Rec1527-89991375 4.377 4.814 1.533 86Int J Plasticity0749-64195276 4.356 4.70.862 87J Chem Inf Model1549-959611250 4.304 4.0670.795 88Langmuir0743-7463106920 4.187 4.4160.793 89Organometallics0276-733339735 4.145 3.653 1.004 90Ultraschall Med0172-46141247 4.116 2.723 1.286 91J Flow Chem2062-249X61 4.091 4.091 1.143 92Curr Med Chem0929-867312773 4.07 4.4710.627 93Struct Bond0081-59931899 4.068 4.24894Mar Drugs1660-33971871 3.978 3.9110.428 95Analyst0003-265416152 3.969 3.9040.785 96Acta Mater1359-645434860 3.941 4.3950.781 97Soft Matter1744-683X15943 3.909 4.35 1.013 98Crystengcomm1466-803312988 3.879 4.0690.863 99Acs Chem Neurosci1948-7193634 3.871 3.9570.729 100Nanotechnology0957-448434133 3.842 3.8380.697 101Org Electron1566-11995856 3.836 4.0210.63 102J Comput Chem0192-865124682 3.835 4.4010.847 103Phys Chem Chem Phys1463-907640969 3.829 3.976 1.052 104Faraday Discuss1359-66405758 3.821 4.148 2.369 105Dalton T1477-922638660 3.806 3.8890.947 106Catal Sci Technol2044-47531079 3.753 3.753 1.024 107Sci Technol Adv Mat1468-69962352 3.752 3.6440.235 108Chembiochem1439-42279616 3.74 3.670.78 109Curr Top Med Chem1568-02664850 3.702 3.8850.655 110Chem Res Toxicol0893-228X10785 3.667 4.0130.807 111Anal Bioanal Chem1618-264221971 3.659 3.7560.727 112Acs Comb Sci2156-8952379 3.636 3.6360.596 113Corros Sci0010-938X18210 3.615 4.0160.757 114J Phys Chem B1520-6106119722 3.607 3.7020.66 115J Am Soc Mass Spectr1044-03058760 3.592 3.5030.925 116J Cheminformatics1758-2946315 3.59 3.6710.314 117Org Biomol Chem1477-052018355 3.568 3.490.952 118Ultrasound Obst Gyn0960-76928490 3.557 3.7080.756 119Colloid Surface B0927-776510312 3.554 3.4170.707 120Int J Hydrogen Energ0360-319933119 3.548 4.0860.584 121Sensor Actuat B-Chem0925-400529909 3.535 3.6680.527 122Dyes Pigments0143-72087664 3.532 3.4330.897 123Expert Opin Ther Pat1354-37761742 3.525 2.5230.6 124Ultrason Sonochem1350-41775008 3.516 3.708 1.234125J Phys Chem Ref Data0047-26894996 3.5 3.7790.391 126Eur J Med Chem0223-523414818 3.499 3.8490.541 127Talanta0039-914026966 3.498 3.7330.531 128Food Hydrocolloid0268-005X6307 3.494 3.5250.953 129Drug Des Dev Ther1177-8881377 3.4860.256 130Carbohyd Polym0144-861718471 3.479 3.9420.665 131Microchim Acta0026-36724743 3.434 2.8830.5 132Appl Catal A-Gen0926-860X27726 3.41 3.910.506 133J Mech Phys Solids0022-509610460 3.406 3.7460.784 134Pure Appl Chem0033-454513333 3.386 3.1150.382 135Micropor Mesopor Mat1387-181115134 3.365 3.4140.869 136J Biol Inorg Chem0949-82574011 3.353 3.3550.713 137Chemphyschem1439-423511279 3.349 3.3880.768 138Eur J Org Chem1434-193X19346 3.344 3.1370.746 139Food Chem0308-814641375 3.334 4.0720.589 140Acs Med Chem Lett1948-58751087 3.311 3.3180.699 141Future Med Chem1756-89191081 3.31 3.2450.933 142J Nat Prod0163-386419898 3.285 3.2670.787 143Electrophoresis0173-083516985 3.261 2.8690.479 144Bioanalysis1757-61801318 3.253 3.0440.723 145J Mass Spectrom1076-51745573 3.214 3.2270.495 146J Inorg Biochem0162-013410014 3.197 3.430.672 147J Mol Catal A-Chem1381-116917999 3.187 3.3190.496 148J Colloid Interf Sci0021-979744929 3.172 3.390.747 149J Anal Atom Spectrom0267-94777362 3.155 2.9530.725 150Sep Purif Rev1542-2119222 3.154 3.5430.3 151J Pharm Sci-Us0022-354917986 3.13 3.3850.6 152Eur J Inorg Chem1434-194816310 3.12 2.9510.728 153Cement Concrete Res0008-884613854 3.112 3.7460.441 154Curr Org Chem1385-27283953 3.039 3.2220.253 155Isr J Chem0021-21481263 3.025 1.9460.478 156Mar Chem0304-420368723 3.3150.466 157Catal Today0920-586123325 2.98 3.4640.515 158Phytomedicine0944-71135244 2.972 3.2580.401 159New J Chem1144-054610014 2.966 2.920.698 160J Pharmaceut Biomed0731-708514648 2.947 2.8530.562 161Photoch Photobio Sci1474-905X4541 2.923 2.810.538 162Catal Commun1566-73679190 2.915 3.3280.535 163Mater Design0261-30699587 2.913 2.8050.703 164J Agr Food Chem0021-856176046 2.906 3.2880.417 165Bioorgan Med Chem0968-089624911 2.903 3.1510.617 166Crit Rev Anal Chem1040-8347979 2.892 3.690.591 167Microchem J0026-265X3428 2.879 2.85 1.048 168Mini-Rev Med Chem1389-55752893 2.865 2.9210.496 169Mol Divers1381-19911289 2.861 3.090.44170Chemmedchem1860-71793723 2.835 3.0750.788 171J Mol Catal B-Enzym1381-11774943 2.823 2.8050.542 172Scripta Mater1359-646219677 2.821 3.1450.534 173Adv Phys Org Chem0065-3160391 2.818 2.609174Electroanal1040-039710646 2.817 2.8620.462 175Fuel Process Technol0378-******** 2.816 3.4930.49 176Tetrahedron0040-402052981 2.803 2.8990.636 177Beilstein J Org Chem1860-53971405 2.801 2.4750.386 178J Phys Chem A1089-563955641 2.771 2.8560.658 179J Ethnopharmacol0378-874121278 2.755 3.3220.519 180Org Process Res Dev1083-61604311 2.739 2.6670.808 181J Supercrit Fluid0896-84466044 2.732 3.1380.472 182Medchemcomm2040-2503690 2.722 2.7220.603 183Adv Inorg Chem0898-******** 2.714 3.3780.25 184J Electroanal Chem1572-665721687 2.672 2.6760.545 185Int J Photoenergy1110-662X945 2.663 2.240.671 186Synlett0936-521417034 2.655 2.4520.604 187Environ Chem1448-25171390 2.652 2.701 1.075 188Opt Mater Express2159-3930593 2.616 2.6220.815 189Anti-Cancer Agent Me1871-52061386 2.610.239 190Top Catal1022-55284896 2.608 2.7080.238 191J Sep Sci1615-93068094 2.591 2.6380.296 192Anal Biochem0003-269739746 2.582 2.9690.558 193Rsc Adv2046-20691816 2.562 2.5670.695 194J Anal Appl Pyrol0165-23705102 2.56 3.0740.447 195Nanoscale Res Lett1931-75733998 2.524 2.7830.2980951-419813285 2.509 2.6110.49 196Rapid Commun Mass Sp196Sci Adv Mater1947-2935553 2.509 2.5610.301 198React Funct Polym1381-51484515 2.505 2.6530.282 199J Chem Technol Biot0268-25756796 2.504 2.4790.573 200Synthesis-Stuttgart0039-788117999 2.5 2.3840.615 201Microsc Microanal1431-92762043 2.495 3.0770.276 202J Chromatogr B1570-023220776 2.487 2.90.319 203Phytochem Analysis0958-03441895 2.48 2.280.292 204Adv Heterocycl Chem0065-2725888 2.478 2.6170.455 205Chem Biol Drug Des1747-02771940 2.469 2.4090.511 206Int J Mol Sci1422-00674706 2.464 2.7320.313 207Ultrasound Med Biol0301-56297839 2.455 2.8440.251 208Gold Bull0017-15571073 2.434 3.1220.04 209Molecules1420-30497552 2.428 2.6790.329 210Plasmonics1557-1955877 2.425 2.9880.525 211J Photoch Photobio A1010-603013061 2.416 2.6910.299 212Tetrahedron Lett0040-403973763 2.397 2.3760.561 213J Alloy Compd0925-838839264 2.39 2.1610.629 214Phys Status Solidi-R1862-62541437 2.388 2.4320.527215Fluid Phase Equilibr0378-******** 2.379 2.3380.544 216Solvent Extr Ion Exc0736-******** 2.375 2.6090.345 217Beilstein J Nanotech2190-4286309 2.374 2.3740.573 218Plant Food Hum Nutr0921-96681569 2.358 2.7620.25 219Acta Pharmacol Sin1671-40835577 2.354 2.5210.596 220Planta Med0032-094311009 2.348 2.4620.304 221Appl Clay Sci0169-13175590 2.342 2.7980.337 222Bioorg Med Chem Lett0960-894X33460 2.338 2.4270.584 222Macromol Mater Eng1438-74922983 2.338 2.3920.509 222Mol Inform1868-1743344 2.338 2.3460.347 225Russ Chem Rev+0036-021X3092 2.299 2.8130.204 226J Chem Thermodyn0021-96145905 2.297 2.2560.735 227Constr Build Mater0950-06187337 2.293 2.8180.391 228Chemometr Intell Lab0169-74394880 2.291 2.4320.253 229Biophys Chem0301-46224822 2.283 2.0940.649 230Arab J Chem1878-5352299 2.2660.343 231J Ginseng Res1226-8453349 2.2590.34 232Materials1996-19441176 2.247 2.3380.222 233Catal Lett1011-372X9373 2.244 2.2610.351 234Theor Chem Acc1432-881X5484 2.233 3.1510.812 235Fitoterapia0367-326X4706 2.231 2.1390.349 236Mater Lett0167-577X23419 2.224 2.3220.489 237Food Addit Contam A1944-00495142 2.22 2.4420.508 238J Biomol Screen1087-05712357 2.207 2.0890.719 239Platin Met Rev0032-1400735 2.194 2.4760.579 240J Nanopart Res1388-07645724 2.175 2.7210.222 241J Mater Sci0022-246131538 2.163 2.10.543 242Adv Quantum Chem0065-3276869 2.161 1.6940.308 242Colloid Polym Sci0303-402X5657 2.161 2.1140.433 244Chem Phys Lett0009-261455163 2.145 2.150.485 244J Ind Eng Chem1226-086X2264 2.145 1.9550.329 246Tetrahedron-Asymmetr0957-416611707 2.115 2.1430.384 247Appl Surf Sci0169-433231193 2.112 2.0990.33 248Synthetic Met0379-677913916 2.109 2.1020.334 249Colloid Surface A0927-775718414 2.108 2.3330.333 249Mat Sci Eng A-Struct0921-509340513 2.108 2.3490.307 251J Anal Toxicol0146-47602660 2.107 1.7580.429 252Solid State Nucl Mag0926-20401202 2.1 2.0570.711 253J Food Compos Anal0889-15753737 2.088 2.7430.19 254J Environ Monitor1464-03254378 2.085 2.1370.322 255Mater Chem Phys0254-058417174 2.072 2.3950.286 256J Pept Sci1075-26171942 2.071 1.8280.434 257Phytother Res0951-418X8059 2.068 2.4380.444 258Nano-Micro Lett2150-5551205 2.057 1.910.35 259Solid State Ionics0167-273820728 2.046 2.5640.26260Carbohyd Res0008-621514176 2.044 2.1780.357 261J Solid State Chem0022-459618166 2.04 2.2950.392 262Curr Org Synth1570-1794673 2.038 2.9140.76 263Ultrasonics0041-624X3651 2.028 2.0540.456 264Smart Mater Struct0964-17267120 2.024 2.3770.289 265Inorg Chem Commun1387-70036416 2.016 1.8810.434 266Appl Organomet Chem0268-26052866 2.011 1.9220.272 267Electrochem Solid St1099-00628883 2.01 2.0260.596 268J Chem Eng Data0021-956815169 2.004 2.1150.295 269Comb Chem High T Scr1386-207314532 1.9750.268 269J Organomet Chem0022-328X217932 1.9920.527 271Thermochim Acta0040-603111408 1.989 2.0460.327 272J Mol Model1610-29403157 1.984 2.3010.378 273J Therm Anal Calorim1388-61509934 1.982 1.7420.243 274Int J Fatigue0142-11235248 1.976 1.9740.352 275J Vib Control1077-54631649 1.966 1.7360.672 276Chem Phys0301-010412935 1.957 2.0590.592 277J Mater Process Tech0924-013618426 1.953 2.1760.332 277Sensors-Basel1424-82207082 1.953 2.3950.321 279Biomed Chromatogr0269-38792861 1.945 1.8150.385 280J Fluorine Chem0022-11394998 1.939 1.9490.465ArticlesCited Half-life Eigenfactor ®Metrics ScoreArticleInfluenc e® Score8699.6 1.5750820.8448329.7 1.3598717.71238018 1.55663 4.8921768.20.2266114.294141 5.20.2278819.481121 3.70.1543615.607390 3.50.178418.8542370.015018.643126 2.40.054018.927207 6.50.108177.90837 3.20.01489 5.62318>10.00.013647.55389.40.008828.821867 5.10.27819 4.24368.40.00872 6.542227 5.50.53637 3.497299.90.01888.1211078 4.40.37491 5.1891191 2.40.20333 4.012473 1.80.05534 3.461368.20.03818 3.07130997.70.83183 2.99465 5.90.01757 3.141169 1.40.00944 3.344569 4.20.12336 3.006377.20.00647 2.73229 1.90.00155 3.27323 3.90.00716 4.441557.70.01045 2.068458 1.40.02121 2.833576 6.70.15215 2.032167.90.00342 3.148457 3.60.07856 2.503112 4.60.01202 1.97501 6.90.05008 1.9062220.00197 2.73916>10.00.003282.792Eigenfactor®Metrics286 2.60.0208 2.0046>10.00.00142 1.13896 2.80.01517 2.301119.20.00397 3.974>10.00.00086 1.894 44940.03848 1.5233>10.00.00076 2.537437.60.01013 2.301 63220.04884 2.393 2150.003483173 4.80.28954 1.5629>10.00.00211 3.009 138 5.80.01695 1.794 1015 1.70.02966 1.597 508.20.01226 2.37 160850.18182 1.40244 5.40.01448 3.213188.30.00431 2.295 107.10.00156 2.34 674 6.40.06437 1.6 1916 4.10.1766 1.469 480 5.40.05059 1.419 284>10.00.04014 1.60356 1.20.00194 1.703 617 3.70.05475 1.603 14797.80.17236 1.533 8907.80.09182 1.35 404 4.40.04405 1.352 239.20.00403 2.705 448 4.10.05929 1.252 507 3.50.04948 1.865 480 5.30.05919 1.416 315 1.30.00585 1.6362>10.00.00057 1.543 255 2.10.01222 1.495 116 6.20.01779 2.376 953 2.30.03563 1.2844>10.00.00016 2.2160 5.10.006940.708137.90.00319 2.303 3283 3.40.34957 1.353 2798.70.02859 1.299 773 3.90.057280.943 493 5.30.04212 1.304 11447.10.090460.92115617.50.124780.98 259 5.70.03346 1.295 372 2.80.02555 1.219 1289>10.00.12482 1.03 7277.10.06585 1.03830 5.80.00343 1.39794 6.80.01439 1.685 303 6.60.019140.892 2119 6.70.21487 1.164 9987.10.059110.76263 3.40.002370.436140.000190.962 475 5.50.03101 1.1898.60.00225 1.225 194 2.50.00680.908 817 6.60.031350.985 68170.09849 1.714 1358 2.50.073 1.334 1194 2.60.032550.749 10720.00309 1.26 1021 4.40.12987 1.194 459 3.20.02416 1.192 2618.90.03446 1.396 1804 3.70.15067 1.296 1418.60.01298 1.609 1709 4.50.08330.842 327 1.30.00320.94181 4.90.00904 1.23 327 4.70.03579 1.258 17750.0131 1.044 2597.10.02119 1.078 877 4.30.068460.99799 1.40.001410.915 449 6.30.028910.786 16157.50.18356 1.081 22860.020170.96535 1.90.00176 1.224 1160 3.80.055440.914 201 6.30.017610.949 426 4.10.025740.772 2120 3.70.074070.708 1016 5.80.054290.757 321 5.50.013530.63690 3.30.006210.726 184 4.80.010070.73123>10.00.00228 1.513 610 3.80.032170.72 842 5.40.057730.829 213 5.40.01270.77439 2.40.001731002 4.80.033670.729 208 4.30.00930.537 5247.60.043570.93 116>10.00.02104 1.812 173>10.00.013330.936 589 5.30.032210.807 108 6.10.009190.931 487 4.70.03951 1.142 76450.047770.779 1666 5.20.091160.854 193 1.80.004620.921 120 2.20.004670.894 3338.70.027140.756 4327.10.027040.625 19520.003610.533 194 6.90.011710.909 2597.30.013360.716 266 6.80.026850.752 87080.070140.869 2187.70.01310.75610 5.60.00062 1.012 4189.20.026590.815 622 5.20.034970.666 1709.90.01712 1.222 178 5.80.009120.836 907.70.00320.65258>10.00.01221 1.371 4877.30.041850.896 187 5.60.009820.662 338 6.50.019280.711 443 6.30.026840.651 20850.01330.806 409 4.50.027060.735 774 3.20.034350.762 15287.90.107180.745 739 5.40.053960.715 227.60.001180.774 105 5.10.007480.647 129 4.90.00780.74475 3.50.002990.639212 3.70.013920.784 201 5.60.009290.599 500 6.30.06138 1.238 >10.00.00042 1.049 290 6.10.019640.625 251 5.60.017330.895 12498.40.075460.68 241 2.20.005560.628 1341 6.70.120620.829 651 6.90.026690.572 224 5.10.010210.672 290 5.30.013220.702 194 1.60.002660.7278>10.00.000620.792 314>10.00.019270.649 32830.001390.381 520 6.30.034550.62153 5.20.004350.904 200 1.30.002460.856 109 3.70.00563143 5.80.013670.825 439 4.20.025470.66 441>10.00.03210.853 17290.80.002020.494 1527.40.009010.741 624 2.40.016810.714 337 6.40.02850.71 173 1.90.002010.614 124 6.30.008010.593 2207.60.010810.602 4708.20.028730.596 221 5.20.00853 1.29 4957.50.036490.714 967.20.003150.52311>10.00.000450.543 184 3.50.006730.588 1064 2.80.017560.666 2437.60.014140.744 257.10.002380.868 105130.022770.574 10130.003550.834 2518.20.017620.63 16669.90.081330.508 1478 4.10.10840.548 14830.008560.8753427.80.015630.587 559.80.002470.6496 1.50.001250.668648.10.002270.561 178 6.20.010850.6 247>10.00.011750.526 202 5.20.013550.701 1484 5.40.069550.546 116 6.40.005740.60572 2.10.00140.58954>10.00.004010.856 351 6.60.011110.57 960 3.90.021660.681 1549.50.006150.597 5790.008180.646 6720.0008353 2.30.00061176 2.60.005320.633 1947.80.016490.569 223 5.60.014960.968 25280.006360.457 1626 5.50.049760.537 1977.10.008730.585 128 5.40.00580.57919>10.00.001080.734 650 3.70.017210.673 9419.10.050830.59113>10.00.00130.598 21090.007690.475 906>10.00.066080.687 350 3.90.005590.396 2247.80.01640.479 1789 5.50.074220.539 419>10.00.014550.491 579 6.80.034280.585 11717.30.084950.73 918.60.003920.471 458.60.00220.655 100 6.20.007370.666 335 5.10.012850.644 964 5.50.038290.59199 5.20.004810.478 302 6.60.012460.48640 2.10.000610.345 392>10.00.021350.719339>10.00.01560.489 5138.90.025150.615 50 4.50.001620.659 1368.50.005580.639 353 5.90.018730.713 495 4.80.011170.321 1147.60.003960.391 156 6.70.022520.706 508 6.50.027410.476 82 5.10.003380.472 410>10.00.020760.421 400>10.00.012770.531 463 4.70.008250.53 728 5.70.012880.253 2367.10.012090.669 180 3.80.004490.45 360>10.00.018270.679 2957.80.035210.653 950 3.40.026580.586 218 5.20.006840.446 2547.50.007820.487。
抗体公司

赛信通(上海)生物试剂有限公司
上海市浦东南路1101号远东大厦514室,200120 info@cst www.cst 2158356288 公司总部: 美国
Established in Beverly, MA in 1999, Cell Signaling Technology (CST) is a privatelyowned company with over 400 employees worldwide. We are dedicated to providing innovative research tools that are used to help define mechanisms underlying cell function and disease. Since its inception, CST has become the world leader in the production of the highest quality activationstate and total protein antibodies utilized to expand knowledge of cell signaling pathways. Our mission is to deliver the world's highest quality research tools that accelerate progress in biological research and personalized medicine. 总引用数为4670,来自于1966篇文章。最常引用的试剂包括: Akt, ERK2, ERK1, p38, Akt1。
AbD Serotec (BioRad)
高效液相色谱-串联质谱法测定贻贝中腹泻性贝类毒素的含量

p r r a c i i crm t rp ytne asset me y ( P C MS ) S ef hsmpew set c d b 0 e om n el ud ho a gah - dm m s pc o t f q o a r r H L - / . hl s a l a x at y8 % MS l i r e
f. hj n i e e uly et g et , aghu hj n 3 0 1,h a 1Z ei g s r s at T sn n e H nzo , ea ,10 2 i ; a F hi Q i i C r Z i C n
2 C l g Fo c n e n i eh o g nier gZ ea g ogh n nvrt, a ghuZ e a, 10 5C i ) . oee f odSi c dBo c nl y g e n, hj n nsag ie i H nzo hj n 30 3 ,hn l o e a t o E n i i G U sy i a
Li a u g r o m a e Li ui nn e s by Hi h Pe f r nc q d Chr m a o r p y Ta e M a s o t g a h nd m s
Sp c r m e r e to ty
Zh n i q He Xi 。 Zh n o g y n a g Ha - i , n , e g Ch n - i g
8 74251 种贝类毒素在2 ~ 0 g 范 围内线性 良好 ;在4 1. 5 .。2 / 080 / L 个添加水平下O A的 回收率为7 . ~8 . , S 95 % 86 R D为 % 84 %~1.%; T 一 的回收率为8 .%~9 .%, S 为42 % ~ . %。 .3 04 D X 1 3 8 1 2 R D .2 65 4 方法灵 敏度高 , 定量 限为00 m /g 来 自市 . g 。 2 k 场 和产地的4 个贻贝样 品残 留分析发现 , 个样 品检 出腹泻性贝类毒 素, 5 有4 检出率为8 %。 . 9
211132698_猪δ冠状病毒辅助蛋白研究进展

·综述·Chinese Journal of Animal Infectious Diseases中国动物传染病学报摘 要:猪δ冠状病毒(PDCoV )是一种新发现的猪肠道冠状病毒,可引起新生仔猪的腹泻、呕吐、脱水、死亡,而且具有跨种感染潜能,给养猪业和公共卫生构成了巨大威胁。
冠状病毒的辅助蛋白(accessory protein )是一类功能特殊的蛋白,不同属甚至同属不同种的冠状病毒编码的辅助蛋白数量、同源性均存在较大差异,具有属或种特异性。
虽然辅助蛋白是冠状病毒增殖非必需的,但在调控宿主天然免疫、病毒复制与致病性中发挥重要作用。
目前,已证实PDCoV 至少编码3个辅助蛋白,而且近年来在PDCoV 辅助蛋白的功能研究方面取得了显著进展。
本文简要介绍了PDCoV 辅助蛋白的鉴定及其在免疫调控和致病性中的作用,并对今后的研究进行了展望。
关键词:猪δ冠状病毒;辅助蛋白;感染;免疫抑制;致病性中图分类号: S852.651文献标志码: A文章编号:1674-6422(2023)01-0213-09Research Progress on A ccessory Proteins of Porcine DeltacoronavirusZHANG Huichang 1,2, FANG Puxian 1,2, FANG Liurong 1,2, XIAO Shaobo 1,2(1. State Key Laboratory of Agricultural Microbiology, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan 430070,China; 2. The Cooperative Innovation Center for Sustainable Pig Production, Wuhan 430070, China)收稿日期:2020-09-03基金项目:国家自然科学基金重点项目(31730095)作者简介:张蕙畅,女,硕士研究生,预防兽医学专业通信作者:肖少波,E-mail:*************猪δ冠状病毒辅助蛋白研究进展张蕙畅1,2,方谱县1,2,方六荣1,2,肖少波1,2(1.华中农业大学动物医学院 农业微生物学国家重点实验室,武汉430070;2.湖北省生猪健康养殖协同创新中心,武汉430070)2023,31(1):213-221Abstract: Porcine deltacoronavirus (PDCoV) is a newly discovered Porcine intestinal coronavirus, which causes diarrhea, vomit, dehydration and death in newborn piglets. Importantly, PDCoV has the potential of cross-species infection, posing a huge threat to public health. Accessory proteins encoded by Coronavirus are a unique set of proteins with special functions. These accessory proteins encoded by different genera, or even by the same genus and different species of Coronavirus vary in number and homology, making them genus or species specifi c. Though accessory proteins are dispensable for replication of Coronavirus in vitro , they play important roles in regulating innate immunity, viral proliferation and pathogenicity. PDCoV has been confi rmed to encode three accessory proteins, and remarkable progress has been achieved on their functions during the past few years. In this review, we briefl y introduced the research progress on PDCoV accessory proteins, including their identifi cation, functions on regulating antiviral innate immune and pathogenicity. We also discussed the future research on PDCoV accessory proteins.Key words: Porcine deltacoronavirus; accessory protein; infection; immunosuppression; pathogenicity· 214 ·中国动物传染病学报2023年2月猪δ冠状病毒(Porcine deltacoronavirus,PDCoV)是一种有囊膜、不分节段的单股正链RN A 病毒,属于套式病毒目冠状病毒科δ冠状病毒属成员,病毒粒子呈球形,直径约为120~180 nm,表面分布有放射状的纤突[1]。
广藿香DXS_基因克隆及表达分析

㊀山东农业科学㊀2024ꎬ56(4):28~35ShandongAgriculturalSciences㊀DOI:10.14083/j.issn.1001-4942.2024.04.004收稿日期:2023-11-08基金项目:广东省中医药局科研项目(20241177)ꎻ广东省重点领域研发计划项目(2020B020221002)作者简介:曾晴(2000 )ꎬ女ꎬ湖南益阳人ꎬ硕士研究生ꎬ研究方向为中药资源开发与品质评价ꎮE-mail:2719041453@qq.com通信作者:张宏意(1977 )ꎬ女ꎬ浙江舟山人ꎬ博士ꎬ副教授ꎬ主要从事药用植物资源与育种研究ꎮE-mail:drizzlezhy@163.com广藿香DXS基因克隆及表达分析曾晴ꎬ严雅玲ꎬ严寒静ꎬ何梦玲ꎬ张宏意(广东药科大学中药学院ꎬ广东广州㊀510006)㊀㊀摘要:1-脱氧-D-木酮糖-5-磷酸合酶(DXS)是调控萜类合成途径中MEP途径的第一个关键酶ꎬ为探究广藿香DXS基因参与其主要萜类成分广藿香醇合成调控的分子机制ꎬ本研究依据本课题组前期获得的转录组DXS基因序列ꎬ以广藿香cDNA为模板ꎬ克隆得到PcDXS基因ꎬ对其进行生物信息学分析ꎬ构建pET-28a-PcDXS原核表达载体诱导蛋白表达ꎬ并采用荧光实时定量PCR法检测广藿香根㊁茎㊁叶㊁芽中DXS基因表达情况ꎮ结果表明ꎬ从广藿香叶中克隆到开放阅读框全长为2151bp和1908bp的PcDXS1㊁PcDXS2基因序列ꎬ分别编码716㊁635个氨基酸ꎮ预测PcDXS1蛋白分子量78.33kDaꎬ为稳定非跨膜非分泌蛋白ꎬ主要定位于叶绿体ꎻPcDXS2蛋白分子量67.92kDaꎬ为不稳定非跨膜非分泌蛋白ꎬ主要定位于叶绿体ꎮPcDXS1㊁PcDXS2均含有DXP_synthase_N㊁Transket_pyr和Transketolase_C结构域模块ꎬ属于DXS超家族ꎮPcDXS1㊁PcDXS2分别与半枝莲㊁糙苏的DXS基因序列具有较高同源性ꎮ使用异丙基-β-D-硫代半乳糖苷(IPTG)诱导的PcDXS1㊁PcDXS2融合蛋白主要在沉淀中表达ꎮPcDXS1㊁PcDXS2基因表达量均在根中最低ꎬPcDXS1基因在叶中显著高表达ꎬPcDXS2基因在叶㊁芽㊁茎中高表达ꎮ本研究结果可为后续深入研究DXS基因在广藿香萜类化合物合成途径中的生物学功能及广藿香萜类代谢途径的基因调控奠定基础ꎮ关键词:广藿香ꎻ1-脱氧-D-木酮糖-5-磷酸合酶(DXS)ꎻ基因克隆ꎻ生物信息学分析ꎻ表达分析中图分类号:S567.2:Q781㊀㊀文献标识号:A㊀㊀文章编号:1001-4942(2024)04-0028-08CloningandExpressionAnalysisofDXSGenefromPogostemoncablin(Blanco)Benth.ZengQingꎬYanYalingꎬYanHanjingꎬHeMenglingꎬZhangHongyi(SchoolofTraditionalChineseMedicineꎬGuangdongPharmaceuticalUniversityꎬGuangzhou510006ꎬChina)Abstract㊀1 ̄Deoxy ̄D ̄xylulose ̄5 ̄phosphatesynthase(DXS)isthefirstkeyenzymetoregulatetheMEPpathwayintheterpenesynthesispathway.InordertoillustratethemolecularmechanismofDXSgenepartici ̄patinginsynthesisandregulationofthemainterpenoidcomponentꎬpatchoulialcoholꎬinpatchouli(Pogoste ̄moncablin)ꎬtheresearchwasconductedbasedonthetranscriptomeDXSgenesequenceobtainedpreviouslybyourresearchgroup.PcDXSgenewasclonedbyusingpatchoulicDNAastemplateꎬandthenanalyzedbybioinformaticsmethod.TheprokaryoticexpressionvectorpET ̄28a ̄PcDXSwasconstructedtoinduceproteinexpressionꎬandreal ̄timefluorescencequantitativePCRwasusedtodetecttheexpressionofDXSgeneinrootsꎬstemsꎬleavesandbudsofpatchouli.TheresultsshowedthatPcDXS1andPcDXS2genesequenceswithopenreadingframelengthof2151bpand1908bpwereclonedfromleavesofpatchouliꎬencoding716and635aminoacidsꎬrespectively.PcDXS1proteinwaspredictedtohave78.33kDaofmolecularweightꎬandwasastableꎬnon ̄transmembraneandnon ̄secretedproteinprimarilylocalizedinchloroplasts.PcDXS2proteinhad67.92kDaofmolecularweightꎬwhichwasanunstableꎬnon ̄transmembraneandnon ̄secretedproteinmainlylocalizedinchloroplasts.PcDXS1andPcDXS2containedDXP̠synthase̠NꎬTransket̠pyrandTransketolase̠CdomainmodulesrespectivelyꎬbelongingtoDXSsuperfamily.PcDXS1andPcDXS2hadhighhomologyingenesequencewithDXSgenesofScutellariabarbataandPhlomisumbrosa.PcDXS1andPcDXS2fusionproteinsinducedbyisopropyl ̄β ̄D ̄thiogalactoside(IPTG)weremainlyexpressedinprecipitation.Theexpressionlev ̄elsofPcDXS1andPcDXS2geneswerethelowestinroots.PcDXS1genewassignificantlyhigherexpressedinleavesꎬwhilePcDXS2genewasrelativelyhigherinleavesꎬbudsandstemscomparedtoroots.TheresultscouldlayfoundationsforfurtherstudiesonbiologicalfunctionsofDXSgeneinterpenoidcompoundsynthesispathwayandgeneregulationmechanismofterpenoidmetabolismpathwayinpatchouli.Keywords㊀Pogostemoncablinꎻ1 ̄Deoxy ̄D ̄xylose ̄5 ̄phosphatesynthase(DXS)ꎻGenecloningꎻBioin ̄formaticsanalysisꎻExpressionanalysis㊀㊀广藿香[Pogostemoncablin(Blanco)Benth.]是唇形科刺蕊草属草本植物ꎬ原产于东南亚ꎬ引种至我国成为岭南道地药材ꎬ是一种具有重要药用价值和广泛工业生产应用价值的经济作物ꎬ具有芳香化浊㊁和中解暑的功效[1]ꎬ是藿香正气丸㊁二妙丸㊁午时茶颗粒等中成药的主要原料ꎮ已检测到广藿香地上部分含有174种化学成分ꎬ其中萜类最多ꎬ达66种ꎬ另有黄酮㊁类固醇㊁吡喃酮㊁多糖等多种生物活性成分[2]ꎬ因而具有抗菌㊁抗炎㊁抗诱变㊁抗细胞凋亡㊁抗HepG19癌细胞增殖等多种药理活性[3-4]ꎮ广藿香提取物广藿香油全球年产可达2000吨[5]ꎬ常被用于各类芳香疗法以舒缓压力㊁减轻疲劳㊁改善失眠㊁缓解焦虑等ꎬ还因气味宜人被广泛应用于香水㊁肥皂㊁化妆品等化工领域[6]ꎻ另外ꎬ在畜禽领域ꎬ广藿香提取物作为饲料添加剂也展现出巨大潜力[7]ꎮ广藿香醇是广藿香挥发油的首要成分ꎬ其生物合成途径有两条ꎬ分别是定位于细胞质的甲羟戊酸(mevalonateꎬMVA)途径和定位于叶绿体的甲基赤藓糖-4-磷酸(methylerythritol ̄4 ̄phosphateꎬMEP)途径[8-9]ꎮMVA途径主要负责倍半萜㊁三萜和橄榄苦苷甾体的合成ꎬ而广藿香醇为三环倍半萜类化合物ꎬ因此在关于广藿香醇的分子调控和生物合成中ꎬ针对MVA途径关键基因的作用已经进行了较多研究[10]ꎮMEP途径除合成单萜㊁二萜㊁四萜还合成植物激素㊁光合色素[11-12]ꎮMVA和MEP途径均可产生戊烯基焦磷酸(IPP)及其异构体二甲基烯丙基焦磷酸(DMAPP)ꎬ这两种物质是合成萜类化合物的共同前体物质ꎮ1-脱氧-D-木酮糖-5-磷酸合酶(DXS)是MEP途径的第一个酶ꎬ催化丙酮酸和3-磷酸甘油醛生成1-脱氧木酮糖-5-磷酸(DXP)[12]ꎮDXS家族可分为3个不同的亚家族 DXS1㊁DXS2和DXS3ꎬ这3种类型的DXS蛋白在保守基序组成㊁二级结构和三级结构方面具有高度相似性[13]ꎮDXS不仅在植物萜类生物合成过程中发挥着重要调控作用ꎬ还在许多生理过程中起着重要作用ꎬ如光合作用㊁植物激素调节㊁逆境抗性和病原体防御等ꎮ过表达马尾松PmDXS可以显著提高其类胡萝卜素㊁叶绿素a㊁叶绿素b含量和DXS活性[14]ꎬ瞬时转化过表达天竺葵的GrDXS基因能增加次生代谢物天竺葵精油中单萜和倍半萜化合物含量[15]ꎬ提示DXS基因在萜类生物合成途径中具有重要调控作用ꎮ迄今ꎬDXS基因已经在银杏[16]㊁乌头[17]㊁穿心莲[18]㊁桔梗[19]㊁荆芥[20]等多种植物中完成克隆和初步研究ꎬ而从广藿香中探究DXS基因的内容鲜见报道ꎮ本研究基于课题组前期获得的转录组数据克隆了广藿香DXS基因ꎬ并对其进行生物信息学分析及在广藿香不同部位的表达分析ꎬ以期为今后深入研究该基因在萜类化合物代谢中的功能奠定基础ꎮ1㊀材料与方法1.1㊀试验材料本研究所用植物材料经广东药科大学中药学院严寒静教授鉴定为唇形科刺蕊草属植物广藿香[Pogostemoncablin(Blanco)Benth.]ꎬ培育于广东药科大学中药学院ꎮ采集生长6个月的广藿香叶片液氮速冻ꎬ于-80ħ超低温冰箱保存ꎬ用于RNA提取与基因克隆ꎮ选取生长6个月状态良好的广藿香根㊁茎(第2㊁3对叶之间)㊁叶(第2对叶)㊁芽ꎬ经液氮速冻后置于-80ħ冰箱ꎬ用于RNA提取及组织特异性表达qRT-PCR分析ꎮDH5α感受态细胞㊁BL21(DE3)表达菌株㊁pET28a原核表达载体均购自北京庄盟国际生物92㊀第4期㊀㊀㊀㊀㊀㊀㊀㊀㊀曾晴ꎬ等:广藿香DXS基因克隆及表达分析基因科技有限公司ꎮ1.2㊀试验方法1.2.1㊀RNA提取与cDNA链合成㊀将广藿香叶片样品经液氮研磨后ꎬ采用天根植物总RNA提取试剂盒提取总RNAꎬ使用超微量紫外分光光度计UV2450测定其浓度和纯度ꎬ用1%琼脂糖凝胶电泳检测RNA完整性ꎮ使用TaKaRa的PrimeScriptRTreagentKitwithgDNAEraser反转录试剂盒合成cDNA第一链ꎮ1.2.2㊀基因克隆㊀根据课题组前期获得的转录组数据得到广藿香PcDXS基因的CDS序列ꎬ设计特异性引物(表1)ꎬ以反转录得到的cDNA为模板进行PCR扩增ꎮPCR反应体系:cDNA1μLꎬ上游引物PcDXS-F1μLꎬ下游引物PcDXS-R1μLꎬPrimeSTAR MaxDNAPolymerase25μL㊁ddH2O补足至50μLꎮPCR反应程序:98ħ10sꎬ55ħ5sꎬ72ħ12sꎬ35个循环ꎻ72ħ2minꎮPCR产物经1%琼脂糖凝胶电泳鉴定后切胶回收ꎬ并检测浓度及纯度ꎬ使用庄盟ZTOPO-Blunt/TA快速克隆试剂盒进行TA克隆ꎻ转化大肠杆菌DH5α感受态细胞ꎬ经菌液PCR鉴定ꎬ挑取阳性克隆ꎬ送广州擎科生物科技股份有限公司测序ꎮ表1㊀引物序列引物名称用途引物序列(5ᶄ-3ᶄ)PcDXS1-F基因克隆ATGGCTTCTGTTTCTTGCCAGAACCPcDXS1-RTCAGAGCATCAATTGAAGAGCGTCGPcDXS2-FATGAAAAACCTCACTGCTAAGGPcDXS2-RTTAGGACATTATCTCTAGGGCCpET-28a-PcDXS1-F构建原核表达载体GACAGCAAATGGGTCGCGGAATGGCTTCT ̄GTTTCTTGCCAGAACCpET-28a-PcDXS1-RCGACGGAGCTCGAATTCGGAGAGCAT ̄CAATTGAAGAGCGTCGCGpET-28a-PcDXS2-FGACAGCAAATGGGTCGCGGAATGAAAAAC ̄CTCACTGCTAAGGAACTGAAACApET-28a-PcDXS2-RCGACGGAGCTCGAATTCGGAGGACAT ̄TATCTCTAGGGCCTCCCTAG18S-FqRT-PCRTCAACCATAAACGATGCCGACC18S-RTTTCAGCCTTGCGACCATACTCCq-PcDXS1-FCGAGACATTTCCCTCCTTGCq-PcDXS1-RGACCACCACAACTTCTTCATCGq-PcDXS2-FGTGATTATGATTGCTTCGGTGCTq-PcDXS2-RATGGCTTCGTATGCTTGACCTG1.2.3㊀生物信息学分析㊀依照对应的生物信息学在线工具(表2)对广藿香的PcDXS1㊁PcDXS2进行序列分析ꎮ从NCBI上进行Blast比对ꎬ获得PcDXS同源序列ꎬ利用MEGA.11邻接法构建进化树ꎬBootstrap值设置为1000次重复ꎮ表2㊀在线工具、用途及网址在线工具用途网址ExPASyProtParam蛋白质理化性质分析https://web.expasy.org/protparam/ExPASyProtScale疏水性分析https://web.expasy.org/protscale/Plant-mPLoc亚细胞定位预测http://www.csbio.sjtu.edu.cn/bioinf/plant ̄multi/DeepTMHMM跨膜结构预测https://dtu.biolib.com/DeepTM ̄HMMSignalP-6.0信号肽预测https://services.healthtech.dtu.dk/services/SignalP ̄6.0/NCBICDSearch保守结构域分析https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgiSOPMA二级结构预测https://npsa ̄prabi.ibcp.fr/cgi ̄bin/npsa_automat.pl?page=npsa%20_sopma.htmlSWISS-MODEL三级结构预测https://swissmodel.expasy.org/1.2.4㊀原核表达㊀设计PcDXS基因与包含pET-28a载体酶切位点的同源臂引物(表1)ꎬ构建pET-28a-PcDXS原核表达载体ꎮPcDXS基因引入pET-28a载体同源臂反应体系为:TA-PcDXS重组质粒1μLꎬ上游引物pET-28a-PcDXS-F1μLꎬ下游引物pET-28a-PcDXS-R1μLꎬPrimeSTAR MaxDNAPolymerase25μLꎬddH2O补足至50μLꎮPCR反应程序:98ħ10sꎬ55ħ5sꎬ72ħ12sꎬ35个循环ꎻ72ħ2minꎮ采用全式金BamHⅠ酶将pET-28a载体进行单酶切线性化ꎮPCR产物及酶切产物经1%琼脂糖凝胶电泳鉴定后切胶回收ꎬ将产物根据庄盟SE无缝克隆和组装试剂盒连接转化DH5α感受态细胞ꎬ菌液经PCR鉴定ꎬ挑取阳性克隆ꎬ送广州擎科生物科技股份有限公司测序ꎮ将pET-28a-PcDXS质粒转化蛋白表达菌株BL21(DE3)ꎬ经PCR鉴定ꎬ挑取阳性菌液摇菌后ꎬ在5mL菌液中添加异丙基-β-D-硫代半乳糖苷(IPTG)至浓度分别为0.25㊁0.5㊁0.75㊁1.0mmol L-1ꎬ160r min-1摇床㊁16ħ培养12hꎬ使用细胞破碎仪(AMP25%ꎬ开5sꎬ关5s)破碎2minꎬ离心收集上清液和沉淀ꎬ进行SDS-PAGE凝胶电泳检测PcDXS蛋白表达情况ꎮ1.2.5㊀荧光定量PCR表达分析㊀以转录合成的cDNA为模板ꎬ18S为内参基因ꎮ根据PcDXS基因的测序结果ꎬ使用Premier5.0设计qRT-PCR引物(表1)ꎮ实时荧光定量PCR采用20μL反应体系:2ˑTBGreenPremixExTaqⅡ10μLꎬcD ̄NA1.5μLꎬ上㊁下游引物各0.4μLꎬddH2O7.7μLꎮ扩增程序:95ħꎬ30sꎻ95ħ5sꎬ60ħ30sꎬ03山东农业科学㊀㊀㊀㊀㊀㊀㊀㊀㊀㊀㊀㊀㊀㊀第56卷㊀40个循环ꎮ每个部位进行3次生物学重复ꎬ每个样品3个技术重复ꎮ采用2-әәCt法计算PcDXS基因在广藿香不同组织部位根㊁茎㊁叶㊁芽中的相对表达量ꎮ采用GraphPadPrism9.5软件进行单向方差多重比较分析数据ꎬ利用MicrosoftExcel作图并用字母标记法标记差异显著性ꎮ2㊀结果与分析2.1㊀PcDXS1㊁PcDXS2基因克隆提取广藿香总RNAꎬ琼脂糖凝胶电泳显示有清晰条带ꎬ经超微量紫外分光光度计UV2450测定浓度和纯度合格ꎬ可用于后续实验ꎮ通过使用基因特异性引物ꎬ成功克隆获得两条序列长度为2151bp和1908bp的序列(图1)ꎬ测序结果与转录组序列相似度分别高达99.95%和100%ꎬ分别编码716个和635个氨基酸ꎬ命名为PcDXS1和PcDXS2ꎮM:DL2000DNAMarkerꎻ1:PcDXS1基因PCR扩增产物ꎻ2:PcDXS2基因PCR扩增产物ꎮ图1㊀PcDXS1㊁PcDXS2基因克隆2.2㊀PcDXS1㊁PcDXS2蛋白理化性质分析经ProtParam预测分析ꎬPcDXS1蛋白分子式为C3473H5496N840O1038S39ꎬ为亲水且稳定的弱酸性蛋白ꎻPcDXS2蛋白分子式为C3009H4790N840O899S25ꎬ为亲水不稳定的弱酸性蛋白(表3)ꎬProtScale分析也表明PcDXS1㊁PcDXS2均为亲水性蛋白(图2)ꎮ两蛋白均定位在叶绿体ꎮ表3㊀PcDXS1㊁PcDXS1蛋白理化性质蛋白氨基酸数分子量/kDa正电残基个数负电残基个数等电点亲水性总平均值(GRAVY)脂肪系数不稳定系数跨膜区域信号肽PcDXS171678.3384665.78-0.02892.3539.35无无PcDXS263567.9271656.36-0.04491.4542.84无无正值表示疏水ꎻ负值表示亲水ꎮ图2㊀PcDXS1(A)㊁PcDXS2(B)蛋白亲疏水性预测2.3㊀PcDXS1㊁PcDXS2保守结构域及二级和三级结构分析PcDXS1㊁PcDXS2蛋白保守结构域分析结果显示ꎬ两者都属于DXS超家族ꎬ含有从N端到C端的DXP_sythase_N㊁Transket_pyr和Transketo ̄lase_C结构域模块(图3)ꎮPcDXS1蛋白二级结构中无规则卷曲占比最高ꎬ为39.80%ꎬ还含有37.85%的α-螺旋㊁15.92%的折叠延伸链㊁6.42%的β-折叠ꎻPcDXS2蛋白中占比最高的是α-螺旋ꎬ为41.89%ꎬ还含有15.59%的折叠延伸链㊁8.03%的β-折叠㊁34.49%的无规则卷曲(图4)ꎮ通过SWISSMODEL构建了两蛋白的三级结构ꎬ对PcDXS1蛋白ꎬ以叶绿体DXPS为模板ꎬ序列一致性55.78%ꎬGMQE得分为0.64(0~1范围内数值越大表明预期质量越高)ꎬGMEAN得分为13㊀第4期㊀㊀㊀㊀㊀㊀㊀㊀㊀曾晴ꎬ等:广藿香DXS基因克隆及表达分析0.67(分数趋近于0反映 类似原生 的结构ꎬ低于-4.0的 QMEAN 分数表示模型质量较低)ꎬ可视为获得了良好的PcDXS1蛋白三级结构模型ꎮ对PcDXS2蛋白ꎬ以番茄DXS1为模板ꎬ二者的相似度高达91.02%ꎬGMQE得分为0.90ꎬ该模型可较好地预测PcDXS2蛋白的三级结构(图5)ꎮ图3㊀PcDXS1㊁PcDXS2蛋白保守结构域分析结果2.4㊀PcDXS1㊁PcDXS2基因系统进化分析使用MEGA11构建PcDXS1㊁PcDXS2与其他物种DXS基因的系统发育树ꎬ结果(图6)显示ꎬPcDXS1与半枝莲DXS(MK035040.1)聚为一支ꎬPcDXS2与糙苏DXS(KU317508.1)聚为一支ꎬ亲缘关系较近ꎮPcDXS1和PcDXS2之间存在系统发育距离ꎬ可能存在功能差异ꎮ蓝色表示α-螺旋ꎻ红色表示β-折叠ꎻ绿色表示β-转角ꎻ紫色表示无规则卷曲ꎮ图4㊀PcDXS1(A)㊁PcDXS2(B)蛋白的二级结构图5㊀PcDXS1(A)㊁PcDXS2(B)蛋白的三级结构Camelliasinensis:山茶花ꎻActinidiachinensis:猕猴桃ꎻPrunellavulgaris:夏枯草ꎻGardeniajasminoides:栀子ꎻPinusmassoniana:马尾松ꎻThujaplicata:北美乔柏ꎻAquilariasinensis:土沉香ꎻPhlomisumbrosa:糙苏ꎻTaraxacumkok ̄saghyz:橡胶草ꎻWithaniasomnifera:南非醉茄ꎻScutellariabarbata:半枝莲ꎻLyciumruthenicum:枸杞ꎻPlectranthusbarbatus:左手香ꎮ图6㊀PcDXS1㊁PcDXS2基因系统发育进化树23山东农业科学㊀㊀㊀㊀㊀㊀㊀㊀㊀㊀㊀㊀㊀㊀第56卷㊀2.5㊀PcDXS1㊁PcDXS2原核表达分析将PcDXS1㊁PcDXS2基因引入同源臂与pET-28a线性化载体连接ꎬ测序分析结果同克隆序列一致ꎬ表明成功构建pET-28a-PcDXS1㊁pET-28a-PcDXS2原核表达载体ꎮPcDXS1㊁PcDXS2蛋白分子量分别为78.33kDa和67.92kDaꎬ加上His标签后蛋白大小分别约为84.6kDa和76.1kDaꎮ将重组质粒转化BL21(DE3)菌株ꎬ挑取阳性菌在16ħ条件下分别用0.25㊁0.50㊁0.75㊁1.0mmol L-1IPTG诱导表达12hꎬ经细胞破碎仪破碎㊁离心㊁SDS-PAGE凝胶电泳检测ꎬ结果(图7)显示ꎬ在沉淀中分别有84.6kDa和76.1kDa的蛋白条带ꎬ说明PcDXS1㊁PcDXS2重组蛋白被成功诱导表达ꎬPcDXS2重组蛋白表达较少ꎬ主要在沉淀中表达ꎬIPTG浓度变化对蛋白表达无明显影响ꎮM:BluePlus®IIProteinMarker(14~120kDa)ꎻ1㊁3㊁5㊁7㊁9:分别为0㊁0.25㊁0.50㊁0.75㊁1.00mmol L-1IPTG诱导重组蛋白表达后的破碎菌体上清ꎻ2㊁4㊁6㊁8㊁10:分别为0㊁0.25㊁0.50㊁0.75㊁1.0mmol L-1IPTG诱导重组蛋白表达后的破碎菌体沉淀ꎮ图7㊀PcDXS1㊁PcDXS2原核表达分析的SDS-PAGE检测结果2.6㊀PcDXS1㊁PcDXS2表达的组织特异性分析结果(图8)显示ꎬPcDXS1㊁PcDXS2在广藿香根㊁茎㊁叶㊁芽中均有表达ꎮ其中ꎬPcDXS1在叶中表达量最高ꎬ在芽和茎中表达量也较高ꎬ均显著高于在根中的表达量ꎻPcDXS2在茎㊁叶㊁芽中的表达量相当ꎬ且均显著高于在根中的表达量ꎮ柱上不同小写字母表示组织间差异显著(P<0.05)ꎮ图8㊀PcDXS1㊁PcDXS2在广藿香不同部位中的表达差异3㊀讨论目前已从多种植物中挖掘出DXS基因ꎬ并对其在萜类化合物生物合成过程中的功能进行了研究ꎬ但DXS基因对广藿香萜类代谢调控的影响尚未见研究报道ꎮDXS基因家族具有较高的保守性[21]ꎬ多含有DXP_sythase_N㊁Transket_pyr和Transketolase_C结构域模块[22]ꎮ本研究从广藿香中克隆出两个DXS基因(PcDXS1㊁PcDXS2)ꎬ均包含该3个保守结构域模块ꎬ符合DXS基因家族结构特征ꎮDXS亚家族中ꎬDXS1被认为是管家基因[23]ꎬDXS1蛋白主要在叶绿体中表达ꎬ是合成光合色素和植物激素所必需的[24]ꎬ在植物叶片和茎的叶绿体以及果实的有色质体中大量表达[12]ꎻDXS2蛋白位于非光合作用质体中ꎬ参与一些防御反应特异的萜类化合物的合成ꎬ优先在植物根的成熟体中表达[12]ꎻDXS3蛋白的表达与参与胚胎后发33㊀第4期㊀㊀㊀㊀㊀㊀㊀㊀㊀曾晴ꎬ等:广藿香DXS基因克隆及表达分析育和繁殖的基因相关[25]ꎮ本研究结果显示ꎬPcDXS1㊁PcDXS2在广藿香根㊁茎㊁叶㊁芽中均有表达ꎬ且主要在地上部表达ꎬ在根中的表达量较低ꎬ尤其PcDXS1在叶中显著高表达ꎬ符合DXS1在光合组织中活跃的特征ꎻ亚细胞定位预测显示主要定位在叶绿体ꎬ说明PcDXS1和PcDXS2可能在叶绿体中发挥作用ꎮ原核表达是常见的表达外源蛋白的分子生物技术ꎬ大肠杆菌(Escherichiacoli)具有生长周期短㊁易于培养及遗传操纵的特点ꎬ是产生重组蛋白的重要宿主ꎬ优化重组蛋白的生物合成至关重要ꎬ常通过改变菌株的培养参数增加蛋白的表达和溶解度ꎬ用于生产重组蛋白[26-27]ꎮ对毛果杨DXS蛋白进行诱导表达ꎬ在沉淀物中检测到PtDXS靶蛋白[28]ꎮ陈文意等[29]考察了20㊁30ħ下ꎬ0.25mmol L-1和0.5mmol L-1IPTG诱导的广藿香Pc ̄JAR1融合蛋白表达ꎬ发现20ħ㊁0.25mmol L-1IPTG诱导下表达菌诱导蛋白在沉淀中更高表达ꎬ说明温度和IPTG浓度影响蛋白表达ꎮ本研究利用0.25㊁0.5㊁0.75㊁1.0mmol L-1IPTG诱导PcDXS1㊁PcDXS2重组蛋白在BL21(DE3)中表达ꎬ结果表明两蛋白主要在沉淀中表达ꎬ表达量与IPTG浓度无明显关联ꎬPcDXS2重组蛋白在同样条件下表达量较少还可能与诱导温度和时间有关ꎮ研究发现ꎬDXS基因的表达水平一般与MEP途径直接调控的萜类含量呈正相关ꎬ过表达薰衣草DXS基因显著促进薰衣草精油中单萜化合物的积累[30]ꎻ二萜类丹参酮是丹参的主要生物活性成分ꎬ丹参SmDXS1和SmDXS2的过表达显著增强其根系中丹参酮的积累[31]ꎮ同时ꎬDXS基因也调控MVA途径萜类化合物合成ꎬ其表达水平会影响MVA途径萜类含量水平的高低ꎬ如沉默甘草DXS基因促进了甘草毛状根中三萜类物质甘草酸的合成ꎬ而过表达DXS基因则导致甘草酸含量下降[32]ꎻ使用DXS基因抑制剂氯马松(CLO)处理檀香幼苗ꎬ倍半萜檀香醇的含量没有降低[33]ꎮ4㊀结论本研究从广藿香中克隆得到PcDXS1和PcDXS2基因ꎬ其全长分别为2151bp和1908bpꎬ分别编码716个和635个氨基酸ꎬ均定位于叶绿体ꎬ均为亲水性㊁非跨膜㊁非分泌蛋白ꎻPcDXS1蛋白为稳定蛋白ꎬPcDXS2蛋白为不稳定蛋白ꎬ均具有DXS基因家族典型特征ꎮ原核表达分析发现两蛋白均受IPTG诱导表达ꎬ且主要在沉淀中表达ꎬ表达水平在不同IPTG浓度间无明显差异ꎮ两基因在广藿香根㊁茎㊁叶㊁芽中均有表达ꎬ但在根中的表达量均较低ꎬPcDXS1主要在叶中表达ꎬPcDXS2在茎㊁叶㊁芽中的表达量均较高且相当ꎮ这为深入研究DXS基因在调控广藿香萜类化合物合成中的作用奠定基础ꎮ为此ꎬ本课题组已构建DXS基因的过表达载体和沉默载体ꎬ期望通过进一步研究揭示其表达与广藿香有效成分积累之间的关系ꎮ参㊀考㊀文㊀献:[1]㊀国家药典委员会.中华人民共和国药典[M].北京:中国医药科技出版社ꎬ2020:46.[2]㊀XuFFꎬCaiWꎬMaTꎬetal.Traditionalusesꎬphytochemistryꎬpharmacologyꎬqualitycontrolꎬindustrialapplicationꎬpharma ̄cokineticsandnetworkpharmacologyofpogostemoncablin:acomprehensivereview[J].TheAmericanJournalofChineseMedicineꎬ2022ꎬ50(3):691-721.[3]㊀PengXJꎬAngSꎬZhangYZꎬetal.ChemicalconstituentswithantiproliferativeactivityfromPogostemoncablin(Blanco)Benth[J].FrontiersinChemistryꎬ2022ꎬ10:938851. [4]㊀FatimaSꎬFarzeenIꎬAshrafAꎬetal.Acomprehensivereviewonpharmacologicalactivitiesofpachypodol:abioactivecompoundofanaromaticmedicinalplantPogostemoncablinBenth.[J].Moleculesꎬ2023ꎬ28(8):3469.[5]㊀VanBeekTAꎬJoulainD.TheessentialoilofpatchouliꎬPog ̄ostemoncablin:areview[J].FlavourandFragranceJournalꎬ2018ꎬ33(1):6-51.[6]㊀SwamyKMꎬSinniahRU.Acomprehensivereviewonthephy ̄tochemicalconstituentsandpharmacologicalactivitiesofPog ̄ostemoncablinBenth.:anaromaticmedicinalplantofindustrialimportance[J].Moleculesꎬ2015ꎬ20(5):8521-8547. [7]㊀高阳ꎬ杜鑫ꎬ马雪ꎬ等.广藿香提取物在动物生产中的应用[J].黑龙江畜牧兽医ꎬ2023(21):49-53.[8]㊀ChappellJ.Thebiochemistryandmolecularbiologyofisopre ̄noidmetabolism[J].PlantPhysiologyꎬ1995ꎬ107(1):1-6. [9]㊀RohmerM.Thediscoveryofamevalonate ̄independentpathwayforisoprenoidbiosynthesisinbacteriaꎬalgaeandhigherplants[J].NaturalProductReportsꎬ1999ꎬ16(5):565-574. [10]张婵ꎬ姚广龙ꎬ张军锋ꎬ等.广藿香百秋李醇分子调控及合成生物学研究进展[J].生物技术通报ꎬ2021ꎬ37(8):55-64.[11]ArendtPꎬPollierJꎬCallewaertNꎬetal.Syntheticbiologyforproductionofnaturalandnew ̄to ̄natureterpenoidsinphotosyn ̄theticorganisms[J].ThePlantJournalꎬ2016ꎬ87(1):16-37.43山东农业科学㊀㊀㊀㊀㊀㊀㊀㊀㊀㊀㊀㊀㊀㊀第56卷㊀[12]GanjewalaDꎬKumarSꎬLuthraR.Anaccountofclonedgenesofmethyl ̄erythritol ̄4 ̄phosphatepathwayofisoprenoidbiosyn ̄thesisinplants[J].CurrentIssuesinMolecularBiologyꎬ2009ꎬ11(s1):35-45.[13]TianSKꎬWangDDꎬYangLꎬetal.Asystematicreviewof1 ̄Deoxy ̄D ̄xylulose ̄5 ̄phosphatesynthaseinterpenoidbiosynthe ̄sisinplants[J].PlantGrowthRegulationꎬ2022ꎬ96:221-235. [14]LiRꎬChenPZꎬZhuLZꎬetal.Characterizationandfunctionofthe1 ̄deoxy ̄D ̄xylose ̄5 ̄phosphatesynthase(DXS)generelat ̄edtoterpenoidsynthesisinPinusmassoniana[J].InternationalJournalofMolecularSciencesꎬ2021ꎬ22(2):848.[15]JadaunJSꎬSangwanNSꎬNarnoliyaLKꎬetal.Over ̄expressionofDXSgeneenhancesterpenoidalsecondarymetaboliteaccu ̄mulationinrose ̄scentedgeraniumandWithaniasomnifera:ac ̄tiveinvolvementofplastidisoprenogenicpathwayintheirbio ̄synthesis[J].PhysiologiaPlantarumꎬ2017ꎬ159(4):381-400. [16]GongYFꎬLiaoZHꎬGuoBHꎬetal.Molecularcloningandex ̄pressionprofileanalysisofGinkgobilobaDXSgeneencoding1 ̄deoxy ̄D ̄xylulose5 ̄phosphatesynthaseꎬthefirstcommitteden ̄zymeofthe2 ̄C ̄methyl ̄D ̄erythritol4 ̄phosphatepathway[J].PlantaMedicaꎬ2006ꎬ72(4):329-335.[17]SharmaEꎬPandeySꎬGaurAK.Identificationandexpressiona ̄nalysisofDXS1geneisolatedfromAconitumbalfouriiStapf[J].ActaPhysiologiaePlantarumꎬ2016ꎬ38:233.[18]SrinathMꎬShailajiAꎬBinduBBVꎬetal.Molecularcloninganddifferentialgeneexpressionanalysisof1 ̄Deoxy ̄D ̄xylulose5 ̄phosphatesynthase(DXS)inAndrographispaniculata(Burm.f)Nees[J].Mol.Biotechnol.ꎬ2021ꎬ63:109-124. [19]董楠ꎬ余函纹ꎬ刘梦丽ꎬ等.桔梗DXS基因的原核表达㊁亚细胞定位和酶活性分析[J].药学学报ꎬ2023ꎬ58(4):1059-1068.[20]林谷音ꎬ周佩娜ꎬ尹梦娇ꎬ等.荆芥1-脱氧-D-木酮糖-5-磷酸合成酶基因克隆及生物信息学分析[J].中草药ꎬ2021ꎬ52(2):527-537.[21]毛积鹏ꎬ黄林旺ꎬ郝静ꎬ等.植物DXS基因的系统发育和分子进化分析[J].生物学杂志ꎬ2022ꎬ39(2):23-28. [22]陈国德ꎬ饶丹丹ꎬ韩豫ꎬ等.土沉香萜类合成相关DXS和DXR基因鉴定㊁进化和表达分析[J/OL].分子植物育种ꎬ2023:1-11[2023-04-17].http://kns.cnki.net/kcms/de ̄tail/46.1068.S.20230417.1520.016.html.[23]WalterMHꎬHansJꎬStrackD.Twodistantlyrelatedgenesen ̄coding1 ̄deoxy ̄d ̄xylulose5 ̄phosphatesynthases:differentialregulationinshootsandapocarotenoid ̄accumulatingmycorrhizalroots[J].ThePlantJournalꎬ2002ꎬ31(3):243-254. [24]KimBRꎬKimSUꎬChangYJ.Differentialexpressionofthree1 ̄deoxy ̄D ̄xylulose ̄5 ̄phosphatesynthasegenesinrice[J].Bi ̄otechnologyLettersꎬ2005ꎬ27(14):997-1001.[25]Luna ̄ValdezLꎬChenge ̄EspinosaMꎬHernández ̄MuñozAꎬetal.Reassessingtheevolutionofthe1 ̄deoxy ̄D ̄xylulose5 ̄phos ̄phatesynthasefamilysuggestsapossiblenovelfunctionfortheDXSclass3proteins[J].PlantScienceꎬ2021ꎬ310:110960. [26]GopalGJꎬKumarA.Strategiesfortheproductionofrecombi ̄nantproteininEscherichiacoli[J].TheProteinJournalꎬ2013ꎬ32:419-425.[27]PouresmaeilMꎬAzizi ̄DargahlouS.Factorsinvolvedinheterolo ̄gousexpressionofproteinsinE.colihost[J].ArchivesofMi ̄crobiologyꎬ2023ꎬ205(5):212.[28]WeiHꎬMovahediAꎬXuCꎬetal.OverexpressionofPtDXSen ̄hancesstressresistanceinPoplars[J].InternationalJournalofMolecularSciencesꎬ2019ꎬ20(7):1669.[29]陈文意ꎬ邓文静ꎬ严寒静ꎬ等.广藿香JAR1基因的克隆及原核表达[J].时珍国医国药ꎬ2022ꎬ33(6):1451-1455. [30]Muñoz ̄BertomeuJꎬArrillageIꎬRosRꎬetal.Up ̄regulationof1 ̄deoxy ̄D ̄xylulose ̄5 ̄phosphatesynthaseenhancesproductionofessentialoilsintransgenicspikelavender[J].PlantPhysiolo ̄gyꎬ2006ꎬ142(3):890-900.[31]ZhouWꎬHuangFꎬLiSꎬetal.Molecularcloningandcharacter ̄izationoftwo1 ̄deoxy ̄D ̄xylulose ̄5 ̄phosphatesynthasegenesinvolvedintanshinonebiosynthesisinSalviamiltiorrhiza[J].MolecularBreedingꎬ2016ꎬ36:124.[32]杨林ꎬ汪逗逗ꎬ田少凯ꎬ等.甘草DXS基因过表达及表达沉默对甘草酸生物合成的影响研究[J].药学学报ꎬ2021ꎬ56(7):2025-2032.[33]ChenXHꎬZhangYYꎬYanHFꎬetal.Cloningandfunctionalanalysisof1 ̄deoxy ̄D ̄xylulose ̄5 ̄phosphatesynthase(DXS)inSantalumalbumL.[J].Geneꎬ2023ꎬ851(30):146762.53㊀第4期㊀㊀㊀㊀㊀㊀㊀㊀㊀曾晴ꎬ等:广藿香DXS基因克隆及表达分析。
GSK2606414_Cell Cycle_PERK_CAS号1337531-36-8说明书_AbMole中国

分子量451.44溶解性(25°C )
DMSO 90 mg/mL 分子式C H F N O Water <1 mg/mL CAS 号1337531-36-8Ethanol 19 mg/mL
储存条件
3年 -20°C 粉末状
生物活性
GSK2606414是一种可口服的、有效的选择性PERK 抑制剂,IC50为0.4 nM ,对PERK 的选择性比对其它EIF2AK 的作用至少高出100倍。
GSK2606414抑制A459细胞中的PERK 自磷酸化,IC50<0.3 μM 。
不同实验动物依据体表面积的等效剂量转换表(数据来源于FDA 指南)
小鼠
大鼠兔豚鼠仓鼠狗重量 (kg)0.020.15 1.80.40.0810体表面积 (m )0.0070.0250.150.050.020.5K 系数
3
6
12
8
5
20
动物 A (mg/kg) = 动物 B (mg/kg) ×
动物 B 的K 系数动物 A 的K 系数
例如,依据体表面积折算法,将白藜芦醇用于小鼠的剂量22.4 mg/kg 换算成大鼠的剂量,需要将22.4 mg/kg 乘以小鼠的K 系数(3),再除以大鼠的K 系数(6),得到白藜芦醇用于大鼠的等效剂量为11.2 mg/kg 。
GSK2606414 目录号M2734
化学数据
2420352m m m m m。
教你看懂基因检测中的那些变异

教你看懂基因检测中的那些变异随着基因检测技术的迅速发展和普及应⽤,越来越多的⼈开始接触到了基因检测。
报告中成堆成串的字母数字专业名词,单个看都认识,合着⼀起看就不认识了。
那么这期我们就从这个点来切⼊,教你看懂基因变异。
学会了这期,看懂报告中的变异内容就轻⽽易举了。
前⾔“突变是指核苷酸序列永久性改变,多态性是指⼈群频率超过1%的变异。
这两个术语已经错误地与致病性和良性结果关联起来,因此,建议使⽤“变异”加以下五个修饰词替代上述两个术语:致病性的、可能致病性的、意义不明确的、可能良性的或良性的。
”——ACMG指南根据HGVS(⼈类基因组变异协会)变异命名法以及ACMG指南,建议使⽤“变异”这个中性词来描述核苷酸的改变。
正确完整的变异结果描述应该包含基因名称,变异的位置,转录本及外显⼦,还有核苷酸的改变以及氨基酸改变。
01变异前缀变异的前缀⽤于指出变异位于哪种序列中:“g.”表⽰基因组序列,如g.455G>T。
“c.”表⽰Coding(编码)DNA序列,如c.455G>A。
“m.”表⽰线粒体DNA序列,如m.766T>C。
“n.”表⽰⾮编码RNA序列。
“r.”表⽰RNA序列,如r.76a>u。
“p.”表⽰蛋⽩质序列,如p.Lys76Asn。
3’规则对于突变的所有描述,最靠近参考序列3'端的描述优先考虑;应⽤于所有关于基因组,基因,转录本,蛋⽩的相关突变描述。
这句话怎么理解呢?序列从5’端向3’端读取,描述靠近3’端的变化。
例如:CTAGAGGTC这段序列变异为CTAGGTC,我们优先描述为缺失后⾯的AG,⽽不是前⾯的AG。
通俗地讲就是“能往下读就往下读,读不动了再说”。
02变异描述的总体规范1、表述符号“>”(⼤于号)表⽰碱基替换,如c.123G>A。
“del”表⽰缺失,如c.76delA。
“dup”表⽰重复,如c.76dupA。
“ins”表⽰插⼊,如c.76_77insG。
