genetic recombinant expression and characterization of human augmenter of liver regeneration
mmlv逆转录酶作用原理

mmlv逆转录酶作用原理MMLV reverse transcriptase is a key enzyme in molecular biology research, particularly in the field of genetic engineering and gene expression analysis. It plays a crucial role in the process of reverse transcription, which is the synthesis of DNA from an RNA template.MMLV reverse transcriptase works by utilizing its inherent ribonuclease H activity to degrade the RNA strand of the RNA-DNA hybrid, leaving a single-stranded DNA molecule. This single-stranded DNA molecule then serves as a template for the synthesis of the complementary DNA strand by the reverse transcriptase, resulting in the formation of a double-stranded DNA molecule.MMLV逆转录酶通过利用其固有的核糖核酸酶H活性来降解RNA-DNA杂交体的RNA链,留下单链DNA分子。
然后,这个单链DNA分子成为逆转录酶合成互补DNA链的模板,从而形成双链DNA分子。
The reverse transcription process has several important applications, including the generation of complementary DNA (cDNA) from messenger RNA (mRNA) for gene cloning and expression analysis.This process is essential for the study of gene regulation, as well as the development of recombinant DNA technology.逆转录过程有几个重要的应用,包括通过从信使RNA(mRNA)中合成互补DNA(cDNA)来进行基因克隆和表达分析。
同源重组和基因工程

同源重组(Homologus Recombination) 是指发生在染色体之间或同一染色体上含有同源序列的DNA分子之间或分子之内的重新组合,同源重组需要一系列的酶催化。
!!!是最基本的DNA重组方式,通过链的断裂和再连接,在两个DNA分子同源序列间进行单链或双链片段的交换。
Holliday中间体切开并修复,形成两个双链重组体DNA,分别为:片段重组体(patch recombinant)拼接重组体(splice recombinant)片段重组体:切开的链与原来断裂的是同一条链,重组体含有一段异源双链区,其两侧来自同一亲本DNA。
拼接重组体:切开的链并非原来断裂的链,重组体异源双链区的两侧来自不同亲本DNA。
基因克隆或重组DNA (recombinant DNA) :应用酶学的方法,在体外将各种来源的遗传物质与载体DNA接合成一具有自我复制能力的DNA分子——复制子(replicon),继而通过转化或转染宿主细胞,筛选出含有目的基因的转化子细胞,再进行扩增提取获得大量同一DNA分子。
基因工程(genetic engineering) :实现基因克隆所用的方法及相关的工作称基因工程。
工具酶功能限制性核酸内切酶识别特异序列,切割DNADNA连接酶催化DNA中相邻的5´磷酸基和3´羟基末端之间形成磷酸二酯键,使DNA切口封合或使两个DNA分子或片段连接DNA聚合酶Ⅰ①合成双链cDNA分子或片段连接②缺口平移制作高比活探针③DNA序列分析④填补3´末端Klenow片段又名DNA聚合酶I大片段,具有完整DNA聚合酶I的5'→3'聚合、3'→5'外切活性,而无5'→3'外切活性。
常用于cDNA第二链合成,双链DNA 3'末端标记等反转录酶①合成cDNA②替代DNA聚合酶I进行填补,标记或DNA序列分析多聚核苷酸激酶催化多聚核苷酸5´羟基末端磷酸化,或标记探针末端转移酶在3´羟基末端进行同质多聚物加尾碱性磷酸酶切除末端磷酸基基因载体:定义:能携带目的基因,具有自我复制能力,实现其无性繁殖或表达有意义的蛋白质的一些DNA分子。
Epigenetics and Gene Expression

Epigenetics and Gene ExpressionEpigenetics and gene expression are two interconnected fields of study that have been gaining a lot of attention in recent years. Epigenetics refers to the study of changes in gene expression that occur without any changes to the underlying DNA sequence. Gene expression, on the other hand, refers to the process by which genetic information is used to produce proteins and other molecules that are essential for life. In this response, I will explore the relationship between epigenetics and gene expression from multiple perspectives.From a scientific perspective, epigenetics and gene expression are intimately linked. Epigenetic modifications, such as DNA methylation and histone modification, can have a profound impact on gene expression. For example, DNA methylation can prevent the binding of transcription factors to DNA, which in turn can prevent the transcription of specific genes. Similarly, histone modifications can alter the structure of chromatin, making it more or less accessible to transcription factors. These modifications can be passed down from one generation to the next, and can even be influenced by environmental factors such as diet and stress.From a medical perspective, the study of epigenetics and gene expression has the potential to revolutionize our understanding of disease. Many diseases, such as cancer and Alzheimer's, are thought to be caused by changes in gene expression. By understanding the epigenetic mechanisms that regulate gene expression, we may be able to develop new treatments for these diseases. For example, drugs that target specific epigenetic modifications may be able to restore normal gene expression patterns in cancer cells, leading to their death. Similarly, drugs that target epigenetic modifications associated with Alzheimer's may be able to slow or even reverse the progression of the disease.From a social perspective, the study of epigenetics and gene expression raises important ethical questions. For example, if epigenetic modifications can be passed down from one generation to the next, what are the implications for future generations? Could environmental factors such as pollution or stress have long-lasting effects on the health of future generations? Similarly, if epigenetic modifications can be influenced by diet andlifestyle, what are the implications for public health policy? Should governments be investing in programs that promote healthy lifestyles in order to prevent epigenetic modifications that could lead to disease?From a personal perspective, the study of epigenetics and gene expression has the potential to change the way we think about ourselves and our place in the world. For example, if epigenetic modifications can be influenced by environmental factors such as diet and stress, then our health may be more within our control than we previously thought. This could be empowering for individuals who are looking to take control of their health. Similarly, if epigenetic modifications can be passed down from one generation to the next, then our actions today may have long-lasting effects on the health of future generations. This could be a sobering realization for individuals who are concerned about the impact of their actions on the world around them.From a philosophical perspective, the study of epigenetics and gene expression raises important questions about the nature of life and the role of genetics in shaping our identities. For example, if epigenetic modifications can have a profound impact on gene expression, then what does this say about the role of genetics in shaping our identities? Are we more than the sum of our genes, or are we simply the products of our genetic makeup and the epigenetic modifications that occur throughout our lives? Similarly, if epigenetic modifications can be influenced by environmental factors such as diet and stress, then what does this say about the nature of life itself? Are we simply passive recipients of environmental influences, or do we have the power to shape our own destinies?In conclusion, the study of epigenetics and gene expression is a complex and multifaceted field that has the potential to revolutionize our understanding of disease, raise important ethical questions, change the way we think about ourselves and our place in the world, and challenge our fundamental assumptions about the nature of life and the role of genetics in shaping our identities. As scientists, medical professionals, policymakers, and individuals, it is our responsibility to engage with these questions in a thoughtful and nuanced way, in order to ensure that we are making the most of this exciting and rapidly evolving field of study.。
生物化学英语

生物化学英语Introduction to BiochemistryBiochemistry is a fascinating interdisciplinary fieldthat combines biology and chemistry to study the chemical processes and molecules that occur within living organisms.It focuses on understanding the molecular mechanisms underlying biological processes and how they are regulated. In this document, we will provide an overview of important concepts and techniques in biochemistry.1. Structure and Function of BiomoleculesBiochemistry studies the structure and function of biomolecules, which include proteins, nucleic acids, carbohydrates, and lipids. Proteins are crucial for various cellular functions, such as enzyme catalysis, cell signaling, and structural support. Nucleic acids, including DNA and RNA, carry genetic information and are involved in protein synthesis. Carbohydrates are important energy sources, while lipids serve as components of cell membranes and energy storage molecules.2. Enzymes and MetabolismEnzymes are proteins that catalyze biochemical reactions, playing a vital role in metabolism. They lower the activation energy required for reactions to occur, thus speeding up the rate of chemical reactions within cells. Metabolism involves a series of interconnected biochemical reactions that convert nutrients into energy and building blocks for cellular processes. An understanding of enzyme kinetics and metabolic pathways is essential in biochemistry.3. Biochemical TechniquesVarious techniques are used in biochemistry to study biomolecules and their functions. These include spectroscopy, chromatography, electrophoresis, centrifugation, and molecular cloning. Spectroscopy allows the analysis of biomolecule structures by using light absorption, emission, or scattering. Chromatography separates mixtures into their individual components. Electrophoresis separates charged molecules based on their size and charge. Centrifugation separates particles based on their size and density. Molecular cloning allows for the replication and manipulation of DNA.4. Gene Expression and RegulationBiochemistry also encompasses the study of gene expression and regulation. Gene expression refers to the process by which information from a gene is used to produce a functional protein or RNA molecule. Regulation of gene expression ensures that the right genes are turned on or off at the appropriate times and in specific cell types. Understanding gene expression and regulation is crucial in understanding development, cell differentiation, and disease.5. Applications of BiochemistryBiochemistry has numerous applications in various fields, including medicine, agriculture, and biotechnology. In medicine, biochemistry is essential for understanding diseases at the molecular level and developing new drugs and therapies. In agriculture, biochemistry is used to improve crop yields and develop genetically modified organisms. Biotechnology relies heavily on biochemistry for geneticengineering, production of recombinant proteins, and designing new biofuels.ConclusionBiochemistry is a vast and dynamic field that plays a critical role in advancing our understanding of life processes and their applications. It provides a foundation for various other branches of biology and chemistry, contributing to fields such as molecular biology, genetics, and pharmacology. By studying the structure and function of biomolecules, enzymes, and metabolic pathways, biochemists continue to unravel the complexities of life.。
生物芯片技术

FGR
FES
ABL
INT2
PIK3CA
NMYC
AKT2
FGFR1
JUNB
AKT1
KRAS2
CDK4
AR
RDA Protocol
RNA extraction and cDNA preparation from archived tissue specimens(tester and driver) Generation of amplified cDNA fragments (‘amplicons’) Subtractive hybridization of amplicons Enrichment of cDNA fragments from differentially expressed genes
DNA Chip Technology
Solid support (glass, plastic, metal, silicon) Miniaturized array of DNA (genetic material) Work on the biochemical principle of DNA/DNA hybridization Hybridized probes (DNA molecules) are fluorescently labeled
应用之一 基因表达谱(gene expression pattern)
Research Use. Clinical Diagnostic Use.
Biological Sample
Functional Information
One Disease——One Gene Expression Pattern
Prototype AmpliOnc™ I Biochip
DNA的重组

有些细菌在自然条件下不发生转化或转化 效率很低,但在实验室中可以人工促使转化。 例如大肠杆菌,用高浓度Ca2+处理,可诱导细 胞成为感受态,重组质粒得以高效转化。
转化过程涉及细菌染色体上10多个基因编码的功能。 转化过程涉及细菌染色体上10多个基因编码的功能。如:感受态因、 10多个基因编码的功能 感受态因、 与膜结合的DNA结合蛋白、自溶素、多种核酸酶等 DNA结合蛋白 与膜结合的DNA结合蛋白、自溶素、多种核酸酶等。感受态因子诱导与 感受态有关的蛋白质表达,自溶素使与膜结合的DNA结合蛋白、 DNA结合蛋白 感受态有关的蛋白质表达,自溶素使与膜结合的DNA结合蛋白、核酸酶 裸露,当游离DNA与膜结合的DNA结合蛋白结合后, DNA与膜结合的DNA结合蛋白结合后 裸露,当游离DNA与膜结合的DNA结合蛋白结合后,核酸酶使其中一条 链降解,另一条链被吸收,与染色体DNA重组。 DNA重组 链降解,另一条链被吸收,与染色体DNA重组。
3.细菌的转导 转导(transduction)是通过噬菌体将细菌基因 从供体转移到受体细胞的过程。 转导有两种类型 普遍性转导(generalized transduction):是指宿 主基因组任意位置的DNA成为成熟噬菌体颗 DNA 粒DNA的一部分而被带入受体菌。 局限性转导(specialized transduction):某些温 和噬菌体在装配病毒颗粒时将宿主染色体整合 部位的DNA切割下来取代病毒DNA。
小鼠: J λ C λ3 小鼠:Vλ2 人: Vλ~ 300 J λ C λ6 λ 小鼠重链基因族( 号染色体 号染色体) 小鼠重链基因族(12号染色体)
细菌的结合作用
traS和traT基因编码表面排斥蛋白,阻止F+细胞之间的转移,F+细胞的性菌毛与F-细胞 基因编码表面排斥蛋白,阻止F 细胞之间的转移, 细胞的性菌毛与F
遗传育种专业英语

Plant Genetic Engineering植物基因工程Key Notes: 要点The concept: 概念Genetic manipulation involves inserting foreign genes or modifying the activity of existing genes. Methods to insert foreign genes are coupled with the methods of plant tissue culture to regenerate identical populations of plants with novel characteristics.基因操作涉及插入外源基因或者改变原有基因的活性。
插入外源基因和植物组织培养技术相结合可再生出具有新特征的纯一的植物群体。
Basic genetic manipulation methods: 基因操作的基本方法Agrobacterium tumifaciens is a soil bacterium with a plasmid that inserts foreign DNA into a plant. The plasmid contains a T-DNA transferred into the plant and a VIR region that facilitates transfer of the T-DNA. Binary vectors for genetic engineering consist of one plasmid containing the VIR region and a second containing the T-DNA including the foreign DNA. Where the Agrobacterium system cannot be used, direct gene transfer techniques may be employed, for instance using a DNA particle gun.农杆菌是一种土壤细菌,它含有一个将外源DNA插人植物体的质粒。
基因工程大肠杆菌发酵生产重组人胰岛素[宝典]
![基因工程大肠杆菌发酵生产重组人胰岛素[宝典]](https://img.taocdn.com/s3/m/90a698430242a8956aece443.png)
基因工程大肠杆菌发酵生产重组人胰岛素[宝典] 基因工程大肠杆菌发酵生产重组人胰岛素的工艺一,背景知识1,基因工程科技名词定义中文名称:基因工程英文名称:genetic engineering;gene engineering其他名称:重组脱氧核糖核酸技术(recombinant DNA technique) 定义1:狭义的基因工程仅指用体外重组DNA技术去获得新的重组基因;广义的基因工程则指按人们意愿设计,通过改造基因或基因组而改变生物的遗传特性。
如用重组DNA技术,将外源基因转入大肠杆菌中表达,使大肠杆菌能够生产人所需要的产品;将外源基因转入动物,构建具有新遗传特性的转基因动物;用基因敲除手段,获得有遗传缺陷的动物等。
定义2:将在体外进行修饰、改造的脱氧核糖核酸分子导入受体细胞中进行复制和表达的技术。
扩充:基因工程是指重组DNA技术的产业化设计与应用,包括上游技术和下游技术两大组成部分。
上游技术指的是基因重组、克隆和表达的设计与构建(即重组DNA技术);而下游技术则涉及到基因工程菌或细胞的大规模培养以及基因产物的分离纯化过程。
一个完整的、用于生产目的的基因工程技术程序包括的基本内容有:(1)外源目标基因的分离、克隆以及目标基因的结构与功能研究。
这一部分的工作是整个基因工程的基础,因此又称为基因工程的上游部分;(2)适合转移、表达载体的构建或目标基因的表达调控结构重组;(3)外源基因的导入;(4)外源基因在宿主基因组上的整合、表达及检测与转基因生物的筛选;(5)外源基因表达产物的生理功能的核实;(6)转基因新品系的选育和建立,以及转基因新品系的效益分析;(7)生态与进化安全保障机制的建立;(8)消费安全评价。
基本操作步骤(上游技术)提取目的基因获取目的基因是实施基因工程的第一步。
如植物的抗病(抗病毒抗细菌)基因,种子的贮藏蛋白的基因,以及人的胰岛素基因干扰素基因等,都是目的基因。
要从浩瀚的“基因海洋”中获得特定的目的基因,是十分不易的。
分子克隆英文版

分子克隆英文版Molecular CloningMolecular cloning is a fundamental technique in modern molecular biology and biotechnology, allowing for the isolation, amplification, and manipulation of specific DNA sequences. This process involves the transfer of a DNA fragment from one organism to a self-replicating genetic element, such as a plasmid or a virus, which can then be introduced into a host cell. The host cell, typically a bacterium or a eukaryotic cell, then replicates the inserted DNA, producing multiple copies of the desired genetic material.The primary motivation behind molecular cloning is the need to obtain large quantities of a specific DNA sequence for various applications, including genetic research, disease diagnosis, and the production of recombinant proteins. By cloning a gene of interest, researchers can generate an unlimited supply of the target DNA, enabling further analysis, modification, and utilization.The process of molecular cloning can be divided into several key steps. The first step involves the isolation of the desired DNA fragment, which can be obtained from a variety of sources, such asgenomic DNA, complementary DNA (cDNA), or synthetic DNA. This DNA fragment is then inserted into a suitable vector, such as a plasmid or a viral genome, using specialized enzymes called restriction endonucleases and DNA ligase.The resulting recombinant DNA molecule is then introduced into a host cell, typically a bacterial cell like Escherichia coli, through a process called transformation. The transformed host cells are then cultured, and the cells containing the desired recombinant DNA are selected and amplified. This amplification process allows for the production of large quantities of the target DNA sequence.One of the most important applications of molecular cloning is the production of recombinant proteins. By cloning a gene encoding a specific protein, researchers can express and purify the protein in a host cell, such as a bacterium or a eukaryotic cell line. This technique has been instrumental in the development of numerous therapeutic proteins, including insulin, growth hormones, and monoclonal antibodies, which have revolutionized the field of medicine.Molecular cloning is also essential for the study of gene function and regulation. By cloning a gene and introducing it into a host cell, researchers can investigate the expression, localization, and interactions of the encoded protein, as well as the regulatory mechanisms that control its activity. This knowledge is crucial forunderstanding the complex biological processes that underlie human health and disease.In addition to its applications in research, molecular cloning has also found widespread use in the field of genetic engineering. By modifying the genetic material of organisms, scientists can create genetically modified organisms (GMOs) with desirable traits, such as increased crop yield, improved nutritional value, or resistance to pests and diseases. This technology has significant implications for agriculture, environmental conservation, and the development of new therapeutic strategies.Despite its many benefits, molecular cloning is not without its challenges and ethical considerations. The potential for misuse or unintended consequences of genetic manipulation has sparked ongoing debates and the need for robust regulatory frameworks to ensure the responsible and ethical use of this powerful technology.In conclusion, molecular cloning is a fundamental technique in modern molecular biology and biotechnology, enabling the isolation, amplification, and manipulation of specific DNA sequences. This technology has revolutionized numerous fields, from genetic research and medicine to agriculture and environmental conservation. As the field of molecular biology continues to evolve, the applications and implications of molecular cloning willundoubtedly continue to expand, posing both exciting opportunities and important ethical considerations for the future.。
分子生物学名词解释(Molecularbiologicalterms)

分子生物学名词解释(Molecular biological terms)The beanmail polar tropical fish is set to exitDouban-douban-douban-douban-douban-douban-douban-douban-dou ban-douban-douban-douban stationDouban searchThe home page of my douban my group of the city browse discoveryExplanation of molecular biology terms.Confused の detectiveThe 2010-05-21 20:43:54 from: confused の detective (if you want to let me live please give me happy pain)Title: molecular biology noun explanation (personal arrangement, only a lot of ~ exams are risky, review should be cautious ~ true love life, far away from the point of birth)Meristematic name solutionProbe: molecular hybridization and the tagged nucleotide chain with specific sequences of nucleotide nucleotide nucleotides can be used to detect specific genes in nucleic acid samples.Molecular hybridization: a technique for qualitative or quantitative analysis of DNA or RNA using the basic properties of DNA degeneration and renature.Gene chip: the support of a specific piece of DNA that is closely aligned in a unit area.Gene library: a clone group that contains the entire DNA sequence of an object in a lifetime.CDNA library: it is contain a tissue cells under certain conditions all mRNA expression by the reverse transcription and synthesis of cDNA sequence of clone population, it is stored in the form of cDNA fragments with the tissue cell gene expression information.Genomic DNA library: a clone population stored in the form of fragments of DNA (including all coding and non-coding regions) of the genome of a living organism.Transgenic technology: gene transfer technology is used to integrate the target genes into the fertilized egg cells or embryonic stem cells, then the cells are imported into the animal's uterus to develop into individual technology.Transgenic: the gene that is being imported in transgenic technologyTransgenic animals: the receptor animals that are genetically engineered to be genetically engineeredNuclear transfer technique: an individual cell nucleus of an animal is introduced into the activated egg cell of another individual to develop into an individual, namely, clone.Gene elimination: a technique for removing certain genes in animals based on homologous recombination.Functional cloning: cloning the pathogenic gene by understanding the function of a pathogenic gene.Location cloning: gradually narrowing the range from the chromosomal localization of a pathogenic gene and finally cloning the gene.Gene diagnosis: direct detection of gene structure and its expression level is normal, so as to diagnose the disease.Gene therapy: an exogenous gene that functions as a defective cell can be used to correct or compensate for its genetic defects to achieve therapeutic purposes.Viral oncogene: a type of gene that is present in tumor viruses (mostly retroviruses) that can cause malignant transformation of the target cell.Proto-oncogene: is the oncogene in normal cells, and its expression products regulate the normal growth and differentiation of cells. When activated, can cause cell growth differentiation abnormality, form tumour. Also called cell carcinoma genes.Oncogene: a normal gene in a living organism or in a cell, which controls the growth and differentiation of cells. Cell carcinogenesis can only be caused when its structure changes or expresses an abnormality.Tumor suppressor gene: inhibits the proliferation and proliferation of cells and thus inhibits the genes of tumor formation.Transformation action: by automatically obtaining or artificially supplying exogenous DNA, the cells obtain a new genetic phenotype, which is called transformation.Conjugation: when a cell or bacterium interacts with the bacteria, the plasmid DNA can be transferred from one cell (bacteria) to another (bacteria). This DNA transfer is called the conjugation.Transduction function: when the release of the virus from infected cells, infected again another cell, occurred in donor cells and DNA transfer and recombination between receptor cells is called transduction.Plasmid: small ring double stranded DNA molecule outside the bacterial chromosomeHomologous recombination: a recombination between the homologous sequences, which is also called fundamental recombinationSite specific recombination: the integration of integrase catalysis between the specific sites of two DNA sequences.Transposition: the translocation or rearrangement of genes mediated by insertion sequence and transposon is referred toas transpositionTransposon: a discrete sequence of repeated sequences that can be transferred from one chromosome site to another.Clone: a collection of identical copies or copies from the same ancestor.Cloning: the process of obtaining the same copy, i.e. asexual reproduction.DNA cloning: the method of application enzymology to reorganize the target gene and carrier DNA in vitro, transform or transfect host cells, and gain a large number of genes. It is also called gene cloning and recombinant DNAGenetic engineering: the methods and related work for gene cloning are described as genetic engineering.Compatibility terminal: some of the restriction enzyme recognition sequences are not exactly the same, but after cutting DNA, they produce the same sticky end, which is called the compatibility terminal.Restriction endonuclease: a specific sequence of DNA that identifies a DNA, and an enzyme that cuts double strand DNA around the identification site or its surroundings.CDNA: transcriptional synthesis of single stranded DNA that complements mRNA. Double stranded cDNA can be synthesized with single stranded cDNA as template and polymerized.Genomic DNA: a complete set of genetic information (chromosomes and mitochondria) of a cell or organism.Gene carrier: some of the DNA molecules used to reproduce or express a meaningful protein for the purpose of carrying a target gene.Gene: a genetic base unit located on a chromosome that carries a DNA fragment of a specific genetic information that can encode a single biological product, including RNA and polypeptide chains.Genome: a complete set of genetic information from a living organism, which is the entire genetic information or whole gene that a cell or virus carries.Gene expression: the process of transcription and translation of genesTime specificity: the expression of a specific gene takes place in a certain chronological order according to the function, which is called the time specificity of gene expressionSpace specificity: in the whole process of individual growth, a certain gene product appears in the order of individual tissue space, which is called the spatial specificity of gene expressionHousekeeping genes: some genes continue to be expressed in almost all cells of an individual, often referred to ashousekeeping genesConstitutive gene expression: usually a gene expression similar to the expression of a butler gene, also known as basic expression.Coordinate expression: under certain mechanism control,A set of genes related to function, no matter how they are expressed, should be coordinated and expressed in a coordinated manner.Trans action: the protein factor expressed by one gene interacts with the specific cis-acting component of another gene to regulate its expression. This conditioning is known as the trans action.Cis: protein factor can recognize, regulate the sequence of its own genes, regulate the expression of its own genes, and call it cis.Self - control: regulation of protein is generally used in automrna, inhibiting the synthesis of itself, and self-controlMonocistron: a coding gene transcribed to generate an mRNA molecule that translates into a polypeptide chain.Protein biosynthesis is the process of synthesizing proteins in the sequence of nucleotides in mRNA molecules.S - D sequence: in prokaryotes initiation codon upstream 8 to13 AUG nucleotide site there are 4-9 consensus sequence, high in purine bases, is with small ribosome binding sites of mRNA, known as the S - D sequence.Ribosomal circulation: the peptide chain is extended continuously in the nucleoprotein body, also known as prolongation. This includes carry, peptide and peptide.Polyribosome: the polymer that mRNA forms with multiple nucleosomes is called polyribosomeMolecular partner: the molecular partner is a nonnatural conformation that identifies a type of protein in the cell that can recognize the right folding of the functional domain and the whole protein.Cistron: a genetic unit that codes for a polypeptide.Signal sequence: all sorting signals exist in the targeted delivery of protein structure, mainly was the specific N terminal amino acid sequence, may guide protein metastasize to the appropriate target cells, the sequences are called signal sequence.Open reading framework: the sequence of nucleotide sequences from the mRNA initiation codon AUG to the termination codon.The degeneracy of the genetic code: an amino acid can have two or more codons coded for it, a feature known as the degeneracy of the genetic code.Enhancers: a sequence of DNA that binds specific gene regulation proteins to promote the expression of specific genes near or far away. The distance of the enhancement subscriptional start point varies greatly, but it always ACTS on the most recent promoter.Transcription is the process by which an organism USES DNA as a template to synthesize RNAStructural genes: segments of RNA that are transcribed from DNA molecules called structural genesAsymmetric transcription: in the genome, the genes are transcribed only by the genes of different developmental timing, conditions and physiological needs of the cell. In the DNA molecule double strand, a strand is used as a template for transcription, and the other strand is not transcribed. The template chain is not always on the same chain.Manipulation: transcription is not continuous. Each transcriptional block can be considered as a transcriptional unit, called the operator. The manipulators include several structural genes and their upstream regulatory sequences.Cis-acting elements: the sequence of DNA that is involved in transcriptional regulation at the beginning of the transcription starting point, consisting of promoters, enhancers, and silences.Anti-type action factor: the ability to recognize and combine the homeopathic components, and reverse the transcriptionaleukaryotic proteins that are transcribed by other genes.Transcription factor: in the trans action factor,The direct or indirect combination of RNA polymerase is called transcription factor.Exon: a sequence of nucleic acid sequences of mature rnas in eukaryotic organisms that appear in the fault gene and its primary transcription products.Intron: linear expression of the partition gene in eukaryotes and the sequence of nucleic acids removed during the splicing process.Nuclease: RNA that has enzymatic activity is called a nucleaseReproduction: refers to the generation of genetic material, the process of synthesizing subchain DNA by the mother chain DNA.Semi - reserved replication: when DNA biosynthesis, the mother chain DNA is unwound into two single strands, each acting as a template by the base pairing rule, and the subchain complemented by the template. The DNA of the daughter cell, a single strand is fully accepted from the parent, and the other single strand is completely resynthesized. The DNA of the two subcells is identical to the parental DNA base sequence. This replication method is called semi-retained replication.Bidirectional replication: when the prokaryote replicates, the DNA dissolves the chain from the starting point to the twodirections, forming the opposite of the two directions of the replication fork, called bidirectional replication.Initiator: a compound structure formed at the beginning of DNA replication, containing helicases, DnaC proteins, primers, and DNA replication initiation regions.Replicator: the unit that completes the replication independently, from the replication point to the replication endpoint.Okazaki fragment: a discontinuous fragment formed in the following chain replication during DNA replication.The double strand of DNA is divided into two segments, each acting as a template, and the subchain lengthens the formed y-font structure along the template to be called a replication fork.Half-discontinuous replication: the lead chain replicates continuously and the attendant chain discontinuous replication is called semi-discontinuous replication.Reverse transcription: the process of synthesizing DNA with RNA as a template for reverse transcriptase.Telomere: the structure of the end of the linear DNA molecule of the eukaryotic biosomatic chromosome, which makes the ends of chromosomes become granule.Transcriptional initiation complex: a compound formed by thebinding of the prokaryotic RNA polymerase, the transcriptional pppGpN - product and template DNA.Boundary sequence: the eukaryotic introns start with GU at 5 'end and AG is 3' end. 5 '- GU... Ag-oh3 is called a boundary sequence, also known as a splice interface.Splicing: removing introns from RNA molecules so that exons can be connected together.Promoter: a sequence of DNA in the upstream of the transcription starting point of RNA polymerase. If the RNA polymerase is combined with it, it can initiate transcription.Central rule: the law of transmission of genetic information from DNA to RNA to protein.Transcriptase: transcriptional complex, which is composed of the nuclease of RNA polymerase and the product of its template DNA, transcribed.Silencing: the negative regulating element in the eukaryotic element, which ACTS as a repressor when it binds the specific protein factor.Gene: the structure of eukaryotic gene, by a number of coding and non-coding area interval with each other, but embedded in a row, after connected to again unless the coding regions, can translate the continuous complete protein amino acid composition. These genes are called broken genes.[figure]DavidDavid (I'm looking for the book direction)It is also the common touch of biochemistryYour response? Who? Who? Who? Who? Who? , ah ha... Good thing ~ plus go> medical student's home groupLatest topic:Molecular biology noun explanation (individual, only a lot more to test a risk... (confused の detective)N reasons you attended medical school (breba)I was depressed. (hh)How microbes learn?Everybody work first or take the exam first. (MaoMaoQ)My man is studying medicine (summer)The division room chooses the direction, especially lost... (ssu xiaawn)Are there any graduate students in Concorde? Seek counsel (summercool)> to report bad information2005-2010 , all rights reserved about , contact us · disclaimer · help center · douban service (API) · mobile phone douban · brand club。
基因工程在英语作文中的应用

基因工程在英语作文中的应用英文回答:Genetic engineering, a powerful technology that enables scientists to manipulate the genetic material of living organisms, has revolutionized various fields of science, including medicine, agriculture, and biotechnology. This groundbreaking technique allows us to modify genes, insert new ones, or even create entirely new genetic constructs.Medical Applications of Genetic Engineering.Genetic engineering has played a crucial role in advancing medical research and treatment. It has led to the development of targeted therapies for genetic diseases, such as sickle cell anemia and cystic fibrosis. By correcting or replacing faulty genes, genetic engineering has the potential to cure or significantly improve the prognosis of debilitating conditions.Moreover, genetic engineering has enabled the production of recombinant proteins, including hormones and enzymes, which are used to treat a wide range of diseases. These proteins can be mass-produced through genetically modified organisms (GMOs), providing affordable and accessible treatments for patients worldwide.Agricultural Applications of Genetic Engineering.In agriculture, genetic engineering has brought about numerous benefits. GMO crops, engineered to possess desirable traits, have increased crop yields, resistance to pests and diseases, and improved nutritional value. This has contributed to global food security, reducing hunger and malnutrition in vulnerable populations.Furthermore, genetic engineering has facilitated the development of crops that are more tolerant to environmental stresses, such as drought and salinity. This technology has the potential to enhance agricultural sustainability and ensure food production in challenging climates.Biotechnological Applications of Genetic Engineering.Genetic engineering has also found extensive applications in biotechnology. Microorganisms, such as bacteria and yeast, are genetically engineered to produce valuable compounds, including enzymes, vitamins, and biofuels. These engineered organisms serve as cost-effective and efficient biofactories, providing renewable and sustainable alternatives to traditional chemical processes.Additionally, genetic engineering has fueled advancements in DNA fingerprinting, DNA sequencing, and other diagnostic techniques. These tools have revolutionized forensic science, enabled personalized medicine, and provided insights into human evolution and genetic diversity.Ethical and Societal Considerations.As with any powerful technology, genetic engineeringraises ethical and societal concerns that must be carefully considered. Issues such as the potential risks to the environment, the unintended consequences of genetic modifications, and the equitable distribution of benefits and risks are important considerations.Ongoing research and thorough risk assessments are essential to ensure the responsible and ethical application of genetic engineering. Open and transparent public dialogue is also crucial to address societal concerns and build trust in this transformative technology.中文回答:基因工程在医学领域的应用。
【双语植物育种14】Genetic_Transformation_and_Production_of_Transgenic_Plants

•Wild type plasmid of A.tumefaciens is responsible for inducing tumour and hence termed as tumor inducing (Ti) plasmid.
Genetic Transformation and Production of Transgenic Plants
Contents
Gene transfer methods
Transient and stable gene expression
Genetic markers in transformation
1. Gene transfer methods
Vector Mediated Gene Transfer
Agro bacterium-mediated Transformation
Method of transformation Intact plants and seedling explants such as cotyledons, hypocotyls, roots, calli and protoplasts can be used for co-cultivation with Agrobacterium cells When Agrobacterium rhizogenes infects plants, adventitious roots rather than containing recombinant plasmids. tumour are formed at the site of infection. This is mediated by Ri (root inducing) However, leaf disc method has been widely used where surface sterilized plasmid. leaf discs are infected with the appropriate strain of Agrobacterium carrying the Vectorsof choice pRiAu which are on regeneration medium for two or three days. vector such as and co-cultured based on Ri plasmid have been developed. The Ri vectorstime,particularly useful for in the bacteria are induced, the bacteria During this are the virulence genes studying nodulation and manipulating root cultures for secondary metabolite and Vasicular Arbuscular Mycorrhiza (丛枝菌 bind to the plant cells around the wounded site and the gene transfer occurs. 根 VAM) production. The leaf discs are then transferred to regeneration/ selection medium which contains 500 (µg/ml carbenicillin to kill the Agrobacteria and the appropriate antibiotic, usually kanamycin, to inhibit the growth of untransformed plant cells. During next 4-5 weeks, the transformed shoots are obtained which are rooted and transferred to soil. Transgenic plants regenerated from various tissues are called T0 plants whereas their subsequent generations are called as T1,T2,T3 etc.
大肠埃希菌严谨反应蛋白RelA的表达载体构建及生物信息学分析

中国病原生物学杂志2020年12月第15卷第12期Journal of Pathogen Biology Dec.2020.Vol.15,No.12・1427・DOI:10.13350/j.cjpb.201212•论著*大肠埃希菌严谨反应蛋白RelA的表达载体构建及生物信息学分析*徐本锦…,刘玲,宋彬裕,杜淼,宣炭,寇妍祺(山西医科大学汾阳学院医学检验系.山西汾阳032200)目的构建大肠埃希菌严谨反应蛋白编码基因relA的原核表达载体.通过生物信息学方法分析RelA蛋白的结构和功能,为阐明RelA介导细菌严谨反应的分子机制奠定基础。
方法利用分子克隆技术得到relA的CDS序列,利用基因工程手段将CDS序列连接到原核表达载体pET-22b中,通过抗性基因筛选和双向测序验证,得到重组表达载体pET-22b-relA…采用Protparam、TMHMM、ProtScale ExPASy>PSORT II、SignalP、InterPro、UniProt、NetPhos 3.NGlyc 1.0、Net()Glyc4.0,Blast等生物信息学软件和数据库对大肠埃希菌RelA蛋白的理化性质、功能位点、翻译后修饰、三维结构及蛋白相互作用网络等生物学特性进行系统分析;利用Clustal X2和MEGA7.0软件对RelA蛋白进行基于氨基酸序列的同源性分析和系统进化分析。
结果成功构建relA基因的原核表达载体pET-22b-relA o生物信息学分析RelA蛋白由744个氨基酸组成.相对分子质量84X103Da,等电点6.29,有两个卷曲螺旋结构,无跨膜螺旋区.为亲水性蛋白。
RelA的N-末端结构域包括PH和SYNTH,C末端结构域包括TGS、a螺旋区、CC及ACT亚结构域;该蛋白主要在细胞质发挥功能.含有10个NTP结合位点,2个金属离子结合位点,16个合成酶活性位点,53个潜在的磷酸化位点和8个可能的糖基化位点;RelA在细菌之间高度保守,与大肠埃希菌RelA亲缘关系最近的是福氏志贺菌,其次是柯氏柠檬酸杆菌。
高中生物第十一节:基因重组与基因转位——基因重组概念
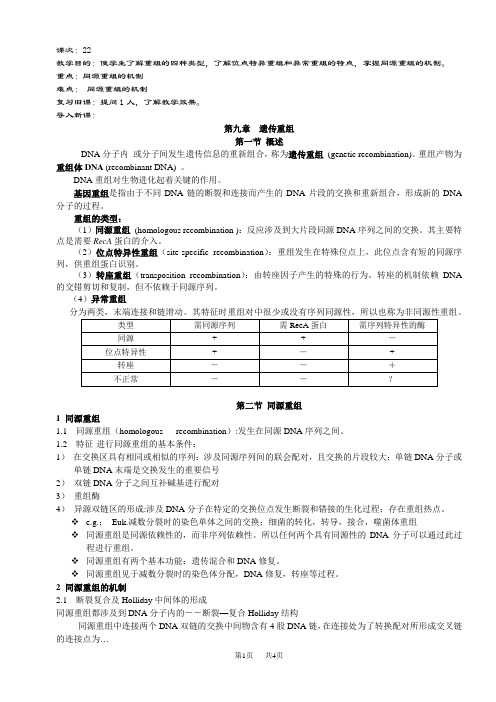
课次:22教学目的:使学生了解重组的四种类型,了解位点特异重组和异常重组的特点,掌握同源重组的机制。
重点:同源重组的机制难点:同源重组的机制复习旧课:提问1人,了解教学效果。
导入新课:第九章遗传重组第一节概述DNA分子内或分子间发生遗传信息的重新组合,称为遗传重组(genetic recombination)。
重组产物为重组体DNA (recombinant DNA) 。
DNA重组对生物进化起着关键的作用。
基因重组是指由于不同DNA链的断裂和连接而产生的DNA片段的交换和重新组合,形成新的DNA 分子的过程。
重组的类型:(1)同源重组(homologous recombination ):反应涉及到大片段同源DNA序列之间的交换。
其主要特点是需要RecA蛋白的介入。
(2)位点特异性重组(site-specific recombination):重组发生在特殊位点上,此位点含有短的同源序列,供重组蛋白识别。
(3)转座重组(transposition recombination):由转座因子产生的特殊的行为。
转座的机制依赖DNA 的交错剪切和复制,但不依赖于同源序列。
(4)异常重组分为两类,末端连接和链滑动。
其特征时重组对中很少或没有序列同源性,所以也称为非同源性重组。
第二节同源重组1 同源重组1.1 同源重组(homologous recombination):发生在同源DNA序列之间。
1.2 特征-进行同源重组的基本条件:1)在交换区具有相同或相似的序列:涉及同源序列间的联会配对,且交换的片段较大;单链DNA分子或单链DNA末端是交换发生的重要信号2)双链DNA分子之间互补碱基进行配对3)重组酶4)异源双链区的形成:涉及DNA分子在特定的交换位点发生断裂和错接的生化过程;存在重组热点。
e.g.:Euk.减数分裂时的染色单体之间的交换;细菌的转化,转导,接合,噬菌体重组同源重组是同源依赖性的,而非序列依赖性。
专业英语遗传学的基础

Dominant and recessive genes: In Mendelian inheritance, there is a distinction between dominant and recessive genes. Dominant genes can express their characteristics, while recessive genes are only expressed when there are no dominant genes present.
02
Genetic material foundation
Discovery
DNA was first isolated from white blood cells in 1869 by Friedrich Miesecher However, its significance was not realized until the 1950s when Watson and Crick proposed its double helix structure
Heterozygous and homozygous: Heterozygous individuals refer to individuals with one dominant gene and one recessive gene in an allele, while homozygous individuals refer to individuals with the same genotype in all alleles.
重组大肠杆菌发酵表达及代谢调控研究进展

食品与药品Food and Drug2021年第23卷第1期85重组大肠杆菌发酵表达及代谢调控研究进展张言慧「,高先岭「,黄魁2,郭青青「,袁建国2*(1.山东国力生物科技有限公司,山东济南250014;2.山东国力生物技术研究院,山东济南250101)摘要:作为应用最为普遍的蛋白质表达系统,重组大肠杆菌表达系统具有其他表达系统无法比拟的优越性。
本文综述了大肠杆菌作为表达系统的特征,釆用重组大肠杆菌表达的重组蛋白及生物化学产品概况,概述了表达载体中营养源诱导启动子、温敏型启动子及乳糖启动子3种不同类型启动子特点。
另外分析了不同大肠杆菌宿主对碳源的代谢特征,乙酸代谢对大肠杆菌生长及重组蛋白表达的影响,丙酮酸代谢及乙醛酸循环的特征,最后介绍了发酵过程调控对重组大肠杆菌表达系统的影响。
关键词:大肠杆菌;发酵;代谢;启动子;重组蛋白中图分类号:Q939.97文献标识码:A文章编号:1672-979X(2021)01-0085-07DOI:10.3969/j.issn.l672-979X.2021.01.018Research Progress on Fermentation Expression and Metabolism Regulation of RecombinantEscherichia ColiZHANG Yan-hui1,GAO Xian-ling1,HUANG Kui2,GUO Qing-qing1,YUAN Jian-guo2收稿日期:2020-10-29基金项目:山东省重大科技创新工程项目(2019JZZY010520)作者简介:张言慧,硕士,工程师,研究方向为微生物发酵过程优化"通讯作者:袁建国,研究员,研究方向为食品与药品生物技术E-mail:******************collagen-induced arthritis[J].Arthritis Rheum,2000,43(8):16981709.[36]Sadallah S,Lach E,Lutz H U,et al.CD35IN synovial fluid frompatients with inflammatory joint diseases[J].Arthritis Rheum, 1997,40(3):520-526.[37]Nilsson S C,Sim R B,Lea S M,et plement factor I inhealth and disease[J].Mol Immunol,2011,48(14):1611-1620. [38]Mastellos D C,Ricklin D,Yancopoulou D,et plementin paroxysmal nocturnal hemoglobinuria:exploiting our current knowledge to improve the treatment landscape[J].Expert Rev Hematol,2014,7(5):583-598.[39]SjOberg A P,Manderson G A,Morgelin M,et al.Short leucine-richglycoproteins of the extracellular matrix display diverse patterns of complement interaction and activation[J].Mol Immunol,2008, 46(5):830-839.[40]Banda N K,Levitt B,Glogowska M J,et al.Targeted inhibitionof the complement alternative pathway with complement receptor 2and factor H attenuates collagen antibody-induced arthritis in mice[J].J Immunol,2009,183(9):5928-5937.[41]Song H,He C,Knaak C,et plement receptor2-mediatedtargeting of complement inhibitors to sites of complement activation[J].J Clin Investig,2003,111(12):1875-1885.[42]Molina H.Distinct receptor and regulatory properties ofrecombinant mouse complement receptor1(CR1)and Crry,the two genetic homologues of human CR1[J].J Exp Med,1992, 175(1):121-129.[43]Michelfelder S,Fischer F,Waldin A,et al.The MFHR1FusionProtein Is a Novel Synthetic Multitarget Complement Inhibitor with Therapeutic Potential[J].J Am Soc Nephrol,2018,29(4): 1141-1153.[44]Noris M,Remuzzi G.Glomerular diseases dependent oncomplement activation,incl-uding atypical hemolytic uremic syndrome,membranoproliferative glomerulonephritis,and C3 glomerulopathy:core curriculum2015[J].Am J Kidney Dis,2015, 66(2):359-375.[45]Melis J PM,Strumane K,Ruuls S R,et plement in therapyand disease:Regul-ating the complement system with antibodybased therapeutics[J].Mol Immunol,2015,67(2):117-130.86食品与药品Food and Drug2021年第23卷第1期(1.Shandong Guoli Biotechnology Limited Company,Jinan250014,China;2.Shandong Guoli BiotechnologyResearch Institute,Jinan250101,China)Abstract:As the most widely used protein expression system,the recombinant Escherichia coli expression system has more advantages than other expression systems.In this paper,the characteristics of Escherichia coli as an expression system were reviewed,and recombinant proteins and biochemical products expressed in recombinant Escherichia coli were introduced.The characteristics of three different types of promoters including nutrition-induced promoters, temperature-sensitive promoters and lactose promoters in expression vectors were summarized.In addition,the metabolic characteristics of carbon sources in different Escherichia coli hosts,the eflect of acetic acid metabolism on the growth of Escherichia coli and expression of recombinant protein,the characteristics of pyruvate metabolism and glyoxylic acid cycle were analyzed.Finally,the effect of fermentation regulation on the expression system of recombinant Escherichia coli was introduced.Key Words:Escherichia coli;fermentation;metabolism;promoter;recombinant protein由于大肠杆菌遗传学背景较清楚,生长速度快,培养基条件要求较低,容易实现高密度培养等特点,长期以来是商业生产重要目的基因的表达系统为了获得高水平的基因表达产物,综合考虑控制转录、翻译、蛋白质稳定性及向胞外分泌等多方面因素,设计了许多具有不同特点的表达载体,以满足表达不同性质、不同要求的目的蛋白需要冈。
生物制药专业英语

生物制药专业英语English:Biopharmaceuticals, also known as biologic drugs, are medicinal products derived from living organisms or their components. These drugs are produced through biotechnological processes involving genetic manipulation, recombinant DNA technology, and cell culture techniques. Biopharmaceuticals encompass a wide range of products, including therapeutic proteins, vaccines, monoclonal antibodies, and gene therapies. Due to their complex nature and manufacturing process, biopharmaceuticals often require stringent regulation and specialized facilities for production. The development of biopharmaceuticals involves multiple stages, including discovery, preclinical research, clinical trials, regulatory approval, and commercialization. These drugs have revolutionized the treatment of various diseases, offering targeted therapies with high efficacy and reduced side effects compared to traditional pharmaceuticals.中文翻译:生物制药品,又称生物制剂,是从活体生物或其组成部分衍生出来的药品。
筛选单克隆细胞株流程

筛选单克隆细胞株流程英文回答:Screening for monoclonal cell lines is an essential process in biotechnology and biomedical research. It involves the identification and selection of individual cells that produce a specific protein or exhibit a desired phenotype. This process is crucial for the development of therapeutic antibodies, recombinant proteins, and cell-based assays.To begin the screening process, I first culture a heterogeneous population of cells, such as hybridoma cells or transfected cells. These cells are typically derived from a parent cell line and express a diverse range of proteins or phenotypes. I then treat the cells with a selective agent or apply a specific assay to identify the desired cells.One common method for screening monoclonal cell linesis fluorescence-activated cell sorting (FACS). In this technique, cells are labeled with fluorescently tagged antibodies or markers that specifically bind to the protein of interest. The cells are then passed through a flow cytometer, which detects and sorts the labeled cells based on their fluorescence intensity. The sorted cells can be collected and further analyzed or expanded for downstream applications.Another approach for screening monoclonal cell lines is limiting dilution. This method involves serially diluting the cell population to a low density, such that each wellin a microplate contains only one cell. The plates are then incubated, and wells with single-cell colonies areidentified and expanded. These colonies can be screened for the desired protein expression or phenotype using immunostaining, enzyme-linked immunosorbent assays (ELISAs), or other specific assays.In addition to these methods, genetic screening techniques such as CRISPR/Cas9-mediated knockout or knock-in can be employed to generate monoclonal cell lines withspecific genetic modifications. These genetically modified cells can then be screened using molecular techniques like polymerase chain reaction (PCR) or sequencing to confirm the desired genetic alteration.Once the monoclonal cell lines are identified and selected, they can be further characterized and validated for their stability, productivity, and functionality. This may involve assessing their growth characteristics, protein expression levels, and functional assays specific to the desired phenotype.Overall, the process of screening for monoclonal cell lines is a crucial step in biotechnology and biomedical research. It requires careful planning, optimization, and validation to ensure the selection of high-quality cell lines that meet the desired criteria.中文回答:筛选单克隆细胞株是生物技术和生物医学研究中的一个重要过程。
遗传学英文词汇

Genetics Glossaries Week 1Heredity 遗传Variation 变异Preformation 先成论Epigenesist 后生说Mendel’s law 孟德尔定律Law of segregation 分离定律Law of independent assortment 自由组合定律Inheritance 遗传特征Trait 特征Full/Constrict 饱满/收缩Pod 荚Axial/Terminal 轴生/顶生Stem 茎Monohybrid cross 单基因杂交Postulate 假说Dominance/Recessiveness 显性/隐形Gamete 配子Likelihood 可能性Punnett square 旁那特方格/棋盘法Genetype 基因型Allele 等位基因Homozygote/ Heterozygous 纯合子/杂合子Phenotype 表现型Test cross 测交Dihybrid cross 双因子杂交Chi-square test 卡方测验Week 2Pedigree 系谱Huntington disease 亨廷顿舞蹈症Cystic fibrosis 囊性纤维化(胰腺病)Vertical inheritance 垂直遗传特性Horizontal inheritance 水平遗传特性Incomplete dominance 不完全显性Semidominance 半显性Codominance 共显性Multiple alleles 复等位基因Self-incompatibility 自交不亲和Pleiotropy 基因多效性Lethal gene 致死基因Cytogenetics 细胞遗传学Chromatin 染色质Chromosome 染色体Haploid 单倍体Diploid 二倍体Karyotype 核型Sex chromosome/Autosome 性/常染色体Moths 蛾Alligator 短吻鳄Parental 亲本的Maternal 母系的Subsequent 随后的Meiosis 减数分裂Drosophila 果蝇Drosophila melanogaster 黑腹果蝇Fruit fly 果蝇1Prolific 多产的Nomenclature 命名法Hemizygous 半合子的Color-blindnenss 色盲Descendant 后代Hormone 荷尔蒙Pattern baldness 模型斑秃Week 3Mitosis 有丝分裂Complement 互补Cytokinesis 胞质分裂Ongoing 持续的Synthesis 合成Telophase 末期Anaphase 后期Aligned 对齐的Metaphase 中期Prophase 前期Duplicated 复制的Duplication 复制Centrosome 中心体Meiosis I/II 减数分裂I/II期Nondisjunction 不分离Red-green colorblindness 红绿色盲Sex-linked 伴性的Hemophilia 血友病Hypophosphatemia 低磷血症Deoxyribonucleic acid 脱氧核糖核酸(DNA)Nuclei 核Principle 组分Ultracentrifugation 超速离心法Predominance 优势Phage 噬菌体Host cell well 宿主细胞壁Double helix 双螺旋Complementary pairing 互补配对Central dogma 中心法则Prokaryote 原核生物Eukaryote 真核生物Week 4Nucleotide 核苷酸Phosphate 磷酸盐Quagga 斑驴Skull 颅骨Neanderthal 穴居人的Uracil 尿嘧啶Thymine 胸腺嘧啶Adenine 腺嘌呤Guanine 鸟嘌呤Cytosine 胞嘧啶Ribonucleotide 核糖核苷酸Tobacco mosaic virus 烟草花叶病毒(TMV)Semiconservative 半保留的Methylate 使甲基化Splicing 剪接Alternative splicing 选择性剪接Reverse transcription 反转录Retrovirus 逆转录病毒2Immunodeficiency 免疫缺陷Matrix 基质Bilipid outer layer 双脂质外层Viral particle 病毒颗粒Disintegrate 破裂Week 5Correlation 相关性Polarity 极性Nonoverlapping 不重叠的Degenerate 简并的Incorporation 编入Nickel hydride 镍氢Wobble rule 摆动法则Peptidyl 肽基Aminoacyl 氨酰基Polyribosome 多核糖体Elongation 延长Termination 终止Multimeric protein 多亚基蛋白质Posttranslational 翻译后Prion 阮病毒Spongiform encephalopathy 海绵状脑病Spongy 海绵似的Proteinaceous 蛋白质的Deposit 沉淀物Incubation 潜伏期Progressive 渐进的Neurodegeneration 神经性退行性病变Infectious 传染的Forward/reverse mutation 正向/反向突变Rearrangement 重排Spontaneous mutation 自发突变Haploid 单倍体Susceptibility 敏感性Mutagen 诱变剂Bactericide 杀菌剂Fluctuation 波动Polymerase 聚合酶Proofreading 校对Crossing-over 互换Transposon 转座子Base analog 碱基类似物Intercalator 插入剂Alkyltransferase 烷基Homology-dependent 同源依赖Excision 切除Methyl 甲基Mismatch 错配Error-prone 易错的Nonhomologous end-joining 非同源末端接合Xeroderma pigmentosum 着色性干皮病Alkaptonuria 尿黑酸症Hypothesis 假说Neurospora 脉胞菌Mold 霉菌Nutritional mutant 营养突变体Auxotroph 营养缺陷型Prototroph 原养型Modulate 调节3Genetics 20104Perception 感觉Week 6-7Transgenic 转基因 Recombinant 重组的 Donor 供体Restriction enzyme 限制性内切酶 Fragment 碎片 Vector 载体 Transformation 转导 Amplification 扩增 Endonuclease 核酸内切酶 Cornerstone 基础 Degrade 降解 Palindrome 回文 Overhang 悬突体 Isoschizomer 同切酶 Isocaudarner 同尾酶 Gel electrophoresis 凝胶电泳 Partial digestion 部分消化 Infer 推断Selectable marker 可选标记 Drug resistance 抗药性 Ligation 连接反应 Ligase 连接酶 Sticky end 粘性末端 Blunt end 平整末端 Cosmid 粘性质粒YAC 酵母人工染色体(yeast artificial chromosome ) Autonomous 自主的Subcloning 亚克隆化β-galactosidase β半乳糖苷酶 Gal 标准编号 Blue dye 蓝色染料 Plaque 噬菌斑Shuttle vector 穿梭载体 Intron 内含子 Probing 探测Southern blotting DNA 印迹 Reverse genetics 反向遗传学 Transgenic 转基因的 Metabolity 代谢物 Gene knockout 基因敲除 Ectopic expression 异位表达Week 8Embryo 胚胎Genetic linkage 遗传连锁 Chiasmata 复交叉Chromosome breakage 染色体断裂 Cytological 细胞学的 Abnormality 异常Keep track of 与……保持联系 Genetic marker 遗传标记 Progeny 子代 Discontinuity 不连续的 Parental class 亲本 Assort 分配 Tracing 追踪 Correction 修正Chromosomal interference 染色体干扰Orient 定向Homologous chromosome 同源染色体Coefficient of coincidence 并发系数Linkage group 连锁群Interchangeable 相互可交换Week 9HGP (Human Genome Project) 人类基因组计划Proposed 被提议Draft 草稿Skepticism 怀疑论Computational biology 计算生物学Ethics 伦理学Legislation 法律Arabidopsis thaliana 拟南芥Facilitate 促进Manipulation 操作-omics 各种组学Transcriptomics 转录组学Proteomics 蛋白质组学Phenomics 表型组学Accuracy 精确性Polymorphism 多态性Heterochromatic DNA 异染色DNAHybridization 杂种Identifying 标记Estimating error 估计误差SNP (Single Nucleotide Polymorphism) 单核苷酸多态性SSR (Simple Sequence Repeat) 简单重复序列Microsatellite 微卫星Genomewide 全基因组Constellation 构象Span 跨度Counterpart 副本Bottom-up approach 自下而上模式STS (Sequence Tagged Site) 标志序列位点Top-down approach自上而下模式Fluorescent 荧光的In situ hybridization 原位杂交Loci (locus复数) 位点Resolution 分辨率Hierarchical shotgun approach 分层散弹枪策略Shearing 剪切Throughput 吞吐量/生产量Distinct 不同的Lateral transfer 横向迁移Complexity 复杂性Shuffling 慢慢移动Module 模块Paralogs 种内同源基因Pseudogene 假基因Duplication 重复Telomere 端粒Orthologous gene 种间/直系同源基因Paralogous gene 种内/旁系同源基因Week 10Organelle 细胞器Saccharomyces cerevisiae 酿酒酵母5Preserve 保护Integrity 完整性Shortening 缩短Fusion 融合Degradation 降解Germ-line cell 生殖细胞Somatic cell 体细胞Histone 组蛋白Heterogeneous 不均匀的,多样的Uneven 不均匀Supercoiling 超螺旋Radial loop 桡箕/反箕Scaffold 支架结构Heterochromatin 异染色质Staining 着色Transcription 转录Inactive 失活Constitutive 组成性的Facultative 兼性的Euchromatin 常染色质Condense 浓缩Dosage compensation 剂量补偿Barr body 巴氏小体/X染色质Deletion 删除Inversion 倒位Translocation 易位Transposition 转置Polytene 多线型Giant chromosome 巨染色体Salivary gland cell 唾液腺细胞Inversion loop 倒位环Chromatid 染色单体Centromere 着丝点Suppressor 抑制物/抑制基因Disruption 分裂Speciation 物种形成Transposable element 转位因子Retroposon 反转录子LINE (Long interspersed element)长散在序列SINE (short interspersed element)短散在序列Relocate 迁移Euploid 整倍体Aneuploid 非整倍体Monosomy 单倍体Trisomy 三倍体Tetrasomy 四倍体Polyploidy 多倍体Colchicines 秋水仙碱Down's syndrome 唐氏综合征Inactivation 失活Mosaic 嵌合体Diploid 二倍体Vigor 活力Sterile 不育的Odd-number 奇数Allopolyploid 异源多倍体Raphanobrassica 萝卜属Week 11Prokaryotic 原核的6Proliferating 增生的ORF (open reading frame) 阅读框架Operon 操纵子Spontaneous 自发的Transformation 转化Conjugation 结合Transduction 转导Recipient 接受者Hfr 高频重组Integrate 融入Excision 切除Reverting 回复Non-Mendelian 非孟德尔式Four-o-clock 紫茉莉Mitochondria 线粒体Polypeptide-encoding 多肽编码Compact 压缩Intron 内含子Liverwort 地钱Protozoan 原生动物Parasite 寄生虫Apparatus 组织/器官Exception 例外mtDNA 线粒体DNA Chloroplast 叶绿体cpDNA 胞质DNAResponsive 回应的Heteroplasmic 异质的Homoplasmic 同质的Bioreactor 生物反应器Week 12Developmental genetics 发育遗传学Manipulation 操纵Species-specific 特种异性的Cell formation 细胞形成Mutant 突变体Loss-of-function 功能性缺失Null 失效的Hypomorphic 亚效等位基因Dominant-negative 显性失活的Gain-of-function 功能性获得Overexpression 超量表达Ectopic expression 异位表达Null mutation 无效突变Leaky 有漏洞的Permissive temp 允许温度Restrictive temp 限制温度Haploinsufficiency 单倍剂量不足Subcellular localization 亚细胞定位Epistasis 上位/异位显性Sepal 萼片Petal 花瓣Stamen 雄蕊Carpel 心皮EMS (Ethylmethane Sulphonate) 乙基甲磺酸Irradiation 放射T-DNA 转运DNAsiRNA 小干扰RNAmiRNA = microRNA 微小RNA7Functional genomics 功能基因组学Adenosine deaminase 腺苷脱氨酶Embryonic 胚胎的Totipotent (细胞)全能的Pluripotent 多能性的Blastocyst 胚泡Multipotent 多能干细胞Hematopoietic 造血的Bone marrow 骨髓Week 13Anterior-posterior 后前位的Syncytium 多核体Cortex 皮层Pole cell 极细胞Blastoderm 胎盘Fertilization 受精Segmentation gene 分节基因Homeotic gene 同源框基因Cellularization 细胞化Gastrulation 原肠胚形成Germ layer 胚层Mesoderm 中胚叶Endoderm 内胚层Ectoderm 外胚层Maternal gene 母体基因Gap gene 裂隙基因Pair-rule gene 成对规则基因Segment-polarity 体节极性基因Maternal-effect 母体影响bicoid (bcd) 果蝇中控制头胸发育的一个关键母体基因Morphogen 成形素Repressor 阻遏物Zygotic gene 合子基因Hierarchy 层次结构Promoter 启动子Affinity 亲和力Regulating 调节Subdivide 细分Mirror-image 镜像Intra-segmental 节内的Patterning 图样Ligand 配合体Transcription factor 转录因子Regulatory cascade 级联调节系统Gene cluster 基因群Biothorax complexHomeodomain 同源域Penetrance 外显率Expressivity 表现度Imprinting 印迹Insulin-like 胰岛素样Epigenetic 表观遗传的Methylation 甲基化作用Prader-Willi syndrome 普拉德-威利综合征Angelman syndrome 天使综合征Haig hypothesis 海格假说Down-regulation 减量调节Sequential 连续的Asymmetric 不对称的8Genetics 20109Intrinsic 固有的 Juxtacrine 邻分泌 Paracrine 旁分泌 Mediated 介导的Week 14Population genetics 种群遗传学 Gene pool 基因库 Microevolution 微观进化 Macroevolution 宏观进化Hardy-Weinberg law 哈代-温伯格定律 Infinite number 无穷 Migration 迁移 Equilibrium 平衡 Correlate 相关 Albino 白化病者 Genetic drift 遗传漂变 Nonrandom mating 选择性交配 Fitness 适合度Natural selection 自然选择 Artificial selection 人工选择 Antibiotic 抗生素 Preexisting 预成 Viability 生存能力 Counteract 抵消 Confer 授予 Persist 保持Heterozygous advantage 杂种优势 Eugenics 优生学Geographically 地理学上的Fluctuation 波动Founder effect 创建者效应 Pathogen 病菌 Insecticide 杀虫剂 Inbreeding 近亲交配 Self-fertilization 自体受精 Hybrid vigor 杂种优势 Deleterious 有害的 Overdominance 超显性Week 15Pre-existing 之前就存在的 Chimpanzee 黑猩猩 Subtle 微妙的 Complexity 复杂度 Transposition 转置 Diversification 多样化 Divergence 分歧Fibrinopeptide 血纤维蛋白肽 Phylogeny tree 系统树。
转基因生物英语

转基因生物英语
转基因生物是指通过基因工程技术对生物体进行基因改造,使其具有特定的性状或功能。
转基因生物广泛应用于农业、医药、工业等领域,但同时也引起了广泛的争议和担忧。
以下是一些转基因生物的英语词汇:
1. genetically modified organism (GMO):转基因生物
2. genetic engineering:基因工程
3. gene splicing:基因剪接
4. gene transfer:基因转移
5. transgene:外源基因
6. genetic modification:基因改造
7. recombinant DNA technology:重组 DNA 技术
8. biotechnology:生物技术
9. genetic diversity:遗传多样性
10. biosafety:生物安全
以上词汇可以帮助人们更好地了解转基因生物及其相关技术和问题。
- 1 -。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
ORIGINAL ARTICLEGenetic Recombinant Expression and Characterization of Human Augmenter of Liver RegenerationChun-Fang Gao ÆFei Guo Zhou ÆHao Wang ÆYing-Feng Huang ÆQiang Ji ÆJie ChenReceived:17March 2008/Accepted:3June 2008/Published online:9July 2008ÓSpringer Science+Business Media,LLC 2008Abstract Aims To establish a highly effective prokary-otic recombinant expression system for human augmenter of liver regeneration (hALR)and to characterize the recombinant hALR both in vitro and in vivo.Methods ALR cDNA was synthesized and inserted into expression vector pET28a +,the recombinant plasmid was transformed into BL21,and expression of hALR was induced by IPTG.Recombinant hALR (rhALR)was purified by sequential detergent wash,enterokinase (EK)digestion,gel-filtration,and chelating chromatography.The rhALR was identified by SDS-PAGE,immunoblotting,MALDI-TOF-MS,and N-terminal sequencer.Cell proliferative effect of rhALR on human hepatocytes was analyzed by MTT.The pro-tective effect of rhALR on liver function was observed on CCl 4-induced intoxicated mice.Results Recombinant expression plasmid of ALR [pET28(a +)-hALR]was con-firmed by restriction enzyme digestion and DNAsequencing.The expressed rhALR constituted 30%of total bacterial protein.Molecular weight was 15,029for mono-mer and 30,136for dimer by mass determination.N-terminal was M-R-T-Q-Q,exactly the same as antici-pated for hALR.The purified protein migrating at about 15KD showed excellent antigenicity in immunoblotting.The rhALR also showed a strong stimulative effect on hepa-tocyte proliferation.ALT and AST levels,liver histological structure,as well as the survival rate of CCl 4-intoxicated mice were significantly improved when rALR was administrated at 40l g/kg or 200l g/kg.Conclusions The rhALR is successfully expressed highly effectively with anticipated MW,N-terminal,and antigenicity.It could play an important role in relieving acute hepatic injury and hepatic failure by promoting hepatic cell proliferation and improving liver function in CCl 4-intoxicated mice.Keywords Gene ÁAugmenter of liver regeneration (ALR)ÁRecombinant ÁHepatocytes ÁLiver functionAbbreviationsALR Augmenter of liver regeneration IPTG Isopropyl-b -galactopyranoside pET Plasmid for expression by T7RNA polymerase MW Molecular weightIntroductionLiver possesses the capacity to restore its tissue mass and attain optimal volume in response to physical,infectious,and toxic injury.The extraordinary ability of the liver to regenerate is regarded to be the effect of cross-talk among growth factors,cytokines,matrix components,and many other factors [1].Among the growth factors promotingChun-Fang Gao,Fei Guo Zhou:the authors contributed equally to this work.C.-F.Gao (&)ÁQ.JiDepartment of Laboratory Medicine,Eastern Hepatobiliary Hospital,Second Military Medical University,225Changhai Road,Shanghai 200438,Chinae-mail:gaocf1115@;wanggaob@ F.G.ZhouDepartment of Hepatic Surgery,Eastern Hepatobiliary Hospital,Second Military Medical University,Shanghai 200438,China H.WangDepartment of Laboratory Medicine,Changzheng Hospital,Second Military Medical University,Shanghai 200003,China Y.-F.Huang ÁJ.ChenLiver Fibrosis Research Center,Changzheng Hospital,Second Military Medical University,200003Shanghai,ChinaDig Dis Sci (2009)54:530–537DOI 10.1007/s10620-008-0372-1hepatocyte growth,augmenter of liver regeneration(ALR) is identified in regenerating rat and canine livers following partial hepatectomy and in the hyperplastic livers of weanling rats,but not in resting adult livers[2].Though initially recognized as a liver-specific,but not species-specific growth factor,ALR has been proved to distribute in many organs and tissues besides liver[3].Quantitative mRNA analysis,immunoblotting,and immunohistological studies revealed increased ALR expression in cirrhosis, hepatocellular carcinoma(HCC),and cholangiocellular carcinoma(CCC),indicating an important role for ALR in hepatocellular regeneration in liver cirrhosis,hepatocarci-nogenesis,and therefore of potential value in the clinical diagnosis of liver cirrhosis and cancer[4].Recently,ALR was identified as aflavin-dependent sulfhydryl oxidase with cytochrome c reductase activity and immune regula-tor,belonging to the new Erv/Alr family[5–7].It could control mitochondrial gene expression and the lytic activ-ity of liver-resident natural killer(NK)cells through regulating interferon-c(IFN c)[8,9].This secondary he-patotrophic protein,also known as hepatopoietin(HPO), can be obtained by gradient purification from hepatic stimulator substance(HSS)or genetic recombinant expression[10–12].However,none of these studies has characterized the rhALR both in vivo and vitron in detail. In this study,a highly effective prokaryotic recombinant human ALR(rhALR)expression system was established, and the biochemical features as well as functional char-acters of the rhALR were analyzed so as to provide experimental bases for developing it into a new potential product as hepatic growth factor for rescuing acute liver failure.Materials and MethodsConstruction and Recombinant Expression of rALRA378-bp coding region for rhALR was optimized for the prokaryotic expression system and avoiding unfavorable features,such as high GC content and undesirable folding domains,in reference to published data and the NCBI website(/molbio/)(sequence patented in China pending,No200710107893.X)[13].This dsDNA was synthesized by Genecore Company,Shanghai, China.The coding fragment with Eco RI,Xho I(underlined) adaptor,and enterokinase(EK)site(bold)was then obtained by PCR.The primers were:50TTG GAA TTC GAC GAC GAC GAC AAG ATG CGT ACC CAG CAG AAA30(upstream)and50TTG CTC GAG TTA GTC GCA AGA ACC GTC30(downstream).The Eco RI-and Xho I-treated fragment was ligated to linearized pET28a(+) vector.The recombinant plasmid was transformed into DH5a and identified with restriction enzyme digestion and DNA sequencing.The correct recombinant plasmid was transformed into BL21(DE3).Cells were grown to an optical density of0.5at600nm and were induced to express hALR by the addition of isopropyl-b-D-thiogalactoside (IPTG)(Amesco,America)to afinal concentration of1mM and incubation for4h.For gel-electrophoretic analysis of successful protein induction,bacteria from0.5-ml aliquots of the induced and uninduced cultures were pelleted and resuspended in100l l of protein sample buffer.Purification and Preparation of rALRBacterial cells were collected by centrifugation(10min at 4,000g),washed once with ice cold PBS and resuspended in PBS,and then frozen at-80°C.The preparation of the inclusion body was performed as described before[14]. Briefly,bacteria were thawed at room temperature,and the mixture was sheared on ice.After centrifugation for30min at10,000g at4°C,the pellet was resuspended in10ml of TED-buffer containing50mM Tris–HCl(pH8.0),2mM EDTA,0.1mM dithiothreitl,and5%glycerol.The sus-pension was sonicated on ice four times for15s.After the addition of sodium deoxycholate to afinal concentration of 0.05%(w/v),the mixture was incubated for15min at4°C and then centrifuged for30min at10,000g.Thefinal pellet was solubilized in50mM Tris(pH8.0)containing8.0M urea and2mM EDTA.The insoluble pellets were removed by centrifugation at15,000g for20min at4°C.The solubilized inclusion bodies were purified by FPLC pro-cedure(AKTA Explorer100,GE).Fusion protein was loaded onto a chelating Sepharose FF column with aflow rate of1–2ml/min.The refolding of the bound protein on the column was achieved by the use of a linear urea gra-dient treatment with aflow rate of0.1ml/min.The refolded protein was then eluted using renaturation buffer with a linear imidazole gradient from20to250mM.The imid-azole and salt were removed by the Sephadex G-25 column,and the fusion protein was cleaved by recombinant enterokinase(EK)(Shanghai Haiyi Biotech,China).His-tag was removed by chelating the Sepharose FF column. After being further purified with Q Sepharose FF column, the purified rhALR was subjected to SDS-PAGE,Western blot,molecular weight(MW)determination,and N-termi-nal amino acid analysis.Characterization of Recombinant rALRSDS-Polyacrylamide Gel Electrophoresis and ImmunoblottingSDS-polyacrylamide gel electrophoresis was performed using14%(w/v)tris–glycine gels under reducingconditions.Briefly,sheared bacterial pellets with or with-out induction were subjected to gel electrophoresis. Analytic gels were stained in a solution containing30%(v/ v)methanol,10%(v/v)acetic acid,and0.5%(w/v)Coo-massie brilliant blue,and distained in a solution containing 30%(v/v)methanol and10%(v/v)acetic acid.For immu-noblotting,the purified proteins were transferred to a positively charged NC membrane(Roche)according to standard protocols.The blotted membrane was blocked overnight with5%(w/v)non-fat dry milk in TBST[10mM Tris,pH7.6;150mM NaCl;0.1%(v/v)Tween20]at4°C. The membrane was subsequently incubated at room tem-perature with a1:2,000diluted rabbit polyclonal anti-ALR antibody(multiBIND biotech GmbH,Dortmund,Ger-many)in1%(w/v)non-fat dry milk in TBST for2h and then with a1:1,000diluted peroxidase(POD)-labeled anti-rabbit IgG as second antibody(Pierce)for45min.The membrane was rigorously washed after each incubation. Color development of POD-labeled secondary antibody was performed using DAB substrate(Pierce).MW Determination and N-SequencingSequentially purified rhALR was ionizedfirstly and entered into the mass analyzer(MALDI-TOF-MS,BrukerAutoflex II).The target protein was identified,and MW was deter-mined according to the difference of mass-to-charge(m/z) ratio.The N-terminal amino acid sequencing of the protein is performed based on the Edman degradation method with two steps.Thefirst step is to cut the amino acids one by one from the N-terminal by a chemical method;the second step is to separate protein by HPLC through separating and analyzing the amino acid.Both of the above determinations were accomplished by the Research Center for Proteome Analysis(RCPA),the Chinese Academy of Sciences,Shanghai.Human Hepatocyte Proliferation MTT Assay Immortalized human hepatocytes(L-02)were obtained from the cell bank of the Chinese Academy of Sciences, Shanghai.Cells were maintained with DMEM(Gibco) supplemented with10%FCS at378C with5%CO2 atmosphere.Hepatocytes were seeded into96-well plates at 59104/ml(100l l/well).The culture medium was replaced with a fresh medium containing0.5%FCS when 60%of cells were confluent.Sixteen hours later,fresh culture medium supplemented with different concentra-tions of rhALR ranging from0.16l g/ml to20l g/ml in double dilution was used for24h.Then cell culture was continued for4h together with10l l/well3-(4,5-dim-ethylthiazolyl-2)-2,5-diphenyltetrazolium bromide(MTT) (Amesco)solution(5mg/ml).The medium was replaced with DMSO(Sigma)100l l/well,and the plate was vor-texed for15s.Absorbance of each well was then detected at wavelength550nm.The experiment was triplicated and repeated three times.Rescue of CCl4Induced Acute Hepatic Injury in Mice Totally60male Chinese Kunming mice(age:6weeks; body weight:25–30g each,Shanghai Experimental Ani-mal Center)were randomly subdivided into four groups (n=15/group),i.e.,group A1:CCl4-induced injury with rhALR treatment(200l g/kg);group A2:CCl4-induced injury with rhALR treatment(40l g/kg);group A3:CCl4-induced injury without rhALR,but with same volume of normal saline instead;group A4:model mock group(with the same volume of olive oil instead of10%CCl4).For the CCl4-induced injury model,CCl4was injected intra-peritoneally at10ml/kg CCl4in10%CCl4olive oil solution.Four hours later,different dosages of rhALR or control normal saline were administrated intraperitoneally every12h.Eight hours after the fourth administration, blood samples were collected,and sera were obtained for alanine aminotransferase(ALT)and aspartate amino-transferase(AST)determination with a routine automatic biochemistry analyzer(model7600,Hitachi,Japan).His-tological study of slices of10%formaldehyde-treated liver tissue was performed with standard hematoxulin and eosin(HE)staining protocols.The study was repeated three times.Protective Effect of rhALR on CCl4-Induced Lethal Injury in MiceAnother60male Chinese Kunming mice(age:6weeks; body weight:25–30g each,Shanghai Experimental Ani-mal Center)were randomly subdivided into four groups (n=15/group):group B1:CCl4-induced lethal injury with rhALR treatment(200l g/kg);group B2:CCl4-induced lethal injury with rhALR treatment(40l g/kg);group B3: CCl4-induced lethal injury without rhALR treatment,but with the same volume of normal saline treatment instead; group B4:model mock group(with the same volume of olive oil instead of50%CCl4).CCl4was injected intra-peritoneally at10ml/kg CCl4in50%CCl4olive oil solution.The rhALR or control normal saline at the same volume was administrated once intraperitoneally1h later. The number of survival mice was recorded for each group over the following7days.Statistical AnalysisResults were expressed as mean±SD for MTT assay and liver function test.Statistical analysis was performedusing SPSS software(SPSS10.0).Student’s t-test was used to determine whether there were any statistical differences between the rhALR-treated group and con-trol.A P value of less than0.05was considered significant.ResultsConstruction of a pET28(a+)Plasmid Encoding Recombinant Human ALRWe cloned a378-bp human ALR coding region corre-sponding to126amino acids into the six His-tagged pET28(a+)vector.By our cloning strategy,an EK diges-tion site immediately before N-terminal of ALR was generated so as to remove the His-tag away from the His-ALR fusion protein.The pET28(a+)vector is a pBR322-based plasmid-containing promoter and transcription ter-minator signal from bacteriophage T7.The nucleotide sequence of the pET28(a+)derivative was confirmed by DNA sequencing.The encoded126-amino acid peptide has a calculated MW of15,027.9.Purification and Identification of Recombinant hALRFor expression of the rhALR,plasmid pET28(a+)-hALR was transformed into the bacterial host strain BL21 (DE3).T7RNA polymerase was induced by IPTG,and recombinant protein synthesis was monitored by SDA-PAGE under reducing conditions.The T7RNA poly-merase directed the synthesis of a fusion protein containing some purification tag with an apparent molecular weight of20,000.The synthesized fusion pro-tein was obtained largely in the insoluble fraction of the bacterial extract,which constitutes30%of total bacterial protein(Fig.1).Purification of rhALR was achieved by sequential detergent washes,Sepharose FF,and Sephadex G-25chromatography,EK digestion,and chelating chro-matography.The purified rhALR was subject to SDS-PAGE,indicating over90%purity,and had a molecular weight of around15,000after removing the unrequired tag in the target protein under reducing conditions (Fig.2).The accurate MW was15,029for monomer and 30,136for dimer by MALDI-TOF-MS determination.The MW for monomer is quite similar to the calculated one (Fig.3).Five N-terminal amino acids were M-R-T-Q-Q, exactly the same as anticipated for hALR.To demonstrate the immunological identity of the protein,an immuno-blotting analysis was performed(Fig.4).As expected,the expressed protein migrating at about15,000showed excellent antigenicity,indicating the excellent immuno-logical identify of the recombinant protein.Biological Assay of rhALR In Vitro and In VivoEffect of rhALR in Vitro on Human Hepatocyte Proliferation(MTT Assay)Recombinant hALR was found to have a strong stimulus on normal human hepatocyte proliferation as measured by MTT assay(see Fig.5).Also,a dose-dependent effect of rhALR on hepatocyte proliferation was observed.The significant stimulative effects appeared at10l g/ml and20 l g/ml.In Vivo Protective Effect of rhALR on Acute Hepatic Failure and Lethal Injury in CCl4-Induced MiceALT and AST were significantly elevated in all10%CCl4-treated mice compared to control,indicating acute liver injury occurred because of the toxic effect of CCl4on liver. Administration of rhALR at40l g/kg could decrease the ALR from441.7U/l to215U/l(P\0.01)and AST from 824U/l to478U/l(P\0.05).Application of rhALR at higher dosage(200l g/kg)could decrease the ALT level significantly,too(P\0.05).So rhALR at low(40l g/kg) or high dosage(200l g/kg)showed the positive effects on hepatocyte function improvement(Fig.6).Upon histolog-ical study,HE staining of the liver tissue section indicated heavy necrotic and degenerative change of hepatocytes in 10%CCl4-induced intoxication.In therhALR-treated Fig.1SDS-polyacrylamide gel electrophoresis of total protein extracts from BL21(DE3)transformed with pET28(a+)-hALR.The bacteria were harvested3h after induction of the T7RNA polymerase.Fusion protein prior to(1)and after induction(2)with IPTG was fractionated on a14%SDS-polyacrylamide gel under reducing conditions and detected by staining with Coomassie brilliant blue.Sizes of protein marker(KD)are given in the left margingroup,fewer necrotic hepatocyes were found,and some regenerative hepatocytes appeared (Fig.7).Among 50%CCl 4-intoxicated mice,40%of them survived after 7days without rhALR treatment,indicating the lethal toxic effect of CCl 4occurred for more than half of the mice,while a protective effect was observed since 80%of mice survived after administration of rhALR at 40l g/kg or at 200l g/kg (Fig.8).DiscussionMammalian augmenter of liver regeneration protein (ALR)was initially identified as a secondary growth factor involved in liver regeneration [1,10].The human gene was found to map to human chromosome 16[13].Its sulfhydryl oxidase activity and involvement in iron homeostasis have been recently demonstrated [3,6,7].ALR is expressed in a broad range of peripheral organs [3].The hepatocyte was the predominant liver cell in which ALR was synthesized and stored [2].ALR appears to be constitutively expressed in hepatocytes in an inactive form and released from the cells in an active form in response to partialhepatectomyFig.2SDS-polyacrylamide gel electrophoresis of rhALR during the process of purification.Sizes of protein marker (KD)are given in the left margin (from upper to lower:97,66,44,29,20,and 14).Lane 1–3:Samples of bacteria lysate (1),inclusion body (2),and washed inclusion body solution (3);lane 4:flow-through liquid from chelating Sepharose FF ne 5:Elution liquid from chelating Sepharose FF ne 6:Final purified sample after EKdigestionFig.3Molecular weight (MW)determination of rhALR by MALDI-TOF-MS.Sequentially purified rhALR was ionized firstly and entered in the Mass Analyzer (BrukerAutoflex II).Target protein was identified,and MW was determined according to the difference of mass-to-charge (m/z)ratio.The MW was 15,029for monomer and 30,136for dimer,with the homodimer predominatingnativelyFig.4Immunoblot of purified rhALR.Purified protein was prepared from IPTG-induced (lane 1–4)bacteria harboring expression plasmid pET28(a +)-hALR.Non-induced (lane 5)bacterial extract was used as negative control.Rabbit polyclonal anti-hALR antibody (multiBIND biotech GmbH,Dortmund,Germany)was used in 1%(w/v)non-fat dry milk in TBST for 2h and then with a 1:1,000diluted peroxidase(POD)-labeled anti-rabbit IgG as second antibody (Pierce)for 45min.Color development of POD-labeled secondary antibody was per-formed using DAB substrate (Pierce).Sizes of prestained protein markers (New England Biolabs)are on the left margin (upper:20KD,lower:14KD)and under other circumstances of liver maturation (as in weanling rats)or regeneration [2].Previous study supports that recombinant ALR has the ability to stimulate DNA synthesis of adult rat hepatocytes in primary culture and HepG2cells in vitro [15,16].Recombinant ALR plasmid DNA injection was helpful in relieving acute hepatic injury and hepatic failure by promoting hepatic cell proliferation and reducing the level of AST and ALT in CCl 4-intoxi-cated rats [17].Hepatic ALR mRNA expression was higher in liver disease patients than in non-liver disease controls,and a correlation was found between serum ALR values and hepatic levels of ALR mRNA [4].ALR was producedby and released from the liver at the time of hepatic injury.Thus,circulation of ALR is also regarded as the markers for liver cirrhosis and HCC [4].Besides the effect as hepatocyte mitogen or potential marker for liver cirrhosis and HCC,ALR induces hepatocyte regeneration via inhi-bition of liver NK cells,since cytotoxicity of liver NK cells against regenerating hepatocytes has been reported as a possible mechanism of regeneration failure in fulminant hepatitis [8,9].The inhibition of the liver-resident NK cells by ALR is achieved via reduction of IFN c by liver resident NK.The administration of IFN c in 70%hepatectomized rats is followed by a significant reduction both of the mitochondrial transcription factor A expression and of liver regeneration,indicating ALR as a growth factor and as an immunoregulator by controlling,through IFN c levels,the mitochondrial transcription factor A expression and the lytic activity of liver-resident NK cells [8,9,18,19].Though ALR was first identified and cloned by Hagiya et al.in 1994[10],and since then more and more studies have focused on the distribution [3,4],functional identi-fication [5,8,9],crystal structure analysis [6],as well as recombinant expression as mentioned above [12,15–17],the low expression efficiency,incomplete characterization of recombinant human ALR (rhALR),and the extraordi-nary effect of rhALR on liver regeneration motivated us to establish a high effective expression system and then per-form a serial of identification and functional studies so as to provide a solid basis for future clinical application of rhALR.The cNDA sequence of human ALR used in this study had been especially optimized to prokaryotic expression,the purification system,and its biological activity withtheFig.5The effect of rhALR on hepatocyte proliferation by MTT.Immortalized human hepatocytes (L-02)were obtained from the cell bank of the Chinese Academy of Sciences,Shanghai.Cells were maintained under standard condition and seeded into 96-well plates at 59104/ml (100l l/well).The culture medium was replaced with a fresh medium containing 0.5%FCS when 60%cells were confluent,and fresh culture medium supplemented with different concentrationsof rhALR ranging from 0.16l g/ml to 20l g/ml in double dilution were used 16h later.Then cell culture was continued for 4h together with 10l l/well MTT (Amesco)solution 24h later.The medium was replaced with DMSO (Sigma),and the plate was vortexed for 15s.Absorbance of each well was then detected at wavelength 550nm.The experiment was triplicated and repeated threetimesFig.6ALT and AST levels of CCl 4-intoxicated mice with or without rhALR administration.Mice were injected intraperitoneally at 10ml/kg CCl 4with 10%CCl 4olive oil solution.Four hours later,different dosages of rhALR or control normal saline were administrated intraperitoneally every 12h.Eight hours after the fourth administra-tion,blood samples were collected,and sera were obtained for alanine aminotransferase (ALT)and aspartate aminotransferase (AST)deter-mination with routine automatic biochemistry analyzer (model 7600,Hitachi,Japan)addition of EK digestion sites for cutting six His-tags away from the recombinant fusion protein (patent pending).The N-terminal amino acid sequence by HPLC,molecular weight by MALDI-TOF-MS,and antigenicity identity by immunoblotting analysis confirmed the identity of the recombinant human ALR as expected.Also,as anticipated,native rhALR was a homodimer since two MWs (15,029and 30,136)were obtained with 30,136(homodimer)pre-dominating (Fig.3),similar to the result by Giorda and Hagiya [13].Sequential detergent washes and gel-filtration as well as chelating chromatography helped to obtain rhALR with high purity (more than 90%),as was shown by SDS-PAGE under reducing conditions (Fig.2).The recombinant ALR could stimulate the proliferation of the human hepatocyte dose dependently,similar to the research on rat hepatocytes [15].The significant stimulation effects appeared at 10l g/ml and 20l g/ml,with proliferation activity more than doubled,indicating the strong mitogen-like effect of rhALR on hepatocytes in vitro.We used 10%CCl 4and 50%CCl 4injections to induce acute liverinjuryFig.7HE staining of the liver tissue from CCl 4-intoxicated mice with or without rhALR administration.Mice were injected intraper-itoneally at 10ml/kg CCl 4with 10%CCl 4olive oil solution.Four hours later,different dosages of rhALR or control normal saline were administrated intraperitoneally every 12h.Eight hours after the fourth administration,liver sections were prepared from the mice and then HE staining was performed.In CCl 4-intoxicated mice administrated normal saline,there was evident and extended centrilobular necrosis with the hepatocellular degeneration of the surrounding zones (a )(9100).The sinusoidal collagen reticulum framework around thecentrilobule collapsed,and the hepatocytes had disappeared with infiltration of abundant inflammatory cells and hyperplasia of Kupffer cells (b )(9400).In CCl 4-intoxicated mice administrated rhALR (40l g/kg),centrilobular necrosis was limited to individual lobules (c )(9100);hepatocyte degeneration of surrounding zones was not prominent,and regeneration of some hepatocytes was revealed (d )(9400).The effect of administration of rhALR at 200l g/kg was similar to that at 40l g/kg.The results indicate that rhALR improves liver structurehistologicallyFig.8Survival curve of 50%CCl 4-intoxicated mice with or without rhALR administration.Mice were injected intraperitoneally at 10ml/kg CCl 4with 50%CCl 4olive oil solution.The rhALR or control normal saline at the same volume was administrated once intraper-itoneally 1h later.The number of survival mice was recorded for each group over the following 7days.The results reveal that rhALR improves the survival rate of CCl 4-intoxicated miceand lethal intoxication,respectively,in mice.Two doses of rhALR were administrated in vivo.The administrations of rhALR intraperitoneally further confirmed that rhALR could decrease ALT(ALR at40l g/kg or200l g/kg)and AST(ALR at40l g/kg)significantly in10%CCl4-induced intoxicated mice;liver function improvement was also confirmed histologically.In50%CCl4-induced lethal intoxication,rhALR could improve the survival of intoxi-cated mice from40to80%,indicating the recombinant human ALR obtained in this study is obviously effective in rescuing CCl4-intoxicated mice via improving liver func-tion and stimulating hepatocyte proliferation.The initial1–3days were especially important for the survival in lethal intoxication,so the administration of the rhALR should be in the very early stage of the injury according to the sur-vival curve(Fig.8).In conclusion,we constructed a highly effective pro-karyotic recombinant expression system pET28(a+)-hALR.The recombinant hALR was effectly purified with serial detergent wash,gel-filtration,EK digestion,and chelating chromotography without His-tag.The rhALR had an anticipated molecular weight of15,029for monomers and30,136for homodimers,with the latter predominating natively.The rhALR was further identified and confirmed by N-terminal sequencing and immunoblotting.Human hepatocytes proliferated dose dependently in response to the rhALR in vitro.In CCl4-intoxicated mice,acute hepatic injury and lethal hepatic failure may be rescued by improving hepatocyte function when rhALR is adminis-trated.The application of recombinant protein has great potential with regard to the improvement of liver function failure upon acute liver injury.Acknowledgments We are especially grateful for the technical support of Dr.Tong-Hai Dou from Fu-Dan University regarding protein purification.References1.Pawlowski R,Jura J(2006)ALR and liver regeneration.Mol CellBiochem288(1–2):159–169.doi:10.1007/s11010-006-9133-7 2.Gandhi CR,Kuddus R,Subbotin VM,Prelich J,Murase N,RaoAS et al(1999)A fresh look at augmenter of liver regeneration in rats.Hepatology29(5):1435–45.doi:10.1002/hep.510290522 3.Tury A,Mairet-Coello G,Lisowsky T,Griffond B,Fellmann D(2005)Expression of the sulfhydryl oxidase ALR(Augmenter of Liver Regeneration)in adult rat brain.Brain Res1048(1–2):87–97.doi:10.1016/j.brainres.2005.04.0504.Thasler WE,Schlott T,Thelen P,Hellerbrand C,Bataille F,Lichtenauer M et al(2005)Expression of augmenter of liver regeneration(ALR)in human liver cirrhosis and carcinoma.His-topathology47(1):57–66.doi:10.1111/j.1365-2559.2005.02172.x5.Farrell SR,Thorpe C(2005)Augmenter of liver regeneration:aflavin-dependent sulfhydryl oxidase with cytochrome c reductase activity.Biochemistry44(5):1532–15416.Wu CK,Dailey TA,Dailey HA,Wang BC,Rose JP(2003)Thecrystal structure of augmenter of liver regeneration:a mammalian FAD-dependent sulfhydryl oxidase.Protein Sci12(5):1109–1118.doi:10.1110/ps.02381037.Lisowsky T,Lee JE,Polimeno L,Francavilla A,Hofhaus G(2001)Mammalian augmenter of liver regeneration protein is a sulfhydryl oxidase.Dig Liver Dis33(2):173–180.doi:10.1016/ S1590-8658(01)80074-88.Polimeno L,Margiotta M,Marangi L,Lisowsky T,Azzarone A,Ierardi E et al(2000)Molecular mechanisms of augmenter of liver regeneration as immunoregulator:its effect on interferon-gamma expression in rat liver.Dig Liver Dis32(3):217–225.doi:10.1016/S1590-8658(00)80824-59.Tanigawa K,Sakaida I,Masuhara M,Hagiya M,Okita K(2000)Augmenter of liver regeneration(ALR)may promote liver regen-eration by reducing natural killer(NK)cell activity in human liver diseases.J Gastroenterol35(2):112–119.doi:10.1007/ s00535005002310.Hagiya M,Francavilla A,Polimeno L,Ihara I,Sakai H,Seki Tet al(1994)Cloning and sequence analysis of the rat augmenter of liver regeneration(ALR)gene:expression of biologically active recombinant ALR and demonstration of tissue distribution.Proc Natl Acad Sci USA91(17):8142–8146.doi:10.1073/pnas.91.17.814211.Gatzidou E,Kouraklis G,Theocharis S(2006)Insights on aug-menter of liver regeneration cloning and function.World J Gastroenterol12(31):4951–495812.Sheng J,Yu H,Li J,Sheng G,Zhou L,Lu Y(2007)Cloning andexpression of the human augmenter of liver regeneration at low temperature in Escherichia coli.J Biochem Biophys Methods 70(3):465–470.doi:10.1016/j.jbbm.2006.11.00913.Giorda R,Hagiya MBB,Seki T,Shimonishi M,Sakai H,Mich-aelson J et al(1996)Analysis of the structure and expression of the augmenter of liver regeneration(ALR)gene.Mol Med 2(1):97–10814.Gao CF,Kong XT,Gressner AM,Weiskirchen R(2001)Theexpression and antigenicity identification of recombinant rat TGF-b1in bacteria.Cell Res11(2):95–100.doi:10.1038/sj.cr.7290073 15.Zhang YD,Zhou J,Zhao JF,Peng J,Liu XD,Liu XS et al(2006)Expression,purification and bioactivity of human augmenter of liver regeneration.World J Gastroenterol12(27):4401–4405 16.Liu Q,Yu HF,Sun H,Ma HF(2004)Expression of humanaugmenter of liver regeneration in pichia pastoris yeast and its bioactivity in vitro.World J Gastroenterol10(21):3188–3190 17.Zhang LM,Liu DW,Liu JB,Zhang XL,Wang XB,Tang LMet al(2005)Effect of naked eukaryotic expression plasmid encoding rat augmenter of liver regeneration on acute hepatic injury and hepatic failure in rats.World J Gastroenterol 11(24):3680–368518.Francavilla A,Vujanovic NL,Polimeno L,Azzarone A,Iaco-bellis A,Deleo A et al(1997)The in vivo effect of hepatotrophic factors augmenter of liver regeneration,hepatocyte growth factor, and insulin-like growth factor-II on liver natural killer cell functions.Hepatology25(2):411–41519.Polimeno L,Capuano F,Marangi LC,Margiotta M,Lisowsky T,Ierardi E et al(2000)The augmenter of liver regeneration induces mitochondrial gene expression in rat liver and enhances oxidative phosphorylation capacity of liver mitochondria.Dig Liver Dis 32(6):510–517.doi:10.1016/S1590-8658(00)80009-2。
