RCA_errorProne
RCA报告范例
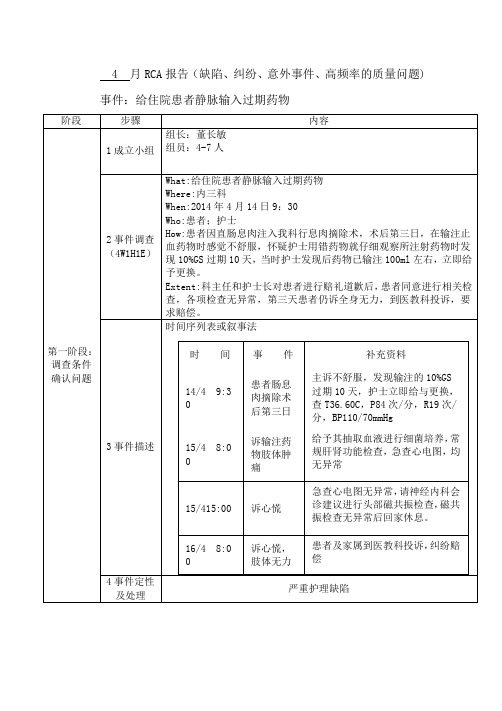
4 月RCA报告(缺陷、纠纷、意外事件、高频率的质量问题) 事件:给住院患者静脉输入过期药物
第二阶段:找出近端
原因确认操作程
序有无问题
为什么会这么做?别人是不是这样做的?操作者是否按程序执行?操
作程序本身有无问题?其他的环境情况、系统有无缺点?
1、护士责任心不足,未按规范操作执行三查七对
2、药房配送液体离有效期太近,护士没注意查对,清点
3、科室未建立近效期药品的管理机制
4、科室忽视大型液体管理,存放量、存放要求无具体说明
第三阶段:确认根本原因1头脑风暴
2鱼骨图
3原因树
4确认根本
原因
根据主要近端原因,反复问是什么情况让这种问题发生?一直追溯到
能找到从根本上解决问题的方案为止,关键是找系统和管理的问题
1、未规范药品物品的检查机制,护理部及护士长督查力度不够
2、未规定所有药品物品按先入先出的顺序摆放,优先使用近效期药品
3、未指定专人负责所有药品的效期审查;
4、护士安全意识差,责任心不强。
unre content-type error

未知的内容类型错误是指在网络通信过程中,服务器无法识别或不支持客户端所请求的内容类型。
这种错误可能会导致用户无法正常获取或显示所需的内容,给全球信息站的正常运行和用户体验带来不便。
为了更好地理解和解决未知的内容类型错误,本文将从以下几个方面进行讨论和分析:1. 未知的内容类型错误的原因2. 未知的内容类型错误对全球信息站和用户的影响3. 解决未知的内容类型错误的方法接下来,我们将逐一对以上三个方面展开阐述,希望能够帮助读者更好地理解和解决未知的内容类型错误问题。
1. 未知的内容类型错误的原因未知的内容类型错误通常是由以下几个原因所导致的:1) 服务器未配置正确的 MIME 类型:MIME 类型是指互联网媒体类型,它告诉浏览器如何处理特定类型的文件。
如果服务器未正确配置所请求内容的 MIME 类型,就会导致未知的内容类型错误。
2) 客户端请求的内容类型不受服务器支持:有时客户端请求的内容类型可能是服务器不支持的,这也会导致未知的内容类型错误。
3) 网络传输中出现错误:在网络传输过程中,数据包可能会丢失、损坏或被篡改,导致服务器无法正常识别所请求的内容类型。
2. 未知的内容类型错误对全球信息站和用户的影响未知的内容类型错误会对全球信息站和用户产生以下几方面的影响:1) 全球信息站排版错乱:未知的内容类型错误可能导致全球信息站排版错乱,使页面无法正常显示,影响用户的浏览体验。
2) 用户无法获取所需的内容:如果用户无法正常获取所需的内容,就会影响其对全球信息站的满意度和使用体验,甚至可能导致用户流失。
3) 全球信息站声誉受损:频繁出现未知的内容类型错误会影响全球信息站的信誉和声誉,降低用户对全球信息站的信任度。
3. 解决未知的内容类型错误的方法针对未知的内容类型错误,我们可以采取以下几种方法来解决:1) 检查服务器配置:我们需要检查服务器的 MIME 类型配置是否正确,确保服务器能够正确识别和处理客户端请求的内容类型。
易错PCR技术在淀粉酶定向进化中的应用

易错PCR技术应用于酶体外进化的研究进展沈思军1*,马春萍2,哈杰提2,马全磊1(1. 石河子大学动物科技学院,新疆,石河子,832003;2. 新疆西部牧业股份有限公司,新疆,石河子,832000)摘要:易错PCR技术是通过调整PCR反应条件,产生随机错配而引入多个突变。
由于该技术操作简单易行,广泛应用于酶工程改造研究中。
本文介绍了易错PCR技术的影响因素及近几年在酶工程改造中的应用进展。
关键词:易错PCR技术;影响因素;酶工程改造在体外进行酶分子改造的常用采用分子体外定向进化技术。
常用的方法有易错PCR (error-prone PCR)、DNA重排(DNA shuffling)、交错延伸法(staggered extension process, StEP)、随机引物体外重组(random-priming in vitro recombination, RPR)、易错滚环扩增法(error-prone rolling circle amplification, EP-RCA)、序列饱和诱变(sequence saturation mutagenesis, SeSaM)、随机插入/删除的链交换突变(random insertion/deletion strand exchange mutagenesis, RAISE)等[1]。
基本原理都是通过导入突变获得大量含有不同突变的酶,在不同环境条件下定向筛选有益突变菌株。
易错PCR是通过改变PCR条件,提高扩增产物的三大错配率,从而获得与原来不同的DNA序列或基因[2],其操作简便易行,成为体外酶工程应用较为广泛的技术之一。
本文就易错PCR 过程中影响突变率的因素和易错PCR技术在酶分子进化中的应用这两个方面,综述国内近几年的研究进展。
1. 影响易错PCR突变率的因素易错PCR技术是结合随机突变与定向筛选,利用Taq DNA 多聚酶不具有3`→5`校对功能的特点,在PCR反应中通过调节离子浓度等方法引入随机突变,在特定培养基或培养条件下定向选择所需酶的方法。
CAPMS使用说明

简介 .................................................................................................................................................... 错误!未定义书签。
目的 ................................................................................................................................................ 错误!未定义书签。
特点 ................................................................................................................................................ 错误!未定义书签。
运行环境......................................................................................................................................... 错误!未定义书签。
CAPMS各模块简介 ..................................................................................................................... 错误!未定义书签。
Common Data公用信息模块 .................................................................................................... 错误!未定义书签。
非平衡及平衡信号

关于非平衡(RCA)输出和平衡(XLR)输出两种输出模式究竟孰优孰劣的问题,从音响厂商一直到音响发烧友的人群里都存在着广泛的不同意见。
支持非平衡输出的人们认为这种输出方式声音最是细腻温柔,最有音乐味,XLR根本不是采用最顶级制作的RCA的对手,像JADIS,AUDIO NOTE,MBL,GOLDMUND这些Hi-End品牌持的就是这种看法,他们的产品都不甚支持平衡,许多机型只设RCA输出,音乐传真的态度则更是显明,连他家的旗舰级产品,售价在80万左右的KW前后级都没有平衡输入。
换作另一边,支持平衡输出的人们则认为这种输出方式声音最是饱满、动态更大、延展更宽,更有音乐味,像贵丰、FM,BOULDER,PASS,MARK,KRELL,CELLO,金嗓子等许多Hi-End 品牌都更多持这种看法,他们的许多产品都建议采用平衡输出方式,甚至只设平衡输出。
我觉得专业器材和非专业器材大可不必共享同一种标准。
专业器材比如一些监听器材要求在长距离传输时不受到外界杂波讯号的干扰,要求有更高的信噪比,平衡输出模式会更具有优势、更合适一些。
但在非专业领域,尤其在家庭使用当中,传输距离并不是一个突出问题,平衡输出的优势也就随之淡化了,平衡和非平衡输出,二者处于同一个基准之上,人们大可各取所需。
当然,在实际使用过程当中还是比较偏重于使用带RCA莲花头的线材而不是带XLR卡侬头的,这纯粹是由惯性使然的。
非平衡又叫单端输入或单端输出。
一个信号端和一个参考端(地)。
平衡又叫双端输入或双端输出。
两个信号端其中一个正向另一个反向。
电子平衡中还有“ 地” 。
平衡电路有两种:1、变压器平衡:它是真正意义上的平衡。
它有极高的共摸抑制比、输入输出完全隔离、无直流、无地线引起的交流声、接成非平衡时,反向输出端接地,增益无变化。
它的缺点是平衡变压器造价昂贵,频响较难做到平直。
2、电子平衡:用电子线路做成的平衡。
它的共摸抑制比一般不会高于集成电路的供电电压(约正负15 伏)。
RCA应用于抽血错误事件的分析报告

RCA应用于抽血错误事件的分析报告Abstract摘要Introduction引言The process of blood collection plays a crucial role in patient diagnosis and treatment. However, errors in this procedure can lead to adverse effects on patient care. Root Cause Analysis (RCA) is a structured approach widely used in various industries, including healthcare, to identify the underlying causes of errors and develop effective preventive measures. This report aims to analyze a specific incident of blood collection error using RCA and provide recommendations for improvement.Incident Description事件描述On [Date], a blood collection error occurred at [Hospital Name]. A 46-year-old female patient was scheduled for routine blood tests in the morning. The phlebotomist assigned to collect the blood sample misplaced the patient's identification label and mistakenly drew blood from another patient with a similar name. The error was discovered during the laboratory analysis, and the correct patient was quickly identified and notified. Fortunately, there were no immediate adverse consequences for either patient, but the incident raised concerns about the hospital's blood collection process.RCA ProcessRCA过程1. Team Formation团队组建To conduct the RCA, a multidisciplinary team consisting of a healthcare quality manager, the phlebotomist involved, the laboratory supervisor, and a representative from the hospital's information technology department was formed. The team had expertise in blood collection processes, quality management, and technology systems.2. Problem Statement问题陈述The team defined the problem as follows: "The misplacement of a patient's identification label during blood collection resulted in a sample being drawn from the wrong patient, indicating a flaw in the hospital's blood collection process."3. Data Collection数据收集The team collected relevant data, including incident reports, interviews with the phlebotomist and laboratory staff, and a review of the hospital's blood collection protocols and systems.4. Root Cause Analysis根本原因分析Based on the collected data, the team performed a thorough analysis using various techniques such as the 5 Whys, fishbone diagrams, and fault tree analysis. The RCA identified the following root causes of the blood collection error:a. Lack of Standard Operating Procedures (SOPs)The hospital did not have standardized SOPs for blood sample collection, including proper verification processes and labeling techniques. This lack of guidelines and training increased the risk of errors.b. Inadequate Staff Training and CommunicationThe phlebotomist involved in the incident had not received comprehensive training on blood collection protocols and lacked awareness of the importance of accurate identification and labeling. Additionally, there was a lack of effective communication channels between the phlebotomy department and the laboratory, making it difficult to rectify errors promptly.c. Technological LimitationsThe hospital's existing information technology system for patient identification and labeling was outdated, prone to errors, and did not provide real-time alerts when potential mismatches occur.5. Recommendations for Improvement改进建议Based on the identified root causes, the RCA team put forward the following recommendations:a. Development and Implementation of SOPsThe hospital should establish standardized SOPs for blood collection, emphasizing the importance of correct patient identification and proper labeling techniques. Regular training sessions should be conducted to educate healthcare staff on these procedures.b. Enhanced Staff Training and CommunicationThe phlebotomy department should provide comprehensive training to all phlebotomists, ensuring they understand and adhere to the SOPs. Additionally, effective communication channels should be established between the phlebotomy department and the laboratory to facilitate immediate error rectification.c. Upgrade of Information Technology SystemThe hospital should invest in an advanced information technology system capable of accurate patient identification and real-time alerts to prevent potential mismatch errors. The new system should integrate with the existing laboratory system and be user-friendly for healthcare professionals.Conclusion结论The RCA process was successfully applied to analyze a blood collection error incident at [Hospital Name]. By identifying the root causes, the report provides recommendations to prevent similar errors in the future.Implementing these recommendations will enhance patient safety, improve the quality of blood collection processes, and ensure accurate diagnostic results. Continuous monitoring, training, and communication are essential to maintain a high standard of care in blood collection procedures.。
病毒转基因技术原理 腺相关病毒

病毒转基因技术原理腺相关病毒
Addtheauthorandtheaccompanyingtitle
第一节腺相关病毒简介
l生物学特性 l致病性与免疫性
第一部分生物学特性
l血清型 l病毒结构 l病毒复制 l对理化因素的抵抗力
血清型
lAAV是从腺病毒的污染物1965年、人群或非人灵长类动物等 的组织中分离鉴定到的, l共鉴定了11个AAV血清型以及108个AAV变株variants, l通过签名PCRsignaturePCR技术,利用高度保守序列扩增Cap 基因的一小段可变区的DNA序列,以筛检是否是新的AAV分离 株,然后利用PCR技术获得新的AAV分离株的Cap或和Rep基 因的全长序列,在人类、非人灵长类动物、马、猪、牛、绵 羊、山羊和蛇等的不同组织器官,发现了大量的具有多样性的 AAV的基因组,但新分离的病毒株,尚未进行血清学分型,通称 为变株, lAAV不同血清型和变株的基因组结构相对较为保守,与AAV-2 型较为类似,不同血清型的衣壳蛋白结构中表位的差异, 50~80%AAV-2抗体在人群中检测出,
第二节腺相关病毒载体 转基因技术原理
l蛋白表达型腺相关病毒载体
―三成分包装系统 ―自我互补型AAV载体self-complementaryAAVvector,scAAV l基―因打反靶式型剪腺接相型关A病AV毒载载体体trans-splicingAAVvector,tsAAV lrA―AV衣的壳纯蛋化和白定修量饰型AAV载体
rca成果报告书模板 -回复

rca成果报告书模板-回复下面是一个基本的RCA成果报告书模板,根据你的要求,我将按照此模板回答问题:1. 引言引言部分介绍了RCA的目标和背景,包括为什么进行RCA以及对组织或项目的重要性。
2. 问题描述在这一部分,详细描述问题以及对组织或项目的影响。
可以提供数据或实际案例来支持问题描述。
3. 数据收集本节描述了收集的数据以及收集数据的方法。
可以包括调查问卷、观察、访谈等方法。
4. 问题分析这一部分分析了导致问题产生的根本原因。
使用工具如因果图、鱼骨图等来帮助分析。
5. 解决方案基于问题分析,提出解决方案。
可以包括具体的行动计划、时间表和责任分配。
6. 实施这一节描述了实施解决方案的步骤和过程。
可以包括培训、沟通和监督等。
7. 结果评估对实施解决方案后的结果进行评估。
可以使用数据和指标来评估解决方案的有效性。
8. 总结和建议在这一部分,总结报告结果,并提出对未来改进的建议。
以下是根据这个模板,对中括号内内容的一步一步回答:1. 引言引言:介绍RCA的目标和背景,包括为什么进行RCA以及对组织或项目的重要性。
在引言中,我们将介绍我们进行RCA的目标和背景。
我们意识到在我们的组织中存在一个重大问题,这个问题严重影响了我们的运营效率和客户满意度。
因此,我们决定进行RCA以找出导致这个问题的根本原因,并提出解决方案。
2. 问题描述问题描述:详细描述问题以及对组织或项目的影响。
可以提供数据或实际案例来支持问题描述。
我们的组织一直面临订单交货延迟的问题。
这个问题导致了客户不满意度的增加,严重影响了我们的声誉和销售业绩。
我们分析了过去一年的订单数据,发现近30的订单无法按时交付,这是一个令人担忧的数字。
这个问题使我们的供应链受到了严重打击,成本也因为返工和补偿付费而大幅增加。
3. 数据收集数据收集:描述收集到的数据和数据收集的方法。
可以包括调查问卷、观察、访谈等方法。
为了深入了解问题的根本原因,我们进行了一系列的数据收集。
rca滚环复制原理

rca滚环复制原理The RCA (Radio Corporation of America) drum is a unique and innovative mechanism used in early television and computer systems for the purpose of storing and reproducing images and data. The drum consists of a rotating cylinder with a magnetic coating, onto which the information is written and read. The principle behind the RCA drum's operation is based on the concept of magnetic recording and playback. When information is written onto the drum, the magnetic particles on the surface of the drum become aligned in a specific pattern, representing the data. When the drum rotates and the information is read, the magnetic particles' alignment is detected and converted back into the original data.The process of duplicating information using the RCA drum involves several key steps. First, the original data is written onto the drum using a magnetic recording head, which applies a magnetic field to the drum's surface, aligning the magnetic particles according to the data. Oncethe information is stored on the drum, it can be reproduced by rotating the drum and using a magnetic playback head to detect the alignment of the magnetic particles and convert it back into the original data. This process allows for the replication of the original information, as the magnetic alignment on the drum's surface can be read and used to recreate the data.One of the key advantages of the RCA drum's replication principle is its ability to store and reproduce information in a relatively compact and efficient manner. Unlike other storage and replication methods of the time, such as tape-based systems, the RCA drum offered a higher data density and faster access times, making it a popular choice for early television and computer systems. Additionally, the ability to duplicate information using the drum allowed for the creation of multiple copies of the same data, enabling widespread distribution and sharing of content.From a technical perspective, the RCA drum'sreplication principle relies on the precise alignment and detection of magnetic particles on the drum's surface. Theaccuracy and consistency of the magnetic recording and playback heads are crucial to ensuring the faithful reproduction of the original data. Additionally, the speed and stability of the drum's rotation play a critical rolein the successful replication of information. Anydeviations or inconsistencies in these factors can resultin errors or data corruption during the duplication process.In conclusion, the RCA drum's replication principle isa fundamental concept in the storage and reproduction of information in early television and computer systems. By leveraging the principles of magnetic recording and playback, the drum allows for the efficient duplication of data, enabling widespread distribution and sharing of content. The technical aspects of the replication process, including the precision of the recording and playback heads, as well as the stability of the drum's rotation, arecrucial to ensuring the accurate reproduction of theoriginal information. Overall, the RCA drum's replication principle represents a significant advancement in thehistory of data storage and replication technology.。
(完整word版)第一季度给药错误RCA报告

措施
方法
时间
责任人
高度重视
警示教育
1.召开质量安全会议,分析发生的原因;
2.用案例进行警示教育,强调医疗安全。
23/2
何莉莉
周丽梅
完善制度
规范流程
1.修订口头医嘱执行流程并进行培训学习;
2.争取医生配合,制定并启用急诊医嘱单,并定期检查。
4/3
何莉莉
第三阶段确认根本原因
鱼骨图
第三阶段确认根本原因
因果关联图
根据主要近端原因,反复问是什么情况让这种问题发生?一直追溯到能找到从根本上解决问题的方案为止,关键是找系统和管理的问题。
1.培训考核力度不够;2.护士不知晓药理知识;
3.使用前未经双人核对药物剂量;4.护士不知晓剂量单位换算;
第四阶
段
:
改进与评价
3月20日
何莉莉
王冬梅
加强督查力度
1.护士长随机检查带教工作完成情况;
2.护士长定期检查新入科及低年资护士工作落实情况并进行相应指导,提高其工作能力。
3-12月
何莉莉
效果评价
1.全科护士知晓口头医嘱执行流程,掌握急救药品的相关知识;
2.启用了急诊医嘱单;
3.改良了科内摆药盒,标识清楚,使用方便。
3.为什么护士会把鲁米那规格记错? 年轻护士急救药品知识缺乏,对药品规格、剂量、用法、注意事项及不良反应掌握不够; 护士不知晓计量换算常识; 急救药品标识不够醒目; 儿科抢救病人较少,儿科抢救用药使用频率低; 医生下口头医嘱不规范,经常无剂量、规格和用法。
4.为什么年轻护士急救药品知识缺乏? 培训考核力度不够,未制定培训目标; 培训方式及效果不佳,护士长督导不够; 急诊科年轻护士经验不足,急救药品使用频率低; 急诊科准入制度不健全,入科培训不到位。
LCR.常见PCHR错误码处理指导书
LCR 5.0 常见PCHR错误码处理指导书TD TECH Communication Technologies Co., Ltd.鼎桥通信技术有限公司x 侵权必究All rights reservedPrepared by拟制吴军、夏仕军拟制Date日期2010-12-30 Reviewed by审核XXXDate日期2010-12-30 Reviewed by审核XXXDate日期2010-12-30 Approved by批准XXXDate日期2010-12-30Revision record 修订记录系统维护部 产品版本 产品补丁集密级 V005R001 内部公开产品名称:TD-SCDMA RAN共48页 Date日期Revision Version 修订 版本 CR ID / Defect ID CR 号 Section Number 修改 章节 Change Description 修改描述 Author 作者2010-12-30 V1.0 第三章 重点局常见错误码处理建议,共包含30个常见错误码。
吴军 2011-01-10V2.0 按照维护文档模板进行修改 夏仕军 2011-01-17V3.0 全篇 根据评审意见修改,补充8个错误码。
夏仕军目录1.概述 .................................................... 错误!未定义书签。
1.1 使用对象 (1)1.2 适用范围 (1)1.3 背景知识 (2)1.4 PCHR日志分析工具 (2)1.5 文档主要内容 (3)2.LCR 5.0 PCHR日志分析及过滤方法 (4)2.1 单点故障PCHR日志分析方法 (4)2.2 KPI专题PCHR日志分析方法 (7)2.3 几种常见的PCHR日志过滤方法及规则 (9)2.3.1 Transdata工具PCHR日志导入过滤 (9)2.3.2 OMSTAR工具PCHR日志分析过滤 (9)2.3.3 Insight工具PCHR日志过滤保存 (12)2.3.4 几种常见的PCHR日志过滤规则 (15)3.LCR 5.0 PCHR错误码处理方法 (16)3.1 RR_ERR_RNCAP_RLC_FAILURE_SRB_RST (16)3.2 RR_ERR_RNCAP_RLC_FAILURE_TRB_RST (17)3.3 RR_ERR_IUB_INTERFACE_PERMANENT_RL_FAILURE (18)3.4 RR_ERR_RNCAP_RB_WAIT_UE_RB_CFG_TIMEOUT (19)3.5 RR_ERR_RNCAP_CU_WAIT_UE_RSP_TIMEOUT (20)3.6 RR_ERR_RNCAP_ALCFG_IUB_AAL2_FAILURE (21)3.7 RR_ERR_RNCAP_RC_REL_MACD_STATUS_ERR (22)3.8 RR_ERR_RNCAP_DEL_OLD_CCB (23)3.9 RR_ERR_RNCAP_RRC_UE_RSP_TIMEOUT (24)3.10 RR_ERR_IU_INTERFACE_RELOC_CANCELLED (25)3.11 RR_ERR_IU_INTERFACE_FAIL_IN_RADIO_INTERF_PROC (25)3.12 RR_ERR_RNCAP_RELOC_PHY_CH_RECFG_CMP_TIMEOUT (26)3.一三 RR_ERR_IU_INTERFACE_UNSPECIFIED_FAIL (26)3.14 RR_ERR_IU_INTERFACE_TIMER_RELOC_CMP_EXPIRY (27)3.一五 RR_ERR_RNCAP_RL_CAUSE_NODEB_TIMEOUT (27)3.16 RR_ERR_IUB_INTERFACE_CAUSE_RADIO_NW_UNSPECIFIED (29)3.17 RR_ERR_UU_INTERFACE_INVALID_CFG_ERR_NULL_TYPE (30)3.一八 NBM_CRA_CELL_RR_CRM_FAIL (31)3.19 RR_ERR_RNCAP_RB_CU_OVERLAP_BACK (31)3.20 RR_ERR_RNCAP_RRC_MAIN_ABNORMAL_ERR (32)3.21RR_ERR_IUB_INTERFACE_REQUESTED_CONFIGURATION_NOT_SUPPORTED . 333.22 RR_ERR_RNCAP_HHO_PHYCH_RECFG_TIMEOUT (34)3.23 RR_ERR_UU_INTERFACE_PH_CH_FAIL_ERR_NULL_TYPE (35)3.24 RR_ERR_IU_INTERFACE_NO_RSRC_AVAIL (36)3.25 RR_ERR_RNCAP_ALCFG_IU_AAL2_FAILURE (36)3.26 RR_ERR_RNCAP_ALCFG_IUB_AAL2_FAILURE (37)3.27 RR_ERR_IUB_INTERFACE_TRANSP_RESOURCE_UNVAILABLE (37)3.28 L2ERR_FPMDC_TR_SYN_NO_RESPONSE_AFTER_MAX_RETRY (38)3.29 RR_ERR_UU_IRCFC_PROTCL_ERROR_ERR_BEGIN (38)3.30 RR_ERR_UU_IRHFC_PROTCL_ERR_BEGIN (39)3.31 RR_ERR_IU_INTERFACE_REL_DUE_TO_UE_GEN_SIG_CONN_REL (40)3.32 RR_ERR_IU_INTERFACE_OM_INTERVENTION (40)3.33 RR_ERR_IU_INTERFACE_REQUESTED_INFO_NOT_AVAIL (41)3.34 RR_ERR_IU_INTERFACE_UNKNOWN_TARGET_RNC (41)3.35RR_ERR_IU_INTERFACE_RELOC_FAIL_IN_TARGET_CN_RNC_OR_TARGET_SYS (42)3.36 NBM_CRA_CELL_RR_FAIL (42)3.37 NBM_IUB_RESET (43)3.38 RR_ERR_RNCAP_CU_CELLFACH_NOT_FIND_PROC (43)图目录图1-1提交PCHR错误码技术咨询单............................................................ 错误!未定义书签。
RCA根本原因分析法

概念
• 根本原因:导致医疗护理执行失效,或其 结果不如预期最基本的原因。 • 根本原因分析:找出造成潜在执行偏差的 最基本或有因果关系原因的流程。 • 执行偏差: • 包括造成非预期及非如所愿的不良结果, 包括警讯事件(sentinel events),也可用于 探索迹近错失(near miss)发生的原因, 作为改善措施重新设计时的依据。
RCA与品质改善工具结合程序图
事故发生
事件发生
品质工具运用
WHAT 界定问题 ?
WHY找原因 ? <人为因素><系统缺乏>
问题描述法
防 止
流程图/鱼骨图 /因果树
HOW
•剖析原因拟定对策
流程解构法( SIPOC )
ACTION
•执行改善对策与稽核管制
管制表
第一阶段 :进行RCA前的准备
• • • • • • • • • 对于严重之异常事件或警讯事件 步骤一: 组织一个小组(Organize a team) 相关流程之一线工作人员 审慎考虑是否接收与事件最直接的关系人 最好不超过十人,必要时可多加开放 成员要求-具批判性观点,并有优秀的分析技巧 ‧组织者: RCA运作的主要负责人 ‧负责人:与事件相关之专业知识且能主导团队运 作 • 组织RCA小组
进行RCA的时机-异常事件严重度评估 准则
结果
死亡 数周 一年 数次 极重度伤 害 重度伤害 中度伤害 无伤害或 轻度伤害
1 1
1 1
2 2
3 3
3 4
频 率
1-2年 一次 2-5年 一次 5年以 上
1 1
2
2 2
3
2 3
3
3 4
4
4 4
CS8900A中文数据手册 中文部分翻译

Байду номын сангаас 目录
4.10.11 I/O 模式下轮询 CS8900A....................................................................... 15 5.2 基本接收操作........................................................................................................ 17
5.2.1.1 数据包................................................................................................ 17 5.2.1.2 帧........................................................................................................ 17 5.2.1.3 传送.................................................................................................... 18 5.2.2 接收配置...................................................................................................... 18 5.2.2.1 配置物理接口.................................................................................... 19
asserrionerror的使用方法

Asserrionerror的使用方法一、什么是AssertionError?AssertionError是Python中的一种异常类型,通常在使用assert语句时发生。
assert语句用于检查程序的某个条件是否为真,如果条件为假,则会引发AssertionError异常。
AssertionError通常用于程序中的断言检查,用于检查程序中的逻辑错误。
二、assert语句的基本语法在Python中,assert语句的基本语法如下所示:assert expression [, arguments]其中,expression是一个返回布尔值的表达式,如果表达式的值为True,则程序会继续执行;如果表达式的值为False,则会引发AssertionError异常。
arguments是可选的,用于指定当assertion 失败时输出的信息。
三、使用示例下面是一个简单的示例,演示了assertion的基本用法:1. 使用assert语句检查一个条件:```pythonx = 10assert x == 10, "x不等于10"print("x的值为10")```在这个例子中,如果x的值不等于10,那么会引发AssertionError异常,同时会输出 "x不等于10";如果x的值等于10,那么程序会继续执行,输出"x的值为10"。
2. 使用assert语句在函数中进行逻辑检查:```pythondef divide(x, y):assert y != 0, "除数不能为0"return x / y```在这个例子中,如果y的值为0,那么会引发AssertionError异常,同时会输出 "除数不能为0";如果y的值不为0,那么程序会继续执行,返回x/y的值。
3. assert语句的启用禁用在Python中,可以通过命令行参数 -O 来禁用assert语句,例如:```shpython -O test.py```这会导致Python忽略所有的assert语句。
贴片机常见错误代码

贴片机常见错误代码Emergency stop SW pccurred.元件废弃异常。
Failed in the Lar recog angle error.激光识别角度发生异常。
R1-Head failed to relea component.在R1-Head中,带回了贴片元件。
The gotten nozzle width does not meet that of le.测定的吸嘴宽度与设置的吸嘴宽度不一致。
Feeder Flow Seneor detected.E051001E109132开倒车在线翻译英译汉E109134The part abandonment is abnormal.Error was detected in checking if any nozzles attached.E242508E249001E291004Axis movement error (Z axis driver error)轴浮动错误。
(Z轴驱动错误)The origin point return is non-completion,plea operate again after compe E109220橙子的英文检查吸嘴安装,发生错误。
Return to home not yet been completed.没有执行返回原点。
Failde to originate,becau of Feeder-Floating.Plea retry.因传送带浮动,返回原点失败,请再次执行。
Failde t北方民族大学怎么样o detect the home of Theta-axis,plea retry origination.因为查出θ轴原点错误,返回原点失败,请再次执行。
E500302E500602E501999E509606E301028E301012merrychristmas是什么意思中文E301015E541502E610026藏语翻译E610200E611030E509701E520231E520238E541132E615065E616049发生紧急停止。
rca单端转平衡电路

rca单端转平衡电路英文回答:Single-Ended to Balanced Circuit.A single-ended to balanced circuit converts a single-ended signal into a balanced signal. A balanced signal is a differential signal where the signal is carried on two conductors, with the signal inverted on one conductor relative to the other. This type of signal is often used in audio applications to reduce noise and interference.There are several different ways to convert a single-ended signal to a balanced signal. One common method is to use a transformer. A transformer is a passive device that consists of two coils of wire that are magnetically coupled. When a signal is applied to one coil, it creates a magnetic field that induces a signal in the other coil. The ratio of the number of turns on the primary coil to the number of turns on the secondary coil determines the gain of thetransformer.Another method for converting a single-ended signal to a balanced signal is to use an operational amplifier (op-amp). An op-amp is an active device that can be used to amplify and condition signals. Op-amps can be used to create a variety of circuits, including single-ended to balanced converters.Single-ended to balanced circuits are used in a variety of applications, including audio, video, and data transmission. They are an important tool for reducing noise and interference in these applications.中文回答:单端转平衡电路。
RCA分析+网络安全事件RCA

RCA分析+网络安全事件RCA简介RCA(Root Cause Analysis)是指通过逐步分析和追溯,找到一个问题最根本的原因。
网络安全事件RCA特指对网络安全事件进行根本原因分析。
1. 事件背景网络安全事件是指发生在网络环境中的各种安全问题,包括但不限于黑客攻击、数据泄露、恶意软件传播等。
这些事件对个人、组织以及整个社会造成了严重的影响。
进行网络安全事件的RCA 有助于找出问题的根本原因,并采取相应的对策来预防未来的类似事件。
2. RCA过程RCA过程通常包括以下几个步骤:2.1 收集数据和证据首先需要收集与事件相关的全部数据和证据,包括网络日志、系统日志、设备配置等。
这些数据和证据可以帮助我们更全面地了解事件发生的过程和细节。
2.2 分析事件在收集到足够的数据和证据后,需要对事件进行仔细分析。
这包括分析攻击的方式、攻击者所使用的工具和技术、目标系统的漏洞等。
通过分析这些信息,可以更好地理解事件是如何发生的。
2.3 确定根本原因在分析事件的基础上,需要进一步确定事件的根本原因。
根本原因通常是导致事件发生的一个或多个核心问题。
比如,一个网络安全事件的根本原因可能是网络设备配置不当、员工的安全意识培训不足等。
2.4 制定对策确定根本原因后,需要制定相应的对策来避免类似事件再次发生。
这包括修补系统漏洞、加强网络安全设备的配置、提高员工的安全意识等。
2.5 监控和评估制定对策后,需要持续监控和评估网络安全状况,确保对策的有效性。
同时,也需要不断研究和研究最新的安全威胁和攻击技术,以便及时更新对策。
3. 结论通过进行RCA分析网络安全事件,可以帮助我们找出问题的根本原因,并制定相应的对策来预防未来的类似事件。
一个系统的网络安全需要持续的监控和改进,只有不断地进行RCA分析并采取相应的措施,才能保证网络环境的安全性。
注:该文档为RCA分析网络安全事件的基本框架和流程,并不涵盖具体的技术细节和实例分析。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
One-step random mutagenesis by error-prone rolling circle amplificationRyota Fujii,Motomitsu Kitaoka*and Kiyoshi HayashiNational Food Research Institute,2-1-12Kannondai,Tsukuba,Ibaraki,305-8642,JapanReceived August 18,2004;Revised and Accepted October 10,2004ABSTRACTIn vitro random mutagenesis is a powerful tool for altering properties of enzymes.We describe here a novel random mutagenesis method using rolling circle amplification,named error-prone RCA.This method consists of only one DNA amplification step followed by transformation of the host strain,without treatment with any restriction enzymes or DNA ligases,and results in a randomly mutated plasmid library with 3–4mutations per kilobase.Specific pri-mers or special equipment,such as a thermal-cycler,are not required.This method permits rapid prepara-tion of randomly mutated plasmid libraries,enabling random mutagenesis to become a more commonly used technique.INTRODUCTIONRandom mutagenesis,coupled with genetic selection or high-throughput screening,is a technique for developing enzymes with novel properties,including altered substrate specificity,enantioselectivity,stability and reaction specificity (1,2).This technique has the advantage of enabling the development of new enzymatic properties without a structural understanding of the targeted enzyme,and often yields unique mutations that could not have been predicted.In addition,further improvements can be expected by repeating the mutagenesis and selection (screening)processes in a manner mimicking Darwinian evolution.This approach,called directed evolution,is a principle method for biomolecular engineering (1–3).The most commonly used random mutagenesis method is error-pronePCR(4),whichintroducesrandommutationsduring PCR by reducing the fidelity of DNA polymerase.The fidelity of DNA polymerase can be reduced by adding manganese ions or by biasing the dNTP e of the compromised DNA polymerase causes mis-incorporation of incorrect nucleo-tides during the PCR reaction,yielding randomly mutated pro-ducts.To convert the product to a suitable form for transforma-tion of a host strain,at least three steps are required:digestion of the product with restriction enzymes,separation of the frag-ments by agarose gel electrophoresis and ligation into a vector.Although these steps do not constitute special techniques,they require almost an entire day of handling time.Further,the liga-tion step can sometimes be troublesome because low ligationefficiency can cause loss of the library.For these reasons,it is desirable to simplify these steps.Another useful random mutagenesis method is the bacterial mutator strain method (5).The most popular mutator strain is Escherichia coli XL1-Red (Stratagene,La Jolla,CA),which lacks three of the primary DNA repair pathways,MutS ,MutD and MutT ,resulting in a random mutation rate $5000-fold higher than in wild type.The protocol for using the mutator strain is composed of two steps:transformation of the mutator strain and recovery of the mutant from the transformant.This protocol is much simpler than error-prone PCR,and a ligation step is unnecessary.However,the mutation frequency is low under the standard conditions (0.5mutations per kilobase)(5),and a cultivation period longer than 24h is often required for introducing multiple mutations.Rolling circle amplification (RCA)(6–8)is an isothermal method that amplifies circular DNA by a rolling circle mechan-ism (9),yielding linear DNA composed of tandem repeats of the circular DNA sequence.This method has several advan-tages over conventional methods for amplifying DNA,such as PCR.For example,it does not require specific primers because random hexamers can be used as a universal primer for any template (10),nor does it require a thermal-cycler because the amplification reaction proceeds at a constant temperature.In addition to the ease of amplifying circular DNA,RCA products have a unique feature in that they can be used for direct trans-formation of E.coli (Fujii,R.,Kitaoka,M.and Hayashi,K.,manuscript submitted)and yeast (11),yielding re-circularized template DNA in the transformants.Therefore,the amplified product can be used directly to transform a host strain.We here describe the ‘simplest’random mutagenesis method using RCA,named error-prone RCA.This method consists of only one RCA step followed by direct transformation of the host strain,and yields mutants with an adequate mutation frequency for in vitro evolution experiments (3–4mutations per kilobase).No restriction enzymes,ligases,specific primers or special equipment such as a thermal-cycler are required.Therefore,this method is much more convenient than any other random mutagenesis methods.This method will enable random muta-genesis to become a more commonly used technique.MATERIALS AND METHODS MaterialsE.coli strains TOP10[F ÀmcrA D (mrr -hsdRMS -mcrBC )f 80lacZ D M15D lacX 74recA 1deoR araD 139D (ala -leu )7697*To whom correspondence should be addressed.Tel:+81298388071;Fax:+81298387321;Email:mkitaoka@nfri.affrc.go.jpNucleic Acids Research,Vol.32No.19ªOxford University Press 2004;all rights reservedNucleic Acids Research,2004,Vol.32,No.19e145doi:10.1093/nar/gnh147Published online October 26, 2004 at :: on May 19, 2011 Downloaded fromgalU galK rpsL (Str R )endA 1nupG ]and DH5a [F Àf 80lacZ D M15D (lacZYA-argF )U169deoR recA 1endA 1hsdR 17(r K Àm K +)phoA supE 44l Àthi -1gyrA 96relA 1]were purchased from Invitrogen (Carlsbad,CA)and TaKaRa (Shiga,Japan),respectively.f 29DNA polymerase was purchased from New England Biolabs (Beverly,MA),and the restriction enzyme BamHI was purchased from TaKaRa.TempliPhi 100DNA amplification kit was purchased from Amersham Biosciences (Piscataway,NJ).Ampicillin sodium salt and ceftazidime pentahydrate were purchased from Nacalai Tesque (Kyoto,Japan)and Sigma (St Louis,MO),respectively.The MinElute Reaction Cleanup and QIAprep miniprep kits were purchased from QIAGEN (Hilden,Germany).Error-prone rolling circle amplificationRCA was performed using the TempliPhi 100DNA amplifica-tion kit,which has a sample buffer containing random hexa-mers that prime DNA synthesis nonspecifically;an enzyme mix containing f 29DNA polymerase and random hexamers,and a reaction buffer containing deoxyribonucleotides.Although the exact composition of the TempliPhi kit is not known,the RCA reaction can be reproduced using f 29DNA polymerase and exonuclease-resistant random hexamers with thiophosphate linkages for the two 30terminal nucleotides (10).In a total volume of 10m l,the final concentrations were 1U/m l of f 29DNA polymerase and 4pmol/m l of exonuclease-resistant random hexamers in 50mM Tris–HCl buffer (pH 7.5),containing 10mM MgCl 2,10mM (NH 4)2SO 4,200ng/m l BSA,4mM DTT,0.2mM dNTP and template DNA (data not shown).As a template for the RCA reaction,we used purified pUC19dissolved in water or E.coli TOP10harboring pUC19,a colony of which was picked from a Luria–Bertani (LB)medium plate using a pipette tip and suspended in 50m l of 10mM Tris–HCl buffer (pH 8.0)containing 1mM EDTA.An aliquot (0.5m l)of the latter was mixed with 5m l of sample buffer,and the mixture was heated at 95 C for 3min to denature the plasmid and to lyse the cells.The sample was cooled to room temperature immediately.The amplification reaction was started by adding a premix from the TempliPhi kit of 5m l of reaction buffer,0.2m l of enzyme mix and 1m l of MnCl 2(0–20mM),followed by incubation at 30 C.The mix-ture was subsequently heated at 65 C for 10min to inactivate the enzyme,and a 5m l aliquot of the product was purified by using the MinElute Reaction Cleanup kit.The amount of amplified DNA was estimated by measuring its absorbance at 260nm with a NanoDrop ND-1000spectrophotometer (NanoDrop Technologies,Rockland,DE).Transformation of E.coli with RCA productElectrocompetent E.coli (50m l)were electroporated with a 1m l aliquot of the RCA product using an E.coli Pulser (Bio-Rad,Hercules,CA)in a 0.1cm electrode cuvette at a voltage setting of 1.8kV.The cells were incubated in 1ml of SOC medium at 37 C for 1h while reciprocal shaking at 160r.p.m.and then plated onto a LB plate containing 20ng/m l ampicillin sodium salt.The plate was incubated at 37 C for 16h.Characterization of plasmid in the transformantColonies on the plate were inoculated into LB medium containing 20m g/ml ampicillin sodium salt,and were incu-bated at 37 C overnight.Plasmid DNA was isolated from the culture medium using a QIAprep miniprep kit.A 1m l aliquot of the isolated plasmid (approx.100ng/m l)was digested with BamHI,and both the digested and undigested plasmids were analyzed by agarose gel electrophoresis.All plasmids of correct size on the agarose gels were sequenced using a CEQ-2000DNA Analysis System with a DTCS quick start kit (Beckman Coulter,Fullerton,CA).To measure mutation frequency in the recovered plasmid,plasmid DNA sequence was determined using an M13(À47)primer.DNA sequences of the other regions were determined using forward primers corresponding to DNA positions 841,1358,1750,2351and 2661.Improvement of ceftazidime resistance of TEM-1b -lactamase using error-prone RCAPlasmid pUC19,which contains TEM-1b -lactamase as a mar-ker for ampicillin,was amplified by error-prone RCA in buffer containing 1.5mM MnCl 2in a volume of 10m l.The product was precipitated with 70%ethanol and used to transform E.coli DH5a in 1ml medium.To estimate the total number of trans-formants,a 5m l aliquot of medium was spread on a LB plate containing 20ng/m l ampicillin sodium salt,and the residual mediumwasspreadonaLB plate containing1ng/m lceftazidime pentahydrate,which completely inhibits the growth of E.coli containing wild-typepUC19(12).After 16hincubationat 37 C,colonies formed on the ceftazidime plate were inoculated into LB liquid medium containing 1ng/m l ceftazidime.The recovered plasmids were subjected to agarose gel electrophor-esis,and the DNA sequence of the TEM-1b -lactamase gene in each transformant was analyzed.RESULTSError-prone rolling circle amplificationRCA is a laboratory method to amplify circular DNA by the rolling circle mechanism,yielding linear DNA composed of tandem repeats of the circular DNA sequence (10).The RCA product has a unique feature in that it can be used for the direct transformation of E.coli ,yielding transformants containing a plasmid identical to the RCA template (Fujii,R.,Kitaoka,M.and Hayashi,K.,manuscript submitted).We therefore con-structed a simple and rapid method for introducing random mutations into plasmid DNA.Plasmid DNA was amplified by the rolling circle mechanism in the presence of manganese ions,which has been shown to reduce the fidelity of DNA polymerases and cause random mutagenesis during RCA.E.coli was directly transformed with the RCA product,resulting in colonies containing a randomly mutated plasmid library.Although the band mobility of the recovered plasmids on agarose gel electrophoresis was mostly identical with pUC19,there were some plasmids with lower mobility than pUC19.These plasmids with lower mobility could be multi-mers,which are circular DNAs having two or more repeated sequences of pUC19(Fujii,R.,Kitaoka,M.and Hayashi,K.,e145Nucleic Acids Research,2004,Vol.32,No.19P AGE 2OF5at :: on May 19, 2011 Downloaded frommanuscript submitted).We analyzed a total of 174clones by agarose gel electrophoresis,resulting in 25clones (14%)being identified as multimers.The proportion of the monomer to multimer was almost constantly independent of mutation frequency (data not shown).When the multimers were sequenced,the mutated residues were often overlapped wild-type residues.This is probably due to the multimeric structure,which contains at least two different plasmid DNA sequences.Therefore,we used only monomers in the following experi-ments characterizing mutations.Mutation frequencyWe found that the mutation frequency increased when the concentration of manganese increased or when the initial amount of template decreased (Table 1).A too high mangan-ese concentration,however,was found to reduce the efficiency of amplification (2mM MnCl 2for 25pg pUC19).The max-imum mutation frequency was 3.5–1.0mutations/kilobase.When we examined the relationship between mutation frequency and amplification rate,we found that,as the amplification reaction proceeded,the mutation frequency increased (Table 2).A considerable number of mutations were introduced after 8h of incubation,and the highest muta-tion frequency was obtained after 24h of incubation.Library sizeIn determining the size of the mutant library produced by the error-prone RCA method (Table 3),we found that,in the presence of 1and 1.5mM MnCl 2,2200and 8900colonies,respectively,were obtained from 1m l of the RCA product.These values were lower than that obtained under error-free conditions (38000),indicating that increasing the concentra-tion of MnCl 2decreased the numbers of colonies.This was due to a decrease in the yield of the RCA product and was inde-pendent of the transformation efficiencies (Table 3).This indi-cates that the effect of mutagenesis on the genes of the plasmid replication system may be low.Characterization of mutationsTo analyze the distribution and variation of mutations caused by error-prone RCA,we isolated seven mutated plasmids from the mutant library constructed in the presence of 1.5mM MnCl 2,and each was sequenced.When we examined the distribution of mutations (Figure 1and Table 4),we found that the mutation frequency in the region from nucleotides 800to 2500was slightly lower than that in other regions.Because this region encodes genes critical for the plasmid (nucleotides 867–1455is the origin of replication,and nucleo-tides 1626–2486is the ampicillin-resistance gene),some mutations in this region may have been lethal and wereTable 1.Effect of RCA conditions on mutation frequency pUC19(pg)MnCl 2(mM)Sequenced base pair Number of mutations Mutation frequency (mutations/kb)a 250505800250.5597610.2–0.225161168 1.3–0.525 1.5376613 3.5–1.0252—b —b —b 2.50.5727210.1–0.12.51440012 2.7–0.82501488410.2–0.22502279031.1–0.6Plasmid pUC19(2.5,25or 250pg)was amplified by TempliPhi 100DNA amplification kit with varying concentrations of MnCl 2(0–2mM)for 24h.The reaction mixture was heated at 65 C for 10min to stop the reaction,and E.coli TOP10was transformed with the amplified product.Plasmids were recovered from the isolated transformants,and their DNA sequence was analyzed using the M13(À47)forward primer.aStandard errors were calculated assuming that the values follow a Poisson distribution.bNo transformants obtained.Table 2.Correlation between reaction time and mutation frequency Reaction time (h)Sequenced base pair Number of mutations Mutation frequency (mutations/kb)a 0—b —b —b4601330.5–0.38696811 1.6–0.5243766133.5–1.0Plasmid pUC19(25pg)was amplified by TempliPhi 100DNA amplification kit with 1.5mM MnCl 2for 0–24h.The reaction mixture was heated at 65 C for 10min to stop the reaction,and E.coli TOP10was transformed with the ampli-fied product.Plasmids were recovered from the isolated transformants,and their DNA sequence was analyzed using the M13(À47)forward primer.aStandard errors were calculated assuming that the values follow a Poisson distribution.bNo transformants obtained.Table 3.Library size MnCl 2(mM)RCA product (ng/m l)Number of colonies Transformation efficiency (c.f.u./m g)0120380003·10512189004·1051.5922002·105Plasmid pUC19(25pg)was amplified by TempliPhi 100DNA amplification kit with varying concentrations of MnCl 2(0,1and 1.5mM)for 24h.The reaction mixture was heated at 65 C for 10min to stop the reaction,and E.coli TOP10was transformed with 1m l of the amplified product.Table 4.Types of mutation Mutation type Number of mutations Percentage in substitutions G to A,C to T 4366G to C,C to G 46G to T,C to A 711A to T,T to A 58A to G,T to C 58A to C,T to G 12Insertion 6Deletion 0Total71100Figure 1.Distribution of mutations.The seven clones produced by RCA in the presence of 1.5mM MnCl 2from 25pg pUC19were sequenced.Lines indicate mutations in the pUC19sequence.Types of mutations are described in Table 4.P AGE 3OF5Nucleic Acids Research,2004,Vol.32,No.19e145at :: on May 19, 2011 Downloaded fromtherefore excluded from the library.The transformation efficiency did not decrease much under error-prone conditions (Table3),however,indicating that the influence of the lethality was trivial for the mutant library.When we examined the diversity of mutagenesis(Table4), we found that the direction of the mutations was biased in favor of C to T and G to A mutations(66%),and that the transition/transversion ratio was2.7.Improvement of ceftazidime resistance of TEM-1b-lactamaseTo verify that the error-prone RCA method can be used for evolutionary experiments,we altered the substrate specificity of TEM-1b-lactamase using error-prone RCA.TEM-1 b-lactamase is an enzyme that hydrolyzes the b-lactam ring and is generally used as a marker for b-lactam antibiotics such as ampicillin.In contrast,this enzyme works poorly against third-generation cephalosporins,such as ceftazidime.We introduced random mutations into the TEM-1b-lactamase gene of pUC19using error-prone RCA in the presence of1.5mM MnCl2.The RCA product was used to transformE.coli DH5a,and mutants with high ceftazidime-hydrolyzing activity were selected.We found that10colonies grew on the LB plate containing1m g/ml ceftazidime,compared with 10000on the ampicillin plate.Of the10plasmids recovered from the ceftazidime plate,agarose gel electrophoresis showed that7had bands of the same size as pUC19,whereas the other 3showed lower mobility than pUC19,indicating that they were probably multimeric plasmid DNA(data not shown). The seven plasmids of the same size as pUC19were sequenced to identify mutations in the b-lactamase gene[Table5;(13)]. All seven plasmids had mutations at R164(to H,G or C)or at D179(to G),all of which are known to increase the ceftazi-dime resistance of TEM-1b-lactamase(14,15).Therefore, we have successfully used error-prone RCA to introduce mutations that altered the substrate specificity of b-lactamase,indicating the applicability of this method for in vitro evolution experiments.DISCUSSIONRandom mutagenesis is a powerful tool for altering the proper-ties of enzymes(1,2).In this study,we have developed a random mutagenesis method using the RCA technique.This method is composed of only one DNA amplification step, followed by direct transformation of the host strain.Target DNA was amplified by the RCA technique in the presence of manganese ions,which reduces thefidelity of DNA polymer-ase and causes the mis-incorporation of nucleic acids(4).This product is used to transform E.coli,yielding colonies that contain randomly mutated plasmids.Although we used E.coli as a host strain in this report,other hosts,including yeast(11)and probably any other host having a DNA recomb-ination system,may also be used.The prime advantage of error-prone RCA is its rapidity. This method consists of only one RCA step,indicating that it is much quicker than the conventional random mutagenesis methods,such as error-prone PCR or mutator strain method (Figure2).The error-prone RCA reaction mixture can be prepared within several minutes,followed by isothermal incu-bation,to yield an amplified DNA suitable for direct transfor-mation of a host strain.Specific primers for target DNA are not necessary because random hexamers can be used as the universal primer for any plasmid.Furthermore,setting of ther-mal-cycling conditions,as in PCR,is not necessary,because the RCA reaction proceeds under isothermal conditions. Therefore,it is no exaggeration to say that error-prone RCA is the simplest of all the random mutagenesis methods. Using error-prone RCA,we introduced random mutations into plasmid DNA at a frequency of up to3.5–1.0mutations per kilobase.This mutation frequency corresponds to almost one amino acid mutation per kilobase and is therefore appro-priate for in vitro evolution experiments(3).In addition,the mutation frequency could be controlled by varying the con-centration of manganese ions.This concentration,however, was limited to below2mM because excess MnCl2decreasedTable5.Mutationsof theTEM-1b-lactamasesequencein ceftazidime-resistant variants produced by error-prone RCAColony no.MutationDNA a Amino acid b1G485A R164H2C82T,G485A,C674T silent,R164H,A227V3T113G,C484G,G852A L40W,R164G,silent4C484T R164C5C273T,G485A,A511G,C526Tsilent,R164H,I173V,R178C6A530G D179G7A530G D179GPlasmid pUC19(25pg)was amplified by TempliPhi100DNA amplification kit with1.5mM MnCl2in10m l volume.After24h incubation at30 C, the reaction was stopped by heating at65 C for10min.The product was precipitated with70%ethanol,and each precipitate was dissolved in1m l water and used to transform E.coli DH5a by electroporation.The transformants were spread on a LB plate containing1ng/m l of ceftazidime and incubated for 16h at37 C.The plasmids were recovered from the colonies,their size was analyzed by agarose gel electrophoresis,and plasmids of the correct size were sequenced to identify mutations in the TEM-1b-lactamase gene.a DNA position based on the TEM-1b-lactamase sequence.b Amino acid position based on the standard numbering for class A b-lactamase(13). at :: on May 19, 2011 Downloaded fromthe RCA e of a modified f 29DNA polymerase without 30–50exonuclease activity may increase the mutation frequency (16).For example,introduction of an H61R muta-tion into f 29DNA polymerase was found to result in a 16-fold increase in mis-incorporation efficiency,as well as a 6-to 23-fold increase in polymerization efficiency.These findings indicate that both mutation frequency and yield may be improved by using the H61R mutant of f 29DNA polymerase.While all types of substitution mutations were found in error-prone RCA variants,the mutation direction of error-prone RCA with f 29DNA polymerase was biased in favor of C to T and G to A mutations (66%).In contrast,these mutations are less favored in error-prone PCR using Taq DNA polymerase (14%)(17).These findings indicate that error-prone RCA could give rise to a wide range of amino acid substitutions not observed in error-prone PCR.Mutations are introduced throughout the entire plasmid by error-prone RCA,as well as in mutator strain mutagenesis (5),and mutations in regions other than the targeted gene might cause unexpected effects.The transformation efficiencies of varying concentrations of MnCl 2were almost constant,however,indicating that these mutations did not have a dele-terious effect on the plasmid replication system.There are many successful reports for improving enzymatic properties by mutating the entire region of the plasmid DNA by mutator strain mutagenesis (18–20),and therefore,the influence of mutations in other regions was probably negligible.To verify that error-prone RCA can be used to alter enzy-matic properties,we used this method to increase the cefta-zidime resistance of TEM-1b -lactamase.The plasmid pUC19,which has TEM-1b -lactamase gene,was mutated by error-prone RCA,and E.coli DH5a transformed with the RCA product was cultured on a ceftazidime plate.Of the seven mutant pUC19plasmids with improved ceftazidime resist-ance,all had mutations at R164(to H,G or C)or D179(to G).Both of these amino acids are located at the root of the V -loop of the TEM-1b -lactamase structure,which forms part of the substrate-binding domain,and mutations in these residues are known to improve ceftazidime resistance (14,15).There-fore,we have demonstrated that error-prone RCA can be used for altering enzymatic properties,indicating the applicability of this method for in vitro evolution experiments.In conclusion,we have developed a simple method for constructing a randomly mutated plasmid library using RCA (error-proneRCA).ThismethodwascomposedofoneRCAstep followed by direct transformation of a host strain.A target plas-mid was amplified by fidelity-reduced RCA,and the host strain was transformed directly with the product to give a mutant library with up to 3.5–1.0mutations per e of this method will save considerable labor in the introduction of random mutants.This is the simplest protocol for the prepara-tionofarandomlymutated plasmid librarytoourknowledgeand will make random mutagenesis more common.ACKNOWLEDGEMENTSThis study was supported by a grant from the Bio-oriented Technology Research Advancement Institution.REFERENCES1.Arnold,F.H.,Wintrode,P.L.,Miyazaki,K.and Gershenson,A.(2001)How enzymes adapt:lessons from directed evolution.Trends Biochem.Sci.,26,100–106.2.Brakmann,S.(2001)Discovery of superior enzymes by directed molecular evolution.Chembiochem.,2,865–871.3.Reetz,M.T.and Jaeger,K.E.(1999)Biocatalysis—From Discovery to Application .Springer-Verlag Berlin,Berlin,Vol.200,pp.31–57.4.Leung,D.W.,Chen,E.and Goeddel,D.W.(1989)A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction.Techniques ,1,11–15.5.Greener,A.,Callahan,M.and Jerpseth,B.(1996)In Vitro Mutagenesis Protocols .Humana press,NJ.6.Fire,A.and Xu,S.Q.(1995)Rolling replication of short DNA circles.Proc.Natl A ,92,4641–4645.7.Liu,D.Y.,Daubendiek,S.L.,Zillman,M.A.,Ryan,K.and Kool,E.T.(1996)Rolling circle DNA synthesis:small circular oligonucleotides as efficient templates for DNA polymerases.J.Am.Chem.Soc.,118,1587–1594.8.Lizardi,P.M.,Huang,X.,Zhu,Z.,Bray-Ward,P.,Thomas,D.C.and Ward,D.C.(1998)Mutation detection and single-molecule counting using isothermal rolling-circle amplification.Nature Genet.,19,225–232.9.Kornberg,A.and Baker,T.(1992)DNA Replication,2nd edn .W.H.Freeman &Company,NY.10.Dean,F.B.,Nelson,J.R.,Giesler,T.L.and Lasken,R.S.(2001)Rapidamplification of plasmid and phage DNA using Phi 29DNA polymerase and multiply-primed rolling circle amplification.Genome Res.,11,1095–1099.11.Ding,X.,Snyder,A.K.,Shaw,R.,Farmerie,W.G.and Song,W.Y.(2003)Direct retransformation of yeast with plasmid DNA isolated fromsingle yeast colonies using rolling circle amplification.BioTechniques ,35,774–779.12.Gaytan,P.,Osuna,J.and Soberon,X.(2002)Novel ceftazidime-resistanceb -lactamases generated by a codon-based mutagenesis method and selection.Nucleic Acids Res.,30,e84.13.Ambler,R.P.,Coulson,A.F.W.,Frere,J.M.,Ghuysen,J.M.,Joris,B.,Forsman,M.,Levesque,R.C.,Tiraby,G.and Waley,S.G.(1991)A standard numbering scheme for the class A b -lactamases.Biochem.J.,276,269–270.14.Vakulenko,S.B.,Taibi-Tronche,P.,Toth,M.,Massova,I.,Lerner,S.A.andMobashery,S.(1999)Effects on substrate profile by mutational substitutions at positions 164and 179of the class A TEM pUC19b -lactamase from Escherichia coli .J.Biol.Chem.,274,23052–23060.15.Vakulenko,S.B.,Toth,M.,Taibi,P.,Mobashery,S.and Lerner,S.A.(1995)Effects of Asp-179mutations in TEM pUC19b -lactamase on susceptibility to b -lactams.Antimicrob.Agents Chemother.,39,1878–1880.16.de Vega,M.,Lazaro,J.M.and Salas,M.(2000)Phage f 29DNApolymerase residues involved in the proper stabilisation of the primer-terminus at the 30–50exonuclease active site.J.Mol.Biol.,304,1–9.17.Shafikhani,S.,Siegel,R.A.,Ferrari,E.and Schellenberger,V.(1997)Generation of large libraries of random mutants in Bacillus subtilis by PCR-based plasmid multimerization.BioTechniques ,23,304–310.18.Camps,M.,Naukkarinen,J.,Johnson,B.P.and Loeb,L.A.(2003)Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I.Proc.Natl A ,100,9727–9732.19.Henke,E.and Bornscheuer,U.T.(1999)Directed evolution of an esterasefrom Psueudomonas fluorescens .Random mutagenesis by error-prone PCR or a mutator strain and identification of mutants showing enhanced enantioselectivity by a resorufin-based fluorescence assay.Biol.Chem.,380,1029–1033.20.Bornscheuer,U.T.,Altenbuchner,J.and Meyer,H.H.(1998)Directedevolution of an esterase for the stereoselective resolution of a key intermediate in the synthesis of epothilones.Biotechnol.Bioeng.,58,554–559.P AGE 5OF5Nucleic Acids Research,2004,Vol.32,No.19e145at :: on May 19, 2011 Downloaded from。
