M6U3-advice_from_grandad_
从国自然放榜的结果来看,m6A真很热

从国自然放榜的结果来看,m6A真很热前几天国自然已经放榜了,中标的肯定很开心,没有中标的心情可能不太好。
但是不管怎么样,大家应该继续努力前进。
中标的应该好好计划开展项目工作,没有中标的需要好好修改,继续申请。
今年准备申请课题的是时候开始准备了。
国自然想不到热点?那就来看看m6A,无论是SCI论文还是国自然标书,都是可以看出m6A是比较热的,就刚放榜的国自然中,就有非常多m6A相关的课题中标了。
感兴趣的临床医生、博士、研究生可以考虑一下m6A, 同时准备开题的研究生也可以考虑一下研究m6A,说不定下一个中标的就是你。
如何用m6A为国自然、博硕研究生开题?利用m6A开题二继续领取6-甲基腺嘌呤(N6-methyladenosine,m6A)是指腺嘌呤(A)的第6 位氮(N)原子上发生的甲基化修饰。
m6A修饰是mRNA与lncRNA最常见的核苷酸修饰。
m6A修饰广泛分布于真核生物的RNA 中,m6A修饰调节RNA 成熟、剪切、转运、降解及翻译等代谢过程。
m6A修饰蛋白或者m6A结合蛋白的表达异常或m6A 修饰水平异常,可导致m6A 修饰相关RNA 代谢的异常,影响基因的表达,与很多疾病发生发展有密切关系,m6A成为了近年来的研究热点,很多国自然或者博士课题都想搭上该热点。
m6A修饰及代谢过程由以下三个部分完成。
甲基转移酶复合物(Writers):METTL3,METTL14,WTAP,RBM15,KIAA1429,HAKAI,ZC3H13去甲基化酶(Erasers):FTO,ALKBH5,ALKBH3结合蛋白(Readers):YTHDF1,YTHDF2, YTHDF3, YTHDC1, YTHDC2, HNRNPA2B1, HNRNPC, HNRNPG, IGF2BP1, IGF2BP2,IGF2BP3参考文献:The role of m6A RNA methylation in cancer (/10.1016/j.biopha.2019.108613)●快速抢发3+SCI●如何用m6A为国自然、博硕研究生开题?●利用m6A开题二继续领取● SCI狂人团队VIP会员●关于答疑时间的通知●如何通过数据挖掘,发现癌症治疗的靶标?●想不到课题?不存在的●论文修回后是否再次回到审稿人手中?取决于你的回复●数据挖掘特训上线●这样的文章,谁都可以做到呀● R代码很容得到,关键是数据不会准备,遇到问题看不懂●如何快速找到值得深入研究的分子?●一大波科研软件免费下载●作为临床医生,不能总靠生信发文章吧?●公共数据库找不到自己需要的数据,该咋办?●躺着也可以轻轻松松得到5篇SCI●学员报喜,再度发表两篇SCI●火山图的高级玩法● VIP会员专属课程再次上线●在GEO数据库随便找一张芯片就可以发SCI?●手把手教你利用WGCNA发文章● Look, 搞一篇SCI so easy●如何快速找到值得深入研究的分子?● SEER数据挖掘这样玩才过瘾的●急!急!急!快来抢发这个数据库●只会用TCGA构建ceRNA你就out了● WB一图多用,有多么严重?●这10个期刊也许是你想要的●甲基化跑不动,我们帮你跑●如何快速学习贝叶斯网状meta分析?●如何搞一张漂亮的列线图?●如何快速学习GEO数据挖掘?●这种发文套路,完全是看数据吃饭的●多张芯片数据合并视频(免费送)●环状RNA数据挖掘课程上线了● TCGA数据下载再也不会难到你●如何绘制两个模型的ROC,并且进行比较?●临床预测模型快速入门课程● GO富集分析这种图,你也可以呀● GEO数据挖掘的深度不够,有没有提高GEO数据挖掘的深度的方法呢?● 10篇SCI还不如一个英语成绩好的临床医生好找工作?●批量挖掘TCGA让发文速度更加快●批量挖掘TCGA临床数据的机会来了●批量挖掘TCGA miRNA●医生朋友,非肿瘤方向也可以进行临床预测模型呀● SEER数据新玩法,你还不来看看。
e-Freight Standard Operating Procedure BRU说明书

e-Freight Standard Operating Procedure BRU 0)IntroductionThis standard operating procedure is applicable for all export, import and transit shipments at Brussels Airport. No limitation is made in product types.1)Inventory of documents in the pouch2)Inventory minimum elements transmitted through e-MessagingFWB version 16 or higher. Mandatory & correct AWB & eAWB fields (include the rules from cargo IMP)FHL is not mandatory for eFreightFSU milestones⇨FSU FOH: Freight on hand / ready for check (goods were received at handlers warehouse where he takes liability for the goods but goods are not yet accepted for carriage) o GHA: is capable to send FOHo Not mandatory for eFreight, but will be required by the FF⇨FSU RCS: Received from shipper / transfer of responsibility at acceptance time, when goods are delivered ‘ready for carriage’o In the Netherlands, a specific paper for acceptance of eFreight export goods needs to be submitted by the GHA to the FF.Suggest: not to make a specific template for it⇨FSU DEP: Departure confirmationo At later stage in scopeo Currently not every forwarder’s system is able to display DEP contento Departure confirmations are sent via e-mail3)Export shipments2.2. Day-to-day operations2.3. Exceptional case: no electronic shipment data in GH A’s system upon delivery2.4. Exceptional case: discrepancy between goods delivered and dataFollow procedure RfC4)Import shipments 1.1.Pre-conditions1.2.Daily operations1.3.Exceptional case 1: electronic shipment data not found in GH A’s system1.4.Exceptional case 2: request for a paper copy of the AWBIn case a particular party needs a paper version; Party itself provides paper version.5)Transit shipments (Transfer)6)Abbreviations & definitionsFF: Freight ForwarderseExport licence: is an advantage but not mandatory. E-Export exists for (non) AEO (Authorized Economic Operator) and it allows FF to present the AWB copy on a later moment then before departure of the goods. The list of the e-Export licensed FF can be found on the ACB website … . Single Process:。
-朱玉贤分子生物学习题题库,DOC

】D.E. 一般需要11.需要A.DNA复制B.DNAC.DNAD.RNAE.RNA12.下列关于A.B.C.由于DNA 复制D.在双螺旋5’-E.连接二个13.关于A.此酶能从B.在DNAC.DNAD.是DNAE.14.A.B.DNAC.D.E.DNA15.A.B.C.需GTPD.需ATPE.16.DNA A.制过程B.需ATPC.D.使相邻的E.催化RNA17.DNAA.作用物为B.C.以NAD+D.E.需要DNA18.DNA A.B.需合成C.D.E.合成DNA19.关于A.DNA pol IB.DNA pol IDNA聚(端粒)全标记15N-DNA是半保)严格遵聚合2)具外切聚合DNADNA连接α通过–OHA.RNAB.RNAC.DNAD.3‘→5’E.5‘→3’2.A.RNAB.RNAC.DNAD.有3‘→5’E.产物是3.下列属于A.ATPD.UTP4.A.dNTPB.RNAC.DNAD.DNA模板5.DNAA.需要DNAB.需要DNAC.D.E.需要RNA6.真核细胞A.有3B.C.D.需Mg2+7.RNAA.A-T8.A.全酶由5B.C.D.σE.σ亚基使9.A.B.D.10.参与A.引物酶D.RNA11.A.C.E.12.A.C.逆转录酶E.DNA13.tRNAA.转录C.翻译E.DNA复制14.tRNAA.剪切5’和B.D.碱基修饰C12.B .B23.C33序l,σ因RNA③对15. 1 2(3 4 12(3(((4物,((1 2 3 1在的,,。
、()。
%,聚合酶和DNA 和rRNAα2ββˊ,聚合,的相RNA聚RNA聚ρ因子ρ变构不DNA模“转录泡”A.140 28A.丝氨酸11.B 2.D 3其中61密码),3 C、E码。
6.C 7阶段需要时,氨基酸14.见page133第142 28.D密码都是二、1A.C.23A.在80S C.E.在70S4A. N-端修饰C. 5.tRNA碱基配对A.U B.C 6A.B.C.mRNAD.E.7ABC、mRNA)上,酶识别。
In-Fusion

Please read the In-Fusion HD Cloning Kit User Manual before using this Protocol-At-A-Glance. This abbreviated protocol is provided for your convenience, but is not intended for first-time users.Cloning more than two fragments at once (e.g, multiple inserts simultaneously into one linearized vector) requires adherence to specific considerations in experimental design and overall cloning protocol. This Protocol-At-A-Glance details these considerations and recommended modifications to ensure cloning success.Please note the following materials are required but not supplied:•Ampicillin (100 mg/ml stock) or other antibiotic required for plating the In-Fusion reaction •LB (Luria-Bertani) medium (pH 7.0) •LB/antibiotic plates • SOC mediumThe table below is a general outline of the protocol used for the In-Fusion HD Cloning Kits. Please refer to the specified User Manual pages for further details on performing each step.Table I. In-Fusion HD Protocol OutlineStepAction User Manual Pages2 Design PCR primers for your sequence(s) of interest with 20-bpextensions (5’) that are complementary to the ends of adjacentsequences (the linearized vector or another insert).6–8 3 Amplify your sequence(s) of interest with CloneAmp™ DNApolymerase. Verify on an agarose gel that your targets have beenamplified and confirm the integrity of the PCR products.8–9 4 Spin-column purify your PCR products OR treat with CloningEnhancer.Spin-Column Protocol I (p. 9–11) OR Cloning Enhancer Protocol II (p. 11) 5 Set up your In-Fusion cloning reaction:2 μl 5X In -Fusion HD Enzyme PremixX μl Linearized vectorX μl Each insertX μl dH 20 to a total reaction v olume of 10 μl. Mix well.6Incubate the reaction for 15 min at 50°C, then place on ice.I. PCR and Experimental Preparation (Section IV of the User Manual)A. Preparation of a Linearized Vector by Restriction DigestionFor vector linearization via PCR, please see primer design recommendations in the User Manual,Section IV.B.Complete, efficient digestion will reduce the amount of cloning background. Generally speaking, twoenzymes cut better than any single enzyme. Digestion efficiency will always be better if the restrictionsites are as far apart as possible.1.Incubate your restriction digest as directed by the restriction enzyme supplier. Longer reactiontimes can increase linearization and reduce background.2.After digestion, purify the linearized vector using a PCR purification kit. We recommend gelpurification using the NucleoSpin Gel and PCR Clean-Up kit (Cat. No. 740609.50).3.[Control] Check the background of your vector by transforming competent cells with 5–10 ng ofthe linearized and purified vector. If background is high, add more restriction enzyme(s) andcontinue digesting the vector (2 hr to overnight). Gel purify the remainder of the vector andtransform again.B. PCR Primer DesignWhen designing In-Fusion PCR primers, consider the following:1.Every PCR primer for multi-insert cloning must be designed in such a way that it generatesproducts containing 5’ ends with 20 bp of homology to the ends of the neighboring cloningfragments (either the linearized vector or other inserts).2.The 3’ portion of each primer should:∙be specific to your template∙be between 18–25 bases in length, with GC-content between 40–60%∙have a T m between 58–65°C; with the difference between the forward and reverse primers≤4°C. T m should be calculated based upon the 3’ (gene-specific) end of the primer, NOT theentire primer.∙not contain identical runs of nucleotides; the last five nucleotides at the 3’ end of eachprimer should not have more than two guanines (G) or cytosines (C)3.Avoid complementarity within each primer and between primer pairs4.Online tools are available to help with primer design:∙BLAST searches can determine specificity and uniqueness of the 3’ end (at/BLAST/)∙Our online primer design tool simplifies PCR primer design for In-Fusion reactions (at/US/Products/Cloning_and_Competent_Cells/Selection_Guides/Online_In-Fusion_Tools)5.Desalted oligonucleotide primers are generally recommended for PCR reactions. However,PAGE purification may be needed for primers of poor quality or longer than 45 nucleotides.C. PCR Amplification of Target Fragment(s)The In-Fusion method is not affected by the presence or absence of A-overhangs, so you can use anythermostable DNA polymerase for amplification, including proofreading enzymes. We recommend using our CloneAmp HiFi PCR Premix (included in every In-Fusion HD Cloning Plus system, and soldseparately as Cat. No. 639298). If you are using a different po lymerase, please refer to the manufacturer’s instructions. If using CloneAmp HiFi PCR Premix, please read the Protocol-at-a-Glance and follow the guidelines below:Table II. Recommendations for PCR with CloneAmp HiFi PCR PremixTemplate Type Template Amount Product Size Extension TimeE. coli genomic DNA 100 pg–100 ng up to 10 kb 5 sec/kbλ DNA10 pg–100 ng up to 15 kb 5 sec/kbPlasmid DNA 10 pg–1 ng up to 15 kb 5 sec/kbcDNA ≤ the equivalent of25–125 ng total RNAup to 6 kb 5–10 sec/kbWhen PCR cycling is complete, confirm your product(s) on an agarose gel.II. In-Fusion Cloning Procedure (Section VI of the User Manual)Both protocols below are appropriate for PCR that produces a single band of the desired size. If non-specificbands are visible on your gel, use Protocol I.A. Protocol I: In-Fusion Cloning Procedure w/Spin-Column Purification1.Isolate each target fragment (insert or linearized vector) by gel extraction followed by spin-columnpurification using a silica-based purification system, such as the NucleoSpin Gel and PCR Clean-Upkit.2.Plan the In-Fusion cloning reaction. Good cloning efficiency is achieved when using 50–200 ng ofvector and inserts, respectively. More is not better. Use the table below for reactionrecommendations.Table III. Recommended In-Fusion Reactions for Purified FragmentsRxn Component Cloning Rxn Negative Control Rxn Positive Control RxnLinearized vector 50–200 ng** 1 μl 1 μl of pUC19 controlvector5X In-Fusion HD EnzymePremix2 μl 2 μl 2 μlDeionized Water to 10 μl to 10 μl to 10 μl *<0.5 kb: 10–50 ng, 0.5 to 10 kb: 50–100 ng, >10 kb: 50–200 ng**<10 kb: 50–100 ng, >10 kb: 50–200 ngMolar Ratio RecommendationsGenerally, the molar ratio of each of the multiple inserts should be 2:1 with regards to the linearized vector, i.e., two moles of each insert for each mole of linearized vector. The molar ratio of two inserts with one vector should be 2:2:1. Specific exceptions are listed below:∙If an insert is large with respect to your linearized vector, we recommend a molar ratio of 1:1 ∙For cloning small DNA fragments (150–350 bp), the suggested insert-to-vector molar ratio is 3–5:1∙For cloning of short synthetic oligos (50–150 bp), the suggested oligo to vector molar ratio is 5–15:1. Depending on the oligo length, the optimal molar ratio must be determined empirically.3.Set up the In-Fusion cloning reaction:2 μl5X In-Fusion HD Enzyme Premixx μl*Linearized vectorx μl*Purified PCR insertx μl*Purified PCR insertx μl dH2O (as needed)10 μl Total volume* For reactions with larger combined volumes of vector and PCR insert (>7 μl of vector + insert), double the amount of enzyme premix, and add dH20 for a total volume of 20 μl.4.Adjust the total reaction volume to 10 µl using deionized H2O, and mix.5.Incubate the reaction for 15 min at 50°C, then place on ice.6.Continue to the Transformation Procedure (Section III). You can store the cloning reactions at –20°Cuntil you are ready.B. Protocol II: In-Fusion Cloning Procedure w/Cloning Enhancer Treatment1.Add 2 µl of Cloning Enhancer to 5 µl of each PCR reaction (insert or linearized vector).e a thermal cycler to incubate at 37°C for 15 min, then at 80°C for 15 min. If you used more than100 ng of DNA template, extend the 37°C incubation to 20 min. If you are using a water bath or heatblock rather than a thermal cycler, extend each incubation to 20–25 min.NOTE: If you cannot proceed immediately to the cloning reaction, store Cloning Enhancer-treatedPCR reactions at –20°C until you are ready.3.Set up the In-Fusion cloning reaction:2 μl5X In-Fusion HD Enzyme Premixx μl*Linearized vectorx μl** Treated PCR insertx μl** Treated PCR insertx μl dH2O (as needed)10 μl Total volume* Use 50–200 ng of linearized vector.** Use 1–2 μl of Cloning Enhancer-treated fragments, regardless of their length. The total volume ofCloning Enhancer-t reated PCR fragments should be up to 4 μl per 10-μl reaction. If you obtain a lowproduct yield from your PCR reaction, we recommend purification of PCR fragments instead ofCloning Enhancer treatment.4.Adjust the total reaction volume to 10 µl using deionized H2O, and mix.5.Incubate the reaction for 15 min at 50°C, then place on ice.6.Continue to the Transformation Procedure (Section III). You can store the cloning reactions at –20°Cuntil you are ready.III. Transformation Procedure Using Stellar™ Competent Cells(Section VIII of the User Manual)This transformation protocol has been optimized for transformation using Stellar Competent Cells, sold inIn-Fusion kits and separately in several formats. If you are not using Stellar Competent Cells, follow the protocol provided by the manufacturer. We strongly recommend the use of competent cells with a transformation efficiency ≥1 x 108 cfu/ug.For complete information on the handling of Stellar Competent Cells, please see the Protocol.1.Thaw Stellar Competent Cells on ice just before use. After thawing, mix gently to ensure even distribution,and then move 50 µl of competent cells to a 14-ml round-bottom tube (Falcon tube). Do not vortex.2.Add 2.5 µl of the In-Fusion cloning reaction to the competent cells.3.Place the tubes on ice for 30 min.4.Heat shock the cells for exactly 45 sec at 42°C.5.Place the tubes on ice for 1–2 min.6.Add SOC medium to bring the final volume to 500 µl. SOC medium should be warmed to 37°C before using.7.Incubate with shaking (160–225 rpm) for 1 hr at 37°C.8.Plate 1/5–1/3 of each transformation reaction into separate tubes and bring the volume to 100 µl with SOCmedium. Spread each diluted transformation reaction on a separate LB plate containing an antibioticappropriate for the cloning vector (e.g., the control vector included with the kit requires 100 µg/ml ofampicillin.)9.Centrifuge the remainder of each transformation reaction at 6,000 rpm x 5 min. Discard the supernatant andresuspend each pellet in 100 µl fresh SOC medium. Spread each sample on a separate antibiotic LB plate.Incubate all plates overnight at 37°C.10.The next day, pick individual isolated colonies from each experimental plate. Isolate plasmid DNA using astandard method of your choice (e.g., miniprep). To determine the presence of inserts, analyze the DNA byrestriction digest or PCR screening.IV. Expected Results (Section IX of the User Manual)The positive control plates typically develop several hundred white colonies when using cells with a minimum transformation efficiency of 1 x 108cfu/μg. The negative control plates should have few colonies.The number of colonies on your experimental plates will depend on the amount and purity of the PCR products and linearized vector used for the In-Fusion cloning reaction.∙The presence of a low number of colonies on both the experimental plate and positive control plate (typically,a few dozen colonies) is indicative of either low transformation efficiency or low quality DNA fragments.∙The presence of many (hundreds) of colonies on the negative control is indicative of incomplete vector linearization.If you do not obtain the expected results, use the guide in Section X of the User Manual to troubleshoot yourexperiment. To confirm that your kit is working properly, perform the control reactions detailed in Section IV.D of the User Manual.NOTE: Many troubleshooting topics are covered in our online In-Fusion Cloning FAQs and Tips.Notice to PurchaserOur products are to be used for research purposes only. They may not be used for any other purpose, including, but not limited to, use in drugs, in vitro diagnostic purposes, therapeutics, or in humans. Our products may not be transferred to third parties, resold, modified for resale, or used to manufacture commercial products or to provide a service to third parties without prior written approval of Takara Bio USA, Inc.Your use of this product is also subject to compliance with any applicable licensing re quirements described on the product’s web page at . It is your responsibility to review, understand and adhere to any restrictions imposed by such statements.©2016 Takara Bio Inc. All Rights Reserved.All trademarks are the property of Takara Bio Inc. or its affiliate(s) in the U.S. and/or other countries or their respective owners. Certain trademarks may not be registered in all jurisdictions. Additional product, intellectual property, and restricted use information is available at .This document has been reviewed and approved by the Quality Department.。
Ovation I O Reference Manual

This publication adds the Eight Channel RTD module to the Ovation I/O Reference Manual. It should be placed between Sections 19 and 20.Date: 04/03IPU No.243Ovation ® Interim Publication UpdatePUBLICATION TITLEOvation I/O Reference ManualPublication No. R3-1150Revision 3, March 2003Section 19A. Eight Channel RTDModule19A-1. DescriptionThe Eight (8) channel RTD module is used to convert inputs from Resistance Temperature Detectors (RTDs) to digital data. The digitized data is transmitted to the Controller.19A-2. Module Groups19A-2.1. Electronics ModulesThere is one Electronics module group for the 8 channel RTD Module:n5X00119G01 converts inputs for all ranges and is compatible only with Personality module 5X00121G01 (not applicable for CE Mark certified systems).19A-2.2. Personality ModulesThere is one Personality module groups for the 8 channel RTD Module:n5X00121G01 converts inputs for all ranges and is compatible only with Electronics module 5x00119G01 (not applicable for CE Mark certified systems).19A-2.3. Module Block Diagram and Field Connection WiringDiagramThe Ovation 8 Channel RTD module consists of two modules an electronics module contains a logic printed circuit board (LIA) and a printed circuit board (FTD). The electronics module is used in conjunction with a personalty module, which contains a single printed circuit board (PTD). The block diagram for the 8 channel RTD moduleis shown in Figure 19A-1.Table 19A-1. 8 Channel RTD Module Subsystem ChannelsElectronic Module Personality Module85X00119G015X00121G01Figure 19A-1. 8 Channel RTD Module Block Diagram and Field Connection Wiring Diagram19A-3. SpecificationsElectronics Module (5X00119)Personality Module (5X00121)Table 19A-2. 8 Channel RTD Module SpecificationsDescription ValueNumber of channels8Sampling rate50 HZ mode: 16.67/sec. normally. In 3 wire mode, leadresistance measurement occurs once every 6.45 sec.during which the rate drops to 3/sec.60 HZ mode: 20/sec. normally. In 3 wire mode, leadresistance measurement occurs once every 6.45 sec.during which the rate drops to 2/sec.Self Calibration Mode: Occurs on demand only. The ratedrops to 1/sec. once during each self calibration cycle.RTD ranges Refer to Table 19A-3.Resolution12 bitsGuaranteed accuracy (@25°C)0.10% ±[0.045 (Rcold/Rspan)]% ± [((Rcold + Rspan)/4096 OHM)]% ± [0.5 OHM/Rspan]% ±10 m V ± 1/2LSBwhere:Rcold and Rspan are in Ohms.Temperature coefficient 10ppm/°CDielectric isolation:Channel to channel Channel to logic 200V AC/DC 1000 V AC/DCInput impedance100 M OHM50 K OHM in power downModule power 3.6 W typical; 4.2 W maximumOperating temperature range0 to 60°C (32°F to 140°F)Storage temperature range-40°C to 85°C (-40°F to 185°F)Humidity (non-condensing)0 to 95%Self Calibration On Demand by Ovation ControllerCommon Mode Rejection120 dB @ DC and nominal power line frequency+/- 1/2%Normal Mode Rejection100 dB @ DC and nominal power line frequency+/- 1/2%Table 19A-3. 8 Channel RTD RangesScale #(HEX)Wires Type Tempo FTempo CRcold(ohm)Rhot(ohm)Excitationcurrent(ma)Accuracy± ±countsAccuracy± ±% ofSPAN1310OhmPL0 to1200–18 t o6496106.3 1.090.222310OhmCU 0 to302–18 t o1508.516.5 1.0 130.32D350OhmCU 32 to2840 to1405080 1.0110.2711350OhmCU 32 to2300 to1105378 1.0120.30193100Ohm PL –4 to334–16 t o16892163.671.0110.27223100Ohm PL 32 to5200 to269100200 1.0100.25233100Ohm PL 32 to10400 to561100301 1.0100.25253120Ohm NI –12 t o464–11 t o240109360 1.0100.25263120Ohm NI 32 to1500 to70120170 1.0130.32283120Ohm NI 32 to2780 to122120225 1.0110.27804100Ohm PL 32 to5440 to290100 208 1.0100.25814100Ohm PL 356 t o446180 t o230168 186 1.0300.74824200Ohm PL 32 to6980 to370200 473 1.0120.30834200Ohm PL 514 t o648268 t o342402452 1.0290.71844100Ohm PL 32 to1240 to51100120 1.0190.47854100Ohm PL 32 to2170 to103100 140 1.0130.3286 4100Ohm PL 32 to4120 to211100 180 1.0110.27874100Ohm PL 32 to7140 to379100 240 1.0100.25884120Ohm PL 511 t o662266 t o350200230 1.0240.5919A-4. 8 Channel RTD Terminal Block Wiring Information19A-4.1. Systems Using Personality Module 5X00121G01 Each Personality module has a simplified wiring diagram label on its side, which appears above the terminal block. This diagram indicates how the wiring from the field is to beconnected to the terminal block in the base unit. The following table lists and defines the abbreviations used in this diagram.Table 19A-4. Abbreviations Used in the DiagramAbbreviation Definition+IN, -IN Positive and negative sense input connectionEarth ground terminal. Used for landing shields when the shield is to begrounded at the module.PS+, PS-Auxiliary power supply terminals.RTN Return for current source connection.SH Shield connector. used for landing shields when the shield is to begrounded at the RTD.SRC Current source connection.Note:PS+ and PS- are not used by this module.19A-5. 8 Channel RTD Module Address Locations19A-5.1. Configuration and Status RegisterWord address 13 (D in Hex) is used for both module configuration and module status. The Module Status Register has both status and diagnostic information. The bit information contained within these words is shown in Table 19A-5.Definitions for the Configuration/Module Status Register bits:Bit 0:This bit configures the module (write) or indicates the configuration state of the module (read). A “1” indicates that the module is configured. Note that until the module is configured, reading from addresses #0 through #11 (B in Hex) will produce an attention status.Bit 1:This bit (write “1”) forces the module into the error state, resulting in the error LED being lit. The read of bit “1” indicates that there is an internal module error,or the controller has forced the module into the error state. The state of this bit is always reflected by the module’s Internal Error LED. Whenever this bit is set,an attention status is returned to the controller when address #0 through #11(B in Hex) are read.Table 19A-5. 8 Channel RTD Configuration/Status Register (Address 13 0xD in Hex)Bit Data Description -Configuration Register (Write)Data Description -Status Register (Read)0Configure Module Module Configured(1 = configured; 0 = unconfigured)1Force errorInternal or forced error(1 = forced error; 0 = no forced error)250/60 Hz select (0 = 60Hz, 1 = 50Hz)50/60 Hz System (1 = 50Hz) d(read back)3SELF_CAL (Initiates Self Calibration)Warming bit (set during power up or configuration)40050060Module Not Calibrated 708CH.1 _ 3/4 Wire.CH.1 _ 3/4 Wire - Configuration (read back)9CH.2 _ 3/4 Wire.CH.2 _ 3/4 Wire - Configuration (read back)10CH.3 _ 3/4 Wire.CH.3 _ 3/4 Wire - Configuration (read back)11CH.4 _ 3/4 Wire.CH.4 _ 3/4 Wire - Configuration (read back)12CH.5 _ 3/4 Wire.CH.5 _ 3/4 Wire - Configuration (read back)13CH.6 _ 3/4 Wire.CH.6 _ 3/4 Wire - Configuration (read back)14CH.7 _ 3/4 Wire.CH.7 _ 3/4 Wire - Configuration (read back)15CH.8 _ 3/4 Wire.CH.8 _ 3/4 Wire - Configuration (read back)Bit 2:The status of this bit (read) indicates the conversion rate of the module, write to this bit configures the conversion rate of A/D converters as shown below.see Table 19A-6.Bit3:Write: This bit is used to initiate self-calibration. Read: This bit indicates that the module is in the “Warming” state. this state exists after power up and ter-minates after 8.16 seconds. the module will be in the error condition during the warm up period.Bit4 & 5:These bits are not used and read as “0” under normal operation.Bit 6:This bit (read) is the result of a checksum test of the EEPROM. A failure of this test can indicate a bad EEPROM, but it typically indicates that the module has not been calibrated. A “0” indicates that there is no error condition. If an error is present, the internal error LED is lit and attention status will be returned for all address offsets 0-11 (0x0 - 0xB). The “1” state of this bit indicates an unre-coverable error condition in the field.Bit 7:This bits is not used and read as “0” under normal operation.Bit 8 - 15:These bits are used to configure channels 1 - 8 respectively for 3 or 4 wire op-eration. A “0” indicates 3 wire and a “1” indicates 4 wire operation, see Table 19A-7 and Table 19A-8).Word address 12 (0xC) is used to configure the appropriate scales for Channels 1 - 4 (refer to Table 19A-7 and Table 19A-8).Table 19A-6. Conversion Rate Conversion Rate (1/sec.)Bit 260 (for 60Hz systems)050 (for 50Hz systems)1Table 19A-7. Data Format for the Channel Scale Configuration Register(0xC)Bit Data Description Configuration (Write)Data Description Status (Read)0 Configure Channel #1scale - Bit 0Channel #1 scale configuration (read back) - Bit 01Configure Channel #1scale - Bit 1Channel #1 scale configuration (read back) - Bit 12Configure Channel #1scale - Bit 2Channel #1 scale configuration (read back) - Bit 23Configure Channel #1scale - Bit 3Channel #1 scale configuration (read back) - Bit 34Configure Channel #2 scale - Bit 0Channel #2 scale configuration (read back) - Bit 05Configure Channel #2 scale - Bit 1Channel #2 scale configuration (read back) - Bit 16Configure Channel #2 scale - Bit 2Channel #2 scale configuration (read back) - Bit 27Configure Channel #2 scale - Bit 3Channel #2 scale configuration (read back) - Bit 38Configure Channel #3 scale - Bit 0Channel #3 scale configuration (read back) - Bit 09Configure Channel #3 scale - Bit 1Channel #3 scale configuration (read back) - Bit 1Caution:Configuring any or all channel scales while the system is running will cause all channels to return attention status for up to two seconds following the reconfiguration.Caution:Configuring any or all channel scales while the system is running will cause all channels to return attention status for up to two seconds following the reconfiguration.10Configure Channel #3 scale - Bit 2Channel #3 scale configuration (read back) - Bit 211Configure Channel #3 scale - Bit 3Channel #3 scale configuration (read back) - Bit 312Configure Channel #4 scale - Bit 0Channel #4 scale configuration (read back) - Bit 013Configure Channel #4 scale - Bit 1Channel #4 scale configuration (read back) - Bit 114Configure Channel #4 scale - Bit 2Channel #4 scale configuration (read back) - Bit 215Configure Channel #4 scale - Bit 3Channel #4 scale configuration (read back) - Bit 3Table 19A-8. Data Format for the Channel Scale Configuration Register(0xE)Bit Data Description Configuration (Write)Data Description Status (Read)0 Configure Channel #5 scale - Bit 0Channel #5 scale configuration (read back) - Bit 01Configure Channel #5 scale - Bit 1Channel #5 scale configuration (read back) - Bit 12Configure Channel #5 scale - Bit 2Channel #5 scale configuration (read back) - Bit 23Configure Channel #5 scale - Bit 3Channel #5 scale configuration (read back) - Bit 34Configure Channel #6 scale - Bit 0Channel #6 scale configuration (read back) - Bit 05Configure Channel #6 scale - Bit 1Channel #6 scale configuration (read back) - Bit 16Configure Channel #6 scale - Bit 2Channel #6 scale configuration (read back) - Bit 27Configure Channel #6 scale - Bit 3Channel #6 scale configuration (read back) - Bit 38Configure Channel #7 scale - Bit 0Channel #7 scale configuration (read back) - Bit 09Configure Channel #7 scale - Bit 1Channel #7 scale configuration (read back) - Bit 110Configure Channel #7 scale - Bit 2Channel #7 scale configuration (read back) - Bit 211Configure Channel #7 scale - Bit 3Channel #7 scale configuration (read back) - Bit 312Configure Channel #8 scale - Bit 0Channel #8 scale configuration (read back) - Bit 013Configure Channel #8 scale - Bit 1Channel #8 scale configuration (read back) - Bit 114Configure Channel #8 scale - Bit 2Channel #8 scale configuration (read back) - Bit 215Configure Channel #8 scale - Bit 3Channel #8 scale configuration (read back) - Bit 3Table 19A-7. Data Format for the Channel Scale Configuration Register(0xC)19A-6. Diagnostic LEDsTable 19A-9. 8 Channel RTD Diagnostic LEDsLED DescriptionP (Green)Power OK LED. Lit when the +5V power is OK.C (Green)Communications OK LED. Lit when the Controller is communicatingwith the module.I (Red)Internal Fault LED. Lit whenever there is any type of error with themodule except to a loss of power. Possible causes are:n - Module initialization is in progress.n - I/O Bus time-out has occurred.n - Register, static RAM, or FLASH checksum error.n - Module resetn - Module is uncalibrated.n - Forced error has been received from the Controllern - Communication between the Field and Logic boards failedCH1 - CH 8 (Red)Channel error. Lit whenever there is an error associated with a channel or channels. Possible causes are:n - Positive overrangen - Negative overrangen Communication with the channel has failed。
219389911_莱茵衣藻碳酸酐酶基因家族的生物信息学分析

1.
1 莱茵衣藻 CA 基因家族成员鉴定
//p
在 Ens
emb
l
eP
l
an
t(
h
t
t
l
an
t
s.
ens
emb
l.
o
rg/
i
ndex.
h
tml)数据库中下载物种莱茵衣藻的全基因组数
ps:
//www.
据,包括 CDS、
Pr
o
t
a
i
ns
equenc
e和 GFF3 文件。从 TAIR(
h
t
t
a
r
ab
i
dops
/L 的 HCO3-为实
实验以 TAP(
Tr
i
sAc
e
t
a
t
e
-Pho
spha
t
e)培养基培养为对照组,在对照组中添加5mmo
l
验组,固定时间间隔进行取样,测定 680nm 处的吸光度值,并对 CA 基因进行表征。
2 结果与分析
2.
1 莱茵衣藻 CA 家族成员鉴定及理化性质分析
按照 1.
1 所 述 方 法 检 索 莱 茵 衣 藻 CA 蛋 白 序 列,进 一 步 通 过 CDD、Pf
-CA 是 一 种 单 体 酶,具 有 高 度 保 守 性,在 不
[
5]
同种藻类中表现出相似的结构和功能特征 。β
-CA 是一种大多数情况下以二聚体形式存在的酶,其基因组
分布较为广泛,具有更多样化的结构和功能 [6]。藻类中 CA 的 活 性 受 到 光 照、温 度、营 养 素 等 环 境 因 素 的 调
m6A文献解读

m6A文献解读相xhog0.8842020.05.10 11:11:23字数 1,442阅读 763前几天我们一直在介绍m6A相关的数据库,正好好久没之前看过一篇关于m6A的文献。
这里就来做一下这个文献的抄读吧。
今天抄读的这篇文献是2019年来自于中山大学发表的一篇m6A 相关的文献。
imageMETTL3在结肠癌当中的基本结果为了评价m6A在大肠癌当中的作用,作者首先评价了m6A的相关基因在TCGA数据库当中的结肠癌的差异表达的情况。
同时利用自己的医院的样本来检验这些差异表达结果的情况。
同时发现METTL3在验证的结果当中存在表达差异。
image一般做组织样本分析的除了做癌和正常的分析,也会进一步的表达和各种临床信息有没有差异的。
所以作者就做了关于METTL3和各种临床信息的分析。
这种一半肯定会做很多临床数据的分析,至于结果的话,可能就是哪个有意义就上哪个了。
imageSOX2收到METTL3介导的m6A甲基化的调控在上面进行各个细胞系表达验证的时候,作者发明METTL3在SW480以及SW620之间的蛋白表达存在差异。
这一对细胞系是从单个患者的腹部转移灶和原发肿瘤中分离出的一对细胞系,两种细胞系表现出不同的转移能力可能说明METTL3和肿瘤的转移存在关系。
image对于m6A测序方面的分析,就是使用MeRIP-seq了。
所以作者做了两组不同的MeRIP-seq的分析。
一组是SW480 vs SW620。
另外一组由于METTL3在SW620当中表达高。
所以就做了SW620 VS 敲除METTL3的SW620细胞系(这个分组的SW620估计就是之前的第一组的那个吧)。
由于我们也介绍过MeRIP-seq只能说明m6A和哪个位置结合,但是并不能说明影响表达,对于相同的分组,作者也做了RNA-seq的分析。
通过最后的交叉分析,最后可以得到158个基因是是两个分组重叠出来的结果。
image进一步对这158个基因进行了富集分析,说明这158个基因主要是和干细胞分化有关系,说明METTL3介导的转移有可能是通过干细胞分析通路来形式作用的。
m6a修饰酶相关关系__概述说明以及解释

m6a修饰酶相关关系概述说明以及解释1. 引言部分的内容:1.1 概述:m6A修饰酶是一类关键的酶,参与了RNA分子上的m6A(甲基化腺嘌呤)修饰反应。
这种修饰是一种重要的RNA表观遗传标记方式,能够影响RNA的稳定性、翻译效率和转运过程。
近年来,随着对m6A修饰的研究深入进行,人们发现了许多与m6A修饰酶相关的功能和调控机制。
本文旨在概述和解释m6A 修饰酶及其相关关系,以加深对这一领域的理解。
1.2 文章结构:本文共包括五个主要部分:引言、M6A修饰酶的相关关系、M6A修饰酶在疾病中的重要性、M6A修饰酶调控策略与应用前景以及结论。
引言部分将介绍本文涉及到的研究领域及其重要性,并给出文章整体结构。
1.3 目的:本文旨在全面介绍M6A修饰酶及其相关关系,并探讨其在疾病发展中的重要性。
此外,我们将讨论利用M6A修饰酶作为靶点进行药物治疗以及RNA编辑技术在M6A修饰调控中的应用前景。
最后,我们会对未来该领域的研究方向进行展望。
通过本文的撰写,我们旨在为读者提供一个清晰而全面的视角,促进对M6A修饰酶相关关系的理解和研究进展。
以上是"1. 引言"部分内容的详细说明,请根据需求进行相应调整和补充。
2. M6A修饰酶的相关关系2.1 M6A修饰酶的定义及作用M6A修饰是一种常见的RNA表观遗传修饰方式,指的是在RNA分子中甲基化腺嘌呤(adenosine)碱基的过程。
M6A修饰是一种高度保守且广泛存在于真核生物细胞中的修饰方式,在多个生物过程中发挥着重要作用。
M6A修饰主要通过M6A修饰酶来进行介导。
目前已知的主要M6A修饰酶包括"Methyltransferase-like 3" (METTL3)、"Methyltransferase-like 14" (METTL14)和"Wilms' tumor 1-associating protein"(WTAP)等。
spyglass-高级lint-check

Richest set of built-in syntax checks2350+ built-in rules for Verilog, VHDL, V2K, SystemVerilog and mixed languageLint, Openmore, Morelint standards575+ lint, coding style, IP-reuse rulesSTARC, STARC2002, STARCAD21, STARC 2005Recognized industry standard rules from consortium of top 11 semiconductor companies in Japan500+ rules for best practices, coding guidelines, IP-reuse, etc.
Source: I.B.S. Inc.
Source: Gartner
SoC Development Cost
Typical Impact Of Poorly Coded RTL
Chip killer bugs/escapesFixing the problems late in design cycle delays scheduleCombinational loops, non tri-state nets cause functional failureIncorrect FSM behavior causes functional failureLong time to complete verificationIncomplete coverage - Only as complete as your set of simulation vectorsCreating test vectors/assertions is time-consuming and complexSynthesis/simulation mismatch, incomplete initialization cause verification delaysPoor area, power, testability and timing of the chipUnintended redundant logic, use of complex FSM’s lead to poor QoRNo predictability in the design process to hit the market windowDesign reviews are manual, ad-hoc and subjectiveA chip company identified 10+ structural issues that would take 2 to 4 days each to identify and fix in the normal implementation flow
EX245系列产品用户操作手册说明书

TroubleshootingRefer to the LED Display. Refer to the operation manual to obtain more detailed information about troubleshooting.Outline with DimensionsRefer to the product catalog or operation manual to obtain more detailed information about outline dimensions.SpecificationRefer to the product catalog or operation manual to obtain more detailed information about product specifications.Note: Specifications are subject to change without prior notice and any obligation on the part of the manufacturer.© 2021 SMC Corporation All Rights Reserved Akihabara UDX 15F, 4-14-1, Sotokanda, Chiyoda-ku, Tokyo 101-0021, JAPAN Phone: +81 3-5207-8249 Fax: +81 3-5298-5362URL https://EX ※※-OMY0028•LED Indicators 2•LED indicators 3Wiring•Power connectorsFieldbus systemInstruction ManualEX245-FPS1/EX245-FPS2/EX245-FPS3Please read this manual carefully before operating the product and make sure you understand its capabilities and limitations.Safety Instructions•FE terminal•The SI Unit must be connected to FE (Functional Earth) to divert electromagnetic interference. The FE terminal and the FE pin of the two power connectors (XD1/XD2)are internally connected. Please connect at least one of these three FEs to ground potential. For maximum protection the FE cable should be as thick and short as reasonably possible. If it is difficult to shorten the power cable, it is recommended to use the FE terminal screw.•FE terminal screw tightening torque = 1.5 Nm.OperatorNOTE•For UL conformity a DC power supply meeting the requirements of UL1310Class 2 must be used. For other applications a SELV or PELV DC power supply should be used.Safety InstructionsPush Pull connetor (24 Volt)LED DisplaySW3SW2These safety instructions are intended to prevent hazardous situations and/or equipment damage.These instructions indicate the level of potential hazard with the labels of"Caution", "Warning" or "Danger". They are all important notes for safety and must be followed in addition to International Standards (ISO/IEC) and other safetyregulations.•EX245-FPS1, EX245-FPS27/8 inch 5-pins plug•EX245-FPS3Installation•Valve manifold connectionConnect the valve manifold with the 2 screws on the SI Unit.(hexagonal socket wrench size 2.5)For torque value, refer to valve manifold catalogue.Mounting and Installation•Module connectionConnect the SI Unit, the I/O modules and the End plate using the 2 modular adaptor assemblies and a joint assembly. These are supplied together in the Joint pack.①1 x Joint assembly②2 x Modular adaptor assembly (hexagonal socket wrench size 2.5 mm,torque = 1.3 Nm)MountingTo prevent the manifold components being damaged, apply the recommended tightening torque.Mount the manifold using the 6 base mounting positions with screws.Required screws are as follows:①2 x M5 (End plate: torque = 1.5 Nm)②2 x M5 (SI unit: torque = 1.5 Nm)③4 x M ∗(Valve manifold: refer to valve manifold catalogue)•PROFINET communication connectors•Safety input connectorsMaintenance•Maintenance should be performed according to the operation manual.•Turn off the power supply, stop the supplied air, exhaust the residual pressure and verify the release of air before performing maintenance.There is risk of unexpected malfunction.34521Mounting•PROFIsafe address switchA ten bit DIP-Swtich is provided for the safety address setting. The switch setting is only checked at power-up. Any changes made during operation are ignored and may lead to problems during the next power-up sequence.Setting•A two bit DIP-switch and a six bit DIP-SwitchTwo DIP-switches SW2 and SW3 are under the M12 safe input connector box. To access the switches remove the retaining screws as shown below.When the DIP-Switches have been set ensure the M12 connector block and all retaining screws are refitted. (torque = 0.4 Nm) The module must be in a fully assembled state with all parts securely fastened before using the product.Refer to the operation manual to obtain more detailed information about DIP-Switch specification.∗∗: Only EX245-FPS1 has this function."0x01F1" will be generated.•LED Indicators 1。
RNA碱基修饰以及m6A的前世今生——m6A专题
RNA碱基修饰以及m6A的前世今生| m6A专题N6-methyladenosine也叫m6A,是一种广泛存在于mRNA上的碱基修饰行为,成为近几年大热的研究方向。
但是早在50年前,人们已经在RNA中发现了多种碱基修饰现象。
除了传统的ACGU四种碱基外,Cohn等人已经在RNA上发现了全新的碱基位点修饰。
Holley等人于1965年,首次在酵母的tRNA中鉴定了包括假尿苷(pseudouridine)在内的十余种不同的RNA修饰。
当然最初这些碱基修饰大多发现于非编码RNA上如tRNA、rRNA等,后来人们发现mRNA中也存在大量的碱基修饰行为。
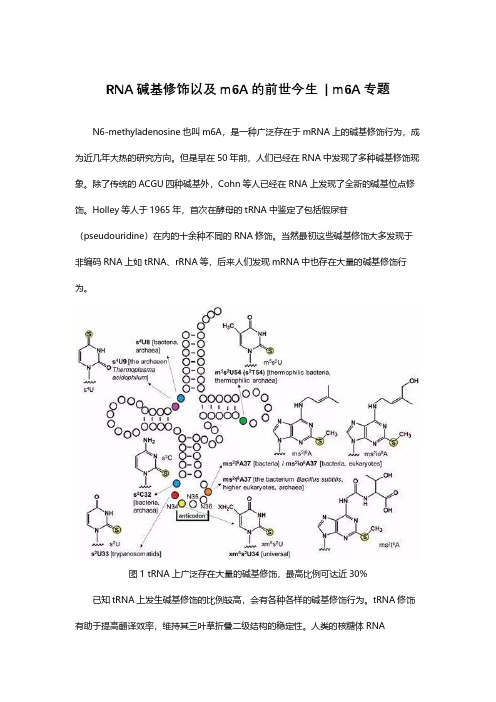
图1 tRNA上广泛存在大量的碱基修饰,最高比例可达近30%已知tRNA上发生碱基修饰的比例较高,会有各种各样的碱基修饰行为。
tRNA修饰有助于提高翻译效率,维持其三叶草折叠二级结构的稳定性。
人类的核糖体RNA(rRNA)上有超过200个碱基修饰位点,而剪切体RNA(spliceosomal RNA)上也有超过50个碱基修饰位点。
图2 真核生物mRNA上存在的各种碱基修饰行为,在5’和3’的UTR以及中间的coding region都有碱基发生修饰目前科学家已经在RNA中鉴定了超过100种不同类型的碱基修饰行为。
在真核生物中,5’端的Cap以及3’的ployA修饰在转录调控中起到了十分重要的作用,而mRNA 的内部修饰则用于维持mRNA的稳定性。
mRNA最常见的内部修饰包括了N6-腺苷酸甲基化(m6A)、N1-腺苷酸甲基化(m1A)、胞嘧啶羟基化(m5C)等。
对于大热的m6A,截止当前,全球的科学家已经鉴定了参与m6A的许多酶,包括去甲基化酶、甲基化酶和甲基化识别酶等(将在下期进行详细介绍)。
图3 m6A甲基化修饰和m6Am超甲基化修饰的概念我们先来了解下什么叫m6A修饰。
从图3中我们可以看到,右侧存在m6A甲基化修饰和m6Am超甲基化修饰。
重复一下N6-methyladenosine就是m6A,一共分为2个大的结构。
中山大学生科院 细胞生物学试卷 (11)

中山大学生科院细胞生物学试卷
填空:
1.呼吸链中的机制叫做———模型(假说)
2.尿素循环是以——的合成开始的
3.参加三羧酸循环的酶中,在线粒体膜上的是——
4.proteasome 是分解——蛋白的
5.当年的诺贝尔生理医学奖和化学奖
判断:
1.肌红蛋白和细胞色素C中的血红素分子是一样的
问答:
1.什么是无效循环?举例说明机体内是如何避免或利用无效循环的
2.CO2是脂肪酸合成的必需原料。
假如用肝脏的可溶部分与14CO2以及其他脂肪酸合成
所需成分保温,生成的软脂酸中是否含有14C?为什么?(5分)
3.The cholesterol in the form of LDL is so-called “bad cholesterol”, and the
cholesterol in the form of HDL is so-called “good cholesterol”. Why?
4.肿瘤为什么酵解速率和葡萄糖利用率都增加?
5.各种核苷酸的合成是如何调节的?
6.下图是有关RNA polymerase 对promoter结合的一个实验,解释实验结果。
(7分)
7.如何识别起始密码子AUG的?。
202 003抗体丨SYSY丨m6A抗体202 003问题使用汇总

202 003抗体丨SYSY丨m6A抗体202 003问题使用汇总202 003,这里并不是单纯的阿拉伯数字,也不是记数系统,数字是一种用来表示数的书写符号,计数也可以是任何环境下的一种特指,而202 003在这里所表达的意思,是生物科研抗体都非常为之耳熟能详的抗体,发表过Cell、Nature,真的超能打!接下来一起来看看这个SYSY 202 003 m6A抗体。
m6A(N6-甲基腺苷)是一种转录后的RNA修饰,在所有种属中都有发现,例如在脊椎动物的snRNAsU2、U4、U6中,在病毒和真核生物的mRNAs中,以及在大肠杆菌的16SrRNA中。
最近的研究发现,mRNA主要是在终止密码子和长的内部外显子处进行m6A修饰,这在小鼠和人类之间是保守的。
所谓的RNA甲基组可能在基因表达的调控中起着重要作用。
在大肠杆菌中,Dam甲基酶在DNA水平上引入5'-GATC-3'座的m6A修饰。
这使得细胞在错配修复过程中能够区分亲本链和子链。
m6A抗体,#202 003:检测RNA和DNA的N6-甲基腺苷(m6A)修饰。
已验证的反应种属有人,小鼠,大鼠,真核细胞,原核细胞等等,用一句话概括就是广泛适用所有的种属;它的应用类型包括WB、IP、IHC、ELISA;免疫原:N6-methyladenosine偶联的BSA,另外,m6A 抗体,#202 003发表文章超过200篇,厉害了!最后,关于SYSY 202 003 m6A抗体如何保存的问题,做科研的都懂这是非常重要的一步,但也存在很多问题,那么接下来就汇总几个比较常见的问题吧~Q1:如何重组(Reconstitution,溶解)202 003 m6A抗体?A1:1.所有抗体均由PBS冻干。
2.制作重悬溶液时请小心打开盖子。
3.为抑制细菌生长,可以添加少量的叠氮化物或硫柳汞作为防腐剂。
4.荧光标记抗体重悬后,添加1:1比例(体积)的甘油至50%最终浓度,从而降低冰点并在-20℃保持液体状态。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
When he was taken off the school football team. When his girl friend told him she wouldn’t go out with him agaoung, I didn’t know…. I didn’t know … I certainly didn’t know…. However, what I know was that … What was Grandad‟s feeling by saying so? If he had known the bad effects of smoking , would not have started smoking he ___________________________. He felt very regretful. What kind of picture did Grandad want to describe in this part? Circle the adjectives. harmful… terrible… difficult… abnormal… breathless…unfit…
Break the habit.
Relax.
Get help if you need it. Keep trying.
Read the advice and fill in the blanks Decided on __________a day that is not ________to stressful quit smoking. Make a list of all the _______ benefits of reread it when you stopping smoking, and ______ ________ feel like smoking. Develop some other habits to busy . If you feel ______ nervous or keep yourself_____ stressed try some ________ relaxation exercises like ________, deep breathing . When you feel really bad, ask chemist for a doctor or _______for help. Don’t feel disappointed if you weaken, and try many times __________ give up and never _______, you will succeed eventually.
To make a comparison?
Para 2 1. Did he scold(责备) James when he knew his
grandson smoked? 2. What did he say?
Believe me, I know how easy it is to begin smoking and how tough it is to stop. You se, during my adolescence, I also smoked and became addicted to cigarettes.
What was his tone of writing the letter? A. a friend‟s B. a grandfather‟s C. a teacher‟s
The advice for quitting smoking
Prepare yourself.
Be determined.
_____ ways to become addicted
physically addicted to nicotine. 1. become _________ habit habitually 2. become addicted through _____. mentally addicted. 3. become ________ 1. What did Gradad feel when he get withdrawal symptoms? bad-tempered, in pain 2. How did Grandad addicted ? How did he know? In all three ways. He was happier and more relaxed after having a cigarette. He could only feel good when he smoked.
Why did Grandad write about his own experiences in this letter?
•to his •to •to shorten the distance between him and grandson arise his grandson‟s resonance(共鸣) make his letter more persuasive
What harmful effects didn’t Grandad know? What did he know? When did he know it was time to quit smoking?
When I was young, I didn’t know…. I didn’t know … I certainly didn’t know…. However, what I did know was that …
1. Why did the grandad use „you” instead of “he” or “one” ? To make his grandson believe that smoking is the case which has something to do with him. 2. What‟s the function of saying “But I did finally manage.”? To give James hope that he could also quit smoking as his grandad did. 3. How does he organize this paragraph? First, … you can…, which …. This means … Secondly,… Lastly,… Tip While writing, you can use first, …; secondly; lastly,… to make your composition more orderly.
To non-smokers The babies of smoking couples may have smaller birth weight or even ____________________________ be abnormal in some way. _______________________
Currently, the mainland has more than 300 million smokers, statistics from the World Health Organization show, and about 1.2 million people die from smokingrelated diseases every year, accounting for one-fifth of the world's total. Meanwhile, an estimated 740 million people are exposed to secondhand smoke, mostly in places like restaurants, bars and workplaces, statistics from the Ministry of Health show.
3. Why did he say that?
He once had the same situation with James.
And he knew his feeling very well, so as to shorten the distance between them.
Para 3
Para 4
Harmful effects of smoking
To smokers themselves lungs •Do terrible damage to hearts ______ and _________ smoking couples to become •Be difficult for _____________ pregnant ________. breath and _______ •_________ clothes smell terrible •The ends of fingers _______________ are turning yellow •Become breathless _______________ quickly sports as much •Not enjoy _____________
Normally, the first part of a suggestion letter is stating the purpose of writing the letter. Why did Grandad describe his active and long life to begin his letter? What was his purpose?
1. Does anybody smoke in your family? 2. Have you ever advised him/her to quit smoking? 3. How did you advise him/her?
