dig RNA labeling kit 罗氏
Roche(罗氏)-地高辛系统产品选择指南

Labeling
PCR DIG Probe Synthesis Kit DIG Northern Starter Kit In Vitro Transcription DIG RNA Labeling Kit (SP6/T7) DIG RNA Labeling Mix
according to the current quality procedures. With DIG-labeled probes, you can easily detect single-copy genes on Southern blots, unique mRNAs on northern blots, or rare recombinants in bacterial colonies or viral plaques.
The DIG System
Labeling and Detection of Nucleic Acids
The DIG System Specifically Label and Detect Nucleic Acids
Publishable results require high level specific detection and low background. Do your hybridizations have nonspecific signals and high background? The DIG System is ideal for nucleic acid labeling. Flexibly use colorimetric, luminescent or fluorescent signal detection. Achieve high sensitivity and low background in very short exposure times
翻译好的罗氏公司Tunel试剂盒操作说明书

罗氏 (Roche)公司 Tunel 试剂盒操作说明书(In situ cell death detection kit-POD法)一、原理:TUNEL (TdT-mediated dUTP nick end labeling)细胞凋亡检测试剂盒是用来检测组织细胞在凋亡早期过程中细胞核DNA 的断裂情况。
其原理是荧光素( fluorescein)标记的 dUTP 在脱氧核糖核苷酸末端转移酶( TdT Enzyme)的作用下,可以连接到凋亡细胞中断裂 DNA 的3’-OH 末端,并与连接辣根过氧化酶(HRP,horse-radish peroxidase)的荧光素抗体特异性结合,后者又与 HRP 底物二氨基联苯胺(DAB )反应产生很强的颜色反应(呈深棕色),特异准确地定位正在凋亡的细胞,因而在光学显微镜下即可观察凋亡细胞;由于正常的或正在增殖的细胞几乎没有 DNA 断裂,因而没有 3‘-OH 形成,很少能够被染色。
本试剂盒适用于组织样本(石蜡包埋、冰冻和超薄切片)和细胞样本(细胞涂片)在单细胞水平上的凋亡原位检测。
还可应用于抗肿瘤药的药效评价,以及通过双色法确定细胞死亡类型和分化阶段。
二、器材与试剂器材:光学显微镜及其成像系统、小型染色缸、湿盒(塑料饭盒与纱布)、塑料盖玻片或封口膜、吸管、各种规格的加样器及枪头等;试剂:试剂盒含:1 号(蓝盖) Enzyme Solution 酶溶液: TdT 10×、2号(紫盖) Label Solution 标记液:荧光素标记的 dUTP 1×、3号(棕瓶) Converter-POD:标记荧光素抗体的 HRP;自备试剂: PBS、双蒸水、二甲苯、梯度乙醇(100、95、90、80、70%)、DAB 工作液(临用前配制, 5 μl 20 ×DAB+1 μL 30%H2O2+94 μl PBS)、Proteinase K工作液( 10-20 μg/ml in 10 mM Tris/HCl ,pH 7.4-8)或细胞通透液(0.1% Triton X-100 溶于 0.1% 柠檬酸钠,临用前配制)、苏木素或甲基绿、 DNase 1(3000 U/ml– 3 U/ml in 50 mM Tris-HCl ,pH 7.5, 10 mM MgCl2 ,1 mg/ml BSA )等。
罗氏RNA提取试剂盒使用指南说明书

Step 1: Sample Preparation & Nucleic Acid IsolationFor great results, use (click product names to learn more):Roche High Pure RNA Isolation KitRoche High Pure FFPET RNA Isolation KitRoche High Pure miRNA Isolation KitRoche RealTime ready Cell Lysis KitFrom which source (animal, organ, tissue) does the examined material originally come from? Which volume or mass or cell number was used for nucleic acid preparation?My MIQE Guide*Empowering results that matter Sponsored by Roche Applied Science Experiment title:Performed by:Date:Institution:Experimental design: How did you choose and set up your study (number of treated samplesand controls)Handling: Which tools or methods were used to obtain and process the primary samples (e.g., micro-dissection, macrodissection)?Method of processing and preservation: How was the sample treated and stored?If frozen – how and how quickly?If fixed – with what, and how quickly?If stored for longer: how and how long? (especially for FFPE samples)Extraction method:Which kit or instrument was used to extract/isolate the DNA/RNA from the starting material? Roche High Pure RNA Isolation Kit, High Pure FFPET RNA Isolation Kit, High Pure miRNA Isolation Kit, RealTime ready Cell Lysis Kit, or other (Please specify)Was the vendor’s protocol modified (If Yes, when, and how? e.g. by using additives)Did you do a DNAse or RNAse treatment? (If Yes, when?)Did you check for nucleic acid purity and integrity? If Yes: By using which instrument and method? What was the resulting purity (A260/A280)? What was the resulting yield? If No: Why not?Did you check for the presence of PCR inhibitors? If Yes: By using what (e.g. Cq dilutions, spike or other (please specify)If No: Why not?Final storage solution (e.g., buffer, H2O) for the purified total RNA:Storage time and temperature of the purified total RNA before use in RT-qPCR:Step 2:Reverse TranscriptionFor optimal results, use:Roche Transcriptor First Strand cDNA Synthesis KitRoche Transcriptor Universal cDNA MasterAmount of RNA and reaction volume:Priming oligonucleotide (if using gene specific primers) and concentration: Reaction temperature and time:Manufacturer of reverse transcription reagent(s) and catalogue number(s): Reverse transcriptase type and used concentration:Storage conditions of cDNA:Step 3:PCR Amplification and AnalysisFor best results, use:LightCycler® 480 Probes MasterFastStart Essential DNA Probes MasterFastStart Universal Probe Master (Rox)Target sequence and amplicon information: Target gene database sequence accession number:Location of amplicon:Amplicon length:Result of in silico specificity screen (BLAST, etc.):Information on pseudogenes, retropseudogenes or other homologs: Secondary structure analysis of amplicon:Determined by which method?Location of each primer relative to exons or introns (if applicable): Targeted splice variants:RTPrimerDB Identification Numbers: Manufacturer of oligonucleotides: Purification method:For probe-based assays: Probe type:qPCR reaction conditionsReaction volume and amount of cDNA/DNA per reaction: Primer, (probe), Mg2+ and dNTP concentrations: Polymerase identity:Buffer/kit manufacturer and identity (e.g., catalog number)Manufacturer and catalog number of plates or tubes and catalog number:Complete thermocycling parameters:Reaction setup: Was it manual or robotic? If robotic: Using which robot?Equipment: Which Real-Time PCR instrument was used? (Which Roche LightCycler® System or other (please specify)?)Validation of qPCR runs:Are you running a multiplex assay? If yes, please describe efficiency and limit of detection foreach assay:How did you check for specificity of amplification for each target (e.g., on a gel, by sequencing, melt-ing curve analysis or digest):For SYBR Green I assays: Cq of the non-template control reaction:Standard curve characteristics (slope and y-intercept):How many replicates did you use to establish the standard curve?(xx replicates per standard concentration)What was the lower and the upper limit of the standard curve?PCR efficiency calculated from slope:Confidence interval for PCR efficiency or standard error:r2 of standard curve:Information on linear dynamic range:Cq variation at lower limit: Confidence intervals throughout range:Evidence for limit of detection:How many reactions per run were used for controls? (please specify positive and negative controls, controls without template and No RT controls, e.g. Positive controls: 3 reactions in 5 replicates per 96 well plate)Data analysis:Vendor software: Which software type, version and algorithm provided by the PCR machine supplier was used to analyze the data?Specialist software: Which (if any) additional software was used? Self-developed algorithms,or other (please specify)Normalisation: Which reference gene(s) were used to calculate the relative expression of the studied genes?What was the reason for choosing these particular genes?Which algorithm (e.g., geNorm, bestkeeper, normfinder) was used to normalize for reference gene(s)Which principle was used for Cq calling?What was the number and of biological replicates used?How was their concordance?How many technical replicates were used, and at which step (RT or qPCR)? What was the observed repeatability (intra-assay variation)?What was the observed reproducibility (inter-assay variation, %CV)The MIQE guidelines empower results that truly matter. And so does Roche.Visit to discover all the materials you need for truly remarkable research results.* modified based on the list in the original MIQE guidelines publication with permission of the MIQE authors.For life science research only. Not for use in diagnostic procedures. LIGHTCYCLER and FASTSTART are trademarks of Roche.All other product names and trademarks are the property of their respective owners. NOTICE: This product may be subject to certain use restrictions. Before using this product, please refer to the Online Technical Support page () and search under the product number or the product name, whether this product is subject to a license disclaimer containing use restrictions.Published byRoche Diagnostics GmbH Sandhofer Straße 116 68305 Mannheim Germany© 2013 Roche Diagnostics. All rights reserved.*********** 1012。
翻译好的罗氏公司Tunel试剂盒操作说明书

罗氏 (Roche)公司 Tunel 试剂盒操作说明书(In situ cell death detection kit-POD法)一、原理:TUNEL (TdT-mediated dUTP nick end labeling)细胞凋亡检测试剂盒是用来检测组织细胞在凋亡早期过程中细胞核DNA 的断裂情况。
其原理是荧光素( fluorescein)标记的 dUTP 在脱氧核糖核苷酸末端转移酶( TdT Enzyme)的作用下,可以连接到凋亡细胞中断裂 DNA 的3’-OH 末端,并与连接辣根过氧化酶(HRP,horse-radish peroxidase)的荧光素抗体特异性结合,后者又与 HRP 底物二氨基联苯胺(DAB )反应产生很强的颜色反应(呈深棕色),特异准确地定位正在凋亡的细胞,因而在光学显微镜下即可观察凋亡细胞;由于正常的或正在增殖的细胞几乎没有 DNA 断裂,因而没有 3‘-OH 形成,很少能够被染色。
本试剂盒适用于组织样本(石蜡包埋、冰冻和超薄切片)和细胞样本(细胞涂片)在单细胞水平上的凋亡原位检测。
还可应用于抗肿瘤药的药效评价,以及通过双色法确定细胞死亡类型和分化阶段。
二、器材与试剂器材:光学显微镜及其成像系统、小型染色缸、湿盒(塑料饭盒与纱布)、塑料盖玻片或封口膜、吸管、各种规格的加样器及枪头等;试剂:试剂盒含:1 号(蓝盖) Enzyme Solution 酶溶液: TdT 10×、2号(紫盖) Label Solution 标记液:荧光素标记的 dUTP 1×、3号(棕瓶) Converter-POD:标记荧光素抗体的 HRP;自备试剂: PBS、双蒸水、二甲苯、梯度乙醇(100、95、90、80、70%)、DAB 工作液(临用前配制, 5 μl 20 ×DAB+1 μL 30%H2O2+94 μl PBS)、Proteinase K工作液( 10-20 μg/ml in 10 mM Tris/HCl ,pH 7.4-8)或细胞通透液(0.1% Triton X-100 溶于 0.1% 柠檬酸钠,临用前配制)、苏木素或甲基绿、 DNase 1(3000 U/ml– 3 U/ml in 50 mM Tris-HCl ,pH 7.5, 10 mM MgCl2 ,1 mg/ml BSA )等。
翻译好的 罗氏公司Tunel试剂盒操作说明书

罗氏(Roche)公司Tunel试剂盒操作说明书(In situ cell death detection kit-POD法)一、原理:TUNEL(TdT-mediated dUTP nick end labeling)细胞凋亡检测试剂盒是用来检测组织细胞在凋亡早期过程中细胞核DNA的断裂情况。
其原理是荧光素(fluorescein)标记的dUTP在脱氧核糖核苷酸末端转移酶(TdT Enzyme)的作用下,可以连接到凋亡细胞中断裂DNA的3’-OH末端,并与连接辣根过氧化酶(HRP,horse-radish peroxidase)的荧光素抗体特异性结合,后者又与HRP底物二氨基联苯胺(DAB)反应产生很强的颜色反应(呈深棕色),特异准确地定位正在凋亡的细胞,因而在光学显微镜下即可观察凋亡细胞;由于正常的或正在增殖的细胞几乎没有DNA断裂,因而没有3‘-OH形成,很少能够被染色。
本试剂盒适用于组织样本(石蜡包埋、冰冻和超薄切片)和细胞样本(细胞涂片)在单细胞水平上的凋亡原位检测。
还可应用于抗肿瘤药的药效评价,以及通过双色法确定细胞死亡类型和分化阶段。
二、器材与试剂器材:光学显微镜及其成像系统、小型染色缸、湿盒(塑料饭盒与纱布)、塑料盖玻片或封口膜、吸管、各种规格的加样器及枪头等;试剂:试剂盒含:1号(蓝盖)Enzyme Solution 酶溶液:TdT 10×、2号(紫盖)Label Solution标记液:荧光素标记的dUTP 1×、3号(棕瓶)Converter-POD:标记荧光素抗体的HRP;自备试剂:PBS、双蒸水、二甲苯、梯度乙醇(100、95、90、80、70%)、DAB工作液(临用前配制,5 μl 20×DAB+1μL 30%H2O2+94 μl PBS)、Proteinase K工作液(10-20 μg/ml in 10 mM Tris/HCl,pH 7.4-8)或细胞通透液(0.1% Triton X-100 溶于0.1% 柠檬酸钠,临用前配制)、苏木素或甲基绿、DNase 1(3000 U/ml–3 U/ml in 50 mM Tris-HCl,pH 7.5,10 mM MgCl2,1 mg/ml BSA)等。
罗氏实时荧光定量PCR仪课件

02
罗氏实时荧光定量PCR仪 介绍
仪器的基本结构
仪器外观
操作界面
罗氏实时荧光定量PCR仪外观为长方 体,尺寸适中,便于实验室放置。
仪器配备彩色触摸屏,操作界面友好 ,方便用户进行参数设置和实验操作 。
内部结构
仪器内部包括加热模块、光学检测系 统、电脑控制系统等部分,各部分协 同工作完成PCR扩增和荧光检测。
制定仪器的维护保养计划,按照 计划进行保养,保证仪器的正常
运行和使用寿命。
THANKS
感谢观看
通过比较不同样本之间的基因表达数据,可以筛选出差异表达的基 因,进一步研究其在生物学过程中的作用。
基因表达调控机制研究
通过实时监测基因转录水平的变化,有助于深入了解基因表达的调 控机制,为相关疾病的研究和治疗提供线索。
在病毒检测中的应用
1 2 3
病毒载量测定
利用罗氏实时荧光定量PCR仪,可以快速、准确 地测定病毒载量,为临床诊断和治疗提供依据。
02
根据错误提示查找相关资料或联系技术支持解决。
仪器运行结果不准确
03
检查样品是否符合要求,运行程序是否正确,仪器是否经过校
正等。
仪器的保养与校正
定期清洗仪器内部
根据仪器使用情况定期清洗仪器 内部,包括清洗反应管、更换滤
芯等。
校正仪器
定期邀请专业技术人员对仪器进 行校正,确保仪器准确性。
建立维护保养计划
详细记录实验数据,包括荧光信号的 变化、扩增曲线等,并对数据进行整 理和分析。
实验后处理
仪器清洁与保养
数据审核与报告撰写
实验结束据进行审核,撰写详细的实验报 告,包括实验目的、方法、结果和结论等 。
样品处理
罗氏地高辛标记说明书 英文版

For life science research only. Not for use in diagnostic procedures.FOR IN VITRO USE ONLY.DIG High Prime DNA Labeling and Detection Starter Kit IFor color detection with NBT/BCIP.Random primed DNA labeling with digoxigenin-dUTP,alkali-labile and detection of hybrids by enzyme immunoassayCat. No. 11 745 832 910Store at Ϫ15 to Ϫ25° C Kit for 12 labeling reactions of 10 ng-3 g DNAand detection of 24 blots of 100 cm²Instruction ManualVersion November 20091.Preface1.1Table of Contents1.Preface (2)1.1Table of Contents (2)1.2Kit contents (3)2.Introduction (5)2.1Product overview (5)3.Procedures and required materials (8)3.1Before you begin (8)3.2DIG-DNA Labeling (9)3.3Determination of labeling efficiency (12)3.4DNA transfer and fixation (15)3.5Hybridization (17)3.6Immunological detection (19)3.7Stripping and reprobing of DNA blots (21)4.Results (22)4.1Typical results (22)5.Appendix (23)5.1Trouble shooting (23)5.2References (24)5.3Ordering Information (25)1.2Kit contentsBottle/ Cap Label Contentincluding function1DIG-HighPrime •50 l DIG-High Prime• 5 × conc. labeling mixture containingoptimal concentrations of random primers, nucle-otides, DIG-dUTP (alkali-labile),Klenow enzyme and buffer components •ready-to-use•clear, viscous solution•for efficient random primed labeling of DNA2DIG-labeledControl DNA •20 l•[5 g/ml] pBR328 DNA(linearized with Bam HI)•clear solution•determination of labeling efficiency3DNA DilutionBuffer •3×1ml•[50 g/ml fish sperm DNA in 10 mM Tris-HCl, 1 mM EDTA; pH 8.0 at +25°C]•clear solution4Anti-Digoxigenin-AP Conjugate •100 l•[750 U/ml]•from sheep, Fab-fragments,conjugated to alkaline phosphatase •clear solution5NBT/BCIP• 6 × 1 ml•50x conc. stock solution [18.75 mg/ml nitrobluetetrazolium chloride and 9.4 mg/ml 5-bromo-4-chloro-3-indolyl-phosphate in67% (v/v) DMSO]•can vary between light yellow and brown, clearsolution•reacts with alkaline phosphatase.6Blockingsolution • 4 × 100 ml•10 × conc.•yellow, viscous solution7DIG Easy HybGranules •to give 4 × 100 ml (preparation page 17)•Hybridization solutionAdditional equipment and reagents requiredIn addition to the reagents listed above, you have to prepare several solutions.In the table you will find an overview about the equipment which is needed for the different procedures.Detailed information is given in front of each procedure.Procedure Equipment Reagents3.2 DIG-DNA labeling Water bath•H2O, sterile, doubledistilled water•EDTA, 0.2 M, pH 8.0,sterile3.3 Semi-quantitativedetermination oflabeling efficiencyNylon membranes positivelycharged*•DIG Wash andBlock Buffer Set*•TE-bufferor•Washing buffer•Maleic acid buffer•Detection buffer•TE-buffer3.4 DNA transferand fixation•UV-transilluminator•commercial availableUV-cross linker• 2 × SSCor•10 × SSC3.5 Hybridization•Nylon membranes, positivelycharged*•Hybridization bags*or•Temperature resistant,sealable plastic bags or rollerbottlesNote: Do not use open trays whenworking with DIG Easy Hyb buffer3.6 Immunologicaldetection•Container of appropriate size inrelation to filter size•Hybridization bags*•DIG Wash andBlock Buffer Set*•TE-bufferor•Washing buffer•Maleic acid buffer•Detection buffer•TE-buffer3.7 Stripping andreprobing of DNAblots•Large tray•Water bath•10 × SSC•10% SDS•0.2 M NaOH2.Introduction2.1Product overviewTest principle The DIG High Prime DNA Labeling and Detection Starter Kit I uses digoxigenin(DIG), a steroid hapten, to label DNA probes for hybridization and subsequentcolor detection by enzyme immunoassay [1].Stage DescriptionDNA labeling DIG-labeled DNA probes are generated with DIG-High Primeaccording to the random primed labeling technique. DIG-High Primeis a specially developed reaction mixture containing digoxigenin-dUTP, alkali-labile (Fig. 2) and all reagents, including enzymenecessary for random primed labeling, premixed in an optimized5 × concentrated reaction buffer.Hybridization DIG-labeled probes are used for hybridization to membraneblotted nucleic acids according to standard methods. The use of thealkali-labile form of DIG-11-dUTP enables easier and more efficientstripping of blots for rehybridization with a secondDIG-labeled probe.Immunological detection The hybridized probes are immunodetected with anti-digoxigenin-AP, Fab fragments and are then visualized with the colorimetric substrates NBT/BCIP.ApplicationDIG-labeled DNA probes can be used:•for all types of filter hybridization•for single copy gene detection in total genomic DNA, even from organisms with high complexity, e.g. human, barley, and wheat.Sample material•DNA fragments of at least 100 bp •linearized plasmid, cosmid or DNA •supercoiled DNAAssay timeThis table lists the reaction time of the single steps Number of tests1 kit is sufficient for•12 standard labeling reactions of up to 3 g template DNA and detection of•24 blots of 100 cm 2.Quality ControlUsing unlabeled control-DNA (pBR 328), labeled as described in the protocol, 0.1 pg of homologous DNA is detected in a dot blot after 16 h color develop-ment (1 pg of homologous DNA can be detected after 1h color development).Step Reaction time DNA labeling 1 h-O/N Hybridization6 h or O/N Immunological detection 1.5 h Color development0.5 - 16 hKit storage/ stabilityThe unopened kit is stable at -15 to -25° C until the expiration date printed on the label. Shipping conditions on dry ice.Once opened, please refer to the following table for proper storage.Sensitivity and specificityA single copy human gene is detected in a Southern blot of 1 g digested pla-centa DNA.Note: Sensitivity depends both on the concentration of labeled DNA in the hybridization and on the time of color reaction.AdvantagesThis table describes benefits and features of the kit.Kit componentStorageAnti-Digoxigenin-AP Conjugate cap 42-8° C, stableBlocking solution bottle 6•unopened, stable at 15-25° C•once opened, it should be aliquoted and stored at -15 to -25° C or at 2-8° C up to one month when keeping sterile•working solution should always be prepared fresh NBT/BCIP cap 6•2-8° C,stable•or at least 4 weeks at 15-25° CNote: During shipment of the kit on dry ice, a precipitate may occur which is easily dissolved by briefly warming to 37°CBenefitFeatureAccurate and fastThe use of premixed DIG-High Prime mini-mizes the hands-on-time required to label DNA probes and increases yields and reproducibility.Sensitive Single-copy genes can be detected in total human DNA complex and plant genomes.Time-savingDIG-labeled probes can be stored for at least one year. Hybridization solutions can be reused 3 – 5 times, depending on the amount of labeled probe used for signal generation in each hybridization.3.Procedures and required materials3.1Before you beginGeneral handling rec-ommendationsThis table describes general hints for DIG labeling and detection.OverviewRecommendation GuidelineWork under clean conditions•Autoclave DIG System solutions•Filter-sterilize solutions containing SDS•Tween 20 should be added to previouslysterilized solutionsUse clean incubation trays Rigorously clean and rinse laboratory traysbefore each use.Membrane handling requirements•Wear powder-free gloves•Handle membrane only on the edges andwith clean forcepsSection 3.2DIG-DNA labeling↓Section 3.3Quantification of labeling efficiency↓Section 3.4DNA fixation↓Section 3.5Hybridization↓Section 3.6Immunological detection↓Section 3.7Stripping and reprobing of DNA blots3.2DIG-DNA LabelingIntroduction DNA is random primed labeled with Digoxigenin-11-dUTP using DIG-HighPrime, a 5x concentrated labeling mixture of random hexamers, dNTP mix con-taining alkali-labile Digoxigenin-11-dUTP, labeling grade Klenow enzyme andan optimized reaction buffer.Additional equipment and reagents required •water bath•ice/waterThis table lists composition, storage and use of the required reagents in addi-tion to kit componentsTemplate DNA The following table lists the recommended features of the template DNALabeling of DNA isolated from agaroseIf you intend to perform genomic Southern blotting, you should separate thetemplate insert DNA from the vector by agarose gel electrophoresis.Excise the DNA fragment from an agarose gel. For best results use either our High Pure PCR Product Purification Kit * or our Agarose Gel DNA ExtractionKit* to separate the DNA from the agarose.Solution Composition Storage Use Water Autoclaved, double distilled water15-25° C,stableDilution of DNAEDTA0.2 M ethylenediamino- tetraacetic acid,pH 8.015-25° C,stableStopping thelabeling reactionFeature DetailPurity For plasmid DNA use the High Pure Plasmid Isolation Kit * for purification.When other commercially available purification kits are used, we recommendto do an additional phenol/chloroform extraction to remove residual protein.This step is also necessary when templates have been treated with restrictionor other modifying enzymes before labeling.Size To obtain optimal results, template DNA should be linearized andshould have a size of 100 or larger. Template DNA >5 kb should be restric-tion-digested using a 4 bp cutter (e.g. Hae III) prior to labeling.Amount With the procedure described below principally 10 ng – 3 g of template can be labeled, however, please check in the given table the necessaryamount of probe needed for your size of blot. By scaling up of all volumes andcomponents accordingly this procedure can be used for labeling of largeramounts. If single-copy gene detection in complex genomes is performed atleast 300 ng of template DNA (probe concentration: 25 ng/ml hybridizationsolution) should be labeled.Procedure This procedure is designed for 10 ng-3 g of DNA. Larger amounts (up to 10g) can be labeled by scaling up of all components and volumes.Yield of labeling reac-tion Table 1:This table shows you the yield of DIG-High Prime labeling under optimal condi-tions.In the standard reaction with 1 g DNA per assay approx. 15% of the nucle-otides are incorporated into about 0.8 g of newly synthesized DIG-labeled DNA within 1 h and approx. 38% of the nucleotides into about 2 g after 20 h. Using DIG-High Prime solution, reactions were performed with increasingamountsof different template DNAs for 1 h and 20 h. The yield of DIG-labeled DNA was determined by incorporation of a radioactive tracer and confirmed by a dot blot (Average of 10 independent labeling assays)Step Action1Add1g template DNA (linear or supercoiled) and autoclaved, double distilled water to a final volume of 16 l to a reaction vial.2Denature the DNA by heating in a boiling water bath for 10 min and quickly chilling in an ice/water bath.Note: Complete denaturation is essential for efficient labeling.3•Mix DIG-High Prime (vial 1) thoroughly and add 4 l to the denatured DNA, mix and centrifuge briefly.•Incubate for 1 h or O/N at 37° C.Note: Longer incubations (up to 20 h) will increase the yield of DIG-labeledDNA (see table below ).4Stop the reaction by adding 2 l 0.2 M EDTA (pH 8.0) and/or by heating to 65°C for 10 min.Note: The length of the DIG-labeled fragments obtained with DIG-High Prime range from 200 bp to 1000 bp or larger, depending on the length of theoriginal template.T emplate DNA 1 h20 h10 ng 45 ng 600 ng30 ng 130 ng1050 ng100 ng 270 ng1500 ng300 ng 450 ng2000 ng 1000 ng 850 ng2300 ng3000 ng1350 ng2650 ng.Fig. 2: Yield of DIG-labeled DNA from different amounts of template DNA after 1 and 20 h incu-bation of the DIG-High Prime reaction at 37°C.3.3Determination of labeling efficiencyIntroduction Determination of the yield of DIG-labeled DNA is most important for optimaland reproducible hybridization results. Too high of a probe concentration in thehybridization mix causes background, while too low of a concentration leads toweak signals.Test principle The preferred method for quantification of labeled probes is the direct detec-tion method.Preparation of additional solutions requiredPlease find in the following table composition and preparation of additionalreagents required. The following buffers are also available in the DIG Wash and Block Buffer Set, DNase and RNase free.*Please note: These solutions are also used in the detection procedure ofchapter 3.6. and can be prepared in larger quantities.Stage Description1• A series of dilutions of DIG-labeled DNA is applied to a small strip of nylon membrane positively charged*.•Part of the nylon membrane is preloaded with defined dilutions of DIG-labeled control DNA (vial 2) which are used as standards.2•The nylon membrane is subjected to immunological detection with anti-digoxigenin-AP conjugate (vial 4) and the premixed stock solutionof NBT/BCIP (vial 5).•The intensities of the dilution series of DIG-labeled DNA and control DNA are compared.Solution Composition / Preparation Storage andstabilityUseWashingbuffer0.1 M Maleic acid, 0.15 M NaCl;pH 7.5 (20° C); 0.3% (v/v) Tween 2015-25° C,stableRemovalof unboundantibody Maleic acidbuffer0.1 M Maleic acid, 0.15 M NaCl;adjust with NaOH (solid) to pH 7.5(20° C)15-25° C,stableDilution ofBlocking solutionDetectionbuffer0.1 M T ris-HCl, 0.1 M NaCl,pH 9.5 (20° C)15-25° C,stableAdjustment ofpH to 9.5TE-buffer10 mM T ris-HCl, 1 mM EDTA,pH 8.015-25° C,stableStopping colorreactionPreparation of kit working solutionsThe following table shows the preparation of kit working solutions ProbequantificationLabeled probes and the DIG-labeled control DNA (vial 2) must be diluted to 1 ng/l, according to the expected yield of synthesized nucleic acid to start the dilution series below. The expected yield of DIG-labeled DNA in your probe can best be estimated by using the table 1 in chapter 3.2. The yield depends on the starting amount of template and incubation time.Note: The yields given in table 1 were achieved under optimal conditions with highly purified template DNA.Dilution seriesPrepare a dilution series of your labeled probe and your control DNA as described in the table:Solution Composition/preparationStorage and stability Use Blocking solutionPrepare a 1 × working solution by diluting the 10x Blocking solution (vial 6) 1:10 in Maleic acid buffer.Always prepare fresh Blocking of unspecific binding sites on the membrane Antibody solutionCentrifuge Anti-Digoxigenin-AP (vial 4) for 5 min at 10 000 rpm in the original vial prior to each use, and pipet the necessary amount carefully from the surface. Dilute Anti-Digoxigenin-AP 1: 5 000 (150 mU/ml) in Blocking solution.12 h at 2-8° CBinding to the DIG-labeled probeColor-substrate solutionAdd 40 l of NBT/BCIP stock solution (vial 5) to 2 ml of Detection buffer.Note: Store protected from light!always prepare freshVisualization of antibody-binding TubeDNA (l)From tube #DNA Dilution Buffer (vial 3)(l)DilutionFinal concentration1diluted original1 ng/l 2211981:10010 pg/l 3152351:3.3 3 pg/l 452451:101 pg/l 553451:100.3 pg/l 654451:100.1 pg/l 755451:100.03 pg/l 856451:100.01 pg/l9-50-0Procedure The following procedure describes the direct detection.Note: Use sufficient buffer volumes to cover the membrane completely duringall steps.Analyzing the resultsCompare the intensity of the spots out of your labeling reaction to the control and calculate the amount of DIG-labeled DNA. If the 0.1 pg dilution spots of your probe and of the control are visible, then the labeled probe has reached the expected labeling efficiency (pls. see table 1 in 3.2.) and can be used in the recommended concentration in the hybridization.The following spots should be visibleStep Action1Apply a1l spot of tubes 2-9 from your labeled probes and the labeled control to the nylon membrane.2Fix the nucleic acid to the membrane by cross linking with UV-light or baking for 30 min at 120 ° C.3•Transfer the membrane into a plastic container with 20 mlMaleic acid buffer.•Incubate under shaking for 2 min at 15-25° C.4Incubate for 30 min in 10 ml Blocking solution.5Incubate for 30 min in 10 ml Antibody solution.6Wash with 10 ml Washing buffer, 2 × 15 min.7Equilibrate 2-5 min in 10 ml Detection buffer.8Incubate membrane in 2 ml freshly prepared color substrate solution in a appropriate container in the dark. Do not shake during color development.Note: The color precipitate starts to form within a few minutes. The membrane can be exposed to light for short time periods to monitor color development.9Stop the reaction, when desired spot or band intensities are achieved, by washing the membrane for 5 min with 50 ml of sterile double dist. wateror with TE-buffer.Results can be documented by photocopying the wet filter or by photography.Incubation time Appearance5-10 min30 pg spot30 min 3 pg spot3.4DNA transfer and fixationTransfer methods and membranes Standard protocols for gel electrophoresis, denaturation and neutralization of the gel are described in Sambrook et al. (2). Gels lacking ethidium bromide are preferred, because ethidium can cause uneven background problems. All com-mon types ofDNA transfer methods are suitable for subsequent DIG hybridization (4,5).In our experience, best results are obtained when gels are blotted by capillary transfer with 20 × SSC on nylon membranes*, positively charged.Note: Alkali transfer (e.g., in 0.4 M NaOH) is not suitable for the transfer of DIG-labeled molecular weight markers*.Fixation procedure Fix the DNA to the membrane by any of the following procedures:Storage of the membranePlease refer to the following table.IF you want to...THEN...UV-crosslinking(nylon membrane)•Place the membrane on Whatman 3MM-paper soakedwith 10 × SSC.•UV-crosslink the wet membrane without priorwashing.•After the UV-crosslinking, rinse the membrane brieflyin double distilled water and allow to air-dry.bake at 120° C(nylon membrane)•Wash the membrane briefly in 2 × SSC.•Bake the nylon membrane at 120° C for 30 min oraccording to the manufacturer`s instructions.bake at 80° C(nylon membrane)•Wash the membrane briefly in 2 × SSC.•Bake at 80° C for 2 h under vacuum.IF...THEN...you want to go e the membrane immediately for prehybridization. you want to work later on store the membrane dry at 2-8° C.3.5HybridizationAdditional equipment required •ice/water•shaking water-bath•or hybridization oven•temperature resistant plastic or glass boxes, petri dishes, roller bottles or sealable plastic bags.Note: Do not use open containers with DIG Easy Hyb buffer.Preparation ofDIG Easy Hyb working solution Add carefully 64 ml sterile double distilled water in two portions to the DIG Easy Hyb Granules (bottle 7), dissolve by stirring immediately for 5 min at 37° C.Hybridization temperature The appropriate hybridization temperature is calculated according to GC con-tent and percent homology of probe to target according to the following equa-tion:Tm= 49.82 + 0.41 (% G + C) - (600/l) [l = length of hybrid in base pairs]Topt.=Tm–20 to 25° C(The given numbers of the equation were calculated according to a standard equation for hybridization solutions containing formamide, 50%.) The actual hybridization temperature T opt. for hybridization with DIG Easy Hyb is 20-25° C below the calculated T m value. T opt. can be regarded as a stringent hybridization temperature allows up to 18% mismatches between probe and target. When the degree of homology of your probe to template is less than 80%, you should lower Topt. accordingly (approx. 1.4°C below Tmper 1 % mismatch) and also adjust the stringent washing steps accordingly (i.e. increase SSC concentration and lower washing temperature).Procedure Please refer to the following table.Storage of hybridization solution DIG Easy Hyb containing DIG-labeled probe can be stored at Ϫ15 to Ϫ25° C and be reused several times when freshly denatured at 68°C for 10 min before use.Note: Do not boil DIG Easy Hyb.Stringency washes For most DNA:DNA applications, a stringency wash with 0.5 × SSC is sufficient. The correct post washing conditions have to be determined empirically for each probe.•For human genomic DNA use 0.5 × SSC and 65° C.•Probes > 150 bp and with a high G/C content should be washed at 68° C.•For shorter probes around 100 bp or shorter, the wash temperature must belowered.This table describes how to perform post-hybridization washes.Step Action1•Pre-heat an appropriate volume of DIG Easy Hyb (10 ml/100 cm2 filter) to hybridization temperature (37 - 42° C).•Prehybridize filter for 30 min with gentle agitation in an appropri-ate container.Note: Membranes should move freely, especially if you use severalmembranes in the same prehybridization solution.2Denature DIG-labeled DNA probe (about 25 ng/ml) by boiling for5 min and rapidly cooling in ice/water.Note: As DIG-11-dUTP is alkali-labile, DNA probes cannot be dena-tured by alkali treatment (NaOH).3Add denatured DIG-labeled DNA probe to pre-heated DIG Easy Hyb(3.5 ml/100 cm2 membrane) and mix well but avoid foaming(bubbles may lead to background).4•Pour off prehybridization solution and add probe/hybridization mixture to membrane.•Incubate 4 hours - O/N with gentle agitation.Step Action1Wash 2 × 5 min in ample 2x SSC, 0.1% SDS at 15-25° C under con-stant agitation.2Wash 2 × 15 min in 0.5x SSC, 0.1% SDS (prewarmed to wash temper-ature) at 65-68° C under constant agitation.3.6Immunological detectionAdditional reagents required Please find in the following table composition and preparation of additional reagents required. The following buffers are also available in the DIG Wash and Block Buffer Set, DNase and RNase free*.Preparation of kit working solutions In the following table the preparation of kit working solutions is described.Solution Composition / Preparation Storage andstabilityUseWashingbuffer0.1 M Maleic acid, 0.15 M NaCl;pH 7.5 (20° C); 0.3% (v/v) Tween2015-25° C,stableWashing ofmembraneMaleic acidbuffer0.1 M Maleic acid, 0.15 M NaCl;adjust with NaOH (solid) to pH7.5 (20° C)15-25° C,stableDilution ofBlockingsolution Detectionbuffer0.1 M Tris-HCl, 0.1 M NaCl,pH 9.5 (20° C)15-25° C,stableAlkalinephosphatasebufferTE-buffer10 mM Tris-HCl, 1 mM EDTA, pH8.015-25° C,stableStoppingcolor reactionSolution Composition / Preparation Storage andstabilityUseBlockingsolutionPrepare a 1x working solution bydiluting 10 × Blocking solution(vial 6) 1:10 with Maleic acidbuffer.Alwaysprepare freshBlocking ofunspecificbinding sitesAntibodysolutionCentrifuge Anti-Digoxigenin-AP(vial 4) for 5 min at 10 000 rpm inthe original vial prior to each use,and pipet the necessary amountcarefully from the surface. DiluteAnti-Digoxigenin-AP 1:5000 (150mU/ml) in Blocking solution.12 h at 2-8° C Binding to theDIG-labeledprobeColor-substratesolutionAdd 200 l of NBT/BCIP stocksolution (vial 5) to 10 ml of Detec-tion buffer.Note: Store protected from light!Alwaysprepare freshVisualizationof antibody-bindingProcedure This table describes how to perform the immunological detection ona 100 cm2 membrane.Note: All incubations should be performed at 15-25° C with agitation. If themembrane is to be reprobed, do not allow the membrane to dry at any time.Storage of membrane Please refer to the following table.Step Action1After hybridization and stringency washes, rinse membrane briefly (1-5) min in Washing buffer.2Incubate for 30 min in 100 ml Blocking solution.3Incubate for 30 min in 20 ml Antibody solution.4Wash 2 x 15 min in 100 ml Washing buffer.5Equilibrate 2-5 min in 20 ml Detection buffer.6Incubate membrane in 10 ml freshly prepared color substrate solu-tion in a appropriate container in the dark. Do not shake duringcolor development.Note: The color precipitate starts to form within a few minutes andthe reaction is usually complete after 16 h. The membrane can beexposed to light for short time periods to monitor color development.7Stop the reaction, when desired spot or band intensities are achieved, by washing the membrane for 5 min with 50 ml of steriledouble dist. water or with TE-buffer.Results can be documented by photocopying the wet filter or by pho-tography.IF...THEN...you want to reprobe the membrane the membrane should not dry off at anytime, store in sealed plastic bag.Note: If you want to maintain the color,store membranes in TE buffer do notallow the membrane to dry.you don´t want to reprobe dry the membrane at 15 to 25°C for stor-age.Note: Color fades upon drying, to revita-lize the color, wet the membrane in TEbuffer.3.7Stripping and reprobing of DNA blotsGeneral The alkali-labile form of DIG-11-dUTP enables easier and more efficient strip-ping of blots for rehybridization experiment.Additional reagents required •Dimethylformamid (DMF)•0.2 N NaOH, 0.1% SDS (w/v)• 2 × SSCProtocolStorage of stripped membrane Please refer to the following table.Note: When stripping and rehybridization of blots is planned, the membrane should not dry off at any time.CAUTION: Work under a fume hoodStep Action1•Heat DMF in a large glass beaker in a water bath under a fume hood to 50-60° C.•Incubate the membranes in the heated DMF until the blue color precipitate is removed from the filter.CAUTION: DMF is volatile and can be ignited above 67° C.2Rinse membrane briefly in double distilled water.3Wash for 2 × 15 min in 0.2 N NaOH, SDS, 0.1% (w/v) at 37° C under constant agitation.4Equilibrate briefly in 2 × SSC.5Prehybridize and hybridize with a second probe.Once the membrane is stripped, it can be stored in Maleic acid buffer or2 × SSC until used again.4.Results4.1Typical resultsGenomic Southern blotFigure 3: This figure shows you the detection of a single-copy gene (ß-actin) in total human DNA using the standard protocol.Size (kb)1234215.1/5.04.33.52.01.91.61.40.950.830.562.551.0 。
栉孔扇贝sox9基因的cDNA克隆及其在不同发育时期性腺中的表达特征

栉孔扇贝sox9基因的cDNA克隆及其在不同发育时期性腺中的表达特征梁少帅;于潇含;杨丹丹;秦贞奎【摘要】SOX9 is a member of the SOXE group in the SOX family, which plays an important role in sex deter-mination and differentiation of vertebrates. sox9 is specifically expressed in the testes of mammals, birds, and rep-tiles, and is expressed in both the testes and ovaries in amphibians and fishes. To measure the expression of sox9 in the gonads of invertebrates, we cloned a 2835 bp full-length cDNA of the scallop Chlamys farreri sox9 (Cf-sox9) which contains a 1413 bp open reading frame (ORF) encoding a 470-amino acid sequence with the HMG-box of the SOX family and the highly conserved region of the SOXE group, although there is no Pro-Gln- Ser-rich region at the C-terminus of vertebrate SOXE proteins. Cf-sox9 DIG-labeled RNA probes and Cf-SOX9 polyclonal anti-bodies were produced based on the Cf-sox9 full-length cDNA sequence. ISH and immunohistochemistry detection showed that Cf-sox9 transcripts and Cf-SOX9 proteins were located in all germ cells of C. farreri testes and ova-ries at different developmental stages. The expression patterns between sox9 mRNA and SOX9 protein were simi-lar. In the testes, the intensity of positive signals was highest in the spermatocytes and lowest in the spermatozoa. In the ovaries, the intensity of positive signals gradually decreased across the oogonia, oocytes, and mature oo-cytes. The expression characteristics of Cf-sox9 in C. farrerigonads were different from that most of vertebrates, which show variability in expression between sex, suggesting its function of gonad development and gametogene-sis might be different from vertebrates.%SOX9是SOX家族的SOXE亚族成员,在脊椎动物骨骼发育、胰腺发育、性别决定与分化以及肿瘤形成中发挥重要的作用.sox9在不同种类脊椎动物性腺中的表达存在差异,在无脊椎动物性腺中的表达特征尚不清楚.本研究利用RACE技术克隆了栉孔扇贝(Chlamys farreri)sox9(Cf-sox9)全长的cDNA序列,其长度为2835 bp,开放阅读框为1413 bp,编码470个氨基酸.预测的氨基酸序列具有SOX家族的HMG-box和SOXE亚族的高保守区域,但没有脊椎动物羧基端的Pro-Gln-Ser rich区域.原位杂交和免疫组织化学技术确定Cf-sox9 mRNA和Cf-SOX9蛋白均定位在栉孔扇贝精巢和卵巢的所有生殖细胞中,并且在不同发育时期性腺中呈现了类似的表达规律,即在精巢中,阳性信号在精母细胞中最强,在精子中最弱;在卵巢中,阳性信号在卵原细胞、卵母细胞以及成熟卵中呈现逐渐减弱的趋势.这一表达特征与脊椎动物性腺中多样的性别差异表达特征不同,提示sox9在贝类性腺发育和配子发生中的作用可能与大多数脊椎动物不同.【期刊名称】《中国水产科学》【年(卷),期】2017(024)006【总页数】9页(P1184-1192)【关键词】栉孔扇贝;sox9;性腺;配子发生【作者】梁少帅;于潇含;杨丹丹;秦贞奎【作者单位】中国海洋大学, 海洋生物遗传学与育种教育部重点实验室, 山东青岛266003;中国海洋大学, 海洋生物遗传学与育种教育部重点实验室, 山东青岛266003;中国海洋大学, 海洋生物遗传学与育种教育部重点实验室, 山东青岛266003;中国海洋大学, 海洋生物遗传学与育种教育部重点实验室, 山东青岛266003【正文语种】中文【中图分类】Q96SOX(SRY-related HMG-box genes)是一类含有 HMG-box的转录因子, 主要与脊椎动物胚胎发生、性别决定、组织器官形成以及疾病发生等有关。
翻译好的罗氏公司Tunel试剂盒操作说明方案

罗氏(R o c h e)公司T u n e l试剂盒操作说明书(Insitucelldeathdetectionkit-POD法)一、原理:TUNEL(TdT-mediateddUTPnickendlabeling)细胞凋亡检测试剂盒是用来检测组织细胞在凋亡早期过程中细胞核DNA的断裂情况。
其原理是荧光素(fluorescein)标记的dUTP在脱氧核糖核苷酸末端转移酶(TdTEnzyme)的作用下,可以连接到凋亡细胞中断裂DNA的3’-OH末端,并与连接辣根过氧化酶(HRP,horse-radishperoxidase)的荧光素抗体特异性结合,后者又与HRP底物二氨基联苯胺(DAB)反应产生很强的颜色反应(呈深棕色),特异准确地定位正在凋亡的细胞,因而在光学显微镜下即可观察凋亡细胞;由于正常的或正在增殖的细胞几乎没有DNA断裂,因而没有3‘-OH形成,很少能够被染色。
本试剂盒适用于组织样本(石蜡包埋、冰冻和超薄切片)和细胞样本(细胞涂片)在单细胞水平上的凋亡原位检测。
还可应用于抗肿瘤药的药效评价,以及通过双色法确定细胞死亡类型和分化阶段。
二、器材与试剂器材:光学显微镜及其成像系统、小型染色缸、湿盒(塑料饭盒与纱布)、塑料盖玻片或封口膜、吸管、各种规格的加样器及枪头等;试剂:试剂盒含:1号(蓝盖)EnzymeSolution酶溶液:TdT10×、2号(紫盖)LabelSolution标记液:荧光素标记的dUTP1×、3号(棕瓶)Converter-POD:标记荧光素抗体的HRP;自备试剂:PBS、双蒸水、二甲苯、梯度乙醇(100、95、90、80、70%)、DAB工作液(临用前配制,5μl20×DAB+1μL30%H2O2+94μlPBS)、ProteinaseK工作液(10-20μg/mlin10mMTris/HCl,pH7.4-8)或细胞通透液(0.1%TritonX-100溶于0.1%柠檬酸钠,临用前配制)、苏木素或甲基绿、DNase1(3000U/ml–3U/mlin50mMTris-HCl,pH7.5,10mMMgCl2,1mg/mlBSA)等。
DIG RNA LABELING KIT(SP6T7)
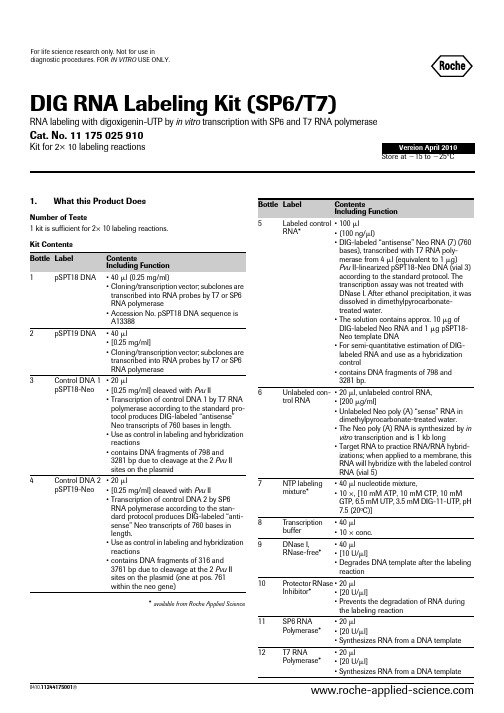
Size
Amount
2.3 Determina- • nylon membranes tion of labeling positively charged* efficiency • UV-transilluminator or • UV cross-linker
Applications DIG-labeled RNA is used for hybridization to • Northern blots • Southern blots • in situ hybridization • plaque or colony lifts • RNase protection experiments L As the linkage between DIG and UTP is resistant to alkali, DIGlabeled RNA can be fragmented by alkaline treatment. Slightly reducing the size of the DIG-labeled RNA probe may make it more suitable for certain applications in in situ hybridization (5, 6).
11
12
0410.11244175001
Storage/Stability The unopened kit is stable at Ϫ15 to Ϫ25°C through the expiration date printed on the label. The kit is shipped on dry ice. Additional Equipment/Reagents Required In addition to the reagents listed above, you must prepare several solutions. In the table you will find an overview of the equipment and reagents needed for the different procedures. Detailed information is given at the beginning of each procedure. Procedure 2.2 DIG RNA labeling Equipment water bath Reagents • H2O, sterile, double distilled water* treated with DMPC • EDTA, 0.2 M, pH 8.0 • RNA dilution buffer • DIG Wash and Block Buffer Set* • Anti-digoxigenin-AP* • CSPD* or CDP-Star* • DIG Luminescent Detection Kit*
TUNEL细胞凋亡检测试剂盒 罗氏

For research purposes only. Not for use for in vitro diagnosticprocedures for clinical diagnosis.In Situ Cell Death Detection Kit, PODKit for immunohistochemical detection and quantification of apop-tosis (programmed cell death) at single cell level, based on labeling of DNA strand breaks (TUNEL technology): Analysis by light microscopy.Cat. No. 1 684 817Store at Ϫ15 to Ϫ25°C 1 Kit (50 tests)Instruction ManualVersion 3, January 20031. Preface1.1Table of contentsP reface (2)1.1.1Table of contents (2) (3)1.2 Kitcontents (5)2. Introduction2.1Product overview (5) (8)2.2 Backgroundinformation3. Procedures and required materials (10)3.1Flow chart (10)3.2Preparation of sample material (11)3.2.1Adherent cells, cell smears and cytospin preparations (11) (12)sections3.2.2 Tissue3.2.2.1 Treatment of paraffin-embedded tissue (12)3.2.2.2Treatment of cryopreserved tissue (14)3.3Labeling protocol (15)3.3.1 Before you begin (15)3.3.2Labeling protocol for adherent cells, cell smears, cytospin preparations,and tissues (16)3.3.3 Labeling protocol for difficult tissue (17) (18)conversion3.4 Signal (19)4. Appendix (19)4.1 Trouble-shooting (22)4.2 References (23)4.3 Relatedproducts1.2 KitcontentsCaution The Label solution contains cacodylate, toxic by inhalation and swal-lowed, and cobalt dichloride, which may cause cancer by inhalation.Avoid exposure and obtain special instructions before use.When using do not eat, drink or smoke. After contact with skin, washimmediately with plenty of water. In case of accident or if you feelunwell seek medical advice immediately (show label where possible).Collect the supernatants from the labeling reactions in a tightly closed,non-breakable container and indicate contents. Discard as regulatedfor toxic waste.Kit contents Please refer to the following table for the contents of the kit.Vial/CapLabel Contents1 blue Enzyme Solution•Terminal deoxynucleotidyl transferasefrom calf thymus (EC 2.7.7.31), recom-binant in E. coli, in storage buffer•10× conc.•5×50l2 violet Label Solution•Nucleotide mixture in reaction buffer•1×conc.• 5 × 550 l3 yellow Converter-POD•Anti-fluorescein antibody, Fab frag-ment from sheep, conjugated withhorse-radish peroxidase (POD)•Ready-to-use• 3.5mlAdditional equipment required In addition to the reagents listed above, you have to prepare several solutions. In the table you will find an overview about the equipment which is needed for the different procedures.Detailed information is given in front of each procedure.Procedure Equipment Reagents Preparation of sample material (section 3.2)•Adherent cells, cell smears and cytospinpreparations (section3.2.1)•Cryopreserved tissue (section 3.2.2.2)•Washing buffer: Phosphate buffered saline(PBS*)•Blocking solution: 3% H2O2 in methanol•Fixation solution: 4% Paraformaldehyde inPBS, pH 7.4, freshly prepared •Permeabilisation solution: 0.1% Triton X-100 in 0.1% sodium citrate, freshly pre-pared (6)Paraffin-embedded tissue (section 3.2.2.1)•Xylene and ethanol (absolute, 95%, 90%, 80%, 70%, diluted in double distilled water)•Washing buffer: PBS*•Proteinase K*, nuclease, working solution: [10-20 µg/ml in 10 mM Tris/HCl, pH 7.4-8] Alternative treatments•Permeabilisation solution: (0.1% Triton1) X–100, 0.1% sodium citrate) , freshly prepared •Pepsin* (0.25% - 0.5% in HCl, pH 2) or trypsin*, 0.01 N HCl, nuclease free•0.1 M Citrate buffer, pH 6 for microwave irradiationLabeling protocol (section 3.3)Positive control (section 3.3.1)•Micrococcal nuclease or •DNase I, grade I *Adherent cells, cell smears, cytospin preparations, and tissues (section 3.3.2) •Parafilm orcoverslips•HumidifiedchamberWashing buffer: PBS*Difficult tissue (section 3.3.3)•Plastic jar•Microwave•Humidifiedchamber•Citrate buffer, 0.1 M, pH 6.0.•Washing buffer: PBS*•Tris-HCl, 0.1 M pH 7.5, containing 3% BSA*and 20% normal bovine serumSignal conversion (section 3.4)•Humidified chamber •Parafilm or coverslip •Washing buffer: PBS*•DAB Metal Enhanced Substrate Set* or alternative POD substrates •Mounting medium for light microscopy1.2 Kitcontents,continued2. Introduction2.1Product overviewTest principle Cleavage of genomic DNA during apoptosis may yield double-stranded, low molecular weight DNA fragments (mono- and oligonu-cleosomes) as well as single strand breaks (“nicks”) in high molecularweight DNA.Those DNA strand breaks can be identified by labeling free 3’-OH ter-mini with modified nucleotides in an enzymatic reaction.Fig. 1: Test principleApplication The In Situ Cell Death Detection Kit is designed as a precise, fast and simple, non-radioactive technique to detect and quantify apoptotic celldeath at single cell level in cells and tissues. Thus, the In Situ CellDeath Detection Kit can be used in many different assay systems.Examples are:•Detection of individual apoptotic cells in frozen and formalin fixedtissue sections in basic research and routine pathology.•Determination of sensitivity of malignant cells to drug induced apo-ptosis in cancer research and clinical oncology.•Typing of cells undergoing cell death in heterogeneous populationsby double staining procedures (6).Specificity The TUNEL reaction preferentially labels DNA strand breaks gener-ated during apoptosis. This allows discrimination of apoptosis fromnecrosis and from primary DNA strand breaks induced by cytostaticdrugs or irradiation (3, 4).Test interference False negative results: DNA cleavage can be absent or incomplete in some forms of apoptotic cell death (37). Sterical hindrance such asextracellular matrix components can prevent access of TdT to DNAstrand breaks. In either case false negative results can be obtained.False positive results: Extensive DNA fragmentation may occur in latestages of necrosis (4, 38).DNA strand breaks may also be prominent in cell populations withhigh proliferative or metabolic activity. In either case false positiveresults may be obtained. To confirm apoptotic mode of cell death, themorphology of respective cells should be examined very carefully.Morphological changes during apoptosis have a characteristic pattern.Therefore evaluation of cell morphology is an important parameter insituations where there is any ambiguity regarding interpretation ofresults.Sample material•Cytospin and cell smear preparations•Adherent cells cultured on chamber slides (31)•Frozen or formalin-fixed, paraffin-embedded tissue sections (1, 25,26, 29, 30, 32–34, 36, 39)Assay time2–3 hours, excluding culture, fixation and permeabilisation of cells and preparation of tissue sections.Number of tests The kit is designed for 50 tests.Kit storage/ stability The unopened kit is stable at Ϫ15 to Ϫ25°C through the expiration date printed on the label.Reagent Storage and stabilityTUNEL reaction mixture The TUNEL reaction mixture should be pre-pared immediately before use and shouldnot be stored.Keep TUNEL reaction mixture on ice untiluse.Converter-POD Once thawed the Converter-POD solutionshould be stored at 2–8°C (maximum stabil-ity 6 months).Note: Do not freeze!Advantage Please refer to the following table.Benefit FeatureSensitive Detection of apoptotic cell death at singlecell level at very early stages (1, 2, 6).Specific Preferential labeling of apoptosis versusnecrosis (3, 4).Fast Short assay time (2-3 h).Convenient•Reagents are provided in stable, opti-mized form.•No dilution steps required.Flexible•Suitable for fixed cells and tissue. Thisallows accumulation, storage and trans-port of samples (2, 5).•Double staining enables identification oftype and differentiation state of cellsundergoing apoptosis (6).Function-tested Every lot is function-tested on apoptoticcells in comparison to a master lot.2.2 BackgroundinformationCell death Two distinct modes of cell death, apoptosis and necrosis, can be distin-guished based on differences in morphological, biochemical andmolecular changes of dying cells.Programmed cell death or apoptosis is the most common form ofeukaryotic cell death. It is a physiological suicide mechanism that pre-serves homeostasis, in which cell death naturally occurs during normaltissue turnover (8, 9). In general, cells undergoing apoptosis display acharacteristic pattern of structural changes in nucleus and cytoplasm,including rapid blebbing of plasma membrane and nuclear disintegra-tion. The nuclear collapse is associated with extensive damage tochromatin and DNA-cleavage into oligonucleosomal length DNA frag-ments after activation of a calcium-dependent endogenous endonu-clease (10, 11). However, very rare exceptions have been describedwhere morphological features of apoptosis are not accompanied witholigonucleosomal DNA cleavage (37).Apoptosis Apoptosis is essential in many physiological processes, includingmaturation and effector mechanisms of the immune system (12, 13),embryonic development of tissue, organs and limbs (14), developmentof the nervous system (15, 16) and hormone-dependent tissueremodeling (17). Inappropriate regulation of apoptosis may play animportant role in many pathological conditions like ischemia, stroke,heart disease, cancer, AIDS, autoimmunity, hepatotoxicity and degen-erative diseases of the central nervous system (18–20).In oncology, extensive interest in apoptosis comes from the observa-tion, that this mode of cell death is triggered by a variety of antitumordrugs, radiation and hyperthermia, and that the intrinsic propensity oftumor cells to respond by apoptosis is modulated by expression ofseveral oncogenes and may be a prognostic marker for cancer treat-ment (21).Identification of apoptosis Several methods have been described to identify apoptotic cells (22– 24). Endonucleolysis is considered as the key biochemical event of apoptosis, resulting in cleavage of nuclear DNA into oligonucleosome-sized fragments. Therefore, this process is commonly used for detec-tion of apoptosis by the typical “DNA ladder“ on agarose gels during electrophoresis. This method, however, can not provide information regarding apoptosis in individual cells nor relate cellular apoptosis to histological localization or cell differentiation.This can be done by enzymatic in situ labeling of apoptosis induced DNA strand breaks. DNA polymerase as well as terminal deoxynucle-otidyl transferase (TdT) (1-6, 25-36, 41) have been used for the incor-poration of labeled nucleotides to DNA strand breaks in situ. The tailing reaction using TdT, which was also described as ISEL (in situ end labeling) (5, 35) or TUNEL (TdT-mediated dUTP nick end labeling) (1, 6, 31, 33) technique, has several advantages in comparison to the in situ nick translation (ISNT) using DNA polymerase:•Label intensity of apoptotic cells is higher with TUNEL compared to ISNT, resulting in an increased sensitivity (2, 4).•Kinetics of nucleotide incorporation is very rapid with TUNEL com-pared to the ISNT (2, 4).•TUNEL preferentially labels apoptosis in comparison to necrosis, thereby discriminating apoptosis from necrosis and from primary DNA strand breaks induced by antitumor drugs or radiation (3, 4).2.2 Backgroundinformation,continued3. Procedures and required materialsThe working procedure described below has been developed andpublished by R. Sgonc and colleagues (6). The main advantage of thissimple and rapid procedure is the use of fluorescein-dUTP to labelDNA strand breaks. This allows the detection of DNA fragmentationby fluorescence microscopy directly after the TUNEL reaction priorto the addition of the secondary anti-fluorescein-POD-conjugate.3.1Flow chartAssay procedure The assay procedure is explained in the following flow chart.Adherent cells, cell smears and cytospin preparations Cryopreservedtissue sectionsParaffin-embeddedtissue sections↓↓↓Fixation •Dewaxation •Rehydration •ProteasetreatmentPermeabilisation of samples↓Addition of TUNEL reaction mixtureOPTIONAL: Analysis of samples by fluorescence microscopy↓Addition of Converter-PODAddition of Substrate solution↓Analysis of samples by light microscopy3.2Preparation of sample material3.2.1Adherent cells, cell smears and cytospin preparationsAdditional solutions required •Washing buffer: Phosphate buffered saline (PBS)•Blocking solution: 3% H2O2 in methanol•Fixation solution: 4% Paraformaldehyde in PBS, pH 7.4, freshly pre-pared•Permeabilisation solution: 0.1% Triton1) X-100 in 0.1% sodium citrate, freshly prepared (6)Procedure In the following table describes the fixation of cells, blocking of endo-genous peroxidase and cell permeabilisation.Note: Fix and permeabilisate two additional cell samples for the nega-tive and positive labeling controls.Step Action1Fix air dried cell samples with a freshly prepared Fixationsolution for 1 h at 15-25°C.2Rinse slides with PBS.3Incubate with Blocking solution for 10 min at 15-25°C.4Rinse slides with PBS.5Incubate in Permeabilisation solution for 2 min on ice (2-8°C).6Proceed as described under 3.3.3.2.2 Tissue sections3.2.2.1 Treatment of paraffin-embedded tissuePretreatment of paraffin embedded tissue Tissue sections can be pretreated in 4 different ways. If you use Pro-teinase K the concentration, incubation time and temperature have to be optimized for each type of tissue (1, 29, 33, 36, 40, 42).Note: Use Proteinase K only from Roche Applied Science, because it is tested for absence of nucleases which might lead to false-positive results!The other 3 alternative procedures are also described in the following table (step 2).Additional solutions required •Xylene and ethanol (absolute, 95%, 90%, 80%, 70%, diluted in dou-ble distilled water)•Washing buffer: PBS•Proteinase K, nuclease free (Cat. No. 745 723), working solution: [10-20 g/ml in 10 mM Tris/HCl, pH 7.4-8]Alternative treatments•Permeabilisation solution: 0.1% Triton1) X–100, 0.1% sodium citrate, freshly prepared•Pepsin* (0.25% - 0.5% in HCl, pH 2) or trypsin*, 0.01 N HCl, nuclease free•0.1 M Citrate buffer, pH 6 for the microwave irradiationProcedure In the following table the pretreatment of paraffin-embedded tissue with Proteinase K treatment and 3 alternative procedures aredescribed.Note: Add additional tissue sections for the negative and positivelabeling controls.Step Action1Dewax and rehydrate tissue section according to standardprotocols (e.g. by heating at 60°C followed by washing inxylene and rehydration through a graded series of ethanoland double dist. water) (1, 33, 36).2Incubate tissue section for 15-30 min at 21–37°C with Pro-teinase K working solution.Alternatives:Treatment:1. Permeabilisa-tion solutionIncubate slides for 8 min.2. Pepsin* (30, 40)or trypsin*15-60 min at 37°C.3. Microwave irradiation •Place the slide(s) in a plastic jar containing 200 ml 0.1 M Citrate buffer, pH6.0.•Apply 350 W microwave irradiation for 5 min.3Rinse slide(s) twice with PBS.4Proceed as described under 3.3.3.2.2.1 Treatment of paraffin-embedded tissue, continued3.2.2.2Treatment of cryopreserved tissueAdditional solutions required •Fixation solution: 4% Paraformaldehyde in PBS, pH 7.4, freshly pre-pared•Washing buffer: PBS•Blocking solution: 3% H2O2 in methanol•Permeabilisation solution (0.1% Triton1) X–100, 0.1% sodium citrate), freshly preparedCryopreserved tissue In the following table the pretreatment of cryopreserved tissue is described.Note: Fix and permeabilisate two additional samples for the negative and positive labeling controls.Step Action1Fix tissue section with Fixation solution for 20 min at 15–25°C.2Wash 30 min with PBS.Note:For storage, dehydrate fixed tissue sections 2 min inabsolute ethanol and store at Ϫ15 to Ϫ25°C.3Incubate with Blocking solution for 10 min at 15–25°C.4Rinse slides with PBS.5Incubate in Permeabilisation solution for 2 min on ice (2–8°C).6Proceed as described under 3.3.3.3Labeling protocol 3.3.1Before you beginPreparation of TUNEL reaction mixtureOne pair of tubes (vial 1: Enzyme Solution, and vial 2: Label Solution) is sufficient for staining 10 samples by using 50 l TUNEL reaction mix-ture per sample and 2 negative controls by using 50 l Label Solution per control.Note : The TUNEL reaction mixture should be prepared immediately before use and should not be stored. Keep TUNEL reaction mixture on ice until use.Additionalreagents required •Micrococcal nuclease or •DNase I, grade I (Cat. No. 104 132)ControlsTwo negative controls and a positive control should be included in each experimental set up.Step Action1Remove 100 l Label Solution (vial 2) for two negative con-trols.2Add total volume (50 l) of Enzyme solution (vial 1) to the remaining 450 l Label Solution in vial 2 to obtain 500 l TUNEL reaction mixture.3Mix well to equilibrate components.Negative control:Incubate fixed and permeabilized cells in 50 l/well Label Solution (without terminal transferase) instead of TUNEL reaction mixture.Positive control:Incubate fixed and permeabilized cells with micro-coccal nuclease or DNase I, grade I (3000 U/ml– 3 U/ml in 50 mM T ris-HCl, pH 7.5, 10 mM MgCl 2 1mg/ml BSA) for 10 min at 15-25°C to induce DNA strand breaks, prior to labeling procedures.3.3.2Labeling protocol for adherent cells, cell smears, cytospin preparations andtissuesAdditional equipment and solutions required •Washing buffer: PBS •Humidified chamber •Parafilm or coverslipProcedure Please refer to the following table.Step Action1Rinse slides twice with PBS.2Dry area around sample.3Add50l TUNEL reaction mixture on sample.Note: For the negative control add 50 l Label solution each.To ensure a homogeneous spread of TUNEL reaction mixtureacross cell monolayer and to avoid evaporative loss, samplesshould be covered with parafilm or coverslip during incuba-tion.4Add lid and incubate for 60 min at 37°C in a humidified atmo-sphere in the dark.5Rinse slide 3 times with PBS.6Samples can be analyzed in a drop of PBS under a fluores-cence microscope at this state. Use an excitation wavelengthin the range of 450–500 nm and detection in the range of515–565 nm (green).3.3.3 Labeling protocol for difficult tissueAdditional equipment and solutions required •Citrate buffer, 0.1 M, pH 6.0.•Washing buffer: PBS•Tris-HCl, 0.1 M pH 7.5, containing 3% BSA and 20% normal bovine serum•Plastic jar•Microwave•Humidified chamberProcedure Please refer to the following table.Step Action1Dewax paraformaldehyde- or formalin-fixed tissue sectionsaccording to standard procedures.2Place the slide(s) in a plastic jar containing 200 ml 0.1 MCitrate buffer, pH 6.0.3•Apply 750 W (high) microwave irradiation for 1 min.•Cool rapidly by immediately adding 80 ml double dist.water (20–25°C).•Transfer the slide(s) into PBS (20–25°C).DO NOT perform a proteinase K treatment!4Immerse the slide(s) for 30 min at 15–25°C in Tris-HCl, 0.1 MpH 7.5, containing 3% BSA and 20% normal bovineserum.5Rinse the slide(s) twice with PBS at 15–25°C.Let excess fluid drain off.6Add50µl of TUNEL reaction mixture on the section and.Note: For the negative control add 50 µl Label solution.7Incubate for 60 min at 37°C in a humidified atmosphere in thedark.8•Rinse slide(s) three times in PBS for 5 min each.•Samples can be analyzed in a drop of PBS under a fluores-cence microscope at this state. Use an excitation wave-length in the range of 450–500 nm and detection in therange of 515–565 nm (green).3.4 SignalconversionAdditional equipment and solutions required •Washing buffer: PBS•Humidified chamber•Parafilm or coverslip•DAB Substrate* (Cat. No. 1 718 096) or alternative POD substrate •Mounting medium for light microscopyProcedure Please refer to the following table.Step Action1Dry area around sample.2Add50l Converter-POD (vial 3) on sample.Note: To ensure a homogeneous spread of Converter-PODacross cell monolayer and to avoid evaporative loss, samplesshould be covered with parafilm or cover slip during incuba-tion.3Incubate slide in a humidified chamber for 30 min at 37°C.4Rinse slide 3× with PBS.5Add 50–100 l DAB Substrate or alternative POD substrates.6Incubate slide for 10 min at 15–25°C.7Rinse slide 3× with PBS.8Mount under glass coverslip (e.g. with PBS/glycerol) and ana-lyze under light microscope.Alternative: Samples can be counterstained prior to analysisby light microscope.4. Appendix4.1 Trouble-shootingThis table describes various troubleshooting parameters. Problem Step/Reagent ofProcedurePossible cause RecommendationNonspecific labeling Embedding of tissue UV-irradiation forpolymerization ofembedding material(e.g. methacrylate)leads to DNA strandbreaksTry different embedding materialor different polymerizationreagent.Fixation Acidic fixatives (e.g.methacarn, Carnoy’sfixative)•Try 4% buffered paraformal-dehyde.•Try formalin or glutaralde-hyde.TUNEL reaction TdT concentration toohighReduce concentration of TdT bydiluting it 1:2 up to 1:10 withTUNEL Dilution Buffer (Cat. No.1 966 06).Converter solution Endogenous PODactivityBlock endogenous POD byimmersing for 10 min in 3%H2O2 in methanol prior to cellpermeabilisation.Non-specific bindingof anti-fluorescein-POD•Block with normal anti-sheepserum.•Block for 20 min with PBScontaining 3% BSA.•Reduce concentration ofconverter solution to 50%. Nucleases Some tissues (e.g.smooth muscles)show DNA strandbreaks very soon aftertissue preparation•Fix tissue immediately afterorgan preparation.•Perfuse fixative through livervein.Some enzymes arestill activeBlock with a solution containingddUTP and dATP.continued on next pageHigh back-ground Fixation Formalin fixation leadsto a yellowish stainingof cells containingmelanin precursorsTry methanol for fixation buttake into account that this mightlead to reduced sensitivity.TUNEL reaction Concentration oflabeling mix is toohigh for mamma car-cinomaReduce concentration of label-ing mix to 50% by diluting withTUNEL Dilution Buffer (Cat. No.1 966 006).Converter solution Endogenous PODactivityBlock endogenous POD byimmersing for 10 min in 3%H2O2 in methanol prior to cellpermeabilisation.Non-specific bindingof anti-fluorescein-POD•Block with normal anti-sheepserum.•Block for 20 min with PBScontaining 3% BSA.•Reduce concentration ofconverter solution to 50%. Sample Mycoplasma contami-nationMycoplasma detection Kit (Cat.No. 1 296 7449).Highly proliferatingcellsDouble staining e.g. withAnnexin-V-Fluos (Cat. No. 1 828681).Note: Measuring via microplatereader not possible because oftoo high background.Low labeling Fixation Ethanol and methanolcan lead to low label-ing (nucleosomes arenot cross-linked withproteins during fixa-tion and are lost dur-ing the proceduresteps)•Try 4% buffered paraformal-dehyde.•Try formalin or glutaralde-hyde.Extensive fixationleads to excessivecrosslinking of pro-teins•Reduce fixation time.•Try 2% buffered paraformal-dehyde.Permeabilisation Permeabilisation tooshort so that reagentscan’t reach their tar-get molecules•Increase incubation time.•Incubate at higher tempera-ture (e.g. 15–25°C).•Try Proteinase K (concentra-tion and time has to be opti-mized for each type oftissue).•Try 0.1 M sodium citrate at70°C for 30 min.continued on next pageProblem Step/Reagent ofProcedure Possible cause Recommendation4.1Trouble-shooting, continuedParaffin-embedding Accessibility forreagents is too low •Treat tissue sections afterdewaxing with Proteinase K (concentration, time andtemperature have to be opti-mized for each type of tis-sue).•Try microwave irradiation at370 W (low) for 5 min in200ml 0.1 M Citrate bufferpH 6.0 (has to be optimizedfor each type of tissue).No signal on positive control DNase treatment Concentration ofDNase is too low•For cryosections apply 3 U/mlDNase I, grade I.•For paraffin-embedded tissuesections apply 1500 U/mlDNase I, grade I.•In general, use 1 U/mlDNase I, grade I, dissolved in10 mM Tris-HCl pH 7.4 con-taining 10 mM NaCl, 5 mMMnCl2, 0.1 mM CaCl2, 25 mMKCl and incubate 30 min at37°C.•Alternative buffer 50 mMTris- HCl pH 7.5 containing1mM MgCl2 and 1 mg/mlBSA.Weak sig-nals Counterstaining Not suitable dye•Counterstaining with 5%methyl green in 0,1 M veronalacetate, pH 4.0 or Hematoxi-lin is possible (43).•Double-staining with propid-ium iodide is possible butonly for detection of morpho-logical cell changes.Problem Step/Reagent ofProcedure Possible cause Recommendation4.1Trouble-shooting, continued4.2 References1Gavrieli, Y., Sherman, Y. & Ben-Sasson, S. A. (1992) J. Cell Biol. 119, 493–501.2Gorczyca, W., Gong, J. & Darzynkiewicz, Z. (1993) Cancer Res. 53, 1945–1951.3Gorczyca, W. et al. (1993) Leukemi a 7, 659–670.4Gold, R. et al. (1994) Lab. Invest. 71, 219.5Gorczyca, W. et al. (1994) Cytometry 15, 169–175.6Sgonc, R. et al. (1994) Trends Genetics 10, 41–42.7Schmied, M. et al. (1993) Am. J. Pathol. 143, 446–452.8Wyllie, A. H. et al. (1980) Int. Rev. Cytol. 68, 251.9Kerr, J. F. R. et al. (1972) Br. J. Cancer 26, 239–257.10Duvall, E. & Wyllie, A. H. (1986) Immunol. To day 7, 115.11Compton, M. M. (1992) Canc. Metastasis Rev. 11, 105–119.12Allen, P. D., Bustin, S. A. & Newland, A. C. (1993) Blood Reviews 7, 63–73.13Cohen, J. J. & Duke, R. C. (1992) Annu. Rev. Immunol. 10, 267–293.14Clarke, P. G. H. (1990) Anat. Embryol. 181, 195–213.15Johnson, E. M. & Deckwerth, T. L. (1993) Annu. Rev. Neurosci. 16, 31–46.16Batistatou, A. & Greene, L. A. (1993) J. Cell Biol. 122, 523–532.17Strange, R. et al. (1992) Development 115, 49–58.18Carson, D. A. & Ribeiro, J. M. (1993) Lancet 341, 1251–1254.19Edgington, S. M. (1993) Biotechnology 11, 787–792.20Gougeon. M.-L. & Montagnier, L. (1993) Science 260, 1269–1270.21Hickman, J. A. (1992) Cancer Metastasis Rev. 11, 121–139.22Afanasyev, V. N. et al. (1993) Cytometry 14, 603–609.23Bryson, G. J., Harmon, B. V. & Collins, R. J. (1994) Immunology Cell Biology 72,35–4124Darzynkiewicz, Z. et al. (1992) Cytometry 13, 795–808.25Ando, K. et al. (1994) J. Immunol. 152, 3245–3253.26Berges, R. R. et al. (1993) Proc. Natl. Acad. Sci. USA 90, 8910– 8914.27Gorczyca, W. et al. (1992) Int. J. Oncol. 1, 639–648.28Gorczyca, W. et al. (1993) Exp. Cell Res. 207, 202–205.29Billig, H., Furuta, I. & Hsueh, A. J. W. (1994) Endocrinology 134, 245–252.30MacManus, J. P. et al. (1993) Neurosci. Lett. 164, 89–92.31Mochizuki, H. et al. (1994) Neurosci. Lett. 170, 191–194.32Oberhammer, F. et al. (1993) Hepatology 18, 1238–1246.33Portera-Cailliau, C. (1994) Proc. Natl. Acad. Sci. USA 91, 974 –978.34Preston, G. A. et al. (1994) Cancer Res. 54, 4214–4223.35Weller, M. et al. (1994) Eur. J. Immunol. 24, 1293–1300.36Zager, R.A. et al. (1994) J. Am. Soc. Nephrol. 4, 1588–1597.37Cohen, G. M. et al. (1992) Biochem. J. 286, 331–334.38Collins, R. J. et al. (1992) Int. J. Rad. Biol. 61, 451–453.39Sei, Y. et al. (1994) Neurosci. Lett. 171, 179–182.40Ansari, B. et al. (1993) J. Pathol. 170, 1–8.41Gold, R. et al. (1993) J. Histochem. Cytochem. 41, 1023–1030.42Negoescu, A. et.al. (1998) Biochemica3, 34-41.43Umermura, S. et al. (1996) J. Histochem. Cytochem. 44, 125-132 .。
DIG标记与检测

D i a g n o s t i c s绿色环保的DIG 检测技术邹 嵘, Ph.D资深技术顾问, Roche 亚太技术支持中心 免费服务电话:800 820 0577电子邮箱:asc.support@D i a g n o s t i c s内容概要•DIG 系统介绍•DIG 系统的应用•DIG 标记试剂盒的选用•DIG 检测产品的选用•DIG 实验结果D i a g n o s t i c sSouthern/Northern 实验中同位素应用•同位素辐射,损害健康 •探针有效时间短由同位素半衰期决定,不能反复使用•专用同位素实验室,配备安全防护设备,缺点D i a g n o s t i c s非同位素标记法 ---地高辛系统介绍独特性:只存在于洋地黄植物花与叶片中的类 固醇物质,抗原性:可产生抗体检测方法;光学显色法, 荧光或者化学发光法实用性:DIG 修饰后的UTP , dUTP ,ddUTP可被RNA 转录酶,DNA 聚合酶,末端转移酶用于核酸链的合成 D. purpurea & D. lanataDIG 系统特点Biotin 特点几乎在所有的组织和细胞中内源性表达Biotin 的结合物Streptavidin ,经常与固体支撑物如 MTP 或膜非特异性结合,产生背景---特异性差D i a g n o ng Neo Poly (A) RNA loaded per laneA. probeDIG-labeled RNA 120 ng/mlFilm exposure: 30 min1 0.1 0.01 0.001 C. probeDIG-labeled DNA 50ng/mlFilm exposure: 30 min10 1 0.1 0.01 B. probe32P-labeled RNA 2 x 106 cpm/ml Film exposure: 16 hours1 0.1 0.01 0.001 D. probe32P-labeled DNA 2 x 106 cpm/ml Film exposure: 24 hours10 1 0.1 0.01显色时间短更敏感D i a g n o A S K i 80006990332P-labeled probe, 10 µg total RNA eachlane, exposure: 4hDIG-labeled probe, PCR derived.Exposure: 10 min.DIG-labeled probe, PCR derived.Exposure: 10 min.(Courtesy of U. Stoeckl, University of Regensburg, Germany) Panel A and B : Blots prepared according to protocol of U. Stoeckl Panel C : Blot prepared according to standard conditions,recommended by ROCHE Application LabCBA1 µg5 µg2,5 µg2,5 µg1 µg5 µg2,5 µg2,5 µgD i a g n o s t i c s(Courtesy of Dr. Peter Hloch, Univ. Homburg, Germany)9.5 7.5 4.4 2.41.3 kb9.5 7.5 4.4 2.41.3kbp68-cDNA probe (radioactive 1 day)p68-cDNA probe (DIG 5min)p68-intron 11 probe (DIG) b -actin probe (DIG)1st rehybridization 2nd rehybridization 3rd rehybridizationMTN 膜,2ug 总RNA膜最多可以反复使用20多次 !!!重复性: DIG 标记的探针D i a g n o s t i c sDIG 标记系统放射性同位素系统安全性 安全存在健康损害风险检测灵敏度 0.1pg for Southern blot好 检测曝光时间几分钟1天到几周探针稳定性> 1 year/半衰期 经济性探针能可以反复使用,好不能反复使用 / 差便利性操作说明完备,技术支持充分,不须特殊实验设备 无技术支持,专用实验室,人员培训上岗,废弃物专项管理DIG 系统的优势D i a g n o s t i c sDIG 标记系统的Southern-Blot 实验流程X-Ray Film32PTarget DNA MembraneX-Ray Film- C - G - T - G - A - T - A - G - C -A - C - U - A - T P PChemiluminescent DetectionCSPD / CDP-StarAntibody-Conjugate(AP 、POD)Labeled-ProbeDigoxigenin AlkalinePhosphataseSubstrateSubstrate32P 标记系统的Southern-Blot 实验流程D i a g n o s t i c s内容概要•DIG 系统介绍•DIG 系统的应用•DIG 标记试剂盒的选用•DIG 检测产品的选用•DIG 实验结果D i a g n o s t i c sDIG 系统的主要应用In Situ Hybridization•检测基因易位,扩增,缺失•基因定位,作图•诊断肿瘤、遗传疾病膜杂交Southern BlotsNorthern Blots Colony Hybr .• 基因组扫描• cDNA 文库筛选• 药物残留检测• 转基因效果检测• 临床疾病监测 (Fragile X)• mRNA 表达分析• mRNA 剪切形式分析D i a g n o s t i c s内容概要•DIG 系统介绍•DIG 系统的应用•DIG 标记试剂盒的选用•DIG 检测产品的选用•DIG 实验结果D i a g n o DNA 探针:稳定,操作方便 随机引物法( Klenow 酶)缺口平移法 (DNA Polymerase I / DNase I )PCR 扩增法 (Taq DNA Polymerase, Pwo DNA Polymerase or Expand HF polymerase )RNA 探针: 灵敏度高,特异性好体外转录法 (SP6-, T3- or T7 RNA Polymerases )Oligonucleotide 探针:也可化学合成 3´-末端标记或加尾法 (Terminal Transferase )方法 1:探针的类型D i a g n o In Situ Hybridization• 缺口平移法• 体外转录法• 随机引物标记法• 寡核苷酸3…末端/加尾标记• PCR 方法膜杂交Southern BlotsNorthern BlotsColony Hybr .• PCR 方法标记• 随机引物标记法• PCR 方法• 寡核苷酸3…末端标记• 体外转录法• 寡核苷酸3… 末端标记• 寡核苷酸3‟加尾标记方法 2:探针的用途D i a g n o 模板: 目标基因片段 量:10ng~3ugKlenow 酶六碱基随机引物标记的探针片段(size range: 300-800-1500 bp )未标记的目标片段优点:产量高,灵敏度高(0.10~0.03pg)应用:Southern blot 、Northern blot 、文库筛选、点杂交DIG High Prime DIG Standard R.P.L a b e l e d D N A (n g )2500 2000 1500 1000 500 0 0200400 6008001000min产品名称Cat. No. DIG High Prime DNA Labeling and Detection Starter Kit I11 745 832 910 DIG High Prime DNA Labeling and Detection Starter Kit II11 585 614 910D i a g n o 模板: 质粒或基因组DNA 量:质粒 10pg~100pg , 基因组DNA 1pg~50pgTaq Polymerase特异性引物 或 多克隆位点的通用引物标记好的探针 “Full -Size”初级延伸产物 (出现于线性扩增阶段!!!)优点:• 模板用量少,模板纯度要求低 • 实验优化简单• 产量高 (2ul / 102cm 膜杂交) • 适用于小片段探针的标记 (<100 bp)应用:Southern blot 、Northern blot 、 文库筛选、点杂交产品名称Cat. No. PCR DIG Probe Synthesis Kit11 636 090 910D i a g n o s t i c sDNA 探针合成-缺口平移法优点:• 能控制探针的长度 (对 ISH 实验特别重要) • 含有全模板序列的探针应用:ISH模板:质粒DNA 或 纯化后的大片段DNA 量:1ug产品名称 Cat. No.DIG-Nick Translation Mix 11 745 816 910不完全的DNaseI 酶切OH OHHOHOHOHO3’3’5’ HO5’ 3’ 5’OH3’ 5’OHDNA 聚合酶+DIG-dUTP3’ 5’OH3’5’ HO15 C 孵育1.5小时后,变性标记结束后,反应体系内探针量为1ug DNAD i a g n o s t i c sRNA 探针合成-体外转录法REpSPT 18/19insertpromoter包含目的DNA 的转录载体, 通过酶切进行线性化 模板量:质粒:1ug含启动子的片段:100~200ngSP6, T3 or T 7 RNA pol 。
罗氏地高辛(DIG)系统产品选择指南设计

罗氏地高辛(DIG)系统产品选择指南发布日期:2011-02-15 来源:浩然生物技术浏览次数:374罗氏应用科学部是最早致力于向用户提供非放射标记技术的公司之一,让更多的科研工作者们可以避免使用危险的放射性同位素。
罗氏专利的地高辛产品上市近二十年,尽管有许多出色的竞争者涉足这一领域,尽管有定量PCR技术的应用,DIG系统仍然是非放射性高效标记&检测技术的首选。
Ø 关于非放射性标记技术罗氏应用科学部是最早致力于向用户提供非放射标记技术的公司之一,让更多的科研工作者们可以避免使用危险的放射性同位素。
罗氏专利的地高辛产品上市近二十年,尽管有许多出色的竞争者涉足这一领域,尽管有定量PCR技术的应用,DIG系统仍然是非放射性高效标记&检测技术的首选。
Ø 地高辛技术优势地高辛标记技术源于一种从洋地黄类植物(毛地黄和毛花毛地黄)中提取的类固醇物质——Digoxigenin (DIG)。
由于洋地黄植物的花和叶片Digoxigenin在自然界中的唯一来源,因此抗DIG 的抗体不会与其他的生物物质结合,从而可以满足特异性标记的需要。
这一点,正是DIG胜于生物素(Biotin)的地方——同样是小分子标记物,生物素广泛存在于各种组织中,对于灵敏度很高的标记检测实验来说,样品自身含有的源生物素,就会对结果产生干扰。
地高辛就能够很好地避免这个问题。
Ø 地高辛技术特点ü可用于标记核苷酸或蛋白质ü检测方法:显色法、荧光或化学发光法ü安全环保,不接触放射性物质,不会对环境造成污染ü曝光时间短,结果显示时间以分钟计算,无需几小时甚至几天的自显影过程ü地高辛标记的探针至少可贮存一年以上,可一次大量制备ü可轻松进行探针剥离和重探Ø 应用领域地高辛系统能够安全高效地标记DNA、RNA或是寡聚核苷酸探针,这些探针可以被广泛应用在:üSouthern blot'' Northern blot'' dot blot ü菌斑杂交üMicroarrayüELISAü原位杂交Ø 根据标记方法选择探针更多产品选择请参考https://.roche-applied-science./sis/lad/index.jsp?id=lad_010100 Ø 根据实验方法选择产品üSouthern blottingüNorthern blottingüColony and plaque hybridizationüIn situ hybridizationhttps://.roche-applied-science./sis/lad/index.jsp?id=lad_010300 Ø 根据检测方法选择产品ü灵敏度更高的化学发光法ü方便直观的显色法。
罗氏10大最具潜力创新药

罗氏拥有丰富的研发管线储备,根据罗氏官网统计,截止2019年10月16日,有多达131个研发管线项目正在进行中,至少处于III期临床的有50个,其中新分子实体有14个,新适应症项目36个。
我们今天就来综合临床研究项目数量、创新性、市场前景等方面盘点一下罗氏10个最具潜力的新药。
10 GantenerumabGantenerumab是一种完全人源化IgG1单克隆抗体,可与大脑中的β-淀粉样蛋白特异性结合,用于治疗老年性痴呆症。
科学家们通过30多年的研究认为β-淀粉样蛋白与老年性痴呆存在着紧密的关系,但是近年来的多个临床药物大部分可以减少β-淀粉样蛋白,却并不能缓解老年性痴呆的疾病进程。
2014年罗氏也搁置了Gantenerumab的研究,直到2015年,Biogen的Aducanumab在1b期临床结果显示,Aducanumab能够降低前驱期或轻微阿尔茨海默病患者的淀粉样蛋白斑,同时可以延缓认知功能的下降,此研究结果佐证了目前科学家关于β淀粉样蛋白导致阿尔兹海默症的假说,给包括Gantenerumab在内的多个临床研究药物带来了希望,同时后续的生物标志物和疗效信号分析表明,在疾病进展最快的患者中,高剂量的Gantenerumab表现出一定疗效,这些促使罗氏又重新启动Gantenerumab的阿尔兹海默症临床III期试验,目前有4个临床III期项目正在进行中。
9 RG6042RG6042是一种用于治疗亨廷顿病(HD)的第二代修饰反义寡核苷酸。
HD是一种罕见的遗传性、进行性疾病,HD的病因是编码亨廷顿蛋白(huntington protein,HTT)的基因出现三核苷酸序列CAG重复超过36次,从而编码结构异常的HTT,在功能上显现出能够杀死神经元细胞的毒性,严重影响患者的行动和思维能力,估计全球约有50万HD患者。
RG6042通过靶向人HTT mRNA来降低突变型HTT(mHTT)蛋白的表达水平,从而减缓或阻止HD患者的疾病进展。
northern和southern探针合成步骤

合成DNA探针步骤:(具体参考Roche DIG DNA Labeling and Detection Kit)1. PCR、凝胶电泳分离目标片段,克隆后测序或者直接把胶回收产物取一部分送测以检验序列是否准确。
(注:假如以前已经测定过该片段,不必反复送测)2. 20μl体系10 ng至3 μg DNA,加双蒸水至15μl,盖紧用蜡封好。
沸水浴10分钟以使DNA变性,迅速置于冰中。
Note: Complete denaturation is essential for efficient labeling. 3 加入以下试剂:Hexanucleotide Mix, 10 × (vial 5) 2 μl dNTP Labeling Mix (vial 6) 2 μl Klenow enzyme labeling grade (vial 7) 1 μl 混匀,短暂离心。
37° C孵育 1 h 到20 h (overnight),一般为16-18小时 4加入 2 μl 0.2 M EDTA (pH 8.0) 或于65° C 加热10 min 以终止反应。
Note: The length of the DIG-labeled fragments range from 200 to 1000 bp.合成RNA探针步骤:(具体参考Roche DIG RNA Labeling Mix, 10 × conc.)过程应保证无RNA酶的环境。
1. PCR、凝胶电泳分离目标片段,克隆后测序,需向公司说明测序结果要带上SP6、T7的序列,RNA探针是单链的,选择使用哪个启动子至关重要。
假如northern blot的对象是单链RNA,合成的RNA探针序列就必须与它互补。
呼肠孤病毒基因组为双链RNA,因此不需要考虑这个问题。
2. 提纯质粒,酶切以进行线性化3. 转录:在冰上加入以下试剂:(20μl体系)1 μg 线性化的质粒DNA x μl DIG RNA labeling mix, 10 ×2 μl Transcription buffer, 10 × 2 μl 加无RNA酶的水至总体积为 18 μl RNA polymerase (SP6, T7 or T3) 20 U/μl 2 μl 混匀,短暂离心。
罗氏荧光定量pcr

罗氏荧光定量 PCR 是一种高灵敏度、高精确度的多重荧光定量 PCR 技术,可以用于定量检测 DNA 或 RNA 中的特定序列。
该技术不仅可以用于基础研究,还可以应用于临床诊断和药物研发等领域。
一、基本原理罗氏荧光定量 PCR 基于 PCR 扩增技术,在 PCR 反应过程中,使用荧光标记的探针来定量检测扩增产物的数量。
该技术采用两个荧光染料:SYBR Green 和 TaqMan 探针。
SYBR Green 是一种结合双链 DNA 的荧光染料,可以在 PCR 反应过程中不断累积,并发出荧光信号;而 TaqMan 探针是一种在 PCR 反应过程中被水解释放的荧光探针,可以在特异性结合目标序列后发出荧光信号。
通过测量这些荧光信号的强度,可以确定 PCR 反应产物的数量。
二、实验步骤1. 样品制备首先需要从样品中提取出目标 DNA 或 RNA,常用的提取方法包括酚-氯仿提取法、磁珠法等。
提取后需要进行纯化和定量,确保样品质量和浓度符合要求。
2. PCR 反应将样品DNA 或RNA 与特异性引物、荧光探针和PCR Master Mix 混合,加入 PCR 反应管中,进行 PCR 扩增反应。
PCR 扩增条件需根据具体引物和模板 DNA/RNA 来确定,一般包括以下步骤:(1) 热变性:将反应体系加热至 95℃,使 DNA/RNA 双链分离为单链。
(2) 退火:将反应体系降温至引物的退火温度,使引物与模板DNA/RNA 特异性结合。
(3) 延伸:利用DNA/RNA 聚合酶将引物延伸,合成新的DNA 单链。
(4) 荧光检测:在延伸过程中,荧光探针与目标序列特异性结合,发出荧光信号。
荧光信号的强度与产物数量成正比,可以通过检测荧光信号来定量 PCR 扩增产物。
3. 数据分析利用荧光数据分析软件对荧光信号进行分析,得出 PCR 扩增产物的数量。
根据标准曲线,可以将荧光信号转化为目标序列的数量,从而定量检测样品中的目标 DNA 或 RNA。
原位杂交实验

原位杂交实验1 探针的设计与合成1)根据实验室已有的p8基因cDNA全长序列,用premier primer5.0设计引物p81和p82,以卤虫cDNA为模板,PCR扩增得到346bp的产物,用T akara胶回收试剂盒回收纯化。
引物编号引物序列长度p81 TGCGGACGAAACAGGAAG 18 bpp82 GCTCAAACAGTGATGCCAGT 20 bp2)目的片段克隆a. 在无菌离心管中加入连接载体的各种成分,载体与片段的摩尔比控制在1:3-1:8,根据凝胶电泳检测后的浓度及载体与片段分子大小来计算摩尔比。
加入成分及比例如下:目的PCR片段 5 μlpGM-T载体(约50ng/uL) 1 μl10×T4 DNA Ligation Buffer 1 μlT4 DNA Ligase(3U/uL) 1 μl无菌去离子水 3 μl总体积10 μlb. 轻轻弹动离心管以混合内容物,短暂离心。
置于PCR仪中16℃过夜连接,反应结束后将离心管置于冰上。
c. 向铺好的含有氨苄青霉素的固体平板表面加入16 μl的IPTG(50mg/ml)、40 μl的X-gal (20mg/ml),使用无菌的弯头玻璃棒将其均匀的涂开,避光置于37℃培养箱1-3小时,使溶解X-gal的二甲基甲酰挥发干净。
d. 将10 μl的连接产物加到100 μl DH5 感受态细胞中,轻弹混匀,冰浴半小时,将离心管置于42℃水浴90秒,取出管后立即置于冰浴上放置2-3分钟,其间不要摇动离心管。
向离心管加入500 μl 37℃预热的LB(不含抗生素)培养基,150rpm摇床37℃振荡培养45分钟。
目的是使质粒上相关的抗性标记基因表达,使菌体复苏。
将菌液于4000g下离心10分钟,去掉上清,加入100 μl培养液重溶并加入到配制好的LB固体培养基上,用无菌的弯头玻璃棒轻轻将细胞均匀涂开。
待平板表面干燥后,倒置平板,37℃培养12-16小时。
DIG Northern标记及杂交检测试剂盒中文说明书

DIG Northern Starter KitDIG Northern标记及杂交检测试剂盒说明书版本:Version 8.0 (2010年8月)利用体外转录法进行地高辛标记的RNA探针合成反应,采用酶联免疫方法及即用型化学发光底物CDP-Star进行杂交分子的检测。
利用本试剂盒可进行10次标记反应,和10次杂交检测反应(10×10cm2杂交膜)。
产品目录号:12 039 672 910试剂盒于-15 ~ -25℃保存1. 概况1.1 说明书内容1. 概况 (2)1.1说明书内容 (2)1.2试剂盒组成 (3)2. 介绍 (5)2.1 产品概述 (5)3. 实验步骤和需要的材料 (7)3.1 实验开始前准备 (7)3.2 实验流程 (7)3.3 DNA模板制备 (7)3.4 DIG-RNA标记 (9)3.5定量标记反应效率 (10)3.6 适用于DIG Northern bolts的凝胶系统 (12)3.7 RNA转膜和固定 (13)3.8 杂交 (14)3.9 免疫检测 (15)3.10探针洗脱和重探 (17)4 附录 (18)4.1 疑难问题解决 (18)4.2参考文献 (19)1.2 试剂盒组成杂交反应所需的仪器和其他反应试剂请根据您的实验操作流程准备杂交反应所需的仪器和其他试剂,参见下表,请注意所有试剂应使用DMPC或DEPC处理的水配置:带*的试剂为罗氏应用科学部提供的试剂。
2.1 产品概述反应原理DIG Northern标记及杂交检测试剂盒采用体外转录法,以地高辛(DIG,一种甾类半抗原)标记的UTP为底物,进行RNA探针的标记合成,所获得的单链RNA探针用于后续的杂交反应,带有地高辛分子的杂交子通过酶联免疫反应和CDP-Star底物进行化学发光检测。
应用尼龙膜上的Northern blots杂交反应样本•线性化质粒,质粒上含相应的RNA聚合酶启动子序列(SP6,T3,T7)注意:质粒上待转录的目标片段长度应在200-1000 bp。
罗氏荧光定量

罗氏荧光定量1. 简介罗氏荧光定量是一种常用的生物化学分析技术,用于测定样品中特定物质的含量。
该技术基于罗氏荧光染料与目标物质之间的特异性结合,通过测量荧光信号的强度来计算目标物质的浓度。
2. 原理罗氏荧光定量的原理基于荧光共振能量转移(FRET)现象。
FRET是一种非辐射能量传递的过程,其中一个荧光染料(受体)吸收能量并将其传递给另一个荧光染料(供体),从而引起供体的荧光发射。
在罗氏荧光定量中,供体和受体分别与目标物质的特定结构域结合。
当目标物质存在时,供体和受体之间的距离会发生改变,从而影响FRET的效率。
通过测量供体的荧光强度变化,可以计算目标物质的浓度。
3. 实验步骤3.1 样品准备首先,需要准备样品,样品可以是生物体、细胞、蛋白质等。
样品应根据实验的要求进行处理和处理,以确保准确测量目标物质的浓度。
3.2 罗氏染料标记接下来,需要将罗氏染料标记在样品中的目标物质上。
这可以通过化学反应或特定的结合方式实现。
标记过程需要确保染料与目标物质之间的特异性结合,并且不影响目标物质的功能和性质。
3.3 荧光测量完成标记后,可以进行荧光测量。
通常使用荧光分光光度计或荧光显微镜来测量样品中的荧光强度。
荧光测量需要选择适当的激发波长和发射波长,并根据实验要求进行多次测量以确保结果的准确性。
3.4 数据处理与分析最后,需要进行数据处理和分析。
根据荧光测量结果,可以绘制荧光强度与目标物质浓度之间的标准曲线。
通过与标准曲线的比较,可以计算样品中目标物质的浓度。
4. 应用领域罗氏荧光定量广泛应用于生物医学研究和临床诊断中。
以下是一些常见的应用领域:4.1 蛋白质定量罗氏荧光定量可用于测定蛋白质样品中特定蛋白质的浓度。
这对于研究蛋白质相互作用、酶促反应和蛋白质表达水平的变化非常有用。
4.2 DNA/RNA定量罗氏荧光定量也可以用于测定DNA或RNA样品中的目标序列的浓度。
这对于分子生物学研究、基因表达分析和疾病诊断非常重要。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Number of Tests 1 kit is sufficient for 2× 10 labeling reactions. Kit Contents Bottle Label 1 pSPT18 DNA Contents Including Function • 40 l (0.25 mg/ml) • Cloning/transcription vector; subclones are transcribed into RNA probes by T7 or SP6 RNA polymerase • 40 l • [0.25 mg/ml] • Cloning/transcription vector; subclones are transcribed into RNA probes by T7 or SP6 RNA polymerase
Labeled control • 100 l RNA* • (100 ng/l) • DIG-labeled “antisense” Neo RNA (7) (760 bases), transcribed with T7 RNA polymerase from 4 l (equivalent to 1 g) Pvu II-linearized pSPT18-Neo DNA (vial 3) according to the standard protocol. The transcription assay was not treated with DNase I. After ethanol precipitation, it was dissolved in dimethylpyrocarbonatetreated water. • The solution contains approx. 10 g of DIG-labeled Neo RNA and 1 g pSPT18Neo template DNA • For semi-quantitative estimation of DIGlabeled RNA and use as a hybridization control • contains DNA fragments of 798 and 3,281 bp. Unlabeled con- • 20 l, unlabeled control RNA, trol RNA • [200 g/ml] • Unlabeled Neo poly (A) “sense” RNA in dimethylpyrocarbonate-treated water. • The Neo poly (A) RNA is synthesized by in vitro transcription and is 1 kb long • Target RNA to practice RNA/RNA hybridizations; when applied to a membrane, this RNA will hybridize with the labeled control RNA (vial 5) NTP labeling mixture* • 40 l nucleotide mixture, • 10 ×, [10 mM ATP, 10 mM CTP, 10 mM GTP, 6.5 mM UTP, 3.5 mM DIG-11-UTP, pH 7.5 (20oC)] • 40 l • 10 × conc. • 40 l • [10 U/l] • Degrades DNA template after the labeling reaction
Cat. No. 11 175 025 910
Kit for 2× 10 labeling reactions
y Version 12 Content version: January 2013 Store at Ϫ15 to Ϫ25°C
1.
What this Product Does
Bottle Label 5
6
4
7
8 9
Transcription buffer DNase I, RNase-free*
10 * available from Roche Applied Science 11
Protector RNase • 20 l Inhibitor* • [20 U/l] • Prevents the degradation of RNA during the labeling reaction SP6 RNA Polymerase* T7 RNA Polymerase* • 20 l • [20 U/l] • Synthesizes RNA from a DNA template • 20 l • [20 U/l] • Synthesizes RNA from a DNA template
12
0113.112441750012
www.roche-appliedity The unopened kit is stable at Ϫ15 to Ϫ25°C until the expiration date printed on the label. Additional Equipment/Reagents Required In addition to the reagents listed above, you must prepare several solutions. In the table you will find an overview of the equipment and reagents needed for the different procedures. Detailed information is given at the beginning of each procedure. Procedure 2.2 DIG RNA labeling Equipment water bath Reagents • Water, PCR Grade treated with DMPC • EDTA, 0.2 M, pH 8.0 • RNA dilution buffer • DIG Wash and Block Buffer Set* • Anti-digoxigenin-AP* • CSPD* or CDP-Star* • DIG Luminescent Detection Kit*
Contents Including Function
2
pSPT19 DNA
3
Control DNA 1 • 20 l pSPT18-Neo • [0.25 mg/ml] cleaved with Pvu II • Transcription of control DNA 1 by T7 RNA polymerase according to the standard protocol produces DIG-labeled “antisense” Neo transcripts of 760 bases in length. • Use as control in labeling and hybridization reactions • contains DNA fragments of 798 and 3,281 bp due to cleavage at the 2 Pvu II sites on the plasmid Control DNA 2 • 20 l pSPT19-Neo • [0.25 mg/ml] cleaved with Pvu II • Transcription of control DNA 2 by SP6 RNA polymerase according to the standard protocol produces DIG-labeled “antisense” Neo transcripts of 760 bases in length. • Use as control in labeling and hybridization reactions • contains DNA fragments of 316 and 3,761 bp due to cleavage at the 2 Pvu II sites on the plasmid (one at pos. 761 within the neo gene)
Size
Amount
2.3 Determina- • nylon membranes tion of labeling positively charged* efficiency • UV-transilluminator or • UV cross-linker
Applications DIG-labeled RNA is used for hybridization to • Northern blots • Southern blots • in situ hybridization • plaque or colony lifts • RNase protection experiments As the linkage between DIG and UTP is resistant to alkali, DIGlabeled RNA can be fragmented by alkaline treatment. Slightly reducing the size of the DIG-labeled RNA probe may make it more suitable for certain applications in in situ hybridization (5, 6).
For life science research only. Not for use in diagnostic procedures.
DIG RNA Labeling Kit (SP6/T7)
