质粒图谱
☆如何阅读质粒图谱

如何阅读质粒图谱最近由于实验需要,需要查阅载体图谱,到园子里搜罗一番,发现虽然有人问载体图谱阅读的问题,也有前辈回答,但都不详细,借自己也在琢磨这个问题的机会,将我学到的东西整理一下,于大家分享。
载体主要有病毒和非病毒两大类,其中质粒DNA是一种新的非病毒转基因载体。
一、一个合格质粒的组成要素#复制起始位点Oril 即控制复制起始的位点。
原核生物DNA分子中只有一个复制起始点。
而真核生物DNA分子有多个复制起始位点。
#抗生素抗性基因可以便于加以检测,如Amp+l ,Kan+#多克隆位点MCS 克隆携带外源基因片段l#P/E 启动子/增强子l#Termsl 终止信号#加poly(A)信号l 可以起到稳定mRNA作用二、如何阅读质粒图谱第一步:首先看Ori的位置,了解质粒的类型(原核/真核/穿梭质粒)第二步:再看筛选标记,如抗性,决定使用什么筛选标记。
(1)Ampr 水解β-内酰胺环,解除氨苄的毒性。
(2)tetr 可以阻止四环素进入细胞。
(3)camr 生成氯霉素羟乙酰基衍生物,使之失去毒性。
(4)neor(kanr)氨基糖苷磷酸转移酶使G418(卡那霉素衍生物)失活(5)hygr 使潮霉素β失活。
第三步:看多克隆位点(MCS)。
它具有多个限制酶的单一切点。
便于外源基因的插入。
如果在这些位点外有外源基因的插入,会导致某种标志基因的失活,而便于筛选。
决定能不能放目的基因以及如何放置目的基因。
第四步:再看外源DNA插入片段大小。
质粒一般只能容纳小于10Kb的外源DNA片段。
一般来说,外源DNA片段越长,越难插入,越不稳定,转化效率越低。
第五步:是否含有表达系统元件,即启动子-核糖体结合位点-克隆位点-转录终止信号。
这是用来区别克隆载体与表达载体。
克隆载体中加入一些与表达调控有关的元件即成为表达载体。
选用那种载体,还是要以实验目的为准绳。
启动子-核糖体结合位点-克隆位点-转录终止信号#启动子-促进DNA转录的DNA顺序,这个DNA区域常在基因或操纵子编码顺序的上游,是DNA分子上可以与RNApol特异性结合并使之开始转录的部位,但启动子本身不被转录。
质粒图谱查询方法

3.google scholar: / 有些质粒是经过改造的,所以通过上述方法不能查询到相应信息。这时,可以在google scholar中输入质粒名称,可以直观地看哪些学者在何文章中使用了该质粒,从而可了解到质粒的来源;或者籍此向作者咨询或索取。 4.尝试从各大生物公司,例如invitrogen网站查询. 5. 这个网站收录了大量图谱: http://www.embl-hamburg.de/~geerlof/webPP/vectordb/bact_vectors/table.html
Pe
te
rX u
file:///D|/中科院/Selective Serotonin Transporter/质粒信息/质粒图谱查询方法.txt
file:///D|/中科院/Selective Serotonin Transporter/质粒信息/质粒图谱查询方法.txt(第 2/6 页)[2011/8/4 18:39:52]
By
0099--pGE-1—Stratagene--RNAi载体 0100--pSUPER.p53—OligoEngine--RNAi载体 0101--palter-ex1--promega 0102--pACYCDuet-1--NOVAGEN 0103--pEX lox(+) Vector—NOVAGEN--原核表达 0104--质粒名称:pBACgus-8 Transfer Plasmid—NOVAGEN--CHUANSUO 0105--pSCREEN?-1b(+) Vector Map—novagen--筛选 0106--PGEX-2T--BD Co--pDsRed2--Clontech 0107--pbgal-Basic—Clontech--mammalian reporter vector 0108—pBI—Clontech--express two genes of interest from a bidirectional tet-responsive promoter 0109--质粒名称:pbgal-Control—Clontech--mammalian reporter vector 0110-- pGEX-5X-1--原核表达 0111--pBI-EGFP—Clontech--pBI-EGFP-- coexpress 0112--pBI-G—Clontech--pBI-G--express b-galactosidase 0113--pBI-GL—Clontech--pBI-GL --express luciferase and b-galactosidase 0114--pCMS-EGFP—Clontech--mammalian expression vector 0115--pd2EYFP-1—Clontech--启动子测定 0116--质粒名称--pd2EYFP-N1—Clontech--融合表达 0117--pd4EGFP-Bid—Clontech--融合表达 Bid 0118--pDNR-CMV—Clontech--pDNR-CMV 0119--pDNR-EGFP Vector—Clontech 0120--pDNR-LacZ –Clontech 0121--pECFP-Endo—Clontech--真核表达0122--pECFP-ER—Clontech--真核表达0123--pEGFP-Actin—Clontech--真核表达0124--pGAD GH--Clontech--酵母表达 0125--pGADT7-Rec –Clontech--酵母表达 0126--pGADT7-RecAB—Clontech--酵母表达 0127--pGADT7-Rec2—Clontech--酵母表达 0128--pGBKT7—Clontech--酵母表达 0129--pHAT 10/11/12—Clontech 0130--pHAT20—Clontech 0131—pHygEGFP—Clontech 0132—pLacZi—Clontech 0133—pM—Clontech--pM is used to generate a fusion of the GAL4 DNA-BD 0134--pPKCa-EGFP—Clontech 0135--pPKCb-EGFP—Clontec 0136--pSIREN-DNR Vector—Clontech--RNAi 0137--pSIREN-DNR-DsRed-Express Vector—Clontech--RNAi 0138--pSIREN-RetroQ—Clontech--RNAi 0139--pIRES-EYFP—Clontech--RNAi 0140--pSRE-Luc—Clontech--RNAi 0141--pTK-neo—novagen--原核表达 0142--pZsGreen Vector—Clontech--pZsGreen is a pUC19-derived prokaryotic expression vector 0143--pTandem-1—novagen--原核表达 0144--pZsGreen1-C1Vector—Clontech----真核表达 0145--质粒名称:M13mp18—novagen--原核表达 0146--pZsGreen1-DR Vector—Clontech--真核表达 0147--PZsGreen1-N1 Vector—Clontech --真核表达 0148--T7Select415-1b—novagen----真核表达 0149--pZsYellow Vector—Clontech --真核表达 0150—pTimer—Clontech --真核表达 0151--pTA-Luc—Clontech --真核表达 0152--pTAL-Luc—Clontech --真核表达 0153--pTA-SEAP—Clontech --真核表达 0154--pTAL-SEAP—Clontech --真核表达 0155--pTet-On—Clontech --真核表达 0156--pTet-Off—Clontech --真核表达 0157--pTet-ATF—Clontech --真核表达 0158--pTet-CREB—Clontech --真核表达
质粒图谱的阅读

第四步,看多克隆位点的序列。
多克隆位点一般会有上图这样的序列,也就是三联体密码加上酶切位点标示的序列。这里的三联体密码,主 要是为了提示你,插入的表达的片段,需要按照这样的三联体进行插入,不要有移码突变。5`末端如果有差 异的话,可以把基因的ATG(甲硫氨酸)的5`末端替换1-2个碱基(变成GTG或者GCG这样),从第二个氨 基酸序列开始完全一致即可。而配合扩增和酶切的话,插入片段是允许有3`末端的冗余。为了可以应付3`末 端的冗余碱基,在多克隆位点序列后,会有译码的终止TAA密码,即使插入的片段使得3`末端产生了移码突
2.密码子阅读与翻译具有一定的方向性:从5'端到3'端。
问题:
1.如何了解质粒的类型(原核/真核/穿梭质粒)?看Ori的位置,原核生物DNA分子中只有一个复制起始
点。而真核生物DNA分子有多个复制起始位点。所谓穿梭质粒是指一类人工构建的具有两种不同复制起 点和选择标记,因而可以在两种不同类群宿主中存活和复制的质粒载体。
第一点是要注意:酶切位点边标注了*的一般是指并不 仅仅存在一个位点,也就不能用来作为构建质粒的酶切
位点。
第二点需要注意:有的酶切位点,在序列中只有一个, 但它上面也会标注一个*或者(dam),这说明可能这 个酶切位点会有CpG岛的甲基化修饰[常见的是XbaI, TCTAG(6m)A中的TC无法切开],一般也是不能用的。 但是非要用这个酶切位点的话,就需要用非甲基化的感 受态细胞,如JM109或者JM110。
变,照样能使表达的蛋白正常终止掉(在酵母双杂的AD质粒中,由于插入的是随机cDNA,所以AD的载体上
会常见这样的结构)。 注:1.起始密码子有两种,一种是甲硫氨酸(AUG),一种是缬氨酸(GUG),而终止密码子(有3个,分 别是UAA、UAG、UGA)没有相应的转运核糖核酸(tRNA)存在,只供释放因子识别来实现翻译的终止。
质粒图谱大全

(转载)一. 九种表达载体Pllp-OmpA, pllp-STII, pMBP-P, pMBP-C,pET-GST, pET-Trx, pET-His, pET-CKS, pET-DsbA二. 克隆载体pTZ19RDNApUC57DNAPMD18TPQE30pUC18pUC19pTrcHisApTrxFuspRSET-ApRSET-BpVAX1PBR322pbv220pBluescriptIIKS( )L4440pCAMBIA-1301pMAL-p2XpGD926三.PET 系列表达载体ProteinExpression?ProkaryoticExpression?pETDsbFusionSystems39band40b ProteinExpression?ProkaryoticExpression?pETExpressionSystem33b ProteinExpression?ProkaryoticExpression?pETExpressionSystems ProteinExpression?ProkaryoticExpression?pETExpressionSystemsplusCompetentCells ProteinExpression?ProkaryoticExpression?pETGSTFusionSystems41and42 ProteinExpression?ProkaryoticExpression?pETNusAFusionSystems43.1and44 ProteinExpression?ProkaryoticExpression?pETVectorDNAProteinPurification?PurificationSystems?Strep?TactinResinsandPurificationKits四.PGEX 系列表达载体TEcoR?pGEX-1I/BAPpGEX-2TpGEX-2TKpGEX-3XpGEX-4T-1pGEX-4T-2pGEX-4T-3pGEX-5X-1pGEX-5X-2pGEX-5X-3pGEX-6P-1pGEX-6P-2pGEX-6P-3五.PTYBsystemPTYB1PTYB2PTYB11PTYB12六. 真核表达载体pCDNA3.1(-)pCDNA3.1( )pPICZalphaApGAPZα APYES2.0pBI121pEGFP-N1pEGFP-C1pPIC9KpPIC3.5K如何阅读分析质粒图谱载体主要有病毒和非病毒两大类, 其中质粒DNA是一种新的非病毒转基因载体。
质粒

pCMV-C-Flag质粒图谱pCMV-C-Flag质粒图谱pCMV-C-Flag的MCS:XmaI PstISacI Small BamHI HindIII651 GAGCTCCACC GCGGTGGCGG CCGCTCTAGC CCGGGCGGAT CCAAGCTTCTCTCGAGGTGG CGCCACCGCC GGCGAGATCG GGCCCGCCTA GGTTCGAAGAFlagEcoRI EcoRV SacI BglII XhoI XbaI D Y K D701 GCAGGAATTC GATATCGTCG ACAGATCTCT CGAGTCTAGA GATTACAAGGCGTCCTTAAG CTATAGCAGC TGTCTAGAGA GCTCAGATCT CTAATGTTCCtagD D D K ApaI751 ATGACGACGA TAAGTAAGGG CCCGGTACCT TAATTAATTA AGGTACCAGGTACTGCTGCT ATTCATTCCC GGGCCATGGA ATTAATTAAT TCCATGGTCC说明:1.该质粒的大小为4317bp2. Ori控制复制起始的位点,该质粒有两个复制位点,分别是:pUCori f1ori,是真核质粒。
3.该质粒图谱上的抗生素抗性基因有: Neor r/kanr4.该质粒的多克隆位点是MCS Flag tag,克隆携带外源基因片段。
5.该质粒有启动子pSV40,p bla,pCMV,可以使目的基因在增殖的细胞中稳定表达。
6.该质粒TkpA、SV40pA信号是加polyA信号,可以起到稳定mRNA的作用。
综上所述,该质粒有质粒所需的组成元素,是一个合格的质粒。
基因工程技术流程图。
pcDNA3.1质粒图谱

pcDNA3.1(+)pcDNA3.1(-)Catalog nos. V790-20 and V795-20, respectivelyVersion I08140128-0104tech_service@iiTable of ContentsTable of Contents (iii)Important Information (v)Purchaser Notification (vi)Methods (1)Overview (1)Cloning into pcDNA3.1 (2)Transfection (6)Creation of Stable Cell Lines (7)Appendix (10)pcDNA3.1 Vectors (10)pcDNA3.1/CAT (12)Technical Service (13)References (15)iiiivImportant InformationContents pcDNA3.1 is supplied as follows:Catalog no.ContentsV790-2020 µg pcDNA3.1(+), lyophilized in TE, pH 8.020 µg pcDNA3.1/CAT, lyophilized in TE, pH 8.0V795-2020 µg pcDNA3.1(-), lyophilized in TE, pH 8.020 µg pcDNA3.1/CAT, lyophilized in TE, pH 8.0 Shipping/Storage Lyophilized plasmids are shipped at room temperature and should be stored at -20°C.Product Qualification Each of the pcDNA3.1 vectors is qualified by restriction enzyme digestion with specific restriction enzymes as listed below. Restriction digests must demonstrate the correct banding pattern when electrophoresed on an agarose gel. The table below lists the restriction enzymes and the expected fragments.Vector Restriction Enzyme Expected Fragments (bp) pcDNA3.1(+)Nhe IPst ISac I54281356, 4072109, 5319pcDNA3.1(-)Nhe IPst ISac I54271363, 4064169, 5258pcDNA3.1/CAT Nhe IPst ISac I62172145, 4072109, 6008vPurchaser NotificationIntroduction Use of pcDNA3.1 is covered under a number of different licenses as described below.CMV Promoter Use of the CMV promoter is covered under U.S. Patent Nos. 5,168,062 and 5,385,839owned and licensed by the University of Iowa Research Foundation and may be used forresearch purposes only. Commercial users must obtain a license to these patents directlyfrom the University of Iowa Research Foundation. Inquiries for commercial use should bedirected to:Brenda AkinsUniversity of Iowa Research Foundation (UIRF)214 Technology Innovation CenterIowa City, IA 52242Phone:319-335-4549BGH Polyadenylation Signal The bovine growth hormone (BGH) polyadenylation sequence is licensed under U.S. Patent No. 5,122,458 for research purposes only. “Research purposes” means uses directed to the identification of useful recombinant proteins and the investigation of the recombinant expression of proteins, which uses shall in no event include any of the following:a.any use in humans of a CLAIMED DNA or CLAIMED CELL;b.any use in human of protein or other substance expressed or made at any stage of itsproduction with the use of a CLAIMED DNA or a CLAIMED CELL;c.any use in which a CLAIMED DNA or CLAIMED CELL would be sold ortransferred to another party other than Invitrogen, its AFFILIATE, or itsSUBLICENSEE;d.any use in connection with the expression or production of a product intended forsale or commercial use; ore.any use for drug screening or drug development.Inquiries for commercial use should be directed to:Bennett Cohen, Ph.D.Research Corporation Technologies101 North Wilmot Road, Suite 600Tucson, AZ 85711-3335Tel: 1-520-748-4400Fax: 1-520-748-0025viMethodsOverviewIntroduction pcDNA3.1(+) and pcDNA3.1(-) are 5.4 kb vectors derived from pcDNA3 and designed for high-level stable and transient expression in mammalian hosts. High-level stable andnon-replicative transient expression can be carried out in most mammalian cells. Thevectors contain the following elements:•Human cytomegalovirus immediate-early (CMV) promoter for high-level expressionin a wide range of mammalian cells•Multiple cloning sites in the forward (+) and reverse (-) orientations to facilitatecloning•Neomycin resistance gene for selection of stable cell lines•Episomal replication in cells lines that are latently infected with SV40 or that expressthe SV40 large T antigen (e.g. COS-1, COS-7)The control plasmid, pcDNA3.1/CAT, is included for use as a positive control fortransfection and expression in the cell line of choice.Experimental Outline Use the following outline to clone and express your gene of interest in pcDNA3.1.1.Consult the multiple cloning sites described on pages 3-4 to design a strategy to cloneyour gene into pcDNA3.1.2.Ligate your insert into the appropriate vector and transform into E. coli. Selecttransformants on LB plates containing 50 to 100 µg/ml ampicillin.3.Analyze your transformants for the presence of insert by restriction digestion.4.Select a transformant with the correct restriction pattern and use sequencing toconfirm that your gene is cloned in the proper orientation.5.Transfect your construct into the mammalian cell line of interest using your ownmethod of choice. Generate a stable cell line, if desired.6.Test for expression of your recombinant gene by western blot analysis or functionalassay.1Cloning into pcDNA3.1Introduction Diagrams are provided on pages 3-4 to help you design a cloning strategy for ligating your gene of interest into pcDNA3.1. General considerations for cloning and transformation arelisted below.General Molecular Biology Techniques For help with DNA ligations, E. coli transformations, restriction enzyme analysis, purification of single-stranded DNA, DNA sequencing, and DNA biochemistry, please refer to Molecular Cloning: A Laboratory Manual (Sambrook et al., 1989) or Current Protocols in Molecular Biology (Ausubel et al., 1994).E. coli Strain Many E. coli strains are suitable for the propagation of this vector including TOP10F´,DH5α™-T1R, and TOP10. We recommend that you propagate vectors containing inserts inE. coli strains that are recombination deficient (rec A) and endonuclease A-deficient(end A).For your convenience, TOP10F´ is available as chemically competent or electrocompetentcells from Invitrogen.Item Quantity Catalog no.One Shot® TOP10F´ (chemically competent cells)21 x 50 µl C3030-03Electrocomp™ TOP10F´ 5 x 80 µl C665-55Ultracomp™ TOP10F´ (chemically competent cells) 5 x 300 µl C665-03Transformation Method You may use any method of your choice for transformation. Chemical transformation is the most convenient for most researchers. Electroporation is the most efficient and the method of choice for large plasmids.Maintenance of pcDNA3.1To propagate and maintain pcDNA3.1, we recommend resuspending the vector in 20 µl sterile water to make a 1 µg/µl stock solution. Store the stock solution at -20°C.Use this stock solution to transform a rec A, end A E. coli strain like TOP10F´, DH5α™-T1R, TOP10, or equivalent. Select transformants on LB plates containing 50 to100 µg/ml ampicillin. Be sure to prepare a glycerol stock of your plasmid-containing E. coli strain for long-term storage (see page 5).Cloning Considerations pcDNA3.1(+) and pcDNA3.1(-) are nonfusion vectors. Your insert must contain a Kozak translation initiation sequence and an ATG start codon for proper initiation of translation (Kozak, 1987; Kozak, 1991; Kozak, 1990). An example of a Kozak consensus sequence is provided below. Please note that other sequences are possible (see references above), but the G or A at position -3 and the G at position +4 are the most critical for function (shown in bold). The ATG initiation codon is shown underlined.(G/A)NNATG GYour insert must also contain a stop codon for proper termination of your gene. Please note that the Xba I site contains an internal stop codon (TCTAGA).continued on next page23Multiple Cloning Site of pcDNA3.1(+)Below is the multiple cloning site for pcDNA3.1(+). Restriction sites are labeled to indicate the cleavage site. The Xba I site contains an internal stop codon (TCTAGA). The multiple cloning site has been confirmed by sequencing and functional testing. Thecomplete sequence of pcDNA3.1(+) is available for downloading from our web site( ) or from Technical Service (see page 13). For a map and adescription of the features of pcDNA3.1(+), please refer to the Appendix , pages 10-11.1109TCCTTTCCTA ATAAAATGAG GAAATTGCAT CAAT TA TABGH poly (A) siteCATTGACGTC AATGGGAGTT TGTTTTGGCA CCAAAATCAA CGGGACTTTC CAAAATGTCGTAACAACTCC GCCCCATTGA CGCAAATGGG CGGTAGGCGT GTACGGTGGG AGGTCTATATAGTCTAGAGG GCCCGTTTAA ACCCGCTGAT CAGCCTCGAC TGTGCCTTCT AGTTGCCAGC CATCTGTTGT TTGCCCCTCC CCCGTGCCTT CCTTGACCCT GGAAGGTGCC ACTCCCACTG6897498098699299891049enhancer region (3´ end)*Please note that there are two Bst X I sites in the polylinker.continued on next page4Multiple CloningSite ofpcDNA3.1(-)Below is the multiple cloning site for pcDNA3.1(-). Restriction sites are labeled to indicate the cleavage site. The Xba I site contains an internal stop codon (TCTAGA). The multiple cloning site has been confirmed by sequencing and functional testing. Thecomplete sequence of pcDNA3.1(-) is available for downloading from our web site ( ) or from Technical Service (see page 13). For a map and a description of the features of pcDNA3.1(-), please see the Appendix , pages 10-11.Hin d III CAAT TA TA Bam H I Bst X I*Eco R I Eco R V Bst X I*BGH poly (A) site Kpn I Afl II Pme I Asp 718 I pcDNA3.1/BGH reverse priming site CCTTTCCTAA TAAAATGAGG AAATTGCATC ATCTGTTGTT TGCCCCTCCC CCGTGCCTTC CTTGACCCTG GAAGGTGCCA CTCCCACTGT GGTACCAAGC TTAAGTTTAA ACCGCTGATC AGCCTCGACT GTGCCTTCTA GTTGCCAGCC GCCGCCACTG TGCTGGATAT CTGCAGAATT CCACCACACT GGACTAGTGG ATCCGAGCTC TAACAACTCC GCCCCATTGA CGCAAATGGG CGGTAGGCGT GTACGGTGGG AGGTCTATAT CATTGACGTC AATGGGAGTT TGTTTTGGCA CCAAAATCAA CGGGACTTTC CAAAATGTCG 11091049989929869809749689enhancer region (3´ end)*Please note that there are two Bst X I sites in the polylinker.continued on next pageCloning into pcDNA3.1, continuedE. coli Transformation Transform your ligation mixtures into a competent rec A, end A E. coli strain (e.g. TOP10F´, DH5α™-T1R, TOP10) and select transformants on LB plates containing 50 to 100 µg/ml ampicillin. Select 10-20 clones and analyze for the presence and orientation of your insert.We recommend that you sequence your construct with the T7 Promoter and BGH Reverseprimers (Catalog nos. N560-02 and N575-02, respectively) to confirm that your gene is in thecorrect orientation for expression and contains an ATG and a stop codon. Please refer to thediagrams on pages 3-4 for the sequences and location of the priming sites. The primers areavailable separately from Invitrogen in 2 µg aliquots.Preparing aGlycerol StockOnce you have identified the correct clone, purify the colony and make a glycerol stock forlong-term storage. You should keep a DNA stock of your plasmid at -20°C.•Streak the original colony out on an LB plate containing 50 µg/ml ampicillin. Incubatethe plate at 37°C overnight.•Isolate a single colony and inoculate into 1-2 ml of LB containing 50 µg/ml ampicillin.•Grow the culture to mid-log phase (OD600 = 0.5-0.7).•Mix 0.85 ml of culture with 0.15 ml of sterile glycerol and transfer to a cryovial.•Store at -80°C.TransfectionIntroduction Once you have verified that your gene is cloned in the correct orientation and contains an initiation ATG and a stop codon, you are ready to transfect your cell line of choice. Werecommend that you include the positive control vector and a mock transfection (negativecontrol) to evaluate your results.Plasmid Preparation Plasmid DNA for transfection into eukaryotic cells must be clean and free from phenol and sodium chloride. Contaminants will kill the cells, and salt will interfere with lipids decreasing transfection efficiency. We recommend isolating plasmid DNA using the S.N.A.P.™ MiniPrep Kit (10-15 µg DNA, Catalog no. K1900-01), the S.N.A.P. ™MidiPrep Kit (10-200 µg DNA, Catalog no. K1910-01), or CsCl gradient centrifugation.Methods of Transfection For established cell lines (e.g. HeLa), please consult original references or the supplier of your cell line for the optimal method of transfection. We recommend that you follow exactly the protocol for your cell line. Pay particular attention to medium requirements, when to pass the cells, and at what dilution to split the cells. Further information is provided in Current Protocols in Molecular Biology (Ausubel et al., 1994).Methods for transfection include calcium phosphate (Chen and Okayama, 1987; Wigler et al., 1977), lipid-mediated (Felgner et al., 1989; Felgner and Ringold, 1989) and electroporation (Chu et al., 1987; Shigekawa and Dower, 1988). Invitrogen offers the Calcium Phosphate Transfection Kit (Catalog no. K2780-01) and a large selection of reagents for transfection. For more information, please refer to our World Wide Web site () or call Technical Service (see page 13).Positive Control pcDNA3.1/CAT is provided as a positive control vector for mammalian transfection and expression (see page 12) and may be used to optimize transfection conditions for yourcell line. The gene encoding chloramphenicol acetyl transferase (CAT) is expressed inmammalian cells under the control of the CMV promoter. A successful transfection willresult in CAT expression that can be easily assayed (see below).Assay for CAT Protein You may assay for CAT expression by ELISA assay, western blot analysis, fluorometric assay, or radioactive assay (Ausubel et al., 1994; Neumann et al., 1987). If you wish to detect CAT protein using western blot analysis, you may use the Anti-CAT Antiserum (Catalog no. R902-25) available from Invitrogen. Other kits to assay for CAT protein using ELISA assay are available from Roche Molecular Biochemicals (Catalog no. 1 363 727) and Molecular Probes (Catalog no. F-2900).Creation of Stable Cell LinesIntroduction The pcDNA3.1(+) and pcDNA3.1(-) vectors contain the neomycin resistance gene forselection of stable cell lines using neomycin (Geneticin®). We recommend that you testthe sensitivity of your mammalian host cell to Geneticin® as natural resistance variesamong cell lines. General information and guidelines are provided in this section for yourconvenience.Geneticin®Selective Antibiotic Geneticin® Selective Antibiotic blocks protein synthesis in mammalian cells by interfering with ribosomal function. It is an aminoglycoside, similar in structure to neomycin, gentamycin, and kanamycin. Expression of the bacterial aminoglycoside phosphotransferase gene (APH), derived from Tn5, in mammalian cells results in detoxification of Geneticin®(Southern and Berg, 1982).Geneticin®Selection Guidelines Geneticin® Selective Antibiotic is available from Invitrogen (Catalog no. 10486-025). Use as follows:•Prepare Geneticin® in a buffered solution (e.g. 100 mM HEPES, pH 7.3).•Use 100 to 800 µg/ml of Geneticin® in complete medium.•Calculate concentration based on the amount of active drug (check the lot label).•Test varying concentrations of Geneticin® on your cell line to determine theconcentration that kills your cells (see below). Cells differ in their susceptibility to Geneticin®.Cells will divide once or twice in the presence of lethal doses of Geneticin®, so the effects of the drug take several days to become apparent. Complete selection can take up to 3 weeks of growth in selective media.Determination of Antibiotic Sensitivity To successfully generate a stable cell line expressing your gene of interest frompcDNA3.1, you need to determine the minimum concentration of Geneticin® required to kill your untransfected host cell line. We recommend that you test a range of concentrations to ensure that you determine the minimum concentration necessary for your host cell line.1.Plate or split a confluent plate so the cells will be approximately 25% confluent.Prepare a set of 7 plates. Allow cells to adhere overnight.2.The next day, substitute culture medium with medium containing varyingconcentrations of Geneticin® (0, 50, 100, 200, 400, 600, 800 µg/ml Geneticin®).3.Replenish the selective media every 3-4 days, and observe the percentage of survivingcells.4.Count the number of viable cells at regular intervals to determine the appropriateconcentration of Geneticin® that prevents growth within 2-3 weeks after addition of Geneticin®.continued on next pagePossible Sites for Linearization of pcDNA3.1(+)Prior to transfection, we recommend that you linearize the pcDNA3.1(+) vector. Linearizing pcDNA3.1(+) will decrease the likelihood of the vector integrating into the genome in a way that disrupts the gene of interest or other elements required for expression in mammalian cells. The table below lists unique restriction sites that may be used to linearize your construct prior to transfection. Other unique restriction sites are possible. Be sure that your insert does not contain the restriction enzyme site you wish to use to linearize your vector.Enzyme Restriction Site (bp)Location SupplierBgl II12Upstream of CMV promoter Invitrogen, Catalog no. 15213-028 Mfe I161Upstream of CMV promoter New England BiolabsBst1107 I3236End of SV40 polyA AGS*, Fermentas, Takara, RocheMol. BiochemicalsEam1105 I4505Ampicillin gene AGS*, Fermentas, TakaraPvu I4875Ampicillin gene Invitrogen, Catalog no. 25420-019 Sca I4985Ampicillin gene Invitrogen, Catalog no. 15436-017 Ssp I5309bla promoter Invitrogen, Catalog no. 15458-011 *Angewandte Gentechnologie SystemePossible Sites for Linearization of pcDNA3.1(-)The table below lists unique restriction sites that may be used to linearize yourpcDNA3.1(-) construct prior to transfection. Other unique restriction sites are possible. Be sure that your insert does not contain the restriction enzyme site you wish to use to linearize your vector.Enzyme Restriction Site (bp)Location SupplierBgl II12Upstream of CMV promoter Invitrogen, Catalog no. 15213-028 Mfe I161Upstream of CMV promoter New England BiolabsBst1107 I3235End of SV40 polyA AGS*, Fermentas, Takara, RocheMol. BiochemicalsEam1105 I4504Ampicillin gene AGS*, Fermentas, TakaraPvu I4874Ampicillin gene Invitrogen, Catalog no. 25420-019 Sca I4984Ampicillin gene Invitrogen, Catalog no. 15436-017 Ssp I5308bla promoter Invitrogen, Catalog no. 15458-011 *Angewandte Gentechnologie Systemecontinued on next pageSelection of Stable Integrants Once you have determined the appropriate Geneticin® concentration to use for selection in your host cell line, you can generate a stable cell line expressing your gene of interest.1.Transfect your mammalian host cell line with your pcDNA3.1 construct using thedesired protocol. Remember to include a plate of untransfected cells as a negativecontrol and the pcDNA3.1/CAT plasmid as a positive control.2.24 hours after transfection, wash the cells and add fresh medium to the cells.3.48 hours after transfection, split the cells into fresh medium containing Geneticin® atthe pre-determined concentration required for your cell line. Split the cells such that they are no more than 25% confluent.4.Feed the cells with selective medium every 3-4 days until Geneticin®-resistant foci canbe identified.5.Pick and expand colonies in 96- or 48-well plates.AppendixpcDNA3.1 VectorsMap ofpcDNA3.1(+) and pcDNA3.1(-)The figure below summarizes the features of the pcDNA3.1(+) and pcDNA3.1(-) vectors.The complete sequences for pcDNA3.1(+) and pcDNA3.1(-) are available for down-loading from our World Wide Web site ( ) or from Technical Service (see page 13). Details of the multiple cloning sites are shown on page 3 for pcDNA3.1(+) and page 4 for pcDNA3.1(-).Comments for pcDNA3.1 (+) 5428 nucleotidesCMV promoter: bases 232-819T7 promoter/priming site: bases 863-882Multiple cloning site: bases 895-1010BGH polyadenylation sequence: bases 1028-1252f1 origin: bases 1298-1726SV40 early promoter and origin: bases 1731-2074Neomycin resistance gene (ORF): bases 2136-2930SV40 early polyadenylation signal: bases 3104-3234pUC origin: bases 3617-4287 (complementary strand)Ampicillin resistance gene (bla ): bases 4432-5428 (complementary strand) ORF: bases 4432-5292 (complementary strand)Ribosome binding site: bases 5300-5304 (complementary strand) bla promoter (P3): bases 5327-5333 (complementary strand)I I (+)( )continued on next pagepcDNA3.1 Vectors, continuedFeatures of pcDNA3.1(+) and pcDNA3.1(-)pcDNA3.1(+) (5428 bp) and pcDNA3.1(-) (5427 bp) contain the following elements. All features have been functionally tested.Feature BenefitHuman cytomegalovirus (CMV)immediate-early promoter/enhancerPermits efficient, high-level expression ofyour recombinant protein (Andersson et al.,1989; Boshart et al., 1985; Nelson et al.,1987)T7 promoter/priming site Allows for in vitro transcription in the senseorientation and sequencing through theinsertMultiple cloning site in forward orreverse orientationAllows insertion of your gene andfacilitates cloningBovine growth hormone (BGH)polyadenylation signalEfficient transcription termination andpolyadenylation of mRNA (Goodwin andRottman, 1992)f1 origin Allows rescue of single-stranded DNASV40 early promoter and origin Allows efficient, high-level expression ofthe neomycin resistance gene and episomalreplication in cells expressing SV40 large TantigenNeomycin resistance gene Selection of stable transfectants inmammalian cells (Southern and Berg,1982)SV40 early polyadenylation signal Efficient transcription termination andpolyadenylation of mRNApUC origin High-copy number replication and growthin E. coliAmpicillin resistance gene (β-lactamase)Selection of vector in E. colipcDNA3.1/CATDescription pcDNA3.1/CAT is a 6217 bp control vector containing the gene for CAT. It wasconstructed by digesting pcDNA3.1(+) with Xho I and Xba I and treating with Klenow.An 800 bp Hin d III fragment containing the CAT gene was treated with Klenow and thenligated into pcDNA3.1(+).Map of Control Vector The figure below summarizes the features of the pcDNA3.1/CAT vector. The complete nucleotide sequence for pcDNA3.1/CAT is available for downloading from our World Wide Web site () or by contacting Technical Service (see page 13).Comments for pcDNA3.1(+)/CAT6217 nucleotidesCMV promoter: bases 232-819CAT ORF: bases 1027-1686pcDNA3.1/BGH reverse priming site: bases 1811-1828BGH polyadenylation sequence: bases 1817-2041f1 origin: bases 2087-2515SV40 early promoter and origin: bases 2520-2863Neomycin resistance gene (ORF): bases 2925-3719SV40 early polyadenylation sequence: bases 3893-4023pUC origin: bases 4406-5076 (complementary strand)Ampicillin resistance gene (ORF): bases 5221-6081 (complementary strand)Technical ServiceVisit the Invitrogen Web Resource using your World Wide Web browser. At the site, youcan:•Get the scoop on our hot new products and special product offers•View and download vector maps and sequences•Download manuals in Adobe® Acrobat® (PDF) format•Explore our catalog with full color graphics•Obtain citations for Invitrogen products•Request catalog and product literatureOnce connected to the Internet, launch your web browser (Internet Explorer 5.0 or neweror Netscape 4.0 or newer), then enter the following location (or URL):...and the program will connect directly. Click on underlined text or outlined graphics toexplore. Don't forget to put a bookmark at our site for easy reference!Contact us For more information or technical assistance, please call, write, fax, or email. Additionalinternational offices are listed on our web page ().United States Headquarters:Japanese Headquarters European Headquarters:Invitrogen Corporation Invitrogen Japan K.K.Invitrogen Ltd1600 Faraday Avenue Nihonbashi Hama-Cho Park Bldg. 4F 3 Fountain DriveCarlsbad, CA 92008 USA2-35-4, Hama-Cho, Nihonbashi Inchinnan Business ParkTel: 1 760 603 7200Tel: 81 3 3663 7972Paisley PA4 9RF, UKTel (Toll Free): 1 800 955 6288Fax: 81 3 3663 8242Tel (Free Phone Orders): 0800 269 210 Fax: 1 760 602 6500E-mail: jpinfo@ Tel (General Enquiries): 0800 5345 5345 E-mail:Fax: +44 (0) 141 814 6287tech_service@ E-mail: eurotech@MSDS Requests To request an MSDS, please visit our web site () and follow theinstructions below.1.On the home page, go to the left-hand column under ‘Technical Resources’ andselect ‘MSDS Requests’.2.Follow instructions on the page and fill out all the required fields.3.To request additional MSDSs, click the ‘Add Another’ button.4.All requests will be faxed unless another method is selected.5.When you are finished entering information, click the ‘Submit’ button. Your MSDSwill be sent within 24 hours.continued on next pageTechnical Service, continuedEmergency Information In the event of an emergency, customers of Invitrogen can call the 3E Company, 24 hours a day, 7 days a week for disposal or spill information. The 3E Company can also connect the customer with poison control or with the University of California at San Diego Medical Center doctors.3E CompanyVoice: 1-760-602-8700Limited Warranty Invitrogen is committed to providing our customers with high-quality goods and services. Our goal is to ensure that every customer is 100% satisfied with our products and our service. If you shouldhave any questions or concerns about an Invitrogen product or service, please contact ourTechnical Service Representatives.Invitrogen warrants that all of its products will perform according to the specifications stated on thecertificate of analysis. The company will replace, free of charge, any product that does not meetthose specifications. This warranty limits Invitrogen Corporation’s liability only to the cost of theproduct. No warranty is granted for products beyond their listed expiration date. No warranty isapplicable unless all product components are stored in accordance with instructions. Invitrogenreserves the right to select the method(s) used to analyze a product unless Invitrogen agrees to aspecified method in writing prior to acceptance of the order.Invitrogen makes every effort to ensure the accuracy of its publications, but realizes that theoccasional typographical or other error is inevitable. Therefore Invitrogen makes no warranty ofany kind regarding the contents of any publications or documentation. If you discover an error inany of our publications, please report it to our Technical Service Representatives.Invitrogen assumes no responsibility or liability for any special, incidental, indirect orconsequential loss or damage whatsoever. The above limited warranty is sole and exclusive.No other warranty is made, whether expressed or implied, including any warranty ofmerchantability or fitness for a particular purpose.ReferencesAndersson, S., Davis, D. L., Dahlbäck, H., Jörnvall, H., and Russell, D. W. (1989). Cloning, Structure, andExpression of the Mitochondrial Cytochrome P-450 Sterol 26-Hydroxylase, a Bile Acid Biosynthetic Enzyme. J.Biol. Chem. 264, 8222-8229.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. (1994).Current Protocols in Molecular Biology (New York: Greene Publishing Associates and Wiley-Interscience).Boshart, M., Weber, F., Jahn, G., Dorsch-Häsler, K., Fleckenstein, B., and Schaffner, W. (1985). A Very Strong Enhancer is Located Upstream of an Immediate Early Gene of Human Cytomegalovirus. Cell 41, 521-530.Chen, C., and Okayama, H. (1987). High-Efficiency Transformation of Mammalian Cells by Plasmid DNA. Mol.Cell. Biol. 7, 2745-2752.Chu, G., Hayakawa, H., and Berg, P. (1987). Electroporation for the Efficient Transfection of Mammalian Cells with DNA. Nuc. Acids Res. 15, 1311-1326.Felgner, P. L., Holm, M., and Chan, H. (1989). Cationic Liposome Mediated Transfection. Proc. West. Pharmacol.Soc. 32, 115-121.Felgner, P. L., and Ringold, G. M. (1989). Cationic Liposome-Mediated Transfection. Nature 337, 387-388.Goodwin, E. C., and Rottman, F. M. (1992). The 3´-Flanking Sequence of the Bovine Growth Hormone GeneContains Novel Elements Required for Efficient and Accurate Polyadenylation. J. Biol. Chem. 267, 16330-16334. Kozak, M. (1987). An Analysis of 5´-Noncoding Sequences from 699 Vertebrate Messenger RNAs. Nuc. Acids Res.15, 8125-8148.Kozak, M. (1991). An Analysis of Vertebrate mRNA Sequences: Intimations of Translational Control. J. Cell Biol.115, 887-903.Kozak, M. (1990). Downstream Secondary Structure Facilitates Recognition of Initiator Codons by Eukaryotic Ribosomes. Proc. Natl. Acad. Sci. USA 87, 8301-8305.Nelson, J. A., Reynolds-Kohler, C., and Smith, B. A. (1987). Negative and Positive Regulation by a Short Segmentin the 5´-Flanking Region of the Human Cytomegalovirus Major Immediate-Early Gene. Mol. Cell. Biol. 7, 4125-4129.Neumann, J. R., Morency, C. A., and Russian, K. O. (1987). A Novel Rapid Assay for Chloramphenicol Acetyltransferase Gene Expression. BioTechniques 5, 444-447.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, Second Edition (Plainview, New York: Cold Spring Harbor Laboratory Press).Shigekawa, K., and Dower, W. J. (1988). Electroporation of Eukaryotes and Prokaryotes: A General Approach tothe Introduction of Macromolecules into Cells. BioTechniques 6, 742-751.Southern, P. J., and Berg, P. (1982). Transformation of Mammalian Cells to Antibiotic Resistance with a BacterialGene Under Control of the SV40 Early Region Promoter. J. Molec. Appl. Gen. 1, 327-339.Wigler, M., Silverstein, S., Lee, L.-S., Pellicer, A., Cheng, Y.-C., and Axel, R. (1977). Transfer of Purified HerpesVirus Thymidine Kinase Gene to Cultured Mouse Cells. Cell 11, 223-232.©1997-2001 Invitrogen Corporation. All rights reserved.15。
质粒pECFP-N1图谱
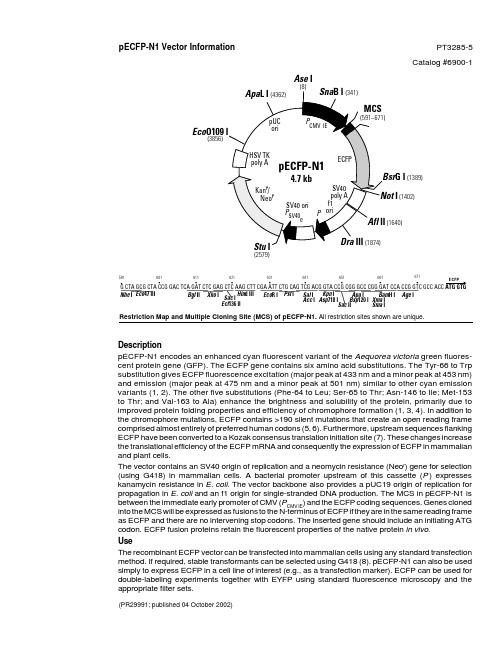
Restriction Map and Multiple Cloning Site (MCS) of pECFP-N1. All restriction sites shown are unique.pECFP-N1 Vector Information PT3285-5Catalog #6900-1(PR29991; published 04 October 2002)DescriptionpECFP-N1 encodes an enhanced cyan fluorescent variant of the Aequorea victoria green fluores-cent protein gene (GFP). The ECFP gene contains six amino acid substitutions. The Tyr-66 to Trp substitution gives ECFP fluorescence excitation (major peak at 433 nm and a minor peak at 453 nm)and emission (major peak at 475 nm and a minor peak at 501 nm) similar to other cyan emission variants (1, 2). The other five substitutions (Phe-64 to Leu; Ser-65 to Thr; Asn-146 to Ile; Met-153to Thr; and Val-163 to Ala) enhance the brightness and solubility of the protein, primarily due to improved protein folding properties and efficiency of chromophore formation (1, 3, 4). In addition to the chromophore mutations, ECFP contains >190 silent mutations that create an open reading frame comprised almost entirely of preferred human codons (5, 6). Furthermore, upstream sequences flanking ECFP have been converted to a Kozak consensus translation initiation site (7). These changes increase the translational efficiency of the ECFP mRNA and consequently the expression of ECFP in mammalian and plant cells.The vector contains an SV40 origin of replication and a neomycin resistance (Neo r ) gene for selection (using G418) in mammalian cells. A bacterial promoter upstream of this cassette (P ) expresses kanamycin resistance in E. coli . The vector backbone also provides a pUC19 origin of replication for propagation in E. coli and an f1 origin for single-stranded DNA production. The MCS in pECFP-N1 is between the immediate early promoter of CMV (P CMV IE ) and the ECFP coding sequences. Genes cloned into the MCS will be expressed as fusions to the N-terminus of ECFP if they are in the same reading frame as ECFP and there are no intervening stop codons. The inserted gene should include an initiating ATG codon. ECFP fusion proteins retain the fluorescent properties of the native protein in vivo .UseThe recombinant ECFP vector can be transfected into mammalian cells using any standard transfection method. If required, stable transformants can be selected using G418 (8). pECFP-N1 can also be used simply to express ECFP in a cell line of interest (e.g., as a transfection marker). ECFP can be used for double-labeling experiments together with EYFP using standard fluorescence microscopy and the appropriate filter sets.G CTA GCG CTA CCG GAC TCA GAT CTC GAG CTC AAG CTT CGA ATT CTG CAG TCG ACG GTA CCG CGG GCC CGG GAT CCA CCG GTC GCC ACC 591•611•601•621•631•641•661•651•671•Hin d III Xho I Apa I Bsp 120 I Kpn I Asp 718 I Bam H I ECFPAge I Xma I Sma ISac II Sal I Acc I Sac I Ecl 136 II Eco R I Pst I Eco 47 IIIBgl II Nhe II (1402) (1640)Eco (3856)(2579)Ase IG I (1389)Location of features•Human cytomegalovirus (CMV) immediate early promoter: 1–589Enhancer region: 59–465TATA box: 554–560Transcription start point: 583C→G mutation to remove Sac I site: 569•MCS: 591–671•Enhanced cyan fluorescent protein (ECFP) geneKozak consensus translation initiation site: 672–682Start codon (ATG): 679–681; stop codon: 1396–1398Insertion of Val at position 2: 682–684ECFP mutations:Phe-64 to Leu, Ser-65 to Thr, and Tyr-66 to Trp: 871–879Asn-146 to Ile: 1117–1119Met-153 to Thr: 1138–1140Val-163 to Ala: 1168–1170His-231 to Leu mutation (A→T): 1373•SV40 early mRNA polyadenylation signalPolyadenylation signals: 1552–1557 & 1581–1586mRNA 3' ends: 1590 & 1602•f1 single-strand DNA origin: 1649–2104(Packages the noncoding strand of ECFP.)•Bacterial promoter for expression of Kan r gene:–35 region: 2166–2171; –10 region: 2189–2194Transcription start point: 2201•SV40 origin of replication: 2445–2580•SV40 early promoterEnhancer (72-bp tandem repeats): 2278–2349 & 2350–242121-bp repeats: 2425–2445, 2446–2466 & 2468–2488Early promoter element: 2501–2507Major transcription start points: 2497, 2535, 2541 & 2546•Kanamycin/neomycin resistance geneNeomycin phosphotransferase coding sequences:Start codon (ATG): 2629–2631; stop codon: 3421–3423G→A mutation to remove Pst I site: 2811C→A (Arg to Ser) mutation to remove Bss H II site: 3157•Herpes simplex virus (HSV) thymidine kinase (TK) polyadenylation signalPolyadenylation signals: 3659–3664 & 3672–3677•pUC plasmid replication origin: 4008–4651Primer Locations•EGFP-N Sequencing Primer (#6479-1): 745–724•EGFP-C Sequencing Primer (#6478-1): 1332–1353Propagation in E. coli•Suitable host strains: DH5α, HB101, and other general-purpose strains. Single-stranded DNA production requires a host containing an F plasmid such as JM101 or XL1-Blue.•Selectable marker: plasmid confers resistance to kanamycin (30 µg/ml) in E. coli hosts.• E. coli replication origin: pUC;Copy number: ~500•Plasmid incompatibility group: pMB1/Col E1References1.Heim, R.,& Tsien, R. Y. (1996) Curr. Biol. 6:178–182.2.Heim, R., et al. (1994) Proc. Natl. Acad. Sci. USA 91:12501–12504.3.Cormack, B., et al. (1996) Gene 173:33–38.4.Heim, R., et al. (1995) Nature373:663–664.5.Yang, T. T., et al. (1996) Nucleic Acids Res.24:4592–4593.6.Haas, J., et al. (1996) Curr. Biol.6:315–324.7.Kozak, M. (1987) Nucleic Acids Res.15:8125–8148.8.Gorman, C. (1985) In DNA cloning: A practical approach, vol. II. Ed. D. M. Glover. (IRL Press, Oxford, U.K.), pp. 143–190.BD Biosciences Clontech Protocol # PT3285-5 2Version # PR29991Protocol # PT3285-5 BD Biosciences ClontechVersion # PR299913Notice to PurchaserUse of BD Biosciences Clontech’s Living Colors™ products containing DNA sequences coding for mutant Aequorea victoria green fluorescent protein (GFP) variants or proteins thereof requires a license from Amersham Biosciences under U.S. Patent Nos. 5,625,048; 5,777,079; 6,054,321and other pending U.S. and foreign patent applications. In addition, certain BD Biosciences Clontech products are made under U.S. Patent No.5,804,387 licensed from Stanford University.Not-For-Profit research institutes or entities are granted an automatic license with the purchase of this product for use in non-commercial internal research purposes, the terms of which are disclosed in detail in the license that accompanies the shipment of this product. Such license specifi-cally excludes the right to sell or otherwise transfer this product or its components to third parties.For-Profit research institutes or entities must obtain a license from Amersham Biosciences. E-mail: gfp@Please contact BD Biosciences Clontech directly for any other assistance, including purchasing and technical support. All companies and institutions purchasing Living Colors™ products will be included in a quarterly report to Aurora Biosciences, as required by the BD Biosciences Clontech/Aurora Biosciences license agreement.This product is intended to be used for research purposes only. It is not to be used for drug or diagnostic purposes nor is it intended for human use. BD Biosciences Clontech products may not be resold, modified for resale, or used to manufacture commercial products without written approval of BD Biosciences Clontech.© 2002, Becton, Dickinson and Company Note: The attached sequence file has been compiled from information in the sequence databases, published literature, and other sources, together with partial sequences obtained by BD Biosciences Clontech. This vector has not been completely sequenced.。
PMD19T 质粒图谱
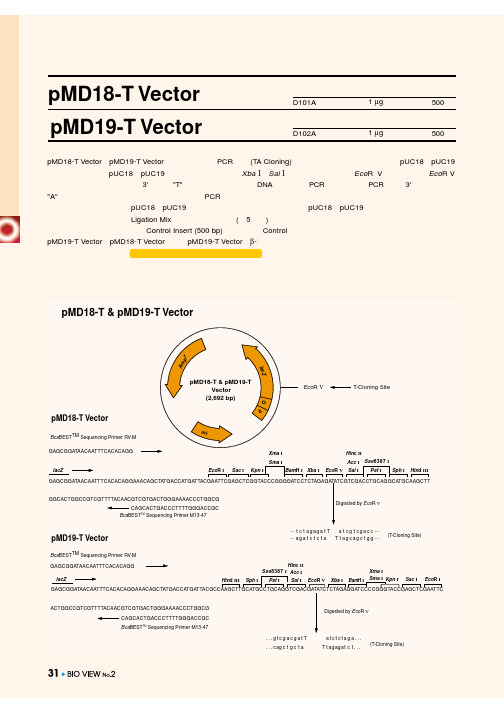
Digested by EcoR V
...gtcgacgatT . . . c ag c t g c t a
atctctaga... T t a g ag a t c t . . .
(T-Cloning Site)
■ 制品内容
pMD18-T or pMD19-T Vector (50 ng/µl) Control Insert (50 ng/µl) Ligation Mix*
852 ~ 1466
Ampr
1626 ~ 2486
【pMD19-T Vector相关位点说明】
Cloning site
431
BcaBEST Sequencing Primer M13-47 binding site 347 ~ 370
BcaBEST Sequencing Primer RV-M binding site 478 ~ 500
上培养,形成单菌落。计数白色、蓝色菌落。 8)挑选白色菌落,使用PCR法确认载体中插入片段的
长度大小。
*1 pMD18-T Vector or pMD19-T Vector的使用量 取0.5 µl实验也可得到满意的结果。实际操作时,可按实验需要确 定T载体的使用量。T Vector 1 µl(50 ng)的摩尔数约为0.03 pmol。
BcaBESTTM Sequencing Primer RV-M
GAGCGGATAACAATTTCACACAGG lacZ
Hind III
Hinc II Sse8387 I Acc I
Sph I Pst I Sal I EcoR V
Xba I
BamH I
Xma I Sma I Kpn I
Sac I
质粒图谱查询方法

3.google scholar: / 有些质粒是经过改造的,所以通过上述方法不能查询到相应信息。这时,可以在google scholar中输入质粒名称,可以直观地看哪些学者在何文章中使用了该质粒,从而可了解到质粒的来源;或者籍此向作者咨询或索取。 4.尝试从各大生物公司,例如invitrogen网站查询. 5. 这个网站收录了大量图谱: http://www.embl-hamburg.de/~geerlof/webPP/vectordb/bact_vectors/table.html
By
0039--pET-24a--Novagene--原核表达--yanBaggio 0040--pGBKT7 和pGADT7--CLOTE--酵母表达--小迷糊 0041--PTXB1--BIOLABS--原核表达--小迷糊 0042--pET-43.1 Ek/LIC--Novagene--原核表达--mcli 0043--PIRES--BD BIOSCIENCES--穿梭载体---joeys008 0044--pBAD/Thio-TOPO--invitrogen--原核表达--mcli 0045--pESC-HIS--Stratagene--酵母表达--zhangqiongyu82 0046--pcDNA3.1/V5-His-TOPO--invitrogen--真核表达--mcli 0047--pMSCVneo--Clontech--真核表达--intron 0048--pCDNA6 /V5 HisB--invitrogen--真核表达--linct97 0049--pcDNA3.1(+)--invitrogen--真核表达--kinase 0050--pSecTag2--invitrogen--真核表达--sssusu 0051--pIRESneo--Clontech--真核表达--sssusu 0052--pBudCE4.1--invitrogen--真核表达--barbie 0053--pGEX-4T-3--amershambio--原核表达--linct97 0054--PESC-TRP--startgene--真核表达--zhangqiongyu82 0055--pPICZ--invitrogen--毕赤酵母表达--yshu3507 0056--pTrc99a--novogen--原核表达--Gmail 0057--pGEX-6p-1--amershambio--原核表达--erik 0058--pSFV1--invitrogen--真核表达--syfnet 0059--pSCA1--赠送--真核表达--syfnet 0060--pUC18--/--克隆--syfnet 0061--pshuttleCMV--KRACKELER--穿梭质粒--renke3333 0062--pdc315--microbix--shuttle--renke3333 0063--pBluescript SK(+)--tianwei--原核表达--fenqinzhuhe 0064--pPIC3.5K--Invitrogen--真核表达--mcli 0065--pFB-hrGFP--Stratagene--真核表达--zhangqiongyu82 0066--pSG5--Stratagene--真核表达--zhangqiongyu82 0067--pCMV-Tag4B--Stratagene--真核表达--小迷糊 0068--pET11a--invitrogen--原核表达--pinghw 0069--pET30a--invitrogen--原核表达--seasider 0070--pCI-NEO--PREMOGA--真核表达--syfnet 0071--phRL-null--PREMOGA--真核表达--kenmed 0072--pBC1--invitrogen--真核表达--竹影烟雨 0073--pGEMEX-1--Promega--体外转录--palmyard 0074--pGEM-T Easy--Promega--克隆--laohu200381 0075--pGEX-5x-1--Pharmacia--原核表达--laohu200381 0076--pshuttle--qbiogene--穿梭质粒--renke3333 0077--pet9c--promega--原核表达--renke3333 0078--trans-vector--qbiogene--穿梭质粒--renke3333 0079--PQBI PGK--QBIOGENE--真核表达--renke3333 0080--PQBI T7 GFP--QBIOGENE--真核表达--renke3333 0081--pcDNA5/FRT/CAT—INVITROGEN--真核表达--renke3333 0082--pBABE Hygro—Geron--真核表达--Jeffrey88 0083--pBABE puro--Geron--Jeffrey88 0084--pMSCV puro—Clontech—RNAi--tuterhu 0085--质粒名--PEGFPN3—Clontech--真核 GFP--renke3333 0086--质粒名称—PSHTULLE—Clontech--穿梭---renke3333 0087--pkk223-3--哈佛大学Brosius等构建--原核表达--kingtsh 0088--pcdna3-c-myc—invitroge--真核表达--giyon 0089—pSG-cmv--新基因--stevenvin 0090--PGEX-3x--BD Co--融合型蛋白原核表达载体--kingtsh 0091--PEGFPc2—Clontech--真核 GFP 0092--PEGFP-N2—Clontech--真核 GFP 0093—PMECA--不详--克隆载体 0094--PGEX 4T-2--不详--克隆载体 0095--siSTRIKE? U6—PROMEGA--RNAi载体 0096--pSilencer? neo—Ambion--RNAi载体 0097--pSilencer? hygro—Ambion--RNAi载体 0098--pSilencer? puro—Ambion--RNAi载体
质粒图谱怎么看

一、质粒(plasmid):存在于许多细菌以及酵母菌等生物中,是染色体外能够自主复制的很小的环状DNA分子。
载体(Vector):简单的来说就是把一个有用的目的DNA片段通过重组DNA技术,送进受体细胞中去进行繁殖和表达的工具叫载体,分为病毒类和非病毒类两种。
我们平时常说的载体是在天然质粒的基础上为适应实验室操作而人工构建的,通常带有一个或一个以上的选择性标记基因(如抗生素抗性基因)和一个人工合成的含有多个限制性内切酶识别位点的多克隆位点序列,并去掉了大部分非必需序列,使分子量尽可能减少,以便于基因工程操作,同时加入一些多用途的辅助序列。
二、如何阅读质粒图谱看懂一个质粒图谱我们需要关注以下几点:1)弄清楚质粒的方向看质粒图谱的时候我们会发现大多数质粒都会有顺时针和逆时针两个方向的箭头,箭头的方向:一个方向是复制起始位点的方向,即该质粒在细菌或真菌中DNA复制的一个方向,有时图谱中还会有f1 ori,这个代表的是噬菌体的复制起始方向,只能复制出单链的DNA,但是可以用来测序。
另一个就是转录方向(一般是正向),主要是从启动子开始,即启动子-核糖体结合位点-克隆位点-转录终止信号。
看懂转录的方向,这样就方便设计插入片段的位置和方向性。
2)复制子复制子又称复制起始区,它控制着质粒DNA的复制,并决定了质粒的宿主和拷贝数。
复制子可分为严谨型复制子与松弛型复制子,分别对应低拷贝数的严紧型质粒和高拷贝数的松弛型质粒。
3)筛选标记:了解筛选标记类型(如抗生素抗性标记),方便后续确定筛选重组质粒载体。
常见抗菌素抗性标记有氨苄青霉素抗性(Amp)、卡那霉素抗性(Kan)、四环素抗性(Tet)、链霉素抗性(Str)、氯霉素(Cmr, 某些酵母表达质粒)、潮霉素(Hyg, 农杆菌里常用的)。
4)多克隆位点MCS区:既外源基因的插入位点。
具有多个限制酶酶切位点,外源性的DNA一般可通过酶切/连接的方式插入质粒。
)其他元件,如表达系统元件和蛋白标签等常见的启动子有:CMV、EF1(常规表达,一般启动长片段的),H1、U6(启动短片段的,比如shRNA),CAMV35S、Ubi(植物表达常用),T7(常用原核系统表达启动子)。
质粒图谱——精选推荐

1)质粒图谱登记号:00012)质粒名称:pIRES3)来源:BD Co4)用途:真核双表达5)是否可以提供更详细资料:可以6)是否可以共享:7)联系方式:PM)质粒图谱登记号:00022)质粒名称:pECFP-C13)来源:BD Co4)用途:检测真核表达5)是否可以提供更详细资料:可以6)是否可以共享:7)联系方式:pm1)质粒图谱登记号:00032)质粒名称:pShuttle3)来源:4)用途:5)是否可以提供更详细资料:6)是否可以共享:7)联系方式:2)pShuttle MCS很多人用pEGFP-C1,我也来发一个)质粒图谱登记号:00042)质粒名称:pS BR322、pUC183)来源:4)用途:5)是否可以提供更详细资料:6)是否可以共享:7)联系方式:1)质粒图谱登记号:00052)质粒名称:pcDNA3.1(+)/CAT3)来源:invitrogen4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:无偿7)联系方式:PM1)质粒图谱登记号:00062)质粒名称:pQEx3)来源:Qiagen4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM2)1)质粒图谱登记号:00072)质粒名称:pIVEX2.33)来源:Rocho4)用途:体外转录翻译5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM质粒图谱登记号:0008质粒名称:pIRES-EGFP来源:用途:是否可以提供更详细资料:是否可以共享:否联系方式:1)质粒图谱登记号:00092)质粒名称:pET-28a(+)3)来源:Novagen4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM1)质粒图谱登记号:00112)质粒名称:pET-32a(+)3)来源:novagen4)用途:原核表达5)是否可以提供更详细资料:6)是否可以共享:7)联系方式:pm1)质粒图谱登记号:00132)质粒名称:pcDNA3.1/Zeo (+)3)来源:invitrogen4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)质粒图谱登记号:00132)质粒名称:pEGFP-N33)来源:clontech4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)质粒图谱登记号:00142)质粒名称:pcDNA33)来源:invitrogen4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)质粒图谱登记号:00152)质粒名称:pfastbac13)来源:invitrogene4)用途:昆虫表达5)是否可以提供更详细资料:不可以6)是否可以共享:不可以7)联系方式:PM)质粒图谱登记号:00162)质粒名称:pEGFP-C33)来源:clontech4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PMwangjun2002274 edited on 2004-06-22 00:041)质粒图谱登记号:00172)质粒名称:pSecTag23)来源:Invitrogen4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)1质粒图谱登记号:00192)质粒名称:pET20b3)来源:NOVAGEN4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM1)质粒图谱登记号:00222)质粒名称:pThioHisA3)来源:invitrogen4)用途:原核表达5)是否可以提供更详细资料:4.365kb , HP-thioredoxin fusion proteinexpressionvector, trc promoter, Ampr, a EK cleavage site lies between HP-thioredoxin and MCS宿主菌TOP10(基因型为:F-mcrA △(mrr-hsd RMS-mcrBC)Ф80 lacZ M15 △lacX74 deoR recAl araD139 △(ara-leu)7697 galU galK rpsL endAl nupG6)是否可以共享:交换或其它7)联系方式:PM2)3)1)质粒图谱登记号:00242)质粒名称:pcDNA3.1-Myc-His-A-3)来源:invitrogen4)用途:真核核表达4))质粒图谱登记号:00252)质粒名称:pS UPER.neo3)来源:4)用途:siRNA5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)质粒图谱登记号:00312)质粒名称:pSilencer1.0-siRNA3)来源:Ambion4)用途:RNAi5)是否可以提供更详细资料:/techlib/Documents.html?fkResSxn=7&fkSub Sxn=236)是否可以共享:实验结束后,可提供含shRNA模板的质粒7)联系方式:pm5))质粒图谱登记号:00322)质粒名称:pSilencer2.0-U6siRNA3)来源:Ambion4)用途:RNAi,与1.0相比,可以建立稳转株5)更详细资料:/techlib/prot/fm_7209.pdf6)是否可以共享:交换7)联系方式:pm6)会员名:mlluoE-mail:*************可提供试验资源名称和简要介绍:pSilencer 3.1-H1 neo Vector,是Ambion公司目前最高版本的shRNA 载体,为扩增此载体我已插入目的片段,如有战友需要,将此片段双酶切再连上自己的片段即可。
pmd18-19-T 质粒图谱

GAGCGGATAACAATTTCACACAGGAAACAGCTATGACCATGATTACGCCAAGCTTGCATGCCTGCAGGTCGACGATATCTCTAGAGGATCCCCGGGTACCGAGCTCGAATTC
ACTGGCCGTCGTTTTACAACGTCGTGACTGGGAAAACCCTGGCG CAGCACTGACCCTTTTGGGACCGC BcaBESTTM Sequencing Primer M13-47
LacZ operator
146 ~ 469
ColE1 ori
852 ~ 1466
Ampr
1626 ~ 2486
■ 实验操作 Control DNA片段的克隆实验 A.操作方法
1)在微量离心管中配制下列DNA溶液,全量为5 µl。
pMD18-T or pMD19-T Vector*1
1 µl
Control Insert*2
■ 相关说明 1.感受态细胞的选择。
PCR产物克隆系列载体
PCR产物克隆系列载体
转化时请使用高效的热转化感受态细胞(转化效率≥1
×108 cfu/µg pUC19),这样才可能得到比较理想的阳
性克隆。 如果需要进行蓝白筛选时,宿主细胞必须具
有正确的基因型(F'编码的[ △M15])产生 ω
Fragment,才可能和载体DNA产生的
ng DNA/ml),将上述培养液稀释10倍后(0.01 ng
DNA/ml)取100 µl涂布平板(0.001 ng DNA/100
µl),记录菌落数。以得到200个克隆体为例计算转化效 率,此时的转化效率=200 cfu/0.001 ng=2×105
cfu/ng=2×108 cfu/µg pUC19 DNA。
质粒图谱大全

质粒图谱大全质粒图谱大全质粒图谱大全(转载)一.九种表达载体Pllp-OmpA, pllp-STII, pMBP-P, pMBP-C, pET-GST, pET-Trx, pET-His, pET-CKS, pET-DsbA二.克隆载体pTZ19RDNApUC57DNAPMD18TPQE30pUC18pUC19pTrcHisApTrxFuspRSET-ApRSET-BpVAX1PBR322pbv220pBluescriptIIKS( )L4440pCAMBIA-1301pMAL-p2XpGD926三.PET系列表达载体ProteinExpression?ProkaryoticExpression?p ETDsbFusionSystems39band40bProteinExpression?ProkaryoticExpression?p ETExpressionSystem33bProteinExpression?ProkaryoticExpression?p ETExpressionSystemsProteinExpression?ProkaryoticExpression?p ETExpressionSystemsplusCompetentCells ProteinExpression?ProkaryoticExpression?p ETGSTFusionSystems41and42ProteinExpression?ProkaryoticExpression?p ETNusAFusionSystems43.1and44 ProteinExpression?ProkaryoticExpression?p ETVectorDNA ProteinPurification?PurificationSystems?Strep?TactinResinsandPurificationKits四.PGEX系列表达载体TEcoR?pGEX-1I/BAP pGEX-2TpGEX-2TKpGEX-3XpGEX-4T-1pGEX-4T-2pGEX-4T-3pGEX-5X-1pGEX-5X-2pGEX-5X-3pGEX-6P-1pGEX-6P-2pGEX-6P-3五.PTYBsystem PTYB1PTYB2PTYB11PTYB12六.真核表达载体pCDNA3.1(-) pCDNA3.1( )pPICZalphaApGAPZαAPYES2.0pBI121pEGFP-N1pEGFP-C1pPIC9KpPIC3.5K如何阅读分析质粒图谱载体主要有病毒和非病毒两大类,其中质粒DNA 是一种新的非病毒转基因载体。
质粒图谱大全之欧阳歌谷创编

(转载)欧阳歌谷(2021.02.01)一.九种表达载体Pllp-OmpA,pllp-STII,pMBP-P,pMBP-C,pET-GST,pET-Trx,pET-His,pET-CKS,pET-DsbA 二.克隆载体pTZ19RDNApUC57DNAPMD18TPQE30pUC18pUC19pTrcHisApTrxFuspRSET-ApRSET-BpVAX1PBR322pbv220pBluescriptIIKS( )L4440pCAMBIA-1301pMAL-p2XpGD926三.PET系列表达载体ProteinExpression?ProkaryoticExpression?pETDsbFusionSystems39b and40bProteinExpression?ProkaryoticExpression?pETExpressionSystem33b ProteinExpression?ProkaryoticExpression?pETExpressionSystems ProteinExpression?ProkaryoticExpression?pETExpressionSystemsplus CompetentCellsProteinExpression?ProkaryoticExpression?pETGSTFusionSystems41a nd42ProteinExpression?ProkaryoticExpression?pETNusAFusionSystems43 .1and44ProteinExpression?ProkaryoticExpression?pETVectorDNA ProteinPurification?PurificationSystems?Strep?TactinResinsandPurific ationKits四.PGEX系列表达载体TEcoR?pGEX-1I/BAP pGEX-2TpGEX-2TKpGEX-4T-1 pGEX-4T-2 pGEX-4T-3 pGEX-5X-1 pGEX-5X-2 pGEX-5X-3 pGEX-6P-1 pGEX-6P-2 pGEX-6P-3五.PTYBsystem PTYB1PTYB2PTYB11PTYB12六.真核表达载体pCDNA3.1(-) pCDNA3.1( ) pPICZalphaA pGAPZαA PYES2.0pBI121pEGFP-C1pPIC9KpPIC3.5K如何阅读分析质粒图谱载体主要有病毒和非病毒两大类,其中质粒DNA是一种新的非病毒转基因载体。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
)质粒图谱登记号:00012)质粒名称:pIRES3)来源:BD Co4)用途:真核双表达5)是否可以提供更详细资料:可以6)是否可以共享:7)联系方式:PM)质粒图谱登记号:00022)质粒名称:pECFP-C13)来源:BD Co4)用途:检测真核表达5)是否可以提供更详细资料:可以6)是否可以共享:7)联系方式:pm1)质粒图谱登记号:00032)质粒名称:pShuttle3)来源:4)用途:5)是否可以提供更详细资料:6)是否可以共享:7)联系方式:2)pShuttle MCS很多人用pEGFP-C1,我也来发一个)质粒图谱登记号:00042)质粒名称:pSBR322、pUC183)来源:4)用途:5)是否可以提供更详细资料:6)是否可以共享:7)联系方式:1)质粒图谱登记号:00052)质粒名称:pcDNA3.1(+)/CAT3)来源:invitrogen4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:无偿7)联系方式:PM1)质粒图谱登记号:00062)质粒名称:pQEx3)来源:Qiagen4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM2)1)质粒图谱登记号:00072)质粒名称:pIVEX2.33)来源:Rocho4)用途:体外转录翻译5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM质粒图谱登记号:0008质粒名称:pIRES-EGFP来源:用途:是否可以提供更详细资料:是否可以共享:否联系方式:1)质粒图谱登记号:00092)质粒名称:pET-28a(+)3)来源:Novagen4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM1)质粒图谱登记号:00112)质粒名称:pET-32a(+)3)来源:novagen4)用途:原核表达5)是否可以提供更详细资料:6)是否可以共享:7)联系方式:pm1)质粒图谱登记号:00132)质粒名称:pcDNA3.1/Zeo (+)3)来源:invitrogen4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)质粒图谱登记号:00132)质粒名称:pEGFP-N33)来源:clontech4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)质粒图谱登记号:00142)质粒名称:pcDNA33)来源:invitrogen4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)质粒图谱登记号:00152)质粒名称:pfastbac13)来源:invitrogene4)用途:昆虫表达5)是否可以提供更详细资料:不可以6)是否可以共享:不可以7)联系方式:PM)质粒图谱登记号:00162)质粒名称:pEGFP-C33)来源:clontech4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PMwangjun2002274 edited on 2004-06-22 00:041)质粒图谱登记号:00172)质粒名称:pSecTag23)来源:Invitrogen4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)1质粒图谱登记号:00192)质粒名称:pET20b3)来源:NOVAGEN4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM1)质粒图谱登记号:00222)质粒名称:pThioHisA3)来源:invitrogen4)用途:原核表达5)是否可以提供更详细资料:4.365kb , HP-thioredoxin fusion proteinexpressionvector, trc promoter, Ampr, a EK cleavage site lies between HP-thioredoxin and MCS宿主菌TOP10(基因型为:F-mcrA △(mrr-hsd RMS-mcrBC)Ф80 lacZ M15 △lacX74 deoR recAl araD139 △(ara-leu)7697 galU galK rpsL endAl nupG6)是否可以共享:交换或其它7)联系方式:PM2)3)1)质粒图谱登记号:00242)质粒名称:pcDNA3.1-Myc-His-A-3)来源:invitrogen4)用途:真核核表达4))质粒图谱登记号:00252)质粒名称:pSUPER.neo3)来源:4)用途:siRNA5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM)质粒图谱登记号:00312)质粒名称:pSilencer1.0-siRNA3)来源:Ambion4)用途:RNAi5)是否可以提供更详细资料:/techlib/Documents.html?fkResSxn=7&fkSub Sxn=236)是否可以共享:实验结束后,可提供含shRNA模板的质粒7)联系方式:pm5) )质粒图谱登记号:00322)质粒名称:pSilencer2.0-U6siRNA 3)来源:Ambion4)用途:RNAi ,与1.0相比,可以建立稳转株5)更详细资料:/techlib/prot/fm_7209.pdf 6)是否可以共享:交换 7)联系方式:pm6)会员名:mlluoE-mail:*************可提供试验资源名称和简要介绍:pSilencer 3.1-H1 neo Vector,是Ambion公司目前最高版本的shRNA 载体,为扩增此载体我已插入目的片段,如有战友需要,将此片段双酶切再连上自己的片段即可。
QQ:无1)质粒图谱登记号:00252)质粒名称:pSilencer 3.1-H1 neo Vector3)来源:Ambion4)用途:RNAi5)是否可以提供更详细资料:可以6)是否可以共享:可交换7)联系方式:E-mail7)1)质粒图谱登记号:0035(后面的会员按此顺序号排)2)质粒名称:pOK123)来源:构建4)用途:克隆大片段,高保真,低突变g/ml)μ片段N端,可以利用蓝白斑筛选重组质粒。
外源片段的插入会减弱质粒的稳定性,特别是当插入片段编码毒性蛋白时,质粒的稳定性更差,所以通过基因修饰减弱了lacZ启动子的活性。
pOK12的复制子来源于P15A,使它成为一个低拷贝的pUC质粒。
pOK12为卡那霉素抗性(10-15α5)是否可以提供更详细资料:低拷贝质粒pOK12(GenBank 登录号:AF223639)具有28个多克隆位点,应用广泛。
多克隆位点位于lacZ6)是否可以共享:7)联系方式:8)1)质粒图谱登记号:00362)质粒名称:pET22b(+)3)来源:Novagene4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:无偿、交换7)联系方式:PM9)1)质粒图谱登记号:00372)质粒名称:pQE93)来源:Qiagen4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:7)联系方式:PM10)1)质粒图谱登记号:00382)质粒名称:pCANTAB5E3)来源:Phamcia4)用途:构建噬菌体单链抗体库5)是否可以提供更详细资料:6)是否可以共享7)联系方式:11))质粒图谱登记号:00392)质粒名称:pET-24a3)来源:Novagen4)用途:原核表达5)是否可以提供更详细资料:/html/NVG/AllTables.html6)是否可以共享:7)联系方式12)1)质粒图谱登记号:00422)质粒名称:pET-43.1 Ek/LIC3)来源:Novagen4)用途:原核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换或其它7)联系方式:PM13)1)质粒图谱登记号:00442)质粒名称:pBAD/Thio-TOPO3)来源:invitrogen4)用途:原核表达5)是否可以提供更详细资料:6)是否可以共享:交换或其它7)联系方式:PM14))质粒图谱登记号:00452)质粒名称:pcDNA3.1/V5-His-TOPO3)来源:invitrogen4)用途:真核表达5)是否可以提供更详细资料:可以6)是否可以共享:交换或其它7)联系方式:PM15)1)质粒图谱登记号:00462)质粒名称:pMSCVneo3)来源:Clontech4)用途:逆转录病毒载体5)是否可以提供更详细资料:可以6)是否可以共享:交换或其它7)联系方式:PM16)1)质粒图谱登记号:00472)质粒名称:pcDNA3.1(+)3)来源:invitrogen4)用途:真核表达5)是否可以提供更详细资料:可以/content/sfs/manuals/pcdna3.1_man.pdf6)是否可以共享:交换或其它7)联系方式:PM17))质粒图谱登记号:00482)质粒名称:pCDNA6 /V5 HisB3)来源:Invitrogen4)用途:真核表达载体5)是否可以提供更详细资料:可以6)是否可以共享7)联系方式:PM18))质粒图谱登记号:00522)质粒名称:pBudCE4.13)来源:Invitrogen4)用途:真核表达载体5)是否可以提供更详细资料6)是否可以共享7)联系方式:PM/content/sfs/manuals/pbudce4_1_man.pdf19)1)质粒图谱登记号:00552)质粒名称:pPICZ A, B, and C3)来源:invitrogen4)用途:毕赤酵母表达载体5)是否可以提供更详细资料:可以6)是否可以共享:7)联系方式:PM20)1)质粒图谱登记号:00602)质粒名称:pUC183)来源:4)用途:克隆5)是否可以提供更详细资料:大家都很清楚了6)是否可以共享:7)联系方式:PM21))质粒图谱登记号:00612)质粒名称:pshuttleCMV3)来源:KRACKELER4)用途:穿梭质粒5)是否可以提供更详细资料:可以6)是否可以共享:交换7)联系方式:PM22)23)24)。
