cikm09-lujh
CIK细胞使用说明书

CIK细胞使用说明书
1.物品准备:输血器、棉棒、碘伏;
2.CIK细胞在室温下(18-22℃)运输;
3.打开运输箱,取出小瓶 (细胞悬液体积约50ml/瓶),共2瓶;
4.检查小瓶外观,注意观察有无漏液、破损。
用腕力震荡小瓶10次,使细胞充分混匀(注:因细胞悬液含有人血白蛋白,若震荡后细胞悬液泡沫增多为正常现象);
5.CIK细胞回输前20分钟遵医嘱给予异丙嗪(非那根)25mg肌肉注射或地塞米松5mg静脉注射(次选);
6.CIK细胞回输前应用输血器建立良好静脉通道,0.9%氯化钠溶液充分冲管;
7.CIK细胞回输时前15分钟滴注速度为30滴/min,15-30分钟滴注速度可增加为45滴/min,如患者无不适可逐渐提高滴速,最高不超过70滴/min,在1小时内输完;
8.滴注过程中每5~10 min轻轻晃动输液瓶,避免细胞沉降堆积,为了防止CIK细胞堵塞输血器的管腔或黏附管壁,应轻弹输血器的管壁并保持输液通畅,直至滴完,输注后再用生理盐水冲洗容器及管路,避免浪费制剂内的免疫细胞。
回输结束拔针后按压穿刺点10 min以上;
9.CIK细胞回输时加强巡视,注意倾听患者主诉,密切观察有无寒战、发热、心慌、胸闷及皮肤黏膜有无皮疹瘙痒等症状;
10.CIK细胞回输后2-6小时,患者可能出现发热,指导患者多饮水,松开被服等,一般持续3~6 h,自然减退,若体温高于39℃,可口服布洛芬、消炎痛或配合物理降温,若回输过程中病人出现休克、喉头痉挛可立即静脉注射激素;
11.CIK细胞治疗期间禁用免疫抑制剂;
12.CIK细胞治疗期间避免使用易引起过敏的药物,同时尽量少食易引起过敏的食物。
cik细胞标准

cik细胞标准CIK细胞,全称为细胞因子诱导的杀伤细胞,是一种新型的免疫疗法,近年来在肿瘤治疗领域备受关注。
CIK细胞的制备和使用需要遵循一定的标准,以确保其质量和安全性。
以下是对CIK细胞标准的详细说明:一、细胞来源CIK细胞的标准来源是健康成年志愿者的外周血单个核细胞(PBMC)。
这些细胞可以通过分离自体或异体输血的方式获得。
在采集前,需要对志愿者的健康状况进行评估,确保其不患有任何严重的疾病,并签署知情同意书。
二、细胞培养CIK细胞的制备需要进行细胞培养。
在培养过程中,需要使用适当的细胞因子和抗体,以诱导细胞因子产生和促进细胞的增殖。
常用的细胞因子包括IL-2、IFN-γ、IL-1等。
在培养过程中,需要对细胞进行监测和记录,确保细胞的生长和分化符合预期。
三、细胞质量CIK细胞的质量是保证其治疗效果的关键因素之一。
因此,在制备和使用CIK细胞时,需要对细胞的质量进行严格的控制。
常用的质量控制指标包括细胞的形态、增殖能力、表面标志物等。
此外,还需要对细胞的活性进行检测,确保其具有较高的细胞毒性。
四、安全性CIK细胞作为一种免疫疗法,其安全性是至关重要的。
因此,在制备和使用CIK细胞时,需要对其安全性进行评估和控制。
常用的安全性指标包括细胞的毒性和致瘤性等。
此外,还需要对细胞的病毒污染情况进行检测,确保其不含有任何潜在的致病因子。
五、临床试验在应用CIK细胞治疗肿瘤之前,需要进行临床试验以评估其安全性和有效性。
在临床试验中,需要对患者的健康状况、肿瘤类型和分期等进行评估,并制定合适的治疗方案。
同时,需要对患者的生命体征和不良反应进行监测和记录,及时处理任何可能的并发症。
六、标准化制备流程为了确保CIK细胞的质量和安全性,需要建立标准化的制备流程。
这包括对细胞的采集、分离、培养、扩增和储存等环节进行严格的控制和管理。
同时,需要对制备过程中的环境和设备进行消毒和清洁,以避免任何潜在的污染风险。
七、质量控制体系为了确保CIK细胞的质量和安全性,需要建立完善的质量控制体系。
智能门锁说明书

Create a Wireless Access System simply, with Trilogy Networx™ Locks via 802.11 or ethernet networkDL6100 & prox/PIN version PDL6100Trilogy Networx Locks eliminate door-to-door programming and audit trail retrieval by com-municating wirelessly via Ethernet or 802.11B/G, while providing all original Trilogy standalone lock’s functionality. They provide a quick, easy, economical access system with advanced features & functions, like automatic schedules, event logs and support for 2000 doors and 5000 existing Prox. ID Cards/Badges, but with no wires to run, and no access panels or power supplies to buy or install. At its simplest it’s one Grade 1 durable wireless lock per door, installed in about an hour, all communicating to a single Gateway (each gateway controls up to 63 locks), networked to any non-dedicated Windows-based PC. In addition to Wireless Trilogy Networx’ unsurpassed battery-life, exceeding 5 years, they feature new network-wide capabilities, executed from any lock or the network’s PC, including emergency priority global lockdown deployed in seconds, and you can activate (non-priority) system-wide free-passage or locking modes from the PC. For existing Trilogy installations, original standalone and new Trilogy Networx locks can seamlessly share one common database hosted on free Alarm Lock DL-WINDOWS software.•Trilogy Networx Locks communicate wirelessly via Ethernet, Bluetooth LE or 802.11B/G for a comprehensive wireless networked door access control system, eliminating door-to-door programming and audit trail retrieval and providing all standalone Trilogy lock functionality plus new network-wide and emergency lockdown features•Unsurpassed 5-year+ battery life, typical, (Automatic battery status reports at the PC; visual and audible low-battery status indicators at lock(s).•Trilogy Networx and gateway modules have an integral transceiver for highly-efficient 900MHz bidirectional communications, for longer range and less interference than with more common 2.4GHz spread spectrum devices. Alarm Lock devices require NO wiring; NO splitters or repeaters; NO hardwiring to the host/server; NO extra power supplies; and NO access controller/panels.•In 10 seconds, activate emergency global lockdown from the Networx PC, any Networx lock or Keyfob RR-4BUTTON. (This priority emergency command is lightning-fast system-wide and supercedes all scheduled programs or updates, etc. However, if desired, locks may be selectively programmed to be excluded from emergency global lockdown function, i.e., they will ignore the command. )•System-wide free-passage enable or disable, can likewise be activated from the Networx PC • Locks support up to 5000 users with 3-6 digit numeric PIN codes and PDL-Series also support prox ID card users (virtually all HID™ format proximity technology ID cards, fobs, credentials.)• On demand, in seconds, wirelessly change users, schedules, lock features or retrieve audit trails of up to 40,000 events/lock from any networked PC. (Note: Transaction processing time is dependent upon system capacity, the more locks/gateways and the more data to be up/ downloaded, the longer the time.)• Unattended programming updates or event log retrievals can be scheduled to run automatically, saving manpower and productive time.• Seamless upgrade path for original Trilogy users: Intermix with existing Trilogy standalone lock installations and share one database and familiar Trilogy lock and software functions: Trilogy Wireless Networx Locks seamlessly integrate in an existing Alarm Lock Trilogy standalone door access system and share the same single DL-WINDOWS database (requires v4.1.x or higher). Existing Trilogy users can easily add one wireless Networx Lock, or as many as two thousand, to an existing system seamlessly with no downtime.• Secure 128bit AES PC encryption and proprietary RF-link lock communication encryption protects the system from interference or tampering (WEP or WPA compatible with wireless modem).• Remote System Management: VPN-connection /remote capability for accessing the Networx System and controlling up to all 2000 locks, with all the security and peace-of-mind of your IT network. (No special modems required.)• Wireless locks are fully keypad programmable and can be up and functioning even before their wireless network is configured, so IT departments don’t have to coordinate with physical security departments or locksmiths• Free Alarm Lock DL-WINDOWS software doesn’t require a dedicated computer or a special computer network or costly server software licenses, it simply runs as a secure Microsoft Windows-based application, on any computer system (as basic as a desktop or laptop PC), a wired or wireless router, and a gateway module or on an internal LAN with static IP add• Minimizing hardware & installation costs, a single gateway module can interface between the PC network and up to 63 Networx locks , with a range of up to 900’ clear field range or up to 200’ depending on building construction. At its maximum, a Trilogy Networx system can comprise up to 32 gateways for the networking of up to 2000 locks or access controlled doors. New Gen 2 Gateways & Expanders for easiest networking• Optional Enterprise Software Integration with Continental, Lenel & Software Housewireless accesscontrol solutions…about 1/2 the cost,on any doorEASY INSTALL Replaces any door lock, on any door type, neatly, quicklyMORE AFFORDABLELowest labor & equipmentcosts without sacrificingtop electronic accessfeaturesEASY NETWORKNo wires to run to doors.Uses customers’ existingnetwork or Ethernet &multi-lock gateways& optional repeatersCENTRALLY MANAGEDAuto-Schedule programupdates, queries, or freeaccess by time,by door & moreUSERSSupports thousandsof PIN, ID or iLock Appusers. Easilyadded/removedlocal or remotelyGLOBAL LOCKDOWNor unlock in secondsfrom the server orany lock345 Bayview Ave, Amityville, New York 117011-800-ALA-LOCK • Fax 631-789-3383Trilogy and Networx is a trademark of Alarm Lock.© NAPCO 11/2017 ALA489EAL-IME2 Gen. 2 GatewayThe Trilogy Networx AL Generation Gateway, interfaces Networx locks with a PC network via a hardwire Ethernet connection. It is very similar in function to the original hardwired NetworxGateway Modules, but is enhanced with additional extended potential range, supporting up to seven(7) Expanders (model AL-IME2-EXP Expanders). Each Expander can extend the AL-IME2 Gateway’s coverage area by approximately 100 feet. Also both Gen. 2 Gateways and matching Expanders have a new modern styling, andthree LED indicators, blending into any décor.AL-IM2-80211 Gen. 2 802.11 GatewaySimilar to model above, but this 802.11 Gateway Module includes a bi-directional radio, andinterfaces Networx locks with a PC network with hardwire Ethernet and wireless 802.11. It is very similar in function to the original 802.11 Networx Gateway Modules, but is enhanced with additional extended potential range, supporting up to seven (7) Expanders (model AL-IME2-EXP Expanders). Each Expander can extend the AL-IM2-80211 Gateway’s coverage area by approximately 100 feet.AL-IME2-POE Gen. 2 GatewayThe Trilogy Networx AL-IME2-POE (Power Over Ethernet)Hardwired Gateway interfaces Networx locks with a PC network with hardwire Power-Over-Ethernet. It is very similar in function to the original hardwired Networx Gateway Modules, but is enhanced with alternative power, and additional extended potential range, supporting up to seven (7) Expanders (model AL-IME2-EXP Expanders).AL-IME2-EXP ExpanderTrilogy ® Networx ™ Expander extends the coverage area of a Gen 2 Series Gateways to Trilogy Networx Locks within a PC network, allowing greater distances for the placement of up to a maximum rated capacity of 63 Networx locks per Version 2 Gateway.AL-IME2-PIE Plug-in ExpanderThe Trilogy ® Networx ™ Plug-In Expander extends the coverage area of a Generation 2 Series AL-IME2 Gateway to Trilogy Networx Locks within a PC network, allowing greater 802.11 distances for the placement of up to a maximum rated capacity of 63 networx locks per Version 2 Gateway.ORIGINAL GATEWAYS AVAILABLE ‡‡AL-IM80211 Hardwire/Wireless Gateway, interfaces up to 63 Networx locks with PC network with hardwired Ethernet and wireless 802.11. AC Xformer supplied; ceiling/wall mount.AL-IME Hardwire Gateway similar to above, but using hardwired connection to the network via RJ45 Ethernet cable. AL-IME-USB Portable USB Gateway.AL-IMEPOE Hardwire Gateway Interface Module + POE, as above, but using hardwired Ethernet and Power Over Ethernet (POE). Connects directly to a network via Ethernet cable. ‡‡Important: For enterprise integration consult technical documentation to match proper AL hardware (Gateways, Expanders,etc.) with enterprisesoftware Versions supported.Trilogy, Networx, ArchiTech & iLock and ** Continental CardAccess or CA4K™ CardAccess are trademarks of Alarm Lock, a division of Napco.**Other marks remain the intellectual properties of their respective companies.• Wall- or ceiling-mountable, compact gateway modules are available in wireless/hardwired and wired-only versions (with Power Over Ethernet (POE) option). The hybrid model connects using either 802.11, Ethernet cable or 100Base-TX (using RJ45 Jack); the wired versions use an RJ-45 Ethernet cable. Networx communications support both wired Ethernet 10BaseT and wireless 802.11.• For the fastest most reliable networked system, in discovery setup mode, the system will locate and DL-WINDOWS will display each lock’s signal strength at each installed gateway, so you can select it and optimize reliable communications and eliminate guesswork.• Lock programming, user- and usage-data are stored locally at the battery-powered lock for uninterrupted door access usage in a power outage or server offline situation.Enterprise Networks, Real-Time Performance• Networx locks can be used as wireless, single-door controllers in real time too• Use with Contintental Access ® CA3000 , CA4K ®, or Lenel OnGuard ® Integrated Access Control & VideoSecurity and Software House Connected Partner ® Program Platforms ‡‡Trilogy Networx Lock Specifications:• Weatherproof locks for indoor/outdoor use with wide operating temperature range (-31 to 151˚ degrees F).• Cylindrical models boast industrial strength BHMA Grade 1 durability and all models feature vandal-resistant all-metal keypad for PIN codes.• PDL versions, HID Prox. Reader. • ADA-compliant leversets.• Easy-to-install modified ANSI A115 Series door prep; easily and neatly retrofit standard cylindrical knobsets and mechanical standalone locks, in under an hour. Non-handed • 3 hour UL fire-rated deadlatch.• Supports 1-5/8 to 1-7/8” door thickness; 2-3/4” backset (2-3/8” available.)• Manual key override standard; most IC cores alss supported on interchangeable core models.• Wide choice of 3 Architectural finishes (US3 polished brass, 26D satin chrome,10B duronodic) and IC core preps (6-7pin). Ordering Information:PDL6100 Cylindrical Trilogy Networx ™ PIN/Prox. Wireless Access Control Lock with built in HID proximity ID card reader, full-metal digital keypad, integral bi-directional radio, 4-C cell battery-operated (batts. supplied), serial number ID card, standard format SCI keyway for manual key override, 4-7/8” ASA Strike (incldd).DL6100 Cylindrical Trilogy ® Networx PIN-Code Wireless Access Control Lock, as above, with full-metal digital keypad only.PDL6500 Mortise version Networx PIN/Prox. lock, similar to 6100 Series above.DL6500 Mortise version Networx PIN-code only lock, similar to 6100 Series above.DL-WINDOWS Free Alarm Lock Trilogy Microsoft Windows-based software application, v4.0.1 or higher, supports Trilogy Networx and Trilogy standalone locks, with single database. No-charge, downloadable online at Network InterfaceINTERFACE: Ethernet 10Base-T or 100Base-TX (using RJ45 Jack)PROTOCOLS: TCP/IP , UDP/IP , DHCP 802.11 Wireless Specifications WIRELESS STANDARDS : IEEE 802.11b; 802.11g FREQUENCY RANGE : 2.412 - 2.484 GHzOUTPUT POWER: 14 dBm + 1.5 dBm/-1.0 dBmMAXIMUM RECEIVE LEVEL: -10dBm (with PER < 8%)DATA RATES WITH AUTOMATIC FALLBACK: 54Mbps - 1Mbps RANGE: Up to 328 feet indoorsMODULATION TECHNIQUES: OFDM, DSSS, CCK, DQPSK, DBPSK, 64 QAM, 16 QAMINTERFACE: Wireless 802.11b, 802.11g and 10/100 Ethernet PROTOCOLS: TCP/IP , UDP/IP , DHCP SECURITY• IEEE 802.11 - PSK with AES Encryption 128-bit Encryption • 128-bit AES Rijndael encryption AL Radio Link• 900 MHz GFSK, 50 channels, 10mW power output VOLTAGE: 5 - 6 volts AC/DCOPERATING TEMPERATURE: -20˚ to 60˚C (-4˚ to 140˚F)STORAGE: -40˚ to 85˚C (-40˚ to 185˚F)AVERAGE POWER CONSUMPTION:• 1300 mW (WLAN mode; maximum data rate)• 300 mW (WLAN mode; idle)• 750 mW (Ethernet Mode)PEAK SUPPLY CURRENT: 650 mA INPUT VOLTAGE: 5 - 6 volts AC/DCOPERATING TEMPERATURE: -31 to 151˚ degrees F STORAGE TEMPERATURE: -40˚ to 85˚C (-40˚ to 185˚F)‡‡Important: For enterprise integration consult technical documentation to match proper AL hardware (Gateways, Expanders,etc.) with enterprise software Versions supported.Up to 2000 locks and a PC communicate wirelessly through bidirectional Gateways – eliminating the need for door-to-doorprogramming and audit trail retrieval. System Management with free DL-WINDOWS or Enterprise Solutions:In seconds, globallockdown or unlock can be implemented from any Trilogy lock, even if the DL-WINDOWS computer is inaccessible or totallyoffline!。
Extract-N-Amp Tissue PCR Kit 产品说明书

Product InformationExtract-N-Amp™ Tissue PCR KitXNAT2, XNAT2RProduct DescriptionThe Extract-N-Amp™ Tissue PCR Kit for direct PCR contains the reagents needed to rapidly extract and amplify genomic DNA from mouse tails and other animal tissues, buccal swabs, hair shafts, and saliva. Briefly, the DNA is released from the starting material by incubating the sample with a mixture of the Extraction Solution and the Tissue Preparation Solution at room temperature for 10 minutes. There is no need for mechanical disruption, organic extraction, column purification, or precipitation of the DNA.After adding Neutralization Solution B, the extract is ready for PCR. An aliquot of the neutralized extract is then combined with the Extract-N-Amp™ PCR Reaction Mix and user-provided PCR primers to amplify target DNA. The Extract-N-Amp™ PCR Reaction Mix is a 2X ready mix containing buffer, salts, dNTPs, and Taq polymerase. It is optimized specifically for use with the extraction reagents. It also contains the JumpStart Taq antibody for hot start PCR to enhance specificity but does not contain the inert red dye found in the REDExtract-N-Amp™ PCR Reaction Mix.Reagents Provided Cat. No. XNAT2 100 Preps,100 PCRsXNAT2R 1000 Preps, 1000 PCRsExtraction SolutionE7526 24 mL 240 mL Tissue Preparation Solution T3073 3 mL 30 mL Neutralization Solution BN391024 mL240 mLExtract-N-Amp™ PCR Reaction Mix This is a 2X PCR reaction mix containing buffer, salts, dNTPs, Taq polymerase, and JumpStart™ Taq antibody.E30041.2 mL12 mLReagents and Equipment Required(Not Provided)•Microcentrifuge tubes (1.5 or 2 mL) or multi-well plate for extractions (200 μL minimal well volume) • Small dissecting scissors• Forceps (small to medium in size)• Buccal swab - Sterile foam tipped applicator (Cat. No. WHAWB100032)•Sample collection card - Bloodstain card (Cat. No. WHAWB100014)• Tubes or plate for PCR• Heat block or thermal cycler at 95 °C • PCR Primers (Cat. No. OLIGO) • Thermal cycler•Water, PCR Reagent (Cat. No. W1754)Precautions and DisclaimerThis product is for R&D use only. Not for drug, household, or other uses. Please consult the Safety Data Sheet for information regarding hazards and safe handling practices.StorageThe Extract-N-Amp™ Tissue PCR Kit can be stored at 2 to 8 °C for up to 3 weeks. For long-term storage, greater than 3 weeks, -20 °C is recommended. Do not store in a "frost-free" freezer.ProcedureAll steps are carried out at room temperature unless otherwise noted.DNA Extraction from Mouse Tails, Animal Tissues, Hair, or Saliva1.Pipette 100 μL of Extraction Solution into amicrocentrifuge tube or well of a multi-well plate.Add 25 μL of Tissue Preparation Solution to thetube or well and pipette up and down to mix.Note: If several extractions will be performed,sufficient volumes of Extraction and TissuePreparation Solutions may be pre-mixed in a ratio of 4:1 up to 2 hours before use.2.For fresh or frozen mouse tails: Rinse thescissors and forceps in 70% ethanol prior to useand between different samples. Place a 0.5–1 cm piece of mouse tail tip (cut end down) into thesolution. Mix thoroughly by vortexing or pipetting.Ensure the mouse tail is in solution.Note: For fresh mouse tails, perform extractions within 30 minutes of snipping the tail.For animal tissues: Rinse the scissors or scalpel and forceps in 70% ethanol prior to use andbetween different samples. Place a 2–10 mgpiece of tissue into the solution. Mix thoroughlyby vortexing or pipetting. Ensure the tissue is inthe solution.For hair shafts: Rinse the scissors and forceps in 70% ethanol prior to use and between differentsamples. Trim excess off of the hair shaft leaving the root and place sample (root end down) intosolution. Only one hair shaft, with root, isrequired per extraction.For Saliva: Pipette 10 μL of saliva into thesolution. Mix thoroughly by vortexing or pipetting.For saliva dried on card: Pipette 50 μL of saliva onto collection card and allow the card to dry.Rinse the punch in 70% ethanol prior to use andbetween different samples. Punch a disk(preferably 1/8 inch or 3 mm) out of the cardfrom the area with the dried saliva sample. Place disk into the solution. Tap tube or plate on hardsurface to ensure disk is in solution forincubation period.3.Incubate sample at room temperature for10 minutes.4.Incubate sample at 95 °C for 3 minutes.Note: Tissues will not be completely digested atthe end of the incubations. This is normal and will not affect performance.5.Add 100 μL of Neutralization Solution B to sampleand mix by vortexing.6.Store the neutralized tissue extract at 4 °C oruse immediately in PCR amplification.Note: For long term storage, remove theundigested tissue or transfer the extracts tonew tubes or wells. Extracts may now be storedat 4 °C for at least 6 months without notable loss in most cases.DNA Extraction for Buccal Swabs1.Collect buccal cells on swab and allow theswab to dry. Drying time is approximately10 to 15 minutes.Note: Due to the low volume of solution used for DNA extraction, a foam tipped swab should beused. Swabs with fibrous tips, such as cotton orDacron®, should be avoided because the solution cannot be recovered efficiently.2.Pipette 200 μL of Extraction Solution into amicrocentrifuge tube. Add 25 μL of TissuePreparation Solution to the tube and pipette upand down to mix.Note: If several extractions will be performed,sufficient volumes of Extraction and TissuePreparation Solutions may be pre-mixed ina ratio of 8:1 up to 2 hours before use.3.Place dried buccal swab into solution and incubateat room temperature for 1 minute.4.Twirl swab in solution 10 times and then removeexcess solution from the swab into the tube bytwirling swab firmly against the side of the tube.Discard the swab. Close the tube andvortex briefly.5.Incubate sample at room temperature for10 minutes.6.Incubate sample at 95 °C for 3 minutes.7.Add 200 μL of Neutralization Solution B to sampleand mix by vortexing.8.Store the neutralized extract at 4 °C or useimmediately in PCR. Continue to PCRamplification.Note: Extracts may be stored at 4 °C for at least6 months without notable loss in most cases. PCR AmplificationThe Extract-N-Amp™ PCR Reaction Mix contains JumpStart™ Taq antibody for specific hot start amplification. Therefore, PCR mixtures can be assembled at room temperature without premature Taq DNA polymerase activity.Typical final primer concentrations are approximately 0.4 μM each. The optimal primer concentration and cycling parameters will depend on the system being used.1.Add the following reagents to a thin-walled PCRmicrocentrifuge tube or plate:Reagent VolumeWater, PCR grade VariableExtract-N-Amp™ PCRreaction mix 10 μLForward primer VariableReverse primer VariableTissue extract 4 μL*Total volume 20 μL*The Extract-N-Amp™ PCR Reaction Mix isformulated to compensate for components in the Extraction, Tissue Preparation, and Neutralization Solutions. If less than 4 µL of tissue extract isadded to the PCR reaction volume, use a 50:50mixture of Extraction and Neutralization BSolutions to bring the volume of tissue extract upto 4 μL.2.Mix gently.3.For thermal cyclers without a heated lid, add20 μL of mineral oil on top of the mixture in eachtube to prevent evaporation.4.Perform thermal cycling. The amplificationparameters should be optimized for individualprimers, template, and thermal cycler.Common cycling parameters:Step Temperature Time Cycles InitialDenaturation 94 °C 3 minutes 1 Denaturation 94 °C 30 seconds Annealing 45 to 68 °C 30 seconds 30-35 Extension 72 °C 1-2 minutes(1 min/kb)FinalExtension 72 °C 10 minutes 1 Hold 4 °C Indefinitely5.The amplified DNA can be loaded onto an agarosegel after the PCR is completed with the addition ofa separate loading buffer/tracking dye such as GelLoading Solution, Cat. No. G2526.Note: PCR products can be purified, if desired, fordownstream applications such as sequencing withthe GenElute PCR Clean-Up Kit, Cat. No.NA1020.Troubleshooting GuideProblem Cause SolutionLittle or no PCR product is detected. PCR reaction may beinhibited due tocontaminants in thetissue extract.Dilute the tissue extract with a 50:50 mix of Extractionand Neutralization Solutions. To test for inhibition, includea DNA control and/or spike a known amount of template(100-500 copies) into the PCR along with the tissue extract. Extraction isinsufficient.Incubate samples at 55 °C for 10 minutes instead ofroom temperature.A PCR component maybe missing or degraded.Run a positive control to ensure that componentsare functioning. A checklist is also recommendedwhen assembling reactions.There may be too fewcycles performed. Increase the number of cycles (5-10 additional cycles at a time). The annealingtemperature maybe too high.Decrease the annealing temperature in 2-4 °C increments.The primers may notbe designed optimally.Confirm the accuracy of the sequence information. If theprimers are less than 22 nucleotides long, try to lengthen theprimer to 25-30 nucleotides. If the primer has a GC contentof less than 45%, try to redesign the primer with a GCcontent of 45-60%.The extension timemay be too short.Increase the extension time in 1-minute increments, especiallyfor long templates.Target templateis difficult.In most cases, inherently difficult targets are due to unusuallyhigh GC content and/or secondary structure. Betaine, Cat. No.B0300, has been reported to help amplification of high GCcontent templates at a concentration of 1.0-1.7 M.Multiple products JumpStart™ Taqantibody is notworking correctly.Do not use DMSO or formamide with Extract-N-Amp™ PCRReaction Mix. It can interfere with the enzyme-antibodycomplex. Other cosolvents, solutes (e.g., salts), and extremesin pH or other reaction conditions may reduce the affinity ofthe JumpStart™ Taq antibody for Taq polymerase and therebycompromise its effectiveness.TouchdownPCR maybe needed.“Touchdown” PCR significantly improves the specificity of manyPCR reactions in various applications. Touchdown PCR involvesusing an annealing/extension temperature that is higher thanthe TM of the primers during the initial PCR cycles. Theannealing/extension temperature is then reduced to the primerTM for the remaining PCR cycles. The change can be performedin a single step or in increments over several cycles.Negative control shows a PCR product or “false positive” result. Reagents arecontaminated.Include a reagent blank without DNA template be included asa control in every PCR run to determine if the reagents used inextraction or PCR are contaminated with a template froma previous reaction.Tissue is not digested after incubations. Tissue is not expectedto be completelydigested.The REDExtract-N-Amp™ Tissue PCR Kit does not require thetissue to be completely digested. Sufficient DNA is released forPCR without completely digesting the tissue.Buccal swab absorbed all the solution. The recommended typeof swab was not used.Due to the low volume of solution used for DNA extraction, afoam tipped swab should be used. Swabs with fibrous tips, suchas cotton or Dacron®, should be avoided because the solutioncannot be recovered efficiently.References1.Dieffenbach, C.W., and Dveksler, G.S. (Eds.), PCRPrimer: A Laboratory Manual, 2nd ed., Cold Spring Harbor Laboratory Press, New York (1995).2.Don, R.H. et al., ‘Touchdown' PCR to circumventspurious priming during gene amplification.Nucleic Acids Res., 19, 4008 (1991).3.Erlich, H.A. (Ed.), PCR Technology: Principles andApplications for DNA Amplification, StocktonPress, New York (1989).4.Griffin, H.G., and Griffin, A.M. (Eds.), PCRTechnology: Current Innovations, CRC Press,Boca Raton, FL (1994).5.Innis, M.A., et al., (Eds.), PCR Strategies,Academic Press, New York (1995).6.Innis, M., et al., (Eds.), PCR Protocols: A Guide toMethods and Applications, Academic Press, SanDiego, California (1990).7.McPherson, M.J. et al., (Eds.), PCR 2: A PracticalApproach, IRL Press, New York (1995).8.Newton, C.R. (Ed.), PCR: Essential Data, JohnWiley & Sons, New York (1995).9.Roux, K.H. Optimization and troubleshooting inPCR. PCR Methods Appl., 4, 5185-5194 (1995).10.Saiki, R., PCR Technology: Principles andApplications for DNA Amplification, Stockton, New York (1989). Product OrderingOrder products online at Related Products Cat. No.Ethanol E7148; E7023; 459836 Forceps,micro-dissecting F4267PCR Marker P9577PCR microtubes Z374873; Z374962;Z374881PCR multi-well plates Z374903Precast Agarose Gels P6097Sealing mats & tapes Z374938; A2350TBE Buffer T4415, T6400, T9525The life science business of Merck operatesas MilliporeSigma in the U.S. and Canada.Merck, Extract-N-Amp, REDExtract-N-Amp, JumpStart, GenElute and Sigma-Aldrich are trademarks of Merck KGaA, Darmstadt, Germany or its affiliates. All other trademarks are theproperty of their respective owners. Detailed information on trademarks is available via publicly accessible resources.NoticeWe provide information and advice to our customers on application technologies and regulatory matters to the best of our knowledge and ability, but without obligation or liability. Existing laws and regulations are to be observed in all cases by our customers. This also applies in respect to any rights of third parties. Our information and advice do not relieve our customers of their own responsibility for checking the suitability of our products for the envisaged purpose. The information in this document is subject to change without notice and should not be construed as a commitment by the manufacturing or selling entity, or an affiliate. We assume no responsibility for any errors that may appear in this document. Technical AssistanceVisit the tech service page at/techservice.Terms and Conditions of SaleWarranty, use restrictions, and other conditions of sale may be found at /terms. Contact InformationFor the location of the office nearest you, go to /offices.。
UVP, LLC Ultra-Violet Products Ltd. White UV Trans

White/UV Transilluminators Instruction ManualUVP, LLC Ultra-Violet Products Ltd. 2066 W 11th Street, Upland, CA 91786 Unit 1,Trinity Hall Farm Estate, Tel: (800) 452-6788 / (909) 946-3197 Nuffield Road Cambridge CB4 1TG UK Fax: (909) 946-3597 Tel: +44(0)1223-420022 / Fax: +44(0)1223-420561Web site: 81-0286-01 Rev DIntroductionThe UVP Transilluminator offers the researcher uniform and intense sources of ultraviolet light(radiation) and white light in one unit. UV and white light 20x20cm filter areas side by side.The special UV design emits high intensity excitation UV wavelength for back-illumination oftransparent fluorescent materials. The UV back-illumination provides a highly sensitive method todetect double-stranded nucleic acids that have been labeled with fluorescent dyes such asethidium bromide or acridine orange. Single stranded nucleic acids may be detected, but withexcitation wavelength sensitive for nucleic acid visualization.White light is used to illuminate coommassie blue, silver stain and protein gels.The transilluminator is uniquely designed with increased UV intensity and uniformity, instant oncapabilities, no lamp flicker and reduced electrical consumption.NOTE:Though the manual refers to the midrange UV waveband as 302nm, others refer to thisregion as 300nm or 312nm. The spectral output of all these regions is the same.A Word of Caution: UV Transilluminators are powerful sources of UV radiation that will causedamage to unprotected eyes and skin. Before operating any unit, be sure all personnel in the areaare properly protected. If not using the transilluminator with an imaging system darkroom, a UVBlocking Cover should be attached to the transilluminator. Even though this cover blocks theultraviolet radiation emitted by the unit, UV Blocking Eyewear should be worn as well.The White/UV Transilluminator features:SpecificationsWhite/UV TransilluminatorsWhite/UV Transilluminator models are designed with white light and one or two UVwavelengths. Units are equipped with a high efficiency electronic ballast.Dimensions: 13.25D x 19.13W x 5.64H in. (337 x 486 x 143mm)No. of Tubes Model Part Number Volt/Hz Wavelength Filter Size /WattsTLW-20 95-0415-01 100-115/60 365nm 20 x 20cm 4 x 8W95-0415-02 230/50 White 20 x 20cm 2 x 8WTMW-20 95-0415-04 100-115/60 302nm 15 x 15cm 4 x 8W95-0415-05 230/50 White 20 x 20cm 2 x 8WLMW-20 95-0418-01 100-115/60 302/365nm 20 x 20cm 4 x 8W95-0418-02 230/50 White 20 x 20cm 2 x 8W Transilluminator OperationSafety PrecautionsWhen the UV Blocking Cover is not beingused, UV light may escape through theholes dedicated to accepting the bracketpins of the UV Blocking Cover.▪Remove the black safety plugs fromtheir package▪Insert the safety plugs through theholes as shown.Safety PlugSetting Up the Transilluminator▪Place the transilluminator on a level work surface. Be sure that an air space exists around the bottom of the work surface. This space allows for the proper air circulation through the unit.▪Plug the female end of the power cord into the transilluminator. For 230 volt models, or those requiring special power cord connectors, ensure that the proper configuration of maleconnector or plug has been properly connected to the power cord.▪Plug the male end of the power cord into a properly grounded electrical outlet. The proper voltage of the transilluminator is found on the product information label. If using thetransilluminator with an imaging system, a jumper cable is required for connecting to thedarkroom. Refer to the imaging system documentation for additional instructions.▪The transilluminator is equipped with a UV Blocking Cover. Remove the brown protective paper from the cover. Insert the bracket pins on the cover into the holes on the front of thetransilluminator. The cover is adjustable to varying angles for access to the filter surface.NOTE: Do not operate the unit without securing the cover. If the cover is missing, a UV Blocking Faceshield must be worn to avoid UV exposure to the skin. UV Blocking Eyewear should be worn even with the cover in place to avoid accidental UV exposure.Permanently Installing the Safety Cover (Optional)Your UVP transilluminator includes a UV-blocking safety shield which can be installed temporarily (as described elsewhere in this manual) or permanently, if desired. The following instructions will explain the process of permanently installing the UVP-supplied UV blocking cover onto the UVP transilluminator.To complete this procedure, the following tools and parts will be required:• Phillips-head screwdriver • 5/16” nut driver• 3/8” narrow walled nut driver• (4) hollow 3/8” nuts (supplied with safety cover)CAUTION: This procedure requires a moderate level of technical competence. If you are not comfortable working with electronics, tools and/or related components, contact UVP for assistance.Note: The transilluminator images shown in this procedure may differ in appearance from your transilluminator. However, the procedure is the same.Removal ProcedureWhen performing the following procedure, place all components (screws, nuts, etc.) in a secure location, as some will be reused for installation.1. Turn off the transilluminator by placing the power switch on the front of the unit in the OFF position. Then, unplug the unit from the wall power.2. Remove the four Phillips-head screws and washers securing the transilluminator filter frame to the body of the transilluminator. Lifting up, remove the transilluminator filter frame from the transilluminator body and place it upside-down on a flat, smooth surface to avoid scratching the filter glass.Two of Four Phillips-Head Screws and WashersTransilluminator Filter FrameTransilluminatorBody3. Locate the two sets of lock nuts and three metal spacers on the inside of the transilluminator filter frame, as shown in the image below. Use the 5/16” nut driver to remove the two sets of lock nuts and spacers.4. Lift the transilluminator filter frame and place the UV blocking cover below the frame. Insert the black tabs, located on the UV blocking cover’s hinges, through the filter frame as shown.Black Tabs5. Use the narrow walled 3/8” nut driver to secure the four supplied 3/8” hollow nuts to the black UV blocking cover hinge tabs.Installation Procedure1. With the UV blocking cover hinges at the front of the unit, replace the transilluminator filter frame on top of the body of the transilluminator.2. Reinstall the four Phillips-head screws and washers on the sides of the transilluminator to secure the transilluminator filter frame to the transilluminator body.Using the TransilluminatorPlace gel/sample on the filter area. It is recommended that researchers place the gels on a Gel-Tray to protect the filter surface from cuts and scratches. It is recommended that gloves be worn to prevent skin contact with gel and staining agents.▪ The transilluminator can be turned on with the wavelength selector knob pointing in anyposition. Press the ON/OFF switch to ON. The tubes within the unit will begin glowing beneath the filter.▪Dial the knob to the appropriate wavelength setting. After viewing the sample, turn the transilluminator off.Phillips-Head Screw and WasherUV Blocking CoverTransilluminatorBodyMaintenance/Repair/Technical AssistanceCleaning and Care of the TransilluminatorClean unit surface with a damp soft cloth or sponge. Never use abrasive cleaners (can damagethe filter surface). To protect the filter glass and minimize moisture and liquids on the glass, it isrecommended that you use a UV transmitting Gel-Tray. Refer to the Replacement Parts forordering information.Replacing Tubes in the Transilluminator▪Disconnect the transilluminator for the electrical supply.▪Remove the filter cover: Use a Phillips head screwdriver to remove the four screws on the sides of the unit. Lift the filter cover off the unit.▪Remove the reflectors on the left and right side. Slide the reflectors up out of the unit.▪Remove the tube: Carefully rotate the tube and slide out of the socket. Replace with a new tube by sliding the tube into the socket and rotating into place. Insert the reflectors back intoplace and reattach the filter cover.Replacement Parts/AccessoriesFor replacement parts or components not shown here, please call UVP Customer Service or placeof purchase. Please have the transilluminator model number available when you call.Replacement Part Description Part NumberFilter Assembly (LMW-20, TMW-20) 38-0189-05Filter Assembly (TLW-20) 38-0189-08Cover, UV blocking 19-0112-01Tube, 8 Watt, 302nm midrange UV (FL8E) 34-0042-01 (Qty. 4 Required)Tube, 8 Watt, 365nm longwave UV (F8T5/BL) 34-0006-01 (Qty. 4 Required)Tube, 8 Watt, cool white (T8T5/CW) 34-0056-01 (Qty. 2 Required)White/UV Selector Switch 53-0196-02Accessories Description Part NumberGel-Cutter 85-0002-01Gel-Scooper 85-0006-01Gel-Tray, UV Transmitting, Lg. 16.5”W x 10.5”D (42 x 27cm) 85-0005-01Gel-Tray, High Performance UV Transmitting (25 x 26cm) 38-0296-03Gel-Ruler, UV Fluorescing 85-0003-01Spectacles, UV Blocking 98-0002-01Goggles, UV Blocking 98-0002-02Faceshield, UV Blocking 98-0002-04Technical SupportUVP offers technical support for all of its products. If you have any questions about the product’suse or, operation, please contact UVP’s offices at the following locations.If you are in North America, South America, If you are in Europe, Africa, the Middle East,East Asia or Australia: Western Asia:⏹ Call(800) 452-6788 or (909) 946-3197⏹ Call+44(0) 1223-420022CustomerCustomer Service regular business regular business days, between 9 amdays, between 7 am and 5 pm PST and 5:30 pm⏹ E-Mail:************⏹ E-Mail:**********.uk⏹ Fax Customer Service: (909) 946-3597⏹ Fax Customer Service: +44(0)1223-420561⏹ Write to: UVP, LLC ⏹ Write to: Ultra-Violet Products Ltd.2066 W. 11th Street Unit 1, Trinity Hall Farm Est., Nuffield Rd.Upland, CA 91786 USA Cambridge CB4 1TG UKNote: A Returned Goods Authorization (RGA) number must be obtained from UVP CustomerService before returning any product.WarrantyUVP, LLC warrants its Ultraviolet Transilluminators to be free of detects in materials and workmanship for a period of two (2) years from date of purchase. The foregoing warranty of UVP shall be of no force and effect if buyer has modified or damaged the product. Tubes and filters are warranted for 90 days.All warranties or merchantability and fitness for any purpose and all other warranties, expressed or implied, except those expressly set forth herein, are deemed waived and excluded.UVP’s duty under the warranty is limited to replacement and/or repair of the defective part at the option of UVP, LLC. UVP shall not be liable for any expenses or damages incurred by the purchaser except as expressly set forth herein, and in no event shall UVP be liable for any special, incidental or consequential damages of any kind. This warranty does not supersede any statutory rights that may be available in certain countries.。
达科为 细胞因子套装(CIK专用)说明书

深圳市达科为生物工程有限公司 地址:深圳市南山区桃源街道光前工业区20栋4C 邮编:518055 Tel: 0755-******** Fax: 0755-******** Web: 细胞因子套装(CIK 专用)说明书CIK Activated Agent (CIK-CYT )DKW35-CYT-CIK001产品描述:细胞因子套装(CIK 专用)是本公司在对CIK 和T 淋巴细胞培养进行大量摸索和实验的基础上,根据优选出的细胞培养所需要的刺激因子的种类及剂量配制而成,主要作为人CIK 细胞培养的诱导活化剂。
本产品批间质量稳定,可搭配各种无血清或有血清培养基用于诱导活化CIK 细胞,使用时按照1:100的比例加入培养基。
包装规格:使用方法:培养细胞时,将组分I (CIK Activated Agent I )和组分II (CIK Activated Agent II )分别按照1:100的比例加入总体系中使用。
培养基总体积组分I组分II 10 mL 100 μL 100 μL 20 mL 200 μL 200 μL 50 mL500 μL500 μL具体培养方法:1. 采血并分离PBMCs 。
2. 使用完全培养基将分离获得的PBMCs 按2-2.5×106个/ml 的浓度接种于T75或T175细胞培养瓶中,按照1:100的比例加入CIK Activated Agent I ,37℃,5%CO 2培养箱中培养。
3. 24hr 后,即第1天按照1:100的比例加入CIK Activated Agent II ,刺激货号名称组分规格 DKW35-CYT-CIK001细胞因子套装(CIK 专用) CIK Activated Agent KitCIK Activated Agent I 500μL CIK Activated Agent II500μL深圳市达科为生物工程有限公司 地址:深圳市南山区桃源街道光前工业区20栋4C 邮编:518055 Tel: 0755-******** Fax: 0755-******** Web: E-mail:*************CIK 细胞的生长和增殖。
外资药厂

CORPORATE SHOCORPORATE DESCMANUFACT. DESCMANUFACT. SHOTOT SEL SELECTED TOTAL#N/AA5Z ASTRAZENECA GROUP#N/AAZN ASTRAZENECA阿斯利康公司AZX ASTRAZENECA WUXI阿斯利康(无锡)制药有限公司PZG PFIZER GROUP#N/APZD PFIZER-DALIAN大连辉瑞制药有限公司PFZ PFIZER美国辉瑞制药公司PHA PHARMACIA CORP法玛西亚普强大药厂-B/S BHC GROUP#N/ABAY BAYER德国拜耳药厂BY6 BAYER BEIJING北京拜耳医药保健品有限公司SGN SCHERING NANJING南京先灵制药有限公司SHG SCHERING AG先灵制药公司LEI LEIRAS芬兰利拉斯药厂SA9 SANOFI-AVENTIS GP.#N/ASG9 SANOFI-SYN.M-SHENG杭州赛诺菲民生制药有限公司AVS AVENTIS PHARMA德国赫司特SPM SANOFIPASTEUR#N/AS-V SANOFI-AVEN DERMIK美国德美克制药公司SSY SANOFI-SYNTHELABO法国赛诺菲-圣德拉堡制药厂RCG ROCHE GROUP#N/AROC ROCHE瑞士罗氏药品有限公司RS- ROCHE-SHANGHAI上海罗氏制药有限公司CHG CHUGAI中外制药公司R-D ROCHE DIAGNOSTICS罗氏诊断公司NVU NOVARTIS GROUP#N/ANVR NOVARTIS瑞士诺华制药有限公司NBJ NOVARTIS BEIJING北京诺华制药有限公司HEX HEXAL德国赫素大药厂S/Z SANDOZ AUSTRIA奥地利山度士生物化学股份有限公司 LEK LEK斯洛文尼亚莱柯医药化学制品有限公司 CTR CHIRON S.P.A.凯龙疫苗公司CVS CIBA VISION诺华视康HXT HEXAL-TIANJIN天津赫素制药有限公司G-S GLAXOSMITHKL GROUP#N/AGSK GLAXOSMITHKLINE PH葛兰素史克公司GX7 GLAXOSMITHKLINE SZ葛兰素史克制药(苏州)有限公司TJS SKF-TIANJIN中美天津史克制药有限公司GTJ GLAXOSMITHKLINE TJ葛兰素史克(天津)有限公司SB- SK-BEECHAM英国比切姆GS1 GLAXOSMITHKLINE SH上海葛兰素史克生物制品有限公司 GCQ GLAXO W.CHONGQING重庆葛兰素制药有限公司NNG NOVO NORDISK GROUP#N/ANKT NOVO NORDISK T/JIN诺和诺德(天津)生物技术有限公司 N-N NOVO-NORDISK丹麦诺和诺德有限公司MSG M.S.D. GROUP#N/AMHU M.S.D.-HANGZHOU杭州默沙东制药有限公司MSD MERCK SHARP&DOHME默沙东J&J J & J GROUP#N/AXJS JANSSEN-XIAN西安杨森制药有限公司JAN JANSSEN比利时杨森制药有限公司JJS J & J SHANGHAI上海强生制药MCN MCNEIL PHARM McNEIL PHARMACEUTICALJJC J&J CHINA强生(中国)有限公司MDX LABORATOIRE MEDIX法国梅迪克斯制药厂MC9 MCNEIL SWEDEN AB#N/AJHC J & J MEDICAL强生(中国)医疗器材有限公司LYG ELI LILLY GROUP#N/ALLY ELI LILLY英国礼来药厂ELU ELI LILLY SUZHOU礼来苏州制药有限公司B6G BAXTER GROUP#N/ABX9 BAXTER - SH.上海百特医疗用品有限公司BTG BAXTER-GZ.广州百特医疗用品有限公司BAX BAXTER HEALTHCARE美国百特医疗用品有限公司BX7 BAXTER TIANJIN天津百特医疗用品有限公司BXG BAXTER-ONCOLOGY德国百特医疗产品股份有限公司 SSW SINO-SWED PHARM无锡华瑞制药有限公司SSW SINO-SWED PHARM无锡华瑞制药有限公司BSG B.MYERS SQUIBB GRP#N/ASSB SQUIBB-SHANGHAI中美上海施贵宝制药有限公司BMS B.MYERS-SQUIBB美国百时美施贵宝公司M-S MERCKSERONO CORP#N/ASER SERONO瑞士雪兰诺大药厂MCK MERCK LIPHA德国默克公司TRM THERAMEX LAB法国赛若美制药厂BAI BEIJING TAIDE PH.北京泰德制药有限公司BAI BEIJING TAIDE PH.北京泰德制药有限公司FG/ FRESENIUS GROUP#N/AFRS FRESENIUS德国费森尤斯卡比有限公司F/B FRESENIUS BEIJING北京费森尤斯卡比医药有限公司 A/U ASTELLAS GROUP#N/AFUJ FUJISAWA日本藤泽药品工业株式会社AES ASTELLAS日本安斯泰来制药AS+ ASTELLAS CHINA安斯泰来制药F.S FUJISAWA-SICHUAN重庆藤泽制药有限公司B.I B.INGELHEIM北京第六制药厂/德勃林格殷格翰 B.I B.INGELHEIM北京第六制药厂/德勃林格殷格翰E-G EISAI GROUP#N/AEI2 EISAI CHINA INC卫材(中国)药业有限公司SG7 SCHERING PLOUGH GP上海先灵葆雅SHC SCHERING PLOUGH美国先灵葆雅药厂ORG ORGANON南京欧加农医药用品有限公司 SP. SCHERING PL-S/HAI上海先灵葆雅制药有限公司SG7 SCHERING PLOUGH GP上海先灵葆雅SVU SERVIER GROUP#N/ATSV SERVIER-TIANJIN施维雅(天津)制药有限公司SVR SERVIER法国施维雅药厂OTU OTSUKA GROUP#N/AOKC OTSUKA-CHINA中国大冢制药有限公司O-G OTSUKA GUANGDONG广东大冢制药有限公司OSU OTSUKA ZHEJIANG浙江大冢制药有限公司SL3 SC. OTSUKA PHARM四川锡成大冢制药有限公司DCG DAIICHI SANKYO GRO#N/ADJG DAIICHI SANKYO BJ#N/ADCH DAIICHI SANKYO SH#N/ADCS DAIICHI SANKYO#N/AWYG WYETH GROUP#N/AWYE WYETH PHARM AYERSTWHZ WYETH OTC SZ惠氏OTC制药有限公司WUZ WYETH SUZHOU惠氏制药有限公司TFS TAIWAN FANSHENG台湾泛生制药厂股份有限公司 TFS TAIWAN FANSHENG台湾泛生制药厂股份有限公司 A5G ABBOTT GROUP#N/AMRI MARUISHI日本丸石株式会社KNO KNOLL德国基诺药厂ABT ABBOTT意大利雅培制药公司SC8 SCICLONE PHARM香港赛生药品股份国际有限公司 SC8 SCICLONE PHARM香港赛生药品股份国际有限公司 U.L THE UNITED LAB香港联邦制药有限公司U.L THE UNITED LAB香港联邦制药有限公司CSL CSL#N/ACSB CSL BEHRING AG#N/AA9E AMERSHAM HEALTH GR#N/ASAH SH.GE HEALTHCARE通用电气药业AM. AMERSHAM HEALTH安盛药业有限公司DSU DAINIPPON SUMITOMO#N/ADSU DAINIPPON SUMITOMO#N/AIPS IPSEN法国博福--益普生制药集团IPS IPSEN法国博福--益普生制药集团UCC UCB CORP#N/AUCB UCB比利时联合化学公司SWR SCHWARZ德国许瓦兹大药厂S-Z SCHWARZ ZHUHAI珠海许瓦兹制药有限公司BF3 BRIGHT FUTURE PH.香港澳美制药BF3 BRIGHT FUTURE PH.香港澳美制药AAA ALTANA PHARMA AG德国安达制药有限公司AAA ALTANA PHARMA AG德国安达制药有限公司RXG RANBAXY GROUP#N/AGXN RANBAXY-GUANGZHOU广州南新制药有限公司RBY RANBAXY印度兰伯西制药有限公司TSD TAIWAN STANDARD CO台湾生达化学制药公司TSD TAIWAN STANDARD CO台湾生达化学制药公司MEI MEIJI SEIKA明治制果公司MEI MEIJI SEIKA明治制果公司BRA BRACCO意大利博莱科化学工业公司BRA BRACCO意大利博莱科化学工业公司DWS DR.WILLMAR SCHWABE德国威玛舒培大药厂DWS DR.WILLMAR SCHWABE德国威玛舒培大药厂N8G NUTRICIA GROUP#N/ANUT NUTRICIA荷兰纽迪希亚NUW NUTRICIA-WUXI纽迪希亚制药DWN KOREAN DAEWOONG PH韩国大熊株式会社DWN KOREAN DAEWOONG PH韩国大熊株式会社B8F BAUSCHELOMB FREDA山东博士伦福瑞达制药有限公司 B8F BAUSCHELOMB FREDA山东博士伦福瑞达制药有限公司 KIR KIRIN GROUP#N/AKRP KIRIN PHARM.CO麒麟麦酒株式会社K4U KIRIN KUNPENG BIO麒麟鲲鹏(中国)生物药业有限公司 SVY SOLVAY PHARM比利时苏威制药SVY SOLVAY PHARM比利时苏威制药STN SANTEN日本参天制药株式会社STN SANTEN日本参天制药株式会社HGP HANMI GROUP#N/AHNM HANMI PHARM韩美药品工业株式会社H3A HANMEI-BJ.北京韩美药品公司ALC ALCON爱尔康眼科产品有限公司ALC ALCON爱尔康眼科产品有限公司TAG TAKEDA GROUP#N/ATAK TAKEDA日本武田药品工业株式会社T/T TAKEDA TIANJIN天津武田药品有限公司SLC SOLCO BASLE瑞士素高·巴塞尔大药厂SLC SOLCO BASLE瑞士素高·巴塞尔大药厂MDO MEDOCHEMIE塞浦路斯麦道甘美公司MDO MEDOCHEMIE塞浦路斯麦道甘美公司BMU BJ.MUNDIPHARM北京萌蒂制药有限公司BMU BJ.MUNDIPHARM北京萌蒂制药有限公司M7B MITSUBISHI GUANGZH三菱制药(广州)有限公司M7B MITSUBISHI GUANGZH三菱制药(广州)有限公司FLK DR.FALK福克大药厂FLK DR.FALK福克大药厂DWO DAEWON KOREA韩国大元制药株式会社DWO DAEWON KOREA韩国大元制药株式会社LUN LUNDBECK丹麦灵北药厂LUN LUNDBECK丹麦灵北药厂BME B.BRAUN瑞士贝朗医药公司BME B.BRAUN瑞士贝朗医药公司CT8 CT意大利斯德大药厂CT8 CT意大利斯德大药厂RED JULIUS-REDEL德国礼达大药厂RED JULIUS-REDEL德国礼达大药厂ESS ESSETI S.A.意大利伊赛特大药厂ESS ESSETI S.A.意大利伊赛特大药厂ASC ASAHI CHEMICAL旭化成工业株式会社ASC ASAHI CHEMICAL旭化成工业株式会社T-C TRB CHEMEDICA#N/ATPQ TRB PHARMA巴西 TRB 制药厂TQI TRB PHARMA IND QUI#N/AOCT OCTAPHARMA AG奥地利奥克特珐玛公司OCT OCTAPHARMA AG奥地利奥克特珐玛公司FER FERRING瑞典辉凌制药有限公司FER FERRING瑞典辉凌制药有限公司H1A HOSPIRA AUSTRALIA澳大利亚科鼎有限公司H1A HOSPIRA AUSTRALIA澳大利亚科鼎有限公司SMT SIGMA-TAU SIGMA-TAUSMT SIGMA-TAU SIGMA-TAUG.R GEDEON RICHTER匈牙利吉瑞大药厂G.R GEDEON RICHTER匈牙利吉瑞大药厂SY. SEIKO EIYO YAKUHIM日本生晃荣养药品株式会社SY. SEIKO EIYO YAKUHIM日本生晃荣养药品株式会社SRB SH.RAAS BLOOD PROD上海莱士血制品有限公司SRB SH.RAAS BLOOD PROD上海莱士血制品有限公司TJ7 TANABE SEIYAKU-TJ天津田边制药有限公司TJ7 TANABE SEIYAKU-TJ天津田边制药有限公司ST6 SHANXI TONGLING PH阿拉宾度同领ST6 SHANXI TONGLING PH阿拉宾度同领K.K KAKEN PHARM生化学工业株式会社K.K KAKEN PHARM生化学工业株式会社MKR MALLINCKRODT加拿大麦林克罗公司MKR MALLINCKRODT加拿大麦林克罗公司HKU HK.UNION PHARM珠海联邦制药股份有限公司中山分公司 HKU HK.UNION PHARM珠海联邦制药股份有限公司中山分公司 MAD MADAUS德国马博士大药厂MAD MADAUS德国马博士大药厂ALW ALFA WASSERMANN意大利阿尔法韦士曼制药公司ALW ALFA WASSERMANN意大利阿尔法韦士曼制药公司A/Z A & Z PHARM美国安士制药有限公司A/Z A & Z PHARM美国安士制药有限公司TS. TOBISHI日本东菱药品工业株式会社TS. TOBISHI日本东菱药品工业株式会社 CHS CHIESI#N/ACH& CHIESI FARM S.P.A.意大利凯西制药公司FND FOURNIER法国科尼(利博福尼)大药厂 FND FOURNIER法国科尼(利博福尼)大药厂 TEV TEVA PHARM以色列梯瓦制药TEV TEVA PHARM以色列梯瓦制药POH G POHL-BOSKAMP德国保时佳大药厂POH G POHL-BOSKAMP德国保时佳大药厂GUE LAB.GUERBET法国加柏大药厂GUE LAB.GUERBET法国加柏大药厂PII POLI CHIMICA意大利普利化学工业股份公司 PII POLI CHIMICA意大利普利化学工业股份公司 T3V TEVA#N/ATJL TJ.HUALIDA BIO.ENG天津华立达生物工程有限公司 BYU BORYUNG PHARM韩国保宁制药有限公司BYU BORYUNG PHARM韩国保宁制药有限公司CHW CHOONGWAE KORE韩国中外制药株式会社CHW CHOONGWAE KORE韩国中外制药株式会社DYK DAIICHI YAKUHIN日本第一药品产业株式会社 DYK DAIICHI YAKUHIN日本第一药品产业株式会社 DPO DUPROMISE MED CO#N/ADPO DUPROMISE MED CO#N/ARTT ROTTA意大利罗达药厂RTT ROTTA意大利罗达药厂EBE EBEWE奥地利依比威药厂EBE EBEWE奥地利依比威药厂N/G NYCOMED GROUP#N/ANYE NYCOMED挪威奈科明有限公司KJE KUKJE PHARM韩国国际药品工业株式会社 KJE KUKJE PHARM韩国国际药品工业株式会社 MRZ MERZ德国麦氏制药有限公司MRZ MERZ德国麦氏制药有限公司ZBG ZAMBON GROUP#N/AZAM ZAMBON法国纳化制药厂H7Z HN.ZAMBON PHARM海南赞邦制药有限公司INP INPHARZAM SA瑞士INPHARZAM SAHL- HANLIN PHARM韩林制药株式会社HL- HANLIN PHARM韩林制药株式会社GEZ GENZYME POLYCLONAL#N/AGEZ GENZYME POLYCLONAL#N/ACKD CHONG KUN DANG韩国钟根堂公司CKD CHONG KUN DANG韩国钟根堂公司BAG BAGO PHARM阿根廷霸科制药厂BAG BAGO PHARM阿根廷霸科制药厂GZY GENZYME CORP#N/AGZY GENZYME CORP#N/AAT4 AMT LAB美国矿维公司AT4 AMT LAB美国矿维公司R7R ROTAM REDDY-K/SHAN昆山龙灯瑞迪制药有限公司 R7R ROTAM REDDY-K/SHAN昆山龙灯瑞迪制药有限公司 LEO LEO丹麦利昂制药有限公司LEO LEO丹麦利昂制药有限公司MN& MINOPHAGEN-JAPAN#N/A MN& MINOPHAGEN-JAPAN#N/A NJM NIN JIOM MEDICINE香港京都念慈庵制药厂NJM NIN JIOM MEDICINE香港京都念慈庵制药厂B&I BOEHRINGER INGEL#N/A BVE BEN VENUE LABORATO#N/A SKO SANKYO PHARM GMBH德国慕尼黑路易坡大药厂 SKO SANKYO PHARM GMBH德国慕尼黑路易坡大药厂 YGS YUNG SHIN GROUP#N/A Y2U YUNG SHIN-KUN SHAN永信药品工业ALL ALLERGAN美国眼力健公司ALL ALLERGAN美国眼力健公司WAT WATSON CHANGZHOU常州华生制药有限公司WAT WATSON CHANGZHOU常州华生制药有限公司HEU HEUMANN德国赫曼大药厂HEU HEUMANN德国赫曼大药厂P.F PIERRE FABRE法国皮尔法伯药物研制公司 P.F PIERRE FABRE法国皮尔法伯药物研制公司 SGI SHIONOGI日本盐野义制药株式会社 SGI SHIONOGI日本盐野义制药株式会社 NP1 NIPPON PHARM日本脏器制药株式会社NP1 NIPPON PHARM日本脏器制药株式会社AKB ALK ABELLO丹麦爱尔开-阿贝优公司AKB ALK ABELLO丹麦爱尔开-阿贝优公司GCR GREEN CROSS GROUP#N/A G/A GREEN CROSS-ANHUI韩国绿十字制药公司G.C GREEN CROSS日本三菱制药ORN ORION芬兰 FARMOS GROUPORN ORION芬兰 FARMOS GROUPMYS MIYARISAN PHARM日本米雅利桑株式会社MYS MIYARISAN PHARM日本米雅利桑株式会社KBN KUNMING BK.NORTON昆明贝克诺顿制药有限公司 KBN KUNMING BK.NORTON昆明贝克诺顿制药有限公司 KHK KYOWA HAKKO日本协和发酵工业株式会社 KHK KYOWA HAKKO日本协和发酵工业株式会社 ND6 NARCO-MED GMBH德国NARCO-MED GMBHND6 NARCO-MED GMBH德国NARCO-MED GMBHGL8 GENELEUK BIOTECH格兰百克生物制药有限公司 GL8 GENELEUK BIOTECH格兰百克生物制药有限公司 GFL LAB.GRIFOLS西班牙基立福药厂GFL LAB.GRIFOLS西班牙基立福药厂KTB KOTOBUKI日本寿制药株式会社KTB KOTOBUKI日本寿制药株式会社P-T POLFA TARCHOMIN波兰波尔法-塔赫敏制药股份公司P-T POLFA TARCHOMIN波兰波尔法-塔赫敏制药股份公司KLI KLINGE PHARM德国克林格制药公司KLI KLINGE PHARM德国克林格制药公司SNJ SENJU日本千寿制药株式会社SNJ SENJU日本千寿制药株式会社S.U STULLN德国施图伦制药公司S.U STULLN德国施图伦制药公司B-C BERLIN-CHEMIE柏林-化学股份有限公司B-C BERLIN-CHEMIE柏林-化学股份有限公司N.K NIPPON KAYAKU日本化药株式会社N.K NIPPON KAYAKU日本化药株式会社S+B SCHAPER & BRUMMER德国沙佩尔-布吕默尔S+B SCHAPER & BRUMMER德国沙佩尔-布吕默尔CIJ CHEIL.JEDANG韩国希杰有限公司CIJ CHEIL.JEDANG韩国希杰有限公司TWX TAIWAN XINDONG台湾信东化学工业股份有限公司TWX TAIWAN XINDONG台湾信东化学工业股份有限公司NSC NESTLE CORPORATION#N/ANSC NESTLE CORPORATION#N/AS9M SH.ETHYPHARM北京第四制药厂/法国爱得发制药公司 S9M SH.ETHYPHARM北京第四制药厂/法国爱得发制药公司 PWP PO WO TONG PHARMA香港保和堂制药有限公司PWP PO WO TONG PHARMA香港保和堂制药有限公司LRQ LAB.LEURQUIN法国乐康大药厂LRQ LAB.LEURQUIN法国乐康大药厂LWI LOTUS-WEIHAI威海路坦制药有限公司LWI LOTUS-WEIHAI威海路坦制药有限公司GLD GALDERMA法国高德美制药公司GLD GALDERMA法国高德美制药公司PZP PLIVA ZAGREB PHARM普利瓦大药厂PZP PLIVA ZAGREB PHARM普利瓦大药厂BMF BIOMEDICA FOSCAMA意大利福斯卡玛生化制药有限公司BMF BIOMEDICA FOSCAMA意大利福斯卡玛生化制药有限公司CFE C.B. FLEET CO.美国辉力公司CFE C.B. FLEET CO.美国辉力公司LPM LISAPHARMA意大利利沙生化大药厂LPM LISAPHARMA意大利利沙生化大药厂BNC BIONORICA AG德国比奥罗历加制药有限公司BNC BIONORICA AG德国比奥罗历加制药有限公司BAD BJ.STADA PHARM史达德药业BAD BJ.STADA PHARM史达德药业AST ASTA德国爱斯达药厂AST ASTA德国爱斯达药厂TJI TEIJIN日本蒂人株式会社TJI TEIJIN日本蒂人株式会社HW9 HN.WANTE PHARM万特制药(海南)有限公司HW9 HN.WANTE PHARM万特制药(海南)有限公司ATC ATLANTIC香港大西洋制药厂ATC ATLANTIC香港大西洋制药厂LO9 LOTUS PHARMA台湾美时化学制药股份有限公司LO9 LOTUS PHARMA台湾美时化学制药股份有限公司DEM DEMO S.A. PHARM希腊 Demo S.A.PharmDEM DEMO S.A. PHARM希腊 Demo S.A.PharmSPI SUN PHARMA.IND.印度太阳药业有限公司SPI SUN PHARMA.IND.印度太阳药业有限公司CMV CHEMINOVA西班牙国际新化学药厂CMV CHEMINOVA西班牙国际新化学药厂C4T C.T.S.英国CONTROLLED THERAPEUTICS公司 C4T C.T.S.英国CONTROLLED THERAPEUTICS公司 BI6 BI SHENG-HK香港毕盛医药国际有限公司BI6 BI SHENG-HK香港毕盛医药国际有限公司ITF ITALFARMACO意大泛马克ITF ITALFARMACO意大泛马克KIS KISSEI PHARMA日本橘生药品工业株式会社KIS KISSEI PHARMA日本橘生药品工业株式会社MBH MITSUBISHI CHEM.日本三菱化学株式会社MBH MITSUBISHI CHEM.日本三菱化学株式会社TIS TAISHO日本大正药品工业株式会社TIS TAISHO日本大正药品工业株式会社SDI SAITAMA DAIICH日本埼玉第一制药株式会社SDI SAITAMA DAIICH日本埼玉第一制药株式会社B.B B.B.MOHAN PHARMACE#N/AB.B B.B.MOHAN PHARMACE#N/AAF+ ACTAVIS FOSHAN PH阿特维斯(佛山)制药有限公司AF+ ACTAVIS FOSHAN PH阿特维斯(佛山)制药有限公司RAT RATIOPHARM德国通益制药有限责任公司RAT RATIOPHARM德国通益制药有限责任公司REC RECORDATI意大利米兰利康大药厂REC RECORDATI意大利米兰利康大药厂BH& BAUSCH & LOMB GROU#N/AMAN DR.GERHARD MANN CH德国博士曼大药厂P9Y PIERRE ROLLLAND FY法国赛特力—碧兰公司P9Y PIERRE ROLLLAND FY法国赛特力—碧兰公司GL& GD.ST.LUSTRE PHARM汕头保税区洛斯特制药有限公司GL& GD.ST.LUSTRE PHARM汕头保税区洛斯特制药有限公司BCX BIOCODEX法国百科达药厂BCX BIOCODEX法国百科达药厂VIF VIFOR瑞士维福国际公司VIF VIFOR瑞士维福国际公司FGM FERRING GMBH德国辉凌制药有限公司FGM FERRING GMBH德国辉凌制药有限公司P2C PACIFIC KOREA韩国太平洋制药有限公司P2C PACIFIC KOREA韩国太平洋制药有限公司JPD JP.DIGUO PHARM日本帝国制药株式会社JPD JP.DIGUO PHARM日本帝国制药株式会社POR POLIFARMA意大利宝利化大药厂POR POLIFARMA意大利宝利化大药厂CIH CHIRONBEHRING GMBH德国凯龙-贝林公司CIH CHIRONBEHRING GMBH德国凯龙-贝林公司LTL LACTEOL法国乐托尔大药厂LTL LACTEOL法国乐托尔大药厂M5A MERCK APOTEC默克-雅柏M5A MERCK APOTEC默克-雅柏DDN DR.DUNNER瑞士端纳博士药厂DDN DR.DUNNER瑞士端纳博士药厂AGP ALLERGOPHARMA JOAC#N/AAGP ALLERGOPHARMA JOAC#N/ALG5 LG LIFE SCIENCES韩国LG生命科学有限公司LG5 LG LIFE SCIENCES韩国LG生命科学有限公司APH ALPHAPHARM澳大利亚艾华大药厂APH ALPHAPHARM澳大利亚艾华大药厂TP5 TAIWAN PEILI PHARM台湾培力药品工业股份有限公司 TP5 TAIWAN PEILI PHARM台湾培力药品工业股份有限公司 .SN NIPPON SHINYAKU日本新药株式会社.SN NIPPON SHINYAKU日本新药株式会社BTT BRUSCHETTINI意大利贝斯迪大药厂BTT BRUSCHETTINI意大利贝斯迪大药厂CDP CD.PASHUN PHARM成都东洋百信制药有限公司CDP CD.PASHUN PHARM成都东洋百信制药有限公司BHL BAUSCH & LOMB博士伦药业BHL BAUSCH & LOMB博士伦药业CKT CHANG KANG TANG-HK香港长康堂制药CKT CHANG KANG TANG-HK香港长康堂制药TYK TOA SEIYAKE INC日本东亚药品工业株式会社TYK TOA SEIYAKE INC日本东亚药品工业株式会社AP5 AMERICAN PHAR PART#N/AAP5 AMERICAN PHAR PART#N/AWE4 WEIDAR CHEM TAIWAN台湾卫达化学制药股份有限公司 WE4 WEIDAR CHEM TAIWAN台湾卫达化学制药股份有限公司 EPH EUROPHARM香港欧化药业有限公司EPH EUROPHARM香港欧化药业有限公司MPH MEPHA PHARM瑞士美化药厂MPH MEPHA PHARM瑞士美化药厂MI6 MINRAD INC.美国MINRAD药厂MI6 MINRAD INC.美国MINRAD药厂HEB DR HERBRAND KG赫尔布兰德KG化学-药物制药厂 HEB DR HERBRAND KG赫尔布兰德KG化学-药物制药厂 PG3 PHAFAG AG瑞士科发大药厂PG3 PHAFAG AG瑞士科发大药厂RHQ RHODIA ORGANIQUE英国罗地亚精细化工有限公司 RHQ RHODIA ORGANIQUE英国罗地亚精细化工有限公司 Y2P YUNGJIN PHARM. CO.韩国永进药业有限公司Y2P YUNGJIN PHARM. CO.韩国永进药业有限公司M-C MERCK APOTEC-CHINA默克雅柏药业M-C MERCK APOTEC-CHINA默克雅柏药业TPU TAIWAN PUTAOWANG台湾葡萄王企业股份有限公司 TPU TAIWAN PUTAOWANG台湾葡萄王企业股份有限公司 EUY EUYANSANG INTERNA余仁生(香港)有限公司EUY EUYANSANG INTERNA余仁生(香港)有限公司AHA ATLANTIC-HAINAN海南大西洋制药有限公司AHA ATLANTIC-HAINAN海南大西洋制药有限公司S4L SK CHEMICALS GROUP#N/ASK6 SK CHEMICALS CO.韩国SK化工株式会社TAN TANABE SEIYAKU日本田边制药株式会社TAN TANABE SEIYAKU日本田边制药株式会社WBG WASSERBURGER G德国米奥大药厂WBG WASSERBURGER G德国米奥大药厂HK5 HK.CONCORD MED.同济药业(香港)有限公司HK5 HK.CONCORD MED.同济药业(香港)有限公司EVE EVERS德国爱活大药厂EVE EVERS德国爱活大药厂AP8 ADVANCE PHARMA香港乐信药业有限公司AP8 ADVANCE PHARMA香港乐信药业有限公司CDS CODAL SYNTO塞浦路斯高特制药有限公司CDS CODAL SYNTO塞浦路斯高特制药有限公司A9N ACTELION PHARMA爱可泰隆药品有限公司(瑞士) A9N ACTELION PHARMA爱可泰隆药品有限公司(瑞士) BSI BESINS-ISCOVESCO法国法杏大药厂BSI BESINS-ISCOVESCO法国法杏大药厂3MH 3M HEALTH CARE法国3M公司3MH 3M HEALTH CARE法国3M公司LOG JACQUES-LOGEAIS法国洛斯大药厂LOG JACQUES-LOGEAIS法国洛斯大药厂ZHP ZHONGXIN PH-JAPAN日本中新药业株式会社ZHP ZHONGXIN PH-JAPAN日本中新药业株式会社S3V SERUM-VACCINE INST瑞士伯尔尼血清疫苗研究所S3V SERUM-VACCINE INST瑞士伯尔尼血清疫苗研究所LD6 LABORATORIOS DALME达尔玛实验室有限公司LD6 LABORATORIOS DALME达尔玛实验室有限公司KSS KYUSHIN SEIYAKU日本救心制药株式会社KSS KYUSHIN SEIYAKU日本救心制药株式会社A.L ALPHARMA APS丹麦阿尔法玛公司A.L ALPHARMA APS丹麦阿尔法玛公司A-T ALPHA THERAPEUTIC美国ALPHA THERAPEUTICA-T ALPHA THERAPEUTIC美国ALPHA THERAPEUTICA&Z A & Z-ZHONGSHAN安士制药(中山)有限公司A&Z A & Z-ZHONGSHAN安士制药(中山)有限公司T-N TAIWAN NANGUANG台湾南光化学制药股份有限公司T-N TAIWAN NANGUANG台湾南光化学制药股份有限公司AWD AWD.PHARMA GMBH德国AWD公司AWD AWD.PHARMA GMBH德国AWD公司IBS IBSA INSTITUTE BIO#N/AIBS IBSA INSTITUTE BIO#N/AJ.I JEIL PHARM韩国第一药品株式会社J.I JEIL PHARM韩国第一药品株式会社ATX APOTEX INC.加拿大奥贝泰克制药有限公司ATX APOTEX INC.加拿大奥贝泰克制药有限公司FSX FIDIA-SINAX意大利菲地亚药厂FSX FIDIA-SINAX意大利菲地亚药厂NID NITTO DENKO COR.日东电工株式会社东北事业所NID NITTO DENKO COR.日东电工株式会社东北事业所J/M JEAN-MARIE PHARMA香港正美药品有限公司J/M JEAN-MARIE PHARMA香港正美药品有限公司SLE SG.LEUNG KAI FOOK新加坡梁介福药业有限公司SLE SG.LEUNG KAI FOOK新加坡梁介福药业有限公司HO& HOSPIRA#N/AHO8 HOSPIRA INC美国赫士睿公司TW2 TW.JINGTIAN PHARM台湾井田制药工业股份有限公司TW2 TW.JINGTIAN PHARM台湾井田制药工业股份有限公司DN4 DANIPHARM APS丹麦丹尼制药厂DN4 DANIPHARM APS丹麦丹尼制药厂M.S MAYOLY SPINDLER法国优德大药厂M.S MAYOLY SPINDLER法国优德大药厂STF STIEFEL-IRELAND施泰福(爱尔兰)大药厂STF STIEFEL-IRELAND施泰福(爱尔兰)大药厂GXD GD.NEXMED PHARMACE尼克美制药(中山)有限公司GXD GD.NEXMED PHARMACE尼克美制药(中山)有限公司SN& SUN STAR日本新时代株式会社SN& SUN STAR日本新时代株式会社LGI LABORATOR GUIDOTT意大利Laboratori Guidotti S.P.A LGI LABORATOR GUIDOTT意大利Laboratori Guidotti S.P.A RHT ROHTO曼秀雷敦(中山)药业有限公司RHT ROHTO曼秀雷敦(中山)药业有限公司D.R DRUG RESEARCH意大利D.R药品拓展制造有限公司 D.R DRUG RESEARCH意大利D.R药品拓展制造有限公司 H5T HK.TONGYITANG PH香港同溢堂中药制药厂H5T HK.TONGYITANG PH香港同溢堂中药制药厂SDO SEPTODONT法国赛特多公司SDO SEPTODONT法国赛特多公司FR& FRANKLIN PHARM香港法健化学制药厂有限公司FR& FRANKLIN PHARM香港法健化学制药厂有限公司ARM ARMOUR PHARM美国亚玛制药公司ARM ARMOUR PHARM美国亚玛制药公司HG5 HONG KONG WONG TO香港黄道益医药控股有限公司HG5 HONG KONG WONG TO香港黄道益医药控股有限公司Y2K YING KUT LEE MEDIC香港英吉利制药厂Y2K YING KUT LEE MEDIC香港英吉利制药厂SGS SHIGAKEN日本滋贺县制药工业株式会社SGS SHIGAKEN日本滋贺县制药工业株式会社ISI ISEI COMPANY INCO.日本点眼药研究所ISI ISEI COMPANY INCO.日本点眼药研究所H/P HK.LIANHUA PHARM香港联华药业H/P HK.LIANHUA PHARM香港联华药业TAI TAIHO PHARMACEUTIC#N/ATAI TAIHO PHARMACEUTIC#N/ATGJ TONGJI TAILEQI PH武汉同济泰乐奇医药有限公司TGJ TONGJI TAILEQI PH武汉同济泰乐奇医药有限公司CED CEDONA荷兰圣多纳药厂CED CEDONA荷兰圣多纳药厂MAC HEINRICH MACK德国马赫大药厂MAC HEINRICH MACK德国马赫大药厂TAD TAD德国泰德制药有限公司TAD TAD德国泰德制药有限公司KW. KOWA PHARM日本兴和株式会社KW. KOWA PHARM日本兴和株式会社HKH HK.HONGLI PHARM香港宏利药厂HKH HK.HONGLI PHARM香港宏利药厂H3D HK.YUANDA PHARMACE香港远大制药有限公司H3D HK.YUANDA PHARMACE香港远大制药有限公司AD2 ADVANCED NUTRITION美国AN药业AD2 ADVANCED NUTRITION美国AN药业C2W CHAI TIN SOW MEDIC香港齐天寿药厂有限公司C2W CHAI TIN SOW MEDIC香港齐天寿药厂有限公司JJ- JX.JIEZHONG BIOCHE江西捷众生物化学有限公司JJ- JX.JIEZHONG BIOCHE江西捷众生物化学有限公司LFS LIFESCAN INTERNATI美国理康国际有限公司LFS LIFESCAN INTERNATI美国理康国际有限公司TBT TAIWAN BIOTECH CO.#N/ATBT TAIWAN BIOTECH CO.#N/ATWB TW.PANION & BF台湾宝龄富锦制药厂股份有限公司 TWB TW.PANION & BF台湾宝龄富锦制药厂股份有限公司 LOP LABORATOIRES SOPHA Laboratoires SophartexLOP LABORATOIRES SOPHA Laboratoires SophartexP7O PO SUM ON MEDICINE香港保心安药厂有限公司P7O PO SUM ON MEDICINE香港保心安药厂有限公司N/C NIPPON CHEMIPHAR日本化学药品株式会社茨城分厂 N/C NIPPON CHEMIPHAR日本化学药品株式会社茨城分厂 B-D BECTON DICKINSON#N/AB-D BECTON DICKINSON#N/AH2F HOE HIN PAK FAH YE香港和兴白花油药厂有限公司 H2F HOE HIN PAK FAH YE香港和兴白花油药厂有限公司 BER BERNA BIOTECH AG瑞士博尔纳生物技术有限公司 BER BERNA BIOTECH AG瑞士博尔纳生物技术有限公司 TQC TAIWAN QUANFU CHEM台湾全福生物制药有限公司TQC TAIWAN QUANFU CHEM台湾全福生物制药有限公司A6Y ARKRAY INC.日本爱科来株式会社A6Y ARKRAY INC.日本爱科来株式会社GRT GRUNENTHAL德国格兰泰公司GRT GRUNENTHAL德国格兰泰公司DRL DR.REDDYS印度瑞迪博士大药厂DRL DR.REDDYS印度瑞迪博士大药厂JIU JINGDU YANGSHENGTA京都养生堂制药厂有限公司JIU JINGDU YANGSHENGTA京都养生堂制药厂有限公司J5T JAPAN TAIKO PHARM日本大幸药品株式会社J5T JAPAN TAIKO PHARM日本大幸药品株式会社FRR FERRER FERRER INTERNACIONAL S.A.FRR FERRER FERRER INTERNACIONAL S.A.AJM AJINOMOTO日本味之素株式会社AJM AJINOMOTO日本味之素株式会社MT7 TAIWAN MIN TONG PH台湾明通化学制药股份有限公司 MT7 TAIWAN MIN TONG PH台湾明通化学制药股份有限公司 RDM RADIUMFARMA意大利 Radiumfarma 国际公司 RDM RADIUMFARMA意大利 Radiumfarma 国际公司 LF/ LIFEPHARM意大利利福法玛公司LF/ LIFEPHARM意大利利福法玛公司SAD STADAPHARM德国史达德大药厂SAD STADAPHARM德国史达德大药厂LAF LAFON法国拉丰制药厂LAF LAFON法国拉丰制药厂B3B BERNA BIOTECH KORE Berna Biotech Korea Corp.B3B BERNA BIOTECH KORE Berna Biotech Korea Corp.TWK TAIWAN KINGDOM CO台湾景德制药有限公司TWK TAIWAN KINGDOM CO台湾景德制药有限公司FC8 FDC LTD印度 FDCFC8 FDC LTD印度 FDCFTU FORTUNE PHARMACAL香港幸福医药有限公司FTU FORTUNE PHARMACAL香港幸福医药有限公司MSH MORISHITA日本罗素-森下株式会社MSH MORISHITA日本罗素-森下株式会社TTS TW.TEH SENG PHARM台湾得生制药股份有限公司TTS TW.TEH SENG PHARM台湾得生制药股份有限公司KER KREUSSLER德国Fabrik Kreussler公司KER KREUSSLER德国Fabrik Kreussler公司BS8 BIOSTIME FRANCE法国合生元公司BS8 BIOSTIME FRANCE法国合生元公司GEI GEISTLICH PHARM瑞士Geistlich药厂GEI GEISTLICH PHARM瑞士Geistlich药厂HK6 HK.TIANFU PH香港天符医药有限公司HK6 HK.TIANFU PH香港天符医药有限公司LUH LOTUS HEALTHCARE美国路坦医药有限公司LUH LOTUS HEALTHCARE美国路坦医药有限公司P8I PHARMASCIENCE加拿大博爱药厂P8I PHARMASCIENCE加拿大博爱药厂PS+ PASHUN GROUP#N/AHKP HK.PA SHUN MED.香港东洋百信药业公司HG- HIGH CHANGE GROUP法国汉臣氏集团(中国)有限公司HG- HIGH CHANGE GROUP法国汉臣氏集团(中国)有限公司LSN LOSAN PHARMA GMBH#N/ALSN LOSAN PHARMA GMBH#N/AGN5 GENTLE PHARMACEUTI#N/AGN5 GENTLE PHARMACEUTI#N/ATEU TERUMO CORPORATION泰尔茂株式会社TEU TERUMO CORPORATION泰尔茂株式会社TNT TORRENT PHARM印度托兰特药业公司TNT TORRENT PHARM印度托兰特药业公司APS APS PHARM英国新百利APS APS PHARM英国新百利LOZ LAB.DE LOZOTHINE法国LABORATOIRES DE L'OZOTHINELOZ LAB.DE LOZOTHINE法国LABORATOIRES DE L'OZOTHINETLW TH.LIWANSHAN PH泰国李万山药厂TLW TH.LIWANSHAN PH泰国李万山药厂LXB LUXEMBERG MEDICAL卢森堡大药厂LXB LUXEMBERG MEDICAL卢森堡大药厂HIG HK.LING NAN MED FA岭南药厂(香港)有限公司HIG HK.LING NAN MED FA岭南药厂(香港)有限公司G2W GD.XM.HAW PAR新加坡虎豹医保有限公司厦门虎标医药有 G2W GD.XM.HAW PAR新加坡虎豹医保有限公司厦门虎标医药有 NU3 NUTRICHEM德国NUTRICHEM DIAT + PHARMANU3 NUTRICHEM德国NUTRICHEM DIAT + PHARMAPFB PIERRE FABRE LABOR法国皮尔法伯雅漾护肤化妆品研制公PFB PIERRE FABRE LABOR法国皮尔法伯雅漾护肤化妆品研制公HKM HK.SHANYUANTANG ME香港善元堂医药生物集团有限公司HKM HK.SHANYUANTANG ME香港善元堂医药生物集团有限公司ZGM ZHONGMEI AIMO PH中美埃默药业有限公司ZGM ZHONGMEI AIMO PH中美埃默药业有限公司LAK LAKESIDE墨西哥 Lakeside, S.A.LAK LAKESIDE墨西哥 Lakeside, S.A.PCH PHARMACHEMIE荷兰 Phamaceuticals B.V.PCH PHARMACHEMIE荷兰 Phamaceuticals B.V.TRD TROMMSDORFF AG德国中央大药厂TRD TROMMSDORFF AG德国中央大药厂WP. WATERFORD PHARM爱尔兰 WATERFORDWP. WATERFORD PHARM爱尔兰 WATERFORDWJ2 W.J RENDELL LTD.英国连德尔大药厂有限公司WJ2 W.J RENDELL LTD.英国连德尔大药厂有限公司SWA SCHWAB & CO SCHWABSWA SCHWAB & CO SCHWABOIS OISHIKOSEIDO CO.日本株式会社大石膏盛堂OIS OISHIKOSEIDO CO.日本株式会社大石膏盛堂C4B CIMAB S.A.古巴分子免疫学中心C4B CIMAB S.A.古巴分子免疫学中心HP4 HANSEO PHARM韩西制药株式会社HP4 HANSEO PHARM韩西制药株式会社L2M LEE MAN SHAN PHARM香港李万山药厂有限公司L2M LEE MAN SHAN PHARM香港李万山药厂有限公司HLX HELIXOR HEILMITTEL#N/AHLX HELIXOR HEILMITTEL#N/ASSH SEVEN SEA LIMITED英国七海有限公司SSH SEVEN SEA LIMITED英国七海有限公司ASN ASTRAPIN德国荷夫曼大药厂ASN ASTRAPIN德国荷夫曼大药厂CPL CIPLA印度悉普拉大药房CPL CIPLA印度悉普拉大药房JPK JAPAN KAWAI日本河合制药株式会社JPK JAPAN KAWAI日本河合制药株式会社HKY HONGKONG YIZHOU CO香港一洲制药有限公司HKY HONGKONG YIZHOU CO香港一洲制药有限公司I5O INFOPIA CO.LTD.韩国Infopia 公司I5O INFOPIA CO.LTD.韩国Infopia 公司OMR OMRON DALIAN CO.欧姆龙(大连)有限公司OMR OMRON DALIAN CO.欧姆龙(大连)有限公司A&T ANTIBIOTICOS S.A西班牙抗生素厂A&T ANTIBIOTICOS S.A西班牙抗生素厂AAI AMGEN美国安进公司AAI AMGEN美国安进公司AVM ADVANCED MAGNETICS美国先进磁性公司AVM ADVANCED MAGNETICS美国先进磁性公司BOO BOOTS英国普施制药公司BOO BOOTS英国普施制药公司BTS BIOTEST德国生物实验制药厂BTS BIOTEST德国生物实验制药厂CEF CEFAK德国彼勒尔公司CEF CEFAK德国彼勒尔公司CN. CLONMEL爱尔兰CLONMEL化学制药有限公司 CN. CLONMEL爱尔兰CLONMEL化学制药有限公司 CVX COVEX西班牙考维克斯公司CVX COVEX西班牙考维克斯公司DDK DONG KOOK KOREA韩国东国制药株式会社DDK DONG KOOK KOREA韩国东国制药株式会社DEL DELALANDE法国巴黎狄朗药厂。
c-Jun N-terminal kinases
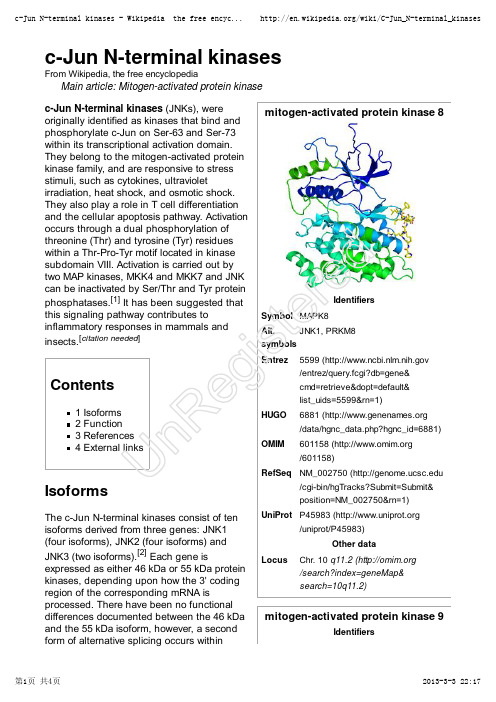
mitogen-activated protein kinase 8IdentifiersSymbol MAPK8Alt.symbols JNK1, PRKM8Entrez5599 ( /entrez/query.fcgi?db=gene&cmd=retrieve&dopt=default&list_uids=5599&rn=1)HUGO 6881 ( /data/hgnc_data.php?hgnc_id=6881)OMIM 601158 ( /601158)RefSeq NM_002750 (/cgi-bin/hgTracks?Submit=Submit&position=NM_002750&rn=1)UniProt P45983 (/uniprot/P45983)Other dataLocusChr. 10 q11.2 ( /search?index=geneMap&search=10q11.2)mitogen-activated protein kinase 9Identifiersc-Jun N-terminal kinasesFrom Wikipedia, the free encyclopediaMain article: Mitogen-activated protein kinase c-Jun N-terminal kinases (JNKs), were originally identified as kinases that bind and phosphorylate c-Jun on Ser-63 and Ser-73within its transcriptional activation domain.They belong to the mitogen-activated protein kinase family, and are responsive to stress stimuli, such as cytokines, ultravioletirradiation, heat shock, and osmotic shock.They also play a role in T cell differentiation and the cellular apoptosis pathway. Activation occurs through a dual phosphorylation of threonine (Thr) and tyrosine (Tyr) residues within a Thr-Pro-Tyr motif located in kinase subdomain VIII. Activation is carried out by two MAP kinases, MKK4 and MKK7 and JNK can be inactivated by Ser/Thr and Tyr protein phosphatases.[1] It has been suggested that this signaling pathway contributes toinflammatory responses in mammals and insects.[citation needed ]Contents1 Isoforms2 Function3 References4 External linksIsoformsThe c-Jun N-terminal kinases consist of ten isoforms derived from three genes: JNK1(four isoforms), JNK2 (four isoforms) and JNK3 (two isoforms).[2] Each gene isexpressed as either 46 kDa or 55 kDa protein kinases, depending upon how the 3' coding region of the corresponding mRNA isprocessed. There have been no functional differences documented between the 46 kDa and the 55 kDa isoform, however, a second form of alternative splicing occurs withinUn Re gi st er edSymbol MAPK9Alt.symbols JNK2, PRKM9Entrez5601 ( /entrez/query.fcgi?db=gene&cmd=retrieve&dopt=default&list_uids=5601&rn=1)HUGO 6886 ( /data/hgnc_data.php?hgnc_id=6886)OMIM602896 ( /602896)RefSeq NM_002752 (/cgi-bin/hgTracks?Submit=Submit&position=NM_002752&rn=1)UniProt P45984 (/uniprot/P45984)Other dataLocusChr. 5 q35 ( /search?index=geneMap&search=5q35)mitogen-activated protein kinase 10IdentifiersSymbol MAPK10Alt.symbols JNK3, PRKM10Entrez 5602 ( /entrez/query.fcgi?db=gene&cmd=retrieve&dopt=default&list_uids=5602&rn=1)HUGO 6872 ( /data/hgnc_data.php?hgnc_id=6872)OMIM602897 ( /602897)RefSeq NM_002753 (/cgi-bin/hgTracks?Submit=Submit&position=NM_002753&rn=1)UniProt P53779 (/uniprot/P53779)Other datatranscripts of JNK1 and JNK2, yielding JNK1-α, JNK2-α and JNK1-β and JNK2-β.Differences in interactions with protein substrates arise because of the mutually exclusive utilization of two exons within the kinase domain.[1]c-Jun N-terminal kinase isoforms have the following tissue distribution:JNK1 and JNK2 are found in all cells and tissues.[3]JNK3 is found mainly in the brain, but is also found in the heart and the testes.[3]FunctionInflammatory signals, changes in levels of reactive oxygen species, ultraviolet radiation,protein synthesis inhibitors, and a variety of stress stimuli can activate JNK. One way this activation may occur is through disruption of the conformation of sensitive protein phosphatase enzymes; specificphosphatases normally inhibit the activity of JNK itself and the activity of proteins linked to JNK activation.[4]JNKs can associate with scaffold proteins JNK interacting proteins as well as their upstream kinases JNKK1 and JNKK2following their activation.JNK, by phosphorylation, modifies the activity of numerous proteins that reside at the mitochondria or act in the nucleus.Downstream molecules that are activated by JNK include c-Jun, ATF2, ELK1, SMAD4, p53and HSF1. The downstream molecules that are inhibited by JNK activation include NFAT4, NFATC1 and STAT3. By activating and inhibiting other small molecules in this way, JNK activity regulates several important cellular functions including cell growth,differentiation, survival and apoptosis.JNK1 is involved in apoptosis,neurodegeneration, cell differentiation andproliferation, inflammatory conditions and cytokine production mediated by AP-1 (activationUn Re gi st er edprotein 1) such as RANTES, IL-8 and GM-CSF.[5]Recently, JNK1 has been found to regulate Jun protein turnover by phosphorylation and activation of the ubiquitin ligase Itch.References^ a b Ip YT, Davis RJ (April 1998). "Signal transduction by the c-Jun N-terminal kinase (JNK)--from inflammation to development". Curr. Opin. Cell Biol. 10 (2): 205–19.doi:10.1016/S0955-0674(98)80143-9 (/10.1016%2FS0955-0674%2898%2980143-9) . PMID 9561845 (// /pubmed/9561845) .1.^ Waetzig V, Herdegen T (2005). "Context-specific inhibition of JNKs: overcoming the dilemma of protection and damage". Br. J. Pharmacol 26 (9): 455–61. doi:10.1016/j.tips.2005.07.006(/10.1016%2Fj.tips.2005.07.006) . PMID 16054242 (// /pubmed/16054242) .2.^ a b Bode AM, Dong Z (August 2007). "The Functional Contrariety of JNK"(///pmc/articles/PMC2832829/) . Mol. Carcinog. 46 (8): 591–8.doi:10.1002/mc.20348 (/10.1002%2Fmc.20348) . PMC 2832829(///pmc/articles/PMC2832829) . PMID 17538955 (// /pubmed/17538955) . ///pmc/articles/PMC2832829/. "The protein products of jnk1 and jnk2 are believed to be expressed in every cell and tissue type, whereas the JNK3protein is found primarily in brain and to a lesser extent in heart and testis"3.^ Vlahopoulos S, Zoumpourlis VC (August 2004). "JNK: a key modulator of intracellular signaling". Biochemistry Mosc. 69 (8): 844–54. doi:10.1023/B:BIRY .0000040215.02460.45(/10.1023%2FB%3ABIRY .0000040215.02460.45) . PMID 15377263(///pubmed/15377263) .4.^ Oltmanns U, Issa R, Sukkar MB, John M, Chung KF (July 2003). "Role of c-jun N-terminal kinase in the induced release of GM-CSF , RANTES and IL-8 from human airway smoothmuscle cells" (///pmc/articles/PMC1573939/) . Br. J. Pharmacol. 139 (6):1228–34. doi:10.1038/sj.bjp.0705345 (/10.1038%2Fsj.bjp.0705345) .PMC 1573939 (///pmc/articles/PMC1573939) . PMID 12871843(///pubmed/12871843) . ///pmc/articles /PMC1573939/.5.External linksJNK+Mitogen-Activated+Protein+Kinases (/cgi/mesh/2011/MB_cgi?mode=&term=JNK+Mitogen-Activated+Protein+Kinases) at the US National Library of Medicine Medical Subject Headings (MeSH)Getting a Handle on Cellular JNK (/?p=1026) (from Beaker Blog)MAP Kinase Resource (http://www.mapkinases.eu)Retrieved from "/w/index.php?title=C-Jun_N-terminal_kinases&oldid=541721429"Categories: Genes on chromosome 10Genes on chromosome 5Cell signaling Signal transduction EC 2.7.11This page was last modified on 2 March 2013 at 13:46.T ext is available under the Creative Commons Attribution-ShareAlike License;Un Re gi st er edadditional terms may apply. See T erms of Use for details.Wikipedia® is a registered trademark of the Wikimedia Foundation, Inc., a non-profit organization.deretsigeRnU。
拓普龙9卡AI服务器机箱规格书说明书

1.6 硬盘背板详解
结构示意图
5/7
深圳拓普龙科技有限公司 Shenzhen Toploong Technology Co.,Ltd.
机箱安装 1 片 12 盘位 12Gb 直连背板,尺寸为 2.5mm*425mm*77.5mm,支持 12 个 SAS/SATA 硬盘, 也可选配 4 个 U.2 NVME 接口,背板设计有 3 个 Mini-sas HD 接口,支持 12*SAS/SATA 硬盘或 8*SAS/SATA+4*SAS/SATA/NVME,背板供电采用 2 个方 8pin 和 3 个大 4pin 电源接口,供电稳定可靠。
深圳拓普龙科技有限公司 Shenzhen Toploong Technology Co.,Ltd.
目录
目 录 ……………………………………………………………………………………1 1.1 产品类型………………………………………………………………………………2 1.2 产品简介………………………………………………………………………………2 1.3 产品特点………………………………………………………………………………3 1.4 产品规格………………………………………………………………………………3 1.5 GPU 底板详解 ……………………………………………………………………4、5 1.6 背板详解 ………………………………………………………………………………6 1.7 选购配件 ………………………………………………………………………………7
标配
标配 选配 选配 标配
标配 标配 标配 标配 标配 选配 选配 选配
6/7
4/7
深圳拓普龙科技有限公司 Shenzhen Toploong Technology Co.,Ltd.
K-9可选配件,微维克斯硬度试验机剪子订单号型号及描述说明书
G
steel ball 1471N(150kgf) beryllium copper, phosphor bronze
H
1/8”
588.4N(60kgf) Bearing metal
E
diameter 980.7N(100kgf)
K
steel ball 1471N(150kgf)
L
1/4”
588.4N(60kgf) Plastics, lead
50x50mm travel stage
Dimensions: 4.92x4.92”(125x125mm) Minimum reading: 0.01mm 810-012
Clamping devices (Vises)
Vise Max. opening: 1.77”(45mm) Standard for the MH series. 810-016
Indenters
Order No. 19BAA061 19BAA058 19BAA062 19BAA059 19BAA060
Type Knoop Indenter Vickers Indenter Knoop Indenter Vickers Indenter Vickers Indenter
Model H, HM Standard Series H, HM Standard Series MVK-H2, H3, HM114 MVK-H2, H3, HM114 HV, AVK-C Series
Micro-Vickers/Vickers Hardness Testing Machine
Test Blocks
Order No. 64BAA173 64BAA174 64BAA175 64BAA176 64BAA177 64BAA178 64BAA179 64BAA180 64BAA181 64BAA182 64BAA183 64BAA184 64BAA185 64BAA186 64BAA187 64BAA188 64BAA189 64BAA190 64BAA191 64BAA192 64BAA193 64BAA194 64BAA195 64BAA196 64BAA197 64BAA198 64BAA199 64BAA200 64BAA201 64BAA202 64BAA203 64BAA204 64BAA205 64BAA206 64BAA207 64BAA208 64BAA209 64BAA210 64BAA211 64BAA212 64BAA213 64BAA214 64BAA215 64BAA216 64BAA217 64BAA218 64BAA219 64BAA220
ртфм 2007 - 2011 Ford Expedition 2011 Ford Explore

Page 1©2007 Whelen Engineering Company Inc.Form No.14105A (020911)For warranty information regarding this product, visit /warrantyA u t o m o t i v e : Installation Guide:Lightbar Roof Rack Mount 2007 - 2011 Ford Expedition2011 Ford Explorer®ENGINEERING COMPANY INC.Internet: Sales e-mail:*******************Customer Service e-mail:*******************51 Winthrop Road,Chester, Connecticut 06412-0684Phone: (860) 526-9504Set Screw (QTY 4)Stop Nut (Qty 6)Installation:IMPORTANT! The lightbar should be a minimum of 16" from any radio antennas!1.Remove the cover from roof rack (see photos), insert the 3 slide mounts into the track in the roof rack and replace the cover (Fig. 1).2.Snap the nylon retaining washers onto the slide mounts then secure the mounting bracket to the slide mounts with the supplied hardware.3.Insert the 3 slide mounts into the tracks on the vehicle roof rack then snap the “nylon retaining washers” onto the slide locks. This will hold them in place while you are doing the installation.4.Slide both mounting brackets into their track in the bottom of the lightbar base (Fig. 2) and secure them there with the supplied set screws.5.When you have positioned the mounting slides where you wish to mount the lightbar, secure the bracket to the slides using the supplied 14-20elastic stop nuts and 1/4” flat washers (Fig. 3) and installation is complete.©2007 Whelen Engineering Company Inc.Form No.14105A (020911)For warranty information regarding this product, visit /warrantyA u t o m o t i v e : Installation Guide:Lightbar Roof Rack Mount 2007 - 2011 Ford Expedition2011 Ford Explorer®ENGINEERING COMPANY INC.Internet: Sales e-mail:*******************Customer Service e-mail:*******************51 Winthrop Road,Chester, Connecticut 06412-0684Phone: (860) 526-9504Set Screw (QTY 4)Stop Nut (Qty 6)Installation:IMPORTANT! The lightbar should be a minimum of 16" from any radio antennas!1.Remove the cover from roof rack (see photos), insert the 3 slide mounts into the track in the roof rack and replace the cover (Fig. 1).2.Snap the nylon retaining washers onto the slide mounts then secure the mounting bracket to the slide mounts with the supplied hardware.3.Insert the 3 slide mounts into the tracks on the vehicle roof rack then snap the “nylon retaining washers” onto the slide locks. This will hold them in place while you are doing the installation.4.Slide both mounting brackets into their track in the bottom of the lightbar base (Fig. 2) and secure them there with the supplied set screws.5.When you have positioned the mounting slides where you wish to mount the lightbar, secure the bracket to the slides using the supplied 14-20elastic stop nuts and 1/4” flat washers (Fig. 3) and installation is complete.。
CIK细胞的特点及其在肿瘤生物治疗中的作用

2. 2 _ 干扰素 _干扰素 ( IFN_ ) 是由 CD4+ 或 CD8+ 细胞产生的同源二聚体 糖蛋白, 可通过多种途径直接或间接发挥抗肿瘤作用, 增强自然 杀伤细胞 ( NK) 、巨噬细胞 ( M #) 以及细胞毒性 T 细胞 ( CTL) 的活 性。在诱导 CIK 细胞形成过程中加入 IFN_ 可降低 IL_2 用量, 同时 IFN_ 加入 的顺序与细胞毒活 性密切相关, 先加入 IFN_ 后加入 IL_2 可提高细胞毒活性, 这是因为先加入 IFN_ , 可促使 PBMC 上 IL_2 受体数量增加, 从而有效地激活效应细胞。 2. 3 白细胞介素 _1 白细胞介素 _1( IL_1) 主要由激活的单核巨噬细胞产生。 IL_1 的 体内抗肿瘤作用机 制是增强 T 细胞 及增加肿瘤浸润淋 巴细胞 的 数量和功 能, IL_1 可以介 导外周血 单个核细胞 上表达 IL_2 受 体, 与 IFN_ 、CD3McAb 合用时可以明显提高 CIK 的细胞毒效应, 但 IL_1 对 CIK 细胞扩增不起作用 [3]。 2. 4 白细胞介素 _2 白细胞介素 _2 ( IL_2) 是 T 淋巴细胞分泌的一种细胞因子 ( 由 133 个氨基酸组成的多肽, 分子量约为 15 kD) 。它是人体免疫应 答的核心物质, 具有免疫增强、抗肿瘤和抗感染等作用。临床上已 用于肾癌、黑色素瘤、淋巴瘤、肺癌、胃癌、乳腺癌、卵巢癌、肠癌、 膀胱癌、头颈部肿瘤、白血病及癌性胸腹水等病症的治疗, 与放 疗、化疗比较, IL_2 对正常细胞毒副作用轻微, 能减轻肿瘤患者的 疼痛, 提高其生活质量。此外结核病、肝炎、爱滋病、性病、成瘾性 吸毒者及其它免疫功能低下病人均可使用 IL_2。CIK 细胞的体外 增殖需要外源性细胞因子如 IL_2、 IL_7、IL_12 等的辅助, 外源性 IL_2、IL_7、IL_12 可以显著促进淋巴细胞的生长, 而对 CIK 细胞的 细胞毒作用无影响 [4]。 CIK 细胞是多种细胞因子共同诱导培养的细胞, 多种细胞因 子的作用是相互协同, 单一因子对效应细胞的增殖及细胞毒活性 无作用或者小于多种细胞因子的联合作用, 联合作用的机制是最 终共同激活静止 T 细胞, 提高细胞表达 IL_2 受体和产生 IL_2 的能 力, 启动自分泌途径 IL_2 依赖的 T 细胞激活反应。
Philips 电子刮胡刀 S9031 21 产品说明书
Shaver series 9000V-Track Precision Blades8-directionContourDetectHeadsSmartClick precision trimmerS9031/21Perfection in every passCuts up to 20% more hair* in a single passThe Shaver 9000 is our most advanced shaver yet. The unique contour detecttechnology offers exceptional coverage over every contour of your face, and theV-Track system guides hairs into the best cutting position for the closest results.Designed for perfectionBlades perfectly guide hairs into position for a close shaveHeads flex in 8 different directions for a superb resultA comfortable shaveGet a comfortable dry or refreshing wet shave with AquatecEasy to useIntuitive icons make the functions easy to use50 minutes cordless shaving after a one-hour chargeShaver can be rinsed clean under the tapWith 2 year guaranteeGet the most of your shaverClick-on trimmer for perfect mustache and sideburn trimmingHighlightsV-Track precision blade systemGet the prefect close shave. The V-TrackPrecision Blades gently positions each hair in the best cutting position, even the flat laying and different length of hairs. Cuts 30% closer in less strokes leaving your skin in great condition.8-direction ContourDetectHeadsFollow every contour of your face and neck with 8-directional ContourDetect heads. You'll catch 20% more hairs with every pass.Resulting in an extremely close, smooth shave.Aquatec Wet & DryChoose how you prefer to shave. With the Aquatec Wet & Dry seal, you can opt for a quick yet comfortable dry shave. Or you can shave wet – with gel or foam – even under the shower.SmartClick precision trimmerClick on our skin-friendly Precision Trimmer to finish your look. It’s ideal for maintaining your mustache and trimming your sideburns.3 level LED displayThe intuitive display shows relevantinformation, enabling you to get the best performance out of your shaver: - 3-level battery and travel lock indicators - Cleaning Indicator - Battery Low Indicator -Replacement Head Indicator50 minutes of cordless shavingOur advanced charging system gives you twoconvenient options: charge for one hour and you’ll get 50 minutes of running time, or do a quick charge for one full shave. All 9000Series Shavers contain a powerful energy-efficient, long-lasting lithium-ion battery. They are designed to operate only in cordless mode,to ensure you’ll always be safe when shaving with water, even under the shower.Fully washable shaverSimply open the shaver head to rinse itthoroughly under the tap.iF DESIGN AWARD 2015Shaver series 9000Precision, control and maneuverability are the defining features of the 9000 shaver. The V-Track system guides hairs into the best cutting position for the closest result, while fully flexible eightdirectional heads catch more hairs for a clean shave in fewer strokes. The “AquaTec Wet &Dry” seal of this shaver allows for acomfortable dry shave or a refreshing wet shave, using a shaving gel or foam for extraskin comfort.Philips Green LogoPhilips Green Products can reduce costs,energy consumption and CO2 emissions. How?They offer a significant environmentalimprovement in one or more of the Philips Green Focal Areas – Energy efficiency,Packaging, Hazardous substances, Weight,Recycling and disposal and Lifetime reliability.SpecificationsShaving PerformanceShaving system: V-Track Precision Blade System, Super Lift & Cut Action Contour following: 8-direction ContourDetectHeadsSkinComfort: AquaTec Wet & Dry AccessoriesSmartClick: Precision trimmerPouch: Travel pouch Ease of useDisplay: 3 level battery indicator, Cleaningindicator, Battery low indicator, Replaceshaving heads indicator, Travel lock indicatorCleaning: Fully washableDesignColor: Black SatinHandle: Ergonomic grip & handlingPowerBattery Type: Lithium-ionRun time: 50 min / 17 shavesCharging: 1 hour full charge, Quick charge for 1shaveAutomatic voltage: 100-240 VMax power consumption: 9 WStand-by power: 0.1 WService2-year guaranteeReplacement head: Replace every 2 yrs withSH90* Cuts up to 20% more hair - versus SensoTouch© 2019 Koninklijke Philips N.V.All Rights reserved.Specifications are subject to change without notice. Trademarks are the property of Koninklijke Philips N.V. or their respective owners.Issue date 2019‑08‑01 Version: 4.0.1。
CIK细胞制备SOP

CIK细胞制备操作SOP一、样本接收1.观察运输箱的温度是否符合要求,采血管或采血袋有无渗漏。
2.查看采血管或采血袋上客户信息是否与交接表一致。
3.样本外表面喷酒精擦拭消毒,并粘贴样本编码,填写样本接收记录,传入洁净区准备制备。
二、PBMC分离、接种1.准备耗材试剂:离心管(50ml、15ml)、离心管架、移液管(25ml、10ml)、包被好的T75、无尘布、EP管、枪头(1000uml)无菌棉球、封口膜、肝素钠管、5ml注射器、50ml注射器针头、止血钳、废液缸(放入安全柜中照紫外)、DPBS、75%酒精、生理盐水、淋巴细胞分离液、血培养皿、CIK细胞培养基常温复温、CIK细胞诱导激活因子常温复温。
2.仪器设备准备及预热:电动移液枪、1000uml手动移液枪、生物安全柜(照紫外30min、通风10min)、离心机开机、水浴锅37℃、全血分析仪开机预热、倒置显微镜、热合仪开机预热(若样本通过采血袋运输)、二氧化碳培养箱(37℃5%CO2)。
3.提前包被T75瓶,2-8℃冰箱过夜或37℃培养箱不少于2H。
包被方法:使用25ml的移液管吸取19mlDPBS加入到T75瓶中,使用1000uml 的移液枪吸取包被因子(0.5mlLT-F-01、0.5mlLT-F-02)加入到该T75瓶中,并吸取瓶中少许DPBS清洗装有因子的西林瓶,在将清洗液加入到T75瓶中,然后从中吸取10ml包被液到另一T75瓶中,一同放入到2-8℃冰箱过夜或37℃培养箱不少于2H。
注意事项:①检查培养瓶的完整性,检查培养瓶是否出现破损。
②包装袋中剩余培养瓶拧紧盖子,袋子重新密封。
③培养瓶避光暂存。
④包被液混匀充分,平铺在培养瓶的培养面上。
⑤不要将包被液碰到培养瓶盖上。
4.分离流程1)将75%酒精喷到台面,用无尘布擦拭台面,包括侧面及玻璃。
2)将样本喷酒精擦拭,放入生物安全柜中,电动移液枪、移液管、DPBS、生理盐水、血培养皿、离心管架喷酒精擦拭,放入生物安全柜中,无菌棉球放入到广口瓶中加75%酒精放入生物安全柜中。
FURUNO GP-1670 1670F 产品说明书

GP-1670/1670F features• Bright 800 cd/m2 LCD gives excellent readability even in direct sunlight • The LCD and AR glass are bonded together to ensure no fogging issues • Clear visibility even when wearing polarized sunglasses• Internal GPS antenna for simple and easy installation• Standard C-Map 4D chart available on SD card• RotoKey TM revolving menu and familiar point-and-click operation• Internal memory: Waypoint/Track 30,000 points, Route 1,000 routesGP-1670F features• Equipped with FURUNO’s latest digital fish finder technology- Bottom Discrimination Function* * Thru-hull transom transducer mount required.- ACCU-FISH TM — A unique fish size analyzer• Post-processing Gain Control applied to all echoes displayed on the screen • White Line function — Discriminates fish lying near the bottomC-MAP 4DDigital Charts that grow with your boating needs. Dual range chart displayPlotter + Fish Finder* (with ACCU-FISH and Bottom Discrimination function ON) *GP-1670F onlyModelModel5.7" GPS/WAAS CHART PLOTTERSPECIFICATIONS SUBJECT TO CHANGE WITHOUT NOTICEPLEASE READ MANUAL BEFORE USING THE UNIT12093SS Printed in JapanCatalogue No. N-872aAll brand and product names are registered trademarks, trademarks or service marks of their respective holders.GPA-017/017S (Option)Internal GPS Antennaor orScreen Size5.7"Screen Resolution 640 x 480 pixels (VGA)Screen Brightness 800 cd/m 2 (typical)LanguageEnglish (US & UK), French, Spanish, German, Italian, Portuguese, Danish, Swedish, Norwegian, Finnish, Greek, Japanese, Chinese, Russian, Thai, Vietnamese,Polish, Bahase Malaysia, Bahasa Indonesia Display ModesCourse plot, Nav Data, Instruments*, Engine monitor*, Anemometer*, Fuel level gauge*, GPS status,Fish nder** *External sensors required **GP-1670F onlyMemory Capacity30,000 points for ship's track and wayponts 1,000 planned routes (Max. 50 points per route)5,000 quickpoints InterfaceCAN bus: 1 portMini USB*: 1 port (2.0) *for maintenance onlySD card slot: 1 slot (up to 32 GB)ChartC-MAP 4DOther Functions AIS target display* *AIS sensor requiredTemperature -15˚C to +55˚CWaterproo ng IP56Power Supply12-24 VDCConsumption CurrentGP-1670: 0.5 - 0.3 AGP-1670F: 0.9 - 0.5 A (When 600 W transducer used)GPS / WAAS Receive TypeGPS: 50 channels WAAS: 1 channelReceiving Frequency L1 (1575.42 MHz)Accuracy Internal antenna : GPS 10 m Max / MSAS 7 m MaxExternal antenna*: GPS 10 m Max / MSAS 7 m Max*GPA-017 (Option)Time to First FIX 90 s (cold start)Tracking Velocity 999 KnTransmission 600 W or 1 kW* *Matching box MB-1100 required for certain transducers Display Range 5 - 1,200 m, shift: 0 - 1,200 m Extension Mode ACCU-FISH*, Auto (Fishing/Cruising/Manual), A-Scope,Marker Zoom, Bottom Zoom, Bottom Lock, Bottom Discrimination*Picture Advance 7 steps: 2/1, 1/1, 1/2, 1/4, 1/8, 1/16, stop*In some instances, bottom component or sh size indicated on the GP-1670F may differ from its actual size or structure. Please carefully read the operation manual prior to using this feature. Equipment List Display unit GP-1670/1670F, Installation materials and standard spare partsOptions • GPS antenna GPA-017/017S • Antenna cable assembly • NMEA2000 heavy cable assembly • Mast mounting kit • Junction box • In-line Terminator • NMEA2000interface unit • NMEA data converter • Waterproof cap • Termination Resistor• NMEA2000 light cable assembly • Cable assembly • Mini/micro T-connector• L-angle mounting base • Handrail mounting base • Rectifier • T-connector • Inner hull mounting kit* • Matching box* • Speed/Temperature sensor*• Temperature sensor* *for GP-1670F only Transducer Options (for GP-1670F only)600 W 1 kW* *Matching box MB-1100 required for certain transducers • 520-5MSD (Bronze) • 50/200-1T • 50B-6B • 520-5PSD (Plastic) • 50/200-12M • 200B-5S • 525-5PWD (Plastic) • 50B-6ViewableConventional GP-1670Easy Replacement from legacy modelsNMEA DATA CONVERTER < IF-NMEA2K2 >IF-NMEA2K2Furuno’s NMEA DATA CONVERTER is a compact interface unit, that converts NMEA 0183 data into CAN bus/NMEA2000 data and vice-versa. Control data such as Heading and Rate-of Turn can be translated in high speed conversion rate.Black out。
cik细胞制备流程

cik细胞制备流程下载温馨提示:该文档是我店铺精心编制而成,希望大家下载以后,能够帮助大家解决实际的问题。
文档下载后可定制随意修改,请根据实际需要进行相应的调整和使用,谢谢!并且,本店铺为大家提供各种各样类型的实用资料,如教育随笔、日记赏析、句子摘抄、古诗大全、经典美文、话题作文、工作总结、词语解析、文案摘录、其他资料等等,如想了解不同资料格式和写法,敬请关注!Download tips: This document is carefully compiled by theeditor. I hope that after you download them,they can help yousolve practical problems. The document can be customized andmodified after downloading,please adjust and use it according toactual needs, thank you!In addition, our shop provides you with various types ofpractical materials,such as educational essays, diaryappreciation,sentence excerpts,ancient poems,classicarticles,topic composition,work summary,word parsing,copy excerpts,other materials and so on,want to know different data formats andwriting methods,please pay attention!CIK 细胞制备流程1. 外周血单核细胞 (PBMC) 分离从健康供者或患者中采集外周血。
CIK细胞培养_CIK细胞扩增_CIK细胞使用说明

GMP级CIK细胞扩增试剂盒
CIK Cell Expansion Kit
作用机理:
同立海源生物GMP级CIK细胞扩增试剂盒操作简便,适用于人CIK细胞培养,细胞总量大、纯度高、存活率高、增殖快,同时效应细胞的表型也较为稳定。
规格参数:
货号:AS-08 规格:套装(CIK试剂A一支、CIK试剂B一支、Human IL-2两支)
产品特点:
1、操作简便,表型稳定;
2、无血清、无动物源成分;
3、ISO9001、ISO13485质量体系认证;
4、GMP制药级生产环境,批次稳定性高;
生产厂家:同立海源生物
标准操作规程:
实验案例:
案例一:人外周血50mL
案例二:人外周血50mL
案例三:人外周血50mL
案例四:人外周血50mL
CIK细胞培养试剂推荐:
CD3单抗、重组人干扰素γ、重组人IL-2、CD28单抗、重组人IL-1α、重组人纤维连接蛋白FN。
CIK培养技术流程

C I K培养技术流程SANY GROUP system office room 【SANYUA16H-SANYHUASANYUA8Q8-C I K细胞免疫治疗方案(一)外周血单个核细胞(PBMC)分离方法1.取患者抗凝全血50ml,加入等体积PBS,充分混匀。
2.分装到4个50ml提前各加入12.5ml淋巴细胞分离液(Ficoll)的离心管中。
3.700G,室温离心20min,缓升缓降(withoutbreaking)。
4.抽取白膜细胞层,加入到装有5mlRPMI1640的50ml离心管中。
5.加入45mlRPMI1640,260G,4℃离心10min,废弃上清液。
6.加入50mlRPMI1640,180G,4℃离心10min,废弃上清液。
7.加入50mlRPMI1640,110G,4℃离心10min,废弃上清液。
8.加入30ml淋巴细胞无血清培养基(Quantum007),备用。
注:Eppendorfcentrifuge5810:水平转子AT-4-62,离心半径16cm。
(二)CIK细胞的培养及扩增1.患者50ml全血经淋巴细胞分离液分离,获得约5×107个单个核细胞。
2.将细胞悬浮于30ml淋巴细胞无血清培养液(Quantum007),细胞浓度为约1.5-2.0×106/ml,加入IFNγ2000IU/ml,将培养瓶置于37℃,5%CO2培养箱条件下培养。
3.培养24小时后(也可在培养开始),加入IL-21000IU/ml,单克隆抗CD3抗体(MabCD3)100ng/ml。
4.在3-4天,加入30ml新鲜淋巴细胞无血清培养液(总体积60ml),并加入IL-21000IU/ml。
5.在5-6天,加入60ml新鲜淋巴细胞无血清培养液(总体积120ml),并加入IL-21000IU/ml。
6.在7-14天,每天根据培养基颜色变化,加入新鲜淋巴细胞无血清培养液将培养体积加倍,并相应加入IL-21000IU/ml,可以多个培养瓶,也可加入培养袋培养。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Efficient Algorithms for Approximate Member Extraction Using Signature-based Inverted ListsJiaheng Lu Jialong Han Xiaofeng MengSchool of Information, Renmin University of China Beijing 100872, China Key Laboratory of Data Engineering and Knowledge Engineering, Ministry of Education, China jiahenglu@ jialonghan@ xfmeng@ABSTRACTWe study the problem of approximate membership extraction (AME), i.e., how to efficiently extract substrings in a text document that approximately match some strings in a given dictionary. This problem is important in a variety of applications such as named entity recognition and data cleaning. We solve this problem in two steps. In the first step, for each substring in the text, we filter away the strings in the dictionary that are very different from the substring. In the second step, each candidate string is verified to decide whether the substring should be extracted. We develop an incremental algorithm using signature-based inverted lists to minimize the duplicate list-scan operations of overlapping windows in the text. Our experimental study of the proposed algorithms on real and synthetic datasets showed that our solutions significantly outperform existing methods in the literature. Categories and Subject DescriptorsH.2.4 [Database Management]: Systems – Textual Databases General TermsAlgorithmsKeywordsApproximate Member Extraction, Filtration-verification, Approximate string matching, Incremental Computation1.INTRODUCTIONIn this paper we study the problem of finding substrings in a text document M that approximately match (e.g. having similarity scores above a given thresholdδ) some strings in a given dictionary R of strings. This problem, called AME (short for A pproximate M ember E xtraction), arises in many applications, as illustrated by the following examples.Named Entity Recognition: With a given document, we want to locate pre-defined entities such as person names, conference names, and company names. We want this extraction to be appro- ximate, i.e., we allow slight mismatches in the substrings. For instance, as shown in Figure 1, suppose we have a collection of conference names, such as “ACM SIGMOD” and “CIKM Conference.”Figure 1. Approximate member extractionWe want to extract all conference names from a given document. We want to find matches such as “CIKM 2009 Conference” and “CIKM International Conference”, even though they do not match the string “CIKM Conference” in the dictionary exactly.Data Cleaning: Documents in many applications could be “dirty” when it contains inconsistencies. Often we need to clean the inconsistencies. We need to perform data cleaning and integration by identifying the dirty words based on an existing dictionary.One naive way to solve the AME problem is to enumerate each substring m of M and check if m matches strings in R approximately. Several algorithms have been proposed for doing the checking efficiently using two steps. In the first step, we filter the dictionary strings that are very different from m. In the second step, we compute the similarity between the remaining candidate strings and the string m to verify its approximate membership.There are two methods to do the filtration in the first step. One is based on an inverted index on the dictionary R. In this method, strings are regarded as collections of tokens. For each token, the index stores the ids of the strings that include this token. For a given string m, we can find candidate similar strings in the dictionary by accessing the lists of the tokens in m and finding those string ids that have enough occurrences on the lists. The second method is based on signatures generation [1, 4]. This method focuses on exploring signature schemes that convert a string to a set of hash codes and filtering irrelevant strings using their signatures.Recently, researchers have been trying to combine these two methods. For instance, Wang et al. propose a method called NGPP [13] to solve the AME problem by assuming the edit-distance similarity function. They shift and extend the partitions of dictionary strings to obtain an inverted index on all the partition variations (an implicit signature). Then by generating the neighborhood of partitions of document substrings and probing the index, the algorithm filters most irrelevant strings and performs verifications for the remaining strings. Chakrabarti et al. [2] studied how to solve the AME problem with other similarity scorePermission to make digital or hard copies of all or part of this work for personal or classroom use is granted without fee provided that copies are not made or distributed for profit or commercial advantage and that copies bear this notice and the full citation on the first page. To copy otherwise, or republish, to post on servers or to redistribute to lists, requires prior specific permission and/or a fee.CIKM’09, November 2-6, 2009, Hong Kong, China.Copyright 2009 ACM 978-1-60558-512-3/09/11...$10.00.functions such as Jaccard coefficient using a method called ISH (Inverted Signature-based Hashtable), which encodes a string and its signatures into a 0-1 matrix. After generating a 0-1 matrix by computing the “bitwise-or” of all small matrices encoded with dictionary strings, ISH converts the problem of searching possible evidence into finding a certain 0-1 submatrix in the big matrix. Our work in this paper is motivated by the following observations: (1) The ISH method generates signatures for strings and prune strings that share signatures with a total weight under a certain lower bound (see Para 8, Page 4 of [2]). Unfortunately, the lower bound may be too high, causing false negatives (see Example 1 for a counter-example). (2) The filtration processing of ISH involves an NP-Complete problem requiring finding solid submatrix (containing only 1) from a given 0-1 matrix under certain constraints. We give the reduction proof in this paper from an NP-Complete problem: Balanced Complete Bipartite Subgraph (see Appendix). Due to the intrinsic complexity, [2] solves this problem by a simple heuristics, which significantly increases the number of false positives, and thus incurs numerous I/Os in the verification phase and deteriorates overall performance. But our research in this paper shows that we can avoid the NP-complete complexity to efficiently solve AME problem by adopting a novel index structure, which effectively controls the number of false positives and consequently improves the overall performance. (3) NGPP[13] and ISH[2] both require that δis provided when preprocessing the data and generating an index for the filter. Once the threshold is fixed, these filters no longer support queries with thresholds other than δunless the index is re-generated.E XAMPLE 1. (Example to illustrate the false negative in [2]) Under Jaccard similarity measurement, with weight assignment {(a, 6), (b, 3.52), (c, 3.51), (d, 3.49), (e, 3.48), (f, 1)}, string r=”bcde” matches m=“abef” under the similarity threshold δ=1/3. But from the following table we see that [2] incorrectly determines that m and r are impossible to match under the threshold 1/3, though their real similarity is 1/3. StringSignatures (k=3)τ valueSharedSignaturesLower BoundRequired byISH Filter r =“bcde” {b,c,d} τ(r)=1.187 m =”abef” {a,b,e} τ(m)=3.67{(b, 3.52)}≥3.67We summarize our contributions as follows:y We adopt the prefix filtering technique [4], and propose new theorems which are all strictly proved and well applied as filtering conditions in our method. These theorems convert approximate matching to prefix signatures sharing, and give a tighter bound. More importantly, this filtering technique brings no false negatives. y We utilize an inverted-list-based filtering index SIL andpropose corresponding algorithm called EvSCAN. By performing inverted list scanning instead of introducing matrix-based combinatorial problem, EvSCAN naturally avoids solving NP-Complete problem. We also apply incremental optimizations on EvSCAN and propose EvITER, which effectively reduces the duplicate list-scanning operations when the substring window shifts over a large document M. Compared with prior solutions, our method produces far less false evidences, thus achieves betterfiltration-verification balance and consequently improves the overall performance significantly.y We modify our SIL to answer queries with dynamic similaritythresholds. Specifically, once initialized with a lower bound δ0, our filter works for any query with a threshold δ≥δ0. However previous filters only allow static similarity threshold and need to be re-initialized (i.e., the indices in filter has to be generated again) once the query threshold is changed. y We provide detailed and accurate experimental results tosupport our argument. We show that SIL is significantly efficient than ISH, both in filtering power and overall running time, and explain the intrinsic reason by analyzing their runtime statistics in details. We also compare EvSCAN and EvITER under various situations, and show that our incremental optimization is effective.The following sections are organized as follows:Section 2 formally defines AME and gives some preliminaries. Section 3 complements the theory of prefix filtering and applies them to build the SIL structure and EvSCAN algorithm. Section 4 introduces the EvITER incremental algorithm. Section 5 shows how to support dynamic thresholds. Section 6 reports our experimental results. Section 7 discusses related work. Section 8 is the conclusion of our study.2. PRELIMINARIES AND PROBLEM STATEMENT2.1 Some Notations and Problem StatementIn this paper, we use the letter “t” to denote a token, and the other lower-case letters are used for strings. We regard strings as sets of tokens. For any token t, we denote wt(t) as the weight of t (e.g. IDF weight), and for any string s, we define wt(s) as ∑∈st t wt )(, i.e. the sum of weights of all tokens in s. Based onthe above notations we define Jaccard similarity of any strings s 1 and s2 as:)()(),(212121s s wt s s wt s s J ∪∩=. E XAMPLE 2. (Reusing the string and weight configuration of Example 1)Under weighted Jaccard similarity, the two strings m=“abef”, r=”bcde” have a similarity of wt({b, e})/wt({a, b, c, d, e, f})=(3.52+3.48)/(6+3.52+3.51+3.49+3.48+1)=1/3.With all notations introduced, we present the formal description of AME as follow:is up to a length threshold L, so we may as well require that |m| ≤ L. Here any extracted m is called approximate member of R, and corresponding evidence for r in R. Note that our algorithms we present later can handle any similarity function that satisfies the following properties:y Sim (m,r) is symmetric, i.e., Sim (m,r) = Sim (r,m).y )()(),(m wt r m wt r m Sim ∩≤.(Symmetrically we have )()(),(r wt r m wt r m Sim ∩≤.)The first property is natural, and the second is also shared by many similarity functions [2]. For example, for any m and r, we have)()()()(),(m wt r m wt r m wt r m wt r m J ∩≤∪∩=because wt(m ∪r) ≥wt(m), so the Jaccard similarity is capable of serving as asimilarity function in our discussion.2.2 The Filtration-Verification FrameworkTo efficiently solve AME and other related problems, researchers have been designing methods following two phases of filtration and verification [1, 2, 4, 7, 13]. In the AME problem, employing this framework usually requires building an indexing structure for the dictionary R. Recall that for each approximate member m extracted, we define the string r in R that is similar enough with m to be m’s evidence. Thus, the task of extracting all approximate members from M can be simply reduced to determining whether there exists any evidence for each substring of M, and filtration-verification is actually referred to as evidence filtration and evidence verification.Generally, our foundation of filtration is based on some necessary condition (denoted as NC) of our matching criterion Sim ≥δ, that is, if some candidate evidence is real evidence, it must satisfy NC. With the dictionary R given offline, we build an index that quickly recommends for a query m ALL potential evidence that meets NC, so that true evidence is never missed. Then the evidence is verified against the actual matching criterion to determine whether the string m is a true approximate member. Note that NC plays a key role in our whole framework. It ensures the correctness of the whole algorithm. Moreover, it determines how balanced our framework is. We can evaluate it through:y How powerful is it? That is, does it eliminate as much falseevidence as possible? y Is it easy-going? That is, can we build a quick index to test itat low cost? There is a tradeoff in the cost between two phases of filtration-verification. For example, with all dictionary strings being potentially possible evidence, the most easy-going filtration approach for them is obviously “no filtration”, which leads to a brute-force method of scanning the whole dictionary. On the other hand, if we try to make our filtration the most powerful, i.e., it produces no false positive and achieves the smallest verification time; it will be expensive to perform such filtering. As a matter of fact, we need to obtain a beneficial compromise between two phases. In the following sections, we will intentionally highlight this issue through our theoretical and experimental analysis.2.3 The K-Signature SchemeThe k-signature scheme is first demonstrated by Chakrabarti et al. in [2]. It is an extension of the prefix signature idea [1]. Here we briefly introduce some key definitions as follows:D EFINITION 1. For a given string s and similarity threshold δ, we sort all its tokens by their weight in descending order (if two tokens appear with the same weight, we sort them lexicographically), and choose the first few tokens to get a subset Sig(s), such that τ(s)=wt(Sig(s))-(1-δ)wt(s)≥0. We call Sig(s) a prefix signature set of string s. For convenience we call it signature set from now on.E XAMPLE 3. (Reusing the string and weight configuration of Example 1) Let δ=0.6, then {a,b} is a signature set of m because τ(m)= wt({a,b})-(1-0.6)*wt({a,b,e,f})=9.52-5.6=3.92≥0. Based on its definition, the following is some facts about prefix signature set:y For any string s, Sig(s) always exists because we can letSig(s)=s, thus making τ(s)=wt(s)-(1-δ)wt(s)=δ*wt(s)≥0, i.e., we choose itself to be its signature set. y A string may have more than one signature set with differentsizes. For instance in Example 3, we see that m has another different signature set {a,b} besides itself. One way to ensure the uniqueness of signature set is to use a parameter k to determine which set we choose for a string s, where k is a positive integer. We call this unique signature set k-signature set, denoted as Sig k (s). When k is fixed, we select Sig k (s) among all available signature sets as follows:y If all signature sets’ sizes are bigger than k, choose thesmallest one. y Else if there is any signature set whose size is exactly k,choose it. y Else, choose the largest one, i.e. the string itself.In some special case, when we set k=1, we call the derived signature scheme as min-signature scheme. When k is set to be ∞, we will get the string itself to be its signature set.E XAMPLE 4. (Following the configurations of Example 3) We list the different Sig(m) under different k settings as below:K 1 2 3 4 5 Sig k (m) {a} {a,b} {a,b,e} {a,b,e,f} {a,b,e,f}Table 1. Signature sets controlled by KNote that parameter k has nothing to do with the requirement of signature set, so we may randomly set k for our filter, and even choose different k for different strings. As a matter of fact, if we regard signature set as a compression of information in strings, then k is the parameter for global compression rate tuning. That is, k only influences the performance of our filter. We will further discuss its role in Section 3.3.With the above description, we find that for any string s, the signature set is actually controlled by δand k, thus should be written as Sig k (s,δ). In fact, the parameter k is fixed to SIL and before Section 5 we also consider that δis static, so when the context is clear, we still use Sig(s) to denote the signature set of s. In Section 5, we use Sig(s,δ) because we start discussing how to support dynamic δduring query processing.3. FILTRATION VIA SIGNATURE-BASED INVERTED LISTSIn this subsection, we show some nice properties about the signature set which will be used in our algorithms. For instance, ifString m and r meet the matching condition Sim(m,r) ≥δ, m must contain at least one of r’s signatures. This is apparently a necessary condition for matching and can be utilized to build a filter. In the following discussion, we’ll further explore the property of signature sets and show that there are better filtering conditions.3.1 The Property of Signature SetsL EMMA 1. For any string s and its selected signatures, we useminsigwt(s) = )}({min )(t wt s sig t ∈ to denote the smallest weight of all s’s signature tokens. Then for any string s:A token t ∈Sig(s) if t ∈s and wt(t) ≥minsigwt(s).This is apparent because the selected signature tokens must have larger weight than unselected tokens.L EMMA 2. (P ROPERTY O F P REFIX S IGNATURES ). For any string m and r, if minsigwt(m) ≥ minsigwt(r), then wt(Sig(m)∩Sig(r)) ≥ wt(Sig(m))- wt(m-r). Here m-r refers to the minus set of m and r.Proof : We transform it to an equivalent form as wt(m-r) ≥ wt(Sig(m)-Sig(r)), and prove this by showing that Sig(m)-Sig(r) is a subset of m-r .For any t ∈Sig(m)-Sig(r) , we have t ∈Sig(m) , so t ∈m .Now we prove that t ∉r. Suppose that t ∈r. because t ∈Sig(m), we know that wt(t) ≥minsigwt(m) ≥ minsigwt(r) . From t ∈r , wt(t) ≥minsigwt(r) and Lemma 1, we conclude that t ∈Sig(r). This is inconsistent with the fact that t ∈Sig(m)-Sig(r) . So t ∉r .From t ∉r and t ∈m , it’s obvious t ∈m-r. So Sig(m)-Sig(r) is a subset of m-r , easily leading to the result that wt(m-r) ≥ wt(Sig(m)-Sig(r)) . ▉ In Lemma 2, we illustrate the signature set overlapping relationship between matching strings. Intuitively this inequality condition is tighter than other conditions proposed in [1], and we believe this property is also useful in other researches involving prefix signatures. However, the set minus operator seems costly to handle, so we need to make this condition more easy-going. We solve this by introducing Theorem 1 as follow:T HEOREM 1 (F ILTERING C ONDITION ). For any m and r that satisfy Sim(m,r) ≥δ, wt(Sig(m)∩Sig(r)) ≥ min{τ(m),τ(r) }. Proof: If minsigwt(m)) ≥ minsigwt(r), according to Lemma 2, we have wt(Sig(m)∩Sig(r)) ≥ wt(Sig(m))- wt(m-m ∩r)= wt(Sig(m))- wt(m)+wt(m ∩r) ≥wt(Sig(m))- wt(m)+δ*wt(m ∪r) ≥wt(Sig(m))- wt(m)+δ*wt(m)= wt(Sig(m))-(1-δ)wt(m)=τ(m) ≥ min{τ(m),τ(r) }.If minsigwt(m) ≤ minsigwt(r), based on the symmetry of Sim() we have the same result. So in conclusion we have wt(Sig(m)∩ Sig(r)) ≥ min{τ(m),τ(r) }. ▉ E XAMPLE 5. (Following the configurations of Example 3) Suppose k=2, we have Sig(m)={a,b},τ(m)= 3.92 and Sig(r) ={b,c},τ(r)=(3.52+3.51)-0.4*14=1.43. So wt(Sig(m)∩Sig(r))= wt({b})=3.52 ≥ min{τ(m),τ(r) } = min{3.92, 1.43}=1.43. We obtain the foundation of filtration phase of SIL so far. It’s easy to discover that when the threshold τ(r) is computed offline, this filtering condition only involves the signature set of all strings,indicating the fact that the time and space requirement of our filter is tightly related to the average signature set size of all strings in the dictionary R, which is controlled by the parameter k. Moreover, different k provides different filtering conditions. Among them we need to decide which one to choose.3.2 Filtration via SILSince for any matched m and r, their signature sets overlaps, it’s easy to come up with the idea of building an inverted index structure for the dictionary R, and filtering by merging inverted lists and accumulating weights. After this index is built up offline, by visiting list[t], we can quickly retrieve for any token t a list of rid of all r in R, who contains token t as a signature. Due to the fact that these lists only involves signatures of all strings in R, we call this index SIL in short for signature-based inverted lists , and Algorithm 1 below shows the method to generate an SIL index. E XAMPLE 6. Suppose we have a dictionary R={r[1]=“SIL’s filtering power”, r[2]=”the power of filtering by SIL”}, the weight of each tokens are {<SIL’s, 5>, <filtering, 4>, <power, 3.5>, <SIL, 3>, <by 2>, <the, 1>, <of, 1>}. We set k=1 and δ=0.55, then we have each strings’ signature set in Table 2 and the SIL built as Figure 2.rid String Signature Set 1 “SIL’s filtering power” {“SIL’s”,”filtering”}2”the power of filtering by SIL”{“filtering”,”power”} Table 2. Signature sets of R’s stringsSignature →String ridsridwt(r)τ(r)“SIL’s” → (1) 1 12.5 3.375 “filtering” → (1), (2) 2 14.5 0.975 “power”→ (2)Figure 2. SIL and additional information for dictionary R When a string m’s membership needs to be checked, wesimply compute the signature set of m, denoted as {t 1,t 2…t n }. Then we scan all n lists that is indexed by list[t 1], list[t 2]…list[t n ], while aggregating the weight of t i to all rid whose record contains t i as one of its signature. To record the aggregated weight, we may use an array Sum[] for convenience or a hash table to save memory space. With all lists scanned, the aggregated weight of any rid is exactly the value of wt(Sig(m)∩Sig(r)). In fact, if any rid appears satisfying the filtering condition of Theorem 1, i.e. with an aggregated weight larger than min{τ(m),τ(r)}, we can store it for later verification to determine whether it is the one that makes m a true member.A LGORITHM 1: BuildSIL ( R, δ, k) 1 for each r ∈R do2Sig ←GenSig (r, δ, k);/*The function GenSig (r, δ, k) generates signature for r under k-signature scheme. */ 3 for each t ∈Sig do4list[t]=list[t]∪{rid (r)};//insert rid of r into list5return list;Note thatτ(r) and wt(r) for any dictionary string r is computed in the signature generating step of Algorithm 1 and they are stored in the main memory for the later use of our algorithms (See Figure2).3.3Additional DiscussionIn the above discussion, one may notice that we didn’t involve the parameter k. This again proves the fact that with any assigned k, our algorithm will run correctly. Since the signature set is a compression of information in a string, we will certainly get more information if we choose a relatively large k, through which we can target potential matching evidences to a smaller scope, thus reducing the cost of verifying these evidences.However, larger k causes longer inverted list length, i.e. more cost on targeting possible evidences. For instance, if we set k = ∞, that is, for all strings s, we set Sig(s) to be s itself, andτ(s)=δ*wt(s), we interestingly find that our method degrades into a common inverted-list based solution. Chakrabarti [2] first analyzed this problem and showed that k=3 is good on average situation, which is also proved in our experimental study.4.OPTIMIZATION BY PROGRESSIVE COMPUTATION4.1Reducing Duplicate ComputationsAlthough EvSCAN algorithm efficiently checks the approximate membership of each single substring m in a document M, it ignores the overlapping between shifting substring windows and consequently takes a lot of time on duplicate computations. Another way to solve AME is to reduce this problem to set similarity join, which is already well studied by researchers [1, 4, 6]. In set similarity join, we are given SA and SB - two columns of sets, a similarity function Sim, and a threshold δ. The task of set similarity join is to join the two columns, where the joining condition is Sim(SA, SB)≥δ. In AME, if we set SA=R, SB={all substrings of M}, run set similarity join between SA and SB and project the result set along SB, we will get the result of AME. Though the problem of set similarity join is explored and optimized in many papers, this method still doesn’t notice the fact that the records in SB are quite similar with each other – they are substrings of a long text.In this section, based on the above observations, we believe that the unique property of AME should be exploited separately, and optimized method could be designed accordingly. So we study the incremental property of Theorem 1, and demonstrate Theorem 2, in purpose of decreasing the duplicate list-scanning when examining all substrings of M.4.2Optimization by Progressive ComputationAssume m⊕t denoting the string which we get by concatenating token t to the tail of string m. Consider the process of checking m and m⊕t: we compute the signature set of m and verify the condition in Theorem 1, then we do the same job for m⊕t. Intuitively the signature set of m and m⊕t are much alike. We observe that: if some r cannot match m and does not contain t, it is not likely to match m⊕t. Before we formalize our intuition into new theorem and algorithm, we introduce Evidence Superset by the lemma and definition below:L EMMA 3. For any m and r that satisfy Sim(m,r) ≥δ, wt(m∩Sig(r))≥min{δ*wt(m),τ(r)}}.This lemma is an inference of Theorem 1. Recall that in previous sections we mention that Theorem 1 remains correct even if we set different k for different strings, Lemma 3 is in fact obtained by setting k=∞ for m in Theorem 1 (so Sig(m) is replaced by m andτ(m) byδ*wt(m)). With Lemma 3 we define Evidence Superset for any query substring m as follow:D EFINITION 2. Suppose δ and k are fixed, for any string m, let ES(m)= {r∈R| wt(m∩Sig(r))≥min{δ*wt(m),τ(r)}}, we call ES(m) an Evidence Superset of m. Based on Lemma 3 it’s obvious that any true evidence for m must be contained in ES(m).From its definition, we see that ES(m) is useful since any evidence matching m will be included in ES(m). If we can efficiently compute ES(m) for any substring m, we can further filter elements in ES(m) to pick out all true evidences and check m’s approximate membership. Our intuition is formalized below.L EMMA 4. For any string m and token t, if a dictionary r∉ES(m) and t∉Sig(r), then r∉ES(m⊕t).Proof: We prove r∉ES(m⊕t) by showing that wt((m⊕t)∩Sig(r)) <min{δ*wt(m⊕t),τ(r)}: wt((m⊕t)∩Sig(r))=wt(m∩Sig(r)) (Because t∉Sig(r)) <min{δ*wt(m),τ(r)}(because r∉ES(m))< min{δ*wt(m⊕t),τ(r)}.▉This lemma states a fact that if a dictionary string is far from being evidence of current substring m and it is not a signature of the coming token t, then it cannot be evidence when the substring window moves to m⊕t. However this lemma still cannot serve as a method for efficiently computing ES(m), we further generalizeLemma 4 to obtain Theorem 2 to demonstrate the incremental property of ES(m).A LGORITHM 2: EvSCAN( M, δ, k, L)1ResultSet←Φ;// for storing approximate members2for each m of M’s substrings (|m| ≤ L) do3 Sig←GenSig(m, δ, k);4Initialize Sum[];//for weight aggregating5 CandSet←Φ;// for storing candidate evidence6 foreach t∈Sig do7for each rid∈list[t] do8Sum[rid]+=wt(rid);//aggregating weight9if Sum[rid]>=min{τ(m),τ(rid)} then10CandSet←CandSet∪{rid};11 foreach rid∈CandSet do//verification12if Sim(m, r(rid))≥δthen//true evidence found 13ResultSet←ResultSet∪{m};14break;15return ResultSet;T HEOREM 2. (I NCREMENTAL P ROPERTY). Suppose δ, k are fixed, it holds for any string m and token t that ES(m⊕t)⊆ES(m)∪list[t].Notice that the approach we build SIL determines that t∈Sig(r) means r∈list[t], so Theorem 2 is obviously based on Lemma4. This theorem indicates an efficient iterative approach of maintaining ES() for the varying substring when the right boundary of the substring windows moves by a token. That is, we check all elements in ES(m)∪list[t] and pick out proper ones into ES(m⊕t). Note that this process requires maintaining another field recording wt(m∩Sig(r)) in the summing table sum[], so it can be combined with the process of filtering by SIL.Now we get a new algorithm of incrementally checking all substrings, that is, we fix the left boundary of the substring window and shift the other boundary to the right. While the substring varies we iteratively maintain corresponding ES() to filter and verify all evidence in it. Figure 3 shows this iteration process, with an instance of M=t1⊕t2. For convenience, we denote this incremental filtering algorithm as EvITER (Evidence Iterating), while EcSCAN in Section 3.2 is short for Evidence Scanning. 4.3Combining Other Filtering ConditionsIn Algorithm 1 and 2, we use “VerifyAllCandidate()” to denote the process of verification, by which one determine if m is really a true member. Since not all r’s in CandSet are the ones that make m a true member, we may as well make “VerifyAllCandidate()” a small filter-verification process, via introducing some other simple yet effective filtering conditions. The following is exactly one of such conditions we want:If Sim(m,r) ≥δ, then wt(m) *δ≤wt(r) ≤ wt(m)/δ.Intuitively, under the non-weighted situation, this condition states that if any two strings have too much difference in length, they are not likely to match each other. To apply this filtering condition, we need only store in memory the weight value of all strings in R. By checking this condition for all r in CandSet, we can quickly narrow our scope to fewer possible r, thus avoiding more disk accessing and making computation more efficient.4.4Algorithm AnalysisIn this section we analyze the time and memory cost of our algorithms, and demonstrate the advantages of EvITER over EvSCAN. The notations to be used are listed in Table 3.|M| The length of text used as the input of AME|R| Dictionarysize L r Average length of dictionary stringsL m Average substring lengthL list Average length of inverted listsδSimilarity thresholdE Total number of evidence that passes the filterC v Time cost of verifying an evidence (including diskaccessing and similarity score computing)In addition to the above notations, we make two assumptionsin order to simplify our discussion: (1) the length of all strings is longer than k. (2) all tokens have the same weight (e.g. 1). Therefore, we obtain an upper bound for the signature set size of any string.L EMMA 5. Suppose k and δ are fixed, then for any string m with length L, |Sig(m)| ≤ max{k,(1-δ)L}.Proof: Because for any string m, |Sig(m)|=wt(Sig(m))≥(1-δ)*wt(m)= (1-δ)L, the smallest signature set size is (1-δ)L.If (1-δ)L≤ k, from the definition of k-signature set we choosethe first k tokens as signatures, so |Sig(m)|=k, else we choose the smallest signature set of size (1-δ)L. Therefore we have |Sig(m)|≤ max{k,(1-δ)L}. ▉A LGORITHM 3: EvITER( M={t[1],t[2],…,t[n]}, δ, k, L)1ResultSet←Φ;// for storing approximate members2for i =1 to n do3 Initialize Sum[];/*Sum[rid].s1 records wt(Sig(m)∩Sig(r(rid))) andSum[rid].s2 for wt(m∩Sig(r(rid))) */4 LastES←Φ;//for iterating ES5for j=i to min{n,i+L-1} do //current m= t[i]…t[j]6ES←Φ;7CandSet←Φ;// for storing candidate evidence8update Sig(m) and maintain sum[];/*because m and Sig(m) changes */9for each rid∈list[t]∪LastES do10if Sum[rid].s2≥min{δ*wt(m),τ(r(rid))} then11ES←ES∪{rid};12if Sum[rid].s1≥min{τ(m),τ(rid)} thenCandSet←CandSet∪{rid};13 VerifyAllCandidate(); //verification14LastES←ES; //iteration for the next window15return ResultSet;Figure 3. Flow of dictionary strings in EvITER。
