ThermoScientific限制性内切酶一览表
Thermo Scientific蛋白酶抑制剂

100mg
1515
78432
抑肽酶(Aprotinin)
25mg
1678
78433
抑氨肽酶(Bestatin)N-[(2S,3R)-3-氨基-2-羟基-4-苯基丁酰]-L-亮氨酸
(N-[(2S,3R)-3-amino-2-hydroxy-4-phenylbutyryl]-L-leucine)
可逆
1mg/ml(水)
胃酶抑素A(PepstatinA)
685.9
天冬氨酸蛋白酶
可逆
1mg/ml(甲醇)
PMSF (苯甲磺酰基氟化物)
174.2
丝氨酸蛋白酶
可逆
18mg/ml(甲醇)
订购消息:
单独包装的蛋白酶抑制剂
产品
货号
产品描述
包装
价格¥
78431
AEBSF [4-(2-氨乙基)氟化苯磺酰•HCl]
10mg
2801
78435
亮抑蛋白酶肽(Leupeptin)
[(乙酰基-亮氨酰-亮氨酰-argininal) •半硫酸盐]
50mg
4317
78436
胃酶抑素A(PepstatinA)
25mg
2118
36978
PMSF (氟化苯基甲磺酰氟)
5mg
911
焦磷酸钠(Sodium
Pyrophosphate)
221.9
丝氨酸/苏氨酸磷酸酶
不可逆
65mg/ml(水)
E-64
357.4
半胱氨酸蛋白酶
不可逆
20mg/ml(1:1乙醇/水)
限制性内切酶

Bam HⅠ
GGATCC CCTAGG
GTC CAG
+
GAC CTG
平端切口
G+
CCTAG
GATCC G
粘端切口
每一种限制酶能辨认特定的限制位点(restriction site) 。
多数限制酶不在两股的同一位置(即限制位点中央)切割,产生两个单股尾巴,
称粘性末端(cohesive/sticky end),因为它容易与另一个DNA片段的互补黏端相结合。
5’NNNNNG
pAATTCNNNNNN3'
3’NNNNNCTTAAp
GNNNNNN5’
EcoRⅠ的识别序列和酶切切口(粘端)
16/35
5’NNNNNNGTTAACNNNNN3‘ 3’NNNNNNCAATTGNNNNN5‘
5’NNNNNGTT 3‘NNNNNCAAp
pAACNNNNN3’ TTGNNNNN5’
甲基化酶 :在专一位点上甲基化,与限制性内切酶相对应 (1)dam甲基化酶
此酶在5'GATC3'序列中的腺嘌呤N6位置上引入甲基,使一些识别顺 序中含有5'GATC3'的限制性内切酶不能切割来自大肠杆菌的DNA。
(2)dcm甲基化酶
此酶在序列5‘CCAGG3’或5‘CCTGG3’中的胞嘧啶C5上引入甲基.
6/35
限制性核酸内切酶
限制性核酸内切酶(restriction endonuclease, RE) 是识别DNA的特异序列, 并在识别位点或其周围切割 双链DNA的一类内切酶。
Bam HⅠ
GGATCC CCTAGG
GCCTAG+
GATCC G
7/35
RESTRICTION ENZYMES
限制性内切酶 MspI_er0541

PRODUCT INFORMATIONMspI (HpaII)#ER05413000 uLot:Expiry Date:5'...C ↓C G G …3'3'...G G C ↑C ...5'Concentration: 10 u/µlSource:Moraxella speciesSupplied with: 2 x 1 ml of 10X Buffer TangoStore at -20°CIn total 3 vials. BSA included/fermentasRECOMMENDATIONS1X Thermo Scientific Tango Buffer (for 100% MspI digestion)33 mM Tris-acetate (pH 7.9), 10 mM magnesium acetate, 66 mM potassium acetate, 0.1 mg/ml BSA. Incubation temperature 37°C.Unit DefinitionOne unit is defined as the amount of MspI required to digest 1 µg lambda DNA in 1 hour at 37°C in 50 µl of recommended reaction buffer. DilutionDilute with the Dilution Buffer (#B19): 10 mM Tris-HCl (pH 7.4 at 25°C), 100 mM KCl, 1 mM EDTA, 1 mM DTT, 0.2 mg/ml BSA and 50% glycerol. Double DigestsTango ™Buffer provided simplifies buffer selection for double digests. 98% of Thermo Scientific restrictionenzymes are active in a 1X or 2X concentration of Tango Buffer. Please go to /doubledigest to choose the best buffer for your experiments. Storage BufferMspI is supplied in: 10 mM potassium phosphate (pH 7.5 at 25°C), 200 mM NaCl, 1 mM DTT, 1 mM EDTA, 0.2 mg/ml BSA and 50% glycerol.Rev.9VRecommended Protocol for Digestion•Add:nuclease-free water 16 µl10X Buffer Tango 2 µlDNA (0.5-1 µg/µl) 1 µlMspI 0.5-2 µl•Mix gently and spin down for a few seconds. •Incubate at 37°C for 1-16 hours.The digestion reaction may be scaled either up or down. Recommended Protocol for Digestion of PCR Products Directly after Amplification•Add:PCR reaction mixture 10 µl (~0.1-0.5 µg of DNA) nuclease-free water 18 µl10X Buffer Tango 2 µlMspI 1-2 µl•Mix gently and spin down for a few seconds. •Incubate at 37°C for 1-16 hours.Thermal InactivationMspI is inactivated by incubation at 80°C for 20 min. ENZYME PROPERTIESEnzyme Activity in Thermo Scientific REase Buffers, %B G O R Tango 2X Tango 50-100 50-100 0-20 0-20 100 50-100 Methylation Effects on DigestionDam: never overlaps – no effect.Dcm: never overlaps – no effect.CpG: completely overlaps – no effect.EcoKI: never overlaps – no effect.EcoBI: never overlaps – no effect.Stability during Prolonged IncubationA minimum of 0.3 units of the enzyme is required forcomplete digestion of 1 µg of lambda DNA in 16 hours at 37°C.Compatible EndsBsp119I, Bsu15I, Hin1I, Hin6I, HpaII, MaeII, NarI,Psp1406I, SsiI, TaqI, XmiI.Number of Recognition Sites in DNAλΦX174pBR322 pUC57 pUC18/19 pTZ19R/U M13mp18/19 328 5 26 13 13 12 18 NoteMspI is an isoshizomer of HpaII. When the external C in the sequence CCGG is methylated, MspI and HpaIIcannot cleave. However, unlike HpaII, MspI can cleave the sequence when the internal C residue is methylated.For CERTIFICATE OF ANALYSIS see back pageCERTIFICATE OF ANALYSISOverdigestion AssayNo detectable change in the specific fragmentationpattern is observed after a 160-fold overdigestion with MspI (10 u/µg lambda DNA x 16 hours).Ligation/Recutting AssayAfter a 50-fold overdigestion (3 u/µg DNA x 17 hours)with MspI, more than 95% of the digested DNAfragments can be ligated at a5'-termini concentration of3.0 µM. More than 95% of these sites can be recut. Labeled Oligonucleotide (LO) AssayNo detectable degradation of single-stranded or double-stranded labeled oligonucleotides occurred duringincubation with 10 units of MspI for 4 hours.Quality authorized by: Jurgita ZilinskienePRODUCT USE LIMITATIONThis product is developed, designed and sold exclusively for research purposes and in vitro use only. The product was not tested for use in diagnostics or for drug development, nor is it suitable for administration to humans or animals.Please refer to /fermentas for Material Safety Data Sheet of the product.© 2011 Thermo Fisher Scientific Inc. All rights reserved. All trademarks are theproperty of Thermo Fisher Scientific Inc. and its subsidiaries.。
常见限制性内切酶识别序列

常见限制性内切酶识别序列(酶切位点)(BamHI、EcoRI、HindIII、NdeI、XhoI等)Time:2009-10-22 PM 15:38Author:bioer Hits: 7681 times在分子克隆实验中,限制性内切酶是必不可少的工具酶。
无论是构建克隆载体还是表达载体,要根据载体选择合适的内切酶(当然,使用T 载就不必考虑了)。
先将引物设计好,然后添加酶切识别序列到引物5' 端。
常用的内切酶比如BamHI、EcoRI、HindIII、NdeI、XhoI等可能你都已经记住了它们的识别序列,不过为了保险起见,还是得查证一下。
下面是一些常用的II型内切酶的识别序列,仅供参考。
先介绍一下什么是II型内切酶吧。
The Type II restriction systems typically contain individual restriction enzymes and modification enzymes encoded by separate genes. The Type II restriction enzymes typically recognize specific DNA sequences and cleave at constant positions at or close to that sequence to produce 5-phosphates and 3-hydroxyls. Usually they require Mg 2+ ions as a cofactor, although some have more exotic requirements. The methyltransferases usually recognize the same sequence although some are more promiscuous. Three types of DNA methyltransferases have been found as part of Type II R-M systems forming either C5-methylcytosine, N4-methylcytosine or N6-methyladenine.酶类型识别序列ApaIType II restrictionenzyme5'GGGCC^C 3'BamHIType II restrictionenzyme5' G^GATCC 3'BglIIType II restrictionenzyme5' A^GATCT 3'EcoRIType II restrictionenzyme5' G^AATTC 3'HindIIIType II restrictionenzyme5' A^AGCTT 3'KpnIType II restrictionenzyme5' GGTAC^C 3'NcoIType II restrictionenzyme5' C^CATGG 3'NdeIType II restrictionenzyme5' CA^TATG 3'NheIType II restrictionenzyme5' G^CTAGC 3'NotIType II restrictionenzyme5' GC^GGCCGC 3'SacIType II restrictionenzyme5' GAGCT^C 3'SalIType II restrictionenzyme5' G^TCGAC 3'SphIType II restrictionenzyme5' GCATG^C 3'XbaIType II restrictionenzyme5' T^CTAGA 3'XhoIType II restrictionenzyme5' C^TCGAG 3'要查找更多内切酶的识别序列,你还可以选择下面几种方法:1. 查你所使用的内切酶的公司的目录或者网站;2. 用软件如:Primer Premier5.0或Bioedit等,这些软件均提供了内切酶识别序列的信息;3. 推荐到NEB的REBASE数据库去查(网址:/rebase/rebase.html)当你设计好引物,添加上了内切酶识别序列,下一步或许是添加保护碱基了,可以参考:NEB公司网站提供的关于设计PCR引物保护碱基参考表下载(也可见图片)双酶切buffer的选择(MBI、罗氏、NEB、Promega、Takara)再给大家推荐一种新的不需要连接反应的分子克隆方法,优点包括:①设计引物不必考虑选择什么酶切位点;②不必考虑保护碱基的问题;③不必每次都选择合适的酶来酶切质粒制备载体;④而且不需要DNA连接酶;⑤假阳性几率低(因为没有连接反应这一步,载体自连的问题没有了)。
常见分子试验限制性内切酶酶切位点大全

Alul 识别位点5: . AG T CT ...T 3:. TCJSA...^Aflll 识别位点Acil 识别位点孑...此GC (3)p J . ..GGG^G. ,.5JAcll 识别位点$…从七US杠…TTGC^AA …hAlwl 识别位点5 ..GljATC<Nl7 3 3 . ,. C C T A G (N),A 5'Acul 识别位点5. . CTGA AGfN), ▼…h3:」GACT TCfN)^.,. 5fAlwNl 识别位点e^XAGMN^TG,, 3 3:..GTC b MNNGAC (5)Afel 识别位点..AGCGCT .. .3P ..TCGCGA b ..5JApal 识别位点亍…GGGC 此.33\,CCCGGG...5JApaLl 识别位点3:..CAGGT i G...SAfllll 识别位点 片…M G R¥GT,・3b : - .TG^RQA …5"ApeKl 识别位点F …GtwGC …可 3\.CGWCJ5.._5]Aatll 识别位点Agel 识别位点员・ ^/T CCGGT ^.O -3\. TGGCC^A ...S^Acc65l 识别位点*:・.GT W KAC 」■丁 P :.. GA KMJG. ..5H Ahdl 识别位点5; ,G ACNN IvfNNGTC 3:..CTGNh!i NNNCAG (5)Accl 识别位点 *:・・ GTW KA C _・3 忆…GA KMJG. ..5Alel 识别位点5 ,TxACNffflNQTO- 7?3H 3 .GTGN\MNCAC 5"Bell识别位点Banl识别位点S^.CCRY^G.-.S^ Banll识别位点Bael识别位点Bcgl识别位点5:爲㈣CKgTGGW 3J 3:.屮㈣GUT(g2®N城” S BciVl识别位点5. ..GTATCCC帖3 . ,. C A 7 AGG(N}^. 5Apol识别位点&…P TA AT T J.y 3:.. YTTAA^R ..5JAscI识别位点S .^GG^GCGCC ・・・3 3J CCGCGCJ3G. ,.5JA Asel识别位点y…AfT AAT ■二住. T AATJA.,,5^AsiSI识别位点$: GRGGe -3”a^.c^ycGAG.-.s*Bbsl识别位点5. ,.GAAGAC(M).T.3" 3: ..CTTCTG{N}ti. . . 5^BbvCl识别位点亍…CtfTCAGC …IT 『…GGAG.CG…h『…GCGAT^GC..・3 p.. CGtyAGCG. . 5Aval识别位点Bbvl识别位点5 .GC AGC(M)D T. .3^ 3. . CGTCGiM^^-.S'CVCGRG…丁p;. G AGCY.C.,,5'Avail识别位点F…(SfeWGC二3" 『…ccwqp…hAvril识别位点忙,比TdGG ,一鼾..GGATGJD (5)Bccl识别位点5 .CCATC(N}f 3 3; .GGTAG(hl). 5ABceAl识别位点5. . . ACGGCIN)!* .3 3:…TGGCG(M)%f ^(fi>AC<N)J GTAYC㈣「3”P^(N)TG(N)4CATRG(叽圄BamHl 识别位点F…G%ATCC・・8 p:,.CCT AG^S.. .5'Bmtl 识别F …TG ATCA ・..3” P J ... ACTAGJ...5JBfal 识别位点&... <Tl AG ….3T 3:,.GAT ;C. ,.5JBfuAI 识别位点5 ACCTGC(N)7 3 :T ,.TGG ACG(N)… . . .5「ABgll 识别位点3 CGGI^NNNNCCG.. .5Bglll 识别位点罚 AT GT …丁 b …TCT*磬…甲Blpl 识别位点|5 …GC T TNAGC …IT 0- CG AN LCG .5'*Bme1580l 识别位点IS. ..GKGCM't , ..3_P^. . <^MCGKG .6BmgBl 识别位点|5\ ,.CAC T GTC.,.3J 忙…GTOf AO …hBmrl 识别位点. ACTGGG(N} T y3: .. TGACCC(N),^..^5;.,,GCTAGC,.・宙 3:. CJ J ATCG...5*Bpml 识别位点5:…CTGEAG(W …3" 3 . ..GACC1. .5'Bpu10l 识别位点酣…GC T TFJ AGC ■…丁3GG Ahl 匚8 …5,BpuEl 识别位点5. t CTTGAQ(M),7.3 3. .. G AACTC(M),^ hBsaAl 识别位点5; , YAC^GTR, ..3' 3: RT(\CAY. . 5^BsaBl 识别位点5; ,G^TNN>JNATC, -33J . CTANNJNNTAG .5'BsaHl 识别位点…GFTtG¥C 、・.3‘ :T.KYGQRG …5’Bsal 识别位点5r . .GGTCTCCM)/ 33. ^CCAGAGCM)^. . ,5HBsaJl 识别位点5:.任NNGG …了3 GGN忖Cf …5「Bmtl识别BsmA I 识别BsaXI 识别位点b J ..\(N)AC(N)^CTCC(«)|, 3y . /NjTGfN^GAGGCN). . 5rBseRI 识别位点|5\ .G AGGAG(N)r T .富 p j . ,.CTCCTC(N)lk ..5-BseYI 识别位点 厂氏G g C …3 卜…GGGT^p …£Bsgl 识别位点..GTGCAG(N}(7 3 3 . . C ACGTciN},^. , .5BsiEI 识别位点CGRv'fcG. .3 3 GCJfRGC. ,5JBsiHKAI 识别位点5:..GWGCWt ・・ + 3" 茁…妄WCGWG.…扩BsiWI 识别位点 耳讯 T R CG 773P'.. GC ATfi/:…h5: . GTCTC (谢一…才 3 . . .C AG AG Nh ■…亍BsmBI 识别位点5 ..CGTCTC(N)7 .3^ 3 . ..GCAGAG (NL ^ …5rBsmFI 识别位点亍…GGG AC^N),7 -3* 書…CCCTG^^. .,5rBsmI 识别位点k ;lh 0AATQCN T \3 T.. XTT ACGN■BsoBI 识别位点F …CGG4G …丁 3:..GRGC^C..5JBsp1286l 识别位点^TTGDGCrffc.rTr3: 一 ・ qHGGDG …5’BspCNI 识别位点5., .CTCAG(N)T 5 ,.y anf1 & ..CTCAGCNJ/7 3: . .G AG TC (N)^ _.53. . ,GAGTCfN>_ . 5_BspDI 识别位点t ;,T AftGAT (ip 3^TAGCJA.-.S^BslI识别位点P:, CC NN NWhT(4NGG. ..3^ 3d GGNN NNNMNCC . ..5F BspEI识别位点5;.. ffcCGGA …丁BssKI识别位点BstZ17l识别位点|5•…ft A T G A . h3^(3: . . AGTAGJ . . 5JBspMI识别位点MCCTGC(N)/. -T4TGG ACG(N\,^,.^ BspQI识别位点S\..GCTCTTC*^T..r S:.XS AGAAG(N^..&'BsrBI识别位点5 - cc(stfcr^33J. .. GGC^AG..・hBsrDI识别位点5 GC AATGHN T,,3^}…CGTTAqNN …hBsrFI识别位点fc;. R'tCGGV..^' 护…YGGcqp…yBsrGI识别位点|sT7 f^ifACA f +.3l p:-, AGATGJ ..5 ・BsrI识别位点S:. . AC TGG . 3*3:. TG ACJ3N …hBssHII识别位点p:. R CNGG…;r3:.i GGNCq t...5,BssSI识别位点S…WZGZ、、S3:_GTGG冬…5’BstAPI识别位点5;. r GCAMNNbTNlGC ,, 3J 3J,..CGTN L NNNNACG....5,JkBstBI识别位点F…TTtGAA …3" 卜…AAGCJT..6BstEII识别位点y…Gt TH AG G …丁扩……5’BstNI识别位点匚ftfwGG …丁3'. GG\\CC. 5^BstUI识别位点GGtG,…丁3:.,GqpCJ …5’BstXI识别位点6;, .CCANNMN^NTGG.T.a 3. .GGTN^NMNNNACC 5BstYI识别位点BssKI识别位点BstZ17l识别位点E:「F T GATCJ■了卜…¥0"初…亍BssKI识别位点BstZ17l识别位点p•…GT"T AC…科A…W冉TO…5"Bsu36l识别位点^.-CCfTNAGG...3rBtgl识别位点Is;.. (7c RY Ge...3" 卜…GG¥RQCBtgZI识别位点p:一GC(3 ATG(N】「33 ..CGC TAC(N)…k..5^BtsCI识别位点p•…GSGTG Wd p\.CGTC AC^hlN,. 5Btsl识别位点p•...G C A G TG 卜...CGTCghIN (5)Cac8l识别位点5•…GC砾IGC…丁『…CG卧X…割Clal识别位点5\ .. AT'feGAT , .,3TAGCJA (51)CspCl识别位点p- :ori(N)CAA(N^GTGG{N>i£l^ 3「(3\^.lJ(N)GTT(N):CACC(N)1frr;. 5*t: ~C^TG ,..3* 買…GT也p…h CviKl-1识别位点S...RC7CY ..*3J …5’CviQl识别位点5:. (frAC …:T3’…C仲…亍Ddel识别位点(7T N A G…3「…GANT卫….亍Dpnl识别位点CH,I;5 G A T C 3^3」CT AG . 5"rC%Dpnll识别位点F" t n T C…雷『…G T A £\一-亍Dral识别位点TT A …丁3 ■…AAAJTT …5’Dralll识别位点『…CACNMNtTG …丁扩…GTG*N 忖NCAG. hDrdl识别位点5\ GACNNNN T N NGTC ,3'3\.CTGNNNNNNCAG. ,.5JCviAll识别位点EcoRV 识别位点fc ;.. CCTNI^NNAGG,.,3^ p ;. r GG AN NNJN NTCC 5^EcoO109lHaell 识别位点F …RGCGE-丁 常…空GCGFt …时b ; , , AT TC ,-3 卜…GTTA Af …亍Eael 识别位点|5:…丹 GCCR,3 p\.RCCG<5^. .5'Eael 识别位点|5 ...丹 GCCR,S 卜...RCCGGy (5)Eagl 识别位点fc. V&GCCG 31 P : . GCGGGJC...5'Earl 识别位点5V T :GA TX TC 7731 3■…CT AJAG …品Fatl 识别位点F …tATG …3「GTAC A ...5JFaul 识别位点犷…CCCGC (N)7. . .3* ,GGGCGtN\^ ,5「Fnu4HI 识别位点6;.. GCTNGC ・・4 3 .. GGN£G (5)识别位点&…RG3CC¥・・:$Fspl 识别位点5;., TG<ft5CA,, 3EcoP15l 识别位点|5\ .C AGCAGfN^^ 3 p : ..GTCGIC(N)^. ,.5^HaelllEcoRl 识别位点Hgal 识别位点5 .GACGC (N )5T .3.CTCTTC(N)7 GAG AAG(N\Ecil 识别位点 B . GGC GG A(N)n T .3" 3 仁.CCGCCTeO 注…h Fokl 识别位点5 GG ATG(M)…T .3^ 3;. .CCT AC{M)nk ..5^Fsel 识别位点EcoNl 识别位点5 …GGCCGG 七 C …3一 3: . CCJ J GGCGG ..5*3. . ,CTGCG{M),k ..5-EcoRV识别位点Hhal识别位点$...GCG七 (3)3. .. QPGG ,.5MHin cll识别位点杖…GTYlH AC…3 歯…GA蔺TG…寸Hin dlll识别位点5;r. A>GCT T・、・3”『…TTCG《…5"Hinfl识别位点F…G鼻NTC…3"P' . C7 . 5"HinP1l识别位点航GG…了b:.tGqp …5*Hpal识别位点|5;.GTr^AC,-3'P:.. CA AJTG.-.5Hpall识别位点$...此GG・・S 卜...GGqp (5)Hphl识别位点6+<. GGTGA ㈣3 忙…GGACT 叫血*6Hpy188l识别位点TC N'GA…3”p j.., A C T …5”Hpy188lll识别位点亍…TCfNNGA…歹31.鼻GNN卫T..hHpy99l识别位点3'..1-h GCWGC. ,5・HpyAV识别位点5...,CCTTCtN)7,. 3* 3.r GGAAG(N)t..亍HpyCH4lll识别位点5;., ACN^T . .*3. , rt^NC A . . 5 HpyCH4lVHpyCH4\识别位点TGt A…3"3J AC fc GT...5JKasl识别位点亍…代CgC…丁3..4CCGCGp....5* Kpnl识别位点6;GGT AC T C f,■歹C£ATGG.…印Mboll识别位点Mbol识别位点SvT^MTrc …歹A…GTAG「HMboll识别位点MspAII 识别位点Nhel 识别位点 Mspl 识别位点3■…GGqC …5’Mlul 识别位点Nael 识别位点Mmel 识别位点Msel 识别位点賓…F T A A …科3:…―匸T …引NgoMIV 识别位点了…GtCGGC ■二歹3:「GGGUQT.Gfi* . G A AG A (N)J .3 P :, + CTTCT (N)7A . + ,5*Mfel 识别位点,…內ATTG …3”& ..GT TA A 卫…£Mwol 识别位点6;. . GCNNNNNT M NG C ・・.33J . . CGNN^NN NHNCG ..5Mlyl 识别位点G AGTC (rj ;7 了 :,CTCA G(N)「…5* h …GC 比GC …3「3:..CGG t CCG.…5JNarl 识别位点『,G 比GCG …丁3: . CCGqGG.^5*Mnll 识别位点5: .,CCTC 侔人二 3” $…GSG 讯厂$Ncil 识别位点&…CC T SGG .,3即…GGS^CC. ..5-Mscl 识别位点5: ,. TGGtCA^.S 3 ... ACCpGT ...5JNcol 识别位点5: ., Ct ATGG , , a^-GGTACp . . .5*Ndel 识别位点5 ..ATQ .3^3:..GT AlAC …5’Msll识别位点5 ...CMNNllNRTG (3)3 GTANN^NNY A C 5MspAII识别位点Nhel识别位点PshAI 识别位点 PaeR7l 识别位点Nlalll 识别位点 石GAT 贰+・玄 扩…少T AC "Pcil 识别位点 F U TGT …扩3 . ..TGTA^A.^5'NotI 识别位点TAGC, ..3p :・"CCGg •3眷…CGATQG 佔3\ GAGCI ;C. ..5*NmeAIII 识别位点5"…GC CG AG 呱二3PflMI 识别位点5;.,CCANNNN T NT&G <1,3 3:「GGT 轉NNNACC ・.,5'PhoI 识别位点5…GGtG …b$…GCGGCCGC …3 fa :.. CGGCGG_CG..,5^NruI 识别位点TCbfcG 昌…3 P :.. AGCpGT ..5PleI 识别位点S ;. . GAGTC IN)T 3 3:., CTCAG (N),A .,.5NsiI 识别位点$…ATGC 肝一团 卜….1;ACGTA .,5'PmeI 识别位点$…GTTT 工冉AG.S 3J ..CAAAJTTG..J 5^NspI 识别位点$…RCAI ■計…tr b ■…百GTMR一$PacI 识别位点b J ... TT AA'Tr AA・「3P J ... A A TJ AA TT (5)PmlI 识别位点CAC'GTG..,3 3: .GTC^CAC . 5JPpuMI 识别位点6;tl RG T GWC CY.^3^ 3: ..YCC W GjGR .. .5F GG 晶g …歹6:..GAGhTNN<5TC ,B …CC 轉GG …h S^.CTGNN^NGAG .NlalV 识别位点PflFI 识别位点P^.CTGNN A NNCAG...5*Psil识别位点胃…TTkTAA,■罔卜…从公T T…因PspGI识别位点卩…丸CWGG…y 卜…GGWCQ…5"PspOMI识别位点|5:・・bGGCCC:・h 忙…CGgcys…hPspXI识别位点6:.. V<fTCGAG6 ...3H 3 .BGAGCT;CV r5^Pstl识别位点$…CTGG帀…3". G^A0GT C .5・Pvul识别位点$…CGiATtG …丁P J...GCJAGC.,.5* Pvull识别位点5:CA^TG.,3-击...GTQJ3AG (5)Rsal识别位点b:.7GT\C...3l 卜…gjTd…戲Rsrll识别位点卧…C西WCCG…歹,…GCCWGjpg…扩Sacl识别位点时…GAGCe…丁扩…CJCGAG.^S^Sacll识别位点6;., GCQd^G,…y 3:.. GG^CGGC -.S^Sall识别位点<fT C G A C. *. 3^ 3J...CAGCT;G...5JSapl识别位点5:…GCTCTTC 啊人=*3「3「・CGAGAAG{Nhv5”Sau3AI识别位点b;..T GATC .313:.. GT AG^ ..5*Sau96l识别位点亍…品NCC…丁3\NG J p...5JSbfl识别位点3」GGf CGTGC …5”Seal识别位点5;, t AGT^CT < ,r3TCAJGA ...S^ ScrFI识别位点SnaBI 识别位点 Tfil 识别位点歛:・TAC'feTA .・.歹3: .. ATGJSAT .. S'Spel 识别位点5:二 Xt TAGT …丁 3J ... TGATC^A .-S^Sphl 识别位点GC ATGfc …3「 『…qpTACG …亍Sspl 识别位点6: ., AA T>TT , 3J ... TTAJAA...5JStul 识别位点AGC^CCT ••謁 3: . TCqSGA …hStyD4l 识别位点6:JCGNGG …孑 3’…GGNCq . SStyl 识别位点E :・,GPWWGG,・・:3”GGWWC&—WSwal 识别位点6*,.. A T T iTi A A T r ,. 3 3: ..T A A Aj T TA (5)Taqa I 识别位点■…fbGA …;r b : ■ • A Gb -CCfhGG . .3 p J . ..GGN^CC •.生SexAl 识别位点,…心GWGGT …了3\..TGGWCqA (5)SfaNI 识别位点fe : . . GC A JC (N)J. ,3' 忙 GGTAG (N);>1 + ,5*Sfcl 识别位点F …疔TRYAG …:T 卜…—ysSfil 识别位点b:. .GGCCNNNr7NGGCC. ..3H p.-CCGGN^NWUCCGG .5Sfol 识别位点S^.GGC'teCC,-^' 3 …CCI^pG 右•一一5"SgrAI 识别位点p •…CF?t CGG¥G …丁 p\.GYGGGC^RC .S^Smal 识别位点『…右右右gUC …hSmll 识别位点b, ,C T T Y RAG …3 P . r ,G AH YT^C .. 5Tlil 识别位点Xhol 识别位点(7TCGAG.-・33;.. G A G C T^C (5)Tsel 识别位点Xmal 识别位点賢…衣CGGG …丁3■…GGGgC …5’Xmnl 识别位点橫一上T£直匚…3",G AANN >IMTTC ,p ; ..CASTG^3:. XT TN N^N MA AG < …5"Zral 识别位点 $…X ATT …wfcC. GAC\STC ・.3rf(5... TT AA M ..5J 3:. CTGJCAG.. .5"TspMl 识别位点 $...比CGGG (3)$…GGGCC£…节与大家共享(非原创)6;ANNNNN^NNNTGG...3J 3; GGTNNNh^NNNNNACC-.STspRl 识别位点|5;-.NNCASTGNM T -.3d忸.&NNGTSACMN …5"Tthllll 识别位点6: , ,G AC hfNNGTC,-・旷 3\.CTGNN i NCAG...5^Xbal 识别位点ftT AGA (3)P ; .. AG A TCJ (5)Tsp45l 识别位点 Tsp509l 识别位点Xcml识别位点SnaBI识别位点Tfil识别位点。
史上最全限制性内切酶酶切位点汇总情况

A系列AatII识别位点Acc65I识别位点AccI识别位点AciI识别位点AclI识别位点AcuI识别位点AfeI识别位点AflII识别位点AflIII识别位点AgeI识别位点AhdI识别位点AleI识别位点AluI识别位点AlwI识别位点AlwNI识别位点ApaI识别位点ApaLI识别位点ApeKI识别位点ApoI识别位点AscI识别位点AseI识别位点AsiSI识别位点AvaI识别位点AvaII识别位点AvrII识别位点BaeI识别位点BamHI 识别位点BanI识别位点BanII识别位点BbvCI识别位点BbvI识别位点BccI识别位点BceAI识别位点BcgI识别位点BciVI识别位点BclI识别位点BfaI识别位点BfuAI识别位点BglI识别位点BglII识别位点BlpI识别位点Bme1580I识别位点BmgBI识别位点BmrI识别位点BmtI识别位点BpmI识别位点Bpu10I识别位点BpuEI识别位点BsaAI识别位点BsaBI识别位点BsaHI识别位点BsaI识别位点BsaJI识别位点BsaWI识别位点BsaXI识别位点BseRI识别位点BseYI识别位点BsiEI识别位点BsiHKAI识别位点BsiWI识别位点BslI识别位点BsmAI识别位点BsmBI识别位点BsmFI识别位点BsmI识别位点BsoBI识别位点Bsp1286I识别位点BspCNI识别位点BspDI识别位点BspEI识别位点BspHI识别位点BspMI识别位点BspQI识别位点BsrBI识别位点BsrDI识别位点BsrFI识别位点BsrGI识别位点BsrI识别位点BssHII识别位点BssKI识别位点BssSI识别位点BstAPI识别位点BstBI识别位点BstEII识别位点BstNI识别位点BstXI识别位点BstYI识别位点BstZ17I识别位点Bsu36I识别位点BtgI识别位点BtgZI识别位点BtsCI识别位点BtsI识别位点Cac8I识别位点ClaI识别位点CspCI识别位点CviAII识别位点CviKI-1识别位点CviQI识别位点DdeI识别位点DpnI识别位点DpnII识别位点DraI识别位点DraIII识别位点DrdI识别位点EaeI识别位点EaeI识别位点EagI识别位点EarI识别位点EciI识别位点EcoNI识别位点EcoO109I 识别位点EcoRI识别位点EcoRV识别位点FatI识别位点FauI识别位点Fnu4HI识别位点FokI识别位点FseI识别位点FspI识别位点HaeII识别位点HaeIIIHgaI识别位点HhaI识别位点HincII识别位点HindIII识别位点HinfI识别位点HinP1I识别位点HpaI识别位点HpaII识别位点HphI识别位点Hpy188I识别位点Hpy188III识别位点Hpy99I识别位点HpyAV识别位点HpyCH4III识别位点HpyCH4IV HpyCH4V识别位点KasI识别位点KpnI识别位点MboI识别位点MboII识别位点MfeI识别位点MluI识别位点MlyI识别位点MmeI识别位点MnlI识别位点MscI识别位点MseI识别位点MslI识别位点MspA1I识别位点MspI识别位点MwoI识别位点NaeI识别位点NarI识别位点NciI识别位点NdeI识别位点NgoMIV识别位点NheI识别位点NlaIII识别位点NlaIV识别位点NmeAIII识别位点NotI识别位点NruI识别位点NsiI识别位点NspI识别位点PacI识别位点PaeR7I识别位点PciI识别位点PflFI识别位点PflMI识别位点PhoI识别位点PleI识别位点PmeI识别位点PmlI识别位点PpuMI识别位点PshAI识别位点PsiI识别位点PspGI识别位点PspOMI识别位点PspXI识别位点PstI识别位点PvuI识别位点PvuII识别位点RsrII识别位点SacI识别位点SacII识别位点SalI识别位点SapI识别位点Sau3AI识别位点Sau96I识别位点SbfI识别位点ScaI识别位点ScrFI识别位点SexAI识别位点SfaNI识别位点SfcI识别位点SfiI识别位点SfoI识别位点SgrAI识别位点SmaI识别位点SmlI识别位点SnaBI识别位点SpeI识别位点SphI识别位点SspI识别位点StuI识别位点StyD4I识别位点StyI识别位点SwaI识别位点TaqαI识别位点TfiI识别位点TseI识别位点Tsp45I识别位点Tsp509I识别位点TspMI识别位点TspRI识别位点Tth111I识别位点XbaI识别位点XcmI识别位点XhoI识别位点XmaI识别位点XmnI识别位点ZraI识别位点。
泰克科学FastDigest Eco31I (IIs类) 限制性内切酶说明书
FastDigest Eco31I (IIs class)Catalog Number FD0293, FD0294Pub. No. MAN0012440 Rev. B.005'...G G T C T C (N)1↓…3' 3'...C C A G A G (N)5↑...5'Contents and storageDescriptionFastDigest Eco31I (IIs class) belongs to specific class of restriction enzymes which recognize asymmetric DNA sequences and cleave outside of their recognition sequence. IIs class enzymes are useful for many applications, including their use in seamless cloning of multiple fragments in a predefined order (1, 2, 3).Thermo Scientific FastDigest enzymes are an advanced line of restriction enzymes for rapid DNA digestion. All FastDigest enzymes are 100 % active in the universal FastDigest and FastDigest Green buffers and are able to digest DNA in5-15 minutes. This enables any combination of restriction enzymes to work simultaneously in one reaction tube and eliminates the need for sequential digestions.∙ FastDigest enzymes are optimized to digest plasmid, genomic and viral DNA as well as PCR products and do not show star activity even in prolonged incubations∙ Enzymes used in common downstream applications such as ligation, blunting and dephosphorylation reactions also have100 % activity in FastDigest and FastDigest Green Buffer.∙ FastDigest Green Buffer includes a density reagent along with blue and yellow tracking dyes that allow for direct loading of the reaction mixtures on a gel.The blue dye of the FastDigest Green Buffer migrates with 3-5 kb DNA fragments in a 1 % agarose gel and has an excitation peak at 424 nm. The yellow dye of the FastDigest Green Buffer migrates faster than 10 bp DNA fragments in a 1 % agarose gel and has an excitation peak at 615 nm.For applications that require analysis by fluorescence excitation FastDigest Buffer is recommended, as the dyes of the FastDigest Green Buffer may interfere with some fluorescence measurements.For Research Use Only. Not for use in diagnostic procedures.WARNING! Read the Safety Data Sheets (SDSs) and follow the handling instructions. Wear appropriate protective eyewear, clothing, and gloves. Safety Data Sheets (SDSs) are available from /support.FastDigest Eco31I (IIs class) cleavage is impaired by overlapping dcm methylation. To avoid dcm methylation, use a dam–, dcm–strains.Protocol for fast digestion of different DNAbine the following reaction components at room temperature in the order indicated:2. Mix gently and spin down.3. Incubate at 37 °C in a heat block or water thermostat for 5 min. Optional: Inactivate the enzyme by heating for 5 min at 65 °C4.If the FastDigest Green Buffer was used in the reaction, load an aliquot of the reaction mixture directly on a gel.Note: The FastDigest Green Buffer can be used as an electrophoresis loading buffer for any DNA sample at a final 1X concentration. Higher concentrations of FastDigest Green Buffer in the sample supply excess salt concentration which may alter DNA mobility.Double and multiple digestion of DNA∙The combined volume of the enzymes in the reaction mixture should not exceed 1/10 of the total reaction volume.∙Use 1 µL of each enzyme and scale up the reaction conditions appropriately.∙If the enzymes require different reaction temperatures, start with the enzyme that requires a lower temperature, then add the second enzyme and incubate at the higher temperature.Note: Increase the incubation time by 3-5 min if the total reaction volume exceeds 20 µL. Use water thermostat, air thermostats are not recommended due to the slow transfer of heat to the reaction mixture.Recommendations for PCR product digestion∙When introducing restriction enzyme sites into primers for subsequent digestion and cloning of a PCR product, refer to/fd, Reaction Conditions Guide, to define the number of extra bases required for efficient cleavage.∙Use Thermo Scientific GeneJET PCR Purification Kit, #K0701 to purify PCR product prior digestion in following cases:-When PCR additives such as DMSO or glycerol where used, as they may affect the cleavage efficiency or cause star activity.-When PCR Product will be used for cloning. Active thermophilic DNA polymerase still present in PCR mixture may alter the ends of the cleaved DNA and reduce the ligation efficiency.* 0.5 mM ATP (#R0441) is required for T4 DNA Ligase activity.2 FastDigest Eco31I (IIs class)Reference1. Engler, C., Gruetzner, R., Kandzia, R., and Marillonnet, S. (2009) Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One 4, e5553.2. Engler, C., Kandzia, R., and Marillonnet, S. (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS One 3, e3647.3. Engler, C., and Marillonnet, S. (2013) Combinatorial DNA assembly using Golden Gate cloning. Methods Mol Biol 1073, 141-156.Limited product warrantyLife Technologies Corporation and/or it affiliate(s) warrant their products as set forth in the Life Technologies’ General Te rms and Conditions of Sale at /us/en/home/global/terms-and-conditions.html. If you have any questions, please contact Life Technologies at /support .The information in this guide is subject to change without notice.DISCLAIMER: TO THE EXTENT ALLOWED BY LAW, THERMO FISHER SCIENTIFIC INC. AND/OR ITS AFFILIATE(S) WILL NOT BE LIABLE FOR SPECIAL, INCIDENTAL, INDIRECT, PUNITIVE, MULTIPLE, OR CONSEQUENTIAL DAMAGES IN CONNECTION WITH OR ARISING FROM THIS DOCUMENT, INCLUDING YOUR USE OF IT. Important Licensing Information : These products may be covered by one or more Limited Use Label Licenses. By use of this product, you accept the terms and conditions of all applicable Limited Use Label Licenses.©2020 Thermo Fisher Scientific Inc. All rights reserved. All trademarks are the property of Thermo Fisher Scientific and its subsidiaries unless otherwise specified./support | /askaquestion Thermo Fisher Scientific Baltics UAB | V.A. Graiciuno 8, LT-02241 Vilnius, LithuaniaFor descriptions of symbols on product labels or product documents, go to /symbols-definition.02 April 2020。
限制性内切酶的热失活温度表
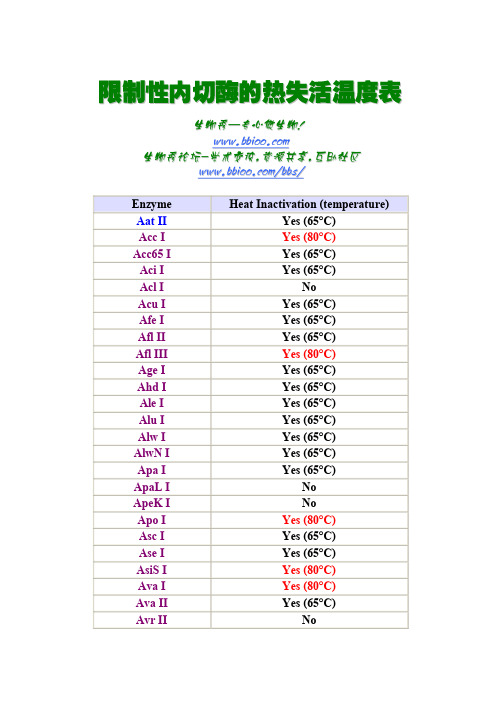
限 限制 制性 性内 内切 切酶 酶的 的热 热失 失活 活温 温度 度表表 生 生物 物秀 秀— —专 专心 心做 做生 生物物! w w w w w w . .b b b b i i o o o o . .c c oo m 生 生物 物秀 秀论 论坛 坛- -学 学术 术交 交流 流, ,资 资源 源共 共享 享, ,互 互助 助社社区 w w w w w w . .b b b b i i o o o o . .c c o o m m / /b b b b s s // EnzymeHeat Inactivation (temperature)Aat II Yes (65°C) Acc I Yes (80°C) Acc65 I Yes (65°C) Aci I Yes (65°C)Acl I No Acu I Yes (65°C) Afe I Yes (65°C) Afl II Yes (65°C) Afl III Yes (80°C) Age I Yes (65°C) Ahd I Yes (65°C) Ale I Yes (65°C) Alu I Yes (65°C) Alw I Yes (65°C) AlwN I Yes (65°C) Apa I Yes (65°C)ApaL I No ApeK I No Apo I Yes (80°C) Asc I Yes (65°C) Ase I Yes (65°C) AsiS I Yes (80°C) Ava I Yes (80°C) Ava II Yes (65°C)Avr IINoBae I Yes (65°C) BamH I NoBan I Yes (65°C) Ban II Yes (65°C) Bbs I Yes (65°C) Bbv I Yes (65°C) BbvC I Yes (80°C) Bcc I Yes (65°C) BceA I Yes (65°C) Bcg I Yes (65°C) BciV I Yes (65°C) Bcl I NoBfa I Yes (80°C) BfrB I Yes (80°C) BfuA I Yes (65°C) BfuC I Yes (80°C) Bgl I Yes (65°C) Bgl II NoBlp I No Bme1580 I Yes (80°C) BmgB I Yes (65°C) Bmr I Yes (65°C) Bmt I Yes (65°C) Bpm I Yes (65°C) Bpu10I Yes (80°C) BpuE I Yes (65°C) Bsa I Yes (65°C) BsaA I Yes (80°C) BsaB I Yes (80°C) BsaH I Yes (80°C) BsaJ I Yes (80°C) BsaW I Yes (80°C) BsaX I NoBseR I Yes (65°C)BseY I Yes (65°C) Bsg I Yes (65°C) BsiE I Yes (80°C) BsiHKA I Yes (80°C) BsiW I Yes (80°C) Bsl I Yes (80°C) Bsm I Yes (80°C) BsmA I Yes (80°C) BsmB I Yes (80°C) BsmF I Yes (80°C) BsoB I No Bsp1286 I Yes (65°C) BspCN I Yes (80°C) BspD I Yes (65°C) BspE I Yes (80°C) BspH I Yes (65°C) BspM I Yes (65°C) Bsr I Yes (80°C) BsrB I Yes (80°C) BsrD I Yes (80°C) BsrF I No BsrG I Yes (80°C) BssH II Yes (80°C) BssK I Yes (80°C) BssS I Yes (80°C) BstAP I Yes (80°C) BstB I No BstE II No BstF5 I No BstN I No BstU I No BstX I Yes (65°C) BstY I Yes (80°C) BstZ17 I NoBsu36 I Yes (80°C) Btg I Yes (80°C) BtgZ I Yes (80°C) Bts I Yes (80°C) Cac8 I Yes (65°C) Cla I Yes (65°C) CviA II Yes (65°C) Dde I Yes (65°C) Dpn I Yes (80°C) Dpn II Yes (65°C) Dra I Yes (65°C) Dra III Yes (65°C) Drd I Yes (65°C) Eae I Yes (65°C) Eag I Yes (65°C) Ear I Yes (65°C) Eci I Yes (65°C) EcoN I Yes (65°C) EcoO109 I Yes (65°C) EcoR I Yes (65°C) EcoR V Yes (80°C) Fat I Yes (65°C) Fau I Yes (65°C) Fnu4H I Yes (65°C) Fok I Yes (65°C) Fse I Yes (65°C) Fsp I Yes (65°C) Hae II Yes (80°C) Hae III Yes (80°C) Hga I Yes (65°C) Hha I Yes (65°C) HinP1 I Yes (65°C) Hinc II Yes (65°C) Hind III Yes (65°C)Hinf I Yes (80°C) Hpa I NoHpa II Yes (65°C) Hph I Yes (65°C) Hpy99 I Yes (65°C) Hpy188 I Yes (65°C) Hpy188 III Yes (65°C) HpyCH4 III Yes (80°C) HpyCH4 IV Yes (65°C) HpyCH4 V Yes (65°C) ICeu I Yes (65°C) ISce I Yes (65°C) Kas I Yes (65°C) Kpn I NoMbo I Yes (65°C) Mbo II Yes (65°C) McrBC Yes (65°C) Mfe I Yes (65°C) Mlu I Yes (65°C) Mly I Yes (65°C) Mme I Yes (80°C) Mnl I Yes (65°C) Msc I Yes (65°C) Mse I Yes (65°C) Msl I Yes (65°C) Msp I Yes (65°C) MspA1 I Yes (65°C) Mwo I NoNae I Yes (65°C) N.Alw I Yes (80°C) Nar I Yes (65°C) N.BbvC IA Yes (80°C) N.BbvC IB Yes (80°C) N.BstNB I Yes (80°C)Nci I No Nco I Yes (65°C) Nde I Yes (65°C) NgoM IV Yes (80°C) Nhe I Yes (65°C) Nla III Yes (65°C) Nla IV Yes (65°C) Not I Yes (65°C) Nru I Yes (65°C) Nsi I Yes (65°C) Nsp I Yes (65°C) Pac I Yes (65°C) PaeR7 I No Pci I Yes (65°C) PflF I Yes (65°C) PflM I Yes (65°C) Pho I NoPIPsp I NoPlSce I Yes (65°C) Ple I Yes (65°C) Pme I Yes (65°C) Pml I Yes (65°C) PpuM I No PshA I Yes (65°C) Psi I Yes (65°C) PspG I No PspOM I Yes (65°C) Pst I Yes (80°C) Pvu I Yes (80°C) Pvu II No Rsa I Yes (65°C) Rsr II Yes (65°C) Sac I Yes (65°C) Sac II Yes (65°C)Sal I Yes (65°C) Sap I Yes (65°C) Sau96 I Yes (80°C) Sau3A I Yes (65°C) Sbf I Yes (65°C) Sca I Yes (80°C) ScrF I Yes (65°C) SexA I Yes (65°C) SfaN I Yes (65°C) Sfc I Yes (65°C) Sfi I No Sfo I Yes (65°C) SgrA I Yes (65°C) Sma I Yes (65°C) Sml I No SnaB I Yes (80°C) Spe I Yes (65°C) Sph I Yes (65°C) Ssp I Yes (65°C) Stu I Yes (65°C) Sty I Yes (65°C) StyD4 I Yes (65°C) Swa I Yes (65°C) Taq I Yes (80°C) Tfi I NoTli I No Tse I No Tsp45 I No Tsp509 I No TspR I No Tth111 I No Xba I Yes (65°C) Xcm I Yes (65°C)Xho I Yes (65°C) Xma I Yes (65°C) Xmn I Yes (65°C) Zra I Yes (80°C)。
常见限制性内切酶识别序列

常见限制性内切酶识别序列(酶切位点)(BamHI、EcoRI、HindII I、NdeI、XhoI等)Time:2009-10-22 PM 15:38Author:bioerHits: 7681 times在分子克隆实验中,限制性内切酶是必不可少的工具酶。
无论是构建克隆载体还是表达载体,要根据载体选择合适的内切酶(当然,使用T 载就不必考虑了)。
先将引物设计好,然后添加酶切识别序列到引物5' 端。
常用的内切酶比如Bam HI、EcoRI、HindII I、NdeI、XhoI等可能你都已经记住了它们的识别序列,不过为了保险起见,还是得查证一下。
下面是一些常用的II型内切酶的识别序列,仅供参考。
先介绍一下什么是II型内切酶吧。
The Type II restri ction system s typica lly contai n indivi dualrestri ction enzyme s and modifi catio n enzyme s encode d by separa te genes. The Type II restri ction enzyme s typica lly recogn ize specif ic DNA sequen ces and cleave at consta nt positi ons at or closeto that sequen ce to produc e 5-phosph atesand 3-hydrox yls. Usuall y they requir e Mg 2+ ions as a cofact or, althou gh some have more exotic requir ement s. The methyl trans feras es usuall y recogn ize the same sequen ce althou gh some are more promis cuous. Threetypesof DNA methyl trans feras es have been foundas part of Type II R-M system s formin g either C5-methyl cytos ine, N4-methyl cytos ine or N6-methyl adeni ne.酶类型识别序列ApaIType II restri ctionenzyme5'GGGCC^C 3'BamHIType II restri ctionenzyme5' G^GATCC3'BglIIType II restri ctionenzyme5' A^GATCT3'EcoRIType II restri ctionenzyme5' G^AATTC3'HindII IType II restri ctionenzyme5' A^AGCTT3'KpnIType II restri ctionenzyme5' GGTAC^C 3'NcoIType II restri ctionenzyme5' C^CATGG3' NdeIType II restri ctionenzyme5' CA^TATG 3'NheIType II restri ctionenzyme5' G^CTAGC3'NotIType II restri ctionenzyme5' GC^GGCCGC 3'SacIType II restri ctionenzyme5' GAGCT^C 3'SalIType II restri ctionenzyme5' G^TCGAC3' SphIType II restri ctionenzyme5' GCATG^C 3'XbaIType II restri ctionenzyme5' T^CTAGA3'XhoIType II restri ctionenzyme5' C^TCGAG3'要查找更多内切酶的识别序列,你还可以选择下面几种方法:1. 查你所使用的内切酶的公司的目录或者网站;2. 用软件如:Primer Premie r5.0或Bioe dit等,这些软件均提供了内切酶识别序列的信息;3. 推荐到NEB的REBA SE数据库去查(网址:http://rebas/rebase/rebase.html)当你设计好引物,添加上了内切酶识别序列,下一步或许是添加保护碱基了,可以参考:NEB公司网站提供的关于设计PC R引物保护碱基参考表下载(也可见图片)双酶切buf fer的选择(MBI、罗氏、NEB、Prom eg a、Takara)再给大家推荐一种新的不需要连接反应的分子克隆方法,优点包括:①设计引物不必考虑选择什么酶切位点;②不必考虑保护碱基的问题;③不必每次都选择合适的酶来酶切质粒制备载体;④而且不需要D NA连接酶;⑤假阳性几率低(因为没有连接反应这一步,载体自连的问题没有了)。
限制性内切酶的说明

限制酶使用说明一、分类目前,已被发现的限制酶,根据其反应的必须因子和切断点等特性,被分为以下三大类:类别反应必须因子切点酶例 I 型 s-腺苷基蛋氨酸、ATP、Mg2+识别部位和切点不同,切断部位不定Eco B、Eco KII 型 Mg2+切断识别部位或其附近的特定部位Eco R I、Bam H I III 型 ATP、Mg2+识别部位和切点不同,但切断特定部位Eco P I、Hin f III 应用于基因工程研究用的限制酶,一般全是II型酶,现在市场上销售的酶都属于II型酶, 这些限制酶由于其反应条件和底物DNA种类的不同,其切断状况及出现Star活性的频率等各有不同,并且其程度也根据酶的不同而千差万别。
因而在使用限制酶时,必须对这些要素充分注意,确保目标序列的切断反应能顺利进行,下面具体介绍一下使用限制酶时的一些注意点。
二、注意事项1. 甲基化的影响从带有DNA甲基化酶基因的宿主菌中制备的DNA,其碱基的一部分已经被甲基化,因此即便使用能够识别、切断被甲基化部分的序列的限制酶,也几乎无法切断被甲基化的部分。
被甲基化的部位,根据底物DNA及宿主种类的不同而不同。
例如宿主菌为大肠杆菌的情况下,根据宿主的种类有以下两种情况:在进行转化时,通常使用的菌株为C600、HB101、JM109等,因为都带有dam、dcm甲基化酶,所以使用这些菌株制备的DNA时,必须注意。
另外,动物由来的DNA,CG序列多为5m CG;植物由来的DNA,CG及CNG序列多为5m CG和5m CNG。
2. Star活性限制酶在一些特定条件下使用时,对于底物DNA的特异性可能降低。
即可以把与原来识别的特定的DNA序列不同的碱基序列切断,这个现象叫Star活性。
Star活性出现的频率,根据酶、底物DNA、反应条件的不同而不同,可以说几乎所有的限制酶都具有Star活性。
并且,它们除了识别序列的范围增大之外,还发现了在DNA的一条链上加入切口的单链切口活性,所以为了极力抑制Star活性,一般情况下,即使会降低反应性能,我们也提倡在低甘油浓度、中性pH、高盐浓度条件下进行反应。
限制性核酸内切酶详细资料大全

限制性核酸内切酶详细资料大全限制性核酸内切酶是可以识别并附着特定的脱氧核苷酸序列,并对在每条链中特定部位的两个脱氧核糖核苷酸之间的磷酸二酯键进行切割的一类酶,简称限制酶。
根据限制酶的结构,辅因子的需求切位与作用方式,可将限制酶分为三种类型,分别是第一型(Type I)、第二型(Type II)及第三型(Type III)。
Ⅰ型限制性内切酶既能催化宿主DNA的甲基化,又催化非甲基化的DNA的水解;而Ⅱ型限制性内切酶只催化非甲基化的DNA的水解。
III型限制性内切酶同时具有修饰及认知切割的作用。
基本介绍•中文名:限制性核酸内切酶•外文名:Restriction endonuclease•作用:切割DNA•别名:限制酶限制性内切酶定义,由来,分布区域,分类性质,用途,命名,类型,第一型限制酶,第二型限制酶,第三型限制酶,生理意义,定义是识别特定的核苷酸序列,并在每条链中特定部位的两个核苷酸之间的磷酸二酯键进行切割的一类酶。
由来一般是以微生物属名的第一个字母和种名的前两个字母组成,第四个字母表示菌株(品系)。
例如,从Bacillus amylolique faciens H 中提取的限制性内切酶称为Bam H,在同一品系细菌中得到的识别不同碱基顺序的几种不同特异性的酶,可以编成不同的号,如HindⅡ、HindⅢ,HpaI、HpaⅡ,MboI、MboⅡ等。
限制性核酸内切酶别名:Endodeoxyribonuclease简称限制酶酶反应限制性内切酶能分裂DNA分子在一限定数目的专一部位上。
它能识别外源DNA并将其降解。
单位定义:在指明pH与37℃,在0.05mL反应混合物中,1小时消化1μg的λDNA的酶量为1单位。
性状制品不含非专一的核酸水解酶(由10单位内切酶与1μg λDNA,保温16小时所得的凝胶电泳图谱的稳定性表示),这类酶主要是从原核生物中分离出来的,迄今已经从近300多种不同的微生物中分离出约4000种限制酶。
4.1.1 限制性内切酶

限制性内切酶LOGOE. coli KE. coli C EOP=1EOP=1EOP=1EOP=10-4E.coli K E.coli B E.coli CλC11110-410-410-410-411λK λB1963年以来,Arber及其同事发现 ,经同位素标记证明,噬菌体在新品系中的损害伴随着其DNA的降解,但宿主自身的DNA不被降解。
Arber的解释:通过噬菌体感染进入细菌细胞的DNA分子能被细菌识别而分解,细菌本身的DNA则由于已被自己所修饰而免于被分解。
1965 年,Arber预言有限制性内切酶的存在发现1968年Messelson首次从E.coli K中分离限制性内切酶 Eco K①需要ATP,S-腺苷甲硫氨酸(SAM)等②有特定的识别位点 ,但没有特定的切割位点③切割位点远离识别位点1000bp以上1970年 美国约翰·霍布金斯大学 H.Smith 偶然发现 流感嗜血杆菌(Haemophilus influenzae) 细胞提取液能迅速降解外源的噬菌体DNA ,但不能降解自身DNA ,这就是Hin dII酶。
Nathans等人立即查明了限制内切酶的有效性,并将其用来分析 DNA 的结构。
1978 H.Smith, W Arber和D Nathans由于限制性内切酶发现和应用的研究得到诺贝尔奖。
命名限制酶的命名最初由Smith和Nathans于1973年提出,1980年再由Roberts进行系统化和分类,人们今天使用的就是这个体系的一个简化版。
1、来源菌属名的首字母和其种名的头两个字母形成的三个字母缩写来表示。
2、用一个字母表示菌株或类型。
3、用罗马字母表示不同的限制与修饰体系。
如Eco RI(Escherichia coli )种类根据限制酶识别切割特性,催化条件,及是否有修饰酶活性分三类(I, II ,III) 。
也有分四类,IIs类,即II 亚类。
type Ⅰ(数量少,约占1%)特点:(1)属复合酶类,兼具修饰和切割DNA两种特性。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
GTGCAC
FastDigest® AscI
GGCGCGCC
FastDigest® AseI
ATTAAT
FastDigest®
AsiSI
GCGATCGC
FastDigest®
AvaI
CYCGRG
FastDigest® AvaII
GGWCC
FastDigest®
AvrII
CCTAGG
37 °C
37 °C
Bpu1102I
FastDigest® BsaAI FastDigest® BsaBI FastDigest® BsaHI FastDigest® BsaJI
BseDI
CCTNAGC GCTNAGC YACGTR GATNNNNA
TC GRCGYC
CCNNGG CCNNGG
BseGI FastDigest® BseGI BseJI
ACTGGG
BfmI
CTRYAG
BfuI
GTATCC
BglI
FastDigest® BglI
BglII FastDigest® BglII FastDigest® BlpI Bme1390I
GCCNNNNN GGC
GCCNNNNN GGC
AGATCT
AGATCT
GCTNAGC
CCNGG
FastDigest® Bme1580I
50-100% 50-100% 20-50% 20-50%
100%
20-50%
F
F
F
T
50-100% 20-50% 0-20%
0-20%
100% 50-100%
T
20-50% 50-100% 20-50% 20-50%
100%
20-50%
F
O
NR
100%
100%
NR
NR
100%
T
20-50%
100% 50-100% 20-50%
Cfr13I
Cfr42I FastDigest® ClaI CpoI
GGNCC CCGCGG ATCGAT CGGWCCG
37 °C
37 °C 37 °C 37 °C 37 °C 37 °C
37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C
37 °C
50-100% 50-100%
0-20% 0-20% 0-20%
100% 100% 0-20%
0-20% 20-50%
100%
20-50% 100%
20-50%
100% 50-100% 20-50% 50-100%
0-20%
0-20%
100%
100%
0-20% 50-100% 100% 50-100%
37 °C
37 °C
37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C
R + SAM
F + SAM
F
R + SAM
R T F O F T F T F B F F F F F F F F
0-20% 100% 20-50%
0-20% 20-50%
100%
20-50%
50-100% 50-100% 50-100% 50-100%
0-20%
0-20%
100%
20-50%
F + SAM
F Buffer Bpu10I
F
T
F
0-20% 20-50% 50-100% 100% 50-100% 100%
GKGCMC
FastDigest® BmtI BoxI
BpiI
GCTAGC
GACNNNNG TC
GAAGAC
BplI
FastDigest® BplI FastDigest® BpmI
GAGNNNNN CTC
GAGNNNNN CTC
CTGGAG
Bpu10I
CCTNAGC
FastDigest® Bpu10I
Bsp1286I
GDGCHC
Bsp1407I
TGTACA
FastDigest® Bsp1407I
FastDigest® BspCNI FastDigest® BspHI BspLI FastDigest® BspMI BspOI
TGTACA
CTCAG TCATGA GGNNCC ACCTGC GCTAGC
BspPI
GGATC
BspTI FastDigest® BsrBI FastDigest® BsrDI FastDigest® BsrFI FastDigest® BssHII Bst1107I
BstXI
FastDigest® BstXI FastDigest® BstZ17I Bsu15I FastDigest® Bsu36I BsuRI
AGCT
Alw21I
GWGCWC
FastDigest®
Alw21I
GWGCWC
Alw26I
GTCTC
FastDigest® Alw26I
GTCTC
Alw44I
GTGCAC
FastDigest® CAGNNNCT
AlwNI
G
ApaI
GGGCCC
FastDigest® ApaI
GGGCCC
FastDigest® ApaLI
37 °C
37 °C
37 °C 37 °C 37 °C 37 °C 37 °C 55 °C 37 °C 37 °C 55 °C 37 °C 37 °C 37 °C 55 °C
100% 100%
FastDigest® BbsI FastDigest® BbvI
BclI
FastDigest® BclI
BcnI
GAAGAC GCAGC TGATCA TGATCA CCSGG
BcuI
ACTAGT
BdaI
FastDigest® BfaI BfiI
TGANNNNN NTCA CTAG
0-20%
0-20% 20-50% 0-20%
20-50% 50-100% 100% 50-100% 0-20% 50-100% 100% 50-100%
F
100% 100% 100% 100% 100% 100%
100% 100% 100%
100% 100%
100% 100% 100% 100% 100% 100% 100% 100% 100% 100% 100% 100%
50-100%
50-100% NR
50-100% 50-100% 20-50%
0-20% NR
F
R
0-20%
0-20% 50-100% 100%
20-50% 50-100%
100% 100%
100%
100% 100% 100% 100% 100%
100% 100% 100% 100% 100% 100% 100% 100%
G
0-20%
F
F
F
Buffer AjiI
NR
AjuI
FastDigest® AjuI FastDigest® AleI
GAANNNNN NNTTGG
GAANNNNN NNTTGG
CACNNNNG TG
AlfI
GCANNNNN
NTGC
AloI
GAACNNNN
NNTCC
AluI
AGCT
FastDigest® AluI
37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C 37 °C
T
50-100%
Buffer AarI + oligo
NR
B
100%
T
50-100%
F
F
O
0-20%
F
F
F
F + SAM
0-20%
0-20% 0-20% 50-100% 0-20% 50-100% 50-100% 100%
BamHI
FastDigest® BamHI FastDigest® BanI BauI
GGATCC GGATCC GGYRCC CACGAG
37 °C Buffer BamHI 20-50%
37 °C
BseLI
BseMI
GGATG
GGATG
GATNNNNA TC
CCNNNNNN NGG
GCAATG
BseMII
CTCAG
BseNI FastDigest® BseNI BseSI
ACTGG ACTGG GKGCMC
BseXI
FastDigest® BseXI Bsh1236I
GCAGC GCAGC CGCG
100%
100% 100%
NR
0-20%
0-20% 20-50% 50-100%
100%
F
O
0-20% 20-50%
100% 50-100% 0-20%
-100% 100% 50-100% 50-100% 50-100%
F
F
T G T + SAM
0-20% 20-50% 0-20%
内切酶
识别位点 孵育温度 推荐缓冲液 B
