TBA CAT RESIDUE EVAP. START UP-614
TBACAT.RES.EVAP.SYST.EMERG.SD-626

CATALYST RESIDUE EVAPORATOR SYSTEM EMERGENCY SHUTDOWN GUIDELINE
NOTE: This is for Training only, and is not intended to be used as a step by step procedure. PO/MTBE
• Isolate the leak. • Depressure the affected area. • Ensure the fire fighting equipment is available for fire fighting, vapor dispersal, or equipment cooling. Utilities failure:
转录因子E2F1及其突变体真核表达质粒的构建及稳定表达细胞株的建立

行检测,如图 2 所示,Flag-E2F1 单克隆的 #6 细胞
株 和 Flag1-60E2F11E71032 单18克0 隆 的190 #2 细20胞0 株2分10 别 稳220定 表
达 Flag-1E602F1 1和70 Fla1g80-E2F1190E132 蛋2白00 且免210疫荧2光20 染色
(1. 天津师范大学生命科学学院,天津 300387;2. 天津师范大学天津市动植物抗性重点实验室,天津 300387)
摘 要:通过细胞转染、药物筛选等方法,获得 Flag-E2F1 和 Flag-E2F1E132 稳定表达的 HeLa 细胞株,用蛋白印迹杂交和细 胞免疫荧光染色的方法对其进行鉴定。结果表明,本研究构建了 Flag-E2F1 和 Flag-E2F1E132 稳定表达的 HeLa 细胞株,这些稳 筛细胞株对于进一步寻找 E2F1 的关键下游靶基因十分重要。
·45·
E2F1 的关键下游靶基因的筛选具有重要意义。
同时,分别将对应细胞转移至 10 cm 培养皿,用 G418
1 材料与方法
1.1 材料
(750 μg/mL)进行筛选,挑取单克隆。通过 Western blot 检 测,将 能 表 达 Flag-E2F1 和 Flag-E2F1E132 的
HeLa 细 胞 和 pFlag 真 核 细 胞 表 达 载 体 由 天 津 师 范 大 学 分 子 细 胞 系 统 生 物 学 重 点 实 验 室 提 供;
Steri-Cycle 直 热 式 Forma CO2 细 胞 培 养 箱,Thermo 公 司;Mini P4 小 型 蛋 白 电 泳 仪,Bio-Rad 公 司;
亮氨酸(L)和 133 位天冬酰胺(N)分别替换为谷氨 酸(E)和丝氨酸(S),使其失去 DNA 结合能力 [6]。
XDR话单

控制面信息 / 用户面信息 控制面信息 / 用户面信息 控制面信息 / 用户面信息 控制面信息 / 用户面信息 基础信息 基础信息 基础信息 基础信息 基础信息 基础信息 基础信息 基础信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息
字段说明onlineid基础信息sessionid基础信息startdatetime基础信息enddatetime基础信息sgsnip基础信息sgsn基础信息bscip基础信息bsc基础信息lac基础信息ci基础信息rac基础信息bvci基础信息imsi基础信息imei基础信息prefiximei基础信息brand基础信息type基础信息apn基础信息eventtype基础信息eventdirection基础信息protocol业务信息iplenul业务信息iplendl业务信息countpacketul业务信息countpacketdl业务信息useriplenul业务信息useriplendl业务信息weightrateul业务信息weightratedl业务信息weightratetotal业务信息duration业务信息sourceip业务信息destip业务信息sourceport业务信息destport业务信息uri业务信息urimain业务信息contenttypepart1业务信息contenttypepart2业务信息useragentmain业务信息servicetype业务信息subservicetype业务信息sedservice业务信息isclient业务信息isprivateservice业务信息isreassemble业务信息request控制面信息用户面信息repeat控制面信息用户面信息reject控制面信息rejectcause控制面信息accept控制面信息acceptdelayfirst控制面信息acceptrepeat控制面信息complete控制面信息completedelayfirst控制面信息rautype控制面信息rauattr控制面信息opplac控制面信息opprac控制面信息bvciold控制面信息bvcinew控制面信息depdpacc控制面信息用户面信息depdpaccdirection控制面信息用户面信息detachacc控制面信息用户面信息detachaccdirection控制面信息用户面信息decause控制面信息用户面信息abnormalreason控制面信息用户面信息abnormaltype控制面信息用户面信息resp用户面信息respdelayfirst用户面信息respcause用户面信息mmscause用户面信息result用/ 用户面信息 控制面信息 / 用户面信息 控制面信息 控制面信息 控制面信息 控制面信息 控制面信息 控制面信息 控制面信息 控制面信息 控制面信息 控制面信息 控制面信息 控制面信息 控制面信息 控制面信息 / 用户面信息 控制面信息 / 用户面信息 控制面信息 / 用户面信息 控制面信息 / 用户面信息 控制面信息 / 用户面信息 控制面信息 / 用户面信息 控制面信息 / 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 用户面信息 控制面信息 / 用户面信息
基于改进YOLOv8s的大田甘蓝移栽状态检测算法

基于改进YOLOv8s的大田甘蓝移栽状态检测算法吴小燕;郭威;朱轶萍;朱华吉;吴华瑞【期刊名称】《智慧农业(中英文)》【年(卷),期】2024(6)2【摘要】[目的/意义]借助智能化识别及图像处理等技术来实现对移栽后蔬菜状态的识别和分析,将会极大提高识别效率。
为了实现甘蓝大田移栽情况的实时监测和统计,提高甘蓝移栽后的成活率以及制定后续工作方案,减少人力和物力的浪费,研究一种自然环境下高效识别甘蓝移栽状态的算法。
[方法]采集移栽后的甘蓝图像,利用数据增强方式对数据进行处理,输入YOLOv8s(You Only Look Once Version 8s)算法中进行识别,通过结合可变形卷积,提高算法特征提取和目标定位能力,捕获更多有用的目标信息,提高对目标的识别效果;通过嵌入多尺度注意力机制,降低背景因素干扰,增加算法对目标区域的关注,提高模型对不同尺寸的甘蓝的检测能力,降低漏检率;通过引入Focal-EIoU Loss(Focal Extended Intersection over Union Loss),优化算法定位精度,提高算法的收敛速度和定位精度。
[结果和讨论]提出的算法经过测试,对甘蓝移栽状态的召回率R值和平均精度均值(Mean Average Precision,mAP)分别达到92.2%和96.2%,传输速率为146帧/s,可满足实际甘蓝移栽工作对移栽状态识别精度和速度的要求。
[结论]提出的甘蓝移栽状态检测方法能够实现对甘蓝移栽状态识别的准确识别,可以提升移栽质量测量效率,减少时间和人力投入,提高大田移栽质量调查的自动化程度。
【总页数】11页(P107-117)【作者】吴小燕;郭威;朱轶萍;朱华吉;吴华瑞【作者单位】广西大学计算机与电子信息学院;国家农业信息化工程技术研究中心;北京市农林科学院信息技术研究中心;农业农村部数字乡村技术重点实验室;农业农村部农业信息技术重点实验室【正文语种】中文【中图分类】TP391.41【相关文献】1.基于改进YOLOv8的森林火灾检测算法研究2.基于改进YOLOv8的交通标志检测算法3.基于改进YOLOv8的火灾检测算法研究4.基于改进YOLOv8的火情智能检测算法5.基于改进Yolov8的摔倒行为检测算法因版权原因,仅展示原文概要,查看原文内容请购买。
残差基数缩放法

残差基数缩放法1. Introduction这篇文章主要的应用背景是根据振动信号(vibration signals)对旋转机械机构进行故障诊断,文中提到目前常见的故障诊断方法包括:signal analysis-based methods (基于信号分析的经典方法)和 machine-learning-powered methods(基于机器学习的智能方法),但经典方法存在问题。
因为对于故障诊断其实主要是对数据进行特征提取并分析,所以采用卷积神经网络(CNN)在合适不过,于是引出Deep residual networks(Res Nets,深度残差网络),它是CNN的一个变种,可以减少训练的复杂程度。
Deep residual networks(Res Nets) are an attractive variant of Conv Nets, which use identity shortcuts to ease the difficulty of parameter optimization.本文有三个贡献:① 在网络结构中加入 Soft thresholding(软阈值)用于过滤噪声相关的feature② 将thresholds(阈值)作为子网络的hyperparameter,并自动训练出来,同时每个震动信号有自己的thresholds③ 提出两种改进的深度残差网络:DRSN-CS 和 DRSN-CW2. Theory of the Developed DRSNsA. Res Nets先介绍一下Res Nets基本结构,然后再介绍其改进结构。
残差模块(Residual Building Unit, RBU)是深度残差网络(Res Nets)的基本组成部分。
如下图所示,长方体表示通道数为C、宽度为W、高为1的特征图;一个残差模块可以包含两个批标准化(Batch Normalization, BN)、两个整流线性单元激活函数(Rectifier Linear Unit activation function, Re LU)、两个卷积层(Convolutional layer)和恒等映射(Identity shortcut)。
SIMATIC Energy Manager PRO V7.2 - Operation Operat

2 Energy Manager PRO Client................................................................................................................. 19
2.1 2.1.1 2.1.2 2.1.3 2.1.4 2.1.5 2.1.5.1 2.1.5.2 2.1.6
Basics ................................................................................................................................ 19 Start Energy Manager ........................................................................................................ 19 Client as navigation tool..................................................................................................... 23 Basic configuration ............................................................................................................ 25 Search for object................................................................................................................ 31 Quicklinks.......................................................................................................................... 33 Create Quicklinks ............................................................................................................... 33 Editing Quicklinks .............................................................................................................. 35 Help .................................................................................................................................. 38
Brassinosteroid, gibberellin and phytochrome impinge on a common

A R T I C L E SBrassinosteroid,gibberellin and phytochrome impinge on a common transcription module in ArabidopsisMing-Yi Bai 1,4,Jian-Xiu Shang 1,2,4,Eunkyoo Oh 1,Min Fan 1,Yang Bai 1,Rodolfo Zentella 3,Tai-ping Sun 3and Zhi-Yong Wang 1,5Brassinosteroid and gibberellin promote many similar developmental responses in plants;however,their relationship remains unclear.Here we show that BR and GA act interdependently through a direct interaction between the BR-activated BZR1and GA-inactivated DELLA transcription regulators.GA promotion of cell elongation required BR signalling,whereas BR or active BZR1suppressed the GA-deficient dwarf phenotype.DELLAs directly interacted with BZR1and inhibited BZR1–DNA binding both in vitro and in vivo .Genome-wide analysis defined a BZR1-dependent GA-regulated transcriptome,which is enriched with light-regulated genes and genes involved in cell wall synthesis and photosynthesis/chloroplast function.GA promotion of hypocotyl elongation requires both BZR1and the phytochrome-interacting factors (PIFs),as well as their common downstream targetsencoding the PRE-family helix–loop–helix factors.The results demonstrate that GA releases DELLA-mediated inhibition of BZR1,and that the DELLA–BZR1–PIF4interaction defines a core transcription module that mediates coordinated growth regulation by GA,BR and light signals.The remarkable plasticity of plant development is believed to rely on networks of interconnected signal transduction pathways that integrate multiple hormonal and environmental signals coordinately regulating common cellular activities and developmental processes 1.However,direct crosstalk between hormone pathways has rarely been observed in plants,although many signalling pathways have been characterized in detail 2.BR and GA are two major growth-promoting hormones that have similar effects on a wide range of developmental processes 1.Mutants deficient in either BR or GA show various degrees of similar phenotypes,including dwarfism,reduced seed germination,de-etiolation in the dark and delayed flowering 2–6.Despite the physiological evidence for overlapping actions on development 7,the relationship between the BR and GA signalling pathways has remained unclear.Both BR and GA signalling pathways have been studied extensively 8,9.BR is perceived by the receptor kinase Brassinosteroid-Insensitive 1(BRI1),and downstream signal transduction leads to activation of the Brassinozale-Resistant 1(BZR1)family transcription factors,which control BR-responsive gene expression 8.When BR levels are low,BZR1is phosphorylated by the GSK3-like kinase Brassinosteroid-Insensitive 2(BIN2),and as a result,loses its DNA-binding activity and becomes retained in the cytoplasm 8.When BR levels are high,activation of BRI11Department of Plant Biology,Carnegie Institution for Science,Stanford,California 94305,USA.2Institute of Molecular Cell Biology,College of Life Science,Hebei Normal University,Shijiazhuang,Hebei,050016,China.3Department of Biology,Duke University,Durham,North Carolina 27708,USA.4These authors contributed equally to this work.5Correspondence should be addressed to Z-Y.W.(e-mail:zywang24@)Received 13January 2012;accepted 13June 2012;published online 22July 2012;DOI:10.1038/ncb2546leads to sequential phosphorylation and activation of the BR-signalling kinases (BSKs and CDG1)and members of the BRI1Suppressor 1(BSU1)family phosphatases 8,10,which inactivates the GSK3s through tyrosine dephosphorylation 11,allowing BZR1activation by PP2A-mediated dephosphorylation 12.Dephosphorylated BZR1translocates into the nucleus to regulate over a thousand target genes 13.GA promotes plant growth by removing the DELLA proteins.Binding of GA to its receptor GA INSENSITIVE DWARF 1(GID1)induces GID1–DELLA interaction and association with the E3ubiquitin ligase SCF SLY1/GID2,leading to polyubiquitylation and degradation of DELLAs (refs 9,14).When GA levels are low,DELLAs accumulate and directly inactivate a number of transcription factors 9,15–21,including the bHLH factor PIF4,which promotes cell elongation when plants are in the dark,shade or high temperature 22.Despite their overlapping physiological functions and the extensive knowledge about each signalling pathway,little is known about how BR and GA interact at the molecular level.Previous genome expression analysis of BR-and GA-responsive genes identified largely non-overlapping transcriptional responses 23;however,such meta-analysis might have biased the results 1.Therefore,it is unclear whether and how the actions of BR and GA are coordinated in regulating common developmental processes 1,2.Here we demonstrate an interdependentNATURE CELL BIOLOGY ADVANCE ONLINE PUBLICATION1© 2012Macmillan Publishers Limited. All rights reserved.A R T I C L E SH y p o c o t y l l e n g t h (m m )det2 +BL Lergai-1dellaGA BL–BL +++–+++–+++BL (nM)ae bc dFigure 1BR signalling and BZR1activity are required for GA promotion of hypocotyl elongation.(a )Wild-type (Col)and BR mutant seedlings were grown under constant light for seven days on medium with or without 1µM GA 3and 1µM brassinolide (BL,the most active brassinosteroid).(b )BR-deficient and insensitive mutants show a GA-insensitive phenotype,which is suppressed by bzr1-1D .Wild-type (Col)and BR mutant seedlings were grown in the dark for six days on medium containing 1µM PAC and different concentrations of GA 3,and with 10nM BL where indicated (+BL).Error bars,s.e.m.(n =32plants).(c )bzr1-1D partly suppressed the ga1-3phenotype.(d )BR promotes cell elongation in GA mutants.Wild-type (Ler),ga1-3and sly1-10seedlings were grown under light for seven days on medium containing 1µM PAC or 1µM BL as indicated.(e )GA enhances the BR sensitivity.Wild-type and det2-1seedlings were grown under constant light for seven days on medium with or without 1µM GA 3and different concentrations of BL.Error bars,s.e.m.(n =35plants).(f ,g )Removing DELLAs enhances BR sensitivity.Ler,gai-1and della seedlings were grown for seven days under constant light on medium containing 2µM brassinazole (BRZ,an inhibitor of BR synthesis)and 10nM (+),1µM (++)(f ),or different concentrations of BL (g ).Error bars,s.e.m.(n =25plants).relationship between BR and GA,mediated by direct interaction between DELLAs and BZR1,and identify a BZR1-dependent GA-regulated transcriptome controlling cell elongation and photosynthesis.These results provide evidence for a central growth-regulating transcription module that integrates BR,GA and environmental signals for regulating cell elongation and seedling etiolation.RESULTSBR or active BZR1/BZR2is required for GA promotion of cell elongationTo understand the relationship between BR and GA,we examined how defects in one hormone pathway influence the sensitivity to the other in hypocotyl elongation.We found that GA increased hypocotyl length in wild-type plants but not in the BR-deficient mutant det2-1or BR-insensitive mutant bri1-5and bri1-119(Fig.1a,b).BR restored GA response to det2-1(Fig.1a,b),and increased the GA sensitivity in a dose-dependent manner (Supplementary Fig.S1a).However,BR cannot restore the GA responsiveness of the BR-insensitive mutants bri1-5and bri1-119(Fig.1a,b),indicating that BR signalling is required for GA-induced hypocotyl elongation.The GA-insensitive phenotypes of bri1-119and bri1-116were suppressed by the dominant gain-of-function mutant bzr1-1D (Fig.1b and Supplementary Fig.S1b,c),in which active BZR1accumulates as a result of increased interaction with PP2A phosphatase 12.The bzr1-1D mutation also partially suppressed the short-hypocotyl phenotype of the GA-deficient ga1-3mutant and wild-type plants treated with a GA biosynthesisinhibitor paclobutrazol (PAC;Fig.1c and Supplementary Fig.S1d –f).BZR2(also known as BES1)is a homologue of BZR1and they have 88%protein identity;its gain-of-function mutant bes1-D showed resistance to PAC and partially suppressed the GA insensitivity of BR-deficient plants (Supplementary Fig.S1g –i).These results suggest that BR or BR-activated BZR1/BZR2is required for GA promotion of hypocotyl elongation.GA-induced DELLA degradation enhances BR responseIn contrast to the GA insensitivity of BR mutants,the GA-deficient mutant ga1-3,GA-insensitive mutant sly1-10and wild-type plants treated with PAC were responsive to BR and partly rescued by a high concentration of BR (Fig.1d and Supplementary Fig.S1j).GA and PAC had no effect on hypocotyl elongation of det2-1and bri1-5mutants,but enhanced and reduced,respectively,the BR-induced hypocotyl elongation in det2-1(Fig.1e and Supplementary Fig.S1k,l),suggesting that GA promotes cell elongation by enhancing the BR-induced response.Such an essential role for BR and an enhancing role for GA are consistent with the stronger dwarf/de-etiolation phenotypes of BR-deficient than GA-deficient mutants.GA is known to promote growth by degradation of the growth-repressor proteins DELLAs (ref.9).The della pentuple mutant lacking all five members of the DELLA family genes showed a markedly enhanced BR response,whereas the GA-insensitive mutant gai-1,which accumulates high levels of GAI (one of the five DELLA proteins in Arabidopsis ;refs 9,15),showed a slightly reduced BR response2NATURE CELL BIOLOGY ADVANCE ONLINE PUBLICATION© 2012Macmillan Publishers Limited. All rights reserved.A R T I C L E Sbzr1-1D BZR1BZR1bzr1-1DBZR1DELLAdomain GRAS domainPoly S/TDELLA/HYNP LHRI VHIIDLHRIPFYRE SAW5871RGA ΔN RGAC RGA ΔSAWADT7RGAN RGA ΔLHRIRGA ADBD–BZR1-His +His-His +HisBD–RGA ΔNpBZ R 1B Z R 1InputPulldownM B P B Z R 1B Z R 1N B Z R 1CAnti-YFPIPAnti-YFPCo lB Z R 1–C F PAnti-RGA R G A –Y F P Anti-Myc InputBL––+Anti-YFPB Z R 1 b i n d i n g705513070551307055CN X 5P P 2A P R E 1P R E 5S a u r -A C 1I A A 19D W F 4PAC GAL U C /R E NL U C /R E N∗∗c pPRE5–LUC∗∗∗∗∗∗AD BD -His +His/B Z R 1–M y cB Z R 1–M y c Input+++––––+++–+MBP MBP–RGA MBP–BZR1024680510pPRE5–LUC12345G F P G F P + b z r 1-D G F P + r g a -Δ17G F P + R G A ΔS A W R G A ΔS A W + b z r 1-D222-G F P B Z R 1B ZR 1 + R G A B Z R 1 + G A I B Z R 1 + R G L 2r g a -Δ17 + b z r 1-D a hijkcd fegbbzr1-1D ADT7 ADT7 RGA RGArga-Δ17rga-Δ17I n p u tP u l l d o w nAnti-YFPIP 130M r (K)pIAA19 Pulldown pSaur-AC1 PulldownFigure 2RGA interacts with BZR1and inhibits the DNA-binding activity of BZR1in vitro and in vivo .(a )Wild-type and dominant mutant forms of BZR1and RGA interact in yeast.(b )A diagram of the structure of RGA.(c )The LHR1domain is necessary for both RGA homodimerization and the interaction with BZR1;the SAW domain is also required for interaction with BZR1.(d ,e )MBP and MBP fusions with the BZR1protein were incubated with GST–RGA bound to glutathione–agarose beads and then eluted and analysed by anti-MBP immunoblotting.(f ,g )BZR1and RGA interact in plants.(f )The seedlings of Col and pBZR1:BZR1–CFP grown in medium containing 100nM PAC under light for seven days were treated with 100nM BL for 1h,and then co-immunoprecipitation was performed using anti-YFP antibody and immunoblotted using anti-RGA and anti-YFP antibodies.(g )Immunoprecipitation was performed using anti-YFP antibody on transgenic Arabidopsis plants expressing 35S::BZR1–Myc only or co-expressing 35S::BZR1–Myc and 35S::RGA–YFP ,and immunoblotted using anti-Myc or anti-YFP antibodies.(h )RGA inhibits BZR1DNA binding in vitro .MBP–BZR1pre-incubated with MBP or MBP–RGA was incubated with biotinylated DNA fragments from the IAA19and Saur-AC1promoters immobilized on streptavidin beads.The DNA-bound proteins were immunoblotted using anti-MBP antibody.(i )GA increases the level of BZR1–DNA binding in vivo .ChIP was performed using anti-YFP antibodies followed by qPCR analysis.The level of BZR1binding was calculated as the ratio between BZR1–CFP and the 35S::YFP control,normalized to that of the control gene CNX5.Error bars,s.d.of three biological repeats.Significant differences between GA and mock treatment are marked by asterisks (P <0.01).(j ,k )Transient reporter gene assays show RGA inhibition of BZR1transcription activity.Arabidopsis protoplasts were transformed with the dual luciferase reporter construct containing pPRE5::LUC (luciferase)and 35S::REN (renilla luciferase),and constructs overexpressing the indicated effectors.The LUC activity was normalized to REN.Error bars,s.d.of three biological repeats.∗:Significant difference between BZR1and BZR1+RGA samples (P <0.01).b:Significant difference between BZR1and BZR1+GAI samples.(P <0.05).#:No Significant difference between BZR1and BZR1+RGL2samples (P >0.05).Uncropped images of blots are shown in Supplementary Fig.S5.(Fig.1f,g).The DELLA protein RGA was degraded normally in response to GA treatment in BR mutants (det2-1and bri1-116)and BR-treated plants (Supplementary Fig.S2a,b),suggesting that BR is not required for GA-induced DELLA degradation and that degradation of DELLA is not sufficient for promoting hypocotyl elongation in the absence of BR.These genetic and physiological results suggest that DELLAs may repress a BR-activated component,probably BZR1or a downstream component,and GA-induced DELLA degradation releases this repression.RGA interacts with BZR1and inhibits BZR1–DNA binding ability DELLAs are known to inhibit several transcription factors through protein –protein interactions 16,17,19,21.The requirement of BR-activated BZR1for GA/DELLA regulation of hypocotyl elongation raises a possibility that DELLAs may directly repress BZR1.Indeed yeast two-hybrid assays showed direct interaction between BZR1and the DELLA protein RGA.Further yeast two-hybrid assays showed that both BZR1and bzr1-1D interacted with RGA (Fig.2a).RGA contains the amino-terminal DELLA domain,which is required for GA-induced degradation 9and possesses transactivation activity 24,and the carboxy-terminal GRAS domain,which is important for its repressor function 25–27(Fig.2b).Deletion of the DELLA domain had no effect on the BZR1interaction,indicating that BZR1interacts with the GRAS domain,but not the DELLA domain,of RGA (Fig.2a,c).The GRAS domain can be subdivided into five distinct sequence motifs:leucine heptad repeat I (LHRI),the VHIID motif,leucineNATURE CELL BIOLOGY ADVANCE ONLINE PUBLICATION3© 2012Macmillan Publishers Limited. All rights reserved.A R T I C L E Sheptad repeat II(LHRII),the PFYRE motif and the SAW motif25 (Fig.2b).Deletion of either the LHRI or SAW motif abolished the interaction with BZR1(Fig.2c).The LHRI domain is required for RGA homodimerization(Fig.2c),and both the LHRI and SAW domains are required for growth-suppression function of DELLAs(refs24, 26,27).RGA can also heterodimerize with other DELLA proteins (Supplementary Fig.S3a,b).These results thus support a possibility that both dimerization and the SAW domain are required for RGA binding to BZR1and suppressing plant growth.The GRAS domain is highly conserved in all members of the DELLA family,and both BZR1 and BZR2interacted with RGA,GAI,RGL1and RGL3,but not RGL2, in yeast(Supplementary Fig.S2a).In vitro pulldown assays showed that GST–RGA interacted strongly with MBP-tagged full-length BZR1and the N-terminal part of BZR1(BZR1N),and weakly with the C-terminal part of BZR1 (BZR1C),but not MBP alone(Fig.2d),suggesting that the N-terminal DNA-binding domain of BZR1has a high affinity for RGA.Interestingly,RGA binds only to unphosphorylated BZR1but not the BIN2-phosphorylated MBP–BZR1(Fig.2e).Consistent with the in vitro data,co-immunoprecipitation assays and bimolecular fluorescence complementation assays showed that BZR1interacts with RGA in vivo and BR-induced BZR1dephosphorylation increased the interaction(Fig.2f,g and Supplementary Fig.S3b). These results demonstrate that RGA binds specifically to the BR-activated form of BZR1.DELLAs are known to inhibit the DNA-binding of transcription factors16,17,19.We thus investigated whether DELLAs block BZR1–DNA binding.MBP–BZR1can be pulled down by biotinylated DNA fragments of the promoters of the BZR1target genes IAA19or SAUR-AC1,but not by the non-target CNX5promoter13(Fig.2h and Supplementary Fig.S3c),confirming the specific interaction between BZR1and target promoters.Incubation of MBP–BZR1with MBP–RGA markedly reduced the level of BZR1binding to DNA,whereas incubation with MBP alone had no effect(Fig.2h and Supplementary Fig.S3c,d),indicating that RGA inhibits BZR1–DNA binding in vitro. To determine whether DELLA proteins inhibit BZR1–DNA binding in vivo,we performed chromatin immunoprecipitation followed by quantitative real-time PCR(ChIP–qPCR)assays.The ChIP–qPCR results show that BZR1binding to the promoters of five BZR1 target genes(PRE1,PRE5,IAA19,SAUR-AC1and DWF4;ref.13) was enhanced by GA treatment,presumably owing to GA-induced degradation of the DELLA proteins(Fig.2i).GA treatment and GA-signalling mutants(rga-24/gai-t6and spy-3)did not affect the abundance or phosphorylation status of BZR1protein(Supplementary Fig.S2c,d),consistent with DELLAs directly blocking DNA binding.In protoplast transient assays,the expression level of luciferase driven by the BR-responsive PRE5promoter was increased by BZR1and bzr1-1D, but this increase was abolished by co-expression of RGA,GAI and rga- 17,but not by RGL2and RGA SAW(Fig.2j,k).These results indicate that RGA specifically interacts with BZR1to inhibit its abilities to bind DNA and regulate transcription.GA and BR regulate overlapping genomic targets involved in photomorphogenesis and cell elongationIf DELLAs inhibit BZR1activity in vivo and GA releases DELLA inhibition,GA should affect the expression of BZR1target genes in similar manners as BR.Indeed,the previously identified microarray data sets of genes affected in the BR-insensitive mutant bri1-116and GA-deficient mutant ga1-3overlap significantly13,28(Fig.3a).Of the 1,194genes differentially expressed in ga1-3when compared with the wild type,419genes(35%)were also affected in the bri1-116mutant,of which296were also affected by light(Fig.3a and Supplementary Table S1).Among these co-regulated genes,387genes(92.3%)were affected in the same way by bri1-116and ga1-3,with a correlation coefficient R=0.76(Fig.3b).The effects of bri1and ga1are also similar to that of light on these genes,consistent with BR and GA repressing light responses(Fig.3b).For276(71%)of these genes,the effects of bri1-116 were reversed by the bzr1-1D mutation(compare bzr1-1D/bri1-116 and bri1-116)and the effects of ga1-3were reversed by loss of DELLA proteins(compare della/ga1-3and ga1-3)(Fig.3c).These results show that GA and BR exert similar effects on a large number of common genes through DELLA and BZR1activities.To further define the BR/BZR1-dependent genomic targets of GA signalling,we analysed the effects of BR-deficiency and bzr1-1D mutation on GA-induced gene expression changes using RNA-sequencing(RNA-Seq).RNA-Seq analysis identified3,570genes affected>1.5-fold by GA treatment in wild-type plants grown without propiconazole(PPZ,a specific inhibitor of BR biosynthesis29),1,629 genes affected by GA in wild-type plants grown on PPZ medium (BR-deficient plants),and4,306genes affected in bzr1-1D when compared with wild-type plants when grown on the PAC+PPZ medium(deficient for both GA and BR;Fig.3d).Of the3,570genes regulated by GA in BR-sufficient plants,only1,187genes(33%) were affected by GA in the BR-deficient plants,suggesting that GA-responsive expression of most genes requires BR.About half(549 genes)of the BR-independent GA-regulated genes were not affected by bzr1-1D in any significant amount or pattern(Class A,Fig.3d,e), whereas the other half(638genes)were affected by bzr1-1D,mostly in similar manners as GA treatment(Class B,Fig.3d,f).It is likely that Class A genes are regulated by other DELLA-interacting transcription regulators,such as JAZ1and EIN3(refs18,19),independently of BR/BZR1,and the Class B genes are regulated through both BR/BZR1-dependent and-independent mechanisms.Of the BR-dependent GA-responsive genes,1,027were affected by bzr1-1D(Class C,Fig.3d), mostly in similar manners as GA treatment(Fig.3g),and their GA responses were quantitatively reduced in the BR-deficient plants (Fig.3g);these genes apparently are regulated by GA through a BR/BZR1-dependent mechanism.Gene Ontology analyses showed that cell-wall-related genes are markedly enriched in the GA-induced Class B(10%)and Class C genes(10%)when compared with the random control(3%; Fig.3h),suggesting that GA promotes cell elongation preferentially through the BZR1-dependent mechanism but also through BR-independent mechanisms.In contrast,photosynthesis/chloroplast genes are markedly enriched in the GA-repressed Class B(44%) and C(62%)but not in Class A or D(13–17%)when compared with the control(15%;Fig.3h),indicating that GA represses photosynthetic genes mainly through a BZR1-dependent mechanism. These genomic data thus provide direct evidence for the important role of BZR1in GA regulation of genome expression,particularly the transcriptomes that promote cell elongation and repress photosynthetic development.4NATURE CELL BIOLOGY ADVANCE ONLINE PUBLICATION©2012Macmillan Publishers Limited. All rights reserved.A R T I C L ESClass C down 629 genesClass D down807 genes1%2% 3% 1% 5% 2%24%WT L versus D2,8784553209692,851296123b r i 1-116 D v e r s u s W T Dga1-3 D versus WT DGA +/–Col GA +/–1,356(D)1,027(C)2,469549 (A)208638(B)172versus Col_PPZ Random15%3%6%4%3%11%5%53%Photosynthesis/chloroplast Cell wallHormone/signalling Growth/development Stress/defence Cell metabolic Transport OtherClass A up 231 genesClass B up 293 genesClass C up 398 genesClass D up 549 genesClass A down 318 genes11%7%11% 4% 3%13%4%47%1%8% 3% 2%11%4% Col_PPZ bzr1-1D _PPZ13%3% 11%8% 4%10%7%44%8%10%14% 5% 2%13%6% 42%7%10%3%5%12%6%46%11% 9%5% 4% 5%13%5%11%48%Class B down 345 genes44%27%62%63%17%1% 2% 5% 2% 8% 2%b z r 1-1D /C o l _GA +/–C o l _GA +/-C ol_P P Z G A +/–C o l_P P Z G A +/-b z r 1-1D _P P Z b z r 1-1D _P P Z v e r s u s C o l _P P Zv e r s u s C o l _P P ZC o l _GA +/-C o l_P P Z G A +/-b z r 1-1D _P P Z v e r s u s C o l _P P Zb r i 1-116b r i 1-116g a 1-3g a 1-3d e l l a /–303–303–303–33Class AClass BClass Cad hefgbcbri1-116 D ga1-3 D versus WT Dversus WT DFigure 3GA and BR co-regulate a large number of genes through DELLAs and BZR1.(a )Venn diagram showing the overlap between sets of genes differentially expressed in dark-grown bri1-116versus wild type (bri1-116D versus WT D),ga1-3versus wild type (ga1-3D versus WT D),and light-grown versus dark-grown wild type (WT L versus D).(b )Scatter plot of log 2fold change values for 419genes differentially expressed in ga1-3D versus WT D and bri1-116D versus WT D.Red and blue colours indicate light-activated and light-repressed genes,respectively;black colour indicates the genes are not regulated by light.(c )Hierarchical clusters analysis of the genes differentially expressed in bzr1-1D/bri1-116versus bri1-116(bzr1-1D/bri1-116),bri1-116versus WT (bri1-116),ga1-3versus Ler (ga1-3),and della/ga1-3versus ga1-3(della/ga1-3).The gradient bar represents the log 2of the ratio.Genes are listed in Supplementary Table S1.(d –h )RNA-Seq analyses of genes affected by GA treatments or by bzr1-1D in BR-deficient plants (grown on 2µM PPZ medium).(d )Venn diagram showing overlaps between sets of genes affected by GA treatment in wild-type (Col)plants grown on medium containing PAC (Col GA +/−)or on medium containing PAC and PPZ (Col_PPZ GA +/−),and genes affected by bzr1-1D in the presence of PAC and PPZ (bzr1-1D _PPZ versus Col_PPZ).(e –g )Hierarchical cluster analysis of the expression data of the genes in Class A (e ),B (f )and C (g )in d .(h )Gene Ontology analysis of cellular functions represented by GA-upregulated and -downregulated genes in each gene Class (A–D)shown in d .All genes detected in RNA-Seq samples were used as control (random).GA-promotion of hypocotyl elongation requires BZR1,PIF4and their common downstream targets paclobutrazol resistance factors (PREs)Both DELLAs and BZR1also interact with PIF4(refs 16,17,30),and PIF4and BZR1together bind to a large number of common promoters in the genome 30.To determine whether the common targets of both BZR1and PIF4were preferentially regulated by GA,we grouped genes on the basis of the ChIP data of BZR1and PIF4and the microarray expression profiling data of pifq and bzr1-1D/bri1mutants 13,30,31,and we calculated the percentage of GA-regulated genes (on the basis of expression data for the ga1-3mutant 28)of each group.The genes regulated by PIFs and/or BZR1included higher percentages of GA-regulated genes than genes unregulated by BZR1and PIFs,and the genes that are common targets co-regulated by BZR1and PIF4NATURE CELL BIOLOGY ADVANCE ONLINE PUBLICATION5© 2012Macmillan Publishers Limited. All rights reserved.A R T I C L E SH y p o c o t y l l e n g t h (m m )PPZH y p o c o t y l l e n g t h (m m )Co l b z r1-1DP IF 4-O xb zr 1-1D /P I P 4-O xbzr1-1Dbzr1-1D bzr1-1D/PIF4-OxBZR1GA-regulated genes (ga1-3)–++++++++–PPZ GA PPZ GA ColColbzr1-1D PIF4-Ox 051015051015abce––++++++++ –+–+–+–+–+–+–+–+–+–+––++++++++–++++++++–ColCol bzr1-1D pifq/pifqCol bzr1-1D pifq/pifqFigure 4BZR1and PIF4are required for the GA promotion of hypocotyl elongation.(a )Venn diagram showing the percentage of GA-regulated genes among the gene sets that BZR1and PIF4bind to and/or regulate.PIF4and BZR1targets were identified by PIF4ChIP-Seq and BZR1ChIP-chip,respectively;PIFs-and BZR1-regulated genes were differentially expressed in pifq versus the wild type and in bzr1-1D/bri1-116versus bri1-116grown in the dark.GA-regulated genes were differentially expressed in ga1-3versus the wild type in the dark.The numbers show the percentages of each gene set that are GA-regulated genes.(b ,c )PIFs are required for BZR1-mediated GA promotion of hypocotyl elongation.Seedlings were grown in the dark for five days on medium containing 0.5µM PAC and 10µM PPZ,with or without 1µM GA 3.Error bars,s.d.(n =10plants).The asterisks mark significant differences between GA and mock treatments (P <0.01).(d ,e )Both BZR1and PIFs are required for GA promotion of hypocotyl elongation in light.Seedlings were grown under red light for five days on medium containing 0.1µM PAC and 2µM PPZ,with 0(M)or 1µM GA 3(GA).Error bars,s.d.(n =10plants).The asterisks mark significant differences between GA and mock treatments (P <0.01).included the highest percentage of GA-regulated genes (Fig.4a and Supplementary Fig.S4),indicating that the genome targets shared by BZR1and PIF4tend to be regulated by GA.We then examined whether GA-induced cell elongation also requires both BZR1and PIFs.The dominant bzr1-1D mutation rescued the GA response in the wild-type background but not in the pifq background when seedlings were grown on the BR biosynthesis inhibitor PPZ in the dark (Fig.4b,c),indicating that PIFs are required for the BZR1-mediated GA response.When grown on medium containing PPZ under light (which causes degradation of PIFs),only bzr1-1D/PIF4-Ox plants showed a robust GA response,whereas PIF4-Ox ,bzr1-1D and wild-type plants were all insensitive to GA (Fig.4d,e).These results indicate that GA promotion of hypocotyl elongation requires both BZR1and PIF4,and is thus probably mediated by the BZR1–PIF4heterodimer.Previous studies have shown that GA,BR and auxin induce the expression of the PRE family helix –loop –helix factors 32–34,which promote cell elongation by antagonizing several inhibitory HLH factors 33,35,36.Several PRE family members,including PRE1,PRE5and PRE6/KIDARI ,are direct targets of both BZR1and PIF4(ref.30).Consistent with GA acting through the DELLA –BZR1–PIF4module,the expression of PRE1,PRE2,PRE5and PRE6was induced by GA treatment in the wild type,but their GA induction was decreased in the bri1-119mutant and recovered in the bri1-119bzr1-1D double mutant,indicating that GA induction of these genes requires active BZR1(Fig.5a).Similarly,the BR-induction of PRE1,PRE5and PRE6was reduced in the dominant gain-of-function gai-1mutant when compared with the wild type (Fig.5b),indicating that BR induction was negated by accumulation of GAI.Two genes,EXP1and EXP8,encoding expansins,cell wall proteins that loosen the cell wall 37,are affected similarly to PREs by bri1-119and gai-1(Fig.5a,b),suggestingthat these expansins might mediate the downstream response in cell elongation.The role of PREs in the GA response was confirmed using the pre-amiR transgenic line,in which four PRE members are suppressed by artificial microRNA 30,and a transgenic line that overexpresses IBH1,the antagonistic partner of PRE1(ref.33).Both pre-amiR and IBH1-overexpression lines had GA-insensitive hypocotyls when compared with the wild type (Fig.5c).Consistent with the GA-insensitivity of pre-amiR ,PRE1overexpression reduced plants’sensitivity to the biosynthesis inhibitors of GA and BR (Fig.5d,e).ChIP –reChIP with transgenic Arabidopsis expressing both BZR1–myc and PIF4–YFP showed that BZR1and PIF4co-occupy the promoter of PRE1,PRE6,EXP1and EXP8(Fig.5f).These results demonstrate that PREs are essential positive regulators for GA-promoted hypocotyl elongation,acting downstream of the DELLA –BZR1–PIF4module.DISCUSSIONHow plant growth is controlled by the wide range of environmental signals and endogenous cues is a fundamental question in plant biology.This study demonstrates a major mechanism of crosstalk between two hormonal signals and illustrates a central network that integrates signalling and growth regulation in plants.Whereas previous studies suggested that BR and GA act independently on highly overlapping developmental responses 1,this study reveals a much closer relationship between these two hormones.Strong evidence from genetic,biochemi-cal and genomic analyses support a model in which GA and BR crosstalk through direct interaction between the key components of each pathway,DELLAs and BZR1(Fig.5g).BR signalling through the BRI1receptor kinase pathway leads to dephosphorylation and subsequent nuclear accumulation of BZR1(ref.8).However,the activity of BZR1is attenuated by DELLA proteins when GA levels are low.GA-induced6NATURE CELL BIOLOGY ADVANCE ONLINE PUBLICATION© 2012Macmillan Publishers Limited. All rights reserved.。
qiime2物种注释缺失值

qiime2物种注释缺失值物种注释是微生物组学研究中的重要步骤,它帮助我们理解不同微生物的功能和相互作用。
然而,在进行物种注释时,我们常常会遇到缺失值的情况。
缺失值的出现可能是由于实验条件、测序技术或者数据处理过程中的错误所导致的。
当我们遇到缺失值时,首先要做的是识别和记录这些缺失值。
在进行物种注释之前,我们需要对数据进行预处理,包括去除低质量序列、去除嵌合序列、去除噪音和伪序列等。
这样可以减少数据中的缺失值,提高注释的准确性和可靠性。
在进行物种注释时,我们可以使用一些常见的工具和数据库,如Greengenes、Silva等。
这些工具和数据库包含了大量的参考序列和物种注释信息,可以帮助我们将未注释的序列与已知物种进行比对和匹配。
通过比对和匹配,我们可以确定未注释序列的物种归属。
然而,在进行物种注释时,由于样本来源复杂多样,可能会出现一些物种无法注释的情况。
这些无法注释的物种可能是新物种、未知物种或者是数据库中缺少相关信息的物种。
在遇到这些情况时,我们可以尝试使用其他工具和数据库进行注释,或者进行进一步的实验和研究,以获取更多的信息。
除了缺失值外,物种注释中还可能出现误注释的情况。
误注释可能是由于数据库中的错误或者序列相似度较高导致的。
为了避免误注释,我们可以使用多个工具和数据库进行注释,并对结果进行比较和验证。
此外,我们还可以利用功能注释等其他信息来帮助判断物种注释的准确性。
物种注释是微生物组学研究中的重要步骤,但在实际操作中可能会遇到缺失值的情况。
我们需要认识到缺失值的存在,并采取合适的方法来处理和解决这些问题。
通过不断的努力和探索,我们可以更准确地注释微生物组中的物种,从而更好地理解微生物的功能和相互作用。
matlab中鲁棒工具箱gap指令

MATLAB中的鲁棒工具箱是一个非常有用的工具,能够帮助用户实现对数据集的鲁棒性分析和鲁棒参数估计。
在鲁棒统计中,gap指令是一个非常重要的工具,它可以帮助用户根据数据集的离裙值情况来选择合适的估计方法。
1. gap指令的作用gap指令是MATLAB中鲁棒工具箱中的一个函数,它的作用是帮助用户确定具有鲁棒性的估计方法。
在统计分析中,数据集中常常存在离裙值,这些离裙值会对参数估计造成较大的影响。
gap指令可以帮助用户根据数据集的离裙值情况选择合适的估计方法,从而提高参数估计的鲁棒性。
2. gap指令的使用方法使用gap指令非常简单,用户只需将数据集作为输入参数传入函数中即可。
gap指令会根据数据集的离裙值情况,选择出具有鲁棒性的估计方法,并返回相应的结果。
用户可以根据返回的结果进行进一步的统计分析和数据处理。
3. gap指令的优势与传统的参数估计方法相比,使用gap指令的优势在于其具有鲁棒性。
在处理含有离裙值的数据集时,传统的参数估计方法往往会出现较大偏差,而使用gap指令选择的估计方法通常能够更好地适应数据集的特点,提高估计的准确性和稳定性。
4. gap指令的应用实例为了更好地理解和使用gap指令,我们可以举一个简单的应用实例来说明其具体的作用。
假设我们有一个含有离裙值的数据集,我们希望对其进行参数估计。
我们可以使用传统的参数估计方法来计算参数估计值,然后再利用gap指令来选择具有鲁棒性的估计方法。
我们可以将两种方法得到的结果进行比较,从而评估gap指令的作用和效果。
5. 总结通过对MATLAB中鲁棒工具箱中的gap指令进行简要介绍和分析,我们可以看到其在处理含有离裙值的数据集时具有明显的优势。
使用gap指令可以帮助用户选择更加鲁棒的参数估计方法,从而提高估计的准确性和稳定性。
在实际的数据分析和统计建模中,gap指令无疑是一个非常有价值的工具,值得研究和应用。
在实际的统计分析和数据建模中,有时候我们会遇到数据集中存在离裙值的情况。
Scaling of virtual machine addresses in datacenter
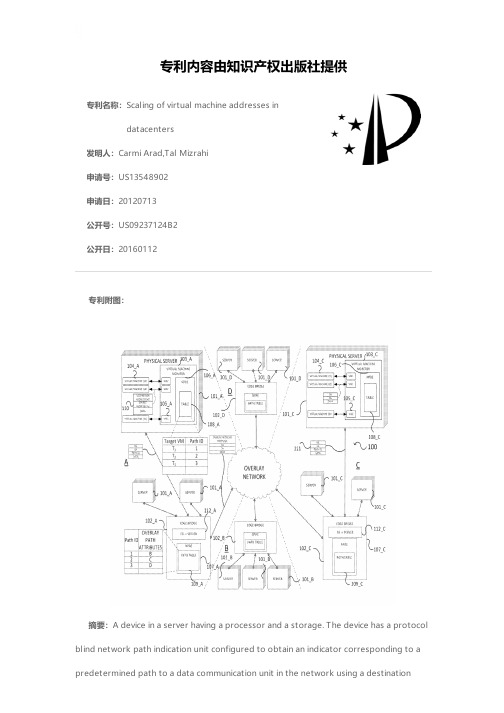
专利名称:Scaling of virtual machine addresses indatacenters发明人:Carmi Arad,Tal Mizrahi申请号:US13548902申请日:20120713公开号:US09237124B2公开日:20160112专利内容由知识产权出版社提供专利附图:摘要:A device in a server having a processor and a storage. The device has a protocol blind network path indication unit configured to obtain an indicator corresponding to a predetermined path to a data communication unit in the network using a destinationaddress of a received data packet, an upstream communication unit configured to transmit a network protocol blind packet including the data packet and the indicator corresponding to the predetermined data path to the data communication unit in the network, a combiner configured to bind the indicator to the data packet received by the downstream communication unit, and a protocol blind correlation storage unit configured to provide information related to target addresses and indicators corresponding to a plurality of predetermined data paths in the network. The protocol blind network path indication unit obtains the indicator corresponding to a predetermined path by accessing the protocol blind correlation structure.申请人:Carmi Arad,Tal Mizrahi地址:Nofit IL,Haifa IL国籍:IL,IL更多信息请下载全文后查看。
ONLINE MACHINE LEARNING-BASED MODEL FOR DECISION R
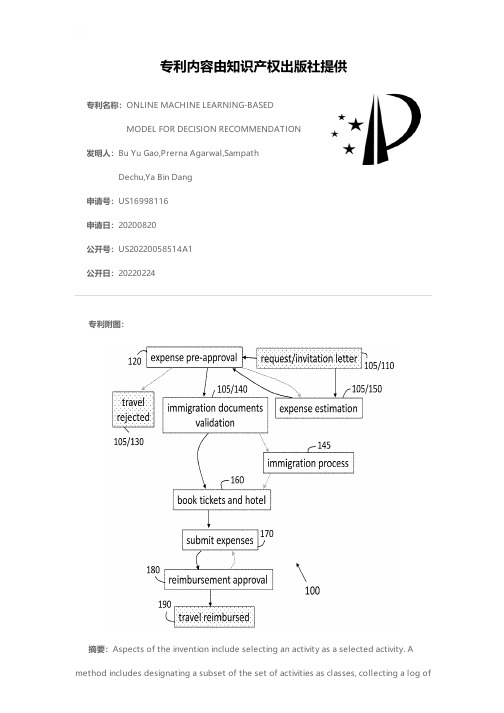
专利名称:ONLINE MACHINE LEARNING-BASEDMODEL FOR DECISION RECOMMENDATION发明人:Bu Yu Gao,Prerna Agarwal,SampathDechu,Ya Bin Dang申请号:US16998116申请日:20200820公开号:US20220058514A1公开日:20220224专利内容由知识产权出版社提供专利附图:摘要:Aspects of the invention include selecting an activity as a selected activity. Amethod includes designating a subset of the set of activities as classes, collecting a log ofinputs and outputs of each encountered activity as a data point each time the process is implemented, and extracting features from each data point that is collected to generate a feature vector from each data point. A teacher model is initialized with a first data point and updated with each data point subsequent to the first data point. A student model is initialized with a set of data points including the first data point such that every one of the classes is encountered at least once. The student model is updated with the teacher model. A set of features is input to the student model to obtain a prediction of the outcome of the selected activity.申请人:International Business Machines Corporation地址:Armonk NY US国籍:US更多信息请下载全文后查看。
如何使用堆叠自动编码器进行异常值检测(十)

在数据科学和机器学习领域,异常值检测是一个重要的问题。
异常值指的是与其他数据点明显不同的数据点,它们可能是由于错误、噪声或者其他原因而产生的。
使用堆叠自动编码器(stacked autoencoder)是一种有效的方法来进行异常值检测。
堆叠自动编码器是一种人工神经网络,用于学习数据的紧凑表示。
它由多个自动编码器组成,每个自动编码器都负责学习数据的一层表示。
通过在多个自动编码器之间传递数据,堆叠自动编码器可以学习数据的层次化表示,从而使得数据的特征更加丰富和抽象。
在使用堆叠自动编码器进行异常值检测时,首先需要对数据进行预处理。
这包括数据清洗、特征选择和标准化等步骤。
然后,我们可以使用堆叠自动编码器来学习数据的表示。
在学习过程中,我们可以利用无监督学习的方法来检测异常值。
堆叠自动编码器的一个常见方法是使用重构误差来进行异常值检测。
在学习过程中,堆叠自动编码器会尝试重构输入数据,然后计算重构误差。
如果某个数据点的重构误差远高于其他数据点,那么它很可能是一个异常值。
通过设置阈值,我们可以将重构误差高于阈值的数据点标记为异常值。
另一个方法是使用编码层的表示来进行异常值检测。
在学习过程中,堆叠自动编码器会学习数据的表示,这些表示可以用来区分正常数据和异常数据。
通过计算编码层表示的距离或相似度,我们可以对数据进行聚类和分类,从而检测异常值。
除了使用堆叠自动编码器进行异常值检测,我们还可以结合其他技术来提高检测效果。
比如,我们可以使用深度学习的方法来学习更加复杂和抽象的表示,从而提高异常值检测的准确性。
此外,我们还可以使用集成学习的方法来结合多个异常值检测器的结果,从而减少误报率和漏报率。
在实际应用中,使用堆叠自动编码器进行异常值检测需要注意一些问题。
首先,我们需要选择合适的网络结构和超参数,以及合适的损失函数和优化算法。
其次,我们需要进行模型评估和调参,以确保检测结果的准确性和稳定性。
最后,我们还需要考虑应用场景和数据特点,从而选择合适的异常值检测方法和技术。
基于LabVIEW的数字通信系统EVM和ACPR全自动化扫描测试

基于LabVIEW的数字通信系统EVM和ACPR全自动化扫
描测试
高风;王占仓
【期刊名称】《现代电子技术》
【年(卷),期】2008(31)15
【摘要】在虚拟仪器开发平台LabVIEW下,提出了一种远程控制安捷伦频谱仪和矢量信号分析仪89600实现对数字通信系统发射链路EVM和ACPR参数进行全自动化扫描的方法.将此方法应用到数字通信集成电路测试系统中,实现了对系统发射链路增益变化对性能参数影响的快速、高精度评估.对TD-SCDMA系统发射链路的测试结果表明,此方法较手动测试有着不可比拟的优势,将产品测试效率提高了60%.
【总页数】4页(P117-119,126)
【作者】高风;王占仓
【作者单位】北京工业大学电子信息与控制工程学院,北京,100022;北京工业大学电子信息与控制工程学院,北京,100022
【正文语种】中文
【中图分类】TP274
【相关文献】
1.基于LabVIEW的机械系统频率特性谐波扫描法测试实验系统 [J], 王珍;祁贵兵;赵洪健
2.基于LabVIEW的某探测组件自动化测试平台设计 [J], 阎俊;费月玲;张敏芳
3.基于TestStand及LabVIEW的航电通用故障监控软件自动化测试研究 [J], 叶颖樑;张铁军
4.基于TestStand及LabVIEW的航电通用故障监控软件自动化测试研究 [J], 叶颖樑;张铁军
5.基于LabVIEW的通用飞机低电平扫描场测试系统设计 [J], 石国昌;廖意;胡雅涵;陈亚南;包贵浩
因版权原因,仅展示原文概要,查看原文内容请购买。
捕获分隔分组接入

捕获分隔分组接入
王玲珑
【期刊名称】《通信技术与制造》
【年(卷),期】1998(000)001
【摘要】本文介绍一种新的蜂窝结构-捕获分隔分组接入(CDPA)这是一种分组结构。
可支持恒定比特速率业务并适用于多媒体的可变带宽业务。
这种方法将多址接入和信道重复使用进行综合,可实现高的频谱效率,即使用于时延受限制的电路业务也显示出许多优点。
不同于CDMA和TDMA,在那里每个连接的有效数据速率仅是分配给PCN(个人通信网)的全部无线信道的一小部分,而CDPA在必要时可使每个用户接入整个信道于一短暂的时间(
【总页数】3页(P27-29)
【作者】王玲珑
【作者单位】无
【正文语种】中文
【中图分类】TN929.5
【相关文献】
1.铁路分组交换网网络技术(三)——铁路分组交换网用户接入技术研究 [J], 汪齐贤
2.IPv6高速网络环境下数据分组捕获技术的研究 [J], 王相林;沈清姿;朱晨;刘立朋
3.分组式终端通过公用电话交换网接入公用分组交换数据网 [J], 郭德明
4.贵州联通分组传送网建设探索--分组系统解决县乡综合业务接入案 [J], 冯军;彭
力;阳林广;陈苛
5.基于平均分组和叠加相关的GPS信号捕获方法 [J], 杨久东;吴风华;王文军因版权原因,仅展示原文概要,查看原文内容请购买。
确认平滑:一种改善无线链路上的TCP性能的实现

确认平滑:一种改善无线链路上的TCP性能的实现
铁玲;诸鸿文;蒋铃鸽;周天翔
【期刊名称】《小型微型计算机系统》
【年(卷),期】2003(024)006
【摘要】许多研究者已经发现,在无线链路上使用链路层自动请求重传(ARQ)来恢复无线链路差错,会在无线链路上和TCP源端产生大量的突发性流,从而使路由器拥塞,导致分组丢失和吞吐量下降.本文提出了一种解决方案,在ARQ的接收端,利用漏桶对接收确认分组进行平滑,使分组的发送能保持一定的间隔.仿真结果表明,平滑能提供更好的吞吐量性能和更低的丢失率.
【总页数】2页(P1002-1003)
【作者】铁玲;诸鸿文;蒋铃鸽;周天翔
【作者单位】上海交通大学,现代通信研究所,上海,200030;上海交通大学,现代通信研究所,上海,200030;上海交通大学,现代通信研究所,上海,200030;上海交通大学,现代通信研究所,上海,200030
【正文语种】中文
【中图分类】TP393.3
【相关文献】
1.一种基于主动网络技术的改善无线链路上TCP性能的方案 [J], 廖小飞;李津生;洪佩琳;孙卫强;袁巍
2.使用改进SNOOP协议改善无线链路TCP的性能 [J], 柏溢;王民北;丁大钊
3.一种改善TCP在无线网络上性能的方法 [J], 朱杰杰;胡维华
4.无线链路的TCP性能问题及其改善 [J], 叶敏华;刘雨;张惠民
5.使用改进的Snoop改善无线链路TCP性能 [J], 刘雅凡
因版权原因,仅展示原文概要,查看原文内容请购买。
基于生物信息学筛选与胶质母细胞瘤预后及免疫相关的铁死亡基因

基于生物信息学筛选与胶质母细胞瘤预后及免疫相关的铁死亡基因孙皓;赵志娟;孟莲;刘春霞【期刊名称】《安徽医科大学学报》【年(卷),期】2024(59)3【摘要】目的探讨胶质母细胞瘤(GBM)中铁死亡的分子机制,为确定新的治疗靶点提供思路。
方法从基因表达综合数据库(GEO)中选取数据集GSE108474并通过GEO2R获取GBM差异表达基因,与铁死亡数据库(FerrDb)中基因集对比,获得铁死亡相关差异基因;利用DAVID数据库进行GO和KEGG富集分析;利用String网站创建蛋白互作(PPI)网络;利用Cytoscape软件确认网络中连接度高的枢纽基因;利用TIMER网站进行预后和免疫浸润分析;利用GEPIA网站进行RNA表达量和基因相关性分析;利用HPA数据库分析枢纽基因蛋白表达差异;利用TISIDB数据库进行肿瘤免疫特征相关性分析;运用实时荧光定量PCR在GBM细胞A172和U251MG 与正常星形胶质细胞HA1800间比较枢纽基因的mRNA差异。
结果5331个差异表达基因中有114个铁死亡相关基因;GO和KEGG富集分析显示114个基因可能在正调控基因表达等方面发挥作用,并通过铁死亡和自噬-动物等途径影响肿瘤进展;114个基因构成的PPI网络中确定了10个枢纽基因,其中细胞黏附分子44(CD44)、鼠双微体基因2(MDM2)和信号转导子和转录激活子3(STAT3)高表达的GBM患者生存率较低;GBM细胞中CD44、MDM2和STAT3的mRNA表达高于正常星形胶质细胞;高级别胶质瘤组织中CD44、MDM2和STAT3的蛋白表达高于正常脑组织;GBM中3个基因的表达均与铁死亡呈负相关;免疫浸润分析显示,GBM中CD44、STAT3与噬中性粒细胞、CD4^(+)T细胞和树突状细胞的浸润相关,MDM2与树突状细胞和CD8^(+)T细胞的浸润相关,并且3个基因与多种趋化因子以及趋化因子受体的表达相关。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Catalyst Residue Evaporator System Startup from T&I
Purpose The purpose of this procedure is to prepare the Catalyst Residue Evaporator System to receive feed from upstream equipment following maintenance. The system will need to initially accept enough feed material from upstream equipment to establish a total reflux condition of the equipment being started up. The act of placing the system in total reflux is to apply the proper amount of reboiler steam flow to create enough vapors in the overhead system to maintain a steady and continuous reflux flow rate while the reflux drum level controller is in Automatic control. Conditions • The Catalyst Residue Evaporator System has been repaired and the equipment is ready to return to service.
NOTE: This is for Training only, and is not intended to be used as a step by step procedure. PO/MTBE
NOTE: This is for Training only, and is not intended to be used as a step by step procedure. PO/MTBE
Catalyst Residue Evaporator System Startup from T&I
NOTE: This is for Training only, and is not intended to be used as a step by step procedure. PO/MTBE
Catalyst Residue Evaporator System Startup from T&I
NOTE: Air or oxygen can form a potentially explosive mixture with hydrocarbons, therefore, nitrogen purge all process lines and equipment to atmosphere until process lines and equipment is less than 3% oxygen. Do not nitrogen purge to the blowdown system or the flare system.
NOTE: This is for Training only, and is not intended to be used as a step by step procedure. PO/MTBE
Catalyst Residue Evaporator System Startup from T&I
NOTE: TBA may freeze and solidify at temperatures below 25˚C. As equipment is shut-down or blocked-in or when flow is stopped, ensure TBA is removed from lines and equipment if not heat traced. This condition can cause unnecessary down time or damage to the equipment. The overhead lines are not heat traced.
• Column and associated equipment has been pressure tested as per the "Tightness Testing of Process Equipment" procedure. • Line up the cooling water on the overhead condenser 01E0368. • Ensure the bypass valve around the overhead vent valve PV39011 is closed. • Ensure the vacuum package control valve PV-39011 is closed. • Line up the vacuum package control valve PV-39011. • Ensure vent line from the overhead condenser to PV-39011 is closed. • Line up the vent line from the reflux drum to PV-39011. • Ensure the bypass valve around the overhead distillate control valve LV-39017B is closed. • Ensure the overhead distillate control valve LV-39017B is closed. • Line up overhead distillate control valve LV-39017B.
NOTE: This is for Training only, and is not intended to be used as a step by step procedure. PO/MTBE
Catalyst Residue Evaporator System Startup from T&I
CATALYST RESIDUE EVAPORATOR SYSTEM STARTUP FROM T&I
NOTEonly, and is not intended to be used as a step by step procedure. PO/MTBE
NOTE: This is for Training only, and is not intended to be used as a step by step procedure. PO/MTBE
Catalyst Residue Evaporator System Startup from T&I
• Ensure the feed control valve FV-34218 is closed. • Line up the feed control valve FV-34218. • Ensure that 40 mm valve on the discharge of 01P0364-A/B that feeds into the Catalyst Recycle Surge Drum is closed. • Notify Console Operator Catalyst Residue Evaporator System is ready for feed.
• Line up the overhead distillate control valve LV-39017B to process fuel. • Ensure the block valve on the discharge side of the TBA Purification Overhead Pump 01P0357-A/B used for acetone flush of the Residue Evaporator Flash Drum level indicator LT-39017A is closed at discharge of TBA Purification Overhead Pump. • Line up the Residue Evaporator Distillate Pumps 01P0365-A/B as required. • Ensure the bypass valve around the bottoms control valve LV39017A is closed. • Ensure the bottoms control valve LV-39017A is closed. • Ensure the bypass valve around the bottoms effluent flow element FE-03049 is closed. • Line up the bottoms effluent flow element FE-39019. • Line up bottoms pumps 01P0364-A/B as required. • Drain the steam condensate from the S4 steam header upstream of Residue Evaporator Exchanger 01E0371-A/B manual block valve (one exchanger is used as spare).
