分子生物学实验-Molecular Biology Practical
课程英文1

本科生课程简介ART1100G 声乐与合唱(Vocality and Chorus)合唱发展的历史梗概;当今合唱用声的理念;歌唱发声基本原理及方法;简谱及五线谱视谱训练;学唱若干首合唱作品;欣赏世界顶级合唱团的演唱。
文法学院全年开课。
ART1101G 西方音乐史与作品欣赏(History and Appreciation of Western Music)本课程是本科学生的公共选修理课。
概括讲授西方音乐的发展历史及欣赏各历史时期的主要作品。
主要内容包括:1.古代音乐; 2.中世纪音乐;3.文艺复兴音乐; 4.巴洛克音乐; 5.古典主义音乐; 6.浪漫主义音乐; 7.20世纪音乐。
文法学院全年开课。
ART1102G 音乐欣赏(Music Appreciation)本课程是本课生的公共选修课。
课程安排是按照音乐的不同体裁对音乐的不同样式进行介绍,其中主要包括:民歌、中外艺术歌曲、中外歌剧、大型声乐作品、交响乐等。
通过本课使学生感受音乐文化的绚丽多姿及它在表现人类精神生活的不同样式。
文法学院全年开课。
ART1200G 花鸟画基本技法(Basic Skills of Drawing Flowers and Birds) 该课先学工笔花鸟画的基本技法,再学写意花鸟画的基本技法。
在讲清基本理论的同时,主要让学生掌握花鸟画的绘画技法。
教师课堂示范与学生课堂练习相结合,课上指导与课下作业相结合。
文法学院全年开课。
ART1201G 绘画与艺术欣赏(Appreciation of Painting and Art) 本课程是本科学生的公共选修课。
概括讲授古今中外绘画及艺术的发展历史,欣赏各历史时期绘画及艺术的主要代表作品与作者简介。
讲授人物画技法和网页创意、色彩学、服装的着装搭配艺术。
文法学院全年开课。
ART1202G 漫画(Caricature) 本课程是本科学生的公共选修课。
概括介绍世界各国漫画种类及名家名作,讲授漫画的基本技法;学会创意、绘制漫画。
英文课程库翻译对照(参考)一、学院开课单位基础医学院PreclinicalM

英文课程库翻译对照(参考)一、学院/开课单位基础医学院:Preclinical Medicine School 临床医学院:Clinical Medical College人文学院:College of Humanities中药学院:College of Traditional Chinese Pharmacy针灸学院:College of Acupuncture and Moxibustion管理学院:School of Management护理学院:College of Nursing信息中心:Information Center研究生部:Graduate faculty二、课程类别选修课:Selected Course必修课:Required Course专业基础课:Special Core Course专业课:Professional Course学位课:Degree course公共课:Public Course补本科课程:Supplement Undergraduate Courses三、专业名称1、中医:中医基础理论:Basic Theory of TCM中医临床基础:TCM Clinical Foundation 中医医史文献:TCM History and Document方剂学:Science of Formulae of Chinese Herbs中医诊断学:Diagnostics of TCM中医内科学:Traditional Chinese Internal Medicine中医外科学:Surgery of TCM中医儿科学:Pediatrics of TCM中医妇科学:Gynecology of TCM 中医骨伤科学:Orthopedics & Traumatology of TCM中医五官科学:Otorhinolaryngology of TCM针灸推拿学:Acupuncture & Moxibustion 民族医学:Ethnomedicine/Medicine of Chinese Minorities临床中药学:Clinical Pharmacology of TCM中医养生康复学:Health-preserving& Rehabilitation of TCM中医护理学:Nursing of TCM2、中西医结合:中西医结合基础:The basis of Integrative Medicine中西医结合临床:Clinical Science of Integrative Medicine3、药学:药物分析学:Pharmacoanalysis微生物与生化药学:Microbial and Biochemical Pharmacy4、中药学:临床中药学:Clinical Pharmacology of TCM中药化学:Chemistry of Chinese Meteria Medica中药药理学:Pharmacology of Chinese Meteria Medica中药制药学:Pharmaceutics of Chinese Meteria Medica中药生药学:Pharmacognostics of Chinese Meteria Medica5、公共管理:社会医学与卫生事业管理:Social Medical and Health Service Management四、学位类型科学学位:Science degree临床专业学位:Clinical professional degree五、学位层次:本科学位:Bachelor degree硕士学位:Master's degree博士学位:Doctor's degree/ Ph.D六、课程名称:1、公共课科学社会主义理论:Theories of Scientific Socialism自然辩证法:Nature Dialectics现代科技革命与马克思主义:Modern Science and Technology Revolution and Marxism硕士英语:English for Master’s Degree博士英语:English for Doctor’s Degree博士日语:Japanese for Doctor’s Degree 博士二外英语:Second Foreign Language for Doctor’s Degree (English)计算机应用:Application of Computer医用统计学:Medical Statistics名师大讲堂:Academician Lectures科研思路与方法:Scientific Ideas and Methods/ Research Courses汉语水平考试:HSK博士专业课:Doctor’s Professional Course2、专业课/专基课/选修课/学位课(1)基础医学院课程内经专题讲座:Forums and Lectures of Internal Classic 难经学术思想:Academic Thoughts of NanJing中国古代哲学:Ancient Chinese Philosophy中医基础专论:Monography of Basic TCM Theories伤寒论专题讲座:Treatise on Cold Diseases金匮要略专题讲座:Treatise on Golden Chamber温病学专题讲座:Treatise on Warm Diseases中医训诂考据学:TCM Textology Exegesis 中国医学史:History of TCM中医文献学:Philology of TCM文献检索:Literature Retrieval各家学说:Various Schools of TCM中医医案:Medical Records of TCM中医处方方法学:Prescription Methodology of TCM中医辨证学:TCM Syndrome Differentiation中医诊断古籍选读:Selected ancient readings of TCM Diagnosis中医内科学(中诊专业):Traditional Chinese Internal Medicine临床中药学:Clinical Pharmacology of TCM神经生理学:Neurophysiology生物化学:Biochemistry生物化学实验:Biochemistry Experiment 医学分子生物学:Medical Molecular Biology分子生物学实验:Molecular Biology Experiment实验动物学:Experimental Zoology神经解剖学:Neuroanatomy局部解剖学:Medical Topography头面部局部解剖学:Craniotopography医用细胞学基础:Medical Foundation of Cytology组织细胞分子学实验:Histiocyte Molecular Experiment病理生理学:Pathophysiology医学免疫学:Medical Immunology临床流行病学(DME):Clinical Epidemiology (DME)DME:Design, Measurement and Evaluation中医基础理论(补本科):The Basic Theory of TCM中医养生学概论:Introduction to TCM Health-preserving中医康复学概论:Introduction to TCM Rehabilitation中医饮食营养学:Nutriology of TCM循证医学:Evidence-based Medicine核酸研究技术在中医药学的应用:Application of Nucleic Acid Research Techniques in TCM(2)临床医学院中医内科学专题讲座:Treatises on Traditional Chinese Internal Medicine中医内科杂病研究:Miscellaneous Internal Diseases of TCM中医外科学专题讲座:Treatises on Traditional Chinese Surgery中医妇科学专题讲座:Treatises on TCM Gynecology中医儿科学专题讲座:Treatises on TCM Pediatrics西医内科学:Internal Medicine中西医结合内科学专题讲座:Treatises on Integrative Internal Medicine中西医结合外科学专题讲座:Treatises on Integrative Surgery中西医结合妇科学专题讲座:Treatises on Integrative Gynecology中西医结合五官科学专题讲座:Treatises on Integrative Otorhinolaryngology针灸推拿学专题讲座:Treatises on Acupuncture and Massage中西医结合骨伤科专题讲座:Treatises on Integrative Orthopedics and Traumatology 中西医结合儿科学专题讲座:Treatises on Integrative Pediatrics西医外科学:Surgery临床病理学基础:Basic Theories of Clinical Pathology(3)针灸学院针灸学:Acupuncture中医推拿学:Chinese Massage实验针灸学:Experimental Acupuncture针灸医籍各家学说:Various Schools of Acupuncture masterpieces中医气功学:Qigong of TCM针灸现代研究进展(博士):Modern Research Progress of Acupuncture and Moxibustion针刀疗法:Acupotomology therapy(4)管理学院高级统计学(一):Advanced Statistics(1)卫生经济理论与方法:Health Economic Theory and Method社会医学:Social Medicine现代医院管理:Modern Hospital Management卫生改革与卫生经济政策:Health Reform and Health Economic Policy卫生机构会计实务:Accounting Practice in Health Institution高等教育学:Higher Pedagogy教育管理学:Science of Educational Management药事管理学:Science of Pharmacy Administration药品知识产权实务:Pharmaceutical Intellectual Property Practice高级数据库开发:Advanced Database Development高级网络技术:Advanced Network TechnologyQOL(生存质量)测量与评价:QOL Measurement and Evaluation社会与发展心理学:Social and Developmental Psychology宏微观经济分析:Macro and Micro Economic Analysis现代企业管理:Modern Enterprise Management医院质量与标准化管理:Hospital Quality and Standardized Management财务分析与管理:Financial Analysis and Management高级统计学(二):Advanced Statistics(1)卫生事业管理:Health Service Management流行病学:Epidemiology卫生统计数据库分析与应用:Health Statistics Database Analysis and Application经济法律通论(民法、诉讼法、合同法):General Theory of Economic Laws(Civil law, procedural law and contract law)知识产权法:Intellectual Property Law药品质量管理:Drug Quality Control药事法学:Law of Pharmaceutical Affairs 计算机统计软件应用:Computer statistics software applications 组织行为学:Science of Organizational Behavior人力资源管理:Human Resource Management技术经济学:Technical Economics医药营销:Pharmaceutical Marketing企业战略管理:Enterprise Strategic Management决策支持系统(信息管理):Decision support systems (information management)教育心理学:Education Psychology电子商务:E-commerce项目管理:Project Management古代管理思想研究:Study on Ancient Management Thought论文写作:Essay writing健康教育与健康促进:Health Education and Health Promotion现代管理理论:Modern Management Theory公共关系与危机管理:Public Relations and Crisis Management卫生服务成本研究与应用:Research and Application on Cost of Health Services(5)中药学中药学专论:Monography of Science of Chinese Materia Medica分析测试技术:Analysis and Test Technology中药化学专论:Monography of Chinese Pharmaceutical Chemistry中药药理学专论:Monography of Traditional Chinese Pharmacology中药制药学专论:Monography of Traditional Chinese Pharmaceutics中药生药学专论:Monography ofTraditional Chinese Pharmacognostics临床中药学专论Monography of Clinical Science of Chinese Materia Medica结构有机化学:Structure of Organic Chemistry波谱分析:Spectrum Analysis药性导论:Introduction of Drug Property 本草文献学:Philology of Chinese Materia Medica中医学选读:TCM Selected Readings中药成分分析:Analysis of Chinese Materia Medica Components中药信息学:Informatics of Chinese Materia Medica中药炮制学专论:Monography of Processing Chinese Materia Medica中成药学专论:Monography of Science of Chinese Patent Drug生物药剂学:Biological Pharmacology药用植物学专论:Monography of Medicinal Botany分子生药学:Molecular Pharmacognostics植物化学分类:Classification of Plant Chemistry中药资源学专论:Monography of Chinese Materia Medica Resources中药显微鉴定:Microscopic Identification of Chinese Materia Medica中药品种论述:Chinese Varieties Treatise 中药生物技术:Chinese Biotechnology生物制药专题:Special Subject on Biopharmaceutics药理学进展:Advancements of Pharmacology中药药效毒理研究思路与方法:Research Ideas and methods of Chinese Materia Medica Effect and Toxicology 物理药剂学:Physical Pharmacology科研思路与方法(中药专业) :Scientific ideas and Methods (for Chinese Materia Medica Profession)微生物与生化药学专论:Monography of Microbial and Biochemical Pharmacology 分子细胞生物学专论:Monography of Molecular Cell Biology中药生物技术应用:Biotechnology Applications in Chinese Materia Medica蛋白质工程:Protein Engineering计算机辅助药物设计:Computer-aided drug design微生物与生化药学研究方法:Microbial and Biochemical Pharmaceutical Research Methods药物分析专论:Monography of Drug Analysis药品质量控制:Drug Quality Control中药成分体内代谢与分析:Vivo Metabolism and Analysis of Chinese Materia Medica Components计算药物分析:Analysis of Drug Calculation生物药物分析:Biopharmaceutical Analysis 新药设计学:Science of New Drug Designing中药药理学:Chinese Pharmacology(6)护理学院护理研究进展:Nursing Research Progress 护理理论:Nursing Theory护理教育:Nursing Education中医临床护理概论:Introduction to TCM Clinical Nursing护理心理学:Nursing Psychology本翻译有不尽之处,欢迎各位老师同学提供更好的翻译建议!研究生院培养办。
PCR实验报告(分子生物学实验)

PCRLin Chengyu Bio 04 2010030007; Cooperator: Yuan XiaowenExperiment date: 2012-03-22 Submit Date: 2012-03-231Introduction1.1Background informationPCR (polymerase chain reaction) is a technique for amplifying DNA sequences in vitro.It was invented by Kary Mullis and his colleagues in 1985. It is widely used in genecloning, mutation, sequencing and detection.1.2Objectives(1)Learn the principle and method of PCR (Polymerase Chain Reaction).(2)Comprehend the significance of PCR technique in DNA manipulation.1.3Major principlesOne typical PCR cycle consists of three steps: denaturation, annealing and extension.(1)DenaturationFigure 1 Step 1: denaturationAs shown in Figure 1, the target double-strand DNA can be separated intosingle-strand DNA under high temperature (about 50 ℃).(2)AnnealingFigure 2 Step 2: annealingAs shown in Figure 2, when temperature is lower, the forward and reverse primers will bind to the single strand DNA, which are about 20 nt long, designed to be complementary to the both end of the target gene and often has restriction enzyme recognition sites at the 5’ ends.(3)ExtensionFigure 3 Step 3: extensionAs shown in Figure 3, when temperature rise to about 72 ℃, which is the optimal temperature of Taq, one kind of DNA polymerase working in high temperature. The discovery of Taq DNA polymerase is fundamental to PCR. As a result, new DNAstrand complementary to the target DNA will be synthesize at the end of 3’ ends ofthe primer until temperature rise to denature the double strand again.2Experiment Operation2.1Material(1)pCMV-Myc-T10 (SIPAR), 10 ng;Figure 4 Plasmid profile (pCMV – Myc)(2)Forward and reverse primer, 1 µM;Figure 5 Forward primerFigure 6 Reverse primer(3)DNA polymerase: r Taq, 5 U / µl.2.2Chemicals and apparatus2.2.1SolutionsTable 1 Solutions2.2.2Apparatus(1)Pipettes;(2)PCR systems;(3)Eppendorf tubes;(4)Centrifuge;(5)Gel electrophoresis apparatus;(6)UV gel imaging system;(7)Biodev PCR purification kit.2.3Procedure2.3.1PCR(1)Add all 7 kinds of materials or solutions into PCR tubes following Table 2;Table 2 50 µl system for PCR(2)Put the tubes into the instrument for PCR, set program as Table 3;(3)Take out the tubes when program stops.2.3.2Agarose gel electrophoresis(1) During PCR, place a clean electrophoresis mold on a horizontal surface,carefully insert a comb (15 µl slots) into the mold;(2) Pour the agarose gel solution (contains 1.0 g agarose in 100 ml TAE) intothe mold gently to avoid bubbles. The height of gel shall be slightly higher than the black line on the side face;(3) Wait for approximately 30 min for solidification, when PCR complete aswell;(4) Add 4 µl 3×Loading buffer, 4 µl ddH2O and 4 µl PCR product to a pieceof sealing film, and mix with pipette;(5) Carefully remove the comb, place the mold in the electrophoresischamber, and add 1×TAE buffer in order to cover the gel to a depth of approximately 1 mm;(6) Load 15 µl solution into the slots of the gel: one slot for one tube. Load 4µl of 1kb DNA ladder into the slot beside the sample;(7) Start electrophoresis immediately samples loading at 100 V, and stopwhen Bromophenol blue is 2/3 gel length from the starting line;(8) Place the gel into EB working solution for 20 min, and observe under UVat 310 nm. The red fluorescent bands show where DNA is and their darkness show the quantity. Then take photograph for records;2.3.3PCR product purification(1)Add 50 µl PCR product to 400 µl Binding buffer, and mix with pipette;(2)Transfer the solution to the mini-spin column, and centrifuge at 12,000 rpmfor 30 sec. Discard the waste solution in the collection tube;(3)Add 450 µl Washing buffer to the mini-spin column, and centrifuge at12,000 rpm for 30 sec. Discard the waste solution in the collection tube;(4)Repeat Step 3 once more;(5)Centrifuge at 12,000 rpm for 2 min to get rid of ethanol;(6)Add 40 µl ddH2O to the center of the mini-spin column, put the column in anew Eppendorf tube, and incubate for 1 min. Then centrifuge at 12,000 rpmfor 1 min.(7)Preserve the product under -20 ℃.3ResultFigure 7 PCR product agarose gel electrophoresis:Lane I – Product of Y uanLane II – Product of LinThe target gene, together with restriction enzyme recognition sequence added in theprimer, is about 0.9 kb long, which corresponds with the band in Figure 7.4ConclusionThe PCR product contains the target gene we want to amplify in the template, and its quantity is enough for the following operation.5Reference【1】Liu Jinyuan, Zhang Shuping, Wu Yaoting, Introduction of Molecular Biology Experiment.。
分子生物学实验手册

版权声明:本站几乎所有资源均搜集于网络,仅供学习参考,不得进行任何商业用途,否则产生的一切后 果将由使用者本人承担! 本站仅仅提供一个观摩学习与交流的平台, 将不保证所提供资源的完 整性,也不对任何资源负法律责任。
所有资源请在下载后 24 小时内删除。
如果您觉得满意, 请购买正版,以便更好支持您所喜欢的软件或书籍!☆☆☆☆☆生物秀[]☆☆☆☆☆中国生物科学论坛[/bbs/]☆☆☆☆☆生物秀下载频道[/Soft/]生物秀——倾力打造最大最专业的生物资源下载平台!■■■ 选择生物秀,我秀我精彩!!■■■欢迎到生物秀论坛(中国生物科学论坛)的相关资源、软件版块参与讨论,共享您的资源,获 取更多资源或帮助。
2000/01 Cardiff School of BiosciencesMolecular Biology IBiochemistryLevel II:PracticalManualDr Peter KilleGENE CLONING PRACTICALThe purpose of this practical is to introduce you to the basic techniques of gene cloning. The techniques that you will use include:The preparation of plasmid DNADigestion of DNA with restriction endonucleasesAnalysis of DNA fragments by electrophoresisLigation of DNA fragments to produce recombinant moleculesTransformation of bacterial cellsSelection of clones by insertional inactivation of a geneSelection of clones by expressed phenotypesAmplification of specific DNA fragments by the polymerase chain reaction (PCR)Induction of gene expressionEnzyme activity measurementParticular attention should be paid to the following points:[1] The manual should be used as a guide to assist you with the experiments rather than asa complete recipe book.[2] There are certain safety requirements (in addition to those appropriate to normallaboratory practice) that must be adhered to (see next page). It is a requirement that you read the section on safety before commencing any work.[3] These experiments will work well but only if the advice on page 4 is followed.TIMETABLEPracticals will be held in lab. 101W every Tuesday. Sessions will start promptly at 10.00 am and will continue through until 18.00. If you cannot attend any part of the practical please inform a demonstrator or the lecturer in charge.Week 1[1] Preparation of plasmid DNA[2] Digestion of DNA with restriction endonucleases[3] Analysis of DNA fragments by electrophoresis[4] Ligation of DNA fragments to produce recombinant moleculesWeek 2[5] Transformation of bacterial cells[6] Plating out of bacteria on selective mediaWeek 3* Monday before practical (approx. 1 hour)[7] Plating out of bacteria for insertional and phenotypic screening* Tuesday[8] Selection of clones by insertional inactivation of a gene[9] Selection of clones by expressed phenotype[11] Preparation of plasmid DNA from recombinant plasmids[10] Identification of specific gene by polymerase chain reaction (PCR) and restrictionmapping.Week 4* Monday before practical (approx. 1 hour)[11] Inoculation of cultures for induced gene expression* Tuesday[12] Analysis of induced gene expression by enzyme activity measurements[13] Analysis of PCR and restriction fragments plasmids prepared in [10].SAFETYGeneral PointsAll experiments involving recombinant DNA work must be approved by a local safety committee who are responsible for quantifying the biological risk and designating the level of containment that is required for the work to proceed. The committee has a legal obligation to submit annual reports on all experiments carried out in College to the Health and Safety Executive. This experiment has been approved subject to the work being carried out in accordance with 'good microbiological practice'.Specific PointsIn addition to the safety procedures that are applicable to any laboratory work the following points should be noted.[1] Experiments must be carried out using good microbiological practice. This meansthat the normal safety precautions applicable to the handling of microorganisms apply to the experiments described in this manual.[2] Compounds that interact with DNA, e.g. ethidium bromide, are mutagens andcarcinogens. Do not allow these compounds to come into contact with yourself orother people. Wear disposable gloves at all times.[3] The transilluminator is a powerful source of ultra violet radiation and will causeserious damage to unprotected skin and eyes. Always ensure that there is a uv-opaque material between you and the light source, i.e. wear gloves and use suitable eye/faceprotection.[4] Phenol-containing solutions can cause severe burns. Make sure that you are wearinggloves when handling these solutions.[5] If you have recently been prescribed medicines by your doctor, especially antibiotics,anti-inflammatory drugs or immunosuppresants, please inform Dr P Kille beforestarting the practical.PRACTICAL CONSIDERATIONSEssentially, the basic techniques of gene cloning are straightforward providing that certain protocols are followed. These practical points are important if you are to obtain successful results.[1] Keep solutions on ice at all times. Even short exposure of certain components, e.g.some restriction endonucleases, to room temperature will cause significantinactivation.[2] Nucleases are ubiquitous and some are extremely stable enzymes. It is vital that theseenzymes are not allowed to come into contact with your apparatus or solutions.Always wear disposable gloves - the skin is an excellent source of nucleases and there is enough enzyme in a fingerprint to ruin your experiments. Also, most of yoursolutions and apparatus have been autoclaved to destroy nucleases so avoidunnecessary handling.[3] The key to many of the techniques described in this manual is the ability to pipettesmall volumes carefully and accurately. It is essential when pipetting small volumes to ensure that the tip of the pipette is touching the surface of the container beforeexpelling the solution. Always use clean pipette tips to avoid cross contamination.Pulse spin microfuge tubes to ensure that all of the added components of reactionmixtures are mixed at the bottom of the tube.[4] The transilluminator should be used for the minimum time possible as prolongedexposure of DNA samples to uv radiation causes random strand breakage - also thefilter has a finite life. Please avoid scratching the surface of the transilluminator asthis will destroy the effectiveness of the filter - a new transilluminator costs £1000.A general point about recombinant DNA work is that reagents are extremely expensive. Please use reagents/enzymes as sparingly as possible. In general, expensive components such as enzymes will have been pre-dispensed into tubes. The other components of the reactionmixture should be added to these tubes (see protocols for further details).BACKGROUND TO THE EXPERIMENTSAlginates are polysaccharides comprised of (1-4)-linked β-D-mannuronate andα-L-guluronate. This polysaccharide is produced by marine macroalgae and certain bacteria, and the algal alginate is widely used by the food and pharmaceutical industries (approx. 22000 tonnes/annum).Certain bacteria produce enzymes (alginate lyases or alginases) which degrade alginates. A gene encoding an alginate lyase has been cloned from the bacterium Klebsiella pneumoniae into a cosmid vector to produce the construct pSP1. In this practical the bacterium E. coli strain LE392 will be used as a source of plasmid pSP1. This plasmid contains the aly gene which encodes the alginate lyase (Fig. 1).Objectives of the Practical[1] Both the plasmid vector (pHG327) and the 'foreign DNA' (pSP1) will be isolated andpurified from cultures of E. coli.[2] Both pHG327 (Fig. 2) and pSP1 will be cut with an appropriate restrictionendonuclease (Hin dIII).[3] Fragments of pSP1 will be added to the cut pHG327 and recombinant molecules willbe allowed to form. The recombinant molecules will be treated with DNA ligase to reform the phosphodiester linkages.[4] Bacterial colonies will be screened on MacConkey agar to detect transformants.[5] Recombinants containing the aly gene will be isolated by detecting expression of thealginate lyase enzyme.[6] DNA will be isolated from recombinants, cut with restriction endonucleases and the fragments analysed on agarose gel electrophoresis. This will allow the aly gene to be mapped onto pSP1.[7] The polymerase chain reaction (PCR) will be used to identify the HindIII fragment of the pSP1 that contains the aly gene from crude bacterial extracts.[8] The expression of alginate lyase by recombinants will be quantified by measurement of specific enzyme activities.HindIIIHindIIIFig. 1 Map of pSP1(Positions of restriction sites are approximate)(Low copy number) (High copy number)Fig. 2 Map of pHG327BACTERIA, ENZYMES AND ANTIBIOTICSBacterial strainsYou will use one or more of these strains of Escherichia coli which have the following genotypes:Escherichia coli JM109 ∆(lacpro AB),end A1, rec A1, thi, sbc B15, hsd R4, sup E44, rel A1(F', tra D36, pro AB+, lac I q Z∆M15)Escherichia coli JM107 ∆(lacpro AB), end A1, gyr A96, thi-1, hsd R17, sup E44, rel A1, (F',tra D36, pro AB+, lac I q Z∆M15) (pHG327, Ap r)Escherichia coli LE392 hsdR514 (rk-,mk+), supE44, supF58, lacY1 (pSP1, Km r)Some genetic markers in frequently used E. coli strains (Data from Promega)Symbol Description Effectend A Endonuclease mutation Improves quality of plasmid DNA isolationsF' Host contains F' episome Provides essential functions to host cellgyr DNA gyrase mutation Confers resistance to nalidixic acidhsd R Restriction system mutations Allows cloning without cleavage of plasmid DNAlac repressor protein Inhibits transcription from lac promoterthelac I q Overproduceslac Y Galactose permease mutation Blocks lactose utilizationlac Z β-D-Galactosidase mutation Unable to produce β-D-galactosidase. Strainssuitable for use with lac Z-containing vectorspro AB Mutations in proline metabolism Requires proline for growth in minimal medium rel A Mutation eliminating stringent factor RNA synthesis in the absence of protein synthesis sbcB15Exonuclease I mutation Allows general recombination in rec BC mutants sup E Supressor mutation Suppresses amber (UAG) mutationssup F Supressor mutation Suppresses amber (UAG) mutationstra D Transfer factor mutation Prevents transfer of F' episomerec A1 Mutation in recombinantion Prevents recombination of introduced DNA withhost DNA.Restriction enzymesEnzyme Microorganism SpecificityBam HI Bacillus amyloliquifaciens H G\GATCCEco RI Escherichia coli RY 13 G\AATTCHin dIII Haemophilus influenzae Rd A\AGCTT164 CTGCA\GstuartiiPst I ProvidenciaXho I Xanthomonas holcicola C\TCGAGAntibioticsAp AmpicillinKm KanamycinAp r Ampicillin-resistantphenotypephenotypeAp s Ampicillin-sensitiveMINI-PREPARATION OF DNAExtraction of Plasmid DNA from bacterial cells.Materials/Solutions Required5ml culture of E. coli JM 107 containing pHG3275ml culture of E. coli LE392 containing pSP1Collection Tubes.Sterile water warmed to 70°CSV Mini prep Kit (The SV Miniprep Kit is manufactured by Promega)Cell resuspension solution (50mM-Tris, pH 7.5; 10mM-EDTA; 100µg/ml RNase A) Cell lysis solution (0.2M-NaOH, 1% SDS)Alkaline Protease solutionNeutralisation solution (4.09M Guanidine hydrochloride, 0.759M-Potassium acetate,2.12M Glacial acetic acid)SV spin columnsSV Column wash solution (162.8mM Potassium Acetate, 27.1mM Tris-HCl, pH7.5 -EDTA - Add 1.75 volumes of 95% ethanol before use i.e. 35mls of ethanol to 20mls Wash concentrate).MethodYou will be provided with bacterial cultures that have been grown overnight at 37°C in medium containing appropriate antibiotics.[1] Pellet bacterial cells by centrifuging 5ml of each bacterial culture for 10mins at 3K ina bench top centrifuge.[2] Remove the clear supernatant. Add 250µl of Cell Resuspension Solution andresuspend the cell pellet by brief vortexing.[3] Transfer each cell suspension to separate sterile eppendofs.[4] To each add 250µl of Cell Lysis Solution and mix by inverting the tube four times (donot vortex). Incubate for 1-5 minutes at room temperature until solution clears.10µl of Alkaline Protease solution and mix by inverting the tube 4 times. [5] AddIncubate for a further 5 minutes at room temperature. Do not incubate for longerthan 5 minutes350µl of Neutralisation Solution and immediately mix by inverting the tube [6] Addseveral times.[7] Spin in the microcentrifuge for 10 min.A. Purification with Centrifugation.[8A] Transfer the cleared lysate to a Spin Column by decanting. Avoid disturbing of transferring any of the white precipitate with the supernatant. Insert the Spin Column into a collection tube.[9A] Spin in the microcentrifuge for 1 min. Remove the spin column from the tube and discard the flowthrough from the Collection tube. Reinsert the Spin Column into the Collection Tube.[10B] Add 750µl of Column Wash solution (previously diluted with ethanol) to the Spin Column.[11A] Spin in the microcentrifuge for 1 min. Remove the spin column from the tube and discard the flowthrough from the Collection tube. Reinsert the Spin Column into the Collection Tube.[12A] Add 250µl of Column Wash solution (previously diluted with ethanol) to the Spin Column.[13A] Spin in the microcentrifuge for 2 min.[14A] Transfer Spin Column to a new sterile eppendoff.B. Purification with Vacuum.[8B] Insert the Spin Column into vacuum manifold. Transfer the cleared lysate to a Spin Column by decanting. Avoid disturbing of transferring any of the white precipitatewith the supernatant.[9B] Apply vacuum to pull liquid through the column. When all liquid has been pulled through column, release vacuum.[10B] Add 750µl of Column Wash solution (previously diluted with ethanol) to the Spin Column.[11B] Apply vacuum to pull liquid through the column. When all liquid has been pulled through column, release vacuum.[12B] Add 250µl of Column Wash solution (previously diluted with ethanol) to the Spin Column.[13B] Apply vacuum to pull liquid through the column. When all liquid has been pulled through column, release vacuum. Transfer to Collection tube and spin in themicrocentrifuge for 2 min to remove any residual wash solution.[14B] Transfer Spin Column to a new sterile eppendoff (with the lid cut off).DNA Elution[15] Elute the plasmid DNA by adding 100µl of nuclease-free water (at 65-70°C) to theMinicolumn.[17] Spin the microfuge tube and column for 1 min in a microcentrifuge.[18] Remove and discard the Minicolumn but keep the liquid. The plasmid DNA, which iscontained in the remaining liquid should be transferred with a pipette to a freshmicrofuge tube (with a lid) and may be stored at 4°C or -20°C until required.The rationale behind the various steps in the experimental protocol is as follows:Step 3 Cell resuspension solution contains EDTA which makes the bacterial outer membrane permeable and RNase which degrades RNA.Step 4 The NaOH denatures the proteins and disrupts the chromosome. The detergent, SDS, lyses the bacteria allowing the plasmids to leak out of thecells. Alkaline Protease breaks down bacterial proteases.Step 5 The guanidine hydrochloride and potassium acetate forms a precipitate with the proteins and chromosomal DNA while the plasmid remains in solution.The acetic acid neutralises the alkali used in the previous step.Step 6-7 The plasmid (in solution) is separated from the precipitated proteins,chromosome and cell debris.Step 8- Under high salt conditions Nucleic acids binds (plasmid DNA) to Spin Column.Step 9-13 Contaminating proteins and salts are washed away. The washing solution contains salt ethanol and a small amount of salt so that mono-nucleotides areremoved but plasmid DNA remains bound.Steps 14 Remaining wash solution is removed.Steps 15-18 Water at 65°C causes the DNA to be released from the resin. The plasmid DNA solution is then collected in a fresh eppendoff when the column is spunin the microcentrifuge.RESTRICTION ENDONUCLEASE DIGESTION OF DNApSP1 is digested into a number of fragments using the restriction enzyme Hind III. The expression vector pHG 327 is also cut with Hind III, in a single location, causing it to linearise. The aim is to ligate the pSP1 fragments into the cut pHG 327. However, when the ligation is performed there is also the possiblity that pHG 327 will reform without incorperating any fragments of pSP1. To prevent this we treat the linearised pHG 327 with a Shimp dephosphorylase enzyme which removes the 5’ phosphate groups making it only possible for the pHG 327 to reform if a fragment from pSP 1 is incorrperated.Materials/Solutions Required10X Digest bufferHin dIII enzymeDNA sampleSterile waterMethod[1] Decide a suitable volume of each plasmid DNA and the final volume of digestion.digest Preparativedigest AnalyticalpHG327 pSP1 DNA volume 10µl 15µl 25µl Final volume 20µl 30µl 50µl [2] You will be provided with tubes containing 2µl of restriction enzyme. To each tubeadd 1/10 final volume of 10X Buffer, sample DNA and sufficient sterile water to give the appropriate final volume.[3] Flick the bottom of the tubes to mix the contents (remember that it is easy to denaturethe enzyme) and then microfuge for 2-3 seconds so that all of the contents are at thebottom of the tube.[4] Incubate the tubes at 37°C for 2 hours (see “DEPHOSPHOYLATION OF VECTORDNA” for instructions of what to do with pHG327 digestion) and then transfer thetubes to a 65°C waterbath for 15 min to inactivate the endonuclease anddephosphoylation enzyme.[5] Use a portion (e.g. 7-8µl) of the digest for analysis by gel electrophoresis and store theremainder for use in the ligation reaction.DEPHOSPHOYLATION OF VECTOR DNAMaterials/Solutions RequiredShrimp PhosphataseMethodpHG327 digestion only.[4a] After the pHG327 Vector DNA has been incubating with Hin dIII for 1 hour remove it from the water bath and add 1µl of Shrimp Phosphatase. Flick the bottom of the tubes to mix the contents and then microfuge for 2-3 seconds so that all of the contents are at the bottom of the tube.[4b] Place the digestion mixture in the water bath to incubate at 37°C for an additional1 hour and then continue as for normal digestion’s at step [5].FORMATION OF RECOMBINANT DNA MOLECULESThe digested pSP1 fragments are ligated into the linearised dephosphorylated pHG 327. Before the ligation is performed the digested DNA will be purified with chloroform/isoamyl alcohol and concentrated by precipitation with ethanol.Materials/Solutions RequiredHin dIII digested DNA samplesSterile waterChloroform/iso amyl alcoholTE buffer3M sodium acetate100% ethanol (at -20°C)10X Ligation bufferT4 DNA ligaseMethod[1] Add the following components in a microfuge tube placed on ice:Dephosphorylated Hin dIII digested pHG327 (vector) DNA 20µlHin dIII digested pSP1 (foreign) DNA 40µlSterile diH2O 40µlTotal 100µl 100µl of chloroform/iso amyl alcohol to the tube, vortex and microfuge for [2] Add2 min.[3] Transfer the aqueous (upper) phase to a fresh tube. It is essential that none of thechloroform phase is transferred.10µl of 3M sodium acetate and 250µl of cold ethanol to the tube. Leave in an [4] Addethanol/dry ice bath (or at -70°C) for about 30 min.[5] Microfuge for 15 min, carefully pour off the supernatant and dry the pellet (may notbe visible). Be careful not to lose the pellet.[6] Resuspend the pellet in 14µl of sterile water[7] You will be provided with a tube containing 2µl of T4 DNA ligase. To this, add 2µlof 10X ligation buffer, the resuspended pellet (14µl) and gently mix.[8] Briefly microfuge to bring contents to the bottom of the tube and incubate at 16°Covernight.TRANSFORMATION OF HOST BACTERIA WITH RECOMBINANT DNAThe recombinant DNA molecules form during the ligation are inserted into E. coli cells and the bacteria containing the plasmids are selected by their resistance to Ampicillin.Materials/Solutions Required50µl competent E. coli DH5α on ice.1 ml Sterile S.O.C. Media.Ligated DNA sampl.eLB Agar capsules.100 mg/ml Amp.Method(a) Preparation of Agar plates[1] Put 8 capsules of LB Agar into the provided 500 ml Duran Bottle and add 200 mls ofSterile water. Screw cap on LEAVING IT LOOSE and cover top with tin foil. Adda strip of autoclave tape and place into autoclave for 20 minutes at 120 p.s.i..[2] When autoclave cycle is finished USE TERMAL RESISTANT GLOVES toremove molten agar from autoclave. Tighten lid and swirl to insure capsules are fully dissolved and place in 55°C water bath.[3] Check molten L Agar is at 50°C remove and pour one agar plate ~20 mls (make surethis plate is labled “LB-AGAR NO ANTIBIOTIC”) then to the remainder add Amp toa final concentration of 100 µg/ml.[4] Mix by swirling and place back into 50°C waterbath for 2-3 minutes until majority ofbubbles disappear.[5] Carefully pour media evenly into 6 sterile petri dishes using good aseptic technique.Any excess LB-agar should be washed down the sink immeadiately with lots of warm water.[6] Wait for plates to solidify (~30 minutes) and dry by opening slightly and placingupside down in the 37°C incubator.[7] Once dry close plate and leave on bench ready for transformation.E. coliof(b) Transformation[8] Add the DNA sample (maximum volume 5µl) to 50µl of competent cells and gentlytap the tube to mix.[9] Incubate the tubes on ice for 30 minutes. Then at 37°C for 20 seconds and thenreplace them on ice for an additional 2 minutes.[10] Add 0.95ml of SOC media and incubate at 37°C for 1 hour.[11] *Label the plates before adding bacteria.[12] Spread 150 µl portions of the transformed bacteria onto 6 of your LB Amp plates(prepared in Section a) and the last 100 µl portion on the LB plate containing noantibiotic. *Label remaining LB Amp plate and store in fridge for use in following week.[13] Incubate upright until the agar surface is dry before inverting the plates and leavingovernight at 37°C.SCREENING CLONESRecombinant clones (i.e. those colonies which contain pHG 327 with a pSP1 insert) will be identified by insertional activation (described in rational section). The recombinant clones identified may contain any of the five fragments produced by digestion of pSP1 with Hind III. In order to determine which of these recombinant clones contains the aly gene (encoding alginate lyase), the ability of the colonies to degrade alginate is assessed.Materials/Solutions RequiredMacConkey plate (contain 100µl Ap/ml)Alginate plate (contain 100µl Ap/ml)Sterile applicator sticksCulture dish containing 24 wells each holding 2mls of LBamp media.Method : Monday Evening[1] Examine the plates that you have prepared previously. The LB plate with noantibiotic should show lawn growth whilst you should be able to identify singlecolonies on the LB Amp plates. Only bacteria containing pHG327 or pHG327 plusinsert DNA should grow on LB Amp plates.[2] On back of MacConkey plate and Alginate plate draw a 6 x 4 grid and label eachsquares A1-A6, B1-B6, C1-C6 and D1-D6. Wells in culture dishes are alreadylabelled in the same manner, check dish orientation.[3] Pick selected individual colony from LB Amp plates with Sterile applicator stick.Place gently onto square A1 of alginate plate, then A1 of MacConkey plate and finally into well A1 of culture dish. Repeat this procedure with new collonies until allsquares all filled (total 24 collonies).[4] Incubate the plates overnight at 37°C.Tuesday Morning[5] Count the number of red colonies which are Lac+ bacteria, and the white colonieswhich are Lac- on the MacConkey agar plate. Note location of white collonies.[6] Overlay the alginate plates with 10%(w/v) cetyl pyridinium chloride (CPC) solutionand leave for 5-10 min. Undegraded alginate will appear opaque white and colonies producing alginate lyase will be surrounded by a clear zone. Placing the plates on ablack background will help you to visualise the clearing zones. CPC is bacteriocidal, therefore you will need to utilise the corresponding colonies on the MacConkey agar plates for further study. Note location of colonies that give clearing zones.[7] Identify i) Collony that is white on MacConkey agar plate and gives clearing zone. ii)Collony that is red with no clearing zone. Take spare LB AMP plate prepared inprevious week, divide in half and using sterile applicator sticks streak out these twocollonies. Give plate to demonstrators at end of session.[8] Locate the wells in culture dish that contain the liquid replicate cultures of the thosecollonies identified in [7]. Prepare bacterial DNA from these cultures and anaylse by performing restriction digests (analytical, as for pHG 327) and PCR.The rationale behind each step of the experimental protocol is as follows:Step 1 Lawn growth on L agar indicates that the competent cells were viable. No growth on MacConkey agar (containing ampicillin) indicates that the host cells were sensitive to ampicillin and cannot grow on this medium.Step 2 pHG327 confers antibiotic resistance on the host cells, therefore the transformants should be able to grow on medium containing ampicillin.Step 3 The vector pHG327 contains the lacZ gene encoding β-galactosidase, a gene which has been partially deleted in the particular host cells that we have used. Therefore, it is only the bacteria containing intact pHG327 that are able to metabolise lactose(present in MacConkey agar) to produce acid which turns the indicator phenol red(also present in MacConkey agar) from colourless to red. Those colonies that remain white should contain genuine recombinant DNA molecules. The cloning site inpHG327 is contained within the lacZ gene. Therefore, if a piece of foreign DNA has been inserted into this site the lacZ gene will be disrupted and no β-galactosidase will be produced. This phenomenon is called 'insertional inactivation' of a marker gene.Step 4 Duplicate plates are required of each colony because the alginate lyase detection procedure (step 6) kills the bacterial cells.Step 6 CPC is a cationic detergent which binds to high molecular weight alginates to form an opaque white precipitate. Alginate lyase reduces the molecular weight of the alginate sufficiently to prevent a significant precipitate from forming, hence the appearance of 'clearing zones' around alginate lyase producing colonies.AGAROSE GEL ELECTROPHORESISDNA may be fractionated according to molecular size by electrophoresis through agarose gels. This method will be used to analyse intact plasmids and also DNA fragments produced by restriction endonuclease digestion.Materials/Solutions RequiredAgarose (0.7%) in 1 X TBEElectrophoresis buffer - 5 X TBE (0.445M Tris, 0.445M boric acid and 0.01M EDTA, pH8.0) Electrophoresis apparatusEthidium bromide solution (2µg/ml)Masking tapeMethod[1] Seal the ends of the gel holder with tape.[2] Place comb in the correct orientation across the end of the gel holder.[3] Melt agarose in the autoclave.[4] Check that agarose is no hotter than 55°C and pour into the holder taking care to avoidair bubbles. Allow to set (approx. 20 min). Do this with the gel holder on the bench - not in the electrophoresis apparatus.。
实验七 外源基因在大肠杆菌中的诱导表达
大肠杆菌包涵体的分离与蛋白质纯化
1 细菌的裂解: 常用方法有:① 高温珠磨法;② 高压匀浆;③ 超声破碎法; ④ 酶溶法;⑤ 化学渗透等。前三种方法属机械破碎法,并且方 法① 、② 已在工业生产中得到应用,后三种方法在实验室研究 中应用较为广泛。 下面介绍酶溶法和超声破碎法的实验步骤。
1、酶溶法。常用的溶解酶有溶菌酶;β-1,3 -葡聚糖酶;β-1,6 -葡聚糖酶;蛋白酶;壳 多糖酶;糖昔酶等。溶菌酶主要对细菌类有作用,而其他几种酶对酵母作用显著。 主要步骤为: ① 4 ℃ ,5000rpm 离心,15 min ,收集诱导表达的细菌培养液(100 mL )。弃 上清,约每克湿菌加3 mL 裂解缓冲液,悬浮沉淀。 ② 每克菌加8μL PMSF 及80μL 溶菌酶,搅拌20 min ;边搅拌边每克菌加4 mg 脱 氧胆酸(在冷室中进行)。 ③ 37 ℃ ,玻棒搅拌,溶液变得粘稠时加每克菌20μL DNase I。室温放置至溶液不 再粘稠。 2、超声破碎法。声频为15-20 kHz 的超声波在高强度声能输入下可以进行细胞破碎, 在处理少量样品时操作简便,液体量损失较少,同时还可对染色体DNA 进行剪切 ,大大降低液体的粘稠度。 ① 收集1 L 诱导表达的工程菌,40 ℃ ,5000r pm 离心,15 min ;弃上清,约每 克湿菌加3 mLTE 缓冲液。 ② 按超声处理仪厂家提供的功能参数进行破菌;10 000g 离心,15min ,分别收集 上清液和沉淀。 ③ 分别取少量上清和沉淀,加入等体积的2× 凝胶电泳加样缓冲液,进行SDS PAGE 。 注意事项:超声破碎与声频、声能、处理时间、细胞浓度、菌种类型等因素有关, 应根据具体情况掌握;超声波破菌前,标本经3 -4 次冻溶后更容易破碎。
实验七 外源基因在大肠杆菌中的诱导表达
实验四 PCR扩增目的基因课件PPT
engineering of DNA
• 等位基因序列变化的分析 Analysis of allelic sequence
variations
2021/3/10
16
PCR反应中的成份
1. 引物:PCR反应产物的特异性由一对上下游引物所 决定。引物的好坏往往是PCR成败的关键。
引物设计和选择目的DNA序列区域时可遵循下列原则:
分子生物学实验 Experiments of molecular biology
实验四 PCR扩增目的基因
2021/3/10
1
实验二 PCR扩增目的基因
【目的要求】 1.掌握实验原理和实验基本过程; 2.了解PCR引物设计原则;学会设计PCR引物 ; 3.了解PCR的优化环节,学会如何优化PCR。了
6
3. PCR管内加好上述试剂之后要充分混合,然后离心数 秒,放入PCR仪上。
4. 编辑PCR反应程序,完成后启动PCR仪, 电泳。
PCR完成后,取5μl扩增产物用1%琼脂糖凝胶进行 电泳分析,检查反应产物及长度。
2021/3/10
7
电泳结果如图所示
2021/3/10
8
[注意] 1. PCR非常灵敏, 操作应尽可能在无菌操作台中进行。 2. 吸头、离心管应高压灭菌, 每次吸头用毕应更换, 不要
• PCR是模拟体内DNA复制条件,应用DNA聚合酶 反应,特异性扩增某一DNA片段的技术。
• 体外程序化的DNA合成技术。
devised by Kary Mullis in 1983
Mullis, K.B. (1993) The unusual origin of the polymerase chain reaction. Scientific American. 262 (4) 56-65.
中国海洋大学国家生命科学与技术人才培养基地班人才培养方案

海洋生物技术应用性实验(2 周,2 学分)
任选一门,可相互替代
六、专业特色课程
生七、实践环节
基地综合性大实验(64 课时,2 学分)
(一)必修实践环节(至少 35 学分)
1.毛泽东思想和中国特色社会主义理论体系概论(64 课时,2 学分)
2.军事训练 (2 周,1 学分)
073302101335
*遗传学
Genetics
细胞生物学
073302102335
遗传学实验
Genetics Laboratory
073302101331
生命科学导航
Life Sciences Navigation
073702101201
生命科学前沿讲座
Frontier Seminar on life Science
18. 基地综合大实验 (2 周,2 学分)
10.植物生物学实验(32 课时,1 学分)
19. 大学体育 I-IV (128 课时,4 学分)
20.以下实验任选一门
发育生物学实验 (32 课时,1 学分)
海洋生物学实验 (32 课时,1 学分)
潜水与海底生物调查 (16 课时,0.5 学分)
生物技术生产性实验 (64 课时,2 学分)海洋
Introduction to Maoism and Theoretical System of Chinese Socialism
008101101013 形势与政策 Ⅰ
Current Situation and Policy Ⅰ
先修课程
008101101015 形势与政策 Ⅱ
Current Situation and Policy Ⅱ
15. 藻类学实验(32 课时,1 学分)
分子生物学实验手册 -高校教材资料精

分子生物学实验室实验一 氯化钙法大肠杆菌感受态细胞的制备及转化 实验二 细菌质粒DNA 的提取实验三 琼脂糖凝胶电泳实验四 细菌基因组DNA 的提取与琼脂糖凝胶电泳实验五 质粒DNA 限制性内切酶实验实验六 聚合酶链式反应(PCR)扩增目的基因实验七 胶回收法纯化DNA 与琼脂糖凝胶电泳南京工业大学制药与生命科学学院生物工程与技术基础实验与工程实训中心《分子生物学实验》教学大纲英文名称: Molecular biology学分:1 学时:32教学对象:生物工程专业、制药工程专业、药剂学专业、食品学专业、生物技术专业教学目的:在开设的理论课程的基础上,结合实验课程的教学,使学生能较全面系统的掌握分子生物学的基本操作,拓宽专业知识面,加深对专业知识的理解,强化动手能力。
基本要求:了解分子生物学的实验操作原理,掌握有关实验仪器的使用方法。
实验内容:实验一.细菌质粒DNA的提取及酶切实验 (8学时)基本要求:通过学习碱变性抽提法对大肠杆菌中的质粒的抽提,掌握质粒DNA的小量制备方法,了解碱裂解法制备质粒DNA的原理。
进一步了解限制性内切酶的特性及酶切反应过程,掌握DNA的酶切技术。
重点:掌握质粒DNA的小量制备方法,了解碱裂解法制备质粒DNA的原理。
进一步了解限制性内切酶的特性,掌握DNA的酶切技术。
难点:掌握质粒DNA的小量制备方法及DNA的酶切技术。
实验二. 细菌基因组DNA的提取 (4学时)基本要求:了解细菌基因组DNA的提取的目的,原理,掌握细菌基因组DNA的提取的实验步骤及操作方法重点:掌握细菌基因组DNA的提取的实验步骤及操作方法难点:掌握细菌基因组DNA的提取的实验步骤及操作方法实验三. 大肠杆菌感受态细胞的制备及转化实验(4学时)基本要求:了解大肠杆菌感受态细胞的制备原理及转化反应的目的,应用范围及操作过程,掌握重组DNA质粒转化大肠杆菌的操作技术。
重点:掌握大肠杆菌感受态细胞的制备及转化的操作技术难点:大肠杆菌感受态细胞的制备及转化实验四. 聚合酶链式反应(PCR)扩增目的基因及琼脂糖凝胶电泳 (8学时) 基本要求:熟悉聚合酶链反应(PCR)的基本原理、实验基本条件;了解 PCR反应条件的优化和注意事项和应用范围 掌握聚合酶链反应(PCR)的实验技术:了解核酸琼脂糖凝胶电泳的原理及操作步骤,掌握琼脂糖凝胶电泳的实验方法。
《医学分子生物学实验》
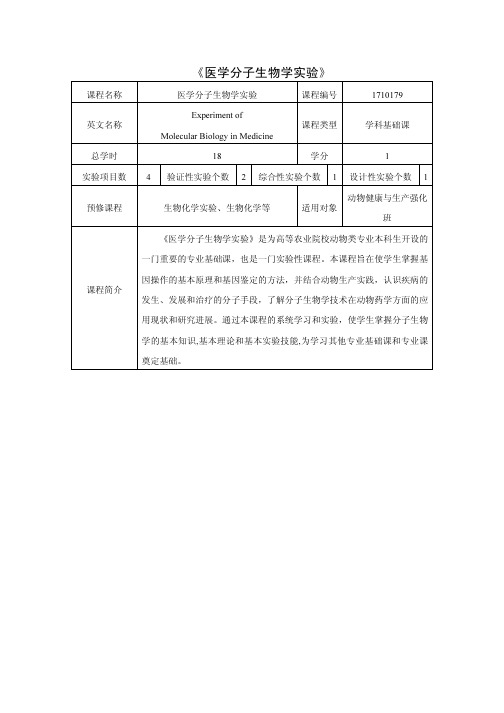
《医学分子生物学实验》
课程名称医学分子生物学实验课程编号1710179
英文名称
Experiment of
Molecular Biology in Medicine
课程类型学科基础课总学时18 学分 1
实验项目数 4 验证性实验个数 2 综合性实验个数 1 设计性实验个数 1 预修课程生物化学实验、生物化学等适用对象
动物健康与生产强化
班
课程简介
《医学分子生物学实验》是为高等农业院校动物类专业本科生开设的一门重要的专业基础课,也是一门实验性课程。
本课程旨在使学生掌握基因操作的基本原理和基因鉴定的方法,并结合动物生产实践,认识疾病的发生、发展和治疗的分子手段,了解分子生物学技术在动物药学方面的应用现状和研究进展。
通过本课程的系统学习和实验,使学生掌握分子生物学的基本知识,基本理论和基本实验技能,为学习其他专业基础课和专业课奠定基础。
(完整)分子生物学实验报告

甘薯adk基因的核心片段的克隆西南大学生命科学学院王丽 222014317011011摘要:甘薯为我国农业主要栽培作物,并且是分子生物学实验室常见材料,本次实验主要以甘薯叶片(部分为茎)为实验材料,第一次实验是用CTAB法提取甘薯DNA,PCR扩增再凝胶电泳检测纯度。
第二次实验从材料中提取检测RNA并反转录cDNA第一链,经PCR扩增得到ADK基因核心片段,将其与T载体相连并导入大肠杆菌DH5a感受态细胞中,扩增后在加有相应抗生素的LB平板上筛选阳性克隆,这对后续生物信息学分析等有重要意义。
关键词:甘薯;PCR技术;cDNA;adk基因 ;感受态大肠杆菌细胞Abstract: Sweet potato for my agricultural main cultivation crop,and is molecular biology laboratory common material,this times experiment main to sweet potato leaves (part for stems)for experiment material。
First times experiment was extracted by CTAB method DNA,PCR amplification and gel electrophoresis and purity of sweet potato。
The second experiment from material in the extraction RNA and reverse recorded cDNA first chain,by PCR spread increased get large ADK gene core fragments,electrophoresis detection rubber recycling ADK gene core fragments,will its and T carrier connected and import Escherichia coli DH5(feel state cell) in ,spread increased in plus has corresponding antibiotics of LB Tablet Shang filter positive clone this importance to subsequent bioinformatics analysis。
生物化学与分子生物学实验原理精选全文

可编辑修改精选全文完整版生物化学与分子生物学实验原理Principles of Biochemistryand Molecular Biology Techniques课程简介本课程主要是介绍常用分子生物学技术测定的原理和机制,以利于研究生了解分子生物学常用技术,并能活用这些技术。
这门课既不同于一般的分子生物学理论课,也不同于实验方法流程的介绍。
该课程分实验技术理论和实验操作两部分。
实验技术理论部分主要通过基因重组技术、目的基因的获得、分子杂交、基因多态性和基因表达调控等,介绍分子生物学实验的方法、设计思路、原理、操作技巧及应用等。
力求培养学生掌握现代分子生物学实验的基础与操作要点,同时邀请校外资深专家介绍分子生物学的新技术及新方法,为今后进一步深入研究奠定良好基础。
实验操作部分另设课程为“分子生物学实验技术”。
This course includes two sections. One section focus on the principles of the techniques used to isolate, identify, modify and analyze three key molecules: DNA, RNA and proteins. The first section includes: DNA recombination technology, molecular hybridization, gene polymorphism, and regulation of gene expression. The second section will be a separate course named the Techniques of Molecular Biology. The goal of this course is to giving students an on-bench training of basic molecular biology techniques.教学大纲一、课程名称:生物化学与分子生物学实验原理二、总学时数及学分:32学时,1.5学分理论课32学时三﹑授课对象:博士生、硕士生预修知识要求:要求有化学、生物学、遗传学、生物化学及微生物学相关知识四、教学目的及要求:目的:通过教学力求培养博士生、硕士生掌握现代生物化学与分子生物学实验的基础理论与基本操作要点,为今后的研究工作奠定良好基础。
研究生分子生物学实验(英文)

Experimental Methods in Molecular BiologySchool of Life ScienceAnhui UniversityDecember,2005ContentsDirectional Cloning into Plasmid Vectors (3)1.Preparation of Plasmid DNA by Alkaline Lysis with SDS:Midipreparation (4)2. Quantitation of DNA and RNA (10)3. Digesting DNA With Restriction Enzymes (12)4. Gel Electrophoresis of DNA (15)5. In Vitro Amplification of DNA by PCR (20)6. Isolation of DNA Fragments from Agarose Gel (26)7. Fresh Competent E.Coli Prepared Using Calcium Chloride (27)8. Ligation Reaction (28)9. Transformation of Recombinant (30)10. Extraction of Total DNA from Plant Tissue (33)11. Fast Protein Liquid Chromatography (FPLC) (35)Directional Cloning into Plasmid Vectors1.Preparation of Plasmid DNA by Alkaline Lysis with SDS:MidipreparationPlasmids as vectorsPlasmids are small,extrachromosomal circular mo1ecules,from 2 to around 200 kb in size,which exist in multiple copies(up to a few hundred)wimhin the host E.coli cell。
从理论到实践:分子生物学实验初学过程中常见错误辨析及相关教学改革建议

从理论到实践:分子生物学实验初学过程中常见错误辨析及相关教学改革建议蒋栋;张玥;陈丹瑛;张剑平;陈晨【期刊名称】《继续医学教育》【年(卷),期】2015(000)011【摘要】分子生物学实验技术是现代生命科学研究工作中最基本的操作技术与实验手段,分子生物学实验所涉及的理论基础非常广泛,而且需要操作者具有一定的动手能力。
而刚进入实验室的学生、研究人员在从理论走向实践的初学过程中常常发生各种“低级”错误。
笔者收集了初学研究者在分子生物学实验过程中发生的一些常见问题,分析其原因,并对分子生物学实验教学提出了针对性的调整建议,以在实践过程中对这些错误及早发现和改正。
%Molecular biology experiment is the basic technic and means of modern life science research work. Molecular biology experiment involves diverse theoretical knowledge and requires the operator to possess certain practical ability. The students and researchers who have just entered the laboratory often make a variety of "low-grade" errors during the course from the theory to the practice. We collected common mistakes of beginner researchers in molecular biology experiment, analyzed its causation, and adjustment advice on molecular biology experiment teaching is proposed to early discover and correct these mistakes in the experiment practice.【总页数】3页(P23-25)【作者】蒋栋;张玥;陈丹瑛;张剑平;陈晨【作者单位】首都医科大学附属北京地坛医院新发突发传染病研究北京市重点实验室,北京 100015;首都医科大学附属北京地坛医院新发突发传染病研究北京市重点实验室,北京 100015;首都医科大学附属北京地坛医院新发突发传染病研究北京市重点实验室,北京 100015;首都医科大学附属北京地坛医院新发突发传染病研究北京市重点实验室,北京 100015;首都医科大学附属北京地坛医院新发突发传染病研究北京市重点实验室,北京 100015【正文语种】中文【中图分类】G642【相关文献】1.高等师范院校本科分子生物学实验教学改革与实践 [J], 冯尚国; 王慧中2.基础医学专业学生分子生物学实验教学改革实践 [J], 宁启兰; 杨旭东; 朱文华; 武丽涛; 蒋小英3.基础医学专业学生分子生物学实验教学改革实践 [J], 宁启兰; 杨旭东; 朱文华; 武丽涛; 蒋小英4.医学生物化学与分子生物学实验教学改革与实践 [J], 李翠萍;杨妍;单琳琳;周贝5.从真理符合论到真理实践论:马克思主义真理观的革命性变革 [J], 吴娜因版权原因,仅展示原文概要,查看原文内容请购买。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
• Lab 4 Total RNA extraction, Reverse transcription and RT-PCR
4.Correctly usethe related experimental instruments such asmicro-sample feeder, high pressure steam sterilizer, constant temperature shaking table, super clean workbench, high speed refrigerated centrifuge, ice maker, PCR instrument, horizontal electrophoresis apparatus, gel imaging systemetc.beskilled in aseptic operation technologyand developed withindependent operation ability and teamwork cooperation spirit.
2.Master the process and basic steps of the experimental technology of bacterial plasmid DNA artificial recombination, as well asthe main experimental techniques of the DNA artificial recombination of animal and plant cells,design the experimentwiththese techniques,observe and analyze the experimental phenomena.
பைடு நூலகம்Pre-requisites
Molecular Biology
Intended Learning Outcomes
The anticipated knowledge, skills and attitude to be developed by the studentsare:
1.Understand the technical system and development status of molecular biology experiment,toenhancethe subjective desire to deeply understand the theoretical knowledge and technical principles of molecular biology, more activelytoexplorerelevant scientific problems in the field of molecular biology and to solve practical problemsusing existing technologies.
Learning Schedule
• Lab 1 Brief introduction, Genomic DNA extraction; PCR amplification
• Lab 2 Construction of recombinant DNA, transformation and selection
3.Independentlyconductgenomic DNA extraction, Plasmid Extraction, total RNA extraction, PCR amplification, RT-PCR amplification, agarose gel electrophoresis;observe and analyze the experimental phenomena;Independentlyconductthe experimental operation of transformingtarget genesinto the competentE.colicellsviaplasmid DNA.
Syllabus ofBiotechnologyat Haide College
Molecular Biology Practical
Description
Molecular Biology Laboratory is a system of molecular biology experiment technology based on biochemistry experiment and microbiology experiment, which is a modern biological technology for research and application of life science. This course is aimed at senior students majoring in biology and biotechnology. It mainly introduces the basic operational techniques and experimental means of molecular biology, including Principles and procedures of experimental projects such as genomic DNA extraction and identification, plasmid DNA extraction and identification, total RNA extraction and identification, PCR amplification, restriction endonuclease digestion, processofmolecular cloning and construction of gene library. Through the course study and operation practice, students are required to master the basic principles and operation skills of molecular biology experiments, and to be able to analyze and solve practical problems in related fields of molecular biologywiththese techniques.
