已发表基因组物种汇总资料2012-9-3
已发表基因组文章的物种汇总资料2012-07-25

豆科苜蓿属 500M 灰胞藻门 70M 423 Mb 禾本科,狗尾草属 茄目、茄科、茄属 760Mb 375Mb 葫芦目、葫芦科、甜瓜属 523Mb 姜目、芭蕉科、芭蕉属 盐芥属 260Mb 多孔菌目、 43.3Mb 灵芝科、灵 302Mb 金虎尾目、 亚麻科、亚
2n=16 2n=18 2n=24 2n=24 2n=22 2n=14
蒺藜苜蓿 蓝载藻(灰胞藻 门) 粟(谷子) 番茄 甜瓜 香蕉 盐芥 灵芝 亚麻
Medicago truncatula Cyanophora paradoxa Setaria italica Solanum lycopersicum Cucumis melo L. Musa acuminata Thellungiella salsuginea lucidum Ganoderma Linum usitatissimum
2011.11 2012.02 2012.05 2012.05 2012.07 2012.07 2012.07 2012.06 2012.06
Nature 34.48 Science 29.747 Nature biotechnology 29.495 Nature 34.48 PNAS 9.432 Nature 34.48 PNAS 9.432 Nature communications The Plant Journal 6.969
刊物 Nature Science Science Science Nature Science Science Nature Nature Nature Science Nature Genetics Nature
IF 34.48 29.747 29.747 29.747 34.48 29.747 29.747 34.48 34.48 34.48 29.747 34.284 34.48 Nature 34.48 Nature 34.48 Science 29.747 Nature Biotechnology 29.495 Plant Cell 9.293 Nature 34.284 Genetics Nature 34.284 Genetics Nature 34.284 Genetics PNAS 9.432 PNAS 9.432 DNA Res. 3.525 Science 29.747 Nature biotechnology 29.495 Nature Genetics 34.284 Nature 34.48 Nature Genetics 34.284 Nature Genetics 34.284 Genome biology 6.89 Nature biotechnology 29.495
已发表所有物种基因组汇总资料 更新至2012 9月

动物基因组(81) 中文名 秀丽隐杆线虫 黑腹果蝇 冈比亚按蚊 小家鼠 红鳍东方豚 玻璃海鞘 线虫 大鼠 黑斑鲀 家蚕 鸡 拟暗果蝇 克氏锥虫 黑猩猩 狗 意蜂 海胆 姥鲨 猕猴 短尾负鼠 青鳉鱼 伊蚊 马来线虫
拉丁名 Caenorhabditis elegans Drosophila melanogaster Anopheles gambiae Mus musculus Fugu rubripes Ciona intestinalis Caenorhabditis briggsae Rattus norvegicus Tetraodon nigroviridis Bombyx mori Gallus sonneratii Drosophila pseudoobscura Trypanosoma cruzi Pan troglodytes Canis familiaris Apis mellifera Strongylocentrotus purpuratus Callorhinchus milii Macaca mulatta Monodelphis domestica Oryzias latipes Aedes aegypti Brugia malayi
刊物 科、属 基因组大小 Science 97M 小杆总科、Caenorhabditis亚属 Science 果蝇科、果蝇属180M Science 蚊科、按蚊属 280M Nature 鼠科 2.5 G Science 鲀科、东方鲀属380M Science 150M 海鞘纲 PLoS Biology Caenorhabditis亚属 104M Nature 啮齿目、鼠科 2.75 G Nature 鲀科 340M Science 鳞翅目、蚕蛾科428.7 M Nature 雉科、原鸡属 1.06G Genome research 果蝇科、果蝇属139M Science 67M 锥虫属 Nature 2.7G 猩猩科、黑猩猩属 Nature 2.5 G 犬科、犬属 Nature 236M 昆虫纲 Science 1G 海胆纲 Plos Biology 软骨鱼纲、银鲛目 0.91G Science 猴科、猕猴属 2.87G Nature 3.4G 负鼠科、短尾负鼠属 Naturer 700M 青鳉属 Science 蚊科、伊蚊属 1376M Science 90M 线虫纲
12个物种miR_302基因簇的生物信息学分析

基因组学与应用生物学,2010年,第30卷,第3期,第379-384页Genomics and Applied Biology,2010,Vol.30,No.3,379-384数据分析An Analysis12个物种miR-302基因簇的生物信息学分析崔亚茹1张坤山2罗玉萍1李思光1,2*1南昌大学生命科学与食品工程学院,南昌,330031;2同济大学医学院,上海,200092*通讯作者,siguangli@摘要很多microRNA(miRNA)基因在基因组上聚集排列形成miRNA基因簇。
miRNA基因簇在进化上较为保守,有其特殊的生物学意义。
miR-302基因簇在胚胎干细胞中特异表达,对胚胎干细胞的自我更新和多潜能维持有重要的作用。
本文采用miRBase数据库查询和BLAST同源搜索方法共搜寻并分析了12个物种的miR-302基因簇。
生物信息学分析表明,这些物种miR-302基因簇均位于LARP7蛋白基因的内含子区,但转录方向与LARP7基因转录方向相反。
序列相似性分析显示,同一物种miR-302簇成员间以及不同物种相应miR-302簇成员间相似性均较高。
进化分析表明,基因复制可能是miR-302基因簇进化的主要驱动力。
关键词基因簇,miR-302,分子进化Bioinformatics Analysis of miR-302Gene Cluster from12Different SpeciesCui Yaru1Zhang Kunshan2Luo Yuping1Li Siguang1,2*1College of Life Sciences and Food Engineering of Nanchang University,Nanchang,330031;2College of Medicine of Tongji University,Shanghai, 200092*Corresponding author,siguangli@DOI:10.3969/gab.030.000379Abstract There are many miRNA genes clustered in genome.miRNA gene cluster is highly conserved in evolu-tion,with its special biological significance.miR-302gene cluster,an embryonic stem cell-specific miRNA clus-ter,plays an important role in maintenance of stem cells self-renewal and pluripotency.In this paper,we analyzed the molecular evolution of miR-302gene cluster from12different species.Bioinformatics analysis showed all of miR-302gene clusters in these species were located in introns of LARP7genes,but transcripted in opposite direc-tion.Sequence identity analysis suggested the high identity of the different members of the miR-302gene cluster of the same specie and the corresponding members of the miR-302gene clusters among different species.The phylogenetic analysis revealed that the miR-302gene cluster originated from gene duplications.Keywords Gene cluster,miR-302,Molecular evolutionmicroRNA(miRNA)是一类长度为19~25个核苷酸的内源非编码小分子RNA,广泛存在于各种后生生物中(Dennis,2002)。
Nature-2012-隼类基因组

l e t t e r sAs top predators, falcons possess unique morphological, physiological and behavioral adaptations that allow them to be successful hunters: for example, the peregrine is renowned as the world’s fastest animal. To examine the evolutionary basis of predatory adaptations, we sequenced the genomes of both the peregrine (Falco peregrinus ) and saker falcon (Falco cherrug ), and we present parallel, genome-wide evidence for evolutionary innovation and selection for a predatory lifestyle. The genomes, assembled using Illumina deep sequencing with greater than 100-fold coverage, are both approximately 1.2 Gb in length, with transcriptome-assisted prediction of approximately 16,200 genes for both species. Analysis of 8,424 orthologs in both falcons, chicken, zebra finch and turkey identified consistent evidence for genome-wide rapid evolution in these raptors. SNP-based inference showed contrasting recent demographic trajectories for the two falcons, and gene-based analysis highlighted falcon-specific evolutionary novelties for beak development and olfaction and specifically for homeostasis-related genes in the arid environment–adapted saker.We carried out next-generation genome sequencing for the peregrine and saker falcon, generating 128.07 Gb and 136.21 Gb of sequence for F. peregrinus and F. cherrug , respectively (Supplementary Tables 1 and 2). Genome size was estimated at 1.2 Gb for both species (Supplementary Fig. 1 and Supplementary Table 3), suggest-ing genome coverage of 106.72× for F. peregrinus and 113.51× for F. cherrug (Supplementary Fig. 2). Assembly using SOAPdenovo 1,2 resulted in contig and scaffold N50 values, respectively, of 28.6 kb and 3.89 Mb for F. peregrinus and 31.2 kb and 4.15 Mb for F. cherrug(Supplementary Table 4). Fosmid-based Sanger sequencing con-firmed >99% (peregrine) and >97% (saker) coverage of euchromatic DNA (Supplementary Table 5). With repeats masked, protein-coding genes were predicted using homology and de novo methods and also with RNA sequencing (RNA-seq) data, employed to refine gene struc-ture and identify novel genes (Supplementary Fig. 3). As a result, 16,263 genes were predicted for F. peregrines , and 16,204 were pre-dicted for F. cherrug (Supplementary Table 6 and Supplementary Note ). Approximately 92% of these genes were functionally annotated using homology-based methods (Supplementary Table 7).Comparative genome analysis was carried out to assess evolution and innovation within falcons using related genomes with comparable assembly quality (Supplementary Table 8). Orthologous genes were identified in the chicken, zebra finch, turkey, peregrine and saker using the program TreeFam 3. For the five genome-enabled avian spe-cies, a maximum-likelihood phylogeny using 861,014 4-fold degen-erate sites from 6,267 single-copy orthologs confirmed that chicken and turkey comprise one evolutionary branch and that zebra finch and the falcons form a second (Fig. 1a ), supporting previous analysis using 19 nuclear sequences 4. Analysis of the peregrine and saker using the same data set (Online Methods) dated the most recent common ancestor of the two falcon species to 2.1 (0.9–4.2) million years ago (Fig. 1a ). More than 99.6% of the peregrine genome was syntenic to the saker genome (Supplementary Table 9).Falcons were found to have less repetitive DNA (Table 1 and Supplementary Table 10), and their transposable element compositionwas most similar to that of zebra finch (with fewer DNA transposable ele-ments and long interspersed nucleotide elements (LINEs) but not short interspersed nucleotide elements (SINES)) (Supplementary Tables 11 and 12). We found fewer large (>1-kb) segmental duplications in fal-cons than in either chicken or zebra finch (Table 1 and Supplementary Table 13), with these elements comprising less than 1% of both falcongenomes. We also found that fewer branch-specific insertions and/ordeletions (indels) have accumulated in falcon genomes over evolutionary time (Table 1 and Supplementary Tables 14 and 15). TreeFam results showed, however, that falcons feature fewer lineage-specific genes than other birds (Fig. 1b and Table 1). It should be noted that our comparative genome analyses are based on alignments from the local to the scaffold level and are not based on whole-chromosome alignments.The olfactory receptor gene repertoire, annotated on the basisof putative functionality and 7-TM (transmembrane) structure, Peregrine and saker falcon genome sequences provide insights into evolution of a predatory lifestyleXiangjiang Zhan 1,7, Shengkai Pan 2,7, Junyi Wang 2,7, Andrew Dixon 3, Jing He 2, Margit G Muller 4,Peixiang Ni 2, Li Hu 2, Yuan Liu 2, Haolong Hou 2, Yuanping Chen 2, Jinquan Xia 2, Qiong Luo 2, Pengwei Xu 2, Ying Chen 2, Shengguang Liao 2, Changchang Cao 2, Shukun Gao 2, Zhaobao Wang 2, Zhen Yue 2, Guoqing Li 2, Ye Yin 2, Nick C Fox 3, Jun Wang 5,6 & Michael W Bruford 11Organisms and Environment Division, Cardiff School of Bioscience, Cardiff University, Cardiff, UK. 2BGI-Shenzhen, Shenzhen, China. 3International WildlifeConsultants, Ltd., Carmarthen, Wales, UK. 4Abu Dhabi Falcon Hospital, Abu Dhabi, United Arab Emirates. 5Department of Biology, University of Copenhagen,Copenhagen, Denmark. 6King Abdulaziz University, Jeddah, Saudi Arabia. 7These authors contributed equally to this work. Correspondence should be addressed to M.W.B. (brufordmw@ ) or Jun Wang (wangj@ ).Received 13 April 2012; accepted 28 February 2013; published online 24 March 2013; doi:10.1038/ng.2588OPEN© 2013 N a t u r e A m e r i c a , I n c . A l l r i g h t s r e s e r v e d .l e t t e r s showed the fewest intact olfactory receptor genes in falcons (n = 28; Supplementary Table 16), even though they have a larger olfactory bulb ratio than the zebra finch and a similar ratio to chicken 5. These two traits have previously been thought to be positively correlated 6 and linked to olfactory function 7. Furthermore, a gene expansion in the olfactory receptor γ-c clade 8 in chicken and zebra finch is not present in falcons (Fig. 1c and Supplementary Table 17), possibly reflecting their reliance on vision for locating prey 5.To compare selection at the gene level, two orthologous gene sets were compiled: the single-copy orthologs for the five avian species and a representative gene set from multiple-copy orthologs (n = 2,157). Analysis of branch-specific Ka /Ks (nonsynonymous -synonymous) substitution ratios (ω) showed that both falcons have higher branch ω values than other birds, independent of the gene set used (Fig. 1a and Supplementary Fig. 4), implying accelerated functional evolution in the falcon lineage. Supporting evidence for this comes from a recent work showing rapid phenotypic evolution and speciation in the falcon family 9. We also calculated the ω value for each orthologous gene and found that the ω values in falcons with mean value of 0.39 were considerably larger than for the other five pairwise avian combina-tions (see distribution in Fig. 2a ). However,these results need to be interpreted in the light of the relative paucity of avian genomes currently sequenced. We further examined both rapidly and slowly evolving gene catego-ries by comparing the two falcons against the galliformes, zebra finch–chicken and zebra finch–turkey pairs, respectively. To account for rapid genome evolution in falcons, each ω value within these categories was normalized using the genome median ω of each species pair. Functional GO (Gene Ontology)10 cat-egories involved in circulation (for example, heme synthesis), the nervous system, olfaction and sodium ion trans-port were found to have evolved rapidly in falcons (Supplementary Fig. 5 and Supplementary Table 18). Notable rapid evolution was also observed in the mitochondrial respiration chain when comparing falcons and galliformes birds but not in the comparisons of falcons with other species (Supplementary Table 18).The gene families defined by TreeFam (Supplementary Table 19) were input into CAFE 11 to examine changes in ortholog cluster sizes between putative ancestors and each species across our phylogeny (Fig. 1a ). We found that net gene loss occurred in all avian genomes, but the rate of loss per unit of time was most rapid in falcons (Fig. 1a ). We then used protein clustering 12 to explore gene number variation in the same protein families between each falcon species and the zebra finch. Loss of protein families within falcons (extinction plus contrac-tion) exceeded gain (innovation plus expansion) (Supplementary Tables 20–24), and contraction was greater than expansion for both the number of protein families and gene number (Fig. 2b ).We identified 879,812 and 761,748 heterozygous SNPs in the per-egrine and saker genomes, respectively. Despite its lower number of heterozygous SNPs, the saker had a higher heterozygous SNP rate than the peregrine (Table 1), although rates for both falcons were lower than for either the chicken 13 or zebra finch 14. Lower genetic diversity might originate from recent population contractions (Fig. 3). A narrow distribution in genome-wide SNP density for the peregrine, unlike the saker, suggests a more heterogeneous SNP distribution, indicating that mutations in the peregrine genome are more evenly distributed (Fig. 3a ). On the basis of local SNP densities, we used the pairwise sequentially Markovian coalescent (PSMC)15 to model the demographic history of both species (Fig. 3b ). For the peregrine,we inferred demographic history from 2 million years ago to 10,000 years ago, whereas, for the saker, the analysis included both the saker and its ancestral hierofalcon 16 because the fossil record indicates that the saker originated less than 34,000 years ago 16. PSMC showed that both falcon species have experienced at least one bottleneck; however,Green anole Chicken TurkeySaker0.63Peregrine Zebra finch0.590.150.150.170.230.1391+129.5+128.1+ 6.1–410.5+13.3+12.7+ 20.8+ 0.8+2.1–391.4– 28.2– 51.4–44.7–49.1–44.8– 57.21009968100M i l l i o n y e a r s a g oac120117558346442461,19043434324465326171381302911837513530303868757588323821701158,944bFigure 1 Comparative genomics in five avian species. (a ) Phylogenetic tree constructed using fourfold degenerate sites from single-copyorthologs, with the branch lengths scaled to estimated divergence time. Branch-specific ω values calculated from concatenated and SATé-aligned single-copy orthologs are shown on the left of each branch, and gene gain (+) and loss (−) per million years are shown on the right. (b ) Venn diagram showing shared and unique gene families in five avian species: red, peregrine; pink, saker; light blue, chicken; orange, turkey; green, zebra finch. (c ) Neighbor-joining tree based on the amino-acid sequences encoded by olfactory receptor genes in the peregrine, saker, chicken and zebra finch. Bootstrap values are shown (clockwise) for the major olfactory receptor clades, namely γ-c, other γ, α, θ and TAAR. Note that both chicken and zebra finch show expansions in the olfactory receptor γ-c clade. Birds are represented by the same colors as in b .table 1 Comparative genome structure summary for the five avian species Species Repetitive DNA (%)Segmental duplications (%)Specific indels per million years Specific gene families (genes)Heterozygous SNP rate per kb Peregrine 6.790.7141.0120 (137)0.7Saker 6.800.6109.0117 (139)0.8Zebra finch 12.83 1.0322.8382 (531) 1.4Turkey 9.62–a –a 170 (182)~1.7Chicken 13.28 5.5278.8115 (174) 4.5Detailed statistics for repetitive DNA are given in supplementary table 10, segmental duplications are listed in supplementary table 13, specific indels are listed in supplementary tables 14 and 15, and gene families identifiedfrom TreeFam analysis are shown in supplementary table 19. Heterozygous SNP rates for non-falcon species havebeen published 12–14.a Excluded from the analysis because of a higher proportion of ambiguous bases in the turkey genome (supplementary Note ).© 2013 N a t u r e A m e r i c a , I n c . A l l r i g h t s r e s e r v e d .l e t t e r swhile this lasted until 100,000 years ago for the peregrine, hierofalcon populations expanded around 300,000 years ago and started to expand again from 60,000 years ago, eventually giving rise to the four spe-cies in the hierofalcon group 16. The peregrine, however, underwent a second bottleneck approximately 20,000 years ago, possibly owing to climate-driven habitat diminution (Supplementary Fig. 6).The word falcon comes from the Latin falco , meaning hook shaped, referring to its beak. The falcon beak is more robust, being longer, wider and deeper, than those of the chicken and zebra finch. We exam-ined the evolution of the genes in two KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways (TGF-β and WNT)17, those in FGF signaling pathways (see URLs) and 98 other genes involved in the development of avian beaks (Online Methods). Bmp4 is a well-studied major-effect gene in beak development 18, and exonization provides a major source of accelerated, lineage-specific evolution 19.We found that falcons have gained two novel exons owing to splice-site mutations in Bmp4 (Supplementary Fig. 7) and have also gained a second copy of TGFB2 (a gene upregulated in curved beaks) and Dkk 2 (a gene related to longer and wider beaks in ducks)20. The two copies originated in different parts of a single ancestral gene (Supplementary Figs. 8 and 9). These changes can both influence gene expression 21,22 and contribute to the regulation of development in falcon beaks (Table 2). Relative to chicken and zebra finch, eight genes in these pathways show evidence of positive selection in falcons (Supplementary Fig. 10), among which TGF βIIr has been shown to be functional in shaping avian beak morphology (Table 2). Although it has been shown that possible errors in multiple-sequence alignment have only a limited impact on estimates of Ka /Ks ratios at the genome level (Supplementary Fig. 11), we caution that the analysis of positive selection remains sensitive to the quality of the multiple-sequence alignments of separate orthologous genes and can be further com-plicated by variance in sequence divergence among them. Analysis of microRNA target sites provided further support for the presence of novel regulatory mechanisms in falcons (Table 2, Supplementary Fig. 10 and Supplementary Table 25).Peregrines and sakers are parapatric breeding species, with the latter confined to the Palearctic and primarily inhabiting arid environments (Supplementary Fig. 6)16,23. Consequently, it is probable that sakers require greater maintenance of osmotic equilibrium and suffer heat stress more than peregrines. We compared the genes from two kidney-expressed KEGG pathways and other related genes involved in home-ostasis in the two species. Rab11a and GNAS have major contributions to the V2R water conservation pathway 17,24, and both have two more copies in saker than in peregrine (Supplementary Table 26).The aldosterone-regulated sodium reabsorption pathway has a major role in determining sodium levels, and the activation of pro-tein kinase C (PKC) inhibits sodium uptake 25. We found that the saker has three more functional copies of PKC β than peregrine. In saker, we also found the exonization of trpv1, a gene involved in pro-moting thermoregulatory cooling by stimulating sweat productiona bN u m b e rlog 2 (Ka/Ks )2004006008001,0001,200log 2 (gene number differences)P r o t e i n f a m i l y n u m b e rFigure 2 Distribution of ω values for each pair of orthologous genes among the five avianspecies and gene gain and loss in falcons.(a ) Distribution of ω values among avian species.The estimates of ω for 8,424 orthologs show a highly significant shift toward larger values in falcons compared with other pairs (P = 1.354 × 10−189, 2.837 × 10−297, 1.621 × 10−284, 7.264 × 10−233 and 4.617 × 10−215 relativeto the other indicated pairs, Mann Whitney test). Genes with Ka = 0 are included in relative frequencies but are not shown. The x axis shows log 2 Ka /Ks , using 0.01 as the unit interval, and the y axis shows the number of orthologs in each interval. (b ) Gene number differencesin contracted and expanded protein families in the falcons compared with zebra finch. Zebra finch genes of unplaced sequence that had more than 97% sequence identity with their closest paralog were excluded to correct for the possible overestimation of zebra finch gene expansion 14.ab3.53.0Peregrine SakerPeregrine Saker2.5E f f e c t i v e p o p u l a t i o n s i z e (×104)P e r c e n t a g e o f w i n d o w s2.01.51.00.50140.0010.0100.100SNP density (%)1.000121086420105Years before the present106104Figure 3 SNP density distribution and demography reconstruction of falcons. (a ) Distribution of SNP density across each falcon genome. Heterozygous SNPs between the two sets of falcon chromosomes were annotated, and heterozygosity density was observed in non-overlapping 50-kb windows. (b ) PSMC inference of falcon population history based on autosomal data. The central bold lines represent inferred population sizes, and the 100 thin curves surrounding each line are the PSMC estimates generated using 100 sequences randomly resampled from the original sequence. The mutation rate on autosomes, which is used in time scaling, was estimated using zebra finch autosome data.© 2013 N a t u r e A m e r i c a , I n c . A l l r i g h t s r e s e r v e d .and preemptive renal water reabsorption through the release of vasopressin (antidiuretic hormone)26 (Supplementary Fig. 12). Supporting evidence for water conservation in sakers comes from the observation that sakers secrete more sodium and chloride in their urine than do peregrines 27. These results, when taken together, suggest a genetic basis by which sakers cope with desert and steppe habitats and heat stress. The functional expression of these genes related to water conservation and sodium secretion warrants future research.The genome sequencing data presented here provide a resource for the future examination of evolution and adaptation in birds and in raptors in particular. Both falcon species are widely distributed across the globe and show a wide variety of local phenotypes and behaviors, and their conservation status varies substantially across their habitat ranges, with the saker falcon being globally classified as vulnerable. Both species have migratory as well as sedentary populations, and understanding the genetic basis of this wide diversity could provide a valuable tool to aid their long-term conservation.URLs. LASTZ and MULTIZ, /miller_lab/; MUSCLE, /muscle ; FGF signaling pathways, /pathway.php?sn=FGF_Signaling .MeThodSMethods and any associated references are available in the online version of the paper .Accession codes. Whole-genome shotgun sequences have been depos-ited at the DNA Data Bank of Japan (DDBJ), the European Molecular Biology Laboratory (EMBL) and GenBank (AKMT00000000 for F. peregrinus and AKMU00000000 for F. cherrug ). The version described in this paper is the first version. Raw DNA, RNA and micro-RNA sequencing reads have been submitted to the NCBI Sequence Read Archive database (SRA054256, SRX225198, SRX225199).Note: Supplementary information is available in the online version of the paper .ACkNoWLeDGMeNtSThis work was supported by the EAAD (Environment Agency of Abu Dhabi). We thank H.E. Mohammed Al Bowardi for his support. We also thankP . Orozco-terWengel and X. Fang for their critical comments on the manuscript, A. Subramanian for suggestions on analysis with DIALIGN-TX, S. Mirarab for suggestions on SATé-II alignment and E. Alm and B.J. Shapiro for advice onKa /Ks normalization. We acknowledge staff at International Wildlife Consultants, EAAD and BGI-Shenzhen who helped conduct this study.AUtHoR CoNtRIBUtIoNSM.W .B. led the UK team. Jun Wang and Junyi Wang led the BGI team. M.W .B., X.Z., A.D. and N.C.F. designed the study. M.G.M., A.D. and X.Z. sampled thefalcons. Q.L., S.G., C.C., P .N., Y.L., S.L., Z.Y., G.L., Z.W . and Y.Y. performed genome sequencing and assembly. X.Z. and Yuanping Chen extracted the DNAand RNA. X.Z., S.P ., J.H., L.H., H.H., J.X., P .X. and Ying Chen analyzed the data. X.Z. and M.W .B. interpreted the results and wrote the manuscript. All authors read and provided input for the manuscript and approved the final version. COMPETING FINANCIAL INTERESTSThe authors declare no competing financial interests.Reprints and permissions information is available online at /reprints/index.html.This work is licensed under a Creative Commons Attribution- NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit /licenses/by-nc-sa/3.0/.1. Kim, E.B. et al. Genome sequencing reveals insights into physiology and longevityof the naked mole rat. Nature 479, 223–227 (2011).2. Li, R. et al. De novo assembly of human genomes with massively parallel shortread sequencing. Genome Res. 20, 265–272 (2010).3. Li, H. et al. TreeFam: a curated database of phylogenetic trees of animal genefamilies. Nucleic Acids Res. 34, D572–D580 (2006).4. Hackett, S.J. et al. A phylogenomic study of birds reveals their evolutionary history.Science 320, 1763–1768 (2008).5. Roper, T.J. Olfaction in birds. Adv. Stud. Behav. 28, 247–332 (1999).6. Steiger, S.S., Fidler, A.E., Valcu, M. & Kempenaers, B. Avian olfactory receptorgene repertoires: evidence for a well-developed sense of smell in birds? Proc. R. Soc. Lond. B 275, 2309–2317 (2008).7. Bang, B.G. Anatomical evidence for olfactory function in some species of birds.Nature 188, 547–549 (1960).8. Steiger, S.S., Kuryshev, V.Y., Stensmyr, M.C., Kempenaers, B. & Mueller, J.C.A comparison of reptilian and avian olfactory receptor gene repertoires: species-specific expansion of group γ genes in birds. BMC Genomics 10, 446 (2009).9. Hugall, A.F . & Stuart-Fox, D. Accelerated speciation in colour-polymorphic birds.Nature 485, 631–634 (2012).10. Ashburner, M. et al. Gene ontology: tool for the unification of biology. The GeneOntology Consortium. Nat. Genet. 25, 25–29 (2000).11. Hahn, M.W., Demuth, J.P . & Han, S.G. Accelerated rate of gene gain and loss inprimates. Genetics 177, 1941–1949 (2007).12. Dalloul, R.A. et al. Multi-platform next-generation sequencing of the domestic turkey(Meleagris gallopavo ): genome assembly and analysis. PLoS Biol. 8, e1000475 (2010).13. Wong, G.K. et al. A genetic variation map for chicken with 2.8 million single-nucleotide polymorphisms. Nature 432, 717–722 (2004).14. Warren, W.C. et al. The genome of a songbird. Nature 464, 757–762 (2010).15. Li, H. & Durbin, R. Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011).16. Nittinger, F ., Haring, E., Pinsker, W., Wink, M. & Gamauf, A. Out of Africa?Phylogenetic relationships between Falco biarmicus and the other hierofalcons (Aves: Falconidae). J. Zoological Syst. Evol. Res. 43, 321–331 (2005).17. Kanehisa, M., Goto, S., Furumichi, M., Tanabe, M. & Hirakawa, M. KEGG forrepresentation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 38, D355–D360 (2010).18. Abzhanov, A., Protas, M., Grant, B.R., Grant, P .R. & Tabin, C.J. Bmp4 andmorphological variation of beaks in Darwin’s finches. Science 305, 1462–1465 (2004).19. Sorek, R. The birth of new exons: mechanisms and evolutionary consequences.RNA 13, 1603–1608 (2007).20. Brugmann, S.A. et al. Comparative gene expression analysis of avian embryonicfacial structures reveals new candidates for human craniofacial disorders. Hum. Mol. Genet. 19, 920–930 (2010).21. Menezes, R.X., Boetzer, M., Sieswerda, M., van Ommen, G.B. & Boer, J.M.Integrated analysis of DNA copy number and gene expression microarray data using gene sets. BMC Bioinformatics 10, 203 (2009).22. Perry, G.H. et al. Diet and evolution of human amylase gene copy number variation.Nat. Genet. 39, 1256–1260 (2007).23. del Hoyo, J., Elliot, A. & Sargatal, J. Handbook of the Birds of the World, Vol. 2(Lynx Edicions, Barcelona, Spain, 1994).24. Tajika, Y. et al. Differential regulation of AQP2 trafficking in endosomes bymicrotubules and actin filaments. Histochem. Cell Biol. 124, 1–12 (2005).25. Hays, S.R., Baum, M. & Kokko, J.P . Effects of protein kinase C activation onsodium, potassium, chloride, and total CO 2 transport in the rabbit cortical collecting tubule. J. Clin. Invest. 80, 1561–1570 (1987).26. Sharif -Naeini, R., Ciura, S. & Bourque, C.W. TRPV1 gene required for thermosensorytransduction and anticipatory secretion from vasopressin neurons during hyperthermia. Neuron 58, 179–185 (2008).27. Cade, T.J. & Greenwald, L. Nasal salt secretion in falconiform birds. Condor 68,338–350 (1966).28. Abzhanov, A. et al. The calmodulin pathway and evolution of elongated beakmorphology in Darwin’s finches. Nature 442, 563–567 (2006).29. Mallarino, R. et al. Two developmental modules establish 3D beak-shape variationin Darwin’s finches. Proc. Natl. Acad. Sci. USA 108, 4057–4062 (2011).table 2 Known functional genes for beak development (length, width and depth) and their innovations in falconsGene LengthWidth Depth Genetic innovation Bmp 4++More exons SCML4+More exons TGFB2+Duplication Dkk 2++DuplicationTGF βIIr ++Positive selection CamKII +Lower copy numberFZD1++Fewer microRNA target sites TGFB3+Fewer microRNA target sites Fgf10−−More microRNA target sites+, upregulation; −, downregulation. The included genes have been shown to be functional in beak development 18,20,28,29.l e t t e r s© 2013 N a t u r e A m e r i c a , I n c . A l l r i g h t s r e s e r v e d .oNLINe MeThodSSampling and DNA extraction. Approximately 2 ml of blood each was taken from one male saker and one male peregrine that were caught in the wild at the Abu Dhabi Falcon Hospital. Genomic DNA was extracted using gravity-flow, anion-exchange tips and buffers (Qiagen Blood & Cell Culture DNA Maxi Kit). Sex was determined using morphology and molecular assays (Supplementary Note ).Genome sequencing and assembly. Sequencing was carried out using an Illumina HiSeq 2000. Paired-end libraries with insert sizes of 170, 500 and 800 bp (short inserts) and 5, 10 and 20 kb (long inserts) were constructed. A detailed description of library construction, sequencing and assembly, genome size estimation, annotation, gene prediction (homology, de novo and RNA-seq) and functional annotation, and RNA (mRNA and microRNA) sequencing is included in the Supplementary Note . Genome assembly quality was assessed using GC content (Supplementary Figs. 13 and 14) and Sanger sequencing of fosmid libraries (Supplementary Note ). The lengths of the repetitive DNA sequences identified are listed in Supplementary Table parative genome analysis. We employed the widely used program TreeFam 3 to define orthologous genes (Supplementary Note ) because it has been inferred that phylogeny-based software is more robust in inferring orthologs 3. The analysis using LASTZ (see URLs) and MCscan 30 in chicken, peregrine and saker showed that approximately 88.9% of TreeFam orthologs were supported by at least one synteny analysis, implying high quality for the orthologs defined (Supplementary Table 28 and Supplementary Note ). On the basis of concatenated 4-fold degenerate sites from 6,267 single-copy orthologs, a maximum-likelihood phylogeny for the falcons, zebra finch, chicken and turkey genomes was constructed using PhyML 31. Ortholog sequences were aligned using MUSCLE (see URLs) with the protein alignment as a guide. The optimal evolutionary model was GTR + I + G, selected using MODELTEST3.7 (ref. 32). The Bayesian relaxed molecular clock approach was used to estimate species divergence time using MCMCTREE in PAML 33, based on the degenerate sites data set used above.Whole-genome synteny analysis was performed for the five bird species. Pairwise alignments were first produced using LASTZ. ChainNet was then applied to merge traditional alignments into larger structures. Scaffolds of <5 kb in size were filtered out to avoid multiple hits for small scaffolds.To detect segmental duplications, an alignment was generated using LASTZ with the parameters T = 2 (no transition), C (chain) = 2, H (inner) = 2,000, Y (ydrop) = 3,400, L (gappedthresh) = 6,000 and K (hspthresh) = 2,200. Before aligning, with repeat sequences masked, the genome assembly was split into 100 subfiles. The maximum simultaneous gap allowed was 100 bp. After align-ing, the blocks obtained were conjoined to obtain larger blocks (chains). We unmasked the repeat sequences in the chains and then selected those with length of >500 bp and identity of >85% and aligned the chains using LASTZ again. Finally, we extracted the aligned chains with length of >1 kb and identity of >90%, considering these predictions to be segmental duplications. After removing the overlapping fragments, we obtained a nonredundant set. Because previous research 34 has shown that highly similar alignments may represent allelic overlaps missed during the assembly process, we removed segmental duplications that were ≥98% identical. Furthermore, to examine the quality of the detected segmental duplications, the sequencing depth distributions of segmental duplications and regions without segmental duplications were compared for peregrine and saker, respectively (Supplementary Fig. 15).For the assessment of lineage-specific indels, MULTIZ (see URLs) was used to integrate all the LASTZ alignments from the synteny analysis to obtain conserved elements among the genomes from the two falcons, zebra finch and chicken. For blocks longer than 1 kb, we identified species-specific short indels using the aligned data. Indels located within 50 bp of the end of the block and pairs of indels with a distance less than 50 bp were filtered out.Avian olfactory receptor genes. The olfactory receptor genes of the five avian species and of green anole, human, cow and dog were annotated using GenBank, Ensembl and published data 8 (Supplementary Note ). Intact avian olfactory receptor genes were aligned using SATé-II (ref. 35), from which a neighbor-joining tree was constructed using MEGA5.03 (ref. 36), with the Poisson modelchosen as the substitution model as previously analyzed 8. The reliability of the phylogenetic tree was evaluated with 1,000 bootstrap replicates.Genome evolution analysis. TreeFam was used to help define both single-copy and multiple-copy orthologs for the five avian species (Supplementary Fig. 16). For multiple-copy orthologs, we used single-copy genes in our out-group (green anole) as a query and selected the best hits in the five birds as the representatives of the ortholog. Similar sequence depth was found for multiple-copy and single-copy genes (Supplementary Fig. 17), suggesting that gene duplications are unlikely to have been misidentified as alleles of the same locus. The two orthologous gene sets were then used for ω ratio calculations as follows.First, on the basis of the concatenated ortholgs, branch-specific ω values for each avian species were calculated using codeml in PAML (Supplementary Note ). Alignment quality is of major importance for ω estimation because errors can lead to the misidentification of synonymous sites as nonsynon-ymous sites 37. To minimize the effect of alignment errors, two algorithms,SATé-II and DIALIGN-TX 38, were chosen to align each ortholog, as they havebeen reported to be robust in dealing with global and local alignments, respec-tively (Supplementary Note ). To further examine the influence of alignment quality on the ω analysis, we randomly selected 421 SATé-aligned orthologs and manually corrected them. All automatic alignment methods and manual corrections produced similar results (Supplementary Fig. 4). Therefore, for the following analysis, we used SATé, except for alignments involving too few taxa (≤3), where we used DIALIGN instead.Second, PAML was used to calculate ω values for each ortholog, and com-parisons were made of the ω distributions of all orthologs between the falcons and other species pairs (chicken and turkey, zebra finch and chicken, zebra finch and turkey, peregrine and zebra finch, and saker and zebra finch).Third, we examined both rapidly and slowly evolving gene categories by comparing the two falcon genomes with the two galliformes genomes, zebra finch and chicken, and zebra finch and turkey. We calculated the median ω values for each gene ontology 10 functional category containing at least ten genes found in the saker-peregrine lineage.Assessment of the possible influence of sequencing errors indicated that they can have little influence on our ω calculations (Supplementary Fig. 11). Detailed information for the above analyses is provided in the Supplementary Note .Gene gain and loss. CAFE based on a random gene birth and death model 11 was used to study gene gain and loss in gene families across our reconstructed phylogenetic tree with six species (Supplementary Fig. 18). The robustness of CAFE was tested by removing one species each time. The results showed that, independent of the species removed, falcons always featured the most rapid gene loss of the studied avian species (Supplementary Fig. 19), although we note that the avian species studied here are more divergent than mammals 11. To further explore gain and loss between each falcon and its closest genome-enabled rela-tive, zebra finch, we used protein clustering 12 to explore gene number variation for the same protein families (Supplementary Note ). Filtering was applied to clean up possible overestimated gene expansion in the zebra finch gene set. However, the patterns of gene turnover in the falcon species were similar with (Fig. 2b ) and without (Supplementary Fig. 20) this filtering.SNP calling and demographic history reconstruction. We use SOAPsnp 39 to detect SNPs between diploid chromosomes of both falcons (Supplementary Note ). The SNP distribution was observed across each genome. To assess whether SNP quality scores or sequencing depth influenced SNP distribution, we compared the results achieved with varied quality scores (20 or 40) and different depth ranges (Supplementary Fig. 21). By comparison with the zebra finch genome, the autosomal mutation rate was estimated to be 1.65 × 10−9 mutations per year for both falcons. Generation time was inferred to be 6.0 and 6.6 years for peregrine and saker, respectively (Supplementary Note ). The demographic history of each falcon was reconstructed using the PSMC model 15 (Supplementary Note ).Beak development. Genes in three signaling pathways (TGF-β, WNT and FGF) and 98 others (Supplementary Table 29) were analyzed in chicken,© 2013 N a t u r e A m e r i c a , I n c . A l l r i g h t s r e s e r v e d .。
已基因组测序物种
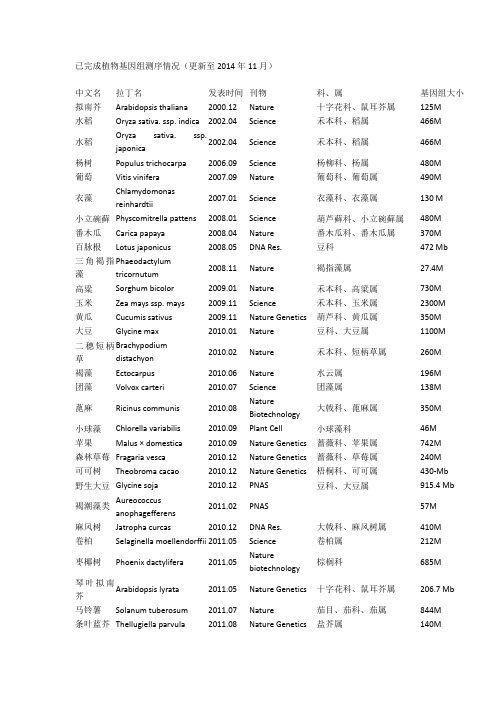
已完成植物基因组测序情况(更新至2014年11月)中文名拉丁名发表时间刊物科、属基因组大小拟南芥Arabidopsis thaliana 2000.12 Nature 十字花科、鼠耳芥属125M水稻Oryza sativa. ssp. indica 2002.04 Science 禾本科、稻属466M水稻Oryza sativa. ssp.japonica2002.04 Science 禾本科、稻属466M杨树Populus trichocarpa 2006.09 Science 杨柳科、杨属480M 葡萄Vitis vinifera 2007.09 Nature 葡萄科、葡萄属490M衣藻Chlamydomonasreinhardtii2007.01 Science 衣藻科、衣藻属130 M小立碗藓Physcomitrella pattens 2008.01 Science 葫芦藓科、小立碗藓属480M 番木瓜Carica papaya 2008.04 Nature 番木瓜科、番木瓜属370M 百脉根Lotus japonicus 2008.05 DNA Res. 豆科472 Mb三角褐指藻Phaeodactylumtricornutum2008.11 Nature 褐指藻属27.4M高粱Sorghum bicolor 2009.01 Nature 禾本科、高粱属730M 玉米Zea mays ssp. mays 2009.11 Science 禾本科、玉米属2300M 黄瓜Cucumis sativus 2009.11 Nature Genetics 葫芦科、黄瓜属350M 大豆Glycine max 2010.01 Nature 豆科、大豆属1100M二穗短柄草Brachypodiumdistachyon2010.02 Nature 禾本科、短柄草属260M褐藻Ectocarpus 2010.06 Nature 水云属196M 团藻Volvox carteri 2010.07 Science 团藻属138M蓖麻Ricinus communis 2010.08 NatureBiotechnology大戟科、蓖麻属350M小球藻Chlorella variabilis 2010.09 Plant Cell 小球藻科46M苹果Malus × domestica 2010.09 Nature Genetics 蔷薇科、苹果属742M森林草莓Fragaria vesca 2010.12 Nature Genetics 蔷薇科、草莓属240M可可树Theobroma cacao 2010.12 Nature Genetics 梧桐科、可可属430-Mb 野生大豆Glycine soja 2010.12 PNAS 豆科、大豆属915.4 Mb褐潮藻类Aureococcusanophagefferens2011.02 PNAS 57M麻风树Jatropha curcas 2010.12 DNA Res. 大戟科、麻风树属410M 卷柏Selaginella moellendorffii 2011.05 Science 卷柏属212M枣椰树Phoenix dactylifera 2011.05 Naturebiotechnology棕榈科685M琴叶拟南芥Arabidopsis lyrata 2011.05 Nature Genetics 十字花科、鼠耳芥属206.7 Mb 马铃薯Solanum tuberosum 2011.07 Nature 茄目、茄科、茄属844M条叶蓝芥Thellugiella parvula 2011.08 Nature Genetics 盐芥属140M白菜Brassica rapa 2011.08 Nature Genetics 十字花科、芸薹属485M 印度大麻Cannabis sativa 2011.1 Genome biology 大麻属534M木豆Cajanus cajan 2011.11 Naturebiotechnology豆科、木豆属833M蒺藜苜蓿Medicago truncatula 2011.11 Nature 豆科苜蓿属500M 蓝载藻Cyanophora paradoxa 2012.02 Science 灰胞藻门70M谷子Setaria italica 2012.05 Naturebiotechnology禾本科、狗尾草属490M谷子Setaria italica 2012.05 Naturebiotechnology禾本科、狗尾草属预估510M,组装出400M番茄Solanum lycopersicum 2012.05 Nature 茄科、茄属900Mb 甜瓜Cucumis melo 2012.07 PNAS 葫芦科、甜瓜属450Mb 亚麻Linum usitatissimum 2012.07 Plant Journal 亚麻科、亚麻属373Mb 盐芥Thellungiella salsuginea 2012.07 PNAS 十字花科、盐芥属260Mb 香蕉Musa acuminata 2012.07 Nature 芭蕉科、芭蕉属523Mb 雷蒙德氏棉Gossypium raimondii 2012.08 Nature Genetics 锦葵科、棉属775.2Mb 大麦Hordeum vulgare 2012.1 Nature 禾本科、大麦属 5.1Gb梨Pyrus bretschneideri 2012.11 Genome Research 蔷薇科、梨属527Mb 西瓜Citrullus lanatus 2012.11 Nature Genetics 葫芦科、西瓜属425 Mb 甜橙Citrus sinensis 2012.11 Nature Genetics 芸香科、柑橘属367 Mb 小麦Triticum aestivum 2012.11 Nature 禾本科、小麦属17Gb两种小型藻Bigelowiella natans,Guillardia theta2012.11 Nature 95Mb 87Mb棉花(雷蒙德氏棉)Gossypium raimondii 2012.12 Nature 锦葵科、棉属761.4Mb梅花Prunus mume 2012.12 NatureCommunications蔷薇科、梨属280M鹰嘴豆Cicer arietinum 2013.01 Naturebiotechnology豆科、鹰嘴豆属738Mb橡胶树Hevea brasiliensis 2013.02 BMC Genomics 大戟科、橡胶树属 2.15Gb 毛竹Phyllostachys heterocycla 2013.02 Nature Genetics 竹科、钢竹属 2.075 Gb短花药野生稻Oryza brachyantha 2013.03NatureCommunications禾本科稻属342Mb-362Mb小麦A Triticum urartu 2013.03 Nature 禾本科、小麦属 4.94 Gb 小麦D grassAegilops tauschii 2013.03 Nature 禾本科、小麦属 4.36Gb 桃树Prunus persica 2013.03 Nature Genetics 蔷薇科、梨属265 Mb 丝叶狸藻Utricularia gibba 2013.05 Nature 狸藻科、狸藻属82Mb中国莲Nelumbo nucifera Gaertn 2013.05 Genome biology 睡莲科、莲属929 Mb 挪威云杉Picea abies 2013.05 Nature 松科、云杉属19.6G海洋球石Emiliania huxleyi 2013.06 Nature 定鞭藻纲141.7Mb藻虫黄藻Symbiodinium minutum 2013.07 Current Biology 甲藻门 1.5G 油棕榈Elaeis guineensis 2013.07 Nature 棕榈科、油棕榈属 1.8G枣椰树Phoenix dactylifera 2013.08 NatureCommunications棕榈科、刺葵属671 Mb醉蝶花Tarenaya hassleriana 2013.08 Plant Cell 醉蝶花科、醉蝶花属290 Mb 莲Nelumbo nucifera 2013.08 Plant Journal 睡莲科、莲属879 Mb桑树Morus notabilis 2013.09 NatureCommunications桑科、桑属357 Mb猕猴桃Actinidia chinensis 2013.10 NatureCommunications猕猴桃属616.1 Mb胡杨Populus euphratica 2013.11 NatureCommunications杨属496.5 Mb八倍体草莓F. x ananassa 2013.12 DNA Research 草莓属698 Mb 康乃馨Dianthus caryophyllus L. 2013.12 DNA Research 石竹属622 Mb 甜菜Beta vulgaris ssp. vulgaris 2013.12 Nature 藜科甜菜属566.6 Mb 无油樟(互叶梅)Amborella trichopoda 2013.12 Science 无油樟属748 Mb辣椒Capsicum annuum(Criolo de Morelos334)2014.1 Nature Genetics 辣椒属 3.48G芝麻Sesamum indicum 2014.2 Genome Biology 胡麻科胡麻属274 Mb辣椒Capsicum annuum(Zunla-1)2014.3 PNAS 辣椒属 3.48G火炬松Pinus taeda(Loblollypine)2014.3 Genome Biology 松属23.2G棉花(亚洲棉)Gossypium arboreum 2014.5 Nature Genetics 锦葵科、棉属1694Mb 萝卜Raphanus sativus L. 2014.5 DNA Research 十字花科、萝卜属402Mb甘蓝Brassica oleracea 2014.5 Naturecommunications十字花科、芸薹属630Mb菜豆Phaseolus vulgaris L.2014.6 Nature Genetics 豆科,菜豆属587Mb野生大豆Glycine soja2014.7Naturecommunications豆科、大豆属868 Mb普通小麦Triticum aestivum 2014.7 Science 禾本科17Gb野生西红柿Solanum pennellii 2014.7 Nature Genetics茄科942 Mb非洲野生稻Oryza glaberrima2014.8 Nature Genetics禾本科316 Mb油菜Brassica napus2014.8 Science十字花科630 Mb中果咖啡Coffea canephora 2014.9 Science 茜草科,咖啡属710 Mb茄子Solanum melongena 2014.9 DNA Research 茄科、茄属1093 Mb多个野生大豆Glycine soja 2014.9Naturebiotechnology豆科、大豆属889.33~1,118.34Mb绿豆Vigna radiata 2014.10 Naturecommunications豆科、豇豆属543 Mb啤酒花Humulus lupulus 2014.11 Plant and CellPhysiology大麻科、葎草属 2.57 Gb蝴蝶兰Phalaenopsis equestris2014.11 Nature Genetics 兰科、蝴蝶兰属 1.16 Gb。
这可能是最全面的已发表基因组物种列表!

这可能是最全面的已发表基因组物种列表!
少罗嗦,直接给网址:
/wiki/Lists_of_sequenced_genomes
点击文末评论区的“阅读原文”可直接在手机上查看,这里建议将链接复制到电脑端的浏览器查看。
打开后的页面是这样的,see,包括了8个列表:分别是动物(animal),古菌(archaea),细菌(bacteria),真菌(fungi),植物(plant),质体(plastid),原生生物(protist)以及真核生物(eukaryote)的总列表。
比如点击植物对应的List,直接进入相应的页面,如下图。
页面包含一个分级目录,类似word的导航窗格,点击对应的分类,比如说点单子叶植物,可快速定位到当前页面相应的位置。
然后可单击列表变量名或者旁边的小三角会进行排序,如下图,比如可按物种名称排序,然后可快速定位到关注的一些物种(真菌和动物的展示方式比较简单)。
将鼠标光标移到物种的拉丁文学名上,会显示该物种相关的简介。
单击物种名称,会跳转到相应的百科词条,如下图。
点击年份的上标,会迅速定位到相应的文献,如下图。
点击引用文献的的doi或PMID号可直接跳转到当前文献的下载页面,如下图。
接着我们就可以下载该文献,进而追踪到基因组数据的下载链接。
当然,你也可以用拉丁名直接到Ensembl,NCBI,UCSC等数据库查找和下载相应物种的基因组数据。
今天的内容就到这里啦~。
已基因组测序植物列表

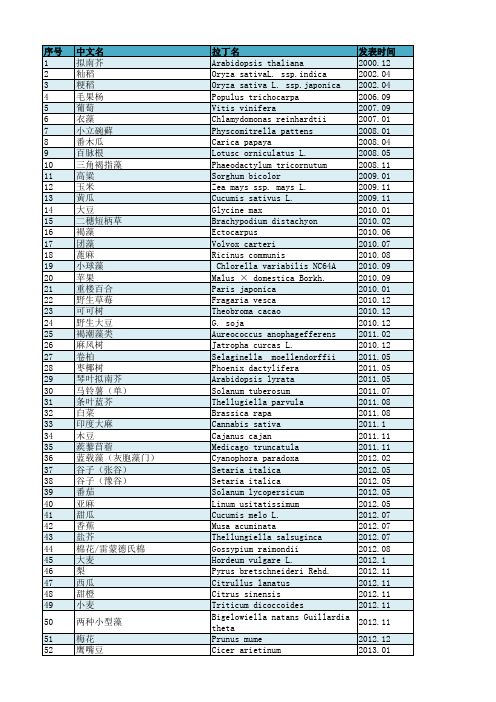
编号中文名拉丁名发表时间 刊物科、属基因组大小1拟南芥Arabidopsis thaliana 2000.12Nature 十字花科、鼠耳芥属125 Mb 2水稻Oryza sativa. ssp. indica 2002.04Science 禾本科、稻属466 Mb 3水稻Oryza sativa. ssp. japonica 2002.04Science 禾本科、稻属466 Mb 4杨树Populus trichocarpa2006.09Science 杨柳科、杨属480 Mb 5葡萄Vitis vinifera 2007.09Nature 葡萄科、葡萄属490 Mb 6衣藻Chlamydomonas reinhardtii 2007.01Science 衣藻科、衣藻属130 Mb 7小立碗藓Physcomitrella pattens2008.01Science 葫芦藓科、小立碗藓属480 Mb 8番木瓜Carica papaya 2008.04Nature 番木瓜科、番木瓜属370 Mb 9百脉根Lotus japonicus 2008.05DNA Research豆科、百脉根属472 Mb 10三角褐指藻Phaeodactylum tricornutum2008.11Nature 褐指藻属27.4 Mb 11高粱Sorghum bicolor2009.01Nature禾本科、高粱属730 Mb 12玉米Zea mays ssp. mays 2009.11Science 禾本科、玉米属2300 Mb 13黄瓜Cucumis sativus 2009.11Nature Genetics葫芦科、黄瓜属350 Mb 14大豆Glycine max 2010.01Nature豆科、大豆属1100 Mb 15二穗短柄草Brachypodium distachyon 2010.02Nature 禾本科、短柄草属260 Mb 16褐藻Ectocarpus 2010.06Nature 水云属 196 Mb 17团藻Volvox carteri2010.07Science团藻属138 Mb 18蓖麻Ricinus communis 2010.08Nature Biotechnology大戟科、蓖麻属350 Mb 19小球藻Chlorella variabilis 2010.09Plant Cell 小球藻科46 Mb 20苹果Malus × domestica 2010.09Nature Genetics 蔷薇科、苹果属742 Mb 21森林草莓Fragaria vesca 2010.12Nature Genetics 蔷薇科、草莓属240 Mb 22可可树Theobroma cacao 2010.12Nature Genetics梧桐科、可可属430- Mb 23野生大豆Glycine soja2010.12PNAS 豆科、大豆属915.4 Mb 24褐潮藻类Aureococcus anophagefferens2011.02PNAS 57 Mb 25麻风树Jatropha curcas2010.12DNA Research大戟科、麻风树属410 Mbs ci en ce ne t.cn : Gr an t l uj i an g26卷柏Selaginella moellendorffii 2011.05Science 卷柏属212 Mb 27枣椰树Phoenix dactylifera 2011.05Nature Biotechnology 棕榈科685 Mb 28琴叶拟南芥Arabidopsis lyrata 2011.05Nature Genetics十字花科、鼠耳芥属206.7 Mb 29马铃薯Solanum tuberosum 2011.07Nature 茄目、茄科、茄属844 Mb 30条叶蓝芥Thellugiella parvula 2011.08Nature Genetics 盐芥属140 Mb 31白菜Brassica rapa 2011.08Nature Genetics 十字花科、芸薹属485 Mb 32印度大麻Cannabis sativa 2011.01Genome Biology 大麻属534 Mb 33木豆Cajanus cajan 2011.11Nature Biotechnology豆科、木豆属833 Mb 34蒺藜苜蓿Medicago truncatula 2011.11Nature 豆科苜蓿属500 Mb 35蓝载藻Cyanophora paradoxa2012.02Science 灰胞藻门70 Mb 36谷子Setaria italica 2012.05Nature Biotechnology 禾本科、狗尾草属490 Mb 37谷子Setaria italica 2012.05Nature Biotechnology禾本科、狗尾草属510 Mb 38番茄Solanum lycopersicum2012.05Nature 茄科、茄属900 Mb 39甜瓜Cucumis melo 2012.07PNAS 葫芦科、甜瓜属450 Mb 40亚麻Linum usitatissimum 2012.07Plant Journal 亚麻科、亚麻属373 Mb 41盐芥Thellungiella salsuginea2012.07PNAS 十字花科、盐芥属260 Mb 42香蕉Musa acuminata 2012.07Nature 芭蕉科、芭蕉属523 Mb 43雷蒙德氏棉 Gossypium raimondii 2012.08Nature Genetics锦葵科、棉属775.2 Mb 44大麦Hordeum vulgare 2012.01Nature禾本科、大麦属 5.1 Gb 45梨Pyrus bretschneideri 2012.11Genome Research 蔷薇科、梨属527 Mb 46西瓜Citrullus lanatus 2012.11Nature Genetics 葫芦科、西瓜属425 Mb 47甜橙Citrus sinensis 2012.11Nature Genetics芸香科、柑橘属367 Mb 48小麦Triticum aestivum 2012.11Nature 禾本科、小麦属17 Gb 49两种小型藻Bigelowiella natans, Guillardiatheta2012.11Nature 95 Mb, 87 Mb 50棉花(雷蒙德氏棉)Gossypium raimondii2012.12Nature 锦葵科、棉属761.4 Mb 51梅花Prunus mume 2012.12Nature Communications 蔷薇科、梨属280 Mb 52鹰嘴豆Cicer arietinum2013.01Nature Biotechnology豆科、鹰嘴豆属738 Mbs ci en ce ne t.cn :Gr an tl uj i an g53橡胶树Hevea brasiliensis 2013.02BMC Genomics 大戟科、橡胶树属2.15 Gb 54毛竹Phyllostachys heterocycla 2013.02Nature Genetics 竹科、钢竹属 2.075 Gb 55短花药野生稻Oryza brachyantha 2013.03Nature Communications禾本科稻属342 Mb-362 Mb56小麦A Triticum urartu 2013.03Nature 禾本科、小麦属 4.94 Gb 57小麦D grassAegilops tauschii 2013.03Nature 禾本科、小麦属 4.36 Gb 58桃树Prunus persica 2013.03Nature Genetics 蔷薇科、梨属265 Mb 59丝叶狸藻Utricularia gibba 2013.05Nature 狸藻科、狸藻属82 Mb 60中国莲Nelumbo nucifera Gaertn2013.05Genome Biology睡莲科、莲属929 Mb 61挪威云杉Picea abies 2013.05Nature 松科、云杉属19.6 Gb 62海洋球石藻Emiliania huxleyi 2013.06Nature 定鞭藻纲141.7 Mb 63虫黄藻Symbiodinium minutum 2013.07Current Biology甲藻门1.5 Gb 64油棕榈Elaeis guineensis 2013.07Nature棕榈科、油棕榈属1.8 Gb 65枣椰树Phoenix dactylifera 2013.08Nature Communications棕榈科、刺葵属671 Mb 66醉蝶花Tarenaya hassleriana 2013.08Plant Cell 醉蝶花科、醉蝶花属290 Mb 67莲Nelumbo nucifera 2013.08Plant Journal 睡莲科、莲属879 Mb 68桑树Morus notabilis 2013.09Nature Communications 桑科、桑属357 Mb 69猕猴桃Actinidia chinensis 2013.01Nature Communications 猕猴桃属616.1 Mb 70胡杨Populus euphratica 2013.11Nature Communications杨属496.5 Mb 71八倍体草莓 F. x ananassa 2013.12DNA Research 草莓属698 Mb 72康乃馨Dianthus caryophyllus L.2013.12DNA Research石竹属622 Mb 73甜菜Beta vulgaris ssp. vulgaris 2013.12Nature 藜科甜菜属566.6 Mb 74无油樟(互叶梅)Amborella trichopoda 2013.12Science 无油樟属748 Mb 75辣椒Capsicum annuum 2014.01Nature Genetics 辣椒属 3.48 Gb 76芝麻Sesamum indicum 2014.02Genome Biology胡麻科胡麻属274 Mb 77辣椒Capsicum annuum (Zunla-1)2014.03PNAS 辣椒属 3.48 Gb 78火炬松Pinus taeda (Loblolly pine)2014.03Genome Biology 松属23.2 Gb 79棉花(亚洲棉)Gossypium arboreum2014.05Nature Genetics锦葵科、棉属1694 Mbs ci en ce ne t.cn : Gr an t l uj i a n g80萝卜Raphanus sativus L. 2014.05DNA Research 十字花科、萝卜属402 Mb 81甘蓝Brassica oleracea 2014.05Nature Communications十字花科、芸薹属630 Mb 82菜豆Phaseolus vulgaris L .2014.06Nature Genetics 豆科,菜豆属587 Mb 83野生大豆Glycine soja 2014.07Nature Communications豆科、大豆属868 Mb 84普通小麦Triticum aestivum 2014.07Science 禾本科17 Gb 85野生西红柿Solanum pennellii 2014.07Nature Genetics茄科942 Mb 86非洲野生稻Oryza glaberrima 2014.08Nature Genetics禾本科316 Mb 87油菜Brassica napus 2014.08Science 十字花科630 Mb 88中果咖啡Coffea canephora 2014.09Science 茜草科,咖啡属710 Mb 89茄子Solanum melongena2014.09DNA Research 茄科、茄属1093 Mb 90多个野生大豆Glycine soja 2014.09Nature Biotechnology 豆科、大豆属889.33~1,118.34 Mb91枣Ziziphus jujuba 2014.10Nature Communications鼠李科、枣属444 Mb92绿豆Vigna radiata 2014.10Nature Communications 豆科、豇豆属543 Mb 93啤酒花Humulus lupulus 2014.11Plant and Cell Physiology 大麻科、葎草属 2.57 Gb 94蝴蝶兰Phalaenopsis equestris 2014.11Nature Genetics 兰科、蝴蝶兰属 1.16 Gb 95铁皮石斛Dendrobium officinale2014.12Molecular Plant 兰科、石斛属 1.35 Gb 96青稞Lasa goumang 2015.01PNAS 禾本科、大麦属 4.5 Gb 97报春花Primula veris2015.01Genome Biology 报春花科、报春花属479 Mb 98陆地棉(南农)Gossypium hirsutum L. acc. TM-12015.03Nature Biotechnology 锦葵科、棉属 2.5 Gb 99陆地棉(棉花所)Gossypium hirsutum L. acc. TM-12015.03Nature Biotechnology 锦葵科、棉属2.173 Gb 100海带Saccharina japonica 2015.04Nature Communications 海带科、海带属537 Mb 101长春花Catharanthus roseus 2015.04The Plant Journal夹竹桃科、长春花属738 Mb 102科民茄Solanum commersor 2015.04Plant cell 茄科、茄属830 Mb 103茭白Zizania latifolia 2015.06The Plant Journal 禾本科、菰属590 Mb 104黑麦草Lolium perenne 2015.09The Plant Journal 禾本科、黑麦草属 2 Gb 105小豆Vigna angularis 2015.10Nature Communications豆科、豇豆属542 Mb 106菠萝Ananas comosus2015.11Nature Genetics 凤梨科、凤梨属526 Mbs ci en ce ne t.cn : Gr an t l uj i an g107复活草Oropetium thomaeum 2015.11Nature 虎尾草亚科245 Mb 108丹参Salvia miltiorrhiza 2015.12GigaScience 唇形科、鼠尾草属641 Mb 109铁皮石斛Dendrobium catenatum 2016.01Scientific Reports 兰科、石斛属 1.11 Gbs ci en ce ne t.cn : Gr an t l uj i a n g。
已发表植物基因组(2018.04)
发表时间 2000.12 2002.04 2002.04 2006.09 2007.09 2007.01 2008.01 2008.04 2008.05 2008.11 2009.01 2009.11 2009.11 2010.01 2010.02 2010.06 2010.07 2010.08 2010.09 2010.09 2010.01 2010.12 2010.12 2010.12 2011.02 2010.12 2011.05 2011.05 2011.05 2011.07 2011.08 2011.08 2011.1 2011.11 2011.11 2012.02 2012.05 2012.05 2012.05 2012.05 2012.07 2012.07 2012.07 2012.08 2012.1 2012.11 2012.11 2012.11 2012.11
Gossypium hirsutum
2015.04
109 海带
Saccharina japonica
2015.04
110 科民茄(南美一种野生马铃薯) Solanum commersonii
2015.04
111 牛耳草
Boea hygrometrica
2015.04
112 长春花
Catharanthus roseus
2016.02
132 落花生
Arachis ipaensis
2016.02
133 鳗草
Zostera marina
2016.02
134 菜豆
Phaseolus vulgaris
2016.02
135 日本结缕草
Zoysia japonica
2016.04
部分植物基因组数据库汇总

部分植物基因组数据库汇总⽜年⼤吉
植物基因组数据库:
1、NCBI中的genome,直接下载NCBI上的基因组⽂件
ftp:///genomes/
2、植物基因组数据库(包含约30个左右的植物,具体查看:)
30个左右植物基因组对应列表下载
3、包含多个物种的基因组数据(如:柑橘、黄⽠、桉树、海棠等约100个左右)100个左右植物基因组对应列表下载
4、Ensembl数据库(植物)
70个左右植物基因组对应列表下载
5、拟南芥数据库
6、另外⼀个综合性的(包含番茄,马铃薯,烟草、茄⼦等)
7、⽔稻
8、马铃薯基因组数据库(较为全⾯)
9、百脉根
10、⾖科植物
11、葡萄基因组
12、⼩麦
13、最全⾯的杨树整合数据(⽬前我看到的)
14、棉花
15、茶
16、苜蓿
17、蔷薇科
18、荞麦
19、各种柑类
20、萝⼘
21、茄⼦
22、各种⽠类(西⽠,甜⽠、南⽠等)
23、梨
24、被⼦植物基因组数据库
25、菠萝
26、⽢草
27、板栗
28、花⽣
29、⽑⽵
30、薄荷
31、蓝莓
32、橡胶草
33、杜仲
33、圣罗勒
34、青蒿
35、向⽇葵
36、胡桃科
37、物种如下图所⽰
38、杨树等
祝⼤家新春快乐,阖家幸福!
⽜年⼤吉,多发Papper,顺利毕业,科研顺利!蚂蚁⽣信和⼤家在新的⼀年⾥⼀起加油!
⽜年更⽜。
小麦族鹅观草属 披碱草属 猬草属和仲彬草 属细胞质基因组

是禾本科 D 3 < 4 < ? 5 ; 5O / F 2 3 4 C 2 ; ? 5 ; 5植 !! 小麦族 收稿日期 ! 修回日期 ! ! ’ ’ % ’ # ) %" ! ’ ’ % ’ & ’ !
物中非常重要的一 个 类 群 $ 它包括了世界上重要的
基金项目 ! 国家自然科学基金 ! 编号 # " $ 教育部长江学者和创新团 队 发 展 计 划 ! $ 和四川省教育厅重点项目% * ’ ! & ’ ’ $ $$ * ’ # & ’ ) * % + , -’ # % *" . / 1 0 $D : 2 C * ’ ! & ’ ’ $ $$ * ’ # & ’ ) * %" 3 2 3 ; FA 2 3B 9 ; = < ; = ? 9 2 > ; 3 H; = 6+ = = 2 I ; 4 < I 5, 5 1 2 3 4 5 67 4 9 5: ; 4 < 2 = ; >. ? < 5 = ? 5@ 2 / = 6 ; 4 < 2 =2 AB 9 < = ;! E G E E. 0 8 " & H 5 ; 3 ? 95 ; F< =J = < I 5 3 H < 4 : 2 C + , -’ # % * ; = 6K 6 / ? ; 4 < 2 =L / 3 5 ; /2 A. < ? 9 / ; =D 3 2 I < = ? 5 8! 作者简介 ! 张 ! 颖! $ 女$ 大连人 $ 硕士 ( 现在东北大学生物技术研究所工作 $ 助教 ) $ & (’" 通讯作者 ! 周永红 ! $ 男$ 教授 $ 博士生导师 $ 研究方向 # 禾本科植物系统与进化 (K # ) $ ( !’" 1 F ; < > M 9 2 / < ? ; / C 5 6 / C ? = 9 "H 8
已发表动植物基因组

已发表动植物基因组在分子生物学领域的“圈内人”,如果还不了解全基因组测序的飞速发展,用娃娃的话说,那真是一个“奥特曼”了。
高通量测序技术出现掀开了生命科学研究的新篇章,促进了许多研究方向的的复苏和蓬勃发展,同时也涌现了一些交叉学科。
2011年德国爆发急性肠出血性流行病疫情——极大困扰着英国剑桥市新生儿护理病房的医护人员。
来自Sanger研究院的研究人员从多个患者身上取样,并采用Illumina MiSeq进行全基因组测序。
通过生物信息学分析得知,在疫情爆发过程中发生了一系列小规模突变,流行病学家据此绘制出一个进化树,追踪疫情逐步靠近可疑的源头。
文章作者Julian Parkhill表示:“基因测序提供了明晰的线索,这是其他方法难以做到的。
”文章发表在流行病学顶级杂志The Lancet Infectious Diseases。
2012年发表在science杂志旗下Science Translational Medicine另外一篇文章也同样表明了全基因组测序技术在流行病学中重要作用。
位于马里兰州贝塞斯达市医疗中心出现了严重的血液传染病事件,在此次事件中,共有17名病人感染了血液传染病,并造成6人死亡。
来自美国NIH人类基因组研究所的研究人员从病人体内和医院一些设备上采集了细菌样本,并对不同菌株的基因组进行测序。
通过对突变菌株进行聚类分析,将病人分组,再结合病例住院的记录(入院时间、所在病房等)画出了最可能的传播路线。
全基因组测序提供的详尽细节让传染病学家们真正地理解了疾病传播途径,详尽的追踪技术将带来更加完善的公共健康领域内的传染病学研究。
所以流行病学家更有可能找到事件发生的真正原因,而非最简单的原因。
这两件事让我们真实的看到了全基因组测序在流行病学的重要作用。
事实上,媲美曼哈顿和登月计划的“人类基因组计划”所带来的巨大作用正在逐渐显现,高通量测序技术走向个性化医疗已是大势所趋。
全基因组测序使得生命可以测量,人们可以更加方便快捷的了解生命的奥秘。
已公布全基因组序列的动植物种类及分类

多房棘球绦虫 (115M;Nature)
微小膜壳绦虫 (141.1M;Nature)
猪蛔虫
(272M;Nature)
南方根瘤线虫(50M;Nat. Biotechnol.)
罗阿丝虫 (91.4M;Nat. Genet.)
海胆
(1G;Science)
旋毛虫 秀丽线虫 线虫 马来丝虫
牡蛎
(16.44MG;;DNNaAt.RGeesenaertc.)h) (97M;Science) (104M;PLoS Biol.) (90M;Science)
家鸽
(1.3G;Science)
游隼 猎隼
(1.2G;Nat. Genet.) (1.2G;Nat. Genet.)
鸭嘴兽 短尾负鼠 袋獾 袋鼠 猛犸象 牛 牦牛
(1.84G;Nature) (3.4G;Nature) (3.3G;PNAS) (2.9G;Genome Biol.) (4.7G;Nature) (2.87G;Science) (2.6G;Nat. Genet.)
黄瓜属 甜瓜属
西瓜属
小立碗藓 (480M;Science)
卷柏
(212M;Science)
枣卷椰柏树
卷(柏685M;Nat. Biotechnol.)
短花卷药柏野生稻 卷(柏281M; Nat. Commum.)
水稻
(466M;Science)
二穗短柄草 (260M;Nature)
高粱
(730M;Nature)
双子叶植物纲
葡萄目
豆目
葡萄科、葡萄属
大豆属
豆科
木豆属 苜蓿属 豌豆属
葡萄
(490M;Nature)
大豆 野生大豆
已完成基因组测序的生物(植物部分)

水稻、玉米、大豆、甘蓝、白菜、高粱、黄瓜、西瓜、马铃薯、番茄、拟南芥、杨树、麻风树、苹果、桃、葡萄、花生拟南芥籼稻粳稻葡萄番木瓜高粱黄瓜玉米栽培大豆苹果蓖麻野草莓马铃薯白菜野生番茄番茄梨甜瓜香蕉亚麻大麦普通小麦西瓜甜橙陆地棉梅毛竹桃芝麻杨树麻风树卷柏狗尾草属花生甘蓝物种基因组大小和开放阅读框文献Sesamum indicum L. Sesame 芝麻(2n = 26)293.7 Mb, 10,656 orfs 1Oryza brachyantha短药野生稻261 Mb, 32,038 orfs 2Chondrus crispus Red seaweed爱尔兰海藻105 Mb, 9,606 orfs 3Pyropia yezoensis susabi-nori海苔43 Mb, 10,327 orfs 4Prunus persica Peach 桃226.6 of 265 Mb 27,852 orfs 5Aegilops tauschii 山羊草(DD)4.23 Gb (97% of the 4.36), 43,150 orfs 6 Triticum urartu 乌拉尔图小麦(AA)4.66 Gb (94.3 % of 4.94 Gb, 34,879 orfs 7 moso bamboo (Phyllostachys heterocycla) 毛竹2.05 Gb (95%) 31,987 orfs 8Cicer arietinum Chickpea鹰嘴豆~738-Mb,28,269 orfs 9 520 Mb (70% of 740 Mb), 27,571 orfs 10Prunus mume 梅280 Mb, 31,390 orfs 11Gossypium hirsutum L.陆地棉2.425 Gb 12Gossypium hirsutum L. 雷蒙德氏棉761.8 Mb 13Citrus sinensis甜橙87.3% of ~367 Mb, 29,445 orfs 14甜橙367 Mb 15Citrullus lanatus watermelon 西瓜353.5 of ~425 Mb (83.2%) 23,440 orfs 16 Betula nana dwarf birch,矮桦450 Mb 17Nannochloropsis oceanica CCMP1779微绿球藻(产油藻类之一)28.7 Mb,11,973 orfs 18Triticum aestivum bread wheat普通小麦17 Gb, 94,000 and 96,000 orfs 19 Hordeum vulgare L. barley 大麦1.13 Gb of 5.1 Gb,26,159 high confidence orfs,53,000 low confidence orfs 20Gossypium raimondii cotton 雷蒙德氏棉D subgenome,88% of 880 Mb 40,976 orfs 21Linum usitatissimum flax 亚麻302 mb (81%), 43,384 orfs 22Musa acuminata banana 香蕉472.2 of 523 Mb, 36,542 orfs 23Cucumis melo L. melon 甜瓜375 Mb(83.3%)27,427 orfs 24Pyrus bretschneideri Rehd. cv. Dangshansuli 梨(砀山酥梨)512.0 Mb (97.1%), 42,812 orfs 25,26Solanum lycopersicum 番茄760/900 Mb,34727 orfs 27S. pimpinellifolium LA1589野生番茄739 MbSetaria 狗尾草属(谷子、青狗尾草)400 Mb,25000-29000 orfs 28,29 Cajanus cajan pigeonpea木豆833 Mb,48,680 orfs 30Nannochloropis gaditana 一种海藻~29 Mb, 9,052 orfs 31Medicago truncatula蒺藜苜蓿350.2 Mb, 62,388 orfs 32Brassica rapa 白菜485 Mb 33Solanum tuberosum 马铃薯0.73 Mb,39031 orfs 34Thellungiella parvula条叶蓝芥13.08 Mb 29,338 orfs 35Arabidopsis lyrata lyrata 玉山筷子芥? 183.7 Mb, 32670 orfs 36Fragaria vesca 野草莓240 Mb,34,809 orfs 37Theobroma cacao 可可76% of 430 Mb, 28,798 orfs 38Aureococcus anophagefferens褐潮藻32 Mb, 11501 orfs 39Selaginella moellendorfii江南卷柏208.5 Mb, 34782 orfs 40Jatropha curcas Palawan麻疯树285.9 Mb, 40929 orfs 41Oryza glaberrima 光稃稻(非洲栽培稻)206.3 Mb (0.6x), 10 080 orfs (>70% coverage) 42Phoenix dactylifera 棕枣380 Mb of 658 Mb, 25,059 orfs 43Chlorella sp. NC64A小球藻属40000 Kb, 9791 orfs 44Ricinus communis蓖麻325 Mb, 31,237 orfs 45Malus domestica (Malus x domestica)苹果742.3 Mb 46Volvox carteri f. nagariensis 69-1b一种团藻120 Mb, 14437 orfs 47 Brachypodium distachyon 短柄草272 Mb,25,532 orfs 48Glycine max cultivar Williams 82栽培大豆1.1 Gb, 46430 orfs 49Zea mays ssp. Mays Zea mays ssp. Parviglumis Zea mays ssp. Mexicana Tripsacum dactyloides var. meridionale 无法下载附表50Zea mays mays cv. B73玉米2.06 Gb, 106046 orfs 51Cucumis sativus 9930 黄瓜243.5 Mb, 63312 orfs 52Micromonas pusilla金藻21.7 Mb, 10248 orfs 53Sorghum bicolor 高粱697.6 Mb, 32886 orfs 54Phaeodactylum tricornutum 三角褐指藻24.6 Mb, 9479 orfs 55Carica papaya L. papaya 番木瓜271 Mb (75%), 28,629 orfs 56 Physcomitrella patens patens小立碗藓454 Mb, 35805 orfs 57Vitis vinifera L. Pinot Noir, clone ENTAV 115葡萄504.6 Mb, 29585 orfs 58 Vitis vinifera PN40024葡萄475 Mb 59Ostreococcus lucimarinus绿色鞭毛藻13.2 Mb, 7640 orfs 60 Chlamydomonas reinhardtii 莱茵衣藻100 Mb, 15256 orfs 61Populus trichocarpa黑三角叶杨550 Mb, 45000 orfs 62Ostreococcus tauri 绿藻12.6 Mb, 7892 orfs 63Oryza sativa ssp. japonica 粳稻360.8 Mb, 37544 orfs 64Thalassiosira pseudonana 硅藻25 Mb, 11242 orfs 65Cyanidioschyzon merolae 10D红藻16.5 Mb, 5331 orfs 66Oryza sativa ssp. japonica粳稻420 Mb, 50000 orfs 67Oryza sativa L. ssp. Indica籼稻420 Mb, 59855 orfs 68Guillardia theta -蓝隐藻,551 Kb, 553 orfs 69Arabidopsis thaliana Columbia拟南芥119.7 Mb, 31392 orfs 70参考文献1 Zhang, H. et al. Genome sequencing of the important oilseed crop Sesamum indicum L. Genome Biology 14, 401 (2013).2 Chen, J. et al. Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nat Commun 4, 1595 (2013).3 Collén, J. et al. Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proceedings of the National Academy of Sciences 110, 5247-5252 (2013).4 Nakamura, Y. et al. The first symbiont-free genome sequence of marine red alga, susabi-nori Pyropia yezoensis. PLoS ONE 8, e57122 (2013).5 Verde, I. et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nature Genetics advance online publication (2013).6 Jia, J. et al. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496, 91-95 (2013).7 Ling, H.-Q. et al. Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496, 87-90 (2013).8 Peng, Z. et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nature Genetics 45, 456-461 (2013).9 Jain, M. et al. A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant Journal, DOI: 10.1111/tpj.12173 (2013).10 Varshney, R. K. et al. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotech 31, 240-246 (2013).11 Zhang, Q. et al. The genome of Prunus mume. Nat Commun 3, 1318 (2012).12 Lee, M.-K. et al. Construction of a plant-transformation-competent BIBAC library and genome sequence analysis of polyploid Upland cotton (Gossypium hirsutum L.). BMC Genomics 14, 208 (2013).13 Paterson, A. H. et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423-427 (2012).14 Xu, Q. et al. The draft genome of sweet orange (Citrus sinensis). Nat Genet 45,59–66 (2013).15 Belknap, W. R. et al. Characterizing the citrus cultivar Carrizo genome through 454 shotgun sequencing. Genome 54, 1005-1015 (2011).16 Guo, S. et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet 45, 51–58 (2013).17 Wang, N. et al. Genome sequence of dwarf birch (Betula nana) and cross-species RAD markers. Mol Ecol Article first published online: 21 NOV 2012 DOI:10.1111/mec.12131 (2012).18 Vieler, A. et al. Genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genet 8, e1003064 (2012).19 Brenchley, R. et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491, 705-710 (2012).20 Consortium, T. I. B. G. S. A physical, genetic and functional sequence assembly of the barley genome. Nature 491, 711–716 (2012).21 Wang, K. et al. The draft genome of a diploid cotton Gossypium raimondii. Nature Genetics 44, 1098–1103 (2012).22 Wang, Z. et al. The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. The Plant Journal 72, 461-473 (2012).23 D'Hont, A. et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488, 213–217 (2012).24 Garcia-Mas, J. et al. The genome of melon (Cucumis melo L.). PNAS 109, 11872-11877 (2012).25 reporter, A. G. s. Consortium releases pear genome data. GenomeWeb Daily News (2012).26 Wu, J. et al. The genome of pear (Pyrus bretschneideri Rehd.). GenomeRes.Published in Advance November 13, 2012, doi:10.1101/gr.144311.112 (2012).27 Consortium, T. T. G. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 (2012).28 Bennetzen, J. L. et al. Reference genome sequence of the model plant Setaria. Nat Biotech 30, 555-561 (2012).29 Zhang, G. et al. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nat Biotech 30, 549-554 (2012).30 Varshney, R. K. et al. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotech 30, 83-89 (2012).31 Radakovits, R. et al. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat Commun 3, 686 (2012).32 Young, N. D. et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524 (2011).33 Wang, X. et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035-1039 (2011).34 Consortium, T. P. G. S. Genome sequence and analysis of the tuber crop potato. Nature 475, 189-195 (2011).35 Dassanayake, M. et al. The genome of the extremophile crucifer Thellungiella parvula. Nat. Genet. 43, 913-918 (2011).36 Hu, T. T. et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43, 476-481 (2011).37 Shulaev, V. et al. The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 43, 109-116 (2011).38 Argout, X. et al. The genome of Theobroma cacao. Nat. Genet. 43, 101-108 (2011).39 Gobler, C. J. et al. Niche of harmful alga Aureococcus anophagefferens revealed through ecogenomics. PNAS 108, 4352-4357 (2011).40 Banks, J. A. et al. The selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960-963 (2011).41 Sato, S. et al. Sequence analysis of the genome of an oil-bearing tree, Jatropha curcas L. DNA Res. 18, 65-76 (2011).42 Sakai, H. et al. Distinct evolutionary patterns of Oryza glaberrima deciphered by genome sequencing and comparative analysis. Plant Journal 66, 796-805 (2011).43 Al-Dous, E. K. et al. De novo genome sequencing and comparative genomics of date palm (Phoenix dactylifera). Nat Biotech 29, 521-527 (2011).44 Blanc, G. et al. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 22, 2943-2955 (2010).45 Chan, A. P. et al. Draft genome sequence of the oilseed species Ricinus communis. Nat Biotech 28(951-956 (2010).46 Velasco, R. et al. The genome of the domesticated apple (Malus x domestica Borkh.). Nat. Genet. 42, 833-839 (2010).47 Prochnik, S. E. et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329, 223-226 (2010).48 Initiative, T. I. B. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763-768 (2010).49 Schmutz, J. et al. Genome sequence of the palaeopolyploid soybean. Nature 463, 178-183 (2010).50 Hufford, M. B. et al. Comparative population genomics of maize domestication and improvement. Nat Genet 44, 808-811 (2012).51 Wei, F. et al. The physical and genetic framework of the maize B73 genome. PLoS Genet 5, e1000715 (2009).52 Huang, S. et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41, 1275-1281 (2009).53 Worden, A. Z. et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324, 268-272 (2009).54 Paterson, A. H. et al. The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551-556 (2009).55 Bowler, C. et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239-244 (2008).56 Ming, R. et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452, 991-996 (2008).57 Rensing, S. A. et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64-69 (2008).58 Velasco, R. et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS One 2, e1326 (2007).59 Jaillon, O. et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463-467 (2007).60 Palenik, B. et al. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. PNAS 104, 7705-7710 (2007).61 Merchant, S. S. et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245-250 (2007).62 Tuskan, G. A. et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596-1604 (2006).63 Derelle, E. et al. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. PNAS 103, 11647-11652 (2006). 64 Project, I. R. G. S. The map-based sequence of the rice genome. Nature 436,793-800 (2005).65 Armbrust, E. V. et al. The genome of the diatom Thalassiosira Pseudonana: ecology, evolution, and metabolism. Science 306, 79-86 (2004).66 Matsuzaki, M. et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653-657 (2004).67 Goff, S. A. et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92-100 (2002).68 Yu, J. et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79-92 (2002).69 Douglas, S. et al. The highly reduced genome of an enslaved algal nucleus. Nature 410, 1091-1096 (2001).70 Kaul, S. et al. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796-815 (2000).。
已测序植物基因组汇总
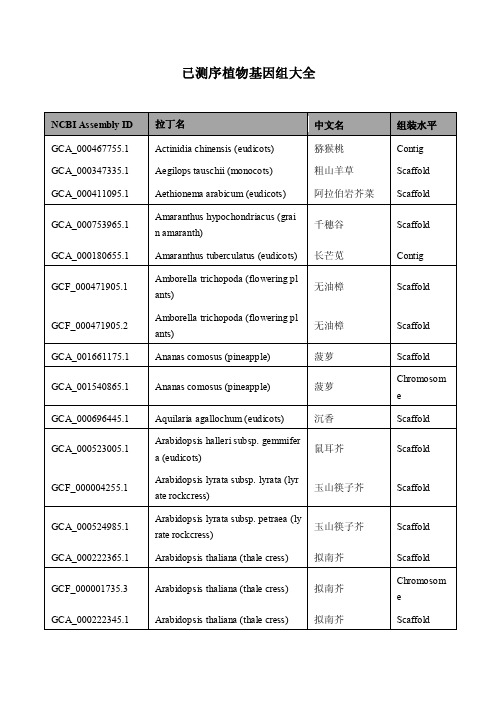
已测序植物基因组大全NCBI Assembly ID 拉丁名中文名组装水平GCA_000467755.1 Actinidia chinensis (eudicots) 猕猴桃Contig GCA_000347335.1 Aegilops tauschii (monocots) 粗山羊草Scaffold GCA_000411095.1 Aethionema arabicum (eudicots) 阿拉伯岩芥菜ScaffoldGCA_000753965.1 Amaranthus hypochondriacus (grain amaranth)千穗谷ScaffoldGCA_000180655.1 Amaranthus tuberculatus (eudicots) 长芒苋ContigGCF_000471905.1 Amborella trichopoda (flowering plants)无油樟ScaffoldGCF_000471905.2 Amborella trichopoda (flowering plants)无油樟ScaffoldGCA_001661175.1 Ananas comosus (pineapple) 菠萝ScaffoldGCA_001540865.1 Ananas comosus (pineapple) 菠萝Chromosom eGCA_000696445.1 Aquilaria agallochum (eudicots) 沉香ScaffoldGCA_000523005.1 Arabidopsis halleri subsp. gemmifera (eudicots)鼠耳芥ScaffoldGCF_000004255.1 Arabidopsis lyrata subsp. lyrata (lyrate rockcress)玉山筷子芥ScaffoldGCA_000524985.1 Arabidopsis lyrata subsp. petraea (lyrate rockcress)玉山筷子芥ScaffoldGCA_000222365.1 Arabidopsis thaliana (thale cress) 拟南芥ScaffoldGCF_000001735.3 Arabidopsis thaliana (thale cress) 拟南芥Chromosom eGCA_000222345.1 Arabidopsis thaliana (thale cress) 拟南芥ScaffoldGCA_000211275.1 Arabidopsis thaliana (thale cress) 拟南芥eGCA_000222385.1 Arabidopsis thaliana (thale cress) 拟南芥ScaffoldGCF_000001735.1 Arabidopsis thaliana (thale cress) 拟南芥Complete G enomeGCF_000001735.2 Arabidopsis thaliana (thale cress) 拟南芥Chromosom eGCA_000835945.1 Arabidopsis thaliana (thale cress) 拟南芥Contig GCA_000222325.1 Arabidopsis thaliana (thale cress) 拟南芥ScaffoldGCA_001651475.1 Arabidopsis thaliana (thale cress) 拟南芥Chromosom eGCA_000612745.1 Arabis alpina (gray rockcress) 小花南芥ContigGCA_000733195.1 Arabis alpina (gray rockcress) 小花南芥Chromosom eGCA_001484125.1 Arabis montbretiana (eudicots) 南芥菜花Contig GCA_001484925.1 Arabis nordmanniana (eudicots) 南芥菜花ScaffoldGCF_000817695.1 Arachis duranensis (eudicots) 蔓花生A亚基因组ChromosomeGCA_001687015.1 Arachis duranensis (eudicots) 蔓花生A亚基因组ScaffoldGCF_000816755.1 Arachis ipaensis (eudicots) 蔓花生B亚基因组ChromosomeGCA_000439995.1 Azadirachta indica (eudicots) 印度苦楝树Contig GCA_000439995.2 Azadirachta indica (eudicots) 印度苦楝树Contig GCA_000439995.3 Azadirachta indica (eudicots) 印度苦楝树Contig GCA_000397105.1 Beta vulgaris subsp. vulgaris (beet) 甜菜ScaffoldGCA_000511025.2 Beta vulgaris subsp. vulgaris (beet) 甜菜Chromosom eGCA_000510975.1 Beta vulgaris subsp. vulgaris (beet) 甜菜eGCA_000510485.1 Beta vulgaris subsp. vulgaris (beet) 甜菜Scaffold GCA_000510875.1 Beta vulgaris subsp. vulgaris (beet) 甜菜Scaffold GCA_000510465.1 Beta vulgaris subsp. vulgaris (beet) 甜菜Scaffold GCA_000729925.1 Beta vulgaris subsp. vulgaris (beet) 甜菜Contig GCA_000510365.1 Beta vulgaris subsp. vulgaris (beet) 甜菜ScaffoldGCF_000511025.1 Beta vulgaris subsp. vulgaris (beet) 甜菜Chromosom eGCA_000327005.1 Betula nana (alpine birch) 高山桦木ScaffoldGCF_000005505.1 Brachypodium distachyon (stiff brome)二穗短柄草ChromosomeGCF_000005505.2 Brachypodium distachyon (stiff brome)二穗短柄草ChromosomeGCF_000686985.1 Brassica napus (rape) 油菜Chromosom eGCA_000751015.1 Brassica napus (rape) 油菜ScaffoldGCA_001682895.1 Brassica nigra (black mustard) 黑芥Chromosom eGCA_000604025.1 Brassica oleracea var. capitata (cabbage)卷心菜ScaffoldGCF_000695525.1 Brassica oleracea var. oleracea (eudicots)卷心菜ChromosomeGCF_000309985.1 Brassica rapa (field mustard) 荠菜Chromosom eGCA_000340665.1 Cajanus cajan (pigeon pea) 木豆Chromosom eGCA_000230855.1 Cajanus cajan (pigeon pea) 木豆ContigGCF_000633955.1 Camelina sativa (false flax) 亚麻荠eGCA_000496875.1 Camelina sativa (false flax) 亚麻荠Scaffold GCA_001509995.1 Cannabis sativa (hemp) 亚麻荠Scaffold GCA_000230575.1 Cannabis sativa (hemp) 亚麻荠ScaffoldGCA_001510005.1 Cannabis sativa subsp. indica (hemp)亚麻荠ContigGCF_000375325.1 Capsella rubella (eudicots) 拟南芥近缘种ScaffoldGCF_000710875.1 Capsicum annuum (eudicots) 辣椒Chromosom eGCA_000512255.1 Capsicum annuum (eudicots) 辣椒ScaffoldGCA_000950795.1 Capsicum annuum var. glabriusculum (eudicots)野生辣椒ChromosomeGCA_000150535.1 Carica papaya (papaya) 木瓜ScaffoldGCA_001310045.1 Carica papaya (papaya) 木瓜Chromosom eGCA_001633085.1 Carthamus tinctorius (safflower) 红花素ScaffoldGCA_000763605.1 Castanea mollissima (Chinese chestnut)板栗ScaffoldGCA_001292525.1 Catharanthus roseus (Madagascar periwinkle)长春花ContigGCA_001292565.1 Catharanthus roseus (Madagascar periwinkle)长春花ContigGCA_000949345.1 Catharanthus roseus (Madagascar periwinkle)长春花ScaffoldGCA_001687005.1 Chenopodium pallidicaule (eudicots)苍白茎藜ScaffoldGCA_001683475.1 Chenopodium quinoa (quinoa) 奎奴亚藜ScaffoldGCA_001687025.1 Chenopodium suecicum (eudicots) 藜ScaffoldGCA_000347275.2 Cicer arietinum (chickpea) 鹰嘴豆Chromosom eGCF_000331145.1 Cicer arietinum (chickpea) 鹰嘴豆Chromosom eGCA_000347275.1 Cicer arietinum (chickpea) 鹰嘴豆Scaffold GCA_000238415.1 Citrullus lanatus (wild melon) 野生甜瓜Contig GCF_000493195.1 Citrus clementina (eudicots) 柑橘Scaffold GCA_000695605.1 Citrus sinensis (sweet orange) 甜橙ScaffoldGCF_000317415.1 Citrus sinensis (sweet orange) 甜橙Chromosom eGCA_000775935.1 Conyza canadensis (horseweed) 小蓬草Contig GCF_000313045.1 Cucumis melo (muskmelon) 甜瓜Scaffold GCF_000004075.1 Cucumis sativus (cucumber) 黄瓜Scaffold GCF_000224045.1 Cucumis sativus (cucumber) 黄瓜Scaffold GCA_001483825.1 Cucumis sativus (cucumber) 黄瓜ContigGCF_000004075.2 Cucumis sativus (cucumber) 黄瓜Chromosom eGCF_001625215.1 Daucus carota subsp. sativus (carrot)胡萝卜ChromosomeGCA_001605985.1 Dendrobium catenatum (monocots) 石斛兰Scaffold GCA_000512335.1 Dianthus caryophyllus (clove pink) 香石竹ScaffoldGCA_001633215.1 Dichanthelium oligosanthes (monocots)* ScaffoldGCA_000774125.1 Diospyros lotus (eudicots) 莲花ScaffoldGCA_001598015.1 Dorcoceras hygrometricum (eudicots)旋蒴苣苔ScaffoldGCA_001647135.1 Eichhornia paniculata (monocots) 凤眼蓝ScaffoldGCF_000442705.1 Elaeis guineensis (African oil palm) 油棕榈Chromosom eGCA_001672495.1 Elaeis guineensis (African oil palm) 油棕榈Scaffold GCA_000441515.1 Elaeis oleifera (American oil palm) 美国油棕ScaffoldGCA_000818735.1 Ensete ventricosum (monocots) 阿比西尼亚红脉蕉ScaffoldGCA_000331365.2 Ensete ventricosum (monocots) 阿比西尼亚红脉蕉ScaffoldGCA_000818735.2 Ensete ventricosum (monocots) 阿比西尼亚红脉蕉ScaffoldGCA_000331365.1 Ensete ventricosum (monocots) 阿比西尼亚红脉蕉ScaffoldGCA_000970635.1 Eragrostis tef (tef) 抗旱粮食作物ScaffoldGCF_000504015.1 Erythranthe guttata (spotted monkey flower)地黄猴面花ScaffoldGCA_000260855.1 Eucalyptus camaldulensis (Murray red gum)赤桉ScaffoldGCF_000612305.1 Eucalyptus grandis (rose gum) 巨桉ScaffoldGCA_000325905.2 Eutrema salsugineum (eudicots) 山嵛菜Chromosom eGCA_000325905.1 Eutrema salsugineum (eudicots) 山嵛菜Scaffold GCF_000478725.1 Eutrema salsugineum (eudicots) 山嵛菜ScaffoldGCA_001661195.1 Fagopyrum esculentum (common buckwheat)普通荞麦ScaffoldGCA_000511975.1 Fragaria iinumae (eudicots) 古二倍体草莓Scaffold GCA_000512025.1 Fragaria nipponica (eudicots) 日本草莓Scaffold GCA_000511995.1 Fragaria nubicola (eudicots) 西藏草莓ScaffoldGCA_000517285.1 Fragaria orientalis (eudicots) 东方草莓ScaffoldGCF_000184155.1 Fragaria vesca subsp. vesca (wild strawberry)野生草莓ChromosomeGCA_000511835.1 Fragaria x ananassa (strawberry) 草莓Scaffold GCA_000511695.1 Fragaria x ananassa (strawberry) 草莓Scaffold GCA_000576895.1 Fraxinus excelsior (European ash) 欧洲草莓Scaffold GCA_000441915.1 Genlisea aurea (eudicots) 螺旋狸藻ScaffoldGCF_000004515.3 Glycine max (soybean) 大豆Chromosom eGCF_000004515.4 Glycine max (soybean) 大豆Chromosom eGCF_000004515.2 Glycine max (soybean) 大豆Chromosom eGCA_001269945.1 Glycine max (soybean) 大豆Contig GCA_001269945.2 Glycine max (soybean) 大豆ContigGCF_000004515.1 Glycine max (soybean) 大豆Chromosom eGCA_000722935.1 Glycine soja (wild soybean) 野生大豆Scaffold GCA_000722935.2 Glycine soja (wild soybean) 野生大豆ScaffoldGCF_000612285.1 Gossypium arboreum (tree cotton) 树棉Chromosom eGCA_000787975.1 Gossypium arboreum (tree cotton) 树棉Scaffold GCA_000612285.1 Gossypium arboreum (tree cotton) 树棉ScaffoldGCF_000987745.1 Gossypium hirsutum (eudicots) 陆地棉Chromosom eGCA_000331045.1 Gossypium raimondii (eudicots) 雷蒙德氏棉Scaffold GCF_000327365.1 Gossypium raimondii (eudicots) 雷蒙德氏棉ChromosomeGCA_001654055.1 Hevea brasiliensis (eudicots) 橡胶树Scaffold GCA_000340545.1 Hevea brasiliensis (eudicots) 橡胶树Scaffold GCA_001696755.1 Hibiscus syriacus (Rose-of-Sharon) 木槿Scaffold GCA_000582825.1 Hordeum pubiflorum (monocots) 大麦Scaffold GCA_000947855.1 Hordeum vulgare (barley) 大麦ContigGCA_000227425.1 Hordeum vulgare subsp. vulgare (domesticated barley)驯化大麦ContigGCA_001077415.1 Hordeum vulgare subsp. vulgare (domesticated barley)驯化大麦ScaffoldGCA_000326125.1 Hordeum vulgare subsp. vulgare (domesticated barley)驯化大麦ScaffoldGCA_000326085.1 Hordeum vulgare subsp. vulgare (domesticated barley)驯化大麦ScaffoldGCA_000830395.1 Humulus lupulus var. cordifolius (European hop)葎草ScaffoldGCA_000831365.1 Humulus lupulus var. lupulus (European hop)葎草ScaffoldGCA_000978395.1 Ipomoea trifida (eudicots) 甘薯近缘野生种Scaffold GCA_000981105.1 Ipomoea trifida (eudicots) 甘薯近缘野生种Scaffold GCA_000208675.1 Jatropha curcas (eudicots) 麻风树Contig GCF_000696525.1 Jatropha curcas (eudicots) 麻风树Scaffold GCA_000208675.2 Jatropha curcas (eudicots) 麻风树Scaffold GCA_001411555.1 Juglans regia (English walnut) 英国胡桃木Scaffold GCA_000227445.1 Lactuca sativa (eudicots) 莴苣ContigGCA_000466325.1 Lagenaria siceraria (white-flowered gourd)葫芦ScaffoldGCA_000411055.1 Leavenworthia alabamica (eudicots) * ScaffoldGCA_000325765.3 Leersia perrieri (monocots) * Chromosom eGCA_000325765.1 Leersia perrieri (monocots) * Contig GCA_000325765.2 Leersia perrieri (monocots) * Scaffold GCA_000224295.1 Linum usitatissimum (flax) 亚麻Contig GCA_000181115.2 Lotus japonicus (eudicots) 百脉根Contig GCA_000181115.1 Lotus japonicus (eudicots) 百脉根ContigGCA_000338175.1 Lupinus angustifolius (narrow-leaved blue lupine)蓝色羽扇豆ScaffoldGCF_000148765.1 Malus domestica (apple) 苹果Chromosom eGCA_000148765.1 Malus domestica (apple) 苹果Chromosom eGCA_000737115.1 Manihot esculenta (cassava) 木薯ScaffoldGCA_001659605.1 Manihot esculenta (cassava) 木薯Chromosom eGCA_000737105.1 Manihot esculenta subsp. flabellifolia (cassava)木薯ScaffoldGCA_001641455.1 Marchantia polymorpha subsp. polymorpha (liverwort)隐花植物地钱ScaffoldGCF_000219495.1 Medicago truncatula (barrel medic) 蒺藜苜蓿Chromosom eGCF_000219495.2 Medicago truncatula (barrel medic) 蒺藜苜蓿Chromosom eGCA_001642375.1 Mentha longifolia (horsemint) 欧薄荷ScaffoldGCA_001662345.1 Metrosideros polymorpha var. glaberrima ('ohi'a lehua)多型铁心木ScaffoldGCF_000414095.1 Morus notabilis (eudicots) 桑树Scaffold GCA_000414095.1 Morus notabilis (eudicots) 桑树ScaffoldGCF_000313855.1 Musa acuminata subsp. malaccensis (wild Malaysian banana)马来西亚野生香蕉ChromosomeGCF_000313855.2 Musa acuminata subsp. malaccensis (wild Malaysian banana)马来西亚野生香蕉ChromosomeGCA_001649415.1 Musa itinerans (monocots) 单子叶植物Scaffold GCF_000365185.1 Nelumbo nucifera (sacred lotus) 莲属椰子Scaffold GCA_000805495.1 Nelumbo nucifera (sacred lotus) 莲属椰子Scaffold GCA_000365185.1 Nelumbo nucifera (sacred lotus) 莲属椰子Scaffold GCA_000723945.1 Nicotiana benthamiana (eudicots) 本氏烟Contig GCA_000715115.1 Nicotiana otophora (eudicots) 烟草Scaffold GCF_000393655.1 Nicotiana sylvestris (wood tobacco) 木烟草ScaffoldGCA_000715095.1 Nicotiana tabacum (common tobacco)普通烟草ScaffoldGCA_000715075.1 Nicotiana tabacum (common tobacco)普通烟草ScaffoldGCF_000715135.1 Nicotiana tabacum (common tobacco)普通烟草ScaffoldGCF_000390325.1 Nicotiana tomentosiformis (eudicots)烟草ScaffoldGCF_000390325.2 Nicotiana tomentosiformis (eudicots)烟草ScaffoldGCA_001278415.1 Ocimum tenuiflorum (holy basil) 圣罗勒Contig GCA_001182835.1 Oropetium thomaeum (monocots) 复活草ContigGCA_000182155.2 Oryza barthii (African wild rice) 短舌野生稻Chromosom eGCA_000182155.3 Oryza barthii (African wild rice) 短舌野生稻Chromosom eGCA_000182155.1 Oryza barthii (African wild rice) 短舌野生稻ContigGCA_000231095.1 Oryza brachyantha (malo sina) 短花药野生稻Chromosom eGCA_000710545.1 Oryza brachyantha (malo sina) 短花药野生稻Chromosom eGCF_000231095.1 Oryza brachyantha (malo sina) 短花药野生稻Chromosom eGCA_000147395.1 Oryza glaberrima (African rice) 非洲栽培稻Scaffold GCA_000147395.2 Oryza glaberrima (African rice) 非洲栽培稻ScaffoldGCA_000576495.1 Oryza glumipatula (monocots) 展颖野生稻Chromosom eGCA_000325645.2 Oryza granulata (monocots) 疣粒稻Scaffold GCA_000325645.1 Oryza granulata (monocots) 疣粒稻ContigGCA_001514335.2 Oryza longistaminata (monocots) 长雄蕊野生稻Chromosom eGCA_000789195.1 Oryza longistaminata (monocots) 长雄蕊野生稻Scaffold GCA_001514335.1 Oryza longistaminata (monocots) 长雄蕊野生稻Scaffold GCA_001551795.1 Oryza meridionalis (monocots) 南方野生稻ContigGCA_000338895.2 Oryza meridionalis (monocots) 南方野生稻Chromosom eGCA_000338895.1 Oryza meridionalis (monocots) 南方野生稻ScaffoldGCA_000632695.1 Oryza minuta (monocots) 异源多倍体水稻(BBCC)ChromosomeGCA_000576065.1 Oryza nivara (monocots) 尼瓦拉野生稻Chromosom eGCA_000710535.1 Oryza nivara (monocots) 尼瓦拉野生稻ChromosomeGCA_000717455.1 Oryza officinalis (monocots) 药用野生稻Chromosom eGCA_000573905.1 Oryza punctata (monocots) 非洲斑点野生稻Chromosom eGCA_000710525.1 Oryza punctata (monocots) 非洲斑点野生稻Chromosom eGCA_000817225.1 Oryza rufipogon (monocots) 普通野生稻Scaffold GCA_001551805.1 Oryza rufipogon (monocots) 普通野生稻ContigGCA_000700045.1 Oryza rufipogon (monocots) 普通野生稻Chromosom eGCA_001648735.1 Oryza sativa (rice) 栽培稻Scaffold GCA_001648745.1 Oryza sativa (rice) 栽培稻ScaffoldGCA_000004655.2 Oryza sativa Indica Group (long-grained rice)栽培稻籼稻ChromosomeGCA_001623365.1 Oryza sativa Indica Group (long-grained rice)栽培稻籼稻ChromosomeGCA_001618795.1 Oryza sativa Indica Group (long-grained rice)栽培稻籼稻ChromosomeGCA_000004655.1 Oryza sativa Indica Group (long-grained rice)栽培稻籼稻ChromosomeGCA_001623345.1 Oryza sativa Indica Group (long-grained rice)栽培稻籼稻ChromosomeGCA_001618785.1 Oryza sativa Indica Group (long-grained rice)栽培稻籼稻ChromosomeGCA_001611235.1 Oryza sativa Indica Group (long-grained rice)栽培稻籼稻ScaffoldGCA_000725085.1 Oryza sativa Indica Group (long-grained rice)栽培稻籼稻ContigGCA_001611195.1 Oryza sativa Indica Group (long-grained rice)栽培稻籼稻ScaffoldGCA_000725085.2 Oryza sativa Indica Group (long-grained rice)栽培稻籼稻ChromosomeGCA_001305255.1 Oryza sativa Indica Group (long-grained rice)栽培稻籼稻ChromosomeGCA_001611255.1 Oryza sativa Indica Group (long-grained rice)栽培稻籼稻ScaffoldGCA_000817615.1 Oryza sativa Japonica Group (Japanese rice)日本晴ChromosomeGCA_000817635.1 Oryza sativa Japonica Group (Japanese rice)日本晴ChromosomeGCA_000321445.1 Oryza sativa Japonica Group (Japanese rice)日本晴ChromosomeGCF_001433935.1 Oryza sativa Japonica Group (Japanese rice)日本晴ChromosomeGCA_000149285.1 Oryza sativa Japonica Group (Japanese rice)日本晴ChromosomeGCA_000164945.1 Oryza sativa Japonica Group (Japanese rice)日本晴ChromosomeGCF_000005425.1 Oryza sativa Japonica Group (Japanese rice)日本晴Complete GenomeGCF_000005425.2 Oryza sativa Japonica Group (Japanese rice)日本晴ChromosomeGCA_000737435.1 Penstemon centranthifolius (eudicots) 大红钓钟柳GCA_000281005.1 Penstemon cyananthus (eudicots) ×GCA_000280985.1 Penstemon davidsonii (eudicots) 钟铃花属的一种蓝紫色小矮花GCA_000280965.1 Penstemon dissectus (eudicots) ×GCA_000281025.1 Penstemon fruticosus (eudicots) ×GCA_000737425.1 Penstemon grinnellii (eudicots) 沙棘GCA_001263595.1 Phalaenopsis equestris (monocots) 小兰屿蝴蝶兰GCF_000499845.1 Phaseolus vulgaris (eudicots) 菜豆GCA_001517995.1 Phaseolus vulgaris (eudicots) 菜豆GCF_000413155.1 Phoenix dactylifera (date palm) 枣椰树GCA_000181215.2 Phoenix dactylifera (date palm) 枣椰树GCA_000181215.1 Phoenix dactylifera (date palm) 枣椰树GCF_000002425.2 Physcomitrella patens (mosses) 小立碗藓GCF_000002425.3 Physcomitrella patens (mosses) 小立碗藓GCA_000411955.3 Picea glauca (white spruce) 白云杉GCA_000411955.5 Picea glauca (white spruce) 白云杉GCA_001687225.1 Picea glauca (white spruce) 白云杉GCA_000411955.4 Picea glauca (white spruce) 白云杉GCA_000411955.1 Picea glauca (white spruce) 白云杉GCA_000966675.1 Picea glauca (white spruce) 白云杉GCA_000411955.2 Picea glauca (white spruce) 白云杉GCA_001447015.2 Pinus lambertiana (sugar pine) 糖松GCA_001447015.1 Pinus lambertiana (sugar pine) 糖松GCA_000404065.1 Pinus taeda (loblolly pine) 厚皮刺果松GCA_000404065.2 Pinus taeda (loblolly pine) 厚皮刺果松GCF_000495115.1 Populus euphratica (Euphrates poplar) 幼发拉底河杨树GCF_000002775.3 Populus trichocarpa (black cottonwood) 黑三角叶杨GCF_000002775.1 Populus trichocarpa (black cottonwood) 黑三角叶杨GCF_000002775.2 Populus trichocarpa (black cottonwood) 黑三角叶杨GCA_000788445.1 Primula veris (cowslip) 黄花九轮草GCA_001077355.1 Primula vulgaris (eudicots) 报春花GCA_001403715.1 Primula vulgaris (eudicots) 报春花GCF_000346735.1 Prunus mume (Japanese apricot) 梅花GCA_000218195.1 Prunus persica (peach) 桃GCA_000218215.1 Prunus persica (peach) 桃GCA_000218175.1 Prunus persica (peach) 桃GCF_000346465.1 Prunus persica (peach) 桃GCA_001517045.1 Pseudotsuga menziesii (Douglas-fir) 花旗松GCF_000315295.1 Pyrus x bretschneideri (Chinese white pear) 中国白梨GCA_001633185.1 Quercus lobata (valley oak) 谷橡木GCA_000769845.1 Raphanus raphanistrum subsp. raphanistrum(eudicots)野萝卜GCA_000801105.2 Raphanus sativus (radish) 萝卜GCA_000801105.1 Raphanus sativus (radish) 萝卜GCA_000715565.1 Raphanus sativus (radish) 萝卜GCA_001047155.1 Raphanus sativus (radish) 萝卜GCF_000151685.1 Ricinus communis (castor bean) 蓖麻GCA_000151685.1 Ricinus communis (castor bean) 蓖麻GCA_001662545.1 Rosa x damascena (damask rose) 玫瑰GCA_000218505.1 Schrenkiella parvula (eudicots) ×GCA_001021135.1 Selaginella kraussiana (club-mosses) 卷柏GCF_000143415.2 Selaginella moellendorffii (club-mosses) 石松类GCF_000143415.1 Selaginella moellendorffii (club-mosses) 石松类GCF_000143415.3 Selaginella moellendorffii (club-mosses) 石松类GCF_000512975.1 Sesamum indicum (sesame) 芝麻GCA_001692995.1 Sesamum indicum (sesame) 芝麻GCA_000975565.1 Sesamum indicum (sesame) 芝麻GCF_000263155.2 Setaria italica (foxtail millet) 谷子GCF_000263155.1 Setaria italica (foxtail millet) 谷子GCA_001652605.1 Setaria italica (foxtail millet) 谷子GCA_001412135.1 Silene latifolia subsp. alba (eudicots) 白花蝇子草GCA_001541825.1 Silybum marianum (eudicots) 水飞蓟GCA_000411075.1 Sisymbrium irio (eudicots) 水蒜芥GCA_000612985.1 Solanum arcanum (eudicots) 野生番茄GCA_001239805.1 Solanum commersonii (Commerson's wildpotato)野生土豆GCA_000577655.1 Solanum habrochaites (eudicots) 多毛番茄GCF_000188115.3 Solanum lycopersicum (tomato) 番茄GCA_000181095.1 Solanum lycopersicum (tomato) 番茄GCA_000325825.1 Solanum lycopersicum (tomato) 番茄GCF_000188115.1 Solanum lycopersicum (tomato) 番茄GCF_000188115.2 Solanum lycopersicum (tomato) 番茄GCA_900008105.1 Solanum lycopersicum (tomato) 番茄GCA_000787875.1 Solanum melongena (eggplant) 茄子GCA_000820945.1 Solanum pennellii (eudicots) 野生西红柿GCA_000577875.1 Solanum pennellii (eudicots) 野生西红柿GCF_001406875.1 Solanum pennellii (eudicots) 野生西红柿GCA_000230315.1 Solanum pimpinellifolium (currant tomato) 醋栗番茄GCA_900004685.1 Solanum tuberosum (potato) 土豆GCF_000226075.1 Solanum tuberosum (potato) 土豆GCA_000003195.2 Sorghum bicolor (sorghum) 高粱GCF_000003195.2 Sorghum bicolor (sorghum) 高粱GCF_000003195.1 Sorghum bicolor (sorghum) 高粱GCA_000236745.2 Sorghum bicolor (sorghum) 高粱GCA_000236765.2 Sorghum bicolor (sorghum) 高粱GCA_000236725.1 Sorghum bicolor (sorghum) 高粱GCA_000236725.2 Sorghum bicolor (sorghum) 高粱GCA_000236745.1 Sorghum bicolor (sorghum) 高粱GCA_000236765.1 Sorghum bicolor (sorghum) 高粱GCA_000510995.2 Spinacia oleracea (spinach) 菠菜GCA_000510995.1 Spinacia oleracea (spinach) 菠菜GCA_000504445.1 Spirodela polyrhiza (great duckweed) 浮萍GCF_000463585.1 Tarenaya hassleriana (spider flower) 醉蝶花GCF_000208745.1 Theobroma cacao (cacao) 可可GCA_000208745.1 Theobroma cacao (cacao) 可可GCF_000403535.1 Theobroma cacao (cacao) 可可GCA_000956625.1 Thlaspi arvense (eudicots) 菥蓂GCA_000583005.2 Trifolium pratense (eudicots) 红花苜蓿GCA_000583005.1 Trifolium pratense (eudicots) 红花苜蓿GCA_900079335.1 Trifolium pratense (eudicots) 红花苜蓿GCA_000210335.1 Triticum aestivum (bread wheat) 面包小麦GCA_000334135.1 Triticum aestivum (bread wheat) 面包小麦GCA_001077335.1 Triticum aestivum (bread wheat) 面包小麦GCA_000334095.1 Triticum aestivum (bread wheat) 面包小麦GCA_000188135.1 Triticum aestivum (bread wheat) 面包小麦GCA_000818885.1 Triticum aestivum (bread wheat) 面包小麦GCA_001485685.1 Triticum aestivum (bread wheat) 面包小麦GCA_000347455.1 Triticum urartu (monocots) 乌拉尔图小麦GCA_000775335.1 V accinium macrocarpon (eudicots) 大果越橘GCA_001375635.1 Vicia faba (fava bean) 蚕豆GCF_001190045.1 Vigna angularis (adzuki bean) 红豆GCA_000465365.1 Vigna angularis var. angularis (adzuki bean) 红豆GCA_001723775.1 Vigna angularis var. angularis (adzuki bean) 红豆GCA_000180895.1 Vigna radiata (mung bean) 绿豆GCA_001584445.1 Vigna radiata var. radiata (mung bean) 绿豆GCA_000741045.1 Vigna radiata var. radiata (mung bean) 绿豆GCF_000741045.1 Vigna radiata var. radiata (mung bean) 绿豆GCA_001687525.1 Vigna unguiculata subsp. unguiculata (cowpea)豇豆GCA_001562795.1 Vitis aestivalis (eudicots) 葡萄-GCA_001282645.1 Vitis cinerea x Vitis riparia (eudicots) 葡萄GCF_000003745.2 Vitis vinifera (wine grape) 葡萄GCF_000003745.3 Vitis vinifera (wine grape) 葡萄GCF_000003745.1 Vitis vinifera (wine grape) 葡萄GCA_000005005.2 Zea mays (maize) 玉米GCA_000223545.1 Zea mays (maize) 玉米GCA_000005005.3 Zea mays (maize) 玉米GCF_000005005.1 Zea mays (maize) 玉米GCA_000005005.1 Zea mays (maize) 玉米GCA_000275765.1 Zea mays (maize) 玉米GCA_000005005.4 Zea mays (maize) 玉米GCA_001644905.1 Zea mays subsp. mays (maize) 玉米GCA_000418225.1 Zizania latifolia (monocots) 野生种“菰GCF_000826755.1 Ziziphus jujuba (eudicots) 枣GCA_001185155.1 Zostera marina (monocots) 鳗草GCA_001602275.1 Zoysia japonica (monocots) 结缕草GCA_001602295.1 Zoysia matrella (Manila grass) 细叶结缕草GCA_001602315.1 Zoysia pacifica (monocots) 太平洋结缕草。
基因组学课件基因组

大肠杆菌
智人
拟南芥
热海栖热袍菌
Buchnerasp. APS 嗜酸热原体
Escherichia coli 家鼠
Homo sapiens 秀丽小杆线虫
Arabidopsis thaliana Thermotoga maritima
大白鼠
疏螺旋体-眼莱姆病
Thermoplasma acidophilum Mus musculus
大肠杆菌(Escherichia coli)
人类研究得最为详尽的模式生物 如:K12菌株,全基因组于1997年测定,长460万bp 长度1.6 m,单细胞原核生物,繁殖快
大肠杆菌及其全基因组
Escherichia coli K12
Escherichia coli O157:H7
基因组学课件基因组
模式生物(Model Organism) 酿酒酵母(Saccharomyces cerevisiae, yeast)
Nature390:580,1997
Welcome Trust资助英、法科学家于 Sanger中心完成,1997, 12宣布 Science281,375,1998
Science 282:2012,1998
Science291,1304,2001 Nature408:796,2000
Science287,2185,2000
基因组大小与人类相近,约30亿个核苷酸对,有19条染色体 2002年
基因组学课件基因组
第1章 什么是基因组
所有生命都具有指令其生长与发育,维持 其结构与功能所必需的遗传信息,生物所 具有的携带遗传信息的遗传物质总和称为 基因组(genome) 基因组(genome)一词出现于80年前,基 因 组 学 (genomics) 则 是 由 美 国 科 学 家 Thomas Roderick在1986年提出的,是指对 所有基因进行基因组作图 (包括遗传图谱、 物理图谱、转录本图谱) ,核苷酸序列分 析,基因定位和基因功能分析的一门科学
已经测序的动物

序号中文名发表时间刊物IF 基因组大小1秀丽隐杆线虫1998.12Science29.74797Mb 2果蝇2000.03Science 29.747120Mb 3冈比亚按蚊2002.1Science 29.747280Mb 4小家鼠2002.12Nature 34.48 2.5 Gb 5红鳍东方豚2002.12Science29.747380Mb 6玻璃海鞘2002.12Science 29.747150Mb 7线虫2003.11PLoSBiology13.501104Mb 8褐家鼠2004.04Nature 34.48 2.75 Gb 9黑斑鲀2004.1Nature 34.48340Mb 10家蚕2004.12Science 29.747428.7 Mb 11鸡2004.12Nature 34.48 1.06Gb 12克氏锥虫2005.07Science 29.74767Mb 13黑猩猩2005.09Nature 34.48 2.7Gb 14狗2005.12Nature 34.48 2.5 Gb 15意蜂2006.1Nature 34.48236Mb 16海胆2006.11Science 29.7471Gb 17姥鲨2007.04PlosBiology13.5010.91Gb 18猕猴2007.04Science 29.747 2.87Gb 19短尾负鼠2007.05Nature 34.48 3.4Gb 20青鳉鱼2007.06Nature 34.48700Mb 21伊蚊2007.06Science 29.7471376Mb 22马来线虫2007.07Science 29.74790Mb 23海葵2007.07Science 29.747357Mb 24猫2007.11GenomeResearch11.2 2.7Gb 25赤拟谷盗2008.04Nature 34.48204Mb 26鸭嘴兽2008.05Nature 34.48 1.84Gb 27文昌鱼2008.06Nature 34.48520Mb 28丝盘虫2008.08Nature 34.48104Mb 29南方根瘤线虫2008.08NatureBiotechno29.49586Mb 30古猛犸象2008.11Nature 34.48 4.7Gb 31牛2009.04Science 29.747 2.87 Gb 32曼氏血吸虫2009.07Nature34.48360Mb 33日本血吸虫2009.07Nature34.48397Mb 34马2009.11Science 29.747 2.7Gb 35熊猫2010.01Nature 34.48 2.25Gb 36金小蜂2010.01Science 29.747295Mb 37豌豆蚜虫2010.02PLoSBiol.13.501517Mb 38水螅2010.03Nature 34.48 1.05Gb 39非洲爪蟾2010.04Science 29.747 1.7 Gb 40珍珠鸟2010.04Nature 34.48 1.2Gb 41人类体虱2010.07PNAS 9.432110Mb 42海绵2010.08Nature 34.48190Mb 43蚂蚁2010.08Science 29.747 C.floridanus2444火鸡2010.09PLoSBiology13.501 1.1 Gbb 45库蚊2010.1Nature 34.48540Mb 46异体住囊虫2010.11Science29.747148Mb 47红毛猩猩2011.01Nature 34.48 3.09Gb 48阿根廷蚁2011.01PNAS 9.432250.8Mb 49红色收割蚁2011.01PNAS9.432250–284 Mb 50火蚁2011.01PNAS 9.432484.2Mb 动物基因组51水蚤2011.02Science 29.747200Mb 52大头切叶蚁2011.02PLoS Genetics 9.532300Mb 53旋毛虫2011.02Nature Genetics 34.28464Mb 54袋獾2011.06PNAS 9.432 3.3Gb 55顶切叶蚁2011.06Genome Research 11.2313Mb 56鹿角珊瑚2011.07Nature 34.48420Mb 57缅甸蟒2011.07Genome Biology 6.89 1.4Gb 58大西洋鳕鱼2011.08Nature 34.48830Mb 59澳大利亚袋鼠2011.08Genome Biology 6.89 2.9Gb 60绿蜥蜴2011.09Nature 34.48 1.78Gb 61裸鼹鼠2011.1Nature 34.48 2.6Gb 62食蟹猴和中国2011.1Nature Biotechno 29.495 2.85Gb 63猪蛔虫2011.1Nature 34.48272Mb 64二斑叶螨(棉红2011.11Nature 34.4890Mb 65帝王蝶2011.11Cell 31.152273Mb 66指猴(夜狐猴)2011.12Genome Biology 2.6743Gb 67埃及血吸虫2012.01Nature Genetics 34.284385Mb 68鳄鱼2012.01Genome Biology 6.89 2.5Gb 69大猩猩2012.03Nature 34.48 3.04Gb 70马氏珠母贝2012.04DNAresea rch 3.525 1.15Gb 71诗神袖蝶2012.05Nature 34.48269Mb 72倭黑猩猩2012.06Nature 34.483Gb 73牦牛2012.07Nature genetics 34.48 2.6Gb 74牡蛎2012.09Nature 34.48559Mb 75双峰骆驼2012.11Nature Communi 7.396 2.38Gb 76猪2012.11Nature 34.48 2.6Gb 77中央狐蝠2012.12Science 31.2012Gb 78大卫鼠耳蝠2012.12Science 31.2012Gb 79山羊2012.12Nature Biotechno 29.425 2.66Gb 80海洋蠕虫2012.12Nature 34.48324Mb 81淡水水蛭2012.12Nature 34.48228Mb 82青螺2012.12Nature 34.48348Mb 83小菜蛾2013.1Nature Genetics 34.284339.4Mb 84家鸽2013.1Science 31.201 1.3Gb85中国树鼩2013.2Nature Communi 7.396 3.2Gb(2.86G bassembled)86海七鳃鳗2013.2Nature Genetics 34.284816Mb 87非洲眼丝虫2013.3Nature Genetics 34.28491.4Mb 88游隼/猎隼2013.3Nature Genetics 34.284 1.2Gb 89西部锦龟2013.3Genome Biology 9.04 2.59Gb 90绦虫2013.3Nature 34.48115Mb-141Mb 91腔棘鱼2013.4Nature 34.48 2.18Gb 92剑尾鱼2013.3.31Nature Genetics 34.284669Mb 93斑马鱼41381Nature 34.48 1.412Gb 94中华鳖和绿海2013.4.28Nature Genetics 34.284 2.21Gb/2.24Gb 95藏羚羊2013.5Nature Communi 7.396 2.75Gb 96鸭2013.6.09Nature Genetics 34.284 1.1GB 97蚊子2013.6.12Nucleic Acids 8.278201Mb 98地山雀2013.7.1Nature Communi 10.015 1.086Gb 99黑线仓鼠2013.7.21Nature Biotechno 32.438 2.7G 100扬子鳄2013.8.06Cell research 10.526 2.3G 101蛭形轮虫2013.8.22Nature 38.597244Mb主要研究单位The C. elegans Sequencing Consortium Celera Genomics等美国塞雷拉基因组公司等Mouse Genome Sequencing Consortium 新加坡分子与细胞生物学研究院等美国能源部JGI等美国冷泉港等美国得克萨斯州贝勒医学院等法国、西班牙、美国西南大学等中国华大基因等美国罗克维尔市基因组研究院等美国哈佛-麻省理工的博德研究所等美国哈佛-麻省理工的博德研究所等人类基因组测序中心等人类基因组测序中心等新加坡分子与细胞生物学研究院等人类基因组测序中心等美国哈佛-麻省理工的博德研究所等日本东京大学等美国MCD基因组研究院等美国匹兹堡大学等美国JGI能源部等马里兰州国立癌症研究所等人类基因组测序中心等美国华盛顿大学等美国JGI能源中心等美国加利福尼亚大学等I法国国家农业研究所等美国宾夕法尼亚州立大学等人类基因组测序中心等Sanger研究院等中国国家人类基因组中心等美国博德研究所等深圳华大基因研究院美国罗切斯特大学等国际蚜虫基因组协会美国JGI能源部等美国JGI能源部等美国华盛顿大学等美国伊利诺斯大学等美国加利福尼亚大学等纽约大学等美国动物家禽科学部等美国加州大学河滨分校等法国CEA等美国华盛顿大学基因组中心等美国旧金山大学等美国里士满厄尔汉学院等瑞士洛桑大学等美国印第安纳大学等美国威斯康星-麦迪逊大学等美国华盛顿大学医学院等澳大利亚罗伊癌症研究中心等哥本哈根大学等日本冲绳县科学与技术研究所等科罗拉多大学医学院等挪威奥斯陆大学等澳大利亚袋鼠基因组研究美国哈佛-麻省理工的博德研究所等韩国梨花女子大学等中山大学、BGI等BGI,墨尔本大学等西安大略大学等马萨诸塞大学医学院、等芝加哥大学、维尔康奈尔医学院BGI、墨尔本大学等加利福尼亚大学、佛罗里达大学等英国韦尔科姆基金会桑格研究所等日本冲绳县科学与技术研究所等伦敦大学学院、剑桥大学等德国马克斯?普朗克人类进化研究所等华大基因中国科学院海洋所等内蒙古大学等国际家猪基因组测序协作组深圳华大基因研究院等深圳华大基因研究院等昆明动物所等莱斯大学等莱斯大学等莱斯大学等福建农林大学等犹他大学等中科院昆明动物所等美国海洋生物学实验室等美国国立卫生研究院等英国卡迪夫大学等美国加州大学等Sanger研究院等美国Benaroya研究所等华盛顿大学医学院等英国桑格研究院深圳华大基因研究院青海大学等中国农业大学等美国加州大学尔湾分校等中科院动物研究所、诺禾致源深圳华大基因研究院浙江大学等美国那慕尔大学等。
常见物种基因组
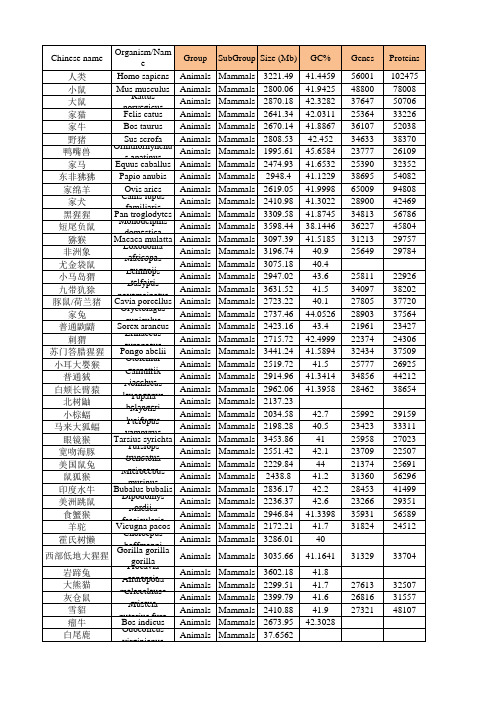
GC% 41.4459 41.9425 42.3282 42.0311 41.8867 42.452 45.6584 41.6532 41.1229 41.9998 41.3022 41.8745 38.1446 41.5185 40.9 40.4 43.6 41.5 40.1 44.0526 43.4 42.4999 41.5894 41.5 41.3414 41.3958 42.7 40.5 41 42.1 44 41.2 42.2 42.6 41.3398 41.7 40 41.1641 41.8 41.7 41.6 41.9 42.3028 -
Group Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals Animals
玻利维亚的灰鼠 猴子 裸鼢鼠 南极须鲸 长鼻猴 金丝猴 美洲野牛 长尾龙猫 威德尔海豹 中华穿山甲 倭黑猩猩 山羊 藏羚羊 非洲跳鼠 东北虎 单峰驼 双峰驼 草原田鼠 星鼻鼹鼠 马铁菊头蝠 大棕蝠 土豚 金毛鼹 八齿鼠 指猴 野骆驼 金仓鼠 缅树鼩 抹香鲸 豚尾猴 白眉猴 太平洋海象 虎鲸 猫头鹰猴 鼠耳蝠 白暨豚 狍 北极熊 布氏鼠耳蝠 野牦牛 黄毛果蝠 印度假吸血蝠 佛罗里达海牛 白犀牛 蒙古野马 天竺鼠 野山羊 旱獭
医学分子生物学基因组
操纵子*
原核基因组特有的结构 数个功能上相关联的结构基因串联在一起,构成信息区,连同其上游的调控区(包括启动子和操纵基因)以及下游的转录终止信号所构成的基因表达单位。 所转录的RNA为多顺反子。 操纵基因并非一个基因,而是一段能被特异阻遏蛋白识别和结合的DNA序列。
Transposon:转座酶+反向重复序列+特定编码基因
Transposable phage: 具备转座功能的溶源噬菌体 Mu噬菌体:温和致突变噬菌体
Ap Kan Tet Hg
细菌转位因子
01
真核生物基因组
模式生物:酵母、线虫、果蝇、小鼠、大 鼠
线状:腺病毒科 、 疱疹病毒科、 痘病毒科、 彩虹病毒科
部分双链:嗜肝DNA病毒 DNA病毒 RNA过程 HBV 基因结构
单击此处可添加副标题
单链RNA 单正链RNA:病毒RNA起类似细胞mRNA作用 帽状结构, PolyA尾, 直接翻译蛋白质 披膜病毒科(Togaviridae): 风疹病毒 黄病毒科(Flaviviridae): 登革病毒 小RNA病毒科(Picornaviridae): 脊髓灰质炎病毒 冠状病毒科(Coronaviridae):人呼吸道冠状病毒 单负链RNA:病毒RNA互补链起类似细胞mRNA作用 正粘病毒科(Orthomyxoviridae):流感病毒(8段) 副粘病毒科(paramyxoviridae): 麻疹病毒、呼吸合胞病毒 弹状病毒(Rhabdoviridae): 狂犬病毒
病毒RNA
01
双链RNA:分节段,分别编码各自蛋白 呼肠病毒科(Reoviridae): 轮状病毒(10段) 逆转录病毒NA:自身配对形成,HDV
单击此处添加小标题
第3章-真核生物基因组资料

主要是由于患者体内少了一条X染色体(45,X)。
第三十一页
二、基因结构与疾病
基因组结构:是指基因组DNA中不同的功能片段 在整个基因组中的分布。
基因组结构的改变不一定导致基因结构的改 变,基因结构的改变也不一定导致基因功能的异 常。只有当缺失、倒位、易位、插入等引起基因 突变,而且这种突变又改变了基因的编码序列或 影响了基因的调控序列时,基因的功能才发生异 常。
内含子与外显子的概念也是相对的.
●间隔区DNA:
真核生物基因之间存在编码空白区或转录的空白区,称之为间
隔区DNA(spacer DNA),这些序列往往在单拷贝的结构基因侧翼,并使
结构基因彼此分开,间隔区DNA也可以存在于rDNA区。
第八页
第九页
四、重复序列
根据复性动力学将真核生物基因组DNA的重复 序列分为三类: (一)高度重复序列:重复频率达百万(106)以上。
可以形成回文结构,既能连接非同源的基因,又可以被参与转位 的特异酶所识别;
④与进化有关:具有种属特异性,但相近种属又有相似性; ⑤与个体特征有关:同一种属中不同个体的高度重复序列的重复次 数不一样,这可以作为每个个体的特征,即DNA指纹; ⑥与染色体减数分裂时染色体配对有关:α卫星DNA成簇分布在染 色体着丝粒附近,同源染色体之间的联会可能依赖于具有染 色体专一性的特定卫星DNA序列。
胞,少于46,称为亚二倍体;多于46,称为超二倍 体;67可称为亚三倍体等。
3.三体性和单体性 体细胞的某号染色体增多一条,称为三体性
(trisomy);
体细胞的某号染色体减少一条,称为单体性 (monosomy)。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
Cajanus cajan
蒺藜苜蓿
Medicago truncatula
蓝载藻(灰胞藻门) 谷子 谷子 番茄 甜瓜 亚麻 盐芥 香蕉 雷蒙德氏棉
Cyanophora paradoxa Setaria italica Setaria italica Solanum lycopersicum Cucumis melo L. Linum usitatissimum Thellungiella salsuginea Musa acuminata Gossypium raimondii
2n=42 2n=38 2n=32
2n=42
2n=48 2n=6 2n=10
海葵
12种果蝇基因组进化 分析(10种新)
猫
Nematostella vectensis D.sechellia, D.simulans , D.yakuba , D.erecta , D.ananassae , D.persimilis , D.willistoni , D.mojavensis , D.virilis and D.grimshawi
Fragaria vesca
Theobroma cacao G. soja
发表时间 刊物
科、属
基因组大小
2000.12
2002.04 2002.04 2006.09 2007.09 2007.01 2008.01
2008.04 2008.11 2009.01 2009.11 2009.11 2010.01 2010.02 2010.06 2010.07 2010.08 2010.09 2010.09
70M 4预9估0M510M,组 装出400M 900Mb 450Mb 373Mb 260Mb 523Mb 775.2Mb
2n=18 2n=18 2n=24 2n = 24 2n = 30 2n = 14 2n = 22 2n = 26
动物基因组(81) 中文名
拉丁名
发表时间 刊物
科、属
基因组大小
染色体 数量
2010.12
2010.12 2010.12
十字花科
Nature
(Brassicaceae) 鼠耳芥属
125M
Science Science Science Nature Science Science
(Arabidopsis)
禾本科Poaceae 稻 禾属 本O科rPyzoaaceae 稻属Oryza
血吸虫属
360M
血吸虫
马 熊猫 金小蜂 豌豆蚜虫 水螅 非洲爪蟾
珍珠鸟 人类体虱 海绵
Schistosoma japonicum
2009.07 Nature
Equus caballus
2009.11
Ailuropoda melanoleura
2010.01
Nasonia vitripennis, N. giraulti, N. longicornis
Rattus norvegicus
Tetraodon nigroviridis Bombyx mori Gallus sonneratii
Drosophila pseudoobscura
Trypanosoma cruzi Pan troglodytes Canis familiaris Apis mellifera Strongylocentrotus purpuratus Callorhinchus milii Macaca mulatta Monodelphis domestica Oryzias latipes Aedes aegypti Brugia malayi
裂吸虫
Schistosoma mansoni
2007.07 Science
海葵目
357M
2007.11 Nature
果蝇科、果蝇 属
2007.11
2008.04 2008.05 2008.06 2008.08 2008.08 2008.11 2009.04
Genome Research
猫科、猫属 2.7G 鞘翅目、拟步
Nature
行虫科、拟谷 204M
Nature
盗 鸭属 嘴兽科、鸭
嘴兽属
1.84G
Nature
文昌鱼科、文
昌鱼属
520M
丝盘虫科、丝
Nature
盘虫属
104M
Nature
线虫属
86M
Biotechnology Nature
象科、猛犸象 属
4.7G
Science
牛科、牛属 2.87 G
2009.07 Nature
鼠科
2.5 G
鲀科、东方鲀 属
380M
海鞘纲
150M
PLoS Biology
Caenorhabditis 亚属
104M
Nature
啮齿目、鼠科 2.75 G
2004.10 2004.12 2004.12 2005.01 2005.07
Nature Science Nature Genome research Science
属 葫芦科、黄瓜 2300M
属
350M
豆科、大豆属 1100M
Nature
禾本科、短柄
草属
260M
Nature
水云属
196M
Science
团藻属
138M
Nature
大戟科、蓖麻
Biotechnology 属
350M
Plant Cell 小球藻科
46M
Nature Genetics
蔷薇科、苹果
属
742M
2005.09 2005.12 2006.10
Nature Nature Nature
鲀科 鳞翅目、蚕蛾 科 雉科、原鸡属
340M 428.7 M 1.06G
果蝇科、果蝇
属
139M
锥虫属
67M
猩猩科、黑猩
猩属
2.7G
犬科、犬属 2.5 G
昆虫纲
236M
2006.11
2007.04
2007.04
2007.05 2007.06 2007.06 2007.07
2n=16 2n=4X=4 8 2n=14 2n=20 2n = 20
2n = 22
2011.11 Nature
豆科苜蓿属 500M
2n=16
2012.02 2012.05 2012.05 2012.05 2012.07 2012.07 2012.07 2012.07 2012.08
Science Nature
2011.02 2010.12 2011.05 2011.05
2011.05
2011.07 2011.08 2011.08 2011.10
2011.11
PNAS
57M
DNA Res.
大戟科、麻风
树属
410M
Science
卷柏属
212M
Nature棕榈科ຫໍສະໝຸດ biotechnology
685M
Nature Genetics
1998.12
2000.03
2002.10 2002.12 2002.12 2002.12 2003.11
2004.04
Science
小杆总科、 Caenorhabditis 97M 亚属
Science
果蝇科、果蝇 属
180M
Science
蚊科、按蚊属 280M
Nature Science Science
Felis catus
赤拟谷盗
Tribolium castaneum
鸭嘴兽
Ornithorhynchus anatinus
文昌鱼
Branchiostoma floridae
丝盘虫
根结线虫 古猛犸象 牛
Trichoplax adhaerens Meloidogyne incognita Mammuthus primigenius Bos taurus
灰胞藻门 禾本科、狗尾
bNiaottuercehnology 草 禾属 本科、狗尾
biotechnology 草属
Nature
茄科、茄属
葫芦科、甜瓜
PNAS Plant Journal PNAS
属 亚麻科、亚麻 属 十字花科、盐 芥 芭属 蕉科、芭蕉
Nature Nature
属 锦葵科、棉属
Genetics
2010.04
2010.07 2010.08
Nature
PNAS Nature
血吸虫属
397M
马科、马属 熊科、大熊猫 属 膜翅目、金小 蜂科
2.7G 2.25G
295M
水螅科、水螅 属 负子蟾科、非 洲爪蟾属
517M 1.05G
1.7 G
文鸟科、梅花 雀属
1.2G
人虱属
110M
多空动物门 190M
Nature Genetics
蔷薇科、草莓
属
240M
Nature Genetics
梧桐科、可可
属
430-Mb
PNAS
豆科、大豆属 915.4 Mb
倍性、 染色体
2n=10
2n=24 2n=24 2n=38 2n=38
2n=18
2n=20 2n=20 2n=14 2n=40 2n=10 36
2n=20 2n=24 2n=34 2n=14 2n=20 2n=40
Science
