pHis2酵母表达载体说明
酵母双杂交原理与实验具体流程.

图2romoters and other cis-acting regulatory elements play a crucial role in yeast-based expression systems and transcriptional assays such as the MATCHMAKER One- and Two-Hybrid Systems. Differences in the promoter region of reporter gene constructs can significantly affect their ability to respond to the DNA-binding domain of specific transcriptional activators; promoter constructs also affect the level of background (or leakiness) of gene expression and the level of induced expression. Furthermore, differences in cloning vector promoters determine the level of protein expression and, in some cases, confer the ability to be regulated by a nutrient (such as g al actose in the case of the G AL 1p romoter) .
酵母双杂交系统原理及具体 操作流程
酵母双杂交系统可进行两个蛋白互作分析,可用
一个已知的蛋白因子(在双杂交系统中称为诱饵蛋白)
白桦E-box元件的载体构建及与BplMYB46转录因子的互作分析

白桦E-box元件的载体构建及与BplMYB46转录因子的互作分析王博;王莲萍;杨春雨;国会艳;魏继承【摘要】MYB转录因子是植物中最大的转录因子家族成员之一,主要参与植物次生代谢调控、激素和环境因子的应答,在植物的生长发育中起着至关重要的作用.已有研究发现,E-box的核心序列为CANNTG(N :A/G/C/T),是一类与光响应和苯丙氨酸生物合成途径相关的元件.在前期研究中,先构建顺式作用元件库,然后以转录因子为中心的酵母单杂交技术筛选发现,白桦的BplMYB46转录因子能够与核心序列为CAAATG 的E-box顺式作用元件结合.但是,是否能与E-box的其他核心序列结合还不清楚.本研究将每一种E-box顺式作用元件的核心序列分别进行双链DNA的复性并连接到pHIS2载体上,然后通过酵母单杂交技术筛选与BplMYB46转录因子能够特异结合的E-box顺式作用元件.结果显示,当E-box的核心序列第3个碱基为A/T/C、第四个碱基为A/G/C时,酵母单菌落能在三缺培养基TDO/3AT上生长,表明与BplMYB46转录因子结合的E-box顺式作用元件的特异性序列为CA(A/T/C)(A/G/C)TG.为后续通过BplMYB46转录因子与E-box顺式作用元件的结合来分析BplMYB46转录因子对下游基因的调控,以及为筛选优良下游基因改良白桦的遗传性状奠定数据基础.【期刊名称】《生物技术通报》【年(卷),期】2018(034)012【总页数】6页(P110-115)【关键词】BplMYB46转录因子;E-box顺式作用元件;酵母单杂交技术【作者】王博;王莲萍;杨春雨;国会艳;魏继承【作者单位】牡丹江师范学院生命科学与技术学院,牡丹江 157011;牡丹江师范学院生命科学与技术学院,牡丹江 157011;牡丹江师范学院生命科学与技术学院,牡丹江 157011;牡丹江师范学院生命科学与技术学院,牡丹江 157011;牡丹江师范学院生命科学与技术学院,牡丹江 157011【正文语种】中文真核生物的基因表达与调控是一系列顺式作用元件(Cis-acting element)与反式作用因子(Transacting factor)相互作用的结果。
酵母表达载体pPICZ手册

pPICZ A, B, and CPichia expression vectors for selection onZeocin™ and purification of recombinant proteins Catalog no. V190-20Rev. Date: 7 July 2010Manual part no. 25-0148MAN00000034User ManualiiTable of ContentsKit Contents and Storage (iv)Accessory Products (v)Introduction (1)Overview (1)Methods (3)Cloning into pPICZ A, B, and C (3)Pichia Transformation (9)Expression in Pichia (13)Purification (15)Appendix (17)Recipes (17)Zeocin™ (19)Map and Features of pPICZ A, B, and C (21)Lithium Chloride Transformation Method (23)Construction of In Vitro Multimers (24)Technical Support (32)Purchaser Notification (33)References (34)iiiKit Contents and StorageContents The following components are included with Catalog no. V190–20. Note that thepPICZ expression vectors are supplied in suspension.Component QuantityCompositionpPICZ A Expression Vector 20 μg 40 μl of 0.5 μg/μl vector in10 mM Tris–HCl, 1 mM EDTA,pH 8.0pPICZ B Expression Vector 20 μg 40 μl of 0.5 μg/μl vector in10 mM Tris–HCl, 1 mM EDTA,pH 8.0pPICZ C Expression Vector 20 μg 40 μl of 0.5 μg/μl vector in10 mM Tris–HCl, 1 mM EDTA,pH 8.0GS115/pPICZ/lacZ Positive1 stab --Control strainShipping/Storage The components included with Catalog no. V190–20 are shipped on wet ice.Upon receipt, store as directed below.For long-term storage of your positive control stab strain, we recommendpreparing a glycerol stock immediately upon receipt and storing at –80°C.Component ShippingStorage pPICZ A Expression Vector Wet ice Store at –20°CpPICZ B Expression Vector Wet ice Store at –20°CpPICZ C Expression Vector Wet ice Store at –20°CGS115/pPICZ/lacZ positive control strain Wet ice Store at 4°CivAccessory ProductsAdditional ProductsThe products listed in this section are intended for use with the pPICZ vectors.For more information, visit our web site at or contactTechnical Support (page 32).Product QuantityCatalogno. X-33 Pichia strain 1 stab C180-00GS115 Pichia strain 1 stab C181-00KM71H Pichia strain 1 stab C182-00SMD1168H Pichia strain 1 stab C184-00pPICZα A, B, and C 20 μg each V195-20pPIC6α A,B, and C 20 μg each V215-20pPIC6 A, B, and C 20 μg each V210-20pPIC6 Starter Kit 1 kit K210-01Original Pichia Expression Kit 1 kit K1710-01EasySelect™Pichia Expression Kit 1 kit K1740-01Pichia EasyComp™ Transformation Kit 1 kit K1730-01Pichia Protocols 1 book G100-01PureLink™ Gel Extraction Kit 50 preps250 prepsK2100–12K2100–25S.N.A.P ™ Gel Purification Kit 25 preps K1999–25PureLink™ Quick Plasmid Miniprep Kit 50 preps250 prepsK2100–10K2100–11PureLink™ HiPure Plasmid Midiprep Kit 25 preps50 prepsK2100–04K2100–13One Shot® TOP10 (chemically competent E. coli) 10 reactions20 reactionsC4040–10C4040–03One Shot® TOP10 Electrocompetent E. Coli 10 reactions20 reactionsC4040-50C4040-52TOP10 Electrocomp™ Kits 20 reactions C664–55Positope™ Control Protein 5 μg R900-50CIAP (Calf Intestinal Alkaline Phosphatase) 1,000 units 18009–019T4 DNA Ligase 100 units500 units15224–01715224–025Zeocin™ 1g5 gR250-01R250-05β-Gal Assay Kit 1 kit K1455-01β-Gal Staining Kit 1 kit K1465-01E-Gel® Agarose Gels E-Gel® Agarose Gels are bufferless, pre-cast agarose gels designed for fast, convenient electrophoresis of DNA samples. E-Gel® agarose gels are available in different agarose percentage and well format for your convenience.For more details on these products, visit our web site at or contact Technical Support (page 32).Continued on next pagevAccessory Products, ContinuedZeocin™Zeocin™ may be obtained from Invitrogen (see above). For your convenience, the drug is prepared in autoclaved, deionized water and available in 1.25 ml aliquotsat a concentration of 100 mg/ml. The stability of Zeocin™ is guaranteed for sixmonths if stored at –20°C.Detection of Fusion Protein A number of antibodies are available from Invitrogen to detect expression ofyour fusion protein from the pPICZ vector. Horseradish peroxidase (HRP)-conjugated antibodies allow one-step detection in Western blots usingcolorimetric or chemiluminescent detection methods. The amount of antibodysupplied is sufficient for 25 Western Blots.Antibody Epitope Catalogno.Anti-myc R950–25 Anti-myc-HRPDetects the 10 amino acid epitopederived from c-myc (Evans et al., 1985):EQKLISEEDLR951–25Anti-His(C-term) R930–25Anti-His(C-term)-HRPDetects the C-terminal polyhistidine(6xHis) tag (requires the free carboxylgroup for detection) (Lindner et al., 1997):HHHHHH-COOHR931–25Purification of Fusion Protein The polyhistidine (6xHis) tag allows purification of the recombinant fusionprotein using metal-chelating resins such as ProBond™. Ordering information forProBond™ resin is provided below.Product QuantityCatalogno. ProBond™ Purification System 6 purifications K850–01ProBond ™ Purification System with Anti-myc-HRP Antibody1 Kit K852–01ProBond ™ Purification System with Anti-His(C-term)-HRP Antibody1 Kit K853–01ProBond™ Nickel-Chelating Resin 50 ml150 mlR801–01R801–15Purification Columns 50 each R640–50viIntroductionOverviewIntroduction pPICZ A, B, and C are 3.3 kb expression vectors used to express recombinantproteins in Pichia pastoris. Recombinant proteins are expressed as fusions to aC-terminal peptide containing the c-myc epitope and a polyhistidine (6xHis) tag.The vector allows high-level, methanol inducible expression of the gene ofinterest in Pichia, and can be used in any Pichia strain including X33, GS115,SMD1168H, and KM71H. pPICZ contains the following elements:•5′ fragment containing the AOX1 promoter for tightly regulated, methanol-induced expression of the gene of interest (Ellis et al., 1985; Koutz et al., 1989;Tschopp et al., 1987a)•Zeocin™ resistance gene for selection in both E. coli and Pichia (Baron et al.,1992; Drocourt et al., 1990)•C-terminal peptide containing the c-myc epitope and a polyhistidine (6xHis)tag for detection and purification of a recombinant fusion protein (if desired)•Three reading frames to facilitate in-frame cloning with the C-terminalpeptideReference Sources The pPICZ A, B, and C expression vectors may be used with the Original Pichia Expression Kit, and are included in the EasySelect™Pichia Expression Kit (see page v for ordering information). Additional general information about recombinant protein expression in Pichia pastoris is provided in the manuals for the Original Pichia Expression Kit and the EasySelect™Pichia Expression Kit. For more information about the Original Pichia Expression Kit, the EasySelect™Pichia Expression Kit, or their manuals, visit our web site at or contact Technical Support (page 32).More detailed information and protocols dealing with Pichia pastoris may also be found in the following general reference:Higgins, D. R., and Cregg, J. M. (1998) Pichia Protocols. In Methods in Molecular Biology, Vol. 103. (J. M. Walker, ed. Humana Press, Totowa, NJ) (see page v for ordering information).Recommended Pichia Host Strain We recommend using the X-33 Pichia strain as the host for expression of recombinant proteins from pPICZ. Other Pichia strains including GS115, KM71H, and SMD1168H are suitable. The X-33 Pichia strain and other strains are available from Invitrogen (see page v for ordering information). The X-33 Pichia strain has the following genotype and phenotype:Genotype: Wild-typePhenotype: Mut+1Overview, ContinuedExperimental Overview The following table describes the basic steps needed to clone and express your gene of interest in pPICZ.Step Action1 Propagate pPICZ A, B, and C by transformation into a rec A, end A1E. coli strain such as TOP10, DH5 , or JM109.2 Develop a cloning strategy and ligate your gene into one of the pPICZvectors in frame with the C-terminal tag.3 TransformintoE. coli and select transformants on Low Salt LB platescontaining 25 μg/ml Zeocin™.4 Analyze 10–20 transformants by restriction mapping or sequencing toconfirm in-frame fusion of your gene with the C-terminal tag.5 Purify and linearize the recombinant plasmid for transformation intoPichia pastoris.6 TransformyourPichia strain and plate onto YPDS plates containing the appropriate concentration of Zeocin™.7 Select for Zeocin™-resistant transformants.8 Optimize expression of your gene.9 Purify your fusion protein on metal-chelating resin (i.e. ProBond™).Continued on next page2MethodsCloning into pPICZ A, B, and CIntroduction The pPICZ vector is supplied with the multiple cloning site in three readingframes (A, B, and C) to facilitate cloning your gene of interest in frame with theC-terminal peptide containing the c-myc epitope and a polyhistidine (6xHis) tag.Use the diagrams provided on pages 5–7 to help you design a strategy to cloneyour gene of interest in frame with the C-terminal peptide. Generalconsiderations for cloning and transformation are discussed in this section.General Molecular Biology Techniques For assistance with E. coli transformations, restriction enzyme analysis, DNA biochemistry, and plasmid preparation, refer to Molecular Cloning: A Laboratory Manual (Sambrook et al., 1989) or Current Protocols in Molecular Biology (Ausubel et al., 1994).E. coli Strain Many E. coli strains are suitable for the propagation of the pPICZ vectorsincluding TOP10, JM109, and DH5 . We recommend that you propagate thepPICZ vectors in E. coli strains that are recombination deficient (rec A) andendonuclease A deficient (end A).For your convenience, TOP10 E. coli are available as chemically competent orelectrocompetent cells from Invitrogen (page v).Transformation Method You may use any method of choice for transformation. Chemical transformation is the most convenient for many researchers. Electroporation is the most efficient and the method of choice for large plasmids.Maintenance of Plasmids The pPICZ vectors contain the Zeocin™ resistance (Sh ble) gene to allow selection of the plasmid using Zeocin™. To propagate and maintain the pPICZ plasmids, we recommend using the following procedure:e 10 ng of your vector to transform a rec A, end A E. coli strain like TOP10,DH5 , JM109, or equivalent (see above).2.Select transformants on Low Salt LB plates containing 25 μg/ml Zeocin™ (seepage 17 for a recipe).3.Prepare a glycerol stock from each transformant containing plasmid forlong-term storage (see page 8).Continued on next page3Cloning into pPICZ A, B, and C, ContinuedGeneral Considerations The following are some general points to consider when using pPICZ to express your gene of interest in Pichia:•The codon usage in Pichia is believed to be similar to Saccharomyces cerevisiae.•Many Saccharomyces genes have proven to be functional in Pichia.•The premature termination of transcripts because of "AT rich regions" has been observed in Pichia and other eukaryotic systems (Henikoff & Cohen, 1984; Irniger et al., 1991; Scorer et al., 1993; Zaret & Sherman, 1984). If you have problems expressing your gene, check for premature termination by northern analysis and check your sequence for AT rich regions. It may be necessary to change the sequence in order to express your gene (Scorer et al., 1993).•The native 5´ end of the AOX1 mRNA is noted in the diagram for each multiple cloning site. This information is needed to calculate the size of the expressed mRNA of the gene of interest if you need to analyze mRNA for any reason.Cloning Considerations For proper initiation of translation, your insert should contain an initiation ATG codon as part of a yeast consensus sequence (Romanos et al., 1992). An example of a yeast consensus sequence is provided below. The ATG initiation codon is shown underlined.(G/A)NNATG GTo express your gene as a recombinant fusion protein, you must clone your gene in frame with the C-terminal peptide containing the c-myc epitope and the polyhistidine tag. The vector is supplied in three reading frames to facilitate cloning. Refer to the diagrams on pages 5–7 to develop a cloning strategy.If you wish to express your protein without the C-terminal peptide, be sure to include a stop codon.Construction of Multimeric Plasmids pPICZ A, B, and C contain unique Bgl II and Bam H I sites to allow construction of plasmids containing multiple copies of your gene. For information on how to construct multimers, refer to pages 24–31.Continued on next page4Multiple CloningSite of pPICZ A Below is the multiple cloning site for pPICZ A. Restriction sites are labeled to indicate the cleavage site. The boxed nucleotides indicate the variable region.The multiple cloning site has been confirmed by sequencing and functionaltesting.You can download the complete sequence of pPICZ A from our web site at or by contacting Technical Support (see page 32).For a map and a description of the features of pPICZ, refer to the Appendix(pages 21–22). AAT AGC GCC GTC GAC CAT CAT CAT CAT CAT CAT TGTTCCTCAG TTCAAGTTGG GCACTTACGA GAAGACCGGT CTTGCTAGAT TCTAATCAAG AGGATGTCAG AATGCCATTT GCCTGAGAGA TGCAGGCTTC ATTTTTGATA CTTTTTTATTTGTAACCTAT ATAGTATAGG ATTTTTTTTG TCATTTTGTT 1218Asn Ser Ala Val Asp His His His His His His ***3´ AOX1 priming site TGA GTTTTAGCCT TAGACATGAC AACCTTTTTT TTTATCATCA TTATTAGCTT ACTTTCATAA TTGCGACTGG TTCCAATTGA CAAGCTTTTG ATTTTAACGA CTTTTAACGA CAACTTGAGA AGATCAAAAA ACAACTAATT ATTCGAAACG AGGAATTCAC GTGGCCCAGC CGGCCGTCTC GGATCGGTAC CTCGAGCCGC GGCGGCCGCC AGCTT GGGCCC GAA CAA AAA CTC ATC TCA GAA GAG GAT CTG 811Glu Gln Lys Leu Ile Ser Glu Glu Asp Leu 5´ AOX1 priming sitemyc epitope3´polyadenylation sitePolyhistidine tag5´ end of AOX1 mRNA Sfu I Eco R I Pml I Sfi I Bsm B I Asp 718 I Kpn I Xho ISac II Not I Apa I 104210981158871931991Continued on next pageMultiple CloningSite of pPICZ B Below is the multiple cloning site for pPICZ B. Restriction sites are labeled to indicate the cleavage site. The boxed nucleotides indicate the variable region.The multiple cloning site has been confirmed by sequencing and functionaltesting.You can download the complete sequence of pPICZ B from our web site at or by contacting Technical Support (see page 32).For a map and a description of the features of pPICZ, refer to the Appendix(pages 21–22). AAT AGC GCC GTC GAC CAT CAT CAT CAT CAT CAT TGA GTTTGTAGCC TTAGACATGA CTGTTCCTCA GTTCAAGTTG GGCACTTACG AGAAGACCGG TCTTGCTAGA TTCTAATCAA GAGGATGTCA GAATGCCATT TGCCTGAGAG ATGCAGGCTT CATTTTTGAT ACTTTTTTAT TTGTAACCTA TATAGTATAG GATTTTTTTT GTCATTTTGT TTC 1216Asn Ser Ala Val Asp His His His His His His ***3´ AOX1 priming site TGA GTTTGTAGCC TTAGACATGA AACCTTTTTT TTTATCATCA TTATTAGCTT ACTTTCATAA TTGCGACTGG TTCCAATTGA CAAGCTTTTG ATTTTAACGA CTTTTAACGA CAACTTGAGA AGATCAAAAA ACAACTAATTATTCGAAACG AGGAATTCAC GTGGCCCAGC CGGCCGTCTC GGATCGGTAC CTCGAGCCGC GGCGGCCGCC AGCTT TCTA GAA CAA AAA CTC ATC TCA GAA GAG GAT CTG 811Glu Gln Lys Leu Ile Ser Glu Glu Asp Leu 5´ AOX1 priming sitemyc epitope3´ polyadenylation site Polyhistidine tag5´ end of AOX1 mRNA Sfu I Eco R I Pml I Sfi I Bsm B I Asp 718 I Kpn I Xho ISac II Not I Xba I 104010961156871931991Continued on next pageMultiple CloningSite of pPICZ C Below is the multiple cloning site for pPICZ C. Restriction sites are labeled to indicate the cleavage site. The boxed nucleotides indicate the variable region.The multiple cloning site has been confirmed by sequencing and functionaltesting.You can download the complete sequence of pPICZ C from our web site at or by contacting Technical Support (see page 32).For a map and a description of the features of pPICZ, refer to the Appendix(pages 21–22). AAT AGC GCC GTC GAC CAT CAT CAT CAT CAT CAT TGA GTTTGTAGCC TTAGACATGA CTGTTCCTCA GTTCAAGTTG GGCACTTACG AGAAGACCGG TCTTGCTAGA TTCTAATCAA GAGGATGTCA GAATGCCATT TGCCTGAGAG ATGCAGGCTT CATTTTTGAT ACTTTTTTAT TTGTAACCTA TATAGTATAG GATTTTTTTT GTCATTTTGT TTC 1217Asn Ser Ala Val Asp His His His His His His ***3´ AOX1 priming siteTGA GTTTGTAGCC TTAGACATGA AACCTTTTTT TTTATCATCA TTATTAGCTT ACTTTCATAA TTGCGACTGG TTCCAATTGA CAAGCTTTTG ATTTTAACGA CTTTTAACGA CAACTTGAGA AGATCAAAAA ACAACTAATT ATTCGAAACG AGGAATTCAC GTGGCCCAGC CGGCCGTCTC GGATCGGTAC CTCGAGCCGC GGCGGCCGCC AGCTT ACGTA GAA CAA AAA CTC ATC TCA GAA GAG GAT CTG 811Glu Gln Lys Leu Ile Ser Glu Glu Asp Leu 5´ AOX1 priming sitemyc epitope3´ polyadenylation site Polyhistidine tag5´ end of AOX1 mRNA Sfu I Eco R I Pml I Sfi I Bsm B I Asp 718 I Kpn I Xho I Sac II Not I Sna B I 104110971157871931991Continued on next pageE. coli Transformation Transform your ligation mixtures into a competent rec A, end A E. coli strain(e.g. TOP10, DH5, JM109) and select on Low Salt LB agar plates containing25 μg/ml Zeocin™ (see below). Note that there is no blue/white screening for the presence of insert with pPICZ A, B, or C. Once you have obtained Zeocin™-resistant colonies, pick 10 transformants and screen for the presence and orientation of your insert.Important To facilitate selection of Zeocin™-resistant E. coli, the salt concentration of the medium must remain low (<90 mM) and the pH must be 7.5. Prepare Low Salt LB broth and plates using the recipe in the Appendix, page 17.Failure to lower the salt content of your LB medium will result in non-selection due to inhibition of the drug.C We recommend that you sequence your construct to confirm that your gene is in the correct orientation for expression and cloned in frame with the C-terminal peptide (if desired). Refer to the diagrams on pages 5–7 for the sequences and location of the priming sites.Preparing a Glycerol Stock Once you have identified the correct clone, be sure to purify the colony and make a glycerol stock for long-term storage. It is also a good idea to keep a DNA stock of your plasmid at –20°C.1.Streak the original colony out on an Low Salt LB plate containing 25 μg/mlZeocin™. Incubate the plate at 37°C overnight.2.Isolate a single colony and inoculate into 1–2 ml of Low Salt LB containing25 μg/ml Zeocin™.3.Grow the culture to mid-log phase (OD600 = 0.5–0.7).4.Mix 0.85 ml of culture with 0.15 ml of sterile glycerol and transfer to acryovial.5.Store at –80°C.Plasmid Preparation Once you have cloned and sequenced your insert, generate enough plasmid DNA to transform Pichia (5–10 μg of each plasmid per transformation). We recommend isolating plasmid DNA using the PureLink™ Quick Plasmid Miniprep Kit or the PureLink™ HiPure Plasmid Midiprep Kit (page v), or CsCl gradient centrifugation.Once you have purified plasmid DNA, proceed to Pichia Transformation, next page.Pichia TransformationIntroduction You should now have your gene cloned into one of the pPICZ vectors. Yourconstruct should be correctly fused to the C-terminal peptide (if desired). Thissection provides general guidelines to prepare plasmid DNA, transform yourPichia strain, and select for Zeocin™-resistant clones.Zeocin™ Selection We generally use 100 μg/ml Zeocin™ to select for transformants when using the X-33 Pichia strain. If you are transforming your pPICZ construct into anotherPichia strain, note that selection conditions may vary. We recommendperforming a dose response curve to determine the appropriate concentration ofZeocin™ to use for selection of transformants in your strain.Method of Transformation We do not recommend spheroplasting for transformation of Pichia with plasmids containing the Zeocin™ resistance marker. Spheroplasting involves removal of the cell wall to allow DNA to enter the cell. Cells must first regenerate the cell wall before they are able to express the Zeocin™ resistance gene. For this reason, plating spheroplasts directly onto selective medium containing Zeocin™ does not yield any transformants.We recommend electroporation for transformation of Pichia with pPICZ A, B, or C. Electroporation yields 103 to 104 transformants per μg of linearized DNA and does not destroy the cell wall of Pichia. If you do not have access to an electroporation device, use the LiCl protocol on page 23 or the Pichia EasyComp™Transformation Kit available from Invitrogen (see below).PichiaEasyComp™Transformation Kit If you wish to perform chemical transformation of your Pichia strain with pPICZ A, B, or C, the Pichia EasyComp™ Transformation Kit is available from Invitrogen (see page v for ordering information). The Pichia EasyComp™ Transformation Kit provides reagents to prepare 6 preparations of competent cells. Each preparation will yield enough competent cells for 20 transformations. Competent cells may be used immediately or frozen and stored for future use. For more information, visit our web site at or contact Technical Support (page 32).Important Since pPICZ does not contain the HIS4 gene, integration can only occur at the AOX1 locus. Vector linearized within the 5´ AOX1 region will integrate by gene insertion into the host 5´ AOX1 region. Therefore, the Pichia host that you use will determine whether the recombinant strain is able to metabolize methanol (Mut+) or not (Mut S). To generate a Mut+ recombinant strain, you must use a Pichia host that contains the native AOX1 gene (e.g. X-33, GS115, SMD1168H). If you wish to generate a Mut S recombinant strain, then use a Pichia host that has a disrupted AOX1 gene (i.e. KM71H).Continued on next pageHis4 Host Strains Host strains containing the his4 allele (e.g. GS115) and transformed with thepPICZ vectors require histidine when grown in minimal media. Add histidine toa final concentration of 0.004% to ensure growth of your transformants.The pPICZ vectors do not contain a yeast origin of replication. Transformantscan only be isolated if recombination occurs between the plasmid and the Pichiagenome.Materials Needed You will need the following items:Note: Inclusion of sorbitol in YPD plates stabilizes electroporated cells as they appear tobe somewhat osmotically sensitive.•5–10 μg pure pPICZ containing your insert•YPD Medium•50 ml conical polypropylene tubes• 1 liter cold (4°C) sterile water (place on ice the day of the experiment)•25 ml cold (4°C) sterile 1 M sorbitol (place on ice the day of the experiment)•30°C incubator•Electroporation device and 0.2 cm cuvettes•YPDS plates containing the appropriate concentration of Zeocin™ (seepage 18 for recipe)Linearizing YourpPICZ ConstructTo promote integration, we recommend that you linearize your pPICZ constructwithin the 5′ AOX1 region. The table below lists unique sites that may be used tolinearize pPICZ prior to transformation. Other restriction sites are possible.Note that for the enzymes listed below, the cleavage site is the same for versionsA, B, and C of pPICZ. Be sure that your insert does not contain the restriction siteyou wish to use to linearize your vector.Enzyme Restriction Site (bp) SupplierSac I 209 ManyPme I 414 New England BiolabsBst X I 707 ManyRestriction Digest 1.Digest ~5–10 μg of plasmid DNA with one of the enzymes listed above.2.Check a small aliquot of your digest by agarose gel electrophoresis forcomplete linearization.3.If the vector is completely linearized, heat inactivate or add EDTA to stopthe reaction, phenol/chloroform extract once, and ethanol precipitate using1/10 volume 3 M sodium acetate and 2.5 volumes of 100% ethanol.4.Centrifuge the solution to pellet the DNA, wash the pellet with 80% ethanol,air-dry, and resuspend in 10 μl sterile, deionized water. Use immediately orstore at –20°C.Continued on next pagePreparation of Pichia for Electroporation Follow the procedure below to prepare your Pichia pastoris strain for electroporation.1. Grow 5 ml of your Pichia pastoris strain in YPD in a 50 ml conical tube at30°C overnight.2. Inoculate 500 ml of fresh medium in a 2 liter flask with 0.1–0.5 ml of theovernight culture. Grow overnight again to an OD600 = 1.3–1.5.3. Centrifuge the cells at 1500 × g for 5 minutes at 4°C. Resuspend the pelletwith 500 ml of ice-cold (0–4°C), sterile water.4. Centrifuge the cells as in Step 3, then resuspend the pellet with 250 ml ofice-cold (0–4°C), sterile water.5. Centrifuge the cells as in Step 3, then resuspend the pellet in 20 ml of ice-cold (0–4°C) 1 M sorbitol.6. Centrifuge the cells as in Step 3, then resuspend the pellet in 1 ml of ice-cold(0–4°C) 1 M sorbitol for a final volume of approximately 1.5 ml. Keep the cells on ice and use that day. Do not store cells.Transformation by Electroporation 1.Mix 80 μl of the cells from Step 6 (above) with 5–10 μg of linearized pPICZDNA (in 5–10 μl sterile water) and transfer them to an ice-cold (0–4°C)0.2 cm electroporation cuvette.2.Incubate the cuvette with the cells on ice for 5 minutes.3.Pulse the cells according to the parameters for yeast (Saccharomycescerevisiae) as suggested by the manufacturer of the specific electroporation device being used.4.Immediately add 1 ml of ice-cold 1 M sorbitol to the cuvette. Transfer thecuvette contents to a sterile 15 ml tube.5.Let the tube incubate at 30°C without shaking for 1 to 2 hours.6.Spread 50-200 μl each on separate, labeled YPDS plates containing theappropriate concentration of Zeocin™.7.Incubate plates for 2–3 days at 30°C until colonies form.8.Pick 10–20 colonies and purify (streak for single colonies) on fresh YPD orYPDS plates containing the appropriate concentration of Zeocin™.Continued on next pageGenerally, several hundred Zeocin™-resistant colonies are generated using theprotocol on the previous page. If more colonies are needed, the protocol may bemodified as described below. Note that you will need ~20, 150 mm plates withYPDS agar containing the appropriate concentration of Zeocin™.1. Set up two transformations per construct and follow Steps 1 through 5 ofthe Transformation by Electroporation protocol, page 11.2. After 1 hour in 1 M sorbitol at 30°C (Step 5, previous page), add 1 ml YPDmedium to each tube.3. Shake (~200 rpm) the cultures at 30°C.4. After 1 hour, take one of the tubes and plate out all of the cells by spreading200 μl on 150 mm plates containing the appropriate concentration ofZeocin™.5. Optional: Continue incubating the other culture for three more hours (for atotal of four hours) and then plate out all of the cells by spreading 200 μl on150 mm plates containing the appropriate concentration of Zeocin™.6. Incubate plates for 2–4 days at 30°C until colonies form.Mut Phenotype If you used a Pichia strain containing a native AOX1 gene (e.g. X-33, GS115,SMD1168H) as the host for your pPICZ construct, your Zeocin™-resistanttransformants will be Mut+. If you used a strain containing a deletion in theAOX1 gene (e.g. KM71H), your transformants will be Mut S.If you wish to verify the Mut phenotype of your Zeocin™-resistant transformants,you may refer to the general guidelines provided in the EasySelect™PichiaExpression Kit manual or the Original Pichia Expression Kit manual or topublished reference sources (Higgins & Cregg, 1998).You are now ready to test your transformants for expression of your gene ofinterest. See Expression in Pichia, next page.。
phis2质粒序列 -回复

phis2质粒序列-回复Phis2质粒序列是一种重要的工具,被广泛应用于分子生物学研究中。
它不仅能够用于基因克隆、表达和转化等方面,还能够用于基因检测和基因治疗等研究领域。
本文将以Phis2质粒序列为主题,一步一步回答相关问题。
第一步:了解Phis2质粒序列的概念和特点Phis2质粒序列是一种常用的基因工程工具,它是由DNA序列组成的环状双链分子。
Phis2质粒序列具有多个特点,包括能够自主复制、具有抗生素抗性基因、能够承载外源基因等。
第二步:探究Phis2质粒序列的应用Phis2质粒序列在分子生物学研究中有广泛的应用。
首先,它可以用于基因克隆,即将感兴趣的DNA片段插入到Phis2质粒序列中,以便后续的研究。
其次,Phis2质粒序列还可以用于基因表达,通过将目标基因插入到Phis2质粒序列中,再将其转化到宿主细胞中,从而实现目标基因的高效表达。
此外,Phis2质粒序列还可以用于基因检测和基因治疗等研究领域。
第三步:详细介绍Phis2质粒序列的结构和构建方法Phis2质粒序列通常由多个功能模块组成,主要包括起始子、多克隆位点、抗生素抗性基因、终止子等。
构建Phis2质粒序列的方法包括PCR 扩增、限制性内切酶切割、连接反应和转化等步骤。
具体而言,首先需要通过PCR扩增得到感兴趣的DNA片段,然后通过限制性内切酶切割将其与Phis2质粒序列中的多克隆位点进行连接。
连接反应可以采用DNA连接酶或Ligase酶进行。
最后,将构建好的Phis2质粒序列转化到宿主细胞中,经过培养和筛选,可以得到目标Phis2质粒序列。
第四步:总结Phis2质粒序列的优势和局限性Phis2质粒序列作为一种重要的分子生物学工具,具有多个优势。
首先,它具有较大的载体容量,能够承载较长的外源基因。
其次,Phis2质粒序列具有自主复制的能力,可以在宿主细胞内独立复制。
此外,Phis2质粒序列还可以通过添加抗生素抗性基因,实现对宿主细胞的筛选。
pHISi使用说明

pHISi编号载体名称北京华越洋VECT76301pHISipHISi载体图谱:pHISi载体简介:pHISi is a yeast integration and reporter vector for use with the MATCHMAKER One-Hybrid System.pHISi contains the yeast HIS3gene downstream of the MCS and the minimal promoter of the HIS3locus(PminHIS).Cis-acting sequences of interest (i.e.,target elements)can be inserted into the MCS.Without activation by a target element,constitutive HIS3expression from PminHIS is very low in yeast,but allows enough growth to select for integration when constructing HIS3reporter strains. During library screening,the leaky expression of HIS3is controlled by adding3-AT to the medium.The yeast URA3and HIS3genes of pHISi can be used as selectable markers for integration into the nonfunctional ura3and his3loci,respectively,of the YM4271 host strain.Before integrating,the vector is linearized at the Xho I or Afl II sites(his3 locus)or at the Apa I site(ura3locus).The Kpn I site cannot be used for integration because it cuts within the coding region of the HIS3gene,and that region is deleted in YM4271.pHISi cannot replicate autonomously in yeast.The plasmid contains a bacterial Col E1origin(ori)and the ampicillin resistance gene(Ampr)for propagation and selection in E.coli.Unique restriction sites are in bold.。
所有质粒载体汇总

pEZZ18 pkk232-8,pkk 233-3,pACYC184,pBR322,pUC119 pTYB1,pTYB2,pTYB4,pTYB11 pBlueScript SK(+),pBlueScript SK(-) pLLP ompA, pINIIIompA, pMBP-P ,pMBP-C, 大肠杆菌冷激质 粒: pColdI pColdII pColdIII pColdTF 原核共表达质粒:pACYCduet1,pETduet-1,pCDFduet-1,pRSFduet-1 Takara公司大肠杆菌分子伴 侣: pG-KJE8 pGro7 pKJE7 pGTf2 pTf16 大肠杆菌宿主细胞: DH5a JM101 JM103 JM105 JM107 JM109 JM110 Top10 Top10F BL21(DE3) HB101 ER2529 E2566 C2566 MG1655 XL-10gold XL blue M15 JF1125 K802 SG1117 BL21(AI) BL21(DE3)plysS TG1 TB1 DH5a(pir) Tuner(DE3) Bl21 codonplusRIPL Novablue(DE3) Rosetta Rosetta(DE3) Rosetta(DE3)plys Rosetta-gami(DE3) RosettagamiB(DE3), Rosetta-gamiB(DE3)plysS Orgami(DE3) OrgamiB(DE3) HMS174(DE3) 植物表达/RNAi载体农杆菌pBI121,pBI121-GFP,pBI101,pBI221,pSN1301, pUN1301,pRTL2 , pRTL2-GFP , pRTL2-CFP, pRTL2-RFP , pRTL2YFP,pCAMBIA 1300, 1301, 1302,1303,1304,1305, 1381Z,1391Z,2300, 2301,3300,3301,pCAMBIA super1300,pCAMBIA super1300GFP,pPZP212,pPZP2121,pPZP212-GFP,pGDG,RNAi载 体pART27,pHANNIBAL,pKANNIBAL, pFGC5941,pTCK303, pTRV1,pTRV2, T-DNA插入载体(随机突变体库)pSKI015,pSKI074,真 菌ATMT载体pBIG2RHPH2-GUS-GFP,pBHt1 枯草芽孢杆菌表达载体pWB980,pHT43,pHP13,pHP43, pBE2,pMUTIN4,pUB110,pE194,pMA5, pMK3,pMK4,pHT304,pHY300PLK, pBest502,pDG1363,pSG1154,pAX01, pSAS144,pDL,pDG148-stu,pDG641, pAL12,pUCX05-bgaB,pHT01, 配套 菌株BS 168,WB600,WB800,WB700, WB800N,1012,FZB42,1A747,广宿主 质粒pVLT33
新-酵母单杂交筛选与启动子pHIS2-A互作蛋白结题报告

目的:筛选与启动子pHIS2-A互作的蛋白。
方法:以启动子A构建到酵母单杂交启动子载体pHIS2,文库质粒为pGADT7-中山杉文库。
材料:1.启动子克隆:pHIS2-A。
根据实验需求进行扩增并酶切构建载体,两端的EcoR I和SacI酶切位点,序列如下:GAATTC AGTTGGAATCACATCTATCATCAGAATGAAAGACCTAAGAAAAAAATGGCA TATAAAAATATGAAAGATTGTTTTTTTTTAACTGATGACCCACATATTTTTTTAATCTGTTCA ACTAAAAAAACTTCTGAAC GAGCTC通过启动子序列两端的酶切位点构建到酵母单杂交启动子载体pHIS2上。
2.启动子载体:pHIS23.prey载体:pGADT74.CLONTECH酵母单杂交系统5.培养基:1). 酵母完全培养基:YPDA:1% Yeast extract,2% Tryptone,2% Glucose, 0.02% Adenine。
2). 酵母缺陷型筛选培养基:参照Clontech公司的PT3024-1/ Yeast Protocols Handbook。
其中各种培养基分别以下列符号表示:SD-T:-trp;SD-TH:-trp,-his;SD-TL:-trp, -leu;SD-TLH:-trp, -leu, -his;1.1酵母转化反应反应AD质粒pHIS2质粒转化平板检测平板检测内容1 2——pGAD53mpHIS2-APHIS2-p53SD-TSD-TLSD-TH+3ATSD-TLH+3AT背景筛选检测&筛库阳性对照1.2 酵母感受态制备及转化方法1. 从YPDA平板挑取Y187单菌落接种于YPDA液体培养基4 ml,30℃,225 rpm,振荡培养18-20 h(过夜),至OD600>1.5,一般为4左右。
2. 转接YPDA液体培养基,培养体积为50 ml,使初始OD600 = 0.2,30℃,225 rpm,振荡培养4-5 h,至OD600 = 0.6。
酵母表达系统

酵母表达系统基因表达是分子生物学领域的重要内容之一,人们利用基因表达技术制备各种目的基因的重组蛋白质,在分析基因的表达与调控、基因的结构与功能、基因治疗以及生物制药等领域均取得了令人振奋的成果。
其中,酵母表达系统拥有转录后加工修饰功能,操作简便,成本低廉,适合于稳定表达有功能的外源蛋白质,而且可大规模发酵,是最理想的重组真核蛋白质生产制备用工具。
1、酵母表达系统的特点酵母是一种单细胞低等真核生物,培养条件普通,生长繁殖速度迅速,能够耐受较高的流体静压,用于表达基因工程产品时,可以大规模生产,有效降低了生产成本。
酵母表达外源基因具有一定的翻译后加工能力,收获的外源蛋白质具有一定程度上的折叠加工和糖基化修饰,性质较原核表达的蛋白质更加稳定,特别适合于表达真核生物基因和制备有功能的表达蛋白质。
某些酵母表达系统具有外分泌信号序列,能够将所表达的外源蛋白质分泌到细胞外,因此很容易纯化。
应用酵母表达系统生产外源基因的蛋白质产物时也有不足之处,如产物蛋白质的不均一、信号肽加工不完全、内部降解、多聚体形成等,造成表达蛋白质在结构上的不一致。
解决内部降解的方法有三:一是在培养基中加入富含氨基酸和多肽的蛋白胨或酪蛋白水解物,通过增加酶作用底物来缓解蛋白水解作用;二是将培养基的pH值调成酸性(酵母可在pH3.0~8.0的范围内生长),以抑制中性蛋白酶的活性;三是利用蛋白酶缺失酵母突变体进行外源基因的表达。
另外,还时常遇到表达产物的过度糖基化情况。
因此,表达系统应根据具体情况作适当的改进。
2、常用酵母表达系统(宿主-载体系统)(1)酿酒酵母(Saccharomyces cerevisiae)表达系统酿酒酵母难于高密度培养,分泌效率低,几乎不分泌分子量大于30 kD的外源蛋白质,也不能使所表达的外源蛋白质正确糖基化,而且表达蛋白质的C端往往被截短。
因此,一般不用酿酒酵母做重组蛋白质表达的宿主菌。
酿酒酵母本身含有质粒,其表达载体可以有自主复制型和整合型两种。
pHIS2
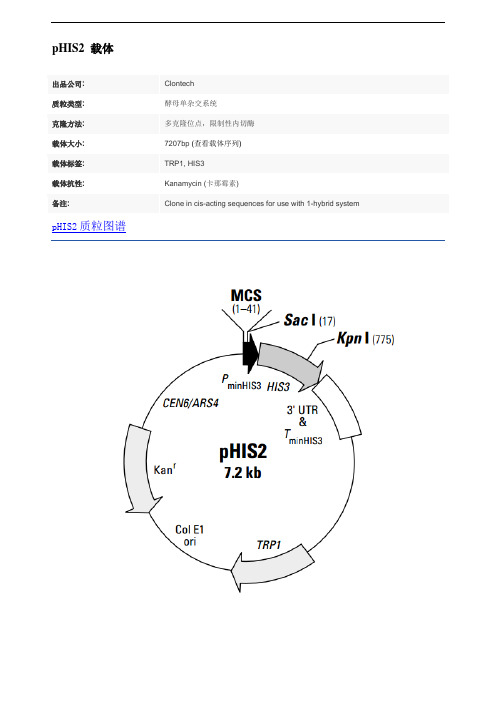
pHIS2 载体pHIS2 is a reporter vector that can be used in yeast one-hybrid assays to identify and characterize DNA-binding proteins. The vector was specifically designed for use with the BD Matchmaker™ One- Hybrid Library Construction & Screening Kit (#K1617-1). It contains a HIS3 nutritional reporter gene, located downstream of a multiple cloning site (MCS) and the minimal promoter of the HIS3 locus (PminHIS3). Cis-acting DNA sequences, or DNA target elements, can be inserted into the MCS and used as baits to screen GAL4 AD/cDNA fusion libraries for proteins that interact with the target sequence.A protein-DNA (or one-hybrid) interaction can be detected by performing the assay in a yeast strain such as Y187 that is auxotrophic for histidine. Positive one-hybrid interactions drive expression of the HIS3 reporter gene, which enables the host cell to grow on histidine-deficient media.In the absence of activation, the constitutive HIS3 expression from PminHIS3 is very low. During library screening, the leaky expression of HIS3 is controlled by adding3-amino-1,2,4-triazole (3-AT) to the medium. The concentration of 3-AT needed to fully suppress leaky HIS3 expression must be determined empirically for each DNA target element.pHIS2 can be maintained in both yeast and bacteria. It contains an autonomous replication sequence (ARS4) and TRP1 nutritional marker for replication and selection in yeast (1, 2); it contains a Col E1 origin and a kanamycin resistance gene (Kanr) for propagation and selection in E. coli. The centromeric sequence CEN6 ensures proper segregation of the plasmid during cell division in yeast (1, 2).载体应用To use pHIS2 in a one-hybrid assay, clone one or more copies of a cis-acting DNA target sequence into the MCS. Then introduce the plasmid into competent yeast cells using the transformation protocol in the BD Matchmaker Library Construction & Screening Kits User Manual (PT3529-1). In contrast to the original BD Matchmaker One- Hybrid System, this reporter vector does not need to be integrated into the yeast genome. Instead, it is maintained as an episome throughout the assay. Inserting your target element may alter the level of background HIS3 expression. Therefore, constructs should be tested for background (leaky) HIS3 expression before you start a one-hybrid analysis. Background growth due to leaky HIS3 expression is controlled by adding 3-AT to the selection medium, as described in the User Manual (PT3529-1).。
pGADT7-Rec2使用说明

pGADT7-Rec2编号载体名称北京华越洋VECT75553pGADT7-Rec2pGADT7-Rec2载体图谱:pGADT7-Rec2载体简介:pGADT7-Rec2is a cloning vector that can be used in yeast to express a protein of interest as a GAL4activation domain(GAL4AD)fusion. Transcription starts with the constitutive ADH1promoter(PADH1)and ends with the ADH1termination signal(TADH1).The GAL4AD sequence includes the SV40nuclear localization signal(SV40NLS;1)so that fusions translocate to the yeast nucleus.GAL4AD fusions also contain a hemagglutinin(HA)epitope tag.The T7promoter in pGADT7-Rec2can be used for in vitro transcription and translation of the HA-tagged fusion protein. It also provides a binding site for sequencing with the T7Promoter Sequencing Primer.pGADT7-Rec2is a shuttle vector;it can be maintained in both yeast and bacteria.It contains an autonomous replication sequence(ARS4) and a LEU2nutritional marker for replication and selection in yeast(2, 3).A centromeric sequence(CEN6)is included to ensure proper segregation of the plasmid during cell division(2,3).For propagation and selection in E.coli,the vector contains a pUC origin of replication(pUC ori)and an ampicillin resistance gene(Ampr).pGADT7-Rec2is derived from pGADT7-Rec,a cloning vector used in the Matchmaker(Two-Hybrid)Library Construction&Screening Kit(Cat.No.630445).It was constructed by replacing the2μori in pGADT7-Rec with the ARS4and CEN6elements.The ARS and CEN elements ensure stable, low-copy propagation of the vector.Unlike pGADT7-Rec,which is able to replicate multiple times during the yeast cell cycle,pGADT7-Rec2,with its ARS and CEN elements,can replicate only once during the cell cycle, so its copy number is restricted.Low-copy plasmids such as pGADT7-Rec2 are preferred for one-hybrid screening because they generate fewer false positives.In the original Matchmaker One-Hybrid System,copy number was restricted not by using low-copy,autonomously replicating plasmids,but by integrating the reporter construct into the yeast genome.With the development of pGADT7-Rec2(and pHIS2,a low-copy reporter vector), integration is no longer necessary because each plasmid,with its CEN and ARS elements,now behaves like a minichromosome,both mitotically and meiotically.pGADT7-Rec2载体其他相关的酵母单杂交载体:p53-ABAIpHIS2pGADT7-RecpGADT7-Rec2pABAI。
毛竹ABCG基因鉴定及其表达模式研究

核农学报2023,37(5):0917~0926Journal of Nuclear Agricultural Sciences毛竹ABCG基因鉴定及其表达模式研究李紫阳杨克彬朱成磊刘燕郭栋肖晓燕高志民 *(国际竹藤中心竹藤资源基因科学与基因产业化研究所,国家林业和草原局/北京市共建竹藤科学与技术重点实验室,北京100102)摘要:ATP结合盒转运蛋白G亚家族(ABCG)在植物体内的物质运输过程中发挥着重要作用。
为探究毛竹(Phyllostachys edulis)中ABCG s的分子特征、表达模式及转录因子对PeABCG15的调控关系,本研究利用生物信息学方法鉴定毛竹ABCG基因家族成员,对其基因结构、编码蛋白的理化性质和系统进化等进行系统分析,通过实时荧光定量PCR(qRT-PCR)技术对毛竹ABCG基因表达模式进行研究,借助酵母单杂交验证转录因子与ABCG基因的调控关系。
结果表明,在毛竹基因组中共鉴定到77个ABCG基因(PeABCG1~PeABCG77),其启动子中含有多种激素和非生物胁迫响应元件,其中脱落酸响应元件ABRE 最多,出现在74个基因的启动子中。
系统进化分析发现,PeABCGs分为白棕色复合体(WBC)和多效性耐药复合体(PDR)两个亚组,与水稻的亲缘关系较近。
qRT-PCR结果显示,在脱落酸(ABA)处理后,7个与ABA运输相关的PeABCG s中有6个呈上调表达趋势,而未检测到PeABCG34表达;在低温处理条件下,7个PeABCG s均受到诱导表达;在干旱处理条件下,有2个PeABCG s的表达受到抑制,其余基因的表达均受到不同程度的诱导。
另外,随着竹笋木质化程度增加,PeABCG15呈持续上调表达趋势,并与木质素合成调控转录因子基因PeKNAT3和PeMYB42的表达趋势一致。
酵母单杂交试验证实,PeKNAT3和PeMYB42均能与PeABCG15的启动子结合。
由此表明,PeABCG s参与毛竹抗逆可能存在ABA介导和非介导两种方式,其中PeABCG15受ABA诱导,且可能通过促进木质素合成参与毛竹抗逆。
酵母单杂phis2实验方法

酵母单杂phis2实验方法一、实验前的准备。
酵母单杂phis2实验呢,可是个很有趣但又有点小复杂的实验哦。
我们得先把各种材料准备好。
首先就是酵母菌株啦,要选择合适的酵母菌株,这就像挑选演员一样,得找对角色呢。
然后是载体,载体就像是酵母的小房子,要确保这个小房子是适合我们要做的实验的。
还有培养基,培养基的成分可得仔细调配。
像碳源啊,氮源啊,这些都是酵母生长的“食物”。
如果这些“食物”没弄好,酵母就会“饿肚子”,不好好工作啦。
各种试剂也不能少,像限制性内切酶、连接酶之类的。
这些试剂就像是建筑工人手里的工具,用来构建我们需要的基因片段呢。
二、构建诱饵载体。
1. 我们要先从基因组中把我们感兴趣的基因片段给找出来。
这就像是在一个大仓库里找一个小零件一样,得有耐心。
可以通过PCR技术来扩增这个基因片段。
PCR的时候呢,要把反应条件设置好,温度啊,时间啊,都得恰到好处。
如果温度不对,可能就扩增不出来我们想要的东西了。
2. 然后把扩增出来的基因片段和载体连接起来。
这个连接过程就像是把两个小积木拼在一起,要靠连接酶这个小“胶水”。
连接的时候要注意比例,基因片段和载体的量要合适,如果一方太多或者太少,都可能连接不成功呢。
3. 连接好之后,我们要把这个重组载体转化到大肠杆菌里面。
大肠杆菌就像是一个小小的工厂,能帮我们大量生产这个重组载体。
转化的时候要用到热激法或者化学转化法,不管用哪种方法,都要按照步骤来,不然大肠杆菌可能就不接受这个外来的“小客人”啦。
三、酵母转化。
1. 把在大肠杆菌里面大量生产的诱饵载体提取出来,这个提取过程要小心,要保证载体的纯度和完整性。
然后把诱饵载体转化到酵母细胞里面。
酵母细胞的细胞壁有点厚,就像一个小城堡一样,要想办法把载体送进去。
可以用醋酸锂法或者电转化法。
醋酸锂法就像是给城堡的大门悄悄送个“小礼物”,让它打开门让载体进去;电转化法就像是给城堡来个小电击,把大门打开,让载体进去。
2. 转化后的酵母细胞要放在合适的培养基上培养。
酵母表达系统步骤

酵母表达系统步骤毕赤酵母表达系统步骤(参考Invitrogen公司说明书):一、pPICZαA、B、C质粒以及DH5α菌株的保存1取0.5μl pPICZα A、B、C质粒,热击转化DH5α,在低盐LB (含有25μg/ml Zeocin)的平板上37℃培养过夜。
2挑取转化子,甘油保存。
二、载体构建1将目的基因构建到pPICZα载体上,转化DH5α,用Zeocin筛选转化子。
2提质粒酶切鉴定或PCR鉴定3载体测序测序可用α-Factor引物或5’AOX1引物,3’AOX1引物三、线性化DNA1提取足够量的质粒DNA(一次转化至少需要5-10μg质粒)2 酶切线性化10μg构建好的载体,同时酶切空载体做对照,根据载体选择线性化酶切位点(样品分管酶切),pPICZα载体在5’AOX1区域有三个酶切位点可选择:SacI、PmeI、BstXI3 取1-2μl酶切产物跑电泳,确定是否酶切完全;4 过柱纯化线性化质粒(用50μl EB洗脱);四、线性化DNA的去磷酸化处理线性化质粒43μlCIAP Buffer 5μlCIAP酶2μl四、总体积为50μl的样品37℃ 1h,过柱纯化,用30μl ddH2O 洗脱;五、感受态细胞的制备实验前准备:无抗性YPD平板一个、无抗生素液体YPD培养基,100μg/ml Zeocin YPD 平板和液体、50ml离心管两个、500ml预冷的无菌水、20ml 1M 山梨醇(灭菌预冷的),0.2cm预冷的电击杯;1YPD平板划线培养菌,30℃培养2-3d;250ml三角瓶中,加入5ml YPD,挑取酵母单菌落,30℃培养过夜;3吸取0.5ml菌液,加入至含有200ml新鲜YPD的1L三角瓶中,30℃,225rpm/min培养至OD值1.3-1.5;41500g,4℃离心5min收集菌体;540ml冰预冷的无菌水重悬沉淀;61500g,4℃,5min;730ml无菌水重悬;81500g,4℃,5min;910ml 1M 山梨醇重悬;101500g,4℃,5min;11加入1ml山梨醇,重悬冰上放置,直接做转化,或加入灭菌甘油每管80ul分装,冻存于-80℃(长时间保存会影响转化效率);六、电击转化15-10μg线性化DNA(20μl<)与80ul上述感受态细胞混合,转移至预冷的0.2cm电击杯中(点击条件:电压1.5kV;电容25μF;电阻200Ω,电击时间为4~10msec);2冰上放置5min3电击(按生产厂商提供的适合酵母用的参数)4迅速加入1ml预冷的1M 山梨醇,转移至1.5ml EP管中530℃静置培养1-2h(如果要增加存活率,获得更多的转化克隆,可在30℃静置培养1h后,加入1mlYPD培养基,30℃200rpm培养1h后取部分涂布与不同浓度抗生素的平板)6取50、100、200ul分别涂布于含有Zeocin的YPD平板,30℃培养2-10 d至有菌落出现;7如果要筛选多拷贝转化子,将转化克隆混合在一起,涂布在Zeocin 浓度为500、1000、2000μg/ml的YPD平板,培养2-3d。
phis2质粒序列 -回复

phis2质粒序列-回复「Phis2质粒序列」为主题的文章:解读构建与应用Phis2质粒序列是在分子生物学研究中广泛应用的一种工具,通过插入特定的DNA序列,可以实现基因的表达、克隆和传递。
本文将一步一步回答关于Phis2质粒序列的构建与应用。
第一步:什么是Phis2质粒序列?以及它的构建原理。
Phis2质粒序列是一种含有DNA片段的人工圆形DNA分子。
它由质粒骨架和DNA片段构成,质粒骨架通常包含起始子、终止子和复制起始位点等功能序列,用于表达和复制所插入的DNA片段。
构建Phis2质粒的过程可以通过以下步骤实现:1. 选择合适的质粒载体:通常从已有的质粒中选择骨架进行改造,选择具有较高复制能力、对外源基因稳定的质粒。
常用的载体有pUC、pBR322等。
2. 得到目标DNA片段:使用PCR或其他方法,在受体DNA或基因组中扩增需要插入的DNA片段。
这个片段可以是一个基因、一个启动子、一个编码序列等。
3. 限制酶消化:使用特定的限制酶切割选择的质粒骨架和目标DNA 片段。
4. 连接:将骨架和目标DNA片段通过DNA连接酶的作用连接起来。
连接的过程中可以使用连接接头如AP拼接。
5. 转化至宿主细胞:将构建好的Phis2质粒通过热激转化、电穿孔等方法导入宿主细胞中。
第二步:Phis2质粒序列的应用领域Phis2质粒序列在分子生物学研究中有着广泛的应用。
以下是几个常见的应用领域:1. 基因表达:通过将目标基因插入Phis2质粒中,可以实现外源基因的表达。
在希望大量表达某个蛋白质的研究中,研究人员可以将该蛋白质的编码序列插入Phis2质粒,并将其导入宿主细胞进行表达。
2. 克隆:Phis2质粒构建方法可以用于分子生物学的克隆操作,如基因克隆等。
通过将目标DNA片段插入Phis2质粒中,可以实现序列的复制和扩增,并进一步进行序列分析和功能验证。
3. 细胞传递:Phis2质粒还可用作载体,将目标基因导入到宿主细胞中。
酵母双杂交原理与实验具体流程
galactosidase. MEL1 is endogenous to both Y187 and AH109. Because αgalactosidase is a secreted enzyme, its activity can be detected by adding X-α-Gal to the selection plate: If MEL1 is active and X-α-Gal is present, the colony will turn blue. lacZ in Y187 exhibits a high level of induced β-galactosidase activity in a poAD通过这个“桥 梁”共同起作用,激活报告基 因(ADE2、HIS3 、 lacZ和 MEL1)的转录。
推荐使用Clontech公司的第三 代载体,pGADT7-Rec 和 pGBKT7进行双杂交筛选,因 为它们产生更少的假阳性。对 于cDNATA regions can be switched to create novel promoters
For GAL4-based systems, either a native GAL UAS or a synthetic UASG 17-mer consensus sequence (Heslot & Gaillardin, 1992) provides the binding site for the GAL4 DNA-BD. If you are putting together your own one- or two-hybrid system, you must make sure that the reporter gene's promoter will be recognized by the DNA-BD moiety encoded in your DNA-BD fusion vector.
pPICZ A酵母表达载体说明

pPICZ A编号 载体名称北京华越洋生物VECT2440 pPICZ ApPICZA载体基本信息出品公司: Invitrogen载体名称: pPICZA, p PICZ A质粒类型: 毕赤酵母蛋白表达载体表达水平: 高拷贝启动子: AOX1克隆方法: 多克隆位点,限制性内切酶载体大小: 3329 b p5' 测序引物及序列: 5´ A OX1:5´-‐GACTGGTTCCAATTGACAAGC-‐3´ 3' 测序引物及序列: 3´ A OX1:5´-‐GCAAATGGCATTCTGACATCC-‐3´ 载体标签: C-‐Myc, C-‐His载体抗性: Zeocin 博来霉素筛选标记: HIS4备注: 插入基因是必需包含起始密码子ATG。
产品目录号: -‐-‐稳定性: 稳定 Stable组成型: 组成型 Constitutive病毒/非病毒: 非病毒pPICZA载体质粒图谱和多克隆位点信息pPICZA多克隆位点pPICZA载体简介pPICZ A,B和C是3.3 k b的毕赤酵母蛋白载体。
表达的重组蛋白是融合蛋白,含有一个C-‐端多肽,多太重含c-‐myc和C-‐His标签。
载体能够在毕赤酵母中利用甲醇诱导的高水平的表达目的蛋白,并且可以用在任何毕赤酵母中,包括X33,GS115菌株,SMD1168H,KM71H。
pPICZ系列载体包含以下元素:•5'片段含有AOX1启动子的严格调控,利用甲醇诱导表达任何感兴趣的基因(Ellis等,1985; Koutz等人,1989;tschopp等人,1987A)。
•Zeocin抗性基因在大肠杆菌和毕赤酵母都能用于筛选(Baron等人,1992; D rocourt等人,1990)。
酵母单杂交系统

酵母单杂交系统应用
1. 鉴别DNA结合位点,并发现潜在的结合蛋白基因,目前对于 酵母单杂交技术的应用主要体现在这方面。 Chew et al (1999)应用酵母单杂交技术证实了在大鼠 脑中存在的COUP-TFⅠ、EAR2和NURR1等蛋白质GRIK5基 因的内含子结合蛋白。 2. 对DNA结合结构域进行分析 如果能得到DNA结合结构域的 结构信息,就可以用酵母单杂交技术对该结构进行分析. Mak et al (1996)运用此技术测试哺乳动物具有基本的 螺旋- 环- 螺旋(bHLH)结构的转录因子,通过对肌调节因子 4(MRF4)的研究,证实其具有转录活性。
基本流程
5. 其他 得到的阳性酵母细行测序和序列比对等后续工作
cDNA插入片段
PADH1
GAL4-AD 转化
TADH1
LEU2
酵母转化体 DRE DRE DRE DRE Pmin LacZ URA3
筛选 测序,进一步验证结合活性 挑取阳性克隆
酵母单杂交是在酵母双杂交的基础上,20世纪9,省略了在酵母双杂交系统中采用 的BD-X蛋白质杂交体,而用特异的DNA序列取代DNAGal4结合位 点
酵母单杂交系统原理
将已知的特定顺式作用原件构建到最基本启动子(Pmin) 上游,把报告基因连接到Pmin下游 编码待测转录因子cDNA与已知酵母转录激活结构域(AD) 融合表达载体导入酵母细胞,该基因产物如果能够和顺式作用 原件结合,就能激活Pmin启动子,使报告基因得到表达
将插入目标元件的pHIS2质粒转化Y187酵母后,在不同浓度的3-AT的SD/-His/-Trp平板进 行培养,获得能抑制组氨酸渗漏的最低3-AT浓度,一般不超过45mmol/L
基本流程2. 构建表达 将样品经过一定处理之后(如胁迫)后,提取mRNA, 经过反转录获得cDNA。
酵母表达简介

酵母表达外源蛋白1、菌株用GS115表达不出蛋白,换KM71H后,大部分克隆能表达。
2、温度:在28度和室温下诱导表达,表达水平可能都不低。
3、pH手册上用6.0,pH提高到6.8,不表达的蛋白可能就表达出来。
BMMY 的pH7.0-7.5比较合适。
国内外做的最好的rHSA,最适pH大概5-6左右。
pH3的时候yeast和peptone好像会沉淀的,可以用磷酸和磷酸二氢钾调,具体比例自己去试试。
4、偏爱密码子codon bias一般不是主要的问题,你要表达的蛋白特性才是主要问题,酵母对分子量大(30KD以上),结构复杂(如一些蛋白酶),二硫键含量多的蛋白往往不能有效表达,尤其是分泌表达。
密码子改造对一些较小的而且结构简单的蛋白表达量的提高可能有一些作用。
比如一位战友用Pichia酵母表达一个单链抗体,29KD,含有2对二硫键,表达量约几毫克每升,选用酵母偏好密码子全基因合成后,表达量没有什么提高。
5、表达时间与空质粒转化对照诱导时间长了以后,是会有很多蛋白分泌出来的,时间越长杂蛋白就越多,且分子量都比较大。
最好做一个空质粒转化的对照,这样就会比较肯定到底是不是自身的蛋白分泌的结果。
6、污染每个样品从G418板上挑10个左右单克隆于2ml BMGY摇菌(30ml 玻璃管,比LB管大一点),纱布一般用8层,一天左右看着比较浑离心,留样1ml,余1ml换2ml BMMY诱导表达,3,4层纱布足够了。
污染一般都是跟瓶口覆盖有关的原因造成的,只盖纱布肯定会污染。
加盖报纸后,就再没遇到过污染。
如果只用6层纱布,污染的可能当然很大,100ml三角瓶,装量10ml培养液,用橡筋把8层纱布和2层报纸拴紧封口,空气浴摇床。
7、不表达蛋白有没有表达就要看你的运气了,一般重复2-3次实验都没有表达菌株,这个蛋白就放弃表达了。
8、表达量30KD,10mg/L表达量已经很高,最直接的方法是发酵,一般提高5-10倍。
大肠杆菌一样出现大团的超表达蛋白。
酵母表达载体是什么?

酵母表达载体是什么?
结构组成及表达元件
来⾃pBR322基本⾻架含有该质粒复制起始位点(ori),lacZ基因、bla或ampr、tetr等基因。
含有URA3、HIS3、LEU2、TRPl、LYS2与特定的宿主营养缺陷突变体互补筛选或利⽤zeocin、basticidin(杀稻瘟菌素)的抗性基因筛选。
启动⼦有GAP、AOXl、AUGl和GAL1等。
特点或优点
GAP是组成型启动⼦。
其余是诱导型启动⼦。
酵母表达载体多为穿梭载体:既可以在⼤肠杆菌中繁殖,获得⾜够的载体DNA进⾏体外操作,⼜可以在酵母中复制、表达和选择。
融合蛋⽩表达载体:于外源基因插⼊位点的C端或N端插⼊了1个6×His标签,或在5’端插⼊了α-因⼦作为分泌信号
备注
由于原核⽣物和真核⽣物存在糖基化、酰基化等翻译后修饰反应机制的差异,真核基因的原核表达难以获得有活性的蛋⽩。
酵母成为真核表达的⾸选系统。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
pHis2编号 载体名称北京华越洋生物VECT2110 pHis2pHis2载体基本信息出品公司: Clontech载体名称: pHis2质粒类型: 酵母单杂交载体高拷贝/低拷贝: -‐-‐启动子: -‐-‐克隆方法: 多克隆位点,限制性内切酶载体大小: 7207bp5' 测序引物及序列: -‐-‐3' 测序引物及序列: -‐-‐载体标签: -‐-‐载体抗性: 卡纳霉素筛选标记: -‐-‐备注: 含HIS3报告基因产品目录号: -‐-‐稳定性: -‐-‐组成型: -‐-‐病毒/非病毒: -‐-‐载体质粒图谱和多克隆位点信息载体序列ORIGIN1 G AATTCCCGG G GAGCTCACG C GTTCGCGAA T CGATCCGCG G TCTAGAAAT T CCTGGCATT 61 A TCACATAAT G AATTATACA T TATATAAAG T AATGTGATT T CTTCGAAGA A TATACTAAA121 A AATGAGCAG G CAAGATAAA C GAAGGCAAA G ATGACAGAG C AGAAAGCCC T AGTAAAGCG181 T ATTACAAAT G AAACCAAGA T TCAGATTGC G ATCTCTTTA A AGGGTGGTC C CCTAGCGAT 241 A GAGCACTCG A TCTTCCCAG A AAAAGAGGC A GAAGCAGTA G CAGAACAGG C CACACAATC 301 G CAAGTGATT A ACGTCCACA C AGGTATAGG G TTTCTGGAC C ATATGATAC A TGCTCTGGC 361 C AAGCATTCC G GCTGGTCGC T AATCGTTGA G TGCATTGGT G ACTTACACA T AGACGACCA 421 T CACACCACT G AAGACTGCG G GATTGCTCT C GGTCAAGCT T TTAAAGAGG C CCTAGGGGC 481 C GTGCGTGGA G TAAAAAGGT T TGGATCAGG A TTTGCGCCT T TGGATGAGG C ACTTTCCAG 541 A GCGGTGGTA G ATCTTTCGA A CAGGCCGTA C GCAGTTGTC G AACTTGGTT T GCAAAGGGA 601 G AAAGTAGGA G ATCTCTCTT G CGAGATGAT C CCGCATTTT C TTGAAAGCT T TGCAGAGGC 661 T AGCAGAATT A CCCTCCACG T TGATTGTCT G CGAGGCAAG A ATGATCATC A CCGTAGTGA 721 G AGTGCGTTC A AGGCTCTTG C GGTTGCCAT A AGAGAAGCC A CCTCGCCCA A TGGTACCAA 781 C GATGTTCCC T CCACCAAAG G TGTTCTTAT G TAGTGACAC C GATTATTTA A AGCTGCAGC 841 A TACGATATA T ATACATGTG T ATATATGTA T ACCTATGAA T GTCAGTAAG T ATGTATACG 901 A ACAGTATGA T ACTGAAGAT G ACAAGGTAA T GCATCATTC T ATACGTGTC A TTCTGAACG 961 A GGCGCGCTT T CCTTTTTTC T TTTTGCTTT T TCTTTTTTT T TCTCTTGAA C TCGAGAAAA 1021 A AAATATAAA A GAGATGGAG G AACGGGAAA A AGTTAGTTG T GGTGATAGG T GGCAAGTGG 1081 T ATTCCGTAA G AACAACAAG A AAAGCATTT C ATATTATGG C TGAACTGAG C GAACAAGTG 1141 C AAAATTTAA G CATCAACGA C AACAACGAG A ATGGTTATG T TCCTCCTCA C TTAAGAGGA 1201 A AACCAAGAA G TGCCAGAAA T AACATGAGC A ACTACAATA A CAACAACGG C GGCTACAAC 1261 G GTGGCCGTG G CGGTGGCAG C TTCTTTAGC A ACAACCGTC G TGGTGGTTA C GGCAACGGT 1321 G GTTTCTTCG G TGGAAACAA C GGTGGCAGC A GATCTAACG G CCGTTCTGG T GGTAGATGG 1381 A TCGATGGCA A ACATGTCCC A GCTCCAAGA A ACGAAAAGG C CGAGATCGC C ATATTTGGT 1441 G TCCCCGAGG A TCCTCTACG C CGGACGCAT C GTGGCCGGC A TCACCGGCG C CACAGGTGC 1501 G GTTGCTGGC G CCTATATCG C CGACATCAC C GATGGGGAA G ATCGGGCTC G CCACTTCGG 1561 G CTCATGAGC G CTTGTTTCG G CGTGGGTAT G GTGGCAGGC C CCGTGGCCG G GGGACTGTT 1621 G GGCGCCATC T CCTTGCATG C ACCATTCCT T GCGGCGGCG G TGCTCAACG G CCTCAACCT 1681 A CTACTGGGC T GCTTCCTAA T GCAGGAGTC G CATAAGGGA G AGCGTCGAC C GATGCCCTT 1741 G AGAGCCTTC A ACCCAGTCA G CTCCTTCCG G TGGGCGCGG G GCATGACTA T CGTCGCCGC 1801 A CTTATGACT G TCTTCTTTA T CATGCAACT C GTAGGACAG G TGCCGGCAG C GCTCTGGGT 1861 C ATTTTCGGC G AGGACCGCT T TCGCTGGAG C GCGACGATG A TCGGCCTGT C GCTTGCGGT 1921 A TTCGGAATC T TGCACGCCC T CGCTCAAGC C TTCGTCACT G GTCCCGCCA C CAAACGTTT 1981 C GGCGAGAAG C AGGCCATTA T CGCCGGCAT G GCGGCCGAC G CGCTGGGCT A CGTCTTGCT 2041 G GCGTTCGCG A CGCGAGGCT G GATGGCCTT C CCCATTATG A TTCTTCTCG C TTCCGGCGG 2101 C ATCGGGATG C CCGCGTTGC A GGCCATGCT G TCCAGGCAG G TAGATGACG A CCATCAGGG 2161 A CAGCTTCAA G GATCGCTCG C GGCTCTTAC C AGCCTAACT T CGATCATTG G ACCGCTGAT 2221 C GTCACGGCG A TTTATGCCG C CTCGGCGAG C ACATGGAAC G GGTTGGCAT G GATTGTAGG 2281 C GCCGCCCTA T ACCTTGTCT G CCTCCCCGC G TTGCGTCGC G GTGCATGGA G CCGGGCCAC2341 C TCGACCTGA A TGGAAGCCG G CGGCACCTC G CTAACGGAT T CACCACTCC A AGAATTGGA 2401 G CCAATCAAT T CTTGCGGAG A ACTGTGAAT G CGCAAACCA A CCCTTGGCA G AACATATCC 2461 A TCGCGTCCG C CATCTCCAG C AGCCGCACG C GGCGCATCG G GGGGGGGGT T TCAATTCAA 2521 T TCATCATTT T TTTTTTATT C TTTTTTTTG A TTTCGGTTT C TTTGAAATT T TTTTAACGA 2581 C ATTACTATA T ATATAATAT A GGAAGCATT T AATAGAACA G CATCGTAAT A TATGTGTAC 2641 T TTGCAGTTA T GACGCCAGA T GGCAGTAGT G GAAGATATT C TTTATTGAA A AATAGCTTG 2701 T CACCTTACG T ACAATCTTG A TCCGGAGCT T TTCTTTTTT T GCCGATTAA G AATTAATTC 2761 G GTCGAAAAA A GAAAAGGAG A GGGCCAAGA G GGAGGGCAT T GGTGACTAT T GAGCACGTG2821 A GTATACGTG A TTAAGCACA C AAAGGCAGC T TGGAGTATG T CTGTTATTA A TTTCACAGG 2881 T AGTTCTGGT C CATTGGTGA A AGTTTGCGG C TTGCAGAGC A CAGAGGCCG C AGAATGTGC 2941 T CTAGATTCC G ATGCTGACT T GCTGGGTAT T ATATGTGTG C CCAATAGAA A GAGAACAAT 3001 T GACCCGGTT A TTGCAAGGA A AATTTCAAG T CTTGTAAAA G CATATAAAA A TAGTTCAGG 3061 C ACTCCGAAA T ACTTGGTTG G CGTGTTTCG T AATCAACCT A AGGAGGATG T TTTGGCTCT 3121 G GTCAATGAT T ACGGCATTG A TATCGTCCA A CTGCATGGA G ATGAGTCGT G GCAAGAATA 3181 C CAAGAGTTC C TCGGTTTGC C AGTTATTAA A AGACTCGTA T TTCCAAAAG A CTGCAACAT 3241 A CTACTCAGT G CAGCTTCAC A GAAACCTCA T TCGTTTATT C CCTTGTTTG A TTCAGAAGC 3301 A GGTGGGACA G GTGAACTTT T GGATTGGAA C TCGATTTCT G ACTGGGTTG G AAGGCAAGA 3361 G AGCCCCGAA A GCTTACATT T TATGTTAGC T GGTGGACTG A CGCCAGAAA A TGTTGGTGA 3421 T GCGCTTAGA T TAAATGGCG T TATTGGTGT T GATGTAAGC G GAGGTGTGG A GACAAATGG 3481 T GTAAAAGAC T CTAACAAAA T AGCAAATTT C GTCAAAAAT G CTAAGAAAT A GGTTATTAC 3541 T GAGTAGTAT T TATTTAAGT A TTGTTTGTG C ACTTGCCCT A GAGTCGACC T GCAGGCATG 3601 C AAGCTTTTG T TCCCTTTAG T GAGGGTTAA T TTCGAGCTT G GCGTAATCA T GGTCATAGC 3661 T GTTTCCTGT G TGAAATTGT T ATCCGCTCA C AATTCCACA C AACATACGA G CCGGAAGCA 3721 T AAAGTGTAA A GCCTGGGGT G CCTAATGAG T GAGCTAACT C ACATTAATT G CGTTGCGCT 3781 C ACTGCCCGC T TTCCAGTCG G GAAACCTGT C GTGCCAGCT G CATTAATGA A TCGGCCAAC 3841 G CGCGGGGAG A GGCGGTTTG C GTATTGGGC G CTCTTCCGC T TCCTCGCTC A CTGACTCGC 3901 T GCGCTCGGT C GTTCGGCTG C GGCGAGCGG T ATCAGCTCA C TCAAAGGCG G TAATACGGT 3961 T ATCCACAGA A TCAGGGGAT A ACGCAGGAA A GAACATGTG A GCAAAAGGC C AGCAAAAGG 4021 C CAGGAACCG T AAAAAGGCC G CGTTGCTGG C GTTTTTCCA T AGGCTCCGC C CCCCTGACG 4081 A GCATCACAA A AATCGACGC T CAAGTCAGA G GTGGCGAAA C CCGACAGGA C TATAAAGAT 4141 A CCAGGCGTT T CCCCCTGGA A GCTCCCTCG T GCGCTCTCC T GTTCCGACC C TGCCGCTTA 4201 C CGGATACCT G TCCGCCTTT C TCCCTTCGG G AAGCGTGGC G CTTTCTCAT A GCTCACGCT 4261 G TAGGTATCT C AGTTCGGTG T AGGTCGTTC G CTCCAAGCT G GGCTGTGTG C ACGAACCCC 4321 C CGTTCAGCC C GACCGCTGC G CCTTATCCG G TAACTATCG T CTTGAGTCC A ACCCGGTAA 4381 G ACACGACTT A TCGCCACTG G CAGCAGCCA C TGGTAACAG G ATTAGCAGA G CGAGGTATG 4441 T AGGCGGTGC T ACAGAGTTC T TGAAGTGGT G GCCTAACTA C GGCTACACT A GAAGGACAG 4501 T ATTTGGTAT C TGCGCTCTG C TGAAGCCAG T TACCTTCGG A AAAAGAGTT G GTAGCTCTT 4561 G ATCCGGCAA A CAAACCACC G CTGGTAGCG G TGGTTTTTT T GTTTGCAAG C AGCAGATTA 4621 C GCGCAGAAA A AAAGGATCT C AAGAAGATC C TTTGATCTT T TCTACGGGG T CTGACGCTC 4681 A GTGGAACGA A AACTCACGT T AAGGGATTT T GGTCATGAG A TTATCAAAA A GGATCTTCA 4741 C CTAGATCCT T TTAAATTAA A AATGAAGTT T TAAATCAAT C TAAAGTATA T ATGAGTAAA 4801 C TTGGTCTGA C AGTCAGAAG A ACTCGTCAA G AAGGCGATA G AAGGCGATG C GCTGCGAAT 4861 C GGGAGCGGC G ATACCGTAA A GCACGAGGA A GCGGTCAGC C CATTCGCCG C CAAGCTCTT 4921 C AGCAATATC A CGGGTAGCC A ACGCTATGT C CTGATAGCG G TCCGCCACA C CCAGCCGGC 4981 C ACAGTCGAT G AATCCAGAA A AGCGGCCAT T TTCCACCAT G ATATTCGGC A AGCAGGCAT 5041 C GTCATGGGT C ACGACGAGA T CCTCGCCGT C GGGCATGCT C GCCTTGAGC C TGGCGAACA 5101 G TTCGGCTGG C GCGAGCCCC T GATGCTCTT C GTCCAGATC A TCCTGATCG A CAAGACCGG 5161 C TTCCATCCG A GTACGTGCT C GCTCGATGC G ATGTTTCGC T TGGTGGTCG A ATGGGCAGG 5221 T AGCCGGATC A AGCGTATGC A GCCGCCGCA T TGCATCAGC C ATGATGGAT A CTTTCTCGG 5281 C AGGAGCAAG G TGAGATGAC A GGAGATCCT G CCCCGGCAC T TCGCCCAAT A GCAGCCAGT 5341 C CCTTCCCGC T TCAGTGACA A CGTCGAGCA C AGCTGCGCA A GGAACGCCC G TCGTGGCCA 5401 G CCACGATAG C CGCGCTGCC T CGTCTTGCA G TTCATTCAG G GCACCGGAC A GGTCGGTCT5461 T GACAAAAAG A ACCGGGCGC C CCTGCGCTG A CAGCCGGAA C ACGGCGGCA T CAGAGCAGC 5521 C GATTGTCTG T TGTGCCCAG T CATAGCCGA A TAGCCTCTC C ACCCAAGCG G CCGGAGAAC 5581 C TGCGTGCAA T CCATCTTGT T CAATCATAC T CTTCCTTTT T CAATATTAT T GAAGCATTT 5641 A TCAGGGTTA T TGTCTCATG A GCGGATACA T ATTTGAATG T ATTTAGAAA A ATAAACAAA 5701 T AGGGGTTCC G CGCACATTT C CCCGAAAAG T GCCACCTGC G GACGGATCG C TTGCCTGTA 5761 A CTTACACGC G CCTCGTATC T TTTAATGAT G GAATAATTT G GGAATTTAC T CTGTGTTTA 5821 T TTATTTTTA T GTTTTGTAT T TGGATTTTA G AAAGTAAAT A AAGAGTAGA A GAGTTACGG 5881 A ATGAAGAAA A AAAAATAAA C AAAGGTTTA A AAAATTTCA A CAAAAAGCG T ACTTTACAT 5941 A TATATTTAT T AGACAAGAA A AGCAGATTA A ATAGATATA C ATTCGATTA A CGATAAGTA 6001 A AATGTAAAA T CACAGGATT T TCGTGTGTG G TCTTCTACA C AGACAAGAT G AAACAATTC 6061 G GCATTAATA C CTGAGAGCA G GAAGAGCAA G ATAAAAGGN A GTATTTGTT G GCGATCCCC 6121 C TAGAGTCTT T TACATCTTC G GAAAACAAA A ACTATTTTT T CTTTAATTT C TTTTTTTAC 6181 T TTCTATTTT T AATTTATAT A TTTATATTA A AAAATTTAA A TTATAATTA T TTTTATAGC 6241 A CGTGATGAA A AGGACCGAC G TCTAAGAAA C CATTATTAT C ATGACATTA A CCTATAAAA 6301 A TAGGCGTAT C ACGAGGCCC T TTCGTCTCG C GCGTTTCGG T GATGACGGT G AAAACCTCT 6361 G ACACATGCA G CTCCCGGAG A CGGTCACAG C TTGTCTGTA A GCGGATGCC G GGAGCAGAC 6421 A AGCCCGTCA G GGCGCGTCA G CGGGTGTTG G CGGGTGTCG G GGCTGGCTT A ACTATGCGG 6481 C ATCAGAGCA G ATTGTACTG A GAGTGCACC A TATGCGGTG T GAAATACCG C ACAGATGCG 6541 T AAGGAGAAA A TACCGCATC A GGAAATTGT A AACGTTAAT A TTTTGTTAA A ATTCGCGTT 6601 A AATTTTTGT T AAATCAGCT C ATTTTTTAA C CAATAGGCC G AAATCGGCA A AATCCCTTA 6661 T AAATCAAAA G AATAGACCG A GATAGGGTT G AGTGTTGTT C CAGTTTGGA A CAAGAGTCC 6721 A CTATTAAAG A ACGTGGACT C CAACGTCAA A GGGCGAAAA A CCGTCTATC A GGGCGATGG 6781 C CCACTACGT G AACCATCAC C CTAATCAAG T TTTTTGGGG T CGAGGTGCC G TAAAGCACT 6841 A AATCGGAAC C CTAAAGGGA G CCCCCGATT T AGAGCTTGA C GGGGAAAGC C GGCGAACGT 6901 G GCGAGAAAG G AAGGGAAGA A AGCGAAAGG A GCGGGCGCT A GGGCGCTGG C AAGTGTAGC 6961 G GTCACGCTG C GCGTAACCA C CACACCCGC C GCGCTTAAT G CGCCGCTAC A GGGCGCGTC 7021 G CGCCATTCG C CATTCAGGC T GCGCAACTG T TGGGAAGGG C GATCGGTGC G GGCCTCTTC 7081 G CTATTACGC C AGCTGGCGA A AGGGGGATG T GCTGCAAGG C GATTAAGTT G GGTAACGCC 7141 A GGGTTTTCC C AGTCACGAC G TTGTAAAAC G ACGGCCAGT G AATTGTAAT A CGACTCACT 7201 A TAGGGC//其他酵母表达载体:p416GFD pPIC9p53blue pPIC9KpACT2-AD pPIC9k-HispAD-GAL4-2.1 pPICZApADH2 pPICZBpAUR123 pPICZCpBridge pPICZαApCL1 pPICZαBpDEST32 pPICZαCpDisplay pPICZαDpDR195 pPICZαFCpESC-His pPICZαGB pESC-Leu pPink-HC pESC-TRP pPink-LC pESC-URA pPinkα-HC pFA6a-FGP(S65T)-kanMX6 pRS316 pFLD pRS403 pFLD/CAT pRS405 pFLDαpRS406 pGADT7-T pRS414 pGAG424 pRS415 pGAPZA pRS416 pGAPZB pRS41H pGAPZαB pRS426 pGAPZαC pRS426gal pGBKT7 pSEP1 pGBKT7-53 pSEP2 pGBKT7-Lam pSEP3pHIC-PI pSospHIL-D2 pSos-MAFB pHIL-S1 pUG66pHis2 pYC2/CT pHisSi-1 pYC2/NTA pMETA pYC2/NTB pMETB pYCP211 pMETC pYEPlac112 pMETαA pYEPlac195 pMETαB pYES2pMETαC pYES2-EGFP pMyr pYES2-kan pPIC3.5 pYES2-NTA pPIC3.5K pYES2-NTB pPIC6B pYES2-NTC pPIC6C pYES3/CT pPIC6αA pYES6/CT pPIC6αB pYES-DEST52 pPIC6αC pYIP211 pYX212 pYIP5 SUMOprotease pYRP7Ycp22lac-EGFP Ycplac33。
