pAcGFP1-N In-Fusion Ready哺乳动物表达载体说明
pEF1α-IRES-DsRed-Express2哺乳动物表达载体说明

GTTAGGCCAG TCTTGGTTCA TCGTGACGCT CGGTACCGCG AGCCGCTTGG TCTTTTGGCA GGTCTTTCCC CCTCTGGAAG CCCCCACCTG AAGGCGGCAC CTCTCCTCAA GGATCTGATC CGTCTAGGCC GGCCACAACC CATGGAGGGC CTACGAGGGC CTGGGACATC CGACATCCCC GAACTTCGAG CTTCATCTAC GAAGAAGACT GAAGGGCGAG CAAGTCAATC CAAGCTGGAC CGAGGCCCGC CCACATTTGT AACATAAAAT AATAAAGCAA GTGGTTTGTC TTAAAATTCG GGCAAAATCC TGGAACAAGA TATCAGGGCG TGCCGTAAAG AAGCCGGCGA CTGGCAAGTG CTACAGGGCG TTTTTCTAAA CAATAATATT AGTTAGGGTG TCAATTAGTC AAAGCATGCA CCCTAACTCC ATGCAGAGGC
GGAGACTGAA TGAGTTTGGA ATTTCAGGTG CTGCAGTCGA TACTGGCCGA CATATTGCCG CATTCCTAGG GGAAGCAGTT GCAGCGGAAC TACACCTGCA AGTCAAATGG CCATTGTATG GTTAAAAAAA ATGATAATAT TCAAGGTGCA AGGGCAAGCC TGCCCTTCGC AGCACCCCGC AGCGCGTGAT AGGACGGCAC CCGTAATGCA ACGGCGTGCT TGGTGGAGTT ACGTGGACTC ACGAGCGCGC ATCAGCCATA CTGAACCTGA AATGGTTACA CATTCTAGTT TAATATTTTG GGCCGAAATC TGTTCCAGTT AAAAACCGTC GGGGTCGAGG TTGACGGGGA CGCTAGGGCG TAATGCGCCG TATTTGTTTA ATAAATGCTT AATGTGTGTC AGCATGCATC AGAAGTATGC CCCATCCCGC TTTTTTATTT
pNFkB-DD-AmCyan1哺乳动物表达载体说明
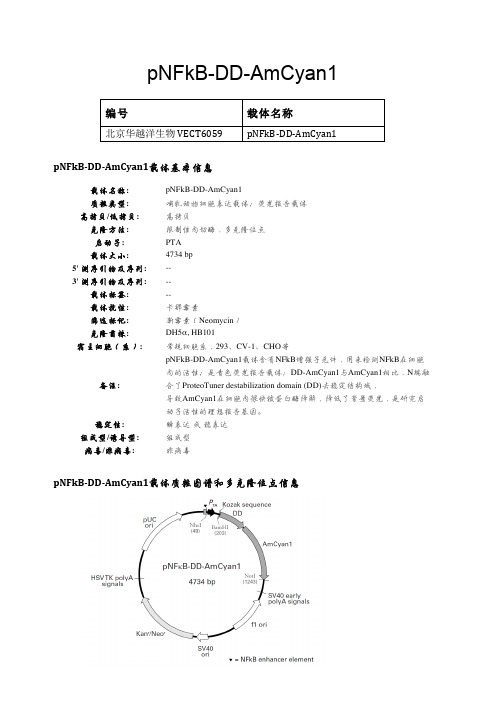
pNFkB-DD-AmCyan1编号载体名称北京华越洋生物VECT6059pNFkB-‐DD-‐AmCyan1pNFkB-‐DD-‐AmCyan1载体基本信息载体名称: pNFkB-DD-AmCyan1质粒类型: 哺乳动物细胞表达载体;荧光报告载体 高拷贝/低拷贝: 高拷贝克隆方法: 限制性内切酶,多克隆位点 启动子: PTA 载体大小: 4734 bp 5' 测序引物及序列: -- 3' 测序引物及序列:-- 载体标签: -- 载体抗性: 卡那霉素筛选标记: 新霉素(Neomycin ) 克隆菌株: DH5α, HB101宿主细胞(系):常规细胞系,293、CV-1、CHO 等备注: pNFkB-DD-AmCyan1载体含有NFkB 增强子元件,用来检测NFkB 在细胞内的活性;是青色荧光报告载体;DD-AmCyan1与AmCyan1相比,N 端融合了ProteoTuner destabilization domain (DD)去稳定结构域,导致AmCyan1在细胞内很快被蛋白酶降解,降低了背景荧光,是研究启动子活性的理想报告基因。
稳定性: 瞬表达 或 稳表达 组成型/诱导型: 组成型 病毒/非病毒:非病毒pNFkB-‐DD-‐AmCyan1载体质粒图谱和多克隆位点信息pNFkB-DD-AmCyan1载体描述pNFkB-DD-AmCyan1 is a reporter vector that allows you to monitor NFkB activation in mammalian cells. The vector contains an NFkB enhancer element (composed of four tandem copies of the NFkB consensus binding sequence; 1) upstream of a minimal TA promoter (PTA), which consists of the TATA box from the herpes simplex virus thymidine kinase (HSV-TK) promoter. The vector encodes the reporter protein DD-AmCyan1, a ligand-dependent, destabilized cyan fluorescent protein that minimizes background fluorescence from leaky promoters.AmCyan1 is a human codon-optimized variant of the wild-type Anemonia majano cyan fluorescent protein (AmCyan) that exhibits enhanced emission characteristics (excitation and emission maxima: 458 and 489, respectively; 2, 3). DD-AmCyan1 is a modified version of AmCyan1 that is tagged on its N-terminus with the ProteoTuner destabilization domain (DD; 4). The presence of this destabilization domain causes rapid, proteasomal degradation of the fluorescent fusion protein; however, when the membrane permeant ligand Shield1 is added to the medium, it binds to the destabilization domain and protects the fusion protein from degradation.In the absence of Shield1, the destabilization domain causes the degradation of any DD-AmCyan1 reporter protein produced prior to promoter activation, thus reducing background fluorescence. In order to analyze NFκB activation, an inducer of choice is added to the medium along with the Shield1 stabilizing ligand, which effectively stabilizes the reporter protein, allowing it to accumulate. As a result, only the reporter molecules expressed during promoter induction will contribute to the fluorescence signal, providing a considerably higher signal-to-noise ratio than thatobtained with non-destabilized or constitutively destabilized reporter systems. The high signal-to-noise ratio also allows the monitoring of NFκB activation during discrete windows of time when Shield1 is added to the cell medium for discrete periods of time.The vector backbone contains an SV40 origin for replication in mammalian cells expressing the SV40 large T antigen, a pUC origin of replication for propagation in E. coli, and an f1 origin for single-stranded DNA production. A neomycin-resistance cassette (Neor) allows stably transfected eukaryotic cells to be selected using G418 (5). This cassette consists of the SV40 early promoter, a Tn5 kanamycin/neomycin resistance gene, and herpes simplex virus thymidine kinase (HSV TK) polyadenylation signals. A bacterial promoter upstream of the cassette expresses kanamycin resistance in E. coli.UseThe pNFkB-DD-AmCyan1 Reporter vector, available as part of the NFkB DD Cyan1 Reporter System (Cat. No. 631083), can be used to monitor NFkB activation in live cells as well as in vivo. pNFkB-DD-AmCyan1 Reporter can be transfected into mammalian cells using any standard transfection method. If required, stable transfectants can be selected using G418.Propagation in E. coliRecommended host strains: DH5α, HB101, and other general purpose strains. Single-stranded DNA productionrequires a host containing an F plasmid such as JM109 or XL1-Blue.Selectable marker: plasmid confers resistance to kanamycin (50 µg/ml) in E. coli hosts.E. coli replication origin: pUCCopy number: highPlasmid incompatibility group: pMB1/ColE1Excitation and emission maxima of AmCyan1Excitation maximum = 458 nmEmission maximum = 489 nm其他哺乳动物表达载体:pCHO1.0 pBApo-CMV-Pur pOPRSVIpcDNA3.1/His C pcDNA5/FRT/V5-His-TOPO pREP4pcDNA3.1/His B pcDNA5/FRT/TO-TOPO pDual-GCpcDNA3.1/His A pcDNA5/TO pBK-RSVpIRESpuro3 pcDNA5/FRT/TO pBK-CMVpIRES2-EGFP pcDNA5/FRT pBI-CMV4pTT5 pFLAG-CMV2 pcDNA4/TO/Myc-His/LacZ pNFkB-DD-tdTomato pcDNA3.1/CT-GFP-TOPO pOPI3CATpBI-CMV5 pcDNA3.1/NT-GFP-TOPO pGene/V5-His B pSEAP2-Basic pOptiVEC-TOPO pSwitchpSEAP2-Control pCMV-MEKK1 pCMVLacIpBI-CMV3 pCMV-MEK1 pVgRxRpBI-CMV2 pCMV-PKA pINDpBI-CMV1 pcDNA6.2/nTC-Tag-DEST pTRE3G-LucpNFκB-MetLuc2-Reporter pcDNA6.2/cTC-Tag-DEST pTRE3GpCRE-MetLuc2-Reporter pcDNA3.2/V5/GW/D-TOPO pTRE2-hygropAcGFP1-Actin pcDNA6.2/V5/GW/D-TOPO pTRE-TightpAcGFP1-N In-Fusion Ready pcDNA6.2/nGeneBLAzer-GW/D-TOPO pTK-hygpAcGFP1-C3 pcDNA6.2/C-YFP-DEST pTet-OnpAcGFP1-C pcDNA6.2/cGeneBLAzer-DEST pTet-OffpAcGFP1-p53 pcDNA6/V5-His A pTet on advanced pAcGFP1-Mito pcDNA6/V5-His B pRevTREpAcGFP1-Mem pcDNA6/V5-His C pRevTet-OnpAcGFP1-Lam pcDNA6/myc-His C pRevTet-OffpAcGFP1-Golgi pcDNA6/myc-His A pCMV-Tet3GpAcGFP1-F pcDNA6/myc-His B pTRE2pAcGFP1-Hyg-C1 pcDNA6.2/nGeneBLAzer-DEST pBD-NF-κBpAsRed2-N1 pcDNA4/HisMax-TOPO pCMV-ADptdTomato-N1 pcDNA6.2/nLumio-DEST pCMV-BDpCMV-tdTomato pcDNA6.2/cLumio-DEST pBIND-Id ControlpCRE-DD-tdTomato pcDNA4/myc-His C pBINDpCMV-DsRed-Express2 pcDNA4/HisMax C pG5 luciferasepEF1α-tdTomato pcDNA4/HisMax A pACT-MyoDpCRE-hrGFP c-Flag pcDNA3 pACTptdTomato-C1 pcDNA4/HisMax B pCMV-SPORT6 pAsRed2-C1 pcDNA4/myc-His B pGL4.13pGL3-Promoter pcDNA4/myc-His A pGL4.19pGL3 basic pcDNA4/His C pGL4.26pAcGFP1-C2 pcDNA4/His B pGL4.20pAcGFP1-C1 pcDNA4/His A pGL4.29pAcGFP1-N3 pcDNA6/TR pGL4.30pAcGFP1-N2 pcDNA4/TO/Myc-His A pGL4.27pAcGFP1-N1 pcDNA4/TO pGL4.75pAcGFP1-C In-Fusion Ready pcDNA4/TO/Myc-His B pGL4.10pCRE-DD-AmCyan1 pcDNA4/TO/Myc-His C pGRN145pNFkB-DD-AmCyan1 pcDNA3.3-TOPO pSecTag2 A pDsRed2-Bid pBudCE4.1 pEBVHis B pDsRED2-Mito pFLAG-CMV-4 pEBVHis ApDD-AmCyan1 Reporter pFLAG-CMV-3 pCMV-Tag 3C pAmCyan1-N1 pFLAG-CMV-2 pCMV-Tag 3A pAmCyan1-C1 pFLAG-CMV-5a pCMV-Tag 3BpEF1α-IRES-DsRed-Express2 p3XFLAG-CMV-9 pCMV-Tag 5CpEF1α-DsRed-Monomer-N1 p3xFLAG-CMV-10 pCMV-Tag 5A pDsRED-Monomer-N1 p3XFLAG-CMV-8 pCMV-Tag 4A pDsRed-Express-N1 p3XFLAG-CMV-7.1 pCMV-Tag 5Bp3XFLAG-CMV-7 pDsRed-Monomer-N In-Fusion Ready pCMV-Tag 4B pDsRed-Express-C1 p3XFLAG-CMV-13 pCMV-Tag 2C pIRES2-ZsGreen1 p3XFLAG-CMV-14 pCMV-Tag 2B pDsRed-Express2-C1 plRES2-ZsGreen1 pCMV-Tag 2A pDsRed-Express2-N1 pBApo-EF1α-pur pCMV-LacZpEF1α-DsRed-Express2 pBApo-EF1α-neo pCMV-MycpIRES2-DsRed-Express pBApo-CMV pEF1α-IRES-AcGFP1 pIRES2-DsRed-Express2 pBApo-CMV-neo pEF1α-IRES-ZsGreen1 pIRES-hrGFP-1a pIRES-EGFP pEF1α-AcGFP1-N1 pIRESneo2 pIRESneo3 pIRES2-DsRed2 pIRESneo pDsRed-Monomer pIRES2-AcGFP1 pIREShyg3 pIRES。
pcDNA4 myc-His C哺乳动物表达载体说明

pcDNA4/myc-His C编号 载体名称北京华越洋生物VECT6123 pcDNA4/myc-‐His C pcDNA4/myc-‐His C载体基本信息载体名称: pcDNA4/myc-His C质粒类型: 哺乳动物表达载体;cDNA表达载体高拷贝/低拷贝: 高拷贝克隆方法: 多克隆位点,限制性内切酶启动子: CMV载体大小: 5071 bp5' 测序引物及序列: T7 Forward: 5’-TAATACGACTCACTATAGGG-3’ 3' 测序引物及序列: BGH Reverse: 5-TAGAAGGCACAGTCGAGG-3 载体标签: His Tag (C-端), c-Myc Epitope Tag(C-端)载体抗性: 氨苄青霉素筛选标记: Zeocin克隆菌株: TOP10F´, DH5a, JM109, TOP10宿主细胞(系): 常规细胞系,如293、Hela等备注: pcDNA4/myc-His C 载体是哺乳动物表达载体,适用于cDNA的表达与克隆;CMV启动子驱动目的基因的高水平表达;pcDNA4/myc-His A,B,C的区别仅在于多克隆位点处。
稳定性: 瞬表达或稳表达组成型/诱导型: 组成型病毒/非病毒: 非病毒pcDNA4/myc-‐His C载体质粒图谱和多克隆位点信息pcDNA4/myc-‐His C载体描述pcDNA4/myc-His A, B, and C are 5.1 kb vectors designed for overproduction of recombinant proteins in mammalian cell lines. Features of the vectors allow purification and detection of expressed proteins (see pages 11-12 for more information). High-level stable and transient expression can be carried out in most mammalian cells. The vectors contain the following elements:Human cytomegalovirus immediate-early (CMV) promoter for high-level expression in a wide range of mammalian cellsThree reading frames to facilitate in-frame cloning with a C-terminal peptide encoding the myc (c-myc) epitope and a polyhistidine (6xHis) metal-binding tagZeocin resistance gene for selection of stable cell lines (Mulsant et al., 1988) (see page 14 for more information).Episomal replication in cell lines that are latently infected with SV40 or that express the SV40 large T antigen (e.g., COS7).The control plasmid, pcDNA4/myc-His/lacZ is included for use as a positive control for transfection, expression, and detection in the cell line of choice.实验流程:Use the following outline to clone and express your gene of interest in pcDNA4/myc-His:1.Consult the multiple cloning sites described on pages 3-4 to determine which vector (A, B, or C) to use for cloning your gene in frame with the C-terminal myc epitope and the polyhistidine tag.2.Ligate your insert into the appropriate vector and transform into E. coli. Select transformants on 50 to 100 μg/mL ampicillin or 25 to 50g/mL Zeocin in Low Salt LB. For more information.3.Analyze your transformants for the presence of insert by restriction digestion.4.Select a transformant with the correct restriction pattern and use sequencing to confirm that your gene is cloned in-frame with the C-terminal peptide.5.Transfect your construct into the cell line of choice using your own method of transfection. Generate a stable cell line, if desired.6.Test for expression of your recombinant gene by western blot analysis or functional assay. For antibodies to the myc epitope or the C-terminal polyhistidine tag.7.To purify your recombinant protein, you may use metal-chelating resin such as ProBond. ProBond resin is available separatelypcDNA4/myc-‐His C载体序列ORIGIN1 GACGGATCGG GAGATCTCCC GATCCCCTAT GGTCGACTCT CAGTACAATC TGCTCTGATG61 CCGCATAGTT AAGCCAGTAT CTGCTCCCTG CTTGTGTGTT GGAGGTCGCT GAGTAGTGCG 121 CGAGCAAAAT TTAAGCTACA ACAAGGCAAG GCTTGACCGA CAATTGCATG AAGAATCTGC 181 TTAGGGTTAG GCGTTTTGCG CTGCTTCGCG ATGTACGGGC CAGATATACG CGTTGACATT 241 GATTATTGAC TAGTTATTAA TAGTAATCAA TTACGGGGTC ATTAGTTCAT AGCCCATATA 301 TGGAGTTCCG CGTTACATAA CTTACGGTAA ATGGCCCGCC TGGCTGACCG CCCAACGACC 361 CCCGCCCATT GACGTCAATA ATGACGTATG TTCCCATAGT AACGCCAATA GGGACTTTCC 421 ATTGACGTCA ATGGGTGGAC TATTTACGGT AAACTGCCCA CTTGGCAGTA CATCAAGTGT 481 ATCATATGCC AAGTACGCCC CCTATTGACG TCAATGACGG TAAATGGCCC GCCTGGCATT 541 ATGCCCAGTA CATGACCTTA TGGGACTTTC CTACTTGGCA GTACATCTAC GTATTAGTCA 601 TCGCTATTAC CATGGTGATG CGGTTTTGGC AGTACATCAA TGGGCGTGGA TAGCGGTTTG 661 ACTCACGGGG ATTTCCAAGT CTCCACCCCA TTGACGTCAA TGGGAGTTTG TTTTGGCACC 721 AAAATCAACG GGACTTTCCA AAATGTCGTA ACAACTCCGC CCCATTGACG CAAATGGGCG 781 GTAGGCGTGT ACGGTGGGAG GTCTATATAA GCAGAGCTCT CTGGCTAACT AGAGAACCCA 841 CTGCTTACTG GCTTATCGAA ATTAATACGA CTCACTATAG GGAGACCCAA GCTGGCTAGT 901 TAAGCTTGGT ACCGAGCTCG GATCCACTAG TCCAGTGTGG TGGAATTCTG CAGATATCCA 961 GCACAGTGGC GGCCGCTCGA GGTCACCCAT TCGAACAAAA ACTCATCTCA GAAGAGGATC 1021 TGAATATGCA TACCGGTCAT CATCACCATC ACCATTGAGT TTAAACCCGC TGATCAGCCT 1081 CGACTGTGCC TTCTAGTTGC CAGCCATCTG TTGTTTGCCC CTCCCCCGTG CCTTCCTTGA 1141 CCCTGGAAGG TGCCACTCCC ACTGTCCTTT CCTAATAAAA TGAGGAAATT GCATCGCATT 1201 GTCTGAGTAG GTGTCATTCT ATTCTGGGGG GTGGGGTGGG GCAGGACAGC AAGGGGGAGG 1261 ATTGGGAAGA CAATAGCAGG CATGCTGGGG ATGCGGTGGG CTCTATGGCT TCTGAGGCGG 1321 AAAGAACCAG CTGGGGCTCT AGGGGGTATC CCCACGCGCC CTGTAGCGGC GCATTAAGCG 1381 CGGCGGGTGT GGTGGTTACG CGCAGCGTGA CCGCTACACT TGCCAGCGCC CTAGCGCCCG 1441 CTCCTTTCGC TTTCTTCCCT TCCTTTCTCG CCACGTTCGC CGGCTTTCCC CGTCAAGCTC 1501 TAAATCGGGG CATCCCTTTA GGGTTCCGAT TTAGTGCTTT ACGGCACCTC GACCCCAAAA 1561 AACTTGATTA GGGTGATGGT TCACGTAGTG GGCCATCGCC CTGATAGACG GTTTTTCGCC 1621 CTTTGACGTT GGAGTCCACG TTCTTTAATA GTGGACTCTT GTTCCAAACT GGAACAACAC 1681 TCAACCCTAT CTCGGTCTAT TCTTTTGATT TATAAGGGAT TTTGGGGATT TCGGCCTATT 1741 GGTTAAAAAA TGAGCTGATT TAACAAAAAT TTAACGCGAA TTAATTCTGT GGAATGTGTG 1801 TCAGTTAGGG TGTGGAAAGT CCCCAGGCTC CCCAGGCAGG CAGAAGTATG CAAAGCATGC 1861 ATCTCAATTA GTCAGCAACC AGGTGTGGAA AGTCCCCAGG CTCCCCAGCA GGCAGAAGTA 1921 TGCAAAGCAT GCATCTCAAT TAGTCAGCAA CCATAGTCCC GCCCCTAACT CCGCCCATCC 1981 CGCCCCTAAC TCCGCCCAGT TCCGCCCATT CTCCGCCCCA TGGCTGACTA ATTTTTTTTA 2041 TTTATGCAGA GGCCGAGGCC GCCTCTGCCT CTGAGCTATT CCAGAAGTAG TGAGGAGGCT 2101 TTTTTGGAGG CCTAGGCTTT TGCAAAAAGC TCCCGGGAGC TTGTATATCC ATTTTCGGAT 2161 CTGATCAGCA CGTGTTGACA ATTAATCATC GGCATAGTAT ATCGGCATAG TATAATACGA 2221 CAAGGTGAGG AACTAAACCA TGGCCAAGTT GACCAGTGCC GTTCCGGTGC TCACCGCGCG 2281 CGACGTCGCC GGAGCGGTCG AGTTCTGGAC CGACCGGCTC GGGTTCTCCC GGGACTTCGT 2341 GGAGGACGAC TTCGCCGGTG TGGTCCGGGA CGACGTGACC CTGTTCATCA GCGCGGTCCA 2401 GGACCAGGTG GTGCCGGACA ACACCCTGGC CTGGGTGTGG GTGCGCGGCC TGGACGAGCT 2461 GTACGCCGAG TGGTCGGAGG TCGTGTCCAC GAACTTCCGG GACGCCTCCG GGCCGGCCAT 2521 GACCGAGATC GGCGAGCAGC CGTGGGGGCG GGAGTTCGCC CTGCGCGACC CGGCCGGCAA 2581 CTGCGTGCAC TTCGTGGCCG AGGAGCAGGA CTGACACGTG CTACGAGATT TCGATTCCAC 2641 CGCCGCCTTC TATGAAAGGT TGGGCTTCGG AATCGTTTTC CGGGACGCCG GCTGGATGAT2701 CCTCCAGCGC GGGGATCTCA TGCTGGAGTT CTTCGCCCAC CCCAACTTGT TTATTGCAGC 2761 TTATAATGGT TACAAATAAA GCAATAGCAT CACAAATTTC ACAAATAAAG CATTTTTTTC 2821 ACTGCATTCT AGTTGTGGTT TGTCCAAACT CATCAATGTA TCTTATCATG TCTGTATACC 2881 GTCGACCTCT AGCTAGAGCT TGGCGTAATC ATGGTCATAG CTGTTTCCTG TGTGAAATTG 2941 TTATCCGCTC ACAATTCCAC ACAACATACG AGCCGGAAGC ATAAAGTGTA AAGCCTGGGG 3001 TGCCTAATGA GTGAGCTAAC TCACATTAAT TGCGTTGCGC TCACTGCCCG CTTTCCAGTC 3061 GGGAAACCTG TCGTGCCAGC TGCATTAATG AATCGGCCAA CGCGCGGGGA GAGGCGGTTT 3121 GCGTATTGGG CGCTCTTCCG CTTCCTCGCT CACTGACTCG CTGCGCTCGG TCGTTCGGCT 3181 GCGGCGAGCG GTATCAGCTC ACTCAAAGGC GGTAATACGG TTATCCACAG AATCAGGGGA 3241 TAACGCAGGA AAGAACATGT GAGCAAAAGG CCAGCAAAAG GCCAGGAACC GTAAAAAGGC 3301 CGCGTTGCTG GCGTTTTTCC ATAGGCTCCG CCCCCCTGAC GAGCATCACA AAAATCGACG 3361 CTCAAGTCAG AGGTGGCGAA ACCCGACAGG ACTATAAAGA TACCAGGCGT TTCCCCCTGG 3421 AAGCTCCCTC GTGCGCTCTC CTGTTCCGAC CCTGCCGCTT ACCGGATACC TGTCCGCCTT 3481 TCTCCCTTCG GGAAGCGTGG CGCTTTCTCA ATGCTCACGC TGTAGGTATC TCAGTTCGGT 3541 GTAGGTCGTT CGCTCCAAGC TGGGCTGTGT GCACGAACCC CCCGTTCAGC CCGACCGCTG 3601 CGCCTTATCC GGTAACTATC GTCTTGAGTC CAACCCGGTA AGACACGACT TATCGCCACT 3661 GGCAGCAGCC ACTGGTAACA GGATTAGCAG AGCGAGGTAT GTAGGCGGTG CTACAGAGTT 3721 CTTGAAGTGG TGGCCTAACT ACGGCTACAC TAGAAGGACA GTATTTGGTA TCTGCGCTCT 3781 GCTGAAGCCA GTTACCTTCG GAAAAAGAGT TGGTAGCTCT TGATCCGGCA AACAAACCAC 3841 CGCTGGTAGC GGTGGTTTTT TTGTTTGCAA GCAGCAGATT ACGCGCAGAA AAAAAGGATC 3901 TCAAGAAGAT CCTTTGATCT TTTCTACGGG GTCTGACGCT CAGTGGAACG AAAACTCACG 3961 TTAAGGGATT TTGGTCATGA GATTATCAAA AAGGATCTTC ACCTAGATCC TTTTAAATTA 4021 AAAATGAAGT TTTAAATCAA TCTAAAGTAT ATATGAGTAA ACTTGGTCTG ACAGTTACCA 4081 ATGCTTAATC AGTGAGGCAC CTATCTCAGC GATCTGTCTA TTTCGTTCAT CCATAGTTGC 4141 CTGACTCCCC GTCGTGTAGA TAACTACGAT ACGGGAGGGC TTACCATCTG GCCCCAGTGC 4201 TGCAATGATA CCGCGAGACC CACGCTCACC GGCTCCAGAT TTATCAGCAA TAAACCAGCC 4261 AGCCGGAAGG GCCGAGCGCA GAAGTGGTCC TGCAACTTTA TCCGCCTCCA TCCAGTCTAT 4321 TAATTGTTGC CGGGAAGCTA GAGTAAGTAG TTCGCCAGTT AATAGTTTGC GCAACGTTGT 4381 TGCCATTGCT ACAGGCATCG TGGTGTCACG CTCGTCGTTT GGTATGGCTT CATTCAGCTC 4441 CGGTTCCCAA CGATCAAGGC GAGTTACATG ATCCCCCATG TTGTGCAAAA AAGCGGTTAG 4501 CTCCTTCGGT CCTCCGATCG TTGTCAGAAG TAAGTTGGCC GCAGTGTTAT CACTCATGGT 4561 TATGGCAGCA CTGCATAATT CTCTTACTGT CATGCCATCC GTAAGATGCT TTTCTGTGAC 4621 TGGTGAGTAC TCAACCAAGT CATTCTGAGA ATAGTGTATG CGGCGACCGA GTTGCTCTTG 4681 CCCGGCGTCA ATACGGGATA ATACCGCGCC ACATAGCAGA ACTTTAAAAG TGCTCATCAT 4741 TGGAAAACGT TCTTCGGGGC GAAAACTCTC AAGGATCTTA CCGCTGTTGA GATCCAGTTC 4801 GATGTAACCC ACTCGTGCAC CCAACTGATC TTCAGCATCT TTTACTTTCA CCAGCGTTTC 4861 TGGGTGAGCA AAAACAGGAA GGCAAAATGC CGCAAAAAAG GGAATAAGGG CGACACGGAA 4921 ATGTTGAATA CTCATACTCT TCCTTTTTCA ATATTATTGA AGCATTTATC AGGGTTATTG 4981 TCTCATGAGC GGATACATAT TTGAATGTAT TTAGAAAAAT AAACAAATAG GGGTTCCGCG 5041 CACATTTCCC CGAAAAGTGC CACCTGACGT C//其他哺乳动物表达载体:pCHO1.0 pBApo-CMV-Pur pOPRSVIpcDNA3.1/His C pcDNA5/FRT/V5-His-TOPO pREP4pcDNA3.1/His B pcDNA5/FRT/TO-TOPO pDual-GCpcDNA3.1/His A pcDNA5/TO pBK-RSVpIRESpuro3 pcDNA5/FRT/TO pBK-CMVpIRES2-EGFP pcDNA5/FRT pBI-CMV4pTT5 pFLAG-CMV2 pcDNA4/TO/Myc-His/LacZ pNFkB-DD-tdTomato pcDNA3.1/CT-GFP-TOPO pOPI3CATpBI-CMV5 pcDNA3.1/NT-GFP-TOPO pGene/V5-His B pSEAP2-Basic pOptiVEC-TOPO pSwitchpSEAP2-Control pCMV-MEKK1 pCMVLacIpBI-CMV3 pCMV-MEK1 pVgRxRpBI-CMV2 pCMV-PKA pINDpBI-CMV1 pcDNA6.2/nTC-Tag-DEST pTRE3G-LucpNFκB-MetLuc2-Reporter pcDNA6.2/cTC-Tag-DEST pTRE3GpCRE-MetLuc2-Reporter pcDNA3.2/V5/GW/D-TOPO pTRE2-hygropAcGFP1-Actin pcDNA6.2/V5/GW/D-TOPO pTRE-TightpAcGFP1-N In-Fusion Ready pcDNA6.2/nGeneBLAzer-GW/D-TOPO pTK-hygpAcGFP1-C3 pcDNA6.2/C-YFP-DEST pTet-OnpAcGFP1-C pcDNA6.2/cGeneBLAzer-DEST pTet-OffpAcGFP1-p53 pcDNA6/V5-His A pTet on advanced pAcGFP1-Mito pcDNA6/V5-His B pRevTREpAcGFP1-Mem pcDNA6/V5-His C pRevTet-OnpAcGFP1-Lam pcDNA6/myc-His C pRevTet-OffpAcGFP1-Golgi pcDNA6/myc-His A pCMV-Tet3GpAcGFP1-F pcDNA6/myc-His B pTRE2pAcGFP1-Hyg-C1 pcDNA6.2/nGeneBLAzer-DEST pBD-NF-κBpAsRed2-N1 pcDNA4/HisMax-TOPO pCMV-ADptdTomato-N1 pcDNA6.2/nLumio-DEST pCMV-BDpCMV-tdTomato pcDNA6.2/cLumio-DEST pBIND-Id ControlpCRE-DD-tdTomato pcDNA4/myc-His C pBINDpCMV-DsRed-Express2 pcDNA4/HisMax C pG5 luciferasepEF1α-tdTomato pcDNA4/HisMax A pACT-MyoDpCRE-hrGFP c-Flag pcDNA3 pACTptdTomato-C1 pcDNA4/HisMax B pCMV-SPORT6 pAsRed2-C1 pcDNA4/myc-His B pGL4.13pGL3-Promoter pcDNA4/myc-His A pGL4.19pGL3 basic pcDNA4/His C pGL4.26pAcGFP1-C2 pcDNA4/His B pGL4.20pAcGFP1-C1 pcDNA4/His A pGL4.29pAcGFP1-N3 pcDNA6/TR pGL4.30pAcGFP1-N2 pcDNA4/TO/Myc-His A pGL4.27pAcGFP1-N1 pcDNA4/TO pGL4.75pAcGFP1-C In-Fusion Ready pcDNA4/TO/Myc-His B pGL4.10pCRE-DD-AmCyan1 pcDNA4/TO/Myc-His C pGRN145pNFkB-DD-AmCyan1 pcDNA3.3-TOPO pSecTag2 A pDsRed2-Bid pBudCE4.1 pEBVHis B pDsRED2-Mito pFLAG-CMV-4 pEBVHis ApDD-AmCyan1 Reporter pFLAG-CMV-3 pCMV-Tag 3C pAmCyan1-N1 pFLAG-CMV-2 pCMV-Tag 3A pAmCyan1-C1 pFLAG-CMV-5a pCMV-Tag 3BpEF1α-IRES-DsRed-Express2 p3XFLAG-CMV-9 pCMV-Tag 5CpEF1α-DsRed-Monomer-N1 p3xFLAG-CMV-10 pCMV-Tag 5A pDsRED-Monomer-N1 p3XFLAG-CMV-8 pCMV-Tag 4A pDsRed-Express-N1 p3XFLAG-CMV-7.1 pCMV-Tag 5Bp3XFLAG-CMV-7 pDsRed-Monomer-N In-Fusion Ready pCMV-Tag 4B pDsRed-Express-C1 p3XFLAG-CMV-13 pCMV-Tag 2C pIRES2-ZsGreen1 p3XFLAG-CMV-14 pCMV-Tag 2B pDsRed-Express2-C1 plRES2-ZsGreen1 pCMV-Tag 2A pDsRed-Express2-N1 pBApo-EF1α-pur pCMV-LacZpEF1α-DsRed-Express2 pBApo-EF1α-neo pCMV-MycpIRES2-DsRed-Express pBApo-CMV pEF1α-IRES-AcGFP1 pIRES2-DsRed-Express2 pBApo-CMV-neo pEF1α-IRES-ZsGreen1 pIRES-hrGFP-1a pIRES-EGFP pEF1α-AcGFP1-N1 pIRESneo2 pIRESneo3 pIRES2-DsRed2 pIRESneo pDsRed-Monomer pIRES2-AcGFP1 pIREShyg3 pIRES。
pLVX-IRES-Neo慢病毒载体使用说明

pLVX-IRES-Neo pLVX-IRES-Neo载体基本信息:载体名称:pLVX-IRES-Neo, pLVX IRES Neo质粒类型: 慢病毒载体;哺乳动物细胞表达载体;双顺反子载体高拷贝/低拷贝: 高拷贝启动子: CMV克隆方法: 多克隆位点,限制性内切酶载体大小: 8269 bp5' 测序引物及序列: CMV-F: CGCAAATGGGCGGTAGGCGTG (Invitrogen)3' 测序引物及序列: IRES-R: CCTCACATTGCCAAAAGACG载体标签: 无标签载体抗性: 氨苄青霉素筛选标记: 新霉素Neomycin克隆菌株: Stbl3 E.coli备注: pLVX-IRES-Neo载体以双顺反子的形式同时表达Neo抗性基因和目的基因;CMV启动子驱动目的基因的过表达。
稳定性: 稳表达组成型: 组成型病毒/非病毒: 慢病毒pLVX-IRES-Neo载体质粒图谱和多克隆位点信息:pLVX-IRES-Neo载体序列:ORIGIN1 TGGAAGGGCT AATTCACTCC CAAAGAAGAC AAGATATCCT TGATCTGTGG ATCTACCACA61 CACAAGGCTA CTTCCCTGAT TAGCAGAACT ACACACCAGG GCCAGGGGTC AGATATCCAC121 TGACCTTTGG ATGGTGCTAC AAGCTAGTAC CAGTTGAGCC AGATAAGGTA GAAGAGGCCA 181 ATAAAGGAGA GAACACCAGC TTGTTACACC CTGTGAGCCT GCATGGGATG GATGACCCGG 241 AGAGAGAAGT GTTAGAGTGG AGGTTTGACA GCCGCCTAGC ATTTCATCAC GTGGCCCGAG 301 AGCTGCATCC GGAGTACTTC AAGAACTGCT GATATCGAGC TTGCTACAAG GGACTTTCCG 361 CTGGGGACTT TCCAGGGAGG CGTGGCCTGG GCGGGACTGG GGAGTGGCGA GCCCTCAGAT 421 CCTGCATATA AGCAGCTGCT TTTTGCCTGT ACTGGGTCTC TCTGGTTAGA CCAGATCTGA481 GCCTGGGAGC TCTCTGGCTA ACTAGGGAAC CCACTGCTTA AGCCTCAATA AAGCTTGCCT 541 TGAGTGCTTC AAGTAGTGTG TGCCCGTCTG TTGTGTGACT CTGGTAACTA GAGATCCCTC601 AGACCCTTTT AGTCAGTGTG GAAAATCTCT AGCAGTGGCG CCCGAACAGG GACTTGAAAG 661 CGAAAGGGAA ACCAGAGGAG CTCTCTCGAC GCAGGACTCG GCTTGCTGAA GCGCGCACGG 721 CAAGAGGCGA GGGGCGGCGA CTGGTGAGTA CGCCAAAAAT TTTGACTAGC GGAGGCTAGA 781 AGGAGAGAGA TGGGTGCGAG AGCGTCAGTA TTAAGCGGGG GAGAATTAGA TCGCGATGGG 841 AAAAAATTCG GTTAAGGCCA GGGGGAAAGA AAAAATATAA ATTAAAACAT ATAGTATGGG 901 CAAGCAGGGA GCTAGAACGA TTCGCAGTTA ATCCTGGCCT GTTAGAAACA TCAGAAGGCT 961 GTAGACAAAT ACTGGGACAG CTACAACCAT CCCTTCAGAC AGGATCAGAA GAACTTAGAT 1021 CATTATATAA TACAGTAGCA ACCCTCTATT GTGTGCATCA AAGGATAGAG ATAAAAGACA 1081 CCAAGGAAGC TTTAGACAAG ATAGAGGAAG AGCAAAACAA AAGTAAGACC ACCGCACAGC 1141 AAGCGGCCGG CCGCTGATCT TCAGACCTGG AGGAGGAGAT ATGAGGGACA ATTGGAGAAG 1201 TGAATTATAT AAATATAAAG TAGTAAAAAT TGAACCATTA GGAGTAGCAC CCACCAAGGC 1261 AAAGAGAAGA GTGGTGCAGA GAGAAAAAAG AGCAGTGGGA ATAGGAGCTT TGTTCCTTGG 1321 GTTCTTGGGA GCAGCAGGAA GCACTATGGG CGCAGCGTCA ATGACGCTGA CGGTACAGGC 1381 CAGACAATTA TTGTCTGGTA TAGTGCAGCA GCAGAACAAT TTGCTGAGGG CTATTGAGGC 1441 GCAACAGCAT CTGTTGCAAC TCACAGTCTG GGGCATCAAG CAGCTCCAGG CAAGAATCCT1501 GGCTGTGGAA AGATACCTAA AGGATCAACA GCTCCTGGGG ATTTGGGGTT GCTCTGGAAA 1561 ACTCATTTGC ACCACTGCTG TGCCTTGGAA TGCTAGTTGG AGTAATAAAT CTCTGGAACA 1621 GATTTGGAAT CACACGACCT GGATGGAGTG GGACAGAGAA ATTAACAATT ACACAAGCTT 1681 AATACACTCC TTAATTGAAG AATCGCAAAA CCAGCAAGAA AAGAATGAAC AAGAATTATT 1741 GGAATTAGAT AAATGGGCAA GTTTGTGGAA TTGGTTTAAC ATAACAAATT GGCTGTGGTA 1801 TATAAAATTA TTCATAATGA TAGTAGGAGG CTTGGTAGGT TTAAGAATAG TTTTTGCTGT 1861 ACTTTCTATA GTGAATAGAG TTAGGCAGGG ATATTCACCA TTATCGTTTC AGACCCACCT 1921 CCCAACCCCG AGGGGACCCG ACAGGCCCGA AGGAATAGAA GAAGAAGGTG GAGAGAGAGA 1981 CAGAGACAGA TCCATTCGAT TAGTGAACGG ATCTCGACGG TATCGCCTTT AAAAGAAAAG 2041 GGGGGATTGG GGGGTACAGT GCAGGGGAAA GAATAGTAGA CATAATAGCA ACAGACATAC 2101 AAACTAAAGA ATTACAAAAA CAAATTACAA AAATTCAAAA TTTTCGGGTT TATTACAGGG 2161 ACAGCAGAGA TCCAGTTTAT CGATAAGCTT GGGAGTTCCG CGTTACATAA CTTACGGTAA 2221 ATGGCCCGCC TGGCTGACCG CCCAACGACC CCCGCCCATT GACGTCAATA ATGACGTATG 2281 TTCCCATAGT AACGCCAATA GGGACTTTCC ATTGACGTCA ATGGGTGGAG TATTTACGGT 2341 AAACTGCCCA CTTGGCAGTA CATCAAGTGT ATCATATGCC AAGTACGCCC CCTATTGACG 2401 TCAATGACGG TAAATGGCCC GCCTGGCATT ATGCCCAGTA CATGACCTTA TGGGACTTTC 2461 CTACTTGGCA GTACATCTAC GTATTAGTCA TCGCTATTAC CATGGTGATG CGGTTTTGGC 2521 AGTACATCAA TGGGCGTGGA TAGCGGTTTG ACTCACGGGG ATTTCCAAGT CTCCACCCCA 2581 TTGACGTCAA TGGGAGTTTG TTTTGGCACC AAAATCAACG GGACTTTCCA AAATGTCGTA 2641 ACAACTCCGC CCCATTGACG CAAATGGGCG GTAGGCGTGT ACGGTGGGAG GTCTATATAA 2701 GCAGAGCTCG TTTAGTGAAC CGTCAGATCG CCTGGAGACG CCATCCACGC TGTTTTGACC 2761 TCCATAGAAG ACACCGACTC TACTAGAGGA TCTATTTCCG GTGAATTCCT CGAGACTAGT 2821 TCTAGAGCGG CCGCGGATCC CGCCCCTCTC CCTCCCCCCC CCCTAACGTT ACTGGCCGAA 2881 GCCGCTTGGA ATAAGGCCGG TGTGCGTTTG TCTATATGTT ATTTTCCACC ATATTGCCGT 2941 CTTTTGGCAA TGTGAGGGCC CGGAAACCTG GCCCTGTCTT CTTGACGAGC ATTCCTAGGG 3001 GTCTTTCCCC TCTCGCCAAA GGAATGCAAG GTCTGTTGAA TGTCGTGAAG GAAGCAGTTC 3061 CTCTGGAAGC TTCTTGAAGA CAAACAACGT CTGTAGCGAC CCTTTGCAGG CAGCGGAACC 3121 CCCCACCTGG CGACAGGTGC CTCTGCGGCC AAAAGCCACG TGTATAAGAT ACACCTGCAA 3181 AGGCGGCACA ACCCCAGTGC CACGTTGTGA GTTGGATAGT TGTGGAAAGA GTCAAATGGC 3241 TCTCCTCAAG CGTATTCAAC AAGGGGCTGA AGGATGCCCA GAAGGTACCC CATTGTATGG 3301 GATCTGATCT GGGGCCTCGG TGCACATGCT TTACATGTGT TTAGTCGAGG TTAAAAAAAC 3361 GTCTAGGCCC CCCGAACCAC GGGGACGTGG TTTTCCTTTG AAAAACACGA TGATAAGCTT 3421 GCCACAACCA TGGCTGAACA AGATGGATTG CACGCAGGTT CTCCGGCCGC TTGGGTGGAG 3481 AGGCTATTCG GCTATGACTG GGCACAACAG ACAATCGGCT GCTCTGATGC CGCCGTGTTC 3541 CGGCTGTCAG CGCAGGGGCG CCCGGTTCTT TTTGTCAAGA CCGACCTGTC CGGTGCCCTG 3601 AATGAACTGC AGGACGAGGC AGCGCGGCTA TCGTGGCTGG CCACGACGGG CGTTCCTTGC 3661 GCAGCTGTGC TCGACGTTGT CACTGAAGCG GGAAGGGACT GGCTGCTATT GGGCGAAGTG 3721 CCGGGGCAGG ATCTCCTGTC ATCTCACCTT GCTCCTGCCG AGAAAGTATC CATCATGGCT 3781 GATGCAATGC GGCGGCTGCA TACGCTTGAT CCGGCTACCT GCCCATTCGA CCACCAAGCG 3841 AAACATCGCA TCGAGCGAGC ACGTACTCGG ATGGAAGCCG GTCTTGTCGA TCAGGATGAT 3901 CTGGACGAAG AGCATCAGGG GCTCGCGCCA GCCGAACTGT TCGCCAGGCT CAAGGCGCGC 3961 ATGCCCGACG GCGAGGATCT CGTCGTGACC CATGGCGATG CCTGCTTGCC GAATATCATG 4021 GTGGAAAATG GCCGCTTTTC TGGATTCATC GACTGTGGCC GGCTGGGTGT GGCGGACCGC 4081 TATCAGGACA TAGCGTTGGC TACCCGTGAT ATTGCTGAAG AGCTTGGCGG CGAATGGGCT4141 GACCGCTTCC TCGTGCTTTA CGGTATCGCC GCTCCCGATT CGCAGCGCAT CGCCTTCTAT 4201 CGCCTTCTTG ACGAGTTCTT CTGAACGCGT CTGGAACAAT CAACCTCTGG ATTACAAAAT 4261 TTGTGAAAGA TTGACTGGTA TTCTTAACTA TGTTGCTCCT TTTACGCTAT GTGGATACGC 4321 TGCTTTAATG CCTTTGTATC ATGCTATTGC TTCCCGTATG GCTTTCATTT TCTCCTCCTT4381 GTATAAATCC TGGTTGCTGT CTCTTTATGA GGAGTTGTGG CCCGTTGTCA GGCAACGTGG 4441 CGTGGTGTGC ACTGTGTTTG CTGACGCAAC CCCCACTGGT TGGGGCATTG CCACCACCTG 4501 TCAGCTCCTT TCCGGGACTT TCGCTTTCCC CCTCCCTATT GCCACGGCGG AACTCATCGC 4561 CGCCTGCCTT GCCCGCTGCT GGACAGGGGC TCGGCTGTTG GGCACTGACA ATTCCGTGGT 4621 GTTGTCGGGG AAGCTGACGT CCTTTCCATG GCTGCTCGCC TGTGTTGCCA CCTGGATTCT 4681 GCGCGGGACG TCCTTCTGCT ACGTCCCTTC GGCCCTCAAT CCAGCGGACC TTCCTTCCCG 4741 CGGCCTGCTG CCGGCTCTGC GGCCTCTTCC GCGTCTTCGC CTTCGCCCTC AGACGAGTCG 4801 GATCTCCCTT TGGGCCGCCT CCCCGCCTGG AATTAATTCT GCAGTCGAGA CCTAGAAAAA 4861 CATGGAGCAA TCACAAGTAG CAATACAGCA GCTACCAATG CTGATTGTGC CTGGCTAGAA 4921 GCACAAGAGG AGGAGGAGGT GGGTTTTCCA GTCACACCTC AGGTACCTTT AAGACCAATG 4981 ACTTACAAGG CAGCTGTAGA TCTTAGCCAC TTTTTAAAAG AAAAGAGGGG ACTGGAAGGG 5041 CTAATTCACT CCCAACGAAG ACAAGATATC CTTGATCTGT GGATCTACCA CACACAAGGC 5101 TACTTCCCTG ATTAGCAGAA CTACACACCA GGGCCAGGGG TCAGATATCC ACTGACCTTT 5161 GGATGGTGCT ACAAGCTAGT ACCAGTTGAG CCAGATAAGG TAGAAGAGGC CAATAAAGGA 5221 GAGAACACCA GCTTGTTACA CCCTGTGAGC CTGCATGGGA TGGATGACCC GGAGAGAGAA 5281 GTGTTAGAGT GGAGGTTTGA CAGCCGCCTA GCATTTCATC ACGTGGCCCG AGAGCTGCAT 5341 CCGGAGTACT TCAAGAACTG CTGATATCGA GCTTGCTACA AGGGACTTTC CGCTGGGGAC 5401 TTTCCAGGGA GGCGTGGCCT GGGCGGGACT GGGGAGTGGC GAGCCCTCAG ATCCTGCATA 5461 TAAGCAGCTG CTTTTTGCCT GTACTGGGTC TCTCTGGTTA GACCAGATCT GAGCCTGGGA 5521 GCTCTCTGGC TAACTAGGGA ACCCACTGCT TAAGCCTCAA TAAAGCTTGC CTTGAGTGCT 5581 TCAAGTAGTG TGTGCCCGTC TGTTGTGTGA CTCTGGTAAC TAGAGATCCC TCAGACCCTT 5641 TTAGTCAGTG TGGAAAATCT CTAGCAGTAG TAGTTCATGT CATCTTATTA TTCAGTATTT 5701 ATAACTTGCA AAGAAATGAA TATCAGAGAG TGAGAGGCCT TGACATTGCT AGCGTTTACC 5761 GTCGACCTCT AGCTAGAGCT TGGCGTAATC ATGGTCATAG CTGTTTCCTG TGTGAAATTG 5821 TTATCCGCTC ACAATTCCAC ACAACATACG AGCCGGAAGC ATAAAGTGTA AAGCCTGGGG 5881 TGCCTAATGA GTGAGCTAAC TCACATTAAT TGCGTTGCGC TCACTGCCCG CTTTCCAGTC 5941 GGGAAACCTG TCGTGCCAGC TGCATTAATG AATCGGCCAA CGCGCGGGGA GAGGCGGTTT 6001 GCGTATTGGG CGCTCTTCCG CTTCCTCGCT CACTGACTCG CTGCGCTCGG TCGTTCGGCT 6061 GCGGCGAGCG GTATCAGCTC ACTCAAAGGC GGTAATACGG TTATCCACAG AATCAGGGGA 6121 TAACGCAGGA AAGAACATGT GAGCAAAAGG CCAGCAAAAG GCCAGGAACC GTAAAAAGGC 6181 CGCGTTGCTG GCGTTTTTCC ATAGGCTCCG CCCCCCTGAC GAGCATCACA AAAATCGACG 6241 CTCAAGTCAG AGGTGGCGAA ACCCGACAGG ACTATAAAGA TACCAGGCGT TTCCCCCTGG 6301 AAGCTCCCTC GTGCGCTCTC CTGTTCCGAC CCTGCCGCTT ACCGGATACC TGTCCGCCTT 6361 TCTCCCTTCG GGAAGCGTGG CGCTTTCTCA TAGCTCACGC TGTAGGTATC TCAGTTCGGT 6421 GTAGGTCGTT CGCTCCAAGC TGGGCTGTGT GCACGAACCC CCCGTTCAGC CCGACCGCTG 6481 CGCCTTATCC GGTAACTATC GTCTTGAGTC CAACCCGGTA AGACACGACT TATCGCCACT 6541 GGCAGCAGCC ACTGGTAACA GGATTAGCAG AGCGAGGTAT GTAGGCGGTG CTACAGAGTT 6601 CTTGAAGTGG TGGCCTAACT ACGGCTACAC TAGAAGAACA GTATTTGGTA TCTGCGCTCT 6661 GCTGAAGCCA GTTACCTTCG GAAAAAGAGT TGGTAGCTCT TGATCCGGCA AACAAACCAC 6721 CGCTGGTAGC GGTGGTTTTT TTGTTTGCAA GCAGCAGATT ACGCGCAGAA AAAAAGGATC6781 TCAAGAAGAT CCTTTGATCT TTTCTACGGG GTCTGACGCT CAGTGGAACG AAAACTCACG6841 TTAAGGGATT TTGGTCATGA GATTATCAAA AAGGATCTTC ACCTAGATCC TTTTAAATTA6901 AAAATGAAGT TTTAAATCAA TCTAAAGTAT ATATGAGTAA ACTTGGTCTG ACAGTTACCA6961 ATGCTTAATC AGTGAGGCAC CTATCTCAGC GATCTGTCTA TTTCGTTCAT CCATAGTTGC7021 CTGACTCCCC GTCGTGTAGA TAACTACGAT ACGGGAGGGC TTACCATCTG GCCCCAGTGC7081 TGCAATGATA CCGCGAGACC CACGCTCACC GGCTCCAGAT TTATCAGCAA TAAACCAGCC7141 AGCCGGAAGG GCCGAGCGCA GAAGTGGTCC TGCAACTTTA TCCGCCTCCA TCCAGTCTAT7201 TAATTGTTGC CGGGAAGCTA GAGTAAGTAG TTCGCCAGTT AATAGTTTGC GCAACGTTGT7261 TGCCATTGCT ACAGGCATCG TGGTGTCACG CTCGTCGTTT GGTATGGCTT CATTCAGCTC7321 CGGTTCCCAA CGATCAAGGC GAGTTACATG ATCCCCCATG TTGTGCAAAA AAGCGGTTAG7381 CTCCTTCGGT CCTCCGATCG TTGTCAGAAG TAAGTTGGCC GCAGTGTTAT CACTCATGGT7441 TATGGCAGCA CTGCATAATT CTCTTACTGT CATGCCATCC GTAAGATGCT TTTCTGTGAC7501 TGGTGAGTAC TCAACCAAGT CATTCTGAGA ATAGTGTATG CGGCGACCGA GTTGCTCTTG7561 CCCGGCGTCA ATACGGGATA ATACCGCGCC ACATAGCAGA ACTTTAAAAG TGCTCATCAT7621 TGGAAAACGT TCTTCGGGGC GAAAACTCTC AAGGATCTTA CCGCTGTTGA GATCCAGTTC7681 GATGTAACCC ACTCGTGCAC CCAACTGATC TTCAGCATCT TTTACTTTCA CCAGCGTTTC7741 TGGGTGAGCA AAAACAGGAA GGCAAAATGC CGCAAAAAAG GGAATAAGGG CGACACGGAA7801 ATGTTGAATA CTCATACTCT TCCTTTTTCA ATATTATTGA AGCATTTATC AGGGTTATTG7861 TCTCATGAGC GGATACATAT TTGAATGTAT TTAGAAAAAT AAACAAATAG GGGTTCCGCG7921 CACATTTCCC CGAAAAGTGC CACCTGACGT CGACGGATCG GGAGATCAAC TTGTTTATTG7981 CAGCTTATAA TGGTTACAAA TAAAGCAATA GCATCACAAA TTTCACAAAT AAAGCATTTT8041 TTTCACTGCA TTCTAGTTGT GGTTTGTCCA AACTCATCAA TGTATCTTAT CATGTCTGGA8101 TCAACTGGAT AACTCAAGCT AACCAAAATC ATCCCAAACT TCCCACCCCA TACCCTATTA8161 CCACTGCCAA TTACCTGTGG TTTCATTTAC TCTAAACCTG TGATTCCTCT GAATTATTTT8221 CATTTTAAAG AAATTGTATT TGTTAAATAT GTACTACAAA CTTAGTAGT//pLVX-IRES-Neo其他相关慢病毒载体:Tet-pLKO-neo Tet-pLKO-puro pPACKH1-GAGpMD2.G pCMV-dR8.2-dvpr pLKO.1-GFP-shRNA pLKO.1-TRC control pLKO.1-hygro pLKO.1-TRCpCDH-MSCV-MCS-EF1-copGFP pCDH-MSCV-MCS-EF1-copGFP-T2A-Puro FUW-tetO-hOKMSFUW-tetO-hOCT4 FUW-tetO-hSOX2 FUW-tetO-hKLF4FUW pLVX-AcGFP1-N1 pLVX-AcGFP1-C1pLVX-AmCyan1-N1 pLVX-DsRed-Express2-C1 pLVX-DsRed-Express2-N1 pLVX-DsRed-Monomer-N1 pLVX-PAmCherry-C1 pLVX-PAmCherry-N1 pLVX-ZsGreen1-N1 pLVX-IRES-ZsGreen1 pLVX-IRES-mCherry pLVX-mCherry-C1 pLVX-mCherry-N1 pLVX-tdTomato-C1 pLKO.1-puro pLentilox 3.7 pLVX-Tet-On-Advanced pLVX-IRES-Puro pLVX-IRES-Neo pLVX-IRES-HygpLVX-EF1α-DsRed-Monomer-C1 pLVX-EF1α-AcGFP1-N1 pLVX-EF1α-AcGFP1-C1 pLVX-EF1α-mCherry-C1 pLVX-EF1α-IRES-mCherry pLVX-EF1α-IRES-ZsGreen1 pLVX-MetLuc Control pLVX-MetLuc pLVX-Hom-Mem1pLVX-Het-2 pLVX-DD-AcGFP1-Actin pPRIME-TET-GFP-FF3pSIH1-H1-CopGFP pCDH-EF1-MCS-T2A-Puro pCDH-CMV-MCS-EF1-Puro pCDF1-MCS2-EF1-copGFP pLOX-CWBmi1 pLOX-CW-CREpRSV-rev pMDLg-pRRE pLL3.7pLVX-DD-AmCyan1 Control pLVX-DD-AmCyan1 Reporter pLVX-DD-tdTomato Reporter pLVX-DD-tdTomato Control pLVX-PTuner-Green pLVX-CherryPicker2pLVX-TetOne-Puro-Luc pLVX-TetOne pLVX-TetOne-PuropLVX-TetOne-Luc pLVX-rHom-Nuc1 pLVX-rHom-Sec1pLVX-rHom-1 pLVX-Hom-Nuc1 pLVX-Het-Nuc1pLVX-PTuner pLVX-PTuner2 pLVX-DD-ZsGreen1 Reporter pLVX-Het-1 pLVX-CherryPicker Control pLVX-Tet3GpCDH-CMV-MCS-EF1-RFP-T2A-Puro pCDH-CMV-MCS-EF1-Hygro pCDH-CMV-MCS-EF1-Neo pCDH-MCS-T2A-Puro-MSCV pCDH1-MCS2-EF1-copGFP pCDF1-MCS2-EF1-Puro pCDH-EF1-MCS-T2A-copGFP pWPXL pLVX-TRE3G-ZsGreen1 pLVX-TRE3G-mCherry pLenti6.3-EmGFP-BveI miR pLenti6/V5-GW/lacZpLenti6.3/V5-GW/EmGFP pLenti6.3-MCS pLenti6.3-DsRed2-BveI miR pLenti6.3-MCS-IRES2-EGFP pLVX-shRNA2 psPAX2VSV-G pSico PGK Puro pcDNA6.2-DsRed2-MCS1 miR pcDNA6.3-EmGFP-NC- II pcDNA6.2-EmGFP-NC- I pcDNA6.2-EmGFP-BsaI miR pLenti6.3-BveI miR pLenti6.3-MCS-IRES2-DsRed2 pLEX-MCSpGIPZ pLP2 pLP1FUGW pFUGW pLOX-Ttag-iresTKpMDLg/pRRE pLentG-KOSM pCMV-dR8.91pLVX-TRE3G-Luc Control pLVX-TRE3G-IRES pCgpvpSico pSicoR pLVTHMpGensil-1 pLVX-EF1α-IRES-Puro pCDF1-MCS2-EF1-copGFP pPACKH1-REV pLVX-Het-Mem1 pLVX-shRNA1pLKO.1-puro-GFP-siRNA pPRIME-TREX-GFP-FF3 pcDNA6.2-DsRed2-BsmBI miR pCDH-MSCV-MCS-EF1-Puro pCDH-CMV-MCS-EF1-copGFP pLVX-TRE3GFUW-tetO-hMYC pLOX-TERT-iresTK pLP/VSVGFUW-M2rtTA pCDH-EF1-MCS-(PGK-Puro) pcDNA6.2-EmGFP-MCS1 miR pLVX-AmCyan1-C1 pLVX-Hom-1 pcDNA6.2-BsaI miRpLVX-DsRed-Monomer-C1 pLVX-mCherry-Actin pTRIPZpLVX-ZsGreen1-C1 pLVX-CherryPicker1 LeGO-iC2pLVX-IRES-tdTomato pCDH-CMV-MCS-EF1-copGFP-T2A-Puro pLKO.3GpLVX-tdTomato-N1 pLVX-PTuner2-C pLVX-PuropLVX-Tight-Puro pLVX-DD-ZsGreen1 Control pSicoR PGK PuropLVX-EF1α-DsRed-Monomer-N1 pCDH-UbC-MCS-EF1-Hygro pLVTHpLVX-EF1α-mCherry-N1 pCDH-CMV-MCS-EF1-RFP。
真核细胞常见表达载体

真核细胞罕见表达载体之马矢奏春创作真核细胞, 表达载体1、pCMVp-NEO-BAN载体特点:该真核细胞表达载体分子量为6600碱基对, 主要由CMVp启动子、兔β-球卵白基因内含子、聚腺嘌呤、氨青霉素抗性基因和抗neo基因以及pBR322骨架构成, 在年夜大都真核细胞内都能高水平稳定地表达外源目的基因.更重要的是, 由于该真核细胞表达载体中抗neo基因存在, 转染细胞后, 用G418筛选, 可建立稳定的、高表达目的基因的细胞株.拔出外源基因的克隆位点包括Sal1、BamH1和EcoR1位点.注意在此载体中有二个EcoR1位点存在.2、pEGFP, 增强型绦色荧光卵白表达载体(Enhanced Fluorecent Protein Vector)特点: pEGFP表达载体中含有绿色荧光卵白, 在PCMV启动子驱动下, 在真核细胞中高水平表达.载体骨架中的SV40origin使该载体在任何表达SV40 T抗原的真核细胞内进行复制.Neo抗性盒由SV40早期启动子、Tn5的neomycin/kanamycin抗性基因以及HSV-TK基因的聚腺嘌呤信号组成, 能应用G418筛选稳定转染的真核细胞株.另外, 载体中的pUC origin 能保证该载体在年夜肠杆菌中的复制, 而位于此表达盒上游的细菌启动子能驱动kanamycin抗性基因在年夜肠杆菌中的表达.用途: 该表达载体EGFP上游有Nde1、Eco47111和Age1克隆位点, 将外源基因扦入这些位点, 将合成外源基因和EGFP的融合基因. 借此可确定外源基因在细胞内的表达和/或组织中的定位.亦可用于检测克隆的启动子活性(取代CMV启动子,Acet1-Nhe1). 3、pEGFT-Actin, 增强型绿色荧光卵白/人肌动卵白表达载体特点:pEGFP-Actin表达载体中含有绿色荧光卵白和人胞浆β-肌动卵白基因, 在PCMV启动子驱动下, 在真核细胞中高水平表达.载体骨架中的SV40origin使该载体在任何表达SV40 T抗原的真核细胞内进行复制.Neo抗性盒由SV40早期启动子、Tn5的neomycin/kanamycin抗性基因以及HSV-TK基因的聚腺嘌呤信号组成, 能应用G418筛选稳定转染的真核细胞株.另外, 载体中的pUC origin 能保证该载体在年夜肠杆菌中的复制, 而位于此表达盒上游的细菌启动子能驱动kanamycin抗性基因在年夜肠杆菌中的表达.用途: pEGFP-Actin载体在真核细胞表达EGFP-Actin融合卵白, 该卵白能整合到胞内正在生的肌动卵白, 因而在活细胞和固定细胞中观察到细胞内含肌动卵白的亚细胞结构.4、pSV2表达载体特点:该表达质粒是以病责SV40启动子驱动在真核细胞目的基因进行表达的, 克隆位点为Hind111.SV40启动子具有组织/细胞的选择特异性.此载体不含neo基因, 故不能用来筛选、建立稳定的表达细胞株.5、CMV4 表达载体特点:该真核细胞表达载体由CMV启动子驱动, 多克隆区域酶切位点选择性较多.含有氨苄青霉素抗性基因和生长基因片段以及SV40复制原点和fl单链复制原点.但值得注意的是, 该表达载体不含有neo基因, 转染細胞后不能用G418筛选稳定的表达细胞株. 其他经常使用克隆Vector:pBluscript II KS DNA 15 ugpUC18 DNA 25 ugpUC19 DNA 25 ug说明: pBluescript II kS、pUC18 &Puc19载体适合于DNA片段的克隆、DNA测序和对外源基因进行表达等.这些载体由于在lacZ基因中含有多克隆位点, 当外源DNA片段扦入, 转化lacZ基因缺乏细胞, 并在含有IPTG和X-gal的培养基上培养时, 含有外源DNA 载体的细胞将为白色菌落, 而无外源DNA片段载体的细胞则为兰色菌落, 由此易于筛选有无外源DNA片段的载体.另外, 这些载体中有便于测序的M13、T7和T3的通用引物进行DNA测序. 保管液: TE BufferTE Buffer组成: 10mM Tris.HCL(Ph 8.0) 1mM EDTA用途:克隆外源DNA片断. 进行基因表达. 使用M13、T7和T3引物进行测序. 存在的问题细胞的生命活动是一个复杂的调控过程, 有些生命活动的机理还不完全清晰, 由于插人的目的基因分歧, 载体构建栗用的顺式组件分歧, 组装的空间位置分歧, 采纳的表达系统分歧, 目的基因表达水平和阳性克隆筛选率城市有很年夜不同.另外, 由于所有的顺式元件存在种属和组织特异性, 所构建的高效表达载体纷歧定在所有细胞株中均高效表达.再者, 细胞生长状态的不同, 转染方法的分歧培养时阉的长短, 筛选药物浓度的高低, 对表达量都有很年夜影响.所以, 需要综合评价一个表达载体和表达系统, 排除一些不定因素, 筛选出最佳组合真核表达载体趋向于既有利于扩增目的基因又有利于筛选阳性克隆的载体的构建.许多有效的应用范围广的强启动子和增强子被人发现并应用于载体中, 年夜年夜提高了目的基因的表达量, 另外一些强的组成型启动子, 如 SV40早期启动子+PHTLv, 已应用于年夜规模生产的细胞系中. 应用以GS为筛选标识表记标帜的表达载体可兼有筛选和扩增目的基因两种功能, 是目前研究的重点.以IRES连接的双顺反子表达载体已成为核表达载体的主体.在同一启动子操纵下, 筛选基因或陈说基因与目的基因转录在同一mRNA上通过IRES的作用, 表达出两种卵白, 因而在理论上可认为, 通过了筛选或表达了陈说基因的克隆必为阳性克隆, 即表达目的基因的克隆, 有报道说这种双顺反子的共表达率为90%.这就年夜年夜提高了筛选率, 简化了实验过程. 将强启动子、增强子和可扩增的筛选标识表记标帜组装在同一载体上, 构建高效表达和筛选的表达载体并将其运用于最合适的表达系统中, 是现今真核表达载体构建研究发展方向.。
pTT5哺乳动物表达载体说明

pTT5编号 载体名称北京华越洋生物VECT6098 pTT5pTT5载体基本信息载体名称: pTT5质粒类型: 哺乳动物细胞载体高拷贝/低拷贝: 高拷贝克隆方法: 限制性内切酶,多克隆位点启动子: CMV载体大小: 4401 bp5' 测序引物及序列: CMV-F:CGCAAATGGGCGGTAGGCGTG 3' 测序引物及序列: --载体标签: 无载体抗性: 氨苄青霉素筛选标记: --克隆菌株: DH5α等宿主细胞(系): 常规细胞系(293、CV-1、CHO等)备注: 利用该质粒构建的基因表达是非融合表达,在构建质粒时需要在基因前面添加Kozak序列来增加蛋白表达效率。
稳定性: 瞬时表达组成型/诱导型: 诱导性病毒/非病毒: 非病毒pTT5载体质粒图谱和多克隆位点信息pTT5载体简介pTT5载体是很好的哺乳细胞瞬时蛋白表达载体,经常用于哺乳细胞的抗体轻链或者重链的表达。
pTT5载体序列ORIGIN1 GTACATTTAT ATTGGCTCAT GTCCAATATG ACCGCCATGT TGACATTGAT TATTGACTAG 61 TTATTAATAG TAATCAATTA CGGGGTCATT AGTTCATAGC CCATATATGG AGTTCCGCGT 121 TACATAACTT ACGGTAAATG GCCCGCCTGG CTGACCGCCC AACGACCCCC GCCCATTGAC 181 GTCAATAATG ACGTATGTTC CCATAGTAAC GCCAATAGGG ACTTTCCATT GACGTCAATG 241 GGTGGAGTAT TTACGGTAAA CTGCCCACTT GGCAGTACAT CAAGTGTATC ATATGCCAAG 301 TCCGCCCCCT ATTGACGTCA ATGACGGTAA ATGGCCCGCC TGGCATTATG CCCAGTACAT 361 GACCTTACGG GACTTTCCTA CTTGGCAGTA CATCTACGTA TTAGTCATCG CTATTACCAT 421 GGTGATGCGG TTTTGGCAGT ACACCAATGG GCGTGGATAG CGGTTTGACT CACGGGGATT 481 TCCAAGTCTC CACCCCATTG ACGTCAATGG GAGTTTGTTT TGGCACCAAA ATCAACGGGA 541 CTTTCCAAAA TGTCGTAATA ACCCCGCCCC GTTGACGCAA ATGGGCGGTA GGCGTGTACG 601 GTGGGAGGTC TATATAAGCA GAGCTCGTTT AGTGAACCGT CAGATCCTCA CTCTCTTCCG 661 CATCGCTGTC TGCGAGGGCC AGCTGTTGGG CTCGCGGTTG AGGACAAACT CTTCGCGGTC721 TTTCCAGTAC TCTTGGATCG GAAACCCGTC GGCCTCCGAA CGGTACTCCG CCACCGAGGG 781 ACCTGAGCGA GTCCGCATCG ACCGGATCGG AAAACCTCTC GAGAAAGGCG TCTAACCAGT 841 CACAGTCGCA AGGTAGGCTG AGCACCGTGG CGGGCGGCAG CGGGTGGCGG TCGGGGTTGT 901 TTCTGGCGGA GGTGCTGCTG ATGATGTAAT TAAAGTAGGC GGTCTTGAGA CGGCGGATGG 961 TCGAGGTGAG GTGTGGCAGG CTTGAGATCC AGCTGTTGGG GTGAGTACTC CCTCTCAAAA 1021 GCGGGCATTA CTTCTGCGCT AAGATTGTCA GTTTCCAAAA ACGAGGAGGA TTTGATATTC 1081 ACCTGGCCCG ATCTGGCCAT ACACTTGAGT GACAATGACA TCCACTTTGC CTTTCTCTCC 1141 ACAGGTGTCC ACTCCCAGGT CCAAGTTTAA ACGGATCTCT AGCGAATTCC CTCTAGAGGG 1201 CCCGTTTCTG CTAGCAAGCT TGCTAGCGGC CGCTCGAGGC CGGCAAGGCC GGATCCCCCG 1261 ACCTCGACCT CTGGCTAATA AAGGAAATTT ATTTTCATTG CAATAGTGTG TTGGAATTTT 1321 TTGTGTCTCT CACTCGGAAG GACATATGGG AGGGCAAATC ATTTGGTCGA GATCCCTCGG 1381 AGATCTCTAG CTAGAGGATC GATCCCCGCC CCGGACGAAC TAAACCTGAC TACGACATCT 1441 CTGCCCCTTC TTCGCGGGGC AGTGCATGTA ATCCCTTCAG TTGGTTGGTA CAACTTGCCA 1501 ACTGAACCCT AAACGGGTAG CATATGCTTC CCGGGTAGTA GTATATACTA TCCAGACTAA 1561 CCCTAATTCA ATAGCATATG TTACCCAACG GGAAGCATAT GCTATCGAAT TAGGGTTAGT 1621 AAAAGGGTCC TAAGGAACAG CGATGTAGGT GGGCGGGCCA AGATAGGGGC GCGATTGCTG 1681 CGATCTGGAG GACAAATTAC ACACACTTGC GCCTGAGCGC CAAGCACAGG GTTGTTGGTC 1741 CTCATATTCA CGAGGTCGCT GAGAGCACGG TGGGCTAATG TTGCCATGGG TAGCATATAC 1801 TACCCAAATA TCTGGATAGC ATATGCTATC CTAATCTATA TCTGGGTAGC ATAGGCTATC 1861 CTAATCTATA TCTGGGTAGC ATATGCTATC CTAATCTATA TCTGGGTAGT ATATGCTATC 1921 CTAATTTATA TCTGGGTAGC ATAGGCTATC CTAATCTATA TCTGGGTAGC ATATGCTATC 1981 CTAATCTATA TCTGGGTAGT ATATGCTATC CTAATCTGTA TCCGGGTAGC ATATGCTATC 2041 CTAATAGAGA TTAGGGTAGT ATATGCTATC CTAATTTATA TCTGGGTAGC ATATACTACC 2101 CAAATATCTG GATAGCATAT GCTATCCTAA TCTATATCTG GGTAGCATAT GCTATCCTAA 2161 TCTATATCTG GGTAGCATAG GCTATCCTAA TCTATATCTG GGTAGCATAT GCTATCCTAA 2221 TCTATATCTG GGTAGTATAT GCTATCCTAA TTTATATCTG GGTAGCATAG GCTATCCTAA 2281 TCTATATCTG GGTAGCATAT GCTATCCTAA TCTATATCTG GGTAGTATAT GCTATCCTAA 2341 TCTGTATCCG GGTAGCATAT GCTATCCTCA TGATAAGCTG TCAAACATGA GAATTAATTC 2401 TTGAAGACGA AAGGGCCTCG TGATACGCCT ATTTTTATAG GTTAATGTCA TGATAATAAT 2461 GGTTTCTTAG ACGTCAGGTG GCACTTTTCG GGGAAATGTG CGCGGAACCC CTATTTGTTT 2521 ATTTTTCTAA ATACATTCAA ATATGTATCC GCTCATGAGA CAATAACCCT GATAAATGCT 2581 TCAATAATAT TGAAAAAGGA AGAGTATGAG TATTCAACAT TTCCGTGTCG CCCTTATTCC 2641 CTTTTTTGCG GCATTTTGCC TTCCTGTTTT TGCTCACCCA GAAACGCTGG TGAAAGTAAA 2701 AGATGCTGAA GATCAGTTGG GTGCACGAGT GGGTTACATC GAACTGGATC TCAACAGCGG 2761 TAAGATCCTT GAGAGTTTTC GCCCCGAAGA ACGTTTTCCA ATGATGAGCA CTTTTAAAGT 2821 TCTGCTATGT GGCGCGGTAT TATCCCGTGT TGACGCCGGG CAAGAGCAAC TCGGTCGCCG 2881 CATACACTAT TCTCAGAATG ACTTGGTTGA GTACTCACCA GTCACAGAAA AGCATCTTAC 2941 GGATGGCATG ACAGTAAGAG AATTATGCAG TGCTGCCATA ACCATGAGTG ATAACACTGC 3001 GGCCAACTTA CTTCTGACAA CGATCGGAGG ACCGAAGGAG CTAACCGCTT TTTTGCACAA 3061 CATGGGGGAT CATGTAACTC GCCTTGATCG TTGGGAACCG GAGCTGAATG AAGCCATACC 3121 AAACGACGAG CGTGACACCA CGATGCCTGC AGCAATGGCA ACAACGTTGC GCAAACTATT 3181 AACTGGCGAA CTACTTACTC TAGCTTCCCG GCAACAATTA ATAGACTGGA TGGAGGCGGA 3241 TAAAGTTGCA GGACCACTTC TGCGCTCGGC CCTTCCGGCT GGCTGGTTTA TTGCTGATAA 3301 ATCTGGAGCC GGTGAGCGTG GGTCTCGCGG TATCATTGCA GCACTGGGGC CAGATGGTAA3361 GCCCTCCCGT ATCGTAGTTA TCTACACGAC GGGGAGTCAG GCAACTATGG ATGAACGAAA3421 TAGACAGATC GCTGAGATAG GTGCCTCACT GATTAAGCAT TGGTAACTGT CAGACCAAGT3481 TTACTCATAT ATACTTTAGA TTGATTTAAA ACTTCATTTT TAATTTAAAA GGATCTAGGT3541 GAAGATCCTT TTTGATAATC TCATGACCAA AATCCCTTAA CGTGAGTTTT CGTTCCACTG3601 AGCGTCAGAC CCCGTAGAAA AGATCAAAGG ATCTTCTTGA GATCCTTTTT TTCTGCGCGT3661 AATCTGCTGC TTGCAAACAA AAAAACCACC GCTACCAGCG GTGGTTTGTT TGCCGGATCA3721 AGAGCTACCA ACTCTTTTTC CGAAGGTAAC TGGCTTCAGC AGAGCGCAGA TACCAAATAC3781 TGTTCTTCTA GTGTAGCCGT AGTTAGGCCA CCACTTCAAG AACTCTGTAG CACCGCCTAC3841 ATACCTCGCT CTGCTAATCC TGTTACCAGT GGCTGCTGCC AGTGGCGATA AGTCGTGTCT3901 TACCGGGTTG GACTCAAGAC GATAGTTACC GGATAAGGCG CAGCGGTCGG GCTGAACGGG3961 GGGTTCGTGC ACACAGCCCA GCTTGGAGCG AACGACCTAC ACCGAACTGA GATACCTACA4021 GCGTGAGCTA TGAGAAAGCG CCACGCTTCC CGAAGGGAGA AAGGCGGACA GGTATCCGGT4081 AAGCGGCAGG GTCGGAACAG GAGAGCGCAC GAGGGAGCTT CCAGGGGGAA ACGCCTGGTA4141 TCTTTATAGT CCTGTCGGGT TTCGCCACCT CTGACTTGAG CGTCGATTTT TGTGATGCTC4201 GTCAGGGGGG CGGAGCCTAT GGAAAAACGC CAGCAACGCG GCCTTTTTAC GGTTCCTGGC4261 CTTTTGCTGG CCTTTTGCTC ACATGTTCTT TCCTGCGTTA TCCCCTGATT CTGTGGATAA4321 CCGTATTACC GCCTTTGAGT GAGCTGATAC CGCTCGCCGC AGCCGAACGA CCGAGCGCAG4381 CGAGTCAGTG AGCGAGGAAG C//其他哺乳动物表达载体:pCHO1.0 pBApo-CMV-Pur pOPRSVIpcDNA3.1/His C pcDNA5/FRT/V5-His-TOPO pREP4pcDNA3.1/His B pcDNA5/FRT/TO-TOPO pDual-GCpcDNA3.1/His A pcDNA5/TO pBK-RSVpIRESpuro3 pcDNA5/FRT/TO pBK-CMVpIRES2-EGFP pcDNA5/FRT pBI-CMV4pTT5 pFLAG-CMV2 pcDNA4/TO/Myc-His/LacZ pNFkB-DD-tdTomato pcDNA3.1/CT-GFP-TOPO pOPI3CATpBI-CMV5 pcDNA3.1/NT-GFP-TOPO pGene/V5-His B pSEAP2-Basic pOptiVEC-TOPO pSwitchpSEAP2-Control pCMV-MEKK1 pCMVLacIpBI-CMV3 pCMV-MEK1 pVgRxRpBI-CMV2 pCMV-PKA pINDpBI-CMV1 pcDNA6.2/nTC-Tag-DEST pTRE3G-LucpNFκB-MetLuc2-Reporter pcDNA6.2/cTC-Tag-DEST pTRE3GpCRE-MetLuc2-Reporter pcDNA3.2/V5/GW/D-TOPO pTRE2-hygropAcGFP1-Actin pcDNA6.2/V5/GW/D-TOPO pTRE-TightpAcGFP1-N In-Fusion Ready pcDNA6.2/nGeneBLAzer-GW/D-TOPO pTK-hygpAcGFP1-C3 pcDNA6.2/C-YFP-DEST pTet-OnpAcGFP1-C pcDNA6.2/cGeneBLAzer-DEST pTet-OffpAcGFP1-p53 pcDNA6/V5-His A pTet on advanced pAcGFP1-Mito pcDNA6/V5-His B pRevTREpAcGFP1-Mem pcDNA6/V5-His C pRevTet-OnpAcGFP1-Lam pcDNA6/myc-His C pRevTet-Off pAcGFP1-Golgi pcDNA6/myc-His A pCMV-Tet3G pAcGFP1-F pcDNA6/myc-His B pTRE2pAcGFP1-Hyg-C1 pcDNA6.2/nGeneBLAzer-DEST pBD-NF-κB pAsRed2-N1 pcDNA4/HisMax-TOPO pCMV-AD ptdTomato-N1 pcDNA6.2/nLumio-DEST pCMV-BDpCMV-tdTomato pcDNA6.2/cLumio-DEST pBIND-Id Control pCRE-DD-tdTomato pcDNA4/myc-His C pBINDpCMV-DsRed-Express2 pcDNA4/HisMax C pG5 luciferasepEF1α-tdTomato pcDNA4/HisMax A pACT-MyoDpCRE-hrGFP c-Flag pcDNA3 pACTptdTomato-C1 pcDNA4/HisMax B pCMV-SPORT6 pAsRed2-C1 pcDNA4/myc-His B pGL4.13pGL3-Promoter pcDNA4/myc-His A pGL4.19pGL3 basic pcDNA4/His C pGL4.26pAcGFP1-C2 pcDNA4/His B pGL4.20pAcGFP1-C1 pcDNA4/His A pGL4.29pAcGFP1-N3 pcDNA6/TR pGL4.30pAcGFP1-N2 pcDNA4/TO/Myc-His A pGL4.27pAcGFP1-N1 pcDNA4/TO pGL4.75pAcGFP1-C In-Fusion Ready pcDNA4/TO/Myc-His B pGL4.10pCRE-DD-AmCyan1 pcDNA4/TO/Myc-His C pGRN145pNFkB-DD-AmCyan1 pcDNA3.3-TOPO pSecTag2 A pDsRed2-Bid pBudCE4.1 pEBVHis B pDsRED2-Mito pFLAG-CMV-4 pEBVHis ApDD-AmCyan1 Reporter pFLAG-CMV-3 pCMV-Tag 3C pAmCyan1-N1 pFLAG-CMV-2 pCMV-Tag 3A pAmCyan1-C1 pFLAG-CMV-5a pCMV-Tag 3BpEF1α-IRES-DsRed-Express2 p3XFLAG-CMV-9 pCMV-Tag 5CpEF1α-DsRed-Monomer-N1 p3xFLAG-CMV-10 pCMV-Tag 5A pDsRED-Monomer-N1 p3XFLAG-CMV-8 pCMV-Tag 4A pDsRed-Express-N1 p3XFLAG-CMV-7.1 pCMV-Tag 5Bp3XFLAG-CMV-7 pDsRed-Monomer-N In-Fusion Ready pCMV-Tag 4B pDsRed-Express-C1 p3XFLAG-CMV-13 pCMV-Tag 2C pIRES2-ZsGreen1 p3XFLAG-CMV-14 pCMV-Tag 2B pDsRed-Express2-C1 plRES2-ZsGreen1 pCMV-Tag 2A pDsRed-Express2-N1 pBApo-EF1α-pur pCMV-LacZpEF1α-DsRed-Express2 pBApo-EF1α-neo pCMV-MycpIRES2-DsRed-Express pBApo-CMV pEF1α-IRES-AcGFP1 pIRES2-DsRed-Express2 pBApo-CMV-neo pEF1α-IRES-ZsGreen1 pIRES-hrGFP-1a pIRES-EGFP pEF1α-AcGFP1-N1 pIRESneo2 pIRESneo3 pIRES2-DsRed2 pIRESneo pDsRed-Monomer pIRES2-AcGFP1 pIREShyg3 pIRES。
pSUMO大肠杆菌表达载体说明

pSUMO编号 载体名称北京华越洋生物VECT4280 pSUMOpSUMO载体基本信息载体名称: pSUMO质粒类型: 大肠杆菌基因表达高拷贝/低拷贝: 高拷贝启动子: T7克隆方法: 多克隆位点,限制性内切酶载体大小: 5598 b p5' 测序引物及序列: T7: 5'-‐TAATACGACTCACTATAGGG-‐3'3' 测序引物及序列: T7t: 5'-‐GCTAGTTATTGCTCAGCGG-‐3'载体标签: N-‐His; N-‐SUMO载体抗性: 卡那霉素备注: -‐-‐稳定性: 瞬时表达组成型/诱导型: 诱导型病毒/非病毒: 非病毒pSUMO载体质粒图谱和多克隆位点信息其他大肠杆菌表达载体:pBV221 ptdTomato pET-52b(+) pAmCyanpDsRed-Express2 pBV220 pCold-GST pColdS-SUMOpCold TF pCold IV pCold III pCold IIpCold I pE-SUMO pCold-ProS2 pBAD102/D-TOPOpBAD202/D-TOPO pACYC184 pBAD/Thio-TOPO pBad/Myc-His CpBad/Myc-His B pBad/Myc-His A pBad/His C pBad/His BpBad/His A pBAD-TOPO pET-23b(+) pET-23a(+)pET-23c(+) pET-23(+) pET-12b(+) pET-12c(+)pET-12a(+) pET-11b(+) pET-11a(+) pET-11c(+)pBad24 pQE-82L pQE-81L pQE-80LpQE-32 pQE-9 pQE-16 pQE-31pQE-60 pQE-70 pQE-40 pET-51b(+)pET-50b(+) pET-49b(+) pET-48b(+) pET-47b(+)pET-26b(+) pET-32a(+) pET-21b(+) pET-22b(+)pET-14b pET-16b pET-15b pET-19bpET-20b(+) pET-21d(+) pET-21c(+) pET-21b(+)pET-21a(+) pET-24a(+) pET-24d(+) pET-25b(+)pET-27b(+) pET-28a(+) pET-30a(+) pET-42a(+) pET-43.1c(+) pET-43.1b(+) pET-43.1a(+) pET-44a(+) pET-44c(+) pET-46 EK/LIC pET-37b(+) pTrcHis2 C pTrcHis2 B pTrcHis2 A pET303/CT-His pET302/NT-His pRSET-CFP pRSET-EmGFP pRSET-BFP pGFPuvpET300/NT-DEST pET301/CT-DEST pGEM-T pBad43pGEX-4T-3 pGEX-5X-2 pBlueScript SK(+) pG-Tf2pG-KJE8 pGro7 pET-SUMO pSE380pET-17b pET102/D-TOPO pCDFDuet-1 pMAL-p5xpTf16 pET-28c(+) pBluescript II SK(+) pET-30b(+) pSUMO pProEX HTc pProEX HTb pProEX HTa pKD3 pKD13 pKD46 pTYB1pTYB2 pTWIN2 pBluescript II KS(-) pTYB12pMAL-p5e pACYCDuet-1 pEGM-11ZF(+) pEGM-7ZF(+) PinPoint Xa-3 PinPoint Xa-2 PinPoint Xa-1 pSP73pSP64 pTWIN1 pTYB11 pTXB1pET-5b(+) pBad/gIII C pBad/gIII B pBad/gIII A pET-5a(+) pMal-p4X pMal-p2G pkk223-3pkk232-8 pCYB1 pEZZ18 pBAD18pMAL-c5x pMal-p2E pMal-p2X pET-44 EK/LIC pET-43.1 EK/LIC pET-41 EK/LIC pMal-c4X pTrcHis BpET-31b(+) pET-3b(+) pET-41a(+) pGEX-3XpGEX-4T-2 pETDuet-1 pGEX-4T-1 pTrc99apET-28b(+) pET-His pALEX a,b,c pACYC177pBR322 pKD4 pKD20 pMXB10pEcoli-6xHN-GFPuv pKJE7 pRSET B pGEX-KGpGEX-2T pRSFDuet-1 pCOLADuet-1 pTrcHis C pTrcHis A pET-41b(+) pET-42b(+) pET-3a(+) pGEX-6P-3 pGEX-6P-2 pGEX-6P-1 pGEX-5X-3 pGEX-5X-1 pGEX-2TK pRSET A pMal-c2GpMal-c2E pMal-c2X pRSET C pQE-30pET-45b(+) pET-44b(+) pET-42c(+) pET-41c(+) pET-40b(+) pET-33b(+) pET-39b(+) pET-32 EK/LIC pET-32 Xa/LIC pET-32c(+) pET-32b(+) pET-30 Xa/LIC pET-30 EK/LIC pET-30c(+) pET-29c(+) pET-29b(+) pET-29a(+) pET-24c(+) pET-24b(+) pET-24(+)pET-23d(+) pET-11d(+) pBad33。
pEGFP-N1哺乳细胞表达载体使用说明

pEGFP-N1哺乳细胞表达载体使用说明
货号:P6460
规格:20μL
保存:-20°C
载体信息:
启动子:CMV
复制子:pUC ori,f1ori
终止子:SV40ploy(A)
载体别名:pEGFPN1,pEGFP N1
载体分类:哺乳细胞表达载体,荧光蛋白报告载体
载体大小:4733bp
载体标签:增强型荧光蛋白报告基因EGFP
原核抗性:卡那霉素(Kanamycin)
筛选标记:新霉素(Neomycin)
克隆菌株:DH5α
表达宿主:哺乳细胞
培养条件:37℃,LB培养基
5'测序引物:CMV-F:5'-CGCAAATGGGCGGTAGGCGTG-3'
3'测序引物:EGFP-N:5'-CGTCGCCGTCCAGCTCGACCAG-3'
备注:载体的MCS多克隆酶切位点,有EcoR I,BamH I等酶切位点。
载体简介:
pEGFP-N1编码野生型GFP的一个红移变体,已被优化,有更亮的荧光,在哺乳细胞有
更高的表达,(最大激发波长=488nm,最大释放波长=507nm)。
EGFP基因编码序列,含有超过190个沉默碱基的变化,这符合人类密码子使用偏好。
EGFP序列的一侧已经转变成和KozaK 一致的翻译起始位点,进一步提高了在真核细胞的翻译效率。
载体图谱:。
pEF1α-DsRed-Monomer-N1 哺乳动物表达载体说明

pEF1α-DsRed-Monomer-N1编号 载体名称北京华越洋生物VECT6014 pEF1α-‐DsRed-‐Monomer-‐N1pEF1α-DsRed-Monomer-N1载体基本信息载体名称: pEF1α-DsRed-Monomer-N1质粒类型: 哺乳动物表达载体;双顺反子载体;荧光报告载体高拷贝/低拷贝: 高拷贝克隆方法: 多克隆位点,限制性内切酶启动子: EF1α载体大小: 5448 bp5' 测序引物及序列: EF1-F: TCAAGCCTCAGACAGTGGTTC3' 测序引物及序列: IRES-R载体标签: 红色荧光蛋白DsRed-Monomer(C-term)载体抗性: 卡那霉素筛选标记: 新霉素(Neomycin)克隆菌株: DH5alpha宿主细胞(系): 大部分细胞类型,包括原代细胞、分化细胞、干细胞。
备注: pEF1α-DsRed-Monomer-N1载体是哺乳动物表达载体;目的基因与报告基因在EF1启动子驱动下转录为双顺反子;二者之间的IRES元件,使得目的基因与报告基因独立翻译;DsRed-Monomer是DsRed的单体突变体,与DsRed相比,编码序列经过了人工优化,适用于在哺乳动物细胞中的高水平表达。
稳定性: 瞬表达或稳表达组成型/诱导型: 组成型病毒/非病毒: 非病毒pEF1α-DsRed-Monomer-N1载体质粒图谱和多克隆位点信息pEF1α-DsRed-Monomer-N载体简介pEF1α-DsRed-Monomer-N1 is designed to express a protein of interest fused to the N-terminus of DsRed-Monomer, a monomeric mutant of the Discosoma sp. red fluorescent protein DsRed (1). The DsRed-Monomer coding sequence has been human-codon-optimized for high expression in mammalian cells (2). The excitation and emission maxima of native DsRed-Monomer are 557 nm and 585 nm, respectively. Expression of fusion proteins that retain the fluorescence properties of unmodified DsRed-Monomer can be monitored by flow cytometry and localized by fluorescence microscopy.The multiple cloning site (MCS) in pEF1α-DsRed-Monomer-N1 is positioned between the EF1 promoter (PEF1α) and the DsRed-Monomer coding sequence. Expression of the fusion protein is driven by the EF1α promoter, which remains constitutively active even after stable integration of the vector into the host cell genome (3). A Kozak consensus sequence, located immediately upstream of the DsRed-Monomer gene, enhances the translational efficiency of DsRed-Monomer in eukaryotic systems (4), and SV40 polyadenylation signals direct proper processing of the 3' end of the DsRed-Monomer mRNA.The vector backbone contains an SV40 origin for replication in mammalian cells expressing the SV40 large T antigen, a pUC origin of replication for propagation in E. coli, and an f1 origin for single-stranded DNA production. A neomycinresistance cassette (Neor) allows stably transfected eukaryotic cells to be selected using G418 (5). This cassette consists of the SV40 early promoter (PSV40 e), the Tn5 neomycin/kanamycin resistance gene, and polyadenylation signals from the herpes simplex virus thymidine kinase (HSV TK) gene. A bacterial promoter upstream of the cassette drives expression of the kanamycin resistance gene in E. coli.Location of FeaturesPEF1α (human elongation factor 1 alpha promoter): 12–1346MCS (multiple cloning site): 1348–1422Kozak consensus sequence: 1429–1439DsRed-Monomer (human-codon-optimized): 1436–2110SV40 polyA signal: 2267–2301f1 origin of replication: 2364–2819 (complementary)PSV40 e (SV40 early promoter and enhancer sequences): 2993–3261SV40 origin of replication: 3160–3295Kanr/Neor (kanamycin/neomycin resistance gene): 3344–4138HSV TK polyA signals: 4374–4392pUC origin of replication: 4723–5366Additional InformationThe gene of interest must be cloned into pEF1α-DsRed-Monomer-N1 so that it isin-frame with the DsRed-Monomer coding sequence. The gene must contain a start codon (ATG), and lack in-frame stop codons. Cells expressing DsRed-Monomer fusions can be detected by flow cytometry or fluorescence microscopy 12–16 hr after transfection. If required, stable transfectants can be selected using G418 (5). pEF1α-DsRed-Monomer-N1 can also be used as a cotransfection marker, as the unmodified vector will express DsRed-Monomer in mammalian cells.Propagation in E. coliSuitable host strains: DH5α, HB101 and other general purpose strains. Single-stranded DNA production requires a host containing an F plasmid, such as theJM109 or XL1-Blue strains.Selectable marker: plasmid confers resistance to kanamycin (50 μg/ml) in E. coli hosts.E. coli replication origin: pUCCopy number: highExcitation and Emission Maxima of DsRed-MonomerExcitation: 557 nmEmission: 585 nm其他哺乳动物表达载体:pCHO1.0 pBApo-CMV-Pur pOPRSVIpcDNA3.1/His C pcDNA5/FRT/V5-His-TOPO pREP4pcDNA3.1/His B pcDNA5/FRT/TO-TOPO pDual-GCpcDNA3.1/His A pcDNA5/TO pBK-RSVpIRESpuro3 pcDNA5/FRT/TO pBK-CMVpIRES2-EGFP pcDNA5/FRT pBI-CMV4pTT5 pFLAG-CMV2 pcDNA4/TO/Myc-His/LacZ pNFkB-DD-tdTomato pcDNA3.1/CT-GFP-TOPO pOPI3CATpBI-CMV5 pcDNA3.1/NT-GFP-TOPO pGene/V5-His B pSEAP2-Basic pOptiVEC-TOPO pSwitchpSEAP2-Control pCMV-MEKK1 pCMVLacIpBI-CMV3 pCMV-MEK1 pVgRxRpBI-CMV2 pCMV-PKA pINDpBI-CMV1 pcDNA6.2/nTC-Tag-DEST pTRE3G-LucpNFκB-MetLuc2-Reporter pcDNA6.2/cTC-Tag-DEST pTRE3GpCRE-MetLuc2-Reporter pcDNA3.2/V5/GW/D-TOPO pTRE2-hygropAcGFP1-Actin pcDNA6.2/V5/GW/D-TOPO pTRE-TightpAcGFP1-N In-Fusion Ready pcDNA6.2/nGeneBLAzer-GW/D-TOPO pTK-hygpAcGFP1-C3 pcDNA6.2/C-YFP-DEST pTet-OnpAcGFP1-C pcDNA6.2/cGeneBLAzer-DEST pTet-OffpAcGFP1-p53 pcDNA6/V5-His A pTet on advanced pAcGFP1-Mito pcDNA6/V5-His B pRevTREpAcGFP1-Mem pcDNA6/V5-His C pRevTet-On pAcGFP1-Lam pcDNA6/myc-His C pRevTet-Off pAcGFP1-Golgi pcDNA6/myc-His A pCMV-Tet3G pAcGFP1-F pcDNA6/myc-His B pTRE2pAcGFP1-Hyg-C1 pcDNA6.2/nGeneBLAzer-DEST pBD-NF-κB pAsRed2-N1 pcDNA4/HisMax-TOPO pCMV-AD ptdTomato-N1 pcDNA6.2/nLumio-DEST pCMV-BDpCMV-tdTomato pcDNA6.2/cLumio-DEST pBIND-Id Control pCRE-DD-tdTomato pcDNA4/myc-His C pBINDpCMV-DsRed-Express2 pcDNA4/HisMax C pG5 luciferasepEF1α-tdTomato pcDNA4/HisMax A pACT-MyoDpCRE-hrGFP c-Flag pcDNA3 pACTptdTomato-C1 pcDNA4/HisMax B pCMV-SPORT6 pAsRed2-C1 pcDNA4/myc-His B pGL4.13pGL3-Promoter pcDNA4/myc-His A pGL4.19pGL3 basic pcDNA4/His C pGL4.26pAcGFP1-C2 pcDNA4/His B pGL4.20pAcGFP1-C1 pcDNA4/His A pGL4.29pAcGFP1-N3 pcDNA6/TR pGL4.30pAcGFP1-N2 pcDNA4/TO/Myc-His A pGL4.27pAcGFP1-N1 pcDNA4/TO pGL4.75pAcGFP1-C In-Fusion Ready pcDNA4/TO/Myc-His B pGL4.10pCRE-DD-AmCyan1 pcDNA4/TO/Myc-His C pGRN145pNFkB-DD-AmCyan1 pcDNA3.3-TOPO pSecTag2 A pDsRed2-Bid pBudCE4.1 pEBVHis B pDsRED2-Mito pFLAG-CMV-4 pEBVHis ApDD-AmCyan1 Reporter pFLAG-CMV-3 pCMV-Tag 3C pAmCyan1-N1 pFLAG-CMV-2 pCMV-Tag 3A pAmCyan1-C1 pFLAG-CMV-5a pCMV-Tag 3BpEF1α-IRES-DsRed-Express2 p3XFLAG-CMV-9 pCMV-Tag 5CpEF1α-DsRed-Monomer-N1 p3xFLAG-CMV-10 pCMV-Tag 5A pDsRED-Monomer-N1 p3XFLAG-CMV-8 pCMV-Tag 4A pDsRed-Express-N1 p3XFLAG-CMV-7.1 pCMV-Tag 5Bp3XFLAG-CMV-7 pDsRed-Monomer-N In-Fusion Ready pCMV-Tag 4B pDsRed-Express-C1 p3XFLAG-CMV-13 pCMV-Tag 2C pIRES2-ZsGreen1 p3XFLAG-CMV-14 pCMV-Tag 2B pDsRed-Express2-C1 plRES2-ZsGreen1 pCMV-Tag 2A pDsRed-Express2-N1 pBApo-EF1α-pur pCMV-LacZpEF1α-DsRed-Express2 pBApo-EF1α-neo pCMV-MycpIRES2-DsRed-Express pBApo-CMV pEF1α-IRES-AcGFP1 pIRES2-DsRed-Express2 pBApo-CMV-neo pEF1α-IRES-ZsGreen1 pIRES-hrGFP-1a pIRES-EGFP pEF1α-AcGFP1-N1pIRESneo2 pIRESneo3 pIRES2-DsRed2 pIRESneo pDsRed-Monomer pIRES2-AcGFP1 pIREShyg3 pIRES。
【VIP专享】pAcGFP1-N1哺乳动物表达载体说明

pAcGFP1-N1编号 载体名称北京华越洋生物VECT6107 pAcGFP1-‐N1pAcGFP1-‐N1载体基本信息载体名称: pAcGFP1-N1质粒类型: 哺乳动物细胞表达载体;荧光报告载体高拷贝/低拷贝: 高拷贝克隆方法: 限制性内切酶,多克隆位点启动子: CMV IE载体大小: 4726 bp5' 测序引物及序列: --3' 测序引物及序列: --载体标签: AcGFP1 (C-端)载体抗性: 卡那霉素筛选标记: 新霉素(Neomycin)克隆菌株: DH5α, HB101宿主细胞(系): 常规细胞系(293、CV-1、CHO等)备注: 哺乳动物载体pAcGFP1-N1组成型表达C端AcGFP融合蛋白;AcGFP1与GFP相比,密码子经过了优化,更适合在哺乳动物细胞中高水平表达,同时也增强了亮度。
稳定性: 稳表达或瞬表达组成型/诱导型: 组成型病毒/非病毒: 非病毒pAcGFP1-‐N1载体质粒图谱和多克隆位点信息pAcGFP1-N1载体描述pAcGFP1-N1 encodes a green fluorescent protein (GFP) from Aequorea coerulescens (excitation maximum = 475 nm; emission maximum = 505 nm). The coding sequence of the AcGFP1 gene contains silent base changes, which correspond to human codon-usage preferences (1). The MCS in pAcGFP1-N1 is between the immediate early promoter of CMV (PCMV IE) and the AcGFP1 coding sequences. Genes cloned into the MCS will be expressed as fusions to the N-terminus of AcGFP1 if they are in the same reading frame as AcGFP1 and there are no intervening stop codons. SV40 polyadenylation signals downstream of the AcGFP1 gene direct proper processing of the 3' end of the AcGFP1 mRNA. The vector backbone also contains an SV40 origin for replication in mammalian cells expressing the SV40 T antigen. A neomycin-resistance cassette (Neor), consisting of the SV40 early promoter, the neomycin/kanamycin resistance gene of Tn5, and polyadenylation signals from the Herpes simplex virus thymidine kinase (HSV TK) gene, allows stably transfected eukaryotic cells to be selected using G418. A bacterial promoter upstream of the gene expresses kanamycin resistance in E. coli. The pAcGFP1-N1 backbone also provides a pUC origin of replication for propagation in E. coli and an f1 origin for single-stranded DNA production.Fusions to the N terminus of AcGFP1 retain the fluorescent properties of the native protein allowing the localization of the fusion protein in vivo . The target gene should be cloned into pAcGFP1-N1 so that it is in frame with the AcGFP1 coding sequences, with no intervening in-frame stop codons. The inserted gene should include the initiating ATG codon. The recombinant AcGFP1 vector can be transfected into mammalian cells using any standard transfection method. If required, stable transformants can be selected using G418 (2). pAcGFP1-N1 can also be used simply to express AcGFP1 in a cell line of interest (e.g., as a transfection marker). Propagation in E. coliSuitable host strains: DH5α, HB101 and other general purpose strains. Single-stranded DNA production requiresa host containing an F plasmid such as JM101 or XL1-Blue.Selectable marker: plasmid confers resistance to kanamycin (30 μg/ml) to E. coli hosts.E. coli replication origin: pUCCopy number: ≈500Plasmid incompatibility group: pMB1/ColE1pAcGFP1-N1载体序列ORIGIN1 TAGTTATTAA TAGTAATCAA TTACGGGGTC ATTAGTTCAT AGCCCATATA TGGAGTTCCG 61 CGTTACATAA CTTACGGTAA ATGGCCCGCC TGGCTGACCG CCCAACGACC CCCGCCCATT 121 GACGTCAATA ATGACGTATG TTCCCATAGT AACGCCAATA GGGACTTTCC ATTGACGTCA 181 ATGGGTGGAG TATTTACGGT AAACTGCCCA CTTGGCAGTA CATCAAGTGT ATCATATGCC 241 AAGTACGCCC CCTATTGACG TCAATGACGG TAAATGGCCC GCCTGGCATT ATGCCCAGTA 301 CATGACCTTA TGGGACTTTC CTACTTGGCA GTACATCTAC GTATTAGTCA TCGCTATTAC 361 CATGGTGATG CGGTTTTGGC AGTACATCAA TGGGCGTGGA TAGCGGTTTG ACTCACGGGG 421 ATTTCCAAGT CTCCACCCCA TTGACGTCAA TGGGAGTTTG TTTTGGCACC AAAATCAACG 481 GGACTTTCCA AAATGTCGTA ACAACTCCGC CCCATTGACG CAAATGGGCG GTAGGCGTGT 541 ACGGTGGGAG GTCTATATAA GCAGAGCTGG TTTAGTGAAC CGTCAGATCC GCTAGCGCTA 601 CCGGACTCAG ATCTCGAGCT CAAGCTTCGA ATTCTGCAGT CGACGGTACC GCGGGCCCGG 661 GATCCACCGG TCATGGTGAG CAAGGGCGCC GAGCTGTTCA CCGGCATCGT GCCCATCCTG 721 ATCGAGCTGA ATGGCGATGT GAATGGCCAC AAGTTCAGCG TGAGCGGCGA GGGCGAGGGC 781 GATGCCACCT ACGGCAAGCT GACCCTGAAG TTCATCTGCA CCACCGGCAA GCTGCCTGTG 841 CCCTGGCCCA CCCTGGTGAC CACCCTGAGC TACGGCGTGC AGTGCTTCTC ACGCTACCCC 901 GATCACATGA AGCAGCACGA CTTCTTCAAG AGCGCCATGC CTGAGGGCTA CATCCAGGAG 961 CGCACCATCT TCTTCGAGGA TGACGGCAAC TACAAGTCGC GCGCCGAGGT GAAGTTCGAG 1021 GGCGATACCC TGGTGAATCG CATCGAGCTG ACCGGCACCG ATTTCAAGGA GGATGGCAAC 1081 ATCCTGGGCA ATAAGATGGA GTACAACTAC AACGCCCACA ATGTGTACAT CATGACCGAC 1141 AAGGCCAAGA ATGGCATCAA GGTGAACTTC AAGATCCGCC ACAACATCGA GGATGGCAGC 1201 GTGCAGCTGG CCGACCACTA CCAGCAGAAT ACCCCCATCG GCGATGGCCC TGTGCTGCTG 1261 CCCGATAACC ACTACCTGTC CACCCAGAGC GCCCTGTCCA AGGACCCCAA CGAGAAGCGC 1321 GATCACATGA TCTACTTCGG CTTCGTGACC GCCGCCGCCA TCACCCACGG CATGGATGAG 1381 CTGTACAAGT GAGCGGCCGC GACTCTAGAT CATAATCAGC CATACCACAT TTGTAGAGGT 1441 TTTACTTGCT TTAAAAAACC TCCCACACCT CCCCCTGAAC CTGAAACATA AAATGAATGC 1501 AATTGTTGTT GTTAACTTGT TTATTGCAGC TTATAATGGT TACAAATAAA GCAATAGCAT 1561 CACAAATTTC ACAAATAAAG CATTTTTTTC ACTGCATTCT AGTTGTGGTT TGTCCAAACT 1621 CATCAATGTA TCTTAAGGCG TAAATTGTAA GCGTTAATAT TTTGTTAAAA TTCGCGTTAA 1681 ATTTTTGTTA AATCAGCTCA TTTTTTAACC AATAGGCCGA AATCGGCAAA ATCCCTTATA 1741 AATCAAAAGA ATAGACCGAG ATAGGGTTGA GTGTTGTTCC AGTTTGGAAC AAGAGTCCAC 1801 TATTAAAGAA CGTGGACTCC AACGTCAAAG GGCGAAAAAC CGTCTATCAG GGCGATGGCC 1861 CACTACGTGA ACCATCACCC TAATCAAGTT TTTTGGGGTC GAGGTGCCGT AAAGCACTAA 1921 ATCGGAACCC TAAAGGGAGC CCCCGATTTA GAGCTTGACG GGGAAAGCCG GCGAACGTGG 1981 CGAGAAAGGA AGGGAAGAAA GCGAAAGGAG CGGGCGCTAG GGCGCTGGCA AGTGTAGCGG 2041 TCACGCTGCG CGTAACCACC ACACCCGCCG CGCTTAATGC GCCGCTACAG GGCGCGTCAG 2101 GTGGCACTTT TCGGGGAAAT GTGCGCGGAA CCCCTATTTG TTTATTTTTC TAAATACATT 2161 CAAATATGTA TCCGCTCATG AGACAATAAC CCTGATAAAT GCTTCAATAA TATTGAAAAA 2221 GGAAGAGTCC TGAGGCGGAA AGAACCAGCT GTGGAATGTG TGTCAGTTAG GGTGTGGAAA 2281 GTCCCCAGGC TCCCCAGCAG GCAGAAGTAT GCAAAGCATG CATCTCAATT AGTCAGCAAC 2341 CAGGTGTGGA AAGTCCCCAG GCTCCCCAGC AGGCAGAAGT ATGCAAAGCA TGCATCTCAA 2401 TTAGTCAGCA ACCATAGTCC CGCCCCTAAC TCCGCCCATC CCGCCCCTAA CTCCGCCCAG 2461 TTCCGCCCAT TCTCCGCCCC ATGGCTGACT AATTTTTTTT ATTTATGCAG AGGCCGAGGC2521 CGCCTCGGCC TCTGAGCTAT TCCAGAAGTA GTGAGGAGGC TTTTTTGGAG GCCTAGGCTT 2581 TTGCAAAGAT CGATCAAGAG ACAGGATGAG GATCGTTTCG CATGATTGAA CAAGATGGAT 2641 TGCACGCAGG TTCTCCGGCC GCTTGGGTGG AGAGGCTATT CGGCTATGAC TGGGCACAAC 2701 AGACAATCGG CTGCTCTGAT GCCGCCGTGT TCCGGCTGTC AGCGCAGGGG CGCCCGGTTC 2761 TTTTTGTCAA GACCGACCTG TCCGGTGCCC TGAATGAACT GCAAGACGAG GCAGCGCGGC 2821 TATCGTGGCT GGCCACGACG GGCGTTCCTT GCGCAGCTGT GCTCGACGTT GTCACTGAAG 2881 CGGGAAGGGA CTGGCTGCTA TTGGGCGAAG TGCCGGGGCA GGATCTCCTG TCATCTCACC 2941 TTGCTCCTGC CGAGAAAGTA TCCATCATGG CTGATGCAAT GCGGCGGCTG CATACGCTTG 3001 ATCCGGCTAC CTGCCCATTC GACCACCAAG CGAAACATCG CATCGAGCGA GCACGTACTC 3061 GGATGGAAGC CGGTCTTGTC GATCAGGATG ATCTGGACGA AGAGCATCAG GGGCTCGCGC 3121 CAGCCGAACT GTTCGCCAGG CTCAAGGCGA GCATGCCCGA CGGCGAGGAT CTCGTCGTGA 3181 CCCATGGCGA TGCCTGCTTG CCGAATATCA TGGTGGAAAA TGGCCGCTTT TCTGGATTCA 3241 TCGACTGTGG CCGGCTGGGT GTGGCGGACC GCTATCAGGA CATAGCGTTG GCTACCCGTG 3301 ATATTGCTGA AGAGCTTGGC GGCGAATGGG CTGACCGCTT CCTCGTGCTT TACGGTATCG 3361 CCGCTCCCGA TTCGCAGCGC ATCGCCTTCT ATCGCCTTCT TGACGAGTTC TTCTGAGCGG 3421 GACTCTGGGG TTCGAAATGA CCGACCAAGC GACGCCCAAC CTGCCATCAC GAGATTTCGA 3481 TTCCACCGCC GCCTTCTATG AAAGGTTGGG CTTCGGAATC GTTTTCCGGG ACGCCGGCTG 3541 GATGATCCTC CAGCGCGGGG ATCTCATGCT GGAGTTCTTC GCCCACCCTA GGGGGAGGCT 3601 AACTGAAACA CGGAAGGAGA CAATACCGGA AGGAACCCGC GCTATGACGG CAATAAAAAG 3661 ACAGAATAAA ACGCACGGTG TTGGGTCGTT TGTTCATAAA CGCGGGGTTC GGTCCCAGGG 3721 CTGGCACTCT GTCGATACCC CACCGAGACC CCATTGGGGC CAATACGCCC GCGTTTCTTC 3781 CTTTTCCCCA CCCCACCCCC CAAGTTCGGG TGAAGGCCCA GGGCTCGCAG CCAACGTCGG 3841 GGCGGCAGGC CCTGCCATAG CCTCAGGTTA CTCATATATA CTTTAGATTG ATTTAAAACT 3901 TCATTTTTAA TTTAAAAGGA TCTAGGTGAA GATCCTTTTT GATAATCTCA TGACCAAAAT 3961 CCCTTAACGT GAGTTTTCGT TCCACTGAGC GTCAGACCCC GTAGAAAAGA TCAAAGGATC 4021 TTCTTGAGAT CCTTTTTTTC TGCGCGTAAT CTGCTGCTTG CAAACAAAAA AACCACCGCT 4081 ACCAGCGGTG GTTTGTTTGC CGGATCAAGA GCTACCAACT CTTTTTCCGA AGGTAACTGG 4141 CTTCAGCAGA GCGCAGATAC CAAATACTGT CCTTCTAGTG TAGCCGTAGT TAGGCCACCA 4201 CTTCAAGAAC TCTGTAGCAC CGCCTACATA CCTCGCTCTG CTAATCCTGT TACCAGTGGC 4261 TGCTGCCAGT GGCGATAAGT CGTGTCTTAC CGGGTTGGAC TCAAGACGAT AGTTACCGGA 4321 TAAGGCGCAG CGGTCGGGCT GAACGGGGGG TTCGTGCACA CAGCCCAGCT TGGAGCGAAC 4381 GACCTACACC GAACTGAGAT ACCTACAGCG TGAGCTATGA GAAAGCGCCA CGCTTCCCGA 4441 AGGGAGAAAG GCGGACAGGT ATCCGGTAAG CGGCAGGGTC GGAACAGGAG AGCGCACGAG 4501 GGAGCTTCCA GGGGGAAACG CCTGGTATCT TTATAGTCCT GTCGGGTTTC GCCACCTCTG 4561 ACTTGAGCGT CGATTTTTGT GATGCTCGTC AGGGGGGCGG AGCCTATGGA AAAACGCCAG 4621 CAACGCGGCC TTTTTACGGT TCCTGGCCTT TTGCTGGCCT TTTGCTCACA TGTTCTTTCC 4681 TGCGTTATCC CCTGATTCTG TGGATAACCG TATTACCGCC ATGCAT//其他哺乳动物表达载体:pCHO1.0 pBApo-CMV-Pur pOPRSVIpcDNA3.1/His C pcDNA5/FRT/V5-His-TOPO pREP4pcDNA3.1/His B pcDNA5/FRT/TO-TOPO pDual-GCpcDNA3.1/His A pcDNA5/TO pBK-RSVpIRESpuro3 pcDNA5/FRT/TO pBK-CMVpIRES2-EGFP pcDNA5/FRT pBI-CMV4pTT5 pFLAG-CMV2 pcDNA4/TO/Myc-His/LacZ pNFkB-DD-tdTomato pcDNA3.1/CT-GFP-TOPO pOPI3CATpBI-CMV5 pcDNA3.1/NT-GFP-TOPO pGene/V5-His B pSEAP2-Basic pOptiVEC-TOPO pSwitchpSEAP2-Control pCMV-MEKK1 pCMVLacIpBI-CMV3 pCMV-MEK1 pVgRxRpBI-CMV2 pCMV-PKA pINDpBI-CMV1 pcDNA6.2/nTC-Tag-DEST pTRE3G-LucpNFκB-MetLuc2-Reporter pcDNA6.2/cTC-Tag-DEST pTRE3GpCRE-MetLuc2-Reporter pcDNA3.2/V5/GW/D-TOPO pTRE2-hygropAcGFP1-Actin pcDNA6.2/V5/GW/D-TOPO pTRE-TightpAcGFP1-N In-Fusion Ready pcDNA6.2/nGeneBLAzer-GW/D-TOPO pTK-hygpAcGFP1-C3 pcDNA6.2/C-YFP-DEST pTet-OnpAcGFP1-C pcDNA6.2/cGeneBLAzer-DEST pTet-OffpAcGFP1-p53 pcDNA6/V5-His A pTet on advanced pAcGFP1-Mito pcDNA6/V5-His B pRevTREpAcGFP1-Mem pcDNA6/V5-His C pRevTet-OnpAcGFP1-Lam pcDNA6/myc-His C pRevTet-OffpAcGFP1-Golgi pcDNA6/myc-His A pCMV-Tet3GpAcGFP1-F pcDNA6/myc-His B pTRE2pAcGFP1-Hyg-C1 pcDNA6.2/nGeneBLAzer-DEST pBD-NF-κBpAsRed2-N1 pcDNA4/HisMax-TOPO pCMV-ADptdTomato-N1 pcDNA6.2/nLumio-DEST pCMV-BDpCMV-tdTomato pcDNA6.2/cLumio-DEST pBIND-Id ControlpCRE-DD-tdTomato pcDNA4/myc-His C pBINDpCMV-DsRed-Express2 pcDNA4/HisMax C pG5 luciferasepEF1α-tdTomato pcDNA4/HisMax A pACT-MyoDpCRE-hrGFP c-Flag pcDNA3 pACTptdTomato-C1 pcDNA4/HisMax B pCMV-SPORT6 pAsRed2-C1 pcDNA4/myc-His B pGL4.13pGL3-Promoter pcDNA4/myc-His A pGL4.19pGL3 basic pcDNA4/His C pGL4.26pAcGFP1-C2 pcDNA4/His B pGL4.20pAcGFP1-C1 pcDNA4/His A pGL4.29pAcGFP1-N3 pcDNA6/TR pGL4.30pAcGFP1-N2 pcDNA4/TO/Myc-His A pGL4.27pAcGFP1-N1 pcDNA4/TO pGL4.75pAcGFP1-C In-Fusion Ready pcDNA4/TO/Myc-His B pGL4.10pCRE-DD-AmCyan1 pcDNA4/TO/Myc-His C pGRN145pNFkB-DD-AmCyan1 pcDNA3.3-TOPO pSecTag2 ApDsRed2-Bid pBudCE4.1 pEBVHis BpDsRED2-Mito pFLAG-CMV-4 pEBVHis ApDD-AmCyan1 Reporter pFLAG-CMV-3 pCMV-Tag 3C pAmCyan1-N1 pFLAG-CMV-2 pCMV-Tag 3A pAmCyan1-C1 pFLAG-CMV-5a pCMV-Tag 3BpEF1α-IRES-DsRed-Express2 p3XFLAG-CMV-9 pCMV-Tag 5CpEF1α-DsRed-Monomer-N1 p3xFLAG-CMV-10 pCMV-Tag 5A pDsRED-Monomer-N1 p3XFLAG-CMV-8 pCMV-Tag 4A pDsRed-Express-N1 p3XFLAG-CMV-7.1 pCMV-Tag 5Bp3XFLAG-CMV-7 pDsRed-Monomer-N In-Fusion Ready pCMV-Tag 4B pDsRed-Express-C1 p3XFLAG-CMV-13 pCMV-Tag 2C pIRES2-ZsGreen1 p3XFLAG-CMV-14 pCMV-Tag 2B pDsRed-Express2-C1 plRES2-ZsGreen1 pCMV-Tag 2A pDsRed-Express2-N1 pBApo-EF1α-pur pCMV-LacZpEF1α-DsRed-Express2 pBApo-EF1α-neo pCMV-MycpIRES2-DsRed-Express pBApo-CMV pEF1α-IRES-AcGFP1 pIRES2-DsRed-Express2 pBApo-CMV-neo pEF1α-IRES-ZsGreen1 pIRES-hrGFP-1a pIRES-EGFP pEF1α-AcGFP1-N1 pIRESneo2 pIRESneo3 pIRES2-DsRed2 pIRESneo pDsRed-Monomer pIRES2-AcGFP1 pIREShyg3 pIRES。
pCMV-LacZ哺乳动物表达载体说明
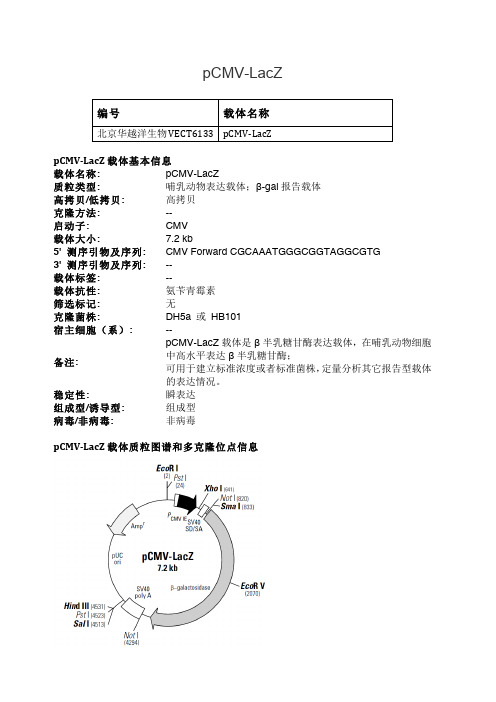
pCMV-LacZ编号 载体名称北京华越洋生物VECT6133 pCMV-‐LacZpCMV-‐LacZ 载体基本信息 载体名称: pCMV-LacZ 质粒类型: 哺乳动物表达载体;β-gal 报告载体 高拷贝/低拷贝: 高拷贝 克隆方法: -- 启动子: CMV 载体大小: 7.2 kb 5' 测序引物及序列: CMV Forward CGCAAATGGGCGGTAGGCGTG 3' 测序引物及序列: -- 载体标签: -- 载体抗性: 氨苄青霉素 筛选标记: 无 克隆菌株: DH5a 或 HB101 宿主细胞(系): --备注:pCMV-LacZ 载体是β半乳糖甘酶表达载体,在哺乳动物细胞中高水平表达β半乳糖甘酶;可用于建立标准浓度或者标准菌株,定量分析其它报告型载体的表达情况。
稳定性: 瞬表达 组成型/诱导型: 组成型 病毒/非病毒: 非病毒pCMV-‐LacZ 载体质粒图谱和多克隆位点信息pCMV-‐LacZ载体简介pCMV-LacZ is a mammalian reporter vector designed to expression β-galactosidasein mammalian cells from the human cytomegalovirus immediate early gene promoter (1). pCMV-LacZ contains an intron (splice donor/splice acceptor; 2) and polyadenylation signal from SV40, and the full-length E. coli β-galactosidase gene with eukaryotic translation initiation signals (3). pCMV-LacZ expresses high levels ofβ-galactosidase and can be used as a reference (control) plasmid when transfecting other reporter gene constructs and can be used to optimize transfection protocols by employing standard assays or stains to assay β-galactosidase activity. Alternatively,the β-galactosidase gene can be excised using the Not I sites at each end to allow other genes to be inserted into the pCMV-LacZ vector backbone for expression in mammalian cells or to insert the β-galactosidase fragment into another expression vector.pCMV-‐LacZ其他哺乳动物表达载体:pCHO1.0 pBApo-CMV-Pur pOPRSVIpcDNA3.1/His C pcDNA5/FRT/V5-His-TOPO pREP4pcDNA3.1/His B pcDNA5/FRT/TO-TOPO pDual-GCpcDNA3.1/His A pcDNA5/TO pBK-RSVpIRESpuro3 pcDNA5/FRT/TO pBK-CMVpIRES2-EGFP pcDNA5/FRT pBI-CMV4pTT5 pFLAG-CMV2 pcDNA4/TO/Myc-His/LacZ pNFkB-DD-tdTomato pcDNA3.1/CT-GFP-TOPO pOPI3CATpBI-CMV5 pcDNA3.1/NT-GFP-TOPO pGene/V5-His B pSEAP2-Basic pOptiVEC-TOPO pSwitchpSEAP2-Control pCMV-MEKK1 pCMVLacIpBI-CMV3 pCMV-MEK1 pVgRxRpBI-CMV2 pCMV-PKA pINDpBI-CMV1 pcDNA6.2/nTC-Tag-DEST pTRE3G-Luc pNFκB-MetLuc2-Reporter pcDNA6.2/cTC-Tag-DEST pTRE3GpCRE-MetLuc2-Reporter pcDNA3.2/V5/GW/D-TOPO pTRE2-hygro pAcGFP1-Actin pcDNA6.2/V5/GW/D-TOPO pTRE-Tight pAcGFP1-N In-Fusion Ready pcDNA6.2/nGeneBLAzer-GW/D-TOPO pTK-hyg pAcGFP1-C3 pcDNA6.2/C-YFP-DEST pTet-On pAcGFP1-C pcDNA6.2/cGeneBLAzer-DEST pTet-Off pAcGFP1-p53 pcDNA6/V5-His A pTet on advanced pAcGFP1-Mito pcDNA6/V5-His B pRevTRE pAcGFP1-Mem pcDNA6/V5-His C pRevTet-On pAcGFP1-Lam pcDNA6/myc-His C pRevTet-Off pAcGFP1-Golgi pcDNA6/myc-His A pCMV-Tet3G pAcGFP1-F pcDNA6/myc-His B pTRE2 pAcGFP1-Hyg-C1 pcDNA6.2/nGeneBLAzer-DEST pBD-NF-κB pAsRed2-N1 pcDNA4/HisMax-TOPO pCMV-AD ptdTomato-N1 pcDNA6.2/nLumio-DEST pCMV-BD pCMV-tdTomato pcDNA6.2/cLumio-DEST pBIND-Id Control pCRE-DD-tdTomato pcDNA4/myc-His C pBINDpCMV-DsRed-Express2 pcDNA4/HisMax C pG5 luciferase pEF1α-tdTomato pcDNA4/HisMax A pACT-MyoD pCRE-hrGFP c-Flag pcDNA3 pACT ptdTomato-C1 pcDNA4/HisMax B pCMV-SPORT6 pAsRed2-C1 pcDNA4/myc-His B pGL4.13pGL3-Promoter pcDNA4/myc-His A pGL4.19pGL3 basic pcDNA4/His C pGL4.26 pAcGFP1-C2 pcDNA4/His B pGL4.20 pAcGFP1-C1 pcDNA4/His A pGL4.29 pAcGFP1-N3 pcDNA6/TR pGL4.30 pAcGFP1-N2 pcDNA4/TO/Myc-His A pGL4.27 pAcGFP1-N1 pcDNA4/TO pGL4.75 pAcGFP1-C In-Fusion Ready pcDNA4/TO/Myc-His B pGL4.10pCRE-DD-AmCyan1 pcDNA4/TO/Myc-His C pGRN145 pNFkB-DD-AmCyan1 pcDNA3.3-TOPO pSecTag2 A pDsRed2-Bid pBudCE4.1 pEBVHis B pDsRED2-Mito pFLAG-CMV-4 pEBVHis ApDD-AmCyan1 Reporter pFLAG-CMV-3 pCMV-Tag 3C pAmCyan1-N1 pFLAG-CMV-2 pCMV-Tag 3A pAmCyan1-C1 pFLAG-CMV-5a pCMV-Tag 3B pEF1α-IRES-DsRed-Express2 p3XFLAG-CMV-9 pCMV-Tag 5C pEF1α-DsRed-Monomer-N1 p3xFLAG-CMV-10 pCMV-Tag 5A pDsRED-Monomer-N1 p3XFLAG-CMV-8 pCMV-Tag 4A pDsRed-Express-N1 p3XFLAG-CMV-7.1 pCMV-Tag 5Bp3XFLAG-CMV-7 pDsRed-Monomer-N In-Fusion Ready pCMV-Tag 4B pDsRed-Express-C1 p3XFLAG-CMV-13 pCMV-Tag 2CpIRES2-ZsGreen1 p3XFLAG-CMV-14 pCMV-Tag 2B pDsRed-Express2-C1 plRES2-ZsGreen1 pCMV-Tag 2A pDsRed-Express2-N1 pBApo-EF1α-pur pCMV-LacZpEF1α-DsRed-Express2 pBApo-EF1α-neo pCMV-MycpIRES2-DsRed-Express pBApo-CMV pEF1α-IRES-AcGFP1 pIRES2-DsRed-Express2 pBApo-CMV-neo pEF1α-IRES-ZsGreen1 pIRES-hrGFP-1a pIRES-EGFP pEF1α-AcGFP1-N1 pIRESneo2 pIRESneo3 pIRES2-DsRed2 pIRESneo pDsRed-Monomer pIRES2-AcGFP1 pIREShyg3 pIRES。
FastDNA SPIN Kit for Soil(mpbio土壤DNA提取试剂盒产品说明书-北京毕特博生物)

Shipping and Storage: The FastDNA® SPIN Kit for Soil is shipped and stored at ambient temperature.
Office Hours:
6:30 am - 5:00 pm P.S.T.(Mon-Fri)
Mailing Address:
FastDNA® SPIN Kit for Soil
Catalog # 6560-200 50 preps
General Information
Qbiogene is a pioneer in developing kits for molecular biology research. We introduced the GENECLEAN® Kits in 1986 and have since been manufacturing products to bring convenience into your research. Our goal is to make your life easier by simplifying the complexities of lab work.
Notes __________________________________________
Call (800) 424-6101 for Technical Support 2 11
Notes __________________________________________
Kit Components 6560-200 (50 preps)
2. Homogenization Reagents
The MT Buffer and Sodium Phosphate Buffer have been carefully selected and prepared to enable complete sample homogenization and protein solubilization. The reagents enable extraction of genomic DNA with minimal RNA contamination.
pPACKH1-REV慢病毒载体使用说明

pPACKH1-REV慢病毒载体使⽤说明pPACKH1-REVpPACKH1-REV 载体基本信息:载体名称:pPACKH1-REV 质粒类型: 慢病毒包装载体⾼拷贝/低拷贝: ⾼拷贝克隆⽅法: 限制性内切酶,多克隆位点启动⼦:CMV 载体⼤⼩:5.5kb 5' 测序引物及序列: CMV Forward : CGCAAATGGGCGGTAGGCGTG3' 测序引物及序列:-- 载体标签:-- 载体抗性: 氨苄青霉素筛选标记:-- 克隆菌株: TOP10等常规菌株宿主细胞(系): 哺乳动物细胞系备注:第三代慢毒包装载体pPACKH1-GAG 与pPACKH1-REV, pVSV-G 是基于HIV-1的慢病毒包装系统。
⽤于第三代慢病毒载体的包装,如: pCDH-CMV-MCS-EF1-puro pCDH-CMV-MCS-EF1-copGFP pCDH-CMV-MCS-EF1-copGFP-T2A-puro pCDH-CMV-MCS-EF1-RFP pCDH-MCS-T2A-puro-MSCV 稳定性: 瞬表达组成型/诱导型: 组成型病毒/⾮病毒: ⾮病毒pPACKH1-REV 载体质粒图谱和多克隆位点信息:pPACKH1-REV载体简介:慢病毒包装质粒pPACKH1-REV使⽤⽅法——慢病毒包装与转染⽅法Production of lentiviral viral stocks requires HIV-1 gag, pol, and rev gene products and vesicular stomatitis virus G (VSV-G) protein encoded by helper plasmids. In general, at least two helper plasmids are required, with one plasmid expressing the Gag-Pol polyprotein and an accessory protein Rev and the other expressing VSV-G as envelop protein to increase cell tropism. Nevertheless, Gag-Pol and Rev proteins can be expressed from separated plasmids as well. Therefore, a three-plasmid system or a four-plasmid system can be used to generate viral stocks, depending on the source and nature of the helper plasmids. In the three-plasmid system, lentiviral transfer vector is cotransfected with two helper plasmids (Gag-Pol + Rev and VSV-G) into cells, while in the four-plasmid system; lentiviral transfer vector is cotransfected with three helper plasmids (Gag-Pol, Rev and VSV-G). It is generally considered to be safer to produce the lentiviral stocks with more helper plasmids, due to the reduced chance of recombination among all vectors that generates replication-competent viruses. The manufacturers of transfection reagents, the suppliers of lentiviral packaging constructs, and many academic laboratories have provided protocols for producing lentiviral stocks. The following procedure is provided as an example only. We produce lentiviral stocks in 293T cells using theBelow we have listed the commonly used vectors for lentiviral packaging.1 For example: pCMV-deltaR8.91 (TRC), psPAX2 (Addgene)2 For example: pMDLg/pRRE (Addgene), pLP1 (Invitrogen), pPACKH1-GAG (SBI)3 For example: pRSV-REV (Addgene), pLP2 (Invitrogen), pPACKH1-REV (SBI)4 For example: pMD.G (TRC), pMD2.G (Addgene), pCMV-VSV-G (Addgene), pVSV-G (SBI), pLP/VSVG (Invitrogen) Note: The transfection reagent Lipofectamine? 2000 (LF2000, Invitrogen) is preferred for transfection. The average lentiviral titers in our preparations are around5 x 106 - 5 x 107 infection units per ml(IU/ml) when titered with 293T cells.Day 0: Seed 6.0 × 106 (1.0 × 106) 293T cells in a 10-cm plate (6-well plate), so that the cell density willbe around 1.0 × 107 (1.7 × 106) at the time of transduction.Day 1: Gently mix 45.0 (7.5) µl LF2000 and 1.5 (0.25) ml Opti-MEM medium and incubate at room temperature for 5 minutes. Meanwhile, gently mix 18.0 (3.0) µg in total of lentiviral transfer vector and helper plasmids mixture into 1.5 (0.25) ml Opti-MEM medium (Invitrogen).Gently mix DNA and LF2000, incubate at room temperature for 20 minutes to allow DNA and lipid to form complexes. In the meantime, replace the overnight culture medium with 5.0 (1.0) ml DMEM + 10% FBS without antibiotics. Add the 3.0 (0.5) ml DNA-LF2000 complexes to 293T cells.Note: We have noticed that fetal bovine serum purchased from different manufacturers may affect the attachment of 293T cells to the bottom of tissue culture plates, resulting in variation in the efficiencyof lentiviral production. We recommend switching to a different brand of FBS if 293T cells disattach from plates during lentiviral production.Day 2: Replace the media containing the DNA-LF2000 complexes with 10.0 (2.0) ml complete medium at 12-16 hours post-transfection.Day4: Collect supernatants at 48 hours post-transfection and transfer media to a polypropylene storage tube. Spin the virus-containing media at 1300 rpm for 5 minutes to pellet any 293T cells that were carried over during collection. Carefully transfer the supernatant to a sterile polypropylene storage tube.Note: Lentiviral stock may be stored at 4 °C for up to 5 days, but should be aliquoted and frozen at-80 °C for long-term storage.Suggestion: To reduce the number of freeze and thaw cycles, aliquot lentiviral stock to smaller tubes before storage at -80 °C. Titering the lentiviral stocksIt is important to titer the lentiviral stocks in the cell line of interest to produce consistent results usingthe equivalent multiplicity of infection (MOI) in experiments. Knowing MOI will help you to control the copy number of lenti-cDNA integrated into the chromosomes of the cells of interest. While titering virus with antibiotic selection, the titering procedure includes selection of stably transduced cells with the corresponding antibiotics and counting the antibiotic-resistant cell colonies. Alternatively, a flow cytometry can be used to determine the viral titer by measuring the number of green (or red) fluorescent cells. If you are using both methods to titer your lentiviruses, please keep in mind that the titers may be different due to sensitivities of FACS machine and cell lines resistant to drug selection. Please be aware the lenti-cDNA virus titer may be underestimated when antibiotic selection or flow cytometry is deployed to determine the titer. For example, the transduced cells with single integration event may not be selected due to low level of selection marker expression by IRES.A. Determination of lentiviral titers by antibiotics selection.Day 0: Seed the cells of your choice in a 6-well plate so that the cell density that will be ~25-50% confluent at the time of transduction.Day 1: Thaw lentiviral stock, gently mix virus and then prepare 2 ml 10-fold serial dilutions ranging from 10-3 to 10-7 in complete medium containing 5-8 µg/ml polybrene.Remove the medium from previous day and add 2 ml fresh dilutions into each well.Suggestion: Leave one well uninfected for mock control. Spin transduction in a desktop centrifuge (e.g. Sorvall RT6000) at 1,000 × g for 30-60 min at room temperature may be necessary if titering virus in cell lines other than 293T.Day 2: Replace the medium containing virus and polybrene with 2 ml of complete medium.Day 3-4: Replace medium with fresh medium containing antibiotic to select for stably transduced cells. Note: At least 48 hours of transduction allows lenti-cDNA to integrate into the host genome. The optimal antibiotic concentration varies from cell line to cell line.Suggestion: A pilot experiment should be performed to determine the minimum concentration of antibiotics required to kill the untransduced cells before this experiment.Day 5-6: Replace medium with 2 ml fresh medium containing antibiotic every 2 days.Day 7-8: Allow antibiotic-resistant colonies to form in dilution wells. No live cells should be growing in the mock control well. Note: The number of days required for the formation of visible colonies may vary among different cell lines.Wash wells twice with 2 ml PBSStain cells with 1 ml 0.5% crystal violet solution in 20% ethanol and incubate for 30 minutes at room temperature.Wash wells with distilled water by submerging the plate in a tray full of water, and repeat the wash one more time.Dry the plate and count the number of blue-stained colonies.The titer should be the average colony number times the dilution factor.Note: The transduction efficiency varies from cell line to cell line. The lentiviral titer in the cell line of your interest may be lower (sometimes more than 10-fold) or higher than the virus titer in commonly used cell lines such as 293T cells.B. Determination of lentiviral titers by flow cytometry.Day 0: Seed the cell of your choice in a 6-well plate so that the cell density that will be ~25-50% confluent at the time of transduction.Note: The number of cells seeded in the well is required to calculate lentiviral titer later.Day 1: Thaw lentiviral stock, gently mix virus and then prepare 2 ml 10-fold serial dilutions ranging from 10-1 to 10-4 in complete medium containing 5-8 µg/ml polybrene.Remove the medium from previous day and add 2 ml fresh dilutions into each well.Suggestion: Leave one well uninfected for mock control. Spin transduction in a desktop centrifuge (e.g. Sorvall RT6000) at 1,000 × g for 30-60 min at room temperature may be necessary if titering virus in cell lines other than 293T.Day 2: Replace the medium containing virus and polybrene with 2 ml of complete medium.Day 3-4: Follow your lab protocol to collect and resuspend cells for flow cytometry to determine the percentage of green or red fluorescent cells.Note: At least 48 hours of transduction allows lenti-cDNA to integrate into the host genome.The titer should be the average number of live fluorescent cells times the dilution factor.Note: The transduction efficiency varies from cell line to cell line. The lentiviral titer in the cell line of your interest may be lower (sometimes more than 10-fold) or higher than the virus titer in commonly used cell lines such as 293T cells. Example: 1 × 105 cells were seeded on Day 0 and the cell doubling time is around 24 hours. The FACS data showed 5% and 40% of green fluorescence cells in the 10-3 and 10-2 dilution wells, respectively. The lentiviral titer is calculated by multiplying the fraction of transduced cells by 2 × 105 (the expected number of cells in the well on Day 1, the time of transduction), and by the dilution factor.0.05 × (2 × 105) × 103 = 1 × 1070.4 × (2 × 105) × 102 = 8 × 106The average lentiviral titer is 9 × 106 IU/mlLentiviral Transduction for Gene Expression.Day 0: Seed cells at appropriate density.Suggestion: Plate cells so that cell density will be ~10-25% confluent at the time of transduction.Day 1: Transduction. Remove the medium from the tissue culture plate by aspiration and replace itwith fresh complete medium containing 5-8 µg/ml polybrene. Gently mix lentivirus with pipette tip,and add appropriate amount of virus to each well.Note: (1) Polybrene may be toxic to some cell lines. (2) The non-concentrated and non-purifiedlentiviral stock collected from 293T supernatant may contain substances affecting the target cell growth, especially when a large volume of low-titer lentiviral stock is added. In case the lentiviral stock inhibits cell growth, concentration and purification of the viruses may be required. (See below for a protocol of virus concentration) Alternatively, incubation time for transduction can be shortened to hours. For example, the virus-containing medium may be replaced with fresh medium after one hourof transduction.Suggestion: Transduce cells at multiplicity of infection (MOI) = 1, 5, 10, 25, and 50 to determine the optimal expression level. Spin transduction in a desktop centrifuge (e.g. Sorvall RT6000) at 1,000 × gfor 30-60 min at room temperature helps increase of transduction efficiency.Day 2: Replace the transduction medium with fresh complete medium to remove lentivirus and polybrene.Day 3-4: Select transduced cells (>50% confluence is recommended) with medium containing appropriate antibiotics or by flow cytometry to sort out fluorescence-positive cells if necessary.Note: The optimal antibiotic concentration varies from cell line to cell line.Suggestion: A pilot experiment should be performed to determine the minimum concentration of antibiotic required to kill the untransduced cells before this experiment.Day 6+: Analysis of transduced cells.Suggestion: Expand the culture of cell lines stably expressing GOI and store the cell line stocks in liquid nitrogen before analyzing the cells.Optional: Concentration of lentivirus by ultracentrifugation1. Filter lentivirus through a 0.45 µm filter.2. Centrifuge at 25,000 rpm for 90 minutes in a SW-28 or SW-41 rotor.3. Discard the supernatant and use a Pasteur pipette with an attached P100 tip to carefully removethe remaining medium.4. Gently resuspend viral pellet in 1/100 volume of DMEM. Let the virus suspension sit for overnightat 4° C.5. On the following day, mix gently, aliquot and freeze virus.pPACKH1-REV载体序列pPACKH1-REV其他相关慢病毒载体:Tet-pLKO-neo Tet-pLKO-puro pPACKH1-GAG pMD2.G pCMV-dR8.2-dvpr pLKO.1-GFP-shRNA pLKO.1-TRC control pLKO.1-hygro pLKO.1-TRC pCDH-MSCV-MCS-EF1-copGFP pCDH-MSCV-MCS-EF1-copGFP-T2A-Puro FUW-tetO-hOKMSFUW-tetO-hOCT4 FUW-tetO-hSOX2 FUW-tetO-hKLF4FUW pLVX-AcGFP1-N1 pLVX-AcGFP1-C1pLVX-AmCyan1-N1 pLVX-DsRed-Express2-C1 pLVX-DsRed-Express2-N1 pLVX-DsRed-Monomer-N1 pLVX-PAmCherry-C1 pLVX-PAmCherry-N1pLVX-ZsGreen1-N1 pLVX-IRES-ZsGreen1 pLVX-IRES-mCherrypLVX-mCherry-C1 pLVX-mCherry-N1 pLVX-tdTomato-C1pLKO.1-puro pLentilox 3.7 pLVX-Tet-On-Advanced pLVX-IRES-Puro pLVX-IRES-Neo pLVX-IRES-HygpLVX-EF1α-DsRed-Monomer-C1 pLVX-EF1α-AcGFP1-N1 pLVX-EF1α-AcGFP1-C1 pLVX-EF1α-mCherry-C1 pLVX-EF1α-IRES-mCherry pLVX-EF1α-IRES-ZsGreen1 pLVX-MetLuc Control pLVX-MetLuc pLVX-Hom-Mem1pLVX-Het-2 pLVX-DD-AcGFP1-Actin pPRIME-TET-GFP-FF3pSIH1-H1-CopGFP pCDH-EF1-MCS-T2A-Puro pCDH-CMV-MCS-EF1-Puro pCDF1-MCS2-EF1-copGFP pLOX-CWBmi1 pLOX-CW-CREpRSV-rev pMDLg-pRRE pLL3.7pLVX-DD-AmCyan1 Control pLVX-DD-AmCyan1 Reporter pLVX-DD-tdTomato Reporter pLVX-DD-tdTomato Control pLVX-PTuner-Green pLVX-CherryPicker2pLVX-TetOne-Puro-Luc pLVX-TetOne pLVX-TetOne-PuropLVX-TetOne-Luc pLVX-rHom-Nuc1 pLVX-rHom-Sec1pLVX-rHom-1 pLVX-Hom-Nuc1 pLVX-Het-Nuc1pLVX-PTuner pLVX-PTuner2 pLVX-DD-ZsGreen1 Reporter pLVX-Het-1 pLVX-CherryPicker Control pLVX-Tet3GpCDH-CMV-MCS-EF1-RFP-T2A-Puro pCDH-CMV-MCS-EF1-Hygro pCDH-CMV-MCS-EF1-Neo pCDH-MCS-T2A-Puro-MSCV pCDH1-MCS2-EF1-copGFP pCDF1-MCS2-EF1-Puro pCDH-EF1-MCS-T2A-copGFP pWPXL pLVX-TRE3G-ZsGreen1 pLVX-TRE3G-mCherry pLenti6.3-EmGFP-BveI miR pLenti6/V5-GW/lacZpLenti6.3/V5-GW/EmGFP pLenti6.3-MCS pLenti6.3-DsRed2-BveI miR pLenti6.3-MCS-IRES2-EGFP pLVX-shRNA2 psPAX2 VSV-G pSico PGK Puro pcDNA6.2-DsRed2-MCS1 miR pcDNA6.3-EmGFP-NC- II pcDNA6.2-EmGFP-NC- I pcDNA6.2-EmGFP-BsaI miR pLenti6.3-BveI miR pLenti6.3-MCS-IRES2-DsRed2 pLEX-MCSpGIPZ pLP2 pLP1FUGW pFUGW pLOX-Ttag-iresTKpMDLg/pRRE pLentG-KOSM pCMV-dR8.91pLVX-TRE3G-Luc Control pLVX-TRE3G-IRES pCgpvpSico pSicoR pLVTHMpGensil-1 pLVX-EF1α-IRES-Puro pCDF1-MCS2-EF1-copGFP pPACKH1-REV pLVX-Het-Mem1 pLVX-shRNA1pLKO.1-puro-GFP-siRNA pPRIME-TREX-GFP-FF3 pcDNA6.2-DsRed2-BsmBI miR pCDH-MSCV-MCS-EF1-Puro pCDH-CMV-MCS-EF1-copGFP pLVX-TRE3GFUW-tetO-hMYC pLOX-TERT-iresTK pLP/VSVGFUW-M2rtTA pCDH-EF1-MCS-(PGK-Puro) pcDNA6.2-EmGFP-MCS1 miR pLVX-AmCyan1-C1 pLVX-Hom-1 pcDNA6.2-BsaI miRpLVX-DsRed-Monomer-C1 pLVX-mCherry-Actin pTRIPZpLVX-ZsGreen1-C1 pLVX-CherryPicker1 LeGO-iC2 pLVX-IRES-tdTomato pCDH-CMV-MCS-EF1-copGFP-T2A-Puro pLKO.3GpLVX-tdTomato-N1 pLVX-PTuner2-C pLVX-Puro pLVX-Tight-Puro pLVX-DD-ZsGreen1 Control pSicoR PGK Puro pLVX-EF1α-DsRed-Monomer-N1 pCDH-UbC-MCS-EF1-Hygro pLVTHpLVX-EF1α-mCherry-N1 pCDH-CMV-MCS-EF1-RFP。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
pAcGFP1-N In-Fusion Ready编号 载体名称北京华越洋生物VECT6052 pAcGFP1-‐N I n-‐Fusion R eady pAcGFP1-‐N I n-‐Fusion R eady载体基本信息载体名称: pAcGFP1-N In-Fusion Ready质粒类型: 哺乳动物细胞表达载体;荧光报告载体高拷贝/低拷贝: 高拷贝克隆方法: 限制性内切酶,多克隆位点启动子: CMV IE载体大小: 4688 bp5' 测序引物及序列: --3' 测序引物及序列: --载体标签: AcGFP1 (C-端)载体抗性: 卡那霉素筛选标记: 新霉素(Neomycin)克隆菌株: DH5α, HB101宿主细胞(系): 常规细胞系(293、CV-1、CHO等)备注: 哺乳动物载体pAcGFP1-N In-FusionReady已经线性化,可以直接用于片段连接;AcGFP1与GFP相比,密码子经过了优化,更适合在哺乳动物细胞中高水平表达,同时也增强了亮度。
稳定性: 稳表达或瞬表达组成型/诱导型: 组成型病毒/非病毒: 非病毒pAcGFP1-‐N I n-‐Fusion R eady载体质粒图谱和多克隆位点信息pAcGFP1-N In-Fusion Ready载体描述pAcGFP1-N In-Fusion Ready vector is a linearized mammalian expression vector that encodes a Green Fluorescent Protein (GFP) from Aequorea coerulescens. This fluorescent protein coding sequence in this construct has been human codon-optimized for efficient expression and enhanced brightness. AcGFP1 protein has an excitation maximum at 475 nm and an emission maximum at 505 nm. The linearized vector allows direct cloning of PCR products without any need for restriction digestion when using Clontech's In-Fusion HD Cloning Plus (638909). This is accomplished by the use of a specific 15 nucleotide long sequence within the sense and antisense primers that overlap with the cut ends created by initial digestion of the vector with Sal I and Hind III.The primers that will be used to amplify In-Fusion Ready PCR products require thefollowing 15 nucleotides on their 5’ends:Sense primer: 5’ AAGGCCTCTGTCGAC followed by sequence of amplification target 3’Antisense primer: 5’ AGAATTCGCAAGCTT followed by sequence of amplification target 3’If the sequence of the gene of interest is added in-frame immediately after the 15 nucleotides mentioned above, this sequence will automatically be in-frame with the AcGFP1 sequence upstream and therefore be expressed as a fusion protein to the N-terminus of AcGFP1.SV40 polyadenylation signals downstream of the AcGFP1 gene direct proper processing of the 3’ end of the mRNA transcript. In addition, the vector also contains a SV40 origin of replication in mammalian cells expressing the SV40 T-antigen. A neomycin resistance cassette (Neor), containing an SV40 early promoter, the neomycin/kanamycin resistance gene of Tn5, and polyadenylation signals from the Herpes simplex virus thymidine kinase (HSV TK) gene, allows selection of stable transformants in eukaryotic cells using G418. A bacterial promoter upstream of the gene allows the plasmid to express kanamycin resistance in E. coli. The pAcGFP1-N In-Fusion Ready Vector also provides a pUC origin of replication for propagation in E. coli and an f1 origin for single-stranded DNA production.Fusions to the N-terminus of AcGFP1 retain the fluorescent properties of the native protein and allow monitoring of fusion protein localization in vivo. The PCR-amplified gene of interest is directly cloned into pAcGFP1-N In-Fusion Ready vector so that it is in-frame with the AcGFP1 coding sequence with no intervening in-frame stop codons. This can be accomplished by using the suggested primers. The inserted gene must include the initiating ATG codon. The recombinant AcGFP1 vector can be transfected into mammalian cells using any standard transfection method. Stable transformants can be selected using G418 (2).Propagation in E. coliSuitable host strains: Stellar Competent Cells.Selectable marker: plasmid confers resistance to kanamycin (50 μg/ml) in E. coli hosts.E. coli replication origin: pUCCopy number: ~ 500Plasmid incompatibility group: pMB1/ColE1pAcGFP1-N In-Fusion Ready载体序列ORIGIN1 AGCTTGCGAA TTCTGGCCTA CCGGTCGCGA GCAAGGGCGC CGAGCTGTTC ACCGGCATCG 61 TGCCCATCCT GATCGAGCTG AATGGCGATG TGAATGGCCA CAAGTTCAGC GTGAGCGGCG 121 AGGGCGAGGG CGATGCCACC TACGGCAAGC TGACCCTGAA GTTCATCTGC ACCACCGGCA 181 AGCTGCCTGT GCCCTGGCCC ACCCTGGTGA CCACCCTGAG CTACGGCGTG CAGTGCTTCT 241 CACGCTACCC CGATCACATG AAGCAGCACG ACTTCTTCAA GAGCGCCATG CCTGAGGGCT301 ACATCCAGGA GCGCACCATC TTCTTCGAGG ATGACGGCAA CTACAAGTCG CGCGCCGAGG 361 TGAAGTTCGA GGGCGATACC CTGGTGAATC GCATCGAGCT GACCGGCACC GATTTCAAGG 421 AGGATGGCAA CATCCTGGGC AATAAGATGG AGTACAACTA CAACGCCCAC AATGTGTACA 481 TCATGACCGA CAAGGCCAAG AATGGCATCA AGGTGAACTT CAAGATCCGC CACAACATCG 541 AGGATGGCAG CGTGCAGCTG GCCGACCACT ACCAGCAGAA TACCCCCATC GGCGATGGCC 601 CTGTGCTGCT GCCCGATAAC CACTACCTGT CCACCCAGAG CGCCCTGTCC AAGGACCCCA 661 ACGAGAAGCG CGATCACATG ATCTACTTCG GCTTCGTGAC CGCCGCCGCC ATCACCCACG 721 GCATGGATGA GCTGTACAAG TGAGCGGCCG CGACTCTAGA TCATAATCAG CCATACCACA 781 TTTGTAGAGG TTTTACTTGC TTTAAAAAAC CTCCCACACC TCCCCCTGAA CCTGAAACAT 841 AAAATGAATG CAATTGTTGT TGTTAACTTG TTTATTGCAG CTTATAATGG TTACAAATAA 901 AGCAATAGCA TCACAAATTT CACAAATAAA GCATTTTTTT CACTGCATTC TAGTTGTGGT 961 TTGTCCAAAC TCATCAATGT ATCTTAAGGC GTAAATTGTA AGCGTTAATA TTTTGTTAAA 1021 ATTCGCGTTA AATTTTTGTT AAATCAGCTC ATTTTTTAAC CAATAGGCCG AAATCGGCAA 1081 AATCCCTTAT AAATCAAAAG AATAGACCGA GATAGGGTTG AGTGTTGTTC CAGTTTGGAA 1141 CAAGAGTCCA CTATTAAAGA ACGTGGACTC CAACGTCAAA GGGCGAAAAA CCGTCTATCA 1201 GGGCGATGGC CCACTACGTG AACCATCACC CTAATCAAGT TTTTTGGGGT CGAGGTGCCG 1261 TAAAGCACTA AATCGGAACC CTAAAGGGAG CCCCCGATTT AGAGCTTGAC GGGGAAAGCC 1321 GGCGAACGTG GCGAGAAAGG AAGGGAAGAA AGCGAAAGGA GCGGGCGCTA GGGCGCTGGC 1381 AAGTGTAGCG GTCACGCTGC GCGTAACCAC CACACCCGCC GCGCTTAATG CGCCGCTACA 1441 GGGCGCGTCA GGTGGCACTT TTCGGGGAAA TGTGCGCGGA ACCCCTATTT GTTTATTTTT 1501 CTAAATACAT TCAAATATGT ATCCGCTCAT GAGACAATAA CCCTGATAAA TGCTTCAATA 1561 ATATTGAAAA AGGAAGAGTC CTGAGGCGGA AAGAACCAGC TGTGGAATGT GTGTCAGTTA 1621 GGGTGTGGAA AGTCCCCAGG CTCCCCAGCA GGCAGAAGTA TGCAAAGCAT GCATCTCAAT 1681 TAGTCAGCAA CCAGGTGTGG AAAGTCCCCA GGCTCCCCAG CAGGCAGAAG TATGCAAAGC 1741 ATGCATCTCA ATTAGTCAGC AACCATAGTC CCGCCCCTAA CTCCGCCCAT CCCGCCCCTA 1801 ACTCCGCCCA GTTCCGCCCA TTCTCCGCCC CATGGCTGAC TAATTTTTTT TATTTATGCA 1861 GAGGCCGAGG CCGCCTCGGC CTCTGAGCTA TTCCAGAAGT AGTGAGGAGG CTTTTTTGGA 1921 GGCCTAGGCT TTTGCAAAGA TCGATCAAGA GACAGGATGA GGATCGTTTC GCATGATTGA 1981 ACAAGATGGA TTGCACGCAG GTTCTCCGGC CGCTTGGGTG GAGAGGCTAT TCGGCTATGA 2041 CTGGGCACAA CAGACAATCG GCTGCTCTGA TGCCGCCGTG TTCCGGCTGT CAGCGCAGGG 2101 GCGCCCGGTT CTTTTTGTCA AGACCGACCT GTCCGGTGCC CTGAATGAAC TGCAAGACGA 2161 GGCAGCGCGG CTATCGTGGC TGGCCACGAC GGGCGTTCCT TGCGCAGCTG TGCTCGACGT 2221 TGTCACTGAA GCGGGAAGGG ACTGGCTGCT ATTGGGCGAA GTGCCGGGGC AGGATCTCCT 2281 GTCATCTCAC CTTGCTCCTG CCGAGAAAGT ATCCATCATG GCTGATGCAA TGCGGCGGCT 2341 GCATACGCTT GATCCGGCTA CCTGCCCATT CGACCACCAA GCGAAACATC GCATCGAGCG 2401 AGCACGTACT CGGATGGAAG CCGGTCTTGT CGATCAGGAT GATCTGGACG AAGAGCATCA 2461 GGGGCTCGCG CCAGCCGAAC TGTTCGCCAG GCTCAAGGCG AGCATGCCCG ACGGCGAGGA 2521 TCTCGTCGTG ACCCATGGCG ATGCCTGCTT GCCGAATATC ATGGTGGAAA ATGGCCGCTT 2581 TTCTGGATTC ATCGACTGTG GCCGGCTGGG TGTGGCGGAC CGCTATCAGG ACATAGCGTT 2641 GGCTACCCGT GATATTGCTG AAGAGCTTGG CGGCGAATGG GCTGACCGCT TCCTCGTGCT 2701 TTACGGTATC GCCGCTCCCG ATTCGCAGCG CATCGCCTTC TATCGCCTTC TTGACGAGTT 2761 CTTCTGAGCG GGACTCTGGG GTTCGAAATG ACCGACCAAG CGACGCCCAA CCTGCCATCA 2821 CGAGATTTCG ATTCCACCGC CGCCTTCTAT GAAAGGTTGG GCTTCGGAAT CGTTTTCCGG 2881 GACGCCGGCT GGATGATCCT CCAGCGCGGG GATCTCATGC TGGAGTTCTT CGCCCACCCT2941 AGGGGGAGGC TAACTGAAAC ACGGAAGGAG ACAATACCGG AAGGAACCCG CGCTATGACG3001 GCAATAAAAA GACAGAATAA AACGCACGGT GTTGGGTCGT TTGTTCATAA ACGCGGGGTT3061 CGGTCCCAGG GCTGGCACTC TGTCGATACC CCACCGAGAC CCCATTGGGG CCAATACGCC3121 CGCGTTTCTT CCTTTTCCCC ACCCCACCCC CCAAGTTCGG GTGAAGGCCC AGGGCTCGCA3181 GCCAACGTCG GGGCGGCAGG CCCTGCCATA GCCTCAGGTT ACTCATATAT ACTTTAGATT3241 GATTTAAAAC TTCATTTTTA ATTTAAAAGG ATCTAGGTGA AGATCCTTTT TGATAATCTC3301 ATGACCAAAA TCCCTTAACG TGAGTTTTCG TTCCACTGAG CGTCAGACCC CGTAGAAAAG3361 ATCAAAGGAT CTTCTTGAGA TCCTTTTTTT CTGCGCGTAA TCTGCTGCTT GCAAACAAAA3421 AAACCACCGC TACCAGCGGT GGTTTGTTTG CCGGATCAAG AGCTACCAAC TCTTTTTCCG3481 AAGGTAACTG GCTTCAGCAG AGCGCAGATA CCAAATACTG TTCTTCTAGT GTAGCCGTAG3541 TTAGGCCACC ACTTCAAGAA CTCTGTAGCA CCGCCTACAT ACCTCGCTCT GCTAATCCTG3601 TTACCAGTGG CTGCTGCCAG TGGCGATAAG TCGTGTCTTA CCGGGTTGGA CTCAAGACGA3661 TAGTTACCGG ATAAGGCGCA GCGGTCGGGC TGAACGGGGG GTTCGTGCAC ACAGCCCAGC3721 TTGGAGCGAA CGACCTACAC CGAACTGAGA TACCTACAGC GTGAGCTATG AGAAAGCGCC3781 ACGCTTCCCG AAGGGAGAAA GGCGGACAGG TATCCGGTAA GCGGCAGGGT CGGAACAGGA3841 GAGCGCACGA GGGAGCTTCC AGGGGGAAAC GCCTGGTATC TTTATAGTCC TGTCGGGTTT3901 CGCCACCTCT GACTTGAGCG TCGATTTTTG TGATGCTCGT CAGGGGGGCG GAGCCTATGG3961 AAAAACGCCA GCAACGCGGC CTTTTTACGG TTCCTGGCCT TTTGCTGGCC TTTTGCTCAC4021 ATGTTCTTTC CTGCGTTATC CCCTGATTCT GTGGATAACC GTATTACCGC CATGCATTAG4081 TTATTAATAG TAATCAATTA CGGGGTCATT AGTTCATAGC CCATATATGG AGTTCCGCGT4141 TACATAACTT ACGGTAAATG GCCCGCCTGG CTGACCGCCC AACGACCCCC GCCCATTGAC4201 GTCAATAATG ACGTATGTTC CCATAGTAAC GCCAATAGGG ACTTTCCATT GACGTCAATG4261 GGTGGAGTAT TTACGGTAAA CTGCCCACTT GGCAGTACAT CAAGTGTATC ATATGCCAAG4321 TACGCCCCCT ATTGACGTCA ATGACGGTAA ATGGCCCGCC TGGCATTATG CCCAGTACAT4381 GACCTTATGG GACTTTCCTA CTTGGCAGTA CATCTACGTA TTAGTCATCG CTATTACCAT4441 GGTGATGCGG TTTTGGCAGT ACATCAATGG GCGTGGATAG CGGTTTGACT CACGGGGATT4501 TCCAAGTCTC CACCCCATTG ACGTCAATGG GAGTTTGTTT TGGCACCAAA ATCAACGGGA4561 CTTTCCAAAA TGTCGTAACA ACTCCGCCCC ATTGACGCAA ATGGGCGGTA GGCGTGTACG4621 GTGGGAGGTC TATATAAGCA GAGCTGGTTT AGTGAACCGT CAGATCCGCT AGCTAAGGCC4681 TCTGTCGA//其他哺乳动物表达载体:pCHO1.0 pBApo-CMV-Pur pOPRSVIpcDNA3.1/His C pcDNA5/FRT/V5-His-TOPO pREP4pcDNA3.1/His B pcDNA5/FRT/TO-TOPO pDual-GCpcDNA3.1/His A pcDNA5/TO pBK-RSVpIRESpuro3 pcDNA5/FRT/TO pBK-CMVpIRES2-EGFP pcDNA5/FRT pBI-CMV4pTT5 pFLAG-CMV2 pcDNA4/TO/Myc-His/LacZ pNFkB-DD-tdTomato pcDNA3.1/CT-GFP-TOPO pOPI3CATpBI-CMV5 pcDNA3.1/NT-GFP-TOPO pGene/V5-His B pSEAP2-Basic pOptiVEC-TOPO pSwitchpSEAP2-Control pCMV-MEKK1 pCMVLacIpBI-CMV3 pCMV-MEK1 pVgRxRpBI-CMV2 pCMV-PKA pINDpBI-CMV1 pcDNA6.2/nTC-Tag-DEST pTRE3G-Luc pNFκB-MetLuc2-Reporter pcDNA6.2/cTC-Tag-DEST pTRE3GpCRE-MetLuc2-Reporter pcDNA3.2/V5/GW/D-TOPO pTRE2-hygro pAcGFP1-Actin pcDNA6.2/V5/GW/D-TOPO pTRE-Tight pAcGFP1-N In-Fusion Ready pcDNA6.2/nGeneBLAzer-GW/D-TOPO pTK-hyg pAcGFP1-C3 pcDNA6.2/C-YFP-DEST pTet-On pAcGFP1-C pcDNA6.2/cGeneBLAzer-DEST pTet-Off pAcGFP1-p53 pcDNA6/V5-His A pTet on advanced pAcGFP1-Mito pcDNA6/V5-His B pRevTRE pAcGFP1-Mem pcDNA6/V5-His C pRevTet-On pAcGFP1-Lam pcDNA6/myc-His C pRevTet-Off pAcGFP1-Golgi pcDNA6/myc-His A pCMV-Tet3G pAcGFP1-F pcDNA6/myc-His B pTRE2 pAcGFP1-Hyg-C1 pcDNA6.2/nGeneBLAzer-DEST pBD-NF-κB pAsRed2-N1 pcDNA4/HisMax-TOPO pCMV-AD ptdTomato-N1 pcDNA6.2/nLumio-DEST pCMV-BD pCMV-tdTomato pcDNA6.2/cLumio-DEST pBIND-Id Control pCRE-DD-tdTomato pcDNA4/myc-His C pBINDpCMV-DsRed-Express2 pcDNA4/HisMax C pG5 luciferase pEF1α-tdTomato pcDNA4/HisMax A pACT-MyoD pCRE-hrGFP c-Flag pcDNA3 pACT ptdTomato-C1 pcDNA4/HisMax B pCMV-SPORT6 pAsRed2-C1 pcDNA4/myc-His B pGL4.13pGL3-Promoter pcDNA4/myc-His A pGL4.19pGL3 basic pcDNA4/His C pGL4.26 pAcGFP1-C2 pcDNA4/His B pGL4.20 pAcGFP1-C1 pcDNA4/His A pGL4.29 pAcGFP1-N3 pcDNA6/TR pGL4.30 pAcGFP1-N2 pcDNA4/TO/Myc-His A pGL4.27 pAcGFP1-N1 pcDNA4/TO pGL4.75 pAcGFP1-C In-Fusion Ready pcDNA4/TO/Myc-His B pGL4.10pCRE-DD-AmCyan1 pcDNA4/TO/Myc-His C pGRN145 pNFkB-DD-AmCyan1 pcDNA3.3-TOPO pSecTag2 A pDsRed2-Bid pBudCE4.1 pEBVHis B pDsRED2-Mito pFLAG-CMV-4 pEBVHis ApDD-AmCyan1 Reporter pFLAG-CMV-3 pCMV-Tag 3C pAmCyan1-N1 pFLAG-CMV-2 pCMV-Tag 3A pAmCyan1-C1 pFLAG-CMV-5a pCMV-Tag 3B pEF1α-IRES-DsRed-Express2 p3XFLAG-CMV-9 pCMV-Tag 5C pEF1α-DsRed-Monomer-N1 p3xFLAG-CMV-10 pCMV-Tag 5A pDsRED-Monomer-N1 p3XFLAG-CMV-8 pCMV-Tag 4A pDsRed-Express-N1 p3XFLAG-CMV-7.1 pCMV-Tag 5Bp3XFLAG-CMV-7 pDsRed-Monomer-N In-Fusion Ready pCMV-Tag 4B pDsRed-Express-C1 p3XFLAG-CMV-13 pCMV-Tag 2C pIRES2-ZsGreen1 p3XFLAG-CMV-14 pCMV-Tag 2B pDsRed-Express2-C1 plRES2-ZsGreen1 pCMV-Tag 2A pDsRed-Express2-N1 pBApo-EF1α-pur pCMV-LacZpEF1α-DsRed-Express2 pBApo-EF1α-neo pCMV-MycpIRES2-DsRed-Express pBApo-CMV pEF1α-IRES-AcGFP1 pIRES2-DsRed-Express2 pBApo-CMV-neo pEF1α-IRES-ZsGreen1 pIRES-hrGFP-1a pIRES-EGFP pEF1α-AcGFP1-N1 pIRESneo2 pIRESneo3 pIRES2-DsRed2 pIRESneo pDsRed-Monomer pIRES2-AcGFP1 pIREShyg3 pIRES。
