pcDNA4 HisMax C哺乳动物表达载体说明
真核细胞常见的表达载体及真核细胞表达外源基因的调控(精)

真核细胞常见表达载体1. pCMVp-NEO-BAN载体特点: 该真核细胞表达载体分子量为6600碱基对,主要由CMVp启动子、兔β-球蛋白基因内含子、聚腺嘌呤、氨青霉素抗性基因和抗neo基因以及pBR322骨架构成,在大多数真核细胞内都能高水平稳定地表达外源目的基因。
更重要的是,由于该真核细胞表达载体中抗neo基因存在,转染细胞后,用G418筛选,可建立稳定的、高表达目的基因的细胞株。
插入外源基因的克隆位点包括Sal1、BamH1和EcoR1位点。
注意在此载体中有二个EcoR1位点存在。
2. pEGFP, 增强型绦色荧光蛋白表达载体(Enhanced Fluorecent Protein Vector特点: pEGFP表达载体中含有绿色荧光蛋白,在PCMV启动子驱动下,在真核细胞中高水平表达。
载体骨架中的SV40 origin使该载体在任何表达SV40 T 抗原的真核细胞内进行复制。
Neo抗性盒由SV40早期启动子、Tn5的neomycin/kanamycin抗性基因以及HSV-TK基因的聚腺嘌呤信号组成,能应用G418筛选稳定转染的真核细胞株。
此外,载体中的pUC origin 能保证该载体在大肠杆菌中的复制,而位于此表达盒上游的细菌启动子能驱动kanamycin抗性基因在大肠杆菌中的表达。
用途: 该表达载体EGFP上游有Nde1、Eco47111和Age1克隆位点,将外源基因扦入这些位点,将合成外源基因和EGFP的融合基因。
借此可确定外源基因在细胞内的表达和/或组织中的定位。
亦可用于检测克隆的启动子活性(取代CMV启动子,Acet1-Nhe1。
Excitation maximum = 488 nm; Emission maximum = 507图示为启动子分泌信号肽和多克隆位点区域:Ase1.pCMV…ccg cta gcg cta ccg gtc gcc acc atg- .EGFP…BamH1…SV40 poly A+Nhe1 Age13. pEGFT-Actin, 增强型绿色荧光蛋白/人肌动蛋白表达载体特点: pEGFP-Actin表达载体中含有绿色荧光蛋白和人胞浆β-肌动蛋白基因,在PCMV启动子驱动下,在真核细胞中高水平表达。
pEF1α-IRES-DsRed-Express2哺乳动物表达载体说明

GTTAGGCCAG TCTTGGTTCA TCGTGACGCT CGGTACCGCG AGCCGCTTGG TCTTTTGGCA GGTCTTTCCC CCTCTGGAAG CCCCCACCTG AAGGCGGCAC CTCTCCTCAA GGATCTGATC CGTCTAGGCC GGCCACAACC CATGGAGGGC CTACGAGGGC CTGGGACATC CGACATCCCC GAACTTCGAG CTTCATCTAC GAAGAAGACT GAAGGGCGAG CAAGTCAATC CAAGCTGGAC CGAGGCCCGC CCACATTTGT AACATAAAAT AATAAAGCAA GTGGTTTGTC TTAAAATTCG GGCAAAATCC TGGAACAAGA TATCAGGGCG TGCCGTAAAG AAGCCGGCGA CTGGCAAGTG CTACAGGGCG TTTTTCTAAA CAATAATATT AGTTAGGGTG TCAATTAGTC AAAGCATGCA CCCTAACTCC ATGCAGAGGC
GGAGACTGAA TGAGTTTGGA ATTTCAGGTG CTGCAGTCGA TACTGGCCGA CATATTGCCG CATTCCTAGG GGAAGCAGTT GCAGCGGAAC TACACCTGCA AGTCAAATGG CCATTGTATG GTTAAAAAAA ATGATAATAT TCAAGGTGCA AGGGCAAGCC TGCCCTTCGC AGCACCCCGC AGCGCGTGAT AGGACGGCAC CCGTAATGCA ACGGCGTGCT TGGTGGAGTT ACGTGGACTC ACGAGCGCGC ATCAGCCATA CTGAACCTGA AATGGTTACA CATTCTAGTT TAATATTTTG GGCCGAAATC TGTTCCAGTT AAAAACCGTC GGGGTCGAGG TTGACGGGGA CGCTAGGGCG TAATGCGCCG TATTTGTTTA ATAAATGCTT AATGTGTGTC AGCATGCATC AGAAGTATGC CCCATCCCGC TTTTTTATTT
常用pGEX载体图谱

常用pGEX载体图谱Rosetta系列的表达菌株可以提供T7 RNA聚合酶,它能表达PET系列载体上的外源基因。
pGEX系列载体上的外源基因不需要T7 RNA 聚合酶,普通的大肠杆菌经IPTG诱导即可表达Tac启动子是一组由Lac和trp启动子人工构建的杂合启动子,受Lac阻遏蛋白的负调节,它的启动能力比Lac和trp都强。
其中Tac 1是由Trp启动子的-35区加上一个合成的46 bp DNA片段(包括Pribnow 盒)和Lac操纵基因构成,Tac 12是由Trp的启动子-35区和Lac 启动子的-10区,加上Lac操纵子中的操纵基因部分和SD序列融合而成蛋白标签:A myc tag is a polypeptide proteintag derived from the c-myc gene product that can be added to a proteinusing recombinant DNA technology. It can be used for affinity chromatography, then used to separate recombinant, overexpressed protein from wild type protein expressed by the host organism. Itcan also be used in the isolation of protein complexes with multiple subunits.A myc tag can be used in many different assays that require recognition byan antibody. If there is no antibody against the studied protein, adding a myc-tag allows one to follow the protein with an antibody against the Myc epitope. Examples are cellular localization studies by immunofluorescence or detectionby Western blotting.The peptide sequence of the myc-tag is (in 1- and 3-letter codes, respectively):N-EQKLISEEDL-C,N-Glu-Gln-Lys-Leu-Ile-Ser-Glu-Glu-Asp-Leu -C, where N stands for Amino-terminus and C stands for Carboxy terminus. The tag is approximately 1202 Daltons in atomic mass and has 10 amino acids.It can be fused to the C-terminus andthe N-terminus of a protein. It is advisablenot to fuse the tag directly behind the signal peptide of a secretory protein, since it can interfere with translocation intothe secretory pathway.A monoclonal antibody against themyc epitope, named 9E10, is available from the non-commercial Developmental Studies Hybridoma BankpGEX4T1载体基本信息出品公司: GE别名: pGEX-4T-1, pGEX4T1, pGEX 4T 1质粒类型: 大肠杆菌蛋白表达载体表达水平: 高拷贝启动子: Tac克隆方法: 多克隆位点,限制性内切酶载体大小: 4969 bp5' 测序引物及序列: pGEX5': GGGCTGGCAAGCCACGTTTGGTG3' 测序引物及序列: pGEX3': CCGGGAGCTGCATGTGTCAGAGG载体标签: N-GST载体抗性: Ampicillin 氨苄备注: 复制子是pMB1产品目录号: 27-4580-01稳定性: 瞬时表达 Transient组成型: 诱导表达病毒/非病毒: 非病毒pGEX4T1载体质粒图谱和多克隆位点信息原核生物DNA复制起始点,是DNA链上独特的具有起始DNA复制功能的碱基序列。
pAD-SV40T使用说明

pAD-SV40T编号载体名称北京华越洋VECT76070pAD-SV40TpAD-SV40T载体图谱:pAD-SV40T载体简介:The pAD-SV40T control plasmid expresses a hybrid protein which contains the NF-κB transcription activation domain fused to amino acids84–708of the SV40large T-antigen.5pAD-SV40T载体相关的哺乳动物表达载体:SuperCos I pDsRed2-Bid pNFκB-MetLuc2-ReporterpYr-adshuttle-3pAcGFP1-N1pEF1α-IRES-DsRed-Express2pVitro2-neo-mcs pSecTag2A pCMV-DsRed-Express2pUB6/V5-His/LacZ pGL4.27pcDNA3.1/NT-GFP-TOPOpUB6/V5-His C pGL4.26pEF1α-IRES-ZsGreen1pUB6/V5-His B pACT pCMV-Tag2ApUB6/V5-His A pBIND-Id Control pCMV-Tag5BpTracer-CMV2pTRE2pAcGFP1-C In-Fusion ReadypSV-β-Galactosidase pRevTRE p3XFLAG-CMV-14pSI pTK-hyg p3XFLAG-CMV-8pSG5pTRE3G-Luc pFLAG-CMV-2pSFV1pSwitch pcDNA3.3-TOPOpSecTag2/Hygro A pcDNA4/His C pcDNA6.2/cLumio-DESTpSecTag B c-Flag pcDNA3pCMV-tdTomatopRluc-N2pcDNA4/TO/Myc-His A pAcGFP1-MitopPICZalpha D pcDNA6/myc-His B pAcGFP1-N In-Fusion Ready pORF-lacZ pcDNA6/V5-His B pDsRed-Monomer-N In-Fusion Ready pORF-HSV1tk pcDNA6.2/nTC-Tag-DEST pcDNA4/TO/Myc-His BpOG44pOptiVEC-TOPO pIRES2-EGFPpNTAP-B pcDNA5/FRT pcDNA3.1/His CpMEP4pGL4.30pcDNA3.1/CT-GFP-TOPOpLVX-ZsGreen-miRNA-Puro pGL4.19pEF1α-IRES-AcGFP1pLVX-IRES-Puro-3xFlag pACT-MyoD pcDNA3.2/V5/GW/D-TOPO pLPCX pCMV-BD pcDNA4/TO/Myc-His/LacZ pLEGFP-N1pCMV-Tet3G pcDNA4/HisMax-TOPOpKH3pTet on advanced p3XFLAG-CMV-13pIRES-puro2pTRE-Tight p3xFLAG-CMV-10phRL-TK pIND pFLAG-CMV-3pG5lac pGene/V5-His B pcDNA4/TO/Myc-His CpFR-luc pOPRSVI pcDNA6.2/C-YFP-DESTpEF6/myc-His C pcDNA4/HisMax A pcDNA6.2/cTC-Tag-DESTpEF4/V5-His A pIRESpuro3pcDNA6.2/nGeneBLAzer-DESTpEF1/myc-His lacZ pIRESneo3pCRE-MetLuc2-ReporterpEF1/myc-His C pIRESneo2pEF1α-DsRed-Express2pEF1/myc-His B pcDNA4/myc-His A pDsRed-Express-C1pEF1/myc-His A pCMV-PKA pEF1α-DsRed-Monomer-N1 pECFP-Mito pAcGFP1-N3pDD-AmCyan1ReporterpECFP-ER pcDNA5/FRT/TO pCRE-DD-AmCyan1pDP8rs pBApo-CMV-Pur pIRES2-DsRed-Express2pDP5rs pBApo-EF1α-pur pDsRed-Express-N1pDP4rs ptdTomato-N1pcDNA6.2/nLumio-DESTpDP3rs pAcGFP1-Golgi pcDNA6/myc-His ApDP2rs pAcGFP1-p53pcDNA6/V5-His ApDP1rs pAcGFP1-Actin pcDNA5/FRT/TO-TOPOpCMV-Myc-C pBI-CMV2pcDNA6.2/V5/GW/D-TOPOpCMV-HA-N pEF1α-tdTomato pcDNA6.2/cGeneBLAzer-DEST pCMV-HA-C pEF1α-tdTomato pcDNA6.2/nGeneBLAzer-GW/D-TOPO pCMV6-XL4pEBVHis A pcDNA5/TOpCMV6-AC-GFP pGL4.10pBApo-CMV-neopCMV5pGL4.29pDsRed-MonomerpCI pGL4.13pIRESpCGN pG5luciferase pIRES-hrGFP-1apcDNA6.2/EmGFP-Bsd/V5-DEST pCMV-AD pDsRed-Express2-N1pcDNA4/V5-His A pRevTet-Off pCMV-Tag3BpcDNA3-mRFP pTet-Off pCRE-hrGFPpcDNA3-CFP pTRE2-hygro pDsRED2-MitopcDNA3.1(+)/myc-His C pVgRxR pAcGFP1-FpcDNA3.1(+)/myc-His A pOPI3CAT pAcGFP1-C1pcDNA3.1(+)/CAT pBK-RSV pAsRed2-C1pcDNA3.1(-)/myc-His C pIRES2-DsRed2pAsRed2-N1pcDNA3.1/Zeo(+)pCMV-Myc pAcGFP1-LampcDNA3.1/Zeo(-)pCMV-Tag2C pAcGFP1-CpcDNA3.1/V5-His C pCMV-Tag5A pSEAP2-BasicpcDNA3.1/Hygro(+)pCMV-Tag3C pBI-CMV3pcDNA3.1/Hygro(-)p3XFLAG-CMV-7pNFkB-DD-tdTomatopcDNA3.1/GS p3XFLAG-CMV-9pcDNA3.1/His ApBS185CMV-Cre pFLAG-CMV-4pEBVHis BpBIND-GFP pBI-CMV4pGL4.75pBiFC-VN155(I152L)pcDNA4/His A pGL4.20pBiFC-CC155pcDNA4/myc-His B pCMV-SPORT6pBiFC-CrN173pcDNA4/HisMax C pCMV-SPORT6pBiFC-bJunVN155(I152L)pCMV-Tag4A pBINDpBD-p53pcDNA6/myc-His C pBD-NF-κBpBD-NF-kB pCMV-Tag2B pRevTet-OnpBC1pGRN145pTet-OnpAD-TRAF pCMV-MEK1pTRE3GpAD-SV40T pCMV-Tag3A pcDNA4/TOpAAV-ZsGreen-miRNA pCMVLacI pcDNA4/His B pAAV-tTS-shRNA pBI-CMV1pcDNA4/HisMax B pIREShyg3pEF1α-AcGFP1-N1pcDNA4/myc-His C pcDNA6/TR pCMV-LacZ pcDNA3.1/His B pDsRed-Express2-C1pCMV-Tag4B pcDNA6/V5-His C pBI-CMV5pCMV-Tag5C pCHO1.0 pAmCyan1-C1plRES2-ZsGreen1pGL3-Promoter pAcGFP1-Mem p3XFLAG-CMV-7.1pCMV-MEKK1 pAcGFP1-C3pFLAG-CMV-5a pFLAG-CMV2 pTT5pBudCE4.1pAcGFP1-C2 pSEAP2-Control pREP4ptdTomato-C1 pIRES2-AcGFP1pBApo-CMV pCRE-DD-tdTomato pAcGFP1-Hyg-C1。
各种表达载体

表达载体一、原核细胞表达载体1. pBAD载体:特点; 该表达质粒含有araBAD(arabinose)操纵子的P BAD启动子和编码该启动子的正负调控子基因araC,具有紧密调控功能和高水平表达外源蛋白质的原核细胞表达载体。
请注意:1. 当要扦入其他信号肽片段,改建此载体时,请不要利用该载体上的Nde1 EcoR1 BamH1 Kpn1和Pst1位点,以免造成重组困难,因为前述内切酶在此载体上均有二个位点,最好使用只有一个酶切位点的Sac1和Hind111位点。
同时记住,如在不含任何信号肽的P BAD表达质粒扦入信号肽,其非编码N-末端要包含核糖体结合位点(RBS)核苷酸序列。
2在使用含Omp A分泌信号肽的P BAD表达质粒时,请应用Omp A分泌信号肽上的Sac1以及载体Hind111酶切位点,这些在载体序列上都是单个酶切位点。
本公司目前有含Omp A分泌信号肽和不含任何信号肽的二种P BAD表达质粒,其多克隆位点区域图谱如下:(a)、含Omp A分泌信号肽的P BAD表达质粒多克隆区域SDP BAD….TACCCGTTTTTTTCC….GCTAGCAGGAGGAAACG ATG AAA AAG ACA GCT ATC GCG ATT GCA GTG GCA CTG GCT GGTA M ATTC GCT ACC GTA GCC ATG GCC GAG CTC GGTACCCGGGGATCCTCTAGAGTCGCCTGCAGGCATCCAAGCTTNco1 Pst1 Hind111 (b)、不含分泌信号肽的P BAD表达质粒多克隆区域EcoR1 Kpn1 BamH1 Pst1P BAD….TACCCGTTTTTTTGG.GCTAGCGAATTCGAGCTCGGTACCCGGGGATCCTCTAGAGTCGCCTGCAGGCATCCAAGCTTNde1 Sac1 Smal 1 Hind111下图显示本公司应用pBAD表达载体完成的实验结果:pBAD载体驱动大分子蛋白质在原核细胞Origami(DE3)内高效表达2. pCAl-n & pCAl-pelB载体特点: 该原核细胞表达载体是来源于以T7 RNA聚合酶为基础的pET载体,含有T7/LacO启动子、编码钙调素结合多肽标鉴和凝血酶切点的核苷酸序列。
pcDNA4 myc-His C哺乳动物表达载体说明

pcDNA4/myc-His C编号 载体名称北京华越洋生物VECT6123 pcDNA4/myc-‐His C pcDNA4/myc-‐His C载体基本信息载体名称: pcDNA4/myc-His C质粒类型: 哺乳动物表达载体;cDNA表达载体高拷贝/低拷贝: 高拷贝克隆方法: 多克隆位点,限制性内切酶启动子: CMV载体大小: 5071 bp5' 测序引物及序列: T7 Forward: 5’-TAATACGACTCACTATAGGG-3’ 3' 测序引物及序列: BGH Reverse: 5-TAGAAGGCACAGTCGAGG-3 载体标签: His Tag (C-端), c-Myc Epitope Tag(C-端)载体抗性: 氨苄青霉素筛选标记: Zeocin克隆菌株: TOP10F´, DH5a, JM109, TOP10宿主细胞(系): 常规细胞系,如293、Hela等备注: pcDNA4/myc-His C 载体是哺乳动物表达载体,适用于cDNA的表达与克隆;CMV启动子驱动目的基因的高水平表达;pcDNA4/myc-His A,B,C的区别仅在于多克隆位点处。
稳定性: 瞬表达或稳表达组成型/诱导型: 组成型病毒/非病毒: 非病毒pcDNA4/myc-‐His C载体质粒图谱和多克隆位点信息pcDNA4/myc-‐His C载体描述pcDNA4/myc-His A, B, and C are 5.1 kb vectors designed for overproduction of recombinant proteins in mammalian cell lines. Features of the vectors allow purification and detection of expressed proteins (see pages 11-12 for more information). High-level stable and transient expression can be carried out in most mammalian cells. The vectors contain the following elements:Human cytomegalovirus immediate-early (CMV) promoter for high-level expression in a wide range of mammalian cellsThree reading frames to facilitate in-frame cloning with a C-terminal peptide encoding the myc (c-myc) epitope and a polyhistidine (6xHis) metal-binding tagZeocin resistance gene for selection of stable cell lines (Mulsant et al., 1988) (see page 14 for more information).Episomal replication in cell lines that are latently infected with SV40 or that express the SV40 large T antigen (e.g., COS7).The control plasmid, pcDNA4/myc-His/lacZ is included for use as a positive control for transfection, expression, and detection in the cell line of choice.实验流程:Use the following outline to clone and express your gene of interest in pcDNA4/myc-His:1.Consult the multiple cloning sites described on pages 3-4 to determine which vector (A, B, or C) to use for cloning your gene in frame with the C-terminal myc epitope and the polyhistidine tag.2.Ligate your insert into the appropriate vector and transform into E. coli. Select transformants on 50 to 100 μg/mL ampicillin or 25 to 50g/mL Zeocin in Low Salt LB. For more information.3.Analyze your transformants for the presence of insert by restriction digestion.4.Select a transformant with the correct restriction pattern and use sequencing to confirm that your gene is cloned in-frame with the C-terminal peptide.5.Transfect your construct into the cell line of choice using your own method of transfection. Generate a stable cell line, if desired.6.Test for expression of your recombinant gene by western blot analysis or functional assay. For antibodies to the myc epitope or the C-terminal polyhistidine tag.7.To purify your recombinant protein, you may use metal-chelating resin such as ProBond. ProBond resin is available separatelypcDNA4/myc-‐His C载体序列ORIGIN1 GACGGATCGG GAGATCTCCC GATCCCCTAT GGTCGACTCT CAGTACAATC TGCTCTGATG61 CCGCATAGTT AAGCCAGTAT CTGCTCCCTG CTTGTGTGTT GGAGGTCGCT GAGTAGTGCG 121 CGAGCAAAAT TTAAGCTACA ACAAGGCAAG GCTTGACCGA CAATTGCATG AAGAATCTGC 181 TTAGGGTTAG GCGTTTTGCG CTGCTTCGCG ATGTACGGGC CAGATATACG CGTTGACATT 241 GATTATTGAC TAGTTATTAA TAGTAATCAA TTACGGGGTC ATTAGTTCAT AGCCCATATA 301 TGGAGTTCCG CGTTACATAA CTTACGGTAA ATGGCCCGCC TGGCTGACCG CCCAACGACC 361 CCCGCCCATT GACGTCAATA ATGACGTATG TTCCCATAGT AACGCCAATA GGGACTTTCC 421 ATTGACGTCA ATGGGTGGAC TATTTACGGT AAACTGCCCA CTTGGCAGTA CATCAAGTGT 481 ATCATATGCC AAGTACGCCC CCTATTGACG TCAATGACGG TAAATGGCCC GCCTGGCATT 541 ATGCCCAGTA CATGACCTTA TGGGACTTTC CTACTTGGCA GTACATCTAC GTATTAGTCA 601 TCGCTATTAC CATGGTGATG CGGTTTTGGC AGTACATCAA TGGGCGTGGA TAGCGGTTTG 661 ACTCACGGGG ATTTCCAAGT CTCCACCCCA TTGACGTCAA TGGGAGTTTG TTTTGGCACC 721 AAAATCAACG GGACTTTCCA AAATGTCGTA ACAACTCCGC CCCATTGACG CAAATGGGCG 781 GTAGGCGTGT ACGGTGGGAG GTCTATATAA GCAGAGCTCT CTGGCTAACT AGAGAACCCA 841 CTGCTTACTG GCTTATCGAA ATTAATACGA CTCACTATAG GGAGACCCAA GCTGGCTAGT 901 TAAGCTTGGT ACCGAGCTCG GATCCACTAG TCCAGTGTGG TGGAATTCTG CAGATATCCA 961 GCACAGTGGC GGCCGCTCGA GGTCACCCAT TCGAACAAAA ACTCATCTCA GAAGAGGATC 1021 TGAATATGCA TACCGGTCAT CATCACCATC ACCATTGAGT TTAAACCCGC TGATCAGCCT 1081 CGACTGTGCC TTCTAGTTGC CAGCCATCTG TTGTTTGCCC CTCCCCCGTG CCTTCCTTGA 1141 CCCTGGAAGG TGCCACTCCC ACTGTCCTTT CCTAATAAAA TGAGGAAATT GCATCGCATT 1201 GTCTGAGTAG GTGTCATTCT ATTCTGGGGG GTGGGGTGGG GCAGGACAGC AAGGGGGAGG 1261 ATTGGGAAGA CAATAGCAGG CATGCTGGGG ATGCGGTGGG CTCTATGGCT TCTGAGGCGG 1321 AAAGAACCAG CTGGGGCTCT AGGGGGTATC CCCACGCGCC CTGTAGCGGC GCATTAAGCG 1381 CGGCGGGTGT GGTGGTTACG CGCAGCGTGA CCGCTACACT TGCCAGCGCC CTAGCGCCCG 1441 CTCCTTTCGC TTTCTTCCCT TCCTTTCTCG CCACGTTCGC CGGCTTTCCC CGTCAAGCTC 1501 TAAATCGGGG CATCCCTTTA GGGTTCCGAT TTAGTGCTTT ACGGCACCTC GACCCCAAAA 1561 AACTTGATTA GGGTGATGGT TCACGTAGTG GGCCATCGCC CTGATAGACG GTTTTTCGCC 1621 CTTTGACGTT GGAGTCCACG TTCTTTAATA GTGGACTCTT GTTCCAAACT GGAACAACAC 1681 TCAACCCTAT CTCGGTCTAT TCTTTTGATT TATAAGGGAT TTTGGGGATT TCGGCCTATT 1741 GGTTAAAAAA TGAGCTGATT TAACAAAAAT TTAACGCGAA TTAATTCTGT GGAATGTGTG 1801 TCAGTTAGGG TGTGGAAAGT CCCCAGGCTC CCCAGGCAGG CAGAAGTATG CAAAGCATGC 1861 ATCTCAATTA GTCAGCAACC AGGTGTGGAA AGTCCCCAGG CTCCCCAGCA GGCAGAAGTA 1921 TGCAAAGCAT GCATCTCAAT TAGTCAGCAA CCATAGTCCC GCCCCTAACT CCGCCCATCC 1981 CGCCCCTAAC TCCGCCCAGT TCCGCCCATT CTCCGCCCCA TGGCTGACTA ATTTTTTTTA 2041 TTTATGCAGA GGCCGAGGCC GCCTCTGCCT CTGAGCTATT CCAGAAGTAG TGAGGAGGCT 2101 TTTTTGGAGG CCTAGGCTTT TGCAAAAAGC TCCCGGGAGC TTGTATATCC ATTTTCGGAT 2161 CTGATCAGCA CGTGTTGACA ATTAATCATC GGCATAGTAT ATCGGCATAG TATAATACGA 2221 CAAGGTGAGG AACTAAACCA TGGCCAAGTT GACCAGTGCC GTTCCGGTGC TCACCGCGCG 2281 CGACGTCGCC GGAGCGGTCG AGTTCTGGAC CGACCGGCTC GGGTTCTCCC GGGACTTCGT 2341 GGAGGACGAC TTCGCCGGTG TGGTCCGGGA CGACGTGACC CTGTTCATCA GCGCGGTCCA 2401 GGACCAGGTG GTGCCGGACA ACACCCTGGC CTGGGTGTGG GTGCGCGGCC TGGACGAGCT 2461 GTACGCCGAG TGGTCGGAGG TCGTGTCCAC GAACTTCCGG GACGCCTCCG GGCCGGCCAT 2521 GACCGAGATC GGCGAGCAGC CGTGGGGGCG GGAGTTCGCC CTGCGCGACC CGGCCGGCAA 2581 CTGCGTGCAC TTCGTGGCCG AGGAGCAGGA CTGACACGTG CTACGAGATT TCGATTCCAC 2641 CGCCGCCTTC TATGAAAGGT TGGGCTTCGG AATCGTTTTC CGGGACGCCG GCTGGATGAT2701 CCTCCAGCGC GGGGATCTCA TGCTGGAGTT CTTCGCCCAC CCCAACTTGT TTATTGCAGC 2761 TTATAATGGT TACAAATAAA GCAATAGCAT CACAAATTTC ACAAATAAAG CATTTTTTTC 2821 ACTGCATTCT AGTTGTGGTT TGTCCAAACT CATCAATGTA TCTTATCATG TCTGTATACC 2881 GTCGACCTCT AGCTAGAGCT TGGCGTAATC ATGGTCATAG CTGTTTCCTG TGTGAAATTG 2941 TTATCCGCTC ACAATTCCAC ACAACATACG AGCCGGAAGC ATAAAGTGTA AAGCCTGGGG 3001 TGCCTAATGA GTGAGCTAAC TCACATTAAT TGCGTTGCGC TCACTGCCCG CTTTCCAGTC 3061 GGGAAACCTG TCGTGCCAGC TGCATTAATG AATCGGCCAA CGCGCGGGGA GAGGCGGTTT 3121 GCGTATTGGG CGCTCTTCCG CTTCCTCGCT CACTGACTCG CTGCGCTCGG TCGTTCGGCT 3181 GCGGCGAGCG GTATCAGCTC ACTCAAAGGC GGTAATACGG TTATCCACAG AATCAGGGGA 3241 TAACGCAGGA AAGAACATGT GAGCAAAAGG CCAGCAAAAG GCCAGGAACC GTAAAAAGGC 3301 CGCGTTGCTG GCGTTTTTCC ATAGGCTCCG CCCCCCTGAC GAGCATCACA AAAATCGACG 3361 CTCAAGTCAG AGGTGGCGAA ACCCGACAGG ACTATAAAGA TACCAGGCGT TTCCCCCTGG 3421 AAGCTCCCTC GTGCGCTCTC CTGTTCCGAC CCTGCCGCTT ACCGGATACC TGTCCGCCTT 3481 TCTCCCTTCG GGAAGCGTGG CGCTTTCTCA ATGCTCACGC TGTAGGTATC TCAGTTCGGT 3541 GTAGGTCGTT CGCTCCAAGC TGGGCTGTGT GCACGAACCC CCCGTTCAGC CCGACCGCTG 3601 CGCCTTATCC GGTAACTATC GTCTTGAGTC CAACCCGGTA AGACACGACT TATCGCCACT 3661 GGCAGCAGCC ACTGGTAACA GGATTAGCAG AGCGAGGTAT GTAGGCGGTG CTACAGAGTT 3721 CTTGAAGTGG TGGCCTAACT ACGGCTACAC TAGAAGGACA GTATTTGGTA TCTGCGCTCT 3781 GCTGAAGCCA GTTACCTTCG GAAAAAGAGT TGGTAGCTCT TGATCCGGCA AACAAACCAC 3841 CGCTGGTAGC GGTGGTTTTT TTGTTTGCAA GCAGCAGATT ACGCGCAGAA AAAAAGGATC 3901 TCAAGAAGAT CCTTTGATCT TTTCTACGGG GTCTGACGCT CAGTGGAACG AAAACTCACG 3961 TTAAGGGATT TTGGTCATGA GATTATCAAA AAGGATCTTC ACCTAGATCC TTTTAAATTA 4021 AAAATGAAGT TTTAAATCAA TCTAAAGTAT ATATGAGTAA ACTTGGTCTG ACAGTTACCA 4081 ATGCTTAATC AGTGAGGCAC CTATCTCAGC GATCTGTCTA TTTCGTTCAT CCATAGTTGC 4141 CTGACTCCCC GTCGTGTAGA TAACTACGAT ACGGGAGGGC TTACCATCTG GCCCCAGTGC 4201 TGCAATGATA CCGCGAGACC CACGCTCACC GGCTCCAGAT TTATCAGCAA TAAACCAGCC 4261 AGCCGGAAGG GCCGAGCGCA GAAGTGGTCC TGCAACTTTA TCCGCCTCCA TCCAGTCTAT 4321 TAATTGTTGC CGGGAAGCTA GAGTAAGTAG TTCGCCAGTT AATAGTTTGC GCAACGTTGT 4381 TGCCATTGCT ACAGGCATCG TGGTGTCACG CTCGTCGTTT GGTATGGCTT CATTCAGCTC 4441 CGGTTCCCAA CGATCAAGGC GAGTTACATG ATCCCCCATG TTGTGCAAAA AAGCGGTTAG 4501 CTCCTTCGGT CCTCCGATCG TTGTCAGAAG TAAGTTGGCC GCAGTGTTAT CACTCATGGT 4561 TATGGCAGCA CTGCATAATT CTCTTACTGT CATGCCATCC GTAAGATGCT TTTCTGTGAC 4621 TGGTGAGTAC TCAACCAAGT CATTCTGAGA ATAGTGTATG CGGCGACCGA GTTGCTCTTG 4681 CCCGGCGTCA ATACGGGATA ATACCGCGCC ACATAGCAGA ACTTTAAAAG TGCTCATCAT 4741 TGGAAAACGT TCTTCGGGGC GAAAACTCTC AAGGATCTTA CCGCTGTTGA GATCCAGTTC 4801 GATGTAACCC ACTCGTGCAC CCAACTGATC TTCAGCATCT TTTACTTTCA CCAGCGTTTC 4861 TGGGTGAGCA AAAACAGGAA GGCAAAATGC CGCAAAAAAG GGAATAAGGG CGACACGGAA 4921 ATGTTGAATA CTCATACTCT TCCTTTTTCA ATATTATTGA AGCATTTATC AGGGTTATTG 4981 TCTCATGAGC GGATACATAT TTGAATGTAT TTAGAAAAAT AAACAAATAG GGGTTCCGCG 5041 CACATTTCCC CGAAAAGTGC CACCTGACGT C//其他哺乳动物表达载体:pCHO1.0 pBApo-CMV-Pur pOPRSVIpcDNA3.1/His C pcDNA5/FRT/V5-His-TOPO pREP4pcDNA3.1/His B pcDNA5/FRT/TO-TOPO pDual-GCpcDNA3.1/His A pcDNA5/TO pBK-RSVpIRESpuro3 pcDNA5/FRT/TO pBK-CMVpIRES2-EGFP pcDNA5/FRT pBI-CMV4pTT5 pFLAG-CMV2 pcDNA4/TO/Myc-His/LacZ pNFkB-DD-tdTomato pcDNA3.1/CT-GFP-TOPO pOPI3CATpBI-CMV5 pcDNA3.1/NT-GFP-TOPO pGene/V5-His B pSEAP2-Basic pOptiVEC-TOPO pSwitchpSEAP2-Control pCMV-MEKK1 pCMVLacIpBI-CMV3 pCMV-MEK1 pVgRxRpBI-CMV2 pCMV-PKA pINDpBI-CMV1 pcDNA6.2/nTC-Tag-DEST pTRE3G-LucpNFκB-MetLuc2-Reporter pcDNA6.2/cTC-Tag-DEST pTRE3GpCRE-MetLuc2-Reporter pcDNA3.2/V5/GW/D-TOPO pTRE2-hygropAcGFP1-Actin pcDNA6.2/V5/GW/D-TOPO pTRE-TightpAcGFP1-N In-Fusion Ready pcDNA6.2/nGeneBLAzer-GW/D-TOPO pTK-hygpAcGFP1-C3 pcDNA6.2/C-YFP-DEST pTet-OnpAcGFP1-C pcDNA6.2/cGeneBLAzer-DEST pTet-OffpAcGFP1-p53 pcDNA6/V5-His A pTet on advanced pAcGFP1-Mito pcDNA6/V5-His B pRevTREpAcGFP1-Mem pcDNA6/V5-His C pRevTet-OnpAcGFP1-Lam pcDNA6/myc-His C pRevTet-OffpAcGFP1-Golgi pcDNA6/myc-His A pCMV-Tet3GpAcGFP1-F pcDNA6/myc-His B pTRE2pAcGFP1-Hyg-C1 pcDNA6.2/nGeneBLAzer-DEST pBD-NF-κBpAsRed2-N1 pcDNA4/HisMax-TOPO pCMV-ADptdTomato-N1 pcDNA6.2/nLumio-DEST pCMV-BDpCMV-tdTomato pcDNA6.2/cLumio-DEST pBIND-Id ControlpCRE-DD-tdTomato pcDNA4/myc-His C pBINDpCMV-DsRed-Express2 pcDNA4/HisMax C pG5 luciferasepEF1α-tdTomato pcDNA4/HisMax A pACT-MyoDpCRE-hrGFP c-Flag pcDNA3 pACTptdTomato-C1 pcDNA4/HisMax B pCMV-SPORT6 pAsRed2-C1 pcDNA4/myc-His B pGL4.13pGL3-Promoter pcDNA4/myc-His A pGL4.19pGL3 basic pcDNA4/His C pGL4.26pAcGFP1-C2 pcDNA4/His B pGL4.20pAcGFP1-C1 pcDNA4/His A pGL4.29pAcGFP1-N3 pcDNA6/TR pGL4.30pAcGFP1-N2 pcDNA4/TO/Myc-His A pGL4.27pAcGFP1-N1 pcDNA4/TO pGL4.75pAcGFP1-C In-Fusion Ready pcDNA4/TO/Myc-His B pGL4.10pCRE-DD-AmCyan1 pcDNA4/TO/Myc-His C pGRN145pNFkB-DD-AmCyan1 pcDNA3.3-TOPO pSecTag2 A pDsRed2-Bid pBudCE4.1 pEBVHis B pDsRED2-Mito pFLAG-CMV-4 pEBVHis ApDD-AmCyan1 Reporter pFLAG-CMV-3 pCMV-Tag 3C pAmCyan1-N1 pFLAG-CMV-2 pCMV-Tag 3A pAmCyan1-C1 pFLAG-CMV-5a pCMV-Tag 3BpEF1α-IRES-DsRed-Express2 p3XFLAG-CMV-9 pCMV-Tag 5CpEF1α-DsRed-Monomer-N1 p3xFLAG-CMV-10 pCMV-Tag 5A pDsRED-Monomer-N1 p3XFLAG-CMV-8 pCMV-Tag 4A pDsRed-Express-N1 p3XFLAG-CMV-7.1 pCMV-Tag 5Bp3XFLAG-CMV-7 pDsRed-Monomer-N In-Fusion Ready pCMV-Tag 4B pDsRed-Express-C1 p3XFLAG-CMV-13 pCMV-Tag 2C pIRES2-ZsGreen1 p3XFLAG-CMV-14 pCMV-Tag 2B pDsRed-Express2-C1 plRES2-ZsGreen1 pCMV-Tag 2A pDsRed-Express2-N1 pBApo-EF1α-pur pCMV-LacZpEF1α-DsRed-Express2 pBApo-EF1α-neo pCMV-MycpIRES2-DsRed-Express pBApo-CMV pEF1α-IRES-AcGFP1 pIRES2-DsRed-Express2 pBApo-CMV-neo pEF1α-IRES-ZsGreen1 pIRES-hrGFP-1a pIRES-EGFP pEF1α-AcGFP1-N1 pIRESneo2 pIRESneo3 pIRES2-DsRed2 pIRESneo pDsRed-Monomer pIRES2-AcGFP1 pIREShyg3 pIRES。
(完整版)所有质粒载体汇总,推荐文档

酿酒酵母表达载体pYES2,pYES2/NT,pYES2/CT,pYES3,pYES6, pYCplac22-GFP,酵母载体pAUR123,pRS303TEF,pRS304, pRS305,pRS306,pY13TEF,pY14TEF,pY15TEF,pY16TEF,酵母基因重组表达载体pUG6, pSH47,酵母单杂载体pHISi,pLacZi,pHIS2, pGAD424, 酵母双杂交系统:酿酒酵母Y187, 酿酒酵母AH109;质粒pGADT7,pGBKT7;对照质粒pGBKT7-53,pGBKT7-lam,pGADT7-T,PCL1,酿酒酵母菌株INVSc1,YM4271, AH109,Y187,Y190,毕赤酵母表达载体pPIC9K,pPIC9K-His,pPIC3.5K,pPICZalphaA,B,C,pPICZA,B,C,pGAPZαA,pAO815,pPIC9k-His,pHIL-S1,pPink hc,配套毕赤酵母Pichiapink,毕赤酵母宿主X33,KM71,KM71H,GS115,原核表达载体pQE30,31,32,40,60,61,62,等原核表达载体,包括pET系列,pET-GST,pGEX系列(含GST标签),pMAL系列pMAL-c2x,-c4x,-c4e,-c5x,-p5x,pBAD,pBADHis,pBADmycHis系列,pQE系列,pTrc99a,pTrcHis系列,pBV220,221,222,pTXB系列,pLLP-ompA,pIN-III-ompA (分泌型表达系列),pQBI63(原核表达带荧光)pET3a, pET 3d, pET 11a, pET 12a, pET 14b, pET 15b, pET 16b, pET 17b, pET 19b, pET 20b, pET 21a,b,d, pET 22b, pET 23a, pET 23b, pET 24a,b, pET 25b, pET 26b, pET 27b, pET 28a,b, pET 29a, pET 30a, pET 31b, pET 32a, pET 35b, pET 38b, pET 39b, pET 40b, pET 41a,b pET 42a, pET 43.1a,b pET 44a, pET 49b pET302,303 pET His,pET Dsb,pET GST,pET Trx pQE2, pQE9 pQE30,31,32, pQE 40 pQE70 pQE80L pQETirs system pRSET-A pRSET-B pRSET-C pGEX4T-1,-2,-3,5x-1,6p-1,6p-2,2tk,3c pBV220,221,222 pTrcHisA,B,C pBAD24,34,43 pBAD HisA,B,C pPinPoint-Xa1,Xa2,Xa3 pMALc2x, p2x pBV220 pGEM Ex1, pGEM7ZF(+), pTrc99A, pTwin1, pEZZ18 pkk232-8,pkk233-3,pACYC184,pBR322,pUC119 pTYB1,pTYB2,pTYB4,pTYB11 pBlueScript SK (+),pBlueScript SK(-)pLLP ompA, pINIIIompA, pMBP-P ,pMBP-C, 大肠杆菌冷激质粒: pColdI pColdII pColdIII pColdTF 原核共表达质粒:pACYCduet-1,pETduet-1,pCDFduet-1,pRSFduet-1 Takara公司大肠杆菌分子伴侣: pG-KJE8 pGro7 pKJE7 pGTf2 pTf16 大肠杆菌宿主细胞: DH5a JM101 JM103JM105 JM107 JM109 JM110 Top10 Top10F BL21(DE3)HB101 ER2529 E2566 C2566 MG1655 XL-10gold XL blue M15 JF1125 K802 SG1117 BL21(AI)BL21(DE3)plysS TG1 TB1 DH5a(pir)Tuner(DE3)Bl21 codonplusRIPL Novablue (DE3)Rosetta Rosetta(DE3)Rosetta(DE3)plys Rosetta-gami(DE3)Rosetta-gamiB(DE3), Rosetta-gamiB(DE3)plysS Orgami(DE3)OrgamiB(DE3)HMS174(DE3)植物表达/RNAi载体农杆菌pBI121,pBI121-GFP,pBI101,pBI221,pSN1301,pUN1301,pRTL2 , pRTL2-GFP , pRTL2-CFP, pRTL2-RFP , pRTL2-YFP,pCAMBIA 1300, 1301, 1302,1303,1304,1305, 1381Z,1391Z,2300, 2301,3300,3301,pCAMBIA super1300,pCAMBIAsuper1300-GFP,pPZP212,pPZP2121,pPZP212-GFP,pGDG,RNAi载体pART27,pHANNIBAL,pKANNIBAL, pFGC5941,pTCK303, pTRV1,pTRV2,T-DNA插入载体(随机突变体库)pSKI015,pSKI074,真菌ATMT载体pBIG2RHPH2-GUS-GFP,pBHt1枯草芽孢杆菌表达载体pWB980,pHT43,pHP13,pHP43,pBE2,pMUTIN4,pUB110,pE194,pMA5, pMK3,pMK4,pHT304,pHY300PLK,pBest502,pDG1363,pSG1154,pAX01, pSAS144,pDL,pDG148-stu,pDG641,pAL12,pUCX05-bgaB,pHT01,配套菌株BS 168,WB600,WB800,WB700,WB800N,1012,FZB42,1A747,广宿主质粒pVLT33RNAi基因沉默干扰敲除载体pSilencer1.0,pSilencer 2.1-U6 hygro, pSilencer 3.1-H1 hygro,pSilencer 3.1-H1 neo, pSilencer 4.1-CMV neo, pSilencer 4.1-CMV puro pMIR-REPORT Luciferase RNAi载体(oligoengine)pSuper-puro RNAi逆转录病毒载体(clontech): RNAi-Ready pSIREN-Retro Q, RNAi-ReadypSIREN-RetroQ-ZsGreen(Luciferase shRNA Annealed Oligonucleotide)RNAi慢病毒载体(addgene): pLKO.1哺乳动物表达载体pcDNA3.1+/-,pcDNA4/HisMax B,pSecTag2 A,pVAX1,pBudCE4.1,pTracer CMV2,pcDNA3.1(-)/myc-His A ,pcDNA6-Myc/His B,pCEP4, pIRES,pIRESneo,pIRES hyg3,pCMV-myc,pCMV-HA,pIRES-puro3,pIRES-neo3,pCAGGS哺乳动物双杂交系统pACT,pBIND,pACT-MyoD,pBIND-Id,pG5luc,pCMV-BD, pCMV-AD, pBD-p53, pFR-luc,Cytotrap Two-Hybrid System:pSos, pSos MAFB, pMyr蜕皮激素诱导系统pIND, pVgRxR,LacSwith II哺乳动物诱导表达系统:pOPRSVI ,pOPI3CAT,pCMVLacI,GeneSwitch System:pSwitch哺乳动物表面展示系统:pDisplay, 四环素调控系统(Invitrogen):pcDNA4/TO/Myc-His A,pcDNA4/TO/Myc-His B,pcDNA4/TO/Myc-His C,pcDNA4/TO/Myc-His/LacZ,pcDNA6/TR四环素调控系统(Clontech):pTet-On,pTet-Off,pTRE2,pRevTRE,pRevTet-On,pRevTet-off信号通路报告载体:pGAS-TA-Luc,pSTAT3-TA-Luc, pISRE-TA-Luc, pTA-Luc,pIκB-EGFP,pNFAT-TA-Luc,pCaspase3-sensor,pAP1(PMA)-Luc;pGL4.26[luc2P/minP/Hygro],pGL4.29[luc2P/CRE/Hygro],pGL4.30[luc2P/NFA T-RE/Hygro],pGL4.75;p53-Luc,pAP-1-Luc, pNF-κB-Luc,pSRE-Luc,pFA2-Elk1,pFC-MEKK,pFR-luc,Gateway系统(invitrogen)pcDNA6.2-GWEmGFP-miR negative, pLenti 6/TR,pcDNA 6.2-GW EmGFP-miR,乳酸菌表达载体及各种乳酸菌乳酸杆菌菌株,pNZ8148,pLEISS,pMG36e,pBBR1MCS-5,pBBR1MCS-6,pRV610,pLEM415,pHY3 00PLK,分泌型乳酸菌表达载体pVE5523,pPG611.1,pPG612.1等和乳酸杆菌菌株宿主菌NZ9000,MG1363,Lactobacillus casei 1.539,Lactobacillus casei,acidophilus NCFM,1.2,Lactobacillus sakei 23K,L.plantarum,L.rhamnosusGG,B.coagulans,Bifidobacterium bifidum,Bifidobacterium infantis,Lactococcus lactis M17,1663,Lactobacillus reuterii广宿主表达载体链球菌表达敲除载体假单胞菌表达载体pVLT33,pBBR1MCS-2,3,4,5,6, pJRD215,pJN105,pME6032,Cos载体pLAFR3,pMP2444(GFP), pHY300PLK,pRT102,pRL1063a, 转座子载体pUT-miniTn5,pMGS100, pWHM10,pKC1139,pSET152,pOJ260,pPG611.1,pPG612.1,腺病毒载体/慢病毒,逆转录病毒表达载体及包装包膜质粒,腺病毒系统(Stratagene): pAdEasy-1,pShuttle-CMV,pShuttle,pAdTrack, pAdTrack-CMV, pShuttle-IRES-hrGFP-1、pShuttle-IRES-hrGFP-2、pShuttle-CMV-lacZ,pShuttle-CMV-EGFP-C,pXC1, pBHGE3, 配套大肠杆菌BJ5183,293,293T cellline 腺相关病毒系统(Stratagene): pAAV-MCS,pAAV-RC,pHelper,pAAV-LacZ,pAAV-IRES-hrGFP,pCMV-MCS,慢病毒载体:pLVX-DsRed-Monomer-N1,pLVX-IRES-ZsGreen1,pLVX-AcGFP1-N1,Lenti6/v 5-EDST-EGFP,pWPXL, FUGW,pLentilox 3.7,RNAi-Ready pSIREN-Retro Q,RNAi-Ready pSIREN-Retro Q-ZsGreen,pSUPER.Retro-GFP/Neo,pSUPER-Retro-Neo, pSUPER.Retro-puro,PLNCX PLNCX2 pMSCV-HYG pMSCV-neo pMSCV-puro pLEGFP-C1 pLOX-CW-CRE pLOX-GFP-IRES-TK pRetroX-IRES-DsRedExpress, pLVX-IRES-mCherry质粒载体。
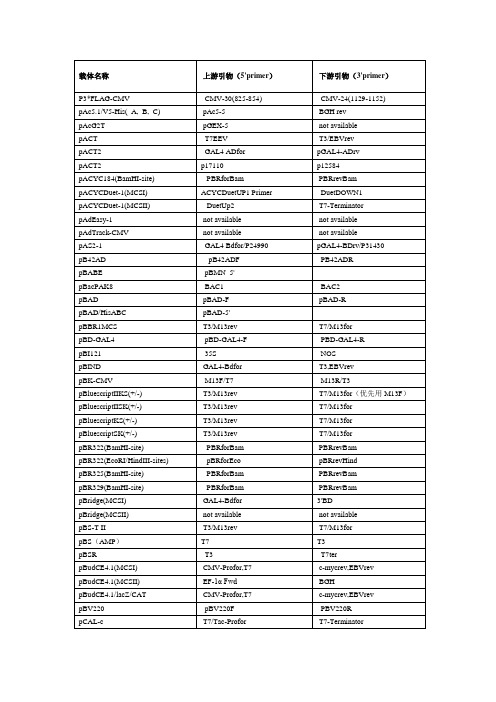
载体及其上下游引物
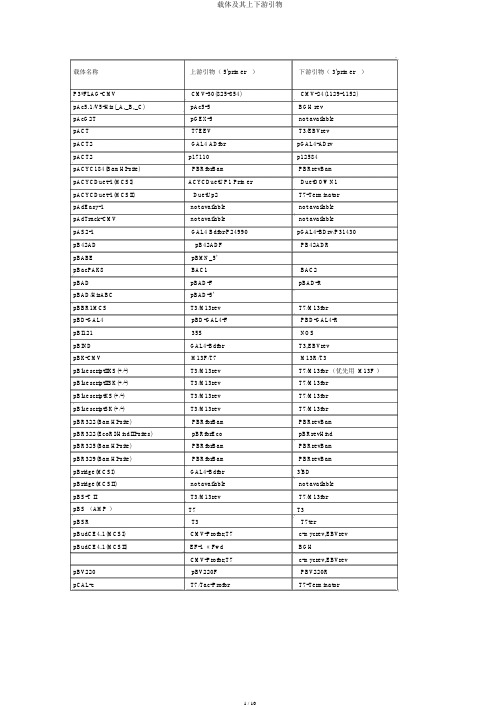
载体名称上游引物( 5'primer)下游引物( 3'primer)P3*FLAG-CMV CMV-30(825-854)CMV-24(1129-1152)pAc5.1/V5-His(_A,_B,_C)pAc5-5BGH revpAcG2T pGEX-5not availablepACT T7EEV T3/EBVrevpACT2GAL4 ADfor pGAL4-ADrvpACT2p17110p12584pACYC184(BamHI-site)PBRforBam PBRrevBampACYCDuet-1(MCSI)ACYCDuetUP1 Primer DuetDOWN1pACYCDuet-1(MCSII)DuetUp2T7-TerminatorpAdEasy-1not available not availablepAdTrack-CMV not available not availablepAS2-1GAL4 Bdfor/P24990pGAL4-BDrv/P31430pB42AD pB42ADF PB42ADRpBABE pBMN_5'pBacPAK8BAC1BAC2pBAD pBAD-F pBAD-RpBAD/HisABC pBAD-5'pBBR1MCS T3/M13rev T7/M13forpBD-GAL4pBD-GAL4-F PBD-GAL4-RpBI12135S NOSpBIND GAL4-Bdfor T3,EBVrevpBK-CMV M13F/T7M13R/T3pBluescriptIIKS(+/-)T3/M13rev T7/M13for (优先用 M13F )pBluescriptIISK(+/-)T3/M13rev T7/M13forpBluescriptKS(+/-)T3/M13rev T7/M13forpBluescriptSK(+/-)T3/M13rev T7/M13forpBR322(BamHI-site)PBRforBam PBRrevBampBR322(EcoRI/HindIII-sites)pBRforEco pBRrevHindpBR325(BamHI-site)PBRforBam PBRrevBampBR329(BamHI-site)PBRforBam PBRrevBampBridge(MCSI)GAL4-Bdfor3'BDpBridge(MCSII)not available not availablepBS-T II T3/M13rev T7/M13forpBS (AMP )T7T3pBSR T3T7terpBudCE4.1(MCSI)CMV-Profor,T7c-mycrev,EBVrevpBudCE4.1(MCSII)EF-1 αFwd BGHCMV-Profor,T7c-mycrev,EBVrevpBV220pBV220F PBV220RpCAL-c T7/Tac-Profor T7-TerminatorpCAL-n T7/Tac-Profor T7-Terminator pCAMBIA1301(1300)pCAMBIA1301-5'(ECORI)/M13R pCAMBIA1301-3'(HindIII) pCAMBIA1304pCAMBIA1301-5'(ECORI)pCAMBIA1301-3'(HindIII) pCAMBIA2300M13R(-48)M13F(-47)/M13F(-49) pCANTB5E S1S6pCAT3-enhancer RVP3此载体无反向引物pCDFDuet-1(MCSI)ACYCDuetUP1 Primer DuetDOWN1 pCDFDuet-1(MCSII)DuetUP2T7TERCMV-Profor,T7pCDM8-3M13for/T7M13rev/Tac-Profor pcDNA3T7/CMV-Profor BGH rev/SP6pcDNA3.1(+/-)BGH revpcDNA3.1/His(_A,_B,_C)T7/CMV-Profor BGH revpcDNA3.1/myc-His(_A,_B,_C)T7/CMV-Profor BGH rev/c-mycrev pcDNA3.1/V5-His(_A,_B,_C)T7/CMV-Profor BGH rev/V5revpcDNA3.1/Hygro(+,-)T7/CMV-Profor BGH rev/V5revpcDNA3.1/Zeo(+.-)T7BGHrevCMV-Profor TKPAreversepcDNA4/HisMax(_A,_B,_C)Xpressfor BGH revpcDNA4/HisMax-TOPO Xpressfor BGH revpcDNA4/HisMax-TOPO/lacZ Xpressfor BGH revpcDNA4/myc-His(_A,_B,_C)T7,CMV-Profor BGH rev,c-mycrev pcDNA4/TO TREfor/CMV-Profor BGH revpcDNA4/V5-His(_A,_B,_C)CMV-Profor,T7BGH rev,V5revpcDNA4-TET/ON/AMP+CMV-ProforpcDNA5/FRT T7/CMV-Profor BGH revpcDNA6/myc-His(_A,_B,_C)CMV-Profor,T7BGH rev,c-mycrev pcDNA6/V5-His(_A,_B,_C)CMV-Profor,T7BGH rev,V5revpCEP4pCEP-F/CMV-Profor EBVrevpCI T7(17BASE )EBVrevpCI-neo T7(17BASE )EBVrev,T3pCMS-EGFP T7T3pCMS-EGFP T7EBVrev,T3pCMV-3Tag-4A T3 /PFLAG-CMV-F T7pCMV5pCMV5F PCMV5RpCMV6CMV-Profor pCMV6-3 、 HGHrev pCMV6-XL4CMV-Profor/T7pCMV6-3 、 HGHrev pCMV-HA PCMV-F,CMV-Profor,TREfor PCMV-R ,EBVrev pCMV-MYC(AMP+)PCMV-F PCMV-R, EBVrev pCMVmyc/nuc CMV-Profor BGH rev,c-mycrev pcmv-scriptEXVector T3T7pCMV-Sport6T7/M13for SP6/M13rev pCOLADuet-1(MCSI)ACYCDuetUP1 Primer DuetDOWN1pCOLADuet-1(MCSII)DuetUP2T7TERT7,M13for M13revM13for,T7M13revT7BGH revT7BGH revpCR4T3/M13rev T7/M13forpCR4-Blunt T3/M13rev T7/M13forpCR4-TOPO M13rev,T3M13for,T7pCR-Blunt M13rev T7,M13forpCR-BluntII M13rev/SP6T7/M13forpCR-BluntII-TOPO M13rev/SP6M13for/T7pCRII M13rev/SP6T7/M13forpCRII-TOPO M13rev/SP6M13for/T7pCR-ScriptSK(+)T7/M13for T3/M13revpCR-XL-TOPO M13rev T7,M13forpCS2+(MCSI)SP6EBVrev,T7?pCS2+(MCSII)T3pSG5-3pDB_GAL4cam T7PpDB_leu PDBLeu-5PDBLeu-3pDEST22DEST22F DEST22RpDEST15pGEX-5T7-TerminatorpDEST20pGEX-5pFastBac-3pDEST26TREfor/CMV-Profor T7/M13forpDisplay T7/CMV-Profor c-mycrevpDNR-LIB(MCS_A)M13for,T7pSG5-3,M13RpDonar M13F M13RpDONR201PDONR-F PDONR-RpDONR207PDONR-F PDONR-RpDONR223M13F M13RpDrive T7/M13rev SP6/M13forpDsRED1-C1(DsRed1-N-f)DsRed1-C-rpDsRed1-N1CMV-Profor RFP-Nrev,CMV-R?pDsRed2-N1(DsRed1-N-f)DsRed1-C-rpDsRed2-C1(DsRed1-N-f)DsRed1-C-rpDsRED2-C1(KAN+)pDsRED-ex-C1-F pEGFP-N-3’pDsRed-Express dsRed1_N_primer/CMV-F dsRed1_C_primer/CMV-R pDSRED-monomer-N1(KAN+)dsRed1_N_primerpDSRED-N1(KAN+)pEGFP-N-5’pDSRED-N-RpEASY-Blunt M13F T7/ M13RT7/ M13R (距离插入位点适合,pEASY-BluntSimple M13F优先安排)M13R (距离插入位点适合,优pEASY-T1 Simple M13F先安排)pEASY-T1pEASY-T3pECFP-N(C)pEF/myc/cyto/ER/mito/nuc pEF1(4,6/myc-His)pEF1(4,6/V5-His)pEF1(4;6/His_A;_B;_C) pEF5/FRT/V5-D(-TOPO) pEGFPpEGFP-1pEGFP-CpEGFP-C1pEGFP-C2pEGFP-C3pEGFP-FpEGFP-NpEGFP-N1pEGFP-N2pEGFP-N3pENTRpET-11(-a,-b,-c,-d)pET-12(-a,-b,-c)pET-14bpET-15bpET-16bpET-17bpET-17xbpET-19bpET-20b(+)pET-21(-a,-b,-c,-d)(+) pET-22b(+)pET-23(-a,-b,-c,-d)(+) pET-24(-a,-b,-c,-d)(+) pET-25b(+)pET-26b(+)pET-27b(+)pET-28a(-b,-c)(+)pET-29a(-b,-c)(+)pET-3(-a,-b,-c,-d)pET-30_Ek/LICpET-30_Xa/LICpET-30a(-b,-c)(+)pET-31b(+)M13F/T7M13F/T7PECFP-N-5.pEF-5T7/pEF-5T7/pEF-5T7/XpressforT7/pEF-5M13revnot availablePEGFP-C-5′EGFP-CforEGFP-CforEGFP-CforEGFP-CforPEGFP-N-5′CMV-ProforCMV-ProforCMV-ProforM13FT7T7T7T7T7T7T7T7T7T7T7T7T7T7T7T7T7T7T7T7not availableM13RM13R/SP6PECFP-N-3,EGFP-NrevEGFP-Nrev/ PEGFP-N-3 ′PEGFP-C-3′SV40-pArevSV40-pArevSV40-pArevSV40-pArevPEGFP-N-3′EGFP-NrevEGFP-NrevEGFP-NrevM13RT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorT7-TerminatorpET-32_Ek/LIC TRXfor T7-TerminatorpET-32_XA/LIC TRXfor T7-TerminatorpET-32a(-b,-c)(+)T7-TerminatorpET-33b(+)T7T7-TerminatorpET-34b(+)T7-TerminatorpET-35b(+)T7-TerminatorpET-36b(+)T7-TerminatorpET-37b(+)T7-TerminatorpET-38b(+)not available T7-TerminatorpET-39b(+)T7-TerminatorpET-40b(+)T7-TerminatorpET-41_Ek/LIC GST-Cfor T7-TerminatorpET-41a(-b,-c)(+)T7-TerminatorpET-42a(-b,-c)(+)T7-TerminatorpET-43.1a(-b,-c)(+)coliDOWNnot availablepET-44a(-b,-c)(+)T7-TerminatorpET-44a_EK/LIC not availablepET-45b(+)T7T7-TerminatorpET-46_Ek/LIC T7T7-TerminatorpET-5(-a,-b,-c)T7T7-TerminatorpET-52b(+)T7T7-TerminatorpET-7T7T7-TerminatorpET-9(-a,-b,-c,-d)T7T7-Terminator pETcoco-1T7T7-Terminator pETDuet-1(MCSI)pETUP1DuetDOWN1 pETDuet-1(MCSII)DuetUp2T7-Terminator pET-T4T7T7-Terminator pEYFP-C1pEGFP-C-5’pEGFP-C-3' pEYFP-N1pEGFP-N-5’ ,CMV-Profor pEGFP-N-3’pFLAG-ATS N26for pTrcHis-rv ; C24rev pFLAG-CTC N26for pTrcHis-rv ; C24rev pFLAG1N26for pTrcHis-rv ; C24rev pFLAG-CMV(-2)CMV-30 ; CMV-Profor CMV-24pFLAG-CMV1CMV-30CMV-24pGAD424GAL4_ADfor3'ADpGADGH GAL4_ADfor3'ADpGADT7T7/GAL4_ADfor3'ADpGADT7-Rec T7/GAL4_ADfor3'ADpGADT-T T73,BDpGAD10GAL4_ADfor3'ADpGAPZa-A a-FACTOR3'AOXpGBKT7T73,BDpGBT9MATCHMAKER5'DNA-BD/GAL4BD MATCHMAKER3'DNA-BDpGEM-1T7SP6pGEM-3T7SP6pGEM-3Zf(+)(-)SP6/M13rev T7/M13forpGEM-4T7SP6pGEM-4Z T7/M13for SP6/M13revpGEM-5Zf(+)(-)SP6/M13rev T7/M13forpGEM-7Zf(+)(-)SP6/M13rev T7/M13forpGEM-9Zf(+)(-)T7/M13for SP6/M13revpGEM-T T7/M13for SP6/M13revpGEM-T_Easy T7/M13for SP6/M13revpGene/V5-His_A(_B,_C)pGENE-5BGH rev,V5revpGEX-2T pGEX-5pGEX-3pGEX-2TK pGEX-5pGEX-3pGEX-3X pGEX-5pGEX-3pGEX-4T-1(-2,-3)pGEX-5pGEX-3pGEX-5X-1(-2,-3)pGEX-5pGEX-3pGEX-6P-1(-2,-3)pGEX-5pGEX-3pGL2GLP1GLP2GLP2(RVP4)( 没有特别说明用pGL3RVP3GLP2)pGL3-Basic(MCSI)RVP3(PGL3+)?GLP2(PGL3-)?pGL3-Basic(MCSII)RVP4,PLXSN-3'?pGL4pGlow(-TOPO)T7pGlow-3pGM-T T7SP6pGWC SP6T7pHook-2T7/CMV-Profor not availablepIND/V5-His_A(_B,_C)not available BGH rev,V5rev PinpointTMVector PinpointPrimer SP6pIRES(MCSI)T7pIRES-3pIRES(MCSII)not available T3,EBVrevpIRES2-EGFP CMV-Profor,pIRES2-EGFP.P5 ’IRESrev,pIRES2-EGFP.P3 ’pIRES-hrGFP-2a CMV-Profor,T3pIRES-hrGFP-3pIRESneo2T7/CMV-Profor pIRES-3pIVEX(all)T7/pBRrevBam T7-TerminatorpLEXA pLEXA-F pLEXA-RpLNCX pLNCX-F pLNCX-RpLNCX2CMV-Profor pRevTRE-3pLVTHM ( H1 启动子下游)H1primer SP6pLXSN pLXSN-F pLXSN-RpMALc MalE primer M13F(-47)pMAL-c2E MalE primer M13F(-47)pMAL-C2x Pmal-c2x-F(P5 ’)Pmal-C2X-R(P3 ’ ) pMALd MalE primer M13F(-47)pMALe MalE primer M13F(-47)pMAL-P4X MalE primer M13F(-47)pMAL-p2X MalE primer M13F(-47)pMET A/B/C AUG1F AUG1RpMD18-T M13R(-48)ECORI M13F(-47)(HIND Ⅲ ) pMD19-T M13F47M13R48pMD20-T M13F47M13R48 pMOSBlue T7/M13rev M13forpMSCV-puro pLXSNfw pMSCVrevpMT/V5-His_A(_B,_C)Mtfor BGH rev,V5rev pNTAP T3T7pCTAP pCTAP5'primer T7pOTB7M13rev/T7M13for/SP6 pOptiVEC-TOPO TA CMV-F/CMV-Profor EMCVIRES reverse pPC86PPC86-F PPC86-RpPC97PBD-GAL4-F PBD-GAL4-R5'AOX3'AOXpPic9k(A+,Z+)5’ AOX(Ppic9k-f)/a-factor3’ AOX(Ppic9k-R) pPICZa5'AOX/PPICZa-F3'AOX pPROEXHTA M13R(-48)无反向引物pProEX-HTa(b,c)M13R48/ Tac-Profor pBADrv/pTrcHis-3M13F20/M13FpQE30or40or60(AMP+)PQE30-F PQE-30RpRC/CMV2/CAT CMV/T7pRECEIVER CMV-F 此载体无反向引物pRECEIVER-M03pRECEIVER-M03F pRECEIVER-M03R pREP4pREP4 forward primer EBV reverse primer pRK5SP6pSG5-3pRSET T7/Xpressfor T7terpRSFDuet-1(MCSI)DuetUP1DuetDOWN1 pRSFDuet-1(MCSII)DuetUP2T7TERpSecTac2T7/CMV-Profor BGH rev/c-mycrev pSG5pSG5-5,/T7PSG5-R(SV40) pSHlox1T7SP6pSHUTTLE-CMV PSHUTTLE-CMV-F PSHUTTLE-CMV-R pSI T7EBVrevT7T3T7M13forT7M13forM13F(-47)pSilencer4_1-CMVneo pSilener4.1-5'CMV-ProforT7pSIneo T7EBVrev,T3Psit T7T7TpSK01-T M13F M13RpSK-CMV(KAN+)T7T3pSOS Psos5'Psos3'pSP64SP6M13revpSP64/65SP6此载体无反向引物pSP65SP6M13revpSP70SP6T7pSP71SP6T7pSP72SP6T7pSP73SP6T7pSport1SP6/M13F(-47)T7/M13RpSPORT1SP6/M13for T7/M13revpSPT18SP6T7pSTBlue T7/M13rev SP6/M13forT7/M13F M13RpSUPER T7T3pSURE-T M13F M13RpSURE-Tsimple M13F-47M13R-48 pSuperCos pBRforEco T7pT3T7M13rev/T3M13for/T7pT7-7T7not availablepT7Blue?T7M13F(-47)pT7T3_18U M13rev/T7M13for/T3pT7T3D-PAC M13rev/T7M13for/T3pTA2M13F/T7( 优先使用 M13F )T3/M13RpT-Adv M13rev T7,M13forpTAg T7/M13rev M13for pTARGETTM pTarget.F( 在 T7 前面) /T7pTG19-T M13F(-47)M13R(-48) pTARGETTM T7/M13F PTARGET-SEQ pThioHisA,B,andC Trx Forward Trx Reverse / M13for pTO-T7T7T7TERpTRC99A PTRC99A-F pTrcHis-rvpTrc99a-c PTRC99C-F PTRC99C-RpTRC99a-c PTRC99C-F PTRC99C-R pTrcHisB PTrcHisA-F/Xpressfor PTrcHisA-R pTrcHisA PTrcHisA-F PTrcHisA-RpT-REX-DEST30TREfor T7pTriEx1T7/TriExup TriExdownT7/TriExup TriExdownT7/TriExup not availableT7/TriExup IRESrevpTriplEX1TriplEx-LD-5T7,TriplEx-LD-3pTriplEX2TriplEx-LD-5 ;pTR5‘T7,TriplEx-LD-3pTWIN-1T7T7TERpTXB1T7MxeInteinrevpTXB2T7MxeInteinrevpTXB3T7MxeInteinrevpTYB1T7InteinrevpTYB2T7InteinrevpTYB3T7InteinrevpTYB4T7InteinrevpTZ_18U T7/M13rev M13forpUB6/V5-His UB-F BGH rev,V5revpUC13M13rev/M13rev(-48)M13for/M13for(-49)pUC18M13rev/M13rev(-48)M13for/M13for(-49)pUC18(19)/118(119)M13F(-47)M13R(-48)/PUC19(HIND Ⅲ ) pUC19M13rev/M13rev(-48)M13for/M13for(-49)pUC57M13rev/M13rev(-48)M13for/M13for(-49)pUC8M13rev/M13rev(-48)M13for/M13for(-49)pUC9M13rev/M13rev(-48)M13for/M13for(-49)pUM-T M13F(-20)M13R(-20)pUCm-T(AMP+)M13F/T7M13RpUNI/V5-His not available BGH rev,V5revpVAX1T7T7/CMV-ProforpVP22/mycHis VP22-Cfor BGH rev,c-mycrevpVP22/mycHis2T7/CMV-Profor VP22-NrevpWE15pBRrevHind T7pYES2T7pYES-DEST52T7/GAL1-Profor V5rev/CYC1-Terminator pYEX_4T_1Clontech pYEX_4T-3pYEX_4T_2pGEX-5pYEX_4T-4pYEX_4T_3pGEX-5pYEX_4T-4pYEX_4T_4pGEX-5pYEX_4T-4pZeoSV2+/hu-Tyr T7not availablepZERO-1M13rev/SP6T7/M13forpZERO-2M13rev/SP6M13for/T7pZome-1-C Zome-F Zome-RpZome-1-N Zome-F Zome-RYCp50(BamHI-site)PBRforBam PBRrevBamYEp13(BamHI-site)PBRforBam PBRrevBamYEp24(BamHI-site)PBRforBam PBRrevBamYIp5(BamHI-site)PBRforBam PBRrevBam RNAi-ReadypSIREN-RetroQ-U6ZsGreen U6菌 PCDB T7BGH天为时代 TA 载体T7T3m13( +)T7?T3?pMSCV-neo pMSCV-5'pMSCV-3'。
禽流感病毒HA基因真核表达质粒的构建与表达

禽流感病毒HA基因真核表达质粒的构建与表达张强哲2) 秦曦明1) 梁 荣1) 邓明俊2) 何宏轩1) 张彦明2) 郑昌学1) 段明星1)3(1)清华大学生物科学与技术系,生物膜与膜生物工程国家重点实验室,北京100084;2)西北农林科技大学动物科技学院,杨凌712100)摘要 血凝素蛋白(HA)基因是禽流感病毒(AIV)重要的保护性抗原基因.为了研究HA基因疫苗,用PCR 扩增H5亚型AIV HA基因,将其克隆到质粒pcDNA4/HisMax和pRc/CMV上得到真核表达质粒pC4H5和pCMVH5.采用Tfx TM220、Superfect转染试剂和电转染法转染HeLa细胞,转染后的HeLa细胞经蛋白质印迹和血凝试验检测HA蛋白及其活性.结果表明,Superfect转染和电转染均能正确表达HA蛋白并具有生物学活性,蛋白质印迹检测到HA和HA裂解的HA1和HA2,与AIV的HA、HA1、HA2蛋白的分子质量一致.从血凝试验结果看,Superfect和电转染表达的HA均具有血凝活性,而经Superfect转染的pC4H5的表达量是pCMVH5的8倍,表明pC4H5是一高效的真核表达质粒.关键词 禽流感病毒,HA基因,真核表达质粒,表达,生物活性学科分类号 S852165915 禽流感(avian influenza,AI)是由A型流感病毒引起的禽类烈性传染病,严重威胁世界各地的养禽业,常常造成巨大的经济损失[1],尤其是H5、H7亚型引起的高致病力禽流感(hightly pathogenic avian influenza,HPAIV)是养禽业的一种灾难性疾病[2].血凝素蛋白(hemagglutinin,HA)在病毒入侵过程中起重要作用,病毒与宿主细胞结合后, HA诱导病毒囊膜与细胞膜结合,使病毒粒子进入细胞.病毒要具有侵染性,必须经过两个过程:a1宿主蛋白酶将HA裂解为HA1和HA2.b1裂解后的HA2暴露出疏水区并与宿主细胞膜脂双层相互结合[3].禽流感病毒HA裂解为HA1和HA2是A IV致病的重要因素,在病毒入侵细胞及决定病毒致病力方面起着关键作用.低致病力禽流感病毒(low pathogenic avian influenza virus,L PA IV)在HA裂解位点上只有一个碱性氨基酸———精氨酸[4],这种结构只能被存在于呼吸道和消化道内的精氨酸特异蛋白酶识别并裂解,因此,L PA IV 感染一般只在呼吸道和消化道内局部繁殖.而HPA IV在HA裂解位点具有多个碱性氨基酸[4],可被机体大多数组织细胞内的蛋白酶识别并裂解,具有广泛的嗜细胞性.HA切割位点处多个碱性氨基酸的插入和HA上糖基化位点的丢失均能使A IV 的致病性增高[5].1997年香港高致病性禽流感H5N1不经过中间宿主而直接由鸡传染给人,导致18人感染,6人死亡[6~8],打破了传统认为的禽流感病毒需在中间宿主体内与人流感重组才能感染人的概念.这引起了许多科学家的高度重视,并正在努力寻找保护率更高,更有效,更便捷的方法,以保护家禽和人类免受HPAIV的攻击[9].我们将对鸡具有高致病性的HPAIV鹅体分离株A/G OOSE/G UAN G DON G/1/96 (H5N1)的HA基因构建到QBI s p163转录增强子和CMV启动子下,在HeLa细胞中进行表达,并检测到表达的HA蛋白,为H5DNA疫苗的后期工作打下了坚实的基础.1 材料和方法111 质粒和菌种含A IV H5基因的质粒PCH5由哈尔滨兽医研究所于康震研究员惠赠;载体pcDNA4/HisMax购自Invtrogen公司;p Rc/CMV质粒由Chember博士惠赠; E.coli DH5α由本室保存;HeLa细胞由清华大学基因组研究所提供.112 工具酶及主要试剂DNA限制性内切酶、T4DNA连接酶、Tfx TM220reagent购自Promega公司;Taq plus DNA聚合酶、dN TP购自上海生工公司;Superfect Transfection Reagent购自Q IA GEN公司;ECF Western blotting kit为Pharmacia公司产品.113 AIV H5基因真核表达质粒的构建11311 A IV H5基因的体外扩增:根据pcDNA4/ HisMax和p Rc/CMV两个载体和H5基因序列分 3通讯联系人. Tel:010*********,E2mail:duanmx@ 收稿日期:2002211220,接受日期:2002212216别设计了两对引物F1、R1和F2、R2,扩增包括H5基因完整的开放阅读框架(ORF)共1707bp.两对引物及包含的酶切位点如下:F15′GC2 GC G A TA TCCA TGG A G A G AA TA GTGCTTCT2T3′(Eco RⅤ),R15′GGCCCTCG A GCTACAA TCG2 TG AACTCCA TAA T3′(XhoⅠ),F25′GCAAA G2 CTTCA TGG A G A G AA TA GTGCTTC3′(Hi n dⅢ), R25′A TA GCGCGCCTACAA TCGTG AACTCCA T2 AA3′(B S S HⅡ).引物由TA KARA公司(大连分公司)合成, PCR反应条件为:95℃预变性5min,94℃1min, 57℃1min,72℃2min,36个循环后72℃延伸10min.11312 A IV H5基因的重组和鉴定:H5基因的PCR扩增产物和pCDNA4/HisMax及p Rc/CMV经双酶切处理后分别进行回收,经T4DNA连接酶连接后,转化入DH5α中经抗性筛选得到阳性重组子.提取质粒对目的基因作限制性酶切鉴定和序列鉴定,构建的真核表达质粒分别命名为pC4H5和pCMV H5.114 AIV H5基因的体外表达11411 Tfx TM220转染HeLa细胞:按Tfx TM220试剂盒说明操作.接种515×105个HeLa细胞于60mm平板上,待细胞达80%融合时,将Tfx TM2 20与pC4H5和pCMV H5以2∶1的电荷比混合转染HeLa细胞,培养48h后收集转染的HeLa细胞于冰预冷的115ml离心管,并作细胞计数.一半用细胞裂解液(400mmol/L KCl,10mmol/L Na2HPO4,1mmol/L ED TA,1mmol/L D TT, 10%甘油,1mmol/L PMSF,015%N P240)裂解,4℃12000g离心10min,回收上清,另一半反复冻融裂解细胞,-20℃保存备用.11412 Superfect转染HeLa细胞:按照Superfect transfect Reagent K it说明操作.接种2×105~8×105细胞于60mm板中,待细胞达40%~80%融合时,将pC4H5、pCMV H5和Superfect试剂以质量与体积比为1∶5的比例转染HeLa细胞,37℃, 5%CO2中培养48h收获细胞.一半用细胞裂解液裂解收获上清,一半反复冻融细胞,-20℃保存备用.11413 电转染法体外转染HeLa细胞:将培养好的HeLa细胞用10%FCS PM EM调至细胞密度5×106/ml,取700μl加入4mm电激杯内,用ECM830电穿孔仪在电压125V,波长为99ms,波数为2个的条件下电击,电击后置于100mm板中培养,待细胞贴壁后,除去死亡细胞,培养48h 收获细胞,一半用细胞裂解液裂解,收获上清,一半反复冻融裂解细胞,-20℃保存备用.115 H5型血凝素的蛋白质印迹检测SDS2聚丙烯酰胺凝胶电泳(SDS2PA GE)按照《分子克隆实验指南》方法进行,积层胶为5%,分离胶为15%,积层胶电压为130V,分离胶为160V.上样量为20μl(每孔约4×105个细胞),设阳性对照,阴性对照,蛋白质低分子质量标准蛋白.电泳结束后立即进行电转移,条件为30V, 90mA,4℃过夜电转,结束后用丽春红染色,标出分子质量标准的位置.用5%脱脂奶粉封闭,一抗为哈尔滨兽医研究所提供抗H5的鸡血清,800倍稀释,二抗为碱性磷酸酶标记的兔抗鸡的IgG,2000倍稀释;底物为ECF试剂盒底物(24μl/cm2),在室温作用10~20min用STORM在Fluorescence mode Blue450nm波长下扫描结果.116 H5型血凝素的活性检测将上述细胞冻融裂解液50μl(1×106个)依次作梯度稀释,再分别加入1%鸡红细胞观察其血凝价.2 结 果211 AIV H5基因真核表达载体的构建用设计的两对引物经PCR扩增、酶切、连接、转化,成功地将1707bp的H5全长基因亚克隆到pcDNA4/HisMax和p Rc/CMV载体中,得到2个含有H5基因的重组真核表达质粒pC4H5和pCMV H5.pC4H5经限制性内切酶Eco RⅤ和XhoⅠ双酶切,pCMV H5经Hi n dⅢ和BSSHⅡ双酶切均获得117kb的目的基因.pC4H5和pCMV H5的序列由TA KARA公司(大连分公司)测定,测序的序列经序列分析软件进行分析,结果表明测序结果与设计结果完全一致,均能找到包含H5基因完整的开放阅读框架,表明两真核表达质粒构建成功.酶切鉴定和测序结果图略.212 pC4H5和pCMVH5体外表达结果用Superfect转染的HeLa细胞,细胞裂解液经蛋白质印迹检测到HA及由HA裂解的HA1和HA2,和A IV的HA、HA1、HA2的分子质量一致(图1a),电转染也检测到与A IV一致的HA(图1b),Tfx TM220转染没有检测到HA(图1c).Fig 11 Western blotting result of expressed HA of H eLa cell(a )Superfect transfection method ;(b )electroporation method ;(c )Tfx TM 220transfection method.1:protein molecular mass marker ;2:AIVHA control ;3:pcDNA4/HisMax ;4:pC4H5;5:pRc/CMV ;6:pCMVH5.213 H eLa 细胞表达的HA 血凝实验结果Superfect (SF )转染和电转染(EP )表达的HA 均检测到血凝价,表达的HA 能使鸡红细胞凝聚,表明HA 具有活性,Tfx TM 220转染(Tfx )的HeLa 细胞和阴性对照(plasmid )检测不到血凝价.具体结果见图2.Fig 12 H emagglutination titer of expressed HA of three plasmids in H eLa cell b y three transfection methods 由图2知Superfect 转染效率最高,电转染效率次之,Tfx TM 220转染效率最差,检测不到血凝价.在Superfect 转染中pC4H5的表达量是pCMV H 5的8倍,而在电转染中pC4H5和pCMV H5的表达量基本相同.3 讨 论HA 是典型的Ⅰ型糖蛋白,即羧基端在囊膜内氨基端在囊膜外.其一级结构含有信号肽(前导序列)、胞浆域、跨膜域、胞外域4个结构域.它的表达涉及到翻译后糖基化的进行、空间构象的折叠及由内质网到高尔基体运送过程中的不断修饰,因此表达比较困难.从图1可看出HeLa 细胞表达的HA 与A IV 自身的HA 分子质量一致,其中图1a第4泳道中部分HA 裂解的HA1和HA2与A IV 裂解的HA1和HA2也基本一致,由图2可知HeLa 细胞表达的HA 与A IV HA 相似能使鸡红细胞凝集,具有血凝活性.这些结果表明A IV HA 基因能在HeLa 细胞中正确表达,因此我们后期的动物试验中pC4H5和pCMV H5取得了良好的保护率(结果在另文报告).图2中Superfect 法转染的HeLa 细胞,pC4H5的表达量是pCMV H5的8倍,原因可能是pcDNA4/HisMax 上游含有QB I sp163转录增强子,所以表达量明显高于不含增强子的p Rc/CMV 质粒.比较图1中蛋白质印迹结果可知,相同的上样量,Superfect 法表达的HA 带浓度最大,电转染次之,Tfx TM 220最差.从图2也可以看出无论是pC4H5还是pCMV H5Superfect 法表达的HA 血凝价最高,电转染次之,Tfx TM 220最差.因此我们认为Superfect 转染法对A IV HA 的表达是一种比较理想的转染方法.K odihalli 等[10]认为A/Hong K ong/156/97(H5N1)(H K/97)和其他A IV 相比明显的特点是HA1的154位氨基酸处的糖基化位点缺失.Matrosovich 等[11]研究发现HA 糖基化位点的增加和NA 氨基酸的缺失会降低A IV 与细胞受体的亲和力和病毒的释放能力.HA1和HA2裂解位点处多个碱性氨基酸的插入和糖基化位点的丢失均能使HA 对蛋白酶切割的敏感性增强,而使致病力提高[5].HPA IV A/GuangDong/1/96(H5N1)(G D/96)的HA 基因与引起香港流感事件的HPA IV A/Hong K ong/156/97(H5N1)(H K/97)的HA 基因核苷酸和氨基酸序列分别为98%和9812%同源率,6个碱性潜在的糖基化位点和2个受体位点完全一致,HA1和HA2裂解位点的6个碱性氨基酸序列(RRR KKR)也完全一致[12,13],G D/96在154位也无糖基化位点,因此我们选用HPA IV G D/96的H5基因作为DNA疫苗的外源基因,以期望它具有更好的抗原性,对HPA IV具有更高的保护率.G oto和Kawaoka[14]研究发现,A/WSN/33 (H1N1)(WSN)的NA对HA的裂解起到关键作用,主要是因为NA羧基端453位是一个赖氨酸残基,146位又缺乏糖基化位点,从而使453位的lysine易于聚集血纤维蛋白溶酶原(plasminogen),致使局部血纤维蛋白溶酶浓度升高,将HA切割成HA1和HA2,增加了病毒的侵袭力.当146位具有糖链时,寡糖链由于空间上的位置结构而阻碍453位lysine与plasminogen结合,使病毒致病性减弱.因此我们下一步将继续构建NA基因的真核表达质粒与HA真核表达质粒联用,或者构建融合表达质粒免疫动物来提高DNA疫苗的保护效果.致谢 感谢湖南正虹科技发展股份有限公司对本研究给予的大力支持.参 考 文 献1 Crawford J,Wilkinson B,Vosnesensky A,et al.Baculovirus2 derived hemagglutinin vaccines protect against lethal influenza infections by avian H5and H7subtypes.Vaccine,1999,17(18):2265~22742 Suarez D L,Schultz2Cherry S.Immunology of avian influenza virus:a review.Developmental&Comparative Immunology, 2000,24(2~3):269~2833 G arten W,K lenk H D.Understanding influenza viruspathogenicity.Trends in Microbiology,1999,7(3):99~100 4 Capua I,Margangon S.The avian influenza epidemic in Italy, 199922000:a review.Avian Pathology,2000,29(3):289~2945 Vogel G.Sequence offers clues to deadly Flu.Science,1998,279 (5349):3246 Hatta M,G ao P,Halfmanm P,et al.Molecular basis for high virulence of Hong K ong H5N1Influenza A virus.Science,2001, 293(5536):1840~18427 G ao P,Watanabe S,Ito T,et al.Biological heterogeneity, including systemic replications in Mice,of H5N1Influenza A virus isolates from humans in Hong K ong.J Virol,1999,73(4): 3184~31898 Ha Y,Stevens D J,Skehel J J,et al.X2ray structures of H5avian and H9swine influenza virus hemagglutinins bound to avian and human receptor analogs.Proc Natl Acad Sci USA,2001,98(20):1181~11869 K odihalli S,K obasa D L,Webster R G.Strategies for inducing protection against avian influenza A virus subtypes with DNA vaccines.Vaccine,2000,18(23):2592~259910K odihalli S,G oto H,K obasa D L,et al.DNA vaccine encoding hemagglutinin provides protective immunity against H5N1influenza virus infection in mice.J Virol,1999,73(3):2094~209811Matrosovich M,Zhou N,K awaoka Y,et al.The surface glycoproteins of H5influenza viruses isolated from humans, chickens,and wild aquatic birds have distinguishable properties.J Virol,1999,73(2):1146~115512陈化兰,于康震,步志高.一株鹅源高致病力禽流感病毒分离株血凝素基因的分析.中国农业科学,1999,32(2):87~92 Chen H L,Yu K Z,Bu Z G.Scientia Agricultura Scienca,1999, 32(2):87~9213Subbarao K,K limov A,K atz J,et al.Characterization of an avian influenza A(H5N1)virus isolated from a child with a fatal respiratory illness.Science,1998,279(5349):393~39614G oto H,K awaoka Y.A novel mechanisms for the acquisition of virulence by a human influenza A virus.Proc Natl Acad Sci USA, 1998,95(17):10224~10228Construction and Expression of Avian Influenza VirusHA G ene Eukaryotic V ectorsZHAN G Qiang2Zhe2),Q IN Xi2Ming1),L IAN G Rong1),DEN G Ming2J un2), HE Hong2Xuan1),ZHAN G Yan2Ming2),ZHEN G Chang2Xue1),DUAN Ming2Xing1)3(1)State Key L aboratory of Biomembrane&Membrane Biotechnology,Depart ment of Biological Sciences&Biotechnology,Tsi nghua U niversity,Beiji ng100084,China;2)College of A nimal Science and T echnology,Northwest Science and T echnology University of A griculture and Forest,Yangling712100,China)Abstract Hemagglutinin(HA)gene is an important gene of protective antigen of Avian Influenza Virus (A IV).In order to study HA gene vaccine,the H5gene of A IV,amplified by PCR,was subcloned into eukaryotic expressing vectors pcDNA4/HisMax and p Rc/CMV.The recombinant plasmids were transfected into HeLa cell by Tfx TM220transfection reagent,Superfect transfection reagent and electroporation respectively.The expressed HA was examined and identified by Western blotting and hemagglutination assay.The results showed that HA was expressed precisely in HeLa cells and had bioactivity.Three special bands(HA,HA1and HA2)were found by Western blotting,which were identical to those of A IV.The quantity of expressed HA from pC4H5,transfected by superfect transfection reagent,was approximately8times as much as that from pCMV H5.These results suggest that pC4H5is a highly efficient eukaryotic expressing vector.K ey w ords avian influenza virus,H5gene,DNA vaccine,expression,bioactivity 3Corresponding author.Tel:86210262773255,E2mail:duanmx@ Received:November20,2002 Accepted:December16,2002一次成功的生物膜学术研讨会———第八次全国暨2003海内外生物膜学术研讨会 由中国生物物理学会、中国生物化学与分子生物学会、中国细胞生物学会、中国植物学会、国家自然科学基金委员会、生物大分子国家重点实验室、生物膜与膜工程国家重点实验室以及上海神经科学开放实验室等联合主办的“第八次全国暨2003海内外生物膜学术研讨会”于2003年3月25~28日在广西北海召开.这是每三年一届的全国生物膜学术讨论会.来自国内外高等院校,科研院所的161位代表参加了这次讨论会,规模超过以往任何一届.代表中有在国内外享有声誉的我国老一辈科学家,也有年富力强的中青年骨干,更令人振奋的是代表中半数以上为研究生,他们是我国生物膜研究的未来.与往届不同,这次研讨会还首次特邀了国外及中国港台地区在生物膜研究中较出色的海外学者做特邀报告.研讨会就A1信号跨膜转导与细胞功能调控;B1生物膜与细胞凋亡;C1生物膜脂筏(lipid raft)和Caveolae;D1细胞内蛋白质的运送;E1膜蛋白与蛋白质组学的研究;F1膜脂与膜蛋白的相互作用;G1转运体和离子通道;H1生物膜研究在医、农中的应用;I1生物膜与膜工程和药物开发等九个方面组织了9个特邀报告,15个专题报告,墙报24份.从中可以看到,我国生物膜研究水平整体上比上届有了较大的提高,特别是在离子通道,信号跨膜转导,生物膜脂筏和细胞凋亡等国际上热门领域投入较多,研究成果突出.但对膜蛋白三维结构、体现物理、化学、生物等多学科的交叉开展重大项目研究,以及利用新的研究技术与手段等的探索有待进一步重视和加大投入的力度.总之,这次研讨会,内容非常丰富,大会特邀报告、专题报告、研究生工作交流和专题座谈会等学术交流形式多样,发言、提问踊跃,讨论热烈,学术交流紧凑活泼,基本上反映了生物膜研究的国际前沿以及国内研究现状和最新研究进展.无论是多次还是初次参加生物膜会议的代表,都觉得本次研讨会开得非常成功,是一次高水平的学术研讨会.本次研讨会对进一步明确在本世纪中我国生物膜研究的方向,瞄准重点,结合我国国情,努力缩短与国际上的差距将会起到积极的推动和促进作用.研讨会期间,还组织了“生物膜与蛋白质组学”和“生物膜研究在21世纪生命科学发展中的作用”的专题座谈会,广泛地探讨了如何开展我国蛋白质组学研究,膜蛋白与蛋白质组学的关系,以及我国生物膜研究的问题和前景.为鼓励研究生进行学术交流,本次研讨会还组织了研究生学术报告会和墙报展讲,并从中评选出了一等奖二名,二等奖五名,颁发了荣誉证书和奖励,收到了良好的效果.这是一次成功的生物膜学术研讨会.相会老朋友,结识新同行,喜看中青年,后继有人在.。
载体——精选推荐
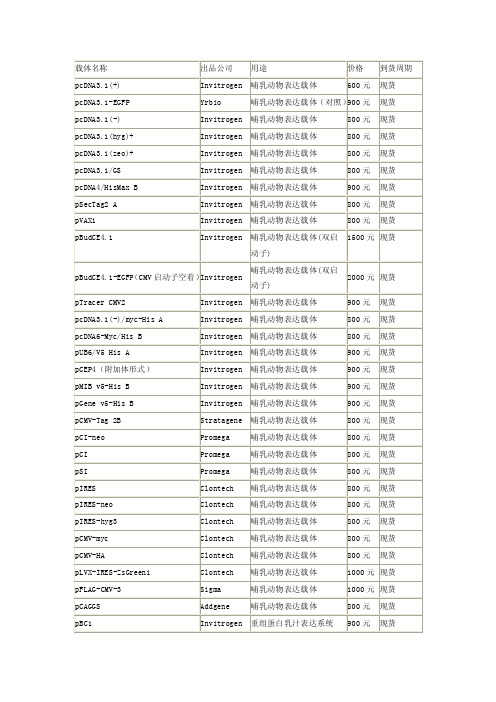
NEB
原核表达 载体
NEB
配套大肠 杆菌
Amersham 原核表达载体
Amersham 原核表达载体
Amersham 原核表达载体
Invitrogen 原核表达载体
Lucigen
原核表达 菌株
Stratagene 原核表达菌株
Qiagen
原核表达 载体
Qiagen
原核表达 载体
Invitrogen 基因载体
700 元 现货 800 元 现货
900 元 现货
800 元 现货 800 元 现货 800 元 现货 800 元 现货 800 元 现货 700 元 现货 800 元 现货 800 元 现货 800 元 现货 800 元 现货 900 元 现货 1000 元 现货 1000 元 现货 1000 元 现货 900 元 现货 1000 元 现货 1500 元 现货 600 元 现货 300 元 现货 300 元 现货 600 元 现货 600 元 现货 800 元 现货 900 元 现货 900 元 现货 900 元 现货
BL21(DE3) BL21(DE3)plysS Rosetta(DE3) Rosetta(DE3)plysS Rosetta-gami(DE3) Rosetta-gami B(DE3)pLysS Orgami(DE3) OrgamiB(DE3) BLR(DE3) pTWIN1 pTYB11 pMXB10 pTXB1 ER2566 pGEX-6P-1 pGEX-4T-2 pGEX-4T-3 pRset-a C41(DE3) BL21 codonplus(DE3)RIPL TAGZyme pQE-2 pQE-Trisystem pOG44 pORF-lacZ pORF-HSV1tk pSP73 pUC18 pUC19 pBR322 pSP64 pBluescript II SK(+) SuperCos 1 pGAPZα A pPICZα A
载体名称
EF-1α Fwd
BGH
pBudCE4.1/lacZ/CAT
CMV-Profor,T7
c-mycrev,EBVrev
pBV220
pBV220F
PBV220R
pCAL-c
T7/Tac-Profor
T7-Terminator
pCAL-n
T7/Tac-Profor
T7-Terminator
pBR322(BamHI-site)
PBRforBam
PBRrevBam
pBR322(EcoRI/HindIII-sites)
pBRforEco
pBRrevHind
pBR325(BamHI-site)
PBRforBam
PBRrevBam
pBR329(BamHI-site)
PBRforBam
PBRrevBam
EBVrev
pCI-neo
T7(17BASE)
EBVrev,T3
pCMS-EG7
EBVrev,T3
pCMV-3Tag-4A
T3 /PFLAG-CMV-F
T7
pCMV5
pCMV5F
PCMV5R
pCMV6
CMV-Profor
pCMV6-3、HGHrev
pCMV6-XL4
CMV-Profor/T7
pcDNA3.3TOPOTA
CMV-Profor
TKPAreverse
pcDNA4/HisMax(_A,_B,_C)
Xpressfor
BGH rev
pcDNA4/HisMax-TOPO
Xpressfor
BGH rev
pcDNA4/HisMax-TOPO/lacZ
载体及通用引物

S1
S6
pCAT3-enhancer
RVP3
此载体无反向引物
pCDFDuet-1(MCSI)
ACYCDuetUP1 Primer
DuetDOWN1
pCDFDuet-1(MCSII)
DuetUP2
T7TER
pcDNA1.1
CMV-Profor,T7
pCDM8-3
pcDNA2.1
M13for/T7
EGFP-Nrev/ PEGFP-N-3′
pEGFP-C
PEGFP-C-5′
PEGFP-C-3′
pEGFP-C1
EGFP-Cfor
SV40-pArev
pEGFP-C2
EGFP-Cfor
SV40-pArev
pEGFP-C3
EGFP-Cfor
SV40-pArev
pEGFP-F
EGFP-Cfor
SV40-pArev
T7
T7-Terminator
pET-25b(+)
T7
T7-Terminator
pET-26b(+)
T7
T7-Terminator
pET-27b(+)
T7
T7-Terminator
pET-28a(-b,-c)(+)
T7
T7-Terminator
pET-29a(-b,-c)(+)
S.tag/T7
T7-Terminator
T7-Terminator
pET-20b(+)
T7
T7-Terminator
pET-21(-a,-b,-c,-d)(+)
T7
真核哺乳动物细胞标签蛋白纯化
真核哺乳动物细胞标签蛋白纯化简介哺乳动物表达系统的突出优点是表达蛋白可正确折叠以及进行糖基化等翻译后修饰,表达产物具有生物活性,已成为表达和生产蛋白质药物,基因工程抗体,疫苗抗原和研究用目的蛋白功能的首选表达体系。
哺乳动物表达体系蛋白表达的主要流程为:PCR扩增目的片段——将基因片段克隆到表达载体上——细胞转染——细胞株的筛选和培养——纯化目的蛋白解决方案1、查找目的基因序列由于引物是根据目的基因序列来设计的,所以查找到目的基因序列是设计引物的第一步。
查找目的基因序列可以在多个网站上查到,也可以在文献中查找。
这里介绍的是比较常用的NCBI网站查找基因序列的方法。
NCBI 是美国国家生物技术信息中心,网站上具有多个生物数据库,是生物学研究常用的网站。
1.1打开NCBI网站,在下拉框选择Gene,输入目的蛋白名称;(这里以p65为例)1.2 找到对应物种,也可以选择右侧框中的物种查找;1.3 页面往下拉,找到mRNA and Protein(s),选择正确的isoform,NMxxx表示mRNA序列,NPxxx表示蛋白质序列;1.4 新页面往下拉,点击CDS,点击右侧FASTA,即可得到目的基因的mRNA序列,复制粘贴到新的文本,以备使用。
2、选择合适的表达载体常用的真核表达载体有pCMV,pcDNA等。
表达载体一般包括启动子,多克隆位点,抗性基因,标签基因等等。
表达载体的选择原则:1)启动子是否为真核启动子;2)标签基因是否是我们需要用的标签。
常用的标签的标签有His,GST,MBP,Strep,Flag标签等,标签选择如下:3、设计引物设计引物也有多个软件可以使用,这里介绍的是常用的Primer 5软件。
设计引物的基本原则是:1)引物长度一般为15-30bp,常用的为18-27bp,但不能大于38bp;2)引物GC含量一般为40%-60%,以45-55%为宜,上下游引物GC含量和Tm值要保持接近;3)引物所对应的模板序列的Tm值最好在72℃左右;4)3'端最好不要是连续碱基,GGG或CCC会导致错误的引发,同时3'端最后一个碱基最好不要是A或T,否则容易导致错配;5)以公式Tm=4*(G+C)+2*(A+T)-5计算Tm值,也就是退火温度。
pTT5哺乳动物表达载体说明

pTT5编号 载体名称北京华越洋生物VECT6098 pTT5pTT5载体基本信息载体名称: pTT5质粒类型: 哺乳动物细胞载体高拷贝/低拷贝: 高拷贝克隆方法: 限制性内切酶,多克隆位点启动子: CMV载体大小: 4401 bp5' 测序引物及序列: CMV-F:CGCAAATGGGCGGTAGGCGTG 3' 测序引物及序列: --载体标签: 无载体抗性: 氨苄青霉素筛选标记: --克隆菌株: DH5α等宿主细胞(系): 常规细胞系(293、CV-1、CHO等)备注: 利用该质粒构建的基因表达是非融合表达,在构建质粒时需要在基因前面添加Kozak序列来增加蛋白表达效率。
稳定性: 瞬时表达组成型/诱导型: 诱导性病毒/非病毒: 非病毒pTT5载体质粒图谱和多克隆位点信息pTT5载体简介pTT5载体是很好的哺乳细胞瞬时蛋白表达载体,经常用于哺乳细胞的抗体轻链或者重链的表达。
pTT5载体序列ORIGIN1 GTACATTTAT ATTGGCTCAT GTCCAATATG ACCGCCATGT TGACATTGAT TATTGACTAG 61 TTATTAATAG TAATCAATTA CGGGGTCATT AGTTCATAGC CCATATATGG AGTTCCGCGT 121 TACATAACTT ACGGTAAATG GCCCGCCTGG CTGACCGCCC AACGACCCCC GCCCATTGAC 181 GTCAATAATG ACGTATGTTC CCATAGTAAC GCCAATAGGG ACTTTCCATT GACGTCAATG 241 GGTGGAGTAT TTACGGTAAA CTGCCCACTT GGCAGTACAT CAAGTGTATC ATATGCCAAG 301 TCCGCCCCCT ATTGACGTCA ATGACGGTAA ATGGCCCGCC TGGCATTATG CCCAGTACAT 361 GACCTTACGG GACTTTCCTA CTTGGCAGTA CATCTACGTA TTAGTCATCG CTATTACCAT 421 GGTGATGCGG TTTTGGCAGT ACACCAATGG GCGTGGATAG CGGTTTGACT CACGGGGATT 481 TCCAAGTCTC CACCCCATTG ACGTCAATGG GAGTTTGTTT TGGCACCAAA ATCAACGGGA 541 CTTTCCAAAA TGTCGTAATA ACCCCGCCCC GTTGACGCAA ATGGGCGGTA GGCGTGTACG 601 GTGGGAGGTC TATATAAGCA GAGCTCGTTT AGTGAACCGT CAGATCCTCA CTCTCTTCCG 661 CATCGCTGTC TGCGAGGGCC AGCTGTTGGG CTCGCGGTTG AGGACAAACT CTTCGCGGTC721 TTTCCAGTAC TCTTGGATCG GAAACCCGTC GGCCTCCGAA CGGTACTCCG CCACCGAGGG 781 ACCTGAGCGA GTCCGCATCG ACCGGATCGG AAAACCTCTC GAGAAAGGCG TCTAACCAGT 841 CACAGTCGCA AGGTAGGCTG AGCACCGTGG CGGGCGGCAG CGGGTGGCGG TCGGGGTTGT 901 TTCTGGCGGA GGTGCTGCTG ATGATGTAAT TAAAGTAGGC GGTCTTGAGA CGGCGGATGG 961 TCGAGGTGAG GTGTGGCAGG CTTGAGATCC AGCTGTTGGG GTGAGTACTC CCTCTCAAAA 1021 GCGGGCATTA CTTCTGCGCT AAGATTGTCA GTTTCCAAAA ACGAGGAGGA TTTGATATTC 1081 ACCTGGCCCG ATCTGGCCAT ACACTTGAGT GACAATGACA TCCACTTTGC CTTTCTCTCC 1141 ACAGGTGTCC ACTCCCAGGT CCAAGTTTAA ACGGATCTCT AGCGAATTCC CTCTAGAGGG 1201 CCCGTTTCTG CTAGCAAGCT TGCTAGCGGC CGCTCGAGGC CGGCAAGGCC GGATCCCCCG 1261 ACCTCGACCT CTGGCTAATA AAGGAAATTT ATTTTCATTG CAATAGTGTG TTGGAATTTT 1321 TTGTGTCTCT CACTCGGAAG GACATATGGG AGGGCAAATC ATTTGGTCGA GATCCCTCGG 1381 AGATCTCTAG CTAGAGGATC GATCCCCGCC CCGGACGAAC TAAACCTGAC TACGACATCT 1441 CTGCCCCTTC TTCGCGGGGC AGTGCATGTA ATCCCTTCAG TTGGTTGGTA CAACTTGCCA 1501 ACTGAACCCT AAACGGGTAG CATATGCTTC CCGGGTAGTA GTATATACTA TCCAGACTAA 1561 CCCTAATTCA ATAGCATATG TTACCCAACG GGAAGCATAT GCTATCGAAT TAGGGTTAGT 1621 AAAAGGGTCC TAAGGAACAG CGATGTAGGT GGGCGGGCCA AGATAGGGGC GCGATTGCTG 1681 CGATCTGGAG GACAAATTAC ACACACTTGC GCCTGAGCGC CAAGCACAGG GTTGTTGGTC 1741 CTCATATTCA CGAGGTCGCT GAGAGCACGG TGGGCTAATG TTGCCATGGG TAGCATATAC 1801 TACCCAAATA TCTGGATAGC ATATGCTATC CTAATCTATA TCTGGGTAGC ATAGGCTATC 1861 CTAATCTATA TCTGGGTAGC ATATGCTATC CTAATCTATA TCTGGGTAGT ATATGCTATC 1921 CTAATTTATA TCTGGGTAGC ATAGGCTATC CTAATCTATA TCTGGGTAGC ATATGCTATC 1981 CTAATCTATA TCTGGGTAGT ATATGCTATC CTAATCTGTA TCCGGGTAGC ATATGCTATC 2041 CTAATAGAGA TTAGGGTAGT ATATGCTATC CTAATTTATA TCTGGGTAGC ATATACTACC 2101 CAAATATCTG GATAGCATAT GCTATCCTAA TCTATATCTG GGTAGCATAT GCTATCCTAA 2161 TCTATATCTG GGTAGCATAG GCTATCCTAA TCTATATCTG GGTAGCATAT GCTATCCTAA 2221 TCTATATCTG GGTAGTATAT GCTATCCTAA TTTATATCTG GGTAGCATAG GCTATCCTAA 2281 TCTATATCTG GGTAGCATAT GCTATCCTAA TCTATATCTG GGTAGTATAT GCTATCCTAA 2341 TCTGTATCCG GGTAGCATAT GCTATCCTCA TGATAAGCTG TCAAACATGA GAATTAATTC 2401 TTGAAGACGA AAGGGCCTCG TGATACGCCT ATTTTTATAG GTTAATGTCA TGATAATAAT 2461 GGTTTCTTAG ACGTCAGGTG GCACTTTTCG GGGAAATGTG CGCGGAACCC CTATTTGTTT 2521 ATTTTTCTAA ATACATTCAA ATATGTATCC GCTCATGAGA CAATAACCCT GATAAATGCT 2581 TCAATAATAT TGAAAAAGGA AGAGTATGAG TATTCAACAT TTCCGTGTCG CCCTTATTCC 2641 CTTTTTTGCG GCATTTTGCC TTCCTGTTTT TGCTCACCCA GAAACGCTGG TGAAAGTAAA 2701 AGATGCTGAA GATCAGTTGG GTGCACGAGT GGGTTACATC GAACTGGATC TCAACAGCGG 2761 TAAGATCCTT GAGAGTTTTC GCCCCGAAGA ACGTTTTCCA ATGATGAGCA CTTTTAAAGT 2821 TCTGCTATGT GGCGCGGTAT TATCCCGTGT TGACGCCGGG CAAGAGCAAC TCGGTCGCCG 2881 CATACACTAT TCTCAGAATG ACTTGGTTGA GTACTCACCA GTCACAGAAA AGCATCTTAC 2941 GGATGGCATG ACAGTAAGAG AATTATGCAG TGCTGCCATA ACCATGAGTG ATAACACTGC 3001 GGCCAACTTA CTTCTGACAA CGATCGGAGG ACCGAAGGAG CTAACCGCTT TTTTGCACAA 3061 CATGGGGGAT CATGTAACTC GCCTTGATCG TTGGGAACCG GAGCTGAATG AAGCCATACC 3121 AAACGACGAG CGTGACACCA CGATGCCTGC AGCAATGGCA ACAACGTTGC GCAAACTATT 3181 AACTGGCGAA CTACTTACTC TAGCTTCCCG GCAACAATTA ATAGACTGGA TGGAGGCGGA 3241 TAAAGTTGCA GGACCACTTC TGCGCTCGGC CCTTCCGGCT GGCTGGTTTA TTGCTGATAA 3301 ATCTGGAGCC GGTGAGCGTG GGTCTCGCGG TATCATTGCA GCACTGGGGC CAGATGGTAA3361 GCCCTCCCGT ATCGTAGTTA TCTACACGAC GGGGAGTCAG GCAACTATGG ATGAACGAAA3421 TAGACAGATC GCTGAGATAG GTGCCTCACT GATTAAGCAT TGGTAACTGT CAGACCAAGT3481 TTACTCATAT ATACTTTAGA TTGATTTAAA ACTTCATTTT TAATTTAAAA GGATCTAGGT3541 GAAGATCCTT TTTGATAATC TCATGACCAA AATCCCTTAA CGTGAGTTTT CGTTCCACTG3601 AGCGTCAGAC CCCGTAGAAA AGATCAAAGG ATCTTCTTGA GATCCTTTTT TTCTGCGCGT3661 AATCTGCTGC TTGCAAACAA AAAAACCACC GCTACCAGCG GTGGTTTGTT TGCCGGATCA3721 AGAGCTACCA ACTCTTTTTC CGAAGGTAAC TGGCTTCAGC AGAGCGCAGA TACCAAATAC3781 TGTTCTTCTA GTGTAGCCGT AGTTAGGCCA CCACTTCAAG AACTCTGTAG CACCGCCTAC3841 ATACCTCGCT CTGCTAATCC TGTTACCAGT GGCTGCTGCC AGTGGCGATA AGTCGTGTCT3901 TACCGGGTTG GACTCAAGAC GATAGTTACC GGATAAGGCG CAGCGGTCGG GCTGAACGGG3961 GGGTTCGTGC ACACAGCCCA GCTTGGAGCG AACGACCTAC ACCGAACTGA GATACCTACA4021 GCGTGAGCTA TGAGAAAGCG CCACGCTTCC CGAAGGGAGA AAGGCGGACA GGTATCCGGT4081 AAGCGGCAGG GTCGGAACAG GAGAGCGCAC GAGGGAGCTT CCAGGGGGAA ACGCCTGGTA4141 TCTTTATAGT CCTGTCGGGT TTCGCCACCT CTGACTTGAG CGTCGATTTT TGTGATGCTC4201 GTCAGGGGGG CGGAGCCTAT GGAAAAACGC CAGCAACGCG GCCTTTTTAC GGTTCCTGGC4261 CTTTTGCTGG CCTTTTGCTC ACATGTTCTT TCCTGCGTTA TCCCCTGATT CTGTGGATAA4321 CCGTATTACC GCCTTTGAGT GAGCTGATAC CGCTCGCCGC AGCCGAACGA CCGAGCGCAG4381 CGAGTCAGTG AGCGAGGAAG C//其他哺乳动物表达载体:pCHO1.0 pBApo-CMV-Pur pOPRSVIpcDNA3.1/His C pcDNA5/FRT/V5-His-TOPO pREP4pcDNA3.1/His B pcDNA5/FRT/TO-TOPO pDual-GCpcDNA3.1/His A pcDNA5/TO pBK-RSVpIRESpuro3 pcDNA5/FRT/TO pBK-CMVpIRES2-EGFP pcDNA5/FRT pBI-CMV4pTT5 pFLAG-CMV2 pcDNA4/TO/Myc-His/LacZ pNFkB-DD-tdTomato pcDNA3.1/CT-GFP-TOPO pOPI3CATpBI-CMV5 pcDNA3.1/NT-GFP-TOPO pGene/V5-His B pSEAP2-Basic pOptiVEC-TOPO pSwitchpSEAP2-Control pCMV-MEKK1 pCMVLacIpBI-CMV3 pCMV-MEK1 pVgRxRpBI-CMV2 pCMV-PKA pINDpBI-CMV1 pcDNA6.2/nTC-Tag-DEST pTRE3G-LucpNFκB-MetLuc2-Reporter pcDNA6.2/cTC-Tag-DEST pTRE3GpCRE-MetLuc2-Reporter pcDNA3.2/V5/GW/D-TOPO pTRE2-hygropAcGFP1-Actin pcDNA6.2/V5/GW/D-TOPO pTRE-TightpAcGFP1-N In-Fusion Ready pcDNA6.2/nGeneBLAzer-GW/D-TOPO pTK-hygpAcGFP1-C3 pcDNA6.2/C-YFP-DEST pTet-OnpAcGFP1-C pcDNA6.2/cGeneBLAzer-DEST pTet-OffpAcGFP1-p53 pcDNA6/V5-His A pTet on advanced pAcGFP1-Mito pcDNA6/V5-His B pRevTREpAcGFP1-Mem pcDNA6/V5-His C pRevTet-OnpAcGFP1-Lam pcDNA6/myc-His C pRevTet-Off pAcGFP1-Golgi pcDNA6/myc-His A pCMV-Tet3G pAcGFP1-F pcDNA6/myc-His B pTRE2pAcGFP1-Hyg-C1 pcDNA6.2/nGeneBLAzer-DEST pBD-NF-κB pAsRed2-N1 pcDNA4/HisMax-TOPO pCMV-AD ptdTomato-N1 pcDNA6.2/nLumio-DEST pCMV-BDpCMV-tdTomato pcDNA6.2/cLumio-DEST pBIND-Id Control pCRE-DD-tdTomato pcDNA4/myc-His C pBINDpCMV-DsRed-Express2 pcDNA4/HisMax C pG5 luciferasepEF1α-tdTomato pcDNA4/HisMax A pACT-MyoDpCRE-hrGFP c-Flag pcDNA3 pACTptdTomato-C1 pcDNA4/HisMax B pCMV-SPORT6 pAsRed2-C1 pcDNA4/myc-His B pGL4.13pGL3-Promoter pcDNA4/myc-His A pGL4.19pGL3 basic pcDNA4/His C pGL4.26pAcGFP1-C2 pcDNA4/His B pGL4.20pAcGFP1-C1 pcDNA4/His A pGL4.29pAcGFP1-N3 pcDNA6/TR pGL4.30pAcGFP1-N2 pcDNA4/TO/Myc-His A pGL4.27pAcGFP1-N1 pcDNA4/TO pGL4.75pAcGFP1-C In-Fusion Ready pcDNA4/TO/Myc-His B pGL4.10pCRE-DD-AmCyan1 pcDNA4/TO/Myc-His C pGRN145pNFkB-DD-AmCyan1 pcDNA3.3-TOPO pSecTag2 A pDsRed2-Bid pBudCE4.1 pEBVHis B pDsRED2-Mito pFLAG-CMV-4 pEBVHis ApDD-AmCyan1 Reporter pFLAG-CMV-3 pCMV-Tag 3C pAmCyan1-N1 pFLAG-CMV-2 pCMV-Tag 3A pAmCyan1-C1 pFLAG-CMV-5a pCMV-Tag 3BpEF1α-IRES-DsRed-Express2 p3XFLAG-CMV-9 pCMV-Tag 5CpEF1α-DsRed-Monomer-N1 p3xFLAG-CMV-10 pCMV-Tag 5A pDsRED-Monomer-N1 p3XFLAG-CMV-8 pCMV-Tag 4A pDsRed-Express-N1 p3XFLAG-CMV-7.1 pCMV-Tag 5Bp3XFLAG-CMV-7 pDsRed-Monomer-N In-Fusion Ready pCMV-Tag 4B pDsRed-Express-C1 p3XFLAG-CMV-13 pCMV-Tag 2C pIRES2-ZsGreen1 p3XFLAG-CMV-14 pCMV-Tag 2B pDsRed-Express2-C1 plRES2-ZsGreen1 pCMV-Tag 2A pDsRed-Express2-N1 pBApo-EF1α-pur pCMV-LacZpEF1α-DsRed-Express2 pBApo-EF1α-neo pCMV-MycpIRES2-DsRed-Express pBApo-CMV pEF1α-IRES-AcGFP1 pIRES2-DsRed-Express2 pBApo-CMV-neo pEF1α-IRES-ZsGreen1 pIRES-hrGFP-1a pIRES-EGFP pEF1α-AcGFP1-N1 pIRESneo2 pIRESneo3 pIRES2-DsRed2 pIRESneo pDsRed-Monomer pIRES2-AcGFP1 pIREShyg3 pIRES。
pCHO1.0哺乳动物表达载体说明

pCHO1.0编号 载体名称北京华越洋生物VECT6001 pCHO1.0pCHO1.0载体基本信息载体名称: pCHO1.0质粒类型: 哺乳动物细胞表达载体, 哺乳动物细胞稳定细胞系建立载体 启动子: CMV表达水平: 高克隆方法: 多克隆位点,限制性内切酶载体大小: 12988 b p5' 测序引物: AB-‐F: G TCTGAGCCTCCTTGTCTTG; EP-‐F:GGTGTCGTGAGGAATTTCAG3' 测序引物: AB-‐R: A GAAGACACGGGAGACTTAG; EP-‐R: G AGGCAGCCGGATCATAATC载体标签: /载体抗性: 卡那筛选标记: Puromycin, DHFR备注: -‐-‐稳定性: 稳表达组成型: 组成型 Constitutive病毒/非病毒: 非病毒pCHO1.0载体质粒图谱和多克隆位点信息pCHO1.0载体简介pCHO 1.0载体是能够表达一个基因或利用载体上面的杂交CMV启动子共表达2个目的基因。
该载体包含有二氢叶酸还原酶(DHFR)选择性标记和嘌呤霉素(puromycin)抗性基因,可以同时使用MTX和嘌呤霉素进行筛选。
其他哺乳动物表达载体:pCHO1.0 pBApo-CMV-Pur pOPRSVIpcDNA3.1/His C pcDNA5/FRT/V5-His-TOPO pREP4pcDNA3.1/His B pcDNA5/FRT/TO-TOPO pDual-GCpcDNA3.1/His A pcDNA5/TO pBK-RSVpIRESpuro3 pcDNA5/FRT/TO pBK-CMVpIRES2-EGFP pcDNA5/FRT pBI-CMV4pTT5 pFLAG-CMV2 pcDNA4/TO/Myc-His/LacZ pNFkB-DD-tdTomato pcDNA3.1/CT-GFP-TOPO pOPI3CATpBI-CMV5 pcDNA3.1/NT-GFP-TOPO pGene/V5-His BpSEAP2-Basic pOptiVEC-TOPO pSwitchpSEAP2-Control pCMV-MEKK1 pCMVLacIpBI-CMV3 pCMV-MEK1 pVgRxRpBI-CMV2 pCMV-PKA pINDpBI-CMV1 pcDNA6.2/nTC-Tag-DEST pTRE3G-LucpNFκB-MetLuc2-Reporter pcDNA6.2/cTC-Tag-DEST pTRE3GpCRE-MetLuc2-Reporter pcDNA3.2/V5/GW/D-TOPO pTRE2-hygropAcGFP1-Actin pcDNA6.2/V5/GW/D-TOPO pTRE-TightpAcGFP1-N In-Fusion Ready pcDNA6.2/nGeneBLAzer-GW/D-TOPO pTK-hygpAcGFP1-C3 pcDNA6.2/C-YFP-DEST pTet-OnpAcGFP1-C pcDNA6.2/cGeneBLAzer-DEST pTet-OffpAcGFP1-p53 pcDNA6/V5-His A pTet on advanced pAcGFP1-Mito pcDNA6/V5-His B pRevTREpAcGFP1-Mem pcDNA6/V5-His C pRevTet-OnpAcGFP1-Lam pcDNA6/myc-His C pRevTet-OffpAcGFP1-Golgi pcDNA6/myc-His A pCMV-Tet3GpAcGFP1-F pcDNA6/myc-His B pTRE2pAcGFP1-Hyg-C1 pcDNA6.2/nGeneBLAzer-DEST pBD-NF-κBpAsRed2-N1 pcDNA4/HisMax-TOPO pCMV-ADptdTomato-N1 pcDNA6.2/nLumio-DEST pCMV-BDpCMV-tdTomato pcDNA6.2/cLumio-DEST pBIND-Id ControlpCRE-DD-tdTomato pcDNA4/myc-His C pBINDpCMV-DsRed-Express2 pcDNA4/HisMax C pG5 luciferasepEF1α-tdTomato pcDNA4/HisMax A pACT-MyoDpCRE-hrGFP c-Flag pcDNA3 pACTptdTomato-C1 pcDNA4/HisMax B pCMV-SPORT6pAsRed2-C1 pcDNA4/myc-His B pGL4.13pGL3-Promoter pcDNA4/myc-His A pGL4.19pGL3 basic pcDNA4/His C pGL4.26pAcGFP1-C2 pcDNA4/His B pGL4.20pAcGFP1-C1 pcDNA4/His A pGL4.29pAcGFP1-N3 pcDNA6/TR pGL4.30pAcGFP1-N2 pcDNA4/TO/Myc-His A pGL4.27pAcGFP1-N1 pcDNA4/TO pGL4.75pAcGFP1-C In-Fusion Ready pcDNA4/TO/Myc-His B pGL4.10pCRE-DD-AmCyan1 pcDNA4/TO/Myc-His C pGRN145pNFkB-DD-AmCyan1 pcDNA3.3-TOPO pSecTag2 A pDsRed2-Bid pBudCE4.1 pEBVHis B pDsRED2-Mito pFLAG-CMV-4 pEBVHis ApDD-AmCyan1 Reporter pFLAG-CMV-3 pCMV-Tag 3C pAmCyan1-N1 pFLAG-CMV-2 pCMV-Tag 3A pAmCyan1-C1 pFLAG-CMV-5a pCMV-Tag 3BpEF1α-IRES-DsRed-Express2 p3XFLAG-CMV-9 pCMV-Tag 5CpEF1α-DsRed-Monomer-N1 p3xFLAG-CMV-10 pCMV-Tag 5A pDsRED-Monomer-N1 p3XFLAG-CMV-8 pCMV-Tag 4A pDsRed-Express-N1 p3XFLAG-CMV-7.1 pCMV-Tag 5Bp3XFLAG-CMV-7 pDsRed-Monomer-N In-Fusion Ready pCMV-Tag 4B pDsRed-Express-C1 p3XFLAG-CMV-13 pCMV-Tag 2C pIRES2-ZsGreen1 p3XFLAG-CMV-14 pCMV-Tag 2B pDsRed-Express2-C1 plRES2-ZsGreen1 pCMV-Tag 2A pDsRed-Express2-N1 pBApo-EF1α-pur pCMV-LacZpEF1α-DsRed-Express2 pBApo-EF1α-neo pCMV-MycpIRES2-DsRed-Express pBApo-CMV pEF1α-IRES-AcGFP1 pIRES2-DsRed-Express2 pBApo-CMV-neo pEF1α-IRES-ZsGreen1 pIRES-hrGFP-1a pIRES-EGFP pEF1α-AcGFP1-N1 pIRESneo2 pIRESneo3 pIRES2-DsRed2 pIRESneo pDsRed-Monomer pIRES2-AcGFP1 pIREShyg3 pIRES。
pCDNA3.1hisabc载体说明书

user guideFor Research Use Only. Not intended for any animal or human therapeutic or diagnostic use.pcDNA ™3.1/His A, B, and CCatalog number V385-20Revision date 2 March 2012Publication Part number 28-0111MAN0000627ContentsKit Contents and Storage (iv)Methods (1)Cloning into pcDNA™3.1/His A, B, and C (1)Transformation and Transfection (6)Appendix (9)Map of pcDNA™3.1/His A, B, and C Vectors (9)Features of pcDNA™3.1/His A, B, and C Vectors (10)Map of pcDNA™3.1/His/lac Z (11)Accessory Products (12)Technical Support (13)Purchaser Notification (14)References (15)Kit Contents and StorageShipping and Storage pcDNA™ 3.1/His vectors are shipped on wet ice. Upon receipt, store vectors at –20ºC.Kit Contents 10 μg each of pcDNA™3.1/His A, B, and C are supplied at 0.5 μg/µL in10 mM Tris-HCl, 1 mM EDTA, pH 8.0 in a total volume of 20 µL.10 μg of pcDNA™3.1/His/lac Z is supplied at 0.5 μg/µL in 10 mM Tris-HCl,1 mM EDTA, pH 8.0 in a total volume of 20 µL.For research use only. Not intended for any human or animal therapeutic ordiagnostic use.Methods Cloning into pcDNA™3.1/His A, B, and CDescription of the System pcDNA™3.1/His A, B, and C are 5.5 kb vectors derived from pcDNA™3.1 anddesigned for high-level expression and purification of recombinant proteins inmammalian hosts. The vectors are supplied in three reading frames to facilitate in frame cloning with a polyhistidine metal-binding tag. The humancytomegalovirus (CMV) immediate-early promoter provides high-levelexpression in a wide range of mammalian cells. In addition, the vector replicatesepisomally in cell lines that are latently infected with SV40 or that express theSV40 large T antigen (e.g. COS7). High-level stable and non-replicative transientexpression can be carried out in most mammalian cells. The control plasmid, pcDNA™3.1/His/lac Z, is the pcDNA™3.1/His B vector with a 3.2 kb fragment containing the β-galactosidase gene cloned in frame with the N-terminal peptide. pcDNA™3.1/His/lac Z is included for use as a positive control for transfection, expression, and purification in the cell line of choice.General Molecular Biology Techniques For help with DNA ligations, E. coli transformations, restriction enzyme analysis, purification of single-stranded DNA, DNA sequencing, and DNA biochemistry, see Molecular Cloning: A Laboratory Manual (Sambrook et al., 1989) or Current Protocols in Molecular Biology (Ausubel et al., 1994).Maintaining pcDNA™3.1/His Many E. coli strains are suitable for the growth of this vector. To propagate and maintain pcDNA™3.1/His A,B, and C, use the supplied stock solution to transform a rec A (recombination deficient), end A (endonuclease A deficient)E. coli strain like TOP10, TOP10F’, DH5α™-T1R, DH10B™, or equivalent (see page12 for ordering information). Select the transformants on LB plates containing50–100 μg/mL ampicillin.For long-term storage, prepare a glycerol stock of your plasmid-containing E. coli strain.Continued on next pageKozak Sequence for Mammalian ExpressionIf you are recombining your entry clone with a destination vector for mammalian expression, your insert should contain a Kozak consensus sequence with an ATG initiation codon for proper initiation of translation (Kozak, 1987; Kozak, 1991; Kozak, 1990). An example of a Kozak consensus sequence is provided below. Other sequences are possible, but the G or A at position –3 and the G at position +4 (shown in bold) illustrates the most commonly occurring sequence with strong consensus. Replacing one of the two bases at these positions provides moderate consensus, while having neither results in weak consensus. The ATG initiation codon is shown underlined. (G/A)NNATG GThe pcDNA ™3.1/His vectors are fusion vectors. To ensure proper expression of your recombinant protein, you must clone your gene in frame with the ATG at base pairs 920–922. This will create a fusion with the N-terminal polyhistidine tag, Xpress ™ epitope, and the enterokinase cleavage site. The vector is supplied in three reading frames to facilitate cloning. See pages 3–5 to develop a cloning strategy. If you wish to clone as close as possible to the enterokinase cleavage site, follow the guidelines below: • Digest pcDNA ™3.1/His A, B, or C with Kpn I • Create blunt ends with T4 DNA polymerase and dNTPs • Clone your blunt-ended insert in frame with the lysine codon (AAG) of the enterokinase recognition sequence. Following enterokinase cleavage, no vector-encoded amino acid residues will be present in your protein. Continued on next pageMultiple Cloning Site ofpcDNA™3.1/His A Below is the multiple cloning site for pcDNA™3.1/His A. Restriction sites are labeled to indicate the cleavage site. The boxed nucleotide indicates the variable region. Note that there is a stop codon after the Xba I site and that the Asp718 I and Kpn I sites are in the same reading frame for all three vectors. The multiple cloning site has been confirmed by sequencing and functional testing. The sequence is available for downloading from or by contacting Technical Support (see page 13).Continued on next pageMultiple Cloning Site ofpcDNA™3.1/His B Below is the multiple cloning site for pcDNA™3.1/His B. Restriction sites are labeled to indicate the cleavage site. The boxed nucleotides indicate the variable region. The multiple cloning site has been confirmed by sequencing and functional testing. Note that the Asp718 I and Kpn I sites are in the same reading frame for all three vectors. The sequence is available for downloading from or by contacting Technical Support (see page 13).Continued on next pageMultiple Cloning Site ofpcDNA™3.1/His C Below is the multiple cloning site for pcDNA™3.1/His C. Restriction sites are labeled to indicate the cleavage site. The multiple cloning site has been confirmed by sequencing and functional testing. Note that the Asp718 I and Kpn I sites are in the same reading frame for all three vectors. The sequence is available for downloading from or by contacting Technical Support (see page 13).Continued on next pageTransformation and TransfectionE. coli Transformation Transform your ligation mixtures into a competent rec A, end A E. coli strain (e.g., TOP10, TOP10F’, DH5α™-T1R, DH10B™, see page 12) and select on LB plates containing 50–100 μg/mL ampicillin. Select 10–20 clones and analyze for the presence and orientation of your insert.Applying Selective Pressure We recommend taking some (if not all) of the following precautions to prevent your clone from being “overrun” by background contaminants:•Use carbenicillin instead of ampicillin. Carbenicillin is more stable than ampicillin, and allows for a longer period of selective pressure, thuspreserving your clones longer.•Increase the antibiotic concentration. More antibiotic means that your clones will not be overwhelmed by β-lactamase buildup.•Periodically refresh plate media. If you suspect that tubes/plates may be beginning to fail, spin them down, remove the old media, and replenish the wells with fresh LB media plus glycerol and antibiotic.Streak clones on selective (preferably carbenicillin) LB agar plates. After about 12 hours, isolate colonies for downstream usage. This will isolate your desired clones from potential background contaminants.We recommend that you sequence your construct with the T7 Forward and BGHReverse primers (see page 12 for ordering information) to confirm that your geneis fused in frame with the N-terminal His tag and the enterokinase site. Refer tothe diagrams on pages 3–5 for the sequences and locations of the priming sites.For your convenience, we offer a custom primer service. For more information,visit or call Technical Support (see page 13).PlasmidPreparationPlasmid DNA for transfection into eukaryotic cells must be very clean and freefrom phenol and sodium chloride. Contaminants will kill the cells and salt willinterfere with lipid complexing, decreasing transfection efficiency. Werecommend isolating DNA using the PureLink® HiPure Miniprep Kit or thePureLink® HiPure Midiprep Kit (see page 12 for ordering information) or CsClgradient centrifugation.Continued on next pageMethods of Transfection For established cell lines (e.g., HeLa), consult original references or the supplier of your cell line for the optimal method of transfection. Precisely follow the protocol for your cell line, paying particular attention to medium requirements, when to pass the cells, and at what dilution to split the cells. Further information is provided in Current Protocols in Molecular Biology (Ausubel et al., 1994). Methods for transfection include calcium phosphate (Chen and Okayama, 1987; Wigler et al., 1977), lipid-mediated (Felgner et al., 1989; Felgner and Ringold, 1989) and electroporation (Chu et al., 1987; Shigekawa and Dower, 1988). For high efficiency transfection in a broad range of mammalian cells, use Lipofectamine®2000 Reagent available for purchase (see page 12). For more information on Lipofectamine® 2000 and other transfection reagents, visit or contact Technical Support (see page 13).Positive Control pcDNA™3.1/His/lac Z is supplied as a positive control vector for mammaliantransfection and expression (see page 11). pcDNA™3.1/His/lac Z may be used tooptimize transfection conditions for your cell line. The gene encodingβ-galactosidase is expressed in mammalian cells under the CMV promoter. Asuccessful transfection results in β-galactosidase expression and can be easilyassayed.Assay forβ-galactosidase Activity You may assay for β-galactosidase expression by activity assay using cell-free lysates (Miller, 1972) or by staining the cells for activity. The β-Gal Assay Kit and the β-Gal Staining Kit are available for purchase (see page 12 for ordering information) for fast, easy detection of β-galactosidase expression.Geneticin®Selective Antibiotic For stable transfection, pcDNA™3.1/His A, B, and C contain the resistance factorto Geneticin®. Geneticin® blocks protein synthesis in mammalian cells byinterfering with ribosomal function. It is an aminoglycoside, similar in structure to neomycin, gentamycin, and kanamycin. Expression in mammalian cells of the bacterial aminoglycoside phosphotransferase gene (APH), derived from Tn5, results in detoxification of Geneticin® Selective Antibiotic (Southern and Berg,1982).Continued on next pageGeneticin®Selection Guidelines Geneticin® is available for purchase (see page 12 for ordering information). Use asfollows:1.Prepare Geneticin® in a buffered solution (e.g., 100 mM HEPES, pH 7.3).e 100 to 1,000 μg/mL of Geneticin® in complete medium.3.Calculate concentration based on the amount of active drug (check the lotlabel).4.Test varying concentrations of Geneticin® on your cell line to determine theconcentration that kills your cells (kill curve). Cells differ in theirsusceptibility to Geneticin®.Cells will divide once or twice in the presence of lethal doses of Geneticin®Selective Antibiotic, so the effects of the drug take several days to become apparent. Complete selection can take from 2 to 4 weeks of growth in selectivemedium.Preparing Cells for Lysis Use the procedure below to prepare cells for lysis prior to purification of your protein on ProBond™ (see page 12 for ordering information). You will need 5 × 106 to 1 × 107 cells for purification of your protein on a 2 mL ProBond™ column (see ProBond™ Protein Purification manual).Note: The N-terminal peptide adds approximately 5 kDa to the size of your recombinant fusion protein.1. Seed cells in five T-75 flasks or 2 to 3 T-150 flasks.2. Grow the cells in selective medium until they are 80–90% confluent.3. Harvest the cells by treating with trypsin-EDTA for 2 to 5 minutes or byscraping the cells in PBS.4. Inactivate the trypsin, if necessary, and transfer the cells to a sterilemicrocentrifuge tube.5. Centrifuge the cells at approximately 250 × g for 5 minutes. You may lyse thecells immediately or freeze in liquid nitrogen and store at −80°C until needed.Lysing Cells If you are using ProBond™ resin, refer to the ProBond™ Protein Purificationmanual for details about sample preparation for chromatography. If you are usingother metal-chelating resin, refer to the manufacturer's instruction.AppendixMap of pcDNA™3.1/His A, B, and C VectorsMap of pcDNA™3.1/His The figure below summarizes the features of the pcDNA™3.1/His vectors. Thesequences for pcDNA™3.1/His A, B, and C are available for downloading from or by contacting Technical Support (see page 13). Details of the multiple cloning sites are shown on page 3 for pcDNA™3.1/His A,page 4 for pcDNA™3.1/His B, and page 5 for pcDNA™3.1/His C.Continued on next pageFeatures of pcDNA™3.1/His A, B, and C VectorsFeatures of pcDNA™3.1/His pcDNA™3.1/His A (5,514 bp), pcDNA™3.1/His B (5,515 bp), andpcDNA™3.1/His C (5,513 bp) contain the following elements. All features have been functionally tested.Map of pcDNA™3.1/His/lac ZDescription pcDNA™3.1/His/lac Z is a 8,577 bp control vector containing the gene forβ-galactosidase. pcDNA™3.1/His B was digested with Bam H I, blunted, anddigested with Eco R I. A 3.2 kb blunt-Eco R I fragment containing theβ-galactosidase gene was then ligated into pcDNA™3.1/His B in frame with theN-terminal peptide. The β-galactosidase protein expressed frompcDNA™3.1/His/lac Z has a size of approximately 120 kDa.Map of Control Vector The figure below summarizes the features of the pcDNA™3.1/His/lac Z vector. The nucleotide sequence for pcDNA™3.1/His/lac Z is available for downloading from or by contacting Technical Support (see page 13).Accessory ProductsAdditional ProductsThe following additional products may be used with the pcDNA ™3.1/His vectors. For more information, visit or contact Technical Support (see page 13).Purifying Fusion ProteinsThe N-terminal polyhistidine tag can be used to purify the recombinant fusion protein with a metal-chelating resin such as ProBond ™. Ordering information for ProBond ™ resin is provided below.Technical SupportObtaining support For the latest services and support information for all locations, go to/support.At the website, you can:•Access worldwide telephone and fax numbers to contact Technical Supportand Sales facilities•Search through frequently asked questions (FAQs)•Submit a question directly to Technical Support (techsupport@)•Search for user documents, SDSs, vector maps and sequences, applicationnotes, formulations, handbooks, certificates of analysis, citations, and otherproduct support documents•Obtain information about customer training•Download software updates and patchesSafety DataSheets (SDS)Safety Data Sheets (SDSs) are available at /support.Certificate of Analysis The Certificate of Analysis provides detailed quality control and product qualification information for each product. Certificates of Analysis are available on our website. Go to /support and search for the Certificate of Analysis by product lot number, which is printed on the box.Limited warranty Life Technologies Corporation is committed to providing our customers withhigh-quality goods and services. Our goal is to ensure that every customer is100% satisfied with our products and our service. If you should have anyquestions or concerns about a Life Technologies product or service, contact ourTechnical Support Representatives.All Life Technologies products are warranted to perform according tospecifications stated on the certificate of analysis. The Company will replace, freeof charge, any product that does not meet those specifications. This warrantylimits the Company’s liability to only the price of the product. No warranty isgranted for products beyond their listed expiration date. No warranty isapplicable unless all product components are stored in accordance withinstructions. The Company reserves the right to select the method(s) used toanalyze a product unless the Company agrees to a specified method in writingprior to acceptance of the order.Life Technologies makes every effort to ensure the accuracy of its publications,but realizes that the occasional typographical or other error is inevitable.Therefore the Company makes no warranty of any kind regarding the contentsof any publications or documentation. If you discover an error in any of ourpublications, report it to our Technical Support Representatives.Life Technologies Corporation shall have no responsibility or liability for anyspecial, incidental, indirect or consequential loss or damage whatsoever. Theabove limited warranty is sole and exclusive. No other warranty is made,whether expressed or implied, including any warranty of merchantability orfitness for a particular purpose.Purchaser NotificationLimited Use Label License No: Research Use Only The purchase of this product conveys to the purchaser the limited, non-transferable right to use the purchased amount of the product only to perform internal research for the sole benefit of the purchaser. No right to resell this product or any of its components is conveyed expressly, by implication, or by estoppel. This product is for internal research purposes only and is not for use in commercial applications of any kind, including, without limitation, quality control and commercial services such as reporting the results of purchaser's activities for a fee or other form of consideration. For information on obtaining additional rights, please contact outlicensing@ or Out Licensing, Life Technologies, 5791 Van Allen Way, Carlsbad, California 92008.Limited Use Label License6x His Tag This product is licensed from Hoffmann-La Roche, Inc., Nutley, NJ and/or Hoffmann-LaRoche Ltd., Basel, Switzerland and is provided only for use in research. Information about licenses for commercial use is available from QIAGEN GmbH, Max-Volmer-Str. 4, D-40724 Hilden, Germany.ReferencesAndersson, S., Davis, D. L., Dahlbäck, H., Jörnvall, H., and Russell, D. W. (1989). Cloning, Structure, and Expression of the Mitochondrial Cytochrome P-450 Sterol 26-Hydroxylase, a Bile Acid Biosynthetic Enzyme. J. Biol. Chem. 264, 8222-8229.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. (1994).Current Protocols in Molecular Biology (New York: Greene Publishing Associates and Wiley-Interscience).Boshart, M., Weber, F., Jahn, G., Dorsch-Häsler, K., Fleckenstein, B., and Schaffner, W. (1985). A Very Strong Enhancer is Located Upstream of an Immediate Early Gene of Human Cytomegalovirus. Cell 41, 521-530.Chen, C., and Okayama, H. (1987). High-Efficiency Transformation of Mammalian Cells by Plasmid DNA.Molec. Cell. Biol. 7, 2745-2752.Chu, G., Hayakawa, H., and Berg, P. (1987). Electroporation for the Efficient Transfection of Mammalian Cells with DNA. Nucleic Acids Res. 15, 1311-1326.Felgner, P. L., Holm, M., and Chan, H. (1989). Cationic Liposome Mediated Transfection. Proc. West.Pharmacol. Soc. 32, 115-121.Felgner, P. L. a., and Ringold, G. M. (1989). Cationic Liposome-Mediated Transfection. Nature 337, 387-388. Goodwin, E. C., and Rottman, F. M. (1992). The 3´-Flanking Sequence of the Bovine Growth Hormone Gene Contains Novel Elements Required for Efficient and Accurate Polyadenylation. J. Biol. Chem. 267, 16330-16334.Miller, J. H. (1972). Experiments in Molecular Genetics (Cold Spring Harbor, New York: Cold Spring Harbor Laboratory).Nelson, J. A., Reynolds-Kohler, C., and Smith, B. A. (1987). Negative and Positive Regulation by a Short Segment in the 5´-Flanking Region of the Human Cytomegalovirus Major Immediate-Early Gene.Molec. Cell. Biol. 7, 4125-4129.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, Second Edition (Plainview, New York: Cold Spring Harbor Laboratory Press).Shigekawa, K., and Dower, W. J. (1988). Electroporation of Eukaryotes and Prokaryotes: A General Approach to the Introduction of Macromolecules into Cells. BioTechniques 6, 742-751.Southern, P. J., and Berg, P. (1982). Transformation of Mammalian Cells to Antibiotic Resistance with a Bacterial Gene Under Control of the SV40 Early Region Promoter. J. Molec. Appl. Gen. 1, 327-339. Wigler, M., Silverstein, S., Lee, L.-S., Pellicer, A., Cheng, Y.-C., and Axel, R. (1977). Transfer of Purified Herpes Virus Thymidine Kinase Gene to Cultured Mouse Cells. Cell 11, 223-232.©2012 Life Technologies Corporation. All rights reserved.The trademarks mentioned herein are the properties of Life Technologies Corporation or their respective owners.Notes1718Headquarters5791 Van Allen Way | Carlsbad, CA 92008 USA | Phone +1 760 603 7200 | Toll Free in USA 800 955 6288 For support visit /support or email techsupport@。
所有质粒载体汇总

酿酒酵母表达载体pYES2,pYES2/NT,pYES2/CT,pYES3,pYES6, pYCplac22-GFP,酵母载体pAUR123,pRS303TEF,pRS304, pRS305,pRS306,pY13TEF,pY14TEF,pY15TEF,pY16TEF,酵母基因重组表达载体pUG6, pSH47,酵母单杂载体pHISi,pLacZi,pHIS2, pGAD424, 酵母双杂交系统:酿酒酵母Y187, 酿酒酵母AH109;质粒pGADT7,pGBKT7;对照质粒pGBKT7-53,pGBKT7-lam,pGADT7-T,PCL1,酿酒酵母菌株INVSc1,YM4271, AH109,Y187,Y190,毕赤酵母表达载体pPIC9K,pPIC9K-His,pPIC3.5K,pPICZalphaA,B,C,pPICZA,B,C,pGAPZαA,pAO815,pPIC9k-His,pHIL-S1,pPink hc,配套毕赤酵母Pichiapink,毕赤酵母宿主X33,KM71,KM71H,GS115,原核表达载体pQE30,31,32,40,60,61,62,等原核表达载体,包括pET系列,pET-GST,pGEX系列(含GST标签),pMAL系列pMAL-c2x,-c4x,-c4e,-c5x,-p5x,pBAD,pBADHis,pBADmycHis系列,pQE系列,pTrc99a,pTrcHis系列,pBV220,221,222,pTXB系列,pLLP-ompA,pIN-III-ompA (分泌型表达系列),pQBI63(原核表达带荧光)pET3a, pET 3d, pET 11a, pET 12a, pET 14b, pET 15b, pET 16b, pET 17b, pET 19b, pET 20b, pET 21a,b,d, pET 22b, pET 23a, pET 23b, pET 24a,b, pET 25b, pET 26b, pET 27b, pET 28a,b, pET 29a, pET 30a, pET 31b, pET 32a, pET 35b, pET 38b, pET 39b, pET 40b, pET 41a,b pET 42a, pET 43.1a,b pET 44a, pET 49b pET302,303 pET His,pET Dsb,pET GST,pET Trx pQE2, pQE9 pQE30,31,32, pQE 40 pQE70 pQE80L pQETirs system pRSET-A pRSET-B pRSET-C pGEX4T-1,-2,-3,5x-1,6p-1,6p-2,2tk,3c pBV220,221,222 pTrcHisA,B,C pBAD24,34,43 pBAD HisA,B,C pPinPoint-Xa1,Xa2,Xa3 pMALc2x, p2x pBV220 pGEM Ex1, pGEM7ZF(+), pTrc99A, pTwin1, pEZZ18 pkk232-8,pkk233-3,pACYC184,pBR322,pUC119 pTYB1,pTYB2,pTYB4,pTYB11 pBlueScript SK (+),pBlueScript SK(-)pLLP ompA, pINIIIompA, pMBP-P ,pMBP-C, 大肠杆菌冷激质粒: pColdI pColdII pColdIII pColdTF 原核共表达质粒:pACYCduet-1,pETduet-1,pCDFduet-1,pRSFduet-1 Takara公司大肠杆菌分子伴侣: pG-KJE8 pGro7 pKJE7 pGTf2 pTf16 大肠杆菌宿主细胞: DH5a JM101 JM103JM105 JM107 JM109 JM110 Top10 Top10F BL21(DE3)HB101 ER2529 E2566 C2566 MG1655 XL-10gold XL blue M15 JF1125 K802 SG1117 BL21(AI)BL21(DE3)plysS TG1 TB1 DH5a(pir)Tuner(DE3)Bl21 codonplusRIPL Novablue (DE3)Rosetta Rosetta(DE3)Rosetta(DE3)plys Rosetta-gami(DE3)Rosetta-gamiB(DE3), Rosetta-gamiB(DE3)plysS Orgami(DE3)OrgamiB(DE3)HMS174(DE3)植物表达/RNAi载体农杆菌pBI121,pBI121-GFP,pBI101,pBI221,pSN1301,pUN1301,pRTL2 , pRTL2-GFP , pRTL2-CFP, pRTL2-RFP , pRTL2-YFP,pCAMBIA 1300, 1301, 1302,1303,1304,1305, 1381Z,1391Z,2300, 2301,3300,3301,pCAMBIA super1300,pCAMBIAsuper1300-GFP,pPZP212,pPZP2121,pPZP212-GFP,pGDG,RNAi载体pART27,pHANNIBAL,pKANNIBAL, pFGC5941,pTCK303, pTRV1,pTRV2,T-DNA插入载体(随机突变体库)pSKI015,pSKI074,真菌ATMT载体pBIG2RHPH2-GUS-GFP,pBHt1枯草芽孢杆菌表达载体pWB980,pHT43,pHP13,pHP43,pBE2,pMUTIN4,pUB110,pE194,pMA5, pMK3,pMK4,pHT304,pHY300PLK,pBest502,pDG1363,pSG1154,pAX01, pSAS144,pDL,pDG148-stu,pDG641,pAL12,pUCX05-bgaB,pHT01,配套菌株BS 168,WB600,WB800,WB700,WB800N,1012,FZB42,1A747,广宿主质粒pVLT33RNAi基因沉默干扰敲除载体pSilencer1.0,pSilencer 2.1-U6 hygro, pSilencer 3.1-H1 hygro,pSilencer 3.1-H1 neo, pSilencer 4.1-CMV neo, pSilencer 4.1-CMV puro pMIR-REPORT Luciferase RNAi载体(oligoengine)pSuper-puro RNAi逆转录病毒载体(clontech): RNAi-Ready pSIREN-Retro Q, RNAi-ReadypSIREN-RetroQ-ZsGreen(Luciferase shRNA Annealed Oligonucleotide)RNAi慢病毒载体(addgene): pLKO.1哺乳动物表达载体pcDNA3.1+/-,pcDNA4/HisMax B,pSecTag2 A,pVAX1,pBudCE4.1,pTracer CMV2,pcDNA3.1(-)/myc-His A ,pcDNA6-Myc/His B,pCEP4, pIRES,pIRESneo,pIRES hyg3,pCMV-myc,pCMV-HA,pIRES-puro3,pIRES-neo3,pCAGGS哺乳动物双杂交系统pACT,pBIND,pACT-MyoD,pBIND-Id,pG5luc,pCMV-BD, pCMV-AD, pBD-p53, pFR-luc,Cytotrap Two-Hybrid System:pSos, pSos MAFB, pMyr蜕皮激素诱导系统pIND, pVgRxR,LacSwith II哺乳动物诱导表达系统:pOPRSVI ,pOPI3CAT,pCMVLacI,GeneSwitch System:pSwitch哺乳动物表面展示系统:pDisplay, 四环素调控系统(Invitrogen):pcDNA4/TO/Myc-His A,pcDNA4/TO/Myc-His B,pcDNA4/TO/Myc-His C,pcDNA4/TO/Myc-His/LacZ,pcDNA6/TR四环素调控系统(Clontech):pTet-On,pTet-Off,pTRE2,pRevTRE,pRevTet-On,pRevTet-off信号通路报告载体:pGAS-TA-Luc,pSTAT3-TA-Luc, pISRE-TA-Luc, pTA-Luc,pIκB-EGFP,pNFAT-TA-Luc,pCaspase3-sensor,pAP1(PMA)-Luc;pGL4.26[luc2P/minP/Hygro],pGL4.29[luc2P/CRE/Hygro],pGL4.30[luc2P/NFA T-RE/Hygro],pGL4.75;p53-Luc,pAP-1-Luc, pNF-κB-Luc,pSRE-Luc,pFA2-Elk1,pFC-MEKK,pFR-luc,Gateway系统(invitrogen)pcDNA6.2-GWEmGFP-miR negative, pLenti 6/TR,pcDNA 6.2-GW EmGFP-miR,乳酸菌表达载体及各种乳酸菌乳酸杆菌菌株,pNZ8148,pLEISS,pMG36e,pBBR1MCS-5,pBBR1MCS-6,pRV610,pLEM415,pHY3 00PLK,分泌型乳酸菌表达载体pVE5523,pPG611.1,pPG612.1等和乳酸杆菌菌株宿主菌NZ9000,MG1363,Lactobacillus casei 1.539,Lactobacillus casei,acidophilus NCFM,1.2,Lactobacillus sakei 23K,L.plantarum,L.rhamnosusGG,B.coagulans,Bifidobacterium bifidum,Bifidobacterium infantis,Lactococcus lactis M17,1663,Lactobacillus reuterii广宿主表达载体链球菌表达敲除载体假单胞菌表达载体pVLT33,pBBR1MCS-2,3,4,5,6, pJRD215,pJN105,pME6032,Cos载体pLAFR3,pMP2444(GFP), pHY300PLK,pRT102,pRL1063a, 转座子载体pUT-miniTn5,pMGS100, pWHM10,pKC1139,pSET152,pOJ260,pPG611.1,pPG612.1,腺病毒载体/慢病毒,逆转录病毒表达载体及包装包膜质粒,腺病毒系统(Stratagene): pAdEasy-1,pShuttle-CMV,pShuttle,pAdTrack, pAdTrack-CMV, pShuttle-IRES-hrGFP-1、pShuttle-IRES-hrGFP-2、pShuttle-CMV-lacZ,pShuttle-CMV-EGFP-C,pXC1, pBHGE3, 配套大肠杆菌BJ5183,293,293T cellline 腺相关病毒系统(Stratagene): pAAV-MCS,pAAV-RC,pHelper,pAAV-LacZ,pAAV-IRES-hrGFP,pCMV-MCS,慢病毒载体:pLVX-DsRed-Monomer-N1,pLVX-IRES-ZsGreen1,pLVX-AcGFP1-N1,Lenti6/v 5-EDST-EGFP,pWPXL, FUGW,pLentilox 3.7,RNAi-Ready pSIREN-Retro Q,RNAi-Ready pSIREN-Retro Q-ZsGreen,pSUPER.Retro-GFP/Neo,pSUPER-Retro-Neo, pSUPER.Retro-puro,PLNCX PLNCX2 pMSCV-HYG pMSCV-neo pMSCV-puro pLEGFP-C1 pLOX-CW-CRE pLOX-GFP-IRES-TK pRetroX-IRES-DsRedExpress, pLVX-IRES-mCherry质粒载体。
GA引物载体对照表1

AmpR AmpR
AmpR ZeocinR ZeocinR ZeocinR ZeocinR ZeocinR ZeocinR
KanaR
pGEM-1 pGEM-11z pGEM-3 pGEM-3z pGEM-3Zf(+)(-) pGEM-4 pGEM-4Z pGEM-4z pGEM-5z pGEM-5Zf(+)(-) pGEM-7z pGEM-7Zf(+)(-) pGEM-9z pGEM-9Zf(+)(-) pGEM-T pGEM-T pGEM-T_Easy pGEM-Teasy pGEX-4T-1(-2,-3) pGEX-5X-1(-2,-3) pGEX-6P-1(-2,-3) pGL2 pGL2-Basic pGL3(MCSI,luc+upstream) pGL3(MCSII,luc+downstream) pGL3-Basic(MCSI) pGL3-Basic(MCSII) pGL4(MCSI,luc+upstream) pGL4(MCSII,luc+downstream) pIRES(MCSI) pIRES(MCSII) pIRESneo2 pLVX-IRES-ZsGreen1 pMALc PMAL-C2X pMALd pMALe pMB438 Pmd18/19-T pPIC3.5K pPIC6A pPIC6alphaA pPIC6alphaB pPIC6alphaC
AmpR&KanaR AmpR&KanaR AmpR AmpR AmpR&KanaR AmpR&KanaR AmpR AmpR&KanaR
pET-3(-a,-b,-c,-d) pET-30_Ek/LIC pET-30_Xa/LIC pET-30a(-b,-c)(+) pET-31b(+) pET-32a(-b,-c)(+) pET-33b(+) pET-34b(+) pET-35b(+) pET-36b(+) pET-37b(+) pET-38b(+) pET-39b(+) pET-40b(+) pET-41a(-b,-c)(+) pET-42a(-b,-c)(+) pET-44a(-b,-c)(+) pET-44a_EK/LIC pET-45b(+) pET-46_Ek/LIC pET-5(-a,-b,-c) pET-7 pET-9(-a,-b,-c,-d) pETDuet-1(MCSI) pETDuet-1(MCSII) pFastBac-Dual pFastBacDual(MCSI,PHpromoter) pFastBacDual(MCSII,p10promoter) pFastBac-GT pFastBac-HT PFLAG-CMV4 pGADT7 pGADT7 pGAD-T7 pGAPZA pGAPZalphaA pGAPZalphaB pGAPZalphaC pGAPZB pGAPZC pGBD-T7 pGBKT7 pGBKT7
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
pcDNA4/HisMax C编号 载体名称北京华越洋生物VECT6173 pcDNA4/HisMax C pcDNA4/HisMax C载体基本信息载体名称: pcDNA4/HisMax C质粒类型: 哺乳动物表达载体;cDNA表达载体高拷贝/低拷贝: 高拷贝克隆方法: 多克隆位点,限制性内切酶启动子: CMV载体大小: 5257 bp5' 测序引物及序列: T7 Forward: 5’-TAATACGACTCACTATAGGG-3’ 3' 测序引物及序列: BGH Reverse: 5-TAGAAGGCACAGTCGAGG-3 载体标签: His Tag (N-端), Xpress Epitope Tag(N-端)载体抗性: 氨苄青霉素筛选标记: Zeocin克隆菌株: TOP10F´, DH5a宿主细胞(系): 常规细胞系,如293、Hela等备注: pcDNA4/HisMax C 载体是哺乳动物表达载体,适用于cDNA的表达与克隆;QBI SP163增强子,使得目的基因的高水平表达提高了3~5倍;pcDNA4/HisMax A,B,C的区别仅在于多克隆位点处;含EK (Enterokinase)切割位点稳定性: 瞬表达或稳表达组成型/诱导型: 组成型病毒/非病毒: 非病毒pcDNA4/HisMax C载体质粒图谱和多克隆位点信息pcDNA4/HisMax A, B, and C载体介绍:pcDNA4/HisMax is specifically designed to maximize protein expression in a variety of mammalian cells. The vector contains the QBI SP163 translational enhancer to increase expression levels two- to five-fold above those seen with the promoter alone . In addition to SP163-enhanced expression, pcDNA4/HisMax includes a cleavable N-terminal Xpress tag for rapid detection of recombinant protein with an Anti-Xpress Antibody. pcDNA4/HisMax is available TOPO Cloning-ready for five-minute cloning of Taqamplified PCR products.pcDNA4/HisMax A, B, and C are 5.3 kb vectors derived from pcDNA4/His and designed for overproduction of recombinant proteins in mammalian cell lines. Features of the vectors allow purification and detection of expressed proteins. High-level stable and transient expression can be carried out in most mammalian cells. The vectors contain the following elements:Human cytomegalovirus immediate-early (CMV) promoter for high-level expression in a wide range of mammalian cellsQBI SP163 translational enhancer for increased levels of recombinant protein expression (Stein et al., 1998) (see page 4 for more information)Three reading frames to facilitate in-frame cloning with an N-terminal peptideencoding the Xpress epitope and a polyhistidine metal-binding tagZeocin resistance gene for selection of stable cell linesEpisomal replication in cell lines that are latently infected with SV40 or that express the SV40 large T antigen (e.g. COS-1, COS-7)The control plasmid, pcDNA4/HisMax/lacZ, is included for use as a positive control for transfection, expression, and detection in the cell line of choice.实验流程:Use the following outline to clone and express your gene of interest in pcDNA4/HisMax.Consult the multiple cloning sites described on pages 5-7 to determine which vector (A, B, or C) should be used to clone your gene in frame with the N-terminal Xpress epitope and the polyhistidine tag.Ligate your insert into the appropriate vector and transform into E. coli. Select transformants on 50 to 100 μg/ml ampicillin or 25-50 μg/ml Zeocin.Analyze your transformants for the presence of insert by restriction digestion. Select a transformant with the correct restriction pattern and use sequencing to confirm that your gene is cloned in frame with the N-terminal peptide.Transfect your construct into the cell line of choice using your own method of transfection. Generate a stable cell line, if desired.Test for expression of your recombinant gene by western blot analysis or functional assay. For antibody to the Xpress epitope, please see the next page.To purify your recombinant protein, you may use metal-chelating resin such as ProBond. ProBond resin is available separately.表达目的基因:We have a wide variety of mammalian expression vectors utilizing the CMV or EF-1αpromoter. Vectors are available with the Xpress (N-terminal), c-myc (C-terminal), or V5 (C-terminal) epitope for detection and either the neomycin, blasticidin, or Zeocin resistance genes. All vectors utilize the polyhistidine tag for purification using ProBond resin.The pcDNA4/HisMax vectors are fusion vectors. To ensure proper expression of your recombinant protein, you must clone your gene in frame with the ATG at base pairs 1080-1082. This will create a fusion with the N-terminal polyhistidine tag, Xpress epitope, and the enterokinase cleavage site. The vector is supplied with the multiple cloning site in three reading frames relative to the N-terminal peptide to facilitate cloning.If you wish to clone your gene as close as possible to the enterokinase cleavage site, follow the guidelines below:Digest pcDNA4/HisMax A, B, or C with Kpn I.Create blunt ends with T4 DNA polymerase and dNTPs.Clone your blunt-ended insert in frame with the lysine codon (AAG) of the enterokinase recognition site.pcDNA4/HisMax C载体序列ORIGIN1 GACGGATCGG GAGATCTCCC GATCCCCTAT GGTCGACTCT CAGTACAATC TGCTCTGATG 61 CCGCATAGTT AAGCCAGTAT CTGCTCCCTG CTTGTGTGTT GGAGGTCGCT GAGTAGTGCG 121 CGAGCAAAAT TTAAGCTACA ACAAGGCAAG GCTTGACCGA CAATTGCATG AAGAATCTGC 181 TTAGGGTTAG GCGTTTTGCG CTGCTTCGCG ATGTACGGGC CAGATATACG CGTTGACATT 241 GATTATTGAC TAGTTATTAA TAGTAATCAA TTACGGGGTC ATTAGTTCAT AGCCCATATA 301 TGGAGTTCCG CGTTACATAA CTTACGGTAA ATGGCCCGCC TGGCTGACCG CCCAACGACC 361 CCCGCCCATT GACGTCAATA ATGACGTATG TTCCCATAGT AACGCCAATA GGGACTTTCC 421 ATTGACGTCA ATGGGTGGAC TATTTACGGT AAACTGCCCA CTTGGCAGTA CATCAAGTGT 481 ATCATATGCC AAGTACGCCC CCTATTGACG TCAATGACGG TAAATGGCCC GCCTGGCATT 541 ATGCCCAGTA CATGACCTTA TGGGACTTTC CTACTTGGCA GTACATCTAC GTATTAGTCA 601 TCGCTATTAC CATGGTGATG CGGTTTTGGC AGTACATCAA TGGGCGTGGA TAGCGGTTTG 661 ACTCACGGGG ATTTCCAAGT CTCCACCCCA TTGACGTCAA TGGGAGTTTG TTTTGGCACC 721 AAAATCAACG GGACTTTCCA AAATGTCGTA ACAACTCCGC CCCATTGACG CAAATGGGCG 781 GTAGGCGTGT ACGGTGGGAG GTCTATATAA GCAGAGCTCT CTGGCTAACT AGAGAACCCA 841 CTGCTTACTG GCTTATCGAA ATTAATACGA CTCACTATAG GGAGACCCAA GCTGGCTAGC 901 GTTTAAACTT AAGCTTAGCG CAGAGGCTTG GGGCAGCCGA GCGGCAGCCA GGCCCCGGCC 961 CGGGCCTCGG TTCCAGAAGG GAGAGGAGCC CGCCAAGGCG CGCAAGAGAG CGGGCTGCCT 1021 CGCAGTCCGA GCCGGAGAGG GAGCGCGAGC CGCGCCGGCC CCGGACGGCC TCCGAAACCA 1081 TGGGGGGTTC TCATCATCAT CATCATCATG GTATGGCTAG CATGACTGGT GGACAGCAAA 1141 TGGGTCGGGA TCTGTACGAC GATGACGATA AGGTACCAGG ATCCAGTGTG GTGGAATTCT 1201 GCAGATATCC AGCACAGTGG CGGCCGCTCG AGTCTAGAGG GCCCGTTTAA ACCCGCTGAT 1261 CAGCCTCGAC TGTGCCTTCT AGTTGCCAGC CATCTGTTGT TTGCCCCTCC CCCGTGCCTT 1321 CCTTGACCCT GGAAGGTGCC ACTCCCACTG TCCTTTCCTA ATAAAATGAG GAAATTGCAT 1381 CGCATTGTCT GAGTAGGTGT CATTCTATTC TGGGGGGTGG GGTGGGGCAG GACAGCAAGG 1441 GGGAGGATTG GGAAGACAAT AGCAGGCATG CTGGGGATGC GGTGGGCTCT ATGGCTTCTG 1501 AGGCGGAAAG AACCAGCTGG GGCTCTAGGG GGTATCCCCA CGCGCCCTGT AGCGGCGCAT 1561 TAAGCGCGGC GGGTGTGGTG GTTACGCGCA GCGTGACCGC TACACTTGCC AGCGCCCTAG 1621 CGCCCGCTCC TTTCGCTTTC TTCCCTTCCT TTCTCGCCAC GTTCGCCGGC TTTCCCCGTC 1681 AAGCTCTAAA TCGGGGCATC CCTTTAGGGT TCCGATTTAG TGCTTTACGG CACCTCGACC 1741 CCAAAAAACT TGATTAGGGT GATGGTTCAC GTAGTGGGCC ATCGCCCTGA TAGACGGTTT 1801 TTCGCCCTTT GACGTTGGAG TCCACGTTCT TTAATAGTGG ACTCTTGTTC CAAACTGGAA 1861 CAACACTCAA CCCTATCTCG GTCTATTCTT TTGATTTATA AGGGATTTTG GGGATTTCGG 1921 CCTATTGGTT AAAAAATGAG CTGATTTAAC AAAAATTTAA CGCGAATTAA TTCTGTGGAA 1981 TGTGTGTCAG TTAGGGTGTG GAAAGTCCCC AGGCTCCCCA GGCAGGCAGA AGTATGCAAA 2041 GCATGCATCT CAATTAGTCA GCAACCAGGT GTGGAAAGTC CCCAGGCTCC CCAGCAGGCA 2101 GAAGTATGCA AAGCATGCAT CTCAATTAGT CAGCAACCAT AGTCCCGCCC CTAACTCCGC 2161 CCATCCCGCC CCTAACTCCG CCCAGTTCCG CCCATTCTCC GCCCCATGGC TGACTAATTT 2221 TTTTTATTTA TGCAGAGGCC GAGGCCGCCT CTGCCTCTGA GCTATTCCAG AAGTAGTGAG2281 GAGGCTTTTT TGGAGGCCTA GGCTTTTGCA AAAAGCTCCC GGGAGCTTGT ATATCCATTT 2341 TCGGATCTGA TCAGCACGTG TTGACAATTA ATCATCGGCA TAGTATATCG GCATAGTATA 2401 ATACGACAAG GTGAGGAACT AAACCATGGC CAAGTTGACC AGTGCCGTTC CGGTGCTCAC 2461 CGCGCGCGAC GTCGCCGGAG CGGTCGAGTT CTGGACCGAC CGGCTCGGGT TCTCCCGGGA 2521 CTTCGTGGAG GACGACTTCG CCGGTGTGGT CCGGGACGAC GTGACCCTGT TCATCAGCGC 2581 GGTCCAGGAC CAGGTGGTGC CGGACAACAC CCTGGCCTGG GTGTGGGTGC GCGGCCTGGA 2641 CGAGCTGTAC GCCGAGTGGT CGGAGGTCGT GTCCACGAAC TTCCGGGACG CCTCCGGGCC 2701 GGCCATGACC GAGATCGGCG AGCAGCCGTG GGGGCGGGAG TTCGCCCTGC GCGACCCGGC 2761 CGGCAACTGC GTGCACTTCG TGGCCGAGGA GCAGGACTGA CACGTGCTAC GAGATTTCGA 2821 TTCCACCGCC GCCTTCTATG AAAGGTTGGG CTTCGGAATC GTTTTCCGGG ACGCCGGCTG 2881 GATGATCCTC CAGCGCGGGG ATCTCATGCT GGAGTTCTTC GCCCACCCCA ACTTGTTTAT 2941 TGCAGCTTAT AATGGTTACA AATAAAGCAA TAGCATCACA AATTTCACAA ATAAAGCATT 3001 TTTTTCACTG CATTCTAGTT GTGGTTTGTC CAAACTCATC AATGTATCTT ATCATGTCTG 3061 TATACCGTCG ACCTCTAGCT AGAGCTTGGC GTAATCATGG TCATAGCTGT TTCCTGTGTG 3121 AAATTGTTAT CCGCTCACAA TTCCACACAA CATACGAGCC GGAAGCATAA AGTGTAAAGC 3181 CTGGGGTGCC TAATGAGTGA GCTAACTCAC ATTAATTGCG TTGCGCTCAC TGCCCGCTTT 3241 CCAGTCGGGA AACCTGTCGT GCCAGCTGCA TTAATGAATC GGCCAACGCG CGGGGAGAGG 3301 CGGTTTGCGT ATTGGGCGCT CTTCCGCTTC CTCGCTCACT GACTCGCTGC GCTCGGTCGT 3361 TCGGCTGCGG CGAGCGGTAT CAGCTCACTC AAAGGCGGTA ATACGGTTAT CCACAGAATC 3421 AGGGGATAAC GCAGGAAAGA ACATGTGAGC AAAAGGCCAG CAAAAGGCCA GGAACCGTAA 3481 AAAGGCCGCG TTGCTGGCGT TTTTCCATAG GCTCCGCCCC CCTGACGAGC ATCACAAAAA 3541 TCGACGCTCA AGTCAGAGGT GGCGAAACCC GACAGGACTA TAAAGATACC AGGCGTTTCC 3601 CCCTGGAAGC TCCCTCGTGC GCTCTCCTGT TCCGACCCTG CCGCTTACCG GATACCTGTC 3661 CGCCTTTCTC CCTTCGGGAA GCGTGGCGCT TTCTCAATGC TCACGCTGTA GGTATCTCAG 3721 TTCGGTGTAG GTCGTTCGCT CCAAGCTGGG CTGTGTGCAC GAACCCCCCG TTCAGCCCGA 3781 CCGCTGCGCC TTATCCGGTA ACTATCGTCT TGAGTCCAAC CCGGTAAGAC ACGACTTATC 3841 GCCACTGGCA GCAGCCACTG GTAACAGGAT TAGCAGAGCG AGGTATGTAG GCGGTGCTAC 3901 AGAGTTCTTG AAGTGGTGGC CTAACTACGG CTACACTAGA AGGACAGTAT TTGGTATCTG 3961 CGCTCTGCTG AAGCCAGTTA CCTTCGGAAA AAGAGTTGGT AGCTCTTGAT CCGGCAAACA 4021 AACCACCGCT GGTAGCGGTG GTTTTTTTGT TTGCAAGCAG CAGATTACGC GCAGAAAAAA 4081 AGGATCTCAA GAAGATCCTT TGATCTTTTC TACGGGGTCT GACGCTCAGT GGAACGAAAA 4141 CTCACGTTAA GGGATTTTGG TCATGAGATT ATCAAAAAGG ATCTTCACCT AGATCCTTTT 4201 AAATTAAAAA TGAAGTTTTA AATCAATCTA AAGTATATAT GAGTAAACTT GGTCTGACAG 4261 TTACCAATGC TTAATCAGTG AGGCACCTAT CTCAGCGATC TGTCTATTTC GTTCATCCAT 4321 AGTTGCCTGA CTCCCCGTCG TGTAGATAAC TACGATACGG GAGGGCTTAC CATCTGGCCC 4381 CAGTGCTGCA ATGATACCGC GAGACCCACG CTCACCGGCT CCAGATTTAT CAGCAATAAA 4441 CCAGCCAGCC GGAAGGGCCG AGCGCAGAAG TGGTCCTGCA ACTTTATCCG CCTCCATCCA 4501 GTCTATTAAT TGTTGCCGGG AAGCTAGAGT AAGTAGTTCG CCAGTTAATA GTTTGCGCAA 4561 CGTTGTTGCC ATTGCTACAG GCATCGTGGT GTCACGCTCG TCGTTTGGTA TGGCTTCATT 4621 CAGCTCCGGT TCCCAACGAT CAAGGCGAGT TACATGATCC CCCATGTTGT GCAAAAAAGC 4681 GGTTAGCTCC TTCGGTCCTC CGATCGTTGT CAGAAGTAAG TTGGCCGCAG TGTTATCACT 4741 CATGGTTATG GCAGCACTGC ATAATTCTCT TACTGTCATG CCATCCGTAA GATGCTTTTC 4801 TGTGACTGGT GAGTACTCAA CCAAGTCATT CTGAGAATAG TGTATGCGGC GACCGAGTTG 4861 CTCTTGCCCG GCGTCAATAC GGGATAATAC CGCGCCACAT AGCAGAACTT TAAAAGTGCT4921 CATCATTGGA AAACGTTCTT CGGGGCGAAA ACTCTCAAGG ATCTTACCGC TGTTGAGATC4981 CAGTTCGATG TAACCCACTC GTGCACCCAA CTGATCTTCA GCATCTTTTA CTTTCACCAG5041 CGTTTCTGGG TGAGCAAAAA CAGGAAGGCA AAATGCCGCA AAAAAGGGAA TAAGGGCGAC5101 ACGGAAATGT TGAATACTCA TACTCTTCCT TTTTCAATAT TATTGAAGCA TTTATCAGGG5161 TTATTGTCTC ATGAGCGGAT ACATATTTGA ATGTATTTAG AAAAATAAAC AAATAGGGGT5221 TCCGCGCACA TTTCCCCGAA AAGTGCCACC TGACGTC//其他哺乳动物表达载体:pCHO1.0 pBApo-CMV-Pur pOPRSVIpcDNA3.1/His C pcDNA5/FRT/V5-His-TOPO pREP4pcDNA3.1/His B pcDNA5/FRT/TO-TOPO pDual-GCpcDNA3.1/His A pcDNA5/TO pBK-RSVpIRESpuro3 pcDNA5/FRT/TO pBK-CMVpIRES2-EGFP pcDNA5/FRT pBI-CMV4pTT5 pFLAG-CMV2 pcDNA4/TO/Myc-His/LacZ pNFkB-DD-tdTomato pcDNA3.1/CT-GFP-TOPO pOPI3CATpBI-CMV5 pcDNA3.1/NT-GFP-TOPO pGene/V5-His B pSEAP2-Basic pOptiVEC-TOPO pSwitchpSEAP2-Control pCMV-MEKK1 pCMVLacIpBI-CMV3 pCMV-MEK1 pVgRxRpBI-CMV2 pCMV-PKA pINDpBI-CMV1 pcDNA6.2/nTC-Tag-DEST pTRE3G-LucpNFκB-MetLuc2-Reporter pcDNA6.2/cTC-Tag-DEST pTRE3GpCRE-MetLuc2-Reporter pcDNA3.2/V5/GW/D-TOPO pTRE2-hygropAcGFP1-Actin pcDNA6.2/V5/GW/D-TOPO pTRE-TightpAcGFP1-N In-Fusion Ready pcDNA6.2/nGeneBLAzer-GW/D-TOPO pTK-hygpAcGFP1-C3 pcDNA6.2/C-YFP-DEST pTet-OnpAcGFP1-C pcDNA6.2/cGeneBLAzer-DEST pTet-OffpAcGFP1-p53 pcDNA6/V5-His A pTet on advanced pAcGFP1-Mito pcDNA6/V5-His B pRevTREpAcGFP1-Mem pcDNA6/V5-His C pRevTet-OnpAcGFP1-Lam pcDNA6/myc-His C pRevTet-OffpAcGFP1-Golgi pcDNA6/myc-His A pCMV-Tet3GpAcGFP1-F pcDNA6/myc-His B pTRE2pAcGFP1-Hyg-C1 pcDNA6.2/nGeneBLAzer-DEST pBD-NF-κBpAsRed2-N1 pcDNA4/HisMax-TOPO pCMV-ADptdTomato-N1 pcDNA6.2/nLumio-DEST pCMV-BDpCMV-tdTomato pcDNA6.2/cLumio-DEST pBIND-Id ControlpCRE-DD-tdTomato pcDNA4/myc-His C pBINDpCMV-DsRed-Express2 pcDNA4/HisMax C pG5 luciferasepEF1α-tdTomato pcDNA4/HisMax A pACT-MyoDpCRE-hrGFP c-Flag pcDNA3 pACTptdTomato-C1 pcDNA4/HisMax B pCMV-SPORT6pAsRed2-C1 pcDNA4/myc-His B pGL4.13pGL3-Promoter pcDNA4/myc-His A pGL4.19pGL3 basic pcDNA4/His C pGL4.26pAcGFP1-C2 pcDNA4/His B pGL4.20pAcGFP1-C1 pcDNA4/His A pGL4.29pAcGFP1-N3 pcDNA6/TR pGL4.30pAcGFP1-N2 pcDNA4/TO/Myc-His A pGL4.27pAcGFP1-N1 pcDNA4/TO pGL4.75pAcGFP1-C In-Fusion Ready pcDNA4/TO/Myc-His B pGL4.10pCRE-DD-AmCyan1 pcDNA4/TO/Myc-His C pGRN145pNFkB-DD-AmCyan1 pcDNA3.3-TOPO pSecTag2 A pDsRed2-Bid pBudCE4.1 pEBVHis B pDsRED2-Mito pFLAG-CMV-4 pEBVHis ApDD-AmCyan1 Reporter pFLAG-CMV-3 pCMV-Tag 3C pAmCyan1-N1 pFLAG-CMV-2 pCMV-Tag 3A pAmCyan1-C1 pFLAG-CMV-5a pCMV-Tag 3BpEF1α-IRES-DsRed-Express2 p3XFLAG-CMV-9 pCMV-Tag 5CpEF1α-DsRed-Monomer-N1 p3xFLAG-CMV-10 pCMV-Tag 5A pDsRED-Monomer-N1 p3XFLAG-CMV-8 pCMV-Tag 4A pDsRed-Express-N1 p3XFLAG-CMV-7.1 pCMV-Tag 5Bp3XFLAG-CMV-7 pDsRed-Monomer-N In-Fusion Ready pCMV-Tag 4B pDsRed-Express-C1 p3XFLAG-CMV-13 pCMV-Tag 2C pIRES2-ZsGreen1 p3XFLAG-CMV-14 pCMV-Tag 2B pDsRed-Express2-C1 plRES2-ZsGreen1 pCMV-Tag 2A pDsRed-Express2-N1 pBApo-EF1α-pur pCMV-LacZpEF1α-DsRed-Express2 pBApo-EF1α-neo pCMV-MycpIRES2-DsRed-Express pBApo-CMV pEF1α-IRES-AcGFP1 pIRES2-DsRed-Express2 pBApo-CMV-neo pEF1α-IRES-ZsGreen1 pIRES-hrGFP-1a pIRES-EGFP pEF1α-AcGFP1-N1 pIRESneo2 pIRESneo3 pIRES2-DsRed2 pIRESneo pDsRed-Monomer pIRES2-AcGFP1 pIREShyg3 pIRES。
