MethylEasy_ Xceed Rapid DNA Bisulphite Modification Kit
甲基胞苷制备方法

甲基胞苷(Methylcytidine)是一种含有甲基化的胞嘧啶核苷。
下面是一种常见的甲基胞苷制备方法:
材料:
●胞苷(Cytidine)
●甲基化试剂(如碘甲烷或N,N-二甲基乙酰胺甲磺酸盐)
步骤:
1.准备反应溶液:将适量的胞苷溶解在无水的有机溶剂中,如二甲基亚砜(DMSO)或二
氯甲烷。
2.添加甲基化试剂:向反应溶液中加入适量的甲基化试剂,如碘甲烷或N,N-二甲基乙酰
胺甲磺酸盐。
通常,使用过量的甲基化试剂可以增加反应的效率。
3.反应条件:根据实验室条件和所用试剂的选择,可以在室温或低温下进行反应。
反应时
间可能需要几小时到数天不等。
4.反应结束后,通过适当的提取和纯化方法(如萃取、结晶或柱层析)分离并纯化产物(甲
基胞苷)。
需要注意的是,以上方法仅提供了一种常见的甲基胞苷制备方法,具体的实验条件和步骤可能因实验室条件、试剂选择和研究目的而有所不同。
在进行化学实验时,请遵循相关安全操作规程,并确保在合适的实验条件下进行。
分子生物学实验室常用有毒药品.

分子生物学实验室有毒常用药品1. 溴化乙锭(EB :具有强诱变致癌性,使用时一定要戴一次性手套,注意操作规范,不要随便触摸别的物品。
2.DEPC(焦碳酸二乙酯 :闻起来香香甜甜的 , 可是害人不眨眼 ! 一种强有力的蛋白质变性剂 , 而且怀疑是致癌剂 . 开瓶时将瓶子远离你 , 内压可导致溅泼 . 操作时戴合适的手套 , 穿工作服 , 并在化学通风橱里进行 .3.PMSF(苯甲基磺酰氟 :老板说是神经毒 !!! 是一种高强度毒性的胆碱酯酶抑制剂 . 它对呼吸道黏膜 , 眼睛和皮肤有非常大的破坏性 . 可因吸入 , 咽下或皮肤吸收而致命 . 戴合适的手套和安全眼镜 , 始终在化学通风橱里使用 . 在接触到的情况下 , 要立即用大量的水冲洗眼镜或皮肤 , 已污染的工作服丢弃掉 .4. 乙腈,易挥发易燃,是一种刺激物和化学窒息剂,通风橱中远离热、火。
5. 放线菌素 D ,是一种致畸剂和致癌剂,通风橱中操作。
6.alpha-鹅膏蕈毒环肽,具有强毒性,可能致命。
7.NN-亚甲双丙烯酰胺,有毒,影响中枢神经系统,切勿吸入粉末。
8. 甲醇,有毒,能引起失明。
9. 乙酸 (浓的 :可能因为吸入或皮肤吸收而受到伤害, 要戴手套和护目镜, 最好在化学通风橱中操作。
10. 过硫酸酸铵:对粘膜和上呼吸道、眼睛和皮肤又较大危害性,吸入可致命。
操作时戴手套、护目镜。
始终在通风橱中操作。
11. 氯化铯:可因吸入、咽下或皮肤吸收而危害健康。
操作时戴手套和护目镜。
12.DTT: 很强的还原剂, 散发难闻的气味。
可因吸入、咽下或皮肤吸收而危害健康。
当使用固体或高浓度储存液时,戴手套和护目镜,在通风橱中操作。
13. 甲醛:毒性较大且易挥发, 也是一种致癌剂, 易通过皮肤吸收, 对眼睛、粘膜和上呼吸道有刺激和损伤作用。
避免一如其挥发的气雾。
戴手套和护目镜。
始终在通风橱中操作。
远离热、火花及明火。
14.TRIzol , 对眼睛有刺激性, 腐蚀皮肤。
DNA甲基化原理
DNA甲基化甲基化检测服务-亚硫酸氢钠处理后测序法(bisulfite genomic sequencing PCR, BSP)是利用未甲基化的胞嘧啶可以被亚硫酸氢钠发生脱氨基变为尿嘧啶的原理,用两一特异性引物扩增后测序。
测序法克服了只能针对单个位点检测,并且这些位点必须是限制性内切酶识别位点的缺点,可以对任何基因序列的甲基化状态进行检测。
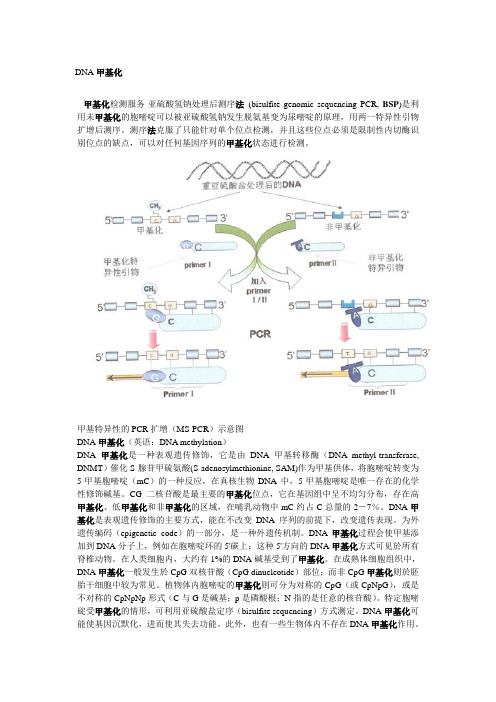
甲基特异性的PCR扩增(MS-PCR)示意图DNA甲基化(英语:DNA methylation)DNA甲基化是一种表观遗传修饰,它是由DNA甲基转移酶(DNA methyl-transferase, DNMT)催化S-腺苷甲硫氨酸(S-adenosylmethionine, SAM)作为甲基供体,将胞嘧啶转变为5-甲基胞嘧啶(mC)的一种反应,在真核生物DNA中,5-甲基胞嘧啶是唯一存在的化学性修饰碱基。
CG二核苷酸是最主要的甲基化位点,它在基因组中呈不均匀分布,存在高甲基化、低甲基化和非甲基化的区域,在哺乳动物中mC约占C总量的2-7%。
DNA甲基化是表观遗传修饰的主要方式,能在不改变DNA序列的前提下,改变遗传表现。
为外遗传编码(epigenetic code)的一部分,是一种外遗传机制。
DNA甲基化过程会使甲基添加到DNA分子上,例如在胞嘧啶环的5'碳上:这种5'方向的DNA甲基化方式可见於所有脊椎动物。
在人类细胞内,大约有1%的DNA碱基受到了甲基化。
在成熟体细胞组织中,DNA甲基化一般发生於CpG双核苷酸(CpG dinucleotide)部位;而非CpG甲基化则於胚胎干细胞中较为常见。
植物体内胞嘧啶的甲基化则可分为对称的CpG(或CpNpG),或是不对称的CpNpNp形式(C与G是碱基;p是磷酸根;N指的是任意的核苷酸)。
特定胞嘧碇受甲基化的情形,可利用亚硫酸盐定序(bisulfite sequencing)方式测定。
DNA甲基化可能使基因沉默化,进而使其失去功能。
Bisulfite Sequencing

Bisulfite Sequencing1. General InformationAs the first discovered mark, DNA methylation plays a vital role in genome dynamics. It is implicated in repression of transcriptional activity and in animals it predominantly involves the addition of a methyl group to the carbon-5 position of residues at cytosine guanine dinucleotide (CpG) sites.Bisulfite sequencing is a powerful technique to study DNA methylation. Treatment of DNA with bisulfite converts cytosine residues to uracil, but leaves 5-methylcytosine residues unaffected. The following PCR amplification treatment converts uracil to thymine. Thus, bisulfite treatment introduces specific changes in the DNA sequence that depends on the methylation status of individual cytosine residues. Coupled with new-generation sequencing technology, it allows for an unbiased genome-wide analysis of DNA methylation and various analyses can be performed on the altered sequence. Bisulfite-seq is the golden standard for DNA methylation analysis. Process includes: treating of DNA with bisulfite to convert cytosine residues to uracil, while leaving 5-methylcytosine residues unaffected; running PCR to convert all the uracil to thymines; in the end, sequencing the PCR product and performingthe bioinformatics analysis compared with the untreated genome to profile quantitive regional methylation pattern.Compared to Sanger sequencing, Bisulfite sequencing is a low-cost method with high reliability and accuracy to determine each cytosine methylation state. Based on new-generation sequencing technology, it avoids mass work of clone sequencing and complicated process. The sequencing primers which will be added to the DNA fragments to process sequencing also can be treated as random amplication primer for DNA samples. Thus, primer designing is not necessary and work of PCR will be decreased when compared to Sanger sequencing to profile genome-wide DNA methylation pattern.2. Experimental PipelineBisulfite treatment of sample DNA and DNA sequencingPerform the bisulfite treatment of qualified DNA and forward to TA clone test. Then the DNA sequencing is carried out using on the new-generation sequencing technology.DNA was extracted and processed by bisulfite sodium and subjected to Illumina GA sequencing with methylated adaptor. The detailed pipeline is showed as following (Figure 6-2-1):3. Bioinformatics AnalysisThe bioinformatics analysis of Bisulfite sequencing is based on the SOAP alignment with C T mismatches tolerated. Ideally, this method would determine the methylation status separately for each allele, even each single strand. The methylation status for each allele is the most important information to detect differentially methylated region as the candidate of imprinting gene. The methylation status for each single strand could be used to describe thehemi-methylation (Figure 6-2-2) .3.1 Basic bioinformatics analysisData production statistics includes image recognition, base calling, filtering adapter sequences and detecting contaminants of sample.3.2 Advanced bioinformatics analysisMap bisulfite-seq reads to referenceGet the methylation profile of the mapped reads.Gather the methylation information of each base of the mapped reads.Get the methylation rate information of all methylated C in CpG of each chromosome.Get the methylation rate information of all dispersed C of each chromosome. Provide the methylated CpG information in different gene regions.Provide the methylated dispersed C information in different gene regions. Provide the methylated CpG information of genome sequence with different features.Provide the methylated dispersed C information of genome sequence with different features.4. Case Studies4.1 Study on an individual human genomeThe match information of all the base sites is gathered and decoded after SOAP alignment. At each base site, tags number which suggests this site to be methylated or not will be given. In the following analysis, the C bases with copy number > 1.5 and quality < 14 are filtered out. The methylated rate of CpG or non-CpG is based on tags number which supports this site to be methylated comparing with and number of all effective tags.5. Frequently Asked Questions1) How many nucleotides are required for Bisulfite sequencing analysis?In order to ensure at least 10X coverage of genome size,we suggest 10 g of DNA to be provided as the mininum amount required.2) How to ensure all the unmethylated C bases to be converted to T bases in the bisulfite treatment of DNA?Our experimental pipeline normally ensures the conversion rate of unmethylated C to T to meet the bioinformatics analysis requirement. Here we used standard DNA and H19 for quality control.3) How to classify the methylation level?We can classify the methylation level through the proportion of total C and total T sequenced in a certain region. For example, if we get 6C and 4T (which in the reference sequencing is C), the methylation level in this region is considered as 6/ (4+6)= 60%.4) Which factors will influence the result of bisulfite-seq?From the scientific view, uncertain factors of DNA methylation research are mainly from undiscovered area, such as methylation difference of individual cell from cell lines, time differences and dynamic changes of methylation during developmental process or pathological process; from the technical view, the length of sample DNA, the conversion rate of DNA during bisulfate treatment and sequencing depth may influence the result of bisulfate-seq.目前, 基因的甲基化研究主要结合亚硫酸氢钠处理和PCR技术,分为甲基化特异性PCR(Methylation specific PCR,MSP)和硫化测序PCR(Bisulfite sequencing PCR, BSP)。
DNA重亚硫酸盐转化
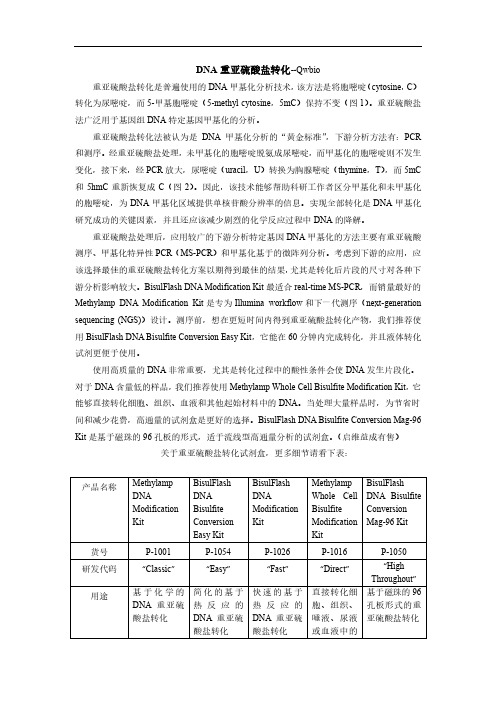
起始材料 最低 DNA 用 量 转化所需总 时间 洗脱体积 转化效率 脱磺酸基作 用/清除 回收率 转化后 DNA 片段尺寸 储存管
> 75% 平 均 : 200-2000 bp 峰值 800 bp 离心管
> 75% 平 均 : 200-2000 bp 峰值 800 bp 离心管
> 75% 平 均 : 100-400 bp 峰值 250 bp 离心管
产品名称methylampdnamodificationkitbisulflashdnabisulfiteconversioneasykitbisulflashdnamodificationkitmethylampwholecellbisulfitemodificationkitbisulflashdnabisulfiteconversionmag96kitp1001p1054p1026p1016p1050研发代码classiceasyfastdirecthighthroughout用途基于化学的dna酸盐转化简化的基于酸盐转化快速的基于酸盐转化直接转化细胞组织唾液尿液或血液中的基于磁珠的96孔板形式的重亚硫酸盐转化dna产品关键词illumina作流程兼容最稳定可靠重亚硫酸盐测序ngs各种msp理想选择illumina作流程兼容热循环变性液体重亚硫酸盐转化试反应快速realtimemsp的理想选择测序最佳选dna分离的步骤dna含量低样品的理想选择illumina工作流程兼容流线型高通量分析专为大量样品或者自动转化设计起始材料dnadnadna细胞组织血液dna最低dna用50pg100pg200pg100个细胞10ng转化所需总时间hour30minuteshour20minutes洗脱体积820转化效率999999999999999脱磺酸基作用清除85转化后dna片段尺寸2002000bp峰值800bp2002000bp峰值800bp100400bp峰值250bp2002000bp峰值800bp2002000bp峰值800bp储存管离心管离心管离心管离心管离心管重亚硫酸盐处理将胞嘧啶c转化为尿嘧啶u5甲基胞嘧啶5mc保持不变
乙酰甲胺磷的合成

乙酰甲胺磷的合成摘要:乙酰甲胺磷(英文通用名:acephate,化学名称:0,S-二甲基一乙酰基一硫代磷酰胺酯)是有机磷农药中的老品种,是一种高效、低毒、低残留、广谱性有机磷杀虫剂。
它是甲胺磷的乙酰化衍生物。
1964年由拜耳公司首先合成。
1972年美国ChevronChemical Co.正式商品化。
而我国在山西省化肥农药研究所与抚顺农药厂协作下,经过近两年的努力,合成了一种新有机磷杀虫剂乙酞甲胺磷,并于于1975年11月份在太原进行了技术鉴定。
一直到2012年由于乙酰甲胺磷易造成甲胺磷残留,才开始被限制使用。
概述:乙酰甲胺磷是有机磷农药中使用历史悠久的杀虫剂,其特点是乙酰甲胺磷为内吸杀虫剂,具有胃毒和触杀作用,并可杀卵,有一定的熏蒸作用,是缓效型杀虫剂,其基本杀虫原理是抑制昆虫乙酰胆碱酯酶。
适用于蔬菜、烟草、果树、棉花、柑橘、水稻、小麦,防治多种咀嚼式、刺吸式口器害虫和害螨。
如果与西维因、乐果等农药混用,有增效作用并可处长持效期。
适用范围适用于蔬菜、茶树、烟草、果树、棉花、水稻、小麦、油菜等作物,防治多种咀嚼式、刺吸式口器害虫和害螨及卫生害虫。
一、乙酰甲胺磷的产品性能及合成1.理化性质:纯品为白色结晶,熔点为91℃。
工业品为白色固体,纯度80-90%,比重1.35,易熔于水、甲醇、乙醇、丙酮等极性溶剂和二氯甲烷、二氯乙烷等卤代烃类。
在苯、甲基苯环与二甲基苯环的混合溶液中溶解度较小。
在碱性介质中极易分解。
英文通用名称acephate 分子式:C4H10NO3PS CAS号:30560-19-1。
分子量:183.1659,其结构式为2.分析方法:可用紫外分光光度计分析,将获得的色谱图与标准色谱图进行对比,波峰数目与波峰状态重合即可。
或者用气相色谱仪测定。
3.乙酰甲胺磷的合成:方法一:生产乙酰甲胺磷的原料有甲基氯化物、氨水、二氯乙烷、乙酐、硫酸二甲酯。
通过胺化、酰化、异构化等反应步骤而得。
1.胺化将甲基氯化物及相应比例量的二氯乙烷分别从高位计量槽加入胺化釜中,开启搅拌和冷冻盐水,当釜内温度降至15℃时,由氨水高位计量槽慢慢滴加氨水于釜中,釜中温度控制在35-40℃,保持35-40 min,滴加氨水完毕后,调节冷冻盐水,使釜内温度降至20-23℃。
磷酸三甲酯为甲基化试剂、氢氧化钙为碱的杂原子甲基化新体系

磷酸三甲酯为甲基化试剂、氢氧化钙为碱的杂原子甲基化新体系下载提示:该文档是本店铺精心编制而成的,希望大家下载后,能够帮助大家解决实际问题。
文档下载后可定制修改,请根据实际需要进行调整和使用,谢谢!本店铺为大家提供各种类型的实用资料,如教育随笔、日记赏析、句子摘抄、古诗大全、经典美文、话题作文、工作总结、词语解析、文案摘录、其他资料等等,想了解不同资料格式和写法,敬请关注!Download tips: This document is carefully compiled by this editor. I hope that after you download it, it can help you solve practical problems. The document can be customized and modified after downloading, please adjust and use it according to actual needs, thank you! In addition, this shop provides you with various types of practical materials, such as educational essays, diary appreciation, sentence excerpts, ancient poems, classic articles, topic composition, work summary, word parsing, copy excerpts, other materials and so on, want to know different data formats and writing methods, please pay attention!探索新型杂原子甲基化体系:磷酸三甲酯与氢氧化钙的结合摘要:在化学合成领域,寻求新型反应体系是一项不断推进的任务。
TaKaRa EpiTaq HS (for bisulfite-treated DNA) 说明书

As template DNA was damaged by bisulfite treatment, amplification efficiency of larger products may be decrease.
VI. Electrophoresis of Amplification Products
Volume/Amount 0.25 μl 5 μl 5 μl 6 μl
< 100 ng 20 pmol 20 pmol to 50 μl
Final Con. 1.25 U / 50 μl
1X 2.5 mM 0.3 mM
0.4 μM 0.4 μM
For a first trial, use the reaction mixture described above. If this reaction mixture fails to result in desired levels of product amplification or results in the appearance of extra bands, adjusting the concentration of MgCl2, dNTP mixture, or primer may improve results. For details, see section X. Troubleshooting.
VII. PCR Products
The majority of PCR products amplified with TaKaRa EpiTaq HS have a single A nucleotide added at 3’ termini, allowing the PCR products to be cloned directly into T-vectors such as pMD20 (Cat. #3270) or pMD19 (Simple) (Cat. #3271).
bisulfite处理原理

bisulfite处理原理
Bisulfite处理原理
Bisulfite处理是一种强有效的用于鉴定DNA甲基化状态的技术。
它利用了甲基化DNA对硫酸盐的特异性敏感性,通过亲和甲基化DNA,将其脱甲基,使其变成被保护的单碱基扩增状态,从而识别出DNA甲基化位点。
Bisulfite处理步骤大致包括:
1)取样:选取DNA样本,把样本加入90%甲醇溶液中,把受试
细胞或组织放入比萨硫酸盐溶液中,并在比萨硫酸盐溶液中加入蛋白酶把DNA片段分解成单个的碱基对。
2)Digestion: 在碱性或中性条件下用碱性聚合酶(DNA Polymerase)对DNA片段进行扩增,形成双链DNA。
3)消除互补序列:把双链DNA片段放入专用的比萨硫酸盐溶液中,经过长时间(一般为一天)的反应,消除DNA片段中未甲基化的互补序列,使甲基化的DNA序列的保持不变。
4)甲基化检测:根据DNA片段的基因型,采用非比较方法或比
较方法,对DNA片段中的甲基化状态进行研究。
- 1 -。
甲磺司特的合成

甲磺司特的合成
简介
甲磺司特(Methanesulfonate)是一种经常用于药物合成的中间体,通常用于合成DNA和RNA的碱基修饰剂。
甲磺司特可以通过甲磺酸的氯化与碳酸二乙酯转化而来,它是一种稳定的、易于纯化和大规模生产的化合物。
原理
甲磺司特的全名为 3-甲基-2-乙氧羰基-1-苯基四氢-1,4-嘧啶并[3,4-c]咪唑-4-乙烯基甲磺酸甲酯。
其化学式为:C10H11NO6S。
甲磺司特的制备方法如下:
材料
•碳酸二乙酯(C5H10O4)
•氯化甲磺酸(ClCH2SO3H)
步骤
1.在室温下,将 30 g 的碳酸二乙酯慢慢加入 20mL 的氯化甲磺酸中,
并用搅拌器搅拌,反应开始就会出现白色沉淀。
2.持续搅拌,反应溶液会变成浑浊的乳白色来,继续搅拌至反应混合物
的溶液完全变成深黄色时即可。
3.冷却反应体系至室温后,用氯化甲磺酸稀释反应混合物,反应液中
甲磺司特便会逐渐析出。
应用
甲磺司特主要用于合成DNA和RNA的碱基修饰剂,具有广泛的应用价值。
总结
同时,甲磺司特的合成方法也是一个重要的有机合成问题,对于学习有机化学的同学而言,掌握甲磺司特的合成方法可以对有机化学实验技能的提高带来很大的帮助。
常用的核酸染料有哪些

EB(溴化乙锭):溴化乙锭是一种高度灵敏的嵌入性荧光染色剂,为强致癌诱变剂,但价格便宜。
它与DNA的结合几乎没有碱基序列特异性。
在高离子强度的饱和溶液中,大约每2.5个碱基插入一个溴化乙锭分子。
EB的这种特性使其成为一种核酸染料,常用于琼脂糖凝胶电泳中的核酸染色。
溴化乙锭在紫外区302nm和366nm处有吸收峰,在紫外光的照射下,溴化乙锭可被激发出橙红色的荧光(590nm)。
结合有DNA的溴化乙锭复合物的荧光强度要比没有结合DNA的染料高出20-30倍,因此溴化乙锭可检测到少至10ng的DNA条带,非常灵敏。
Gel Green和Gel Red:GelRed 和 GelGreen 是两种集高灵敏、低毒性和超稳定性于一身的极佳的荧光核酸凝胶染色试剂。
其特点有:高灵敏度:GelRed 和 GelGreen 是目前市场中最灵敏的凝胶核酸染料之一;稳定性极好:可以使用微波炉加热,可以在室温下保存;更安全:艾姆斯氏试验结果表明,该染料的诱变性远小于溴化乙锭(EB);广泛的适应性:适用于预制凝胶和凝胶电泳后染色;染色过程简单:与EB一样,在预制胶和电泳过程中不必担心染料降解;而电泳后染色过程也只需30分钟且无需脱色或冲洗;对 DNA 和 RNA 的迁移影响小:对核酸迁移的影响小于 SYBR Green 。
与标准凝胶成像系统以及可见光激发的凝胶观察装置完美兼容:使用 312nm 激发的 UV 凝胶成像系统时,GelRed 可以完美的替代EB;使用 254nm 激发的 UV 凝胶成像系统或可见光激发的凝胶观察装置时,GelGreen 足以替代任意一种 SYB染料。
但该染料会使小片段DNA迁移慢一些(与EB相比).Gold View I型/II型:Goldview对于大片段DNA的染色效果良好,但是在对于500bp以下的片段效果不是很好,还有一个很致命的弱点是它的荧光基团在紫外灯下非常容易淬灭,一般5-10min条带就会消失。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
More Sensitive Excellent DNA Preservation
Typical PCR results after MethylEasy™ Xceed conversion of human genomic DNA is shown below. The results demonstrate whole genome conversion via the amplification of 48 different genomic loci from just 50 ng of starting DNA. Unsurpassed sensitivity of MethylEasy™ Xceed is seen below in the amplification of 2 separate PCRs when starting with only 100 pg or 50 pg of mammalian DNA. Either 100 pg or 50 pg of DNA (16 or 8 mammalian cell equivalents respectively) was treated with MethylEasy™ Xceed as per the user manual and eluted in 12 µl. Nested PCR for the Lim2 gene was performed on 4 µl of converted DNA. The second round amplification was elecrophoresed on a 2% agarose gel. Lanes 1 and 2: 50 pg starting DNA; Lanes 3 and 4: 100 pg starting DNA; M: DNA marker.
MethylEasy Xceed Rapid DNA Bisulphite Modification Kit
™
Take a Leap Forward in Methylation Detection
1 2
100 pg of starting DNA
50 pg of starting DNA
Hale Waihona Puke “Loss-less” conversion technology. Lane 1: DNA (2µg) recovered and electrophoresed after using MethylEasy™ conversion technology Lane 2: (2µg) recovered and electrophoresed after using conventional bisulphite conversion.
Easy to Use
MethylEasy™ technology does not require any DNA pre-treatment (i.e restriction digestion) and the DNA is now purified via a simple column. The vastly reduced processing time is demonstrated below: A total of 50 ng of human DNA was converted as per the MethylEasy™ Xceed product manual. The DNA was resuspended in 120 µL and a total of 2 µL was seeded into 48 different PCRs. All amplifications were two rounds, and 10 µL of the second round product was electrophoresed on a 2% agarose gel. Original MethylEasy™
Denature DNA with NaOH and incubate for 15 minutes Add conversion reagents and incubate for 4-16 hours Precipitate and wash the DNA, 45 to 75 minutes Resuspend the DNA and desulphonate for 30 mins to 1 hour. The DNA sample is now fully converted – 6 to 18 hours total time
Conventional bisulphite PCR product The above sequence shows a portion of the Myog gene that was assayed for CpG methylation using either the MethylEasy™ Xceed kit (top panel) or the original MethylEasy™ kit. The sequence was generated using a reverse primer and hence any methylated cytosines are represented as guanines. The above data was generated during beta-testing in the laboratory of Prof Philippe Collas1.
Better DNA Conversion
The sequencing chromatograms below show full conversion in the DNA treated with MethylEasy™ technology, compared to many non-converted sites in the DNA converted with conventional methods (Arrows denote blocked sites.) MethylEasy™ PCR product Original MethylEasy™ (16 hour bisulphite incubation)
Exquisite Sensitivity: Either 100 pg or 50 pg of DNA (16 or 8 mammalian cell equivalents respectively) was treated with MethylEasy™ Xceed as per the user manual and eluted in 12 µl. Nested PCR for the Lim2 gene was performed on 4 µl of converted DNA. The second round was detected in real time on a Corbett Rotor-Gene 6000 using Syto-9 as the detection dye at a final concentration of 1 µM.
Consistent Results as Compared to Original MethylEasy™
The new MethylEasy™ Xceed method achieves the same results in 90 minutes that took 6-18 hours with the original MethylEasy™ method. The figure below compares the sequence data of PCR fragments amplified from DNA that had been treated with either the MethylEasy™ Xceed or Original MethylEasy™ protocols. MethylEasy™ Xceed (45 minute bisulphite incubation)
“The rapid bisulfite conversion time with MethylEasy™ Xceed greatly facilitates and accelerates our research on DNA methylation in stem cells”. Prof. Philippe Collas
*1992, Frommer et al., Proc. Nat. Acad. A, 89,1827-1831.
MethylEasy™ Xceed DNA Bisulphite Modification Kit
Conventional bisulphite treatments result in the loss of between 84% and 96% of the starting DNA2, require pre-treatment with restriction endonuclease digests, the embedding of the DNA in agarose, or multiple tube changes and precipitations. MethylEasy™ Xceed addresses all of these shortcomings and is a simple, sensitive and quick modification method that requires no DNA pre-treatment and as little as 50 pg of starting material. MethylEasy™ “loss-less” sodium bisulphite technology results in virtually no loss of DNA, improved sensitivity, higher amplification efficiency, longer fragment generation and increased stability of the template DNA.
