ImageJ Western 量化 拿走不谢
ImageJ分析western步骤

ImageJ分析western步骤
点击file →ope n→任意一张图
点击再点击,左右鼠标键可改变图片大小,以便分析,图片大小不影响数据。
点击再点倒数第二项,减去背景,保存原设置值。
点击再点invert
点击再点 set measurements,勾选如下
Ok确定
Mean Gray Value:平灰度值
Integrated Density:总灰度值
点击,在区域内可改变大小,最好刚刚圈住每个印记,
不改变方框大小,移到旁边黑色区域,计算背景面积
点击,measure
再把方框移到每个组别,仍点击
所有值将会依次出现,
如果中间改变方框大小,那么必须再重新将方框移到黑色区域,计算背景面积。
所有印记的真实值=系统出现值-同样方框下的背景值。
用imagej进行WB定量分析

ImagJ是一款简单的图像处理与分析软件,可以用来进行WB定量分析。
一、利用ImagJ对WB条带进行灰度分析1、File——》open 打开WB片子2、把图片转化成灰度图片image——》type——》8-bit3、消除背景影响process——》subtract background 选择50基本可以4、设置定量参数analyze——》set measurements,点击面积,平均密度和灰度值及Integrated Density 5、设置单位analyze——》set scale ,在“unit of length”的方框里输入“pixels”6、把图片转换成亮带,Edit——》invert7、选择FreehandSelection,尽量把条带圈起来,点击键盘m,出来IntDen灰度值当测定完所有条带,选结果中的“Edit ”的“SelectAll”,然后复制数据“IntDen”到Excel表即可进行分析8、复制数据IntDen进行分析二、利用ImagJ对WB条带进行密度分析1、File——》open 打开WB片子2、如条带不正,需修正image——》transform——》rotate调节angle值,直到条带水平为止3、选中矩形选项,圈中第一个条带,然后 analyze——》gels——》select firstlane(快捷键ctrl+1),然后移动第一个条带上的矩形到第二个条带上,analyze——》gels——》select secondlane(快捷键 ctrl+2),最后analyze——》gels——》plotlanes4、选中直线工具,将开口的波峰关闭5、选中魔棒工具,点击波峰可以显示波峰下面积,即条带的密度值6、以第一个数值为基数,其他数值与第一个数值的比值为相对密度The protocol of imageJ for quantitative analysis of Western blotting:- open an image- transfer the image to 8-bitsimage → type: 8 bits- Process → substract background 50- analyze → set measurements: pick area, mean gray value, inte grated density. - analyze → set scale, fill “unit of length” with “pixels”- switch the image to bright bandsedit → invert- freehand selection, choose the target area on the image . a band), type a “m” for measurement, then we get the result from a new window.- after measuring all of the bands, pick the “edit” of the result window. “select all”, copy the integrated density values to excel to analyze.The integrated density means that the area value multiplies the mean gray value. For Western blotting analysis the integrated density is very important, because the area and gray value both of them have meaning for Western blotting bands.。
image_j的中文使用方法-值得借鉴哈
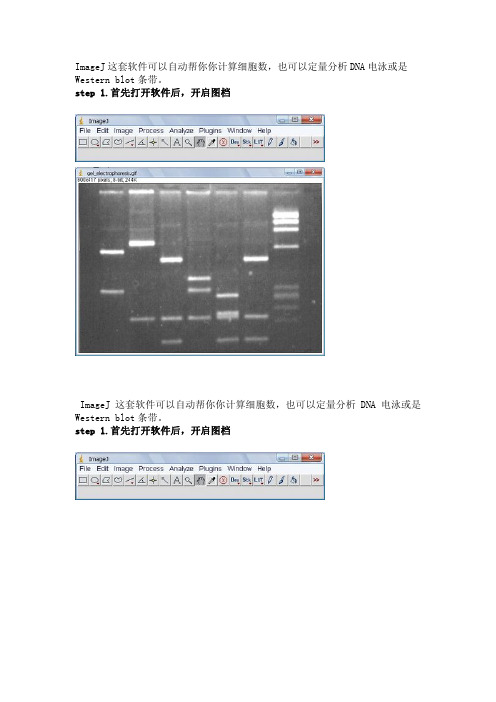
ImageJ这套软件可以自动帮你你计算细胞数,也可以定量分析DNA电泳或是Western blot条带。
step 1.首先打开软件后,开启图档ImageJ这套软件可以自动帮你你计算细胞数,也可以定量分析DNA电泳或是Western blot条带。
step 1.首先打开软件后,开启图档step 2.请先做校正,选择Analyze底下的Calibrate选项,再选择校正的模式,使用Uncalibrate OD,再按ok按下ok之后会出现校正的图形Step 3.在要分析的第一条(first lane)加上一个长型框(工具列第一个选项),再按下Analyze/Gels/select first Lane快速键(Ctr+1),此时框架中会出现一个号码1,之后可以移动框架到第二个lane再选择Analyze/Gels/select second Lane快速键(Ctr+2),当然可以一直加下去,最后按Analyze/Gels/plot Lanes快速键(Ctr +3)。
Step 4.分析以后会出现图型表示你刚选择的框内的影像强度,此时可以看到有几个比较高的区段,就是我们想定量的band,使用直线工具(工具列第五个选项)先将图形中高点为有band的区域和没有band的区域分开再,使用魔术棒工具(工具列第八个选项)点选要分析的区域。
Step 5.当我们点选分析时,在result的对话视窗会出现分析的数据,依序点选就会出现每个band的值。
注:当我们选择分析的条带也可以是横向选取,就可以只比较相同大小的DNA 的含量,同样也可以应用在western blot或其它类似实验条带的分析上。
使用ImageJ 分析图像中的颗粒数[] 原创教程,转载请保留此行1,到本站资料下载-实用小工具栏目下载 ImageJ 并安装。
2,打开ImageJ并打开要分析的图片。
请看演示图片。
3,把图像二值话或者设定阈值。
选择Image - Adjust - Threshold...根据提示设定你需要的阈值。
10分钟Get!大牛教你用imageJ对Westernblot条带进行灰度分析!
10分钟Get!大牛教你用imageJ对Westernblot条带进行
灰度分析!
科研人员做的western blot实验一般需要对其结果扫描后进行灰度分析,一般使用的软件为Image 。
采用Image J 软件对蛋白质印迹实验结果(Western Blot)进行灰度半定量分析,是分子生物学实验中比较常用的方法,可实现对WB实验结果进行准确、简单和快速的分析。
ImageJ分析Westernblot条带图
Step 1:打开Image J, 导入图片→Image→Type→8 bit
Step 2:选择工具框中第一个矩形框选项,选中所有的条带
Step 3:点击Analyze→Gels→Plot Lanes
Step 4:选用工具框中的直线工具,将开口波峰封闭
Step 5:选用工具框中的魔棒工具,点击相应的波峰,相应area值
ImageJ分析Westernblot条带图
Step 1:打开Image J, 导入图片。
Image→Type→8 bit
Step 2:扣除背景P→rocess→Subtract Background→选择50 pixels和Lightbackground
Step 3:设定参数→Analyze→Set Measures→按照下图勾选参数
Step 4:设定参数→Analyze→Set scale→unit of length选项改为pixels
Step 5:图像分割→Edit→Invert→选用椭圆或自定义工具选中条带
Step 6:Analyze→Measure→得到 IntDen值Step 7:重复Step5、6,得到所有条带的IntDen值
以上实用干货来源于课程
30分钟精通Image J图片处理。
imagej教程

关于本手册- 请先阅读ImageJ是一个公共领域的Java图像处理程序由美国国立卫生研究院的Macintosh形象的启发。
它运行在任何一个Java 1.1或更高版本的虚拟机电脑,无论是作为一个applet或网上下载的应用程序作为一个。
笔者,韦恩Rasband(wayne@),是在研究科,国立心理健康,贝塞斯达,马里兰州,美国。
约ImageJ信息的最佳来源,可以发现在ImageJ网页(/ij/)和通过订阅ImageJ邮件列表(在主页上的细节)。
本手册的目的是要介绍给ImageJ光学显微镜- 一个ImageJ的剧目一小部分。
ImageJ的一种方式,一个“插件”现在越来越多的补充本地大量功能(可选需要额外安装)。
的核心职能进行了详细的描述在ImageJ网站(按照单证链接)。
一个插件是一个文件(名为*.类),需要在“插件”子文件夹ImageJ文件夹,否则ImageJ不会加载它。
在本手册中,本机的功能是指在斜体,黑白文本,由插件斜体,深蓝色文本。
ImageJ功能都可以通过键盘快捷键访问,或“热键”。
一些热键是硬到有线ImageJ,而另一些用户定义的。
与大多数其他Windows应用程序,键盘快捷方式不要求“控制”键被按下(在Microsoft Word中,热键“复制”,例如,是按Ctrl + C)后,在ImageJ,这就是“C”的。
除非你已经安装了从赖特细胞成像设备网站(手册ImageJ 和软件的链接),插件本手册中提到的和定制的键盘快捷方式或多或少(“热键”)可能无法工作。
这也仅仅是被插件收集整理和我组织,他们都已经获得摆脱ImageJ网站或其他地方上网。
这项工作的信贷应该去作者的插件(见附件)。
请确保他们得到适当的承认,在任何出版,他们的工作便利。
我试图包括适当引用或联系方式每位作者。
让我知道任何遗漏,我会立即改正。
如果你是一个插件作者,你是不是你的插件的方式描述或包含或快乐引,请让我知道。
我会在必要时更新此手册。
使用ImageJ分析Western
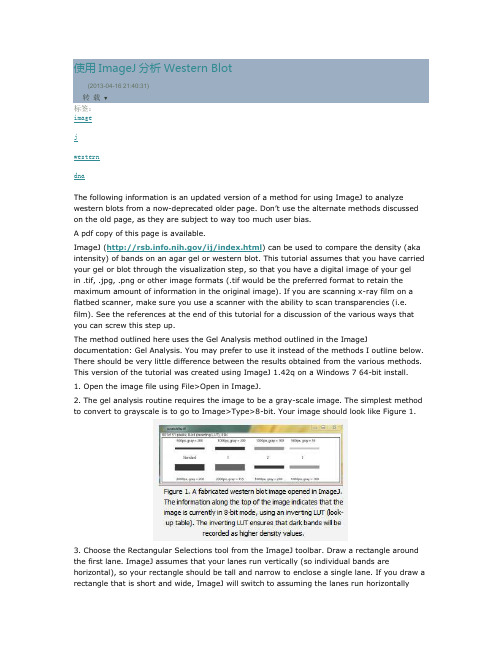
使用ImageJ 分析Western Blot (2013-04-16 21:40:31)转载▼标签: imagejwesterndnaThe following information is an updated version of a method for using ImageJ to analyze western blots from a now-deprecated older page. Don’t use the alternate methods discussed on the old page, as they are subject to way too much user bias.A pdf copy of this page is available.ImageJ (/ij/index.html ) can be used to compare the density (aka intensity) of bands on an agar gel or western blot. This tutorial assumes that you have carried your gel or blot through the visualization step, so that you have a digital image of your gel in .tif, .jpg, .png or other image formats (.tif would be the preferred format to retain the maximum amount of information in the original image). If you are scanning x-ray film on a flatbed scanner, make sure you use a scanner with the ability to scan transparencies (i.e. film). See the references at the end of this tutorial for a discussion of the various ways that you can screw this step up.The method outlined here uses the Gel Analysis method outlined in the ImageJdocumentation: Gel Analysis. You may prefer to use it instead of the methods I outline below. There should be very little difference between the results obtained from the various methods. This version of the tutorial was created using ImageJ 1.42q on a Windows 7 64-bit install.1. Open the image file using File>Open in ImageJ.2. The gel analysis routine requires the image to be a gray-scale image. The simplest method to convert to grayscale is to go to Image>Type>8-bit. Your image should look like Figure 1.3. Choose the Rectangular Selections tool from the ImageJ toolbar. Draw a rectangle around the first lane. ImageJ assumes that your lanes run vertically (so individual bands arehorizontal), so your rectangle should be tall and narrow to enclose a single lane. If you draw a rectangle that is short and wide, ImageJ will switch to assuming the lanes run horizontally(individual bands are vertical), leading to much confusion.4. After drawing the rectangle over your first lane, press the 1 key or go toAnalyze>Gels>Select First Lane to set the rectangle in place. The 1st lane will now be highlighted and have a 1 in the middle of it.5. Use your mouse to click and hold in the middle of the rectangle on the 1st lane and drag it over to the next lane. You can also use the arrow keys to move the rectangle, though this is slower. Center the rectangle over the lane left-to-right, but don’t worry about lining it up perfectly on the same vertical axis. Image-J will automatically align the rectangle on the same vertical axis as the 1st rectangle in the next step.6. Press 2 or go to Analyze>Gels>Select Next Lane to set the rectangle in place over the 2nd lane. A 2 will appear in the lane when the rectangle is placed.7. Repeat Steps 5 + 6 for each subsequent lane on the gel, pressing 2 each time to set the rectangle in place (Figure 3).8. After you have set the rectangle in place on the last lane (by pressing 2), press 3, or go to Analyze>Gels>Plot Lanes to draw a profile plot of each lane.9. The profile plot represents the relative density of the contents of the rectangle over each lane. The rectangles are arranged top to bottom on the profile plot. In the example western blot image, the peaks in the profile plot (Figure 4) correspond to the dark bands in the original image (Figure 3). Because there were four lanes selected, there are four sections in the profile plot. Higher peaks represent darker bands. Wider peaks represent bands that cover a wider size range on the original gel.10. Images of real gels or western blots will always have some background signal, so the peaks don’t reach down to the baseline of the profile plot. Figure 5 shows a peak from a real blot where there was some background noise, so the peak appears to float above the baseline of the profile plot. It will be necessary to close off the peak so that we can measure its size.11. Choose the Straight Line selection tool from the ImageJ toolbar (Figure 6). For each peak you want to analyze in the profile plot, draw a line across the base of the peak to enclose the peak (Figure 5). This step requires some subjective judgment on your part to decide where the peak ends and the background noise begins.12. Note that if you have many lanes highlighted, the later lanes will be hidden at the bottom of the profile plot window. To see these lanes, press and hold the space bar, and use the mouse to click and drag the profile plot upwards.13. When each peak has been closed off at the base with the Straight Line selection tool, select the Wand tool from the ImageJ toolbar (Figure 8).14. Using the spacebar and mouse, drag the profile plot back down until you are back at the first lane. With the Wand tool, click inside the peak (Figure 9). Repeat this for each peak as you go down the profile plot. For each peak that you highlight, measurements should pop up in the Results window that appears.15. When all of the peaks have been highlighted, go to Analyze>Gels>Label Peaks. This labels each peak with its size, expressed as a percentage of the total size of all of the highlighted peaks.16. The values from the Results window (Figure 10) can be moved to a spreadsheet program by selecting Edit>Copy All in the Results window. Paste the values into a spreadsheet.Note: If you accidentally click in the wrong place with the Wand, the program still records that clicked area as a peak, and it will factor into the total area used to calculate the percentage values. Obviously this will skew your results if you click in areas that aren’t peaks. If you do happen to click in the wrong place, simple go to Analyze>Gel>Label Peaks to plot the current results, which displays the incorrect values, but more importantly resets the counter for theResults window. Go back to the profile plot and begin clicking inside the peaks again, starting with the 1st peak of interest. The Results window should clear and begin showing your new values. When you’re sure you’ve click in all of the correct peaks without accidentally clicking in any wrong areas, you can go back to Analyze>Gels>Label Peaks and get the correct results. Data analysisWith your data pasted into a spreadsheet, you can now calculate the relative density of the peaks. As a reminder, the values calculated by ImageJ are essentially arbitrary numbers, they only have meaning within the context of the set of peaks that you selected on the single gel image you’ve been working on. They do not have units of μg of protein or any other real-world units that you can think of. The normal procedure is to express the density of the selected bands relative to some standard band that you also selected during this process.1. Place your data in a spreadsheet. One of the peaks should be your standard. In this example we’ll use the 1st peak as the standard.2. In a new column next to the Percent column, divide the Percent value for each sample by the Percent value for the standard (the 1st peak in this case, 26.666).3. The resulting column of values is a measure of the relative density of each peak, compared to the standard, which will obviously have a relative density of 1.4. In this example, the 2nd lane has a higher Relative Density (1.86), which corresponds well with the size and darkness of that band in the original image (Figure 1). Recall that these data are for the upper row of bands on the original western blot image.5. If you want to compare the density of samples on multiple gels or blots, you will need to use the same standard sample on every gel to provide a common reference when you calculate Relative Density values. See the sections below for more detailed discussion of these requirements.6. In order to test for significant differences between treatments in an experiment, all of your gels or blots will need to be scanned and quantified using this method, and the values will be expressed in terms of Relative Density, or you can treat Relative Density as a fold-change value (i.e. a Relative Density difference of 2 between a control and treatment wouldindicate a 2-fold change in expression). If you will be using analysis of variance techniques to test your data, you may need to ensure that your Relative Density values are normally distributed and that there is homogeneity of variance among the different treatments.7. It should be noted here that some researchers make the extra effort to include a set of serial dilutions of a known standard on each blot. Using the serial dilution curve and the quantification techniques outlined above, it should be possible to express your sample bands in terms of picograms or nanograms of protein.A more involved example using loading-controls.We’ll use Figure 12 as a representative western blot. On this blot, we will pretend that we loaded four replicate samples of protein (four pipette loads out of the same vial of homogenate), so we expect the densities in each lane to be equivalent. The upper row of bars will represent our protein of interest. The lower set of bars will represent our loading-control protein, which is meant to ensure that an equal amount of total protein was loaded in each lane. This loading-control protein is a protein that is presumably expressed at a constant level regardless of the treatment applied to the original organisms, such as actin (though many people will question the assertion that actin will be expressed equivalently across treatments).Looking at Figure 12, we had hoped to load equivalent amounts of total protein in each lane, but after running the western blot, the size and intensity of the lower bars in each lane varies quite a lot. The two left lanes appear equivalent, but the 3rd lane has half the density (gray value) compared to lanes 1+2, while lane 4 has half the density and half the size compared to lanes 1+2. Because our loading controls are so different, the density values of the upper set of bands may not be directly comparable.We’ll use ImageJ’s gel analysis routine to quantify the density and size of the blots, and use the results from our loading-controls (lower bands) to scale the values for our protein of interest (upper bands).1. Open the western blot image in ImageJ.2. Make sure that the image is in 8-bit mode: go to Image>Type>8-bit.3. Use the rectangle tool to draw a box around the entire 1st lane (both upper and lower bars included.4. Press “1″ to set the rectangle. A “1″ should appear in the middle of the rectangle.5. Click and hold in the middle of the rectangle and drag it over the 2nd lane.6. Press “2″ to set the rectangle for lane 2. A “2″ should appear in the middle of the rectangle.7. Repeat steps 5 + 6 for each subsequent lane, pressing “2″ to set the rectangle over each subsequent lane (see Figure 13).8. When you have placed the last lane (and pressed “2″ to set it in place), you can press “3″ to produce a plot of the selected lanes (see Figure 14).9. The profile plot essentially represents the average density value across a set of horizontal slices of each lane. Darker blots will have higher peaks, and blots that cover a larger size range (kD) will have wider peaks. In our example western blot, the bands are perfect rectangles, but you will notice some slope in the profile plot peaks, as ImageJ is applying a bit of averaging of density values as it moves from top to bottom of each lane. As a result, the sharp transition from perfect white to perfect black on the bands of lane 1 is translated into a slight slope on the profile plot due to the averaging.10. On our idealized western blot used here, there is no background noise, so the peak reaches all the way down to the baseline of the profile plot. In real western blots, there will be some background noise (the background will not be perfectly white), so the peaks won’t reach the baseline of the profile plot (see figure 5 above). As a result, each plot will need to have a line drawn across the base of the peak to close it off.11. Choose the Straight Line selection tool from the ImageJ toolbar. For each peak you want to analyze in the profile plot, draw a line across the base of the peak to enclose the peak (Figure 7). This step requires some subjective judgment on your part to decide where the peak ends and the background noise begins.12. When each peak of interest is closed off with the straight line tool, switch to the Wand tool. We will use the wand tool to highlight each peak of interest so that Image-J can calculate its relative area+density.13. We will start by highlighting the loading-control bands (lower row) on our example western blot. Beginning at the top of the profile plot, use the wand to click inside the 1st peak (Figure 15). The peak should be highlighted after you click on it. Continue clicking on the loading-control peaks for the other lanes. If a lane is not visible at the bottom of the profile plot, hold down the space bar and click-and-drag the profile plot upwards to reveal the remaining lanes.14. When the loading control peak for each lane has been highlighted with the wand, go to Analyze>Gel>Label Peaks. Each highlighted peak will be labeled with its relative size expressed as a percentage of the total area of all the highlighted peaks. You can go to the Results window and choose Edit>Copy All to copy the results for placement in a spreadsheet.15. Repeat steps 13 + 14 for the real sample peaks now. We are selecting these peaks separately from the loading-control peaks so that those areas are not factored into the calculation of the density of our proteins-of-interest. As before, use the Wand tool to click inside the area of the peak in the 1st lane, then continue clicking inside the peaks of the remaining lanes. When finished, go to Analyze>Gel>Label Peaks to show the results. Copy the results to a spreadsheet alongside the data for the loading-control bands (Figure 17).Data Analysis with loading-control bands1. With all of the relative density values now in the spreadsheet, we can calculate the relative amounts of protein on the western blot. Remember that the “Area” and “Percent” values returned by ImageJ are expressed as relative values, based only on the peaks that you highlighted on the gel. Start the analysis by calculating Relative Density values for each of the loading-standard bands. In this case, we’ll pretend that Lane 1 is our contr ol that we want to compare the other 3 lanes to. Divide the Percent value for each lane by the Percent value in the control (Lane 1 here) to get a set of density values that is relative to the amount of protein in Lane 1′s loading-control band (Figure 18).2. Next we’ll calculate the Relative Density values for our sample protein bands (upper row on the example western blot). We carry out a similar calculation as step 1, dividing the Percent value in each row by the Percent value of our control’s protein band (Lane 1 here).Note: Recall that because some of our loading-control bands were wildly different on the original western blot, we can’t simply use the Relative Density values from our Samples calculated in Step 2 as the final results. Now it is necessary to scale the Relative Density values for the Samples by the Relative Density of the corresponding loading-control bands for each lane. We do this based on the assumption that the proportional differences in the Relative Densities of the loading-control bands represent the proportional differences in amounts of total protein we loaded on the gel. In our example western blot, we have evidence of massively different amounts of total protein in each sample (poor pipetting practice, probably).3. The final step is to scale our Sample Relative Densities using the Relative Densities of the loading-controls. On the spreadsheet, divide the Sample Relative Density of each lane by the loading-control Relative Density for that same lane.9. The profile plot essentially represents the average density value across a set of horizontal slices of each lane. Darker blots will have higher peaks, and blots that cover a larger size range (kD) will have wider peaks. In our example western blot, the bands are perfect rectangles, but you will notice some slope in the profile plot peaks, as ImageJ is applying a bit of averaging of density values as it moves from top to bottom of each lane. As a result, the sharp transition from perfect white to perfect black on the bands of lane 1 is translated into a slight slope on the profile plot due to the averaging.10. On our idealized western blot used here, there is no background noise, so the peak reaches all the way down to the baseline of the profile plot. In real western blots, there will be some background noise (the background will not be perfectly white), so the peaks won’t reach the baseline of the profile plot (see figure 5 above). As a result, each plot will need to have a line drawn across the base of the peak to close it off.11. Choose the Straight Line selection tool from the ImageJ toolbar. For each peak you want to analyze in the profile plot, draw a line across the base of the peak to enclose the peak (Figure 7). This step requires some subjective judgment on your part to decide where the peak ends and the background noise begins.12. When each peak of interest is closed off with the straight line tool, switch to the Wand tool. We will use the wand tool to highlight each peak of interest so that Image-J can calculate its relative area+density.13. We will start by highlighting the loading-control bands (lower row) on our example western blot. Beginning at the top of the profile plot, use the wand to click inside the 1st peak (Figure 15). The peak should be highlighted after you click on it. Continue clicking on the loading-control peaks for the other lanes. If a lane is not visible at the bottom of the profile plot, hold down the space bar and click-and-drag the profile plot upwards to reveal theremaining lanes.14. When the loading control peak for each lane has been highlighted with the wand, go to Analyze>Gel>Label Peaks. Each highlighted peak will be labeled with its relative size expressed as a percentage of the total area of all the highlighted peaks. You can go to the Results window and choose Edit>Copy All to copy the results for placement in a spreadsheet.15. Repeat steps 13 + 14 for the real sample peaks now. We are selecting these peaks separately from the loading-control peaks so that those areas are not factored into the calculation of the density of our proteins-of-interest. As before, use the Wand tool to click inside the area of the peak in the 1st lane, then continue clicking inside the peaks of the remaining lanes. When finished, go to Analyze>Gel>Label Peaks to show the results. Copy the results to a spreadsheet alongside the data for the loading-control bands (Figure 17).。
干货▎ImageJ分析WesternBlot蛋白条带灰度值

干货▎ImageJ分析WesternBlot蛋白条带灰度值聊点学术,一键关注“你已经是个成熟的条带了,要学会自己分析!”“连Western blot都没学好,又让我学Image J,我......”This is the dividing line.Image J分析蛋白条带灰度值首先解释一下,为什么要分析灰度值?灰度的概念是使用黑色调表示物体,即用黑色为基准色,不同的饱和度的黑色来显示图像。
采用ECL发光时,蛋白条带发出的荧光会曝光在胶片上,留下的黑色条带。
然而胶片扫描后为蓝色背景、黑色条带,这并不是单纯的黑灰白颜色。
因此,我们首先得在Photoshop 中将整张图片去色,使彩色图片变成黑白图片,形成近灰色背景、黑色条带的图片类型。
此时,条带的深浅和面积综合代表着蛋白的量。
灰度值分析自然就成为我们的首选。
延伸说一下,免疫组织化学染色(IHC)结果本身为近白色背景、棕黄色阳性表达,这时我们就不能直接用灰度反映阳性表达强弱了,而是积分吸光度,也就是咱们常说的平均光密度(IOD)。
如果你真的想用灰度计算IHC结果,显然你需要将图片转为灰度图再计算。
此处不表。
分析图文步骤如下:Image J软件界面↓1. 图片转换为黑白图:首先使用Photoshop打开胶片扫描图片,点击“图像>调整>去色”,将彩色图片转化为黑白图片,并使用裁剪工具将目标条带裁剪至合适大小后另存图片。
2.打开Image J软件:2.1.打开图片文件:File>Open;将图片转化为8bit类型:Image>Type>8-bit。
(此步骤的目的是为了将图片的每个像素用8bit表示,这样的话,整个图片的灰度将分为256个级别,即黑、灰、白的像素模式;若保留原始的16bit格式或RGB格式,图片灰度级别会呈指数增长,并有其它颜色混入,造成计算误差。
)2.2.将整张图片背景灰度均一化,消除图片背景影响:Process>Subtract Background,默认数值为50即可。
ImageJ之对免疫组化图片定量评价和自动评分

ImageJ之对免疫组化图片定量评价和自动评分首先这款神器的插件在哪里可以下载?IHC Profiler插件可以在/projects/ihcprofiler/免费获取。
其次这款插件如何安装到自己的Imagj软件中?1、解压下载好的IHC_Profiler.zip文件,得到两个文件:IHC Profiler文件和IHC_Profiler.txt宏文件。
2、复制这两个文件粘贴至Plugins文件夹中:My Computer > C: > Program Files > ImageJ > plugins此外复制IHC_Profiler.txt文件粘贴至macros文件夹中:My Computer > C: > Program Files > ImageJ > macros重启软件即可,我们会在Imagej软件Plugins中发现已经安装好的IHC_Profiler插件。
如何使用该插件?对于胞浆着色的IHC图片,读入图片能够快速得到结果。
对于胞核着色的IHC图片:计算Score的公式为:Score有四个等级,分别为4/3/2/1,High Positve为4分,Positive为3分,Low Positive为2分,negative 为1分。
整个分析的流程如下:现在我们以实际例子来演示整个操作:1、打开Imagej软件,软件界面如下:File,Open,打开胞浆着色的图片:2、点击Plugins,点击IHC Profiler,进入以下界面:询问是否是胞浆着色,下拉可选择核着色,此处选择胞浆着色,点击OK,进入以下界面,选择H DAB点击OK。
得到结果为High Positive:而对于胞核着色,操作略微不同:1、打开Imagej软件,打开胞核着色图片:2、点击Plugins,点击IHC Profiler,进入以下界面,选择胞核着色:同上选择H DAB,点击OK进入以下界面:调节Threshold阈值选中所有核着色,点击Plugins,选择IHC Profiler Macro,得到结果:这个用于临床或者科研组织病理学分析的新工具可以在全球范围内用于评估大多数定位于细胞质或者细胞核的蛋白质,这种方法可以尽量减少不同实验室之间观察者之间差异的问题,为进一步为跨国间的临床试验提供了有效的保证。
你真的用ImageJ测到了WB条带灰度值吗?

你真的用ImageJ测到了WB条带灰度值吗?关于用Image J量化westernblot条带灰度值,在网上一直盛传着两种操作方法,然鹅~小伙伴们却一直不确定到底哪种才是正确的测量灰度方法,甚至将灰度与光密度弄混淆。
今天咱们一起就这个机会学习下,请先看看你是用以下那种方法测量灰度值?以下是两种方法的具体操作。
方法一1、打开Image J软件→左上角file→Open 选择自己的条带图片(事先把条带摆正)2、把图片转化成灰度图片:Image→type→8-bit3、第一个矩形工具→选上所有条带4、analyze→Gels→select first lane→Gels→plot lanes(在这第四步也可以分开选取,如:框选第一个条带→analyze→Gels→select firstlane→将第一个框拖移到余下条带→Gels→select nextlane)5、选中直线工具,将开口波峰关闭6、选中魔棒工具(正数第七个),点击波峰,就得出所有area 值。
方法二1、打开Image J软件→左上角file→Open 选择自己的条带图片(事先把条带弄正)2、把图片转化成灰度图片:Image→type→8-bit3、扣除背景:Process→Subtract Background→50pixels 并勾选Light Background5、设定参数:Analyze→set measurement→勾选Area、Mean gray value、Min & max gray value、Integrated density6、Analyze→set scale→unit of length选项里改为pixels,确定6、图像分割。
Edit→Invert→选中椭圆圈(手残党首选)/不规则圆圈(心灵手巧党义无反顾),手动圈上单个条带7、Analyze→Measurement即得到Intden,重复6、7步就得到所有Intden值小伙伴们,你们是哪种方法呢?首先先把结论说出来,其实两者得出的area和Intden严格来说都不是灰度值,前者是面积而后者是光密度值,但用来量化WB条带都是OK的。
灰度分析和细胞计数神器:ImageJ(附软件下载)
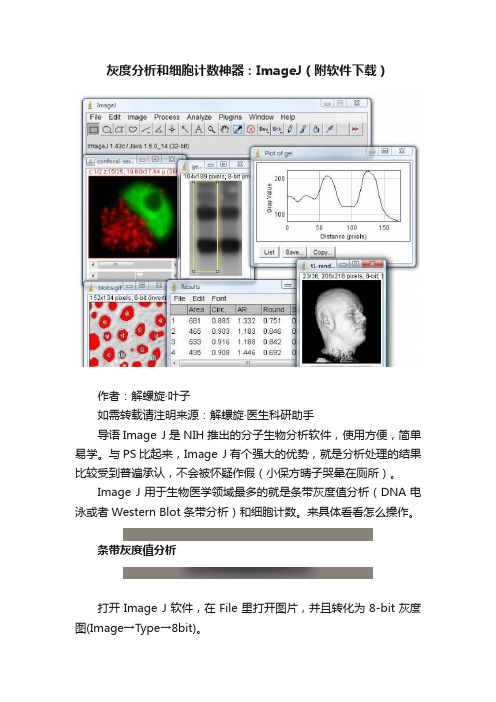
灰度分析和细胞计数神器:ImageJ(附软件下载)作者:解螺旋·叶子如需转载请注明来源:解螺旋·医生科研助手导语Image J是NIH推出的分子生物分析软件,使用方便,简单易学。
与PS比起来,Image J有个强大的优势,就是分析处理的结果比较受到普遍承认,不会被怀疑作假(小保方晴子哭晕在厕所)。
Image J用于生物医学领域最多的就是条带灰度值分析(DNA电泳或者Western Blot条带分析)和细胞计数。
来具体看看怎么操作。
条带灰度值分析打开Image J软件,在File里打开图片,并且转化为8-bit灰度图(Image→Type→8bit)。
方框工具选择并画出条带,然后选择Analyze→Gels→Select First Lane。
最后按Analyze→Gels→plot Lanes,就会出现山峰样图。
但这样是不够的,要把每个山峰分开。
这就用到了直线功能,把下面都封口后点击"魔棒"。
分别点击每个峰下方的区域,在"Results" 里就会出现面积,表示相应条带的灰度值。
如果要测多个条带的灰度,就用在Select First Lane后,点Select Next Lane来复选。
这里注意,每个框的形状大小必须是一样的。
分析图像中的颗粒数(细胞计数)打开Image J软件,在File里打开图片,并且转化为8-bit灰度图(Image→Type→8bit)。
这种黑底的图数起来非常困难,需要反相下变成白底图。
框定区域后,选择Edit→Invert。
比之前清楚多了吧,再设置下阈值(Image→Adjust→Threshold)。
经过调整后对比非常明显。
最后,进入Analyze→Analyze Particles, 键入微粒大小的下限和上限,并且选择显示轮廓(Show outlines)和显示结果(Display Results)。
点击ok,被计数的微粒将显示轮廓和编号。
ImageJ对WesternBlot进行灰度分析

ImageJ对WesternBlot进行灰度分析Western Blotting是生物学领域常用的分析蛋白质的实验技术,它可以提供蛋白质定量和免疫印迹图像。
Western Blot的数据分析需要使用图像处理软件,ImageJ是一款免费的开源软件,是生物医学图像处理分析领域的标准工具之一。
本文将介绍ImageJ如何对Western Blot图像进行灰度分析。
背景Western Blotting技术是分析蛋白质表达及功能的一种常用方法,它可以检测蛋白质的存在、分子量、相对表达量、修饰等信息。
通常利用免疫学方法检测Western Blot膜上的蛋白质,检测后得到的结果是一张黑白图像。
这个图像需要进行处理和分析,以得到蛋白质的数量和变化。
ImageJ是一个开源的图像处理和分析软件,它可以用于各种各样的生物医学图像分析,包括Western Blotting图像分析。
ImageJ提供了灰度分析的功能,可以实现对Western Blotting图像的结果定量分析。
灰度分析可以测量Western Blotting图像的灰度值,这个值可以代表蛋白质的含量,从而实现对蛋白质表达量的分析。
安装ImageJ和导入图像在使用ImageJ进行Western Blotting图像分析前,需要先安装并打开ImageJ。
安装完成后,可以通过以下步骤导入Western Blotting图像:1.打开ImageJ并选择“File”菜单中的“Open”选项。
2.找到Western Blotting图像并选择打开。
3.如果需要对导入的图像进行调整,可以使用ImageJ提供的功能或插件进行操作。
灰度分析灰度分析是Western Blotting图像分析的一个重要步骤,旨在定量分析蛋白质的含量。
以下是使用ImageJ进行灰度分析的步骤:1.进入“Analyze”菜单并选择“Gels”选项。
2.在“Gels”菜单中,选择“Lane Profile”选项。
ImageJ,图像处理与分析的得手利器

本例中,日志文本表明几乎所有像素值都已使用,即使所占总体比例很小。
Brightness Adjustment
亮度调节
The brightness adjustment essentially adds or subtracts a constant to every pixel, causing a shift in the histogram along the x axis, but no change in the distribution
可以将一幅彩色图像看作是由3幅原色图像叠加所合成的。
As we move the cursor over different parts ቤተ መጻሕፍቲ ባይዱf the image, the color values appear in the status bar of the program.
当我们将鼠标移到图像的不同部位,颜色值将显示在程序的状态栏。
However, some images, such as fluorescence micrographs taken as RGB images, can yield surprises.
然而,某些图像,例如RGB图像下获得的荧光显微照片,会产生令人意外的结果。
The reason that the image is so dark is that the routine averages the three channels (rgb) to generate the image. Since there is no data in g or b, the values for the red channel are divided by 3, yielding a dark image.
imageJ

http://fiji.sc/Welcome /ij/
Fiji Is Just ImageJ
Image J 是什么?
• ImageJ是一个基于java的公共的图像处理 软件,它是由美国国立卫生研究院开发的。 可运行于Microsoft Windows,Mac OS, Mac OS X,Linux,和Sharp Zaurus PDA 等多种平台。
首先通过直线工具将 峰值部分封闭,如箭 头处所示。(也可添 加垂直方向的线)
选择魔棒工具,选中封 闭区域,即可显示其面 积,也就是对应条带的 辉度值(图中2峰得出的 值分别为6166.598和 2995.477,所代表的条 带被窗口挡住了。)
测量小鱼面积
(图中黑色金鱼,标尺为原图片自带,正规应该放测量工具实物)
3、方框工具选择并画出条带1, Analyze→Gels→Select First Lane (快捷键Ctrl+1或数字1)
4、在第一个边框边缘左键 拖动移至第二条带, Analyze→Gels→Select Ne xt Lane (或Ctrl+2),重复 该步骤,(快捷键一直都是 Ctrl+2 或数字2)
3、设置阈值 Image →Adjust →Threshold
4、Analyze →Analyze Particles, 键入微粒大小的下 限(为5)和上限,并且选择显 示轮廓(Show outlines)和 显示结果(Display Results)。 点击ok
5、结果,由于下限选择 了5平方厘米,所以只有 大金鱼的面积被测量。( 右上角的小金鱼由于颜色 较浅,面积显示比较小, 不到5)
所有边框选择完毕后, Analyze→Gels→Plot Lanes (快捷键Ctrl+3或3)。 提示:Ctrl+3不能多次按, 可改为 Analyse →Gels →Re plot Lanes
老司机带你解锁ImageJ实用技巧(下)
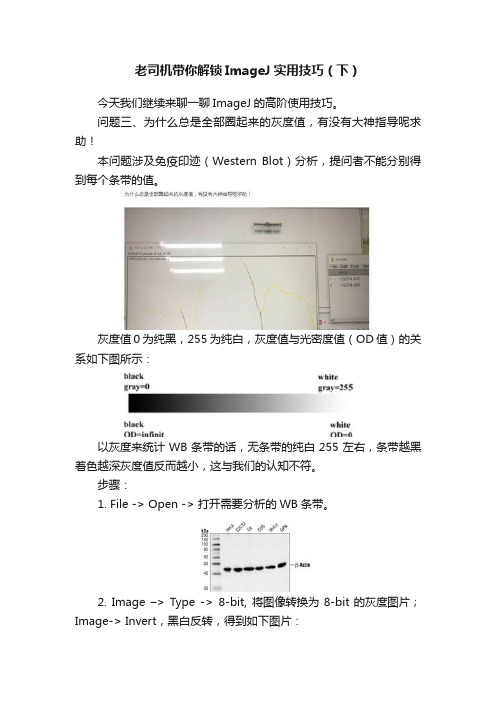
老司机带你解锁ImageJ实用技巧(下)今天我们继续来聊一聊ImageJ的高阶使用技巧。
问题三、为什么总是全部圈起来的灰度值,有没有大神指导呢求助!本问题涉及免疫印迹(Western Blot)分析,提问者不能分别得到每个条带的值。
灰度值0为纯黑,255为纯白,灰度值与光密度值(OD值)的关系如下图所示:以灰度来统计WB条带的话,无条带的纯白255左右,条带越黑着色越深灰度值反而越小,这与我们的认知不符。
步骤:1. File -> Open -> 打开需要分析的WB条带。
2. Image –> Type -> 8-bit, 将图像转换为8-bit的灰度图片;Image-> Invert,黑白反转,得到如下图片:3. Analyze -> Calibrate, 校正光密度值Function中选择Uncalibrated OD,左下方Global calibration,勾选表示有多张图片打开时对所有图片进行此操作。
否则只对当前图片进行此操作。
4. 在工具栏中选择矩形工具——Rectangular, 最左边的为矩形工具,选择条带:键盘上按数字1,弹出如下提示框:点Yes,键盘上按数字3,得到如下:5. 工具栏中选择直线工具——Straight,下图最右边的为直线工具,按住Shift键使用直线工具画竖线将步骤4的峰进行分割:6. 选择魔棒工具——Wand tool,分别点击分割好的峰,即可得到结果,保存结果(File -> Save as…)即可:7. WB统计分析一般将对照组标准化为100%或1,上述结果6个条带前3个为对照组。
在Excel中,先计算对照组3个的平均值(AVERAGE(B2:B4)):然后所有B列的数值除以对照组平均值,此时对照组的均值为1:8. 将所得数据放入Graphpad prism绘图即可:拓展:弯曲的WB条带应如何使用ImageJ拉直?1. 使用Segmented line沿着倾斜条带画间断线段:2. 双击Segmented line,设置线段的宽度至包含所有条带:3. 点击菜单栏Edit -> Selection-> Straighten,倾斜的WB条带即可拉直:问题四、如何统计SEM图片小球数量?步骤:1. 打开ImageJ软件,File -> Open打开SEM图片。
搞定Western的结果图只要简单三步

搞定Western的结果图只要简单三步【前言】辛辛苦苦做完了Western,还在犯愁怎么处理结果么?花了两三天跑Western,难道你还再用大半天来作图么?没钱换新电脑,用着老本儿同时打开PS和AI,都快卡出翔了,你还能忍么?现在就跟着我,简单三步,只用AI,搞定Western的结果!【目的】利用Adobe Illustrator,处理出漂亮美观的Western Blotting的结果图【工具】Adobe Illustrator CS5 (可能有点老,但用着很顺手)【步骤】1)新建文档与打开图片(常用AI者可不看)菜单栏:文件-->新建(快捷键:Ctrl N)-->修改宽度与高度(见下图),常用170mm宽与230mm高-->点击“确定”菜单栏:文件-->打开(快捷键:Ctrl O)-->选择图片-->点击“打开”2)裁剪图片矩形工具(见下图)-->拖拽至覆盖目的条带-->透明度:0%(见下图)-->选择工具(见下图)-->同时选择矩形及其图片或全选-->右键:建立剪切蒙版(快捷键:Ctrl 7)-->加边框(见下图)3)加文字文字工具(见下图)-->选择合适位置拖动并输入文字-->修改字体与大小(见下图),一般为8 pt-12 ptBingo!这样一张Western 结果图就处理好了,多张结果图组合时只要随意的复制(Ctrl C)粘贴(Ctrl V)就可以了。
【小贴士】(1)超常用快捷键双手奉上,望陛下笑纳。
#撤销# Ctrl Z#全选# Ctrl A#水平线# 按住Shift键#连续复制# 按住Alt键(2)若感觉图片较小可利用左下角显示的“100%”进行大小调整(见下图),一般为200%或300%(满屏)更养眼噢。
(3)在选择“矩形工具”或“文字工具”时,记得在选取“选择工具”状态下进行其他操作,需要小小注意一下呦~华丽丽的分割线李莫愁博士:感谢郭影的投稿,如果大家也有什么好的经验和有意思的科研想法也可以给我们投稿哦!希望这些分享出来的经验也能对大家有所帮助,好啦,今天就先策到这里吧。
ImageJ 技能之长度和面积测量
ImageJ技能之细胞长度和面积测量
上两篇小编给大家分享了细胞计数和荧光定量分析的方法。
为响应广大忠粉的需要,今天小编给大家分享一下细胞长度和面积测量方法,最简单的方法就是可以直接在我们Revolve正倒置一体显微镜上做长度和面积测量。
Revolve正倒置一体显微镜
下图就是在显微镜上直接测量长度和面积
如果大家还是觉得不过瘾,小编再给大家分享另外一种办法-ImageJ技能之长度和面积测量。
这是一张我们Revolve正倒置一体显微镜镜头下的微滴图片(与上图为同一倍数图片)。
接下来我们就通过ImageJ来测量长度和面积吧。
Step1:ImageJ打开一张需要测量的图片。
File→Open
Step2:选择直线工具,在标尺上画出单位长度的直线,想要数值更精确可放大图片;
Step3:选择Analyze->Set Scale。
Known distance输入所画直线长度数值(此图为200),Unit of length输入单位(此图为um),Global复选框打钩(将此标准用于全部图片),点击OK
Step4:在需要测量的图片上用直线选择需要测量的长度或者用区域选择工具选择需要测量的面积;
Step5:Ctrl+M(Measurement),结果会显示并记录在Results窗口中。
其中Length显示的即为换算后的长度数值。
与显微镜测量结果基本一致。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
ImageJ实用技巧之Western blot量化
原创2016-05-26 李莫愁博士实验万事屋
ImageJ对医学生来讲最常用的功能是WesternBlot量化和细胞计数,又以前者最常见。
不是所有的WB都需要量化,看审稿人的要求,好了,今天内容比较枯燥,我们一步一步来学习image J量化Western blot的条带吧!
首先下载安装Image J,温馨提示软件在网上有下载。
安装完image J,打开一张要量化的图片如下图:
点击选择方形选框工具,按住左键,在图片上拖动,选择需要分析的条带区域,如图中黄色方框所示。
依次选择,如下图,Select First Lane选项后会出现以上对话框,直接点击Yes 即可:
当选区出现1表示选择成功:
然后选择,Analyze→Gels→Plot Lanes,就会弹出山峰样的图。
峰的面积即灰度值,但是这几个峰连在了一起要先把他们分开,点击直线工具,画直线。
点击选择魔棒工具,然后分别点击每个峰的下方区域,然后就会弹出各个峰面积的结果,分别与六个条带的灰度值相对应,表示六个条带所代表的目的蛋白的表达水平。
使用同样的方法也可以对beta-actin进行量化,最后通过标准化计算得到该Western Blot的量化结果,至于如何将量化结果变成审稿人要求的柱状图,就留给你们自己思考。
