银染protocol概要版 老尚实验室版
银染
1)30%(w/v)Acrylamide 1L,(丙烯酰胺:甲叉双丙烯酰胺=29:1):配制方法:称量丙烯酰胺290g、甲叉双丙烯酰胺10g溶于600ml去离子水中,充分搅拌溶解,定容至1L,用0.45um滤膜滤去杂质,于棕色瓶中4°C保存。
2)5×TBE(1L):配制方法:称量54gTris碱,3.72gNa2EDTA2H2O,27.5g硼酸溶于800ml去离子水中,充分搅拌溶解,定容至1L,高压灭菌后室温保存。
3)10%(W/V)过硫酸铵(10 ml):配制方法:称取1g过硫酸铵,溶于10ml去离子水中搅拌溶解,储存于4℃。
5)10%乙醇(200ml):配制方法:20ml无水乙醇加水溶解,最后定容至200ml。
6)1%的HNO3溶液(200ml):配制方法:2ml硝酸加入到198ml双蒸水中,边加边搅拌。
7)染色液:0.2%的AgNO3溶液(200ml):配制方法:快称0.4克AgNO3倒入200ml双蒸水中,黑暗中搅拌溶解后,快速倒入一棕色瓶中。
8)显色液:2%的Na2CO3及0.4%的甲醛溶液(200ml):配制方法:称取4g Na2CO3精确量取0.8ml甲醛溶于少量双蒸水中,定容至200ml。
9)终显液:4%冰乙酸(200ml):配制方法:量取8ml冰醋酸溶于双蒸水中,定容至200ml。
注:1)---3)为制胶配方5)---9)为银染试剂配方实验步骤:PCR产物的检测(12% 的PAGE电泳检测):洗净玻璃板、胶条及梳子,晾干,然后用夹子夹好制成灌胶板。
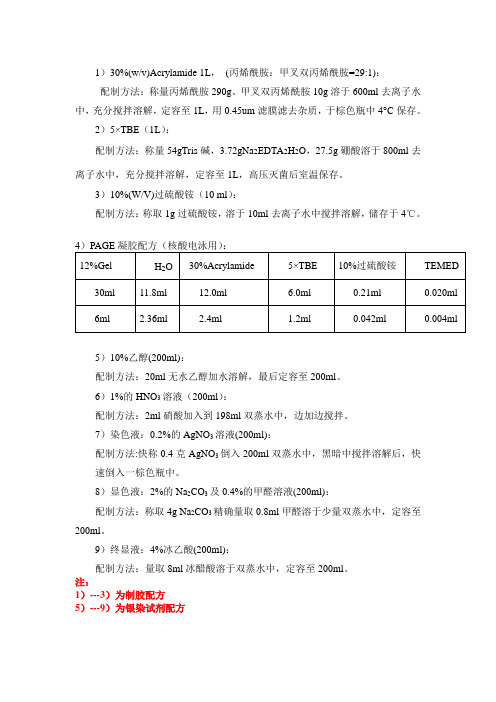
↓用枪吸取 2.36ml三蒸水加入10ml离心管中,再加1.2ml 5×TBE缓冲液,2.4ml 30% Acr,4ul TEMED,最后加42ul APS(过硫酸铵),用移液抢吹打混合混匀,然后缓慢倒入灌胶板中,插上梳子,慢慢放平,让其凝固。
↓待凝胶凝固后,拔掉梳子及玻璃板底部胶条,放入电泳槽,加1×TBE电泳缓冲液并接通电源,60伏,预电泳30min。
蛋白银染

蛋白银染1 固定:50%甲醇,10%冰醋酸浸泡1小时。
2 漂洗:去离子水反复漂洗30分钟。
3 银染:0.8%的银染液(4ml 20% AgNO3滴加到0.36%NaOH与30%氨水的混合液中,不停搅拌,以使沉淀顺速溶解),轻摇15分钟。
4 漂洗:去离子水漂洗2 分钟。
5 显色:1% 柠檬酸和30%甲醛的混合液中震荡直至出现清晰条带6 终止:1%冰醋酸终止反应。
此方法在我们实验室应用效果非常好。
此法出自王家政,蛋白质技术手册P92-97固定:500ml乙醇,100ml冰醋酸,400mldH2O,至少30min浸泡:75ml乙醇,17g醋酸钠,1.25ml25%戊二醛,0.5g硫代硫酸钠·5H2O用dH2O溶解后加至250ml 30min漂洗:用dH2O漂洗3次5min/次银染:0.25g硝酸银,50ul甲醛,用dH2O加至250ml 20min显色:6.25g 碳酸钠,25ul甲醛,用dH2O加至250ml 2-10min视蛋白带显示深棕色终止:3.65gEDTA-Na2·2H2O,用dH2O加至250ml 10min漂洗:用dH2O漂洗3次5min/次保存:25ml甘油用dH2O加至250ml 30min取出晾干以上protocol出自:《蛋白质电泳实验技术》,郭尧君编著,科学出版社出版,p107这是我在郭尧君编著的《蛋白质电泳实验技术》一书上查到的,这是Krause用考玛斯亮蓝染色并干燥后的凝胶再进行的快速银染,供你参考:干胶的5分钟快速银染(Krause)1.配置贮液:溶液A:25gNa2CO3+500ml ddH2O溶液B:1.0g硝酸铵+1.0g硝酸银+5.0gtungstosilisic acid+7ml 37%甲醛,加ddH2O至500ml。
2.使用前混合35mlA和65mlB。
将凝胶放在摇床上振摇,直到蛋白带显色,用ddH2O淋洗。
3.用0.05mol/L甘油终止。
非变性聚丙烯酰胺凝胶电泳银染实验操作指导书(银染) V1.0
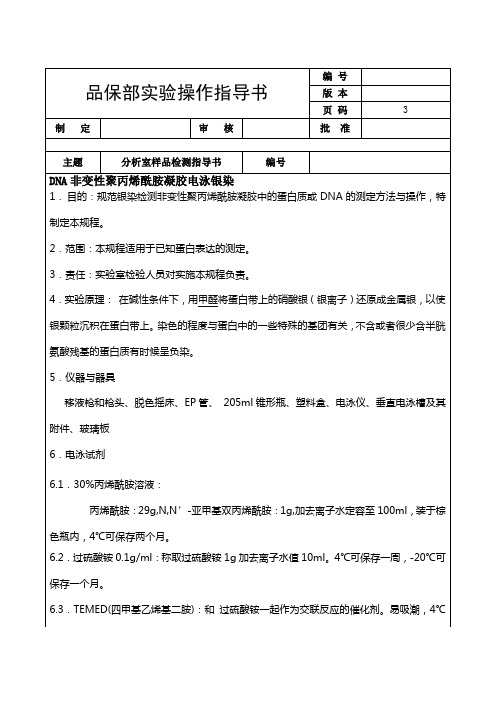
8.1.2红色硅胶条应保持清洁,以保证良好的密封性。然后将玻璃板置于制胶夹上的红色硅胶条中心,轻推夹牢即可,无需下压(按压夹子时应轻柔)。
8.2制胶(6%)
8.2.130%聚丙烯酰胺(29:1)8ml,5*TBE,定容至40ml(每块胶约5ml),加过硫酸铵200ul,TEMED20ul,插入梳子。室温凝固,垂直向上轻轻拔出梳子。避免产生气泡。(若有气泡,用手轻轻弹几下玻璃板,赶尽气泡)
7.银染试剂
7.1固定液:100ml无水乙醇,5ml冰醋酸,定容至1L。
7.2 0.4%硝酸银:硝酸银1克,水250ml。
7.3 1.5%氢氧化钠:氢氧化钠15克,水1L。
7.4 37%甲醛
8.非变性聚丙烯酰胺凝胶的制备
8.1安装玻璃板夹心
8.1.1分别取内板(10cm*7.3cm)、外板(10cm*8.3cm)各一块(注意勿用手接触灌胶面的玻璃),将内板置于外板夹条一侧,使内外玻板底部、两边齐平(否则需重新安装),置于玻璃夹中,将两边固定(夹子应夹在侧边条中央位置)。
5.仪器与器具
移液枪和枪头、脱色摇床、EP管、205ml锥形瓶、塑料盒、电泳仪、液:
丙烯酰胺:29g,N,N’-亚甲基双丙烯酰胺:1g,加去离子水定容至100ml,装于棕色瓶内,4℃可保存两个月。
6.2.过硫酸铵0.1g/ml:称取过硫酸铵1g加去离子水值10ml。4℃可保存一周,-20℃可保存一个月。
6.3.TEMED(四甲基乙烯基二胺):和过硫酸铵一起作为交联反应的催化剂。易吸潮,4℃封闭保存。
6.4. 5*TBE电泳缓冲液
分别称取54克Tris,27.5克硼酸,20ml0.5M/EDTA(pH=8.0)定容至1L
PAGE银染方法

一、实验原理银染是一种重要的PAGE染色方法,由于其成本低,所用试剂安全、快速、灵敏度高而被广泛应用。
银染的原理是银离子在碱性pH 环境下被还原成金属银,沉淀在蛋白质的表面上而显色。
由于银染的灵敏度很高,可染出凝胶上低于1 ng/蛋白质点,故广泛的用在2D凝胶分析上,及极低蛋白含量测定的垂直凝胶中。
这里介绍的是一种实验室常用的银染方法,主要是用于垂直凝胶电泳中低丰度蛋白的检测。
二、实验试剂1. 乙醇、冰醋酸、乙酸钠、硫代硫酸钠、硝酸银、碳酸钠、甘氨酸或EDTA钠盐、去离子水、甲醛。
三、实验步骤1. 固定量取100ml 乙醇,25ml 冰醋酸加去离子水到250ml(视胶板大小情况,适当等比例配制,增加固定液量,以下相同。
),使其终浓度达到 40% 乙醇,10% 冰醋酸。
将凝胶板浸入固定液中,固定30min以上。
2. 水洗去离子水浸泡。
3x 10 min。
2. 致敏生物科研提醒:量取75ml 乙醇,17g 乙酸钠或28.2g三水乙酸钠,0.5g硫代硫酸钠加去离子水到体积250ml。
将固定后的凝胶板拿起,浸入致敏液中,浸泡30 min。
4. 水洗去离子水浸泡。
3x 10 min。
5.银染0.625g AgNO3、100 ul 37%甲醛(在使用前加入)加去离子水到终体积250ml。
将胶板浸入银染液中,浸泡30 min。
6.水洗去离子水浸泡。
3x 20 sec。
注意把握时间,水洗时间长显色速度慢,点的颜色偏黄色。
水洗不充分,背景较深。
7.显色6.25 g Na2CO3、50 ul 37% 甲醛(在使用前加入)加去离子水到终体积250ml。
2-15min,显色时间视凝胶大小、厚薄情况而定。
看到各条带,即可将凝胶捞出。
8.终止3.65g EDTA钠盐或者1g 甘氨酸加去离子水到终体积250ml。
浸泡凝胶10 min。
9.保存1% 冰醋酸,4 ℃。
四、注意事项1.银染主要出现在胶的表面,用薄胶(0.5-0.75mm)可以提高灵敏度。
银染方法

A modified silver staining protocol for visualization of proteins compatible with matrix-assisted laser desorption/ionization and electrospray ionization-mass spectrometryThe growing availability of genomic sequence information,together with improvements in analytical methodology,have enabled high throughput,high sensitivity protein identi-fication.Silver staining remains the most sensitive method for visualization of proteins separated by two-dimensional gel electrophoresis (2-D PAGE).Several silver staining protocols have been developed which offer improved compatibility with subsequent mass spectrometric analysis.We describe a modified silver staining method that is available as a commercial kit (Silver Stain PlusOne;Amersham Pharmacia Biotech,Amersham,UK).The 2-D patterns abtained with this modified protocol are comparable to those from other silver staining methods.Omitting the sensitizing reagent allows higher loading without saturation,which facilitates protein identification and quantita-tion.We show that tryptic digests of proteins visualized by the modified stain afford excellent mass spectra by both matrix-assisted laser desorption/ionization and tandem electrospray ionization.We conclude that the modified silver staining protocol is highly compatible with subsequent mass spectrometric analysis.Keywords:Proteomics /Two-dimensional gel electrophoresis /Silver stain /Mass spectrometry /Protein identification /Matrix assisted laser desorption/ionization ±time of flight /Electrospray ionization ±time of flightEL 4190Jun X.Yan 1Robin Wait 2Tom Berkelman 3Rachel A.Harry 1Jules A.Westbrook 1Colin H.Wheeler 1Michael J.Dunn 11Department of Cardiothoracic Surgery,National Heart and Lung Institute,ImperialCollege School of Medicine,Heart Science Center,Harefield Hospital,Harefield,Middlesex,UK 2Kennedy Institute ofRheumatology,Hammersmith,London,UK 3Amersham Pharmacia Biotech,San Francisco,CA,USAThe increasing availability of genomic sequence informa-tion,together with improvements in protein characteriza-tion by mass spectrometry,have facilitated huge in-creases in the throughput of protein identification.Most commonly,sample components are separated by two-dimensional gel electrophoresis (2-D PAGE)and protein spots are visualized by staining silver,Coomassie blue or SYPRO fluorescent dyes [1±3].Individual spots are then excised from the gel,proteolytically digested,and the masses of the resulting peptides are determined by matrix assisted laser desorption/ionization ±time of flight ±mass spectrometry (MALDI-TOF-MS).The list of peptide masses thus obtained can then be used as a highly spe-cific query to interrogate a protein database [4±9].Recent advances in tandem electrospray ionization-mass spec-trometry (ESI-MS/MS),particularly the development of hybrid quadrupole /orthogonal acceleration TOF instru-ments (Q-TOF),enable routine de novo sequencing of low femtomole levels of peptides [10±13].The ability to determine 10±20amino acid lengths of sequence greatly facilitates cross-species protein identification and retrieval of homologous proteins from genomic and EST data-bases,even when an exactly matching sequence is not present.It is desirable to visualize protein spots in the gel at sensitivities which are roughly comparable to those of the subsequent MALDI-and ESI-MS analyses (usually in the range of nanograms per protein spot).Silver staining has been widely used for this purpose [14,15]since it re-quires relatively inexpensive equipment and reagents and remains one of the most sensitive methods for perma-nently staining proteins in polyacrylamide gels.For protein silver staining,a polyacrylamide gel is soaked in a solution containing soluble silver ions (Ag +)and sub-sequently developed by treatment with a reductant.Pro-tein molecules in the gel promote the reduction of silver ions to metallic silver (Ag 0),which is insoluble and visible.Initial deposition of metallic silver promotes further depo-sition by an autocatalytic process,resulting in exception-ally high sensitivity.There are many published versions of the silver staining process [16,17],which may incorpo-rate,in addition to silver impregnation and development,fixation steps,incubations with sensitivity enhancers (e.g.,glutaraldehyde or formaldehyde),stopping and preservation,and washing steps.The reagents used vary,but the silver reductant is always formaldehyde.The high sensitivity of silver staining comes at the cost of sus-ceptibility to interference from a variety of scources.Correspondence:Dr.Jun X.Yan,Heart Science Center,Hare-field Hospital,Hill End Road,Harefield,Middlesex,UB96JH,UK E-mail:jun.yan@ Fax:+44-(0)1895-828-9003666Electrophoresis 2000,21,3666±3672WILEY-VCH Verlag GmbH,69451Weinheim,20000173-0835/00/1717-3666$17.50+.50/0Exceptional cleanliness must therefore be practiced and reagent and water quality are critical.Silver staining pro-tocols have been developed specifically for visualizingproteins prior to in-gel digestion and mass spectrometric analysis [14,18].Subsequent MS constrains the choice of reagents that can be used during silver staining,because the proteins in the gel must not be chemically modified.Thus many common sensitization reagents (e.g.,glutaraldehyde and strong oxidizing agents)cannot be employed.Since silver staining is a multistep process utilizing numerous reagents,the quality of which is critical,it is often advantageous to purchase a dedicated kit in which the reagents are quality-assured specifically for sil-ver staining.We report here a modified silver staining method that is available as a commercial kit (Silver Stain PlusOne;Amersham Pharmacia Biotech)and we show that it is compatible with subsequent in-gel digestion,MALDI,and ESI analysis.The method is based on that of Heukes-hoven and Dernick [19],but omits the use of glutaralde-hyde in the sensitization step and formaldehyde in the sil-ver impregnation step.The detailed protocol is shown in Table 1.Staining was performed in glass dishes and par-ticular care was taken to avoid contamination by keratin and other extraneous proteins.Electrophoresis 2000,21,3666±3672Silver staining compatible with mass spectrometric analysis 3667Table 1.The modified silver staining protocol using Silver Stain PlusOne kit Step Solution (250mL per gel)Time (min)1.Fix 25mL acetic acid,100mL methanol,125mL milli-Q water 152.Fix25mL acetic acid,100mL methanol,125mL milli-Q water153.Sensitization a)75mL methanol,10ml sodium thiosulfate (5%),17g30sodium acetate 165mL milli-Q water 4.Wash 250mL milli-Q water 55.Wash 250mL milli-Q water 56.Wash 250mL milli-Q water57.Silver a)25mL silver nitrate (2.5%),225mL milli-Q water 208.Wash 250mL milli-Q water 19.Wash 250mL milli-Q water110.Develop6.25g sodium carbonate,100m L formaldehyde,250mL milli-Q water 11.Stop 3.65g EDTA,milli-Q water 1012.Wash 250mL milli-Q water 513.Wash 250mL milli-Q water 514.Wash250mL milli-Q water5a)Omitting the use of glutaraldehyde in the sensitization step and formaldehyde in the sil-ver impregnation step.Working solutions are freshly made immediately prior tostaining.Figure 1.Gel image of normal rat left ventricle using IPG pH 3±10NL 2-D PAGE (12%T)with 100m g total protein loading and silver staining (Owl silver stain kit).P r o t e o m i c s a n d 2-D ENormal human and rat heart left ventricle tissues were used.Sample preparation and2-D PAGE were performed essentially according to Weekes et al.[20].We routinely use100m g total protein loading for analytical gels(pH 3±10NL)and the Owl silver stain kit(Owl Separation Sys-tem,Portsmouth,UK)for visualization.A typical image of one of these gels is shown in Fig.1.To investigate the sensitivity of the PlusOne kit,100,200,300and400m g total protein loading were used.The corresponding gel images are shown in Fig.2.The patterns obtained using the two different kits(Figs.1and2d)are very similar. This,therefore,facilitates comparisons between semipre-parative and analytical gels stained with conventional pro-tocols(e.g.,the Owl kit).Excellent patterns were achieved at higher protein loadings(200,300,and400m g;Fig.2a±c),whereas many spots display negative staining at these loadings when more sensitive staining methods,such as the Owl kit,are used(data not shown).Note that the high-er background obtained from the higher protein loading gels were removed using transform to autoscale the gel image in PDQuest2-D software version6.1(Bio-Rad, Hercules,CA,USA).A400m g total protein loading for IPG3±10strips appears to be optimal in that adequate concentrations of most spots are obtained for MS analy-3668J.X.Yan et al.Electrophoresis2000,21,3666±3672Figure2.Gel images of normal rat left ventricle using IPG pH3±10NL2-D PAGE(12%T)and modi-fied silver staining described in Table1(modified PlusOne silver stain kit)with total protein loading of(a)400m g,(b)300m g,(c)200m g and(d)100m g.sis,while minimizing excessive background and the for-mation of large spot clusters.Higher loadings are possi-ble,however,when using narrow-range IPG strips(data not shown).We investigated the compatibility of the PlusOne modified protocol with ESI-and MALDI-MS.While MALDI is rela-tively tolerant of salts and other contaminants[21,22], the ESI technique is much more susceptible to such inter-ference.Thus,it is necessary to validate the compatibility of the modified stain with both ionization methods.Figure 3shows a gel image of human heart left ventricle(400m g total protein loading)from which20proteins spots were excised for MS characterization.A modified sample prep-aration method was used,which incorporates a destain-ing step[15]to remove silver prior to in-gel digestion with trypsin[14].Aliquots(0.5m L)of the digest supernatant were applied directly to the MALDI target,after which,if necessary,the remainder of the sample was extracted, desalted,and analyzed by ESI-MS/MS.These data are summarized in Table.2.If the results from MALDI mass mapping were ambiguous,ESI-MS/MS was used to gen-erate amino acid sequence data suitable for sequence tag or similarity searching using BLAST.Figure4shows the MALDI mass spectrum of spot19which,when sub-mitted to database searching,retrieved a highly signifi-cant match to ubiquinol cytochrome C reductase.Figure 5a shows a MALDI mass spectrum obtained from spot4, which,when searched,did not find any unambiguous hits.Electrophoresis2000,21,3666±3672Silver staining compatible with mass spectrometric analysis3669Figure3.Gel image of human left ventricle using IPG pH3±10NL2-D PAGE(12%T)with400m gtotal protein loading.The gel was stained using the PlusOne silver stain kit.Protein spots(1±20)labeled on the image were subjected to trypsin digestion and MALDI-TOF-MS or ESI-MS(Table2).This protein was identified by ESI-MS/MS of a doubly charged ion at m/z 777.4,from which 13residues of amino acid sequence were deduced (Fig.5b),which exactly matched the sequence of ATPsynthase a -chain.We conclude that the modified silver staining kit is com-patible with both MALDI-and ESI-MS.Although the sensi-tivity is somewhat lower than other versions,this kit ena-bles higher protein loading,thus facilitating identification by MS.In our laboratory the modified protocol has pro-vided consistent results over a 12-month period and,since the resulting patterns are similar to those produced by the Owl silver stain kit,we have been able to correlate results from semipreparative and analytical gels.Spots of interest are easily located for excision and further charac-terization.A loading of 400m g protein on IPG 3±10strips provides adequate concentrations for successful MALDI analysis of the majority of visible spots.For very low abu-dance proteins,use of the modified stain in conjunction with high protein loadings on narrow pH range IPG strips avoids excessive background staining and spot cluster-ing.We thank the Cardiovascular Disease Group,Rhone-Poulenc Rorer (Collegeville,PA,USA)for providing the rat heart tissue,and Tim Harwood,Amersham PharmaciaElectrophoresis 2000,21,3666±3672Silver staining compatible with mass spectrometric analysis3671T a b l e 2.c o n t i n u e dS p o t E s t i m a t e d I d e n t i f i c a t i o n (a c c e s s i o n n u m b e r )T h e o r e t i c a l A m i n o a c i d P e p t i d e s e q u e n c e i n f o r m a t i o n o b t a i n e d b y E S I -T O F -M S /M S a n a l y s i sN o .p I /M r (D a )p I /M r (D a )c o v e r a g e (%)w i t h M A L D I -T O F -M S 195.7/46200U b i q u i n o l -c y t o c h r o m e c r e d u c t a s e c o m p l e x c o r e p r o t e i n I 5.94/5261939.31.m /z 787.5(3+)A V E L L G D I V Q N C S L E D S Q I E K p r e c u r s o r ,h u m a n (P 31930)2.m /z 901.5(3+)A G Y G P L E Q L P D Y N R3.m /z 1044.3(3+)...F Q G T P L A Q A V E G P S E N V R 4.m /z 1180.7(2+)A V E L L G D I V Q N C S L E D S Q I E K5.m /z 1351.7(2+)Y I I D Q C P A V A G Y Y P I E Q L P D Y N R 205.9/40700A c t i n ,a -c a r d i a c ,h u m a n (P 03996)5.24/4200930.21.m /z 565.9(2+)G Y S F V T T A E R2.m /z 652.7(3+)V A P E E H P T L L T E A P L N P K3.m /z (2+)P y r -E Y D E A G P S I V H RP r o t e i n s p o t s 1±20w e r e e x c i s e d f r o m t h e 2-D g e l s h o w n i n F i g .3a n d a n a l y z e d b y M A L D I -a n d E S I -M S .T h e r e s u l t i n g p e p t i d e m a s s m a p s w e r e s e a r c h e d a g a i n s t S W I S S -P R O T /T r E M B L r e l e a s e 35,u s i n g P r o t e i n P r o b e (M i c r o m a s s ),o r a g a i n s t a n o n r e d u n d a n t d a t a b a s e m a i n t a i n e d b y t h e N a t i o n a l C e n t e r f o r B i o t e c h n o l o g y I n f o r m a t i o n (N C B I )(h t t p ://w w w .n c b i.n l m .n i h .g o v )u s i n g t h e M a s c o t [23]s e a r c h e n g i n e (h t t p ://w w w .m a t r i x s c i e n c e .c o .u k ).A n i n i t i a l m a s s t o l e r a n c e o f 100p p m w a s u s e d ,b u t w a s r e d u c e d t o 50p p m i f e x c e s s i v e n u m b e r s o f h i t s w e r e r e t r i e v e d .A m i n o a c i d s e q u e n c e s o b t a i n e d f r o m E S I -M S /M S w e r e s e a r c h e d a g a i n s t a n o n r e d u n -d a n t d a t a b a s e i n N C B I u s i n g t h e B L A S T p r o g r a m [23].M S /M S s h o w e d t h a t s p o t 1c o n t a i n e d t w o -c o m i g r a t i n g p r o t e i n s .S p o t s 6,8±13,a n d 15±17w e r e n o t a n a l y z e d b y M S /M S b e c a u s e t h e s e a r c h r e s u l t s f r o m t h e M A L D I d a t a w e r e u n a m b i g u o u s .T h e s e q u e n c e c o v e r a g e o f 6,8,9a n d 12a p p e a r s l o w ,b e c a u s e t h e o b s e r v e d p r o -t e i n s p o t s c o r r e s p o n d t o t r u n c a t e d f o r m s ,w h e r e a s t h e c o v e r a g e w a s c a l c u l a t e d f r o m t h e f u l l -l e n g t h s e q u e n c e.Biotech,for conducting this collaboration.JXY acknowl-edges Aventis for their financial support.RAH and JAW thank Proteome Sciences Inc.for their financial support. RW thanks the Wellcome Trust for purchase of the MALDI spectrometer.Work in MJD©s laboratory is sup-ported by the British Heart Foundation.Received May,30,2000References[1]Berggren,K.,Chernolalskaya,E.,Steinberg,T.H.Kemper,C.,Lopez,M.F.,Diwu,Z.,Haugland,R.P.,Patton,W.F.,Electrophoresis2000,21,2509±2521.[2]Steinberg,T.H.,Lauber,W.M.,Berggren,K.,Kemper,C.,Yue,S.,Patton,W.F.,Electrophoresis2000,21,497±508.[3]Steinberg,T.H.,Jones,L.J.,Haugland,R.P.,Singer,V.L.,Anal.Biochem.1996,239,223±237.[4]Henzel,W.J.,Billeci,T.M.,Stults,J.T.,Wong,S.C.,Grim-ley,C.,Watanabe,C.,A1993,90, 5011±5015.[5]Mann,M.,Hojrup,P.,Roeppstorff,P.,Biol,Mass Spectrom.1993,22,338±345.[6]Pappin,D.J.C.,Hojrup,P.,Bleasby,A.J.,Curr.Biol.1993,3,327±332.[7]Liang,X.L.,Bai,J.,Liu,Y.H.,Lubman,D.M.,Anal.Chem.1996,68,1012±1018.[8]Patterson,S.D.,Aebersold,R.,Electrophoresis1995,16,1791±1814.[9]Wheeler,C.H.,Berry,S.L.,Wilkins,M.R.,Corbett,J.M.,Ou,K.,Golley,A.A.,Humphery-Smith,I.,Williams,K.L., Dunn,M.J.,Electrophoresis1996,7,580±587.[10]Morris,H.R.,Paxton,T.,Dell,A.,Langhorne,J.,Berg,M.,Bordoli,R.S.,Hoyes,J.,Bateman,R.H.,Rapid Commun.Mass Spectrom.1996,10,889±896.[11]Shevchenko,A.,Chernushevich,I.,Ens,W.,Standing,K.G.,Thomson,B.,Wilm,M.,Mann,M.,Rapid Commun.Mass Spectrom.1997,11,1015±1024.[12]Borchers,C.,Peter,J.F.,Hall,M.C.,Kunkel,T.A.,Tomer,K.B.,Anal.Chem.2000,72,1163±1168.[13]Kristensen,D.B.,Imamura,K.,Miyamoto,Y.,Yoshizato,K.,Electrophoresis2000,21,430±439.[14]Shevchenko,A.,Wilm,M.,Vorm,O.,Mann,M.,Anal.Chem.1996,68,850±858.[15]Gharahdaghi,F.,Weinberg,C.R.,Meagher,D.A.,Imai,B.S.,Mische,S.M.,Electrophoresis1999,20,601±605.[16]Rabilloud,T.,Electrophoresis1990,11,785±794.[17]Rabilloud,R.,Electrophoresis1992,13,429±439.[18]Arnott,D.,O©Connell,K.L.,King,K.L.,Stults,J.T.,Anal.Biochem.1998,258,1±18.[19]Heukeshoven,J.,Dernick,R.,Electrophoresis1985,6,103±112.[20]Weekes,J.,Wheeler,C.H.,Yan,J.X.,Weil,J.,Eschenha-gen,T.,Scholtysik,G.,Dunn,M.J.,Electrophoresis1999, 20,898±906.[21]Garden,R.W.,Moroz,L.L.,Moroz,T.P.,Shippy,S.A.,Sweedler,J.V.,J.Mass Spectrom.1996,31,1126±1130.[22]Winkler,M.A.,Kundu,S.,Robey,T.E.,Robey,W.G.,J.Chromatogr.A1996,744,177±185.[23]Perkins,D.N.,Pappin,D.J.,Creasy,D.M.,Cottrell,J.S.,Electrophoresis1999,20,3551±3567.[24]Altschul,S.F.,Madden,T.L.,Schäffer,A.A.,Zhang,J.,Zhang,Z.,Miller,W.,Lipman,D.J.,Nuclei Acids Res.1997, 25,3389±3402.[25]Vorm,O.,Mann,M.,J.Am.Soc.Mass Spectrom.1994,5,955±958.3672J.X.Yan et al.Electrophoresis2000,21,3666±3672。
银染 protocol

试剂准备(所有试剂均用超纯水制备):
1)固定液:甲醇50ml,乙酸12ml,甲醛(37%)135ul,加水至100ml;
2)35% 乙醇;
3)2% Na2S2O3:0.1克Na2S2O3用水稀释到5ml使用;
4)染色液(现配现用):AgNO3 0.1g,甲醛(37%)100ul,加水至50ml;
5)显色液(现配现用):Na2CO36g,甲醛(37%)135ul,Na2S2O3(2%)20ul,加水至100ml;
6)终止液:甲醇50ml,乙酸12ml,加水至100ml。
步骤:
1)取下2.7电泳分离蛋白样品后的聚丙烯酰胺凝胶,泡在固定液中,摇床上低速、室温固定2h或过夜(以下步骤均可摇床上低速晃动完成);
2)弃去固定液,加入35%乙醇洗涤,隔20min换新的35%乙醇再次洗涤,共洗三次;
3)弃去35%乙醇,于0.02%的Na2S2O3溶液中激活凝胶,2min;
4)弃去3)的Na2S2O3溶液,用高纯水洗涤凝胶三次,每次5min;
5)弃去高纯水,用染色液浸泡凝胶,染色20min;
6)弃去染色液,用高纯水洗涤凝胶两次,每次1min;
7)弃去高纯水,加入显色液,密切关注显色情况,以清晰看到条带为宜;
8)弃去显色液,加入终止液,5min;
9)弃去终止液,凝胶可泡在1%的乙酸溶液中,4℃保存。
silver stain (protocol)——银染

Staining the SDS-PAGE gel with Silver saltsThe silver stains are capable of detecting as little as 0.1 t o10 ng of polypeptide in a single band, and silver nitrate solutions are easier to prepare.1.The SDS-PAGE gel is fixed in at least 5 gel volumes of a solution ofethanol:glacial acetic acid:water(30:10:60) for 4 -12 h with gentle shaking.2.Discard the fixing solution, and add at least 5 gel volumes of 30% ethanol.Incubate the gel for 30 min at room temperature with gentle shaking.3.Repeat step 2.4.Discard the ethanol and add 10 volumes of deionized water. Incubate the gel for10 min at room temperature with gentle shaking.5.Repeat step 4 twice.6.Discard the last of the water washes, and, wearing gloves, add 5 gel volumes of 10.1% solution of AgNO3(freshly diluted). Incubate the gel for 30 min at roomtemperature with gentle shaking.+7.Discard the AgNO3 solution, and wash both sides of the gel 20 s under a steam ofdeionized water.8.Add 5 gel volume of a freshly made aqueous solution of 2.5% sodium carbonate,0.02% formaldehyde. Incubate the gel at room temperature with gentle agitation.Washing the gel carefully. Stained bands of protein should appear within a few minutes. Continue incuation until desired contrast is obtained.9.Quench the reaction by washing the gel in 1% acetic acid for a few minutes. Thenwash the gel several times with deionized wither (10 min per wash)10.Dry the gel.。
银染流程及注意事项

银染流程及注意事项
银染流程银染 protocol
注意事项:
在最初甲醇固定时就应该先除去甘油、尿素、甘氨酸、Triton X 100和两性电解质这些干扰性物质。
蛋白胶固定之前一定要先用水涮干净!
SDS凝胶中的巯基乙醇会导致在60 KDa或67 KDa处出现两条水平线。
减少巯基乙醇的用量即可避免。
不同蛋白质对银染的反应是不一样的,尤其是碱性蛋白染色效果差。
因此,不宜用银染测定不同蛋白的比例。
染色过程中,缓慢的振荡是必要的,一般选择 40 60 rpm.
对于银染条带不理想可尝试优化:
① 反应溶液都平衡到室温,
② 硝酸银和显色液容易失效,可以现配现用
③ PAGE胶固定之前,用水浸洗掉running buffer
④ 检查用水器具是否都去离子彻底(用新鲜配置50%硝酸润洗)。
PAGE银染方法

TA钠盐或者1g甘氨酸加去离子水到终体积250ml。 浸泡凝胶10min。 9.保存 1%冰醋酸,4℃。 1.银染主要出现在胶的表面,用薄胶(0.5-0.7
5mm)可以提高灵敏度。 2.固定时间较长,则加一步水洗30min,以免胶 太脆而破碎。 3.最好多配制一份显色液,第一次显色到溶液 变混浊时换一份显色液,显色
由于其成本低,所用试剂安全、快速、灵敏度高 而被广泛应用。银染的原理是银离子在碱性pH环 境下被还原成金属银,沉淀在蛋白质的表面上而 显色。由于银染的灵敏度很高,可染
出凝胶上低于1ng/蛋白质点,故广泛的用在2D凝 胶分析上,及极低蛋白含量测定的垂直凝胶中。 这里介绍的是一种实验室常用的银染方法,主要 是用于垂直凝胶电泳中低丰度蛋
.室温操作,温度的波动往往会干扰银染的效果, 恒温水浴可以解决这个问题。 14.当蛋白质中含有核酸或金属时,银染则不会 奏效。改变固定剂和染色之前对胶进行漂洗可以
改进染色效果。 15.据称戊二醛预处理可以使各种蛋白质的染色 提高40倍。在染色效果不佳的情况下可以考虑。 16.银染应尽快照相,随着时间延长,蛋白条带 会变浅
,而背景会加深。 17.不同蛋白质对银染反应不一样,尤其是碱性 蛋白染色效果差。因此不宜用银染测定不同蛋白 的比例。
上海坤肯生物化工有限公司主营分子生物学
试剂、免疫试剂、细胞培养基、生化试剂、实验 室耗材及仪器。公司以专业的技术支持,和周到 快捷的服务,努力成为生物化工领域的领头羊。 公司的经营理念:以专业的产品、专业
致在60KDa或67KDa处出现两条水平线。减少巯 基乙醇的用量即可避免。。 8.染色、水洗等过程中,缓慢的振荡是必要的, 一般选择40-60rpm。没有设备,用
手推轻晃亦可。 9.凝胶表面的裂纹多是由于压力、手印及表面 干燥所致,所以全程操作中都应带手套。 10.凝胶背景呈均一的黑色多是水中的杂质引起 的,所以溶液的配
silverstain(protocol)——银染

silverstain(protocol)——银染Staining the SDS-PAGE gel with Silver saltsThe silver stains are capable of detecting as little as 0.1 t o10 ng of polypeptide in a single band, and silver nitrate solutions are easier to prepare.1.The SDS-PAGE gel is fixed in at least 5 gel volumes of a solution ofethanol:glacial acetic acid:water(30:10:60) for 4 -12 h with gentle shaking.2.Discard the fixing solution, and add at least 5 gel volumes of 30% ethanol.Incubate the gel for 30 min at room temperature with gentle shaking.3.Repeat step 2.4.Discard the ethanol and add 10 volumes of deionized water. Incubate the gel for10 min at room temperature with gentle shaking.5.Repeat step 4 twice.6.Discard the last of the water washes, and, wearing gloves, add 5 gel volumes of 10.1% solution of AgNO3(freshly diluted). Incubate the gel for 30 min at roomtemperature with gentle shaking.+7.Discard the AgNO3 solution, and wash both sides of the gel 20 s under a steam ofdeionized water.8.Add 5 gel volume of a freshly made aqueous solution of 2.5% sodium carbonate,0.02% formaldehyde. Incubate the gel at room temperaturewith gentle agitation.Washing the gel carefully. Stained bands of protein should appear within a few minutes. Continue incuation until desired contrast is obtained.9.Quench the reaction by washing the gel in 1% acetic acid for a few minutes. Thenwash the gel several times with deionized wither (10 min per wash)10.Dry the gel.。
我们实验室用的银染方法

我们实验室用的银染方法1.50%甲醇浸泡15分钟(置于摇摆床上,以下省略,本步骤两次,每15分钟更换一次)2.10%甲醇浸泡10分钟3.水漂洗3次,每次数秒4.2μM DTT溶液浸泡20分钟5.0.1%硝酸银溶液浸泡20分钟6.水漂洗3次,每次不超过15秒(每次约150ML)7.显色液漂洗3次,每次不超过15秒(每次约100ML)8.显色液浸泡显色至看到目的蛋白带(约200ML)9.10G柠檬酸定影显色液:15G无水碳酸钠溶于500ml水,用前加入250μL福尔马林搅拌均匀水用双蒸水就可以,纯度越高越好,其他试剂用分析纯,据书上说甲醇用化学纯的比分析纯更灵敏此方法出自冷泉港蛋白质技术指南(实际上是其蛋白质技术会议的总结)灵敏度稍低于分子克隆上老方法,但是非常方便,也比其他方法快速,只需要1.5小时,我染出来的胶非常漂亮,有需要的话可以上传给大家看看一般来说,高背景是由于杂质产生的,银染的水质很重要,最好要用去离子水或双蒸水,装胶的器皿也一定要洗得很干净。
固定、银染之后均需充分的漂洗。
其实银染就是银镜反应,可能是你的样品浓度高,所以白了,浓度更高还会变成一条能反光的带。
固定时间不需加长,我有时来不及还减少固定时间,银染关键是器皿和溶液干净,显色前胶一定要漂干净大板本来效果就差点,浓度越大,条带形状越差,这也很正常,银染薄点的胶效果要好些,因为染的是表面的蛋白1.取下凝胶,蒸馏水冼胶30秒.2. 0.1% 硝酸银染色10分钟.3. 弃去染色液,蒸馏水洗胶1分钟.4.显色液显色15秒,换用新的显色液,显清条带为止.显色液配方: 10克氢氧化钠+0.2克碳酸钠+2m甲醛溶液,定溶到500ml.这个染色方法挺好的,我用过.注意染色时间不能长.材料没什么要求,玻璃会更好一点,但不好搞到,我们是见到什么大小合适就用什么。
液体的量一般是胶体积的5倍,比如13×13×0.1cm的胶,用一百ml一定够,80也行。
我们实验室用的银染方法

我们实验室用的银染方法1.50%甲醇浸泡15分钟(置于摇摆床上,以下省略,本步骤两次,每15分钟更换一次)2.10%甲醇浸泡10分钟3.水漂洗3次,每次数秒4.2μM DTT溶液浸泡20分钟5.0.1%硝酸银溶液浸泡20分钟6.水漂洗3次,每次不超过15秒(每次约150ML)7.显色液漂洗3次,每次不超过15秒(每次约100ML)8.显色液浸泡显色至看到目的蛋白带(约200ML)9.10G柠檬酸定影显色液:15G无水碳酸钠溶于500ml水,用前加入250μL福尔马林搅拌均匀水用双蒸水就可以,纯度越高越好,其他试剂用分析纯,据书上说甲醇用化学纯的比分析纯更灵敏此方法出自冷泉港蛋白质技术指南(实际上是其蛋白质技术会议的总结)灵敏度稍低于分子克隆上老方法,但是非常方便,也比其他方法快速,只需要1.5小时,我染出来的胶非常漂亮,有需要的话可以上传给大家看看一般来说,高背景是由于杂质产生的,银染的水质很重要,最好要用去离子水或双蒸水,装胶的器皿也一定要洗得很干净。
固定、银染之后均需充分的漂洗。
其实银染就是银镜反应,可能是你的样品浓度高,所以白了,浓度更高还会变成一条能反光的带。
固定时间不需加长,我有时来不及还减少固定时间,银染关键是器皿和溶液干净,显色前胶一定要漂干净大板本来效果就差点,浓度越大,条带形状越差,这也很正常,银染薄点的胶效果要好些,因为染的是表面的蛋白1.取下凝胶,蒸馏水冼胶30秒.2. 0.1% 硝酸银染色10分钟.3. 弃去染色液,蒸馏水洗胶1分钟.4.显色液显色15秒,换用新的显色液,显清条带为止.显色液配方: 10克氢氧化钠+0.2克碳酸钠+2m甲醛溶液,定溶到500ml.这个染色方法挺好的,我用过.注意染色时间不能长.材料没什么要求,玻璃会更好一点,但不好搞到,我们是见到什么大小合适就用什么。
液体的量一般是胶体积的5倍,比如13×13×0.1cm的胶,用一百ml一定够,80也行。
PAGE银染方法

一、实验原理银染是一种重要的PAGE染色方法,由于其成本低,所用试剂安全、快速、灵敏度高而被广泛应用。
银染的原理是银离子在碱性pH 环境下被还原成金属银,沉淀在蛋白质的表面上而显色。
由于银染的灵敏度很高,可染出凝胶上低于1 ng/蛋白质点,故广泛的用在2D凝胶分析上,及极低蛋白含量测定的垂直凝胶中。
这里介绍的是一种实验室常用的银染方法,主要是用于垂直凝胶电泳中低丰度蛋白的检测。
二、实验试剂1. 乙醇、冰醋酸、乙酸钠、硫代硫酸钠、硝酸银、碳酸钠、甘氨酸或EDTA钠盐、去离子水、甲醛。
三、实验步骤1. 固定量取100ml 乙醇,25ml 冰醋酸加去离子水到250ml(视胶板大小情况,适当等比例配制,增加固定液量,以下相同。
),使其终浓度达到 40% 乙醇,10% 冰醋酸。
将凝胶板浸入固定液中,固定30min以上。
2. 水洗去离子水浸泡。
3x 10 min。
2. 致敏生物科研提醒:量取75ml 乙醇,17g 乙酸钠或28.2g三水乙酸钠,0.5g硫代硫酸钠加去离子水到体积250ml。
将固定后的凝胶板拿起,浸入致敏液中,浸泡30 min。
4. 水洗去离子水浸泡。
3x 10 min。
5.银染0.625g AgNO3、100 ul 37%甲醛(在使用前加入)加去离子水到终体积250ml。
将胶板浸入银染液中,浸泡30 min。
6.水洗去离子水浸泡。
3x 20 sec。
注意把握时间,水洗时间长显色速度慢,点的颜色偏黄色。
水洗不充分,背景较深。
7.显色6.25 g Na2CO3、50 ul 37% 甲醛(在使用前加入)加去离子水到终体积250ml。
2-15min,显色时间视凝胶大小、厚薄情况而定。
看到各条带,即可将凝胶捞出。
8.终止3.65g EDTA钠盐或者1g 甘氨酸加去离子水到终体积250ml。
浸泡凝胶10 min。
9.保存1% 冰醋酸,4 ℃。
四、注意事项1.银染主要出现在胶的表面,用薄胶(0.5-0.75mm)可以提高灵敏度。
PAGE银染方法

一、实验原理银染是一种重要的PAGE染色方法,由于其成本低,所用试剂安全、快速、灵敏度高而被广泛应用。
银染的原理是银离子在碱性pH 环境下被还原成金属银,沉淀在蛋白质的表面上而显色。
由于银染的灵敏度很高,可染出凝胶上低于1 ng/蛋白质点,故广泛的用在2D凝胶分析上,及极低蛋白含量测定的垂直凝胶中。
这里介绍的是一种实验室常用的银染方法,主要是用于垂直凝胶电泳中低丰度蛋白的检测。
二、实验试剂1. 乙醇、冰醋酸、乙酸钠、硫代硫酸钠、硝酸银、碳酸钠、甘氨酸或EDTA钠盐、去离子水、甲醛。
三、实验步骤1. 固定量取100ml 乙醇,25ml 冰醋酸加去离子水到250ml(视胶板大小情况,适当等比例配制,增加固定液量,以下相同。
),使其终浓度达到 40% 乙醇,10% 冰醋酸。
将凝胶板浸入固定液中,固定30min以上。
2. 水洗去离子水浸泡。
3x 10 min。
2. 致敏生物科研提醒:量取75ml 乙醇,17g 乙酸钠或28.2g三水乙酸钠,0.5g硫代硫酸钠加去离子水到体积250ml。
将固定后的凝胶板拿起,浸入致敏液中,浸泡30 min。
4. 水洗去离子水浸泡。
3x 10 min。
5.银染0.625g AgNO3、100 ul 37%甲醛(在使用前加入)加去离子水到终体积250ml。
将胶板浸入银染液中,浸泡30 min。
6.水洗去离子水浸泡。
3x 20 sec。
注意把握时间,水洗时间长显色速度慢,点的颜色偏黄色。
水洗不充分,背景较深。
7.显色6.25 g Na2CO3、50 ul 37% 甲醛(在使用前加入)加去离子水到终体积250ml。
2-15min,显色时间视凝胶大小、厚薄情况而定。
看到各条带,即可将凝胶捞出。
8.终止3.65g EDTA钠盐或者1g 甘氨酸加去离子水到终体积250ml。
浸泡凝胶10 min。
9.保存1% 冰醋酸,4 ℃。
四、注意事项1.银染主要出现在胶的表面,用薄胶(0.5-0.75mm)可以提高灵敏度。
银染

银染方法比较生物科学2008-08-16 00:20:42 阅读92 评论1银染步骤:1 改进的Bassam 银染方法固定液配方:100mL冰醋酸(10%),加900mL去离子水;染色液配方:lg AgNO3定容(0.1%)到1L去离子水中;显影液配方:60g Na2CO3定容到lL去离子水中,4~C保存。
用时加入200微升浓度为10g/L 的硫代硫酸钠,37%甲醛1.5mL;终止液配方:同固定液配方。
操作流程固定2Omin 去离子水洗涤lOmin一染色3Omin一去离子水洗涤lOs一显色(至所要的程度)一终止显影。
2 改进的Sanguinetti银染方法染色液配方:lg AgNO3定容到lL去离子水中;显影液配方:16g的NaOH,37%甲醛8mL定容到1L去离子水中;终止液配方:7.5g Na2CO3, 定容到lL 去离子水中。
操作流程去离子水洗涤15S--染色8min--去离子水洗涤l5s---显影至所需程度(3—4min)---终止显影。
从染色效果来看,改进的Bassam方法比改进的Sanguinetti方法颜色背景浅,灵敏度高,能够看到副带染色。
但此方法相比后者又有着染色时间长,所用试剂品种多,价格较贵的缺点。
尽管改进的Sanguinetti方法对条带的鉴别力不如改进的Bassam方法,但因省时省力成本相对较低,更适用作进行大规模引物筛选时的染色方法。
南农时银染步骤:固定:(10%)乙醇,(0.5%)冰乙酸的固定液,每次6min,两次;渗透:(0.2%)AgNO3渗透液,12min;漂洗:ddH2O,清洗2遍,各30s,10%的硫代硫酸钠(Na2S2O3)溶液,漂洗30s;显色:(1.5%)NaOH,0.4%的甲醛显色液显色约15min。
Bioscience博客SSR银染方法(轻轻震荡)1、固定10%乙醇+0.5%冰乙酸6min×2次2、染色0.2% AgNO3 12min3、漂洗(1)蒸馏水1-2min(2)0.002% Na2S2O3 1-2min(3)显色1.5%NaOH + 0.4%甲醛(4)保存0.75%Na2CO3定稿银染步骤如下固定:(10%)乙醇,(0.5%)冰乙酸的固定液,10min;450 ml ddH2O加无水酒精50 ml,冰乙酸2.5 ml。
银染方法总汇及注意事项

ptotocol是这样的,请看哪里不妥:蛋白银染步骤步骤溶液时间固定甲醇100ml冰醋酸25ml加双蒸水至250ml 30min冲洗双蒸水3次敏化甲醇75ml戊二醛(25%w/)1.25ml硫代硫酸钠(5%w/) 10ml醋酸钠(17g)加双蒸水至250ml 30min冲洗双蒸水3次银反应硝酸银溶液(2.5%w/) 25ml甲醛(37%w/) 0.1ml加双蒸水至250ml 20min冲洗双蒸水2次显色碳酸钠(6.25g)甲醛(37%w/) 0.05ml加双蒸水至250ml2-5min终止EDTA-Na22H2O(3.65g)加双蒸水至250ml 10min冲洗双蒸水3次建议使用silia的方法!!简单,快!本人的银染方法:1. 固定:100ml MeOH, 24ml H Ac, 56ml 0.04%HCHO, 20ml ddH2O,洗三次,每次>=20分钟;2. 50%EtOH洗PAGE胶三次,每次20分钟;3. 预处理:0.02%Na2S2O3 处理一分钟;4. 水洗:ddH2O漂洗三次,每次20秒;5. 染色:0.8ml 20%AgNO3+72ml 0.04%HCHO处理PAGE胶20分钟;6. 水洗:同上;7. 显色:25ml 12%Na2CO3, 24ml 0.04%HCHO, 1ml 0.02%Na2S2O3, 漂洗10-15分钟;8. 水洗:同上;9. 终止:50%MeOH, 12%H Ac,10分钟。
以上步骤从中科院生化所学来,过程和silia 的基本一致,不知道是否有出入,请大家指正。
谢谢!!!5.2.2.3 银染色1 银染液1(1000ml):甲醇50%乙酸12%甲醇500ml 冰醋酸120ml2 银染液2(1000ml):甲醇30%,乙酸钠0.4M,戊二醛0.5%,硫代硫酸钠0.1%,(含乙酸1-2ml)甲醇300ml 乙酸钠54.4g 戊二醛10ml 硫代硫酸钠1g 乙酸1-2ml3 显色液(1000ml) 无水碳酸钠2.5% 甲醛0.02%无水碳酸钠25g,甲醛1.5ml4 硝酸银(100ml):20%硝酸银(过滤后待用)染色步骤:1 银染液1 洗2次,每次5分钟2 银染液2 洗1次,每次30分钟3 超纯水洗2次,每次1分钟4 0.5%硝酸银洗30分钟5 超纯水洗5次,每次1分钟(关键步骤,共计5分钟)显色液显色6 看到各条带,则倾去显色液, 1%乙酸中止我感觉浸银后、显色前的水洗非常关键,时间不能太长,次数不能太多,否则可能会染不上颜色。
- 1、下载文档前请自行甄别文档内容的完整性,平台不提供额外的编辑、内容补充、找答案等附加服务。
- 2、"仅部分预览"的文档,不可在线预览部分如存在完整性等问题,可反馈申请退款(可完整预览的文档不适用该条件!)。
- 3、如文档侵犯您的权益,请联系客服反馈,我们会尽快为您处理(人工客服工作时间:9:00-18:30)。
银染protocol概要版
1.接种细胞于6-8个15cm的大板, 次日使用PEI转染(24μg 质粒/板,
质粒:PET约1:5) 细胞, 分别转染vector和含flag的目的基因, 48h 后用RIPA裂解液裂解收集细胞.
2.弃去培养基, 用冷的PBS洗两遍, 每大盘加入2ml IP buffer (加
cocktail), 4℃摇20min.
3.刮下细胞, 4℃,最大转速离心15min.
4.吸取上清转入干净的15 ml离心管中.
5.用IP buffer裂解细胞时, 制备flag-M2 resin. 取130μl M2的
珠子, PBS洗珠子五遍. 每次4℃离心, 500g, 5min, 最后制成50%的悬浮液
6.目的蛋白上清和vector中各加入100μl 珠子. 4℃孵育过夜或
3-4h(较好).然后4℃,500g,5min收珠子, 转移至1.5ml Ep管中.
7.用IP buffer 洗珠子5遍, 4℃,500g,5min离心.
8.25x(见多肽说明书, 4mg溶于PBS 1.6ml)的flag多肽, 用前先用
PBS稀释成1x的. 然后每管加入60μl (体积要大于纯珠子的体积, 上面用50%的100μl,因此应大于50μl)的flag多肽. 4℃, 5min 结合(摩天轮上快摇), 5min离心,收集上清,反复三次, 3xflag配方稍有不同,详见说明书.
9.每次上清收集后加入4X银染专用loading,70度保温10min.(可以
用实验室通用的loading buffer, 可煮沸)
10.珠子用PBS洗三次,加入含叠氮钠的PBS或者50%甘油保存,可反复
使用.
11.得到的蛋白上预制胶,用专用的3μl的sharp Marker或者3μl蛋白
预染marker跑电泳
12.电泳时80v
13.胶用三蒸水洗5min, 两次(用新的25cm的盘子装)
14.用30%无水乙醇: 10%醋酸: 60%的三蒸水固定胶15min, 两遍.
15.用10%乙醇洗胶5min两遍, 然后用三蒸水洗5min洗2遍.
16.准备sensitizer work solution (50μl sensitizer 于25ml水中), 严格增
敏1min, 然后用水洗1min 洗两遍.
17.准备stain working solution (0.5ml enhancer 于25ml stain中). 染胶
30min.
18.准备Developer working solution (0.5ml enhancer 于25ml developer
中), 用三蒸水洗胶20s 两遍, 然后用Developer working solution增敏2-3min, 直至条带出现.
19.终止5%乙酸中. 10min
说明: 胶用Invitrogen公司的NP0335, 是4%-10%的梯度胶.
Flag多肽F3290 sigma公司.。
